Abstract
Background:
A combination of proton-pump inhibitors (PPI) and topical steroids (TS) is used to treat children with eosinophilic esophagitis (EoE). However, a subset of children do not respond to this combination therapy. We aimed to identify the esophageal transcriptional, cell composition, and microbial differences between the non-responders (EoE-PPI-TSnr; n = 7) and responders (EoE-PPI-TSr; n = 7) to the combination therapy for EoE and controls (n = 9) using metatranscriptomics.
Methods:
Differential gene expression analysis was used to identify transcriptional differences, validated using the EoE diagnostic panel (EDP). Deconvolution analysis was performed to identify differences in their cell type composition. Microbiome analysis was conducted from esophageal biopsies RNAseq data, and microbial abundance was correlated with esophageal gene expression.
Results:
In all, 3164 upregulated and 3154 downregulated genes distinguished EoE-PPI-TSnr from EoE-PPI-TSr. Eosinophilic inflammatory response, cytokine signaling, and collagen formation pathways were significantly upregulated in EoE-PPI-TSnr. There was a 56% overlap in dysregulated genes between EoE-PPI-TSnr and EDP, with a perfect agreement in the directionality of modulation. Eosinophils, dendritic cells (DCs), immature DCs, megakaryocytic-erythroid progenitors, and T helper type 1 cells were significantly higher in EoE-PPI-TSnr. There was no significant difference in microbiome diversity. The relative abundance of Fusobacterium sp. and Acinetobacter sp. notably differed in EoE-PPI-TSnr and correlated with the key pathways.
Conclusion:
Our results provide critical insights into the molecular, cellular, and microbial factors associated with the lack of response to PPI and TS combination therapy in children with EoE. This study advances our understanding of the pathobiology of EoE while guiding personalized treatment strategies.
Keywords: eosinophilic esophagitis, gene expression, immune response, Metatranscriptomics, microbiome
Graphical Abstract
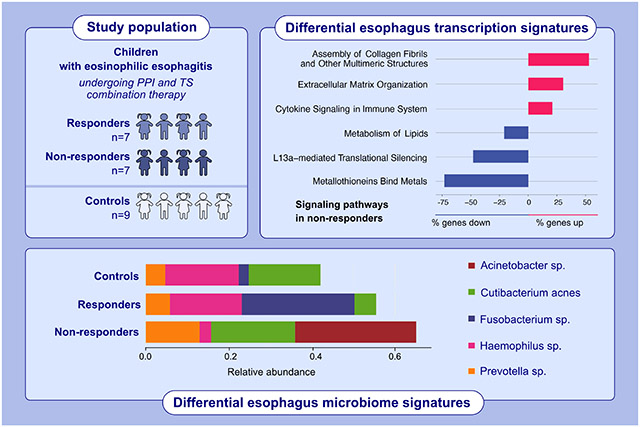
This study aimed to identify the esophageal transcriptional, cell composition, and microbial differences between the non-responders and responders to the combination therapy for EoE and controls using metatranscriptomics. In children with eosinophilic esophagitis and on a combination of proton-pump inhibitors and topical steroids therapy, non-responders show upregulation of cytokine signaling, assembly of collagen fibrils, and extracellular matrix organization, along with downregulation of the L13a-mediated translational silencing and metallothionein pathways. Additionally, there are distinct esophageal microbiome signatures that appear to be linked to the differential transcription signatures.
Abbreviations: EoE, eosinophilic esophagitis; PPI, proton pump inhibitors; TS, topical steroids
1 ∣. INTRODUCTION
Eosinophilic esophagitis (EoE) is a T helper 2 mediated, allergen-driven chronic disease of the esophagus characterized by complex interactions between the host and environmental factors.1 It is estimated to affect 34 per 100,000 children annually.2 Affected children typically present with refusal to feed, swallowing difficulties, vomiting, and abdominal pain. Their diagnosis is confirmed by an intense eosinophilic inflammation [defined as a peak eosinophil count (PEC) of ≥15 eosinophils per high power field (eos/hpf)] in at least one of the multiple esophageal biopsies in the absence of other causes of esophageal eosinophilia.3 Sub-optimal control of the esophageal eosinophilic inflammation, either due to a delay in diagnosis or non-response to therapies, can lead to fibrostenotic complications requiring endoscopic or surgical interventions.4 Therefore, the primary goals of EoE therapy are to ameliorate symptoms, control eosinophilic inflammation, and prevent complications.5
Proton-pump inhibitors (PPIs) are commonly used as the firstline pharmacological agent to manage pediatric EoE.6 However, on average, only about 39% (range 23%–83%) of children with EoE on a PPI achieve histologic remission (defined as a PEC of <15 eos/hpf).7,8 Although there is limited data, in those non-responsive to PPI monotherapy, topical steroids (TSs) can be added off-label (combination therapy) to induce and maintain histologic remission.9 However, some children may not respond to this combination therapy. They continue to experience clinical symptoms while remaining at a higher risk for ensuing complications due to uncontrolled eosinophilic inflammation. The host and environmental factors associated with non-response to combination therapy remain to be thoroughly evaluated.
Esophageal gene expression using microarray analysis and RNA sequencing has been performed to elucidate molecular signatures associated with response to PPI8-11 and TS.12 Likewise, multiple studies using the 16S rRNA-sequencing technique have demonstrated that esophageal microbial dysbiosis can be associated with the pathogenesis and treatment response in EoE.13-16 However, little is known about the molecular and commensal microbial alterations associated with non-response to PPI and TS combination therapy in children with EoE. This represents a critical knowledge gap with significant implications for advancing our understanding of the pathobiology of EoE and personalizing pharmacological management of pediatric EoE.
Recent advances in high-throughput sequencing technologies, such as metatranscriptomics, have enabled a comprehensive analysis of host and non-host gene expression and microbiome through more extensive sequencing and sophisticated bioinformatic analysis compared to conventional approaches (such as microarray analysis, RNA sequencing, 16S rRNA sequencing).17 This technique is uniquely suited for studying the host microbiome as it allows for species-level identification of micro-organisms and their potential roles in disease states.18,19 Furthermore, this versatile approach can enhance our ability to unravel the complex interaction between host cells and microbiome in the pathogenesis of disease state and treatment response. However, metatranscriptomics has not been applied to elucidate the host esophageal and microbial associations with a non-response to the combination therapy in children with EoE.20
To address the knowledge gap, we primarily sought to identify molecular and microbial differences between children with EoE who responded to combination therapy, those who did not, and non-EoE controls using metatranscriptomics. The secondary aims were to validate the differences in gene expression using the EoE diagnostic panel (EDP) and to determine the differences in the cell type composition between responders and non-responders. EDP comprises of representative EoE genes and has been validated for molecular diagnosis of EoE, disease endotyping, and predicting treatment response.10,21 Our exploratory aim was to assess the correlation between the esophageal molecular response and the esophageal microbiome in non-responders. We hypothesized that children with EoE who were non-responsive to combination therapy would have a distinct esophageal gene expression profile and microbial community compared to responders and non-EoE controls.
2 ∣. METHODS
2.1 ∣. Study design
Prospectively collected distal esophageal biopsies and corresponding clinical metadata after obtaining appropriate informed consent and assent under the auspices of the Vanderbilt institutional review board approved protocols (# 151341 and 160785) for previously published studies were analyzed.22,23 Briefly, children 6–18 years old with a previous diagnosis of EoE or upper gastrointestinal symptoms suggestive of EoE and undergoing an esophagogastroduodenoscopy (EGD) at Vanderbilt Children's Hospital were enrolled. During the EGD, 2–3 biopsies, each from the proximal and distal esophagus (4–6 per participant), were collected for clinical care and submitted for histopathologic assessment per our institutional protocol. In addition, two distal esophageal biopsies (≤5 cm from the lower esophageal sphincter) were collected for research purposes and stored in RNA later (Qiagen, catalog number 76154) separately at −80°C. Children with known esophageal injury or surgery, celiac disease, inflammatory bowel disease, and exposure to antibiotics and systemic steroids within the past 30 days were excluded to minimize confounding.
