Abstract
Objective
International Classification of Diseases (ICD) codes recorded in electronic health records (EHRs) are frequently used to create patient cohorts or define phenotypes. Inconsistent assignment of codes may reduce the utility of such cohorts. We assessed the reliability across time and location of the assignment of ICD codes in a US health system at the time of the transition from ICD-9-CM (ICD, 9th Revision, Clinical Modification) to ICD-10-CM (ICD, 10th Revision, Clinical Modification).
Materials and methods
Using clusters of equivalent codes derived from the US Centers for Disease Control and Prevention General Equivalence Mapping (GEM) tables, ICD assignments occurring during the ICD-9-CM to ICD-10-CM transition were investigated in EHR data from the US Veterans Administration Central Data Warehouse using deep learning and statistical models. These models were then used to detect abrupt changes across the transition; additionally, changes at each VA station were examined.
Results
Many of the 687 most-used code clusters had ICD-10-CM assignments differing greatly from that predicted from the codes used in ICD-9-CM. Manual reviews of a random sample found that 66% of the clusters showed problematic changes, with 37% having no apparent explanations. Notably, the observed pattern of changes varied widely across care locations.
Discussion and conclusion
The observed coding variability across time and across location suggests that ICD codes in EHRs are insufficient to establish a semantically reliable cohort or phenotype. While some variations might be expected with a changing in coding structure, the inconsistency across locations suggests other difficulties. Researchers should consider carefully how cohorts and phenotypes of interest are selected and defined.
Keywords: International classification of diseases, clinical coding, data accuracy
Introduction
The adoption of electronic health record (EHR) systems was an effort to improve patient care by improving the accuracy and clarity of medical records and by increasing the availability of the records to patients and physicians. It has been observed that EHR data can be very useful for studying patient outcomes in real-world settings. EHR data has become widely used for observational clinical research to improve post-care surveillance, and to perform comparative effectiveness studies. 1 However, the usefulness of EHR-based research depends centrally on the quality of the data to be found there. 2
Traditionally, EHR data quality is measured along five dimensions: (1) completeness (e.g., the amount of missing data), (2) correctness (e.g., a plausible distribution of values), (3) concordance (e.g., the use of terminology standards), (4) plausibility (e.g., out of range values), and (5) currency (e.g., the timeliness or recency of data). 3 None of these measures looks specifically at the quality of coding.
There are challenges when transforming the raw data into a dataset for analysis. 4 One such challenge is the need to define a cohort, often to ascertain the presence or absence of a disease or phenotype. Most often this is accomplished by using standard codes, particularly the International Classification of Diseases (ICD). Even as researchers have begun using other data (e.g., using free-text notes through natural language processing [NLP]) to augment phenotype definitions,5–7 the ICD codes remain a critical piece in defining cohorts for study.8–11 While we might expect codes to be used consistently over time and across different healthcare organizations and facilities, the degree of consistency has not been established.
The ICD codes are updated roughly every 10–20 years to reflect the changes in medical science and clinical practice. Each update usually introduces new codes, and it takes time for human coders and coding software to make necessary adjustments, discover problems, and develop solutions. The transition, on 1 October 2015, from coding using ICD, 9th Revision, Clinical Modification (ICD-9-CM) to ICD, 10th Revision, Clinical Modification (ICD-10-CM) in the United States, allowed us to conduct a formal investigation of coding quality in large EHR databases. The difficult transition in coding from using ICD-9-CM (with approximately 14,000 diagnostic codes) to using ICD-10-CM (with about 70,000 codes, providing for greater specificity in the codes) was eased somewhat by the General Equivalence Mapping (GEM) tables provided by the Centers for Disease Control and Prevention (CDC). Despite those efforts, however, marked changes in the frequency of certain conditions were observed by some researchers.12–15 Unfortunately, many researchers using EHR data appear unconcerned with the problems associated with the ICD transition, or with other coding quality challenges. Accordingly, we began a systematic investigation into the frequency and causes of problems associated with the ICD transition.
Methods
For our investigation, we used the US Veteran Affairs (VA) Corporate Data Warehouse, with 20 million patients and up to 20 years of follow-up, 4 billion outpatient visits, and 16 million inpatient visits (numbers as of August 2022). The US VA healthcare system has 1298 healthcare facilities, including 171 VA Medical Centers and 1113 outpatient sites.
