Abstract
Escherichia coli leucyl-tRNA synthetase (LeuRS) aminoacylates up to six different class II tRNAleu molecules. Each has a distinct anticodon and varied nucleotides in other regions of the tRNA. Attempts to construct a minihelix RNA that can be aminoacylated with leucine have been unsuccessful. Herein, we describe the smallest tRNAleu analog that has been aminoacylated to a significant extent to date. A series of tRNAleu analogs with various domains and combinations of domains deleted was constructed. The minimal RNA that was efficiently aminoacylated with LeuRS was one in which the anticodon stem–loop and variable arm stem–loop, but neither the D-arm nor T-arm, were deleted. Aminoacylation of this minimal RNA was abolished when the discriminator base A73 was replaced with C73 or when putative tertiary interactions between the D-loop and T-loop were disrupted, suggesting that these identity elements are still functioning in the minimized RNA. The various constructs that were significantly aminoacylated were also tested for amino acid editing by the synthetase. The anticodon and variable stem–loop domains were also dispensable for hydrolysis of the charged tRNAleu mimics. These results suggest that LeuRS may rely on identity elements in overlapping domains of the tRNA for both its aminoacylation and editing activities.
INTRODUCTION
Leucyl-tRNA synthetase (LeuRS) aminoacylates up to six tRNA isoacceptors with different anticodons (1). In most cases, the isoacceptors are class II tRNAs that contain an extra-long variable loop. The primary identity element for leucylation is believed to be the unpaired discriminator base A73 on Escherichia coli, yeast cytoplasmic and human cytoplasmic tRNAleu (2–4). With the exception of the yeast cytoplasmic enzyme, LeuRS is not known to interact with the anticodon stem–loop (2–4). The extra-long variable arm that is common among most class II tRNA molecules, surprisingly, does not appear to be an identity element for E.coli tRNAleu (2,5). However, recent in vitro selection experiments suggest that tertiary interactions between A15:U48 and the location of G18:G19 in the D-loop may play a key role in discriminating tRNAleu from other class II RNA molecules, tRNAser and tRNAtyr (2,6). Consistent with this, even though the discriminator base is the primary recognition determinant, tRNA acceptor stem microhelices from human tRNAleu could not be aminoacylated by LeuRS (7).
Escherichia coli LeuRS also binds tRNA in a separate domain that is distinct from the aminoacylation active site in order to hydrolytically edit misaminoacylated tRNAleu (8–10). Other amino acid editing tRNA synthetases have been shown to be RNA dependent (11) and rely on specific tRNA interactions (11,12). In the best characterized example, isoleucyl-tRNA synthetase (IleRS) relies on the D-loop of tRNAile to confer editing of mischarged tRNAs (12). Substitution of two critical nucleotides abolishes post-transfer editing activity by IleRS. Interestingly, they are separate and distinct from the anticodon and discriminator base identity elements required for IleRS aminoacylation.
Specific regions or nucleotides that are critical to amino acid editing have not been investigated in tRNAleu. In fact, these sorts of investigations are hindered by the difficulties in stably producing mischarged tRNA as a substrate. However, an important amino acid editing mutant of LeuRS (T252A) that hydrolyzes aminoacylated leu-tRNAleu correctly has been reported (9). The alanine mutation prevents discrimination of leucine in the editing pocket. The steric mechanism that blocks a cognate leucine molecule from entering or binding efficiently to the amino acid binding pocket of the editing active site is eliminated by the single amino acid change. We used the LeuRS T252A mutant and correctly charged tRNA analogs to probe specific tRNA domains for their role in amino acid editing.
Herein, we report the successful construction of a minimized RNA that can be aminoacylated with LeuRS. In order to obtain this RNA, we studied a series of E.coli tRNAleu analogs to further characterize which RNA domains were essential for aminoacylation by LeuRS. Our results provide further evidence that the anticodon and extra-long variable loop are dispensable for recognition of tRNAleu. The minimal chargeable tRNAleu analog is one in which the anticodon stem–loop and extra arm were deleted. Within the context of this minimal tRNAleu analog, it was found that the discriminator base and tertiary interactions between the D-loop and T-loop continued to be essential for successful aminoacylation. Modification of the linker regions connecting the D-loop and T-loop in the minimized RNA had a significant effect on aminoacylation efficiency but never abolished activity. Chargeable constructs were also tested for their effect on the hydrolytic editing activity of E.coli LeuRS. The results show that neither the tRNA anticodon stem–loop or variable loop domains impact amino acid editing or aminoacylation activity.
MATERIALS AND METHODS
Materials
Plasmid ptDNAleu that expresses the suppressor tRNAleu (UAA) was kindly provided by Drs Abelson and Tocchini-Valentini (California Institute of Technology). Oligonucleotides were purchased from Genosys Biotechnologies, Inc. (Woodlands, TX) or MWG Biotech, Inc. (High Point, NC). T7 RNA polymerase was purified from E.coli (BL21) harboring the pAR1219 plasmid according to Grodberg and Dunn (13). Escherichia coli LeuRS with a six-histidine N-terminal fusion was purified as described previously (9,14). l-14C-leucine and l-4,5-3H-leucine were purchased from Amersham Pharmacia Biotech. Inc. (Piscataway, NJ). Solutions were prepared with diethyl pyrocarbonate-treated water to inhibit potential RNase activity.
Expression and purification of tRNA molecules
Escherichia coli tRNAleu (UAA) and minimalist tRNAleu molecules were made by in vitro T7 RNA polymerase run-off transcription (15) using a specifically designed ptDNAleu template or a synthetic oligonucleotide with a 5′-T7 promoter. The plasmid DNA template was prepared by digestion with BstNI (New England Biolabs, Beverly, MA) at 60°C for 4 h. Approximately 50 µg of template were incubated for 4 h at 42°C in a 1 ml in vitro transcription reaction mixture, which was optimized for each specific template. The reaction contained 40 mM Tris buffer (pH 8.1), 80 mg/ml polyethylene glycol 8000, 5 mM dl-dithiothreitol (DTT), 30 mM MgCl2, 2 mM spermidine, 50 µg/ml bovine serum albumin, 0.01% Triton X-100 and 0.003 U/ml inorganic pyrophosphatase (Sigma, St. Louis, MO). It was experimentally determined that a final concentration range of 2.0–7.5 mM for each nucleotide (depending on the specific template) and 0.1 mM GMP yielded optimal RNA production. Approximately 100 µg of T7 RNA polymerase were added initially to each 1.0 ml in vitro RNA synthesis mixture followed by a second aliquot after a 2 h incubation.
