Abstract
Recently, it has been shown that blocking the binding of valosin-containing protein (VCP) to mutant huntingtin (mtHtt) can prevent neuronal mitochondrial autophagy in Huntington’s disease (HD) models. Herein, we describe the development and efficacy of a protein-like polymer (PLP) for inhibiting this interaction in cellular and in vivo models of HD. PLPs exhibit bioactivity in HD mouse striatal cells by successfully inhibiting mitochondrial destruction. PLP is notably resilient to in vitro enzyme, serum, and liver microsome stability assays, which render analogous control oligopeptides ineffective. PLP demonstrates a 2000-fold increase in circulation half-life compared to peptides, exhibiting an elimination half-life of 152 hours. In vivo efficacy studies in HD transgenic mice (R6/2) confirm the superior bioactivity of PLP compared to free peptide through behavioral and neuropathological analyses. PLP functions by preventing pathologic VCP/mtHtt binding in HD animal models; exhibits enhanced efficacy over the parent, free peptide; and implicates the PLP as a platform with potential for translational central nervous system therapeutics.
Proteomimetic polymer blocks protein-protein interaction and displays potential as a platform for neurodegenerative therapeutics.
INTRODUCTION
Huntington’s disease (HD) is an autosomal dominant, fatal neurodegenerative disease affecting approximately 41,000 individuals in the United States with at least 200,000 other Americans with a 50% risk of developing the disease (1–5). The disease affects anywhere from 4 to 13 individuals per 100,000 in Western populations (5–9). HD is characterized by motor dysfunction, involuntary movements, dystonia, cognitive decline, intellectual impairment, and emotional disturbances (1, 5, 8–13). Symptoms are usually adult onset, beginning between 30 and 50 years of age with the course of the disease lasting 15 to 20 years depending on the prevalence of CAG repeats (1, 9, 13–15). While immobility complications such as cardiac disease, infection, and pneumonia contribute heavily to the cause of death, 10 to 25% of patients attempt to take their own lives, and 9% commit suicide (1, 5, 9).
HD is terminal upon diagnosis, as the expansion of the CAG codon from the huntingtin (Htt) gene produces mutant huntingtin (mtHtt) proteins and causes mitochondrial dysfunction (1, 6, 8, 10, 11, 14, 16). mtHtt on the mitochondria binds to valosin-containing protein (VCP) erroneously, inducing aberrant mitochondrial VCP accumulation, resulting in excessive mitophagy and subsequent neuronal cell death (6–8, 16). Neuronal loss is particularly prominent in the striatum of the basal ganglia, where atrophy has substantial impact on motor function and motor planning, causing chorea or involuntary movement (1, 6–8, 10, 14, 16). In patients with HD and mouse models of HD, concentrations of dopamine signaling protein, dopamine- and cyclic AMP–regulated phosphoprotein of molecular weight 32,000 (DARPP-32), are decreased in the striatum as a result of the distinct loss of striatal medium spiny neurons (MSN) (17). Although the disease mutant of HD has been identified and a genetic test is available to identify those individuals who carry the mutation and will succumb to the disease, there is currently no therapy to slow or prevent disease progression, only symptomatic treatments with limited impact (1, 5, 9, 10). Recent developments of therapeutics including those targeting the striatum via injection are invasive, have low efficacy, and are not sustainable (11, 13).
Peptides have been explored as biocompatible therapeutics with enhanced target specificity for HD. Recently, the peptide sequence HVLVMCAT (HV3) was shown to target the VCP/mtHtt interaction (16). Lacking cell-penetrative capability, this sequence was coupled to the cell-penetrating, HIV-derived TAT peptide (18–21) to yield the active, HV3-TAT peptide. HV3-TAT peptide blocks mtHtt-induced VCP translocation to the mitochondria, thereby rescuing neurons from mitochondrial destruction. Although this peptide displayed effective target engagement, it was limited by rapid enzymatic clearance. These limitations (poor pharmacokinetic properties and limited cell penetration) generally plague peptide-based therapeutics. In terms of pharmacokinetic limitations, a major factor is that peptides have an inherently low molecular weight, leading to rapid renal clearance (22). This rapid renal clearance is coupled with decreased stability and high susceptibility to rapid degradation by proteolytic enzymes in vivo, leading to overall poor performance for peptide-based drugs (22). In their current state, free peptides, although advantageous for targeting and biocompatibility, generally present substantial hurdles to translation to clinical use (23–27). Hence, there is a demonstrated need for new, enabling platform technologies.
In contrast to the classical, biological, and linear polypeptide configuration, we have developed a class of “polypeptide” wherein peptides form side chains originating from a polymer backbone scaffold (24, 28). These structures, termed protein-like polymers (PLPs), are synthesized via graft-through, living polymerization methods using peptide-modified monomers. The resulting structure, consisting of a hydrophobic polymer, with dense array of hydrophilic side-chain peptides leads to the formation of a globular “protein-like” structure protecting the peptides from proteolytic degradation and providing a means to penetrate cells, with judicious choice of charge, combined with their metaphilic (29) or transiently amphiphilic structure (28, 30–32). In addition, we hypothesized that PLPs, having a higher molecular weight compared to free peptides, would avoid rapid renal clearance. Herein, we demonstrate the utility of the PLP system to overcome the challenges facing peptide-based therapeutics using the HV3 peptide displayed on a PLP backbone as a proof of concept in an HD mouse model. The PLP is shown to penetrate mouse-derived HD striatal neurons (HdhQ111) containing homozygous HTT loci with polyglutamine repeats (33–36), successfully blocking VCP/mtHtt binding, an intracellular protein-protein interaction (PPI) (16, 37–40). This PPI serves as a proof-of-concept target, providing further encouragement to pursue other intracellular disease driving and difficult to drug protein interfaces. Moreover, the PLP demonstrates biocompatibility, low toxicity, and exceptional resistance to enzymatic proteolysis, resulting in extended half-life upon intravenous administration. PLP exhibited therapeutic efficacy, providing neuroprotection in an in vivo HD mouse model.
RESULTS
Synthesis and characterization of PLPs
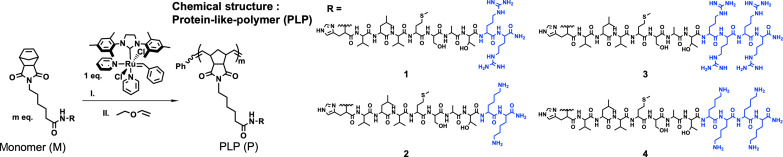
For the synthesis of PLPs, the original HV3 peptide sequence, HVLVMCAT, was modified to prevent the possibility of disulfide bond formation and aggregation between peptide side chains on the polymers by substitution of cysteine to serine to give HVLVMSAT. In addition, two or four positively charged arginine or lysine residues were introduced to facilitate cellular uptake (Fig. 1). The four resulting peptide sequences (HVLVMSATRR, HVLVMSATKK, HVLVMSATRRRR, and HVLVMSATKKKK) were synthesized and conjugated to N-(hexanoic acid)-exo-5-norbornene-2,3-dicarboximide at the N terminus using solid-phase peptide synthesis (fig. S1). Polymerization reactions were performed on high-performance liquid chromatography (HPLC)–purified norbornenyl peptide-monomers via ring-opening metathesis polymerization (ROMP) polymerization with the Grubbs’ ruthenium initiator, (IMesH2)(C5H5N)2(Cl)2Ru = CHPh (41), and were monitored by nuclear magnetic resonance (1H NMR) spectrometry (figs. S2 and S3). The resulting PLPs generated from the four peptide monomers M1 to M4, named P1, P2, P3, and P4, respectively (Fig. 1), were then characterized via size exclusion chromatography with a multi-angle light scattering detector (figs. S4 and S5).
Fig. 1. Structure of PLP.
HV3 peptide analogs with different number of positive charged amino acids (blue) were polymerized into PLPs P1, P2, P3, and P4 using ROMP polymerization technique.
In vitro assessment of PLP efficacy
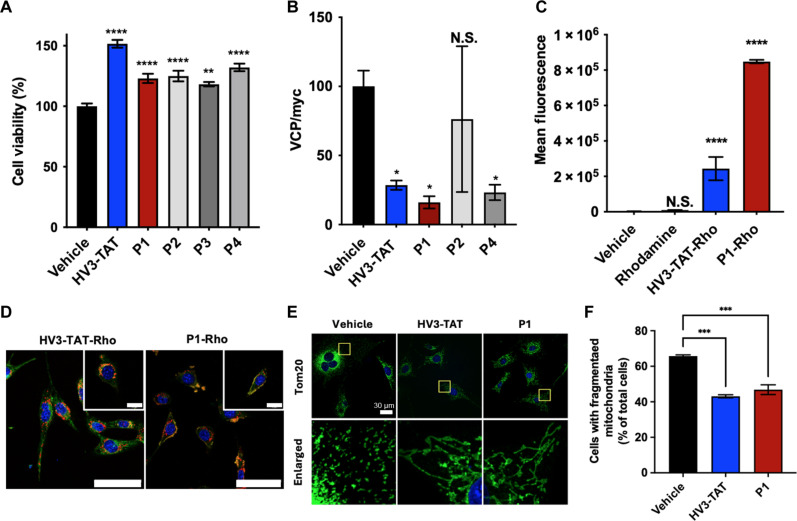
In vitro assays were carried out to examine the bioactivity of PLPs, P1, P2, P3, and P4 and to compare against the known, active HV3-TAT (Fig. 2). First, to demonstrate cell rescue, HdhQ111 cells were serum-starved before treatment with HV3-TAT or P1 and subsequently assessed for cell viability. HdhQ111 cells were immortalized from knock-in mice carrying a humanized exon 1 with 111 CAG repeats in the mouse Htt gene (16, 34). Compared to the vehicle control, both the peptide and PLPs exhibited significant increases in cell viability (Fig. 2A). Subsequently, P1, P2, and P4 were selected for analysis via a VCP association assay as they exhibited higher statistical significance (P1: P < 0.0001, P2: P < 0.0001, P4: P < 0.0001) in comparison to P3 (P = 0.0018) when compared to the vehicle control in the initial cell viability rescue screen.
Fig. 2. In vitro efficacy assays.
