Abstract
Spontaneous hydrolytic deamination of DNA cytosine and 5-methyl-cytosine residues is an abundant source of C/G (5-meC/G) to T/A transition mutations. As a result of this pressure, at least six different families of enzymes have evolved that initiate repair at U/G (T/G) mispairs, the relevant pre-mutagenic intermediates. The necessarily higher rate of the process at elevated temperatures must pose a correspondingly accentuated problem to contemporary thermophilic organisms and may have been a serious bottleneck in early evolution when life passed through a phase of very high ambient temperatures. Here we show that Thermus thermophilus, an aerobic, Gram-negative eubacterium thriving at up to 85°C, harbors two uracil-DNA glycosylases (UDGs), termed TTUDGA and TTUDGB. According to both amino acid sequence and enzymatic properties, TTUDGA clearly belongs to the family of ‘thermostable UDGs’. TTUDGB shares with TTUDGA 23% sequence identity, but differs from it in profound functional aspects. TTUDGB, unlike TTUDGA, does not act upon uracil residues in the context of single-stranded DNA whereas both enzymes process various double-stranded substrates, albeit with different preferences. TTUDGB shows a number of sequence features characteristic of the UDG superfamily, but surprisingly lacks any polar residue within its so-called motif 1 (GLAPG-X10-F). This finding is in conflict with a previously assumed crucial catalytic role of motif 1 in water activation and supports a more recently suggested alternative of a dissociative (‘SN1-type’) reaction mechanism. Together, the characteristics of TTUDGB and its homologs in other organisms define a novel family of UDG repair enzymes.
INTRODUCTION
Water, the major constituent of every metabollically active cell, is a genotoxic agent of considerable potency (1). Among a variety of spontaneous DNA hydrolysis reactions, deamination of cytosine and 5-methyl-cytosine residues to uracil and, respectively, thymine residues is genetically most significant due to the relatively high rate of the process and due to the coding properties of the hydrolysis products, which—unrepaired—direct the formation of C/G (5-meC/G) to T/A transition mutations (1). Not surprisingly, therefore, DNA repair systems counteracting this mutagenic threat are spread throughout the entire realm of life. To date, six classes of enzymes have been identified that initiate repair at DNA U/G (T/G) sites, the relevant pre-mutagenic intermediates. One class, Vsr (2), is a mismatch endonuclease that directly introduces a nick next to the damaged nucleotide, the other five classes comprise specialized DNA glycosylases, i.e. enzymes that remove the damaged base from DNA by hydrolyzing its glycosidic bond, leaving strand incision to a separate, subsequent step. One of these, Mig (3), is a member of the ‘helix–hairpin–helix’ (HhH) superfamily of DNA repair glycosylases (4) the other four classes all belong to the UDG superfamily of uracil-DNA glycosylases (UDGs) (5). In addition to Mig, the HhH superfamily is made up of families of enzymes specialized inter alia for the repair of the following lesions (substrate residue underlined): A/G and A/8-oxoG, MutY (6); 8-oxoG/C, Ogg1 (4); alkylated purines, AlkA (7); and 5,6-saturated pyrimidines, EndoIII (8). The four classes that make up the UDG superfamily are Ung (9), Mug/TDG (10,11), sMUG (12) and ‘thermostable UDG’ (13).
Spontaneous hydrolysis must accelerate with increasing temperature and thermophilic organisms can therefore be expected to counter the correspondingly more pronounced challenge to their genome integrity by any combination of the following measures: (i) minimizing genome size; (ii) protecting DNA from hydrolytic attack; (iii) increasing repair efficiency. With this rationale in mind, we have previously started to investigate the DNA repair status of thermophilic microorganisms and have identified in the archaeon Methanobacterium thermoautotrophicum the first member of the Mig family of T(U)/G glycosylases (3). Here we demonstrate the presence in Thermus thermophilus, a Gram-negative eubacterium, of two UDGs, TTUDGA and TTUDGB, the latter of which constitutes the prototype of a novel class of enzymes within the UDG superfamily.