The RNA product was ethanol precipitated, washed with 70% ethanol and dried in a speed-vac. RNA precipitants were rehydrated directly in 8 M urea/50 mM Tris (pH 8) buffer and loaded onto a 10 or 16% acrylamide:bisacrylamide (19:1) and 8 M urea denaturing gel. RNA bands were visualized by UV shadowing using Whatman 250 µm layer flexible TLC plates (PE SIL G/UV254), cut out of the gel and eluted twice in an equal volume of 0.5 M NH4OAc/1 mM EDTA (15). RNA was concentrated to ∼400 µl via butanol extraction and precipitated with ethanol at –80°C using 62.5 µg/ml glycogen stock as a carrier. The final concentration of each RNA product was determined by UV spectroscopy at 260 nm. Extinction (ɛ) coefficients (M–1 cm–1) for each template were calculated as described previously and are listed in Table 1 (16).
Table 1. Calculated ɛ-coefficients for each minimalist tRNAleu analog.
| tRNAleu molecule | ɛ-Coefficient (M–1 cm–1) |
|---|---|
| tRNAleu |
853660 |
| tRNAleu-ΔAC |
701590 |
| tRNAleu-ΔVL |
755180 |
| tRNAleu-ΔACVL1 |
558990 |
| tRNAleu-ΔACVL1 (A73C) |
553190 |
| tRNAleu-ΔACVL1 (G18A:U55C) |
558520 |
| tRNAleu-ΔACVL1 (G19A:C56U) |
564660 |
| G18A:U55C/G19A:C56U |
562210 |
| tRNAleu-ΔACVL2 |
587070 |
| tRNAleu-ΔACVL3 |
583330 |
| tRNAleu-ΔACDL1 |
501810 |
| tRNAleu-ΔACDL2 |
531840 |
| tRNAleu-ΔACDL3 |
511820 |
| tRNAleu-ΔACDL4 |
521830 |
| Minileu2 |
322080 |
| Minileu3B | 323970 |
Aminoacylation of leu-tRNAleu
The tRNAleu and minimalist tRNA substrates were aminoacylated with 14C-leucine or 3H-leucine using E.coli LeuRS. Purified tRNAleu transcript was diluted in H2O depending on the anticipated final reaction volume (100–250 µl) and denatured at 90°C for 2 min. Addition of MgCl2 to a final concentration of 1 mM facilitated RNA folding under quick cooling conditions on ice. Each reaction mixture contained 100 nM LeuRS, purified tRNAleu (0.7–100 µM) or minimalist tRNA molecules (0.7–60 µM), 20.5 µM 14C-leucine (50 µCi/ml) or 20 µM 3H-leucine (100 µCi/ml), 2 mM ATP, 0.5 mM DTT, 10 mM MgCl2 and 60 mM Tris buffer (pH 7.5), and was incubated at 25°C. The reaction was initiated by addition of LeuRS. Reactions were quenched either by acidification to pH 5 with 10% acetic acid (17), or precipitation onto a filter pad pre-wet with 5% tricholoroacetic acid (TCA). The kinetic constants for tRNAleu, tRNAleu-ΔAC and tRNAleu-ΔVL were derived from Lineweaver–Burke plots representing the average of three independent experiments.
Acid gel analysis of aminoacylated RNAs
Aminoacylation of each RNA substrate with 14C-leucine was visualized by separation on denaturing 16% polyacrylamide acid gels that contained 25 mM NaOAc (pH 5)/8 M urea as described previously (18,19). The RNA was electrophoresed at 10 mA overnight (∼12 h). The polyacrylamide gel was then dried for analysis by phophoroimaging. Approximately 2–3 weeks of imaging using a Fuji MacBAS plate was necessary to detect 14C-leucine aminoacylated RNA products with a Fuji BAS1000 phosphoroimager.
Aminoacylated RNA was also visualized by native acid gel electrophoresis. A portion of each aminoacylation reaction mixture was mixed with 40% sucrose and loaded onto a 20% polyacrylamide (29:1) gel buffered in 100 mM NH4OAc (pH 5). The polyacrylamide gels were stained with ethidium bromide in 25 mM NH4OAc (pH 5) buffer and visualized using the Stratagene Eagle Eye II Still Video System and software. Subsequent to drying they were imaged for 2–4 weeks as described above.
RNA melting properties
Purified RNA samples ranging in concentration from 1 to 60 µM were heated at 90°C for 2 min. MgCl2 was added to a final concentration of 1 mM and the samples were quick cooled on ice to facilitate intramolecular folding. These samples were diluted in 10 mM Tris (pH 7.5) buffer and transferred to 1.0 ml quartz cuvettes for RNA thermal melting assays. Each sample was degassed using a house vacuum and covered with mineral oil to prevent buffer evaporation during data acquisition at high temperatures. Thermal melting curves were carried out on a Cary 3E spectrophotometer equipped with a Peltier 12 cell holder capable of varying temperature from 0 to 95°C. Full-length tRNAleu, tRNAleu-ΔACVL1, tRNAleu-ΔACVL1 mutants and tRNAleu-ΔACDL1 RNA substrates were heated from 0 to 95°C at 2°C/min intervals. The tRNAleu-ΔACDL2 molecules and leucine minihelix (Minileu) were heated from 0 to 90°C at 1°C/min intervals. The absorbance at 260 nm was recorded at every 0.5°C. All data were analyzed with CaryWinUV software.
Hydrolytic editing assays
Each of the minimized tRNAleu variants was aminoacylated with 3H-leucine (100 µCi/ml) and isolated (17). Phenol-extraction (pH 5) was used to remove protein followed by ethanol precipitation. The aminoacylated RNA was rehydrated in 50 mM KH2PO4 buffer (pH 5) prior to use. Each charged minimalist molecule was tested for hydrolytic editing in reaction mixtures consisting of 60 mM Tris (pH 7.0), 10 mM MgCl2, ∼4 µM 3H-leu-tRNAleu or aminoacylated tRNAleu mimic, and 100 nM LeuRS T252A mutant enzyme (9). The reaction was initiated by the addition of enzyme. A 10 µl aliquot was removed at periodic time intervals and quenched by TCA precipitation.
