Abstract
Background
Canadian Arctic summer sea ice has dramatically declined due to global warming, resulting in the rapid opening of the Northwest Passage (NWP), slated to be a major shipping route connecting the Atlantic and Pacific Oceans by 2040. This development elevates the risk of oil spills in Arctic regions, prompting growing concerns over the remediation and minimizing the impact on affected shorelines.
Results
This research aims to assess the viability of nutrient and a surface washing agent addition as potential bioremediation methods for Arctic beaches. To achieve this goal, we conducted two semi-automated mesocosm experiments simulating hydrocarbon contamination in high-Arctic beach tidal sediments: a 32-day experiment at 8 °C and a 92-day experiment at 4 °C. We analyzed the effects of hydrocarbon contamination, biostimulation, and a surface washing agent on the microbial community and its functional capacity using 16S rRNA gene sequencing and metagenomics. Hydrocarbon removal rates were determined through total petroleum hydrocarbon analysis. Biostimulation is commonly considered the most effective strategy for enhancing the bioremediation process in response to oil contamination. However, our findings suggest that nutrient addition has limited effectiveness in facilitating the biodegradation process in Arctic beaches, despite its initial promotion of aliphatic hydrocarbons within a constrained timeframe. Alternatively, our study highlights the promise of a surface washing agent as a potential bioremediation approach. By implementing advanced -omics approaches, we unveiled highly proficient, unconventional hydrocarbon-degrading microorganisms such as Halioglobus and Acidimicrobiales genera.
Conclusions
Given the receding Arctic sea ice and the rising traffic in the NWP, heightened awareness and preparedness for potential oil spills are imperative. While continuously exploring optimal remediation strategies through the integration of microbial and chemical studies, a paramount consideration involves limiting traffic in the NWP and Arctic regions to prevent beach oil contamination, as cleanup in these remote areas proves exceedingly challenging and costly.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-024-00626-w.
Keywords: The Northwest Passage, High Arctic beaches, Oil contamination, Hydrocarbon biodegradation, Hydrocarbon degrading microorganisms, Bioremediation, Biostimulation
Background
The Arctic is experiencing a disproportionate impact from global warming, with a rate of warming more than double the global average [1–3]. Temperatures have increased at an unprecedented rate since the mid-twentieth century, projected to continue warming at a rate of approximately 0.5 degrees per decade [4]. This has resulted in a drastic decline in summer sea ice cover in Canadian Arctic waters, with reductions ranging from 2.9 to 11.3% per decade [5]. The rapidly opening Northwest Passage (NWP) is expected to become a major shipping route connecting the Atlantic and Pacific Oceans by 2040 [6–10], elevating the risk of oil spills [11, 12]. Should an oil spill occur and reach Arctic shorelines, it would have profound impacts on indigenous communities and the surrounding environment [13]. Oil that becomes stranded on the surface of beach sediments can persist for an extended period, leading to long-term environmental problems [14–16]. As a result, there is growing concern regarding the remediation and minimizing impact of spilled oil on contaminated beaches.
Microorganisms are critical to the natural attenuation of oil spills, and bioremediation is a cost-effective approach in various ecosystems, including the Arctic [17, 18]. Supplementing nutrients accelerates the biodegradation process by stimulating hydrocarbon degrading microorganisms that convert aromatic and aliphatic hydrocarbons aerobically and anaerobically into the tricarboxylic acid cycle [19–21]. The degradation of aliphatic and aromatic hydrocarbons is facilitated by enzymes such as alkane monooxygenases (ex. AlkB) and naphthalene dioxygenases (ex. NDO), respectively. Arctic soils, seawater and sea ice contain hydrocarbon degraders and functional genes associated with hydrocarbon degradation, indicating the feasibility of bioremediation in sub-zero environments [20, 22, 23]. Previous research confirms widespread hydrocarbon degrading microorganisms and detectable degradation genes in high Arctic beach sediments [24]. However, degradation efficiency is highly dependent on environmental conditions, especially temperature and nutrient availability [18, 25]. Consequently, the bioremediation of hydrocarbon spills in extreme Arctic environments, such as the NWP, presents a significant challenge due to the very low metabolic rates of microorganisms at cold and sub-zero temperatures combined with nutrient scarcity.
Two commonly used methods to expedite the rate of biodegradation are biostimulation, involving the addition of nutrients to facilitate natural attenuation, and the use of chemical surfactants [18, 19]. The efficacy of inorganic, slow-release, and oleophilic fertilizers, which are frequently employed as nutrients, has been demonstrated in cold conditions [18, 20, 26]. Examples of effective biostimulation include the use of monoammonium phosphate (MAP), an inorganic fertilizer that stimulated the biodegradation process in Arctic soils [20], and S-200 Oilgone (formerly referred to as Inipol EAP22), an oleophilic fertilizer that was found to promote biodegradation on shorelines following the Exxon Valdez oil spill [27]. Surface washing agents (SWAs), such as COREXIT™ EC9580A, containing surfactants and solvents, have also been proposed as a viable approach to clean up oil-contaminated shorelines [28]. The performance of SWAs on shorelines is heavily influenced by environmental factors, such as tidal waves, currents, and temperatures [28], with colder and subzero temperatures resulting in increased oil viscosity and decreased oil bioavailability [30–32]. The implementation of these methods in Arctic beach bioremediation remains limited, and further investigation is necessary to evaluate their efficacy.
Advanced molecular and chemical techniques are increasingly employed in the development of effective bioremediation strategies. Next-generation sequencing and -omics approaches allow for a more comprehensive understanding of changes in microbial community composition and function following oil spills [26–28]. Conventional hydrocarbon-degrading microorganisms, including Pseudomonas, Colwellia, Oleispira, and Cycloclasticus, have been identified in Arctic marine environments such as seawater, sea ice, and sediments, but their distribution varies according to different environmental variables [22, 24, 29–31]. Genome-resolved metagenomics can link specific hydrocarbon-degrading taxa with functional genes [32]. One such example is Oleispira antarctica, a ubiquitous hydrocarbonoclastic bacterium thought to play a crucial role in cold marine environment oil degradation and which possesses a variety of genes related to alkane monooxygenases [33]. Nevertheless, there is a lack of systematic understanding of biodegradation processes in Arctic beaches, and little is known about the microbial community structure and response to oil contamination in these regions. Furthermore, as it is inappropriate to perform in situ studies that introduce high concentrations of oil on pristine Arctic beaches [34, 35], a handful of studies are mainly limited to investigating unpremeditated oil spills in situ, such as the Exxon Valdez and the Deepwater Horizon oil spills [36–38], as well as conducting small-scale microcosm studies using Arctic beach sediments, sea ice or seawater [22, 24, 39, 40]. Few studies have conducted in situ experiments employing materials such as slate tiles and oil-coated adsorbents to investigate degradation processes in Arctic rocky shorelines, seawater, and sea ice under natural conditions [23, 41].
In this study, we aim to address two primary questions: (1) Evaluate the potential of nutrient addition (organic and inorganic fertilizers) and SWAs as bioremediation techniques for Arctic beaches, and (2) assess their impact on the microbial community and its associated functional capacity. To address these research questions, we developed a semi-automated mesocosm system (robo-beach) that mimics the Arctic beach sediment environment. The mesocosm robo-beaches, designed in a column format, simulate the natural tidal cycle by filling and draining every 12 h with artificial seawater. Two separate laboratory mesocosm experiments were conducted to simulate hydrocarbon contamination in high-Arctic beach tidal sediments and test the efficacy of different bioremediation techniques (nutrient/SWA additions) in reducing overall hydrocarbon concentrations. We followed up these experiments by assessing the levels and diversity of hydrocarbon degradation genes and associated microorganisms in the sediments and effluents of the mesocosms using 16S rRNA gene sequencing and metagenomics. Additionally, hydrocarbon biodegradation rates were determined through total petroleum hydrocarbon (TPH) analysis. We hypothesized that (1) nutrient addition would significantly alter the microbial community by promoting the growth of hydrocarbon-degrading microorganisms and increase hydrocarbon biodegradation, and (2) SWAs would improve oil biodegradation from Arctic beach sediments by enhancing oil bioavailability. The results of this study are expected to contribute to the development of effective and sustainable strategies for oil spill remediation in Arctic coastal areas.
Methods
Sampling and setup of the laboratory mesocosm experiments
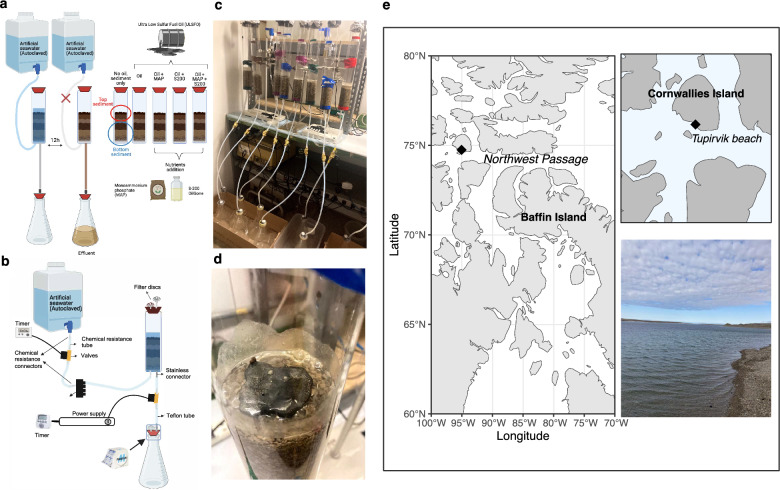
Sediments were collected from Tupirvik beach (74.74542° N, 95.03954° W) located on Cornwallis Island, Nunavut, Canada in July 2019 and Aug 2021 (Fig. 1e). Surface sediments (< 10 cm) were collected using a shovel which was carefully rinsed with ethanol before each sample collection and transferred to sterile Whirl–Pak® bags (Whirl–Pak® Flitration Group). The samples were promptly placed in a cooler and stored at − 20 °C until they were transported to the laboratory for further experiments. Two separate robo-beach mesocosm experiments were performed, with durations of 32 days (2020/9/24 to 2020/10/26) and 92 days (2022/3/1 to 2022/6/1), respectively. The basic setup of the mesocosms was consistent between the two experiments. Briefly, 600 g of sediment was placed in each column mesocosm and was contaminated with 6000 ppm of Ultra Low Sulfur Fuel Oil (ULSFO; Shell Trading Canada, 002D4509) to simulate an oil spill [36]. Sterile artificial seawater was introduced (filled from the bottom and then drained) to the columns every 12 h to simulate tidal cycles (Fig. 1a, b). Detailed information about the ULSFO can be found on the Shell website (https://www.shell.com/business-customers/marine/fuel/marine157safety-data-sheets.html). For the initial mesocosm Column Experiment I (CI), the impact of inorganic (monoammonium phosphate, MAP; Sigma) and oleophilic fertilizers (S-200 OilGone; International Environmental Products, llc) (i.e. nutrients) on hydrocarbon degradation was studied. Five treatments were established: control, oil only, oil with 0.15 g MAP, oil with 4 mL S-200 OilGone, and oil with a combination of both fertilizers (0.15 g MAP and 4 mL S-200 OilGone). Fertilizer concentrations were determined through reference to prior studies on Arctic soil microcosms and a field bioremediation assay for the Prestige oil spill [42, 43]. All treatments were performed in triplicate at 8 °C for 32 days. Time 0 sediment samples were randomly collected from the well-mixed sediments used in each column at the beginning of the experiment. Sediment samples were collected from the top (0–3 cm) and bottom (3–10 cm) of the columns at the end of the experiment. Effluent samples were periodically collected from the flasks connected to the outflow of the columns, as shown in Fig. 1a and 1c, at three specific intervals: T1 (0–14 days), T2 (15–21 days), and T3 (22–32 days). These samples were weighed and transferred to glass bottles for storage until chemical analysis. Biofilm samples were collected from the oil surface on day 32. All samples were proceeded for further microbial community, metagenome and chemical analyses.
Fig. 1.
