Abstract
Mitochondrial DNA (mtDNA) mutations are frequent in cancer, yet their precise role in cancer progression remains debated. To functionally evaluate the impact of mtDNA variants on tumor growth and metastasis, we developed an enhanced cytoplasmic hybrid (cybrid) generation protocol and established isogenic human melanoma cybrid lines with wild-type mtDNA or pathogenic mtDNA mutations with partial or complete loss of mitochondrial oxidative function. Cybrids with homoplasmic levels of pathogenic mtDNA reliably established tumors despite dysfunctional oxidative phosphorylation. However, these mtDNA variants disrupted spontaneous metastasis from primary tumors and reduced the abundance of circulating tumor cells. Migration and invasion of tumor cells were reduced, indicating that entry into circulation is a bottleneck for metastasis amid mtDNA dysfunction. Pathogenic mtDNA did not inhibit organ colonization following intravenous injection. In heteroplasmic cybrid tumors, single-cell analyses revealed selection against pathogenic mtDNA during melanoma growth. Collectively, these findings experimentally demonstrate that functional mtDNA is favored during melanoma growth and supports metastatic entry into the blood.
Pathogenic mtDNA is tolerated in melanoma growth but inhibits metastasis by blocking bloodstream entry.
INTRODUCTION
Pathogenic mutations within mitochondrial DNA (mtDNA) are widely recognized as causative for inherited diseases, yet their role in the pathology of acquired diseases is largely unknown (1–3). Somatic mtDNA mutations commonly occur in human tumors, with an incidence rate greater than 50% (4–6). Certain tumor types such as colorectal, thyroid, and renal cancers exhibit a disproportionately high incidence and allelic burden of deleterious mtDNA mutations (5–9), and are typically associated with “oncocytic” changes secondary to excessive mitochondrial accumulation (10). However, high allelic deleterious mtDNA mutations are atypical in most tumors, and most cancers maintain somatic mtDNA mutations at a low allelic frequency (4–6). In addition, it has been suggested that most cancer types favor functional mtDNA, as indicated by a lower allelic frequency of deleterious mutations relative to nonpathogenic variants (4–6).
Evaluating the impact of somatic mtDNA variants on tumorigenesis is challenged by an inability to precisely edit the mitochondrial genome as current mtDNA editing technologies suffer from a limited scope of editable mutations, off-target editing, and an inability to achieve homoplasmy (11, 12). Previous studies have largely relied on the transplantation of patient mtDNA into mtDNA devoid (ρ0) cells, with most reports using the 143B osteosarcoma cell line (13–19). Two major limitations limit the generality of existing transmitochondrial cytoplasmic hybrid (cybrid) creation protocols (20, 21). First, selection mechanisms are required to remove residual mtDNA donor cells following generation. The frequently used 143B cell line contains a genetic deficiency in thymidine kinase, which confers resistance to bromodeoxyuridine (BrdU), and thus cytotoxic concentrations of BrdU selectively remove residual mtDNA donor cells. Beyond the 143B cybrid system, mtDNA mutations have only been investigated in a few other cancer lines: HeLa, A549, PC3, and murine lung carcinoma (22–26). The generation of these cybrid lines used alternative strategies for removal of contaminating mtDNA donor cells, including inherent resistance to 6-thioguanine treatment, growth in HAT media, or engineered neomycin resistance (24, 25, 27). Second, to remove ρ0 cells that failed to acquire donor mtDNA, pyruvate and uridine nutrients are withdrawn from culture. As these supplements are required to support aspartate and pyrimidine levels during mitochondrial oxidative dysfunction, only cells with residual mitochondrial activity will survive (21, 28–30). As a result, these previous studies are limited to partial loss of mitochondrial oxidative function mtDNA mutations that do not confer pyruvate/uridine auxotrophy. Updated techniques for generating transmitochondrial cells are available; however, they continue to rely on selection mechanisms to isolate cybrid lines (31, 32).
Homoplasmic cybrids produced from these previous generation methods have demonstrated that mtDNA variants can affect tumor progression. However, these studies suffered from a limited number of mtDNA variants and cancer types assessed, a lack of biological replicates, an incompatibility with mtDNA mutations that cause complete loss of oxidative mitochondrial function (due to pyruvate and uridine withdrawal selection), and an inability to establish heteroplasmic pathogenic alleles, a scenario compromising most human cancers. Beyond mtDNA genetics, a number of studies have suggested that mitochondrial activity promotes cancer progression. For example, disseminated cancer cells in melanoma, breast, and renal cancers display increased expression of the mitochondrial biogenesis transcription factor PGC-1α, which promotes mitochondrial mass and oxygen consumption (33–35). In oral squamous cell carcinoma, metastatic cells are observed to enhance mtDNA translation through mt-tRNA modifications (36). From a metabolic standpoint, activity in the tricarboxylic acid (TCA) cycle is elevated in metastatic tumors of triple-negative breast cancer and clear cell renal cell carcinoma when compared to primary tumors (37, 38). Further, pharmacologic inhibition of mitochondrial complex I with IACS-010759 in preclinical models curbed melanoma brain metastases without impeding primary tumor growth (35).
Here, we investigated somatic mtDNA mutations in a cohort of advanced-stage melanomas, observing a rare subset of melanomas with high allele frequency truncating mtDNA mutations. To functionally study the biological consequences of mtDNA mutations, we present a simplified cybrid generation method by incorporating flow cytometric enrichment to enable homoplasmic or heteroplasmic transplantation of mitochondrial genomes into any cell line without requiring selective enrichment dependent on inherent resistances or metabolic deficiencies. We use this method to generate nuclear-isogenic biological replicates for a series of wild-type (WT) and patient-derived dysfunctional mtDNA cybrids in metastatic human melanoma cell lines. In addition to transplanting pathogenic mutations in MT-ND1 and MT-ATP6 genes, we exploit our cybrid generation method for mtDNA derived from a human melanoma with a homoplasmic deletion in MT-CO1, an mtDNA variant not amenable to existing cybrid generation protocols. We characterized the effects of these pathogenic mtDNA alleles on melanoma tumor progression. Despite introduced mtDNA-encoded functional deficits, all homoplasmic cybrid lines invariably established tumors when injected into immunocompromised mice. These mtDNA mutant tumors demonstrated a pronounced reduction in spontaneous metastasis, which we isolated to a specific defect in bloodstream invasion within the tumor interstitial environment. Consistent with this, short-term treatment of mtDNA-functional melanomas with a complex I inhibitor eliminated the presence of circulating tumor cells. We further leveraged our cybrid generation protocol for assessment of selective pressures exerted on the mitochondrial genome in a series of heteroplasmic cybrid lines, revealing that in vivo melanoma growth selects against pathogenic mtDNA variants. These findings provide the first direct in vivo experimental verification of mtDNA selection during tumor growth and demonstrate that functional mtDNA promotes metastatic entry of melanoma cells into the blood.
RESULTS
Somatic mtDNA mutations are a common feature of advanced-stage melanomas
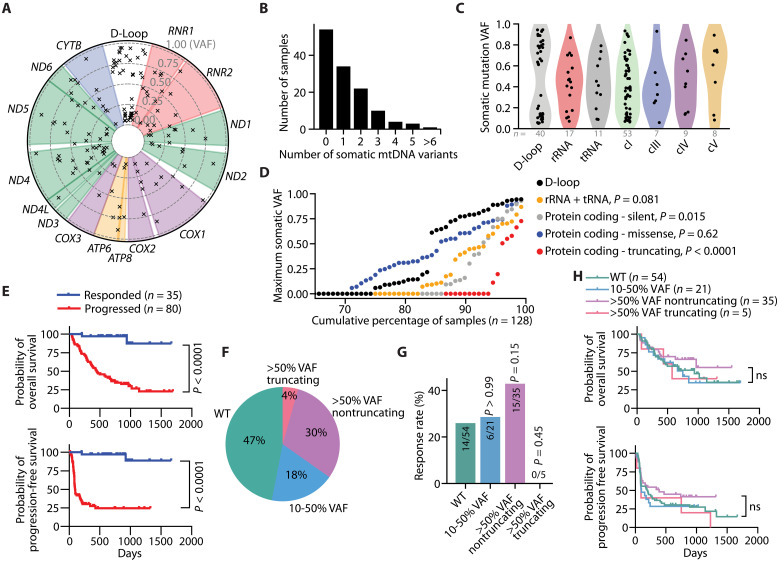
We analyzed the mtDNA status of melanoma and matched healthy tissue samples from a cohort of patients with advanced-stage melanoma (Fig. 1A) (39). Within this cohort, 128 patients exhibited sufficient mtDNA read depth in both normal and melanoma tissues to identify tumor-specific somatic mtDNA mutations (fig. S1, A to E). Notably, 57.8% (74 of 128) of the melanomas displayed at least one detectable tumor somatic mtDNA mutation (Fig. 1B). Most of these mtDNA mutations were C>T or T>C transitions, similar to mtDNA mutational signatures previously identified in other cancers (fig. S2A) (4–6). We identified somatic mtDNA mutations in both noncoding (D-loop) and coding (rRNA, tRNA, cI, cIII, cIV, and cV) regions (Fig. 1, A and C, and data S1), which were present at a variety of allelic frequencies (fig. S2, B to H). Analysis of the maximum variant allele frequency (VAF) for each mtDNA mutational type across the cumulative fraction of samples revealed that noncoding (D-loop) and protein-coding missense mutations were the most prevalent type of mtDNA mutations in individual melanomas (Fig. 1D). Conversely, truncation (protein coding indels and nonsense) mutations were notably infrequent with suppressed VAFs among the melanomas (Fig. 1D).
Fig. 1. Somatic mtDNA variants commonly arise in late-stage melanomas.
(A) Circos plot presenting all somatic mtDNA mutations identified within the melanoma cohort (n = 128 samples). The radial depth of the circos plot, extending from the center to the outer edge, reflects the variant allelic frequency (VAF) of each somatic mutation. (B) The number of somatic mutations identified within each melanoma sample. (C) Variant allele frequencies of somatic mtDNA mutations across different coding regions of mtDNA. (D) The cumulative distribution of the maximum mtDNA variant allele frequency categorized by mutation type. P values reflect comparisons with D-loop. (E) Kaplan-Meier plots showing overall survival (top) and progression-free survival (bottom) in melanomas with sufficient mtDNA read depth coverage. (F) The percentage of samples with annotated clinical outcomes (n = 115 total) for each mtDNA status. The WT group had no detectable somatic mtDNA mutations >0.1 VAF, while the missense and nonsense groups had the described somatic mtDNA mutations >0.5 VAF. (G) The percentage of samples responding to anti-PD1 therapies for each mtDNA status group. (H) Kaplan-Meier plot for overall survival among nonresponders to anti-PD1 therapy across samples with different mtDNA statuses. The number of samples (biological replicates) analyzed per group is indicated. Statistical significance was assessed using Mann-Whitney U test (D), Mantel-Cox (log-rank) [(E) and (H)] or χ2 test (G).
We subsequently explored the impact of mtDNA mutations by analyzing outcomes in samples with annotated clinical data. As expected, overall survival and progression-free survival were significantly associated with immunotherapy response (Fig. 1E) (39). To assess the impact of somatic mtDNA mutations on clinical outcomes, we followed previous analyses (5, 40) and classified samples based on whether they contained somatic mtDNA mutations at 0 to 10% VAF (WT), 10 to 50% VAF, or >50% VAF (Fig. 1F). The >50% VAF group was further divided into samples containing truncating (indel or nonsense mutation) or nontruncating mutations (Fig. 1F). The identified truncating mutations were exclusively in complex I–encoding genes (fig. S3). While there were differences in the anti-PD1 response rate among these groups, they did not rise to statistical significance (Fig. 1G). Further, we did not observe significant differences in overall survival or progression-free survival between these categories (Fig. 1H).
A flow cytometry–based protocol enables transplantation of mitochondrial genomes
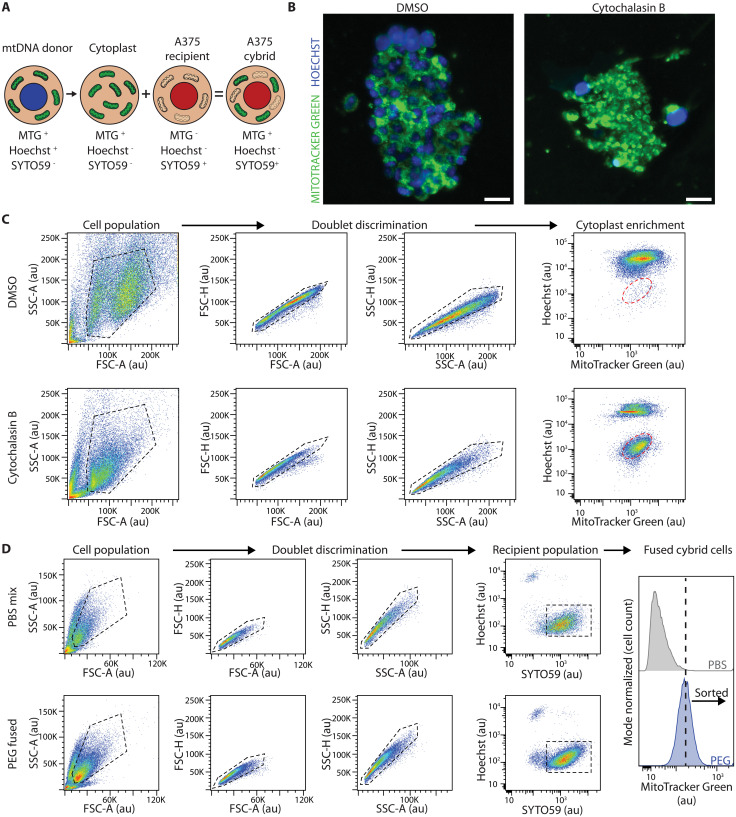
As patient samples and patient-derived tumor grafts and cell lines have differing nuclear genetic backgrounds, we sought to directly evaluate the impact of mtDNA variants on tumorigenesis using nuclear isogenic melanoma lines with mtDNA mutations of known effects. Existing cybrid fusion protocols rely on both small-molecule and metabolic selection to eliminate unfused contaminating donor cells and ρ0 acceptor cells (27, 41). These limitations are due to inefficiencies in both the enucleation and fusion steps and have resulted in most studies being performed in cancer cell lines with engineered or endogenous selection mechanisms. To overcome these limitations and render the protocol more versatile across cell lines, we integrated cellular compartment staining followed by flow cytometry–based enrichment, effectively removing the necessity for antibiotic and metabolic selection (Fig. 2A). Initially, the mitochondria and nuclei of mtDNA donor cells are stained with MitoTracker Green (MTGreen) and Hoechst 33342. Stained donor cells are then incubated with cytochalasin B, an actin polymerization inhibitor, and subjected to a high-speed centrifugation atop a Percoll cushion, thereby generating a mixture of nuclei negative cytoplasts and whole cells (42) (Fig. 2B). Enucleated cytoplasts, identified as the Hoechst−,MTGreen+ population, are enriched through flow cytometry (Fig. 2C). These enriched cytoplasts are subsequently fused with ρ0 cells that have been prestained with the nuclear dye SYTO59 (Fig. 2A). Last, fused cybrid cells, identified as the SYTO59+,MTGreen+,Hoechst− population, are enriched by flow cytometry (Fig. 2D).