(A) Cell viability of HV3-TAT peptide and PLPs. Concentration, 3 μM with respect to peptide (n = 8). (B) Association of VCP to mtHtt (Q73) was quantified by IP followed by Western blot upon treatment of HV3-TAT peptide or PLPs (n = 5). (C) Quantification of mean fluorescence output of flow cytometry cellular uptake in HdhQ111 cells. Treatments: HV3-TAT-Rho peptide, P1-Rho, rhodamine dye, and vehicle (n = 3). (D) Mitochondrial localization in live HdhQ111 cells. HdhQ111 cells were treated with HV3-TAT-Rho peptide, P1-Rho, and rhodamine dye alone at a concentration of 3 μM. Yellow indicates colocalization of green and red channels, signifying localization to the mitochondria. Mitochondria, MitoTracker Green FM (green channel). Material, Rhodamine (red channel). Nuclei, Hoechst 33342 stain (blue channel). Scale bars, 60 μm; 20 μm (inset). (E) Mitochondrial fragmentation assay. Mitochondrial morphology was determined by staining cells with anti-Tom20 antibody (Green). Nuclei, Hoechst 33342 stain (blue channel). Scale bar, 30 μm. The yellow boxes in the top panel have been magnified in the panel below for a more detailed view. (F) Mitochondrial fragmentation assay. The percentage of HdhQ111 cells with fragmented mitochondria relative to the total number of cells was quantitated. At least 95 cells per group were counted (n = 3). One-way analysis of variance (ANOVA) with multiple comparisons of the mean in each group with that of vehicle was used for analysis. Statistical significance was defined as follows: N.S. P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001. Values are means ± SEM.
As noted above, the HV3-TAT peptide and PLPs are designed to selectively inhibit the interaction between VCP and mtHtt as the mtHtt-induced VCP mitochondrial accumulation triggers neuronal mitophagy, ultimately leading to cell death. To test for target engagement, protein-binding assays were conducted. The inhibitory effects of PLPs on mtHtt-VCP binding in cells are examined with immunoprecipitation (IP) (Fig. 2B and figs. S6). Briefly, human embryonic kidney (HEK) 293T cells transfected with full-length human Htt containing 73 polyglutamine repeats (myc-Htt-Q73-FL construct, which was obtained from the CHDI Foundation, CH00039), were treated with HV3-TAT or PLPs. After treatment, total lysates were subjected to IP with anti-myc antibody (recognizes Htt-Q73-FL), and immunoprecipitants were analyzed by Western blot with anti-VCP antibody. Here, HV3-TAT peptide, P1 and P4 exhibited evidence for blocking PPI of VCP/mtHtt, with P2 exhibiting lower performance. With P1 and P4 showing equivalent activity, and to further down select for subsequent cellular and in vivo studies, P1 was chosen. Key factors were the lower molecular weight, presence of arginine residues that have been shown to provide more efficient uptake of PLPs (42), and lower overall charge.
A major drawback for most peptide-based drugs is poor innate cell membrane permeability, such that many peptide-based drugs fail to reach their putative intracellular targets (25, 43). Cellular uptake was evaluated by confocal microscopy and flow cytometry using a rhodamine dye–labeled version of HV3-TAT (HV3-TAT-Rho; fig. S7) and a rhodamine-labeled version of P1 PLP (P1-Rho; fig. S8). Distinct intracellular signals (red) were observed for both dye-labeled treatment groups, confirming the ability of the PLPs to penetrate the cellular membrane (figs. S9 to S11). Furthermore, as demonstrated through flow cytometry analysis (Fig. 2C and fig. S12), P1-Rho exhibited significantly enhanced cell penetration when compared to both the HV3-TAT-Rho peptide and the rhodamine dye control. In addition, we conducted an accumulation assay of the HV3-TAT peptide and PLP within HdhQ111 cells (fig. S13). Here, P1-Rho was detected in the HdhQ111 cells with sustained intensities at 2 and 7 days after one initial treatment as opposed to the decrease in mean fluorescence observed for the peptide alone.
A decisive pathologic hallmark for HdhQ111 cells is mitochondrial fragmentation. Since the mitochondrial membrane is the specific intracellular target location for the designed system, mitochondrial localization of P1-Rho in HdhQ111cells was evaluated (Fig. 2D and fig. S14). To visualize mitochondrial localization, the mitochondria were stained with MitoTracker (shown in green). Both HV3-TAT-Rho and P1-Rho–treated cells exhibited overlapping signals with mitochondria (yellow), confirming localization. This demonstrates that P1-Rho can be delivered intracellularly and subsequently localize to the mitochondrial membrane, which is where the target interaction of mtHtt/VCP is located. In contrast, the rhodamine dye alone did not show overlapping signals with mitochondria (fig. S15), indicating that the dye itself does not localize to the mitochondria and therefore does not contribute to the localization of the conjugated peptide or polymer.
Preventing the fragmentation of the mitochondria is important for maintaining the health of striatal cells in the HD in vitro model. Therefore, to demonstrate the ability of the PLP to prevent mitochondrial fragmentation, confocal fluorescent imaging was used to assess mitochondrial morphology after treatment (Fig. 2E). While mitochondrial fragmentation was observed in the vehicle treatment group, a marked reduction in the number of fragmented mitochondria was evident in both the HV3-TAT and P1 treatment groups. The quantification of mitochondrial fragmentation was based on an analysis of a minimum of 95 cells per group (Fig. 2F).
With cellular VCP engagement verified (Fig. 2B), P1 was further assessed for VCP binding via bio-layer interferometry (BLI) to compare with HV3-TAT peptide. Briefly, immobilized VCP was exposed to aqueous solutions of the HV3-TAT peptide or P1 at various concentrations to determine a dissociation constant (fig. S16). A Kd of 92 nM was determined for P1, driven by slow off-rates (koff) (Table 1). These data reveal a 150-fold decrease in dissociation constant compared to the HV3-TAT peptide at a Kd of 14 μM. The avidity of the multivalent PLP compared to free peptide is an inherent attribute of the system where statistical rebinding of the PLP to the target protein, VCP, drives the significantly decreased off rate observed.
Table 1. Binding constants of BLI.
| HV3-TAT | P1 | |
|---|---|---|
| Kd (M)* | 1.4 × 10−5 | 9.2 × 10−8 |
| kon (1/Ms) | 1.6 × 103 | 2.3 × 103 |
| koff (1/s) | 2.3 × 10−2 | 2.5 × 10−5 |
*Binding affinity of HV3-TAT and P1 measured by BLI. Binding kinetics of HV3-TAT peptide and P1 were measured using VCP protein. kon represents the association rate constant or on-rate, and koff represents the dissociation rate constant or off-rate.
Proteolytic stability of PLP
PLPs have been shown to confer proteolytic resistance on the peptide side chains driven by polymer backbone hydrophobicity and the resulting globular PLP structure (28, 44). To ascertain the extent to which proteolysis is abated by the peptide side chains of HV3-based PLPs, enzyme degradation and serum stability studies were conducted (figs. S17 to S23). First, P1 was treated separately with pronase (30), chymotrypsin (45), and elastase for 1 hour and then assessed for changes in molecular weight using gel electrophoresis (fig. S17). Enzymatically treated P1 maintained molecular weight running together with the untreated P1 control, demonstrating proteolytic stability of P1. To further confirm the enhanced proteolytic resistance of PLPs compared to the free HV3-TAT peptide, each compound was incubated with pepsin and subsequently analyzed using analytical HPLC to evaluate specific peptide fragments and to determine percent cleavage as a function of time (fig. S18). Pepsin is a common gastric enzyme that was selected for its ability to fragment the HV3 peptides at only one cleavage site. Hence, fragmentation could be confirmed by comparing the HPLC traces of the enzyme-treated compounds with that of the synthesized peptide fragment generated by pepsin (sequence = SATRR). The HV3-TAT peptide was 100% cleaved into two fragments in only 50 min, while P1 remained protected, with no cleavage observed for up to 150 min (figs. S18 and S19). Stability of HV3-TAT and P1 were also assessed by incubating materials in serum [10% fetal bovine serum (FBS)–rich media and 25% FBS-rich media]. The HV3-TAT peptide continued to show rapid degradation under both 10 and 25% FBS conditions, in contrast to the stability of P1 seen with serum treatments (figs. S19 and S20).
Furthermore, resistance to enzyme degradation was further confirmed by preparing a donor-acceptor labeled fluorogenic substrate (EDANS-DABCYL pair; figs. S21 to S23). In these experiments, fluorogenic P1 (P1-ED) demonstrated excellent resistance to enzyme degradation with little observed change in fluorescence intensity, compared to clear dequenching of EDANS for the fluorogenic monomer peptide (M1-ED) when treated with the three enzymes, trypsin, thermolysin, and elastase (fig. S23).
This robust stability of P1 over HV3-TAT was also evident in a liver microsome assay (fig. S24). In this assay, P1 or HV3-TAT were exposed to pooled human liver microsomes (0.5 mg/ml) activated by 1 mM reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) cofactor and analyzed via analytical HPLC. While HV3-TAT peptide exhibited rapid degradation of 82% within 1 hour, P1 displayed a decelerated degradation rate.
Furthermore, to verify that the P1 retained bioactivity after being exposed to enzymes or serum treatments, the viability of HD mouse mutant striatal HdhQ111 cells was assessed following treatment with HV3-TAT and P1 following enzymatic exposure (fig. S25). Briefly, HV3-TAT peptide or P1 were pretreated with trypsin, pronase, or 10% FBS for 1 hour, and enzymes were heat-neutralized before incubation with HdhQ111 cells. Cells treated with enzyme or serum-pretreated HV3-TAT peptide showed a dramatic decrease in cell viability relative to those treated with undigested peptide. Notably, P1 did not show a significant change in cell viability between groups with and without pretreatment with enzyme or serum (fig. S25).
In vitro analysis of hemocompatibility and complement activation
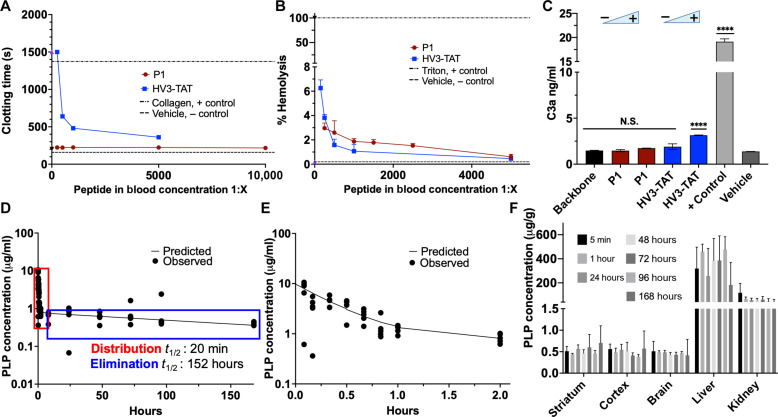
Hemocompatibility of the compounds was assessed via activated clotting time (ACT) and hemolytic activity using a dosing guide previously published (Fig. 3, A and B) (46). In the ACT hemochron assay, a viscosity-based assay to assess the common clotting pathway, P1 did not show any significant change in ACT compared to the vehicle control (lower dashed line) and only the highest concentration of HV3-TAT hindered clotting similar to the Triton X-100 detergent (top dashed line) positive control (Fig. 3A). Hemolytic activity of P1 was also found to be nonsignificant compared to vehicle control (bottom dashed line) (Fig. 3B).