GEM mapping
We identified clusters of codes made up from both versions of ICD that were suggested to be equivalent by the GEM. The GEM tables, consisting of one for mapping ICD-9-CM to ICD-10-CM and one for mapping ICD-10-CM to ICD-9-CM, were provided by the Center for Disease Control to assist in the transition from ICD-9-CM to ICD-10-CM. As the two tables are not symmetric, we ignored the mapping directions and merged the mapped pairs of codes to yield a single combined GEM table; it included four types of ICD-9-CM to ICD-10-CM code mappings: one-to-one, one-to-many, many-to-one, and many-to-many.
We constructed new diagnosis variables (clusters) with GEM mappings (version 2018) as building blocks. If the whole GEM table is a bipartite graph, in which nodes are codes and edges are correspondence relations between the two versions of codes, then the GEM mappings are exactly the connected components (as defined in graph theory). These mappings connected all the components of the two versions. The sets of codes in the fully connected subgraphs are then referred to as clusters. Each code is present in one, and only one, cluster; a total of 6707 clusters were identified.
The frequency of use of the ICD-9/10-CM codes within 1 year of the ICD 9/10 transition (between 1 October 2014 and 30 September 2016) was used to identify the number of patients with each diagnostic code. Limiting the codes for further study to those assigned to at least 0.1% of the observed population resulted in 700 clusters.
Baseline investigations
The frequency with which the clusters were used at each station of the VA on a weekly basis was obtained by summing the frequency of the codes in the cluster. In a similar manner computed the weekly frequency of use of each cluster was summed across the entire VA system.
The data was prepared for multivariate time-series analysis. The weekly visit counts of each cluster at each station were collected from 2001 to 2019, a total of 992 weeks and 2.6 billion visits. Candidate observables included: (1) total weekly inpatient and outpatient visit counts; (2) total patient counts by gender, race, and ethnicity; (3) top 500 weekly lab test counts grouped by analyte group; and (4) weekly drug fill counts grouped by VA drug class (based on pharmacological effects). Because the visit count has seasonal patterns, the number of the week in the year was also added. This resulted in a total of 860 observables to be used as predictors of the ICD code cluster.
To eliminate variation due to seasonal effects and trends, transformer-based deep learning (DL) and statistical (seasonal autoregressive integrated moving average [SARIMA]) models were built on the time-series data of ICD-9-CM usage from 1 January 2001 to 30 September 2015. The performance of the DL models was measured using the mean squared error.
DL model
A two-branch DL model was designed for time series forecasting. The first branch of the model processing temporal data was composed of a linear projection layer, a positional encoding layer, and two transformer blocks resembling those in the original transformer architecture for NLP. For a given week t, the prior N weekly data points Xt−N+1, …, Xt were used as the input data. Each data point Xt was a scaled vector containing both weekly visit count for a given cluster and covariate values. The linear projection layer then projected each data point to a vector of dimension dmodel. The positional encoding layer mapped the corresponding position of each data point in the sequence to a vector of dimension dmodel using sine and cosine functions. The positions were numbered as 1 through N starting from Xt and backward. The element-wise sum of the two sequences of vectors were then fed into the two transformer blocks.
The second branch, processing the input data containing the covariate information of week t + 1 was a feedforward neural network (FFNN) with residual connection. The outputs of the two branches were then combined by element-wise addition. The final output went through another FFNN with residual connection before reaching the output layer. The output layer had only one node with no nonlinear activation functions and output the predicted (re-scaled) visit count for the week t + 1. The loss function for training was the mean squared error function.
For each cluster, a model was trained separately. N = 12 weekly data points predicted the next future week visit count. The stochastic gradient descent with Nesterov momentum method was used as the optimizer. The mean square error of the predicted and the observed counts was calculated for model evaluation. The dropout method was used for regularization.
SARIMA model
Independent SARIMA models were fit to the weekly visit counts for each cluster. With the 860 variables prepared above, laboratory tests and medication fills were linearly combined using principal component (PC) analysis. We used the first 143 PC which explained 95% of the variability implied by the matrix of standardized counts arranged by date. The selected PC, the log count of patients, demographics, and an indicator for ICD-10 week, were later used as covariates during the SARIMA model development and selection phase within each of the GEM clusters. The indicator variable functions in each of the models as global step effect change on the log count due to the change of the ICD system use.