Fig. 2. A flow-cytometry protocol to transplant mitochondrial genomes.
(A) Illustration of the cellular fractionation and staining used during generation of A375 cybrids. Donor mtDNA lines were stained with MitoTracker Green (MTG) and Hoechst before cytoplast generation. Cytoplasts were generated, enriched via flow cytometry, and then fused with mtDNA-depleted A375 ρ0 cells that were prestained with SYTO59. Successful cybrid cells were further enriched based on the presence of the SYTO59 signal and MitoTracker Green signal and on the absence of Hoechst signal. (B) Representative epifluorescence images of stained mtDNA donor 143Bnuclear Wild-typemtDNA cells following treatment with DMSO vehicle (left) and cytochalasin B (right) and centrifugation to generate cytoplasts. Scale bar, 25 μm. (C) Enrichment of cytoplasts following differential centrifugation with treatment of DMSO (top) or cytochalasin B (bottom). Cell gating populations are displayed with black dashed lines and sorted cytoplast population is shown with red dashed lines. SSC-A, side scatter area; SSC-H, side scatter height; FSC-A, forward scatter area; FSC-H, forward scatter height; au, arbitrary intensity units. (D) Flow cytometric enrichment of PBS (phosphate-buffered saline)–mixed (top) and PEG (polyethylene glycol)–fused cybrid cells (bottom). Cell gating populations are displayed with black dashed lines. Fusion positive populations were sorted for an enrichment of MitoTracker Green in the PEG-fused cybrid cells relative to the PBS-mixed cells.
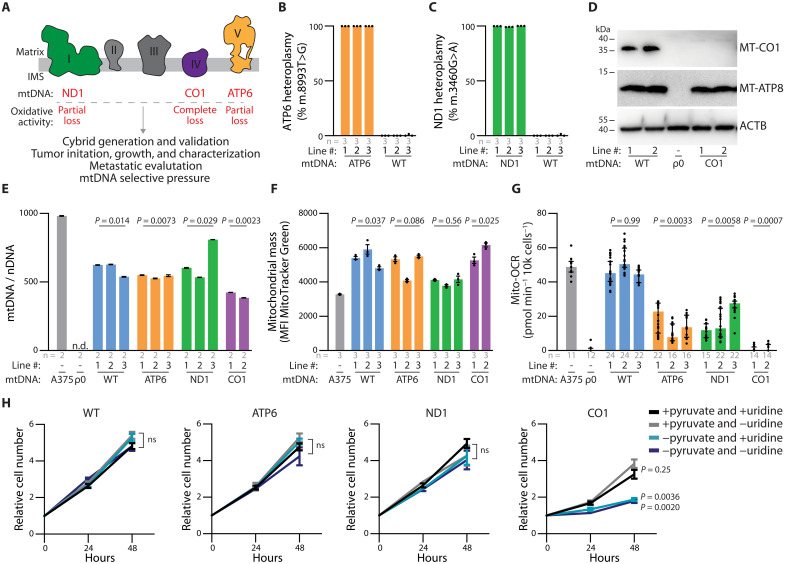
We implemented this protocol to transplant mitochondrial genomes into the A375 human melanoma cell line, enabling a systematic investigation of the impact of mtDNA variants on tumor growth and progression (Fig. 3A). We first generated mtDNA-depleted A375 cells by treating with dideoxycytidine (ddC), an irreversible inhibitor of mtDNA replication (43). Following a 2-week treatment with 5 or 10 μM ddC, we established multiple A375 ρ0 clones in which mtDNA was reduced to undetectable levels (fig. S4A). While the parental line demonstrated functional mitochondrial oxygen consumption, clones from both ddC treatment concentrations exhibited no mitochondrial oxygen consumption (fig. S4, B and C). Clones treated with 5 μM ddC were selected as the ρ0 recipient line and used to generate a panel of A375 homoplasmic cybrids (VAF = 1). These cybrids carried either WT mtDNA (with no pathogenic variants) or pathogenic (partial or complete loss of function) variants associated with human disease (Table 1). We established and validated multiple independently created lines for each cybrid model, ensuring the retention of the A375 nuclear genome (through short tandem repeat analysis) and successful transplantation of the mtDNA genome by Sanger sequencing (fig. S5, A to C). To precisely verify homoplasmic allelic frequency for the partial loss of oxidative mitochondrial function lines, ATP6 and ND1, we used digital droplet polymerase chain reaction (ddPCR) (Fig. 3, B and C, and fig. S5, D to I). The pathologic deletion in the complete loss of oxidative mitochondrial function model, CO1, involves a frameshift within a homopolymeric region, preventing quantitative ddPCR analysis. However, Western blot analysis revealed a loss of MT-CO1 protein expression, while expression of another mtDNA-encoded protein, MT-ATP8, remained intact (Fig. 3D).
Fig. 3. mtDNA pathogenic variants in A375 homoplasmic cybrid lines affect mitochondria ETC activity.
(A) Schematic of electron transport chain with complexes labeled to display subunits with mtDNA variants investigated in this study. (B and C) ddPCR quantitation of allelic fraction for (B) MT-ATP6 mutation m.8993 T>G and (C) MT-ND1 mutation m.3460G>A in homoplasmic cybrid lines. (D) Western blot analysis for the indicated mtDNA-encoded proteins in homoplasmic cybrid cell lines. ACTB levels are shown as loading control. (E) Mitochondrial genome (mtDNA)–to–nuclear genome (nDNA) ratios in the indicated homoplasmic cybrid lines. n.d., not detected. P values reflect comparisons with the A375 parental line. (F) Mitochondrial mass as assessed by flow cytometric analysis of MTGreen staining in the indicated homoplasmic cybrid lines. P values reflect comparisons with the A375 parental line. MFI, mean fluorescence intensity. (G) Mitochondrial oxygen consumption rates (Mito-OCR) in indicated homoplasmic cybrid lines. P values reflect comparisons with the A375 parental line. (H) A375 cybrid in vitro growth rates in culture conditions deplete of pyruvate and/or uridine. P values reflect comparisons with complete media condition (+pyruvate and +uridine) at the 48-hour time point. ns, not significant. The number of samples (biological replicates) analyzed per group is indicated. Data are mean [(B) and (C)], mean ± SEM [(E), (F), and (H)], and median ± interquartile range (G). Statistical significance was assessed using one-way ANOVA with Dunn’s multiple comparison adjustment (D), nested one-way ANOVA with Dunn’s multiple comparison adjustment [(F) and (G)], and two-way ANOVA with Dunn’s multiple comparison adjustment (H).
Table 1. Origin of cybrid donor mtDNA.
| Cybrid | ETC complex | NT change (AA change) | Oxidative activity | Origin of mtDNA |
|---|---|---|---|---|
| WT | – | – | Full activity | Verified healthy donor |
| ATP6 | V | m.8993 T>G (L156R) | Partial | mtDNA disease maternally inherited Leigh syndrome and neuropathy ataxia and retinitis pigmentosa |
| ND1 | I | m.3460G>A (A52T) | Partial | mtDNA disease Leber hereditary optic neuropathy |
| CO1 | IV | m.6692del (truncation) | Absent | Melanoma patient–derived xenograft |
All cybrid lines displayed a restoration of mitochondrial genome content relative to the A375 ρ0 clone, albeit at levels lower than the parental line (Fig. 3E). The mitochondrial mass (based on MTGreen staining) of the cybrid lines was generally elevated relative to the parental line (Fig. 3F). The influence of the mitochondrial genome on oxygen consumption reflected the anticipated functional consequences of the respective pathogenic mtDNA alleles (44, 45). Specifically, the mitochondrial oxygen consumption rate was unchanged between the parental line and the WT cybrids, partially reduced in the ATP6 and ND1 cybrids, and completely ablated in the CO1 cybrids (Fig. 3G). Consistent with these changes, the in vitro growth rates of WT, ATP6, and ND1 cybrids were independent of pyruvate or uridine supplementation, while the CO1 cybrids growth required pyruvate but not uridine supplementation (Fig. 3H).
Tumor growth is sustained in mtDNA dysfunctional cybrids
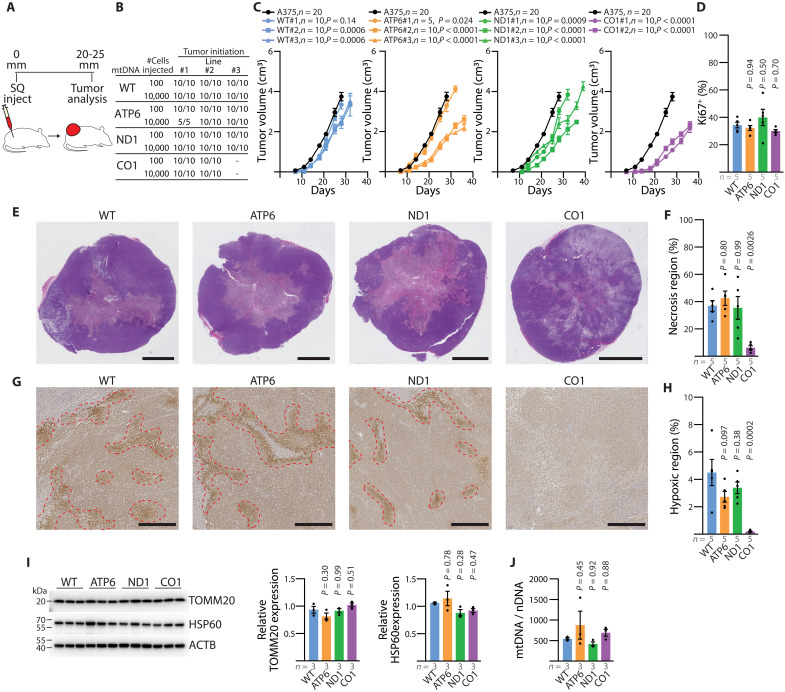
We subcutaneously xenografted each A375 cybrid line into the hind flank of immunocompromised NOD.CB17-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice and monitored tumor growth over time (Fig. 4A). Despite variations in electron transport chain (ETC) capacity, all cybrid lines reliably established tumors at either 100 or 10,000 cell injections (Fig. 4, B and C). Upon tumor harvesting, we confirmed the in vivo stability of the homoplasmic pathogenic mtDNA variants (fig. S5, J to L). Observed growth rates among the subcutaneous cybrid tumors were heterogeneous, with a general reduction in comparison to the parental line—an effect further accentuated in the lines with mtDNA dysfunction (Fig. 4C). Assessment of Ki67 staining across the tumors revealed comparable levels of proliferation among the cybrid lines (Fig. 4D and fig. S6, A to D). Histological analysis indicated substantial areas of tumor necrosis within the WT, ND1, and ATP6 cybrid tumors (Fig. 4, E and F, and fig. S6E). Conversely, the CO1 tumors exhibited decreased tumor necrosis and an increased prevalence of disorganized, or discohesive, regions (Fig. 4, E and F, and fig. S6, E to G).
Fig. 4. Functional mtDNA is dispensable for primary melanoma growth.
(A) Illustration of subcutaneous injection to assess primary tumors of homoplasmic cybrid lines. (B) Frequency of tumor formation for indicated injection counts across homoplasmic cybrid lines. (C) Tumor growth rate of indicated cybrid lines following 10,000 cell subcutaneous injection. Homoplasmic growth data (black circle) are repeated as a reference in all panels. P values reflect comparison with the wild-type (WT) group. (D) Quantitation of the percentage of Ki67+ nuclei across subcutaneous tumors. P values reflect comparison with the WT group. (E) Representative H&E images of subcutaneous tumors. Scale bars, 5000 μm. (F) Quantitation of necrotic region as a percentage of tumor cross-sectional area. P values reflect comparison with the WT group. (G) Representative immunohistochemistry images for pimonidazole staining from subcutaneous tumors of the indicated mtDNA haplotype. Scale bars, 500 μm. (H) Quantitation of pimonidazole positive (hypoxic) regions as a percentage of tumor cross-sectional area. P values reflect comparison with the WT group. (I) Western blot analysis of mitochondrial outer membrane protein TOMM20 and matrix protein HSP60. ACTB expression is shown as loading control. P values reflect comparison with the WT group. (J) Mitochondrial genome content (mtDNA/nDNA) for subcutaneous tumors of the indicated mtDNA haplotype. The number of samples (biological replicates) analyzed per group is indicated. Data are mean ± SEM [(C), (D), (F), (H), (I), and (J)]. Statistical significance was assessed using exponential growth least squares fitting on the mean values of replicates followed by extra sum-of-squares F test with Holm-Sidak’s adjustment (C) and one-way ANOVA with Dunn’s multiple comparison adjustment [(D), (F), (H), (I), and (J)].
We assessed hypoxic regions by pimonidazole staining, which revealed that WT, ATP6, and ND1 tumors contained comparable levels of hypoxia (Fig. 4, G and H). In contrast, no detectable hypoxic regions were observed in CO1 tumors (Fig. 4, G and H). We failed to find significant differences in mitochondrial biomass, assessed by measuring mitochondrial protein expression (TOMM20 on the outer membrane and HSP60 in the matrix), and mitochondrial genome content among the cybrid tumors (Fig. 4, I and J), indicating a lack of oncocytic transformation.