Schematic design of robo-beach mesocosm experiments. The mesocosm Column Experiment I (CI) is depicted, with each treatment performed in triplicate (a). b A detailed illustration of the setup is provided, highlighting the integration of chemical-resistant connectors and tubes at the inlet, along with stainless connectors and Teflon tubes at the outlet to prevent chemical contamination and oil absorbance. Additionally, photographic evidence is displayed in c and d, demonstrating the actual status of columns and the solidification of loaded oil upon sediment contact at 8 °C. e A map illustrating the sampling site, Tupirvik beach, is provided, featuring coordinates and a corresponding photograph of the site
To assess the efficacy of bioremediation techniques other than nutrients, we introduced a surface washing agent (COREXIT™ EC9580A; Corexit Environmental Solutions LLC) and fertilizers (MAP and S-200 OilGone) in Column Experiment II (CII) to study their impact on oil bioremediation in Arctic beaches. Five treatments were prepared: control, oil only, oil with two nutrients (0.15 g MAP and 4 ml S-200 OilGone), oil with 1.15 mL COREXIT™ EC9580A, and a combination of nutrients and surface washing agents (0.15 g MAP, 4 ml S-200 OilGone and 1.15 mL COREXIT™ EC9580A). The concentration of COREXIT™ EC9580A was based on the company’s guidelines. All treatments were performed in triplicate at 4 °C for 92 days. Sediment samples were collected at the end of the experiment, and all samples were analyzed for microbial community and chemical analysis. Furthermore, to investigate whether oil degradation occurred under abiotic conditions, an autoclaved sediment control was established for each column experiment (CI and CII).
DNA extraction and 16S rRNA gene sequencing
DNA was extracted from sediment, effluent, and biofilm samples using the DNeasy PowerLyzer PowerSoil Kit according to the manufacturer's protocol (Qiagen, Germany). Additionally, a negative control that consisted solely of Invitrogen™ Nuclease-Free Water was extracted. The purity and yield of nucleic acids were verified via Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific) and a Qubit 4 fluorometer (Thermo Fisher Scientific). The libraries for 16S rRNA sequencing were prepared based on the Illumina 16S Metagenomic Sequencing Library Preparation protocol (Part # 15044223 Rev. B), with three modifications. First, 2 × HotStarTaq Plus Master Mix from Qiagen was used in the PCR steps. Secondly, the ratio of amplicon PCR reagents was adjusted to 7.5 μL nuclease-free water, 1.5 μL 10 μM forward primer, 1.5 μL 10 μM reverse primer, 12.5 μL HotStarTaq Plus, and 2 μL genomic DNA. Finally, the earth microbiome project primers (515F-Y (5′-GTGYCAGCMGCCGCGGTAA) and 926R (5′-CCGYCAATTYMTTTRAGTTT)) were used [44]. The beach sediment amplicons were indexed, pooled, and sequenced using Illumina Nextera XT kit following manufacturer’s instructions. The libraries were sequenced in-house on an Illumina MiSeq platform using the Illumina V3 600 cycle kit with 300 base pairs end option. Sequencing reads were analyzed using the dada2 package in R [45], with forward reads trimmed to 280 base pairs and reverse reads trimmed to 220 base pairs. The negative control showed no detectable DNA amplification by PCR, indicating negligible contamination. The decontam package in R was applied to further reduce sequence contamination [46]. The Silva v138 databases were utilized to align all sequencing reads for taxonomic classification.
Microbial composition, diversity and functional prediction from 16S rRNA gene sequencing data
R software version 4.2.1 (2022/6/23) was used to execute all data analysis and visualization using the R packages phyloseq, ggplot2, ggpubr, and vegan [47–50]. To assess alpha diversity, the Shannon index was computed, and variations were examined with ANOVA and Tukey's post hoc tests. For the first mesocosm experiment (CI), a total of 98 samples comprising sediment, effluent, and biofilm were analyzed. To analyze beta diversity, sequencing was rarefied at 10,000 per sample, and sequencing depth was determined by balancing sequence richness and sample number using rarefaction curves. Samples with less than 10,000 sequencing depth were excluded from the analysis, leaving 35 sediment samples and 41 effluent and biofilm samples. Weighted UniFrac distances were utilized, and the results were visualized in a PCoA plot. Differences in community composition were analyzed using PERMANOVA and PERMDISP tests. All analyses for the second mesocosm experiment (CII) were conducted using the same method. A total of 29 sediment samples were included in the analyses. For beta diversity of CII, rarefaction was set at 15,000 per sample, and all 29 samples were included in the analysis. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2.0 (PICRUSt2 v2.5.0) was used to predict the functional capacity of sediment samples from the 16S rRNA gene sequencing data [51]. Normalization of the gene abundance data from PICRUSt2 was performed using the cumulative sum scaling (CSS) transformation. We then utilized the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [52] to perform the analysis from the PICRUSt2 output, with specific focus on 13 KEGG Orthology (KO) numbers related to hydrocarbon degradation pathway. These KO numbers are K00496 (alkB), K14338 (cypD_E), K15760 (tmoA), K15761 (tmoB), K15764 (tmoE), K18223 (prmA), K18224 (prmC), K03380 (pheA and pheB), K16245 (dmpO), K14578 (nahAb), K14581(nahAa), K10700 (ebdA), and K07540 (bssA).
Metagenome analysis and functional annotation
Total microbial DNA was extracted from the surface sediment and biofilm samples of CI mesocosm experiment for metagenomic sequencing, using the Illumina Nextera XT kit according to the manufacturer's instructions. Metagenome sequencing was conducted by Genome Quebec using Illumina NovaSeq 6000 SP PE100. PhiX removal, adapter and low-quality base trimming of metagenomes were performed using BBDuk function of BBMap v38.86 [53]. We pooled the DNA from 28 samples to construct one metagenome for metagenomic binning. Clean reads were assembled with MEGAHIT v1.2.9 [54], and BBMap function of BBMap v38.86 was used for mapping reads to assembled contigs. Metagenomic binning was performed using MetaBAT2 v2.15 [55] and MaxBin2 v2.2.7 [56], followed by dereplication using DAS_Tool v1.1.3 [57] and dRep v3.0.0 [58]. Genome UNClutterer v1.0.5 [59] was utilized for chimerism detection, while CheckM v1.2.0 [60] was employed for assessing completeness and contamination of metagenome-assembled genomes. 146 bins were recovered, but only 65 passed the dereplication and quality control and were selected for further analysis. Taxanomy classification of MAGs was aligned with the Genome Taxonomy Database (GTDB-tk v2.1.1) [61] and the Microbial Genomes Atlas (MiGA) webserver [62]. PhyloFlash was used to examine the microbial composition of the metagenomes [63]. For the binned assembled sequences, open reading frames were predicted using Prodigal v2.6.3 [64] and annotated by eggNOG-mapper v2 [65]. KEGG databases [52] was used to search for key metabolic genes including genes of aerobic respiration coxA and ccoN, reductive TCA cycle (aclB), Wood Ljungdahl pathway (acsB), photosynthesis (rbcL), sulfur cycle (sqr, soxB, dsrA and sor), nitrogen cycle (narG, nirK, norB and nosZ), and stress response (nhaA and cspA). To identify crucial genes involved in hydrocarbon degradation both aerobically and anaerobically, we employed the Calgary approach to ANnoTating HYDrocarbon degradation genes (CANT-HYD) with a noise cut-off of E-value at 0.01 [66] for analysis across both binned and unbinned metagenomes. This approach allowed us to annotate the presence of 37 key genes, including genes of propane monooxygenase (prmAC), butane monooxygenase (pBmoABC and sBmoXYZ), alkane hydroxylase (AlkB and CYP153), flavin binding monooxygenase (AlmA), long-chain alkane hydroxylase (LadA and LadB), toluene-4-monooxygenase (TmoA, TmoB and TmoE), monoaromatic dioxygenase (MAH_alpha and MAH_beta), toluene-ortho-monooxygenase/phenol hydroxylase (TomA1, TomA3 and TomA4/DmpO), naphthalene-1,2 dioxygenase (NdoB, NdoC and non-NdoB), dibenzothiophene monooxygenase (DszC), alkylsuccinate synthase (AssA), putative alkane C2 methylene hydroxylase (AhyA), ethylbenzene dehydrogenase (EdbA and CmdA), benzylsuccinate synthase (BssA), 2-naphthylmethyl-succinate synthase (NmsA), benzene carboxylase (AbcA) and naphthalene carboxylase (K27540). For unbinned metagenomic sequences, reads were co-assembled to construct 28 metagenomes using MEGAHIT v1.2.9 9 [54] and co-assembled contigs were predicted by Prodigal v2.6.3 [64]. The number of CANT-HYD HMM hits to hydrocarbon degradation genes in each metagenome was adjusted to account for the variation in the number of protein-coding genes, using normalization of hits per million genes.
Chemical analyses
Hydrocarbons analyses were performed according to the established protocols of SGS Canada (Montreal, Quebec). For aqueous samples, the extraction of semi-volatile organic compounds (SVOCs) was carried out using dichloromethane by liquid–liquid extraction to ensure complete removal of the analytes from the water, and the quantification of SVOCs was conducted using the United States Environmental Protection Agency (EPA) Method 8270E (U.S.EPA, 2014). The analysis of petroleum hydrocarbons, including BTEX and specific PAH compounds, was performed according to the CCME Reference Method for the Canada-Wide Standard for Petroleum Hydrocarbons in Soil—Tier 1. We examined a total of nineteen PAHs, which included acenaphthene, acenaphthylene, anthracene, benzo(a)anthracene, benzo(a)pyrene, benzo(b + j)fluoranthene, benzo(ghi)perylene, benzo(k)fluoranthene, chrysene, dibenzo(a,h)anthracene, fluoranthene, fluorene, indeno(1,2,3-cd)pyrene, 1-methylnaphthalene, 2-methylnaphthalene, methylnaphthalene, 2-(1-), naphthalene, phenanthrene, and pyrene. In addition, we also examined benzene, ethylbenzene, toluene, xylene, and aliphatic hydrocarbons (C6-C50). The initial amount (μg) of each hydrocarbon group in ULSFO was determined and calculated by employing autoclaved controls. Once ULSFO was applied to the surface of the sediment column, it solidified immediately. In the autoclaved control after incubation, all the oil added was recovered from the surface layer where it was initially applied. No oil was detected in either the column effluent or the sediment underlying the surface, indicating that the oil remained intact and did not degrade or disperse into the sediment or effluent. Since the sediment was autoclaved, no biodegradation was expected. While the column was sealed with a rubber cap to minimize evaporation under low-temperature conditions, some loss was inevitable. The only potential loss could be attributed to evaporation, which is discussed further in the discussion section. After measuring the concentration (µg/g) of each group, we multiplied it by the weight of each sample to determine the actual amount (µg) present in the remaining oil and sediment. The hydrocarbon removal rate was determined by subtracting the measured amounts of each group in the remaining oil and sediment from the initial amount in ULSFO. This result was divided by the concentration (μg) of that group in ULSFO and multiplied by 100 to obtain the percentage of hydrocarbon removal.
Results
Hydrocarbon-degrading taxa dominate microbial community after addition of fertilizers and a surface washing agent
In Column Experiment I (CI) and the follow up Column Experiment II (CII), we assessed microbial diversity using alpha and beta diversity based on 16S rRNA gene sequencing after 32 days and 92 days of incubation, respectively. We analyzed sediment, effluent, and biofilm samples in CI. However, due to the limited response of the microbial community in effluent samples and the scarcity of biofilms, we excluded them from the analyses and focused on sediment samples in CII. Alpha diversity was measured by both observed ASVs and the Shannon index, with the latter revealing a significant difference (p < 0.05, one-way ANOVA) among treatments in the surface sediment and biofilm in CI (Fig. S1a, S1b and Table S3). The S-200 and S-200 with MAP addition groups displayed significantly lower Shannon index than the control and oil only groups (Fig. S1a, S1b and Table S3). In contrast, discernible differentiation based on treatment was absent in effluent samples, but temporal differences were observed (Fig. S1b). A considerable decrease in Shannon index was evident in treatments that utilized a combination of two nutrients and a surface washing agent in the surface sediment in CII (Fig. S1c and Table S3).