2.2 ∣. Study groups
Per the 2018 AGREE consensus statement, children with symptoms of esophageal dysfunction and a PEC of ≥15 eos/hpf without other causes of esophageal eosinophilia were diagnosed with EoE.24 Children with EoE and on PPI and TS for at least 12 weeks at their EGD were classified as non-responders (EoE-PPI-TSnr) if their PEC was ≥15 eos/hpf. Participants were considered responders (EoE-PPI-TSr) if their PEC was <15 eos/hpf. In our institution, as a standard of care, EoE patients are typically initiated on a PPI monotherapy, and TS is added to PPI monotherapy non-responders to treat them with PPI-TS combination therapy. The treatment response is typically assessed between 12 weeks and 16 weeks in children with EoE, and post-treatment biopsies were analyzed in this study. The non-EoE control (controls) group comprised of children who did not have a prior diagnosis of EoE and did not meet the histologic criteria for EoE. This group included children with abdominal pain and functional dyspepsia.
2.3 ∣. Clinical data collection
Demographic (age at EGD, sex, ethnicity), clinical (weight, indication for EGD, allergic co-morbidities, PPI and TS exposure and dose, and the number of esophageal biopsies), endoscopic (esophageal mucosal) abnormalities visualized during EGD and rated per the validated endoscopic reference score [EREFS],25 and histologic (PEC) information was gathered from the electronic medical records.
2.4 ∣. Metatranscriptomic sequencing of esophageal biopsies
The total RNA from the esophageal biopsies collected for research and stored at −80°C was extracted.26 Illumina sequencing libraries were made using the NEBNext Ultra II RNA Library Prep Kit (NEB #E7775), with human rRNA removal prior to library preparation.27 The quality of the libraries was assessed using an Agilent Bioanalyzer DNA High Sensitivity chip. The libraries were then sequenced on an Illumina NovaSeq 6000 platform (S4 flow cells run) with 2 × 150 base pair reads, with a sequencing depth of ~40 million paired-end reads per sample (see Data S1 for additional details).
2.5 ∣. Statistical analysis
Descriptive statistics were used to characterize the cohort. Continuous variables are presented as median (interquartile range [IQR]), and frequencies and percentages describe categorical variables.
To assess host gene expression, we mapped reads identified as originating from human transcripts to the human genome (hg19) using HISAT2.28 We quantified the read counts for genomic features using HTseq and combined the feature counts of all samples into a single matrix using a custom R script.29 Differential expression analysis was performed to comparing EoE-PPI-TSr, EoE-PPI-TSnr, and control groups using the DESeq2 package.30 Genes with a significant log2 fold change with an adjusted p-value < 0.05 were considered as differentially expressed. The differentially expressed genes for each group were analyzed for enrichment of Reactome Human Pathways using Enrichr and regarded as significant when false discovery rate (FDR) <0.05.31 Further, to substantiate our findings and to identify new pathways for investigation. We compared the differentially expressed genes' directionality with the previously validated EDP.
The individual cell types that contributed to the gene expression patterns in EoE-PPI-TSnr and EoE-PPI-TSr groups were identified through deconvolution analysis using xCell.32,33 We converted raw gene counts to log counts per million (CPM) with a prior count of 3 using limma (version 3.50.3) and calculated raw enrichment scores, which were then normalized. To improve accuracy, a beta distribution was used to exclude all cell types not in the gene count mixture using a threshold of cell types present in at least three samples. Finally, spillover was calculated with a default alpha of 0.5, and Z-scores demonstrating enrichment were graphed using a complex heatmap (version 2.10.0).34 A Wilcoxon-signed rank test with Benjamin–Hochberg correction for multiple testing was performed to assess the statistical relation between the cell type scores and treatment response status.
For the microbiome analysis, we used microbial reads to profile the composition of microbial communities at the species level using metagenomics operational taxonomic units (mOTUs) profiler.35 The differences in relative abundance (proportion) of bacteria of interest were calculated by using a Kruskal–Wallis test.36 Microbial richness and alpha diversity metrics were analyzed using the Phyloseq R package version 1.30.0.37 The Shannon and Chao1 diversity indices were calculated from the mOTUs profiler counts in the samples to assess the alpha diversity of the microbial communities they represent.35 The Wilcoxon rank sum test assessed the statistical significance of microbial richness and alpha diversity differences.
Next, we explored the correlation between differentially expressed esophageal genes and the microbiome. We performed nonparametric Kendall's tau rank correlations between each microbial species' relative abundance and each differentially expressed gene's normalized expression level in EoE-PPI-TSnr and EoE-PPI-TSr children (excluding controls). Kendall's tau correlation accounted for ties caused by heavily zero-inflated microbial abundances.
Finally, we assessed the power of our sample size by performing nonparametric resampling of the target genes/microbes within the EoE-PPI-TSr and EoE-PPI-TSnr groups. The results from differential expression/abundance analysis on the resampled data using DESeq2 for genes and Kruskal–Wallis tests for microbes were synthesized across 100 resampling iterations to estimate power.
3 ∣. RESULTS
3.1 ∣. Characteristics of the cohort
Esophageal samples from 23 children were analyzed, including 7 with EoE-PPI-TSnr, 7 with EoE-PPI-TSr, and 9 with controls. The cohort's median age was 14 years10-15 at the time of EGD, most of whom were White (91%) and male (65%). Expectedly, children with EoE had a higher burden of atopic co-morbidities than controls, and EoE-PPI-TSnr children had significantly higher PEC than EoE-PPI-TSr and controls. Interestingly, children in the EoE-PPI-TSnr group had significantly higher rates of food allergy than those in the EoE-PPI-TSr group (Table 1).
TABLE 1.
Demographic, clinical, endoscopic, and histologic features of the cohort.
| EoE-PPI-TS (n = 14) | ||||||
|---|---|---|---|---|---|---|
| Non- responder (n = 7) |
Responder (n = 7) |
Control (n = 9) |
p value | |||
| A | B | C | A vs. B | A VS. C | B VS. C | |
| Age (years)a | 12 (10–15) | 14 (8–15) | 14 (11–15) | NS | NS | NS |
| Ethnicityb | ||||||
| White | 6 (86) | 6 (86) | 9 (100) | NS | NS | NS |
| Others | 1 (14) | 1 (14) | 0 (0) | NS | NS | NS |
| Maleb | 6 (86) | 5 (71) | 4 (44) | NS | NS | NS |
| Weight (kg) | 57 (30–67) | 54 (32–77) | 64 (46–67) | NS | NS | NS |
| Indication for EGDb | ||||||
| Nausea | – | 1 (14) | 2 (22) | – | – | NS |
| Abdominal pain | – | 1 (14) | 6 (67) | – | – | 0.005 |
| Swallowing difficulties | – | – | 2 (22) | – | – | – |
| Known EoE | 7 (100) | 7 (100) | – | NS | – | – |
| Atopic comorbidityb | ||||||
| Food allergy | 6 (86) | 3 (43) | 3 (33) | NS | 0.03 | NS |
| Allergic rhinitis | 5 (71) | 7 (100) | 1 (11) | NS | <0.001 | <0.001 |
| Asthma | 3 (43) | 5 (71) | 1 (11) | NS | NS | <0.001 |
| Atopic dermatitis | 3 (43) | 4 (57) | 2 (22) | NS | NS | NS |
| Medicationsb | ||||||
| Exposure | ||||||
| Nasal steroids | 4 (57) | 3 (43) | 2 (22) | NS | NS | NS |
| Proton-pump inhibitors | 7 (100) | 7 (100) | – | NS | – | – |
| Topical steroids | 7 (100) | 7 (100) | – | NS | – | – |
| Dosea | ||||||
| Proton-pump inhibitors (mg/kg/day) | 0.64 (0.56–1.33) | 0.62 (0.52–1.01) | 0 | NS | – | – |
| Topical steroids (mg/day) | 1 (1–2) | 1 (1–2) | 0 | NS | – | – |
| EREFSa | 2 (2–3) | 0 (0–0) | 0 (0–0) | NS | NS | NS |
| Number of biopsiesa | 5 (4–6) | 5 (4–6) | 3 (3–4) | NS | NS | NS |
| Peak eosinophil counta | 81 (65–101) | 0 (0–4) | 0 (0–0) | <0.001 | <0.001 | NS |
Abbreviations: EREFS, endoscopic reference score; NS, not significant.
Median (interquartile range).
Number (percentage).
3.2 ∣. Unique transcriptional changes distinguish non-responders to the combination therapy
Using gene expression data, the PCA plot distinctly segregated EoE-PPI-TSnr from EoE-PPI-TSr and controls. In all, 6318 genes were significantly modulated. Of these, 3164 genes were upregulated, and 3154 were downregulated in EoE-PPI-TSnr children compared to EoE-PPI-TSr children (Figure 1). Only three genes (CCL22, HLA-DRB5, and MT-CO1) were downregulated in EoE-PPI-TSr children compared to controls, indicating that the transcriptional profile of the EoE-PPI-TSr children and controls are substantially similar.