Fig. 3. Biocompatibility and pharmacokinetic profiling for P1.
(A) Viscosity-based assay of ACTs of whole human blood. One-way ANOVA with repeated measures compared to mean of the positive control, collagen (n = 4). (B) % Hemolysis of red blood cells after the addition of P1 and HV3-TAT. One-way ANOVA with repeated measures compared to mean of the vehicle (n = 4). (C) Complement C3a activation ELISA to assess immunogenicity of P1 versus HV3-TAT. Positive control is cobra venom factor. One-way ANOVA performed with respect to vehicle (n = 4). (D) Pharmacokinetic profile of P1-Gd with predicted two-compartment model trend outlined overlayed with pharmacokinetic data. Elimination half-life is 152 hours (n = 5). (E) Distribution phase of the pharmacokinetic profile of P1-Gd from 5 min to 2 hours. Distribution half-life is 20 min (n = 5). (F) Biodistribution of P1-Gd. Micrograms of P1 per gram of organ across main time points, organized by organ (n = 5). One-way ANOVA comparisons of the mean of each group (striatum, cortex, and brain) was used for analysis (F). Statistical significance was defined as follows: N.S. P > 0.05, ****P ≤ 0.0001. Values are means ± SEM.
High doses of P1 and HV3-TAT (10 mg/kg), the therapeutic dose of P1 and HV3-TAT (3 mg/kg), and the PLP backbone were assayed in duplicate using a C3a enzyme-linked immunosorbent assay (ELISA) assay, with results quantified by absorbance (Fig. 3C). In this assay, mouse weight was estimated as 30 g with an average blood volume of 2 ml for dosing preparation conversion of 3 mg/kg to 17.8 μM and 10 mg/kg to 59.4 μM for HV3-TAT, and 9.9 and 33 μM for P1, respectively. The PLP backbone and tested concentrations of P1 did not show a statistically significant difference in activity compared to the vehicle. HV3-TAT did not show significant activation at the therapeutic concentration of 17.8 μM (3 mg/kg), but above 59.4 μM (10 mg/kg), significant activation was observed. The results of these in vitro assays suggest that the anti-HD PLP, P1, can confidently be assessed in vivo at the therapeutic dose of 3 mg/kg head-to-head with HV3-TAT.
In vivo pharmacokinetic profiling and biodistribution
Gadolinium (Gd) is a commonly used metal as an internal standard for inductively coupled plasma mass spectrometry (ICPMS) due to its stability, compatibility, and sensitivity. Consequently, we prepared a Gd-metalated termination agent (fig. S26 to S31) to serve as a labeling tag for PLPs (47). This tag enables in vivo pharmacokinetic analysis using ICPMS for quantification in both blood and tissue samples (fig. S31). Gd-labeled P1 (P1-Gd) was administered via a single intravenous tail vein injection at 5.54 mg/kg (10 mg/kg with respect to peptide) in wild-type (WT) mice. The pharmacokinetics of the PLP followed a two-compartment model. Here, an initial 20-min distribution half-life was observed, followed by an extended 152-hour elimination half-life (Fig. 3, D and E). This represents at least a 2000-fold increase in circulation detection compared to the HV3-TAT peptide, which is not detectable after 5 min.
While most of the compound was detected in the liver and kidney, primary sites for drug elimination, P1-Gd was detectable in the central nervous system (CNS) (Fig. 3F and fig. S32). One-way analysis of variance (ANOVA) showed no significant difference between the concentrations in the brain, striatum, and cortex for all time points, suggesting that an average concentration of 0.6 μg/g is maintained after 5 min, indicating that repeated dosing is needed to reach the target efficacious dose in the brain with in vitro studies as the reference.
Toxicity of P1 in WT mice
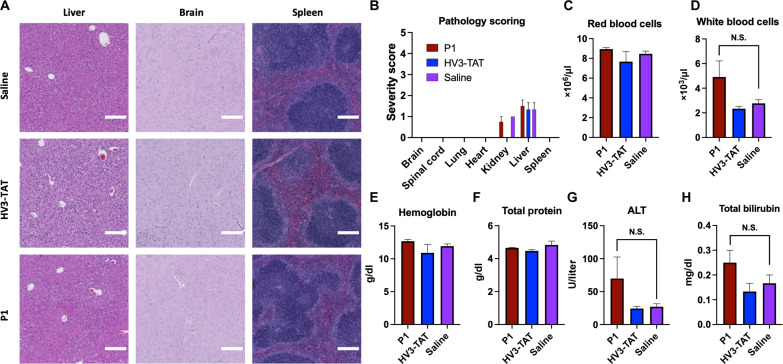
With the efficacy study designed to test P1, HV3-TAT, and saline in mice via a subcutaneously implanted Alzet osmotic pump over the course of 2 months, we next assessed toxic effects in healthy, WT mice. Here, C57BL/6J WT mice were implanted with Alzet osmotic pumps containing saline, HV3-TAT, or P1 at the previously established (16) operating dose of HV3-TAT, 3 mg/kg with respect to peptide. After analyzing the organs, no significant differences in pathology were observed between the saline control group and the HV3-TAT or P1 (blinded, senior pathologist) (Fig. 4, A and B, and table S6). A complete blood count (table S3) and blood chemistry profile (table S4) also revealed no significant deviation of any markers for P1 or HV3-TAT from saline. This overall assessment confirms the nontoxic nature of P1 within the dose range.
Fig. 4. Representative optical micrographs and selected data for toxicity pathology analysis.
(A) WT C57BL/6J mice were dosed with saline or 3 mg kg−1 day−1 (with respect to peptide) of P1 or HV3-TAT from 6 to 13 weeks of age. Continuous dose administered via Alzet osmotic pump. Photomicrographs of hematoxylin and eosin (H&E)–stained tissue showing similar morphology across all groups. Scale bar, 200 μm. (B) Pathology severity scoring of H&E-stained tissue. Full scoring comments in table S6. (C to H) Representative graphs from complete blood count and blood biochemistry panel (remaining graphs in figs. S33 and S34). Group: Vehicle (n = 3), HV3-TAT (n = 3), P1 (n = 4). One-way ANOVA comparisons of the mean of each group were used for analysis [(C) to (H)]. Statistical significance was defined as follows: N.S. P > 0.05. Values are means ± SEM.
In vivo efficacy of P1 as an HD therapeutic
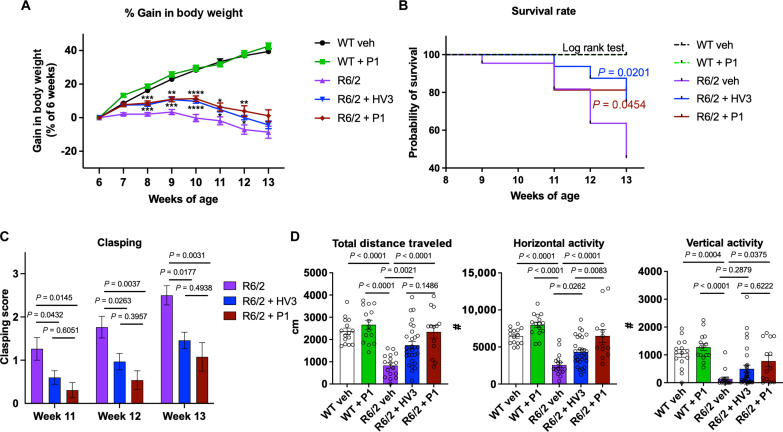
We next examined whether P1 treatment provides neuroprotection in vivo using the R6/2 transgenic mouse model for HD. As a primary screening model for HD drug candidates, the R6/2 mice model was previously used to assess efficacy of the HV3-TAT peptide (35). R6/2 mice aggressively develop HD-associated pathologies exhibiting a significant decrease in motor coordination, general motility and body weight in male mice (48). P1 was administered to HD R6/2 and WT mice via Alzet osmotic pump at a dose of 1.69 mg kg−1 day−1 (3 mg kg−1 day−1 on a per peptide basis, matching HV3-TAT peptide) from 6 to 13 weeks of age. The disease group (saline-treated R6/2 vehicle control) showed a decrease in body weight over the course of the study compared to WT controls (Fig. 5A), with P1 and HV3-TAT significantly inhibiting weight loss and extended survival over the R6/2 saline control (Fig. 5, A and B). Hindlimb clasping was assessed weekly, with R6/2 control mice exhibiting progressively severe motor deficits over time. Treatments of both HV3-TAT peptide and P1 attenuated the clasping behavior of R6/2 mice, with P1-treated mice exhibiting shorter clasping times than HV3-TAT–treated mice (Fig. 5C). Open field locomotor activity was assessed 1 week before euthanasia, at the age of 12 weeks. R6/2 vehicle mice exhibited less total distance traveled as well as decreased horizontal and vertical activities (Fig. 5D). P1 administration markedly corrected these deficits of spontaneous locomotion, significantly improving vertical activity over R6/2 vehicle mice and increasing total distance and horizontal activity to a comparable level with WT mice (Fig. 5D).
Fig. 5. Behavioral analysis of P1 treatments in HD mice.
Male HD R6/2 mice and WT littermates were dosed with 3 mg kg−1 day−1 of HV3-TAT peptide, labeled HV3 in the graphs, and 1.69 mg kg−1 day−1 of P1 (3 mg kg−1 day−1 with respect to peptide) from 6 to 13 weeks of age. Continuous dose administered via Alzet osmotic pump. (A) Body weight was recorded from the age of 6 to 13 weeks (at week 6: WT, n = 15; WT + P1, n = 15; R6/2 veh, n = 22; R6/2 + HV3, n = 32; and R6/2 + P1, n = 16). Statistical significance was defined as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (B) Survival was recorded from the age of 6 to 13 weeks and analyzed by the log-rank (Mantel-Cox) test [same n number of each group with body weight in (A)]. (C) Hindlimb clasping was assessed through tail suspension test once a week from the ages of 11 to 13 weeks (at week 11: R6/2 veh, n = 19; R6/2 + HV3, n = 30; and R6/2 + P1, n = 13). (D) One hour of overall movement activity in R6/2 mice and WT littermates (total traveled distance, horizontal and vertical activities) was determined by locomotion activity chamber at the age of 12 weeks (WT, n = 15; WT + P1, n = 15; R6/2 veh, n = 17; R6/2 + HV3, n = 30; and R6/2 + P1: n = 13). All values are reported as means ± SEM. Data were compared using one-way ANOVA with Tukey’s post hoc test in (A), (C), and (D). Exact P values are shown in the figures. Values are means ± SEM.
