Abstract
Daily behavioral and physiological rhythms are controlled by the brain’s circadian timekeeping system, a synchronized network of neurons that maintains endogenous molecular oscillations. These oscillations are based on transcriptional feedback loops of clock genes, which in Drosophila include the transcriptional activators Clock (Clk) and cycle (cyc). While the mechanisms underlying this molecular clock are very well characterized, the roles that the core clock genes play in neuronal physiology and development are much less understood. The Drosophila timekeeping center is composed of ~150 clock neurons, among which the four small ventral lateral neurons (sLNvs) are the most dominant pacemakers under constant conditions. Here, we show that downregulating the clock gene cyc specifically in the Pdf-expressing neurons leads to decreased fasciculation both in larval and adult brains. This effect is due to a developmental role of cyc, as both knocking down cyc or expressing a dominant negative form of cyc exclusively during development lead to defasciculation phenotypes in adult clock neurons. Clk downregulation also leads to developmental effects on sLNv morphology. Our results reveal a non-circadian role for cyc, shedding light on the additional functions of circadian clock genes in the development of the nervous system.
Author summary
Daily behaviors and physiological processes are governed by the brain’s circadian clock, a network of neurons that regulates internal rhythms through cyclic gene expression. In Drosophila (fruit flies), the circadian clock consists of approximately 150 neurons, with a small subset playing a key role in maintaining these rhythms. While the molecular mechanisms of clock genes, including Clock (Clk) and cycle (cyc), are well understood in terms of timekeeping, their roles in other aspects of neuronal physiology are less explored. In this study, we found that the cyc gene, beyond its function in maintaining circadian rhythms, also influences the development of key clock neurons. Specifically, reducing cyc expression in a group of clock neurons during development leads to defects in the morphology of those neurons in both larval and adult stages and affects the ability of adult flies to maintain behavioral rhythms in the absence of environmental cues. These findings reveal that clock genes like cyc have important roles in brain development, highlighting their broader significance beyond circadian regulation. This work provides new insights into how genetic factors involved in timekeeping also contribute to the formation of neural circuits, expanding our understanding of the diverse functions of circadian clock genes.
Introduction
The proper wiring of neuronal circuits during development is essential for the neuronal control of behavior. Across animal species, sleep/wake cycle rhythms, as well as many other behavioral and physiological rhythms, are controlled by the circadian timekeeping system, a network of neurons that maintains endogenous molecular oscillations and rhythmic behavior with a ~24 hour period [1]. The proper functioning of this circadian network requires the formation of synaptic and peptidergic connections during development [2,3].
The Drosophila circadian clock neuron network comprises ~150 neurons and is the functional equivalent of the mammalian suprachiasmatic nuclei, which contain 20,000 neurons in mice [4–6]. All circadian clock neurons contain an intracellular molecular clock consisting of a transcriptional feedback loop of clock genes[7]. CLOCK (CLK) and CYCLE (CYC) are heterodimeric transcriptional activators that directly activate transcription of the period (per) and timeless (tim) genes. PER and TIM encode repressors that inhibit CLK-CYC function. Subsequently, PER and TIM are degraded, which enables the cycle to reinitiate every morning. CLK and CYC also interact with other genes in a secondary circadian loop by activating the genes vrille (vri), and Par domain proteinε (Pdp1ε) [8, 9]. Clk and cyc expression can be detected in almost all clock neurons even before some of these neurons show molecular oscillations [10], suggesting that these genes serve functions that precede the establishment of molecular rhythms.
Drosophila clock neurons are classified into multiple clusters with distinct patterns of gene expression, anatomy, physiology, and synaptic connectivity [5, 6, 11–16]. Among these clusters, the small ventral lateral neurons (sLNvs) are considered the most dominant pacemakers since they are critical for maintaining behavioral rhythmicity under constant darkness and temperature (DD, or free-running) [17–20]. The sLNvs release the neuropeptide Pigment Dispersing Factor (PDF) [21], a key output signal within the clock neuron network [22]. PDF accumulates rhythmically at the sLNv dorsal termini [23] and can be released from both the neurites and soma [24]. Loss of PDF severely reduces the amplitude of the endogenous circadian rhythm and shortens its free-running period in DD [21]. The large LNvs also produce PDF but do not play a role in maintaining rhythms in DD [17].
The projections of the four sLNvs form a bundle and remain fasciculated as they extend from the ventral to the dorsal brain during development. These four projections are usually difficult to distinguish from each other until they begin to defasciculate in the dorsal protocerebrum [25] and extend their dorsal arborizations toward the area where dorsal clusters of clock neurons are located [26]. In adult flies, the dorsal termini of the sLNv projections show rhythmic structural plasticity [27], which relies on daily and circadian rhythms in outgrowth and fasciculation [28–31]. Both Clk and cyc mutants have lower Pdf RNA levels, and the PDF peptide can barely be detected in the sLNv projections [23, 32].
Cyc is a homolog of the mammalian gene Bmal1, although CYC protein levels do not cycle, unlike BMAL1 and several other Drosophila circadian proteins [33]. There is growing evidence for non-circadian functions of BMAL1. First, its downregulation induces apoptosis and cell-cycle arrest in Glioblastoma Stem Cells (GSC), and it was found to preferentially bind metabolic genes and associate with active chromatin regions in GSCs [34]. Second, brain knockdown of Bmal1 using CRISPR/Cas9 made glioblastomas grow at faster rates than controls [35], and similar effects were observed in B16 melanoma cells. Moreover, Bmal1(-/-) mice exhibit defects in short- and long-term memory formation [36] and show reduced lifespan and multiple symptoms of premature aging [37]. Overall, results from studies in different animal models suggest that Bmal1 plays a role in the development of various neurological disorders [38].
The Drosophila sLNvs offer an excellent model for exploring the non-circadian roles of canonical clock genes such as cycle. To determine if the phenotypes previously observed for cyc mutants are specific to PDF expression or involved a broader, non-circadian effect in the development of PDF- expressing cells, we downregulated cyc specifically in the Pdf-expressing cells and observed pronounced defasciculation of the sLNv projections. Similar phenotypes were observed upon expression of a dominant negative form of cyc. Moreover, we found that cyc downregulation in Pdf+ cells during development is sufficient to prevent the fasciculation of the adult sLNvs and results in the loss of behavioral rhythms in adult flies. Manipulations of Clk expression also affect sLNv morphology, although remarkably, the phenotypes of Clk and cyc manipulation differ. Our results show that cyc plays a role in the development of pacemaker neurons, which is likely independent of its role in the circadian molecular oscillator.
Results
cyc downregulation in circadian pacemaker neurons affects the formation of sLNv axon bundles
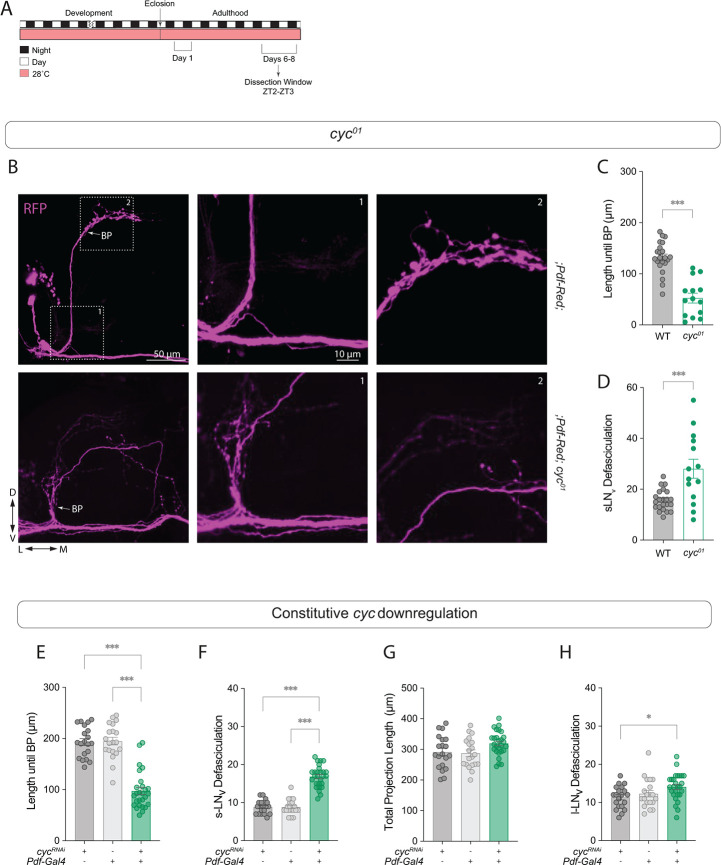
Mutations in both Clk and cyc severely reduce pdf RNA and neuropeptide levels [23, 32]. In cyc null mutants, sLNvs projections are often undetectable in larval and adult brains stained with PDF antibodies [23, 39, 40], although around half of the brains show ‘stunted’ projections [40]. We observed that cyc null mutants (cyc01) showed a substantial reduction in PDF levels at ZT2, consistent with previous studies, but we also noticed the presence of thin, misrouted sLNv projections in cyc01 flies at higher magnification and intensity (S1A Fig). Upon close observation, PDF could often be detected in the sLNv projections. However, these projections did not form the stereotypical bundle observed in control brains when extending from the anterior medulla toward the dorsal area of the brain.
Because highly defasciculated projections might contribute to the weaker PDF levels observed in cyc01 mutants, we examined the structure of the sLNv projections using a Pdf-RFP transgene, in which a cytosolic Red Fluorescent Protein (RFP) is controlled by the Pdf regulatory sequence [41]. Flies were raised at 28°C throughout development, and experiments were conducted at 28°C in 6–8-day old flies (Fig 1A). In control brains, the projections from the four sLNvs remain fasciculated, forming a bundle until reaching the superior medial protocerebrum (SMP). In contrast, the sLNvs of cyc01 mutants often began to defasciculate in the ventral brain, near their cell bodies (Fig 1B). Some sLNv projections were severely misrouted and did not reach the dorsal brain, extending instead toward the midline or other brain regions. As a result, it was possible to distinguish individual projections from each sLNv even in the ventral brain in most cyc01 mutant brains. This is almost never observed in control brains until the sLNv projections reach the main branching point in the SMP. The morphological phenotypes of cyc01 flies are highly variable, and in some instances the projections are barely visible (S1B Fig).
Fig 1. Cyc downregulation in circadian pacemaker neurons prevents the formation of sLNvs axon bundles.
The cyc01 mutant has disrupted sLNv morphology. (A) Representative timeline of the experiments in the figure. Flies were kept at 28°C throughout development and experiments were performed within days 6–8 post-eclosion. (B) Representative confocal images of Pdf-RFP controls and; Pdf-RFP;cyc01 experimental flies stained with anti-RFP (magenta). The branching point (BP) of the dorsal projections is indicated. Scale bar = 25 μm. Boxes with dashed lines indicate the proximal (1) and distal (2) projections, corresponding to the labeled projection images in the center and right panels, respectively. An unpaired t-test was used to quantify the sLNv projection length until the branching point (BP) (C), and the total number of intersections of the sLNv ventral projections (D). Results from two independent experiments, with each dot representing one brain. For each genotype, the number of subjects (n) fall in the range: 13 ≤ n ≤ 22. (E-H) Quantification of the LNv morphology phenotypes of experimental flies in which a cycRNAi transgene was driven by a; Pdf-RFP,Pdf-Gal4;Tub-Gal80ts driver compared to the parental controls. The sLNv projection length until the branching point (BP) (E), the total number of intersections of the sLNv ventral projections (F), the total sLNv projection length (G), and the total number of intersections of the lLNv projections along the optic tract (OT) (H) are shown. Results from three independent experiments, with each dot representing one brain. For each genotype, the n falls in the range: 20 ≤ n ≤ 27. For nonparametric data sets, statistical comparisons were done with Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests. For parametric data sets, statistical comparisons were done with one-way ANOVAs followed by Tukey post hoc tests. Differences that are not significant are not indicated. *p < 0.05, *** p < 0.001. Error bars indicate SEM.
