Abstract
Background
H9N2 avian influenza viruses have been circulating in Bangladesh since 2006, affecting multiple avian species and resulting in economic losses. The recent emergence of tribasic strains, along with co-infections, has increased the risk to poultry health. Therefore, the study aimed to compare the immune responses of Sonali (crossbred) and commercial broiler chickens infected with tribasic H9N2 low pathogenic avian influenza (LPAI) virus.
Methods
Following H9N2 infection, proinflammatory (IL-6, IL-8, IL-1β and TNF-α) and antiviral (IFN-β and IFN-γ) cytokine expressions were observed in the trachea, lungs, intestine, and lymphoid tissues in Sonali and broiler chickens from 1 day post infection (dpi) to 10 dpi by qPCR.
Results
Sonali chickens exhibited significantly higher proinflammatory and antiviral cytokine expressions in the trachea at 3–7 days post infection (dpi), while broiler chickens showed lower immune responses. Broiler chickens displayed prolonged IL-6, IL-8, and IL-1β expression in lungs at 3–10 dpi compared to Sonali chickens. In the intestine, broiler chickens showed higher IL-6 and IL-8 expression that peaks at 1–3 dpi, while in Sonali chickens only IL-1β elevated at 10 dpi. In response to the H9N2 viruses, broiler chickens exhibited a stronger early IFN-β responses and a delayed IFN-γ responses in their lymphoid organs compared to Sonali chickens.
Conclusion
This suggests distinct immune profiles between the chicken types in response to the H9N2 infection. The information sheds light on the function of innate immunity in the pathophysiology of currently circulating tribasic H9N2 virus and could assist in effective controlling of avian influenza virus spread in poultry and designing vaccines.
Keywords: Sonali chickens, broilers, tribasic H9N2, proinflammatory, antiviral cytokines
Background
H9N2 viruses of the G1 lineage have been frequently isolated from backyard and commercial poultry across Bangladesh since 2006 [1–3]. The avian influenza virus (AIV) can cause influenza in chickens, showing a variety of clinical signs. These range from no symptoms or mild respiratory conditions to severe and potentially fatal disease [4, 5]. High mortality rates are typically the result of highly pathogenic AIV (HPAIV) infection, which can spread to various tissues and organs in wide range of avian species [6]. However, no clinical signs or mild respiratory distress are typically the outcome of infection with low pathogenic AIV (LPAIV). These H9N2 viruses remains a threat to the worldwide poultry industry including Bangladesh [4, 5], since co-infection with other respiratory pathogens like Newcastle Disease Virus (NDV), Infectious Bronchitis Virus (IBV), E. coli, Mycoplasma are very common in field. AIV subtypes is influenced by hemagglutinin endoproteolytic cleavage site (HACS) to produce disease in avian hosts. Reportedly, mono and di-basic (PAKSSR*GLF) cleavage site motifs of H9N2 viruses cleaved by trypsin-like proteases that are produced by respiratory and gastrointestinal cells, is common in Bangladesh [7]. However, recent field outbreak showing evidence of a tri-basic (PAKSKR*GLF) HA cleavage site motif with increasing frequency [8]. The tribasic H9N2 viruses increased the viral replication, stability, pathogenicity and transmission in chickens compared to the monobasic H9N2 viruses [9]. Besides, an in ovo pathogenesis study describes the replication of tribasic H9N2 viruses in remarkably wide range of embryonic organs [10]. This H9N2 virus has become important due to its role as a genetic donor in the reassortment of zoonotic AIV, including subtypes H5, H7, and H10 [11] that posing a greater risk for zoonotic transmission and potential pandemic spread.
The interaction between the virus and its host is primarily influenced by the pathogen’s virulence factors and the host’s immune response [12]. The role of host immunity, notably innate immunity, plays a significant role in the pathogenesis of influenza viruses [13]. AIV infection commonly leads to an imbalance in cytokine levels, often termed a “cytokine storm,” marked by increased levels of proinflammatory cytokines and an interferon (IFN) reaction [4, 5]. One of the most important markers of the pathogenicity of the influenza virus is elevated proinflammatory cytokine expression in the respiratory tract [14]. Generally, respiratory and digestive tracts were the main sites of H9N2 virus replication in specific pathogen-free (SPF) chickens, and commercial poultry under experimental condition [4, 5]. According to our previous study, H9N2 LPAI virus infected Sonali and broiler chickens develop mild tracheitis, pneumonia, and enteritis [15]. Although replication of H9N2 LPAIVs is limited to trypsin-expressing epithelial cells lining the respiratory and gastrointestinal tracts, mononuclear cells can also be infected [16–18].
Innate immunity draws immune cells to the site of infection by producing cytokines. The most evident immune cells are alveolar macrophages, which commonly reside on the respiratory tract’s mucosal surface and induce inflammation [19]. Following infection with influenza viruses, there is a prominent surge in cytokine production, including interferon-alpha (IFN-α), interferon-gamma (IFN-γ), tumor necrosis factor-alpha (TNF-α), interleukin-6 (IL-6), interleukin-8 (IL-8), interleukin-10 (IL-10), interleukin-1β (IL-1β), and interleukin-12 (IL-12) across different organs. However, the levels of cytokine induction appear to vary depending on the specific strain of the virus [18, 20–23] and the magnitude of viral replication in affected organs [24].
Prior research has shown that cytokine responses play a role in the immune reaction to H9N2 infection across various avian species [25–27]. Reportedly, there is modulation of various cytokines, including IFN-γ, IFN-α, IL-1β, IL-4, IL-6, IL-8, IL-10, and IL-17, detected in chickens across different organs such as lungs, spleen, and cecal tonsils [18, 20, 25, 27, 28]. In addition, some research has verified that Japanese quails [25] as well as mallards [26] express IFN-α, IL-1β, IL-6, RIG-I and Mx against avian influenza infection. It has been reported that the pathogenicity of influenza viruses in the host is influenced by the levels of proinflammatory cytokines (IL-1β and IL-6) and antiviral cytokines (IFN-γ) [29–31]. Proinflammatory cytokines like interleukin and TNF work to worsen disease, while antiviral cytokines like interferon combat viruses by inhibiting their replication in cells [32, 33].