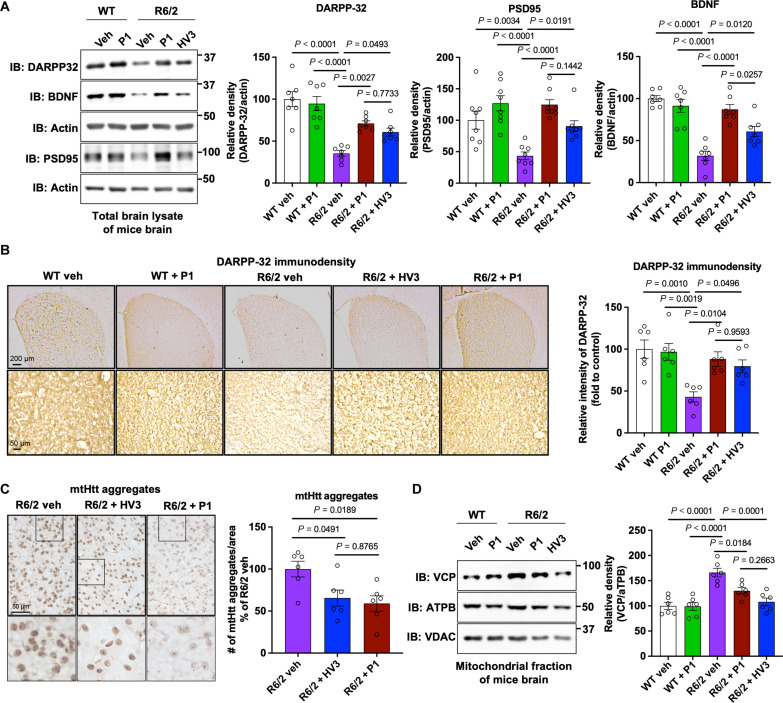
To further assess the efficacy of P1 in modulating HD neuropathology, the protein levels of dopamine- and cyclic adenosine monophosphate–regulated phosphoprotein 32 kDa (DARPP-32), postsynaptic density protein (PSD95), and brain-derived neurotrophic factor (BDNF) were determined, each of which are decreased in the MSN in striatum of HD R6/2 mouse model. MSN is the most susceptible neuron that degenerates in HD, and DARPP-32 is a typical MSN marker (16, 17, 49). BDNF is required for MSN survival, and BDNF decrease is also featured in the HD brain (16, 50). PSD95 is a synapse marker, and its level can generally reflect neuronal function (8). Western blot analysis of striatal extracts revealed a significant reduction of DARPP-32, PSD95, and BDNF in saline-treated R6/2 mice (Fig. 6A). R6/2 mice treated with P1 had significantly increased levels of these proteins over saline control, including a significantly higher level of BDNF as compared to the HV3-TAT treatment group (Fig. 6A). Enhanced levels of DARPP-32, an MSN marker, were consistently observed in immunohistochemical staining (Fig. 6B), indicating striatal neuron rescue after P1 treatment. As the neuropathology hallmark of HD, the formation of mtHtt aggregates can be observed in striatum of HD mouse models, like R6/2 mice. MtHtt aggregation was visualized by immunohistochemical staining by anti-mtHtt antibody (Fig. 6C). Both HV3-TAT peptide and P1 inhibit mtHtt aggregation resulting in a reduced number of mtHtt aggregates in striatum compared to saline-treated R6/2 mice. The treatment had no significant effects on motor activity, body weight, or neuropathology of WT mice, confirming a lack of toxicity of P1 treatment. These results collectively demonstrated that P1 treatment reduced neuropathology and behavioral phenotypes in HD animal models.
Fig. 6. Neuropathology analysis of P1 treatments in HD mice.
Male HD R6/2 mice and WT littermates were dosed with HV3-TAT peptide, labeled HV3 in the graphs for brevity, and P1 from 6 to 13 weeks of age via Alzet osmotic pump. (A) Total lysates from the striatum of 13-week-old R6/2 mice and WT littermates were subjected to WB with the indicated antibodies. Shown blots are representative of independent experiments. Histogram: the relative abundance of DARPP-32, PSD95, and BDNF. Actin was used as a loading control. n = 7 mice for DARPP-32 and BDNF, and n = 8 mice for PSD95. Brain sections of 13-week-old R6/2 mice and WT littermates were stained with (B) anti-DARPP-32 and (C) anti-Htt (clone EM48) antibodies. (B) DARPP-32 immunodensity was examined and quantified in the dorsolateral striatum of mice. n = 6 mice per group. (C) mtHtt aggregate numbers per area were quantified. n = 6 mice per group. (D) Mitochondrial fractions were isolated from the striatum of 13-week-old R6/2 mice and WT littermates and subjected to WB with the indicated antibodies. ATPB was used as a loading control of mitochondria. Left, representative blots from six independent experiments. Right, relative density of VCP in contrast to ATPB. All values are reported as means ± SEM. Data were compared using one-way ANOVA with Tukey’s post hoc test in (A) to (D). Exact P values are shown in the figures. Values are means ± SEM.
Previous work demonstrated that mtHtt is required for VCP accumulation on mitochondria, which causes excessive mitophagy and subsequence neuropathology in HD (16, 39, 51). As noted above, the HV3-TAT peptide was shown to block mtHtt-VCP binding and to inhibit VCP mitochondrial accumulation, which reduces behavioral and neuropathological phenotypes of HD (16). Therefore, mitochondrial levels of VCP were examined to test whether P1 retains the mechanism established for HV3-TAT peptide and recapitulates in vivo what was concluded from in vitro studies (Fig. 2B). Here, mitochondrial fractions of striatum were isolated from mice in each treatment group. Protein levels of VCP on mitochondria were analyzed by Western blotting, using adenosine triphosphate synthase beta subunit (ATPB) as a loading control for mitochondria. These experiments indicate that P1 indeed blocks aberrant VCP mitochondrial translocation in R6/2 mice (Fig. 6D).
DISCUSSION
We describe PLPs for inhibiting the mtHtt-VCP PPI and for blunting neuropathology in the R6/2 HD mouse model. We reasoned that leveraging a peptide with an established mechanism of action would provide a proof-of-concept study to examine a range of features of PLPs as a potential therapeutic platform technology. First, we established that PLPs can be designed for the inhibition of specific intracellular PPIs and, in particular, that PLPs can inhibit such interactions at specific locations within the cell, here at the mitochondria. Moreover, PLPs can enter and inhibit PPIs in neurons without the initial peptide being cell penetrating (i.e., HV3 without the TAT sequence), as modification of the peptide with limited arginine residues, followed by polymerization, leads to uptake (Fig. 2C) (31, 52, 53). Second, PLPs exhibit exceptional stability compared to free peptides with the origin of this stability, with respect to proteolytic degradation, being the dense display of peptides on the hydrophobic polymer backbone, leading to globular structures, inaccessible to proteases (24, 28, 30). This ensures that PLPs maintain their activity in rescuing neurons following serum incubation, in contrast to peptides (fig. S25). Third, PLPs exhibit enhanced multivalency via statistical rebinding (Table 1). Specifically, we observe an on-rate defined by the peptide sequence and a significantly reduced off-rate for the PLP. Therefore, once bound, the reduced dissociation constant is driven by the high probability of reengagement of the target protein due to the high-density display of neighboring peptides. This high-affinity binding coupled with proteolytic stability suggests that the PLPs are flexible and capable of induced fit binding upon engaging target proteins while resisting degradation by enzymes with sterically restricted active sites.
The PLP shows significantly extended elimination half-lives (t1/2 = 152 hours) compared to HV3-TAT, which is undetectable after 5 min, with P1 showing no signs of toxicity despite high and continuous dosing over 8 weeks at 1.7 mg kg−1 day−1. These features of being well-tolerated, while exhibiting long circulation times, will inform future PLP design against other targets and the optimization of the PLP and dosing strategies for tissue-selective PPI inhibition. Notably, the extended half-life of the PLP should enable lower-dose, single intravenous injection of PLP, or inform sustained, low-dose continuous administration in future studies.
Efficacy studies in vivo revealed that P1 is effective in terms of behavioral assessments. This includes significant rescue of the clasping phenotype relative to R6/2, maintaining levels near WT to day 11 in study. Notably, P1-treated animals returned to near WT activity in terms of horizontal activity in contrast to severe deficits in spontaneous locomotion exhibited by untreated R6/2 animals at week 12. Together with these analyses, P1 treatment significantly elevated DARPP-32, BDNF, and PSD95 levels compared to the saline and the HV3-TAT treatment groups. In addition, regarding target engagement, P1 demonstrated on-target selectivity with the capacity to inhibit the aggregation of mtHtt and the translocation of VCP to mitochondria, thereby extending survival compared to saline control groups and overall outperforming the HV3-TAT peptide at approximately half the dose.
In summary, functional group tolerant polymerization methods provide a route to the incorporation of complex monomers into well-defined, discrete polymer chains. Here, the polymerization of peptide-modified norbornenyl monomers to generate peptide brush polymers, results in a functional therapeutic entity that displays serum stability, cell-penetrative capabilities, and on-target disruption of a disease-relevant PPI in the CNS, following intravenous administration. Excitingly, these studies support the idea of “polymers as therapeutics,” rather than polymers as carriers of drug molecules in the typical sense of serving as a payload delivery system. Of course, biologics in the form of antibodies are precisely polymeric therapeutics although not described as such and not, to date, capable of intracellular activity. Instead, antibodies and proteins generally are largely restricted in the clinic to extracellular targets (54). If generalizable, the PLP then serves as a platform technology for taking lead peptides into the cell, with promise as tools for engaging difficult and currently undruggable protein targets, in particular, the troublesome collection of PPIs that exhibit featureless yet disease driving interfaces. Last, given the encouraging performance of P1 in this model system, future studies will include optimization of the platform for blood-brain barrier crossing in vivo to take full advantage of extended half-lives. Moreover, given the efficacy of P1 in cultured cells and animal models, it is plausible that P1 will exhibit similar, if not superior, protective effects in neurons derived from induced pluripotent stem (iPS) cells of patients with HD. These experiments are the subject of ongoing investigations to fully explore the translational therapeutic potential of P1 and the platform technology overall.
MATERIALS AND METHODS
Experimental design
Preparation of peptide monomers via SPPS (general procedure)
All peptide monomers including M1, M2, M3, and M4 were synthesized on rink resin (0.67 mmol/g) using standard 9-fluorenyl methoxycarbonyl (Fmoc) solid-phase peptide synthesis (SPPS) procedures on an AAPPTec Focus XC automated synthesizer or Liberty Blue Automated Microwave Synthesizer. After that, M1 to M4 peptide monomers were prepared via amide coupling to N-(hexanoic acid)-cis-5-norbornene-exo-dicarboximide (3.0 equiv.) in the presence of HBTU (2.9 equiv.) and DIPEA (6.0 equiv.). The peptide monomers were cleaved off the resin by treating the resin with trifluoroacetic acid (TFA)/H2O/triisopropylsilane (TIPS) (95:2.5:2.5 v/v) for 2 hours. The crude products were obtained by precipitation in cold diethyl ether and further purified via preparative HPLC.