RESULTS
RNA substrate design
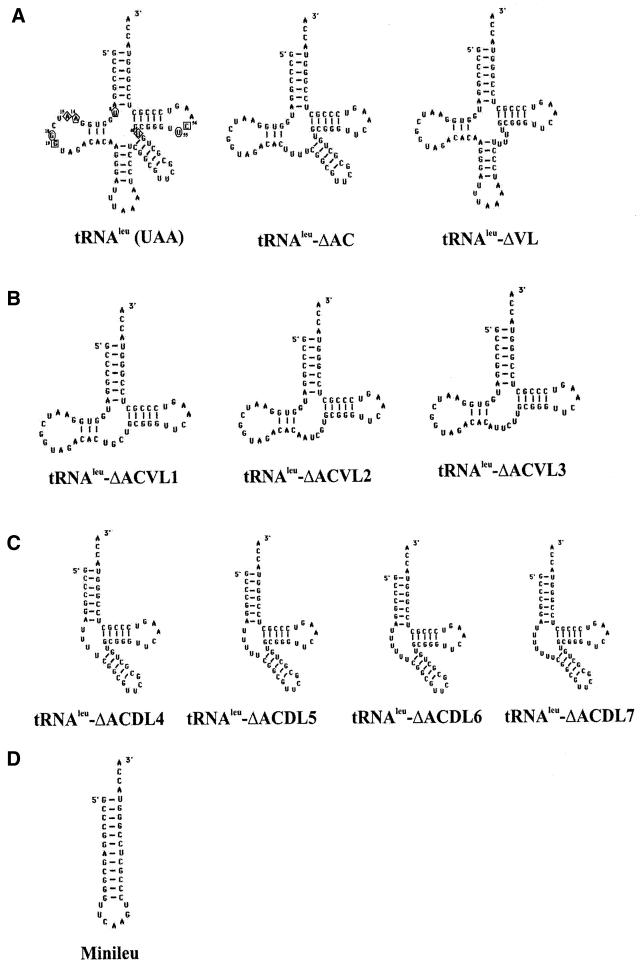
We designed a series of truncated minimalist tRNAleu molecules to test the importance of each discrete domain for LeuRS recognition and aminoacylation. As shown in Figure 1A, the anticodon stem and loop (tRNAleu-ΔAC) or variable arm (tRNAleu-ΔVL) were eliminated. We also deleted both the variable arm and anticodon stem–loop (tRNAleu-ΔACVL), replacing them with linker regions of variable length and sequence (Fig. 1B). These linkers were designed to preserve tertiary interactions in the folded tRNA core based on the tRNAphe crystal structure (20). Specifically, these tRNAleu-ΔACVL variants were expected to maintain the A15:U48, G18:U55, G19:G56 and U8:A14 base pair interactions which are highlighted in Figure 1A. In the tRNAleu-ΔACVL1 variant, the A26:C44 tertiary interaction was eliminated. Longer linker regions for the tRNAleu-ΔACVL2 and tRNAleu-ΔACVL3 mimics were designed to restore this tertiary interaction by incorporating A and C nucleotides into the linker region. The D-stem and loop and anticodon stem and loop were also removed (Fig. 1C) and replaced with varying lengths of polyuridine linkers (4–7 nt) to allow flexibility for tertiary folding. Figure 1D shows that the leucine minihelix (Minileu) was comprised of the acceptor stem and TψC-stem and loop (18).
Figure 1.
Sequence and secondary structures of minimized E.coli tRNAleu variants. (A) tRNAleu and truncated variants lacking either the anticodon domain (ΔAC) or the extra-long variable stem and loop (ΔVL). The symbols used for important tRNAleu tertiary interactions are: squares, G19:C56; circles, G18:U55; diamonds, A15:U48; and pentagons, U8:A14. (B) Truncated variants with combined deletions of the anticodon domain and variable loop (ΔACVL). Each RNA substrate varies only by the number and sequence of nucleotides in the linker region to replace the anticodon and variable loop domains. Specifically, tRNAleu-ΔACVL1, tRNAleu-ΔACVL2 and tRNAleu-ΔACVL3 have 4 nt (UCGU), 6 nt (UGCUAA) and 6 nt (UUCUUA), respectively. (C) Minimalist RNA molecules (ΔACDL4–7) replace the anticodon and D-stem–loops with polyuridine linkers that range from 4 to 7 nt. (D) Leucine minihelix is comprised of the acceptor stem and the TψC stem–loop (Minileu).
Aminoacylation of minimized tRNAleu molecules
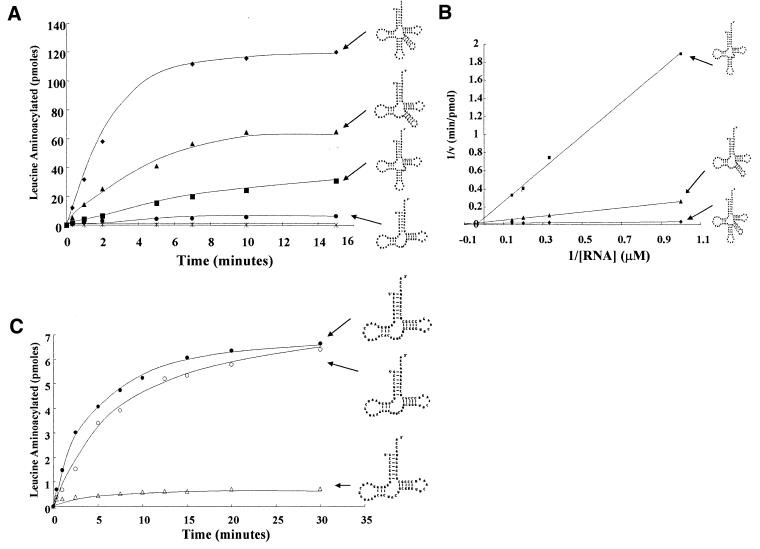
Full-length E.coli tRNAleu and each of the tRNAleu mimics were tested for aminoacylation by E.coli LeuRS. The most efficiently charged minimalist RNA molecules were tRNAleu-ΔAC followed by tRNAleu-ΔVL (Fig. 2A). The relative kcat/KM decreased 17-fold and 125-fold, respectively (Table 2 and Fig. 2B). Changes in both parameters, specifically an increase in KM and decrease in the turnover rate (kcat) contribute to the reduction of aminoacylation efficiency. While the kcat was reduced only 3–5-fold, the KM increased ∼10-fold for tRNAleu-ΔAC and 26-fold for tRNAleu-ΔVL. When both the anticodon and variable loop domains were deleted within a single tRNA analog (tRNAleu-ΔACVL1, tRNAleu-ΔACVL2 and tRNAleu-ΔACVL3) the RNA molecules were still recognized as substrates and aminoacylated by LeuRS (Fig. 2C).
Figure 2.
Aminoacylation activity of various minimalist tRNAleu molecules by E.coli LeuRS. (A) Reactions contained 10 µM tRNA, 20 µM 3H-leucine (100 µCi/ml) and 100 nM LeuRS, and were carried out under the conditions described in Materials and Methods. Diamonds, full-length tRNAleu; triangles, tRNAleu-ΔAC; squares, tRNAleu-ΔVL; circles, tRNAleu-ΔACVL2; stars, no RNA. (B) Lineweaver–Burke analysis of three independent experiments for tRNAleu (diamonds), tRNAleu-ΔAC (triangles), and tRNAleu-ΔVL (squares) using an RNA concentration range of 0.7–7 µM at 25°C. (C) Reactions contained 10 µM tRNA, 20 µM 3H-leucine (100 µCi/ml) and 100 nM LeuRS, and were carried out under the conditions described in Materials and Methods. Closed circles, tRNAleu-ΔACVL2 (UGCUAA linker region); open circles, tRNAleu-ΔACVL3 (UUCUUA linker region); and open triangles, tRNAleu-ΔACVL1 (UCGU linker region).