Several models for each of the 700 ICD clusters to describe the visit count trends were tested and only one chosen for the respective cluster, as we describe below.
The log count series associated with each ICD cluster was sequentially tested for stationarity using the Dickey–Fuller test at the 0.05 alpha level. In this way, we determine whether to model the original log time series, the first or the second differentiation. All models with no more than 15 moving average components (q) and no more than 15 autoregressive components (p) were fitted to the data, starting with a model with no seasonal effects. The model with lower Akaike information criteria with correction for overfitting (AICc) was selected to describe the time series. A similar process was repeated for models with seasonal effects of 52, 5, and 4 weeks, representing possible weekly, monthly, or yearly effects. When including a seasonal effect, the autoregressive (P) and moving average (Q) parameters for the seasonal part in the model need to be considered. Of all the models for each seasonal pattern with P < 2 and Q < 2, p < 10, and q < 10, the model used in further analysis was selected by using the partial autocorrelation plots, and the consistency of predictions depending on the time intervals between training data and outcome. Overfitting was avoided by fitting a similar SARIMA model but removing some PC predictors and checking that the coefficients of the remaining covariates did not change. The final models were checked for goodness of fit. We use the final SARIMA model of each cluster for one-step-ahead predictions with their respective 95% pointwise confidence intervals.
Applying the predictions of the models to the transition
The fitted models were then applied to the fiscal years 2016–2019 to predict the usage of the cluster-equivalent ICD-10-CM codes. We focused on the time period between 1 October 2015 and 30 September 2016 to detect significant changes at the time of the transition. We computed the difference between the predicted value and the actual value of the frequency of use. The standard deviation of the residuals in the training phase (1 January 2001–30 September 2015) provided a measure of the variability of the difference between the prediction and the actual values. We calculated the absolute value of the standardized residual (ASR) (the residual divided by the standard deviation) to measure the significance of changes. For each cluster mode, two sets of ASR values were computed: ASR1, the ASR of the first week after transition (4 October 2015) and ASR2, the averaged ASR of the year after transition. This enabled us to measure both the immediate abrupt change and year trend change. Aberrance was defined as an ASR value greater than 4.
Using each station as its own control, we calculated the rate of change of each cluster frequency at the time of the transition from ICD-9-CM to ICD-10-CM.
Human review
To examine the quality of the predictions and to better review the face validity of the clusters, we performed human review of the results from a randomly selected stratified set of clusters for each model. For each model, the 687 clusters were sorted using the ASR1. The sorted clusters were then placed in 10 equally sized bins. Five clusters and their associated ASR1 were selected at random from each bin for human review. This resulted in a total of 98 clusters (two clusters had been chosen at random from both the DL and SARIMA models).
The lowest ASR for the DL model was 0.28 and the highest 24.25. For the SARIMA model, the lowest was 0.09 and the highest 56.30. Two reviewers judged whether the cluster showed clear changes in frequency after the ICD transition. They also examined GEM-based mapping in the GEM to assess if problematic mappings were potential causes of the changes.
Results
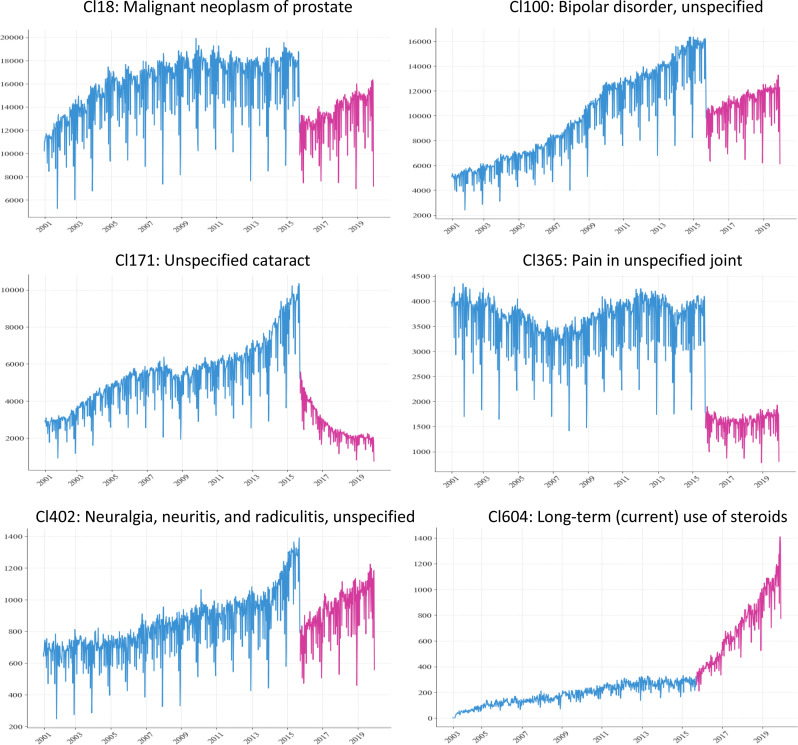
Dramatic code use changes were observed in many clusters after the ICD transition (Figure 1).