Synthesis of P1, P2, P3, and P4 via ROMP
HV3 PLPs were achieved by ROMP under nitrogen gas in a glove box. Norbornene-conjugated peptide monomers (20 mg, 15.0 equiv., 30 mM) were dissolved in degassed N,N′-dimethylformamide (DMF) with 1 M LiCl. Next, the olefin metathesis initiator (IMesH2)(C5H5N)2(Cl)2Ru = CHPh stock solution (1.0 equiv., 20 mg/ml in DMF) was quickly added into the monomer solution. The solution was left to stir for 12 hours until the full consumption of monomers as initially determined by NMR. After the polymerization, the polymer solution was precipitated in diethyl ether and was further purified via dialysis into deionized water. Last, the polymer product was obtained by lyophilization.
VCP protein association assay
HEK293 cells were cultured and plated 1 day before treatment. The cells were pretreated with HV3-TAT or PLPs at 3 μM for 2 hours. The cells were then cotransfected with green fluorescent protein–VCP and myc-Q73 FL plasmids. The cells were treated with a second dose of peptide or PLPs 6 hours after transfection, and a final third dose of peptide or PLPs treatment was administered 24 hours after transfection. Cell total lysate was harvested 48 hours after transfection, and IP analysis was performed to test the binding between mtHtt and VCP [IP myc—Western blot (WB) VCP].
Synthesis of HV3-TAT-Rho
HV3-TAT was synthesized on a peptide synthesizer on rink resin with an additional Mtt protected lysine at the C terminus of the HV3-TAT peptide following general procedure (3.1). The Mtt protection group was deprotected with TFA/TIPS/dichloromethane (DCM) (1:2:97 v/v/v), and Rhodamine B (481 mg, 1.005 mmol) was coupled to HV3-TAT (0.5 mg on resin loaded at 0.67 mmol/g, 0.335 mmol).
Synthesis of P1-Rho
P1-Rho achieved by ROMP under nitrogen gas in a glove box. M1 (51.4 mg, 15.0 equiv., 30 mM) was dissolved in degassed DMF with 1 M LiCl. Next, the olefin metathesis initiator (IMesH2)(C5H5N)2(Cl)2Ru = CHPh stock solution (1.0 equiv., 20 mg/ml in DMF) was quickly added into the monomer solution. The solution was left to stir for 12 hours until the full consumption of monomers. Then, norbornene-coupled rhodamine [fig. S9A, prepared as described in a previous report (55)] was added (1.62 mg, 1 equiv., 10 mM) for 3 hours. After the polymerization, the polymer solution was precipitated in diethyl ether and was further purified via dialysis into deionized water. Last, the polymer product was obtained by lyophilization.
Confocal laser scanning microscopy
Mouse striatal HdhQ111 cells were plated in a 4-chamber 35-mm round glass-bottom dishes at a density of 50,000 per well. The cells were incubated for 24 hours in a 5% CO2 atmosphere at 33°C. HV3-TAT, HV3 PLPs, rhodamine dye, and vehicle control in 10% FBS Dulbecco’s modified Eagle’s medium (DMEM) without phenol red were incubated with the cells for 24 hours, respectively. The cell nuclei (stained with Hoechst) were accomplished using a 405-nm laser with a 15% laser power. The cell membrane (stained with wheat germ agglutinin, Alexa Fluor 488 conjugate) was accomplished using a 488-nm laser with a 12% laser power. The mitochondria (stained with MitoTracker Green FM) were accomplished using 490-nm laser with a 12% laser power. Cell imaging for rhodamine fluorescence was accomplished using a 581-nm laser with an 8% laser power.
Cellular uptake measurement
For cellular uptake measurement, mouse striatal HdhQ111 cells were plated in 12-well plates at a density of 100,000 cells per well and incubated for 24 hours in a 5% CO2 atmosphere at 33°C. HV3-TAT, HV3 PLPs, rhodamine dye, and vehicle control in 10% FBS DMEM without phenol red were incubated with the cells for 24 hours, respectively. After triple washing with Dulbecco’s phosphate-buffered saline (DPBS), 100 μl of 0.25% Trypsin-EDTA was added to each well for 10 min at 33°C. The cell uptake study was analyzed through flow cytometry by using a BD FacsAria IIu 4-Laser flow cytometer (Becton Dickinson Inc., USA). Mean fluorescence intensity was prepared for presentation using FlowJo v10.
Mitochondrial fragmentation
HdhQ111 cells were treated with HV3-TAT or PLPs at 3 μM three times for 3 days. Confocal immunofluorescence imaging of the mitochondrial marker Tom20 were performed after treatments. Briefly, cells grown on coverslips were washed with cold PBS, fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 2% normal goat serum at room temperature. The cells were incubated overnight at 4°C with primary antibodies against Tom20 (1:4000, Proteintech, 11802-1-AP) followed by Alexa Fluor 488–labeled secondary antibodies (2 hours at room temperature). After staining of nuclei with Hoechst dye (1:10,000 dilution), the cells were mounted onto slides and imaged using an Olympus Fluoview FV1000 confocal microscope. The percentage of cells with fragmented mitochondria relative to the total number of cells was quantified. At least 95 cells per group were counted.
BLI (BLItz) binding assay
Binding affinity was measured by using BLItz instrument. The anti-HIS sensor (HIS2) was hydrated before the installation. After the sensor installation to the BLItz instrument, histidine-tagged VCP proteins (50 μg/ml in sodium acetate buffer at pH 6.5) were immobilized on the sensor. HV3-TAT and P1 were dissolved in the tris buffer (pH 8.4) with range of concentration. Using BLItz instrument, the dissociation constant (KD), on-rate (ka), and off-rate (kd) of each analyte were measured.
Evaluation of the stability of PLP against protease
For enzyme-induced cleavage experiments, the molar concentration of enzymes was set to 0.1 μM. The concentration of side-chain peptides (HVLVMSATRR) varied in the range of 50 to 200 μM. For each enzyme, the temperature was set for optimal enzyme activity. In a typical experiment, P1 or HV3-TAT was dissolved in DPBS (pH 7.4) or hydrochloric acid solution (pH 2) to obtain a stock polymer/peptide solution with a peptide concentration of 200 μM. Next, 5 μl of enzyme stock solution (10 μM) was added into 500 μl of the polymer/peptide solution, which was subsequently stirred in a preheated oil bath at 37°C. In this case, the molar ratio of peptide substrate to enzymes was 2000:1. During the cleavage, aliquots were taken for HPLC analysis at predetermined time points. Each degradation experiment was repeated three times. Enzyme pretreated P1 was also run on SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and visualized with Coomassie Blue staining.
Serum stability assay
Polymers/peptide (200 μM with respect to peptide) were incubated in DMEM supplemented with 10 or 25% FBS at 37°C. At each time point, 50 μl of cold acetonitrile with 0.1% TFA was added to 50 μl of aliquot to deproteinize the plasma proteins and subjected to centrifugation at 4°C for 15 min at 10,000 rpm. The supernatant was injected to the HPLC to analyze degradation kinetics.
Synthesis of EDANS-DABCYL monomer
Fmoc-Lys(dabcl)-OH (Sigma-Aldrich) and Fmoc-Glu(EDANS)-OH (Sigma-Aldrich) were coupled on a peptide synthesizer to opposing ends of the peptide sequence of M1 as shown in fig. S12A.
Synthesis of P1- EDANS-DABCYL polymer
P1-ED was achieved by ROMP under nitrogen gas in a glove box. M1-EDANS-DABCYL (8.73 mg, 10.0 equiv.) was dissolved in degassed DMF with 1 M LiCl. Next, the olefin metathesis initiator (IMesH2)(C5H5N)2(Cl)2Ru = CHPh stock solution (0.3 mg, 1.0 equiv) was quickly added into the monomer solution. The solution was left to stir for 12 hours until the full consumption of monomers. After the polymerization, the polymer solution was precipitated in diethyl ether and was further purified via dialysis into deionized water. Last, the polymer product was obtained by lyophilization.
Enzyme resistance of fluorogenic polymer
Different enzymes (0.1 μM) were treated to the EDANS-DABCYL–conjugated monomer (M1-ED) and polymer (P1-ED). The concentration of M1-ED and P1-ED was set to 50 μM with respect to peptide. The fluorescence changes were observed by a plate reader for 300 min.
Liver microsomal stability of P1 and HV3-TAT
P1 or HV3-TAT (2 mg/ml) were incubated in pooled human liver microsome (0.5 mg/ml) activated by 1 mM NADPH cofactor. At each time point, 10 μl of supernatant was diluted to the 90 μl of water with 0.1% TFA. Degradation kinetics of P1 or HV3 were analyzed by analytic HPLC.
Cell viability assay and enzyme pretreatment cell viability assay
Mouse striatal HdQ111 neurons were grown in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were maintained at 33°C and 5% CO2 with a relative humidity of 95%. HdQ111 cells were plated in 96-well plates at a density of 5000 cells per well and then left to attach for 24 hours. The cells were treated with HV3-TAT or PLPs at 3 μM with respect to peptide once a day for 3 days. After the second day of treatment, the cells were washed twice with serum-free medium and then incubated in serum-free medium containing HV3-TAT or PLPs to induce a starvation state. The medium was replaced every day. After 72 hours, the cells were washed three times with PBS buffer. At this point, 20 μl of CellTiter-Blue (CTB) reagent in 80 μl of medium was added, and the cells were incubated for 3 hours at 37°C. Fluorescence was measured at 590 nm with excitation at 560 nm using a PerkinElmer EnSpire plate reader. The average background fluorescence of CTB in media without cells was subtracted from the average fluorescence readings of the experimental wells. Viability was calculated as the average background-subtracted signal in a well compared to that of a negative control well (treatment with vehicle or media). Three replicates were performed for each independent sample. Ten % dimethyl sulfoxide was used as a positive control and untreated cells in complete medium as a negative control. Viability is reported as a percentage of untreated cells.
C3a complement activation evaluation by ELISA
Tested compounds were incubated at a ratio of 1:250 and 1:500 of compound to human serum. Compounds were incubated in human serum for 4 hours before dilution and start of the assay. Using a C3a ELISA (Thermo Fisher Scientific, BMS2089) and following the manufacturer’s protocol, the samples were diluted with sample diluent and C3a control reconstituted with water. Cobra venom factor, structurally similar to C3a, was used as positive control, with DPBS as negative vehicle control. Serum samples and controls were incubated in C3a antibody–coated wells for 2 hours with gentle shaking before they were gently washed three times with washing buffer and primary antibody biotin solution administered to wells. After 1 hour of shaking, the plate was washed three times and streptavidin solution added for 1 hour. After a final wash, developing substrate was added for 30 min before imaging on plate reader at 450 nm.