To test whether the effect of cyc loss on the LNvs is cell-autonomous, we next expressed a UAS-cyc dsRNA transgene (UAS-cycRNAi) using the Pdf-Gal4 driver. We quantified the length of the projections, starting at the point where the projections of the sLNvs intersect with those of the lLNvs (“point of origin”, POI, S1C Fig), until the first branching point (“branching point”, BP). This branching point is where the sLNvs ramify and extend their stereotypical arborizations in the dorsal protocerebrum in control brains, and these arborizations show daily, clock-controlled rhythms in their fasciculation and outgrowth [27]. Downregulating cyc in the Pdf-expressing cells significantly decreased the distance to the branching point (Fig 1E). Using a modified Scholl’s analysis [42], we quantified the degree of branching in the ventral projections starting at the POI. We observed pronounced defasciculation in the sLNv projections in Pdf > cycRNAi flies (Fig 1F). cyc01 mutants also showed decreased distance to BP and sLNv fasciculation (Fig 1C and 1D). The total projection length in Pdf > cycRNAi flies was not different from that of the controls (Fig 1G), and the defasciculation phenotype was not observed in the contralateral projections that extended from the lLNvs (Fig 1H). Since the lLNvs are born later in development during metamorphosis [25], this result suggests that cyc plays a role in neuronal development during an earlier developmental stage, when the sLNvs begin to extend their projections toward the dorsal brain. These experiments were conducted at 28°C to allow subsequent comparisons with adult-specific and development-specific downregulations of cyc using Gal80ts. Similar results were obtained with flies raised at 25°C (S1F–S1I Fig).
Expression of dominant negative forms of Clk and cyc is an effective strategy for preventing circadian molecular oscillations in specific groups of clock neurons [43–45]. In these dominant negative forms, the DNA binding ability is disrupted while the ability to heterodimerize is preserved [43]. Based on the phenotypes induced by cyc downregulation, we asked if expressing a dominant negative form of cyc in the sLNvs also leads to aberrant projection morphology. We found that sLNvs expressing Δ-cyc using Pdf-Gal4 had a significantly shorter distance until the branching point (S2B and S2C Fig) and a greater degree of sLNv projection defasciculation (S2B and S2D Fig), similar to the effects observed in Pdf > cycRNAi flies. The total projection length and the projections of the lLNvs were unaffected (S2E and S2F Fig).
PER levels in Pdf+ neurons are reduced upon cell-specific cyc knockdown
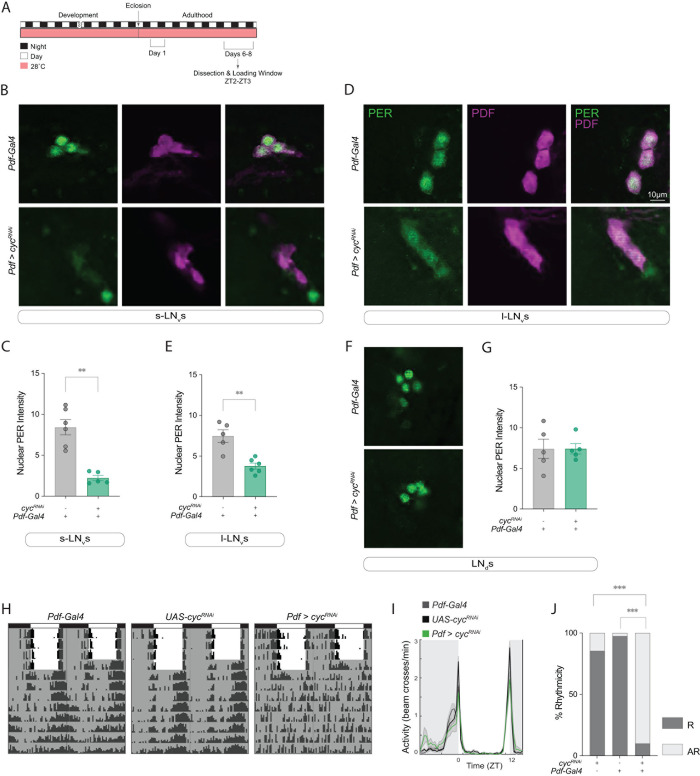
CYC activates per transcription, and thus, PER levels in the brain are significantly reduced in cyc mutants [33]. To test whether the phenotypes of cycRNAi expression in the Pdf-expressing neurons are consistent with what would be expected from cyc downregulation, we compared PER levels in the parental control (Pdf-Gal4, Pdf-RFP/+) with those in Pdf > cycRNAi flies at the end of the night (ZT23), when PER nuclear levels are highest [46]. We found that nuclear PER levels in Pdf > cycRNAi flies were significantly reduced in the sLNvs (Fig 2B and 2C) and lLNvs (Fig 2D and 2E). In contrast, PER levels were unaffected in the Dorsal Lateral Neurons (LNds) (Fig 2E and 2G). These results confirmed that, at least in a light-dark cycle (LD), Pdf > cycRNAi flies have lower PER levels in Pdf+ neurons.
Fig 2. Constitutive cyc downregulation in Pdf+ cells leads to a reduction in PER levels and arrhythmicity under free-running conditions.
(A) Representative timeline of the experiments in the figure. Flies were kept at 28°C for their entire lifespan. Experiments were performed within days 6–8 post-eclosion. Dissections were performed at ZT2-3. (B,D,F) Representative confocal images of PER (green) and PDF (magenta) staining in the sLNvs (B), lLNvs (D), and LNds (F) of Pdf > cycRNAi experimental and Pdf-Gal4 /+ control flies (n = 5–6 brains per clock neuron group). All lines also included a Pdf-RFP transgene. Scale bar = 10 μm. (C,E,G) Mann-Whitney tests were used to compare nuclear PER intensity levels in the sLNvs (C), lLNvs (E), and LNds (G) in flies of the indicated genotypes. Differences that are not significant are not indicated. ** p < 0.01. Error bars indicate SEM. (H) Representative actograms of flies of the indicated genotypes under 5 days of LD entrainment followed by 7 days of free-running (DD). To allow comparison with development-specific cyc downregulation, flies in this experiment were raised at 28°C for their entire lifespan and the experiment was conducted at 28°C. (I) Population activity plots for flies during days 3–5 of the LD cycle at 28°C. (J) Fisher’s exact contingency tests were used to analyze the percentage of rhythmic flies of the indicated genotypes under DD (DD1-7). The driver line also included a tub-Gal80ts transgene. Additional quantifications can be found in Table 1. R = Rhythmic and AR = arrhythmic. Differences that are not significant are not indicated. *** p < 0.001. Behavioral data corresponds to two independent behavior experiments. For each genotype: 40 ≤ n ≤ 48.
cyc null mutant flies have pronounced behavioral phenotypes. Their activity is unimodal instead of bimodal during LD, and they are predominantly nocturnal [47]. Additionally, cyc mutants are largely arrhythmic in DD due to the key role of cyc in circadian molecular oscillations [33]. We conducted behavioral experiments to determine the extent to which downregulating cyc specifically in PDF-expressing neurons recapitulates the phenotype of the cyc mutant. We found that at 28°C, the activity pattern of Pdf > cycRNAi flies was still bimodal in LD (Fig 2H and 2I). However, the majority (~90%) of the experimental flies were arrhythmic in DD (Fig 2J). Pdf > Δ-cyc flies showed similar behavioral phenotypes (S2G and S2H Fig), consistent with what was reported for their free-running behavior at 25°C [43].
cyc acts during development to shape neuronal morphology in adults
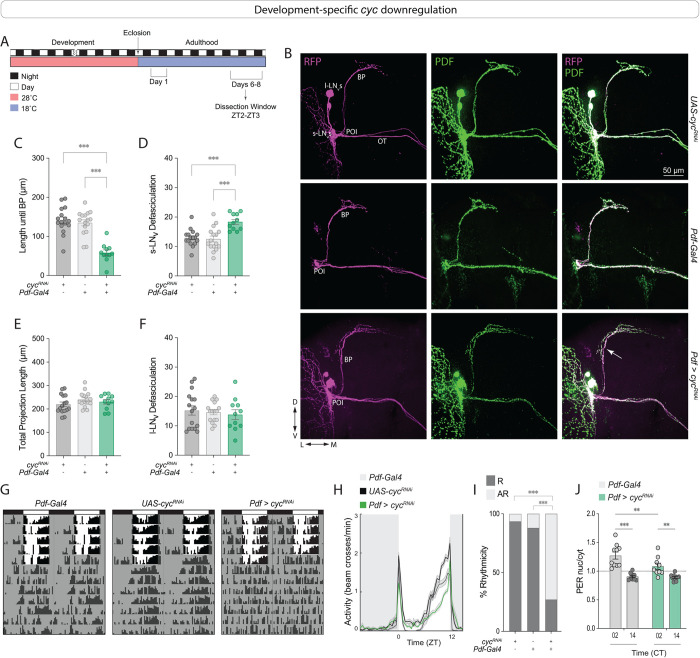
To knock down cyc specifically during development, we employed a temperature-sensitive Gal80 (Gal80ts) variant with ubiquitous expression to conditionally inhibit Gal4-mediated expression of the RNAi [48]. This method enables the temporal regulation of UAS transgenes, as Gal80ts remains active at lower temperatures but becomes inactive at higher temperatures. We raised flies at 28°C to allow cyc downregulation during development then transferred them to 18°C immediately after eclosion (Fig 3A). After 1 week at 18°C, the brains were dissected at ZT2 and stained with PDF and RFP antibodies (see methods section). As shown in Fig 3, downregulating cyc exclusively before eclosion resulted in abnormal morphology of the sLNv axonal projections in adult flies (Fig 3B). The phenotypes resembled those observed with constitutive downregulation, with a significantly shorter distance to the branching point (Fig 3B and 3C) and a greater degree of defasciculation compared to parental controls (Fig 3B–3D). No significant differences were found in the total projection length or the degree of defasciculation of the lLNvs (Fig 3E and 3F).
Fig 3. Development-specific cyc downregulation in Pdf+ cells prevents sLNv fasciculation.
(A) Representative timeline of the experiments in the figure. Flies were raised in LD at 28°C, and transferred to 18°C immediately after eclosion. Dissections were then performed in 6–8 day old adults at ZT2-3. (B) Representative confocal images of anti-PDF (green) and anti-RFP (magenta) staining of adult fly brains in which cyc was downregulated only during development. Each line also included a Pdf-RFP transgene. The white arrow indicates the increased defasciculation in the sLNv projections in experimental flies. Scale bar = 50 μm. (C-F) Quantification of the LNv morphology phenotypes of flies of the indicated genotypes. The driver line also included a tub-Gal80ts transgene. The sLNv projection length until the branching point (BP) (C), the number of intersections of sLNv ventral projections (D), the total sLNv projection length (E), and the total number of intersections of the lLNv projections along the optic tract (OT) (F) are shown for flies in which cyc was downregulated in Pdf+ cells until eclosion. Two independent experiments were conducted. For each genotype: 11 ≤ n ≤ 16. One-way ANOVA tests were used to quantify the LNv morphology. *** p < 0.001. Error bars indicate SEM. Each dot corresponds to one brain. (G-I). Behavioral phenotypes of development-specific cyc knockdown. Flies were raised in LD at 28°C, before being transferred to 18°C upon eclosion. Experiments were conducted at 18°C. (G) Representative actograms of flies of the indicated genotypes under free-running (see Table 1 for n and additional quantifications). (H) Population activity plots for flies during days 3–5 of the LD cycle at 18°C. (I) Percent rhythmicity for the indicated genotypes under DD. R = Rhythmic and AR = arrhythmic. Fisher’s exact contingency tests were used to analyze the percentage of rhythmic flies under DD (DD1-7). *** p < 0.001. Error bars indicate SEM. The data correspond to three independent behavior experiments. For each genotype: 68 ≤ n ≤ 94. (J) Quantification of nuclear over cytoplasmic PER immunosignal within the sLNvs on day 2 of constant darkness at 18°C from brains of Gal4 controls or cyc RNAi-expressing flies. A two-way ANOVA was employed for statistical analysis. ** p < 0.01, *** p < 0.001. Error bars indicate SEM.