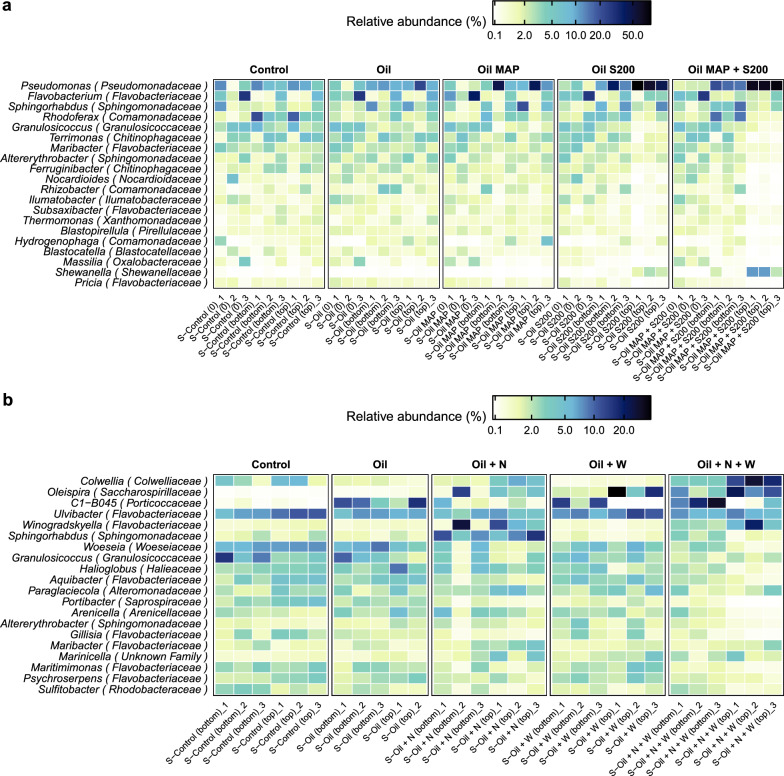
We used weighted Unifrac analysis to assess beta diversity. This revealed significant differences in community structure after treatment. In CI, the surface sediment treated with both fertilizers differed from the other samples (Fig. S1d). In CII, the groups with surface washing agent addition showed notable differences compared to the others (Fig. S1f). These differences were mainly attributed to the presence of hydrocarbon-degrading taxa. In CI, the relative abundance of surface sediment samples revealed that Pseudomonas comprised 72% of the microbial community, followed by Shewanella (8.2%), Flavobacterium (2.8%) and Sphingorhabdus (2.5%) in the S-200 with MAP treatment (Fig. 2a and Table S1). The genus Pseudomonas and Shewanella both exhibited significant increases compared to the control group, where they comprised only 8.7% and 0.15% of the community, respectively. In CII, these dominant hydrocarbon-degrading taxa, primarily Colwellia, Oleispira, and C1 − B045 (a member of Porticoccaceae family) displayed treatment-dependent distribution variations (Fig. 2b). Addition of the surface washing agent (SWA) significantly impacted the community structure, with Oleispira's abundance increasing from 0.1% in the control group to 19% and 14% in the SWA and the combined SWA and nutrient treatments, respectively. Specifically, Colwellia dominated in the group of SWA plus two nutrients, while C1 − B045 was prevalent in the oil-only group (Table S2). Distinct differentiation based on treatments was also evident in biofilm samples in CI, particularly in the groups that treated with S-200 and S-200 with MAP treatments (Fig. S1e). In congruence with sediment samples, Pseudomonas, Flavobacterium, and Sphingorhabdus were the most prevalent genera. Janthinobacterium, despite not being a well-described hydrocarbon-degrading bacterium, was ranked as the fourth dominant genus in the biofilm samples (Table S1). Unlike sediment and biofilm samples, effluent samples showed no treatment differences but varied with sampling dates (Fig. S1e).
Fig. 2.
Microbial composition differences among five treatments in two column experiments following oil amendment are depicted. Heatmaps illustrate the 20 most prevalent genera detected in sediment samples obtained from CI and CII (a, b). 'S' denotes sediment, and the treatments are labeled as follows: no oil control (Control), oil only (Oil), oil with inorganic fertilizer, MAP (Oil MAP), oil with oleophilic fertilizer, S200 (Oil S200), and oil with both fertilizers (Oil MAP + S200) for CI; and no oil control (Control), oil only (Oil), oil with both fertilizers (Oil + N), oil with SWA (Oil + W), and oil with both fertilizers and SWA (Oil + N + W) for CII. '0' represents time 0, 'top' refers to surface sediment, and 'bottom' pertains to bottom sediment. The numbers indicate three biological replicates for each condition
We observed the shift in microbial community dynamics after 32 and 92 days of exposure to treated oil and oil with nutrients. The top 20 microbial taxa significantly changed over 32 and 92 days of incubations in distinct ways. Altererythrobacter, Shingorhabdus, Granulosicoccus, and Maribacter were among the top 20 genera that were present during both the 32 and 92-day incubation periods, but with distinct relative abundance. Over the designated periods, a combination of two fertilizers induced changes in the population distribution of Altererythrobacter, Sphingorhabdus, Granulosicoccus, and Maribacter in the surface sediment compared to the control group. For example, Sphingorhabdus experienced an increase from 2.3 to 2.5% and from 1.5 to 9.4% in the combination of two fertilizers treatment compared to the control group after 32 and 92 days, respectively, while Granulosicoccus exhibited a slight decrease in both time periods, from 4.7 to 0.9% and 3.0 to 1.7% (Table S1 and S2). These taxa were also found in eight different Arctic beaches across four locations in Nunavut, Canada [24], indicating their importance as core members of the arctic marine beach microbial communities. During a 32-day incubation period, we found that four Flavobacteriaceae genera—Flavobacterium, Maribacter, Subsaxibacter, and Pricia—ranked among the top 20 most abundant. In contrast, a greater diversity of Flavobacteriaceae genera—Ulvibacter, Winogradskyella, Aquibacter, Gillisia, Maribacter, Maritimimonas, and Psychroserpens—was observed during the 92-day incubation period. Despite changes in the abundance of specific genera, the Flavobacteriaceae family maintained its position as the dominant microbial family throughout the incubation periods.
The addition of fertilizers and a surface washing agent revealed a moderate level of hydrocarbon removal in Arctic beach sediments
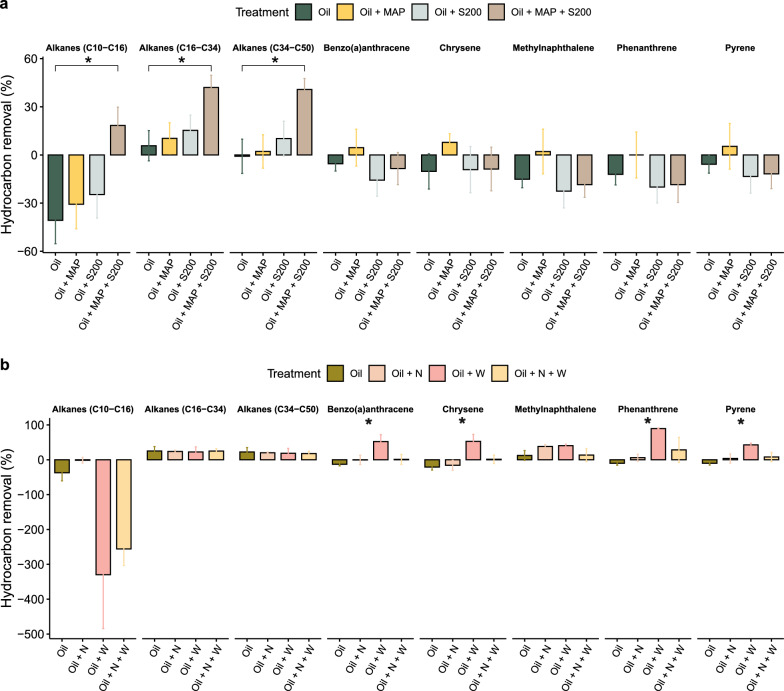
We also investigated the impact of nutrient addition and a surface washing agent (SWA) on the degradation of hydrocarbons in Arctic beach sediments after 32-day and 92-day incubations at 4 °C and 8 °C, respectively. We measured total petroleum hydrocarbons (TPH), as well as different hydrocarbon fraction short- (C10-C16), middle- (C16-C34), and long-chain (C34-C50) alkanes, and various polyaromatic compounds (PAHs). However, we only reported benzo(a)anthracene, chrysene, methylnaphthalene, 2-(1-), phenanthrene, and pyrene among the PAHs as the other compounds (i.e., fluoranthene, fluorene, and naphthalene) were not detectable in our results. The study focused on surface sediments that had direct exposure to ULSFO, while the oil content in bottom sediments was below the detection limit and thus negligible. The effluent samples from CI exhibited a range of 200 to 300 µg of short-chain alkanes (C6–C10) (Fig. S3), significantly lower in comparison to the initial oil amount over 105 µg. This difference may indicate a washing away effect due to the artificial seawater. Notably, these measurements were below the detection limit in CII. Therefore, we discounted the influence of the hydrocarbon concentration in effluents when calculating the hydrocarbon removal rate. In the CI experiment, combining MAP and S-200 fertilizers significantly reduced aliphatic hydrocarbons (p < 0.05) compared to the oil-only treatment after 32 days, but did not significantly accelerate PAH degradation (Fig. 3a). Moreover, in contrast to our findings after a 32-day incubation in CI, middle- and long-chain alkanes consistently demonstrated a removal rate ranging from 17 to 25% across all treatments in the CII experiment (Fig. 3b). The addition of SWA enhanced the degradation of benzo(a)anthracene, chrysene, phenanthrene, and pyrene, with phenanthrene showing a 90% reduction (Fig. 3b and Table S4). However, the combined application of SWA and nutrients did not provide a synergistic enhancement in the removal rate. Overall, alkane removal was observed with or without treatments over a longer period of time at 4 °C on Arctic beaches. Adding a combination of MAP and S-200 enhanced alkane degradation in immediate exposure situations such as 32 days at 8 °C, although this treatment only resulted in 40% of removal (Fig. 3a). The addition of SWA notably improved PAH degradation after 92 days at 4 °C (Fig. 3b and Table S4). These findings provide insights into the effectiveness of remediation strategies for petroleum hydrocarbon-contaminated sediments in the Arctic regions.
Fig. 3.
The assessment of hydrocarbon removal efficiency through total petroleum hydrocarbon analysis in two column experiments. a The bar chart displays a comparison of hydrocarbon removal rates in Column Experiment I (CI) for treatments: oil only (Oil), oil with inorganic fertilizer, MAP (Oil MAP), oil with oleophilic fertilizer, S200 (Oil S200), and oil with both fertilizers (Oil MAP + S200). b The bar chart illustrates hydrocarbon removal rates for treatments in Column Experiment II (CII): oil only (Oil), oil with both fertilizers (Oil + N), oil with SWA (Oil + W), and oil with both fertilizers and SWA (Oil + N + W). In panel a, the " *" indicates a significant difference (p < 0.05, t-test) between the oil only group and the oil with both fertilizers group, while in panel b, the " *" denotes a significant difference across the four different treatments (p < 0.05, ANOVA)
Deciphering the hydrocarbon-degrading potential of Arctic beaches through functional prediction using CANT-HYD and PICRUSt2
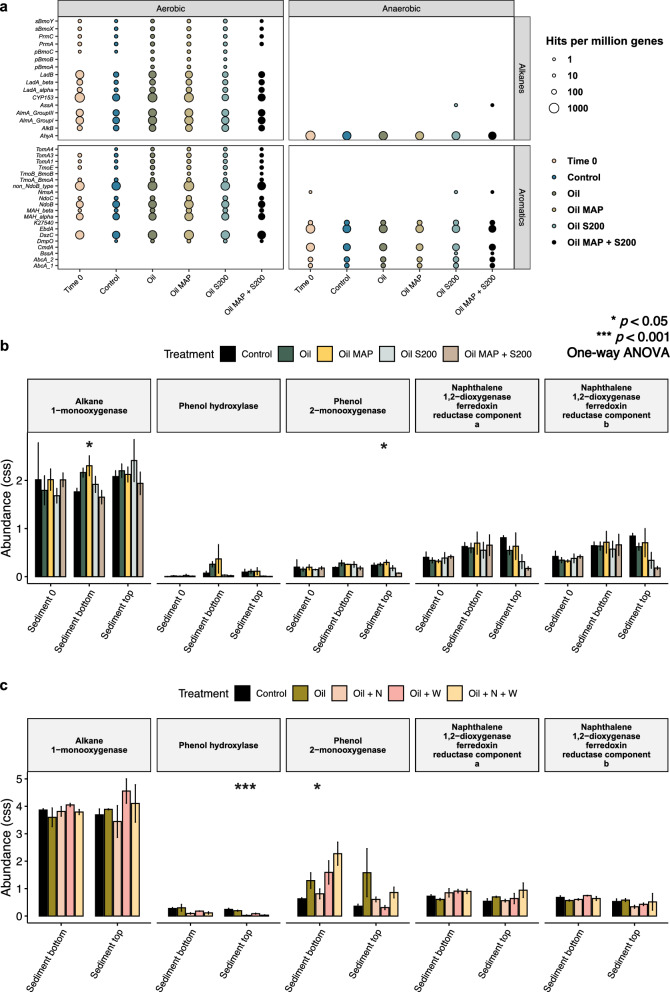
We employed metagenome analyses to investigate the functional gene capacity of Arctic sediments in CI. To validate our microbial composition results obtained from metagenomes, we used PhyloFlash to confirm consistency with 16S rRNA sequencing results at the phylum level (Fig. S4). Subsequently, we used CANT-HYD to analyze the abundance of hydrocarbon degradation genes in each metagenome and PICRUSt2 to predict the functional capacity based on 16S rRNA gene sequencing. Our results showed that hydrocarbon degradation in sediments was predominantly aerobic (Fig. 4a). Sediments across all treatments had high abundances of cytochrome P450 (CYP153) and naphthalene-1,2 dioxygenase (non-NdoB type) genes, which are crucial for aerobic alkane and PAH degradation. Genes of putative alkane C2 methylene hydroxylase (AhyA) and ethylbenzene dehydrogenase (EdbA and CmdA) were also abundant, indicating the possibility of anaerobic degradation, as these enzymes are involved in the degradation of alkanes and ethylbenzene under anaerobic conditions [67, 68]. Interestingly, the MAP and S-200 combination group showed lower levels of key biodegradation genes, including alkane 1-monooxygenase (AlkB) and cytochrome P450 (CYP153), compared to the oil-only group (p < 0.05, one-way ANOVA; Fig. 4a and Table S5), indicating that nutrient additions did not increase the abundance of these genes. Since both genes are vital for aerobic alkane degradation, this finding is unexpected, especially in light of the TPH analysis indicating a reduction in alkane levels in the MAP and S-200 combination group. Additionally, the distribution of key genes was consistent across different treatments in biofilm samples (Fig. S5).