Blood dilutions for hemocompatibility
Peptide-in-blood and polymer-in-blood dilutions were performed at volume ratios of 1:10,000, 1:5000, 1:1000, 1:500, 1:250, and in some cases 1:100 as described previously in our laboratory (46). Given efficient distribution after injection, we could expect a peptide-in-blood or polymer-in-blood dilution to be between 1:1000 and 1:500, whereas concentration immediately after bolus in tail vein could be as high as 1:250 to 1:100, informing higher concentrations. However, we anticipated efficient distribution of compounds (as would be informed by pharmacokinetics), and the injection will take place over the course of minutes, allowing the compounds to diffuse. Efficacy would be performed with compounds released via Alzet pump, resulting in even further blood dilution, and thus our concentration range is sufficient to inform us of hemocompatibility issues even with buildup over time at dilute levels.
Hemocompatibility: ACT ex vivo
Hemocompatibility was performed as previously described in our laboratory (46). Briefly, ACT of whole human blood was measured using a Hemochron 801 instrument calibrated with electronic system verification. To minimize variability in starting time points for clotting in all assays, ACTs were measured using sodium citrate–treated whole human blood with calcium chloride added at a specific time. Controls with type I collagen (0.095 mg/ml, 1:240 dilution) or without calcium both used 1× DPBS as a vehicle to ensure consistent blood dilution in all experiments. Four microliters of CaCl2 (1.1 M) and 36 μl of peptide stock solution (12.2× final blood concentration) was added to each Hemochron P214 tube with glass beads. The samples were mixed, and citrated whole human blood (400 μl) was then added, which marked t = 0 s, mixed by hand for 10 s, and added to the instrument. Clot formation was recorded by the instrument as the time points at which the magnet was displaced by the formed clot. Each experiment was performed n = 4 times with averages and SE plotted. For samples without calcium, blood clotting times exceeded the instrument maximum range (>1500 s).
Hemocompatibility: Hemolysis
As previously described in our laboratory (46), hemolytic assessments required 25 ml of whole human blood mixed with Na citrate anticoagulant to be obtained. To prepare solution of erythrocytes, or red blood cells, whole blood was centrifuged at 500g for 5 min and plasma aspirated. The same volume of plasma removed was replaced with 150 mM NaCl buffer (in Millipore water). The tube was inverted to mix, centrifuged at 500g for 5 min, and the supernatant removed, and this step was repeated twice. Last, red blood cells were resuspended in PBS. Ten microliters of compound was added to 190 μl of diluted red blood cell erythrocyte solution in a 96-well plate (n = 4). Twenty % Triton X-100 was positive control; PBS is negative control/vehicle. Plates were incubated at 37°C for 1 hour. The plates were centrifuged for 5 min at 500g to pellet intact erythrocytes. Using a multichannel pipette, 100 μl of supernatant was transferred from each well into a clear, flat-bottomed 96-well plate. Absorbance of supernatants was measured with a plate reader at 405 nM.
Synthesis and characterization of DOTA terminating agent
First, 51.69 mg (0.10375 mmol) 4,4′-[(2Z)-2-Butene-1,4-diylbis(oxy)]bis[benzeneethanamine] (prepared via methods from a previous report) (56) was deprotected with 3 ml of TFA in 3 ml of DCM for 2 hours and rotovapped to leave a brown oil and then mixed with DOTA-NHS Ester (Macrocyclics, B-280), 158 mg (0.2075 mmol) in DIPEA (5 ml) and DCM (5 ml) and MeOH (1 ml). The reaction mixture was stirred at room temperature for 48 hours. Solvent was removed in vacuo leaving a brown oil. NMR peaks: 1H NMR- D2O- 7.25 to 6.85 (m, 8H), 5.9 (m, 2H), 4.7 (m,4H), 4.1-2.6 (m, 32H), 2 (s, 1H), 1.9 (s, 1H)
Synthesis and characterization of Gd-DOTA terminating agent
DOTA-TA (35.92 mg) was dissolved in DCM (5 ml), and 2 equiv. of Gd(OAc)3 was added (25.87 mg, 0.0656 mmol). The reaction was stirred for 48 hours at room temperature. Solvent was removed in vacuo to yield Gd-DOTA-TA as a white microcrystalline solid. This powder was purified using preparative HPLC on a gradient of 25 to 35% buffer B (MeCN with 0.1% TFA) in buffer A (water with 0.1% TFA). Purified, lyophilized product was characterized via electrospray ionization (ESI) and reversed-phase HPLC. Gd metalation efficiency was confirmed via ICPMS. Briefly, 0.68 mg of Gd-DOTA-TA was weighed and digested overnighted, diluted, and then run on ICPMS where metalation efficiency was determined to be 98.99%.
P1-Gd polymerization
P1-Gd was achieved by ROMP under nitrogen gas in a glove box. M1 (88.2 mg, 15.0 equiv., 30 mM) was dissolved in degassed DMF with 1 M LiCl. Next, the olefin metathesis initiator (IMesH2)(C5H5N)2(Cl)2Ru = CHPh stock solution (3 mg, 1.0 equiv., 10 mM) was quickly added into the monomer solution. The solution was left to stir for 12 hours until the full consumption of monomers. Then, Gd-DOTA-TA was added (8.71 mg, 1.5 equiv., 10 mM). After the polymerization, the polymer solution was precipitated in diethyl ether and was further purified via dialysis into deionized water. Last, the polymer product was obtained by lyophilization. Gd metalation efficiency was confirmed via ICPMS. Briefly, serial dilutions of P1-Gd were prepared in Millipore H2O, digested according to ICPMS protocol, and ICPMS run, confirming metalation efficiency at 95%.
Pharmacokinetics and biodistribution via ICPMS
CD1 mice were purchased from Charles River laboratory and housed in Northwestern University’s CCM core, handled by the DTC. All of the mice were maintained with a 12-hour light/dark cycle (on, 06:00 hours; off, 18:00 hours) and acclimated for 1 week. The mice were dosed intravenously with 10 mg/kg with respect to peptide (5.54 mg/kg of polymer) with a dose for a 30-g mouse being 0.189 mg of polymer in 100 μl of saline. At time points of 5, 10, 20, 30, 40, and 50 min, and 1, 2, 4, 8, 24, 48, 72, 96, and 168 hours, five mice were anesthetized and exsanguinated. Whole blood was collected in preweighed Falcon tubes. Livers, kidneys, striatum, cortex, and the rest of the brain tissue were collected in preweighed tubes. The tubes were weighed again to determine total mass of tissue collected in each tube. To each tube of all samples except liver, 300 μl of conc nitric acid and 30% hydrogen peroxide were added. Liver samples had 500 μl of conc nitric acid and 30% hydrogen peroxide added. All samples were digested in a 65°C water bath for 24 hours until the solutions were transparent. The tubes were diluted to 5 ml of total volume (10 ml for liver) with Millipore water and weighed for a final time. After this point, any excess digested tissue that precipitated was removed by centrifugation and the supernatant saved. Each sample was run on high-resolution ICPMS, the Thermo iCAP Q ICPMS, which, after calibration with known Gd concentrations, resulted in exact Gd concentration in parts per billion (ppb) of each sample. The ppb of Gd were directly related to the calibration curve of known Gd PLP concentrations and was used to calculate the amount of PLP present in each sample. Pharmacokinetics expert M.J.A. was consulted to perform pharmacokinetics analysis and assist in plotting the fit of the two-compartment pharmacokinetic model to the measured blood concentrations.
Toxicity analysis in C57BL/6J mice
To assess the toxicity of P1 at the administrable dose in healthy animals, as compared to HV3-TAT and at its operating dose and saline, male C57BL/6J mice (The Jackson Laboratories) were assigned to three groups (n = 3 to 4 per group) and were administered via osmotic subcutaneous pump (Alzet) either HV3-TAT at 3 mg/kg, P1 at 3 mg/kg, all with respect to peptide concentration, or saline as a negative control for 8 weeks. Animals were checked weekly after drug administration for mortality. At euthanasia, blood was collected from each animal via cardiac puncture using a 25-gauge needle while under isoflurane, and then the animals were euthanized and major organs (brain, lung, heart, liver, spleen, spinal cord, and kidneys) collected, weighed, and stored in 10% buffered formalin followed by paraffin embedment. A complete blood count panel and serum chemical analysis was obtained (University of Illinois Veterinary Diagnostic Laboratory, Buffalo Grove, IL), with no significant difference between groups. All values were not significantly different from saline control values, suggesting no toxicity. The tissues were stained with hematoxylin and eosin (H&E) and scored by a senior pathologist at Charles River Laboratory. No pathological differences between the treated (P1 or HV3-TAT) and untreated (saline) were observed.
Histological scoring
The histologic evaluation of the tissue was performed using a five-grade scoring system to record the severity of histopathology. Each H&E slide was carefully reviewed for changes in cell size, shape, number, and organization, including necrosis, apoptosis, and dysplasia. Disruption of normal tissue structure, such as inflammation, fibrosis, or necrosis, was also screened for. Some tissues have organ-specific changes that are evaluated, such as the presence of fibrosis, mesenchymal cells, hematopoietic cells, basophilia, and inflammation. Severity was scored as follows: 0 = none, 1 = minimal, 2 = mild, 3 = moderate, 4 = marked, and 5 = severe. Specific findings were noted in the microscopic findings in table S6 (Supplementary Materials), which were deemed not notable enough to declare toxicity.
Animal models of HD
All animal studies were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of Case Western Reserve University and were performed on the basis of the National Institutes of Health Guide for the Care and Use of Laboratory Animals (protocol number: 2015-0024; Case Western Reserve University). Sufficient procedures were used for reducing pain or discomfort of mice during the experiments. Male R6/2 mice and their WT littermates (5 weeks old) were purchased from The Jackson Laboratory [Bar Harbor, ME, USA; B6CBA-TgN (HD exon 1); JAX stock number: 006494]. R6/2 mice (C57BL/6 and CBA genetic background) are transgenic for the 5′ end of the human HD gene carrying 100 to 150 glutamine (CAG) repeats. Male R6/2 mice at 6 to 13 weeks were used in the study. All mice were maintained with a 12-hour light/dark cycle (on at 06:00 hours and off at 18:00 hours).
Systemic peptide treatment in HD mice
All randomization and peptide treatments were prepared by an experimenter not associated with behavioral and neuropathology analysis. Male hemizygous R6/2 mice (Tg) and their age-matched WT littermates (6-week-old) were implanted with a 28-day osmotic pump (Alzet, Cupertino CA) containing saline (negative control), HV3-TAT peptide (3 mg kg−1 day−1), or P1 (1.69 mg kg−1 day−1). The first pump was implanted subcutaneously in the back of 6-week-old mice between the shoulders and replaced once after 4 weeks. By the age of 13 weeks, the treatments were terminated and the mouse samples were harvested for analysis.