A previous study showed that panneuronal rescue of cyc expression in a cyc01 mutant exclusively during development was sufficient to partially rescue arrhythmicity in adult flies [40]. Therefore, we asked if downregulating cyc in the Pdf+ cells specifically during development would lead to behavioral phenotypes similar to those seen in the cyc null mutants. We found that under free-running conditions at 18°C, most (~78%) of the Pdf > cycRNAi flies were arrhythmic (Fig 3G–3I). An analysis of PER subcellular localization in DD2 revealed clear cycling with nuclear PER higher at CT2 (Fig 3J). These results indicate that developmental downregulation of cyc specifically in the Pdf+ cells is sufficient to prevent behavioral rhythms in adults.
To determine whether adult-specific cyc knockdown in the Pdf-expressing cells would also lead to morphological phenotypes, we raised flies at 18°C and switched them to 28°C immediately after eclosion (S3A Fig, see the methods section). This manipulation did not result in morphological phenotypes either in terms of the length to the branching point or in the degree of sLNv defasciculation (S3B–S3F Fig). Under free-running 28°C, the majority of the experimental flies were arrhythmic (S3G–S3H Fig), indicating that, as expected, cyc is required in adult clock neurons for proper circadian clock function.
Cyc manipulations lead to aberrant sLNv projections in larval clock neurons
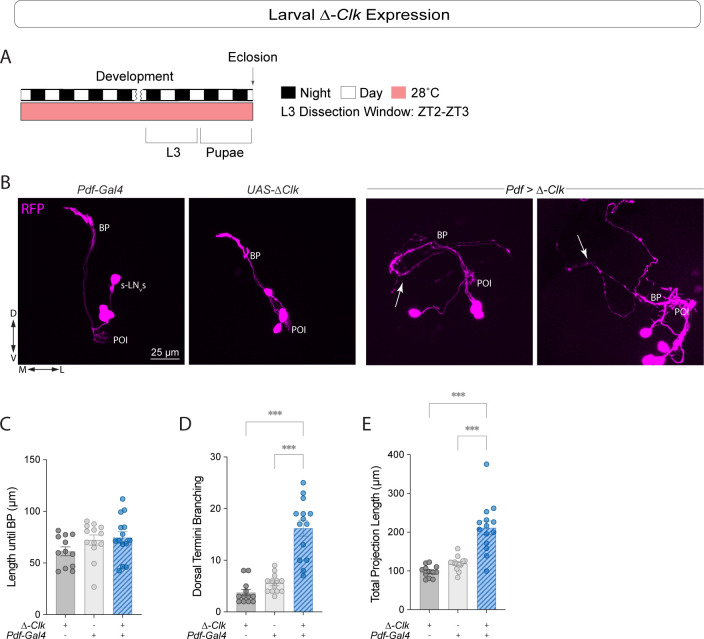
Next, we asked if cyc downregulation results in clock neuron morphology phenotypes during earlier developmental stages. The four larval sLNvs, which modulate the sensitivity of larvae to light and mediate a circadian rhythm in visual sensitivity [49], appear to be identical in their anatomy and synaptic connections [50]. We expressed the cycRNAi transgene under the Pdf-Gal4 driver and dissected third larval instar (L3) brains (Fig 4). In brains of experimental larvae the length to the branching point did not differ from that of the controls (Fig 4B and 4C), but the degree of dorsal termini branching was significantly higher (Fig 4B–4D). This quantification is similar to that previously described when quantifying the arborization of the dorsal projections sLNvs in adults [27], where the concentric circles are centered at the main dorsal branching point (S1B Fig; see methods section). The total sLNv projection length was not affected by the genetic manipulation (Fig 4E).
Fig 4. Cyc manipulations lead to aberrant sLNv projections in larval clock neurons.
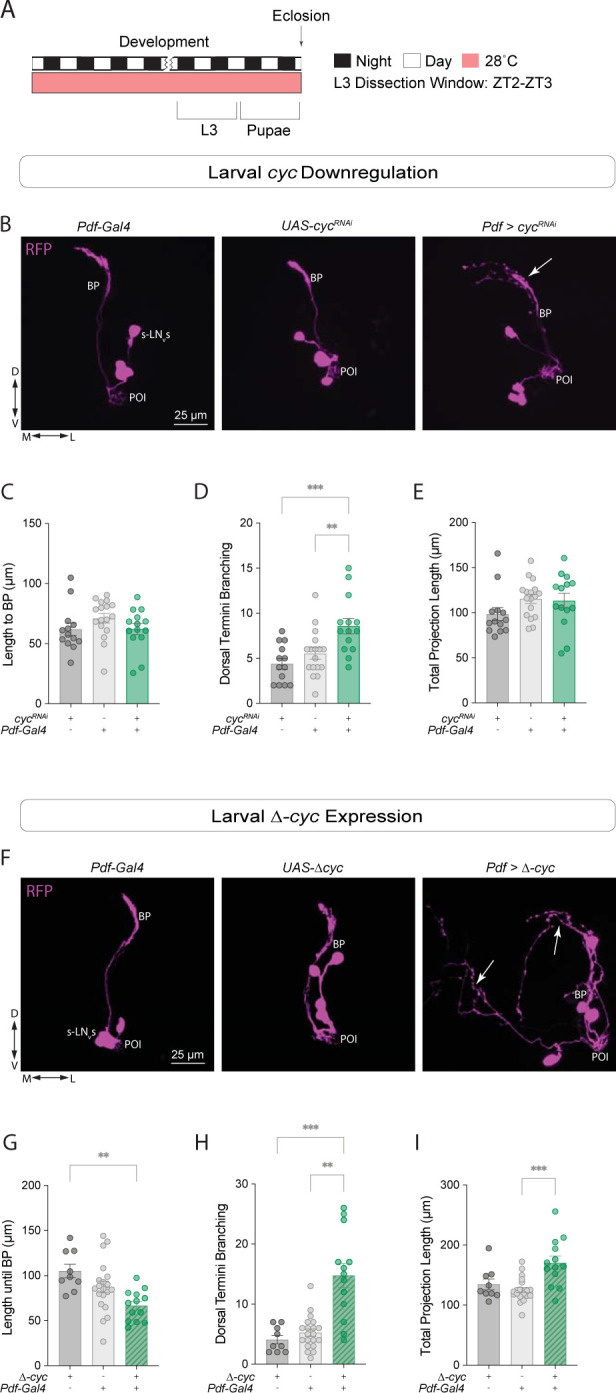
(A) Representative timeline of the experiments in the figure. Larvae were raised in LD at 28°C. Third instar larvae (L3) were dissected at ZT2-3. (B-E) Developmental effects of cyc knockdown in the sLNvs. (B) Representative confocal images of L3 larval brains stained with anti-RFP, labeling the sLNvs. (C-E) The projection length from the POI to the BP (C), the degree of sLNv dorsal termini branching (D), and the total projection length (E) were compared. For each genotype: 13 ≤ n ≤ 17. (F-I) Developmental effects of expressing a dominant-negative form of cyc, Δ-cyc, in the larval sLNvs. (F) Representative confocal images of anti-RFP staining in the sLNvs of L3 larvae. A one-way ANOVA followed by a Tukey’s Multiple Comparisons tests was used to compare the projection length from the POI to the BP (G). A Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests compared the nonparametric data sets: the degree of sLNv dorsal termini branching (H) and the total projection length (I). Each dot corresponds to one brain. For each genotype: 9 ≤ n ≤ 20. ** p < 0.01, *** p < 0.001. Three independent experiments were conducted for each genetic manipulation and each line also included a Pdf-RFP transgene. The driver lines also included a tub-Gal80ts transgene. Error bars indicate SEM.
Since the effects of cyc knockdown via RNAi and the expression of a cyc dominant negative form in adults were similar (Figs 1 and S2), we analyzed the morphology of the sLNv projections in L3 larvae upon Δ-cyc expression. In Pdf > Δ-cyc larval brains, the length to the branching point was significantly lower (Fig 4F and 4G) and the number of branches was significantly greater than that of controls (Fig 4H). The total projection length was not affected (Fig 4I). Taken together, these results suggest that cyc plays a role in the development of the larval sLNv neurons.
Clk downregulation increases sLNv dorsal arborizations
Clk and cyc mutations produce similar effects on the expression pattern of PDF in adult brains [23]. CLK and CYC act as heterodimeric transcriptional activators, and the circadian phenotypes associated with mutations in these core circadian clock genes, both molecular and behavioral, are largely similar [43, 47, 51]. To determine if downregulating Clk in the sLNvs leads to the same defasciculation of the sLNvs observed with cyc manipulations, we performed similar experiments as those described above, in which we expressed ClkRNAi in Pdf+ neurons. We found that Pdf > ClkRNAi flies also showed neuronal morphology phenotypes (Fig 5).
Fig 5. Clk and cyc manipulations result in different morphology phenotypes in clock neurons.