Fig. 4.
Evaluating the hydrocarbon degradation capability of microbial communities from Arctic beach sediments after various treatments. a The metagenomes obtained from sediment samples in CI were analyzed using CANT-HYD to identify significant hydrocarbon degradation genes. The relative abundance of 5 key hydrocarbon degradation genes in CI and CII sediments was predicted using PICRUSt2 based on 16S rRNA gene amplicon sequencing data (b, c). The treatments are labeled as follows: CI—no oil control (Control), oil only (Oil), oil with inorganic fertilizer, MAP (Oil MAP), oil with oleophilic fertilizer, S200 (Oil S200), and oil with both fertilizers (Oil MAP + S200); CII—no oil control (Control), oil only (Oil), oil with both fertilizers (Oil + N), oil with SWA (Oil + W), and oil with both fertilizers and SWA (Oil + N + W). The labeling 'Sediment 0' denotes time 0, 'Sediment top' corresponds to surface sediment, and 'Sediment bottom' signifies bottom sediment. Additionally, the asterisks “*” and “***” indicate significant differences (p < 0.05, ANOVA and p < 0.001, respectively) among the four different treatments
To corroborate CANT-HYD results, PICRUSt2 was employed on 14 key hydrocarbon degradation genes, with 6 genes (alkB, pheA, pheB, dmpO, nahAa, and nahAb) showing higher abundances with an average > 0.1 while the rest were estimated to be low in abundance with an average < 0.1 (Table S6). Interestingly, alkB (alkane 1-monooxygenase), dmpO (phenol hydroxylase), nahAa, and nahAb (naphthalene 1,2-dioxygenase) showed no significant differences in abundance across treatments in surface sediments, while pheA and pheB (phenol 2-monooxygenase) demonstrated a significantly lower abundance (p < 0.05, one-way ANOVA) in the group of a combination of MAP and S-200. These findings indicate that most of the key hydrocarbon degradation genes remained relatively stable, regardless of the presence or absence of treatments. Furthermore, in CII sediments, the abundances of alkB, pheA, pheB, nahAa, and nahAb did not exhibit any significant differences in surface sediments (Fig. 4c), despite TPH analyses revealing an increase in PAH removal. This discrepancy between gene abundance and observed PAH reduction is an interesting aspect of our results that we will elaborate further in the discussion.
It is important to note that the choice of annotation method and interpretation of results should consider the inherent limitations and uncertainties in analysing environmental samples. Although the discrepancy between the CANT-HYD and PICRUSt2 methods is expected due to differences in databases and variations in sensitivity and resolution, some results exhibited a degree of similarity. For instance, when exposed to a combination of MAP and S-200, NodB genes in CANT-HYD showed a trend similar to that of nahAa and nahAb (naphthalene 1,2-dioxygenase) predicted by PICRUSt2, with a slight decrease in abundance compared to the oil-only group (p < 0.05, one-way ANOVA; p < 0.05, t-test). The CANT-HYD approach employs a specialized database for hydrocarbon degradation genes in environmental samples, while PICRUSt2 analysis uses a broader reference genome database, resulting in differences in the accuracy and completeness of gene annotations. Additionally, the CANT-HYD approach employs HMM searches and manual curation while PICRUSt2 infers functional profiles from 16S rRNA gene sequences computationally, which may vary in sensitivity and resolution, resulting in differences in gene identification and annotation.
Metagenome-assembled genomes (MAGs) indicated hydrocarbon-degrading genes are ubiquitous among microorganisms in Arctic beaches
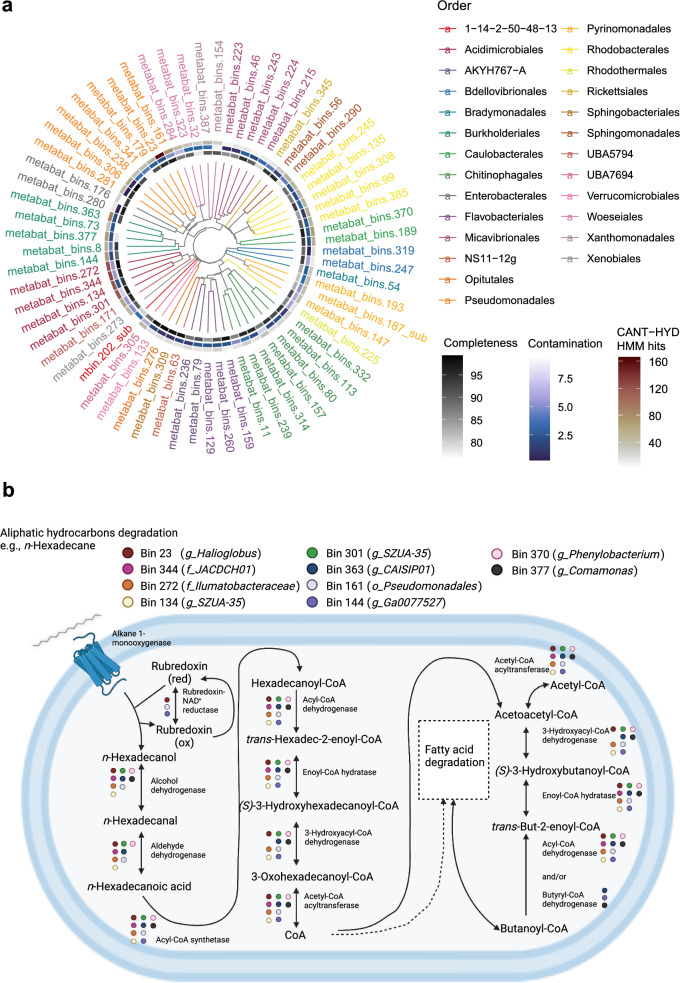
During our metagenomic investigations of key hydrocarbon degradation genes using CANT-HYD, we selected 65 medium to high-quality metagenomic bins from diverse orders (Fig. 5a and Table S7). Of the 65 MAGs, Pseudomonadales and Chitinophagales each had 7 representatives, followed by Micavibrionales, Rhodobacterales, and Burkholderiales with 5 representatives each, and Acidimicrobiales with 4 (Table S7). All All selected MAGs contained at least two key hydrocarbon genes. Pseudomonadales showed potential for both aerobic and anaerobic degradation of alkanes as all seven bins contained genes of AlkB, AlmA, and AhyA (Table S7). However, their ability to degrade PAHs was limited, as most of the MAGs had only a few genes related to PAH degradation, except for Bin 23, Halioglobus, which has 56 key genes related to aerobic PAH degradation (Table 1 and Table S7). Chitinophagales, Rhodobacterales, and Micavibrionales MAGs exhibited limited potential as primary hydrocarbon degraders, with hydrocarbon degradation gene hits around or below the average of 36.5 for 65 MAGs (Fig. 5a and Table S7). Burkholderiales and Acidimicrobiales MAGs showed the most promise as strong hydrocarbon degraders, with three of five Burkholderiales MAGs and all four Acidimicrobiales MAGs containing over 65 and 116 potential hydrocarbon degradation genes, respectively (Fig. 5a and Table S7).
Fig. 5.
Assessing phylogenetic information and hydrocarbon degradation pathways of 65 metagenome-assembled genomes (MAGs). a An unrooted phylogenetic tree was reconstructed to explore MAG diversity, with different colors representing various microbial taxonomic orders. Three inner circles indicate CANT-HYD HMM hits, contamination levels, and completeness. b The top 10 MAGs with abundant hydrocarbon degradation genes were selected to elucidate the aerobic alkane degradation pathway. The metabolic pathway of n-Hexadecane was reconstructed, and key enzymes involved in the degradation process were highlighted
Table 1.
The results of the CANT-HYD Hidden Markov Model (HMM) analysis performed on metagenome-assembled genomes (MAGs)
| Bin 23 GTDB:g_Halioglobus MiGa (AAI):Halioglobus 60.77% |
Bin 344 GTDB:f_JACDCH01 MiGa (AAI):Actinomarinicola 50.83% |
Bin 272 GTDB:f_Ilumatobacteraceae MiGa (AAI):Nocardioides 43.19% |
Bin 134 GTDB:g_SZUA35 MiGa (AAI):Micromonospora 43.60% |
Bin 301 GTDB:g_SZUA35 MiGa (AAI):Actinomarinicola 49.87% |
Bin 363 GTDB:g_CAISIP01 MiGa (AAI):Curvibacter 62.96% |
Bin 161 GTDB:o_Pseudomonadales MiGa (AAI):Spongiibacter 52.79% |
Bin 144 GTDB:g_Ga0077527 MiGa (AAI):Thauera 52.07% |
Bin 370 GTDB:g_Phenylobacterium MiGa (AAI):Phenylobacterium 59.95% |
Bin 377 GTDB:g_Comamonas MiGa (AAI):Acidovorax 83.65% |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Aerobic alkane degradation | ||||||||||
| AlkB | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 0 | 0 |
| AlmA_GroupI | 11 | 10 | 8 | 6 | 7 | 4 | 3 | 2 | 8 | 2 |
| AlmA_GroupIII | 9 | 9 | 5 | 5 | 7 | 6 | 2 | 1 | 7 | 1 |
| CYP153 | 21 | 20 | 8 | 11 | 15 | 5 | 2 | 0 | 11 | 2 |
| LadA_alpha | 10 | 9 | 4 | 9 | 8 | 1 | 2 | 1 | 2 | 4 |
| LadA_beta | 6 | 11 | 11 | 10 | 9 | 0 | 3 | 1 | 2 | 5 |
| LadB | 20 | 23 | 22 | 23 | 26 | 0 | 4 | 1 | 4 | 4 |
| pBmoB | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| pBmoC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PrmA | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| PrmC | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| sBmoX | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| sBmoY | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aerobic aromatic degradation | ||||||||||
| DmpO | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DszC | 25 | 16 | 15 | 15 | 13 | 15 | 9 | 13 | 17 | 11 |
| MAH_alpha | 9 | 3 | 7 | 3 | 2 | 5 | 5 | 1 | 1 | 4 |
| MAH_beta | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 1 |
| NdoB | 9 | 3 | 7 | 4 | 3 | 8 | 6 | 1 | 2 | 8 |
| NdoC | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 1 |
| non_NdoB_type | 11 | 5 | 9 | 6 | 4 | 13 | 9 | 1 | 3 | 9 |
| TmoA_BmoA | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 |
| TmoB_BmoB | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TmoE | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| TomA1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| TomA3 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| TomA4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anaerobic alkane degradation | ||||||||||
| AhyA | 10 | 6 | 8 | 6 | 8 | 7 | 7 | 7 | 2 | 5 |
| AssA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anaerobic aromatic degradation | ||||||||||
| AbcA_1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 1 | 0 |
| AbcA_2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 6 | 1 | 0 |
| CmdA | 9 | 5 | 7 | 5 | 6 | 6 | 6 | 6 | 1 | 4 |
| EbdA | 9 | 6 | 8 | 6 | 8 | 6 | 6 | 8 | 2 | 4 |
| K27540 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 1 | 0 |
Among the analyzed MAGs, the top 10 MAGs with the most abundant key hydrocarbon degradation genes were identified and listed
We ranked MAGs based on the quantity of hydrocarbon degradation genes. The top 10 MAGs with the highest number of essential hydrocarbon degradation genes were identified and selected for further analysis (Tables 1 and S8). Among them, Halioglobus (Pseudomonadales) displayed the highest number of key hydrocarbon degradation genes with a total of 166 genes, including DszC (25 genes), LadB (20 genes), and CYP153 (21 genes) (Table 1). Notably, Bin 134 was unique among the top 10 MAGs for including genes for propane monooxygenase (PrmAC) and butane monooxygenase (sBmoXYZ) (Table 1 and Table S7). Bin 144 showed a strong potential operating anaerobic PAH degradation, harbouring 30 key genes related to this metabolism (Table 1 and Table S7). We then conducted a detailed examination of the key aerobic alkane degradation pathway, key aerobic aromatic degradation pathway and general metabolisms using KEGG databases. This aerobic alkane degradation pathway involves the oxidation of the terminal methyl group to form primary alcohols, aldehydes, and carboxylic acids. Most of the top 10 MAGs contain an almost complete set of genes that encode key enzymes involved in this alkane degradation pathway (Fig. 5b). The search result of key genes and enzymes is not consistent in CANT-HYD and KEGG. For example, 3 out of top 10 MAGs have genes for AlkB in CANT-HYD, but none show Alkane-1-monooxygenase using KEGG. Also, the majority of the top 10 MAGs have few genes for aerobic aromatic degradation, whereas only MAG 363 (a CAISIP01 genus in Burkholderiales), harbors the nahAa and nahAb genes in KEGG; In CANT-HYD, moderate to high levels of NodB and non-NodB type genes can be detected in the top 10 MAGs. This discrepancy highlights the importance of employing a combination of two different search approaches to comprehensively understand the functional repertoire of these MAGs.
In addition to the top 10 MAGs displaying the most prominent presence of key hydrocarbon degradation genes, we have carefully selected 7 additional MAGs based on the abundances from the top 20 genera identified in CI and CII experiments (Figs. 2a, b and S2). This deliberate selection enables a comprehensive assessment of the biodegradation capacity and general metabolic traits of these MAGs in relation to the top 10 MAGs. Notably, our analysis revealed that these MAGs encompass a moderate number of hydrocarbon degradation genes, ranging from 20 to 46 (Table S9). Similar to the top 10 MAGs, these selected MAGs harbor an almost complete repertoire of genes for the alkane degradation pathway, underscoring their capacity for aliphatic hydrocarbon degradation, while exhibiting limited aromatic degradation ability (Table S8).
We systematically screened the MAGs to identify key functional genes involved in both aerobic and anaerobic respiration, nitrogen cycling, sulfur cycling, and stress response providing a comprehensive assessment of their metabolic capabilities. Our analysis also uncovered aerobic respiration as a prominent metabolic pathway, and identified genes associated with denitrification (narG, nirK, norB and nosZ) and sulfide oxidation (soxB and sqr) in these genomes (Table S8). Notably, 7 of the top 10 MAGs contain genes for both denitrification and sulfide oxidation, suggesting potential coupled processes in oxygen-limited environments [69]. A comparative analysis reveals that the 7 additional MAGs exhibit a higher proportion of genes related to nitrogen cycling as opposed to sulfur cycling, with only 28.6% of these MAGs demonstrating the presence of both nitrogen and sulfur cycling genes, in contrast to the 70% observed in the top 10 MAGs (Table S9). This variation underscores the metabolic diversity between the top 10 MAGs and the 7 additional MAGs, reflecting the metabolic versatility of microbial communities. Key genes associated with salinity tolerance (nhaA) and cold shock proteins (cspA) were commonly detected in these 17 selected MAGs, with a prevalence of 58.8% and 82.4% respectively (Table S9).
Discussion
In this study, we examined the bioremediation potential of native microbial communities in Canadian high Arctic tidal beach sediments within the Northwest Passage (NWP). Using semi-automated mesocosm systems to simulate seasonal variations and NWP tidal dynamics, along with metagenomics and hydrocarbon analysis, we found that nutrient additions enhanced alkane biodegradation but had limited effects on aromatic hydrocarbons. The addition of a surface washing agent proved to be a more reliable remediation approach, effectively removing PAHs. The CI experiment simulated summer conditions in Arctic beaches at 8 °C for 32 days, whereas the CII experiment represented spring/fall at 4 °C for 92 days. The varying temperatures and incubation times produced distinct outcomes in alkane degradation. Middle- and long-chain alkane removal was more pronounced with MAP and S200 treatment at 8 °C for 32 days compared to 4 °C for 92 days, indicating that nutrient addition was more effective under warmer conditions and shorter durations. After 92 days at 4 °C, nutrient addition did not enhance alkane degradation and showed comparable removal levels to other treatments, suggesting that nutrient addition was less effective at lower temperatures and over extended periods. Notably, a biofilm layer formed on the oil at 8 °C, whereas no growth was observed at 4 °C. The presence of the biofilm may aid in oil dispersion by reducing oil–water interfacial tension [70]. Pseudomonas, prevalent in our biofilm samples (Fig. S2) and known for hydrocarbon degradation, suggests its role in this process. However, we did not specifically examine whether the biofilm contributes to the degradation in this study.
In the TPH analysis, we recognized an unusual pattern of hydrocarbon removal, where several treatments showed a negative removal rate for some hydrocarbon fractions (Fig. 3a). The autoclaved control, collected alongside other treatments at the end of the column experiment, may have lost some hydrocarbon fractions due to evaporation during the incubation period. This loss in the autoclaved control could result in a negative value when used to standardize removal rates, as evaporation in the other treatments may have been less prevalent. Additionally, the negative removal rate in short-chain alkanes in the CI experiment could result from the breakdown of medium- or long-chain alkanes. In the CII experiment, the negative removal rate in short-chain alkanes in the surface washing agent addition groups might be attributed to the de-aromatized hydrocarbon solvent system in the surface washing agent. However, we have ruled out the possibility of free-phase oil adhering to the sides of the cores and tubing, which could potentially influence the extraction results. Due to the cold setup conditions, the oil solidified immediately upon contact with the sediment surface, with no oil sticking to the device. Moreover, interference from the surface washing agent can be excluded, as the negative removal rates are more prevalent in the CI experiment, which did not include the addition of a surface washing agent.
After a systematic examination in our study, we conclude that nutrient additions promoted alkane degradation but had minimal impact on PAH degradation in Arctic sediments. This finding aligns with the results from our previous microcosm experiment, which similarly showed that nutrient addition did not expedite PAH degradation [24]. A recent microcosm study in Labrador Sea sediments at 4 °C also showed that biostimulation did not enhance PAH biodegradation with high (1%) concentrations of diesel or crude oil [39]. We propose two possible explanations for this phenomenon. First, alkanes are relatively easy degraded and less energy costly for microorganisms. Pseudomonas or other hydrocarbon degrading bacteria can prioritize the usage of aliphatic hydrocarbons over aromatic compounds under such cold conditions. Our MAGs investigation supported the hypothesis that microorganisms in Arctic marine beach sediments often harbor alkane degradation genes, which may serve as a beneficial survival strategy in the nutrient-limited Arctic beach environment. The top 10 MAGs, along with an additional 7 selected MAGs, predominantly feature an almost complete set of genes for aerobic alkane degradation while having fewer genes related to PAH degradation pathways. This highlights their superior capability in alkane degradation compared to PAHs (Table S8). Furthermore, all 65 MAGs were found to harbor a minimum of 2 hydrocarbon-degradation genes associated with alkane degradation, indicating the widespread nature of this trait across various taxa (Table S7). Additionally, the functional prediction results associated with alkane degradation pathways displayed minimal variations across different treatments (Fig. 4c). Chemical analyses supported these genetic observations, indicating that despite 92 days of nutrient addition, there was minimal improvement in alkane degradation. The oil-only group eventually matched the degradation rate of the nutrient addition group in aliphatic hydrocarbons (Fig. 3b). These collective findings indicate that the presence of these genes is a common occurrence, potentially representing a natural adaptation to sustain life in Arctic environments. Despite the scarcity of nutrients and carbon sources in Arctic sediments, an important carbon source can be derived from cyanobacteria and algae, which produce long-chain alkanes as part of the ocean hydrocarbon cycle [71]. Furthermore, marine cold seeps in the Arctic Ocean release abundant methane, sulfide, and other reduced chemicals such as hydrogen sulfide, ammonia, reduced iron, and hydrogen [72]. A study of a cold seep near Scott Inlet in Baffin Bay, Canada, revealed that bacterial communities linked to hydrocarbon and methane oxidation contribute to reducing hydrocarbon emissions from natural geological sources [73]. A recent study in a High Arctic lake system proposed a cryptic hydrocarbon cycle (i.e., the biogenic and short-term hydrocarbon cycle) exhibited in Arctic water ecosystems and suggested that biogenic hydrocarbons may sustain a large fraction of freshwater and oceanic microbiomes in the Arctic [74]. Based on these observations, alkane degradation metabolism may serve as a crucial survival strategy in Arctic marine environments.
The second explanation supporting our finding is based on the premise that supplementing nutrients can enhance bacterial competition, favouring those with superior nutrient uptake efficiency. However, the proliferation of specific bacterial types does not always benefit the overall hydrocarbon degradation process; it can introduce additional competition and selective stress within the community. For example, the addition of nitrogen and phosphorus promotes the proliferation of specific microbial populations such as Pseudomonas and Sphingorhabdus, yet these two taxa only exhibit moderate hydrocarbon degradation ability (Fig. 2a, b, and Table S7). It is worth noting that while some Pseudomonas species can degrade PAHs [75], this function may not be universally present among Arctic Pseudomonas species. More than 50% of Pseudomonas species in our samples remain unidentified and are novel at the species level (Fig. S6). Species commonly known for their ability to degrade PAHs, such as Pseudomonas putida and Pseudomonas aeruginosa, represent only a small fraction of the overall Pseudomonas distribution in our samples (Fig. S6 and Table S10). Microbial species with a broader range of hydrocarbon degradation capabilities (e.g., Halioglobus) fail to outcompete Pseudomonas and Sphingorhabdus, and instead, they experience selective pressures in response to nutrient supplementation. This hypothesis is further supported by the functional analyses (Fig. 4a, b), which reveal that the majority of key hydrocarbon degradation genes exhibit lower abundance in the group of two nutrient additions (Fig. 4a) or maintain relatively constant abundance across various treatments (Fig. 4b, c). Hydrocarbon-degrading taxa with high abundance of key degradation genes were replaced by those with moderate abundance, enriched by nutrients. Though these competitors initially promote alkane degradation, this effect did not persist over a long period of time and did not extend to PAH degradation.
The impact of nutrient addition on bioremediation in Arctic beach sediments remains uncertain, but we cannot rule out the fact that it may still alter microbial metabolism. The study of a High Arctic lake revealed a potential connection between the cryptic hydrocarbon cycle and the sulfur and nitrogen cycles, revealing a co-occurrence pattern of key hydrocarbon degradation genes with key sulfur and nitrogen cycling genes [74]. We also observed a similar trend in our top 10 MAGs with the most abundant hydrocarbon genes. 90% of the top 10 MAGs have genes related to sulfur oxidation and denitrification and 70% of them have genes for both reactions (Table S8). Interestingly, when we looked at the other 7 MAGs with moderate abundance of hydrocarbon degradation genes, only 28.6% of them have genes for both metabolisms (Table S9). Additionally, it is plausible that the introduction of nutrients to oil-contaminated sediments may stimulate the growth of denitrifying bacteria [76]. In our study, the population of Rhodoferax, a typical denitrifier, which had the highest copy number of denitrification genes among our selected MAGs, increased in the bottom sediments after nutrient addition (Table S8 and Fig. 2a). Contrary to prior findings suggesting denitrifiers' capability to aid hydrocarbon degradation under oxygen-limited conditions [76–79], our results did not conclusively show that the increase of Rhodoferax led to increased hydrocarbon degradation. Our recent study demonstrated that adding nutrients did not clearly enhance the degradation of marine diesel, rather it showed a protection effect on maintaining baseline microbial activity when oil is present [80]. However, a comprehensive investigation is required to decipher the intricate relationship between hydrocarbon degradation and nutrient cycles.