Table 2. Kinetic analysis of minimalist tRNAleu substrate leucylation by E.coli LeuRS.
| tRNAleu minimalist substrates | KM(µM) | kcat(min–1) | kcat/KM(µM–1 min–1) | kcat/KM (relative) | Loss |
|---|---|---|---|---|---|
| tRNAleu (UAA) | 0.9 | 0.354 | 3.9 × 10–1 | 1 | 0 |
| tRNAleu -ΔAC | 7.8 | 0.188 | 2.4 × 10–2 | 0.06 | 17 |
| tRNAleu-ΔVL | 23.5 | 0.071 | 3.0 × 10–3 | 0.008 | 125 |
| tRNAleu-ΔACVL1 | NM | 1.56 × 10–3 | NM | NM | NM |
| tRNAleu-ΔACVL2 | NM | 8.80 × 10–3 | NM | NM | NM |
| tRNAleu-ΔACVL3 | NM | 7.45 × 10–3 | NM | NM | NM |
| tRNAleu-ΔACDL4 | - | - | - | - | - |
| tRNAleu-ΔACDL5 | - | - | - | - | - |
| tRNAleu-ΔACDL6 | - | - | - | - | - |
| tRNAleu-ΔACDL7 | - | - | - | - | - |
| Minileu3B | - | - | - | - | - |
NM, aminoacylation occurs but kinetic parameters are not measurable.
The truncated tRNAleu mimics that are missing anticodon and D-stem–loop domains (tRNAleu-ΔACDL4, tRNAleu-ΔACDL5, tRNAleu-ΔACDL6 and tRNAleu-ΔACDL7) and the leucine minihelix (Minileu) were not aminoacylated by LeuRS (data not shown). The RNA and LeuRS concentrations were increased to 100 and 1 µM, respectively, to enhance sensitivity. However, analysis of aminoacylation activity assays using either acid gel electrophoresis or TCA precipitation methods failed to yield detectable aminoacylated RNA products. Since the anticodon stem–loop alone is dispensable for aminoacylation, these experiments suggest that the D-stem and loop is an important tRNA domain for recognition and aminoacylation by E.coli LeuRS.
RNA folding and structure
We probed the structures of the tRNAleu mimics using RNA melting studies (16). Each RNA sample was thermally denatured at varied concentrations to determine whether folding was based on intramolecular or intermolecular interactions. Shifts in the melting temperature as well as the shape of the melting curves for each RNA molecule were dependent on concentration, suggesting that the RNA is, at least initially, folded in a multimeric complex (data not shown). Specifically, full-length tRNAleu and tRNAleu-ΔACVL1 displayed non-two state behavior common to complex RNA folding or indicative of potential RNA aggregation (16). Interestingly, leucine minihelix and truncated tRNAleu-ΔACDL analogs that were not aminoacylated by LeuRS displayed two-state behavior between double stranded and single stranded transitions.
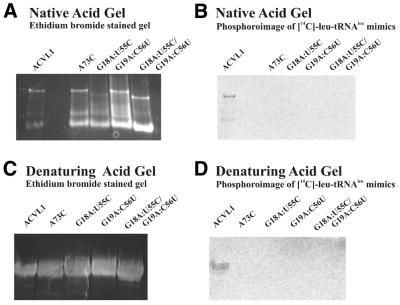
We further probed the structure of the aminoacylatable minimized RNA molecules by analyzing the charged product via acid gel electrophoresis. Acidic conditions (pH 5) are required to stabilize the labile aminoacyl bond between the leucine radiolabel and the RNA substrate (19). Aminoacylated tRNAleu-ΔACVL1, tRNAleu-ΔACVL2 and tRNAleu-ΔACVL3 with concentrations of 10 or 60 µM were separated on a native acid gel by electrophoresis (Fig. 3A and B). Four distinct bands were detected by ethidium staining, suggesting that a mixed population of conformers form for each minimal RNA substrate (Fig. 3A). Under denaturing conditions one primary band was detected, indicating that multimers seen under native conditions originate from a single purified RNA molecule (Fig. 3A and C). Acidic native gels reveal that LeuRS aminoacylates each of the RNA molecules despite the conformational differences (Fig. 3B).
Figure 3.
Native and denaturing acid gels of aminoacylated minimalist tRNAleu-ΔACVL molecules. Each aminoacylation reaction was performed with either 10 or 60 µM RNA, 20.5 µM 14C-leucine (50 µCi/ml) and 100 nM LeuRS under the conditions described in Materials and Methods. (A) RNA bands were detected by ethidium bromide staining of a native acidic gel. (B) RNA aminoacylated with 14C-leucine was visualized by phosphoroimaging the dried acidic gel for two weeks. Arrows in both (A) and (B) emphasize four distinct RNA bands observed using native acid gel electrophoresis. (C) Denatured RNA bands were detected by ethidium bromide staining of a denaturing acidic polyacrylamide gel. (D) Denatured 14C-leucine aminoacylated RNA products were visualized by phosphoroimaging the dried acidic gel for two weeks.
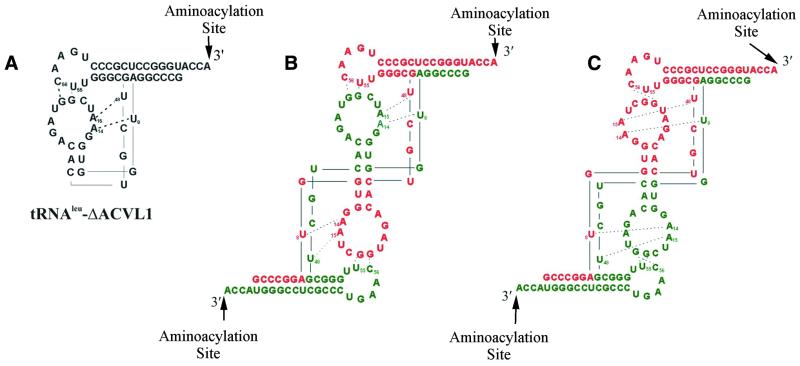
We propose that at least two of the bands represent monomers and dimers based on our thermal denaturation studies as well as previous investigations of RNA folding (21,22). Small RNA oligonucleotides can be difficult to visualize with ethidium bromide staining because the dye can not intercalate efficiently (21). Because the tRNAleu-ΔACVL1 monomer would have less double stranded helix regions available for ethidium bromide intercalation than the dimer, it is probable that the lower band, which is difficult to visualize on the gel, is a monomer (Fig. 3A). Beuning et al. (22) used both NMR structural analysis and native acid gel electrophoresis to show that a decrease in gel mobility of RNA hairpins was consistent with aggregation at high concentrations. One would then expect that a tRNAleu-ΔACVL dimer has retarded gel mobility relative to the monomer. Thus, the presence of a dimer is a more likely explanation for one of the upper bands on the gel (Fig. 3A and B). In addition, dimer formation appears to increase at a higher RNA concentration (60 µM), as seen in Figure 3A. We hypothesize that the multiple bands may be due to the conformational flexibility of the RNA dimers consistent with the description provided by Beuning et al. (22). Figure 4 illustrates a potential monomer and two possible dimer conformations of tRNAleu-ΔACVL1.
Figure 4.