Figure 1.
Examples of weekly visit trend of diagnosis clusters.
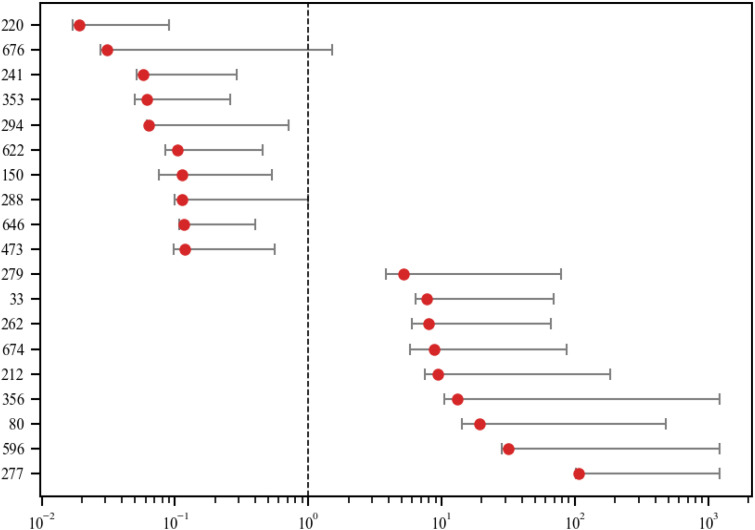
We noted wide variance between differing VA stations in the way the codes were used (Figure 2).
Figure 2.
Variation of code frequency rate changes among VA stations of top 20 most changed clusters. The whisker range covers 95% of all variations and excludes outliers.
Of the 700 clusters studied, 13 clusters were mapped with ICD-10-CM codes effective after 2017 and thus no data was available for the testing phase. With the remaining 687 clusters, 323 (47.0%) of the DL model and 480 (69.9%) of the SARIMA models exhibited a significant abrupt change with ASR1 exceeding 4.
Expanding the analysis to the year after transition, the median ASR2 of the 687 clusters were 9.22 and 3.94 for SARIMA and DL models, respectively, indicating the code usages of more than half of the clusters underwent dramatic shifts in the year after transition, and the aberrant signals were not just limited to 1 week. To confirm the findings, using human review of the 98 clusters for changes at the time of transition as the reference standard, we obtained area under the curve (AUC) values of 81.3% for the SARIMA and 93.6% for the DL models.
We further explored other potential causes of the dramatic changes by searching for evidence of alterations in clinical and coding practice, disease outbreaks, and introduction of new treatments or diagnostic approaches that might explain the changes which coincided with the ICD-9-CM to ICD-10-CM transition. Of the 98 clusters selected for manual review, only 5 clusters had a potential plausible cause induced from the changes in knowledge and practice. Another 23 of the clusters chosen for review were thought to be related to a different cause: problematic mappings in the GEM.
Discussion
Significance
We have observed a wide variation over time and space of ICD coding. This variation suggests that any observational study relying solely on ICD coding for diagnosis is untrustworthy.
Abrupt changes in coding associated with the ICD-9/10-CM transition have been reported in the literature.12,13 The remarkable finding in this investigation is the high prevalence of these changes in a large and diverse EHR dataset despite the attempts to reduce the size of those changes with the GEM mappings. Our analysis found that the use of the codes in the clusters formed from the GEM deviated significantly from what was predicted. A large proportion of the clusters showed a deviance of 4 standard deviations or more in their predicted use. The human review suggests that most data aberrancies are not attributable to the flaws in GEM. There is little evidence of other legitimate causes of abrupt changes such as outbreaks or new diagnostic practices.