Although the addition of nutrients did not exhibit promising results as a bioremediation approach, we propose exploring the following potential solutions for further investigation. The use of a surface washing agent (SWA) has been proposed as a useful approach to improve shoreline clean-up by effectively removing oil adhered to solid surfaces.[81–83]. Our study revealed that the incorporation of a SWA significantly enhanced the removal of PAHs from oil-contaminated sediments (Fig. 3b). This increased rate of removal could be attributed to the proliferation of the well-known cold-adapted marine hydrocarbon degrader, Oleispira [84, 85]. Notably, the abundance of this taxon increased by over tenfold compared to the treatment with oil alone. Considering such a significant augmentation of recognized hydrocarbon-degrading bacterium, one would expect a corresponding elevation in key hydrocarbon degradation genes, including those responsible for naphthalene 1,2-dioxygenase biodegradation. However, no significant increase was observed across treatments (Fig. 4c). The absence of significant changes in hydrocarbon degradation gene abundance does not imply unchanged functionality. Our TPH analysis demonstrates a clear enhancement in PAH degradation upon the introduction of SWA, surpassing the oil-only treatment. One plausible explanation for this activation is that SWA emulsifies and enhances the bioavailability of oil [81, 86], facilitating its accessibility to microorganisms for utilization and enabling the function of their PAH degradation genes. The presence of surfactant molecules at the oil–water interface reduces the interfacial tension and increases the surface tension of the oil [87]. This effect is particularly important in cold environments, as oil aggregates together and decreases the surface tension at cold temperatures [25]. The observed enhancement in PAH degradation following the introduction of SWA may be attributed to changes in the expression levels of degradation genes rather than alterations at the genetic level. Without metatranscriptomic analysis, we can only infer that SWA induces different responses in degradation gene DNA and RNA profiles. In a previous microcosm study, discrepancies between metagenomic and metatranscriptomic data indicated that certain hydrocarbon degradation genes exhibited high expression levels despite low gene abundance [85]. Further research is necessary to elucidate how SWA impact the expression of key hydrocarbon degradation genes. Unfortunately, the addition of nutrients with SWA showed no synergistic effect in hydrocarbon degradation (Fig. 3b). This result further supports our previous assumption that the addition of nutrients introduces more competition and selective stress to the environment, which may not necessarily lead to beneficial outcomes for bioremediation in Arctic beach environments. Although Colwellia, a renowned marine cold-adapted hydrocarbon degrader [30, 88], significantly increased in abundance after nutrient and SWA addition (Fig. 2b), it did not contribute to more PAH degradation compared to the SWA addition group in the TPH analysis (Fig. 3b). However, its potential significance in PAH removal cannot be dismissed, necessitating further study to understand its metabolism when both nutrient and SWA are present.
The second possible solution is to explore the biodegradation capacity of unconventional or novel hydrocarbon-degrading microorganisms. From the observation of the predominant genus from biofilm samples, Janthinobacterium demonstrated potential for crude oil hydrocarbon bioremediation. Janthinobacterium is reported to degrade alkanes, PAHs, and petroleum compounds in oil-contaminated soils and water [65, 66], while also producing biosurfactants that enhance hydrocarbon availability for microbial degradation, making it a promising candidate for oil-contaminated environments bioremediation [67]. Our MAGs result also indicated that Halioglobus and Acidimicrobiales genera could be crucial in decontaminating hydrocarbon pollution in Arctic beaches due to their high presence of key hydrocarbon degradation genes (e.g., CYP153, LadA, and NodB genes) in their genomes (Tables 1 and S7). Lack of sufficient representatives in the existing databases, presenting a challenge in assigning them to the same taxonomy group (especially below the family level) using different taxanomy databases such as GTDB and MiGA. For example, Bin 344, ranked second with a total of 126 potential genes, belongs to the family JACDCH01 (Acidimicrobiales) according to GTDB-tk classification. Nevertheless, through a search in MiGA, Bin 344 was more closely related to Actinomarinicola tropica (Acidimicrobiales), with an average amino acid identity (AAI) of 50.83% (Table 1). Similarly, six other bins exhibited the same inconsistency in family or species levels using different taxonomy classification databases (Table 1). All the aforementioned emphasizes the enigmatic nature of the microbial community within this environment. Moreover, the metabolic processes of the microbiome and their role in hydrocarbon degradation within this specific environment remain unclear. Some limited studies have been conducted on Halioglobus, a halophilic bacteria [89], which was discovered in a hydrocarbon-contaminated Gulf of Mexico sediment sample, strongly suggesting its potential role in hydrocarbon degradation [90]. Most of studies pertaining to Acidimicrobiales and their proficiency in oil degradation have been conducted in soil environments [91, 92]. Several taxa within this order have been found to increase their abundance in hydrocarbon contamination, but only a few studies have been conducted in marine environments [93, 94]. Additionally, Bin 144 Ga0077527 (Burkholderiales) may degrade hydrocarbons in anaerobic conditions based on our MAGs survey (Table 1). Bin 144 exhibits a limited number of genes for aerobic alkane degradation and only a single copy of the essential aerobic respiration gene, coxA, suggesting constrained competitiveness as aerobic hydrocarbon degraders (Table S8). However, upon closer examination of the anaerobic-related hydrocarbon degradation genes in Bin 144, the K27540 gene was found to be relatively abundant; this gene encodes naphthalene carboxylase, a key enzyme in anaerobic naphthalene degradation (Table 1), suggesting that Bin 144 exhibits traits indicative of functioning as a facultative anaerobe and an anaerobic hydrocarbon degrader. Further comprehensive research is needed to fully understand the capabilities of these novel hydrocarbon degraders in the hydrocarbon degradation processes. To further elucidate and confirm the biodegradative capabilities of these bacteria and their potential role in bioremediation in Arctic marine beaches, a combination of culture-dependent (isolating and culturing aerobic/anaerobic hydrocarbon-degrading bacteria) and culture-independent techniques (metatranscriptomic analyses) should be employed. We isolated 22 strains from the CI experiment, with five matching the MAGs identified in this study (Bins 56, 179, 245, 290, and 341) at the genus level. These strains exhibit notable hydrocarbon degradation abilities, confirmed by whole-genome sequencing and TPH analysis [95]. This result signifies the advantages and the effectiveness of employing a combined approach.
Conclusions
Arctic coastal sediments are dynamic environments influenced by extreme cold, oligotrophy, wave energy, sea-ice extent, and coverage duration [96, 97]. Consequently, microorganisms in these sediments exhibit metabolic versatility, performing both aerobic and anaerobic metabolisms to survive fluctuating conditions. To effectively remediate potential oil spills on Arctic beaches, sophisticated research methods combining chemical and advanced molecular approaches are necessary. Examining key functional genes is crucial for understanding hydrocarbon degradation by atypical degraders. More specific and sophisticated targeting databases and bioinformatics tools are needed to unravel complex hydrocarbon degradation processes, given the limitations of current methods. While PICRUSt2 and CANT-HYD show promise in predicting functional genes from 16S rRNA sequencing data and metagenomes, metatranscriptome analyses are necessary for assessing microbial gene expression. To fully evaluate the hydrocarbon degradation capacity of microbial communities, both culture-independent and culture-dependent methods are required. Isolating and culturing unconventional hydrocarbon-degrading microorganisms is also critical to studying their metabolisms and degradation abilities. In Arctic environments, the cold temperatures limit the activity of hydrocarbon-degrading microorganisms, resulting in very slow biodegradation process for oil spills in Arctic beaches. This slow rate underscores the urgency of immediate removal approaches to minimize environmental damage. Mechanical or chemical clean-up methods effectively remove oil spills but are costly and may have adverse environmental effects. Bioremediation is often considered the most cost-effective approach for treating oil contamination. However, our study indicates that adding nutrients, a commonly used biostimulation method, has limited effectiveness in facilitating the biodegradation process. Our research suggests that applying a surface washing agent, an effective shoreline cleaner, shows potential as a bioremediation method. This agent emulsifies oil, aiding its removal from beaches and facilitating its dispersion into seawater. Despite potentially lower toxicity compared to dispersants, the environmental impact of surface washing agents and released oil entering the seawater cannot be ignored. Immediate recovery methods, such as sorbents or skimmers, should be applied to prevent water column contamination. Further in-depth in situ experiments are needed to assess the feasibility of using such chemical reagents. In conclusion, with the inevitable receding of sea ice and the increasing maritime traffic in the Northwest Passage (NWP), it is critical to raise awareness and preparedness for potential oil spills in the Arctic. Developing comprehensive strategies that combine preventive and responsive measures will be crucial for effectively mitigating the environmental impacts of oil spills. Above all, implementing measures to restrain traffic in the NWP and Arctic regions to prevent oil contamination on beaches would be the best approach, as the removal of oil in these remote areas would be extremely difficult and costly.
Supplementary Information
Supplementary Material 1: Figure S1. Alpha and beta diversity were used to analyze the microbial diversity of microorganisms from the sediment samples. Alpha diversity was evaluated by analyzing observed ASVs and the Shannon index for CI's sediment (a), effluent, and biofilm (b), and CII's sediment (c) samples. Beta diversity was assessed using the Weighted-Unifrac metric, presenting PCoA plots for CI's sediment (d), effluent, and biofilm (e), and CII's sediment (f) samples.
Supplementary Material 2: Figure S2. The heatmap depicts the relative abundance of the top 20 most prevalent genera in CI's effluent and biofilm samples, offering a graphical representation of their distribution patterns.
Supplementary Material 3: Figure S3. Total petroleum analysis of effluent samples from CI. Effluent samples were periodically collected on day 14 and 32, enabling quantification and comparison of hydrocarbon concentrations across various treatment conditions.
Supplementary Material 4: Figure S4. Comparative analysis of microbial composition at the phylum level in sediment and biofilm samples using metagenome and 16S rRNA sequencing data. Microbial communities were generated by PhyloFlash from metagenome data (a) and 16S rRNA sequencing data (b). 'S' denotes sediment, ‘Bio’ denotes biofilm and the treatments are labeled as follows: no oil control (Control), oil only (Oil), oil with inorganic fertilizer, MAP (Oil MAP), oil with oleophilic fertilizer, S200 (Oil S200), and oil with both fertilizers (Oil MAP + S200).
Supplementary Material 5: Figure S5. The identification of key hydrocarbon degradation genes in each biofilm metagenome using the CANT-HYD approach for gene search. The treatments are labeled as follows: oil only (Oil), oil with inorganic fertilizer, MAP (Oil MAP), oil with oleophilic fertilizer, S200 (Oil S200), and oil with both fertilizers (Oil MAP + S200).
Supplementary Material 6: Figure S6. Relative abundance of various Pseudomonas species in sediment, biofilm, and effluent samples collected from CI under different treatments. 'R' represents sediment samples; 'E' signifies effluent samples; 'Bio' denotes biofilm samples. 'T0' refers to Day 0, 'T2' refers to the period from 15 to 21 days, and 'T3' from 22 to 32 days. 'T' designates surface sediments, while 'B' indicates bottom sediments.
Supplementary Material 7: Table S1. ASV table, including raw data and relative abundance, in sediment and effluent samples from CI experiment.
Supplementary Material 8: Table S2. ASV table, including raw data and relative abundance, in sediment and effluent samples from CII experiment.
Supplementary Material 9: Table S3. Alpha diversity and statistical analysis of microbial communities in sediment samples from CI and CII experiments.
Supplementary Material 10: Table S4. Quantification of total petroleum hydrocarbons (TPH) and statistical analysis for CI and CII experiment samples.
Supplementary Material 11: Table S5. Identification of 37 key hydrocarbon degradation genes from CANT-HYD search results in 28 metagenomes from CI experiment.
Supplementary Material 12: Table S6. Prediction of key gene abundance through PICRUSt2 in CI and CII experiment samples.
Supplementary Material 13: Table S7. 37 key hydrocarbon degradation genes identified in 65 metagenome-assembled genomes (MAGs) from the CI experiment via CANT-HYD search.
Supplementary Material 14: Table S8. Key metabolic genes, involving respiration, nitrogen cycles, sulfur cycles, and hydrocarbon degradation, in selected MAGs via KEGG database.
Supplementary Material 15: Table S9. Comparative analysis of CANT-HYD and KEGG database search results between top 10 MAGs and 7 other selected MAGs.
Supplementary Material 16: Table S10. Assessment of Pseudomonas relative abundance in sediment samples from the CI experiment.
Supplementary Material 17: Table S11 Metadata information associated with the 16S rRNA sequencing data for the CI and CII column experiments, the metagenome sequencing data, and the metagenome-assembled genomes for the CI column experiment.
Acknowledgements
The surface washing agent, COREXIT™ EC9580A, used in this study was kindly provided by Dr. Chunjiang An at the Department of Building, Civil, and Environmental Engineering at Concordia University.
Abbreviations
- NWP
The Northwest Passage
- MAP
Monoammonium phosphate
- S-200
S-200 OilGone
- ULSFO
Ultra low sulfur fuel oil
- SWA
Surface washing agent
- PAHs
Polyaromatic compounds
- TPH
Total petroleum hydrocarbon
- CANT-HYD
The Calgary approach to ANnoTating HYDrocarbon degradation genes
- KEGG
The Kyoto Encyclopedia of Genes and Genomes database
- PICRUSt2
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2.0
- MAGs
Metagenome-assembled genomes
Author contributions
YJC and IA conceptualized and designed the study. YJC and NJF performed the experiments. YJC, NJF, EG and AL participated in fieldwork and sample collection. YJC analyzed and visualized the data. YJC wrote the first draft of the manuscript. IA and LGW provided comments and revised the manuscript. CWG and LGW supervised the study. LGW administered the funding support. All authors have read and agreed to the final version of the manuscript.
Funding
This study was funded by Fisheries and Oceans Canada under the Multi-Partner Research Initiative and supported by the Polar Continental Shelf Program.