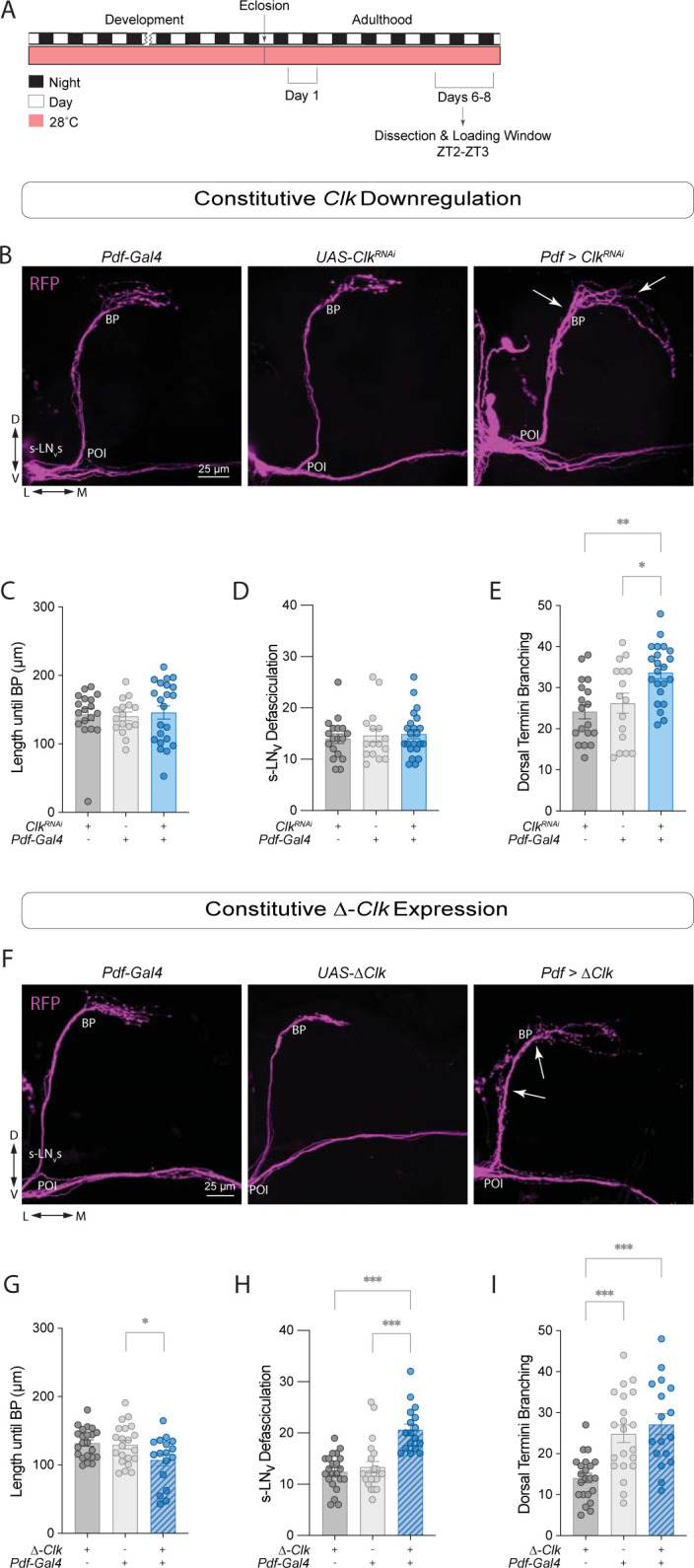
(A) Representative timeline of the experiments in the figure. Flies were kept in LD conditions at 28°C for their entire lifespan. Dissections were performed within Days 6–8 post-eclosion at ZT2-3. (B) Representative confocal images of anti-RFP staining in the sLNvs adult brains of control (ClkRNAi /+ and Pdf-Gal4;tub-Gal80ts/+), and experimental (Pdf > ClkRNAi) flies. White arrows indicate the BP (left) and extension of some of the sLNv dorsal projections (right) in the experimental line. All lines employed also included a Pdf-RFP transgene. Scale bar = 25 μm. (C-E) Quantification of sLNv morphology using Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests for nonparametric datasets, compared the length until the branching point (C) and the total number of axonal crosses of the sLNvs (D). For parametric data, ordinary one-way ANOVA tests followed by Tukey’s Multiple Comparisons tests compared the total number of axonal crosses after the BP (E). Each dot corresponds to one brain. Two independent experiments were conducted. For each genotype: 16 ≤ n ≤ 22. * p < 0.05, *** p < 0.001. Error bars indicate SEM. (F) Representative confocal images of anti-RFP (magenta) staining in the sLNvs adult brains of control (UAS-ΔClk /+ and Pdf-Gal4;tub-Gal80ts/+), and experimental (Pdf > Δ-Clk) flies. White arrows indicate the BP (top) and increased defasciculation along the sLNv projections (bottom). All lines employed also included a Pdf-RFP transgene. Scale bar = 25 μm. (G-I) Quantification of sLNv morphology phenotypes: length until the branching point (G), the total number of axonal crosses of the sLNvs (H), and the total number of axonal crosses after the BP (I). For parametric data, ordinary one-way ANOVA tests followed by Tukey’s Multiple Comparisons tests were employed. For nonparametric data, Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests were employed. See S1 Table for details about statistical analysis. Each dot corresponds to one brain. Two independent experiments were conducted. For each genotype: 17 ≤ n ≤ 22. ** p < 0.01, *** p < 0.001. Error bars indicate SEM.
In a previous study, Clk downregulation resulted in overfasciculation of the sLNv dorsal termini when stained with anti-PDF [52]. However, RFP labeling of the sLNv membrane indicated that these termini were actually more expanded than those of control flies, resulting in significantly higher dorsal termini branching (Figs 5B–5E and S1C). In Pdf > ClkRNAi flies, neither the distance to the branching point (Fig 5C) nor the degree of defasciculation differed from controls (Fig 5D). Neither the sLNv total projection length nor the lLNv projections were affected (S4C and S4D Fig). Only ~48% of the Pdf > ClkRNAi flies were rhythmic, and those that were rhythmic exhibited a lengthening of the free-running period (S4B Fig and Table 1). Nuclear PER levels in Pdf > ClkRNAi flies were significantly reduced in the LNvs (S4H–S4J Fig).
Table 1. Summary of free running activity rhythms.
Related to Figs 2, 3, and 7 and S1–S4. Activity analysis of the above genotypes at 25°C, 28°C, or 18°C. Light conditions for each experiment was 12:12 LD for 5 days followed by DD for at least 8 days. Depending on the experiment, flies were raised at 25°C, 28°C, or 18°C. Flies raised a 25°C were kept at that temperature throughout the behavior experiment. Flies raised at 18°C were transferred to 28°C upon eclosion and behavior experiments were conducted at 28°C. Flies raised at 28°C were either kept at 28°C for behavior experiments (constitutive knockdown) or transferred to 18°C upon eclosion (development-specific knockdown). ClockLab’s χ-square periodogram analysis was used to analyze rhythmicity, rhythmic power, and free-running period for each above genotypes. The % rhythmicity along with the number of rhythmic flies (nR), the period in hours with the SEM, and the rhythmic power with the SEM are indicated. Arrhythmic flies were not included in the analysis of period or power.
| Temperature pre-eclosion: 28°C, Temperature post-eclosion: 28°C | ||||
| Genotype | Number of Flies (n) |
% Rhythmicity (nR) |
Period (h) ± SEM |
Rhythmic Power ± SEM |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts | 40 | 97.50 (39) | 24.44 ± 0.06 | 109.80 ± 6.39 |
| ;UAS-cycRNAi 42563; | 48 | 85.42 (41) | 24.02 ± 0.08 | 85.74 ± 7.91 |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts >; UAS-cycRNAi 42563; | 40 | 10.00 (4) | 25.88 ± 3.03 | 24.32 ± 3.71 |
| ;UAS-ClkRNAi 42566; | 26 | 92.31 (24) | 23.73 ± 0.10 | 97.66 ± 8.90 |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts >; UAS-ClkRNAi 42566; | 21 | 52.38 (11) | 25.36 ± 0.22 | 36.74 ± 6.76 |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts | 24 | 100.00 (24) | 24.5 ± 0.07 | 101.40 ± 8.97 |
| ;UAS-cas9/CyO; UAS-Vrig/TM6b Tb | 16 | 87.50 (14) | 23.86 ± 0.11 | 124.6 ± 10.39 |
|
;Pdf-Red,Pdf-Gal4;Tub-Gal80ts >; UAS-cas9/CyO; UAS-Vrig/TM6b Tb |
18 | 38.89 (7) | 23.50 ± 0.15 | 29.02 ± 3.68 |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts | 31 | 83.87 (26) | 24.98 ± 0.07 | 56.76 ± 6.73 |
| ;UAS-Δcyc; | 24 | 95.83 (23) | 23.54 ± 0.08 | 72.79 ± 9.29 |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts >; UAS-Δcyc; | 32 | 3.13 (1) | 25.50 ± 0.00 | 13.66 ± 0.00 |
| ;UAS-ΔClk | 27 | 100.00 (27) | 23.52 ± 0.06 | 93.52 ± 8.71 |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts >; UAS-ΔClk | 28 | 3.57 (1) | 23.50 ± 0.00 | 17.18 ± 0.00 |
|
Temperature pre-eclosion: 28°C, Temperature post-eclosion: 18°C | ||||
| Genotype | Number of Flies (n) |
% Rhythmicity (nR) |
Period (h) ± SEM |
Rhythmic Power ± SEM |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts | 68 | 88.24 (60) | 24.40 ± 0.17 | 87.80 ± 7.33 |
| ;UAS-cycRNAi 42563; | 94 | 93.62 (88) | 24.18 ± 0.04 | 81.06 ± 4.38 |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts >; UAS-cycRNAi 42563; | 70 | 27.14 (19) | 23.82 ± 0.81 | 21.42 ± 1.78 |
| Temperature pre-eclosion: 18°C, Temperature post-eclosion: 28°C | ||||
| Genotype | Number of Flies (n) |
% Rhythmicity (nR) |
Period (h) ± SEM |
Rhythmic Power ± SEM |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts | 30 | 100 (30) | 24.93 ± 0.04 | 127.8 ± 6.76 |
| ;UAS-cycRNAi 42563; | 25 | 96 (24) | 24.04 ± 0.07 | 150.1 ± 12.44 |
| ;Pdf-Red,Pdf-Gal4;Tub-Gal80ts >; UAS-cycRNAi 42563; | 31 | 38.71 (12) | 23.67 ± 0.61 | 24.27 ± 3.68 |
| Temperature pre-eclosion: 25°C, Temperature post-eclosion: 25°C | ||||
| Genotype | Number of Flies (n) |
% Rhythmicity (nR) |
Period (h) ± SEM |
Rhythmic Power ± SEM |
| ;Pdf-Red,Pdf-Gal4; | 29 | 89.66 (26) | 24.52 ± 0.08 | 152.2 ± 11.53 |
| ;UAS-cycRNAi 42563; | 31 | 83.87 (26) | 24.06 ± 0.08 | 95.44 ± 9.61 |
| ;Pdf-Red,Pdf-Gal4; >; UAS-cycRNAi 42563; | 31 | 12.90 (4) | 23.13 ± 0.24 | 21.55 ± 6.93 |
Expression of Δ-Clk in the Pdf+ cells did not result in changes in the sLNv projection length until branching point (Fig 5F and 5G) or the total length of the projections (S4F Fig). However, the Pdf > Δ-Clk brains had increased defasciculation of the ventral projection (Fig 5H). The degree of dorsal termini branching in the Pdf > Δ-Clk flies was not significant (Fig 5I), not was the degreed of lLNv defasciculation (S4G Fig). Under DD at 28°C, the majority of Δ-Clk expressing flies were arrhythmic (S4E Fig), consistent with what was reported at 25°C [43].
We then examined L3 larval brains to determine if the observed phenotypes were already present at this developmental stage. While expression of ClkRNAi did not result in morphological phenotypes in larval LNvs (S5 Fig), expression of Δ-Clk resulted in pronounced phenotypes (Fig 6A and 6B). We observed a significant increase in sLNv dorsal termini branching (Fig 6D) and total projection length (Fig 6E) in Pdf > Δ-Clk larvae. The length to the branching point for the experimental larvae was not significantly different from that of the control lines (Fig 6C). Expressing Δ-Clk led to more pronounced phenotypes in the larval stage than Clk downregulation, possibly due to incomplete knockdown.
Fig 6. Expressing Δ-Clk in the sLNvs leads to axonal morphology phenotypes in L3 larvae.
(A) Representative timeline of the experiments in the figure. Larvae were raised in LD at 28°C. Third instar larvae (L3) were dissected at ZT2-3. (B) Representative confocal images of anti-RFP (magenta) staining in the sLNvs when Δ-Clk was expressed in Pdf+ neurons in L3 larvae. Each line also included a Pdf-RFP transgene, and the driver line also included a tub-Gal80ts transgene. White arrows indicate misrouting of the sLNv projections in the experimental line. Scale bar = 25 μm. For nonparametric data, Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests were used to compare the length to the BP (C). One way ANOVA tests were used to compare dorsal termini branching (D) and the total projection length (E). Two independent experiments were conducted. Each dot corresponds to one brain. For each genotype: 12 ≤ n ≤ 14. *** p < 0.001. Error bars indicate SEM.