Collectively, these pathogenic mtDNA variants did not preclude the growth of subcutaneous melanoma xenografts, as evidenced by 100% of implants forming tumors, but generally reduced tumor growth rates and affected histological features. To determine whether the molecular and metabolic features of the tumor are affected by mtDNA variants, we transcriptionally and metabolically profiled subcutaneous tumors (data S3 to S7). Hierarchical clustering of differentially expressed genes separated transcriptomes of CO1 tumors from those of WT, ATP6, and ND1 tumors (fig. S7A). Gene set enrichment analysis (GSEA) revealed no significant pathway enrichments between WT and ATP6 tumors (fig. S7B and data S5). A number of hallmark pathways were enriched among differentially expressed genes in ND1 and CO1 tumors relative to WT tumors (fig. S7B and data S5), including hypoxia-related genes that were repressed in CO1 tumors (fig. S7C). Of note, no differential enrichment was identified in the hallmark epithelial-to-mesenchymal transition (EMT) gene set between the WT and mutant cybrid tumors (fig. S7, D to F). Partial least squares discriminant analysis (PLS-DA) of metabolomics profiles also separated CO1 tumors from WT, ATP6, and ND1 tumors (fig. S7G and data S6), and the abundance of a number of metabolites was altered in CO1 tumors (fig. S7H and data S7).
Pathogenic mtDNA variants suppress spontaneous metastasis
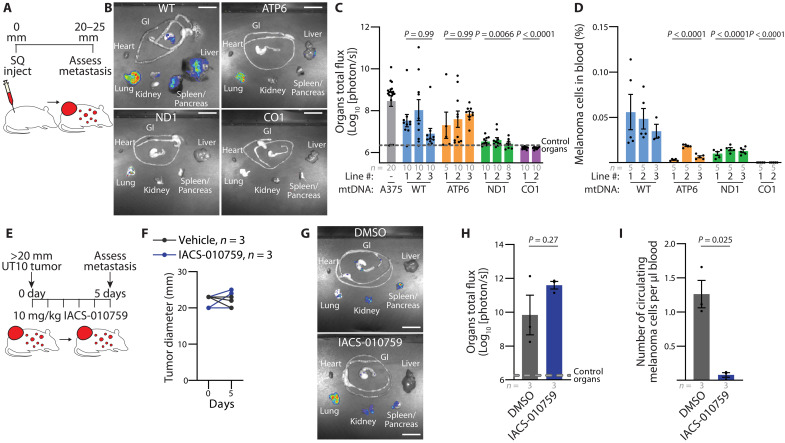
The xenografted A375 cybrid and parental lines were engineered to express luciferase, enabling quantitation of spontaneous metastatic disease burden via bioluminescence imaging of organs. Specifically, the total spontaneous metastatic burden, as measured by bioluminescence of dissected organs, was analyzed when primary subcutaneous tumors attained a size of 20 to 25 mm in diameter (Fig. 5A). Both the WT and ATP6 cybrid tumors exhibited spontaneous metastasis at levels comparable to the parental line (Fig. 5, B and C). In contrast, the ND1 cybrid tumors exhibited a substantial decrease in spontaneous metastatic burden, and no metastasis was detected from the CO1 cybrid tumors (Fig. 5, B and C). Correspondingly, there was a substantial reduction in the frequency of circulating melanoma cells in the blood of all mtDNA mutant cybrid lines (Fig. 5D and fig. S8, A and B).
Fig. 5. Spontaneous metastasis is reduced by dysfunctional mtDNA.
(A) Illustration of subcutaneous injection to assess spontaneous metastasis. (B and C) Representative images (B) and quantitation (C) of metastatic bioluminescence signal of organs from mice with late-stage subcutaneous xenografts of indicated cybrid lines. P values indicate comparisons with the A375 (parental) group. Scale bars, 20 mm. (D) Frequency of circulating melanoma cells from mice (as a percentage of DAPI negative events) with late-stage subcutaneous xenografts of indicated cybrid lines. P values indicate comparisons with the WT group. (E) Illustration of acute pharmacological evaluation of IACS-010759 to assess metastasis in the late-stage PDX model UT10. (F) Tumor diameter of xenografted UT10 mice treated with IACS-010759 or vehicle. (G and H) Representative images (G) and quantitation (H) of organ bioluminescence signal of UT10 xenograft following treatment with IACS-010759 or vehicle. Scale bars, 20 mm. (I) Circulating melanoma cells normalized to total blood volume of xenografted UT10 mice following treatment with IACS-010759 or vehicle. The number of samples (biological replicates) analyzed per group is indicated. Data are mean ± SEM [(C), (D), (H), and (I)]. Statistical significance was assessed using nonparametric Kruskal-Wallis test with Dunn’s multiple comparison adjustment (C), nested one-way ANOVA with Dunn’s multiple comparison adjustment (D), and two-tailed unpaired t test with Welch’s correction [(H) and (I)].
These results indicate that pathogenic mtDNA mutations are well tolerated for primary growth of A375 xenografted tumors but suppress metastatic behavior. To extend these findings, we first investigated the impact of the CO1 mutation in an independent human melanoma cell line (MeWo). After the creation of MeWo ρ0 lines with ddC treatment, we generated and validated MeWo WT and CO1 cybrids using FACS (fluorescence-activated cell sorting)–based enrichment as described above (fig. S9, A and B). Both WT and CO1 cybrids reliably established tumors after transplantation in NSG mice, with growth rates suppressed in the CO1 cybrids (fig. S9C). Histological analysis revealed decreased necrotic regions (fig. S9D), similar to our observations in the A375 background (Fig. 3F). We readily observed metastatic nodules in the livers of mice with WT MeWO tumors, which were suppressed in organs from mice bearing CO1 MeWo tumors (fig. S9E).
We next investigated whether pharmacologic inhibition of mitochondrial ETC function in a tumor with functional mtDNA could suppress the emergence of circulating melanoma cells in the blood. Mice bearing advanced-stage melanoma patient–derived xenograft (PDX) UT10 (46, 47) tumors were subjected to an acute 5-day oral gavage of either IACS-010759 [10 mg/kg; an established bioavailable complex I inhibitor (48)] or 0.5% methylcellulose vehicle control (Fig. 5E). The short-term IACS-010759 treatment did not induce changes in primary tumor size or organ metastatic burden (Fig. 5, F to H). However, IACS-010759 treatment led to a significant decrease in the number of circulating melanoma cells in the blood (Fig. 5I). These findings indicate that either genetic or pharmacologic impairment of mitochondrial ETC activity can inhibit the appearance of melanoma cells in the blood.
Pathogenic mtDNA variants inhibit tumor cell motility and invasion
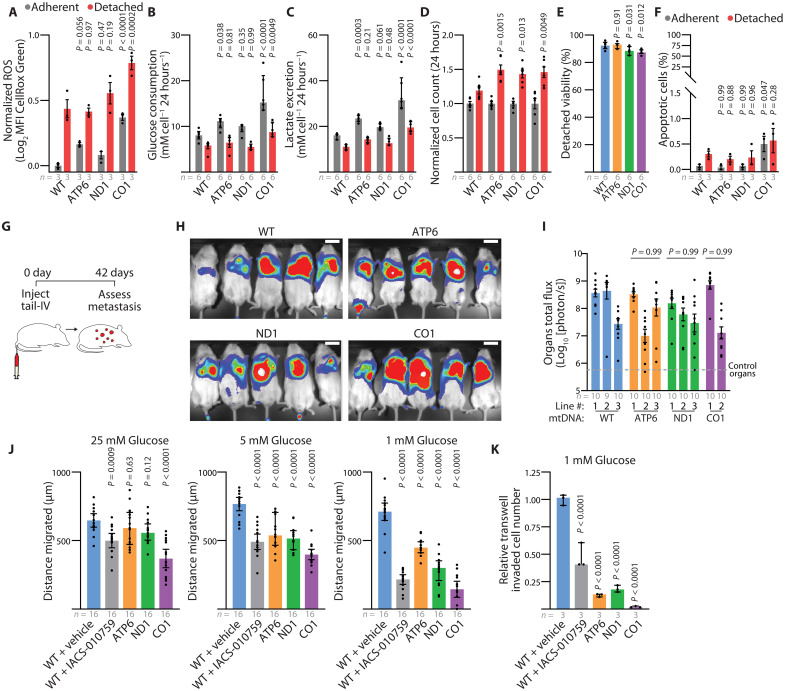
Our transcriptomic analysis from subcutaneous tumors did not identify changes in EMT-related genes (fig. S7, D to F); however, these results do not rule out alternative mechanisms that might limit metastatic properties related to motility and survival after detachment. Considering that the onset of anoikis may limit metastasis (49), we investigated whether the cybrid lines exhibited differential detachment survival potentials. In line with observations of metabolic perturbation induced by detachment (50–53), detached culture of the A375 cybrid lines increased reactive oxygen species (ROS), reduced glucose consumption, and reduced lactate excretion (Fig. 6, A to C). Relative to the WT line, these effects were exacerbated in the CO1 line. Despite these detachment-induced stresses, the mtDNA mutant cybrid lines exhibited significantly elevated cell counts following 24 hours of detached culture relative to the WT line (Fig. 6D). Detachment resulted in only marginal reductions in viability and minimal increases in apoptosis for all cybrid lines (Fig. 6, E and F). To directly investigate metastatic seeding from the bloodstream, we injected 1000 cells of each A375 cybrid line into the tail vein of NSG mice (Fig. 6G). Live bioluminescent imaging showed substantial signal in all cybrid lines, irrespective of ETC capacity (Fig. 6H). Further analysis of dissected organs indicated no significant difference in the total metastatic disease burden, indicating that these mtDNA mutations do not inhibit the ability of melanoma cells to survive detachment or seed metastatic sites following direct bloodstream injections (Fig. 6I).
Fig. 6. Dysfunctional mtDNA disrupts migratory potential in human melanoma.
(A to D) Reactive oxygen species (A), glucose consumption (B), lactate excretion (C), and cell count (D) for indicated cybrid cells after 24-hour adherent and detached culture. P values indicate comparison with WT group for each condition. MFI, mean fluorescence intensity. Same color scheme throughout. (E) Detached viability of indicated cybrid lines following 24 hours of culture. P values indicate comparison with the WT group. (F) Percentage of apoptotic cells following 24-hour culture in adherent or detached conditions. P values indicate comparison with the WT group. (G) Illustration of intravenous injection of cybrid lines to assess organ colonization. (H and I) Representative bioluminescence imaging of live mice (H) and quantitation of metastatic bioluminescence signal of organs (I) following intravenous injection of indicated cybrid lines. P values indicate comparisons with the WT group. Scale bars, 50 mm. (J) Twenty-four–hour wound healing assay quantification for indicated cybrid lines at 25, 5, and 1 mM glucose concentrations. Gap distance was quantified from the difference of the 0- and 24-hour wound gap. P values indicate comparisons with the WT + vehicle group for each glucose concentration. (K) Relative transwell invaded cells at 1 mM glucose concentration. P values indicate comparisons with the WT + vehicle group for each glucose concentration. The number of samples (biological replicates) analyzed per group is indicated. Data are mean ± SEM (A) and median ± interquartile range [(I), (J), and (K)]. Statistical significance was assessed using one-way ANOVA with Dunn’s multiple comparison adjustment (E), nonparametric Kruskal-Wallis test with Dunn’s multiple comparison adjustment (I), two-way ANOVA with Dunn’s multiple comparison adjustment [(A), (B), (C), (D), and (F)], and one-way ANOVA with Dunn’s multiple comparison adjustment [(J) and (K)].
We therefore hypothesized that mtDNA mutations might impede metastatic entry into the blood. The migratory potential of cybrids was examined under various glucose concentrations that correspond to high (25 mM), plasma (5 mM), and tumor interstitial fluid (TIF) (1 mM) levels (54). Under conditions of TIF glucose availability, continuous oxygen consumption analysis revealed a significant increase in the oxygen consumption for the WT, ATP6, and ND1 cybrid lines, indicating that TIF conditions stimulate mitochondrial oxidative activity (fig. S10, A and B). Notably, these differences were not a consequence of altering cellular viability (fig. S10, C to E). Assessment through a scratch-wound assay demonstrated a reduction in migration among the pathogenic mtDNA cybrid lines under conditions simulating TIF glucose availability (Fig. 6J and fig. S10, F and G). Furthermore, a significant decrease in invasion, as evaluated by Boyden transwell assays, was observed in the pathogenic mtDNA cybrid lines under TIF conditions (Fig. 6K and fig. S10H). Pharmacologic inhibition of ETC function with IACS-010759 in the WT cybrid line mirrored the results observed in the mutant mtDNA cybrid lines (Fig. 6, J and K). These findings indicate that these partial and complete loss-of-function mtDNA variants limit migration and invasion under low-glucose conditions similar to the TIF.