Possible secondary and tertiary interactions of the folded truncated tRNAleu-ΔACVL1 molecules. (A) An L-shaped representation of the monomer. Dashed lines indicate hydrogen bonds predicted to play an important role in intramolecular interactions for tRNAleu tertiary folding. (B and C) Two separate strands are distinguished by green and red to depict the two possibilities of monomer assembly into a dimer. Arrows indicate the 3′ end of the tRNA which is aminoacylated.
The putative dimeric tRNAleu-ΔACVL1 molecules (Fig. 4B and C) resemble a full-length tRNA molecule with an extra acceptor stem/TψC stem–loop appendage at one end. Interestingly, these two 3′ ends could potentially be aminoacylated yielding a higher intensity band for phophoroimaging. Indeed, it appears that the visible intensity of the presumed aminoacylated duplex is significantly increased compared with the monomer (Fig. 3B). If such dimers of the tRNAleu mimics exist, recognition of the RNA would not be affected by the distal end of the RNA substrate, because LeuRS does not interact with the anticodon region (23).
Aminoacylation identity elements of minimized tRNAleu molecules
Another explanation for the dramatic decrease in aminoacylation may be a change in the structural environment such that key identity determinants are no longer being used effectively. The most important of these is the discriminator base, A73, in full-length E.coli tRNAleu. Changes at this position dramatically decrease aminoacylation activity in the wild-type tRNA (2,4,24). We therefore investigated whether the discriminator base remained a major identity element for LeuRS recognition by changing A73 to C73 in the tRNAleu-ΔACVL1 construct. This variant failed to yield TCA-precipitated aminoacylated product (data not shown). Potential aminoacylated product of this mutant was also separated via acidic polyacrylamide gel electrophoresis. As Figure 5A shows, the A73C mutant showed multiple conformers on a native acid gel similar to those seen with tRNAleu-ΔACVL1. However, in this case none of the conformers yielded aminoacylated RNA product (Fig. 5B). These results show that the discriminator base continues to play a crucial role in aminoacylation even when other domains of the minimized tRNAleu molecules are no longer available for recognition.
Figure 5.
Acid gel analysis of tRNAleu-ΔACVL1 mutant. A73C depicts the discriminator base change from A73 to C73. G18A:U55C, G19A:G56U and G18A:U55C/G19A:G56U are notations for the mutational changes G18:U55 to A18:C55, G19:C56 to A19:U56 and a double mutant with both G18:U55 and G19:C56 changed to A18:C55 and A19:U56, respectively. Each aminoacylation reaction was performed with 60 µM RNA, 20.5 µM 14C-leucine (50 µCi/ml) and 100 nM LeuRS under the conditions described in Materials and Methods. (A) RNA products were detected by ethidium bromide staining of a native acidic gel. An extra lane without loaded RNA (lane 2) separates assays for tRNAleu-ΔACVL1 from each of its mutant variants. (B) RNA products aminoacylated with 14C-leucine were visualized by phosphoroimaging the dried acidic gel for two weeks. (C) Denatured RNA products were detected by ethidium bromide staining of a denaturing acidic polyacrylamide gel. (D) Denatured 14C-leucine aminoacylated products were visualized by phosphoroimaging the dried acidic gel for two weeks.
Several conserved bases in the D- and T-loops have also been implicated as identity determinants using in vitro selection experiments of a partially randomized E.coli tRNAleu molecule (6). Specifically, Asahara et al. (2,6) showed that the invariant nucleotides G18 and G19 in the D-loop appeared to be a requirement for recognition and aminoacylation by LeuRS. Based on comparison with the crystal structure of tRNAphe, these bases are involved in tertiary interactions with bases of the TψC-loop (20). These interactions likely stabilize the core region of a folded tRNA molecule and, hence, it is not clear whether they are essential for aminoacylation because they maintain structural integrity, or if they might be actual identity elements. Therefore, we sought to test this by introducing rational changes into tRNAleu-ΔACVL1 that would be predicted to maintain the structure of the RNA.
The G18:U55 interaction is an unusual bifurcated base pair (N1-carbonyl; amino-carbonyl) (25). An A18:C55 variant was constructed as a possible replacement because the A–C mismatch can form a one hydrogen bond structure that is very similar to the bifurcated pair. The G19:C56 interaction is of the Watson–Crick type and hence replacement with a perfectly isosteric A19:U56 pair is unlikely to disrupt the structure. Finally, a double mutant was also generated that combined both the A18:C55 and A19:U56 changes.
All three variants were tested for aminoacylation using a range of RNA concentrations (10–60 µM). Both TCA precipitation assays (data not shown) and acid gel analysis failed to detect charged RNA products for the G18A:U55C, G19A:C56U and double mutants (G18A:U55C, G19A:C56U). Figure 5 shows the native acid gel analysis for tRNAleu-ΔACVL1 wild-type and each mutant analog. Several conformers for each of the RNA molecules were observed as distinct and compact bands in the ethidium bromide stained native acid gel (Fig. 5A). Each variant appears to exhibit monomer and dimer formation, comparable with the duplexes discussed previously and diagrammed for the tRNAleu-ΔACVL1 substrate in Figure 4. Although each of the tRNAleu-ΔACVL1 variants appears to have retained critical global structural interactions, leucylation activity was abolished in all of them (Fig. 5B). If the structure has in fact been maintained, as is especially likely for the A19:U56 variant, these results are a significant addition to the evidence establishing these interactions as identity elements for LeuRS aminoacylation of tRNAleu and tRNAleu-ΔACVL1.
tRNA-dependent amino acid editing with aminoacylated minimalist tRNAleu molecules
Escherichia coli LeuRS hydrolytically cleaves mischarged amino acids from tRNAleu in a post-transfer editing mechanism (8–10). Mursinna et al. (9) isolated an editing mutant in which a conserved threonine residue, T252, was changed to an alanine (T252A). This single site mutant enzyme could not discriminate its cognate amino acid, leucine, in the editing active site and hydrolytically cleaved correctly charged leu-tRNAleu. We employed the E.coli LeuRS T252A mutant to test for RNA-dependent effects of post-transfer editing using our charged minimized leu-tRNAleu molecules.
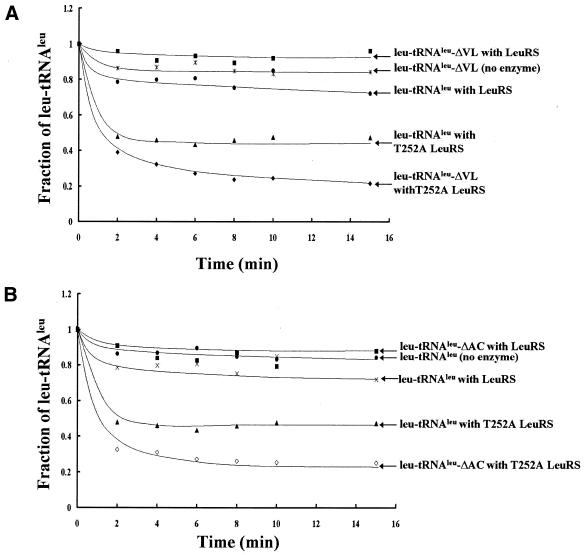
The two most highly aminoacylatable minimized RNAs reported herein were charged with 3H-leucine, recovered and then incubated with the E.coli LeuRS T252A mutant or the wild-type enzyme. Neither deletion of the anticodon stem–loop (3H-leu-tRNAleu-ΔAC) nor variable loop (3H-leu-tRNAleu-ΔVL) eliminated editing (Fig. 6). In fact, these smaller RNAs reproducibly exhibited somewhat faster rates of editing compared with the full-length tRNA. Further investigation of other tRNAleu domains for editing determinants is hampered by the inability to effectively charge smaller or altered tRNA molecules in order to obtain appropriate editing substrates. However, these initial results suggest that at least the anticodon and variable stem–loops do not play a significant role in RNA-dependent post-transfer editing by E.coli LeuRS.
Figure 6.
Hydrolytic amino-acid editing for each of the minimalist variants aminoacylated with 3H-leucine. All reactions contained ∼18 µM aminoacylated substrate, 100 nM of either wild-type LeuRS or T252A mutant enzyme, 60 mM Tris (pH 7.0) and 10 mM MgCl2. Editing activity was tested for tRNAleu-ΔVL (A) and tRNAleu-ΔAC (B).
DISCUSSION
Each aminoacyl-tRNA synthetase (AARS) may be divided into two distinctly folded domains that specifically and separately interact with the two domains of tRNA molecules (26). In particular, the N-terminal conserved catalytic core of the AARS interacts with the acceptor stem and TψC stem and loop domain. A non- or less-conserved C-terminal domain typically interacts with the tRNA anticodon and D-stem and loop domain. Although LeuRS also has a two-domain modular structure, in most cases the LeuRS C-terminal domain does not bind the anticodon. However, the class II tRNAleu has an extra-long variable arm that is found in only two other tRNAs (tRNAser and tRNAtyr). Thus, a priori one might expect that either the anticodon arm or the long variable arm would be key identity elements for leucyl tRNAs.
Although an atomic resolution structure is not yet available for tRNAleu either alone or in complex with LeuRS, considerable effort has been focused on understanding the origin of its identity elements and its interactions with the synthetase. We have compiled investigations that have determined identity elements in E.coli, human (cyt) and yeast (cyt) tRNAleu, which are summarized according to each tRNA domain in Table 3. The anticodon is not important for RNA recognition and aminoacylation for E.coli and human LeuRS. In addition, footprint analysis using bean cytoplasmic LeuRS with Phaseolus vulgaris, yeast and E.coli tRNAleu revealed that the D-arm and hinge region are in contact with the enzyme, but not the anticodon loop or the variable loop (23). The extra-long variable arm also does not appear to be an identity element in E.coli because replacement of the type II variable arm with a shorter class I consensus sequence maintained leucine identity in vivo. In contrast, the long variable arm is absolutely required for aminoacylation of human tRNAleu and becomes an inactive substrate when truncated (24).
Table 3. Compilation of identity determinants for tRNAleu in several organisms.
| Domain/site | Mutations | LeuRS aminoacylation | Reference |
|---|---|---|---|
| E.coli | |||
| Discriminator base (A73) | Replaced with C, U or G | Base preference: A>C>U>G | (2) |
| tRNAser G73 replaced with A | Conferred leucylation | (5) | |
| Replaced with C (ΔACVL1) | Leucylation abolished | This work | |
| Acceptor stem | G1 changed to A1 | No effect | (2) |
| C2:G71 changed to G2:C71 | No effect | ||
| Leucine minihelix | No aminoacylation | This work | |
| D-stem–loop | U16 changed to A16 | No effect | (2) |
| A14 changed to C, U or G | Decreased 100-fold | ||
| tRNAser G18G19 location altered | Conferred leucylation | ||
| tRNAtyr G18G19 location altered | Conferred leucylation | ||
| A15:U48 changed to G15:C48 | Small reduction | ||
| Randomized for in vitro selection | A15, G18, G19, A20a conserved | (6) | |
| G18:G19 fixed position | |||
| tRNAser G15:C48 changed to A15:U48 | In vivo analysis: 82% suppression | (5) | |
| tRNAser G18G19 location altered | 100% leu insertion | ||
| D-stem–loop domain deleted (ΔACDL1–4) | No aminoacylation | This work | |
| G18 changed to A18 (ΔACVL1) | Aminoacylation abolished | ||
| G19 changed to A19 (ΔACVL1) | Aminoacylation abolished | ||
| G18G19 changed to A18A19 (ΔACVL1) | Aminoacylation abolished | ||
| Anticodon stem–loop | G37 changed to U37 | No effect | (2) |
| U32 changed to C32 | No effect | ||
| Anticodon domain deleted (ΔAC) | Reduced 17-fold | This work | |
| Variable arm | G45:C47I changed to C45:G47I | No effect | (2) |
| G47J changed to A47J | Reduced 2-fold | ||
| Two base pair deletions | Reduced 2-fold | ||
| Randomized for in vitro selection | U48 conserved | (6) | |
| Variants are good substrates | |||
| 47 nt inserted in tRNAser | Conferred leucylation | (5) | |
| Variable domain deleted (ΔVL) | Reduced 125-fold | This work | |
| Anticodon stem–loop and variable domains deleted (ΔACVL1–3) | Reduced significantly | ||
| T-stem–loop | N59 randomized for in vitro selection | No effect | (6) |
| U55 changed to C55 (ΔACVL1) | Aminoacylation abolished | This work | |
| C56 changed to U56 (ΔACVL1) | Aminoacylation abolished | ||
| U55C56 changed to C55U56 (ΔACVL1) | Aminoacylation abolished | ||
| Human (cyt) | |||
| Discriminator base (A73) | U73 | Low efficiency | (4) |
| C73 and G73 | Reduced aminoacylation | ||
| Acceptor stem | C3:G70 inserted into tRNAser | Conferred leucylation 50-fold | |
| A4:U69 inserted into tRNAser | |||
| G5:C68 inserted into tRNAser | |||
| Leucine minihelix | No aminoacylation | (7) | |
| D-stem–loop | Removed C20a | Decreased 30-fold | (24) |
| Variable arm | Variable loop inserted into tRNAser | Conferred leucylation | (24) |
| Variable loop replaced with 5 nt arm | Abolished leucylation | ||
| T-stem-loop | U50:A64 inserted into tRNAser | No effect | (24) |
| G51:C63 inserted into tRNAser | No effect | (24) | |
| Yeast (cyt) | |||
| Discriminator base (A73) | A73 (plus A35 and G37 in anticodon) inserted into tRNAser | Conferred leucylation | (3) |
| Anticodon stem–loop | A35 and G37 | Necessary identity determinants | (3) |
Herein, we sought to obtain a minimal RNA substrate for tRNAleu and thereby further refined our understanding of its identity elements. A previous attempt to construct a microhelix that could be aminoacylated by LeuRS failed (7), presumably because essential identity elements suggested by the results summarized in Table 3 were not present. Our strategy was to construct a series of truncated tRNAleu RNA mimics that lacked entire domains of the RNA tertiary structure. The results showed that an E.coli tRNAleu based leucine minihelix and truncated tRNAleu-ΔACDL mimics that lack the anticodon and D-stem–loop failed to be aminoacylated by LeuRS, regardless of the number of connecting bases that we used to bridge the region between the CCA stem and the anticodon stem. However, the tRNAleu-ΔAC substrate with a 4-nt polyuridine linker replacing the complete anticodon domain was efficiently charged with only a 17-fold decrease in leucylation activity compared with the full-length tRNAleu (Table 2). Likewise, tRNAleu-ΔVL that was missing the extra-long variable loop retained significant leucylation activity compared with full-length tRNAleu. The smallest aminoacylatable tRNAleu mimic that we generated (tRNAleu-ΔACVL) incorporated deletions of both the anticodon and variable stem–loop.