Behavioral analysis in HD mice
All behavioral analyses were conducted by a researcher blinded to genotypes and treatment groups. Gross locomotor activity was assessed in R6/2 mice and age-matched WT littermates at the ages of 12 weeks. In an open-field activity chamber (Omnitech Electronics Inc., Columbus, OH, USA), the mice were placed in the center of the chamber and allowed to explore while being tracked by an automated beam system (Vertax, Omnitech Electronics Inc). The analyzer records the time and position of the mice based on beam break information and rapidly analyzes it. The computer software then calculates multiple activity measures, including total distance traveled, horizontal activities (ambulation, horizontally directed movement), and vertical activities (rearing, vertically directed movement). One hour of locomotor activity analysis was conducted for R6/2 mice and WT littermates.
Hindlimb clasping was assessed with the tail suspension test weekly from the ages 7 to 13 weeks in R6/2 mice and WT littermates. Briefly, the mice were suspended by the tail for 20 s, and the latency for the hindlimbs or all four paws to clasp was recorded using the following score system: clasping over 10 s, score 3; 5 to 10 s, score 2; 0 to 5 s, score 1; 0 s, score 0. The body weight and survival rate of R6/2 mice and WT littermates were also recorded throughout the study period.
Preparation of total lysates
Cells were washed in cold PBS (pH 7.4) and incubated on ice for 30 min in total lysis buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, protease inhibitors cocktail, and phosphatase inhibitors cocktail (MilliporeSigma)]. Mouse brains were minced and homogenized in lysis buffer and placed on ice for 30 min. Cells or tissues were centrifuged at 12,000g for 10 min at 4°C to generate total lysate supernatants.
Isolation of mitochondria-enriched fractionations
Mouse brains were minced and homogenized in mitochondrial lysis buffer [250 mM sucrose, 20 mM Hepes-NaOH (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, protease inhibitor cocktail, and phosphatase inhibitor cocktail] to release mitochondria. The mouse brains were disrupted 20 times by repeated aspiration through a 25-gauge needle and then incubated on ice for 30 min. The homogenates were spun at 800g for 10 min at 4°C, and the resulting supernatants were spun at 10,000g for 20 min at 4°C. The pellets were washed with mitochondrial lysis buffer and spun at 10,000g again for 20 min at 4°C. The final pellets of mitochondria were suspended in lysis buffer containing 1% Triton X-100 and were assigned as mitochondrial-rich lysate fractions.
Western blotting–SDS-PAGE
The protein concentration in each sample was determined using protein assay dye reagents (Bio-Rad, 5000006). The proteins were resuspended in Laemmli buffer, separated using SDS gels, and transferred to nitrocellulose membranes. The membranes were probed with the indicated antibodies, and the specific proteins were visualized by electrochemiluminescence. The antibodies used in the study are listed below: anti–DARPP-32 (ab40801, Abcam, Cambridge, UK, 1:3000), anti-huntingtin protein (MAB5374, clone EM48, MilliporeSigma, 1:1000), anti-BDNF (ab108319, Abcam, 1:1000), anti–β-actin (A1978, MilliporeSigma, 1:10,000), anti- PSD95 (2507, Cell Signaling Technology, Danvers, MA, USA, 1:5000), VCP (ab109240, Abcam, Cambridge, UK, 1:5000), anti-ATPB (17247-1- AP, Proteintech, 1:3000), anti-VDAC1 (14734, Abcam, 1:2000), anti–c-myc (sc-40, Santa Cruz Biotechnology, 1:1000), horseradish peroxidase (HRP)–conjugated anti-rabbit, or anti-mouse IgG (31430/31460, Thermo Fisher Scientific, 1:5000).
Immunohistochemistry
Mice were anesthetized with isoflurane and transcardially perfused with PBS. After being postfixed with 4% paraformaldehyde, the brains were sliced and frozen. The brain sections (20 μm, coronal) were used for immunohistochemical localization of DARPP-32 and mtHtt aggregation. Briefly, frozen brain sections were hydrated by tris-buffered saline wash (twice). After antigen retrieval in 0.01 M sodium citrate buffer plus 0.1% Tween 20 (pH 6.0), the slides were incubated with 3% hydrogen peroxide (H2O2) in methanol to quench endogenous peroxidase. They were then treated with 5% normal goat serum (Invitrogen) in tris-buffered saline with Tween 20 (TBST) for 1 hour at room temperature. The sections were then incubated with anti–DARPP-32 (1710-1, Epitomics, Burlingame, CA, USA; 1:500) and anti-huntingtin protein (MAB5374, clone EM48, MilliporeSigma, 1:500) in a humidified chamber overnight at 4°C. The next day, the slides were incubated with biotin-conjugated secondary antibody (goat anti-mouse/rabbit) and streptavidin-conjugated HRP using an immunohistochemistry select HRP/DAB kit (MilliporeSigma, DAB150) and DAB solution following the manufacturer’s instructions. Slides were mounted after sequential dehydration in water, 50% ethanol, 70% ethanol, 95% ethanol, 100% ethanol, and xylene. Images were captured using a digital microscope (VHX-7000, Keyence, Osaka, Japan). Quantitation of DARPP-32 intensity and the number of mtHtt aggregates per area were conducted using the ImageJ software. The same image exposure times and threshold settings were used for all the group sections.
Materials
All materials and reagents, unless otherwise noted, were purchased from Sigma-Aldrich and Fisher Chemicals. All amino acids used to prepare peptides by SPPS were obtained from AAPPTec, Chem-Impex, and NovaBiochem. N-(hexanoic acid)-cis-5-norbornene-exo-dicarboximide and olefin metathesis initiator (IMesH2)(C5H5N)2(Cl)2Ru = CHPh was synthesized according to previous report (32, 56). CellTiter-Blue was purchased from Promega Corporation. DPBS with Ca2+ and Mg2+ was purchased from Corning Cellgro. VCP (His tag) was purchased from Antibodies-online. All the enzymes were purchased from Promega. Protein phosphatase inhibitor cocktail (P5726) and protease inhibitor cocktails (8340) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Statistical analysis
Sample sizes were determined by power analysis based on pilot data from our laboratories or published studies. For animal studies, we used n = 13 to 32 mice per group for behavioral tests, n = 6 to 8 mice per group for neuropathology analysis, and n = 3 to 4 mice per group for pathology studies. In cell culture studies, each experiment was performed with at least three independent replications. All animal studies were conducted with randomization and blinded experimental procedures. For imaging analysis, quantification was conducted by an observer blinded to the experimental groups. No samples or animals were excluded from the analysis.
Data were analyzed using Student’s t test or ANOVA with Tukey’s post hoc test for comparisons between two groups. Survival, behavioral tests, and body weight were analyzed by repeated-measures one-way ANOVA. Data are expressed as means ± SEM. Statistical significance was considered achieved when P < 0.05.
Acknowledgments
We would like to thank the members of the Gianneschi laboratory for all the insightful discussions and support. This work made use of (i) the IMSERC MS facility at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-2025633), the State of Illinois, the International Institute for Nanotechnology (IIN), and Northwestern University; (ii) the IMSERC NMR facility at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-2025633), NSF CHE-1048773, and Northwestern University; (iii) the Keck Biophysics Facility, a shared resource of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University supported in part by the NCI Cancer Center Support Grant #P30 CA060553; (iv) the Biological Imaging Facility at Northwestern University (RRID:SCR_017767), supported by the Chemistry for Life Processes Institute, the NU Office for Research, the Department of Molecular Biosciences. Microscopy was performed at the Northwestern University Center for Advanced Microscopy supported by CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center; and (v) the Quantitative Bio-element Imaging Center, supported by NASA Ames Research Center NNA06CB93G. This work was supported by the Developmental Therapeutics Core at Northwestern University and the Robert H. Lurie Comprehensive Cancer Center support grant (NCI CA060553).
Funding: This work was supported by the International Institute of Nanotechnology Convergence Science Medicine Institute grant (N.C.G.), National Institutes of Health award 1F31AG076334 (M.F.), National Institutes of Health award 1F30AG076317 (K.P.C.), International Institute of Nanotechnology Ryan Fellowship (M.F. and K.P.C.), National Institutes of Health grant R01AG065240 (X.Q.), National Institutes of Health grant R01NS115903 (X.Q.), National Institutes of Health grant R01AG076051 (X.Q.), and National Institutes of Health grant RF1AG074346 (X.Q.).
Author contributions: Conceptualization: N.C.G., X.Q., W.C., M.F., M.P.T., and O.B. Investigation: X.Q., W.C., M.F., M.P.T., Y.S., Z.L., D.H., K.B., and O.B. Visualization: M.F., W.C., Y.S., and K.P.C. Methodology: X.Q., W.C., M.F., M.J.A., K.P.C., and O.B. Formal analysis: W.C., M.F., Y.S., M.J.A., K.P.C., and Z.L. Validation: X.Q., M.F., W.C., and Y.S. Data curation: W.C. Project administration: N.C.G., M.F., M.P.T., and W.C. Funding acquisition: N.C.G. and X.Q. Resources: M.F., M.P.T., W.C., and Z.L. Writing—original draft: W.C., M.F., and Y.S. Writing—review and editing: N.C.G., M.F., W.C., M.J.A., Y.S., K.P.C., K.B., and O.B. Supervision: N.C.G., W.C., and O.B.