Selective pressure during tumor growth favors functional mtDNA
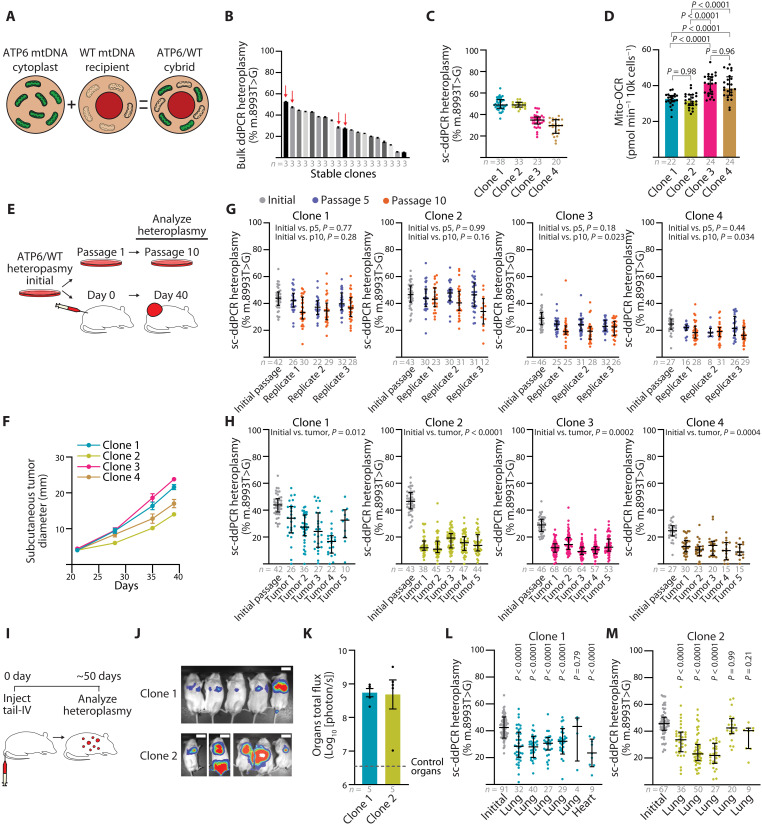
Although sequence analysis of human tumors has suggested that cancers select for WT mitochondrial genomes, our homoplasmic cybrid experiments demonstrate that partial or complete loss of oxidative mitochondrial function mtDNA mutations do not abolish growth of subcutaneous melanoma xenografts. However, these experiments did not address pressures that might restrict the expansion of dysfunctional mtDNA genomes in heteroplasmic scenarios. An advantage of our flow cytometry–based protocol is that it allows the generation of heteroplasmic cybrid lines by skipping the metabolic selection normally used to eliminate unfused recipient (ρ0) cells. We therefore generated A375 heteroplasmic cybrid clones by fusing cytoplasts derived from mtDNA dysfunctional donors into the homoplasmic WT cybrid line (Fig. 7A). Immediately following heteroplasmic cybrid fusion, the MT-ATP6, MT-ND1, and MT-CO1 mutant alleles presented as heteroplasmic with their respective WT alleles (fig. S11, A to C). However, the ND1 and CO1 heteroplasmic lines consistently shifted toward increased VAF of the WT allele, indicating that these heteroplasmic pathogenic alleles were not stable during clonal expansion (fig. S11, D and E). Notably, the MT-ATP6 mutant allele maintained heteroplasmy in culture, as assessed by ddPCR analysis (Fig. 7B). We chose four heteroplasmic ATP6/WT clones for further analysis (Fig. 7B, red arrows). Single-cell analysis of the MT-ATP6 (m.8993 T>G) heteroplasmic frequencies in individual clones demonstrated that these clonal lines contained a distribution of single cells with allelic frequencies centered around the calculated allelic frequencies from bulk analysis (Fig. 7C). We noted that the cybrid lines with a higher m.8993 T>G frequency (~50%, clone 1 and clone 2) exhibited a lower oxygen consumption rate than clones with lower allelic frequency (~30%, clone 3 and clone 4) (Fig. 7D).
Fig. 7. Purifying mtDNA selection is a feature of melanoma growth.
(A) Overview of the ATP6/WT heteroplasmic cybrid generation. (B) Bulk ddPCR analysis of m.8993 T>G heteroplasmic frequency for ATP6/WT cybrids after clonal line establishment. Data from technical replicates are provided. Clones 1 to 4 selected for experiments are indicated with red arrows. (C) Single-cell ddPCR (sc-ddPCR) analysis of m.8993 T>G heteroplasmy for selected ATP6/WT cybrid clones. (D) Mitochondrial oxygen consumption rate of ATP6/WT heteroplasmic clones. (E) Illustration of heteroplasmic selection experiment. Initial cells were passaged for extended in vitro culture or xenografted for subcutaneous tumors in NSG mice. (F) Subcutaneous tumor diameter over time after xenograft of 100 cells from heteroplasmic ATP6/WT clones. (G) Single-cell ddPCR analysis of m.8993 T>G heteroplasmy for ATP6/WT heteroplasmic clones at passage 5 (p5) and passage 10 (p10) of in vitro culture. Three replicates were independently passaged and analyzed for each clone. (H) Single-cell ddPCR analysis of m.8993 T>G heteroplasmy for ATP6/WT heteroplasmic clones of subcutaneous xenograft of 100 cells following tumor growth. (I) Illustration of heteroplasmic selection assay following tail vein intravenous injection of ATP6/WT heteroplasmic clones. (J) Bioluminescence imaging of live mice following intravenous injection of 1000 cybrid cells. Scale bars, 50 mm. (K) Quantification of organ bioluminescence total flux following intravenous injections. (L and M) Single-cell ddPCR analysis of m.8993 T>G heteroplasmy for ATP6/WT heteroplasmic clones’ tumor nodules following intravenous injection. P values reflect comparisons with the initial passage. The number of samples (biological replicates, unless otherwise indicated) analyzed per group is indicated. Data are mean ± SEM [(B) and (F)] and median ± interquartile range [(C), (D), (G), (H), (K), and (L)]. Statistical significance was assessed by one-way ANOVA with Tukey’s multiple comparison adjustment (D). Nested one-way ANOVA with Dunn’s multiple comparison adjustment [(G) and (H)], one-way ANOVA with Dunn’s multiple comparison adjustment (L), and nonparametric Kruskal-Wallis test with Dunn’s multiple comparison adjustment (M).
These four heteroplasmic ATP6/WT clones, two at ~50% m.8993 T>G VAF and two at ~30% m.8993 T>G VAF, were used for concurrent passage in culture and subcutaneous xenografting (Fig. 7E). After subcutaneous xenograft of 100 cells, all clones reached maximal tumor size in ~40 days (Fig. 7F). In culture, there were minimal shifts in the single-cell allelic frequency in these four clones and changes in heteroplasmy were not associated with pyruvate availability (Fig. 7G and fig. S11F). In contrast, we observed that subcutaneous tumors consistently shifted toward increased VAF of the WT allele (Fig. 7H). We observed similar results following subcutaneous injection of 10,000 cells per mouse (fig. S12, A to E). Further, intravenous injection of heteroplasmic cell lines also shifted toward the WT allele in metastatic nodules of visceral organs (Fig. 7, I to M). Therefore, A375 melanoma growth exhibits selection for WT mitochondrial genomes when implanted in mice, irrespective of growth in subcutaneous or visceral space.
DISCUSSION
We identified that isogenic melanoma cybrids transplanted with dysfunctional mitochondrial genomes are capable of sustaining tumor proliferation. The ND1 and CO1 mutant mtDNA cybrid lines displayed a reduction in spontaneous metastatic dissemination and all dysfunctional cybrid lines exhibited a reduction in circulating melanoma cells in the blood. Correspondingly, short-term treatment of severe metastatic disease with IACS-010759 ablated the abundance of melanoma cells within the blood of melanoma PDX UT10. In contrast, when mtDNA mutant cybrid lines were delivered through direct intravenous injections, bypassing the process of metastatic circulation entry, they resulted in uniform metastatic seeding regardless of mtDNA mutational status. These results suggest that ETC dysfunctional subcutaneous tumors fail to disseminate in the blood. Moreover, mutant mtDNA cybrid lines exhibited decreased migration and invasion, particularly at the low glucose availabilities similar to the TIF. Therefore, we propose that limited circulation entry functions as a metastatic bottleneck amidst mtDNA dysfunction.
Genomic sequencing analyses of human tumors have revealed that a small number of tumor types (colorectal/renal/thyroid) exhibit enrichment for loss-of-function mtDNA mutations (5–9). These tumors can exhibit a notable proliferation of mitochondria, resulting in an oncocytic appearance (10). However, we found that dysfunctional mitochondrial genomes were not sufficient to impart oncocytic features in human melanoma, suggesting that tissue specificity and/or alternative alterations drive oncocytoma mitochondrial phenotypes. Further, analysis from pan-cancer mtDNA sequencing studies have suggested that human tumors select against dysfunctional mitochondrial genomes, and here, our results provide the first direct experimental verification of this selective pressure. Inherited mitochondrial diseases often manifest in a state of heteroplasmy; thus, insights into the mechanisms that drive tumor selection for functional mtDNA could potentially unveil innovative treatment strategies for mitochondrial disease.
Before this report, the impact of complete loss of mitochondrial oxidative phosphorylation on tumorigenesis has made use of ρ0 cells, or cells with nuclear deficiencies in complex III genes (55–57). These previous results have indicated that complex III of the mitochondrial ETC is required for tumor growth. We find that complete loss of complex IV function can be compatible with melanoma tumor growth. The m.6692del mutation in MT-CO1 in this paper was derived from a human melanoma PDX model (M405) (45) and has also been reported in human colonic crypts (58, 59), myopathy (60), peripheral blood of a patient with breast cancer (61), and within the PCAWG/TCGA dataset for the following cancer types: bone, breast cancer, prostatic adenocarcinoma, esophageal adenocarcinoma, renal cell carcinoma, glioma, hepatobiliary cancer, and non–small cell lung cancer (5, 6). We previously reported metabolic tracing in the m.6692del M405 PDX model and demonstrated a lack of TCA cycle metabolic activity, as well as minimal metabolic perturbations by treatment with IACS-010759 (45). Here, we build on the effects of m.6692del mutation and demonstrate that these tumors histologically do not present regions of tumor necrosis or hypoxia, yet exhibit a high proportion of discohesive regions. These results indicate that mitochondrial respiration can contribute to tumor necrotic processes, and future studies expanding to more cancer types will be needed to establish the relationship between severe mtDNA impairment and necrosis.
Our heteroplasmic and homoplasmic cybrids robustly establish tumors and, following tail vein injections, proliferate within visceral organs, which confirms that melanomas can tolerate mtDNA mutations. The negative impact of mtDNA mutations on melanoma metastasis contrasts with cybrid studies in other types of cancer, which suggest that mtDNA mutations enhance tumor aggressiveness, potentially via increases in ROS (22, 26, 62–64). In melanomas, elevated ROS levels have been shown to inhibit metastatic behavior in mice (46, 65). Ultimately, these differences may be the result of unique cell-intrinsic factors that modulate the oncogenic response to ROS levels.
Last, the analyzed partial loss of oxidative mitochondrial function cybrid lines, ATP6 and ND1, were derived from well-characterized human mitochondrial diseases. The influence of mitochondrial disease on cancer progression is largely uncharacterized, but will grow in importance as patient survival improves. Within the scope of the studied alleles and cell lines, these findings suggest that mitochondrial disease may not preclude melanoma development but rather attenuate its severity. We note that our findings are currently limited to the specific cell lines tested, and the impact of mtDNA mutations may be dependent not only on the tumor type studied here, but also on the specific oncogenic driver mutations present in these lines. Our flow cytometry–based technique allows cybrid creation of both homoplasmic and heteroplasmic cancer cell lines without the need for engineered or endogenous selection protocols, and will enable similar studies in other cell lines and cancer types to investigate the generality of these results.
MATERIALS AND METHODS
Melanoma and normal tissue mtDNA sequencing and analysis
Clinically annotated late-stage human melanoma whole exome sequencing data from Liu et al. (39) were analyzed to detect mtDNA mutations. These data were previously acquired with informed consent procedures approved by the central Ethics Committee of the University Hospital Essen (12-5152-BO and 11-4715). The sequencing analysis used an in-house pipeline as previously described (66). Briefly, sequencing reads were checked for quality using FastQC 0.11.8 and trimmed and filtered using TrimGalore 0.6.4 to maintain a quality threshold above Q30. These processed reads were then aligned to the Homo sapiens mitochondrial genome (GRCh38) using MapSplice2 2.2.1. After mapping, paired-end reads were further quality filtered using SAMtools 1.9.0. Bam-readcount 0.8.0 was used to assess per-base sequencing depth and to identify point mutations in the quality-filtered mapped reads. Only samples with ≥90% of the mitochondrial genome having a read depth ≥5 in both the normal and tumor sequences were used to call somatic mtDNA mutations. To reduce false-positive results, variant calling (both substitutions and indels) required detection on both the heavy and light strands, with a minimum of four total supporting reads and an allelic frequency ≥5%. For the identification of somatic tumor mutations, corresponding normal samples were analyzed for suspected variants with a lowered detection threshold of one supporting read at any allelic frequency. Consequently, somatic tumor variants were confirmed only in the absence of any evidence of these variants in the healthy tissue.
Experimental models
Immortalized human melanoma cell line A375 (CRL-1619) and MeWo (HTB-65) were obtained from the American Type Culture Collection (ATCC). Melanoma PDX model UT10 was obtained with informed consent according to protocols approved by the Institutional Review Board (IRB) of the University of Texas Southwestern Medical Center (IRB approval 102010-051). Melanoma specimens used in the manuscript are available, either commercially or from the authors, though there are restrictions imposed by IRB requirements and institutional policy on sharing of material from patients.
Immortalized cells were maintained in complete media [Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, 100 μM uridine, and 1% penicillin/streptomycin]. For detached experiments, ultralow attachment surface plates were used (Corning Costar Ultra-Low Attachment Microplates). All cells used in this study were cultivated at 37°C with 5% CO2. The PDX UT10 model was maintained in NOD.CB17-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory, RRID:IMSR_JAX:005557) (details below). Cell culture lines were confirmed as mycoplasma negative using the Universal Mycoplasma Detection Kit (ATCC 30-1012 K). Cell lines were authenticated through small-tandem repeat analysis by the UT Southwestern McDermott Center Sequencing Core facility.
mtDNA depletion and establishment of stable ρ0 lines
Cells were cultured in complete media (DMEM supplemented with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 100 μM uridine, and 1% penicillin/streptomycin) and treated with 5 or 10 μM ddC as indicated. Cells were passaged every 2 days for 2 weeks, at which time serial dilution was used to establish single-cell colonies. The 5 μM clones demonstrated mtDNA depletion and were selected for cybrid generation (see below).
Cybrid generation
Mitochondrial donor cell lines with mtDNA mutations were previous described (44, 45). For cytoplast generation, 1 × 106 to 1 × 107 mitochondrial donors were suspended in 1:1 Percoll (Sigma-Aldrich, 65455-52-9) and cybrid generation buffer [D5030 (Sigma-Aldrich), 25 mM glucose, 2% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 100 μM uridine, and 1% penicillin/streptomycin, pH 7.2–7.4] supplemented with cytochalasin B (10 mg/ml; Cayman Chemical Company, 14930-96-2), 2000 nM MTGreen FM (Invitrogen, M7514), and Hoechst 33342 (40 μg/ml; Thermo Fisher Scientific, H3570). Cells were then subjected to 39,800g centrifugation for 70 min at 37°C. The resulting hazy cellular band located above the Percoll cushion was collected (~5 ml), diluted 10-fold with cybrid generation buffer, and centrifuged at 650g for 10 min at room temperature. The cytoplast containing pellet was resuspended in cybrid generation buffer for FACS isolation of cytoplasts defined as MTGreen+ and Hoechst−.