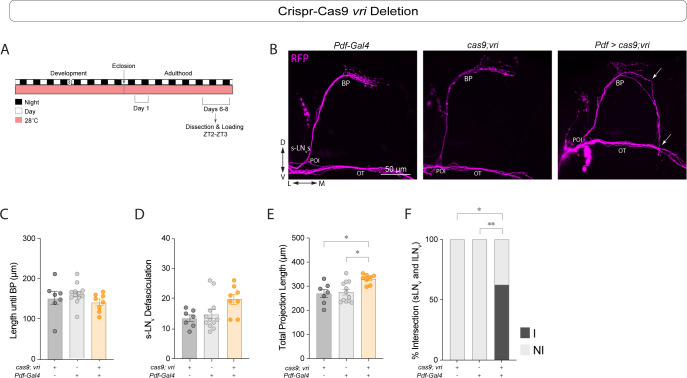
In addition to the main feedback loop, CLK and CYC form a secondary loop by activating vri and Pdp1ε [8, 9], which repress and activate Clk expression, respectively. The low PDF peptide in the sLNvs projections of cyc01 mutants can be rescued by vri overexpression [39]. To determine if vri expression also affects sLNvs morphology we expressed a line with a CRISPR/Cas9-based gRNA targeting the vri gene [53] under the control of the Pdf-Gal4 driver. We found that in Pdf > Cas9 + vri-g flies neither the distance until branching point in the s-LNvs (Fig 7B and 7C) nor the degree of fasciculation of the s-LNvs (Fig 7D) was different from controls. The total projection length was significantly higher than controls (Fig 7E), but in this case due to projections extending ventrally towards the optic tract (Fig 7B). In the majority of the brains, some s-LNv projections extended towards the ventral brain after reaching the SMP (Fig 7B) and in most cases contacted the l-LNv contralateral projections in the optic tract (Fig 7F and 7G), a phenotype that was never observed in control brains.
Fig 7. Vri mutagenesis results in sLNv hyperextension.
(A) Representative timeline of the experiments in the figure. Flies were kept in LD conditions at 28°C for their entire lifespan. Dissections were performed in 6–8 day old adults at ZT2-3. Behavioral experiments were run at constant 28°C. (B) Representative confocal images of anti-RFP (magenta) staining in the sLNvs adult brains of control (cas9;vrig\/+ and Pdf-Gal4;tub-Gal80ts/+), and experimental (Pdf > cas9;vrig) flies. All lines employed also included a Pdf-RFP transgene. White arrows indicate the misrouting of the sLNv dorsal projections (top), and the intersection of the sLNv with the lLNvs at the OT (bottom). Scale bar = 25 μm. (C-E) Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests were used to compare the length until the branching point (C), the total number of intersections of the sLNvs ventral projections (D), and the longest path of the sLNv projections (without including misrouting) (E). (F) Fisher’s exact contingency tests were used to analyze the percentage of brains where the sLNvs intersected with the lLNvs at the optic tract (I = Intersecting, N.I. = Not Intersecting). See Table 1 for additional quantifications. Each dot corresponds to one brain. Two independent experiments were conducted. For each genotype: 7 ≤ n ≤ 12. * p < 0.05, ** p < 0.01, *** p < 0.001. Error bars indicate SEM.
Discussion
Our results reveal a role for the circadian clock gene cyc in establishing the proper cellular morphology of the key clock pacemaker neurons, the sLNvs. Both constitutive cyc knockdown or expression of a dominant negative form of cyc in Pdf+ cells result in increased defasciculation of the sLNvs. In addition, Clk downregulation and expression of a dominant negative form of Clk also result in sLNv morphology phenotypes, although some of those phenotypes appear to be distinct from those caused by cyc manipulations. Expressing the dominant-negative forms of either Clk or cyc has been used in previous studies as an effective way to prevent molecular oscillations in subsets of clock neurons. However, our results indicate that these genetic manipulations lead to additional morphological phenotypes beyond molecular timekeeping that are already detectable during the larval stages.
In addition to anatomical and functional classifications, clock neurons can be divided into early or late developmental groups depending on when circadian oscillations can be detected. In the early groups, which include the sLNvs, per and tim expression rhythms can be detected at the first instar (L1) larval stage, whereas in the late groups, such rhythms cannot be detected until metamorphosis [25, 54]. However, cyc and Clk expression using GFP-cyc and GFP-Clk transgenes can be detected in almost all groups of clock neurons at early developmental stages, even days before per oscillations begin [10]. This suggests that cyc and Clk play additional roles in the development of clock neurons beyond their role in the molecular oscillator.
Both cyc and Clk modulate PDF expression in both larval and adult clock neurons. In Clkjrk mutants, neither PDF nor Pdf mRNA can be detected in most larval [32] or adult sLNvs [23], and similar effects have been observed for the cyc02 mutant [23]. However, around half of the cyc01 brains stained with PDF exhibit ‘stunted’ sLNv projections which appear to lack their dorsal termini [40]. This study by Goda et al. also showed that panneuronal rescue of cyc expression throughout development is sufficient to restore PDF expression in the LNv dorsal projections of cyc01 mutants [40]. Overexpression of vri, a clock gene that is downstream of CLK/CYC and acts as a repressor of CLK transcription [9, 32], causes a severe reduction in PDF levels in larval brains [32], and the low PDF levels in the sLNvs of cyc01 mutants can be rescued by vri overexpression [39]. However, restoring PDF expression in the sLNvs in flies lacking vri expression is not sufficient to rescue activity rhythms.
Unlike in cyc and Clk mutants [23], PDF can be detected in the sLNvs projections in per01 and tim01 mutants, although it no longer shows rhythms in its accumulation in the dorsal termini [23]. In addition, structural plasticity rhythms in the sLNvs are absent in both per01 and tim01 mutants [27]. Downregulation of Clk [52], expression of Δ-cyc [30], and overexpression of vri in Pdf-expressing cells [39] also result in impaired plasticity rhythms [55]. Although the anatomical phenotypes seen in these mutants are milder than those that observed when cyc and Clk are downregulated or when their dominant negative forms are expressed, the sLNv projections of both per and tim null mutants also exhibit altered morphology [27].
Our results suggest that Clk and cyc manipulations produce different phenotypes, however, it is possible that this is partially due to a less effective knockdown of Clk. Behavioral experiments show that cyc knockdown in Pdf+ neurons result in a larger fraction of arrhythmic flies than knockdown of Clk (Table 1). Use of RNAi often reduces gene expression but does not completely eliminate it, and may lead to off-target side effects. In addition, RNAi efficiency may vary over time. Expression of dominant negative alleles was used as an independent approach, but this method has limitations as well: over-expression levels for cyc and Clk may differ, and non-native molecular interactions may occur at high concentrations. However, differential effects of cyc and Clk mutations have been previously described: cyc01 and Clkjrk mutants showed differences in their sleep consolidation during the day and in their ability to recover after sleep deprivation [47].
CYC/CLK may regulate neuronal fasciculation by modulating the expression of genes involved in cell migration or cytoskeletal dynamics. For example, increasing matrix metalloproteinases 1 (MMP1) expression reduces the complexity of the sLNv arborizations along the projections [31]. MMP1 promotes fasciculation in Drosophila motor neuron axons [56]. Clk has been shown to affect sLNv dorsal termini arborization through the activation of Mef2, which negative regulates Fas2 expression [52]. Our results from Clk downregulation show increased rather than decreased sLNv dorsal termini arborization. One important difference is that we used RFP to label the membrane rather than a PDF staining. As for cyc, among the sLNv morphology phenotypes reported in the literature, including those of other clock mutants, PDF/PDFR [57], and Rho GTPases [28], among others, the phenotype most similar to cyc downregulation is the downregulation of the Medea (Med). Med is homolog of the human tumor-suppressor gene DPC4 and is involved in the decapentaplegic (dpp) pathway [58], and its downregulation via RNAi in Pdf+ neurons results in decreased fasciculation along the projections of the sLNvs [57]. In addition, similar to cyc manipulations, developmental specific downregulation of Med leads to morphology phenotypes in adult clock neurons [57].
Expression of Clk outside the clock network leads to the generation of ectopic clocks [59], but they require cyc expression. A study by Liu et al. showed that Clk stabilizes CYC both in cultured Drosophila Schneider 2 (S2) cells and in vivo: upon ectopic Clk expression, GFP-CYC can be detected in additional cells beyond the clock neuron network, suggesting that although with this reporter the CYC protein could be detected in the brain only in clock neurons, cyc mRNA is more broadly expressed [60]. In addition, cyc mRNA was not enriched in the LNvs compared to other elav-expressing neurons in the head [61]. Single cell RNA sequencing data revealed that cyc mRNA is present in non-clock neurons as well as in various tissues throughout the fly’s body, with particularly high expression in the gut, ovaries, and testes [62]. In some instances, cyc mRNA expression levels are very high while Clk mRNA levels are low, such as in intestinal stem cells and the chordotonal organ [62]. The role of cyc mRNA expression in non-clock cells remains unknown. An interesting question for future studies is whether CLK and CYC act as an obligate heterodimer in their neurodevelopmental function and other possible non-circadian roles. In mammals, BMAL1 can dimerize with NPAS2 in addition to CLOCK [63], and a recent study in Drosophila detected co-binding of CYC and FOXO in the promoter region of vrille [64]. Single seq RNA sequencing in the sLNvs and other clock neurons, comparing the effects of cyc vs Clk downregulation, could help clarify the degree to which they function independently. Our results suggest that Clk and cyc are involved in shaping the morphology of clock neurons, and it is possible that they play similar roles in non-clock neurons as well.
Materials and Methods
Fly lines and rearing
Flies were raised on cornmeal-sucrose yeast media in a Percival Incubator under 12:12 LD at different temperature conditions. Depending on the experiment, flies were raised under either 18°C, 25°C, or 28°C (indicated in the figure legends). The lines UAS-cycRNAi (BDSC #42563), UAS-ClkRNAi (BDSC #42566), w1118 (BDSC #3605) and CS (BDSC #64349) were obtained from the Bloomington Drosophila Stock Center. The lines Pdf-RFP,Pdf-Gal4;Tub-gal80ts and w;Pdf-RFP;MKRS/TM6 were donated by Justin Blau (New York University). The cyc01, UAS-Δ-cyc, and UAS-Δ-Clk stocks were donated by Paul Hardin (University of Texas).
Immunohistochemistry
LNv PDF levels and neuronal morphology
Brains of 6–8-day-old adult males or L3 larvae were dissected between ZT2 and ZT3 in ice-cold Schneider’s Drosophila Medium (S2) (Thermo Fisher, #21720024). They were fixed immediately after dissection in 2% Paraformaldehyde (PFA) in S2 for 30 minutes. Brains were then treated with blocking solution (5% goat serum in 0.3% PBS-Tx) for 1 hour at room temperature followed by incubation with primary antibodies at 4°C for 24–48 hr. The primary antibodies used were 1:3000 mouse anti-PDF (Developmental Hybridoma Bank) and 1:1000 rabbit anti-RFP (Rockland, #600-401-379-RTU). After incubation, the brains were rinsed 6 times in 0.3% PBS + Triton X-100 (PBT), after which they were incubated with Alexa-fluor conjugated secondary antibodies for 24-hr at 4°C. The secondary antibodies used were 1:3000 Alexa-488 (Thermo Fisher, #A11029) and 1:1000 Alexa-568 (Thermo Fisher, #A11036). The brain samples were further washed 6 times with 0.3% PBT, cleaned and mounted on a clean glass slide in Vectashield (Vector Laboratories, #H-1000-10) mounting media. A list of reagents can be found on Table 2.