The diversity in the patterns of the changes in code use between VA stations is telling. Bearing in mind that the prevalence of diseases may be variable depending on location, using each station as its own control, and looking at the differences in the rate of change allows the elimination of prevalence as a cause for the diversity. While this finding may not be surprising, the extent of diversity quantifies what may have been only suspected. The way that code use changed from one station to another at the time of transition dramatically illustrates the inconsistency in the manner of assignment of diagnostic codes.
While we cannot completely rule out all other causes, the most likely cause of the majority of aberrancy is a lack of what we call representational semantic (RS) integrity. The interpretation of a given clinical situation might be represented using separate sets of codes, a violation of RS integrity. Clinical factors contributing to RS integrity violations might include unfamiliarity with a new coding system, a change in coding practices, or inconsistent training in and application of coding rules. The coding system itself may be a contributing factor. Ambiguity in the definition of a code, or complex rules regarding when a code should or should not be used make coding systems more difficult to use.
RS integrity can be thought of as a generalization of the Cimino terminology desiderata of avoiding redundancy. 16 While we would hope that all comparable cases would be coded identically, such is not the case, as revealed by our analysis. In this generalization of the desiderata, the concern is not simply the terminology, but also the practical use of codes in the EHR data. The variance between VA stations in their use of codes is an example, one not limited to the transition. Another type of RS integrity violation was induced by the transition, where the changes in the coding systems could have led to confusion or misuse of the codes. Some of the ICD-9-CM and ICD-10-CM codes with the same exact description were used indisputably differently by coders. This finding adds to the concern about coded data, such as that noted by Glynn and Hoffman, 17 who observed that data in the Cerner Real World Database was collected and recorded differently in different places.
With the increasing acceptance of NLP, some may feel that structured data quality issue has become less important. While our team has been active in the development of NLP-based phenotypes, we believe that NLP complements structured data but cannot replace it. One example is the phenotype knowledgebase (PheKB) which contains dozens of phenotypes sourced from eMerge and other projects. About half of these phenotypes involved NLP while almost all of them involved ICD codes and other structured data. We also note that in some large databases (e.g., Cerner Real World Data, IBM Market Scan, and NIH N3C) only structured data is available, precluding the ability to use any NLP techniques.
Implications
The challenge posed by violations of RS integrity should not be taken lightly. Each year, hundreds, if not thousands, of scientific papers are published based on data containing ICD codes to define cohorts and phenotype variables.8–11 Take the VA EHR database for example: it is used by dozens of clinical and services studies each year; with the ICD codes frequently used in the studies.18–24 Phenotyping efforts sometimes employ sophisticated NLP and machine learning methods to improve the sensitivity and specificity by 5–30 percentage points.5–7 Some of the changes in ICD codes we observed in the study are far more dramatic.
Inconsistent coding will inevitably affect the downstream analysis. Ignoring a violation of RS integrity can propagate errors in data analysis during cohort creation, calculations of prevalence or incidence rates, identification of risk factors, and development of predictive models. For example, if two variables corresponding to two distinct ICD codes are included in a Cox survival analysis as independent variables where the two codes are assigned by different coders to the exact same condition, it will inevitably lead to wrong estimates of the hazard ratio and p-value for those variables.
The RS integrity challenge is not unique to the VA, as was observed in the papers noting the major changes in coding.12–15 In a prior study, on the large Cerner EHR dataset, we observed similar abrupt changes in ICD code frequencies. 25 In addition, these coding changes did not just occur during the ICD transition nor are they only associated with ICD codes. Similarly to ICD, Current Procedural Terminology (CPT) and Systematized Nomenclature of Medicine Clinical Terms (SNOMED) are evolving. 26 CPT is updated annually while SNOMED CT is updated every 6 months. RS integrity issues similar to those associated with ICD can be expected to occur in varying degrees in CPT and SNOMED CT, because coding variations using both vocabularies are not uncommon.27–30 As such, our findings and the methods we developed have broader implications.
A limitation of our work is that this study is based on data generated in the United States. The US model of health care is based on fee-for-service reimbursement where the diagnostic code may play some role in the amount of reimbursement. To minimize this effect, we used only VA data, which is based on a single payer. Another concern is that there is no data on who was responsible for assigning a code. In some situations, health information specially trained in coding is responsible; in others, it might be the caregiver or one of the caregiver team; in either case, the coder may or may not have access to a computer program suggesting possible codes. This aspect may play a role in leading to inconsistency.
Given our findings, we would like to advocate for all healthcare organizations and large clinical and claims data repositories that are actively collecting data to conduct regular examinations of RS integrity in their coding. We want to recommend researchers who utilize coded clinical and claims data to check for RS integrity. To facilitate the adaptation of this practice, we deposited our source code into a GitHub repository (https://github.com/GW-BIC/ICD-TimeSeries).
We chose to limit the assessment of aberrancy to the ICD transition period because of the high prevalence of changes during this phase. Given the high AUC achieved by the DL model when using human expert review as the reference, it may be feasible to automatically monitor for aberrant signals at a large scale using DL models. Unlike simple frequency-based monitoring, DL models can take account of seasonal changes and other contextual factors to reduce the potential for false alarms.
How should a researcher approach this problem of data quality? Undisputably, the law of large numbers helps. If most of the diagnostic codes are correct, the larger the cohort, the closer the estimates of the outcomes will be to a reliable finding. Additionally, Blois 31 pointed out that in most disease descriptions, the attributes are neither necessary nor sufficient to establish a diagnosis. However, there may very well be one or more attributes that are more “hard-edged,” e.g., measurements or observations, and thus less susceptible to misinterpretation. Using those attributes, perhaps in addition to the codes, will help define a cohort or phenotype more reliably.
Conclusions
The wide discrepancy between what might be expected to occur and what did occur with the changeover from ICD-9-CM to ICD-10-CM should cause alarm for those who are using historical EHR data in observational studies and for cohort identification. As we anticipate a change to ICD-11, our findings of what happened during the transition from ICD-9-CM to ICD-10-CM should arouse further caution. Without an established model of how the changes in standard terminologies should be applied to existing databases, persons attempting to perform studies over longer periods of time must determine for themselves how to avoid the RS integrity problems in building their cohorts.
This investigation suggests that coding inconsistency is a serious data quality issue in large EHR datasets and can be detected using machine learning and statistical methods. Because most researchers are unaware of the significance of RS integrity; few studies check the longitudinal use of structured data codes for consistency. We believe more efforts need to be invested to monitor and improve the quality of EHR data for health services and clinical research.
Acknowledgements
The authors are grateful for helpful discussions with James Cimino.
Footnotes
Contributorship: SJN and QZT designed and supervised the study. MST served as an unpaid advisor to the study. YY, PM, YJS, and ETR performed the data analysis, built the models, and performed statistical tests. SJN, PM, and JG participated in the manual review. All authors reviewed the data and analysis and contributed to the writing of the manuscript.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: Research using data from the VA was determined by the IRB to be de-identified and exempt.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and publication of this article. This work was supported by the US Veterans Administration Health Services Research Department [Grant No. 1I21HX003278-01A1] and by the Agency for Healthcare Research and Quality [Grant No. R01 HS28450-01A1].
Guarantor: SJN.
ORCID iDs: Stuart J. Nelson https://orcid.org/0000-0002-8756-0179
Ying Yin https://orcid.org/0000-0003-2156-4588
Phillip Ma https://orcid.org/0000-0002-8302-5021
References
- 1.Cowie MR, Blomster JI, Curtis LH, et al. Electronic health records to facilitate clinical research. Clin Res Cardiol 2017; 106: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn MG, Callahan TJ, Barnard J, et al. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. EGEMS (Wash DC) 2016; 4: 1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc 2013; 20: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastarache L, Brown JS, Cimino JJ, et al. Developing real-world evidence from real-world data: transforming raw data into analytical datasets. Learn Health Syst 2022; 6: e10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P, Garza M, Zozus M. Cancer phenotype development: a literature review. Stud Health Technol Inform 2019; 257: 468–472. [PubMed] [Google Scholar]
- 6.Sivakumar K, Nithya NS, Revathy O. Phenotype algorithm based big data analytics for cancer diagnoses. J Med Syst 2019; 43: 264. [DOI] [PubMed] [Google Scholar]
- 7.Liao KP, Ananthakrishnan AN, Kumar V, et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS One 2015; 10: e0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen LV, Thompson WK, Pacheco JA, et al. Design patterns for the development of electronic health record-driven phenotype extraction algorithms. J Biomed Inform 2014; 51: 280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao KP, Cai T, Savova GK, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. Br Med J 2015; 350: h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao KP, Sun J, Cai TA, et al. High-throughput multimodal automated phenotyping (MAP) with application to PheWAS. J Am Med Inform Assoc 2019; 26: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart CC, Lu CY, Yoon TK, et al. Impact of ICD-10-CM transition on mental health diagnoses recording . EGEMS (Wash DC) 2019; 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon J, Chow A. Comparing chronic condition rates using ICD-9 and ICD-10 in VA patients FY2014-2016. BMC Health Serv Res 2017; 17: 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Columbo JA, Kang R, Trooboff SW, et al. Validating publicly available crosswalks for translating ICD-9 to ICD-10 diagnosis codes for cardiovascular outcomes research. Circ Cardiovasc Qual Outcomes 2018; 11: e004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung KW, Richesson R, Smerek M, et al. Preparing for the ICD-10-CM transition: automated methods for translating ICD codes in clinical phenotype definitions. EGEMS (Wash DC) 2016; 4: 1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998; 37: 394–403. [PMC free article] [PubMed] [Google Scholar]
- 17.Glynn EF, Hoffman MA. Heterogeneity induced by EHR system implementation in a de-identified data resource from 100 non-affiliated organizations. J Am Med Inform Assoc Open 2019; 2: 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odden MC, Li Y, Graham LA, et al. Trends in blood pressure diagnosis, treatment, and control among VA nursing home residents, 2007–2018. J Am Geriatr Soc 2022; 70: 2280–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang RDL, Nguyen XM, Peloso GM, et al. Genome-wide and phenome-wide analysis of ideal cardiovascular health in the VA million veteran program. PLoS One 2022; 17: e0267900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swann AC, Graham DP, Wilkinson AV, et al. Suicide risk in a national VA sample: roles of psychiatric diagnosis, behavior regulation, substance use, and smoking. J Clin Psychiatry 2022; 83: 21m14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slobodnick A, Toprover M, Greenberg J, et al. Allopurinol use and type 2 diabetes incidence among patients with gout: a VA retrospective cohort study. Medicine (Baltimore) 2020; 99: e21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta S, Liu L, Patterson OV, et al. A framework for leveraging “big data” to advance epidemiology and improve quality: design of the VA colonoscopy collaborative. EGEMS (Wash DC) 2018; 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Badhwar V, Rankin JS, Thourani VH, et al. The Society of Thoracic Surgeons Adult Cardiac Surgery Database: 2018 update on research: outcomes analysis, quality improvement, and patient safety. Ann Thorac Surg 2018; 106: 8–13. [DOI] [PubMed] [Google Scholar]
- 24.Collett GA, Song K, Jaramillo CA, et al. Prevalence of central nervous system polypharmacy and associations with overdose and suicide-related behaviors in Iraq and Afghanistan war veterans in VA care 2010–2011. Drugs Real World Outcomes 2016; 3: 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miran SM, Nelson SJ, Redd D, et al. Using multivariate long short-term memory neural network to detect aberrant signals in health data for quality assurance. Int J Med Inform 2021; 147: 104368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui L, Zhu W, Tao S, et al. Mining non-lattice subgraphs for detecting missing hierarchical relations and concepts in SNOMED CT. J Am Med Inform Assoc 2017; 24: 788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deeken-Draisey A, Ritchie A, Yang G-Y, et al. Current procedural terminology coding for surgical pathology: a review and one academic center’s experience with pathologist-verified coding. Arch Pathol Lab Med 2018; 142: 1524–1532. [DOI] [PubMed] [Google Scholar]
- 28.Balla F, Garwe T, Motghare P, et al. Evaluating coding accuracy in general surgery Residents’ accreditation council for graduate medical education procedural case logs. J Surg Educ 2016; 73: e59–e63. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Lee JH, Ryu W, et al. Computer-based clinical coding activity analysis for neurosurgical terms. Yeungnam Univ J Med 2019; 36: 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liljeqvist HT, Muscatello D, Sara G, et al. Accuracy of automatic syndromic classification of coded emergency department diagnoses in identifying mental health-related presentations for public health surveillance. BMC Med Inform Decis Mak 2014; 14: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blois MS. Information and medicine: the nature of medical descriptions. Berkeley, CA, USA: University of Califonria Press, 1984. [Google Scholar]