Several research performed real-time RT-PCR or microarray methodologies to gain insights into the interaction between H9N2 and avian cells [4, 5]. Our previous study focused on the gross and microscopic changes, viral distribution in tissues, and viral shedding of two low pathogenic avian influenza (LPAI) H9N2 subtype viruses in Sonali (Rhode Island Red cock x Fayoum hen crossbreed) and Cobb 500 broiler chickens [15] that revealed mild pathological changes including tracheitis, pneumonia and enteritis without showing any clinical signs.
The aim of this study was to evaluate the expression of proinflammatory and antiviral cytokines in Sonali and broiler chickens after infection with the H9N2 avian influenza virus. This is the first comparison of cytokine expressions in response to tribasic H9N2 in commercial broiler (Cobb 500) chickens and local crossbred Sonali chickens. Our investigation will deepen our understanding of the aspects of pathogenesis and host specific immunity by observing quantitative measurement of cytokine levels in different organs at different time points in Sonali and broiler chickens.
Methods
Virus
For the cytokine expression, A/chicken/Bangladesh/2458-LT2/2020 (LT_2) field H9N2 LPAI isolate was recovered from the virus repository of the Department of Pathology, Faculty of Veterinary Science, Bangladesh Agricultural University. The selected isolate A/chicken/Bangladesh/2458-LT2/2020 (LT_2) was propagated and isolated in 10 days old embryonated chicken eggs [15] and then harvested allantoic fluids were collected and stored at -80 °C until use.
Experimental design and sampling
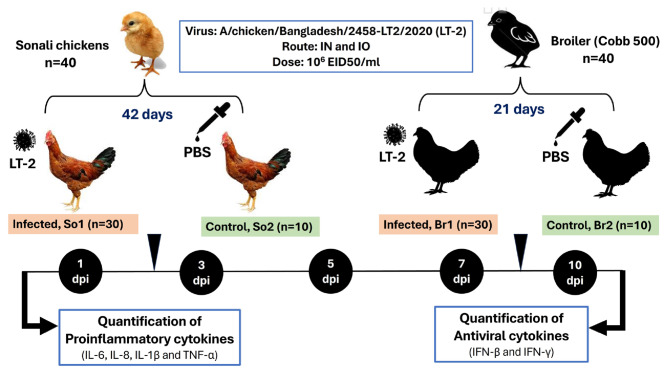
A total of 80 day old chicks consisting of 40 Sonali and 40 broiler chicks, were obtained from a leading and reliable commercial company of Bangladesh and were raised in separate enclosures, providing them with ad libitum access to food and water. At 21 days (broiler chickens) and 42 days (Sonali chickens), birds that tested negative for H9N2 LPAI antibodies were divided into four groups of birds: LT_2 infected Sonali (LT_2-So1) (n = 30), and the control group (So2) (n = 10); similarly, broilers were distributed to LT_2 infected broiler (LT_2-Br1) (n = 30) and the control group (Br2) (n = 10). The different groups were housed separately. Chicks in the infected groups (LT_2-So1 and LT_2-Br1) were intranasally and intraocularly administered 200 µl of 106 EID50/ml of the selected isolate. Meanwhile, control groups (So2 and Br2) received 200 µl of PBS using the same routes. The birds were monitored daily for any clinical signs of infection. Subsequently, six birds from each infected group and two from each control group were euthanized and necropsied at 1, 3, 5, 7, and 10 days post-infection (dpi). All experimental birds were humanely euthanized by cervical dislocation performed by a veterinarian. Tissue samples, including the trachea, lungs, intestine and lymphoid organs (pooled thymus, spleen and bursa) were collected for cytokine expression analysis (Fig. 1).
Fig. 1.
In vivo experimental design of tribasic H9N2 infection in Sonali and Cobb 500 broiler chickens using A/chicken/Bangladesh/2458-LT2/2020 (LT_2) field isolate for measurement of cytokine expression by qPCR
RNA extraction and quantification
Tissue samples were ground and suspensions were prepared at a 20% w/v ratio using PBS supplemented with gentamicin (500 µg/ml) with the help of Tissue Lyser instrument (Qiagen, Hilden, Germany). RNA was extracted from the tissue suspensions using a QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. Then the RNA was quantified by Nanodrop One (Thermo Scientific, Waltham, MA, USA).
cDNA synthesis
Extracted RNA were used for reverse transcriptase using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific™, USA). Reaction mixtures contained 4 µl of Reaction Buffer, RiboLock RNase Inhibitor (20 U/µl) 1 µl, 10 mM dNTP Mix 2 µl, Revert Aid M-MuLV RT (200 U/µl) 1 µl, Oligo(dT)18 Primer 1 µl and nuclease-free water to make 20 µl, incubated in a thermal cycler for 60 min at 42 °C and 5 min at 70 °C with the reaction component.
Quantification of proinflammatory and antiviral cytokines
The synthesized cDNA was diluted (1:10) and subjected to qPCR for the measurement of relative cytokine expression of TNF-α, IL-6, IL-8, IL-1β, IFN-β, IFN-γ and GAPDH for samples from both control and infected birds as changes in SYBR green fluorescence after amplification with specific primer sets. GAPDH was used as housekeeping gene. Details of primers are listed in Table 1. Reactions were done in duplicate on 10 µl reaction mixtures containing 100 ng of cDNA (1 µl), 5 pmol of each primer (1 µl), Luna Universal qPCR Master Mix (5 µl) and nuclease free water (2 µl) using Luna® Universal qPCR Master Mix (New England Biolabs, Inc.). Amplification and detection of specific cytokines were performed using the qPCR with the following cycle profile: one cycle at 95˚C for 2 min, 40 cycles at 95˚C for 15 s and 72˚C for 1 min. One cycle for the dissociation curve for all reactions was added and the melting curve was analyzed.