Table 2. List of reagents.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental Models: Organisms/Strains | ||
| w;Pdf-RFP,Pdf-Gal4;Tub-gal80ts | J. Blau, NYU | |
| w;Pdf-RFP;MKRS/TM6 | J. Blau, NYU | |
| ;UAS-ΔClk #1 | J. Blau, NYU | |
| W;UAS-cas9/CyO;UAS-Vrig;TM6b Tb | M.Rosbash, Brandeis | |
| w;;UAS-cyc RNAi42563 | Bloomington Drosophila Stock Center | BDSC 42563 |
| w;;UAS-Clk RNAi 42566 | Bloomington Drosophila Stock Center | BDSC 42566 |
| w 1118 ;+;+ | Bloomington Drosophila Stock Center | BDSC 3605 |
| Canton-S | Bloomington Drosophila Stock Center | BDSC 64349 |
| cyc 01 | P. Hardin, University of Texas | BDSC 80929 |
| ;UAS-Δcyc; | P. Hardin, University of Texas | |
| ;UAS-ΔClk | P. Hardin, University of Texas; Bloomington Drosophila Stock Center | BDSC 3618 |
| Antibodies | ||
| Rabbi anti-RFP (1:1000) | Rockland | #600-401-379-RTU |
| Mouse anti-PDF (1:3000) | Developmental Hybridoma Bank | |
| Rat anti-PER (1:500) | O. Shafer (ASRC CUNY) | |
| Anti-rabbit Alexa-568 (1:1000) | Thermo Fisher | A11036 |
| Donkey anti-rat Alexa-488 (1:500) | Thermo Fisher | A21208 |
| Anti-mouse Alexa-488 (1:3000) | Thermo Fisher | A11029 |
| Software | ||
| Fiji | http://fiji.sc | RRID: SCR_002285 |
| MATLAB R2022b | MathWorks, Natick | RRID: SCR_001622 |
| GraphPad Prism 9.0 | GraphPad Software | RRID: SCR_002798 |
| DAM FileScan | Trikinetics | |
| ClockLab | Actimetrics | RRID:SCR_014309 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Vectashield Mounting Medium | Vector Laboratories | #H-1000-10 |
| Premix PBS Buffer (10x) | Sigma-Aldrich | Cat# 11666789001 |
| 2% Paraformaldehyde (PFA) | Sigma-Aldrich | 47608-250ML-F |
| Triton- X-100 | Bio Basic | CAS#9002-93-1 |
| Schneider’s Drosophila Medium (S2) | Thermo Fisher | 21720024 |
| Other | ||
| DAM2 Drosophila Activity Monitors | Trikinetics | |
| DAM Drosophila Environmental Monitors | Trikinetics | |
PER Staining
Brains of 6–8-day-old males were dissected one hour before lights-on (ZT23) in ice-cold Schneider’s Drosophila Medium (S2) (Thermo Fisher, #21720024). Immediately after dissection, brains were fixed in 2% paraformaldehyde (PFA) for 30 minutes, stained and mounted as described above. The primary antibodies used were 1:1000 rabbit anti-RFP (Rockland, #600-401-379-RTU) and 1:500 rat anti-PER (donated by Orie Shafer). The secondary antibodies used were 1:1000 Alexa-568 (Thermo Fisher, #A11036) and 1:500 Alexa-488 (Thermo Fisher, #A21208).
For the analysis of PER subcellular localization (Fig 3J), flies were raised at 28C under LD and transferred to 18°C immediately after eclosion. After 5 days under LD 18C, flies were transferred to constant darkness at 18°C and brains were dissected on the second day of DD (DD2).
Imaging, quantification, and statistical analysis
All images were acquired on an Olympus Fluoview 1000 laser-scanning confocal microscope using a 40x/1.10 NA FUMFL N objective (Olympus, Center Valley, PA) at the Advanced Science Research Center (ASRC-CUNY). For all the experiments, only one hemisphere per brain was imaged (the right hemisphere, unless it was damaged, in which case we imaged the left hemisphere).
Quantification of adult LNv morphology (sLNv and lLNv): We quantified 1) sLNv total projection length, 2) sLNv length from the point of origin (POI) until the branching point (BP) (‘length until BP’), 3) the degree of defasciculation of the sLNv ventral projections, 4) the sLNv dorsal termini branching, 5) the degree of defasciculation of the l-LNv projections, and 6) intersections between sLNv projections and lLNv projections along the optic tract.
1-Total projection length: The total length of the dorsal projection was determined by a line drawn from the point of intersection (POI) between the sLNvs and the optic tract until the end of the dorsal termini. If the projection length went past the midline of the brain, the length was measured up to the midline.
2-Length until BP: The partial length of the dorsal projections was determined by a line drawn from the POI until the BP of the sLNvs at the dorsal termini. The projection length and partial projection length of the sLNvs were quantified using Fiji in ImageJ.
3- Defasciculation of the sLNv ventral projections (‘sLNv Defasciculation’): A modified Scholl’s analysis [42], was used to analyze the degree of defasciculation of the ventral area of the sLNv projections, near the cell bodies. Six concentric circles, each 25 μm apart, were placed centered in the POI (S1C Fig). Each intersection between an individual ventral projection and any of the 6 circles was counted. A value of ‘10’ denotes 10 total intersections between any of the projections and any of the circles.
4-sLNv Dorsal termini branching: A modified Scholl’s analysis was used to analyze the degree of defasciculation of the dorsal termini of the sLNv projections. This method is similar to what was previously described to quantify sLNv dorsal termini [27]. In this study, 8 concentric circles, each 12.5 um apart, were centered in the BP (S1E Fig). Each intersection between an individual dorsal projection and any of the 8 circles was counted. A value of ‘10’ denotes 10 total intersections between any of the dorsal projections and any of the circles.
5-Defasciculation of the lLNv optic tract projections. The degree of defasciculation of the lLNvs was determined using the same 6 concentric circles centered in the POI what were used to quantify defasciculation of the sLNv ventral projections (2) (S1C Fig). Each intersection between an individual lLNv projection and any of the 6 circles was counted.
6- Intersections between sLNv projections and lLNv projections along the optic tract. We quantified the percentage of brains in which at least one sLNv dorsal projection turned ventrally and extended towards the optic tract, contacting at least one lLNv projection. This phenotype was not observed in brains of control flies but was present in more than half of the brains of flies in which vri was knocked out (shown in Fig 7F).
Quantification of larval sLNv morphology
We quantified the total projection length of the sLNvs, the axonal projection length until the branching point of the and the degree of branching in the sLNv dorsal projections (S1D Fig). The projection length, partial projection length, and area of the sLNvs were quantified using Fiji. The projection length was measured by a line drawn from a determined first point of intersection (POI) of each of the sLNv cell bodies until the end of the dorsal termini. The partial length of the axonal projections was determined by a line drawn for the same point of intersection until the branching point (BP) of the sLNvs at the dorsal termini. A modified Scholl’s analysis was used to measure the branching of the sLNv projections. Six concentric circles were placed around the same branching point used in the length measurements. The concentric circles were each 12.5 μm away from each other, so that the farthest circle was 75 μm away from the POI. The number of visible neurites of the sLNvs that intersected with each circle were counted and summed, yielding the total number of intersecting neurons for the dorsal projections.
Quantification of PER levels
Single optical sections of either sLNvs, lLNvs or LNds were imaged using the same settings using a 40x/1.10 objective. PDF was used to identify the small and large LNvs. The LNds were identified based on their localization, size, and morphology. PER levels were determined through normalization of nuclear staining within each cell to the background. The average value for each brain within a cluster was computed by averaging the values obtained from multiple cells within that cluster. Quantification was performed using images from 5–6 brains per each cluster at each timepoint. For the analysis of PER subcellular localization (Fig 3J), the ratio of nuclear vs cytoplasmic PER levels was determined for individual sLNvs and compared using a two-way ANOVA.
Locomotor activity rhythm recording and analysis
DAM2 Drosophila Activity Monitors (TriKinetics, Waltham, MA) were used to record the locomotor activity rhythms of adult male flies aged three- to five-days, as previously described [65]. Flies were entrained to 12:12 LD cycles for at least five days, and then transferred to constant darkness (DD) for at least eight days at a constant temperature of 28°C, unless otherwise specified. Free-running activity rhythms were analyzed with ClockLab software from Actimetrics (Wilmette, IL). We employed ClockLab’s χ-square periodogram function, which was integrated into ClockLab software, for the analysis of rhythmicity, rhythmic power, and free-running period in individual flies, using a confidence level of 0.01 [33]. For each of the tested genotypes, only significant periodicities falling within the 14 to 34-hour range were taken into consideration. In instances where an individual fly exhibited multiple periodicities with peaks surpassing the significance threshold, only the period with the highest amplitude was utilized when calculating the average periods presented in Table 1. ClockLab assigns each peak in the χ-square periodogram both a "Power" value and a "Significance" value. The "Rhythmic Power" for each designated rhythmic fly was determined by subtracting the "Significance" value from the "Power" value associated with the predominant peak. Flies that did not exhibit a periodicity peak above the threshold (10) were categorized as "arrhythmic," and their period and rhythmic power were not included in the analysis [65].
Statistical analysis
Pearson’s D’Agostino normality tests were performed for all the datasets. Depending on whether the data were normally distributed, statistical analyses were performed using either a one-way ANOVA with a Tukey’s multiple comparisons test or a Kruskal-Wallis test with a Dunn’s multiple comparisons test for 3 or more groups, or a t-test for comparisons between 2 groups. Fisher’s exact contingency tests were run to analyze the percent rhythmicity for the indicated genotypes under DD.
Supporting information
(A-B) The cyc01 mutant has disrupted sLNv morphology. (A) Representative confocal images of anti-PDF staining in Canton-S control and cyc01 adult male brains. The sLNvs and optic tract (OT) are indicated. Scale bar = 25 μm. The inserts on the right show the sLNv projections with the signal intensity adjusted for visibility in the cyc01 mutants. The top insert shows the distal (dorsal) area and the bottom insert shows the proximal (ventral) area of the sLNv projections. Scale bar = 10 μm. (B) Representative images of eight brains of; Pdf-RFP;cyc01 experimental flies stained with anti-RFP (magenta). Flies were raised at 28°C. Most of cyc01 mutant flies (~78.5%, 11 out of 14 brains) exhibit severe phenotypes in their sLNv morphology compared to the effects of cyc downregulation in Pdf+ neurons. (C-E) Representative confocal images of adult (C, E) and L3 larvae (D) control brains stained with anti-RFP (magenta). (C) To determine the degree of defasciculation of the sLNv ventral projections in adult brains, 6 concentric circles separated by is 25 μm were centered at the point of intersection (POI), where the projections of the sLNvs and those of the lLNvs intersect. The most distant circle does not reach the main branching point (BP) in control brains; therefore, the dorsal termini are not included. The number of intersections between either the sLNvs or the lLNvs and each concentric circle were quantified. (D) Dorsal projection branching in the larval sLNvs was measured by counting the number of intersections the sLNvs had at each of the 6 concentric circles separated by 12.5 μm. (E) Adult sLNv dorsal projection branching was measured by counting the number of intersections the sLNvs had at each of 8 concentric circles separated by 12.5 μm. This was adapted from a previous study [27] to capture the hyperextended projection phenotype of Pdf > Δ-Clk flies. (F-I) Quantification of the LNv morphology phenotypes of experimental flies in which a cycRNAi transgene was driven by a; Pdf-RFP,Pdf-Gal4; driver compared to the parental controls. Flies were raised at 25°C. The sLNv projection length until the branching point (BP) (F), the total number of intersections of the sLNv ventral projections (G), the total sLNv projection length (H), and the total number of intersections of the lLNv projections along the optic tract (OT) (I) are shown. Graphs are representative of two independent experiments, with each dot representing one brain. For each genotype, n falls in the range: 18 ≤ n ≤ 21. (I-J). **p < 0.01, *** p < 0.001. Error bars indicate SEM. See S1 Table for additional quantifications.
(PDF)
(A) Representative timeline of the experiments in the figure. Flies were kept in LD conditions at 28°C for their entire lifespan. Dissections were performed within days 6–8 post-eclosion at ZT2-3. (B) Representative brain confocal images of anti-PDF (green) and anti-RFP (magenta) staining in the sLNvs of flies in which Δ-cyc was constitutively expressed in the Pdf+ cells using a Pdf-Gal4;tub-Gal80ts driver. Each line also included a Pdf-RFP transgene. White arrows indicate branching of sLNv dorsal projections (left), and dorsal termini of the sLNv projections (right) for the experimental genotype. The images are representative of two independent experiments. Scale bar = 50 μm. LNv morphology was quantified by comparing the sLNv projection length until the branching point (C), the total number of intersections of the sLNv ventral projections (D), the full sLNv projection length (E), and the total number of lLNv intersections (F). One-way ANOVA was used to analyze normally distributed data (C, F). For nonparametric data sets, a Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests was used (D,E). * p < 0.05, ** p < 0.01, *** p < 0.001. Error bars indicate SEM. Each dot corresponds to one brain. For each genotype: 9 ≤ n ≤ 12. (G-H) Behavioral phenotypes of constitutive Δ-cyc expression. Experiments were conducted at 28°C. (G) Population Activity (left) plots for flies during days 3–5 of the LD cycle at 28°C (see Table 1 for additional quantifications). (H) Percent rhythmicity for the indicated genotypes under DD. R = Rhythmic and AR = arrhythmic. Fisher’s exact contingency tests were used to analyze the percentage of rhythmic flies under DD (DD1-8). *** p < 0.001. Error bars indicate SEM. For each genotype: 24 ≤ n ≤ 32.
(PDF)
(A) Representative timeline of the experiments in the figure. Flies were raised in LD at 18°C, and transferred to 28°C immediately after eclosion. Dissections were then performed within days 6–8 post-eclosion at ZT2-3. (B) Representative confocal images of anti-PDF (green) and anti-RFP (magenta) staining in the sLNvs when cyc was downregulated exclusively after eclosion using a Pdf-Gal4;tub-Gal80ts driver. The images are representative of two independent experiments. Scale bar = 50 μm. Each line also included a Pdf-RFP transgene. Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests were used to quantify the projection length until BP (C), the total number of intersections of the sLNv ventral projections (D), the full sLNv projection length (E), and the total number of lLNv intersections (F). * p < 0.05. Datasets are nonparametric (C-F). Each dot corresponds to one brain. For each genotype: 17 ≤ n ≤ 24. (G-H) Behavioral phenotypes of adult-specific cyc knockdown. Flies were raised in LD at 18°C, before being transferred to 28°C upon eclosion. Experiments were conducted at 28°C. (G) Population Activity plots for flies during days 3–5 of the LD cycle at 18°C (see Table 1 for additional quantifications). (H) Percent rhythmicity for the indicated genotypes under DD. Fisher’s exact contingency tests were used to analyze the percentage of rhythmic flies under DD (DD1-8). *** p < 0.001. Error bars indicate SEM. For each genotype: 25 ≤ n ≤ 31.
(PDF)
(A) Representative timeline. Flies were raised in LD at 28°C for their entire lifespan. Behavioral assays and dissections were performed within days 6–8 post-eclosion at ZT2-3. Experiments were conducted at 28°C. (B) Fisher’s tests were used to compare the percent of rhythmic flies of each indicated genotype (additional quantifications can be found in Table 1). ** P < 0.01, *** P < 0.001. For each genotype: 21 ≤ n ≤ 26. (C-D) Additional quantifications of effects of ClkRNAi expression in the Pdf+ cells in adult brains. Neither the sLNv total projection length (C) nor the lLNv projections (D) were affected. Datasets were quantified with ordinary one-way ANOVA tests followed by Tukey’s Multiple Comparisons tests. For each genotype: 16 ≤ n ≤ 22. (E) Fisher’s tests were used to compare the percent of rhythmic flies of each indicated genotype (additional quantifications shown in Table 1). For each genotype: 27 ≤ n ≤ 31. (F-G) Effects of Δ-Clk expression in the Pdf+ cells in adult brains. The sLNv total projection length (F) and the lLNv projections (G) were quantified using one-way ANOVA tests followed by Tukey’s Multiple Comparisons tests. Each dot corresponds to one brain. For each genotype: 17 ≤ n ≤ 22. (H-J) Mann-Whitney tests were used to compare nuclear PER intensity levels in the sLNvs (H), lLNvs (J), and LNds (J) in flies of the indicated genotypes. Flies were raised at constant 28°C for their entire lifespan and dissections were performed at ZT2-3 * p < 0.05, ** P < 0.01, *** P < 0.001. Error bars indicate SEM.
(PDF)
(A) Representative timeline of the experiments in the figure. Larvae were raised in LD at 28°C. Third instar larvae (L3) were dissected at ZT2-3. (B) Representative confocal images of anti-RFP (magenta) staining in the sLNvs when ClkRNAi was expressed in L3 larvae. Each line also contains a Pdf-RFP transgene. Scale bar = 25 μm. Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests were used to compare the projection length from the POI to the BP (C), the total number of axonal intersections (D), and the total projection length from the POI (E). * p < 0.05. Error bars indicate SEM. Each dot corresponds to one brain. For each genotype: 4 ≤ n ≤ 9.
(PDF)
Statistical analysis of each experiment, labelled with the corresponding figure, the genotypes used, and the comparisons between genotypes. D’Agostino & Pearson Normality tests were employed to determine if datasets followed a normal distribution. For comparisons between two independent variables unpaired t-tests were used for parametric datasets, while Mann-Whitney tests were employed for nonparametric datasets. For comparisons between three independent variables ordinary one-way ANOVA tests followed by Tukey’s Multiple Comparisons tests or Holm-Šídák’s Multiple Comparisons tests were used for parametric datasets, while Kruskal-Wallis tests followed by Dunn’s multiple comparisons tests were employed for nonparametric datasets. The p value for each test run is indicated, as is each tests corresponding significance. NS indicates results that are not significant. * p < 0.05, ** p < 0.01, *** p < 0.001.
(XLSX)
Acknowledgments
We are very grateful to Justin Blau and Paul Hardin for their valuable feedback on various aspects of this project and for sharing fly lines with us. We are also grateful to M.Fernanda Ceriani, Amanda González-Segarra, Aishwarya Ramakrishnan Iyer, Orie Shafer, and Troy Shirangi for helpful comments on the manuscript, Annika Barber and Troy Shirangi for helpful discussions, Orie Shafer for the rat anti-PER antibody and Michael Rosbash for fly lines. The mouse anti-PDF antibody was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Data Availability
All the data are freely available, without restrictions, and it can be found here: https://github.com/graceb4/cyc-ms-raw-data.
Funding Statement
This work was supported by a National Science Foundation (NSF IOS 2239994) to M.P.F. This grant provided the salary for G.B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8(10):790–802. Epub 2007/09/21. doi: 10.1038/nrn2215 . [DOI] [PubMed] [Google Scholar]
- 2.Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. 2012;76(1):82–97. Epub 2012/10/09. doi: 10.1016/j.neuron.2012.08.035 ; PubMed Central PMCID: PMC3466441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sehgal A, Price J, Young MW. Ontogeny of a biological clock in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1992;89(4):1423–7. Epub 1992/02/15. doi: 10.1073/pnas.89.4.1423 ; PubMed Central PMCID: PMC48463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helfrich-Forster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Tech. 2003;62(2):94–102. Epub 2003/09/11. doi: 10.1002/jemt.10357 . [DOI] [PubMed] [Google Scholar]
- 5.Shafer OT, Helfrich-Forster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498(2):180–93. Epub 2006/07/21. doi: 10.1002/cne.21021 ; PubMed Central PMCID: PMC2596765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schubert FK, Hagedorn N, Yoshii T, Helfrich-Forster C, Rieger D. Neuroanatomical details of the lateral neurons of Drosophila melanogaster support their functional role in the circadian system. J Comp Neurol. 2018;526(7):1209–31. Epub 2018/02/10. doi: 10.1002/cne.24406 ; PubMed Central PMCID: PMC5873451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343(6258):536–40. Epub 1990/02/08. doi: 10.1038/343536a0 . [DOI] [PubMed] [Google Scholar]
- 8.Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112(3):329–41. Epub 2003/02/13. doi: 10.1016/s0092-8674(03)00074-6 . [DOI] [PubMed] [Google Scholar]
- 9.Glossop NRJ, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE Feeds Back to Control Circadian Transcription of Clock in the Drosophila Circadian Oscillator. Neuron. 2003;37(2):249–61. doi: 10.1016/s0896-6273(03)00002-3 [DOI] [PubMed] [Google Scholar]
- 10.Liu T, Mahesh G, Houl JH, Hardin PE. Circadian Activators Are Expressed Days before They Initiate Clock Function in Late Pacemaker Neurons from Drosophila. The Journal of Neuroscience. 2015;35(22):8662–71. doi: 10.1523/JNEUROSCI.0250-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma D, Przybylski D, Abruzzi KC, Schlichting M, Li Q, Long X, et al. A transcriptomic taxonomy of Drosophila circadian neurons around the clock. eLife. 2021;10:e63056. doi: 10.7554/eLife.63056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shafer OT, Gutierrez GJ, Li K, Mildenhall A, Spira D, Marty J, et al. Connectomic analysis of the Drosophila lateral neuron clock cells reveals the synaptic basis of functional pacemaker classes. Elife. 2022;11. Epub 2022/06/30. doi: 10.7554/eLife.79139 ; PubMed Central PMCID: PMC9365390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helfrich-Forster C. Neurobiology of the fruit fly’s circadian clock. Genes Brain Behav. 2005;4(2):65–76. Epub 2005/02/22. doi: 10.1111/j.1601-183X.2004.00092.x . [DOI] [PubMed] [Google Scholar]
- 14.Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, et al. RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet. 2017;13(2):e1006613. Epub 2017/02/10. doi: 10.1371/journal.pgen.1006613 ; PubMed Central PMCID: PMC5325595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rieger D, Shafer OT, Tomioka K, Helfrich-Forster C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J Neurosci. 2006;26(9):2531–43. Epub 2006/03/03. doi: 10.1523/JNEUROSCI.1234-05.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murad A, Emery-Le M, Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron. 2007;53(5):689–701. Epub 2007/03/03. doi: 10.1016/j.neuron.2007.01.034 ; PubMed Central PMCID: PMC1852515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431(7010):869–73. Epub 2004/10/16. doi: 10.1038/nature02935 . [DOI] [PubMed] [Google Scholar]
- 18.Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431(7010):862–8. Epub 2004/10/16. doi: 10.1038/nature02926 . [DOI] [PubMed] [Google Scholar]
