Abstract
Background and Objectives
Subarachnoid neurocysticercosis (SANCC) is the most severe form of Taenia solium CNS infection and accounts for the majority of neurocysticercosis-associated mortality. Inflammation is important in the treatment of SANCC because overactivity can lead to serious complications, but excessive suppression may be counterproductive toward parasite eradication. A relative abundance of CSF IL-10 to IL-12 has been associated with increased treatment duration for patients with SANCC, suggesting that IL-10 plays an important role in this disease process. To better understand SANCC immunology and the major sources of IL-10 during anthelmintic treatment, we took an unbiased and comprehensive approach to phenotype the immune cell populations in the CSF and peripheral blood of patients with SANCC.
Methods
Eight samples of CSF cells collected from 5 patients with SANCC during treatment were evaluated using single-cell RNA sequencing. Matched CSF and peripheral blood mononuclear cells from 4 patients were assessed using flow cytometry. Staining for extracellular and intracellular markers allowed for the characterization of IL-10–producing T cells.
Results
The CSF during SANCC contains a diversity of immune cell populations including multiple myeloid and lymphoid populations. Although there were changes in the composition of CSF cells during treatment, the largest population at both early and late time points was CD4+ T cells. Within this population, we identified 3 sources of IL-10 unique to SANCC CSF compared with controls: natural regulatory T cells (nTregs), induced regulatory T cells (iTregs), and Th17 cells. The abundance and phenotype of these IL-10–producing populations differed between CSF and blood in patients with SANCC, but iTregs were the single most productive population in the CSF. During treatment, these IL-10 producers persisted in consistent proportions despite decreases in parasite antigen over time.
Discussion
This profile of immune cell populations in the CSF provides a comprehensive blueprint of the local and systemic immunology associated with SANCC. The identification of IL-10–producing cells in the CSF and peripheral blood deepens our understanding of the immunosuppressive phenotype that deters SANCC treatment success. Finally, the discovery that these IL-10 producers persist throughout treatment highlights the endurance of these populations in the CNS.
Introduction
Neurocysticercosis (NCC) is an important cause of adult-onset epilepsy and long-term disability in developing areas.1,2 The disease results from infection of the CNS by the larval form of Taenia solium, and symptoms often reflect the anatomical location where the Taenia larvae encyst. However, the most severe form of NCC occurs when larval cysts reside in the subarachnoid space.3 This results in subarachnoid neurocysticercosis (SANCC), which is the primary driver of NCC-associated mortality.4 In these cases, stem cells in the cestode tegument have the ability to proliferate, causing a progressive, relapsing, and treatment-refractory form of the disease.5
It is understood that, in SANCC, anthelmintic treatment must be carefully balanced with immunosuppressive treatment because excessive neuroinflammation can result in serious post-treatment complications. Although the inflammation associated with SANCC has been identified as a major driver of morbidity and mortality,6 it is also beneficial for parasite clearance to maintain some level of bias toward a proinflammatory environment.7,8 In addition, it has been shown that high CSF levels of interleukin-10 (IL-10), an immunosuppressive cytokine, relative to CSF IL-12 levels, are associated with longer time to SANCC cure.8 Together, this suggests that an understanding of the major cytokine producers—especially those producing IL-10—will enable us to more effectively manage the factors that affect treatment success.
Despite our recognition that the immune environment plays a critical role in the treatment of SANCC, we still lack a foundational understanding of the neuroimmunology associated with this disease process. Early studies of immune cell populations in the CNS during SANCC found evidence for B-cell and cytotoxic T-cell populations in the CSF.9,10 However, more recent work has focused on FoxP3+ natural regulatory T cells (nTregs) and described correlations between immune populations in CSF and blood.11 While these data provide important immunologic insights, it is still unclear whether these cells exist simultaneously and whether these data provide a complete picture of the immunology associated with SANCC. Given the importance of IL-10, it is also vital that we identify all major sources of IL-10 and the dynamics of these populations over the course of treatment.
This study aimed to further the understanding of neuroimmunologic factors shaping the treatment of SANCC by identifying important cell populations and primary drivers of IL-10 production. In this study, we obtained a comprehensive, unbiased profile of immune cells found in the CSF of patients with SANCC at the time of initiating treatment, identified the major sources of IL-10 in the CSF and peripheral blood, and described the dynamics of these populations during anthelmintic treatment.
Methods
Samples
All CSF samples used for this study were collected from 7 patients with subarachnoid neurocysticercosis. This cohort was composed of 2 women and 5 men. The geometric mean age of all patients was 35 years with a range of 15–44 years. Putative regions of infection included Guatemala, Honduras, El Salvador, and Mexico.
CSF samples were categorized into 2 groups depending on the patient's treatment status at the time of collection. Early-treatment samples were obtained from patients with a new or recurrent episode of SANCC while off anthelmintics or within 1 month of starting adequate anthelmintic therapy (albendazole 15 mg/kg/d with or without praziquantel). A recurrent episode was defined as a clinical and/or radiologic change consistent with subarachnoid neurocysticercosis in the presence of CSF antigen and qPCR positivity. T. solium qPCR was performed as previously described.12 Taenia antigen relative quantification was determined in real time for most samples, but the particular assay changed over time. Therefore, all samples underwent either capture monoclonal ELISA TsG10,13 commercial apDia Cysticercosis ELISA (Turnhout, Belgium), or Luminex-based platform assay using B60/158 monoclonal antibodies.14 However, repeat samples from the same patient underwent identical Taenia antigen quantification protocols, making them comparable between time points. Sample information including experimental usage, patient clinical presentation, anthelmintic and corticosteroid usage at the time of each lumbar puncture, and qPCR and Taenia antigen results are presented in Table 1. All patients improved with treatment, although some currently remain on therapy. To date, there have been no deaths.