Table 1.
List of primers used in quantification of proinflammatory and antiviral cytokines in this study
| Gene | Name | Cytokine nature | Sequences (5ʹ-3ʹ) | References |
|---|---|---|---|---|
| IL-6 | IL6-F | Proinflammatory | 545-ATCCGGCAGATGGTGATAAA | |
| IL6-R | 707-CCCTCACGGTCTTCTCCATA | |||
| IL-8 | IL8-F | 459-CATCATGAAGCATTCCATCT | Nang et al., 2011 | |
| IL8-R | 663-CTTCCA AGGGATCTTCATTT | |||
| IL-1β | IL1β-F | 134-GGATTCTGAGCACACCACAGT | ||
| IL1β-R | 405-CTGGTTGATGTCGAAGATGTC | |||
| TNF-α | TNFα-F | 57-CTTCTGAGGCATTTGGAAGC | ||
| TNFα-R | 407-ACTGGGCGGTCATAGAACAG | |||
| IFN-β | IFNβ-F | Antiviral | 42-GCTCACCTCAGCATCAACAA | |
| IFNβ-R | 607-GGGTGTTGAGACGTTTGGAT | Nang et al., 2011 | ||
| IFN-γ | IFNγ-F | 320-TGAGCCAGATTGTTTCGATG | ||
| IFNγ-R | 471-CTTGGCCAGGTCCATGATA |
Statistical analysis
Fold change was determined by the -ΔCt method using GAPDH as the reference gene to normalize the level of target gene expression to uninfected control birds of each breed. Data were analyzed by two-way ANOVA followed by the Bonferroni post hoc test (GraphPad Prism 8, USA). The standard error mean was calculated using the fold change values of three replicates for each gene measured. Comparisons were considered significant to control at P ≤ 0.05.
Results
Expression of cytokines in respiratory and gastrointestinal tract
Trachea
Proinflammatory cytokines
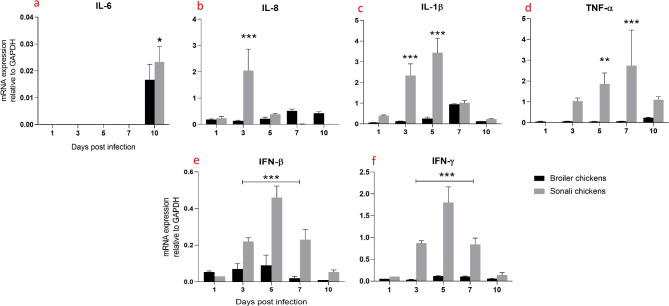
In the trachea, IL-6 expression was significantly increased (*** p ≤ 0.001), (** p ≤ 0.01) and (*p ≤ 0.05) at 10 dpi in Sonali chickens compared to broiler chickens (Fig. 2a). Whereas, a significant increase in IL-8 expression in the Sonali chicken group at 3 dpi compared to broiler. However, IL-8 expression was returned to lower level for Sonali chickens, at 5, 7, and 10 dpi; whereas, it remained at the minimum level at all the time points tested for broiler chickens (Fig. 2b). Sonali chickens demonstrated a notable increase in IL-1β expression at 3 and 5 dpi, suggesting a robust inflammatory reaction to the infection during these days. In contrast, broiler chickens during the same time period (3 dpi and 5 dpi) displayed lower levels of IL-1β expression (Fig. 2c). Sonali chickens showed a significant increase in TNF-α expression at 5 and 7 dpi, whereas, broiler chickens showed consistently low TNF-α expression levels throughout the infection period, suggesting a much weaker inflammatory response in trachea (Fig. 2d).
Fig. 2.
Expression of proinflammatory and antiviral cytokines mRNA expression in trachea of Sonali and broiler chickens, infected with H9N2 LPAI virus. Bar diagram showing relative expression of IL-6 (a), IL-8 (b), IL-1β (c), TNF-α (d), IFN-β (e) and IFN-γ (f) in the trachea of two type of chickens. Error bars represent mean ± SEM. Two-way ANOVA with Bonferroni’s multiple comparison test, *** p ≤ 0.001, ** p ≤ 0.01 and *p ≤ 0.05. Non-significant differences are not indicated
Antiviral cytokines
The antiviral cytokines, both IFN-β and IFN-γ, were expressed at significantly higher levels in the trachea of Sonali chickens at 3, 5, and 7 dpi whereas, broiler chickens exhibited consistently low expression for both cytokines at different days of post infection (Fig. 2e-f).
Lungs
Proinflammatory cytokines
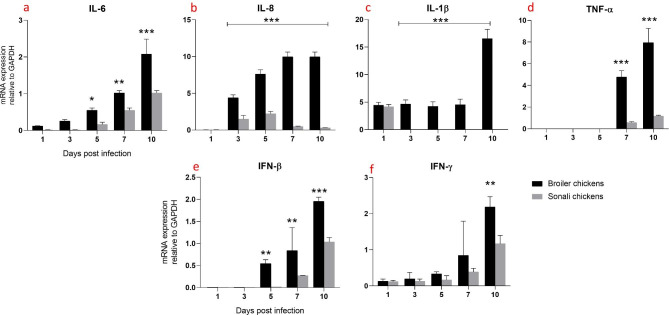
Between 5 and 10 dpi, there was a significant increase in the expression of IL-6 in the lungs of broiler chickens. However, Sonali chickens showed a higher level of IL-6 expression over time (not significantly) (Fig. 3a). In broiler chickens, both IL-8 and IL-1β exhibited comparable patterns and significantly increased expression from 3 dpi to 10 dpi. In contrast, Sonali chickens exhibited slightly lower levels of IL-8 expression over the same periods (Fig. 3b), and IL-1β expression was only observed at 1 dpi (Fig. 3c). Additionally, another proinflammatory cytokine TNF-α was expressed at significantly higher levels in broiler chickens compared to Sonali chickens at 7 and 10 dpi (Fig. 3d).