This minimalist RNA was found to include several conformers under non-denaturing conditions, some of which may be the result of dimerization. Multimers, if present, would have unusual RNA architectures. Notably, several large molecules such as viral RNAs or the tmRNA (10Sa RNA) can be charged with an amino acid by AARSs although they do not have a typical tRNA structure (27). In these cases, one portion of the molecule maintains the canonical acceptor stem and TψC-stem–loop domain, while distal appendages do not interfere with aminoacylation. In the case of LeuRS, we propose that as long as D- and T-stem–loop domain interactions are preserved, the extra appendage of the duplex, such as those shown in Figure 5, did not disrupt recognition and aminoacylation. This was supported by thermal denaturing studies and native acid gel analysis, which revealed that the multimer folding and alternate conformations of the tRNA molecules were efficiently aminoacylated by LeuRS.
The minimalist tRNAleu-ΔACVL RNAs were also examined in detail to determine if putative identity elements were still being recognized by LeuRS. Aminoacylation of this RNA was abolished when the discriminator base, A73, was replaced with C73. This result shows that the discriminator base continues to be recognized by LeuRS when the anticodon or variable arms have been deleted. Therefore, it is likely that the local structural environment of this region of the RNA continues to resemble that of the wild-type tRNAleu.
Two base pair interactions between the D- and T-loops that were suggested as possible identity elements (6) have also been investigated in the context of the minimalist RNA. Variants were designed that would alter the critical interactions in the hinge region with minimal disruption to the structure of the tRNA molecule. Thus, we hypothesized that the Watson–Crick geometry of the G19:C56 interaction could be maintained with an isosteric A19:U56 substitution (25). This variant would lack a hydrogen bond involving the exocyclic amino group of the guanosine residue; however, the overall tertiary structure of the RNA would likely remain intact. The other interaction, G18:U55, was more problematic because it was unclear how to choose an equivalent geometrical substitution for this unusual bifurcated pair. We created a tRNAleu-ΔACVL1 variant with an A18:C55 substitution, which seemed to be the most likely substitution to preserve the intermolecular interactions without disrupting the geometry of base pairs. In both cases the changed bases abolished leucylation as did a double variant carrying both changes. This evidence strongly implicates that E.coli LeuRS is sensitive to interactions in the core region of E.coli tRNAleu for aminoacylation, especially the structural conformation of the D-loop and T-loop. It also greatly strengthens the argument that these residues are key identity determinants in the wild-type tRNAleu and continue to be used in the minimalist RNA. Indeed, this recognition paradigm has been identified in cysteine tRNA synthetase (CysRS) (28,29), which is closely related to LeuRS. CysRS relies on tertiary base pair interactions as critical identity elements for aminoacylation (28,29). It is possible that LeuRS recognizes tertiary elements similar to CysRS. However, since we have not examined the structures at high resolution, the alternative explanation of a general structural requirement cannot be entirely excluded.
The excellent aminoacylation levels obtained with the single domain deletions unequivocably demonstrate that LeuRS does not interact or rely on either the anticodon arm or the variable arm. Although we were successful in constructing a minimal RNA, tRNAleu-ΔACVL1, in which both domains were deleted, there was a much greater loss of aminoacylation activity than seen with the individual domain deletions. It was speculated that this loss of activity was due to disruption of the overall structure rather than the loss of an identity element. We therefore constructed alternative variants, which might better preserve structural elements by modifying the junction sequence that replaced the deleted domains. As expected, tRNAleu-ΔACVL2 and tRNAleu-ΔACVL3 (6-nt linker regions) gave a 5–6-fold increase in detectable aminoacylation compared with tRNAleu-ΔACVL1 (4-nt linker region). The RNAs did, however, continue to form a mixture of conformers.
Sampson and Saks (30) created similar types of minimalist molecules with entire domain deletions for E.coli tRNAser. An insignificant decrease in serylation was detected when the anticodon stem–loop domain was removed, because it was not an essential domain for seryl-tRNA synthetase (SerRS) recognition. However, in contrast with our results with tRNAleu, aminoacylation was drastically reduced when the tRNAser extra-long variable arm was removed. The variable loop of human tRNAser has also been shown to be important for aminoacylation by human SerRS using minimized versions of tRNAser (31). Clearly, SerRSs from various species recognize structural elements in the variable arm of tRNAser (30,32–34). Interestingly, but not surprisingly, the synthetases of these two similar class II tRNAs have evolved significantly different specificities for interaction.
The availability of truncated RNAs that could be aminoacylated allowed us to begin mapping RNA regions that may be important or dispensable for hydrolytic editing. We used an editing mutant (T252A) that hydrolyzes correctly charged leucine from leu-tRNAleu to test substrates for post-transfer editing activity. The hydrolytic editing experiments of aminoacylated tRNAleu that lacked either the anticodon stem–loop or variable loop showed similar editing activities to the full-length tRNAleu. Thus, as found for the aminoacylation reaction, the tRNAleu anticodon stem–loop and variable loop are dispensable for RNA dependent editing.
In the case of IleRS, aminoacylation and editing rely on distinct RNA domains and nucleotide determinants to trigger efficient activities. It is possible that further investigation may reveal that the D-loop of tRNAleu is similarly important to editing since the IleRS and LeuRS editing mechanisms are thought to be closely related. Should this be the case, identity determinants for aminoacylation and editing, or at least the required domains, may overlap in contrast to the IleRS system. Resolution of these hypotheses will rely on the ability to stably generate altered tRNA molecules that can then be used as substrates for editing.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr John Abelson for providing the ptDNAleu plasmid. We are grateful to Dr U. Nagaswamy for help with analyzing and interpreting structural data as well as with manuscript preparation. Special acknowledgement goes to Mr R. Mursinna for sharing his purified LeuRS and T252A and Mr T. Lincecum Jr for assistance with RNA purification. This work was supported by NASA grants NTG5-50182 to D.C.L. and NAG5-8140 to G.E.F., as well as Robert A. Welch Foundation grants E-1404 to S.A.M. and E-1451 to G.E.F.
REFERENCES
- 1.Lincecum T.L. Jr and Martinis,S.A. (2002) Leucyl-transfer ribonucleic acid synthetases. In Cusack,S., Franklyn,C. and Ibba,M. (eds), Aminoacyl-tRNA Synthetases, in press.
- 2.Asahara H., Himeno,H., Tamura,K., Hasegawa,T., Watanabe,K. and Shimizu,M. (1993) Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J. Mol. Biol., 231, 219–229. [DOI] [PubMed] [Google Scholar]
- 3.Soma A., Kumagai,R., Nishikawa,K. and Himeno,H. (1996) The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNALeu. J. Mol. Biol., 263, 707–714. [DOI] [PubMed] [Google Scholar]
- 4.Breitschopf K. and Gross,H.J. (1996) The discriminator bases G73 in human tRNASer and A73 in tRNALeu have significantly different roles in the recognition of aminoacyl-tRNA synthetases. Nucleic Acids Res., 24, 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Normanly J., Ollick,T. and Abelson,J. (1992) Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc. Natl Acad. Sci. USA, 89, 5680–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asahara H., Nameki,N. and Hasegawa,T. (1998) In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J. Mol. Biol., 283, 605–618. [DOI] [PubMed] [Google Scholar]
- 7.Metzger A.U., Heckl,M., Willbold,D., Breitschopf,K., RajBhandary,U.L., Rosch,P. and Gross,H.J. (1997) Structural studies on tRNA acceptor stem microhelices: exchange of the discriminator base A73 for G in human tRNALeu switches the acceptor specificity from leucine to serine possibly by decreasing the stability of the terminal G1-C72 base pair. Nucleic Acids Res., 25, 4551–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J.F., Guo,N.N., Li,T., Wang,E.D. and Wang,Y.L. (2000) CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry, 39, 6726–6731. [DOI] [PubMed] [Google Scholar]
- 9.Mursinna R.S., Lincecum,T.L.,Jr and Martinis,S.A. (2001) A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry, 40, 5376–5381. [DOI] [PubMed] [Google Scholar]
- 10.Englisch S., Englisch,U., von der Haar,F. and Cramer,F. (1986) The proofreading of hydroxy analogues of leucine and isoleucine by leucyl-tRNA synthetases from E. coli and yeast. Nucleic Acids Res., 14, 7529–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schimmel P. and Schmidt,E. (1995) Making connections: RNA-dependent amino acid recognition. Trends Biochem. Sci., 20, 1–2. [DOI] [PubMed] [Google Scholar]
- 12.Hale S.P. and Schimmel,P. (1996) Protein synthesis editing by a DNA aptamer. Proc. Natl Acad. Sci. USA, 93, 2755–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grodberg J. and Dunn,J.J. (1988) ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol., 170, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinis S.A. and Fox,G.E. (1997) Non-standard amino acid recognition by Escherichia coli leucyl-tRNA synthetase. Nucleic Acids Symp. Ser., 36, 125–128. [PubMed] [Google Scholar]
- 15.Sampson J.R. and Uhlenbeck,O.C. (1988) Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA, 85, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puglisi J.D. and Tinoco,I.,Jr (1989) Absorbance melting curves of RNA. Methods Enzymol., 180, 304–325. [DOI] [PubMed] [Google Scholar]
- 17.Schreier A.A. and Schimmel,P.R. (1972) Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry, 11, 1582–1589. [DOI] [PubMed] [Google Scholar]
- 18.Martinis S.A. and Schimmel,P. (1992) Enzymatic aminoacylation of sequence-specific RNA minihelices and hybrid duplexes with methionine. Proc. Natl Acad. Sci. USA, 89, 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varshney U., Lee,C.P. and RajBhandary,U.L. (1991) Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem., 266, 24712–24718. [PubMed] [Google Scholar]
- 20.Kim S.H., Suddath,F.L., Quigley,G.J., McPherson,A., Sussman,J.L., Wang,A.H., Seeman,N.C. and Rich,A. (1974) Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science, 185, 435–440. [DOI] [PubMed] [Google Scholar]
- 21.Liu H. and Musier-Forsyth,K. (1994) Escherichia coli proline tRNA synthetase is sensitive to changes in the core region of tRNAPro. Biochemistry, 33, 12708–12714. [DOI] [PubMed] [Google Scholar]
- 22.Beuning P.J., Tessmer,M.R., Baumann,C.G., Kallick,D.A. and Musier-Forsyth,K. (1999) Sequence-dependent conformational differences of small RNAs revealed by native gel electrophoresis. Anal. Biochem., 273, 284–290. [DOI] [PubMed] [Google Scholar]
- 23.Dietrich A., Romby,P., Marechal-Drouard,L., Guillemaut,P. and Giegé,R. (1990) Solution conformation of several free tRNALeu species from bean, yeast and Escherichia coli and interaction of these tRNAs with bean cytoplasmic leucyl-tRNA synthetase. A phosphate alkylation study with ethylnitrosourea. Nucleic Acids Res., 18, 2589–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breitschopf K., Achsel,T., Busch,K. and Gross,H.J. (1995) Identity elements of human tRNALeu: structural requirements for converting human tRNASer into a leucine acceptor in vitro. Nucleic Acids Res., 23, 3633–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaswamy U., Larios-Sanz,M., Hury,J., Collins,S., Zhang,Z., Zhao,Q. and Fox,G.E. (2002) NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res., 30, 395–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinis S.A. and Schimmel,P. (1995) Small RNA oligonucleotide substrates for specific aminoacylations. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. American Society for Microbiology, Washington, DC, pp. 349–370.
- 27.Giegé R., Sissler,M. and Florentz,C. (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res., 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamann C.S. and Hou,Y.M. (1997) An RNA structural determinant for tRNA recognition. Biochemistry, 36, 7967–7972. [DOI] [PubMed] [Google Scholar]
- 29.Hou Y.M., Westhof,E. and Giegé,R. (1993) An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl Acad. Sci. USA, 90, 6776–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampson J.R. and Saks,M.E. (1993) Contributions of discrete tRNASer domains to aminoacylation by E. coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res., 21, 4467–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heckl M., Busch,K. and Gross,H.J. (1998) Minimal tRNA(Ser) and tRNA(Sec) substrates for human seryl-tRNA synthetase: contribution of tRNA domains to serylation and tertiary structure. FEBS Lett., 427, 315–319. [DOI] [PubMed] [Google Scholar]
- 32.Himeno H., Hasegawa,T., Ueda,T., Watanabe,K. and Shimizu,M. (1990) Conversion of aminoacylation specificity from tRNATyr to tRNASerin vitro. Nucleic Acids Res., 18, 6815–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Himeno H., Yoshida,S., Soma,A. and Nishikawa,K. (1997) Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNALeuin vitro. J. Mol. Biol., 268, 704–711. [DOI] [PubMed] [Google Scholar]
- 34.Achsel T. and Gross,H.J. (1993) Identity determinants of human tRNASer: sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J., 12, 3333–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]