MATERIALS AND METHODS
Bacterial strains and vectors
TOP10 One Shot™: F–, mcrA, Δ(mrr-hsdRMS-mcrBC), ϕ80-dlacZΔM15, ΔlacX74, recA1, deoR, araD139, Δ(ara-leu)7697, galU, galK, rpsL (Strr), endA1, nupG (Invitrogen, Groningen, The Netherlands). BL21(DE3) pLysS-derivative: Escherichia coli B, F–, ompT, hsd, SB (rB– mB–), gal, dcm, λ(DE3), pLysS (Cmr), (Novagen, Madison, WI). pCR-Blunt II-TOPO (Invitrogen). pET-21d (Novagen).
Enzymes
Pfu Turbo DNA polymerase was purchased from Stratagene (La Jolla, CA). Restriction enzymes and T4 DNA ligase were from New England Biolabs (Beverly, MA) and MBI Fermentas (Vilnius, Lithuania). Reagents were of analytical grade and supplied from Merck (Darmstadt, Germany) or Sigma (St Louis, MO).
Cloning of genes
The following 2′-deoxyoligonucleotide primer pairs were used to isolate the TTUDGA and the TTUDGB gene from genomic DNA of T.thermophilus HB27: A_UP, 5′-CCGCAAGCCCCTGCCATGGCCCTGGAACTG; A_LO, 5′-CGCGGGGGCTTACTCGAGGGGCTCCTGGC; B_UP, 5′-CGACAACATCCCCCTGCCATGGACAGGGAAG; B_LO, 5′-TGGACTACGAGGACCTCCTCTCCCGGCCGAAAGCCCGGCGAGGC. PCR products were cloned in pCR-Blunt II vector/‘TOP10 One Shot’ host bacteria. Relevant fragments were re-cloned in pET-21d vector. Cleavage sites for the re-cloning step were provided by the PCR primers as follows: A_UP and B_UP contain an NcoI restriction site, A_LO an XhoI and B_LO an EagI site. Correct DNA sequences of expression constructs were confirmed experimentally.
Production and purification of TTUDGA and TTUDGB
Escherichia coli BL21(DE3) pLysS, harboring pET-21d plasmid with the respective gene insert was grown in 1 l LB medium containing 50 µg/ml ampicillin and 34 µg/ml chloramphenicol at 37°C until an OD600 of 0.6. IPTG was added to a final concentration of 1 mM and the culture was incubated for an additional 3 h at 30°C. Cells were harvested by centrifugation, resuspended in 20 ml of 0.5 M NaCl, 25 mM HEPES–KOH, pH 7.6 and frozen at –70°C. Cell lysis was induced by rapidly thawing in a water bath of ∼30°C, followed by sonication. Crude lysates were clarified by centrifugation at 15 000 g at 4°C for 20 min and applied to a column filled with 3 ml of Chelating Sepharose™ Fast Flow (Amersham Pharmacia Biotech) loaded with Ni2+ (IMAC). TTUDGA and TTUDGB were eluted with 200–300 mM imidazole in 0.5 M NaCl, 25 mM HEPES–KOH, pH 7.6. Eluates were diluted 10-fold with cold 25 mM HEPES–KOH, pH 7.6 and loaded onto an HS cation exchange column (POROS 20, 4.6 × 100 mm). Proteins were eluted in a linear gradient of 0–1.5 M NaCl (BioCad™ Workstation, PerSeptive Biosystems). The main peaks, eluting between 500 and 600 mM NaCl, were pooled, desalted and concentrated 10 times by ultrafiltration (Millipore Centriprep cartridge, molecular weight cut-off ∼3000). The resulting solutions were diluted with an equal amount of glycerol and dithiothreitol was added to a final concentration of 1 mM. The enzymes were stored at –20°C. For both TTUDGA and TTUDGB, yields were in the range of 0.2–0.3 mg per batch with some variation between individual preparations.
Qualitative glycosylase assay
Substrates for qualitative glycosylase assays were prepared from the following 2′-deoxyoligonucleotides (‘[F]’ indicates a 5′-fluorescein moiety; target/mispaired residues underlined; syntheses by Metabion GmbH, München, Germany): SUB1_U, [F]-5′-GGGTACTTGGCTTATCTCCGAGGUCCTTAATCTGTCGCA; SUB1_T, [F]-5′-GGGTACTTGGCTTATCTCCGAGGTCCTTAATCTGTCGCA; SUB2_U, [F]-5′-GGGTACTTGGCTTATCTCCGCCUGGGTTAATCTGTCGCA; COMP1_G, 5′-TGCGACAGATTAAGGGCCTCGGAGATAAGCC; COMP2_A, 5′-TGCGACAGATTAACCCAGGCGGAGATAAGCC. U/G-containing duplex was prepared by hybridization of SUB1_U and COMP1_G as described earlier (3). Likewise, T/G-containing duplex was prepared from SUB1_T and COMP1_G, U/A-containing duplex from SUB2_U and COMP2_A. For assays of single-stranded DNA as substrate, SUB1_U was used. The standard reaction mixture consisted of 40 fmol of substrate and 10 pmol of the respective enzyme in 20 µl of reaction buffer [20 mM Tris–HCl pH 9.0, 20 mM (NH4)2SO4]. The reaction was carried out at 50°C for 30 min. A 2 µl aliquot of 1 M NaOH was added and the mixture kept at 95°C for 7 min. After cooling on ice, 10 µl of gel loading solution (95% formamide, 20 mM EDTA, 3 mg/ml dextran blue) was added. Aliquots of 10 µl were analyzed by gel electrophoresis (Pharmacia A.L.F. sequencer, 10% acrylamide/bisacrylamide gel, 7 M urea, 0.6× TBE running buffer, 25 W, 50°C). When assaying for glycosylase-associated AP lyase activity, the alkali/heat treatment step was omitted.
Multiple substrate kinetics
For multiple substrate kinetics, the following additional 2′-deoxyoligonucleotides were synthesized (Metabion GmbH): SUB3_U, [F]-5′-ACTTGGCTTATCTCCGCCUGGGTTAATCTGTCGCA; COMP2_C, 5′-TGCGACAGATTAACCCCGGCGGAGATAAGCC; COMP2_G, 5′-TGCGACAGATTAACCCGGGCGGAGATAAGCC. Fluorescently labeled substrate duplexes were prepared as follows: the U/G-containing duplex from SUB3_U and COMP2_G, the U/A-containing duplex from SUB2_U and COMP2_A, the U/C-containing duplex from SUB2_U and COMP2_C; the procedure as in the preceding paragraph. Note that SUB2_U is four nucleotide residues longer at its 5′-end than SUB3_U and thus creates correspondingly longer single-stranded protrusions with the U/C- and the U/A-containing duplexes as compared with the U/G-containing duplex. Multiple substrate kinetics were carried out pairwise and in triplicate (U/C and U/G together, then U/A and U/G) and the results plotted together with U/G as a common reference. Reaction mixtures were composed as follows: TTUDGA, 12 pmol of enzymes were mixed with 240 pmol of each substrate, consisting of 480 fmol labeled and 239.5 pmol unlabeled DNA in a total of 240 µl buffer (compare preceding paragraph); TTUDGB, 12 pmol of enzyme and 300 fmol of each labeled substrate in 240 µl buffer. The reactions were incubated at 50°C; aliquots of 20 µl were removed at fixed time points (compare Fig. 4) and worked up as described above. Peak areas were integrated using Fragment Manager V1.2 software (Pharmacia). With the exception of U/G cleavage by TTUDGB, SigmaPlot software was used to fit single exponential decay functions to the experimental data. From these, relative rate constants were derived as ratios of the respective exponents.
Figure 4.
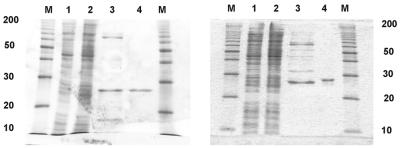
Analysis of enzyme purification by 15% SDS–PAGE; gels are stained with Coomassie brilliant blue. M, marker proteins (Gibco ‘10 kDa protein ladder’ as indicated). Lane 1, crude extract of E.coli cells harboring expression plasmid, before induction. Lane 2, crude extract of cells after induction with IPTG. Lane 3, pooled fractions of Ni2+-IMAC (Pharmacia) column (200–300 mM imidazole). Lane 4, pooled fractions of Poros HS cation exchange column (500–600 mM NaCl). (Left) TTUDGA. (Right) TTUDGB.
RESULTS
Bioinformatics
The genome of T.thermophilus (at >99.5% completion of a genome sequencing project presently pursued at Göttingen Genomics Laboratory) was scanned with representative sequences of all six classes of DNA-U processing enzymes mentioned in the Introduction using the TBLASTN algorithm (14). Only two genes yielded statistically significant hits; both were in the class of ‘thermostable UDGs’; search sequence was TMUDG of Thermotoga maritima, the first enzyme discovered in that class (13). Putative products of the two newly identified genes were termed TTUDGA and TTUDGB. At the level of derived amino acid sequence, TMUDG shares with TTUDGA 42% identity and 55% similarity, and with TTUDGB 29% identity and 55% similarity. The high degree of sequence identity between TTUDGA and TMUDG immediately suggests functional relatedness. TTUDGA and TTUDGB share 23% identity and 35% similarity; for sequence alignment see Figure 1.
Figure 1.
Multiple alignment of amino acid sequences of TTUDGA, TTUDGB and TMUDG of Thermotoga maritima (AAD35596) (13) using the CLUSTAL W algorithm (26). Vertical black bars indicate identity between all three sequences, gray bars either identity between two or similarity between all three sequences. Motif 1 (compare Fig. 2) is underscored. NCBI database accession number is given in parentheses.
A striking feature of the TTUDGB sequence is the absence of any polar amino acid residue in the so-called motif 1 (Fig. 2). Motif 1 is conserved throughout the entire UDG superfamily (5) and, as X-ray crystallographic studies revealed (15,16), makes up part of the active site of these enzymes. It invariably contains an amino acid residue with a side chain ending in a carboxylate or an amide function. This group is considered crucial for catalysis in that it positions and—in the case of aspartate or glutamate—activates by deprotonation a water molecule for nucleophilic attack on the glycosidic sugar center (17)—compare the Discussion. A corresponding conserved aspartate residue is also present in the HhH superfamily of DNA repair glycosylases (4).
Figure 2.
Comparison of motif 1 sequences. MUG (E.coli) (P43342), Ung (E.coli) (P12295), sMUG (Xenopus laevis) (AAD17300). (Left) MUG, Ung, sMUG and TTUDGA represent the four families of DNA repair glycosylases within the UDG superfamily mentioned in the Introduction. TTUDGB lacks the polar residue underlined. (Right) Selection of TTUDGB-like motifs 1 identified in public domain databases. NCBI database accession numbers are given in parentheses—other sources as indicated in the legend to Figure 3.
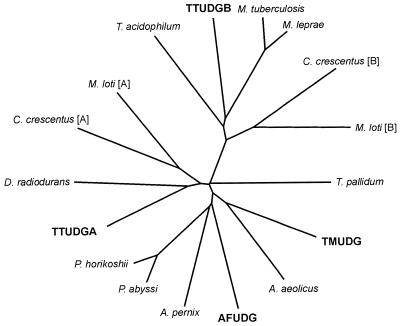
Further database searches revealed that TTUDGB (shown at the 12:00 h position in Fig. 3) branches off deeply from the lineage leading to TTUDGA and to TMUDG. Furthermore, it is part of a group of at least six proteins of phylogenetically distant microorganisms. Significantly, all proteins in that group lack the polar amino acid residue in motif 1 (Fig. 2), whereas it is present in all the others (alignments not shown).
Figure 3.
Tree diagram of sequence relatedness, derived for members of the family of ‘thermostable UDGs’ applying CLUSTAL (26) and TREEVIEW (27) programs (bold face lettering indicates biochemically verified enzymatic activity). Aquifex aeolicus (AAC07559), Archaeoglobus fulgidus (AFUDG) (NP_071102), Aeropyrum pernix (BAA79385), Caulobacter crescentus (AAK23314) [A] and (AAK23528) [B], Deinococcus radiodurans (AAF11304), Pyrococcus abyssi (CAB49606), Pyrococcus horikoshii (BAA30579), Mesorhizobium loti (NP_107967) [A] and (BAB54148) [B], Mycobacterium leprae (CAC31486), Mycobacterium tuberculosis (CAB00912), Thermoplasma acidophilum (CAC11619), Thermotoga maritima (TMUDG) (AAD35596), Treponema pallidum (AAC65215). TTUDGA and TTUDGB, this work.
This immediately ruled out the possibility of the peculiar motif 1 sequence of TTUDGB being the product of a fortuitous deactivating nonsense mutation and the entire reading frame thus being a pseudogene. It also made it clear that the simultaneous presence of both types of genes in some of the genomes listed does not mean that they are related by a recent gene duplication. TTUDGA and TTUDGB, rather, each define their own group of orthologous genes.
Production and purification of TTUDGA and TTUDGB
Even with functional significance of the TTUDGB motif 1 taken for granted, this function need not necessarily be hydrolytic removal of the uracil base from DNA—as long as this essential DNA repair step is performed by TTUDGA or some other still unknown enzyme(s). In order to approach this question biochemically, the TTUDGA and TTUDGB genes were cloned and expressed in E.coli (with six histidine codons added to the 3′-ends of each gene). Crude extracts were fractionated by Ni2+-IMAC chromatography, followed by re-chromatography of enriched fractions on a cation exchange matrix (for details refer to Materials and Methods).
As illustrated in Figure 4, this procedure resulted in essentially homogeneous products. Calculated relative molecular masses (including His6-tag) are 23.7 × 103 for TTUDGA and 25.2 × 103 for TTUDGB.
Qualitative assay of glycosylase activity
For a qualitative survey of enzymatic activities, various single-stranded and double-stranded oligonucleotide substrates were synthesized and incubated with TTUDGA and, respectively, TTUDGB as described in detail in the Materials and Methods section. The assay detects cleavage of substrate DNA by gel electrophoresis in a DNA sequencer (3,18), results are shown in Figure 5.
Figure 5.
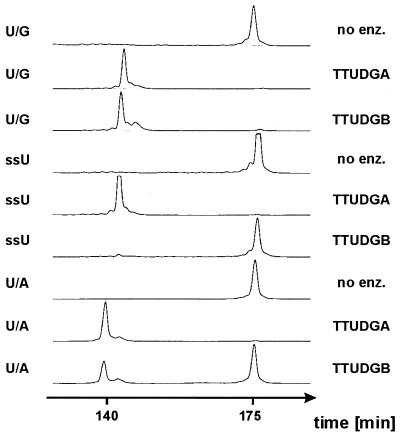
Qualitative assays of enzyme activity. Gel electrophoretic lane tracings of assay products (Pharmacia ‘A.L.F.’ DNA sequencer and ‘Fragment Manager’ software). Substrate DNAs (39mer) pass the laser beam after ∼175 min, products (23mer and 22mer) after ∼140 min. Sodium hydroxide treatment of reaction products was included throughout. For experimental details refer to Materials and Methods.
Clearly, both enzymes efficiently process U/G mispairs. U/A pairs were also substrates, but for TTUDGB reduced efficiency was noticeable even under the conditions of the merely qualitative assay used. The single-stranded substrate was efficiently processed by TTUDGA, whereas it was essentially refractory to the action of TTUDGB. The very small amount of product of the reaction of the single-stranded substrate with TTUDGB became gradually still less upon increasing the assay temperature, in steps of 10°C, from 50 to 90°C (data not shown). Therefore, we tentatively attribute this marginal activity to transient self-annealing of the substrate strand—possibly stabilized by binding to the protein—with formation of cleavable base/base oppositions. Both enzymes also processed U/C and U/T mispairs and neither acted on any opposition of regular DNA bases (data not shown). The enzymatic activity of TTUDGB in combination with the special features of motif 1 define the TTUDGB branch of the tree shown in Figure 3 as a distinct and novel class of enzymes within the UDG superfamily.