Fig. 3.
Expression of proinflammatory and antiviral cytokines mRNA expression in lungs of Sonali and broiler chickens, infected with H9N2 LPAI. Bar diagram showing relative expression of IL-6 (a), IL-8 (b), IL-1β (c), TNF-α (d), IFN-β (e) and IFN-γ (f) in lungs of two type of chickens. Error bars represent mean ± SEM. Two-way ANOVA with Bonferroni’s multiple comparison test, *** p ≤ 0.001, ** p ≤ 0.01 and *p ≤ 0.05. Non-significant differences are not indicated
Antiviral cytokines
In both groups of birds, the expression of antiviral cytokines (IFN-β and IFN-γ) in the lungs increased over time. However, in broiler chickens, the levels of both IFN-β and IFN-γ were significantly higher compared to Sonali chickens. To be more precise, IFN-β significantly increased at 5 dpi and IFN-γ increased at 10 dpi (Fig. 3e-f).
Intestine
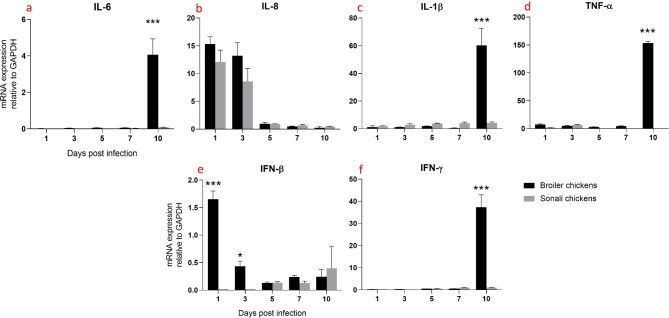
Proinflammatory cytokines
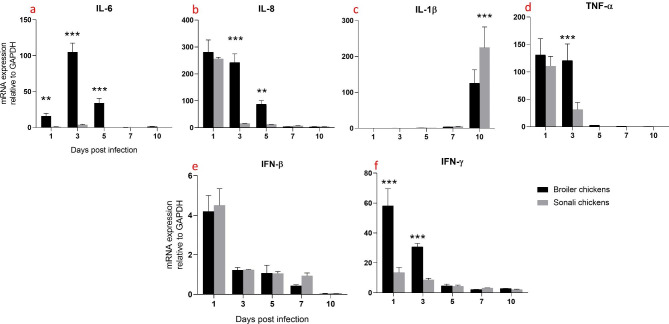
In the intestine of broiler chickens, IL-6 and IL-8 expression levels were significantly higher from 1 dpi to 5 dpi compared to Sonali chickens. IL-6 peaked at 3 dpi in broilers (Fig. 4a), whereas IL-8 peaked at 1 dpi in both broilers and Sonali chickens which decreased over time (Fig. 4b). In contrast, IL-1β expression significantly increased in Sonali chickens at 10 dpi (Fig. 4c). TNF-α expression in both varieties of chickens had a similar pattern, peaking at 1 dpi and then gradually decreasing; however, at 3 dpi, broiler chickens had substantially higher TNF-α expression than Sonali chickens. (Fig. 4d).
Fig. 4.
Expression of proinflammatory and antiviral cytokines mRNA expression in intestines of Sonali and broiler chickens, infected with H9N2 LPAI. Bar diagram showing relative expression of IL-6 (a), IL-8 (b), IL-1β (c), TNF-α (d), IFN-β (e) and IFN-γ (f) in intestine of two type of chickens. Error bars represent mean ± SEM. Two-way ANOVA with Bonferroni’s multiple comparison test, *** p ≤ 0.001, ** p ≤ 0.01 and *p ≤ 0.05. Non-significant differences are not indicated
Antiviral cytokines
Broiler chickens exhibited significantly higher IFN-γ mRNA expression in the intestine compared to Sonali chickens at 1 dpi to 3 dpi. Both chicken types showed low and similar levels of IFN-γ expression with no significant differences between the groups at 5, 7 and 10 dpi (Fig. 4f). In case of IFN-β, both broiler and Sonali chickens exhibited high levels of expression; however, the difference between two groups of birds was not statistically significant. At 3, 5, 7, and 10 dpi, the levels of IFN-β expression decreased gradually over time, with no significant differences between the two types of chickens (Fig. 4e).
Expression of cytokine in lymphoid tissues (pooled thymus, spleen and bursa)
Proinflammatory cytokines
IL-6, IL-1β and TNF-α, all exhibited identical expression patterns and showed higher expression at later times. In broiler chickens, their expression level significantly increased at 10 dpi compared to Sonali chickens (Fig. 5a, c and d). But in both bird groups, IL-8 expressed at high levels at 1 and 3 dpi, with broiler chickens expressing it at a slightly higher level. There were no appreciable variations in the expression levels between the two varieties of chickens at 3, 5, 7, and 10 dpi during these days (Fig. 5b).
Fig. 5.
Expression of proinflammatory and antiviral cytokines mRNA expression in lymphoid tissues of Sonali and broiler chickens, infected with H9N2 LPAI. Bar diagram showing relative expression of IL-6 (a), IL-8 (b), IL-1β (c), TNF-α (d), IFN-β (e) and IFN-γ (f) in lymphoid tissues of two type of chickens. Error bars represent mean ± SEM. Two-way ANOVA with Bonferroni’s multiple comparison test, *** p ≤ 0.001, ** p ≤ 0.01 and *p ≤ 0.05. Non-significant differences are not indicated
Antiviral cytokines
For antiviral cytokines, broiler chickens showed a significant increase in IFN-β mRNA expression at 1 dpi. At 3 dpi, there was a slight increase in IFN-β mRNA expression in broiler chickens. From day 5 to day 10 post-infection, both broiler and Sonali chickens showed low and similar levels of IFN-β mRNA expression (Fig. 5e). Both broiler and Sonali chickens exhibited similar patterns with low levels of IFN-γ mRNA expression at 1, 3, 5 and 7 dpi. However, broiler chickens showed a significant increase in IFN-γ mRNA expression at 10 dpi (Fig. 5f). This suggests that broiler chickens had a stronger early IFN-β response and a delayed but strong IFN-γ response to H9N2 infection compared to Sonali chickens.