FIGURE 1.
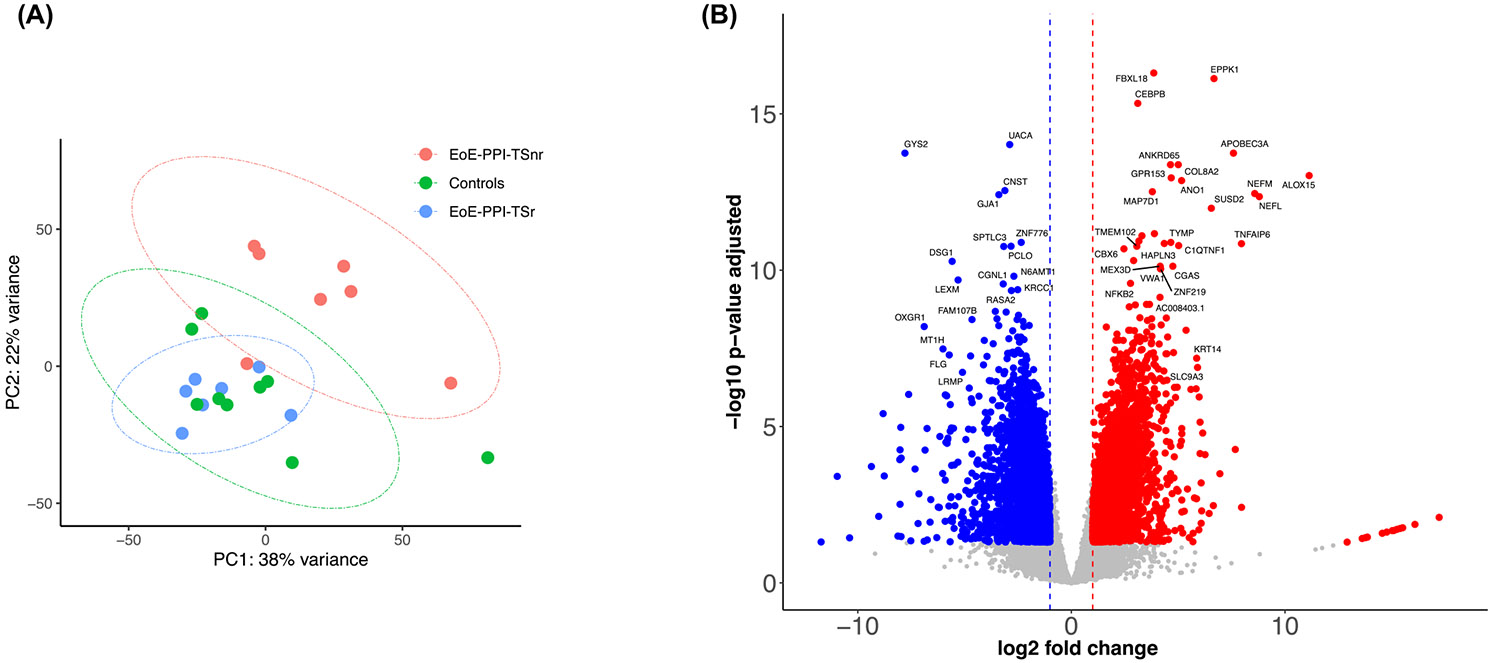
Esophagus tissue transcriptome. (A) PCA plot depicting the EoE-PPI-TSr, EoE-PPI-TSnr, and Control groups. (B) Volcano plot illustrating significantly modulated genes, including upregulated genes 3164 (red) and downregulated genes 3154 (blue) in EoE-PPI-TSnr children compared to EoE-PPI-TSr children, with thresholds set at log2 (FC) of −1 and 1 (x-axis) and −log10(FDR) = 0.05 (y-axis).
In EoE-PPI-TSnr children, the pathways involved in eosinophilic inflammatory response such as cytokine signaling, signaling to RAS, assembly of collagen fibrils and other multimeric structures, extracellular matrix organization, integrin cell surface interactions, signaling by NTRKs, signaling by receptor tyrosine kinases and neutrophil degranulation were notably upregulated compared to EoE-PPI-TSr children. Conversely, expression of several pathways, including metallothioneins that bind metals, cohesin loading onto chromatin, the establishment of sister chromatid cohesion, synthesis of PIPs at the late endosome membrane, defects in cobalamin (B12) metabolism, L13a-mediated translational silencing of ceruloplasmin expression, antigen processing, and lipid metabolism, were downregulated in EoE-PPI-TSnr compared to EoE-PPI-TSr children (Figure 2A). Compared to the EoE-PPI-TSSr group, 150 out of 702 (21%) genes involved in the cytokine signaling pathway were upregulated (Figure 2B), and 52 out of 108 (48%) genes within the L13a-mediated translational silencing of ceruloplasmin expression pathway were downregulated in the EoE-PPI-TSnr group (Figure 2C). Additionally, genes associated with metallothioneins (MT) such as MT2A, MT1A, MT1M, MT1F, MT1G, MT1H, MT1X, and MT1E (Figure 2D), alongside serine peptidase inhibitors (SPINK), including SPINK5, SPINK7, and SPINK8, and key epithelial barrier genes including desmoglein-1 (DSG1) were downregulated in EoE-PPI-TSnr children compared to EoE-PPI-TSr children were downregulated in children with EoE-PPI-TSnr relative to those in the EoE-PPI-TSr group (Figure 2).
FIGURE 2.
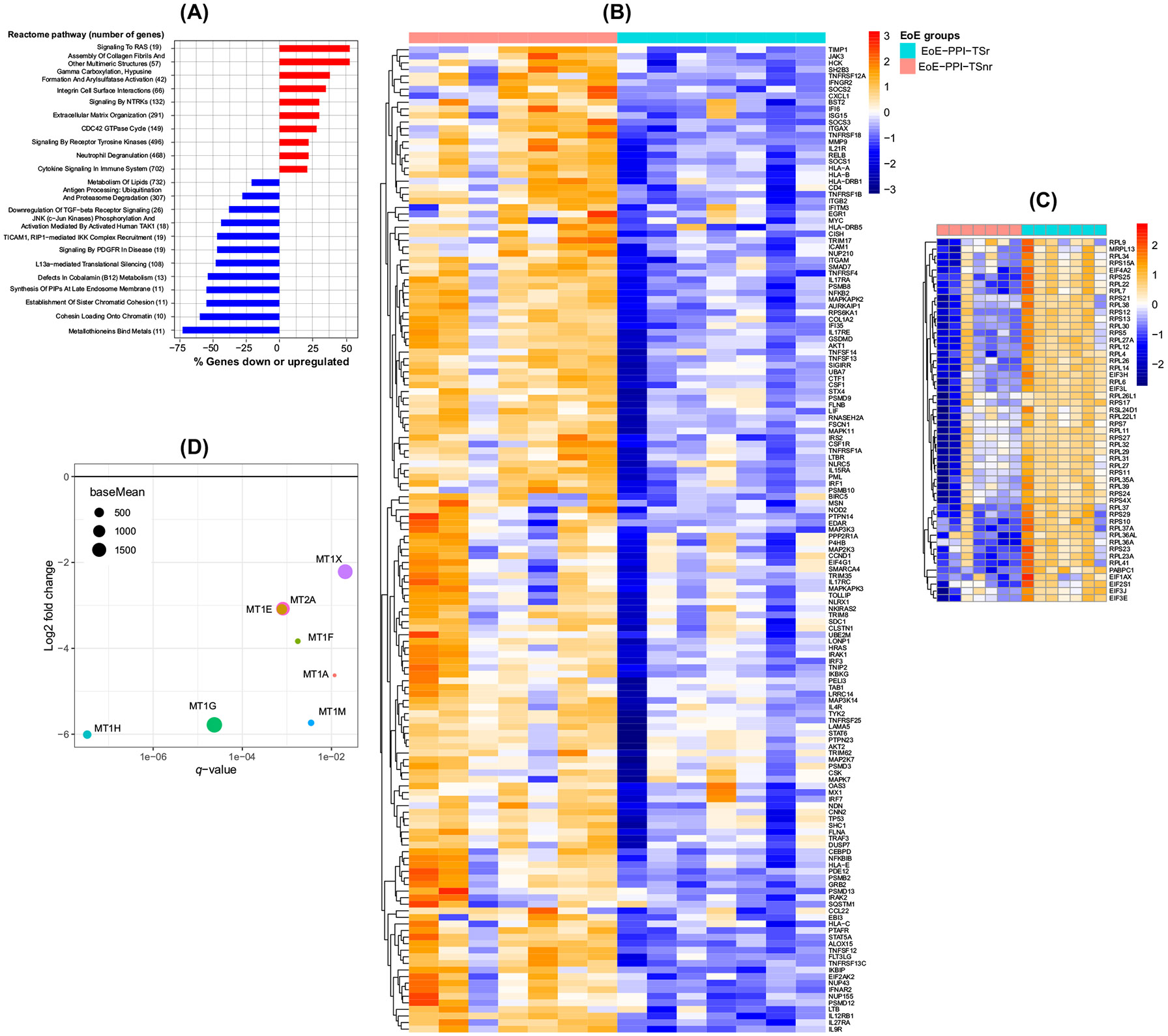
Signaling pathways that are modulated in EoE-PPI-TSnr compared to EoE-PPI-TSr. (A) Reactome human pathways from the differential gene expression analysis. Upregulated pathways are shown in red, and downregulated pathways are shown in blue. On the y-axis, the pathway name and the total number of genes in each pathway are displayed. On the x-axis is displayed the percentage of genes in each pathway that are up- or downregulated; only a subset of significant pathway enrichment with q < 0.05 is shown. (B) The heat map was generated using normalized counts to highlight expression trends of genes that are significantly upregulated in EoE-PPI-TSnr compared to EoE-PPI-TSr, specifically within the cytokine signaling pathways. (C) The heat map was generated using normalized counts to highlight expression trends of genes that are significantly downregulated in EoE-PPI-TSnr compared to EoE-PPI-TSr, specifically within the iL13a-mediated translational silencing of the Ceruloplasmin expression pathway. (D) Plot showing the reactome metallothioneins bind metals pathway genes significantly downregulated in the EoE-PPI-TSnr compared with the EoE-PPI-TSr group. On the x-axis is displayed the q-value for the downregulated genes with q < 0.05. On the y-axis, the log2 fold change for those genes is displayed. The size of the dots represents the base mean, the mean of normalized counts of all samples.