Table 1.
Sample Usage and Clinical Data
| Participant | A | B | C | D | E | F | G | |||||
| CSF collection time point | Early | Late | Early | Late | Early | Late | Early | Late | Early | Early | Late | Late |
| Flow | + | + | + | + | + | + | + | + | — | — | — | — |
| scRNA | — | — | — | — | + | + | + | + | + | + | + | + |
| Clinical presentation | Headaches, communicating hydrocephalus | Communicating hydrocephalus, meningitis | Obstructive hydrocephalus, meningitis | Difficulty walking, spinal cord compression | Seizures | Ischemic stroke | Headaches, communicating hydrocephalus | |||||
| New diagnosis vs. recurrence | New Diagnosis | Recurrence | Reccurence | Recurrence | New Diagnosis | Recurrence | Recurrence | |||||
| On albendazole and/or praziquantel | Yes | Yes | No | Yes | Yes | Yes | No | Yes | No | No | Yes | Yes |
| Time on anthelmintics (mo) | <1 | 4 | — | 3 | 1 | 6 | — | 60 | — | — | 3 | 8 |
| Dexamethasone equivalent (mg) | 7.5 | 9 | 0 | 0.9 | 1.5 | 0.8 | 0 | 0 | 18 | 1.1 | 4 | 0.8 |
| Steroid-sparing agents | — | — | — | — | — | Etanercept | Methotrexate | — | — | — | Etanercept, methotrexate | — |
| T. solium qPCR (Cq) | 19.6 | 25.6 | 32.8 | 28.2 | 22.1 | 24.2 | 35.8 | 29 | 40 | 33.2 | 27.1 | 34.3 |
| Taenia antigena (ng/mL) | 17,104 | 8,207 | >4,000 | 20 | 2,239 | 47 | 6,580 | 527 | 21 | 10 | 0 | 0 |
(—) Indicated field not applicable.
Capture ELISA TsG10, commercial ELISA, or B60/158 Luminex assay.
After collection, CSF was centrifuged at 1,500 rpm for 7 minutes at room temperature and the cell pellet was resuspended in 5 mL of RPMI. After an additional spin at 1,500 rpm, the supernatant was discarded and the pellet was resuspended in 1.8 mL of freezing media (RPMI, 20% FCS, 2 mM glutamine, 1% Pen/Strep, 1 mM HEPES) with 0.2 mL of DMSO. Cells were then cryopreserved in 1.8 mL cryovials (ThermoFisher Scientific, cat#375418) using controlled cryopreservation containers with isopropanol, stored at −80°C overnight. Cells were then transferred to liquid nitrogen freezers until thawed for use.
Sample Preparation for Single-Cell RNA Sequencing
Cryopreserved cells were rapidly thawed in a 37°C water bath for no more than 2 minutes. An equal volume of complete media (RPMI + GlutaMAX-1, 10% FBS, 1% HEPES, 1% Pen/Strep) with 10 μg/mL of DNase was added to the sample dropwise, and the diluted sample was transferred into multiple wells of a 96-well V-bottom tissue culture treated plate (Corning, cat#3894). The plate was spun at 1,500 rpm for 7 minutes at 4°C, and the supernatant was discarded. Each pellet was resuspended in 50 μL of complete media, and samples were each pooled back into a single well. Cells were pelleted again by spinning at 2000 rpm for 3 minutes at room temperature, and the supernatant was again discarded. Cells were resuspended in 100 μL of 1:500 diluted Live/Dead viability marker (Fixable Aqua–UV405, Invitrogen, cat#L34957) and incubated for 15 minutes at room temperature. Cells were then diluted with 200 μL of FACS buffer and pelleted by spinning at 2000 rpm for 3 minutes at room temperature. The supernatant was discarded, and cells were resuspended in 200 μL of FACS buffer. Cells were filtered through a 35-μm cell strainer to achieve a single cell suspension and sorted for only viable cells on BD FACSymphony. Sorted cells were collected in 5-mL polystyrene round-bottom tubes (Falcon, cat#352052).
Cells for which collection and single-cell capture were to occur on the same day were never frozen and thus not subject to cell sorting. Freshly collected samples were centrifuged at 1,500 rpm for 7 minutes at room temperature and separated from the CSF. The cell pellet was resuspended in 1 mL of PBS and transferred into a 5-mL polystyrene round-bottom tube.
Single-Cell Library Construction and Sequencing
Single-cell capture, cDNA amplification, and library construction were completed using Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 (10x Genomics, cat#1000127, 1000128) according to the manufacturer's protocols. SPRIselect Reagent (Beckman Coulter, cat#B23317) was used for cDNA cleanup, and quality control steps were completed using Bioanalyzer High Sensitivity DNA Kits (Agilent, cat#5067-4626).
Two 10x Genomics Single Cell Chromium 3′ mRNA libraries were made (S1 and S2) and sequenced as part of 1 Illumina NovaSeq S1 run. Each sample had a sequencing yield of greater than 399 million reads. The sequencing run was performed using recommended parameters with 28 cycles + 90 cycles of asymmetric run.