Competing interests: N.C.G. is a cofounder of Grove Biopharma to whom intellectual property related to this work is exclusively licensed. N.C.G, W.C., X.Q., and M.F. are all coinventors on a patent filed by Northwestern University on 2023-08-18 (App. Num. US18/452,059, pending) related, in part, to the work described herein. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Instrumentation
Figs. S1 to S35
Tables S1 to S6
REFERENCES AND NOTES
- 1.Frank S., Treatment of Huntington’s disease. Neurotherapeutics 11, 153–160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan L., Feigin A., Huntington’s disease: New frontiers in therapeutics. Curr. Neurol. Neurosci. Rep. 21, 10 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Gupta S., Khan A., Vishwas S., Gulati M., Gurjeet T., Dua K., Kumar S., Najda A., Sayed A. A., Almeer R., Abdel-daim M. M., Demethyleneberberine: A possible treatment for Huntington’s disease. Med. Hypotheses 153, 110639 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Gibson J. S., Isaacs D. A., Claassen D. O., Stovall J. G., Lifetime neuropsychiatric symptoms in Huntington’s disease: Implications for psychiatric nursing. Arch. Psychiatr. Nurs. 35, 284–289 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Ferguson M. W., Kennedy C. J., Palpagama T. H., Waldvogel H. J., Faull R. L. M., Kwakowsky A., Current and possible future therapeutic options for Huntington’s disease. J. Cent. Nerv. Syst. Dis. 14, 117957352210925 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez-Sanchez M., Licitra F., Underwood B. R., Rubinsztein D. C., Huntington’s disease: Mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb. Perspect. Med. 7, a024240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franco-Iborra S., Vila M., Perier C., Mitochondrial quality control in neurodegenerative diseases: Focus on Parkinson’s disease and Huntington’s disease. Front. Neurosci. 12, 342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin X., Manczak M., Reddy P. H., Mitochondria-targeted molecules MitoQ and SS31 reduce mutant huntingtin-induced mitochondrial toxicity and synaptic damage in Huntington’s disease. Hum. Mol. Genet. 25, 1739–1753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahamad S., Bhat S. A., The emerging landscape of small-molecule therapeutics for the treatment of Huntington’s disease. J. Med. Chem. 65, 15993–16032 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Kumar A., Kumar Singh S., Kumar V., Kumar D., Agarwal S., Rana M. K., Huntington’s disease: An update of therapeutic strategies. Gene 556, 91–97 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kordasiewicz H. B., Stanek L. M., Wancewicz E. V., Mazur C., McAlonis M. M., Pytel K. A., Artates J. W., Weiss A., Cheng S. H., Shihabuddin L. S., Hung G., Bennett C. F., Cleveland D. W., Sustained therapeutic reversal of Huntington’s disease by transient repression of Huntingtin synthesis. Neuron 74, 1031–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawlins M. D., Wexler N. S., Wexler A. R., Tabrizi S. J., Douglas I., Evans S. J. W., Smeeth L., The prevalence of Huntington’s disease. Neuroepidemiology 46, 144–153 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Jurcau A., Jurcau M. C., Therapeutic strategies in Huntington’s disease: From genetic defect to gene therapy. Biomedicine 10, 1895 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bano D., Zanetti F., Mende Y., Nicotera P., Neurodegenerative processes in Huntington’s disease. Cell Death Dis. 2, e228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui M., Yoshimori T., Nakamura S., Autophagy system as a potential therapeutic target for neurodegenerative diseases. Neurochem. Int. 155, 105308 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Guo X., Sun X., Hu D., Wang Y. J., Fujioka H., Vyas R., Chakrapani S., Joshi A. U., Luo Y., Mochly-Rosen D., Qi X., VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington’s disease. Nat. Commun. 7, 12646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bibb J. A., Yan Z., Svenningsson P., Snyder G. L., Pieribone V. A., Horiuchi A., Nairn A. C., Messer A., Greengard P., Severe deficiencies in dopamine signaling in presymptomatic Huntington’s disease mice. Proc. Natl. Acad. Sci. U.S.A. 97, 6809–6814 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mäe M., Langel Ü., Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr. Opin. Pharmacol. 6, 509–514 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Zou L., Peng Q., Wang P., Zhou B., Progress in research and application of HIV-1 TAT-derived cell-penetrating peptide. J. Membr. Biol. 250, 115–122 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Feng S., Holland E. C., HIV-1 tat trans-activation requires the loop sequence within tar. Nature 334, 165–167 (1988). [DOI] [PubMed] [Google Scholar]
- 21.Rizzuti M., Nizzardo M., Zanetta C., Ramirez A., Corti S., Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov. Today 20, 76–85 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Muttenthaler M., King G. F., Adams D. J., Alewood P. F., Trends in peptide drug discovery. Nat. Rev. Drug Discov. 20, 309–325 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Otvos L. Jr., Wade J. D., Current challenges in peptide-based drug discovery. Front. Chem. 2, 62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callmann C. E., Thompson M. P., Gianneschi N. C., Poly(peptide): Synthesis, structure, and function of peptide–polymer amphiphiles and protein-like polymers. Acc. Chem. Res. 53, 400–413 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craik D. J., Fairlie D. P., Liras S., Price D., The future of peptide-based drugs. Chem. Biol. Drug Des. 81, 136–147 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Fosgerau K., Hoffmann T., Peptide therapeutics: Current status and future directions. Drug Discov. Today 20, 122–128 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Harris D., Robinson J. R., Bioadhesive polymers in peptide drug delivery. Biomaterials 11, 652–658 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Sun H., Qiao B., Choi W., Hampu N., McCallum N. C., Thompson M. P., Oktawiec J., Weigand S., Ebrahim O. M., De La Cruz M. O., Gianneschi N. C., Origin of proteolytic stability of peptide-brush polymers as globular proteomimetics. ACS Cent. Sci. 7, 2063–2072 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M. W., Han M., Bossa G. V., Snell C., Song Z., Tang H., Yin L., Cheng J., May S., Luijten E., Wong G. C. L., Interactions between membranes and “metaphilic” polypeptide architectures with diverse side-chain populations. ACS Nano 11, 2858–2871 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Blum A. P., Kammeyer J. K., Yin J., Crystal D. T., Rush A. M., Gilson M. K., Gianneschi N. C., Peptides displayed as high density brush polymers resist proteolysis and retain bioactivity. J. Am. Chem. Soc. 136, 15422–15437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blum A. P., Kammeyer J. K., Gianneschi N. C., Activating peptides for cellular uptake via polymerization into high density brushes. Chem. Sci. 7, 989–994 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kammeyer J. K., Blum A. P., Adamiak L., Hahn M. E., Gianneschi N. C., Polymerization of protecting-group-free peptides via ROMP. Polym. Chem. 41, 3929–3933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V. C., Sharp A. H., Persichetti F., Cattaneo E., MacDonald M. E., Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 9, 2799–2809 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Yhnell E., Dunnett S. B., Brooks S. P., A longitudinal operant assessment of cognitive and behavioural changes in the HdhQ111 mouse model of Huntington’s disease. PLOS ONE 11, e0164072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X., Qi X., VCP cooperates with UBXD1 to degrade mitochondrial outer membrane protein MCL1 in model of Huntington’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 552–559 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghose J., Sinha M., Das E., Jana N. R., Bhattacharyya N. P., Regulation of miR-146a by RelA/NFkB and p53 in STHdh q111/Hdh q111 cells, a cell model of Huntington’s disease. PLOS ONE 6, e23837 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori F., Tanji K., Toyoshima Y., Sasaki H., Yoshida M., Kakita A., Takahashi H., Wakabayashi K., Valosin-containing protein immunoreactivity in tauopathies, synucleinopathies, polyglutamine diseases and intranuclear inclusion body disease. Neuropathology 33, 637–644 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Hübbers C. U., Clemen C. S., Kesper K., Böddrich A., Hofmann A., Kämäräinen O., Tolksdorf K., Stumpf M., Reichelt J., Roth U., Krause S., Watts G., Kimonis V., Wattjes M. P., Reimann J., Thal D. R., Biermann K., Evert B. O., Lochmüller H., Wanker E. E., Schoser B. G. H., Noegel A. A., Schröder R., Pathological consequences of VCP mutations on human striated muscle. Brain 130, 381–393 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Yeo B. K., Yu S. W., Valosin-containing protein (VCP): Structure, functions, and implications in neurodegenerative diseases. Anim. Cells Syst. 20, 303–309 (2016). [Google Scholar]
- 40.Wanker E. E., Ast A., Schindler F., Trepte P., Schnoegl S., The pathobiology of perturbed mutant huntingtin protein–protein interactions in Huntington’s disease. J. Neurochem. 151, 507–519 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Sanford M. S., Love J. A., Grubbs R. H., A versatile precursor for the synthesis of new ruthenium olefin metathesis catalysts. Organometallics 20, 5314–5318 (2001). [Google Scholar]
- 42.Su Y., Doherty T., Waring A. J., Ruchala P., Hong M., Roles of arginine and lysine residues in the translocation of a cell-penetrating peptide from13C, 31P, and 19F solid-state NMR. Biochemistry 48, 4587–4595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakase I., Tadokoro A., Kawabata N., Takeuchi T., Katoh H., Hiramoto K., Negishi M., Nomizu M., Sugiura Y., Futaki S., Interaction of arginine-rich peptides with membrane-associated proteoglycans is crucial for induction of actin organization and macropinocytosis. Biochemistry 46, 492–501 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Choi W., Sun H., Battistella C., Berger O., Vratsanos M. A., Wang M. M., Gianneschi N. C., Biomolecular densely grafted brush polymers: Oligonucleotides, oligosaccharides, and oligopeptides. Angew. Chem. Int. Ed. Engl. 59, 19762–19772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blum A. P., Kammeyer J. K., Rush A. M., Callmann C. E., Hahn M. E., Gianneschi N. C., Stimuli-responsive nanomaterials for biomedical applications. J. Am. Chem. Soc. 137, 2140–2154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlini A. S., Choi W., McCallum N. C., Gianneschi N. C., pH-Responsive charge-conversion progelator peptides. Adv. Funct. Mater. 31, 2007733 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caravan P., Ellison J. J., McMurry T. J., Lauffer R. B., Gadolinium(III) chelates as MRI contrast agents: Structure, dynamics, and applications. Chem. Rev. 99, 2293–2352 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Li J. Y., Popovic N., Brundin P., The use of the R6 transgenic mouse models of Huntington’s disease in attempts to develop novel therapeutic strategies. NeuroRx 2, 447–464 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansson O., Petersen A., Leist M., Nicotera P., Castilho R. F., Brundin P., Penney J. B., Bates G. P., Young A. B., Transgenic mice expressing a Huntington’s disease mutation are resistant to quinolinic acid-induced striatal excitotoxicity. Proc. Natl. Acad. Sci. U.S.A. 96, 8727–8732 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Couly S., Paucard A., Bonneaud N., Maurice T., Benigno L., Jourdan C., Cohen-Solal C., Vignes M., Maschat F., Improvement of BDNF signalling by P42 peptide in Huntington’s disease. Hum. Mol. Genet. 27, 3012–3028 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Li X., Zhang Y., Fu Y., Zhang H., Li H., Li Q., Tan R., Jiang C., Jiang W., Li Z., Luo C., Lu B., Dang Y., Autophagic degradation of mutant Huntingtin by enhancement of the complex of VCP / p97-LC3-mHTT. Authorea, 1–17 [preprint] (2020). [Google Scholar]
- 52.Zhu J., Sun H., Callmann C. E., Thompson M. P., Battistella C., Proetto M. T., Carlini A. S., Gianneschi N. C., Paclitaxel-terminated peptide brush polymers. Chem. Commun. 56, 6778–6781 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Blum A. P., Nelles D. A., Hidalgo F. J., Touve M. A., Sim D. S., Madrigal A. A., Yeo G. W., Gianneschi N. C., Peptide brush polymers for efficient delivery of a gene editing protein to stem cells. