Abstract
The eukaryotic Rad51 protein is a structural and functional homolog of Escherichia coli RecA with a role in DNA repair and genetic recombination. Five paralogs of Rad51 have been identified in vertebrates, Rad51B, Rad51C, Rad51D, Xrcc2 and Xrcc3, which are also implicated in recombination and genome stability. Here, we identify a mammalian cell mutant in Rad51C. We show that the Chinese hamster cell mutant, CL-V4B, has a defect in Rad51C. Sequencing of the hamster Rad51C cDNA revealed a 132 bp deletion corresponding to an alternatively spliced transcript with lack of exon 5. CL-V4B was hypersensitive to the interstrand cross-linking agents mitomycin C (MMC) and cisplatinum, the alkylating agent methyl methanesulfonate and the topoisomerase I inhibitor campthotecin and showed impaired Rad51 foci formation in response to DNA damage. The defect in Rad51C also resulted in an increase of spontaneous and MMC-induced chromosomal aberrations as well as a lack of induction of sister chromatid exchanges. However, centrosome formation was not affected. Intriguingly, a reduced level of sister chromatid cohesion was found in CL-V4B cells. These results reveal a role for Rad51C that is unique among the Rad51 paralogs.
INTRODUCTION
Homologous recombination (HR) is a major pathway involved in the repair of double-strand breaks, interstrand cross-links and other types of DNA damage (1–3). HR requires extensive regions of DNA homology and accurately repairs DNA damage using the information of the undamaged sister chromatid or the homologous chromosome. HR has been studied extensively in the yeast Sacharomyces cerevisiae where genes of the Rad52 epistasis group, including RAD51, mediate this process (4,5). The Rad51 protein in eukaryotic cells is a functional homolog of the bacterial RecA protein that forms nucleoprotein filaments on DNA and promotes exchange between homologous sequences (4). Furthermore, Rad51 displays a dynamic redistribution into nuclear foci after treatment with DNA damaging agents (6). These foci are formed at the site of DNA damage (7) and contain additional proteins involved in HR such as Rad52, Rad54 and the single-stranded DNA binding protein RPA (8–10).
Saccharomyces cerevisiae cells contain, in addition to RAD51, two RAD51 paralogs, RAD55 and RAD57, which form a heterodimer that weakly interacts with RAD51 and stimulates RAD51-mediated strand exchange reactions (11). Another RAD51 paralog, Dmc1, has a specialized role in meiosis. Seven members of the Rad51 protein family have been identified in humans: Rad51 (12), Dmc1 (13), XRCC2 (14,15), XRCC3 (15–17), Rad51B (18,19), Rad51C (20) and Rad51D (19,21,22). The Rad51 paralogs share limited sequence homology, which is mainly concentrated in the central part of the proteins and includes the two Walker A and B motifs potentially involved in ATP hydrolysis (23). Physical interactions can occur between human Rad51 and XRCC3, Rad51C and XRCC3, Rad51B and Rad51C, Rad51D and Rad51C as well as between Rad51D and XRCC2 (24–27) suggesting that these paralogs may function as Rad51 accessory factors, comparable with S.cerevisiae RAD55 and RAD57.
Although the functional roles of the Rad51 family members are not understood in human cells, evidence from Chinese hamster cells, chicken DT40 cells and knockout mouse models shows that they contribute to genomic stability and are involved in genetic recombination processes. The Chinese hamster cell mutants, irs1 and irs1SF, defective in XRCC2 and XRCC3, respectively, have been most widely used to study the consequences of a deficiency in a Rad51 paralog. These mutants are phenotypically similar and display sensitivity to diverse DNA damaging agents such as ionizing radiation (IR; ∼3-fold), UV (∼3-fold), monofunctional alkylating agents (2–10-fold) and camptothecin (∼5-fold) as well as an extreme sensitivity to DNA cross-linking agents (60–100-fold) such as MMC (15,28–30). Both mutants exhibit a high incidence of spontaneous and IR-induced chromosomal aberrations (CAs) and show defects in chromosome segregation due to aberrant centrosomes (15,31,32). Moreover, both irs1 and irs1SF show a decreased frequency of DNA double-strand break repair by HR (17,33) and a lack of DNA damage-inducible nuclear Rad51 foci (34,35). The first evidence for a role of Rad51B, C and D in genetic recombination processes came from chicken DT40 cells with deficiencies in those genes (36,37). These chicken cell mutants all exhibited chromosomal instability caused by increased spontaneous CAs and reduced levels of sister chromatid exchanges (SCEs). Furthermore, they showed hampered Rad51 foci formation after IR and were 2–3-fold more sensitive to IR and MMC. The important roles that Rad51 paralogs play in the maintenance of genomic stability during proliferation is further emphasized by the embryonic lethality in Rad51B–/–, Rad51D–/– and Xrcc2–/– knockout mice (38–40).
In this paper, we describe an MMC-hypersensitive Chinese hamster cell mutant, CL-V4B, which was found to be the first mammalian mutant defective in Rad51C. We show that Rad51C plays a pivotal role in protection against the deleterious effects of DNA interstrand cross-links as well as in the maintenance of genome stability. This deficiency in Rad51C affects CA levels as well as the induction of SCEs. Importantly, among the Rad51 paralogs, Rad51C appears to play a unique role in chromatid cohesion.
MATERIALS AND METHODS
Cell culture
The MMC-sensitive mutant CL-V4B was isolated upon ENU treatment of Chinese hamster V79B cells by the replica plating method described previously (41). The MMC-sensitive cell line irs1SF of CHO (AA8) was kindly provided by Dr R. B. Painter (University of California, San Francisco) and irs1TOR of Chinese hamster V79 cells by Dr John Thacker (MRC Radiobiology Unit, Harwell, UK). V-H4, and the parental lines V79 and V79B were described previously (42,43). All cells were cultured in plastic dishes (P94; Greiner) in Ham’s F10 medium (Life Technologies) without hypoxanthine and thymidine or in Dulbecco’s modified Eagle’s medium/F12, supplemented with 10% fetal calf serum (Bodingo) and penicillin (100 U/ml) and streptomycin (0.1 mg/ml). Cells were maintained at 37°C in a 5% CO2 atmosphere, humidified to 95–100%. For subcultures the cells were detached by use of 0.25% trypsin containing 0.02% EDTA.
Chemicals
Polyethylene glycol (PEG; 1450 molecular weight), cytochalasin-B, camptothecin, leupeptin and aprotinin were purchased from Sigma Chemical Co.; Pefabloc (AEBSF) from Boehringer Mannheim; methyl methanesulfonate (MMS) from Merck; mitomycin C (MMC) from Kyowa; colcemid and geneticin (G418) from Gibco BRL; ethylnitrosourea (ENU) from Pfalz and Bauer; bleomycin (BLM) from Dagra Pharma B.V.; cis-diamminedichloroplatinum (cis-DDP) was a gift from Dr J. Brouwer (University of Leiden, The Netherlands).
Irradiation
For X-ray irradiation cells were irradiated in tissue culture medium at a dose rate of 2.8 Gy/min (200 kV, 4 mA, 1 mm Al). For irradiation with UV light of 254 nm, a Philips TUV germicidal lamp was used with a fluence rate of 0.19 W/m2, measured with the IL/770 germicidal radiometer.
Clonogenic survival assays
Cultures in exponential growth were trypsinized and 300–1000 cells were plated in P94 dishes in duplicate (controls in triplicate), left to attach for 4 h, and then irradiated or exposed to MMS or cis-DDP for 1 h, to camptothecin for 24 h and to MMC continuously, in complete medium. After treatment with chemicals the cells were washed twice with PBS, with the exception of MMC, and returned to fresh medium. After 8–10 days the dishes were rinsed with 0.9% NaCl, dried, stained with methylene blue (0.25%) and visible colonies were counted. In all cases, the parental cell line was treated in an identical manner to serve as control. Each survival curve represents the mean of at least three independent experiments. Error bars represent standard errors of the mean (SEM).
Microcell-mediated chromosome transfer
Microcells with a single human chromosome 17 were obtained as described previously (44). The microcell suspension was added to a monolayer of recipient cells (3.0 × 106 mutant cells per P94 dish) and allowed to attach at room temperature for 15 min. The cells were fused by treatment with 2 ml of 47% PEG (in serum-free medium containing 10% DMSO) for 1 min followed by washing in serum-free medium containing 10% DMSO. After 24 h the cells were trypsinized and split to five P94 dishes with selective medium containing G418. The resulting microcell hybrids were isolated after 10–14 days.
Transfections
Human Rad51, Rad51B, Rad51C and Rad51D cDNAs cloned into pcDNA3 with G418 as selectable marker (2 µg/transfection) were transfected into CL-V4B cells (1 × 105 on a P60 dish) using the GenePORTER™ transfection reagent (BIOzym) according to the manufacturer’s instructions. After 48 h the cells were trypsinized divided over four P94 dishes in medium containing G418 (Gibco) at 400 µg/ml. More than 100 G418-resistant clones were pooled from each transfection and these pools were used for further analysis.
Immunofluorescence labeling and microscopy
To examine centrosomes or Rad51 foci, Chinese hamster cells were grown on sterile glass slides for 1 day, giving sub-confluent cells at time of fixation. For centrosome analysis cells were fixed with methanol:acetone (7:3). For Rad51 foci analysis, cells were either mock-treated or treated with MMC (2.4 µg/ml for 1 h) or 12 Gy X-ray irradiation. After a 2, 8 or 24 h recovery period, cells were fixed with 2% paraformaldehyde in PBS, and permeabilized for antibody staining with PBS/0.1% Triton X-100. Subsequently the slides were blocked for 30 min in PBS/BSA (0.5%)/glycin (0.15%) and thereafter incubated with rabbit anti-hRad51 antiserum (FBE2) or with rabbit anti-γ-tubulin antiserum (centrosome staining) for 90 min at 37°C in a humidified atmosphere. The slides were washed three times in PBS/0.1% Triton X-100 and then incubated with Alexa™ 488-conjugated goat anti-rabbit IgG (Molecular Probes) for 1 h at 37°C in a humidified atmosphere. After three washes with PBS/0.1% Triton X-100 the cells were counterstained with 4′,6-diamino-2-phenylindole (DAPI; 0.1 µg/ml) in Vectashield mounting medium (Vector Laboratories). Rad51 foci and centrosomes were examined under a Leitz Axioplan fluorescence microscope.
Immunoblotting
Whole cell extracts (WCE) were made by lysis of 5–10 × 106 exponentially growing hamster cells in 30–50 µl of lysis buffer (10 mM Tris pH 7.4, 150 mM NaCl, 1% Igepal CA-360, 0.5 % sodium deoxycholate, 1 mM EDTA, 1 mM DTT, 0.5 mg/ml Pefabloc, 1 µg/ml aprotinin and 1 µg/ml leupeptin). For western analysis aliquots of extracts were used containing equal amounts of protein as determined with a Bio-Rad protein assay (Bio-Rad). WCE were resolved by SDS–PAGE, transferred to a PVDF membrane (Millipore), and hybridized with rabbit anti-hRad51C antiserum (at a 1:1000 dilution) followed by a peroxidase-labeled anti-rabbit antibody (Amersham). Antibody binding was detected by enhanced chemoluminescence (Amersham). Equality of loading was confirmed by hybridizing with a monoclonal antibody against actin (Santa-Cruz).
RT–PCR analysis
Total RNA was extracted using RNAzolB (Cinna/Biotecx Laboratories). Oligo-dT primed first strand cDNA was reverse transcribed from 2 µg RNA using the Riboclone cDNA synthesis system (Promega) according to the manufacturer’s instructions. Rad51C cDNA was amplified by using primer pairs F1C–R1 spanning exons 2–9, F4–R1 spanning exons 4–9 and F3–R1 spanning exons 5–9 (Gibco BRL) resulting in bands from 990 and 858 bp for the first pair, 325 and 457 bp for the second pair and a 324 bp band with the third pair. The PCR was performed with cycles consisting of denaturation for 1 min at 94°C, primer annealing for 1 min at 49°C and extension for 2.5 min at 72°C in a DNA engine gradient cycler (BIOzym) with AmpliTaq DNA polymerase (Applied Biosystems). The primers used were: F1C, 5′-CAG CAA AGA AGT TGG GAT ATC-3′; F3, 5′-TGA TCA TTT GTT GGG CGA GGC C-3′; F4, 5′-TGC TGG CAC AAG TCT ATC TCC-3′; R1, 5′-GGT TCT CGT GAC CGT TTC CGG-3′.
DNA sequence analysis
The PCR products were generated as described in RT–PCR analysis and gel-purified by using Qiaex columns (Qiagen). Products were sequenced using forward and reverse primers as described above, at the Baseclear facilities (Baseclear)
Nucleotide sequence accession numbers
The Chinese hamster partial Rad51C cDNA sequence has been deposited in the GenBank database under accession number AJ428572. The GenBank accession numbers for human and mouse Rad51C sequences are AF029669 and AF324883, respectively.
Analysis of CAs, SCEs and sister chromatid cohesion
Exponentially growing V79B, CL-V4B and CL-V4B/hRad51C cells were either mock treated or treated for 2 h (SCEs) with MMC or for 24 h (CAs) with 1, 4, 40, 80 and 160 ng MMC/ml (V79B and CL-V4B/Rad51C) or with 0.5, 1.0, 1.5, 2.0 and 4.0 ng MMC/ml for CL-V4B cells. For SCE analysis, subsequently 5-bromo-2′-deoxyuridine (Sigma) was added to the medium (10 µM final concentration) to enable sister chromatid differentiation. The cells were harvested by trypsinization 24–48 h after (mock) treatment, following 2 h incubation with colcemid (1 µg/ml final concentration). For the sister chromatid cohesion analysis, preparations were also made with omission of colcemid in the procedure to serve as controls. The cells were fixed in ethanol:glacial acetic acid (3:1) after treatment with hypotonic solution (0.6% sodium citrate). Air-dried preparations were made and stained with a 5% aqueous Giemsa solution for 5 min to analyze for CAs and with FPG to visualize SCEs (45). For CAs 100 mitotic cells were scored at each dose, SCEs were analyzed in 25 cells and sister chromatid cohesion was scored in 100 cells. Data for CAs, SCEs and sister chromatid cohesion analyses are from at least two independent experiments of which the mean and the SEM were calculated. Statistical analysis was performed using t-tests.
RESULTS
Isolation of mammalian cell mutant, CL-V4B, and its sensitivity to DNA damaging agents
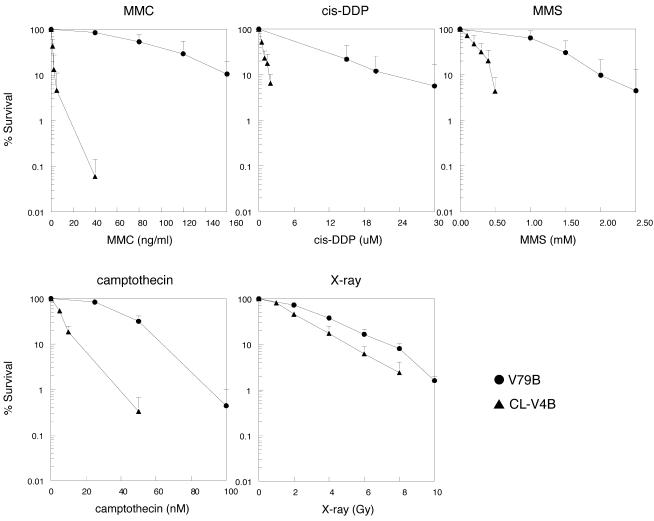
The Chinese hamster cell mutant, CL-V4B, has been isolated from an ENU-mutagenized population of V79B cells on the basis of its hypersensitivity to MMC, by the replica plating method described previously (41). Clonogenic survival experiments indicated that the CL-V4B mutant was very sensitive to DNA cross-linking agents. As shown in Figure 1, CL-V4B cells were ∼32-fold more sensitive to MMC and 14-fold more sensitive to cis-DDP than parental V79B cells, based on the dose required to reduce the survival to 10% (D10). This mutant also showed sensitivity to the mono-functional alkylating agent MMS (∼4-fold) and to the topoisomerase I-inhibitor camptothecin (∼4-fold). However, CL-V4B cells were only slightly sensitive to X-ray irradiation (∼1.3-fold) (Fig. 1), the radiomimetic agent BLM (∼1.3-fold) and to UV irradiation (∼1.5-fold) (data not shown). The generation time of CL-V4B cells was comparable with parental V79B cells (∼15 h), but the cloning efficiency was slightly reduced from 73 ± 9% in V79B to 58 ± 11% for CL-V4B (data not shown).
Figure 1.
Clonogenic survival of CL-V4B and parental V79B cells after exposure to MMC, cis-DDP, MMS, camptothecin and X-rays. Data are the means of at least three experiments. Error bars represent the SEM.
Impaired Rad51 nuclear foci formation after genotoxic treatment in CL-V4B
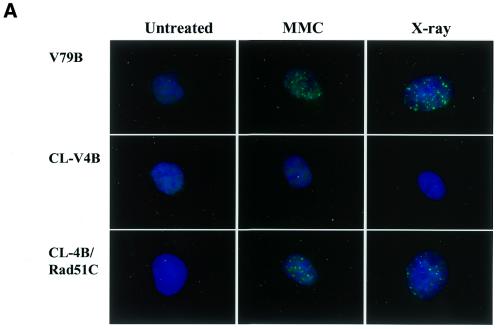
Given the hypersensitivity of CL-V4B to MMC that causes formation of interstrand cross-links, which can be repaired by a process depending on HR (46,47), we determined whether CL-V4B cells were able to form nuclear Rad51 foci after treatment with this agent. We therefore treated CL-V4B cells with MMC, fixed them after 8 h and subsequently immunostained the cells with anti-Rad51 antibodies. CL-V4B cells were severely impaired in the formation of nuclear Rad51 foci following MMC treatment (Fig. 2A), whereas V79B cells readily formed Rad51 foci under the same conditions. Since the nuclear Rad51 foci are also formed after X-ray irradiation (6), we also analyzed Rad51 foci formation in CL-V4B in response to X-rays. Intriguingly, despite the fact that CL-V4B is only slightly sensitive to cell killing by X-rays (Fig. 1), this mutant was also severely impaired in the formation of X-ray induced nuclear Rad51 foci (Fig. 2A). Immunoblot analysis of the Rad51 protein showed normal levels in CL-V4B cells prior to and after treatment with MMC when compared with wild-type (data not shown), indicating that CL-V4B cells are most probably defective in a gene responsible for the assembly and/or stabilization of the Rad51 complex upon the induction of exogenous DNA damage.
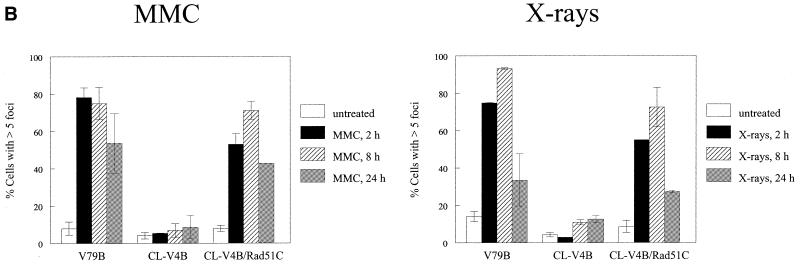
Figure 2.
Impaired DNA-damage induced Rad51 nuclear foci formation in CL-V4B. (A) Immunofluorescent visualization of nuclear Rad51 foci in untreated V79B, CL-V4B and CL-V4B/hRad51C cells, as well as in cells 8 h after treatment with either MMC (2.4 µg/ml for 1 h) or X-ray irradiation (12 Gy). Depicted is the merged picture of the FITC signal used to detect the Rad51 protein and the DAPI signal. (B) Kinetics of Rad51 foci formation in V79B, CL-V4B and CL-V4B/hRad51C cells analyzed 2, 8 and 24 h after treatment with MMC (2.4 µg/ml for 1 h) or after X-ray irradiation (12 Gy). A cell with more than five distinct foci in the nucleus was considered to be positive. Data are the means of at least two experiments. Error bars represent the SEM.
Complementation of MMC sensitivity and Rad51 nuclear foci formation by hRad51C cDNA transfection and by human chromosome 17 transfer
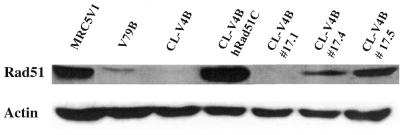
In our search for the gene defective in CL-V4B, we analyzed several genes involved in Rad51 complex assembly for their ability to complement the MMC sensitivity of this mutant. Transfection of CL-V4B cells with hRad51, hRad51B and hRad51D cDNAs did not result in complementation of their MMC sensitivity (Fig. 3). However, transfection of CL-V4B with hRad51C cDNA restored the MMC sensitivity as well as the MMS sensitivity of this mutant to a great extent (Fig. 3; data not shown), suggesting that CL-V4B is defective in Rad51C. Since cDNA transfection resulted in a high expression of hRad51C (Fig. 4), we also transferred a single human chromosome 17 to CL-V4B cells, presumably providing a more physiological level of Rad51C. However, this human chromosome also partially complemented the MMC sensitivity of CL-V4B (Fig. 3). Immunoblot analysis using anti-hRad51C antibodies indicated a high level of Rad51C protein in the CL-V4B/hRad51C transfectants and a lower level of protein in two complementing CL-V4B/#17 hybrids (#17.4 and #17.5) as shown in Figure 4. No protein could be detected in the non-complementing CL-V4B/#17 hybrid (#17.1) indicating that the lack of complementation in CL-V4B/#17.1 is due to absence of hRad51C protein (Fig. 4; data not shown). Microcell-mediated transfer can lead to damage of the transferred chromosome and this could explain the lack of Rad51C expression in the #17.1 hybrid. The endogenous hamster Rad51C was poorly recognized by the antibodies raised against hRad51C. However, while endogenous Rad51C could be detected in V79B cells, it was not detected in CL-V4B cells (Fig. 4).
Figure 3.
Complementation of MMC sensitivity of CL-V4B upon hRad51C cDNA transfection or transfer of human chromosome 17. Survival curves of V79B, CL-V4B and CL-V4B cells transfected with hRad51, hRad51B, hRad51D and hRad51C cDNA or after transfer of a single human chromosome 17 (#17) after treatment with different doses of MMC. Data are the average of at least three independent experiments. Error bars represent the SEM.
Figure 4.
Expression of the Rad51C protein is restored in complementing CL-V4B/#17 and CL-V4B/hRad51C cells. V79B, CL-V4B, CL-V4B/hRad51C, CL-V4B/#17 (hybrids 1, 4, 5) and human immortal fibroblast MRC5V1 cells were lysed and whole cell extracts were electrophoresed on a 12% SDS–PAGE gel and transferred to a membrane. The Rad51C protein was visualized by using polyclonal rabbit anti-hRad51C antibodies and actin by using mouse anti-actin antibodies to confirm equality of loading.
Transfection of CL-V4B with hRad51C cDNA also restored the ability to form nuclear Rad51 foci after treatment with MMC or X-rays (Fig. 2A), with kinetics comparable with parental cells (Fig. 2B). In untreated cells a level of ∼10% Rad51 foci-positive cells could be observed in V79B cells, as well as in CL-V4B/hRad51C transfectants. Two hours after treatment with X-rays or MMC the number of foci-positive cells increased to ∼70–90% in both parental cells and transfectants. These levels of foci positive cells were maintained at 8 h after the treatment and declined after 24 h (Fig. 2B). In contrast, the level of Rad51 foci-positive cells in CL-V4B remained at the level found in untreated cells (Fig. 2B).
The Rad51C cDNA of CL-V4B contains a deletion of 132 bp corresponding to lack of exon 5
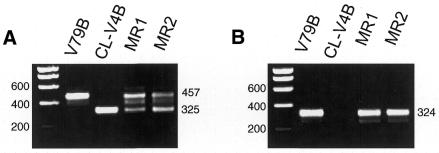
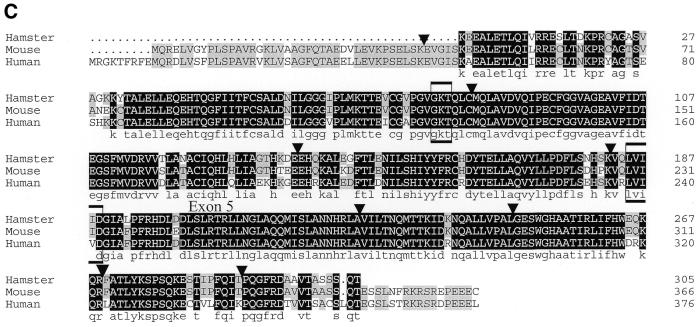
To study the molecular nature of the defect in CL-V4B, we designed primers based on the human and mouse Rad51C cDNA sequences (20,48) to amplify the hamster Rad51C cDNA using RT–PCR. Total RNA of CL-V4B and parental V79B cells was converted to cDNA and amplified using Rad51C-specific primer pairs: F1C–R1, F3–R1 and F4–R1. The RT–PCR with primer pair F1C–R1 showed a major band of 990 bp in V79B, whereas a shorter product of 858 bp was present in CL-V4B (data not shown). Similarly, RT–PCR with primers F4–R1 revealed a major band of 457 bp in V79B cells and a shorter band of 325 bp in CL-V4B cells, whereas MMC-resistant revertants of CL-V4B, MR1 and MR2, revealed the presence of both bands (Fig. 5A). These results indicate that the functional hemizygosity often observed in hamster cells due to silencing of the second allele via methylation (49) or alternative pathways was inverted in these CL-V4B revertants giving rise to one wild-type and one mutant allele. All amplified Rad51C PCR products were sequenced resulting in an almost complete cDNA sequence. Sequence analysis revealed a deletion of 132 bp in the CL-V4B Rad51C sequence from amino acids 183–226 when compared with the sequences of two different wild-type hamster cell lines, V79B and AA8, which showed identical sequences (Fig. 5C). Furthermore, we compared the hamster Rad51C sequence with the recently available human genomic DNA sequence of the 17q23 region which indicated that the 132 bp deletion found in the CL-V4B Rad51C cDNA was due to splicing out of exon 5. This was confirmed by a RT–PCR with primers F3–R1 in which the forward primer was located in exon 5, showing a lack of PCR product in CL-V4B and the predicted 324 bp band in V79B cells and in the revertants, MR1 and MR2 (Fig. 5B). Deletion of exon 5 results in the deletion of the Walker B box, which destroys the ATP binding site and the core domain of the protein.
Figure 5.
CL-V4B has a 132 bp deletion in Rad51C cDNA due to lack of exon 5. RT–PCR of hamster Rad51C from cDNA generated from parental V79B cells, CL-V4B cells and CL-V4B derived MMC-resistant clones, MR1 and MR2, using primer pairs (A) F4–R1 spanning exons 4–9 and (B) F3–R1 spanning exons 5–9. The position of the molecular size markers (bp) is shown on the left of each panel, as well as the size of the PCR products on the right. (C) Alignment of predicted amino acid sequences of Chinese hamster Rad51C (exons 2–9) with mouse and human Rad51C. Identical amino acids are highlighted in black, identical amino acids in two of the three sequences are highlighted in gray. The triangles represents the intron–exon borders as deduced from the human #17q23 sequence, boxed amino acids represent nucleotide binding regions.
Increased levels of spontaneous and MMC-induced CAs in CL-V4B
To further investigate the role of Rad51C in mammalian cells, we analyzed the spontaneous and MMC-induced CAs in the CL-V4B mutant and the parental V79B cells (Tables 1 and 2). This analysis revealed higher levels of spontaneous CAs (∼6-fold) in CL-V4B cells than parental V79B cells. The majority of these aberrations in CL-V4B were of the chromatid type, which are usually lethal and seldom seen in V79B cells. Chromatid breaks as well as exchanges were observed with a normal ratio of inter- versus intra-chromatid changes. After MMC treatment the relative chromosomal sensitivity of CL-V4B further increased to ∼300-fold (Table 2). Transfection of the hRad51C cDNA into CL-V4B restored the spontaneous and MMC-induced CAs to a great extent (Tables 1 and 2), confirming that the genomic instability was due to the defect in Rad51C.
Table 2. Relative sensitivities for chromosomal aberrations.
| Relative sensitivity | |||
|---|---|---|---|
| V79B | CL-V4B | CL-V4B/hRad51C | |
| Spontaneous |
1a |
6 |
1 |
| MMC-induced | 1b | >300 | 11 |
aThe frequency of spontaneous aberrations per 100 cells in parental V79B cells was 7 ± 2, and this was set at 1.
bThe slope of the induction curve of CA after MMC treatment in parental V79B cells was 0.4 aberrations per 100 cells per ng/ml MMC, and this was set as 1. All slopes were determined by using at least three different dose levels.
Table 1. Spontaneous and MMC-induced CAs per 100 cells.
| Cell line | Cells with CA | Chromatid | Chromosome | Total breaksc ± SEM | |||
|---|---|---|---|---|---|---|---|
| Breaks | Exchangesb | Breaks | Exchanges | ||||
| V79B |
Spontaneous |
7 ± 1 |
6 |
0 |
1 |
0 |
7 ± 2 |
| |
80 ng/ml MMCa |
30 ± 8 |
26 |
4 |
7 |
1 |
43 ± 7 |
| CL-V4B |
Spontaneous |
15 ± 3 |
29 |
3 |
5 |
0 |
40 ± 8 |
| |
2 ng/ml MMCa |
48 ± 10 |
47 |
23 |
4 |
1 |
99 ± 24 |
| CL-V4B/hRad51C |
Spontaneous |
12 ± 1 |
8 |
1 |
2 |
0 |
12 ± 2 |
| 80 ng/ml MMCa | 49 ± 3 | 148 | 23 | 2 | 0 | 196 ± 7 | |
aThe indicated frequencies of MMC-induced CA are after treatment with equitoxic doses that give 20–60% survival.
bChromatid exchanges are interchanges, intrachanges, triradials or subchromatid exchanges.
cFor calculations of the total number of breaks the exchanges were counted as two breaks.
Rad51C influences the spontaneous and MMC-induced SCEs
Recent studies in Rad51 paralog-knockout DT40 cells showed a reduced frequency of spontaneous and MMC-induced SCEs (36,37); therefore, we examined the frequency of spontaneous and MMC-induced SCEs in CL-V4B. This analysis revealed slightly reduced spontaneous SCE numbers in CL-V4B (4.9 SCEs/cell) compared with V79B (6.6 SCEs/cell) (Table 3). After treatment with various doses of MMC, V79B cells showed a significant increase of SCE level up to 21 SCEs/cell at a dose of 30 ng/ml (P < 0.01), whereas no clear SCE induction was observed in CL-V4B cells (5.4 SCEs/cell at a dose of 1.5 ng/ml) (Table 3). This was not solely due to the low doses of MMC that could be used in CL-V4B due to the high toxicity, since preliminary data with another MMC-sensitive hamster cell mutant V-H4 (42), which is not affected in Rad51 foci formation, demonstrated that an increase in SCEs can be observed after low doses of MMC (data not shown). Furthermore, SCEs were significantly increased in the CL-V4B/hRad51C transfectant after MMC treatment (P < 0.05) albeit at significantly lower frequency than in V79B cells (P < 0.05; Table 3). These data indicate that CL-V4B cells are hampered in MMC-induced SCE formation and that transfection of hRad51C cDNA partially restores this capacity. Therefore, Rad51C apparently plays a role in the formation of SCEs as part of the cellular response to cross-links.
Table 3. Analysis of spontaneous and MMC-induced SCEs in V79B, CL-V4B and the CL-V4B/hRad51C transfectant.
| Cell line | Concentration MMC (ng/ml) | SCEs/cell ± SEM |
|---|---|---|
| V79B |
0 |
6.6 ± 0.4 |
| |
1 |
5.9 ± 0.4 |
| |
15 |
15.0 ± 0.7 |
| |
30 |
21.3 ± 1.3 |
| CL-V4B |
0 |
4.9 ± 0.4 |
| |
0.5 |
4.7 ± 0.6 |
| |
1.0 |
5.5 ± 0.3 |
| |
1.5 |
5.4 ± 0.3 |
| CL-V4B/hRad51C |
0 |
5.8 ± 0.4 |
| |
5 |
7.2 ± 0.5 |
| 15 | 8.9 ± 0.6 |
Normal centrosome formation but impaired sister chromatid cohesion in CL-V4B
In XRCC2-, XRCC3-, Brca1- and Brca2-deficient rodent cells incorrect chromosome segregation as a result of abnormal centrosomes can be observed which may contribute to the chromosomal instability in these cells (32,50,51). Therefore, we investigated whether a defect in Rad51C would also give rise to abnormal centrosomes. Centrosomes were visualized microscopically after labeling the cells with anti-γ-tubulin antibodies. This analysis demonstrated that CL-V4B cells did not differ from V79B cells with respect to centrosome formation. In 90–95% of the cells centrosomes were visible as clearly defined dot shaped structures in both cell lines (data not shown). These results indicate that the defective Rad51C protein as observed in CL-V4B cells does not influence centrosome formation, which is in contrast to cell lines defective in Brca1, Brca2, XRCC2 and XRCC3.
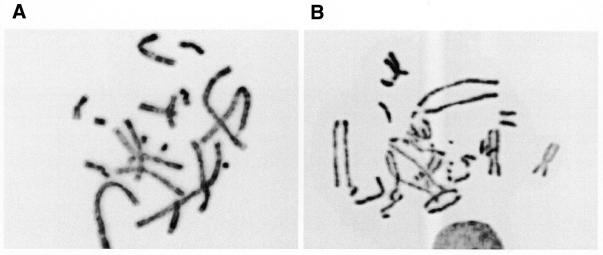
Intriguingly, we observed that a defect in Rad51C affected sister chromatid cohesion. In the colcemid accumulated metaphase spreads of untreated CL-V4B cells, in ∼58% of the cells the sister chromatids were not attached at their centromeres and therefore not lying close to each other (Fig. 6). In contrast, in V79B only 26% of the cells showed separated chromatids (Table 4). After omission of colcemid, CL-V4B still displayed reduced sister chromatid cohesion with 21% separated chromatids compared with 8% in V79B (Table 4), which indicated that the colcemid treatment was not underlying this reduced cohesion. Moreover, transfection of hRad51C cDNA restored the sister chromatid cohesion (11% separated chromatids in the absence of colcemid), suggesting that the impaired cohesion of sister chromatids was indeed due to the defect in Rad51C. Similar analyses in irs1 and in irs1SF, defective in XRCC2 and XRCC3, respectively, did not show any influence of these genes on sister chromatid cohesion when compared with their parental cell lines (Table 4) indicating that this is a unique feature of Rad51C. Moreover, analysis of chromosomes at the G2 phase of the cell cycle by premature chromosome condensation using caliculin A treatment revealed normal cohesion of sister chromatids in both V79B and CL-V4B (data not shown) indicating that cohesion is normal at G2 phase in Rad51C-deficient cells but that sister chromatids separate prematurely around metaphase–anaphase transition.
Figure 6.
Impaired sister chromatid cohesion. Shown are representative pictures of (A) metaphase spread from a V79B cell with normal cohesion and (B) metaphase spread from a CL-V4B cell with impaired sister chromatid cohesion (see Table 4). Both metaphase spreads were generated after treatment with colcemid.
Table 4. Cells with precocious sister chromatid separation per 100 cells.
| + Colcemid ± SEM | – Colcemid ± SEM | |
|---|---|---|
| V79B |
26 ± 7 |
8 ± 1 |
| CL-V4B |
58 ± 5 |
21 ± 2 |
| CL-V4B/hRad51C |
42 ± 5 |
11 ± 2 |
| V79 |
30 ± 8 |
3 ± 1 |
|
irs1 |
18 ± 6 |
4 ± 1 |
| AA8 |
1 ± 1 |
0 |
| irs1SF | 1 ± 1 | 0 |
DISCUSSION
Here, we describe the first mammalian cell mutant defective in Rad51C. The Rad51C cDNA derived from the Chinese hamster cell mutant, CL-V4B, contains a 132 bp deletion that corresponds to a deletion of exon 5. Comparison of human, mouse and hamster Rad51C reveals that the protein is highly conserved in evolution. It contains a bipartite Walker A and B ATPase domain (52) characteristic of the RecA/Rad51 protein family (3). Deletion of exon 5 results in loss of the Walker B box motif, which is located in what is the core domain of the homologous RecA protein (53).
The general importance of Rad51C in protecting genome stability is emphasized by the various DNA damage sensitivities of CL-V4B cells. The sensitivity of the CL-V4B mutant to cross-links, monofunctional alkylating agents and to topoisomerase I inhibitors is in concordance with results obtained in XRCC2- or XRCC3-deficient cell lines (15,28–30). In comparison with the Rad51C–/– chicken DT40 cells (37), CL-V4B hamster cells are much more sensitive to cross-linking agents. Conversely, only a slight sensitivity to X-ray irradiation is found in CL-V4B cells, in contrast to what has been observed in chicken Rad51 paralog knockout cells as well as in XRCC2- and XRCC3-deficient rodent cells that show a 2-fold increase in sensitivity (28,36,37,54). This suggests that Rad51C-dependent pathways of DNA repair might contribute differentially towards repair of cross-links and X-ray induced DNA damage in different cell types or in different species. Finally, the Rad51C-deficient CL-V4B cells differ from XRCC2- and XRCC3-deficient hamster cells as well as from the Rad51 paralog knockout chicken cells in that they are not significantly affected in cell growth and only slightly affected in cloning efficiency (28,36,37,54).
The defect in Rad51C affected the formation of Rad51 nuclear foci in CL-V4B cells after treatment with cross-linking agents as was observed in Brca2- and XRCC3-defective rodent cell mutants (34,51) as well as in the Rad51B-deficient chicken cells (36). All Rad51 paralog-deficient cells identified so far display mild sensitivity to cell killing by X-rays and show severely impaired Rad51 foci formation after X-ray irradiation. CL-V4B cells show similarly impaired Rad51 foci formation after X-ray irradiation but are less sensitive to cell killing by X-rays suggesting that the role of Rad51C in X-ray induced DNA damage repair might be redundant, whereas its role in cross-link repair is not. The impaired DNA damage-induced Rad51 foci formation indicates that Rad51C is involved in a Rad51-dependent HR pathway most probably via a role in assembly and/or stabilization of the Rad51 complex after genotoxic treatment.
CL-V4B cells display an increased frequency of spontaneous and MMC-induced CAs, which is in line with data from Brca2-, XRCC2- and XRCC3-deficient rodent cells and Rad51 paralog knockout chicken cells (15,31,36,37,51,55). This indicates that all these genes, most likely through their function in HR, are involved in maintenance of genomic stability. Intriguingly, the chromosomal instability in CL-V4B does not greatly reduce the cell viability of this mutant, which is in contrast to the Rad51 paralogs knockout chicken cells (36,37) but the cloning efficiency is reduced in CL-V4B cells albeit to a much lower extent. This indicates that a defect in Rad51C influences the CA frequencies but that its effect on cell viability may vary between different species or cell types. This disparity could be caused by differences in apoptotic potential. Because chicken DT40 cells are of lymphoid origin they could possibly undergo apoptosis much more readily than Chinese hamster lung-derived fibroblasts.
SCEs likely reflect post-replicational repair by HR that is associated with crossing-over between sister duplexes (56). The reduced SCE frequency of CL-V4B and the lack of induction of SCEs after MMC treatment suggests that Rad51C is indeed involved in HR. Similar observations have been made for other HR-deficient vertebrate cell lines including Brca2-deficient rodent cells, Rad51 paralog knockout chicken cells and Rad54 knockout mouse embryonic stem cells (36,37,51,57).
An additional aspect of chromosome metabolism that has been found to be aberrant in HR-deficient cells is the number of centrosomes. Abnormalities in centrosome number have been observed in Brca1-, Brca2-, XRCC2- and XRCC3-deficient rodent cells (32,50,51). In contrast, the defect in Rad51C in CL-V4B cells does not result in abnormalities in centrosome number. These observations infer that the chromosomal instability in CL-V4B is not caused by spindle-mediated chromosome missegregation. Intriguingly, the mutated Rad51C in CL-V4B leads to impaired sister chromatid cohesion, which is complemented by Rad51C cDNA transfection. This reduced sister chromatid cohesion seems to be a unique feature of the defective Rad51C because it is only observed in CL-V4B cells and not in XRCC2-, XRCC3- or Brca2-defective (data not shown) hamster cells.
Chromosome maintenance activities such as sister chromatid cohesion and chromosome condensation are regulated by members of the structural maintenance of chromosomes (SMC) family of proteins. In eukaryotes the cohesin multiprotein complex containing SMC1, SMC3, Scc3 (SA) and Scc1 (Rad21) is required for sister chromatid cohesion (58–61). The SMC1/SMC3 heterodimer is also associated with DNA polymerase ɛ and DNA ligase III in the RC-1 complex that promotes DNA recombination (62,63). The RC-1 complex is not the only indication for involvement of SMC proteins in DNA recombination and repair processes. The yeast Rad18 and Spr18 are SMC-like proteins with a role in recombinational repair (64). Recently, it has been shown that cohesin (Smc1, Scc1) as well as factors needed for its generation and subsequent loading onto chromosomes are required for post-replicative double-strand break repair during late S and early G2 phase in yeast (65). Indeed, DNA double-strand breaks are preferentially repaired by HR during late S and early G2 phase when an undamaged sister chromatid is available (2). The present data indicate that Rad51C influences sister chromatid cohesion but whether this is due to a direct interaction with the cohesin complex or with the interacting proteins remains to be investigated.
Our results indicate that the hamster CL-V4B mutant offers a tool to study the functions of the Rad51C gene in mammalian cells, since so far only DT40 chicken cells defective in Rad51C are available (37). The complementation of all the phenotypic characteristics of CL-V4B upon transfection with human Rad51C cDNA indicates that the described mutation in Rad51C is responsible. However, we cannot formally exclude that an additional mutation might also be involved. The observed complex phenotype of CL-V4B might be due to the fact that Rad51C is part of at least two distinct protein complexes. Rad51C and XRCC3 can form a heterodimer, which binds single-stranded DNA (26). Rad51C is also part of a multi-protein complex comprised of at least Rad51B, C, D and XRCC2 that binds single-stranded DNA and single strand gaps in duplex DNA (66). Furthermore, a subcomplex consisting of Rad51B and Rad51C can act as a mediator in the Rad51/RPA-catalyzed DNA strand exchange (27). Thus, Rad51C appears to be the only Rad51 paralog that is a common component of distinct Rad51 paralog-containing complexes.
Defects in HR are thought to play a significant role in promoting tumorigenesis because of their effect on genomic stability (67–69). For example, chromosomal translocations that involve Rad51B are frequently observed in uterine leiomyoma (70) and mutations in Rad54 and its homolog Rad54B are observed in lymphoma, colon and breast cancer (71,72). Since CL-V4B cells display genomic instability with increased levels of CAs, lack of SCE induction after genotoxic treatment as well as impaired sister chromatid cohesion, defects in the Rad51C gene will most probably play an important role in cancer development. In fact, 17q23 amplifications in breast cancer also involve the Rad51C gene (73). Therefore, it would be of interest to screen different tumors including breast cancers for Rad51C mutations.
Acknowledgments
ACKNOWLEDGEMENTS
We thank F. E. Benson and M. Oshimura for providing us with Rad51 antibodies and the A9Neo#17 cells, respectively. We thank N. van Wessel and L. Wetselaar for their skillful technical assistance. This work was supported, in part, by the Netherlands Organization for Scientific Research (NWO, grant 901-01-190).
DDBJ/EMBL/GenBank accession no. AJ428572
REFERENCES
- 1.Kanaar R., Hoeijmakers,J.H.J. and van Gent,D. (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol., 8, 483–489. [DOI] [PubMed] [Google Scholar]
- 2.Karran P. (2000) DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev., 10, 144–150. [DOI] [PubMed] [Google Scholar]
- 3.Thompson L.H. and Schild,D. (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res., 477, 131–153. [DOI] [PubMed] [Google Scholar]
- 4.Baumann P. and West,S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- 5.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Sacharomyces cerevisiae.Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haaf T., Golub,E.I., Reddy,G., Radding,C.M. and Ward,D.C. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA, 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tashiro S., Walter,J., Shinohara,A., Kamada,N. and Cremer,T. (2000) Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol., 150, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu N. and Maizels,N. (2000) Coordinated response of mammalian Rad51 and Rad52 to DNA damage. EMBO Rep., 1, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan T.L.R., Essers,J., Citterio,E., Swagemakers,S.M.A., de Wit,J., Benson,F.E., Hoeijmakers,J.H.J. and Kanaar,R. (1999) Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol., 9, 325–328. [DOI] [PubMed] [Google Scholar]
- 10.Raderschall E., Golub,E.I. and Haaf,T. (1999) Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl Acad. Sci. USA, 96, 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara A.H., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fission yeast genes homologous to RAD51 and recA. Nature Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 13.Habu T., Taki,T., West,A., Nishimune,Y. and Morita,T. (1996) The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res., 24, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cartwright R., Tambini,C.E., Simpson,P.J. and Thacker,J. (1998) The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucleic Acids Res., 26, 3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N., Lamerdin,J.E., Tebbs,R.S., Schild,D., Tucker,J.D., Shen,M.R., Brookman,K.W., Siciliano,M.J., Walter,C.A., Fan,W., Narayana,L.S., Zhou,Z.-Q., Adamson,A.W., Sorensen,K.J., Chen,D.J., Jones,N.J. and Thompson,L.H. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- 16.Tebbs R.S., Zhao,Y., Tucker,J.D., Scheerer,J.B., Siciliano,M.J., Hwang,M., Liu,N., Legerski,R.J. and Thompson,L.H. (1995) Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl Acad. Sci. USA, 92, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce A.J., Johnson,R.D., Thompson,L.H. and Jasin,M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev., 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albala J.S., Thelen,M.P., Prange,C., Fan,W., Christensen,M., Thompson,L.H. and Lennon,G.G. (1997) Identification of a novel human RAD51 homolog, RAD51B. Genomics, 46, 476–479. [DOI] [PubMed] [Google Scholar]
- 19.Cartwright R., Dunn,A.M., Simpson,P.J., Tambini,C.E. and Thacker,J. (1998) Isolation of novel human and mouse genes of the recA/Rad51 recombination-repair gene family. Nucleic Acids Res., 26, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dosanjh M.K., Collins,D.W., Fan,W., Lennon,G.G., Albala,J.S., Shen,Z. and Schild,D. (1998) Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acids Res., 26, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawabata M. and Saeki,K. (1998) Sequence analysis and expression of a novel mouse homolog of Escherichia coli recA gene. Biochim. Biophys. Acta, 1398, 353–358. [DOI] [PubMed] [Google Scholar]
- 22.Pittman D.L., Weinberg,L.R. and Schimenti,J.C. (1998) Identification, characterization, and genetic mapping of Rad51D, a new mouse and human RAD51/recA-related gene. Genomics, 49, 103–111. [DOI] [PubMed] [Google Scholar]
- 23.Thacker J. (1999) A surfeit of RAD51-like genes. Trends Genet., 15, 166–168. [DOI] [PubMed] [Google Scholar]
- 24.Schild D., Lio,Y.-C., Collins,D.W., Tsomondo,T. and Chen,D.J. (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem., 275, 16443–16449. [DOI] [PubMed] [Google Scholar]
- 25.Braybrooke J.P., Spink,K.G., Thacker,J. and Hickson,I.D. (2000) The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J. Biol. Chem., 275, 29100–29106. [DOI] [PubMed] [Google Scholar]
- 26.Masson J.-Y., Stasiak,A.Z., Stasiak,A., Benson,F.E. and West,S.C. (2001) Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl Acad. Sci. USA, 98, 8440–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigurdson S., Van Komen,S., Bussen,W., Schild,D., Albala,J.S. and Sung,P. (2001) Mediator function of the human Rad51B–Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev., 15, 3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones N.J., Cox,R. and Thacker,J. (1987) Isolation and cross-sensitivity of X-ray sensitive mutants of V79-4 hamster cells. Mutat. Res., 183, 279–286. [DOI] [PubMed] [Google Scholar]
- 29.Thacker J. and Ganesh,A.N. (1990) DNA-break repair, radioresistence of DNA synthesis, and camptothecin sensitivity in the radiation-sensitive irs mutants: comparisons to ataxia-telangiectasia cells. Mutat. Res., 235, 49–58. [DOI] [PubMed] [Google Scholar]
- 30.Caldecott K. and Jeggo,P. (1991) Cross-sensitivity of γ-ray-sensitive hamster mutants to cross-linking agents. Mutat. Res., 255, 111–121. [DOI] [PubMed] [Google Scholar]
- 31.Tucker J.D., Jones,N.J., Allen,N.A., Minkler,J.L., Thompson,L.H. and Carrano,A.V. (1991) Cytogenetic characterization of the ionizing radiation-sensitive hamster mutant irs1. Mutat. Res., 254, 143–152. [DOI] [PubMed] [Google Scholar]
- 32.Griffin C.S., Simpson,P.J., Wilson,C.R. and Thacker,J. (2000) Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nature Cell Biol., 2, 757–761. [DOI] [PubMed] [Google Scholar]
- 33.Johnson R.D., Liu,N. and Jasin,M. (1999) Mammalian XRCC2 promotes the repair of DNA double strand breaks by homologous recombination. Nature, 401, 397–399. [DOI] [PubMed] [Google Scholar]
- 34.Bishop D.K., Ear,U., Bhattacharyya,A., Calderone,C., Beckett,M., Weichselbaum,R.R. and Shinohara,A. (1998) Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem., 273, 21482–21488. [DOI] [PubMed] [Google Scholar]
- 35.O’Regan P., Wilson,C., Townsend,S. and Thacker,J. (2001) XRCC2 is a RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem., 276, 22148–22153. [DOI] [PubMed] [Google Scholar]
- 36.Takata M., Sasaki,M.S., Sonoda,E., Fukushima,T., Morrison,C., Albala,J.S., Swagemakers,S.M.A., Kanaar,R., Thompson,L.H. and Takeda,S. (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol., 20, 6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takata M., Sasaki,M.S., Tachiiri,S., Fukushima,T., Sonoda,E., Schild,D., Thompson,L.H. and Takeda,S. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol., 21, 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu Z., Smith,S., Wang,L., Rice,M.C. and Kmiec,E.B. (1999) Disruption of the muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53–/– background. Mol. Cell. Biol., 19, 8686–8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittman D.L. and Schimenti,J.C. (2000) Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51L3. Genesis, 26, 167–173. [DOI] [PubMed]
- 40.Deans B., Griffin,C.S., Maconochie,M. and Thacker,J. (2000) Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J., 19, 6675–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zdzienicka M.Z. and Simons,J. (1987) Mutagen-sensitive cell lines are obtained at high frequency in V79 Chinese hamster cells. Mutat. Res., 178, 235–244. [DOI] [PubMed] [Google Scholar]
- 42.Zdzienicka M.Z., Arwert,F., Neuteboom,I., Rooimans,M. and Simons,J.W.I.M. (1990) The Chinese hamster V79 cell mutant V-H4 is phenotypically like Fanconi anemia cells. Somat. Cell Mol. Genet., 16, 575–581. [DOI] [PubMed] [Google Scholar]
- 43.Telleman P., Overkamp,W.J.I., van Wessel,N., Studzian,K., Wetselaar,L., Natarajan,A.T. and Zdzienicka,M.Z. (1995) A new complementation group of mitomycin C-hypersensitive Chinese hamster cell mutants that closely resembles the phenotype of Fanconi anemia cells. Cancer Res., 55, 3412–3416. [PubMed] [Google Scholar]
- 44.Koi M., Shimizu,M., Morita,H. and Oshimura,M. (1989) Construction of mouse A9 clones containing a single human chromosome tagged with neomycin-resistance gene via microcell fusion. Jpn J. Cancer Res., 80, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry P. and Wolff,S. (1974) New Giemsa staining for the differential staining of sister chromatids. Nature, 251, 156–158. [DOI] [PubMed] [Google Scholar]
- 46.De Silva I.U., McHugh,P.J., Clingen,P.H. and Hartley,J.A. (2000) Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol., 20, 7980–7990.11027268 [Google Scholar]
- 47.Dronkert M.L.G. and Kanaar,R. (2001) Repair of DNA interstrand cross-links. Mutat. Res., 486, 217–247. [DOI] [PubMed] [Google Scholar]
- 48.Leasure C.S., Chandler,J., Gilbert,D.J., Householder,G.D.J., Stephens,R., Copeland,N.G., Jenkins,N.A. and Sharan,S.K. (2001) Sequence, chromosomal location and expression analysis of the murine homologue of human RAD51L2/RAD51C. Gene, 271, 59–67. [DOI] [PubMed] [Google Scholar]
- 49.Jeggo P.A. and Holliday,R. (1986). Azacytidine-induced reactivation of a DNA repair gene in Chinese hamster ovary cells. Mol. Cell. Biol., 6, 2944–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu X., Weaver,Z., Linke,S.P., Li,C., Gotay,J., Wang,C.-X., Harris,C.C., Ried,T. and Deng,C.-X. (1999) Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell, 3, 389–395. [DOI] [PubMed] [Google Scholar]
- 51.Kraakman-van der Zwet M., Overkamp,W.J.I., van Lange,R.E.E., Essers,J., van Duijn-Goedhart,A., Wiggers,I., Swaminathan,S., van Buul,P.P.W., Errami,A., Tan,R.T.L., Jaspers,N.G.J., Sharan,S.K., Kanaar,R. and Zdzienicka,M.Z. (2002) Brca2 (XRCC11) deficiency results in radioresistent DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol., 22, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Story R.M. and Steitz,T.A. (1992) Structure of the recA protein–ADP complex. Nature, 355, 374–376. [DOI] [PubMed] [Google Scholar]
- 54.Fuller L.F. and Painter,R.B. (1988) A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res., 193, 109–121. [DOI] [PubMed] [Google Scholar]
- 55.Tutt A., Bertwistle,D., Valentine,J., Gabriel,A., Swift,S., Ross,G., Griffin,C., Thacker,J. and Ashworth,A. (2001) Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J., 20, 4704–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,T. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dronkert M.L.G., Beverloo,H.B., Johnson,R.D., Hoeijmakers,J.H.J., Jasin,M. and Kanaar,R. (2000) Mouse RAD54 affects double-strand break repair and sister chromatid exchange. Mol. Cell. Biol., 20, 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strunnikov A.V. and Jessberger,R. (1999) Structural maintenance of chromosomes (SMC) proteins conserved molecular properties for multiple biological functions. Eur. J. Biochem., 263, 6–13. [DOI] [PubMed] [Google Scholar]
- 59.McKay M.J., Troelstra,C., van der Spek,P., Kanaar,R., Smit,B., Hagemeijer,A., Bootsma,D. and Hoeijmakers,J. (1996) Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics, 36, 305–315. [DOI] [PubMed] [Google Scholar]
- 60.Hoque Md.T. and Ishikawa,F. (2001) Human chromatid cohesion component hRad21 is phosphorylated in M phase and associated with metaphase centromeres. J. Biol. Chem., 276, 5059–5067. [DOI] [PubMed] [Google Scholar]
- 61.Losada A., Yokochi,T., Kobayashi,R. and Hirano,T. (2000) Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol., 150, 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jessberger R., Riwar,B., Beachtold,H. and Akhmedov,A.T. (1996) SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J., 15, 4061–4068. [PMC free article] [PubMed] [Google Scholar]
- 63.Stursberg S., Riwar,B. and Jessberger,R. (1999) Cloning and characterization of mammalian SMC1 and SMC3 genes and proteins, components of the DNA recombination complexes RC-1. Gene, 228, 1–12. [DOI] [PubMed] [Google Scholar]
- 64.Fousteri M.I. and Lehmann,A.R. (2000) A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J., 19, 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sjögren C. and Nasmyth,K. (2001) Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol., 11, 991–995. [DOI] [PubMed] [Google Scholar]
- 66.Masson J.-Y., Tarsounas,M.C., Stasiak,A.Z., Stasiak,A., Shah,R., McIlwraith,M.J., Benson,F. and West,S.C. (2001) Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev., 15, 3296–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jasin M. (2000) Chromosome breaks and genomic instability. Cancer Invest., 18, 78–86. [DOI] [PubMed] [Google Scholar]
- 68.Pastink A., Eeken,J.C.J. and Lohman,P.H.M. (2001) Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res., 480–481, 37–50. [DOI] [PubMed] [Google Scholar]
- 69.Van Gent D.C., Hoeijmakers,G.H. and Kanaar,R. (2001) Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet., 3, 196–206. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi T., Nagai,N., Oda,H., Kamada,N. and Miyagawa,K. (2001) Evidence for Rad51L1/HMGIC fusion in the pathogenesis of uterine leiomyoma. Genes Chromosom. Cancer, 30, 196–201. [DOI] [PubMed] [Google Scholar]
- 71.Matsuda M., Miyagawa,K., Takahashi,M., Fukuda,T., Kataoka,T., Asahara,T., Inui,H., Watatani,M., Yasutomi,M., Kamada,N., Dohi,K. and Kamiya,K. (1999) Mutations in the RAD54 recombination gene in primary cancers. Oncogene, 18, 3427–3430. [DOI] [PubMed] [Google Scholar]
- 72.Hiramoto T., Nakanishi,T., Sumiyoshi,T., Fukuda,T., Matsuura,S., Tauchi,H., Komatsu,K., Shibasaki,Y., Inui,H., Watatani,M., Yasutomi,M., Sumii,K., Kajiyama,G., Kamada,N., Miyagawa,K. and Kamiya,K. (1999) Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene, 18, 3422–3426. [DOI] [PubMed] [Google Scholar]
- 73.Wu G.-J., Sinclair,C.S., Paape,J., Ingle,J.N., Roche,P.C., James,D. and Couch,F.J. (2000) 17q23 amplification in breast cancer involve the PAT1, RAD51C, PS6K, and SIGMA1B genes. Cancer Res., 60, 5371–5375. [PubMed] [Google Scholar]