Initial processing of the 2 samples included removal of cells with a low number of unique molecular identifier counts using Cellranger v7.1.015 with default parameters. Demuxlet16 was used to call multiplet annotations.
The remainder of the single-cell RNA (scRNA) analysis was performed with Seurat v4.1.0.17 Cell type identification was completed using a combination of SingleR18 with the Novershtern hematopoietic database19 and canonical immune cell markers. Sample S1 was filtered for cells containing greater than 200 and less than 4,000 detected genes, less than 5% total mitochondrial gene expression, and singlet calls by demuxlet.16 Sample S2 was filtered for cells containing greater than 200 and less than 4,000 detected genes, less than 10% total mitochondrial gene expression, and singlet calls by demuxlet.16 The final total numbers of cells in S1 and S2 were 668 and 1,336, respectively.
Whole-exome sequencing data were used to deconvolute the patient samples in S1 and S2 with demuxlet.16 FASTQ files were processed to a joint variant call format (VCF) file as described previously.20 Early and late samples were identified, and samples S1 and S2 were then reintegrated using Seurat. Standard scRNA analysis was completed using 20 principal components to visualize the UMAP diagrams. After unsupervised clustering, induced regulatory T cells (iTregs) were defined as the CD4+ T-cell cluster that contained a majority of cells with 0 FoxP3 expression and greater than 0 CTLA4 expression. nTregs were defined as the CD4+ T-cell cluster that contained a majority of cells with greater than 0 FoxP3 expression.
Control CSF data were downloaded from GEO/SRA (bioproject: PRJNA979258; samples: SAMN35573421, SAMN35573423, SAMN35573425, SAMN35573427, SAMN35573429, SAMN35573431, SAMN35573433) and processed using the same methods as mentioned above with the following exception: control samples were filtered for cells containing greater than 200 and less than 5,000 detected genes and less than 10% total mitochondrial gene expression. These data were collected from the cryopreserved CSF of 7 patients with a variety of noninfectious neurologic processes.
Immunophenotypic Analysis
Cryopreserved cells were thawed as described above and diluted in an equal volume of complete media with 10 µg/mL of DNAse. Cells were then transferred to 5-mL polystyrene round-bottom tubes and centrifuged at 1,000 rpm for 10 minutes at room temperature. The supernatant was discarded, and samples were resuspended in 300 µL of PBS. Cells were spun again at 1,200 rpm for 6 minutes at room temperature, and the supernatant was again discarded. Samples were resuspended in 100 µL of 1:500 diluted Live/Dead viability marker (Fixable Blue – UV450, Invitrogen, cat#L34962) and incubated for at least 10 minutes at room temperature. Cells were then stained for extracellular surface markers including CD294 (CRTH2), CD134 (OX40), TCRγδ, CD25, CD197 (CCR7), CD185 (CXCR5), CD196 (CCR6), CD45RA, CD279 (PD-1), CD4, CD194 (CCR4), CD8a, CD183 (CXCR3), and CD127 (IL-7Rα) and 20 µL of Brilliant Stain Buffer Plus (BD Biosciences, cat#566385). After a 15-minute incubation, samples were washed in 150 µL of FACS buffer (PBS, 2% FBS) and centrifuged at 1,200 rpm for 6 minutes at room temperature. The supernatant was again discarded, and cells were fixed with 4.2% formaldehyde (BD Cytofix, BD Biosciences cat#51-2090KZ), permeabilized (eBioscience, Permeabilization Buffer 10x, cat#00-8333-56), and washed. Then, samples were stained with the intracellular antibody mix containing IL-10, CD69, IL-13, FoxP3, IFN-γ, CD3, IL-5, IL-4, CD154, and TNF and 10 µL of Brilliant Stain Buffer Plus for 30 minutes at 4°C (eTable 1).
Gating and quantification analysis were performed using FlowJoTM v10.9.0 Software (BD Life Sciences). Positive and negative gates for all markers were determined based on the staining of live lymphocytes. This allowed for reasonable gating when cell numbers were too diminished for clear distinction of multiple populations. All gates were identically applied to CSF and peripheral blood mononuclear cell (PBMC) samples and samples of different time points (eFigure 1).
Statistical Analyses
All data was analyzed and plotted using Prism 10.0.1 (Graphpad Software; San Diego, CA). Geometric means were used as the primary measures of centration when comparing populations. Comparisons of cell populations between CSF and PBMC or between time points were assessed using two-way ANOVA with the Sidak multiple comparisons test or two-tailed paired t test. Within each compartment, comparisons of more than 2 cell populations were analyzed using repeated-measures one-way ANOVA with the Tukey multiple comparisons test.
Standard Protocol Approvals, Registrations, and Patient Consents
All samples were collected on the NIH Division of Intramural Research IRB-approved protocol (NCT00001205) after obtaining written informed consent. Whole-exome sequencing of 3 patients was performed as part of the Clinical Sequencing Initiative at the National Institutes of Allergy and Infectious Disease (NCT02417766) and is available as part of a dbGAP submission related to that project (dbGaP accession phs001899.v2.p1)21 (Phenotips IDs: P0004036, P0006831, P0007400). Whole-exome sequencing of the other 2 patients was performed under NCT00001205 in a similar manner.
Data Availability
The scRNA data set in this study can be found in the Gene Expression Omnibus repository using accession GSE260929. Scripts used to process all data are available on request.