3.3 ∣. Gene expression profile in non-responders is validated by the EDP
We validated gene expression changes observed in our EoE-PPI-TSnr children with the validated EDP panel. The EDP panel consists of 96 genes, of which 77 genes are modulated (50 genes were upregulated and 27 genes were downregulated), and there is no change in the expression of 19 genes quantified using PCR. We found that 56% (43 of 77) genes were differentially expressed in the EoE-PPI-TSnr children compared to the EoE-PPI-TSr overlapped with the genes modulated in the EDP panel. Of these, 20 genes were upregulated in our cohort of EoE-PPI-TSnr children, and the same genes were also upregulated in the EDP panel. Likewise, 23 genes downregulated in our cohort of EoE-PPI-TSnr children were also as downregulated in the EDP panel. Importantly, the directionality of gene modulation in the EoE-PPI-TSnr children perfectly correlated with EDP panel gene expression annotations. Similarly, we validated the differentially expressed in the EoE-PPI-TSnr compared to healthy controls. We observed that 52% (40 of 77) of genes differentially expressed in EoE-PPI-TSnr overlapped with the EDP panel. Of these, 21 genes were upregulated and 19 genes downregulated, and the directionality of gene modulation perfectly correlated with EDP panel gene expression annotations (Figure 3).
FIGURE 3.
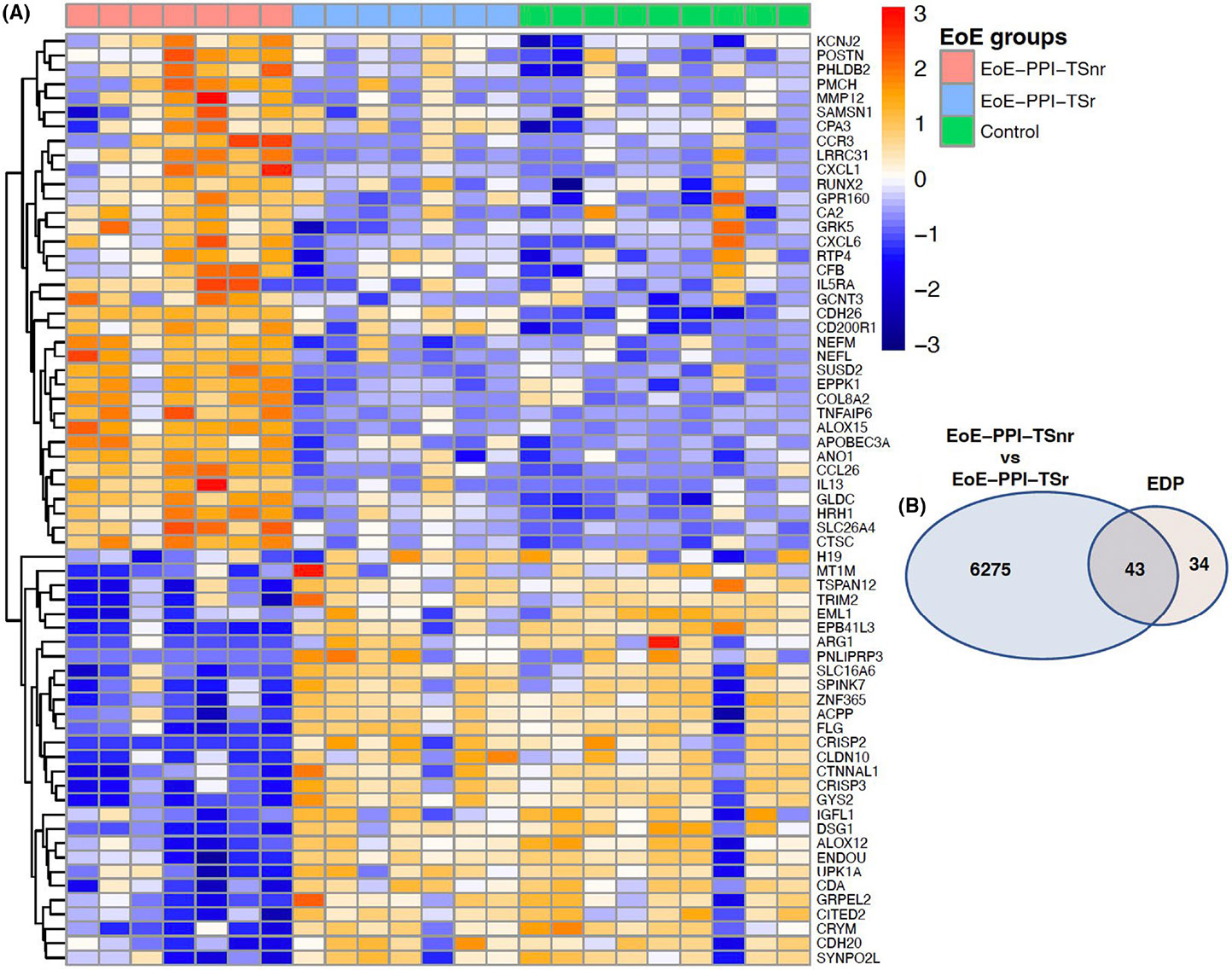
(A) Heatmap showing the expression profile of genes in our cohort of EoE-PPI-TSnr, EoE-PPI-TSr, and controls which are included in the EoE diagnostic panel (EDP). (B) Venn diagram illustrating total number of differentially expressed genes in EoE-PPI-TSnr versus EoE-PPI-TSr and their overlap with the differentially expressed genes in the EDP panel (77 genes).
3.4 ∣. Cell type composition is distinct in non-responders to combination therapy
Deconvolution analysis revealed that cell type composition was significantly different in the EoE-PPI-TSnr group compared to EoE-PPI-TSr and control groups. Specifically, the eosinophils, dendritic cells (DC), immature DCs, megakaryocytic-erythroid progenitors, chondrocytes, mast cells and T helper type 1 cell were significantly higher in EoE-PPI-TSnr children compared to EoE-PPI-TSr and control groups (Figure 4). Given the high relevance of mast cells in the pathophysiology of EoE, we validated mast cell density (CD117) per high power field (40×) on the clinical biopsies. We found that esophageal samples from children with EoE-PPI-TSnr had a significantly higher density of CD117 stained cells compared to EoE-PPI-TSr and controls (Data S1).
FIGURE 4.
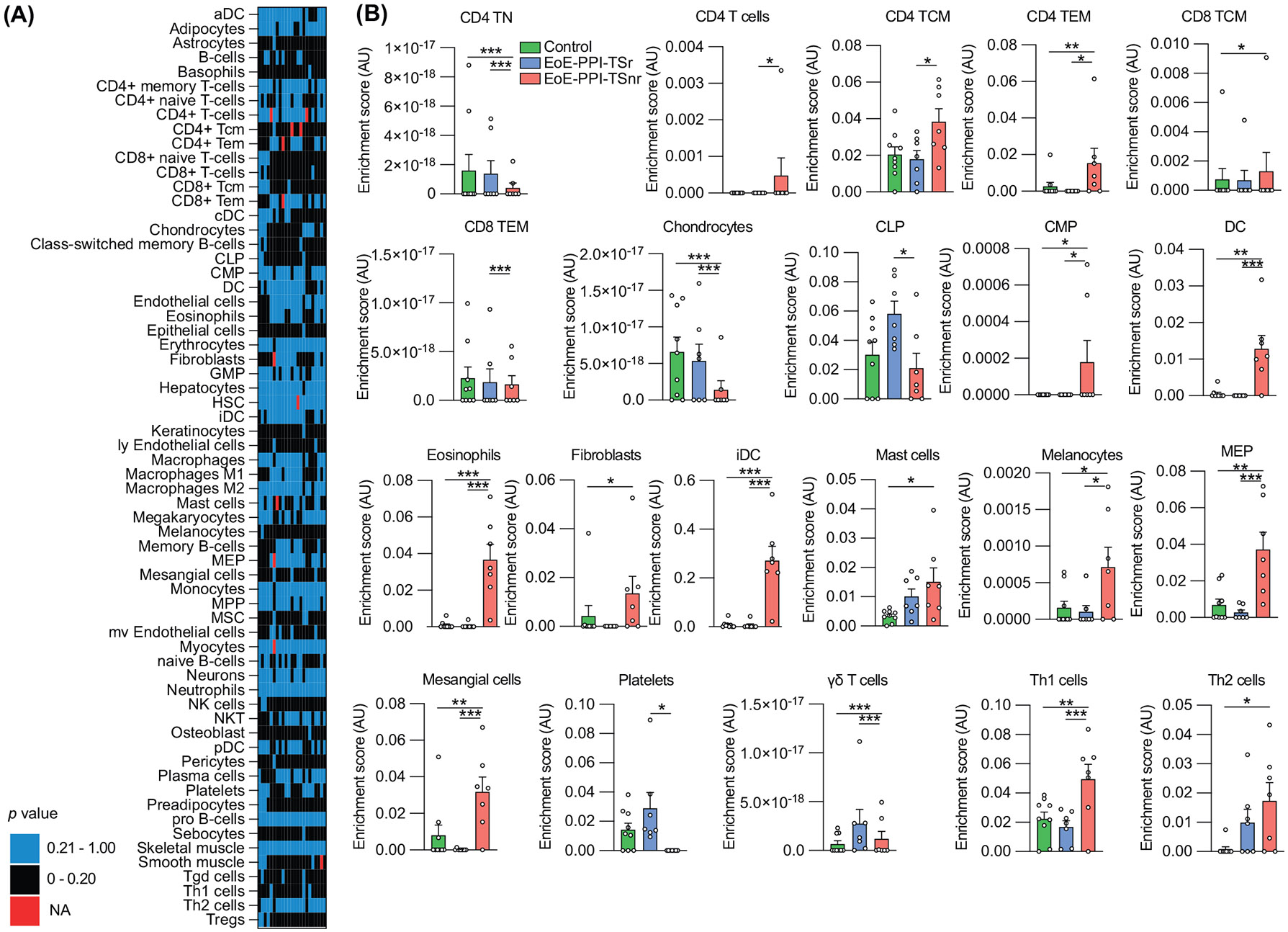
(A) Heatmap and (B) barplots of cell types present in EoE-PPI-TSnr, EoE-PPI-TSr, and controls. The cell type classification was derived using deconvolution of bulk RNA-sequencing data. Absent cell types were excluded to increase the sensitivity of the analysis. The cell types present and the cell types for which at least one comparison was statistically significant are shown. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
3.5 ∣. Esophageal microbiome can be differentially altered in non-responders to combination therapy
Hemophilus sp., Cutibacterium acnes, and Prevotella sp. were the dominant members of the esophageal microbiome in all three groups. We observed no significant differences in the species diversity index (α-diversity) and overall community composition as measured by β-diversity metrics (Figure 5A,B). However, the differences in the abundance of Acinetobacter sp., Fusobacterium sp., and Prevotella sp. between groups were observed. Acinetobacter sp. was notably abundant in EoE-PPI-TSnr (average relative abundance of 27%) compared to the EoE-PPI-TSr children and controls (both at zero presence). However, this difference in abundance did not achieve statistical significance (p = 0.1751, Kruskal–Wallis test). On the other hand, the relative abundance of Fusobacterium sp. was significantly higher in the EoE-PPI-TSr group (27%) compared to the controls (2.5%) and the EoE-PPI-TSnr group (0%), p-value of 0.049 (Figure 5C).
FIGURE 5.
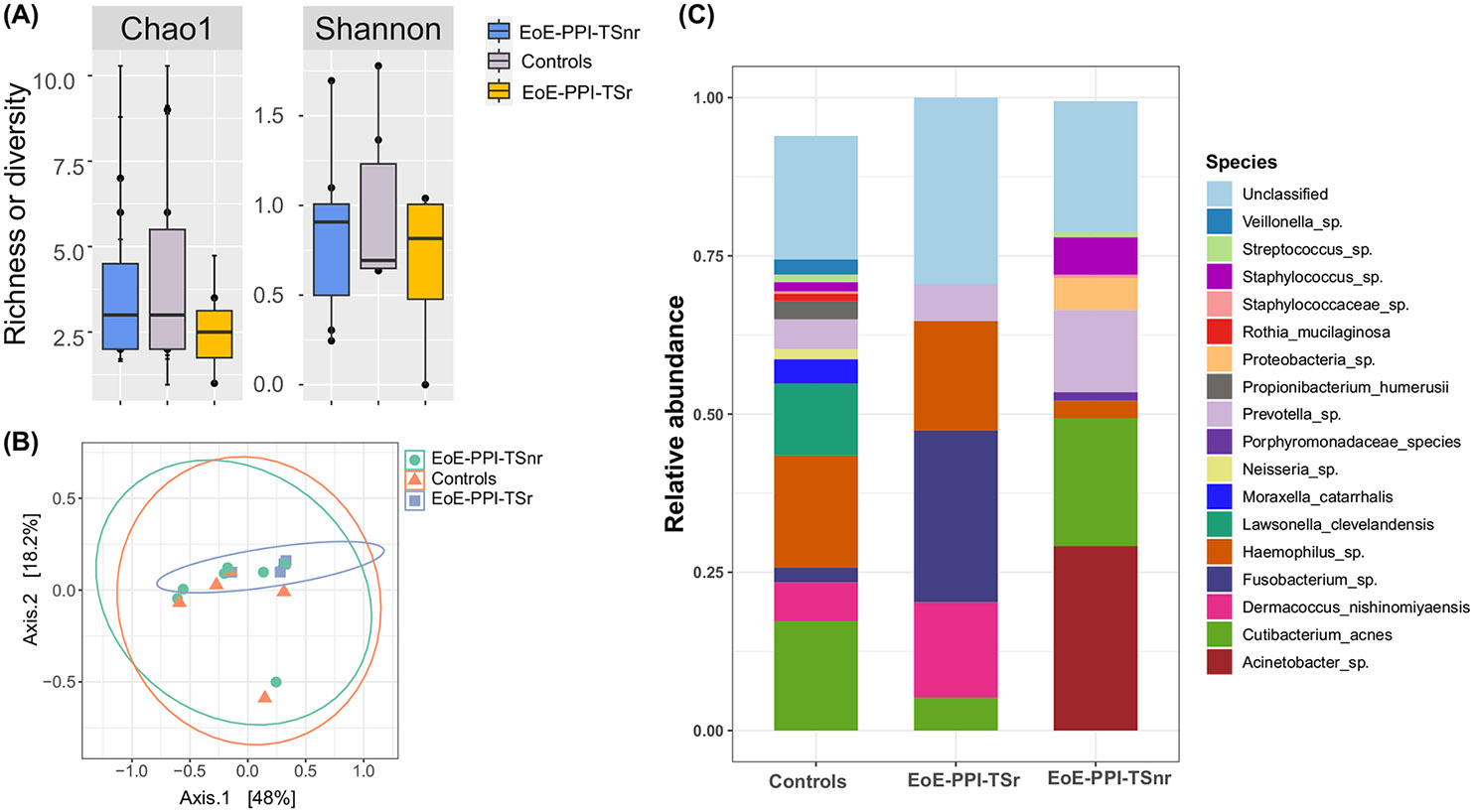
Esophagus microbiome diversity and abundance. (A) The esophagus microbiome alpha diversity and richness were compared between EoE-PPI-TSnr, EoE-PPI-TSr, and control group. (B) A principal coordinate analysis plot of Bray–Curtis dissimilarities (beta-diversity) over the first two-axis is shown. Dots represent individual data points, and the color represents sample groups. The microbiome community composition was not significantly dissimilar among the EoE-PPI-TSnr, EoE-PPI-TSr, and control groups. (C) A color-coded bar plot showing the relative abundance of the microbiome at the species level. The samples are portioned into EoE-PPI-TSnr, EoE-PPI-TSr, and control groups.