Availability of data and materials
The 16S rRNA sequencing data for the CI and CII column experiments were deposited at the National Center for Biotechnology Information (NCBI) under the BioSample accession numbers SAMN40210342 and SAMN40243382, respectively, within the BioProject accession number PRJNA1082469. Additionally, the metagenomic sequence data for the CI column experiment was deposited at NCBI under the SRA accession number from SRX24007725 to SRX24007752, within the BioProject accession number PRJNA1086230. Furthermore, the metagenome-assembled genomes were deposited at NCBI under the BioSample accession numbers ranging from SAMN40570777 to SAMN40570841, all within the BioProject accession number PRJNA1086230. Metadata information associated with the sequencing data has been attached as a supplementary table (Table S11).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bush EaL DS, editors: Canada’s Changing Climate Report. 2019.
- 2.Zhang X, Vincent LA, Hogg WD, Niitsoo A. Temperature and precipitation trends in Canada during the 20th century. Atmos Ocean. 2000;38:395–429. [Google Scholar]
- 3.Academies CoC: Canada’s Top Climate Change Risks, Ottawa (ON): The Expert Panel on Climate Change Risks and Adaptation Potential. 2019.
- 4.Lemmen DS, Warren FJ, Lacroix J, Bush E, editors: From Impacts to Adaptation: Canada in a Changing Climate 2007. 2008.
- 5.Tivy A, Howell SEL, Alt B, McCourt S, Chagnon R, Crocker G, Carrieres T, Yackel JJ. Trends and variability in summer sea ice cover in the Canadian Arctic based on the Canadian Ice Service Digital Archive, 1960–2008 and 1968–2008. J Geophys Res (Oceans). 2011;116:C03007. [Google Scholar]
- 6.Melling H. Sea ice of the northern Canadian Arctic Archipelago. J Geophys Res Oceans. 2002;107:2-1–2-21. [Google Scholar]
- 7.Howell SEL, Duguay CR, Markus T. Sea ice conditions and melt season duration variability within the Canadian Arctic Archipelago: 1979–2008. Geophys Res Lett. 2009;36:L10502. [Google Scholar]
- 8.Smith LC, Stephenson SR. New trans-Arctic shipping routes navigable by midcentury. Proc Natl Acad Sci. 2013;110:E1191–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khon VC, Mokhov II, Latif M, Semenov VA, Park W. Perspectives of Northern Sea Route and Northwest Passage in the twenty-first century. Clim Change. 2010;100:757–68. [Google Scholar]
- 10.Melia N, Haines K, Hawkins E. Sea ice decline and 21st century trans-Arctic shipping routes. Geophys Res Lett. 2016;43:9720–8. [Google Scholar]
- 11.Afenyo M, Ng AKY, Jiang C. A multiperiod model for assessing the socioeconomic impacts of oil spills during arctic shipping. Risk Anal. 2022;42:614–33. [DOI] [PubMed] [Google Scholar]
- 12.Afenyo M, Jiang C, Ng AKY. Climate change and Arctic shipping: a method for assessing the impacts of oil spills in the Arctic. Transp Res Part D Transp Environ. 2019;77:476–90. [Google Scholar]
- 13.Frisso GM. Vulnerability, arctic indigenous groups and oil spills: potential contributions to the work of the Arctic Council. Yearb Int Disaster Law Online. 2022;3:352–73. [Google Scholar]
- 14.Challenger GE, Gmur S, Taylor E. A review of Gulf of Mexico coastal marsh erosion studies following the 2010 Deepwater Horizon oil spill and comparison to over 4 years of shoreline loss data from Fall 2010 to Summer 2015. Mar Pollut Bull. 2021;164: 111983. [DOI] [PubMed] [Google Scholar]
- 15.Asif Z, Chen Z, An C, Dong J. Environmental impacts and challenges associated with oil spills on shorelines. J Mar Sci Eng. 2022;10:762. [Google Scholar]
- 16.Owens EH, Taylor E, Humphrey B. The persistence and character of stranded oil on coarse-sediment beaches. Mar Pollut Bull. 2008;56:14–26. [DOI] [PubMed] [Google Scholar]
- 17.Aislabie J, Saul DJ, Foght JM. Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles. 2006;10:171–9. [DOI] [PubMed] [Google Scholar]
- 18.Góngora E, Chen Y-J, Ellis M, Okshevsky M, Whyte L. Hydrocarbon bioremediation on Arctic shorelines: historic perspective and roadway to the future. Environ Pollut. 2022;305: 119247. [DOI] [PubMed] [Google Scholar]
- 19.Thomassin-Lacroix EJ, Eriksson M, Reimer KJ, Mohn WW. Biostimulation and bioaugmentation for on-site treatment of weathered diesel fuel in Arctic soil. Appl Microbiol Biotechnol. 2002;59:551–6. [DOI] [PubMed] [Google Scholar]
- 20.Yergeau E, Sanschagrin S, Beaumier D, Greer CW. Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high Arctic soils. PLoS ONE. 2012;7: e30058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leahy JG, Colwell RR. Microbial degradation of hydrocarbons in the environment. Microbiol Rev. 1990;54:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garneau M-È, Michel C, Meisterhans G, Fortin N, King TL, Greer CW, Lee K. Hydrocarbon biodegradation by Arctic sea-ice and sub-ice microbial communities during microcosm experiments, Northwest Passage (Nunavut, Canada). FEMS Microbiol Ecol. 2016;92:fiw130. [DOI] [PubMed] [Google Scholar]
- 23.Vergeynst L, Christensen JH, Kjeldsen KU, Meire L, Boone W, Malmquist LMV, Rysgaard S. In situ biodegradation, photooxidation and dissolution of petroleum compounds in Arctic seawater and sea ice. Water Res. 2019;148:459–68. [DOI] [PubMed] [Google Scholar]
- 24.Ellis M, Altshuler I, Schreiber L, Chen YJ, Okshevsky M, Lee K, Greer CW, Whyte LG. Hydrocarbon biodegradation potential of microbial communities from high Arctic beaches in Canada’s Northwest Passage. Mar Pollut Bull. 2022;174: 113288. [DOI] [PubMed] [Google Scholar]
- 25.Das N, Chandran P. Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Int. 2011;2011: 941810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yergeau E, Maynard C, Sanschagrin S, Champagne J, Juck D, Lee K, Greer CW. Microbial community composition, functions, and activities in the Gulf of Mexico 1 year after the deepwater horizon accident. Appl Environ Microbiol. 2015;81:5855–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dombrowski N, Donaho JA, Gutierrez T, Seitz KW, Teske AP, Baker BJ. Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat Microbiol. 2016;1:16057. [DOI] [PubMed] [Google Scholar]
- 28.Atlas RM, Stoeckel DM, Faith SA, Minard-Smith A, Thorn JR, Benotti MJ. Oil biodegradation and oil-degrading microbial populations in marsh sediments impacted by oil from the deepwater horizon well blowout. Environ Sci Technol. 2015;49:8356–66. [DOI] [PubMed] [Google Scholar]
- 29.Ribicic D, Netzer R, Winkler A, Brakstad OG. Microbial communities in seawater from an Arctic and a temperate Norwegian fjord and their potentials for biodegradation of chemically dispersed oil at low seawater temperatures. Mar Pollut Bull. 2018;129:308–17. [DOI] [PubMed] [Google Scholar]
- 30.Peeb A, Dang NP, Truu M, Nõlvak H, Petrich C, Truu J. Assessment of hydrocarbon degradation potential in microbial communities in Arctic Sea Ice. Microorganisms. 2022;10:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzo C, Malavenda R, Gerçe B, Papale M, Syldatk C, Hausmann R, Bruni V, Michaud L, Lo Giudice A, Amalfitano S. Effects of a simulated acute oil spillage on bacterial communities from Arctic and Antarctic marine sediments. Microorganisms. 2019;7:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somee MR, Amoozegar MA, Dastgheib SMM, Shavandi M, Maman LG, Bertilsson S, Mehrshad M. Genome-resolved analyses show an extensive diversification in key aerobic hydrocarbon-degrading enzymes across bacteria and archaea. BMC Genom. 2022;23:690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kube M, Chernikova TN, Al-Ramahi Y, Beloqui A, Lopez-Cortez N, Guazzaroni M-E, Heipieper HJ, Klages S, Kotsyurbenko OR, Langer I, et al. Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica. Nat Commun. 2013;4:2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sergy GA, Blackall PJ. Design and conclusions of the Baffin island oil spill project. Arctic. 1987;40:1–9. [Google Scholar]
- 35.Prince RC, Owens EH, Sergy GA. Weathering of an Arctic oil spill over 20 years: the BIOS experiment revisited. Mar Pollut Bull. 2002;44:1236–42. [DOI] [PubMed] [Google Scholar]
- 36.Bragg JR, Prince RC, Harner EJ, Atlas RM. Effectiveness of bioremediation for the Exxon Valdez oil spill. Nature. 1994;368:413–8. [Google Scholar]
- 37.Pritchard PH, Mueller JG, Rogers JC, Kremer FV, Glaser JA. Oil spill bioremediation: experiences, lessons and results from the Exxon Valdez oil spill in Alaska. Biodegradation. 1992;3:315–35. [Google Scholar]
- 38.Atlas RM, Hazen TC. Oil biodegradation and bioremediation: a tale of the two worst spills in U.S. history. Environ Sci Technol. 2011;45:6709–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy SMC, Bautista MA, Cramm MA, Hubert CRJ. Diesel and crude oil biodegradation by cold-adapted microbial communities in the labrador sea. Appl Environ Microbiol. 2021;87: e0080021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nõlvak H, Dang NP, Truu M, Peeb A, Tiirik K, O’Sadnick M, Truu J. Microbial community dynamics during biodegradation of crude oil and its response to biostimulation in svalbard seawater at low temperature. Microorganisms. 2021;9:2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gustavson K, Hansson SV, van Beest FM, Fritt-Rasmussen J, Lassen P, Geertz-Hansen O, Wegeberg S. Natural removal of crude and heavy fuel oil on rocky shorelines in Arctic climate regimes. Water Air Soil Pollut. 2020;231:479. [Google Scholar]
- 42.Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Greer CW. Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J. 2013;7:1200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiménez N, Viñas M, Sabaté J, Díez S, Bayona JM, Solanas AM, Albaiges J. The prestige oil spill. 2. Enhanced biodegradation of a heavy fuel oil under field conditions by the use of an oleophilic fertilizer. Environ Sci Technol. 2006;40:2578–85. [DOI] [PubMed] [Google Scholar]
- 44.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. [DOI] [PubMed] [Google Scholar]
- 45.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valero-Mora PM. ggplot2: elegant graphics for data analysis. J Stat Softw Book Rev. 2010;35:1–3. [Google Scholar]
- 49.Kassambara A: ggpubr: 'ggplot2' Based Publication Ready Plots. R package version 0.6.0. https://rpkgsdatanovia.com/ggpubr/ 2023.
- 50.Oksanen J: Vegan: community ecology package. R package version 2. 6–4. http://CRANR-project.org/package=vegan 2022.
- 51.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38:685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44:D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bushnell B: BBMap: A Fast, Accurate, Splice-Aware Aligner. https://www.sourceforgenet/projects/bbmap 2014.