Results
CSF Cellular Profiling in SANCC Through scRNA
A total of 8 CSF samples collected from 5 patients were assessed using scRNA. 3 patients provided paired samples at both early and late time points, and there were 2 patients from whom only a single sample at 1 time point (1 early, 1 late) was available.
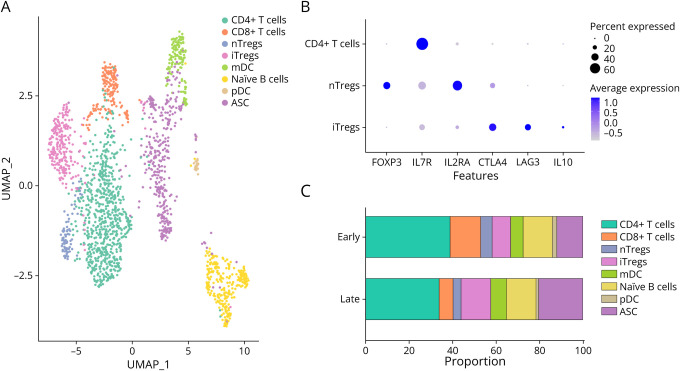
Analysis of all samples, regardless of time points, demonstrated a clear delineation of populations of CD4+ and CD8+ T cells, myeloid and plasmacytoid dendritic cells (mDCs, pDCs), naïve B cells, antibody-secreting B cells (ASCs), and 2 populations of regulatory T cells (Tregs): FoxP3+ natural Tregs (nTregs) and FoxP3- induced Tregs (iTregs) (Figure 1A). The dual presence of both nTregs and iTregs in CSF was unique to SANCC when compared with CSF collected from 7 patients with non-neuroinfectious processes (eFigure 2). Rather, these control CSF samples contained only nTregs with no discernible population of iTregs.
Figure 1. CSF Immune Milieu Is Diverse During SANCC Treatment.
(A) CSF cells collected from 5 patients with SANCC were merged and surveyed using scRNA. Colors denote different cell populations identified using reference databases and canonical markers. (B) 2 populations of CD4+ Tregs were observed in the CSF: nTregs and iTregs. These were isolated as a subset of CD4+ T cells. Canonical Treg-associated features are displayed on the x-axis with percent and average expression portrayed by shape size and color intensity. (C) Samples were collected from 5 patients; 3 patients provided samples at both the early and late time points while 2 provided samples at a single time point. With 4 samples in each group, aggregate scRNA analysis demonstrated shifts in CSF cell populations over the course of treatment with an observed expansion of iTregs, mDCs, and ASCs. Colors parallel those in panel A. ASCs = antibody-secreting B cells; iTregs = induced regulatory T cells; mDC = myeloid dendritic cell; nTregs = natural regulatory T cells; pDC = plasmacytoid dendritic cell; SANCC = subarachnoid neurocysticercosis; scRNA = single-cell RNA sequencing.
In the SANCC samples, both populations of Tregs expressed CD4 and were low in IL7R (CD127). As can be seen in Figure 1B, not only did the Treg populations differ in their expression of FoxP3, but also the population of iTregs had relatively lower expression of IL2RA (CD25) and higher expression of CTLA4, LAG3, and notably IL-10. In fact, most cells producing IL-10 belonged to the subpopulation of iTregs. Meanwhile, production of IL-12 was minimal across all cell populations (eFigure 3).
When we compared the CSF immune milieu between the early and late time points in treatment in aggregate, we were able to see alterations in the cellular composition of the CSF (Figure 1C). Late in treatment, we saw an increase in the proportion of iTregs, mDCs, and ASCs. Of interest, the increase in iTregs at the later time point was accompanied by a decrease in nTregs.
Composition of CD4+ T Cells in CSF Differs From Peripheral Blood
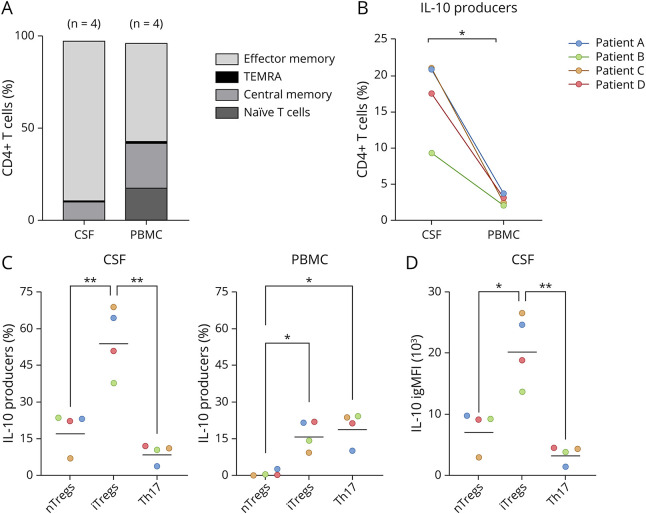
With this new understanding of the immune populations in the CSF during SANCC, we sought to determine whether the inflammation seen in the local CNS environment reflected that seen in peripheral blood. To this end, we used flow cytometry to focus our assessment on T cells present early in SANCC treatment in the CSF and peripheral blood. Using paired CSF and peripheral blood samples from 4 patients early in treatment, we found that the phenotypes of T cells in the peripheral blood were starkly different from those in the CSF (Figure 2A). Across the 4 patients, CSF CD4+ T cells (CD3+TCRgd-CD4+CD8−) were 86.5% effector memory (CD45RA-CCR7-), 9.7% central memory (CD45RA-CCR7+), and only 0.3% naïve (CD45RA+CCR7+). By contrast, CD4+ T cells in the peripheral blood were more evenly distributed with only 53.1% effector memory, 23.9% central memory, and 17.7% naïve. Both CSF and peripheral T-cell populations had very small proportions of terminally differentiated effector CD4+ T cells (TEMRA: CD45RA+CCR7-) with only 0.6% TEMRAs found in CSF and 1.3% in PBMC. Overall, the CSF and peripheral blood differed in their respective proportions of naïve T cells (padj = 0.019) and effector memory T cells (padj = 0.0003).