The major biochemical feature that sets TTUDGB apart from TTUDGA is its specificity for double-stranded substrates, which brings with it the option of dual substrate selectivity, U/G and T/G, as observed for Mig (3,19) and hTDG (11) glycosylases. Correspondingly constructed oligonucleotide duplexes containing an T/G mismatch, however, were not processed by TTUDGB, which thus resembles MUG glycosylase of E.coli (10), rather than Mig (3,19) and hTDG (11) enzymes (data not shown). For obvious reasons, hydrolytic attack on the glycosidic bond of T residues is a forbidden reaction for glycosylases that also process single-stranded substrates. A lack of activity towards T/G mispairs was therefore expected for TTUDGA and was verified experimentally (data not shown).
Up to this point, two-step cleavage assays had been applied throughout, i.e. incubation of substrate with enzyme was followed by treatment with sodium hydroxide. This second step drives strand cleavage at the base-free DNA site by β-elimination (3). Some DNA repair glycosylases are bifunctional in the sense that they can also catalyze the strand cleavage step (4); in these cases the sodium hydroxide treatment can be omitted from the assay. Corresponding one-step assays were performed with the unambiguous result that both TTUDGA and TTUDGB are strictly monofunctional glycosylases (data not shown).
Multiple substrate kinetics
DNA repair glycosylases are generally known to flip the base to be removed from the interior of the DNA double helix to its periphery and to bind it there in a tightly tailored active site pocket (5,17). Clearly, the flipping process itself requires activation energy which conceivably could even be rate-limiting. Rate comparison between different substrates that offer the DNA uracil residue in various forms that differ in the strength of binding of the base within the stack could shed light on this mechanistic problem. We have therefore tested both TTUDGA and TTUDGB in multiple substrate kinetics (19,20) by which three near-identical substrates were compared. These substrates differ from one another in only two respects: they contain three different base/base oppositions (U/G, U/C and U/A) and they have different lengths of the single-stranded protrusion that connects the 5′-fluorescein moiety to the body of the double-stranded part. It was verified separately that, within the limits relevant here, the length of this protrusion has no influence on reaction rates (data not shown).
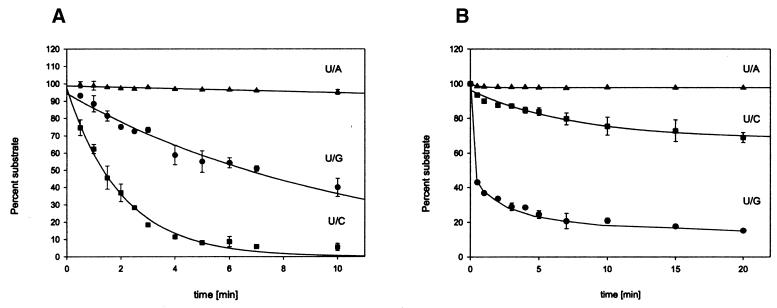
For TTUDGA, the data can be fitted smoothly to simple exponential decays with U/C being the most efficient substrate (Fig. 6). Setting U/G at reference value 1.0, U/C has a relative rate constant of 7.1, and U/A one of 0.05. From these data we derive the tentative hypothesis that base flipping might be rate limiting and the latter be governed by the strength of pairing the substrate uracil residue to the opposite base (nearest neighbors; i.e. stacking environment kept constant). This hypothesis can be tested by straightforward experiments outside the scope of the study presented here. Note that the much smaller rate of processing U/A, as compared with U/G, was completely missed in the qualitative assay (Fig. 5).
Figure 6.
Multiple substrate kinetics. Quantitative comparison of reaction rates of different U/X base/base oppositions. (A) TTUDGA, substrate:enzyme ratio, 40:1. (B) TTUDGB, substrate:enzyme ratio, 1:20. For experimental details refer to Materials and Methods.
A different picture emerges for TTUDGB. Here U/G, the mispair relevant to the presumed natural mutagenesis pathway, is by far the most efficient substrate; the time-course of the reaction, however, is biphasic; a brief period of very fast reaction is followed by levelling off before the reaction reaches completion. At present, this kinetic behavior precludes more precise, quantitative rankings. Still, the pronounced preference for U/G exhibited by TTUDGB seems to hint at a possible process by which the enzyme checks not only the presence of a uracil residue but also the structural nature of the base juxtaposed to it. Clearly, TTUDGA turns over (enzyme:substrate ratio was 1:40); TTUDGB, under conditions of the assay, does not (20-fold excess of enzyme).
DISCUSSION
In summary, both TTUDGA and TTUDGB qualify as tools that can help T.thermophilus counteract the genetic threat of hydrolytic deamination of DNA cytosine residues. The encoding of two such enzymes with similar, but in parts distinctly different catalytic properties in an otherwise minimalistic genome (1.8 Mb chromosome size) can be taken as an indication of an as yet ill understood division of different tasks. TTUDGA displays all the properties expected for a member of the TMUDG family (13,21,22). TTUDGB defines a novel family of UDGs, its most prominent structural feature being the lack of a polar amino acid residue that is otherwise conserved throughout the entire UDG superfamily.
A widely accepted catalytic role of this conserved residue (e.g. Asp145 in hUDG) is (partial) deprotonation of a water molecule, thereby activating it for nucleophilic attack on the glycosidic carbon center. This function is particularly important if one assumes a concerted (‘SN2-type’) mechanism of nucleophilic displacement of the base, with the latter being made a better leaving group by electron pull. This, for example, can be provided by a likewise conserved histidine residue (His268 in hUDG) that is within hydrogen bonding range to O-2 of the uracil ring in an enzyme–product complex (23).
Early indication that the carboxylate function of Asp145 of hUDG is not strictly required for catalysis came from crystallographic studies of a corresponding D145N mutant (23) that could be co-crystallized with a substrate oligonucleotide but had cleaved the uracil glycosidic bond by the time X-ray scattering data were taken (23). The nature of this residual catalytic activity was later followed up by X-ray crystallography (24) performed on a complex of hUDG with a substrate analog containing a non-hydrolyzable ‘C-glycosidic’ bond (i.e. a pseudouridine residue) and by energy calculations (25). Taken together, the result of these studies is that a dissociative (‘SN1-type’) reaction mechanism with breakage of the glycosidic bond preceding the entry of water at carbon center C-1′ is a viable alternative. The role of the enzyme would be to force upon the substrate residue a strongly distorted conformation with the result of destabilization of the glycosidic bond through steric strain and stereoelectronic antibinding forces (24) and/or to electrostatically stabilize the carbocation intermediate through nearby burying in the protein of four phosphodiester groups of the DNA substrate. The latter mechanism would amount to enzyme-mediated substrate autocatalysis (25).
If these latter effects are to claim the lion’s share of catalytic rate enhancement, one should expect to find fully active UDGs with no polar residue within a wild-type motif 1. TTUDGB is the first enzyme identified that meets these criteria. Conceivably, catalysis exerted by TTUDGB rests on substrate distortion and/or electrostatic substrate autocatalysis alone. Alternatively, amino acid residues outside the canonical motif 1 may substitute for the role of the lacking polar group. Clearly, these newly arising questions are amenable to experimental scrutiny.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Alessandro A. Sartori and Josef Jiricny for exchange of unpublished information. The financial support of Göttingen Genomics Laboratory by the state of Lower Saxony is gratefully acknowledged.
REFERENCES
- 1.Lindahl T. (1993) Instability and decay of the primary strucure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 2.Hennecke F., Kolmar,H., Bründl,K. and Fritz,H.-J. (1991) The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature, 353, 776–778. [DOI] [PubMed] [Google Scholar]
- 3.Horst J.-P. and Fritz,H.-J. (1996) Counteracting the mutagenic effect of hydrolytic deamination of DNA 5-methylcytosine residues at high temperature: DNA mismatch N-glycosylase Mig.Mth of the thermophilic archaeon Methanobacterium thermoautotrophicum THF. EMBO J., 15, 5459–5469. [PMC free article] [PubMed] [Google Scholar]
- 4.Nash H.M., Bruner,S.D., Schärer,O.D., Kawate,T., Addona,T.A., Spooner,E., Lane,W.S. and Verdine,G.L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol., 6, 968–980. [DOI] [PubMed] [Google Scholar]
- 5.Pearl L.H. (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res., 460, 165–181. [DOI] [PubMed] [Google Scholar]
- 6.Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,R.S. and Tainer,J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair superfamily. Nature Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- 7.Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) DNA bending and flip-out mechanism for base excision by the helix–hairpin–helix DNA glycosylase, Escherichia coli AlkA. EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) Novel DNA binding motifs in the DNA repair endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindahl T. (1974) An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl Acad. Sci. USA, 71, 3649–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallinari P. and Jiricny,J. (1996) A new class of uracil-DNA glycosylase related to human thymine-DNA glycosylase. Nature, 383, 735–738. [DOI] [PubMed] [Google Scholar]
- 11.Neddermann P., Gallinari,P., Lettieri,T., Schmid,D., Truong,O., Hsuan,J.J., Wiebauer,K. and Jiricny,J. (1996) Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem., 271, 12767–12774. [DOI] [PubMed] [Google Scholar]
- 12.Haushalter K.A., Stukenberg,P.T., Kirschner,M.W. and Verdine,G.L. (1999) Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitores. Curr. Biol., 9, 174–185. [DOI] [PubMed] [Google Scholar]
- 13.Sandigursky M. and Franklin,W.A. (1999) Thermostable uracil-DNA glycosylase from Thermotoga maritima, a member of a novel class of DNA repair enzymes. Curr. Biol., 9, 531–534. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden,T., Schaeffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped BLAST and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res., 27, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parikh S.S. Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett T.E., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed] [Google Scholar]
- 17.Schärer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- 18.Gläsner W., Merkl,R., Schmidt,S., Cech,D. and Fritz,H.-J. (1992) Fast quantitative assay of sequence-specific endonuclease-activity based on DNA sequencer technology. Biol. Chem. Hoppe-Seyler, 373, 1223–1225. [DOI] [PubMed] [Google Scholar]
- 19.Fondufe-Mittendorf Y.N., Härer,C., Kramer,W. and Fritz,H.-J. (2002) Two amino acid replacements change the substrate preference of DNA mismatch glycosylase Mig.MthI from T/G to A/G. Nucleic Acids Res., 30, 614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gläsner W., Merkl,R., Schellenberger,V. and Fritz,H.-J. (1995) Substrate preferences of Vsr DNA mismatch endonuclease and their consequences for the evolution of the Escherichia coli K-12 genome. J. Mol. Biol., 245, 1–7. [DOI] [PubMed] [Google Scholar]
- 21.Sandigursky M. and Franklin,W.A. (2000) Uracil-DNA glycosylase in the extreme thermophile Archeoglobus fulgidus. J. Biol. Chem., 275, 19146–19149. [DOI] [PubMed] [Google Scholar]
- 22.Sartori A.A., Schär,P., Fitz-Gibbons,S., Miller,J.H. and Jiricny,J. (2001) Biochemical characterization of uracil processing activities in the hyperthermophilic Archaeon Pyrobaculum aerophilum. J. Biol. Chem., 276, 29979–29986. [DOI] [PubMed] [Google Scholar]
- 23.Slupphaug G., Mol,C.D., Kavli,B., Arvai,A.S., Krokan,H.E. and Tainer,J.A. (1996) A nucleotide-flipping mechanism from the strucure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- 24.Parikh S.S., Walcher,G., Jones,G.D., Slupphaug,G., Krokan,H.E., Blackburn,G.M. and Tainer,J.A. (2000) Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl Acad. Sci. USA, 97, 5083–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinner A.R., Blackburn,G.M. and Karplus,M. (2001) Uracil-DNA glycosylase acts by substrate autocatalysis. Nature, 413, 752–755. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page R.D.M. (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci., 12, 357–358. [DOI] [PubMed] [Google Scholar]