- 54.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. [DOI] [PubMed] [Google Scholar]
- 55.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7: e7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2015;32:605–7. [DOI] [PubMed] [Google Scholar]
- 57.Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. Isme J. 2017;11:2864–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orakov A, Fullam A, Coelho LP, Khedkar S, Szklarczyk D, Mende DR, Schmidt TSB, Bork P. GUNC: detection of chimerism and contamination in prokaryotic genomes. Genome Biol. 2021;22:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2019;36:1925–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez RL, Gunturu S, Harvey WT, Rosselló-Mora R, Tiedje JM, Cole JR, Konstantinidis KT. The Microbial Genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018;46:W282-w288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gruber-Vodicka HR, Seah BKB, Pruesse E. phyloFlash: rapid small-subunit rRNA profiling and targeted assembly from metagenomes. Systems. 2020;5:e00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 2021;38:5825–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khot V, Zorz J, Gittins DA, Chakraborty A, Bell E, Bautista MA, Paquette AJ, Hawley AK, Novotnik B, Hubert CRJ, et al. CANT-HYD: a curated database of phylogeny-derived hidden Markov models for annotation of marker genes involved in hydrocarbon degradation. Front Microbiol. 2021;12: 764058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spormann AM, Widdel F. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation. 2000;11:85–105. [DOI] [PubMed] [Google Scholar]
- 68.Boll M, Heider J. Anaerobic degradation of hydrocarbons: mechanisms of C-H-bond activation in the absence of oxygen. In: Timmis KN, editor. Handbook of hydrocarbon and lipid microbiology. Berlin: Springer; 2010. p. 1011–24. [Google Scholar]
- 69.Callbeck CM, Lavik G, Ferdelman TG, Fuchs B, Gruber-Vodicka HR, Hach PF, Littmann S, Schoffelen NJ, Kalvelage T, Thomsen S, et al. Oxygen minimum zone cryptic sulfur cycling sustained by offshore transport of key sulfur oxidizing bacteria. Nat Commun. 2018;9:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Omarova M, Swientoniewski LT, Mkam Tsengam IK, Blake DA, John V, McCormick A, Bothun GD, Raghavan SR, Bose A. Biofilm formation by hydrocarbon-degrading marine bacteria and its effects on oil dispersion. ACS Sustain Chem Eng. 2019;7:14490–9. [Google Scholar]
- 71.Love CR, Arrington EC, Gosselin KM, Reddy CM, Van Mooy BAS, Nelson RK, Valentine DL. Microbial production and consumption of hydrocarbons in the global ocean. Nat Microbiol. 2021;6:489–98. [DOI] [PubMed] [Google Scholar]
- 72.Åström EKL, Carroll ML, Ambrose WG Jr, Sen A, Silyakova A, Carroll J. Methane cold seeps as biological oases in the high-Arctic deep sea. Limnol Oceanogr. 2018;63:S209–31. [Google Scholar]
- 73.Cramm MA, Neves BM, Manning CCM, Oldenburg TBP, Archambault P, Chakraborty A, Cyr-Parent A, Edinger EN, Jaggi A, Mort A, et al. Characterization of marine microbial communities around an Arctic seabed hydrocarbon seep at Scott Inlet, Baffin Bay. Sci Total Environ. 2021;762:143961. [DOI] [PubMed] [Google Scholar]
- 74.Vigneron A, Cruaud P, Lovejoy C, Vincent WF. Genomic insights into cryptic cycles of microbial hydrocarbon production and degradation in contiguous freshwater and marine microbiomes. Microbiome. 2023;11:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whyte LG, Bourbonniére L, Greer CW. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl Environ Microbiol. 1997;63:3719–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang K, Hu Z, Zeng F, Yang X, Wang J, Jing R, Zhang H, Li Y, Zhang Z. Biodegradation of petroleum hydrocarbons and changes in microbial community structure in sediment under nitrate-, ferric-, sulfate-reducing and methanogenic conditions. J Environ Manag. 2019;249: 109425. [DOI] [PubMed] [Google Scholar]
- 77.Hutchins SR, Sewell GW, Kovacs DA, Smith GA. Biodegradation of aromatic hydrocarbons by aquifer microorganisms under denitrifying conditions. Environ Sci Technol. 1991;25:68–76. [Google Scholar]
- 78.Wilkes H, Buckel W, Golding BT, Rabus R. Metabolism of hydrocarbons in n-alkane-utilizing anaerobic bacteria. J Mol Microbiol Biotechnol. 2016;26:138–51. [DOI] [PubMed] [Google Scholar]
- 79.Foght J. Anaerobic biodegradation of aromatic hydrocarbons: pathways and prospects. J Mol Microbiol Biotechnol. 2008;15:93–120. [DOI] [PubMed] [Google Scholar]
- 80.Durand M, Touchette D, Chen Y-J, Magnuson E, Wasserscheid J, Greer CW, Whyte LG, Altshuler I. Effects of marine diesel on microbial diversity and activity in high Arctic beach sediments. Mar Pollut Bull. 2023;194: 115226. [DOI] [PubMed] [Google Scholar]
- 81.Chen Z, An C, Boufadel M, Owens E, Chen Z, Lee K, Cao Y, Cai M. Use of surface-washing agents for the treatment of oiled shorelines: research advancements, technical applications and future challenges. Chem Eng J. 2020;391: 123565. [Google Scholar]
- 82.Bi H, An C, Chen X, Owens E, Lee K. Investigation into the oil removal from sand using a surface washing agent under different environmental conditions. J Environ Manag. 2020;275: 111232. [DOI] [PubMed] [Google Scholar]
- 83.Bi H, Mulligan CN, Zhang B, Biagi M, An C, Yang X, Lyu L, Chen X. A review on recent development in the use of surface washing agents for shoreline cleanup after oil spills. Ocean Coast Manag. 2023;245: 106877. [Google Scholar]
- 84.Gregson BH, Metodieva G, Metodiev MV, Golyshin PN, McKew BA. Protein expression in the obligate hydrocarbon-degrading psychrophile Oleispira antarctica RB-8 during alkane degradation and cold tolerance. Environ Microbiol. 2020;22:1870–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tremblay J, Fortin N, Elias M, Wasserscheid J, King TL, Lee K, Greer CW. Metagenomic and metatranscriptomic responses of natural oil degrading bacteria in the presence of dispersants. Environ Microbiol. 2019;21:2307–19. [DOI] [PubMed] [Google Scholar]
- 86.Prince RC, Varadaraj R, Fiocco RJ, Lessard RR. Bioremediation as an oil spill response tool. Environ Technol. 1999;20:891–6. [Google Scholar]
- 87.Maier RM, Gentry TJ. Chapter 17. Microorganisms and organic pollutants. In: Pepper IL, Gerba CP, Gentry TJ, editors. Environmental microbiology (third edition). San Diego: Academic Press; 2015. p. 377–413. [Google Scholar]
- 88.Brakstad OG, Nonstad I, Faksness L-G, Brandvik PJ. Responses of microbial communities in arctic sea ice after contamination by crude petroleum oil. Microb Ecol. 2008;55:540–52. [DOI] [PubMed] [Google Scholar]
- 89.Park S, Yoshizawa S, Inomata K, Kogure K, Yokota A. Halioglobus japonicus gen. nov., sp. Nov. and Halioglobus pacificus sp. Nov., members of the class Gammaproteobacteria isolated from seawater. Int J Syst Evol Microbiol. 2012;62:1784–9. [DOI] [PubMed] [Google Scholar]
- 90.Suárez-Moo P, Lamelas A, Garcia-Bautista I, Barahona-Pérez LF, Sandoval-Flores G, Valdes-Lozano D, Toledano-Thompson T, Polanco-Lugo E, Valdez-Ojeda R. Characterization of sediment microbial communities at two sites with low hydrocarbon pollution in the southeast Gulf of Mexico. PeerJ. 2020;8: e10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pacwa-Płociniczak M, Biniecka P, Bondarczuk K, Piotrowska-Seget Z. Metagenomic functional profiling reveals differences in bacterial composition and function during bioaugmentation of aged petroleum-contaminated soil. Front Microbiol. 2020;11:2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Militon C, Boucher D, Vachelard C, Perchet G, Barra V, Troquet J, Peyretaillade E, Peyret P. Bacterial community changes during bioremediation of aliphatic hydrocarbon-contaminated soil. FEMS Microbiol Ecol. 2010;74:669–81. [DOI] [PubMed] [Google Scholar]
- 93.Rodríguez-Salazar J, Loza A, Ornelas-Ocampo K, Gutierrez-Rios RM, Pardo-López L. Bacteria from the Southern Gulf of Mexico: baseline, diversity, hydrocarbon-degrading potential and future applications. Front Mar Sci. 2021;8:625477. [Google Scholar]
- 94.Lofthus S, Bakke I, Tremblay J, Greer CW, Brakstad OG. Biodegradation of weathered crude oil in seawater with frazil ice. Mar Pollut Bull. 2020;154: 111090. [DOI] [PubMed] [Google Scholar]
- 95.Lirette AO, Chen YJ, Freyria NJ, Góngora E, Greer CW, Whyte LG. Characterization of hydrocarbon degraders from Northwest Passage beach sediments and assessment of their ability for bioremediation. Can J Microbiol. 2024;70:163–77. [DOI] [PubMed] [Google Scholar]
- 96.Irrgang AM, Bendixen M, Farquharson LM, Baranskaya AV, Erikson LH, Gibbs AE, Ogorodov SA, Overduin PP, Lantuit H, Grigoriev MN, Jones BM. Drivers, dynamics and impacts of changing Arctic coasts. Nat Rev Earth Environ. 2022;3:39–54. [Google Scholar]
- 97.Barnhart KR, Overeem I, Anderson RS. The effect of changing sea ice on the physical vulnerability of Arctic coasts. Cryosphere. 2014;8:1777–99. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Figure S1. Alpha and beta diversity were used to analyze the microbial diversity of microorganisms from the sediment samples. Alpha diversity was evaluated by analyzing observed ASVs and the Shannon index for CI's sediment (a), effluent, and biofilm (b), and CII's sediment (c) samples. Beta diversity was assessed using the Weighted-Unifrac metric, presenting PCoA plots for CI's sediment (d), effluent, and biofilm (e), and CII's sediment (f) samples.
Supplementary Material 2: Figure S2. The heatmap depicts the relative abundance of the top 20 most prevalent genera in CI's effluent and biofilm samples, offering a graphical representation of their distribution patterns.
Supplementary Material 3: Figure S3. Total petroleum analysis of effluent samples from CI. Effluent samples were periodically collected on day 14 and 32, enabling quantification and comparison of hydrocarbon concentrations across various treatment conditions.
Supplementary Material 4: Figure S4. Comparative analysis of microbial composition at the phylum level in sediment and biofilm samples using metagenome and 16S rRNA sequencing data. Microbial communities were generated by PhyloFlash from metagenome data (a) and 16S rRNA sequencing data (b). 'S' denotes sediment, ‘Bio’ denotes biofilm and the treatments are labeled as follows: no oil control (Control), oil only (Oil), oil with inorganic fertilizer, MAP (Oil MAP), oil with oleophilic fertilizer, S200 (Oil S200), and oil with both fertilizers (Oil MAP + S200).
Supplementary Material 5: Figure S5. The identification of key hydrocarbon degradation genes in each biofilm metagenome using the CANT-HYD approach for gene search. The treatments are labeled as follows: oil only (Oil), oil with inorganic fertilizer, MAP (Oil MAP), oil with oleophilic fertilizer, S200 (Oil S200), and oil with both fertilizers (Oil MAP + S200).
Supplementary Material 6: Figure S6. Relative abundance of various Pseudomonas species in sediment, biofilm, and effluent samples collected from CI under different treatments. 'R' represents sediment samples; 'E' signifies effluent samples; 'Bio' denotes biofilm samples. 'T0' refers to Day 0, 'T2' refers to the period from 15 to 21 days, and 'T3' from 22 to 32 days. 'T' designates surface sediments, while 'B' indicates bottom sediments.
Supplementary Material 7: Table S1. ASV table, including raw data and relative abundance, in sediment and effluent samples from CI experiment.
Supplementary Material 8: Table S2. ASV table, including raw data and relative abundance, in sediment and effluent samples from CII experiment.
Supplementary Material 9: Table S3. Alpha diversity and statistical analysis of microbial communities in sediment samples from CI and CII experiments.
Supplementary Material 10: Table S4. Quantification of total petroleum hydrocarbons (TPH) and statistical analysis for CI and CII experiment samples.
Supplementary Material 11: Table S5. Identification of 37 key hydrocarbon degradation genes from CANT-HYD search results in 28 metagenomes from CI experiment.
Supplementary Material 12: Table S6. Prediction of key gene abundance through PICRUSt2 in CI and CII experiment samples.
Supplementary Material 13: Table S7. 37 key hydrocarbon degradation genes identified in 65 metagenome-assembled genomes (MAGs) from the CI experiment via CANT-HYD search.
Supplementary Material 14: Table S8. Key metabolic genes, involving respiration, nitrogen cycles, sulfur cycles, and hydrocarbon degradation, in selected MAGs via KEGG database.
Supplementary Material 15: Table S9. Comparative analysis of CANT-HYD and KEGG database search results between top 10 MAGs and 7 other selected MAGs.
Supplementary Material 16: Table S10. Assessment of Pseudomonas relative abundance in sediment samples from the CI experiment.
Supplementary Material 17: Table S11 Metadata information associated with the 16S rRNA sequencing data for the CI and CII column experiments, the metagenome sequencing data, and the metagenome-assembled genomes for the CI column experiment.
Data Availability Statement
The 16S rRNA sequencing data for the CI and CII column experiments were deposited at the National Center for Biotechnology Information (NCBI) under the BioSample accession numbers SAMN40210342 and SAMN40243382, respectively, within the BioProject accession number PRJNA1082469. Additionally, the metagenomic sequence data for the CI column experiment was deposited at NCBI under the SRA accession number from SRX24007725 to SRX24007752, within the BioProject accession number PRJNA1086230. Furthermore, the metagenome-assembled genomes were deposited at NCBI under the BioSample accession numbers ranging from SAMN40570777 to SAMN40570841, all within the BioProject accession number PRJNA1086230. Metadata information associated with the sequencing data has been attached as a supplementary table (Table S11).