Figure 2. CSF and Peripheral Blood Have Distinctly Different Phenotypes.
(A) Flow cytometry analysis of matched CSF and peripheral blood revealed differences in the relative proportions of CD4+ T-cell populations. The colors denote distinct populations with the height of each section representing the geometric mean proportion of that population across the 4 sampled patients. When characterized by CCR7 and CD45RA expression, CSF CD4+ T cells (CD3+TCRgd-CD4+CD8−) were predominantly effector memory (CD45RA-CCR7-) while peripheral blood CD4+ T cells demonstrated higher proportions of naïve (CD45RA+CCR7+) and central memory T (CD45RA-CCR7+) cells. Both compartments harbored small populations of terminally differentiated effector CD4+ T or TEMRA cells (CD45RA+CCR7-). (B) Within CD4+ T cells, IL-10+ cells were identified and named IL-10 producers. The frequency of IL-10 producers differed between the CSF and peripheral blood of paired samples from 4 patients (each represented by a different color, p = 0.011). (C) In the CSF of these 4 patients, repeated-measures one-way ANOVA with the Tukey multiple comparisons test showed that there were more iTregs than both nTregs (padj = 0.009) and Th17 cells (padj = 0.003), whereas there were more iTregs and Th17 cells than nTregs (iTregs [padj = 0.024], Th17 cells [padj = 0.011]) in the peripheral blood. Horizontal bars indicate geometric mean of percent of IL-10 producers, and symbol colors reflect the patient identity of each sample, consistent with those in panel B. (D) Using integrated geometric mean fluorescence intensity (igMFI) to compare the contributions of each IL-10–producing cell population, iTregs were found to produce more IL-10 than nTregs (padj = 0.015) and Th17 cells (padj = 0.004). Horizontal bars indicate the geometric mean of IL-10 igMFI, and symbol colors reflect the patient identity of each sample, consistent with those in panel B. igMFI = integrated geometric mean of fluorescence intensity; iTregs = induced regulatory T cells; nTregs = natural regulatory T cells; PBMC = peripheral blood mononuclear cell.
Sources of IL-10 in the CSF in SANCC
Given the importance of IL-10 in altering treatment success in SANCC and that CD4+ T cells were the major source of IL-10 in our scRNA data, we sought to determine which IL-10–producing CD4+ T cells were critical in driving the IL-10–mediated phenomena observed in SANCC. As seen in Figure 2B, the proportion of these IL-10–producing CD4+ T-cell populations differed between the CSF and peripheral blood with the CSF harboring a higher proportion of these IL-10 producers (p = 0.011).
Of interest, the distribution of phenotypes of IL-10 producers in CSF and PBMC diverged as well (Figure 2C). In both compartments, we observed 3 distinct populations: nTregs (CD127-FoxP3+), iTregs (CD127-FoxP3-), and Th17 cells (CD127+CRTH2-CCR6+CCR4+).22,23 In CSF, iTregs were the dominating population of IL-10 producers compared with nTregs (padj = 0.009) and Th17 cells (padj = 0.003). In the peripheral blood, iTregs and Th17 cells were present in modest proportions with a smaller proportion of nTregs (iTregs [padj = 0.024], Th17 [padj = 0.011]). When directly comparing CSF with PBMC, the abundance of all 3 populations differed between these compartments (nTregs [p = 0.017], iTregs [p = 0.017], Th17 [p = 0.008]). Of interest, the largest population of IL-10 producers in PBMC was a CD127+CRTH2-CCR6+CCR4- population of CD4+ T cells that was not reflected in the CSF.
After determining the primary IL-10–producing populations in the CSF, we used the integrated geometric mean fluorescence intensity (igMFI)24 to determine which lone population produced the most IL-10 in the CNS. As seen in Figure 2D, iTregs were the largest source of IL-10 in the CSF, producing 2.87 times more IL-10 than nTregs (padj = 0.015) and 6.25 times more IL-10 than Th17 cells (padj = 0.004).