Discussion
Sonali and broiler chickens are two different breeds of chickens with distinct physiological characteristics and genetic backgrounds. Sonali chickens, a crossbred recognized for their resilience and adaptability [34], whereas, broiler chickens are mainly for fast growth and high meat purposes [35]. Previous studies have suggested that genetic differences among chicken breeds can influence their immune responses to viral infections, including avian influenza viruses [36]. However, no studies have been conducted on the immune responses to tribasic H9N2 LPAI to date. In this study, we compared the host immune responses of Sonali and broiler chickens from 1 dpi to 10 dpi after infection with tribasic H9N2 LPAI viruses, where we measured the expression of proinflammatory (TNF-α, IL-6, IL-8, and IL-1β) and antiviral (IFN-β and IFN-γ) cytokines relative to GAPDH. In this study, Broiler and Sonali chickens were infected with tribasic H9N2 virus at 21 and 42 days, respectively. Here, chicken breeds of two different ages were used, assuming that 2–3 weeks differences had no or very minimal effect on immune responses against viral diseases of poultry. However, some studies suggest that older animals exhibit lower immune responses [37, 38], though others found no significant age-related differences in cytokine (IFN-α and IL-2) expression in ducks [39]. Furthermore, in our earlier investigation, infection of Broiler and Sonali chickens at different ages (21 days and 42 days respectively) had no significant differences on viral loads and pathogenicity [15].
Reportedly, proinflammatory cytokines have a particular affinity for the upper respiratory tract and lymphoid organs [40, 41]. Viral replication, tissue damage, and fever are all intimately correlated with their expression levels [42]. In our study, Sonali chickens exhibited stronger proinflammatory responses in trachea compared to broiler chickens at 3–7 dpi. This was consistent with higher levels of viral load observed during the same period, as reported in our previous study [15]. Both groups of chickens showed a continuous increase in the expression of proinflammatory cytokines in their lungs. However, broiler chickens showed higher viral loads with significantly increased cytokine expression compared to Sonali chickens [15]. Other studies have shown that the inductions of IL-1β, IL-6, IFN-α, IFN-β, and IFN-γ increase in the lungs and trachea (both in vivo and in vitro) of H9N2-infected chickens, which aligns with our findings [28, 41, 43, 44].
In contrast, in the intestine, the expression of IL-6, IL-8, TNF-α, IFN-β, and IFN-γ decreased over time in both chicken hosts, while IL-1β showed a delayed expression. Broilers had significantly higher IL-6 (at 1–5 dpi), IL-8 (at 3–5 dpi), TNF-α (at 3 dpi) and IFN-γ (from 1 to 3 dpi) expression levels in the intestine. However, viral load in the intestine of both types of breeds was high at 3 and 5 dpi [15]. Additionally, in lymphoid organs, proinflammatory cytokines expression peaked at 10 dpi except IL-8, with broilers exhibiting higher levels compared to Sonali chickens. Another study also reported significantly higher expression of TNF-α and IL-1β in the bursa of Fabricius at 1 and 10 dpi, consistent with the study findings [22]. However, they also noted a significant increase in IL-6 expression at 1–3 dpi, which does not align with the current results.
The interferon system inhibits viral replication and stimulates immune responses that fight viral infection [45]. To avoid the host immune response and spread infection, influenza viruses have evolved complex defense mechanisms that antagonize interferon production and signaling [46].
Like proinflammatory cytokines, Sonali chickens showed a significant antiviral response from 3 to 7 dpi in the trachea than broiler chickens. Another study reported that higher levels of IFN-γ were induced in the trachea on 4 dpi [41]. Over time, antiviral responses increased progressively in the lungs of both chicken hosts. However, broiler chickens showed a higher level of cytokine expression compared to Sonali chickens. Several researches have indicated that the expression of IFN-α, IFN-β and IFN-γ was significantly higher at 4–10 dpi in the lung of chickens infected with H9N2 LPAI [14, 22] which is in line with the study findings.
In intestine, both groups displayed a decreasing trend in antiviral responses over time which is consistent with previous finding [47]. In lymphoid organs, the antiviral cytokine IFN-β was expressed from 1 to 10 dpi, with its peak expression occurring at 1 dpi. In contrast, INF-γ exhibited minimal expression at early time points but reached its peak at 10 dpi. Another study indicates that IFN-γ was expressed from 1 to 7 dpi, with a significant upregulation observed on 4 dpi in chickens inoculated with the H9N2 virus [28].
In our previous study, we noted that infected Sonali and broiler chickens showed no mortality or clinical signs with limited pathological changes including mild tracheitis, pneumonia and enteritis [15]. Despite this, our present study compared significant cytokine induction in both chicken hosts. This finding contrasted with some studies that reported a correlation between clinical signs and the production of inflammatory cytokines [48, 49]. Some studies reported that viral pathogenicity was not simply related to the extent of viral replication [50]. Though the current study found that viral replication [15] and cytokine induction correlated with each other, these differences might be complex interplay between host factors (such as immune response and genetic makeup) and viral characteristics (such as strain virulence and ability to induce cytokine production). These factors collectively influence the severity and manifest clinical disease in different host species.