3.6 ∣. Relationship between differentially expressed esophageal genes and microbiome and EoE activity indices
Finally, we explored the associations between esophageal gene expression and the esophageal microbiome among EoE-PPI-TSnr and EoE-PPI-TSr patients. As such, the identified microbe-gene associations are in concordance with and likely reflect the differences between the two groups. We found a positive correlation between Acinetobacter abundance and cytokine and interleukin signaling. Specifically, cytokines and interleukins such as interleukin (IL)-13, IL5RA, CXCL1, CCL26 and CXCL6 and genes involved in the inflammation process, including CBP, ALOX15, HRH1, and TNFAIP6 were upregulated in samples with a higher Acinetobacter sp abundance. Conversely, the expression of genes involved in epithelial barrier function, such as DSG1 and filaggrin (FLG) were negatively correlated with Acinetobacter abundance. We also observed a negative correlation in the abundance of Fusobacterium sp. and the expression of IL-13, IL5RA, CXCL1, CCL26, CXCL6, and genes associated with inflammation, including CBP, ALOX15, HRH1, and TNFAIP6 (Figure 6). There was a positive association between Acinetobacter and EREFS score (p-value: 0.014, padj: 0.23). Among the bacteria of interest, Acinetobacter positively associated with the PEC; however, the adjusted p-value did not achieve statistical significance (p = 0.086).
FIGURE 6.
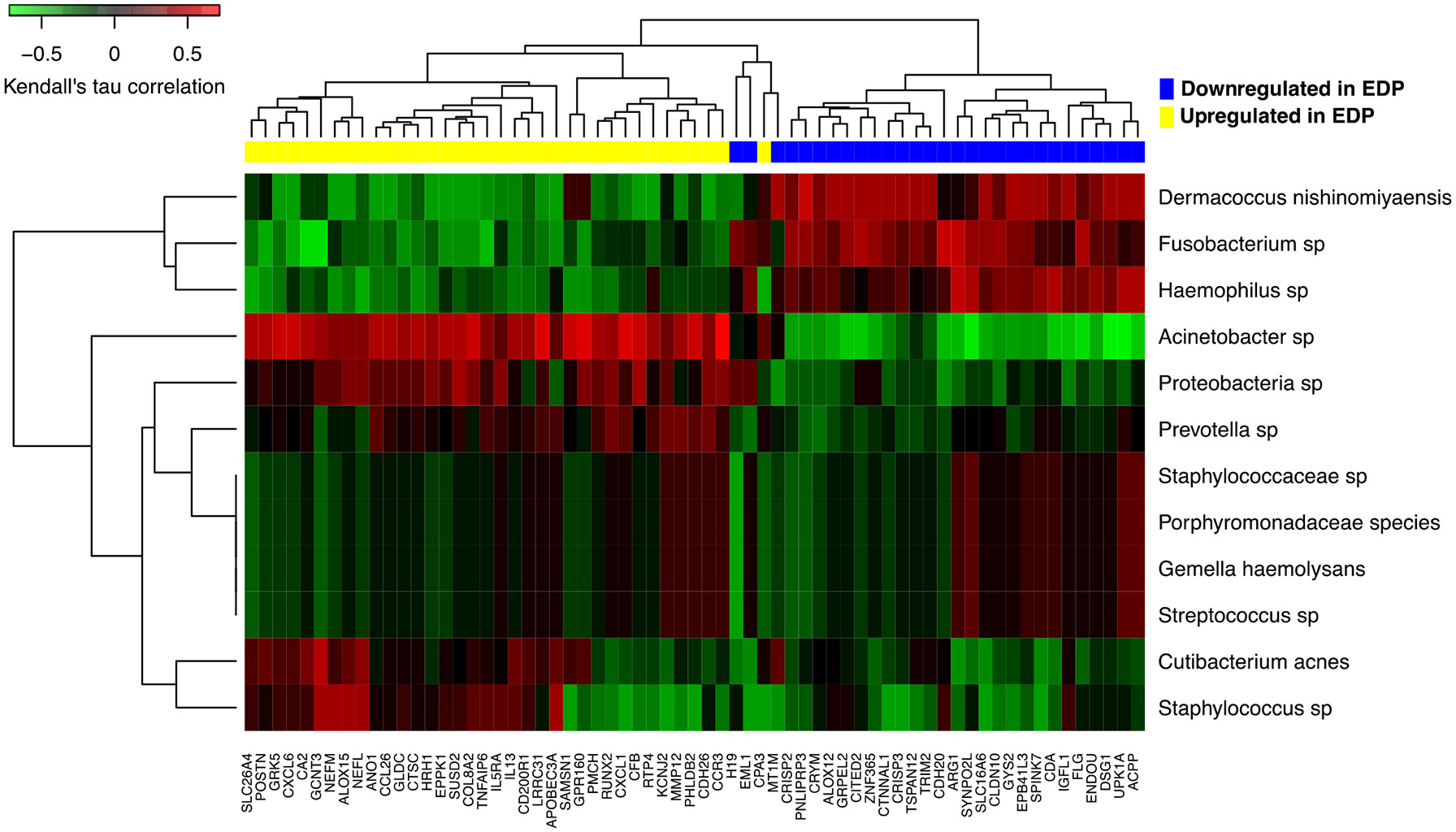
Heatmap illustrating the relationship between microbial abundance and the differentially expressed genes in EoE-PPI-TSnr (compared to EoE-PPI-TSr) included in the EDP. Using EoE-PPI-TSr and EoE-PPI-TSnr (excluding controls) samples, we calculated nonparametric Kendall's tau rank correlations between each microbial species' relative abundance and DE gene's normalized expression level. Microbe-gene correlation coefficients are then visualized through heatmap along with hierarchical clustering, from which we observed distinct correlation patterns between groups of microbial species and DE genes.
3.7 ∣. Power analysis
For gene expression, we selected representative genes upregulated in EoE-PPI-TSnr compared to EoE-PPI-TSr. These have log2 fold changes of 0.87, 1.07, and 3.15, corresponding to the first, second, and third quartiles of all significant genes' log2 fold changes. After correcting for multiple tests, our sample size had >94% power to detect a log2 fold change of ≥1.07. This ensured the reliability of our gene expression findings. On the other hand, we selected Acinetobacter, Prevotella (both enriched in EoE-PPI-TSnr), and Fusobacterium (depleted in EoE-PPI-TSnr) and found that we had inadequate power to detect changes in the esophageal microbiome. These power analysis results agreed with our empirical findings (Data S1).
4 ∣. DISCUSSION
PPI and TS remain the mainstay of the pharmacological approach to managing children with EoE. However, treatment non-response poses a significant challenge. This study used metatranscriptomics to investigate the association between esophageal gene expression and microbiota and the response to PPI and TS combination therapy. We observed that the transcriptional profiles of EoE-PPI-TSr and controls clustered together, suggesting that combination therapy normalizes gene expression in responders. In contrast, we found that the prevalence of food allergy was higher in EoE-PPI-TSnr and the pathways involved in the eosinophilic inflammatory response, cytokine signaling, and extracellular matrix organization were significantly upregulated in EoE-PPI-TSnr. This is consistent with our current understanding that potential immunologic mechanisms, including IL-4 and IL-13 signaling pathways are central in sustaining eosinophilic inflammation in EoE.38,39
Dupilumab is a monoclonal antibody directed against IL-4 receptor alpha, a receptor involved in signaling pathways for both IL-4 and IL-13 known to promote eosinophil recruitment, fibroblast proliferation, and B-cell class switching to IgE production, and was recently approved by the United States Food and Drug Administration for use in children (≥1 year and weighing ≥15 kg) with EoE.40,41 Many unanswered questions exist about the position of this biologic in routine clinical practice. Our observations about the upregulated eosinophilic inflammatory pathways in EoE-PPI-TSnr provide an immunopathologic basis for using Dupilumab as a therapeutic option for those who failed to respond to combination therapy. However, it is important to note that the patient characteristics, clinical presentation, caregivers, and healthcare providers' preferences could substantially influence the initiation of Dupilumab therapy in a pediatric EoE patient.
Next, we found that the several MT genes (MT2A, MT1A, MT1M, MT1F, MT1G, MT1H, MT1X, and MT1E) were significantly decreased in EoE-PPI-TSnr children compared to EoE-PPI-TSr children. MTs are cysteine-rich cytosolic proteins that regulate cell differentiation and proliferation.42 The MT genes are key in modulating innate and adaptive immunologic responses by acting as regulators of zinc metabolism and protecting cells against oxidative stress, and can be induced by PPIs.11 The diminished expression of MT genes in EoE-PPI-TSnr is likely due to the undifferentiated esophageal epithelial cell subpopulations, which can be seen in inflammatory states and inadequate response to PPIs.43 These genes hold potential as a biomarker for predicting response to combination therapy and could provide novel insights into the mechanisms underlying non-response in children with EoE.
Novel to our study is the identification of L13a-mediated translational silencing of the ceruloplasmin expression pathway being downregulated in EoE-PPI-TSnr compared to EoE-PPI-TSr. While the L13a-mediated translational silencing pathway is primarily associated with regulating copper homeostasis, it can also play a pivotal role in modulating allergic inflammation.44 L13a can bind to the mRNA of pro-inflammatory cytokines such as IL-5, which activates and recruits eosinophils and immune cells associated with allergic inflammation. By inhibiting the translation of IL-5, L13a may be involved in modulating reciprocal changes in its downstream targets, such as overexpression of pro-inflammatory cytokine pathways known to contribute to eosinophilic inflammation and regulation of the translation of other genes involved in epithelial barrier integrity and adhesion molecules such as SPINK and DSG1.45,46 Based on our data, L13-a mediated translational silencing of ceruloplasmin holds potential as a novel therapeutic target to manage EoE.
Deconvolution analysis added depth and granularity to our observations by allowing us to characterize the cellular composition and heterogeneity across the study groups. Specifically, it allowed us to infer that the composition of the esophageal cell type is distinct in EoE-PPI-TSnr children and may contribute to the observed transcriptional profiles. The higher levels of eosinophils, DCs, and immature DCs in EoE-PPI-TSnr children reflect ongoing inflammation and immune dysregulation in this subset of patients. Additionally, the higher levels of megakaryocytic-erythroid progenitors, which give rise to megakaryocyte and erythroid lineages, and T helper type 1 cells involved in the immune response against pathogens warrant further investigation. A more focused approach, such as single cell-RNA sequencing, might allow us to confirm these differences and overcome some of the limitations of deconvolution analysis by providing high-resolution insights into cell-to-cell heterogeneity, characterization of different cell states, and unbiased exploration of cell populations.
With growing interest in host-microbiome interactions in the pathogenesis and management of EoE, we explored how the esophageal microbiome might influence the esophageal epithelial gene expression concerning the treatment response. We found a positive correlation between Acinetobacter abundance and the upregulation of genes involved in cytokine and interleukin signaling pathways, including IL-13, IL5RA, CXCL1, CCL26, and CXCL6. In contrast, there was a negative correlation between Acinetobacter abundance and genes involved in epithelial barrier function, such as DSG1 and FLG. This suggests that Acinetobacter may play a role in influencing the response to combination therapy in children with EoE. Animal model studies are being planned to investigate how the microbiome (including Acinetobacter) influences the dysregulated pathways observed in EoE-PPI-TSnr.
There are limitations to our study. This single-center study was conducted in the pediatric population, so the results may not be more generalizable to adults with EoE. A majority of children with EoE presenting to our referral center are already on a PPI, TS, or a combination therapy. This limited our ability to collect and analyze pre- and post-PI-TS combination therapy esophageal samples. As such, it was beyond the scope of this study to determine if the non-response was due to primary or secondary resistance and if the L13a pathway is differentially expressed at baseline or is a feature of lack of treatment response in children with EoE-PPI-TSnr. Likewise, microbiome alterations may have existed prior to treatment initiation, and the temporal relationship between microbiome composition and treatment response needs to be studied further. While our study had sufficient power to identify critical differences in esophageal gene expression between the EoE-PPI-TSnr and the EoE-PPI-TSr groups, it was not powered to detect more moderate gene expression changes, such as that expected between the EoE-PPI-TSr and the control group and in the esophageal microbiome with statistical significance. A multi-center, prospectively study with optimal sample size and pre- and post-treatment esophageal samples from the same subject would enable us to overcome these limitations and also study the impact of other biological variables (such as age, gender, and ethnicity) on the response to combination therapy. We used distal esophageal samples, which may not reflect an individual's global changes in the esophagus. However, it has been previously demonstrated that EDP analysis of the distal esophagus assesses spatial inflammation along the length of the esophagus.47 Lastly, removing host ribosomal RNA from the tissue samples poses a significant challenge with metatranscriptomics to capture microbial signatures from tissue samples precisely. Although our protocol allowed us to capture microbial transcripts successfully, further optimization may be required to capture the complete transcriptome of the microbial community to assess their functional capabilities.
Despite these limitations, our study has several strengths. We used the most recent guidelines to define EoE and the widely accepted threshold to define treatment response, thereby increasing the reproducibility of our study. This is the first study to use metatranscriptomics to investigate molecular mechanisms and microbial changes associated with response to combination therapy in children with EoE. We report two original observations, including identifying L13-a translational silencing of the ceruloplasmin expression pathway and unique cell composition in EoE-PPI-TSnr children. We observed a positive correlation of Acinetobacter abundance with IL-13, IL5RA, CXCL1, CCL26, and CXCL6 gene expression. The functional relevance of these interactions requires further investigation.
In conclusion, this study provides novel insights into the immunobiology of EoE and highlights the underlying molecular mechanisms and microbial changes in non-responders to combination therapy. It also identifies MT genes as potential biomarkers predicting response to combination therapy and the L13a-mediated translational silencing pathway as a novel therapeutic target for managing EoE. The study also suggests that Acinetobacter may have a role in treatment response. These findings provide valuable information to foster future research on EoE pathogenesis and treatment, leading to informed clinical decision-making, personalized care, and identifying novel therapeutic targets.
Supplementary Material
Clinical Implications.
Non-response to pharmacologic therapy in children with eosinophilic esophagitis (EoE) poses a significant burden and is associated with persistent symptoms and impaired quality of life, psychological impact, and disease progression.
Using metatranscriptomics, this study identifies dysregulated genes and pathways in the esophageal tissue in non-responders to the proton-pump inhibitors (PPI) and topical steroids (TS) (combination therapy). It provides invaluable insights into the underlying molecular mechanisms of non-responsiveness to combination therapy in children with EoE. The high concordance between dysregulated genes in non-responders and the EoE diagnostic panel (EDP) suggests the potential utility of the EDP in predicting treatment response.
Differences in esophageal cell type composition, such as increased levels of eosinophils, dendritic cells, and T helper type 1 cells in non-responders, maybe a potential biomarker for predicting response to combination therapy.
The association between the abundance of Fusobacterium sp., Acinetobacter sp., and inflammatory signaling pathways, as well as epithelial barrier dysfunction, highlights the role of microbial factors in treatment response and suggests targeting the microbiome for improved therapeutic outcomes in children with EoE.
ACKNOWLEDGMENTS
The authors acknowledge Ms. Regina Tyree for managing tissue biorepository and database.
FUNDING INFORMATION
G.H. is supported by the NIH 1K23DK131341 and 5R21AI168832. Y.C. is supported by a VA-Career Development Award IK2BX004648, a Vanderbilt Burroughs Welcome Supporting Careers in Research for Interventional Physicians and Surgeons (SCRIPS), and a DDRC Pilot and Feasibility Grant Application P30 058404. S.V.R. is supported by the NIH 5R21AI168832. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations:
- EDP
Eosinophilic esophagitis diagnostic panel
- EGD
Esophagogastroduodenoscopy
- EoE
Eosinophilic esophagitis
- EoE-PPI-TSnr
Eosinophilic esophagitis non-responsive to PPI and TS therapy
- EoE-PPI-TSr
Eosinophilic esophagitis responsive to PPI and TS therapy
- Eos/hpf
Eosinophils per high power field
- EREFS
Endoscopic reference score
- FDR
False Discovery Rate
- PEC
Peak eosinophil count
- PPI
Proton-pump inhibitor
- rRNA
Ribosomal ribonucleic acid
- RT-qPCR
Quantitative reverse transcription PCR
- TS
Topical steroids
Footnotes
CONFLICT OF INTEREST STATEMENT
G.H. serves a consultant to Bristol Myer Squibb, Regeneron, and Sanofi. In addition, he has received speaker fees from Bristol Myer Squibb. All other authors declare that the research was conducted without commercial or financial relationships that could be construed as a potential conflict of interest.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Furuta GT, Katzka DA. Eosinophilic esophagitis. N Engl J Med. 2015;373:1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navarro P, Arias Á, Arias-González L, Laserna-Mendieta EJ, Ruiz-Ponce M, Lucendo AJ. Systematic review with meta-analysis: the growing incidence and prevalence of eosinophilic oesophagitis in children and adults in population-based studies. Aliment Pharmacol Ther. 2019;49:1116–1125. [DOI] [PubMed] [Google Scholar]
- 3.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20.e6. [DOI] [PubMed] [Google Scholar]
- 4.Schoepfer AM, Safroneeva E, Bussmann C, et al. Delay in diagnosis of eosinophilic esophagitis increases risk for stricture formation in a time-dependent manner. Gastroenterology. 2013;145:1230–1236. [DOI] [PubMed] [Google Scholar]
- 5.Rank MA, Sharaf RN, Furuta GT, et al. Technical review on the management of eosinophilic esophagitis: a report from the AGA institute and the joint task force on allergy-immunology practice parameters. Gastroenterology. 2020;158:1789–1810.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marabotto E, Giannini EG, Zentilin P, et al. Pharmacotherapies in eosinophilic esophagitis: state of the art. Minerva Gastroenterol. 2022;68:69–76. [DOI] [PubMed] [Google Scholar]
- 7.Gutiérrez-Junquera C, Fernández-Fernández S, Cilleruelo ML, Rayo A, Román E. The role of proton pump inhibitors in the management of pediatric eosinophilic esophagitis. Front Pediatr. 2018;6:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franciosi JP, Mougey EB, Dellon ES, et al. Proton pump inhibitor therapy for eosinophilic esophagitis: history, mechanisms, efficacy, and future directions. J Asthma Allergy. 2022;15:281–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laserna-Mendieta EJ, Casabona S, Savarino E, et al. Efficacy of therapy for eosinophilic esophagitis in real-world practice. Clin Gastroenterol Hepatol. 2020;18:2903–2911.e4. [DOI] [PubMed] [Google Scholar]
- 10.Wen T, Dellon ES, Moawad FJ, Furuta GT, Aceves SS, Rothenberg ME. Transcriptome analysis of proton pump inhibitor-responsive esophageal eosinophilia reveals PPI-reversible allergic inflammation. J Allergy Clin Immunol. 2015;135:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rochman M, Xie YM, Mack L, et al. Broad transcriptional response of the human esophageal epithelium to proton pump inhibitors. J Allergy Clin Immunol. 2021;147:1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eluri S, Selitsky SR, Perjar I, et al. Clinical and molecular factors associated with histologic response to topical steroid treatment in patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2019;17:1081–1088.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laserna-Mendieta EJ, FitzGerald JA, Arias-Gonzalez L, et al. Esophageal microbiome in active eosinophilic esophagitis and changes induced by different therapies. Sci Rep. 2021;11:7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucendo AJ, de Rezende L, Comas C, Caballero T, Bellón T. Treatment with topical steroids downregulates IL-5, eotaxin-1/CCL11, and eotaxin-3/CCL26 gene expression in eosinophilic esophagitis. Am J Gastroenterol. 2008;103:2184–2193. [DOI] [PubMed] [Google Scholar]
- 15.Benitez AJ, Hoffmann C, Muir AB, et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome. 2015;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park CH, Lee SK. Exploring esophageal microbiomes in esophageal diseases: a systematic review. J Neurogastroenterol Motil. 2020;26:171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shakya M, Lo C-C, Chain PSG. Advances and challenges in Metatranscriptomic analysis. Front Genet. 2019;10:904. doi: 10.3389/fgene.2019.00904. https://www.frontiersin.org/articles/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bikel S, Valdez-Lara A, Cornejo-Granados F, et al. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput Struct Biotechnol J. 2015;13:390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Li L, Butcher J, Stintzi A, Figeys D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome. 2019;7:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busing JD, Buendia M, Choksi Y, Hiremath G, Das SR. Microbiome in eosinophilic esophagitis—metagenomic, Metatranscriptomic, and metabolomic changes: a systematic review. Front Physiol. 2021;12:731034. doi: 10.3389/fphys.2021.731034. https://www.frontiersin.org/articles/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen T, Rothenberg ME. Clinical applications of the eosinophilic esophagitis diagnostic panel. Front Med (Lausanne). 2017;4:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiremath G, Locke A, Thomas G, et al. Novel insights into tissue-specific biochemical alterations in pediatric eosinophilic esophagitis using Raman spectroscopy. Clin Transl Gastroenterol. 2020;11:e00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiremath G, Shilts MH, Boone HH, et al. The salivary microbiome is altered in children with eosinophilic esophagitis and correlates with disease activity. Clin Transl Gastroenterol. 2019;10:e00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dellon ES, Liacouras CA, Molina-Infante J, et al. Updated international consensus diagnostic criteria for eosinophilic esophagitis: proceedings of the AGREE conference. Gastroenterology. 2018;155:1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano I, Moy N, Heckman MG, Thomas CS, Gonsalves N, Achem SR. Endoscopic assessment of the oesophageal features of eosinophilic oesophagitis: validation of a novel classification and grading system. Gut. 2013;62:489–495. [DOI] [PubMed] [Google Scholar]
- 26.Rajagopala SV, Bakhoum NG, Pakala SB, et al. Metatranscriptomics to characterize respiratory virome, microbiome, and host response directly from clinical samples. Cell Rep Methods. 2021;1:100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MI Love, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aran D. Cell-type enrichment analysis of bulk transcriptomes using xCell. In: Boegel S, ed. Bioinformatics for Cancer Immunotherapy: Methods and Protocols. Springer US; 2020:263–276. [DOI] [PubMed] [Google Scholar]
- 33.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu Z, Hübschmann D. Make interactive complex heatmaps in R. Bioinformatics. 2022;38:1460–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunagawa S, Mende DR, Zeller G, et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods. 2013;10:1196–1199. [DOI] [PubMed] [Google Scholar]
- 36.Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc. 1952;47:583–621. [Google Scholar]
- 37.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arias Á, Lucendo AJ. Molecular basis and cellular mechanisms of eosinophilic esophagitis for the clinical practice. Expert Rev Gastroenterol Hepatol. 2019;13:99–117. [DOI] [PubMed] [Google Scholar]
- 39.Davis BP, Rothenberg ME. Mechanisms of disease of eosinophilic esophagitis. Annu Rev Pathol. 2016;11:365–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Press Release. Dupixent® FDA approved as first and only treatment indicated for children aged 1 year and older with eosinophilic esophagitis (EoE). Accessed April 8, 2024. https://www.sanofi.com/en/media-room/press-releases/2024/2024-01-25-19-30-00-2817342
- 41.Aceves SS, Dellon ES, Greenhawt M, Hirano I, Liacouras CA, Spergel JM. Clinical guidance for the use of dupilumab in eosinophilic esophagitis: a yardstick. Ann Allergy Asthma Immunol. 2023;130:371–378. [DOI] [PubMed] [Google Scholar]
- 42.Subramanian Vignesh K, Deepe GS. Metallothioneins: emerging modulators in immunity and infection. Int J Mol Sci. 2017;18:2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rochman M, Wen T, Kotliar M, et al. Single-cell RNA-seq of human esophageal epithelium in homeostasis and allergic inflammation. JCI Insight. 2022;7:e159093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. [DOI] [PubMed] [Google Scholar]
- 45.Mazumder B, Li X, Barik S. Translation control: a multifaceted regulator of inflammatory response. J Immunol. 2010;184:3311–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azouz NP, Ynga-Durand MA, Caldwell JM, et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med. 2018;10:eaap9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min S, Shoda T, Wen T, Rothenberg ME. Diagnostic merits of eosinophilic esophagitis diagnostic panel from a single esophageal biopsy. J Allergy Clin Immunol. 2022;149:782–787.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