IL-10 Producers Persist in the CSF Throughout SANCC Treatment
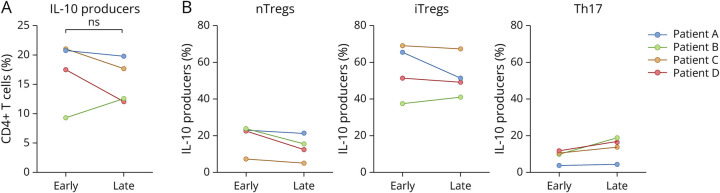
Given that CSF IL-10 levels during SANCC treatment have been shown to positively correlate with time to cure,8 we used flow cytometry analysis to observe changes in the populations of IL-10 producers over the course of treatment in samples longitudinally collected from 4 patients at early and late time points (Figure 3). Despite all samples exhibiting substantial decreases in parasite antigen over the course of treatment (Table 1), we found that there was no change in the proportion of IL-10 producers within CD4+ T cells (p = 0.445) (Figure 3A). Furthermore, we determined that nTregs (p = 0.085), iTregs (p = 0.399), and Th17 cells (p = 0.110) all persisted in similar proportions (Figure 3B). These data largely corroborate the persistence of IL-10 when measured in the CSF of those with SANCC late in treatment.8
Figure 3. IL-10 Producers Persist in the CSF Late in Treatment.
(A) Flow cytometric analysis and paired t test of CSF samples provided by 4 patients (represented by color) early and late in treatment showed no difference in the proportion of IL-10 producers between the 2 treatment time points (p = 0.445). (B) Within these IL-10 producers, paired t tests found no significant changes in relative proportions of nTregs, iTregs, or Th17 cells over the course of treatment (nTregs [p = 0.085], iTregs [p = 0.399], Th17 [p = 0.110]), suggesting that the persistent population of IL-10 producers does not change in composition. Symbol colors reflect the patient identity of each sample, consistent with those in panel B. iTregs = induced regulatory T cells; nTregs = natural regulatory T cells.
Discussion
This study establishes a comprehensive foundation for understanding the neuroimmunology associated with SANCC. In this study, we observed a diversity of expanded lymphocytic and myeloid cell populations including CD4+ and CD8+ T cells, mDCs, pDCs, naïve B cells, and ASCs. Although some of these populations have been reported previously,9,10 this study describes the co-occurrence of these populations, their relative proportions, and the kinetics of these populations over time.
Our identification of 2 dendritic cell populations—mDCs and pDCs—in the local CNS environment supports the long proposed mechanism that dendritic cells (DCs) induce Treg differentiation in the context of NCC.25,26 Before this study, these hypotheses were based on the expression of markers of dendritic cell contact (CTLA-4, LAG-3, GITR, PD-1) on Tregs isolated from the CSF of patients with NCC,26 the observation of DC-mediated Treg induction in vitro, and studies of DCs isolated from PBMC of patients with NCC. Our data provide additional evidence supporting this theory by definitively documenting the presence of DCs in the CSF of those with SANCC. Similarly, this demonstration of both naïve B cells and terminally differentiated ASCs in the CSF during SANCC provides new insights into the local production of antibodies in SANCC and might explain the relationship between the presence of autoantibodies27,28 and the pathology seen in SANCC.
We also identified 2 distinct populations of CD4+ Tregs in the CSF of patients with SANCC: nTregs and iTregs. It is important to note that iTregs were not identified in control CSF from patients with noninfectious neurologic conditions. Although iTregs have been reported in other parasitic diseases, it was previously unknown whether they played a role in NCC. In both Schistosoma mansoni and malaria pathogenesis, iTregs have been reported to control inflammation through an IL-10–dependent mechanism that deters parasite clearance.29,30 This study suggests that iTregs serve a similar immunosuppressive role in SANCC as a major producer of IL-10 in the PBMC and the most abundant source of IL-10 in the CSF.
Our comparison of important T-cell populations between CSF cells and PBMC during SANCC revealed that the local CNS and peripheral blood compartments differ. Previous work had suggested that the immune cells found in the CSF were recruited from the blood.11 However, our data demonstrate disparities between these compartments, including a stronger polarity toward effector memory cells in the CSF and higher proportions of IL-10–producing CD4+ T cells in the CSF compared with PBMC. These data thus support previous findings that cytokines observed in the CSF are locally produced rather than peripherally imported8 and identify the cell populations likely responsible for their production. Our study also found differences in the phenotypes of these IL-10 producers in each compartment. In the CSF, IL-10–producing CD4+ T cells were mainly iTregs with small populations of nTregs and Th17 cells. Meanwhile, the IL-10 producers in PBMC were primarily composed of a population that was CD4+CD127+CRTH2-CCR6+CCR4-.
By profiling CSF cells collected early and late in disease, we assessed the evolution of the CSF immune milieu over the course of treatment. For all patients in this study, the late time point in treatment had a much lower CSF white blood cell count than the earlier time point. However, our scRNA data suggested that this reduction in cell count was not uniform across all cell types. Notably, these data showed an increase in iTregs and a decrease in nTregs at the later time point. Of interest, when characterizing the IL-10–producing cells in the CSF using flow cytometry, we observed no change in the proportion of IL-10 producers in CD4+ T cells, nTregs, or iTregs over the course of treatment. This discrepancy may be due to the difference in platforms and small sample size, limiting the ability to detect small shifts over time.
Limitations affecting this work included small sample size, cell fragility, and the confounding use of corticosteroids with anthelmintics during patient treatment. Because our samples were cryopreserved, the cells were more fragile, preventing us from using stimulation to increase cytokine production in our analyses. This also resulted in an absence of granulocytes because these cells were not able to survive the freeze/thaw process. Despite these challenges, our data provide important insight into the composition and dynamics of immune cell populations within the CSF over the course of SANCC treatment.
Overall, this work describes the neuroimmunology associated with SANCC. We presented an unbiased profile of the immune cell populations within the CSF early and late in anthelmintic treatment. We demonstrated that the immune milieu in the CSF is distinct from that of the periphery, supporting the notion that inflammation found in the CSF is produced locally. Finally, we identified multiple populations of IL-10–producing cells that likely influence treatment responsiveness. It is important to note that we discovered that iTregs are the primary drivers of IL-10 in the CSF.
These findings leave us with many opportunities for future investigations. These include efforts to understand the roles of each of the cell populations identified here within the CSF, their interactions with and effects on co-existent populations, and the mechanism of induction of these newly observed iTregs. Having identified iTregs as the most abundant source of IL-10 in the CSF, future work aimed to specifically target this population could result in increased success and shorter courses of anthelmintic treatment. Ultimately, this work provides a blueprint for us to better understand the complex immunologic mechanisms that aid or deter our abilities to treat SANCC effectively.
Acknowledgment
The authors appreciate the participants in this study and those who supported their care. The authors are grateful to all members of the Laboratory of Parasitic Diseases who contributed to this work. The authors also acknowledge Jason Brenchley and his group at the National Institute of Allergy and Infectious Diseases for their support regarding this project.
Glossary
- ASCs
antibody-secreting B cells
- DC
dendritic cell
- iTregs
induced regulatory T cells
- NCC
neurocysticercosis
- nTregs
natural regulatory T cells
- PBMC
peripheral blood mononuclear cell
- SANCC
subarachnoid neurocysticercosis
- scRNA
single-cell RNA sequencing
Appendix. Authors
| Name | Location | Contribution |
| Nina L. Tang, BA | Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases | Drafting/revision of the manuscript for content, including medical writing for content; major role in the acquisition of data; analysis or interpretation of data |
| Paul Schaughency, PhD | Integrated Data Sciences Section, National Institute of Allergy and Infectious Diseases | Drafting/revision of the manuscript for content, including medical writing for content; analysis or interpretation of data |
| Pedro Gazzinelli-Guimaraes, PhD | Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases | Major role in the acquisition of data; analysis or interpretation of data |
| Justin Lack, PhD | Integrated Data Sciences Section, National Institute of Allergy and Infectious Diseases | Analysis or interpretation of data |
| Lauren Thumm, RN, MSN | Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases; Clinical Monitoring Research Program Directorate, Frederick National Laboratory for Cancer Research | Major role in the acquisition of data |
| Emily Miltenberger, BS | Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases | Major role in the acquisition of data |
| Theodore E. Nash, MD | Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases | Major role in the acquisition of data |
| Thomas B. Nutman, MD | Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design |
| Elise M. O'Connell, MD | Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases | Drafting/revision of the manuscript for content, including medical writing for content; study concept or design |
Study Funding
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases.
Disclosure
The authors report no relevant disclosures. Go to Neurology.org/NN for full disclosures.
References
- 1.Butala C, Brook TM, Majekodunmi AO, Welburn SC. Neurocysticercosis: current perspectives on diagnosis and management. Front Vet Sci. 2021;8:615703. doi: 10.3389/fvets.2021.615703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taeniasis/Cysticercosis. Accessed October 21, 2021. who.int/news-room/fact-sheets/detail/taeniasis-cysticercosis
- 3.Nash TE, O'Connell EM, Hammoud DA, Wetzler L, Ware JM, Mahanty S. Natural history of treated subarachnoid neurocysticercosis. Am J Trop Med Hyg. 2020;102(1):78-89. doi: 10.4269/ajtmh.19-0436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abanto J, Blanco D, Saavedra H, et al. . Mortality in parenchymal and subarachnoid neurocysticercosis. Am J Trop Med Hyg. 2021;105(1):176-180. doi: 10.4269/ajtmh.20-1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orrego MA, Verastegui MR, Vasquez CM, et al. . Identification and culture of proliferative cells in abnormal Taenia solium larvae: role in the development of racemose neurocysticercosis. PLoS Negl Trop Dis. 2021;15(3):e0009303. doi: 10.1371/journal.pntd.0009303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nash TE, O'Connell EM. Subarachnoid neurocysticercosis: emerging concepts and treatment. Curr Opin Infect Dis. 2020;33(5):339-346. doi: 10.1097/QCO.0000000000000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cárdenas G, Fragoso G, Rosetti M, et al. . Neurocysticercosis: the effectiveness of the cysticidal treatment could be influenced by the host immunity. Med Microbiol Immunol. 2014;203(6):373-381. doi: 10.1007/s00430-014-0345-2 [DOI] [PubMed] [Google Scholar]
- 8.Harrison S, Thumm L, Nash TE, Nutman TB, O'Connell EM. The local inflammatory profile and predictors of treatment success in subarachnoid neurocysticercosis. Clin Infect Dis. 2021;72(9):e326-e333. doi: 10.1093/cid/ciaa1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bueno EC, Vaz AJ, Oliveira CA, et al. . Analysis of cells in cerebrospinal fluid from patients with neurocysticercosis by means of flow cytometry. Cytometry. 1999;38(3):106-110. doi: [DOI] [PubMed] [Google Scholar]