- 19.Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci. 1992;12(9):3321–49. Epub 1992/09/01. doi: 10.1523/JNEUROSCI.12-09-03321.1992 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12(3):555–70. Epub 1994/03/01. doi: 10.1016/0896-6273(94)90212-7 . [DOI] [PubMed] [Google Scholar]
- 21.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99(7):791–802. Epub 2000/01/05. doi: 10.1016/s0092-8674(00)81676-1 . [DOI] [PubMed] [Google Scholar]
- 22.Shafer OT, Yao Z. Pigment-Dispersing Factor Signaling and Circadian Rhythms in Insect Locomotor Activity. Curr Opin Insect Sci. 2014;1:73–80. Epub 2014/11/12. doi: 10.1016/j.cois.2014.05.002 ; PubMed Central PMCID: PMC4224320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97(7):3608–13. Epub 2000/03/22. doi: 10.1073/pnas.97.7.3608 ; PubMed Central PMCID: PMC16287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klose MK, Bruchez MP, Deitcher DL, Levitan ES. Temporally and spatially partitioned neuropeptide release from individual clock neurons. Proc Natl Acad Sci U S A. 2021;118(17). Epub 2021/04/21. doi: 10.1073/pnas.2101818118 ; PubMed Central PMCID: PMC8092580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helfrich-Forster C, Shafer OT, Wulbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500(1):47–70. Epub 2006/11/14. doi: 10.1002/cne.21146 . [DOI] [PubMed] [Google Scholar]
- 26.Helfrich-Forster C, Homberg U. Pigment-dispersing hormone-immunoreactive neurons in the nervous system of wild-type Drosophila melanogaster and of several mutants with altered circadian rhythmicity. J Comp Neurol. 1993;337(2):177–90. Epub 1993/11/08. doi: 10.1002/cne.903370202 . [DOI] [PubMed] [Google Scholar]
- 27.Fernandez MP, Berni J, Ceriani MF. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 2008;6(3):e69. Epub 2008/03/28. doi: 10.1371/journal.pbio.0060069 ; PubMed Central PMCID: PMC2270325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petsakou A, Sapsis Themistoklis P, Blau J. Circadian Rhythms in Rho1 Activity Regulate Neuronal Plasticity and Network Hierarchy. Cell. 2015;162(4):823–35. doi: 10.1016/j.cell.2015.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pírez N, Ceriani María F. Circadian Pacemaker Neurons Change Synaptic Contacts across the Day. Current Biology. 2014;24(18):2161–7. doi: 10.1016/j.cub.2014.07.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrero A, Duhart JM, Ceriani MF. Neuronal and Glial Clocks Underlying Structural Remodeling of Pacemaker Neurons in Drosophila. Front Physiol. 2017;8:918. Epub 2017/12/01. doi: 10.3389/fphys.2017.00918 ; PubMed Central PMCID: PMC5694478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Depetris-Chauvin A, Fernández-Gamba Á, Gorostiza EA, Herrero A, Castaño EM, Ceriani MF. Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons. PLOS Genetics. 2014;10(10):e1004700. doi: 10.1371/journal.pgen.1004700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blau J, Young MW. Cycling vrille Expression Is Required for a Functional Drosophila Clock. Cell. 1999;99(6):661–71. doi: 10.1016/s0092-8674(00)81554-8 [DOI] [PubMed] [Google Scholar]
- 33.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93(5):805–14. Epub 1998/06/18. doi: 10.1016/s0092-8674(00)81441-5 . [DOI] [PubMed] [Google Scholar]
- 34.Dong Z, Zhang G, Qu M, Gimple RC, Wu Q, Qiu Z, et al. Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock. Cancer Discov. 2019;9(11):1556–73. Epub 2019/08/29. doi: 10.1158/2159-8290.CD-19-0215 ; PubMed Central PMCID: PMC6983300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner PM, Prucca CG, Velazquez FN, Sosa Alderete LG, Caputto BL, Guido ME. Temporal regulation of tumor growth in nocturnal mammals: In vivo studies and chemotherapeutical potential. FASEB J. 2021;35(2):e21231. Epub 2021/01/12. doi: 10.1096/fj.202001753R . [DOI] [PubMed] [Google Scholar]
- 36.Kondratova AA, Dubrovsky YV, Antoch MP, Kondratov RV. Circadian clock proteins control adaptation to novel environment and memory formation. Aging (Albany NY). 2010;2(5):285–97. Epub 2010/06/04. doi: 10.18632/aging.100142 ; PubMed Central PMCID: PMC2898019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20(14):1868–73. Epub 2006/07/19. doi: 10.1101/gad.1432206 ; PubMed Central PMCID: PMC1522083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng Y, Pan L, Wang F, Yan J, Wang T, Xia Y, et al. Neural function of Bmal1: an overview. Cell & Bioscience. 2023;13(1):1. doi: 10.1186/s13578-022-00947-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunawardhana KL, Hardin PE. VRILLE Controls PDF Neuropeptide Accumulation and Arborization Rhythms in Small Ventrolateral Neurons to Drive Rhythmic Behavior in Drosophila. Curr Biol. 2017;27(22):3442–53 e4. Epub 2017/11/07. doi: 10.1016/j.cub.2017.10.010 . [DOI] [PubMed] [Google Scholar]
- 40.Goda T, Mirowska K, Currie J, Kim MH, Rao NV, Bonilla G, et al. Adult circadian behavior in Drosophila requires developmental expression of cycle, but not period. PLoS Genet. 2011;7(7):e1002167. Epub 20110707. doi: 10.1371/journal.pgen.1002167 ; PubMed Central PMCID: PMC3131292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruben M, Drapeau MD, Mizrak D, Blau J. A mechanism for circadian control of pacemaker neuron excitability. J Biol Rhythms. 2012;27(5):353–64. doi: 10.1177/0748730412455918 ; PubMed Central PMCID: PMC4019749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87(4):387–406. ; PubMed Central PMCID: PMC1244622. [PMC free article] [PubMed] [Google Scholar]
- 43.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14(8):638–49. doi: 10.1016/j.cub.2004.04.009 . [DOI] [PubMed] [Google Scholar]
- 44.Collins B, Kane Elizabeth A, Reeves David C, Akabas Myles H, Blau J. Balance of Activity between LNvs and Glutamatergic Dorsal Clock Neurons Promotes Robust Circadian Rhythms in Drosophila. Neuron. 2012;74(4):706–18. doi: 10.1016/j.neuron.2012.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorber C, Leleux S, Stanewsky R, Lamaze A. Light triggers a network switch between circadian morning and evening oscillators controlling behaviour during daily temperature cycles. PLOS Genetics. 2022;18(11):e1010487. doi: 10.1371/journal.pgen.1010487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1(2):141–50. Epub 1988/04/01. doi: 10.1016/0896-6273(88)90198-5 . [DOI] [PubMed] [Google Scholar]
- 47.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18(1):12–25. Epub 2003/02/06. doi: 10.1177/0748730402239673 . [DOI] [PubMed] [Google Scholar]
- 48.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004(220):pl6. Epub 2004/02/19. doi: 10.1126/stke.2202004pl6 . [DOI] [PubMed] [Google Scholar]
- 49.Mazzoni EO, Desplan C, Blau J. Circadian pacemaker neurons transmit and modulate visual information to control a rapid behavioral response. Neuron. 2005;45(2):293–300. Epub 2005/01/25. doi: 10.1016/j.neuron.2004.12.038 . [DOI] [PubMed] [Google Scholar]
- 50.Larderet I, Fritsch PMJ, Gendre N, Neagu-Maier GL, Fetter RD, Schneider-Mizell CM, et al. Organization of the Drosophila larval visual circuit. eLife. 2017;6:e28387. doi: 10.7554/eLife.28387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keene AC, Duboue ER, McDonald DM, Dus M, Suh GS, Waddell S, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010;20(13):1209–15. Epub 2010/06/15. doi: 10.1016/j.cub.2010.05.029 ; PubMed Central PMCID: PMC2929698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sivachenko A, Li Y, Abruzzi KC, Rosbash M. The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron. 2013;79(2):281–92. Epub 2013/07/31. doi: 10.1016/j.neuron.2013.05.015 ; PubMed Central PMCID: PMC3859024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richhariya S, Shin D, Le JQ, Rosbash M. Dissecting neuron-specific functions of circadian genes using modified cell-specific CRISPR approaches. Proc Natl Acad Sci U S A. 2023;120(29):e2303779120. Epub 2023/07/10. doi: 10.1073/pnas.2303779120 ; PubMed Central PMCID: PMC10629539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houl JH, Ng F, Taylor P, Hardin PE. CLOCK expression identifies developing circadian oscillator neurons in the brains of Drosophila embryos. BMC Neurosci. 2008;9:119. Epub 20081218. doi: 10.1186/1471-2202-9-119 ; PubMed Central PMCID: PMC2628352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herrero A, Yoshii T, Ispizua JI, Colque C, Veenstra JA, Muraro NI, et al. Coupling Neuropeptide Levels to Structural Plasticity in Drosophila Clock Neurons. Curr Biol. 2020;30(16):3154–66 e4. Epub 2020/07/04. doi: 10.1016/j.cub.2020.06.009 . [DOI] [PubMed] [Google Scholar]
- 56.Miller CM, Page-McCaw A, Broihier HT. Matrix metalloproteinases promote motor axon fasciculation in the Drosophila embryo. Development. 2008;135(1):95–109. Epub 2007/11/30. doi: 10.1242/dev.011072 . [DOI] [PubMed] [Google Scholar]
- 57.Gorostiza EA, Ceriani MF. Retrograde Bone Morphogenetic Protein Signaling Shapes a Key Circadian Pacemaker Circuit. The Journal of Neuroscience. 2013;33(2):687–96. doi: 10.1523/JNEUROSCI.3448-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das P, L. Maduzia L, Wang H, L. Finelli A, Cho S-H Smith MM, et al. The Drosophila gene Medea demonstrates the requirement for different classes of Smads in dpp signaling. Development. 1998;125(8):1519–28. doi: 10.1242/dev.125.8.1519 [DOI] [PubMed] [Google Scholar]
- 59.Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, et al. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113(6):755–66. Epub 2003/06/18. doi: 10.1016/s0092-8674(03)00400-8 . [DOI] [PubMed] [Google Scholar]
- 60.Liu T, Mahesh G, Yu W, Hardin PE. CLOCK stabilizes CYCLE to initiate clock function in Drosophila. Proc Natl Acad Sci U S A. 2017;114(41):10972–7. Epub 2017/10/05. doi: 10.1073/pnas.1707143114 ; PubMed Central PMCID: PMC5642697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci U S A. 2010;107(30):13497–502. Epub 2010/07/14. doi: 10.1073/pnas.1002081107 ; PubMed Central PMCID: PMC2922133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patop IL, Anduaga AM, Bussi IL, Ceriani MF, Kadener S. Organismal landscape of clock cells and circadian gene expression in Drosophila. bioRxiv. 2023:2023.05.23.542009. doi: 10.1101/2023.05.23.542009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: An Analog of Clock Operative in the Mammalian Forebrain. Science. 2001;293(5529):506–9. doi: 10.1126/science.1060699 [DOI] [PubMed] [Google Scholar]
- 64.Shokri L, Inukai S, Hafner A, Weinand K, Hens K, Vedenko A, et al. A Comprehensive Drosophila melanogaster Transcription Factor Interactome. Cell Rep. 2019;27(3):955–70.e7. Epub 2019/04/18. doi: 10.1016/j.celrep.2019.03.071 ; PubMed Central PMCID: PMC6485956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandez MP, Pettibone HL, Bogart JT, Roell CJ, Davey CE, Pranevicius A, et al. Sites of Circadian Clock Neuron Plasticity Mediate Sensory Integration and Entrainment. Curr Biol. 2020;30(12):2225–37 e5. Epub 2020/05/11. doi: 10.1016/j.cub.2020.04.025 ; PubMed Central PMCID: PMC7314648. [DOI] [PMC free article] [PubMed] [Google Scholar]