Recipient cells (either ρ0 cells for homoplasmic cybrids or WT cybrids for heteroplasmic cybrids) were prestained with 100 nM SYTO59 (Invitrogen, S11341) for 30 min in complete media at 5% CO2 and 37°C. Cells were washed 3× with phosphate-buffered saline (PBS), trypsin digested, centrifuged at 300g for 5 min at room temperature, and resuspended in cybrid generation buffer. Cytoplasts isolated via FACS were mixed with recipient cells in a 1:1 ratio. A portion of the mixture (1/10 total volume) was set aside as a nonfusion control for FACS gating. The cell/cytoplast mixture was centrifuged at 300g for 5 min at room temperature and all supernatant was removed. The pellet was softly resuspended and mixed with 100 μl of polyethylene glycol Hybri-Max [polyethylene glycol (PEG)] (Sigma-Aldrich, P7306) for 1 min at room temperature. Slowly and with constant mixing, 37°C cybrid generation buffer (no FBS) was added at a rate of 100 μl the first minute, 200 μl the second minute, 300 μl the third minute, and so on up to the seventh minute. Next, over the course of 2 min, 10 ml of cybrid buffer was added to the cells and the postfusion cell suspension was incubated for 10 min at 37°C. The cells were centrifuged at 200g for 7 min and resuspended in cybrid generation buffer for FACS isolation of cybrid cells. Successfully fused cybrid cells were defined as MTGreen+, Hoechst−, and SYTO59+. Cybrids were directly sorted into complete media and plated for cell culture.
Single-cell digital droplet PCR quantification of mtDNA
Single-cell digital droplet PCR analysis was performed as previously described (67) with the following exceptions: Single melanoma cells were sorted into cell lysis solution {Proteinase K (Fisher BioReagents Proteinase K catalog no. BP1700-100) in 10% [v/v] NP-40 (Thermo Fisher Scientific, FNN0021), 4.5% [v/v] TNES (50 mM tris, 0.4 M NaCl, 100 mM EDTA, and 0.5% SDS)}. Proteinase K digestion was performed at 50°C for 30 min, followed by inactivation at 100°C for 10 min, then cooled to 12°C. Next, the region of interest was minimally amplified through a seven-cycle PCR (Phusion High-Fidelity DNA Polymerase, NEB) surrounding the mtDNA regions for ddPCR detection. Following amplification, 15% of the limited PCR product was input for ddPCR probe analysis (Bio-Rad, QX100 droplet reader) following manufacturer instructions. ddPCR data analysis was performed in QuantaSoft Analysis Pro version 1.0.596. To establish a standard curve, regions of mtDNA were cloned from donor cell lines into p-GEM-T Easy vector (Promega, A1360). Samples were then diluted to specific concentrations and analyzed to evaluate probe specificity.
For detection of the m.8993 T>G mutation within the MT-ATP6 gene, a surrounding region was amplified using forward (GAAACCATCAGCCTACTCATTC) and reverse (GCTTCCAATTAGGTGCATGAG) primers. Fluorescent labeled WT probe (HEX; AGCCCTTGGCCGTACGCC) and mutant probe (FAM; AGCCCGGGCCGTACGC) were used for the detection of each variant.
For detection of the m.3460 G>A mutation within the MT-ND1 gene, a surrounding region was amplified using forward (CAACGTTGTAGGCCCCTAC) and reverse (GAGGGTGATGGTAGATGTGG) primers. Fluorescent labeled WT probe (HEX; TGGCGTCAGCGAAGGG) and mutant probe (FAM; TGGTGTCAGCGAAGGGTTG) were used for the detection of each variant.
Quantitative PCR for mtDNA/nDNA
For DNA extraction, cells were lysed by digestion with proteinase K (Fisher BioReagents Proteinase K catalog no. BP1700-100) in digestion buffer (20 mM tris, 100 mM NaCl, 0.5% SDS, and 10 mM EDTA, pH 7.6) at 44°C overnight. After digestion, the samples were supplemented with additional NaCl to reach a final concentration of 2 mM, enhancing DNA extraction yield. Cellular debris was pelleted via centrifugation at 14,000g for 10 min. Total DNA was isolated from the supernatant via phenol-chloroform extraction and ethanol precipitation as previously described (68, 69). For mtDNA/nDNA measurements, 3 ng of total DNA was input, and samples were analyzed with iTAQ Universal SYBR Green Supermix (Bio-Rad, no. 1725120). Quantitative PCR analysis was used to determine the levels of mtDNA (targeting regions 7773 to 7929) and the nuclear genome for histone H4c, as previously described (70). The following primers were used for detection of mtDNA (forward primer: CCGTCTGAACTATCCTGCCC; reverse primer: GCCGTAGTCGGTGTACTCGT) and nuclear DNA (nDNA) (forward primer: GGGATAACATCCAGGGCATT; reverse primer: CCCTGACGTTTTAGGGCATA).
Extracellular flux assay
Cells were seeded in Seahorse XFe96 cell culture plates overnight in complete growth media. The next day, cells were washed twice with 200 μl of assay medium [DMEM (5030, Sigma-Aldrich) supplemented with 10 mM glucose, 2 mM l-glutamine, 1 mM sodium pyruvate, and 1% penicillin/streptomycin]. Subsequently, 150 μl of the assay medium was added to each well. The cells were then placed in a 37°C, CO2-free incubator for 1 hour. Oxygen consumption measurements were conducted on a Seahorse XFe96 instrument, using a 3-min mix and a 3-min measure cycle. Three measurements were recorded at baseline and after injecting each compound. Inhibitors were sequentially administered at the specified final concentrations: 2 μM oligomycin, 3 μM CCCP (carbonyl cyanide m-chlorophenyl hydrazone), and 3 μM antimycin A. Data collection and analysis were performed using WAVE software (v.2.4.1.1). Upon completion of the experiment, cells were fixed with formalin and stained with DAPI (4′,6-diamidino-2-phenylindole), and cell counts per well were determined using a Celigo imaging cytometer (Nexcelom Bioscience, 5.1.0.0). Mitochondrial OCR (oxygen consumption rate) was calculated as follows: basal (pre-oligomycin) OCR − baseline (post-antimycin) OCR. Negative Mito-OCR values were replaced with zero. OCR values were normalized based on cell count per well.
Analysis of mitochondrial mass
To assess intracellular mitochondrial mass, adherent cells were washed with PBS and incubated for 30 min at 5% CO2 and 37°C in staining medium with 20 nM MTGreen FM (Thermo Fisher Scientific, M7514). Cells were then washed with staining medium and resuspended in DAPI (1 μg/ml; Sigma-Aldrich) to eliminate dead cells from sorts and analyses. Cells were examined on a FACS Fusion Cell Sorter (Becton Dickinson).
Mouse studies and xenograft assays
All mouse experiments complied with all relevant ethical regulations and were performed according to protocols approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center (protocol 2016-101360). For transplantation, cell culture lines were trypsin (T4049, Sigma-Aldrich) digested for 5 min at 37°C to dissociate from adherent cultures followed by room temperature centrifugation at 200g for 3 min. Cells were resuspended, at the desired cell count for injection, in staining media [L15 medium containing bovine serum albumin (1 mg/ml), 1% penicillin/streptomycin, and 10 mM Hepes, pH 7.4] with 25% high-protein Matrigel (354248; BD Biosciences). PDX single-cell suspensions were obtained by mechanical dissociation (12-141-363, Thermo Fisher Scientific) on ice followed by enzymatic digestion in collagenase IV (200 U/ml; Worthington), DNase (50 U/ml), and 5 mM CaCl2 for 20 min at 37°C. Cells were filtered through a 40-μm cell strainer to remove clumps, pelleted at 200g for 5 min at 4°C, and resuspended in staining media. Subcutaneous injections were performed in the right flank of NSG mice in a final volume of 50 μl. Estimated three-dimensional subcutaneous tumor volume was calculated using the formula: [length 1 × (length 22)]/2. Four to eight-week-old male and female NSG mice were transplanted with 100 or 10,000 melanoma cells subcutaneously as indicated. Intravenous injections were done via tail vein injection into NSG mice with 1000 melanoma cells in 100 μl of staining media. Mouse cages were randomized between treatments (mice within the same cage received the same treatment). Subcutaneous tumor diameters were measured weekly with calipers until any tumor in the mouse cage reached 2.5 cm in its largest diameter, in agreement with the approved animal protocol. At that point, all mice in the cage were euthanized and spontaneous metastasis was evaluated by gross inspection of visceral organs for macrometastases and bioluminescence imaging of visceral organs to quantify metastatic disease burden (see details below).
For short-term treatment with IACS-010759 (Chemietek), when subcutaneous tumors reached greater than 2.0 cm in diameter, the mice were administered IACS-010759 or control solution by oral gavage daily for 5 days [10 mg/kg body mass in 100 μl of 0.5% methylcellulose and 4% dimethyl sulfoxide (DMSO), adapted from (45, 48)]. On the fifth day and 2 hours following final oral gavage administration, mice were euthanized and spontaneous metastasis was evaluated.
Bioluminescence imaging
Metastatic disease burden was monitored using bioluminescence imaging [all melanomas were tagged with a bicistronic lentiviral (FUW lentiviral expression construct) carrying dsRed2 and luciferase (dsRed2-P2A-Luc)]. Five minutes before performing luminescence imaging, mice were injected intraperitoneally with 100 μl of PBS containing d-luciferin monopotassium salt (40 mg/ml) (Biosynth) and mice were anesthetized with isoflurane 2 min before imaging. All mice were imaged using an IVIS Imaging System 200 Series (Caliper Life Sciences) with Living Image software. Upon completion of whole-body imaging, mice were euthanized and individual organs were surgically removed and imaged. The exposure time ranged from 10 to 60 s, depending on the maximum signal intensity. To measure the background luminescence, a negative control mouse not transplanted with melanoma cells was imaged. The bioluminescence signal (total photon flux) was quantified with “region of interest” measurement tools in Living Image (PerkinElmer) software. Metastatic disease burden was calculated as observed total photon flux across all organs in xenografted mice.
Cell labeling and flow cytometry for circulating melanoma cells
For analysis of circulating melanoma cells, blood was collected from mice by cardiac puncture with a syringe pretreated with citrate-dextrose solution (Sigma-Aldrich) when subcutaneous tumors reached greater than 2.0 cm in diameter. Red blood cells were sedimented using Ficoll, according to the manufacturer’s instructions (Ficoll Paque Plus, GE Healthcare). The remaining cells were washed with Hanks’ balanced salt solution (Invitrogen) before antibody staining and flow cytometry. Melanoma cells were identified via flow cytometry as previously described (46, 47). All antibody staining was performed on ice for 20 min. Cells were stained with directly conjugated antibodies against mouse CD45 (violetFluor 450, eBiosciences), mouse CD31 (390-eFluor450, BioLegend), mouse Ter119 (eFluor450, eBiosciences), and human leukocyte antigen (HLA)-A, B, C (G46-2.6-FITC, BD Biosciences). Cells were washed with staining medium and resuspended in staining media supplemented with DAPI (1 μg/ml; Sigma-Aldrich) to eliminate dead cells from analyses. Human melanoma cells were isolated as events positive for HLA and negative for mouse endothelial and hematopoietic markers.
Continuous oxygen consumption assay
Thirty-thousand cells were seeded in each well of a 96-well plate in complete growth media and cultured overnight to form a monolayer. The next day, cells were washed twice with PBS and replaced with DMEM (5030, Sigma-Aldrich) supplemented with 1 or 25 mM glucose, 2 mM l-glutamine, and 1% penicillin/streptomycin. Continuous oxygen consumption was monitored for 6 hours with a Resipher instrument (Lucid Technologies Inc.) and analyzed with Lucid Labs Software. OCR values were normalized based on final cell count per well.
Histology
Tumors were dissected, fixed in 10% neutral buffered formalin for 48 hours, and paraffin embedded. For pimonidazole staining, 3 hours before collection, mice received an intraperitoneal injection of pimonidazole (60 mg/kg) dissolved in saline. For pimonidazole detection, 4.3.11.3 mouse FITC-Mab (Hypoxiaprobe, 1:100) was used in combination with mouse on mouse blocking immunoglobulin G (IgG) reagent (Vector, MKB-2213-1). The following antibodies were used for Ki67 staining: Ki67 (ab15580, Abcam, 1:100, RRID:AB_443209) and anti-rabbit IgG-biotin conjugated (Vector Laboratories, BA-1000-1.5, 1:500, RRID:AB_2313606). Five-micrometer serial sections were used for hematoxylin and eosin (H&E) staining, Ki67 staining, and pimonidazole staining. Slides were scanned at 40× using a Nanozoomer 2.0HT (Hamamatsu) at the UTSW Whole Brain Microscopy Facility. QuPath (71) was used to quantify all histological sections. Quantification of H&E structural regions was performed through training the QuPath pixel detection tool on annotated regions. Ki67-stained nuclei were detected using the Positive Cell Detection tool in QuPath. Pimonidazole-positive regions were detected by applying a threshold in ImageJ (72).
RNA sequencing analysis
RNA isolation was carried out using the RNEasy Kit in accordance with the manufacturer’s guidelines (Qiagen, Germany). To assess the concentration and quality of RNA, Qubit (Invitrogen, Waltham, MA, USA) and Agilent Bioanalyzer RNA nano chips (Agilent, Santa Clara, CA, USA) were used. Subsequent to this, library preparation was conducted using Lexogen’s QuantSeq 3′ mRNA-seq Library Prep Kit FWD, and sequencing took place on a NextSeq2000 (Illumina, San Diego, CA, USA). The acquired sequences underwent trimming using TrimGalore (v.0.6.0 DOI:10.14806/ej.17.1.200) and alignment with HISAT2 (73) to the GRCh38 reference genome. DESeq2 v1.42.0 R package (74) was used to identify differentially expressed genes across the four experimental conditions (WT, ATP6, ND1, and CO1). Raw read counts were normalized, and differential expression analysis was performed using the likelihood ratio test (LRT), with the WT condition set as the reference. Genes with adjusted P values below 0.05 were considered significantly differentially expressed. GSEA (75, 76) was conducted using the fgsea v1.28.0 R package (77), with hallmark pathways retrieved with the msigdbr v.7.5.1 R package (DOI: 10.18129/B9.bioc.msigdb). Log2 fold change values were ranked and analyzed against predefined gene sets, with pathway enrichment results filtered by adjusted P value (q value <0.25) to identify significant pathways.
Metabolomics analysis
Chemicals
Liquid chromatography–mass spectrometry (LC-MS) grade acetonitrile and methanol were purchased from VWR International (Darmstadt, Germany). LC-MS grade acetic acid and ammonium acetate were purchased from Sigma-Aldrich (Steinheim, Germany). Isotopically labeled compounds, including 13C6-l-arginine hydrochloride (Cambridge Isotope Laboratories, Tewksbury, MA, USA), 13C5-l-valine, 13C2-citric acid, 2H4-succinic acid (Sigma-Aldrich, Steinheim, Germany), and 13C6-fructose-6-phosphate disodium salt hydrate (Omicron Biochemicals Inc., South Bend, USA), were used for correction of suspected target data. Nanosep centrifugal devices with omega membrane were obtained from Thermo Fisher Scientific GmbH (Schwerte, Germany). Ultrapure and desalted water with (18.2 megohms/cm resistivity) was generated by a Sartorius Stedim water purification system (Sartorius, Goettingen, Germany). All other chemicals were purchased by a local distributor in the highest possible grade.
Extraction of metabolites
Metabolites were extracted by a two-step liquid extraction adapted as described before (78). Briefly, 10 to 25 mg of frozen tissue and 500 μl of prechilled methanol (−80°C) were spiked with 25 μl of internal standard (13C6-l-arginine, 13C5-l-valine, 13C2-citric acid, 2H4-succinic acid, and 13C6-fructose-6-phosphate, each 100 μM). The samples were homogenized using a tissue slicer and vortexed for 2 min. Subsequently, the samples were sonicated for 2 min in a chilled (0°C) ultrasonic bath followed by a freezing/thawing cycle at −80°C for 5 min. The thawed samples were vortexed and sonicated for 2 min and centrifuged at 3000g for 5 min. The supernatant was collected and 250 μl of water acidified with 0.1% acetic acid (LC-MS grade) was added. The samples were vortexed and sonicated for 2 min and subjected to another freezing/thawing cycle. The samples were vortexed, sonicated for 2 min, and centrifuged at 3000g for 5 min before the supernatant was collected. The combined supernatants were completely dried in a vacuum centrifuge for 45 min. The dried extracts were resuspended in 250 μl of acetonitrile/water (60/40; v/v) by sonication and vortexed for 2 min. The samples were centrifuged at 14,000 rpm for 5 min and filtered using Nanosep 3 K Omega centrifugal filters. QC samples were generated by pooling 25 μl of each extract. All samples were analyzed in a randomized order with five QC samples after every eighth sample.
LC-MS analysis and quantification
Chromatography was performed using an Agilent 1290 Infinity II Bio LC system (Agilent Technologies Inc., Waldbronn, Germany) coupled to a Thermo Orbitrap Q Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). An AdvanceBio MS Spent Media column (150 × 2.1 mm, 2.7 μm; Agilent Technologies Inc., Waldbronn, Germany) was used for HILIC separation of the metabolites. The elution gradient used water (10 mM ammonium acetate, pH 9) as solvent A and acetonitrile/water (95/5; v/v; 10 mM ammonium acetate, pH 9) as solvent B, at a flow rate of 450 μl/min. The linear gradient was as follows: 0 min, 100% B; 0.5 min, 100% B; 6.5 min, 50% B; 7 min, 50% B; 7.01 min, 100% B. The column temperature was set at 70°C, and the injection volume was 1 μl. Ionization was performed in electrospray ionization negative and positive modes, and MS parameters were optimized for high-resolution scans (MS1) and data-dependent MS2. The spray voltage was 3.5 kV, with S-lens RF level 40, MS inlet capillary 300°C, and aux gas 350°C. The MS1 acquisition was performed in a scan range from 70 to 1050 mass/charge ratio (m/z) at a resolution of 35,000 at m/z 200. MS parameters for MS1 were 1 microscan, the default charge 1, AGC target 1 × 106, max. IT 50 ms, and chromatographic peak width 12 s. Data-dependent MS2 has been used with a resolution of 17,500 at m/z 200. MS2 AGC and AGC target were 8 × 103 and 2 × 105, respectively and max It was 100 ms. A stepped NCE (30, 50, 70) was used for fragmentation with an isolation window of 1 m/z and a loop count of 10. Dynamic exclusion of 4 s isotope exclusion was applied. Internal lock masses were 391.2842 m/z for positive mode and 265.1476 m/z for negative mode.
Data analysis
Data analysis was carried out by a suspect target and a nontarget approach. For the analysis of suspect target data, Skyline Daily was used to generate extracted ion chromatograms based on the known m/z, fragmentation pattern by MS/MS, and retention times for metabolites. Peaks were automatically integrated by Skyline and manually checked. Nontarget data analysis was performed using MSDial 4.90, which includes a series of automated steps (e.g., peak detection, identification, alignment, or gap filling). Identification was performed by matching the measured MS/MS with the spectral library provided by MSDial (“ESI(−)-MS/MS from standards + bio + in silico” and “ESI(+)-MS/MS from standards + bio + in silico” [https://systemsomicslab.github.io/compms/msdial/main.html#MSP (accessed date December 14, 2023)]. The resulting data were filtered based on an existing database match, an RSD of less than 25% in the quality control (QC) samples, and no missing values in the QC. Data were analyzed by PLS-DA using the mixOmics package v6.26.0 in R, with eight components and the defined sample groups as the response variable (79).
Metabolic glucose and lactate consumption and excretion assay
Cells were cultured in adherent culture conditions or detached conditions (Thermo Fisher Scientific, 07-200-601) for 24 hours in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 1% penicillin/streptomycin. Glucose and lactate were measured in culture medium using an automated electrochemical analyzer (BioProfile Basic-4 analyzer, NOVA).
Migration and invasion assays
Wound healing migration assay was performed as previously described (80). Briefly, cells were seeded to form a monolayer and subjected to overnight serum starvation. Scratches were created with a sterile p200 tip and wells were washed with PBS to remove detached cells. DMEM (no glucose) supplemented with 10% FBS, 2 mM l-glutamine, 1% penicillin/streptomycin, the appropriate concentration of glucose (1, 5, or 25 mM), and 100 nM IACS-010759 or DMSO vehicle was added to wells. To prevent consumption of all available glucose at lower availabilities, the medium was replaced every 8 hours. Images were taken at 0- and 24-hour time points with a Celigo image cytometer-4 Channel software version 5.1.0 (Nexcelom Bioscience). The difference in gap length between the 0- and 24-hour time points was reported as distance migrated.
Boyden transwell assay invasion assay was performed as previously described (80). Briefly, cells were seeded in DMEM (no glucose) supplemented with 1 mM glucose, 2 mM l-glutamine, 1% penicillin/streptomycin, and 100 nM IACS-010759 or DMSO vehicle and were added to wells. Following a 6-hour serum starvation, cells were trypsin digested, pelleted at 300g, and washed in the appropriate glucose concentration DMEM, and 50,000 cells per well were plated into the upper chamber of a transwell insert with 8 μm pore size (Celltreat, 2306439). The bottom chamber of the transwell was filled with DMEM (no glucose) supplemented with 10% FBS, 1 mM glucose, 2 mM l-glutamine, 1% penicillin/streptomycin, and 100 nM IACS-010759 or DMSO vehicle. To prevent consumption of all available glucose, the medium was replaced every 8 hours. Following 24 hours, the insert was transferred to a PBS wash and a cotton swab (Q-tip) dipped in PBS was used to remove nonmigrated cells from the upper chamber. The cells were fixed in 10% buffered formalin for 20 min, followed by a 20-min stain in 0.1% crystal violet solution (0.1% crystal in 10% ethanol). The inserts were washed 3× with PBS and the upper chamber was cleaned again with a Q-tip. The inserts were allowed to dry for several hours, after which the membrane was cut and imaged with Primovert ZEISS microscope on a 10× objective. All images were recorded with ZEN 3.1 (Blue ed) software and analyzed with ImageJ.
Analysis of adherent and detached ROS, viability, and apoptosis
Cells were lifted from adherent passage with PBS supplemented with 1 mM EDTA and 1% FBS. Cells were cultured in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 1% penicillin/streptomycin for 24 hours in adherent conditions or detached (Ultra-Low Attachment Microplates, Thermo Fisher Scientific, 07-200-601) conditions.
For ROS measurements, both conditions were washed with PBS; adherent cells were directly washed with PBS and detached cells were centrifuged at 200g for 3 min and resuspended with PBS. Cells were incubated for 30 min at 5% CO2 and 37°C in staining medium [L15 medium containing bovine serum albumin (1 mg ml−1), 1% penicillin/streptomycin, and 10 mM Hepes (pH 7.4)] with 5 μm CellROX Green (Thermo Fisher Scientific, C10444). Cells were washed with staining medium and resuspended in DAPI (1 μg/ml; Sigma-Aldrich) to eliminate dead cells from sorts and analyses. Cells were examined on a FACS Fusion Cell Sorter (Becton Dickinson).
For detached viability, cells were collected and spun down at 200g for 4 min. Cell viability was determined by the percentage of cells that were trypan blue positive (Sigma-Aldrich). For apoptosis analysis, the Apo-Direct TUNEL assay kit (MilliporeSigma) was used following the manufacturer’s protocol, and cells were examined on a FACS Fusion Cell Sorter (Becton Dickinson). For adherent monolayer viability, cells were incubated with DAPI (1 μg/ml) for 20 min at 5% CO2 and 37°C. Afterward, DAPI-positive cell counts were determined using a Celigo image cytometer-4 Channel software version 5.1.0 (Nexcelom Bioscience). For total cell counts, cells were fixed with 10% buffered formalin overnight, followed by staining with DAPI (1 μg/ml) in PBS for 30 min and total cell counts were determined using a Celigo image cytometer-4 Channel software version 5.1.0 (Nexcelom Bioscience).
Western blot analysis
Tumors were excised and rapidly snap frozen in liquid nitrogen. Tumor lysates were prepared by mincing tissue in radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, 89901) supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) and maintained in constant agitation for 2 hours at 4°C. For cells, lysis buffer was added to the dish and cells were scrapped on ice before constant agitation for 30 min at 4°C. Lysates were spun down at 12,000g at 4°C for 10 min. The DC protein assay kit II (Bio-Rad) was used to quantify protein concentrations. Equal amounts of protein (5 μg) were loaded into each lane and separated on 4 to 20% polyacrylamide tris glycine SDS gels (Bio-Rad), then transferred to 0.45-μm PVDF membranes (MilliporeSigma). The membranes were blocked for 1 hour at room temperature with 5% milk in TBS supplemented with 0.1% Tween 20 (TBST) and then incubated with primary antibodies overnight at 4°C. After washing and incubating with horseradish peroxidase–conjugated secondary antibody (Cell Signaling Technology), signals were developed using Immobilon Western Chemiluminescent HRP Substrate (MilliporeSigma). Blot data collection was performed using Amersham ImageQuant 800. Blots were sometimes stripped using Restore PLUS stripping buffer (Thermo Fisher Scientific) and restained with other primary antibodies. The following antibodies were used for Western blots: anti-mtCO1 (ab14705, Abcam, RRID:AB_2084810), anti-ATP8 (26723-1-AP, Proteintech, RRID:AB_2880614), anti-HSP60 (15282-1-AP, Proteintech, RRID:AB_2121440), anti-TOMM20 (11802-1-AP, Proteintech), and anti–β-actin (4970, Cell Signaling, RRID:AB_2223172).
Statistical analysis
Mice were allocated to experiments randomly and samples were processed in an arbitrary order, but formal randomization techniques were not used. Sample sizes were not predetermined based on statistical power calculations but were based on our experience with these assays. For assays in which variability is commonly high, we typically used n > 10. For assays in which variability is commonly low, we typically used n < 10. All data representation is indicated in the figure legend of each figure. No blinding or masking of samples was performed. All represented data are unique biological replicates unless indicated otherwise.
Before analyzing the statistical significance of differences among treatments, we tested whether the data were normally distributed and whether variance was similar among treatments. To test for normal distribution, we performed the Shapiro-Wilk test. To test whether variability significantly differed among treatments, we performed F tests. When the data significantly deviated from normality or variability significantly differed among treatments, we log2 transformed the data and tested again for normality and variability. Fold change data were log2 transformed. If the transformed data no longer significantly deviated from normality and equal variability, we performed parametric tests on the transformed data. If the transformed data remained significantly deviated from normality or equal variability, we performed nonparametric tests on the nontransformed data. For normally distributed data, groups were compared using the two-tailed Student’s t test (for two groups), or one-way analysis of variance (ANOVA) or two-way ANOVA (>2 groups), followed by Dunnett’s or Tukey’s test for multiple comparisons. For data that were not normally distributed, we used nonparametric testing (Kruskal-Wallis test for multiple groups), followed by Dunn’s multiple comparisons adjustment. All statistical analyses were performed with GraphPad Prism 9.5.1 or in R 4.0.2.
Acknowledgments
We thank the Moody Foundation Flow Cytometry Facility, the Children’s Research Institute Next Generation Sequencing Facility, the UT Southwestern Histo Pathology Core, the UT Southwestern Whole Brain Microscopy Facility, and the UT Southwestern BioHPC supercomputing facility. We thank M. Mulkey for mouse colony management. We would like to acknowledge the Quantitative Light Microscopy Core and the Tissue Management Core, a Shared Resource of the Harold C. Simmons Cancer Center, supported in part by an NCI Cancer Center Support Grant, 1P30 CA142543-01. We thank B. Budeus and the team from the Essen University Hospital Genomics & Transcriptomics Facility for RNA sequencing.
Funding: This research was supported by the Cancer Prevention and Research Institute of Texas (RP180778 to S.J.M., R.J.D., and P.M.), the National Institutes of Health (1DP2ES030449-01 from NIEHS to P.M.; 1R01AR073217-01 from NIAMS to P.M.; 1F30DK137407 from NIDDK to C.J.M.; R35CA22044901 from NCI to R.J.D.; K08CA279757 from NCI to J.G.G.; U01CA228608 from NCI to S.J.M.), the UT Southwestern KCP SPORE CEP to P.M. (NIH/NCI P50CA196516), the HHMI Investigator Program (to R.J.D. and S.J.M.), and the Moody Medical Research Institute (research grant to P.M.). S.D.S. was supported by the National Science Foundation (GRF 2019281210), and A.T. was supported by an Emmy Noether Award from the German Research Foundation (DFG, 467788900), the Ministry of Culture and Science of the State of North Rhine-Westphalia (NRW-Nachwuchsgruppenprogramm), and a European Research Council (ERC) Starting Grant (METATARGET, ERC101078355). A.T. holds the Peter Hans Hofschneider of Molecular Medicine endowed professorship by the Stiftung Experimentelle Biomedizin. F.R. is funded by Melanoma Research Alliance and the Wolfgang & Gertrud Boettcher Foundation. D.S. receives funding via institutional grants from Amgen, BMS, MSD, and Pfizer.
Author contributions: Conceptualization: S.D.S., X.W., S.J.M., A.T., and P.M. Methodology: S.D.S., Z.C., X.W., S.S.K.V., S.W.M., S.J.M., A.T., and P.M. Software: P.Z., Z.Z., and P.M. Validation: S.D.S., X.W., J.K., A.T., and P.M. Formal analysis: S.D.S., S.W.M., P.Z., F.R., Z.Z., R.J.D., and P.M. Investigation: S.D.S., S.H., L.M.N.M., V.R., X.W., C.B.L., C.J.M., G.A., J.K., J.R., D.S., and A.T. Resources: S.D.S., L.M.N.M., T.W., X.W., S.S.K.V., D.S., J.G.G., S.J.M., A.T., and P.M. Data curation: S.D.S., S.W.M., A.T., and P.M. Writing—original draft: S.D.S., A.T., and P.M. Writing—review and editing: S.D.S., T.W., D.S., R.J.D., S.J.M., A.T., and P.M. Visualization: S.D.S., A.T., and P.M. Supervision: S.D.S., S.W.M., D.S., S.J.M., A.T., and P.M. Project administration: S.D.S., X.W., A.T., and P.M. Funding acquisition: D.S., S.J.M., A.T., and P.M.
Competing interests: D.S. receives consulting fees from Philogen, lnFlarX, Neracare, Merck Sharp & Dohme, Novartis, Bristol Myers Squibb, Pfizer, Pierre Fabre, Replimune, SunPharma, Daiichi Sanyo, Astra Zeneca, IQVIA, LabCorp, UltimoVacs, Seagen, Immunocore, Immatics, BioNTech, PamGene, BioAlta, Regeneron, Agenus, Erasca, Formycon, NoviGenix, CureVac, and Sanofi; received speakers’ honoraria and/or travel support from Neracare, Merck Sharp & Dohme, Novartis, Bristol Myers Squibb, Pierre Fabre, Replimune, SunPharma, and Sanofi; and is a member or chair of the following advisory or steering boards: EORTC-MG, DeCOG, NVKH, and CCC. R.J.D. is a founder and advisor at Atavistik Bioscience, and an advisor at Agios Pharmaceuticals, Vida Ventures, Droia Ventures, and Faeth Therapeutics. S.J.M. is a founder of Garuda Therapeutics and an advisor at Kojin Therapeutics, Inception Therapeutics, and Conception Biosciences. P.M. is a member of the scientific and medical advisory board for the United Mitochondrial Disease Foundation. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Human melanoma sequencing datasets are available at dbGAP (accession number phs000452.v3.p1). Code used for mtDNA mapping is available at https://doi.org/10.5281/zenodo.10480633. RNA sequencing datasets are available at the NCBI GEO under accession GSE276027. The mtDNA cybrid cell lines can be provided by P.M. pending scientific review and a completed material transfer agreement. Requests for mtDNA cybrid cell lines should be submitted to UTSW Material requests: CopperativeResearch@utsouthwestern.edu.
Supplementary Materials
The PDF file includes:
Figs. S1 to S12
Legends for data S1 to S7
Other Supplementary Material for this manuscript includes the following:
Data S1 to S7
REFERENCES AND NOTES
- 1.Holt I. J., Harding A. E., Morgan-Hughes J. A., Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Tuppen H. A., Blakely E. L., Turnbull D. M., Taylor R. W., Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta 1797, 113–128 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Gorman G. S., Chinnery P. F., DiMauro S., Hirano M., Koga Y., McFarland R., Suomalainen A., Thorburn D. R., Zeviani M., Turnbull D. M., Mitochondrial diseases. Nat. Rev. Dis. Primers. 2, 16080 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ju Y. S., Alexandrov L. B., Gerstung M., Martincorena I., Nik-Zainal S., Ramakrishna M., Davies H. R., Papaemmanuil E., Gundem G., Shlien A., Bolli N., Behjati S., Tarpey P. S., Nangalia J., Massie C. E., Butler A. P., Teague J. W., Vassiliou G. S., Green A. R., Du M.-Q., Unnikrishnan A., Pimanda J. E., Teh B. T., Munshi N., Greaves M., Vyas P., El-Naggar A. K., Santarius T., Collins V. P., Grundy R., Taylor J. A., Hayes D. N., Malkin D.; ICGC Breast Cancer Group; ICGC Chronic Myeloid Disorders Group; ICGC Prostate Cancer Group, Foster C. S., Warren A. Y., Whitaker H. C., Brewer D., Eeles R., Cooper C., Neal D., Visakorpi T., Isaacs W. B., Bova G. S., Flanagan A. M., Futreal P. A., Lynch A. G., Chinnery P. F., Dermott U. M., Stratton M. R., Campbell P. J., Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife 3, e02935 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorelick A. N., Kim M., Chatila W. K., La K., Hakimi A. A., Berger M. F., Taylor B. S., Gammage P. A., Reznik E., Respiratory complex and tissue lineage drive recurrent mutations in tumour mtDNA. Nat. Metab. 3, 558–570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Y., Ju Y. S., Kim Y., Li J., Wang Y., Yoon C. J., Yang Y., Martincorena I., Creighton C. J., Weinstein J. N., Xu Y., Han L., Kim H. L., Nakagawa H., Park K., Campbell P. J., Liang H.; PCAWG Consortium , Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet. 52, 342–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maximo V., Soares P., Lima J., Cameselle-Teijeiro J., Sobrinho-Simoes M., Mitochondrial DNA somatic mutations (point mutations and large deletions) and mitochondrial DNA variants in human thyroid pathology: A study with emphasis on Hurthle cell tumors. Am. J. Pathol. 160, 1857–1865 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis C. F., Ricketts C. J., Wang M., Yang L., Cherniack A. D., Shen H., Buhay C., Kang H., Kim S. C., Fahey C. C., Hacker K. E., Bhanot G., Gordenin D. A., Chu A., Gunaratne P. H., Biehl M., Seth S., Kaipparettu B. A., Bristow C. A., Donehower L. A., Wallen E. M., Smith A. B., Tickoo S. K., Tamboli P., Reuter V., Schmidt L. S., Hsieh J. J., Choueiri T. K., Hakimi A. A.; The Cancer Genome Atlas Research Network, Chin L., Meyerson M., Kucherlapati R., Park W. Y., Robertson A. G., Laird P. W., Henske E. P., Kwiatkowski D. J., Park P. J., Morgan M., Shuch B., Muzny D., Wheeler D. A., Linehan W. M., Gibbs R. A., Rathmell W. K., Creighton C. J., The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gopal R. K., Kubler K., Calvo S. E., Polak P., Livitz D., Rosebrock D., Sadow P. M., Campbell B., Donovan S. E., Amin S., Gigliotti B. J., Grabarek Z., Hess J. M., Stewart C., Braunstein L. Z., Arndt P. F., Mordecai S., Shih A. R., Chaves F., Zhan T., Lubitz C. C., Kim J., Iafrate A. J., Wirth L., Parangi S., Leshchiner I., Daniels G. H., Mootha V. K., Dias-Santagata D., Getz G., McFadden D. G., Widespread chromosomal losses and mitochondrial dna alterations as genetic drivers in hurthle cell carcinoma. Cancer Cell 34, 242–255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tasdogan A., McFadden D. G., Mishra P., Mitochondrial DNA haplotypes as genetic modifiers of cancer. Trends Cancer 6, 1044–1058 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Yi Z., Zhang X., Tang W., Yu Y., Wei X., Zhang X., Wei W., Strand-selective base editing of human mitochondrial DNA using mitoBEs. Nat. Biotechnol. 42, 498–509 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mok B. Y., de Moraes M. H., Zeng J., Bosch D. E., Kotrys A. V., Raguram A., Hsu F., Radey M. C., Peterson S. B., Mootha V. K., Mougous J. D., Liu D. R., A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cruz-Bermudez A., Vallejo C. G., Vicente-Blanco R. J., Gallardo M. E., Fernandez-Moreno M. A., Quintanilla M., Garesse R., Enhanced tumorigenicity by mitochondrial DNA mild mutations. Oncotarget 6, 13628–13643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calabrese C., Iommarini L., Kurelac I., Calvaruso M. A., Capristo M., Lollini P.-L., Nanni P., Bergamini C., Nicoletti G., De Giovanni C., Ghelli A., Giorgio V., Caratozzolo M. F., Marzano F., Manzari C., Betts C. M., Carelli V., Ceccarelli C., Attimonelli M., Romeo G., Fato R., Rugolo M., Tullo A., Gasparre G., Porcelli A. M., Respiratory complex I is essential to induce a Warburg profile in mitochondria-defective tumor cells. Cancer Metab. 1, 11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iommarini L., Kurelac I., Capristo M., Calvaruso M. A., Giorgio V., Bergamini C., Ghelli A., Nanni P., De Giovanni C., Carelli V., Fato R., Lollini P. L., Rugolo M., Gasparre G., Porcelli A. M., Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum. Mol. Genet. 23, 1453–1466 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Gasparre G., Kurelac I., Capristo M., Iommarini L., Ghelli A., Ceccarelli C., Nicoletti G., Nanni P., De Giovanni C., Scotlandi K., Betts C. M., Carelli V., Lollini P. L., Romeo G., Rugolo M., Porcelli A. M., A mutation threshold distinguishes the antitumorigenic effects of the mitochondrial gene MTND1, an oncojanus function. Cancer Res. 71, 6220–6229 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Porcelli A. M., Ghelli A., Ceccarelli C., Lang M., Cenacchi G., Capristo M., Pennisi L. F., Morra I., Ciccarelli E., Melcarne A., Bartoletti-Stella A., Salfi N., Tallini G., Martinuzzi A., Carelli V., Attimonelli M., Rugolo M., Romeo G., Gasparre G., The genetic and metabolic signature of oncocytic transformation implicates HIF1α destabilization. Hum. Mol. Genet. 19, 1019–1032 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Nunes J. B., Peixoto J., Soares P., Maximo V., Carvalho S., Pinho S. S., Vieira A. F., Paredes J., Rego A. C., Ferreira I. L., Gomez-Lazaro M., Sobrinho-Simoes M., Singh K. K., Lima J., OXPHOS dysfunction regulates integrin-beta1 modifications and enhances cell motility and migration. Hum. Mol. Genet. 24, 1977–1990 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kulawiec M., Owens K. M., Singh K. K., Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol. Ther. 8, 1378–1385 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Bacman S. R., Nissanka N., Moraes C. T., Cybrid technology. Methods Cell Biol. 155, 415–439 (2020). [DOI] [PubMed] [Google Scholar]
- 21.King M. P., Attardi G., Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 246, 500–503 (1989). [DOI] [PubMed] [Google Scholar]
- 22.Arnold R. S., Sun C. Q., Richards J. C., Grigoriev G., Coleman I. M., Nelson P. S., Hsieh C. L., Lee J. K., Xu Z., Rogatko A., Osunkoya A. O., Zayzafoon M., Chung L., Petros J. A., Mitochondrial DNA mutation stimulates prostate cancer growth in bone stromal environment. Prostate 69, 1–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shidara Y., Yamagata K., Kanamori T., Nakano K., Kwong J. Q., Manfredi G., Oda H., Ohta S., Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 65, 1655–1663 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Yuan Y., Wang W., Li H., Yu Y., Tao J., Huang S., Zeng Z., Nonsense and missense mutation of mitochondrial ND6 gene promotes cell migration and invasion in human lung adenocarcinoma. BMC Cancer 15, 346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa K., Takenaga K., Akimoto M., Koshikawa N., Yamaguchi A., Imanishi H., Nakada K., Honma Y., Hayashi J., ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 320, 661–664 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Petros J. A., Baumann A. K., Ruiz-Pesini E., Amin M. B., Sun C. Q., Hall J., Lim S., Issa M. M., Flanders W. D., Hosseini S. H., Marshall F. F., Wallace D. C., mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 102, 719–724 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi J. I., Ohta S., Kikuchi A., Takemitsu M., Goto Y., Nonaka I., Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl. Acad. Sci. U.S.A. 88, 10614–10618 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birsoy K., Wang T., Chen W. W., Freinkman E., Abu-Remaileh M., Sabatini D. M., An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan L. B., Gui D. Y., Hosios A. M., Bush L. N., Freinkman E., Vander Heiden M. G., Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loffler M., Jockel J., Schuster G., Becker C., Dihydroorotat-ubiquinone oxidoreductase links mitochondria in the biosynthesis of pyrimidine nucleotides. Mol. Cell. Biochem. 174, 125–129 (1997). [PubMed] [Google Scholar]
- 31.Wada K. I., Hosokawa K., Ito Y., Maeda M., Harada Y., Yonemitsu Y., Generation of transmitochondrial cybrids using a microfluidic device. Exp. Cell Res. 418, 113233 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Patananan A. N., Sercel A. J., Wu T. H., Ahsan F. M., Torres A. Jr., Kennedy S. A. L., Vandiver A., Collier A. J., Mehrabi A., Van Lew J., Zakin L., Rodriguez N., Sixto M., Tadros W., Lazar A., Sieling P. A., Nguyen T. L., Dawson E. R., Braas D., Golovato J., Cisneros L., Vaske C., Plath K., Rabizadeh S., Niazi K. R., Chiou P. Y., Teitell M. A., Pressure-driven mitochondrial transfer pipeline generates mammalian cells of desired genetic combinations and fates. Cell Rep. 33, 108562 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeBleu V. S., O’Connell J. T., Gonzalez Herrera K. N., Wikman H., Pantel K., Haigis M. C., de Carvalho F. M., Damascena A., Domingos Chinen L. T., Rocha R. M., Asara J. M., Kalluri R., PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 16, 992–1003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vazquez F., Lim J. H., Chim H., Bhalla K., Girnun G., Pierce K., Clish C. B., Granter S. R., Widlund H. R., Spiegelman B. M., Puigserver P., PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 23, 287–301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer G. M., Jalali A., Kircher D. A., Lee W. C., McQuade J. L., Haydu L. E., Joon A. Y., Reuben A., de Macedo M. P., Carapeto F. C. L., Yang C., Srivastava A., Ambati C. R., Sreekumar A., Hudgens C. W., Knighton B., Deng W., Ferguson S. D., Tawbi H. A., Glitza I. C., Gershenwald J. E., Vashisht Gopal Y. N., Hwu P., Huse J. T., Wargo J. A., Futreal P. A., Putluri N., Lazar A. J., DeBerardinis R. J., Marszalek J. R., Zhang J., Holmen S. L., Tetzlaff M. T., Davies M. A., Molecular profiling reveals unique immune and metabolic features of melanoma brain metastases. Cancer Discov. 9, 628–645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaunay S., Pascual G., Feng B., Klann K., Behm M., Hotz-Wagenblatt A., Richter K., Zaoui K., Herpel E., Munch C., Dietmann S., Hess J., Benitah S. A., Frye M., Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature 607, 593–603 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartman C. R., Weilandt D. R., Shen Y., Lee W. D., Han Y., TeSlaa T., Jankowski C. S. R., Samarah L., Park N. R., da Silva-Diz V., Aleksandrova M., Gultekin Y., Marishta A., Wang L., Yang L., Roichman A., Bhatt V., Lan T., Hu Z., Xing X., Lu W., Davidson S., Wuhr M., Vander Heiden M. G., Herranz D., Guo J. Y., Kang Y., Rabinowitz J. D., Slow TCA flux and ATP production in primary solid tumours but not metastases. Nature 614, 349–357 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bezwada D., Perelli L., Lesner N. P., Cai L., Brooks B., Wu Z., Vu H. S., Sondhi V., Cassidy D. L., Kasitinon S., Kelekar S., Cai F., Aurora A. B., Patrick M., Leach A., Ghandour R., Zhang Y., Do D., McDaniel P., Sudderth J., Dumesnil D., House S., Rosales T., Poole A. M., Lotan Y., Woldu S., Bagrodia A., Meng X., Cadeddu J. A., Mishra P., Garcia-Bermudez J., Pedrosa I., Kapur P., Courtney K. D., Malloy C. R., Genovese G., Margulis V., DeBerardinis R. J., Mitochondrial complex I promotes kidney cancer metastasis. Nature 633, 923–931 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Arnon L., Zimmer L., Gutzmer R., Satzger I., Loquai C., Grabbe S., Vokes N., Margolis C. A., Conway J., He M. X., Elmarakeby H., Dietlein F., Miao D., Tracy A., Gogas H., Goldinger S. M., Utikal J., Blank C. U., Rauschenberg R., von Bubnoff D., Krackhardt A., Weide B., Haferkamp S., Kiecker F., Izar B., Garraway L., Regev A., Flaherty K., Paschen A., Van Allen E. M., Schadendorf D., Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahmood M., Liu E. M., Shergold A. L., Tolla E., Tait-Mulder J., Huerta-Uribe A., Shokry E., Young A. L., Lilla S., Kim M., Park T., Boscenco S., Manchon J. L., Rodriguez-Antona C., Walters R. C., Springett R. J., Blaza J. N., Mitchell L., Blyth K., Zanivan S., Sumpton D., Roberts E. W., Reznik E., Gammage P. A., Mitochondrial DNA mutations drive aerobic glycolysis to enhance checkpoint blockade response in melanoma. Nat. Cancer 5, 659–672 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soler-Agesta R., Marco-Brualla J., Fernandez-Silva P., Mozas P., Anel A., Moreno Loshuertos R., Transmitochondrial cybrid generation using cancer cell lines. J. Vis. Exp. 193, e65186 (2023). [DOI] [PubMed] [Google Scholar]
- 42.Prescott D. M., Myerson D., Wallace J., Enucleation of mammalian cells with cytochalasin B. Exp. Cell Res. 71, 480–485 (1972). [DOI] [PubMed] [Google Scholar]
- 43.Chen C. H., Cheng Y. C., Delayed cytotoxicity and selective loss of mitochondrial DNA in cells treated with the anti-human immunodeficiency virus compound 2′,3′-dideoxycytidine. J. Biol. Chem. 264, 11934–11937 (1989). [PubMed] [Google Scholar]
- 44.Lesner N. P., Gokhale A. S., Kota K., DeBerardinis R. J., Mishra P., α-Ketobutyrate links alterations in cystine metabolism to glucose oxidation in mtDNA mutant cells. Metab. Eng. 60, 157–167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pachnis P., Wu Z., Faubert B., Tasdogan A., Gu W., Shelton S., Solmonson A., Rao A. D., Kaushik A. K., Rogers T. J., Ubellacker J. M., LaVigne C. A., Yang C., Ko B., Ramesh V., Sudderth J., Zacharias L. G., Martin-Sandoval M. S., Do D., Mathews T. P., Zhao Z., Mishra P., Morrison S. J., DeBerardinis R. J., In vivo isotope tracing reveals a requirement for the electron transport chain in glucose and glutamine metabolism by tumors. Sci. Adv. 8, eabn9550 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piskounova E., Agathocleous M., Murphy M. M., Hu Z., Huddlestun S. E., Zhao Z., Leitch A. M., Johnson T. M., DeBerardinis R. J., Morrison S. J., Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 527, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tasdogan A., Faubert B., Ramesh V., Ubellacker J. M., Shen B., Solmonson A., Murphy M. M., Gu Z., Gu W., Martin M., Kasitinon S. Y., Vandergriff T., Mathews T. P., Zhao Z., Schadendorf D., DeBerardinis R. J., Morrison S. J., Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molina J. R., Sun Y., Protopopova M., Gera S., Bandi M., Bristow C., McAfoos T., Morlacchi P., Ackroyd J., Agip A. A., Al-Atrash G., Asara J., Bardenhagen J., Carrillo C. C., Carroll C., Chang E., Ciurea S., Cross J. B., Czako B., Deem A., Daver N., de Groot J. F., Dong J. W., Feng N., Gao G., Gay J., Do M. G., Greer J., Giuliani V., Han J., Han L., Henry V. K., Hirst J., Huang S., Jiang Y., Kang Z., Khor T., Konoplev S., Lin Y. H., Liu G., Lodi A., Lofton T., Ma H., Mahendra M., Matre P., Mullinax R., Peoples M., Petrocchi A., Rodriguez-Canale J., Serreli R., Shi T., Smith M., Tabe Y., Theroff J., Tiziani S., Xu Q., Zhang Q., Muller F., DePinho R. A., Toniatti C., Draetta G. F., Heffernan T. P., Konopleva M., Jones P., Di Francesco M. E., Marszalek J. R., An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 24, 1036–1046 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Paoli P., Giannoni E., Chiarugi P., Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta 1833, 3481–3498 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Mullen A. R., Wheaton W. W., Jin E. S., Chen P. H., Sullivan L. B., Cheng T., Yang Y. F., Linehan W. M., Chandel N. S., DeBerardinis R. J., Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schafer Z. T., Grassian A. R., Song L., Jiang Z., Gerhart-Hines Z., Irie H. Y., Gao S., Puigserver P., Brugge J. S., Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang L., Shestov A. A., Swain P., Yang C., Parker S. J., Wang Q. A., Terada L. S., Adams N. D., McCabe M. T., Pietrak B., Schmidt S., Metallo C. M., Dranka B. P., Schwartz B., DeBerardinis R. J., Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Labuschagne C. F., Cheung E. C., Blagih J., Domart M. C., Vousden K. H., Cell clustering promotes a metabolic switch that supports metastatic colonization. Cell Metab. 30, 720–734.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lien E. C., Westermark A. M., Zhang Y., Yuan C., Li Z., Lau A. N., Sapp K. M., Wolpin B. M., Vander Heiden M. G., Low glycaemic diets alter lipid metabolism to influence tumour growth. Nature 599, 302–307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinberg F., Hamanaka R., Wheaton W. W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G. M., Budinger G. R., Chandel N. S., Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. U.S.A. 107, 8788–8793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan A. S., Baty J. W., Dong L. F., Bezawork-Geleta A., Endaya B., Goodwin J., Bajzikova M., Kovarova J., Peterka M., Yan B., Pesdar E. A., Sobol M., Filimonenko A., Stuart S., Vondrusova M., Kluckova K., Sachaphibulkij K., Rohlena J., Hozak P., Truksa J., Eccles D., Haupt L. M., Griffiths L. R., Neuzil J., Berridge M. V., Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 21, 81–94 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Martinez-Reyes I., Cardona L. R., Kong H., Vasan K., McElroy G. S., Werner M., Kihshen H., Reczek C. R., Weinberg S. E., Gao P., Steinert E. M., Piseaux R., Budinger G. R. S., Chandel N. S., Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature 585, 288–292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor R. W., Barron M. J., Borthwick G. M., Gospel A., Chinnery P. F., Samuels D. C., Taylor G. A., Plusa S. M., Needham S. J., Greaves L. C., Kirkwood T. B., Turnbull D. M., Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 112, 1351–1360 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutierrez-Gonzalez L., Deheragoda M., Elia G., Leedham S. J., Shankar A., Imber C., Jankowski J. A., Turnbull D. M., Novelli M., Wright N. A., McDonald S. A., Analysis of the clonal architecture of the human small intestinal epithelium establishes a common stem cell for all lineages and reveals a mechanism for the fixation and spread of mutations. J. Pathol. 217, 489–496 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Valente L., Piga D., Lamantea E., Carrara F., Uziel G., Cudia P., Zani A., Farina L., Morandi L., Mora M., Spinazzola A., Zeviani M., Tiranti V., Identification of novel mutations in five patients with mitochondrial encephalomyopathy. Biochim. Biophys. Acta 1787, 491–501 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Li L., Chen L., Li J., Zhang W., Liao Y., Chen J., Sun Z., Correlational study on mitochondrial DNA mutations as potential risk factors in breast cancer. Oncotarget 7, 31270–31283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arnold R. S., Sun Q., Sun C. Q., Richards J. C., O’Hearn S., Osunkoya A. O., Wallace D. C., Petros J. A., An inherited heteroplasmic mutation in mitochondrial gene COI in a patient with prostate cancer alters reactive oxygen, reactive nitrogen and proliferation. Biomed. Res. Int. 2013, 239257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park J. S., Sharma L. K., Li H., Xiang R., Holstein D., Wu J., Lechleiter J., Naylor S. L., Deng J. J., Lu J., Bai Y., A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum. Mol. Genet. 18, 1578–1589 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulawiec M., Owens K. M., Singh K. K., mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. J. Hum. Genet. 54, 647–654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ubellacker J. M., Tasdogan A., Ramesh V., Shen B., Mitchell E. C., Martin-Sandoval M. S., Gu Z., McCormick M. L., Durham A. B., Spitz D. R., Zhao Z., Mathews T. P., Morrison S. J., Lymph protects metastasizing melanoma cells from ferroptosis. Nature 585, 113–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X., Shelton S. D., Bordieanu B., Frank A. R., Yi Y., Venigalla S. S. K., Gu Z., Lenser N. P., Glogauer M., Chandel N. S., Zhao H., Zhao Z., McFadden D. G., Mishra P., Scinderin promotes fusion of electron transport chain dysfunctional muscle stem cells with myofibers. Nat. Aging. 2, 155–169 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Hara R., Tedone E., Ludlow A., Huang E., Arosio B., Mari D., Shay J. W., Quantitative mitochondrial DNA copy number determination using droplet digital PCR with single-cell resolution. Genome Res. 29, 1878–1888 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Green M. R., Sambrook J., Isolation of high-molecular-weight DNA using organic solvents. Cold Spring Harb. Protoc. 2017, 356–359 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Green M. R., Sambrook J., Precipitation of DNA with ethanol. Cold Spring Harb. Protoc. 2016, 1116–1120 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Mao L., Wertzler K. J., Maloney S. C., Wang Z., Magnuson N. S., Reeves R., HMGA1 levels influence mitochondrial function and mitochondrial DNA repair efficiency. Mol. Cell. Biol. 29, 5426–5440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bankhead P., Loughrey M. B., Fernandez J. A., Dombrowski Y., McArt D. G., Dunne P. D., McQuaid S., Gray R. T., Murray L. J., Coleman H. G., James J. A., Salto-Tellez M., Hamilton P. W., QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L., Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C., PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.G. Korotkevich, V. Sukhov, N. Budin, B. Shpak, M. N. Artyomov, A. Sergushichev, Fast gene set enrichment analysis. bioRxiv 060012 [Preprint] (2021). 10.1101/060012. [DOI]
- 78.Rogava M., Aprati T. J., Chi W. Y., Melms J. C., Hug C., Davis S. H., Earlie E. M., Chung C., Deshmukh S. K., Wu S., Sledge G., Tang S., Ho P., Amin A. D., Caprio L., Gurjao C., Tagore S., Ngo B., Lee M. J., Zanetti G., Wang Y., Chen S., Ge W., Melo L. M. N., Allies G., Rosler J., Gibney G. T., Schmitz O. J., Sykes M., Creusot R. J., Tuting T., Schadendorf D., Rocken M., Eigentler T. K., Molotkov A., Mintz A., Bakhoum S. F., Beyaz S., Cantley L. C., Sorger P. K., Meckelmann S. W., Tasdogan A., Liu D., Laughney A. M., Izar B., Loss of Pip4k2c confers liver-metastatic organotropism through insulin-dependent PI3K-AKT pathway activation. Nat. Cancer 5, 433–447 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rohart F., Gautier B., Singh A., Le K. A., Cao, mixOmics: An R package for ’omics feature selection and multiple data integration. PLOS Comput. Biol. 13, e1005752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soflaee M. H., Kesavan R., Sahu U., Tasdogan A., Villa E., Djabari Z., Cai F., Tran D. H., Vu H. S., Ali E. S., Rion H., O’Hara B. P., Kelekar S., Hallett J. H., Martin M., Mathews T. P., Gao P., Asara J. M., Manning B. D., Ben-Sahra I., Hoxhaj G., Purine nucleotide depletion prompts cell migration by stimulating the serine synthesis pathway. Nat. Commun. 13, 2698 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Legends for data S1 to S7
Data S1 to S7