There is some potential limitation in our study as it is restricted to evaluate a limited set of cytokines (IL-6, IL-8, IL-1β, TNF-α, IFN-β, and IFN-γ) expression using only RT-qPCR. The study didn’t explore the adaptive immune responses or involvement of key immune cells (T-cells, B-cells, or NK cells) using modern techniques like flow cytometry or single cell RNA sequencing. Therefore, the study required further an in-depth investigation of immune cell populations or broader immune pathway analysis against tribasic H9N2 infection in chickens that could provide more comprehensive view of the immune response against tribasic H9N2 infection in Broiler and Sonali chickens.
Conclusion
Infection of Sonali and broiler chickens with the tribasic H9N2 LPAI virus induce proinflammatory (IL-6, IL-8, IL-1β and TNF-α) and antiviral (IFN-β and IFN-γ) cytokine expressions in different organs. Sonali chickens exhibited a significant proinflammatory and antiviral response in the trachea until 3 to 7 dpi. However, broiler chickens showed significant proinflammatory and antiviral cytokine responses in lungs, intestine and lymphoid tissues until 3 to 10 dpi. Furthermore, broiler showed stronger early IFN-β response and a delayed IFN-γ response compared to Sonali chickens. Understanding the innate immune response by cytokine profile of tribasic H9N2 LPAI revealed their potential for cross-species transmission, and is crucial for assessing their pandemic potential.
Acknowledgements
Not applicable.
Abbreviations
- AIV
Avian influenza virus
- dpi
Days post infection
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HACS
Hemagglutinin endoproteolytic cleavage site
- HPAIV
Highly pathogenic AIV
- IBV
Infectious Bronchitis Virus
- IFN
interferon
- IFN-α
Interferon alpha
- IFN-γ
Interferon gamma
- IL-10
Interleukin 10
- IL-12
Interleukin 12
- IL-1β
Interleukin 1β
- IL-6
Interleukin 6
- IL-8
Interleukin 8
- IN
Intranasal
- IO
Intraocular
- LPAIV
Low pathogenic AIV
- LPAI
Low pathogenic avian influenza
- NDV
Newcastle Disease Virus
- PBS
Phosphate-buffered saline
- qPCR
Quantitative Polymerase Chain Reaction
- SPF
Specific pathogen-free
- TNF-α
tumor necrosis factor alpha
Author contributions
I.H. wrote the first draft of the manuscript. J.A.B., R.A.S., M.M.U., and I.H. organized samples and data collection. J.A.B., R.P. M.M.U., I.H., and R.A.S. analyzed formally and interpreted the results. R.P., J.A.B., I.H., and E.H.C. edited and critically reviewed the manuscript. J.A.B. and R.P. designed the study and J.A.B. supervised the study. All authors have read and agreed to the published version of the manuscript.
Funding
This research was jointly funded by Bangladesh Agricultural University (BAU), project no. 2020/963/BAU, Ministry of Science and Technology (MoST), project no. 2023/24/MoST/R&D and University Grant Commission (UGC), Project No. 2023/11/UGC.
Data availability
The original contributions presented in the study are included in the manuscript. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All applicable guidelines from national and institutional were followed for the care and use of animals. The present study was carried out in strict accordance with the approvals of the Bangladesh Agricultural University, Mymensingh Ethical Standard of Research Committee. The Ethical Standard of the Research Committee reviewed and approved the protocol and procedures of this experiment (7/BAURES/ESRC/VET-16/22). For the animal trial, day-old chicks of both Sonali and broiler chickens were sourced from a leading and reliable commercial supplier. The supplier provided full consent for their chicks to be used in this animal trial.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gerloff NA, Khan SU, Zanders N, Balish A, Haider N, Islam A, et al. <ArticleTitle Language=“En”>Genetically diverse low pathogenicity avian influenza a virus subtypes co-circulate among poultry in Bangladesh. PLoS ONE. 2016;11(3):e0152131. 10.1371/journal.pone.0152131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shanmuganatham K, Feeroz MM, Jones-Engel L, Smith GJ, Fourment M, Walker D, et al. Antigenic and molecular characterization of avian influenza A (H9N2) viruses, Bangladesh. Emerg Infect Dis. 2013;19(9):1393. https://doi.org/10.3201%2Feid1909.130336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan SU, Gurley ES, Gerloff N, Rahman MZ, Simpson N, Rahman M, et al. Avian influenza surveillance in domestic waterfowl and environment of live bird markets in Bangladesh, 2007–2012. Sci Rep. 2018;8(1):9396. 10.1038/s41598-018-27515-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kariithi HM, Welch CN, Ferreira HL, Pusch EA, Ateya LO, Binepal YS, et al. Genetic characterization and pathogenesis of the first H9N2 low pathogenic avian influenza viruses isolated from chickens in Kenyan live bird markets. Infect Genet Evol. 2020;78:104074. 10.1016/j.meegid.2019.104074. [DOI] [PubMed] [Google Scholar]
- 5.Kye S-J, Park M-J, Kim N-Y, Lee Y-N, Heo G-B, Baek Y-K, et al. Pathogenicity of H9N2 low pathogenic avian influenza viruses of different lineages isolated from live bird markets tested in three animal models: SPF chickens, Korean native chickens, and ducks. Poult Sci. 2021;100(9):101318. 10.1016/j.psj.2021.101318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander DJ. A review of avian influenza in different bird species. Vet microbiol. 2000;74(1–2):3–13. 10.1016/s0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 7.Parvin R, Heenemann K, Halami MY, Chowdhury EH, Islam M, Vahlenkamp TW. Full-genome analysis of avian influenza virus H9N2 from Bangladesh reveals internal gene reassortments with two distinct highly pathogenic avian influenza viruses. Arch virol. 2014;159:1651–61. 10.1007/s00705-014-1976-8. [DOI] [PubMed] [Google Scholar]
- 8.Parvin R, Begum JA, Nooruzzaman M, Chowdhury EH, Islam MR, Vahlenkamp TW. Review analysis and impact of co-circulating H5N1 and H9N2 avian influenza viruses in Bangladesh. Epidemiol Infect. 2018;146(10):1259–66. 10.1017/S0950268818001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Ma K, Li B, Chen Y, Qiu Z, Xing J, et al. A risk marker of tribasic hemagglutinin cleavage site in influenza A (H9N2) virus. Commun Biol. 2021;4(1):71. 10.1038/s42003-020-01589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parvin R, Schinkoethe J, Grund C, Ulrich R, Bönte F, Behr KP, et al. Comparison of pathogenicity of subtype H9 avian influenza wild-type viruses from a wide geographic origin expressing mono-, di-, or tri-basic hemagglutinin cleavage sites. Vet Res. 2020;51:1–12. 10.1186/s13567-020-00771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parvin R, Nooruzzaman M, Kabiraj CK, Begum JA, Chowdhury EH, Islam MR, et al. Controlling avian influenza virus in Bangladesh: challenges and recommendations. Viruses. 2020;12(7):751. 10.3390/v12070751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casadevall A, Pirofski L. Host-pathogen interactions: the attributes of virulence. J Infect Dis. 2001;184(3):337–44. 10.1086/322044. [DOI] [PubMed] [Google Scholar]
- 13.Lednicky JA, Hamilton SB, Tuttle RS, Sosna WA, Daniels DE, Swayne DE. Ferrets develop fatal influenza after inhaling small particle aerosols of highly pathogenic avian influenza virus A/Vietnam/1203/2004 (H5N1). Virol J. 2010;7:1–15. 10.1186/1743-422X-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taye B, Chen H, Myaing MZ, Tan BH, Maurer-Stroh S, Sugrue RJ. Systems-based approach to examine the cytokine responses in primary mouse lung macrophages infected with low pathogenic avian Influenza virus circulating in South East Asia. BMC Genomics. 2017;18:1–16. 10.1186/s12864-017-3803-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begum JA, Hossain I, Nooruzzaman M, King J, Chowdhury EH, Harder TC, et al. Experimental pathogenicity of H9N2 avian influenza viruses harboring a tri-basic hemagglutinin cleavage site in Sonali and broiler chickens. Viruses. 2023;15(2):461. 10.3390/v15020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujisawa H, Tsuru S, Taniguchi M, Zinnaka Y, Nomoto K. Protective mechanisms against pulmonary infection with influenza virus. I. Relative contribution of polymorphonuclear leukocytes and of alveolar macrophages to protection during the early phase of intranasal infection. J Gen Virol. 1987;68(2):425–32. 10.1099/0022-1317-68-2-425. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann A, Salentin R, Meyer RG, Bussfeld D, Pauligk C, Fesq H, et al. Defense against influenza A virus infection: essential role of the chemokine system. Immunobiology. 2001;204(5):603–13. 10.1078/0171-2985-00099. [DOI] [PubMed] [Google Scholar]
- 18.Xing Z, Cardona CJ, Li J, Dao N, Tran T, Andrada J. Modulation of the immune responses in chickens by low-pathogenicity avian influenza virus H9N2. J Gen Virol. 2008;89(5):1288–99. 10.1099/vir.0.83362-0. [DOI] [PubMed] [Google Scholar]
- 19.Gu Y, Hsu ACY, Pang Z, Pan H, Zuo X, Wang G, et al. Role of the innate cytokine storm induced by the influenza A virus. Viral Immunol. 2019;32(6):244–51. 10.1089/vim.2019.0032. [DOI] [PubMed] [Google Scholar]
- 20.Rebel JM, Peeters B, Fijten H, Post J, Cornelissen J, Vervelde L. Highly pathogenic or low pathogenic avian influenza virus subtype H7N1 infection in chicken lungs: small differences in general acute responses. Vet Res. 2011;42:1–11. 10.1186/1297-9716-42-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karpala AJ, Bingham J, Schat KA, Chen L-M, Donis RO, Lowenthal JW, et al. Highly pathogenic (H5N1) avian influenza induces an inflammatory T helper type 1 cytokine response in the chicken. J Interferon Cytokine Res. 2011;31(4):393–400. 10.1089/jir.2010.0069. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Cao Z, Guo X, Zhang Y, Wang D, Xu S, et al. Cytokine expression in three chicken host systems infected with H9N2 influenza viruses with different pathogenicities. Avian Pathol. 2016;45(6):630–9. 10.1080/03079457.2016.1193665. [DOI] [PubMed] [Google Scholar]
- 23.Vervelde L, Reemers SS, van Haarlem DA, Post J, Claassen E, Rebel JM, et al. Chicken dendritic cells are susceptible to highly pathogenic avian influenza viruses which induce strong cytokine responses. Dev Comp Immunol. 2013;39(3):198–206. 10.1016/j.dci.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Mogensen TH, Paludan SR. Molecular pathways in virus-induced cytokine production. Microbiol Mol Biol Rev. 2001;65(1):131–50. 10.1128/MMBR.65.1.131-150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertran K, Pantin-Jackwood MJ, Criado MF, Lee D-H, Balzli CL, Spackman E, et al. Pathobiology and innate immune responses of gallinaceous poultry to clade 2.3. 4.4 A H5Nx highly pathogenic avian influenza virus infection. Vet Res. 2019;50:1–14. 10.1186/s13567-019-0704-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helin AS, Wille M, Atterby C, Järhult JD, Waldenström J, Chapman JR. A rapid and transient innate immune response to avian influenza infection in mallards. Mol Immunol. 2018;95:64–72. 10.1016/j.molimm.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Lee D-H, Yuk S-S, Park J-K, Kwon J-H, Erdene-Ochir T-O, Noh J-Y, et al. Innate immune response gene expression profiles in specific pathogen-free chickens infected with avian influenza virus subtype H9N2. BioChip J. 2013;7:393–8. [Google Scholar]
- 28.Guan J, Fu Q, Sharif S. Replication of an H9N2 avian influenza virus and cytokine gene expression in chickens exposed by aerosol or intranasal routes. Avian Dis. 2015;59(2):263–8. 10.1637/10972-110714-Reg. [DOI] [PubMed] [Google Scholar]
- 29.Adams SC, Xing Z, Li J, Cardona CJ. Immune-related gene expression in response to H11N9 low pathogenic avian influenza virus infection in chicken and Pekin duck peripheral blood mononuclear cells. Mol ImmunoL. 2009;46(8–9):1744–9. 10.1016/j.molimm.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Daviet S, Van Borm S, Habyarimana A, Ahanda M-LE, Morin V, Oudin A, et al. Induction of Mx and PKR failed to protect chickens from H5N1 infection. Viral Immunol. 2009;22(6):467–72. 10.1089/vim.2009.0053. [DOI] [PubMed] [Google Scholar]
- 31.Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, et al. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J Virol. 2007;81(22):12439–49. 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinarello CA. Impact of basic research on tomorrow’s medicine. Proinflammatory cytokines Chest. 2000;118(2):503–8. 10.1378/chest.118.2.503. [DOI] [PubMed] [Google Scholar]
- 33.Ank N, West H, Paludan SR. IFN-λ: novel antiviral cytokines. J Interferon Cytokine Res. 2006;26(6):373–9. 10.1089/jir.2006.26.373. [DOI] [PubMed] [Google Scholar]
- 34.Saleque M, Saha A, editors. Production and economic performance of small scale Sonali bird farming for meat production in Bangladesh. Proceedings of the Semian, 8th International Poultry Show and Seminar, Dhaka, World Poultry Science Association Barach; 2013.