- 10.Bueno EC, dos Ramos Machado L, Livramento JA, Vaz AJ. Cellular immune response of patients with neurocysticercosis (inflammatory and non-inflammatory phases). Acta Trop. 2004;91(2):205-213. doi: 10.1016/j.actatropica.2004.05.010 [DOI] [PubMed] [Google Scholar]
- 11.Adalid-Peralta L, Fleury A, García-Ibarra TM, et al. . Human neurocysticercosis: in vivo expansion of peripheral regulatory T cells and their recruitment in the central nervous system. J Parasitol. 2012;98(1):142-148. doi: 10.1645/GE-2839.1 [DOI] [PubMed] [Google Scholar]
- 12.O'Connell EM, Harrison S, Dahlstrom E, Nash T, Nutman TB. A novel, highly sensitive quantitative polymerase chain reaction assay for the diagnosis of subarachnoid and ventricular neurocysticercosis and for assessing responses to treatment. Clin Infect Dis. 2020;70(9):1875-1881. doi: 10.1093/cid/ciz541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corda M, Sciurba J, Blaha J, et al. . A recombinant monoclonal-based Taenia antigen assay that reflects disease activity in extra-parenchymal neurocysticercosis. PLoS Negl Trop Dis. 2022;16(5):e0010442. doi: 10.1371/journal.pntd.0010442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandt JR, Geerts S, De Deken R, et al. . A monoclonal antibody-based ELISA for the detection of circulating excretory-secretory antigens in Taenia saginata cysticercosis. Int J Parasitol. 1992;22(4):471-477. doi: 10.1016/0020-7519(92)90148-e [DOI] [PubMed] [Google Scholar]
- 15.Stuart T, Butler A, Hoffman P, et al. . Comprehensive integration of single-cell data. Cell. 2019;177(7):1888-1902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang HM, Subramaniam M, Targ S, et al. . Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018;36(1):89-94. doi: 10.1038/nbt.4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Y, Hao S, Andersen-Nissen E, et al. . Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573-3587.e29. doi: 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aran D, Looney AP, Liu L, et al. . Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163-172. doi: 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novershtern N, Subramanian A, Lawton LN, et al. . Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296-309. doi: 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazzinelli-Guimaraes PH, Sanku G, Sette A, et al. . Antigenic determinants of SARS-CoV-2-specific CD4+ T cell lines reveals m protein-driven dysregulation of interferon signaling. Front Immunol. 2022;13:883159. doi: 10.3389/fimmu.2022.883159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Similuk MN, Yan J, Ghosh R, et al. . Clinical exome sequencing of 1000 families with complex immune phenotypes: toward comprehensive genomic evaluations. J Allergy Clin Immunol. 2022;150(4):947-954. doi: 10.1016/j.jaci.2022.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acosta-Rodriguez EV, Rivino L, Geginat J, et al. . Surface phenotype and antigenic specificity of human interleukin 17–producing T helper memory cells. Nat Immunol. 2007;8(6):639-646. doi: 10.1038/ni1467 [DOI] [PubMed] [Google Scholar]
- 23.Becattini S, Latorre D, Mele F, et al. . T cell immunity. Functional heterogeneity of human memory CD4⁺ T cell clones primed by pathogens or vaccines. Science. 2015;347(6220):400-406. doi: 10.1126/science.1260668 [DOI] [PubMed] [Google Scholar]
- 24.Shooshtari P, Fortuno ES, Blimkie D, et al. . Correlation analysis of intracellular and secreted cytokines via the generalized integrated mean fluorescence intensity. Cytometry A . 2010;77(9):873-880. doi: 10.1002/cyto.a.20943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adalid-Peralta L, Arce-Sillas A, Fragoso G, et al. . Cysticerci drive dendritic cells to promote in vitro and in vivo tregs differentiation. Clin Dev Immunol. 2013;2013:981468. doi: 10.1155/2013/981468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arce‐Sillas A, Álvarez‐Luquín DD, Cárdenas G, et al. . Interleukin 10 and dendritic cells are the main suppression mediators of regulatory T cells in human neurocysticercosis. Clin Exp Immunol. 2016;183(2):271-279. doi: 10.1111/cei.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhouse RME, Carpio A, Cortez MM, von Kriegsheim A, Fesel C. Anti-brain protein autoantibodies are detectable in extraparenchymal but not parenchymal neurocysticercosis. J Neuroimmunol. 2020;344:577234. doi: 10.1016/j.jneuroim.2020.577234 [DOI] [PubMed] [Google Scholar]
- 28.Parkhouse RME, Sciutto E, Hernández M, Cortez MM, Carpio A, Fleury A. Extraparenchymal human neurocysticercosis induces autoantibodies against brain tubulin and MOG35-55 in cerebral spinal fluid. J Neuroimmunol. 2020;349:577389. doi: 10.1016/j.jneuroim.2020.577389 [DOI] [PubMed] [Google Scholar]
- 29.Dewals B, Hoving JC, Horsnell WGC, Brombacher F. Control of Schistosoma mansoni egg-induced inflammation by IL-4-responsive CD4(+)CD25(-)CD103(+)Foxp3(-) cells is IL-10-dependent. Eur J Immunol. 2010;40(10):2837-2847. doi: 10.1002/eji.200940075 [DOI] [PubMed] [Google Scholar]
- 30.Couper KN, Blount DG, Wilson MS, et al. . IL-10 from CD4CD25Foxp3CD127 adaptive regulatory T cells modulates parasite clearance and pathology during malaria infection. PLoS Pathog. 2008;4(2):e1000004. doi: 10.1371/journal.ppat.1000004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The scRNA data set in this study can be found in the Gene Expression Omnibus repository using accession GSE260929. Scripts used to process all data are available on request.