- 35.Amin JR, Mercier Y, Iji P, editors. Gross response and meat yield of broile r chickens fed different levels of digestible methionine. Proceedings of the 19t h European Symposium on Poultry Nutrition; 2013.
- 36.Ruiz-Hernandez R, Mwangi W, Peroval M, Sadeyen J-R, Ascough S, Balkissoon D, et al. Host genetics determine susceptibility to avian influenza infection and transmission dynamics. Sci Rep. 2016;6(1):26787. https://www.nature.com/articles/srep26787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renshaw M, Rockwell J, Engleman C, Gewirtz A, Katz J, Sambhara S. Cutting edge: impaired Toll-like receptor expression and function in aging. J Immunol. 2002;169(9):4697–701. 10.4049/jimmunol.169.9.4697. [DOI] [PubMed] [Google Scholar]
- 38.Boehmer ED, Meehan MJ, Cutro BT, Kovacs EJ. Aging negatively skews macrophage TLR2-and TLR4-mediated pro-inflammatory responses without affecting the IL-2-stimulated pathway. Mech Age Dev. 2005;126(12):1305–13. 10.1016/j.mad.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Pantin-Jackwood MJ, Smith DM, Wasilenko JL, Cagle C, Shepherd E, Sarmento L, et al. Effect of age on the pathogenesis and innate immune responses in Pekin ducks infected with different H5N1 highly pathogenic avian influenza viruses. Virus Res. 2012;167(2):196–206. 10.1016/j.virusres.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen GT, Rauw F, Steensels M, Ingrao F, Bonfante F, Davidson I, et al. Study of the underlying mechanisms and consequences of pathogenicity differences between two in vitro selected G1-H9N2 clones originating from a single isolate. Vet Res. 2019;50:1–12. 10.1186/s13567-019-0635-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nang NT, Lee JS, Song BM, Kang YM, Kim HS, Seo SH. Induction of inflammatory cytokines and toll-like receptors in chickens infected with avian H9N2 influenza virus. Vet Res. 2011;42:1–8. 10.1186/1297-9716-42-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser L, Fritz RS, Straus SE, Gubareva L, Hayden FG. Symptom pathogenesis during acute influenza: interleukin-6 and other cytokine responses. J Med Virol. 2001;64(3):262–8. 10.1002/jmv.1045. [DOI] [PubMed] [Google Scholar]
- 43.Reemers SS, van Haarlem DA, Groot Koerkamp MJ, Vervelde L. Differential gene-expression and host-response profiles against avian influenza virus within the chicken lung due to anatomy and airflow. J Gen Virol. 2009;90(9):2134–46. 10.1099/vir.0.012401-0. [DOI] [PubMed] [Google Scholar]
- 44.Jiang H, Yu K, Kapczynski DR. Transcription factor regulation and cytokine expression following in vitro infection of primary chicken cell culture with low pathogenic avian influenza virus. Virol J. 2013;10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westenius V, Mäkelä SM, Ziegler T, Julkunen I, Österlund P. Efficient replication and strong induction of innate immune responses by H9N2 avian influenza virus in human dendritic cells. Virology. 2014;471:38–48. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Liu S, Goraya MU, Maarouf M, Huang S, Chen J-L. Host immune response to influenza A virus infection. Front Immunol. 2018;9:320. 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleming-Canepa X, Aldridge JR Jr, Canniff L, Kobewka M, Jax E, Webster RG, et al. Duck innate immune responses to high and low pathogenicity H5 avian influenza viruses. Vet Microbiol. 2019;228:101–11. 10.1016/j.vetmic.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richard M, Graaf Md, Herfst S. Avian influenza A viruses: from zoonosis to pandemic. Future Virol. 2014;9(5):513–24. 10.2217/fvl.14.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindh E, Ek-Kommonen C, Väänänen V-M, Vaheri A, Vapalahti O, Huovilainen A. Molecular epidemiology of H9N2 influenza viruses in Northern Europe. Vet Microbiol. 2014;172(3–4):548–54. 10.1016/j.vetmic.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 50.Kobasa D, Takada A, Shinya K, Hatta M, Halfmann P, Theriault S, et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431(7009):703–7. 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the manuscript. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.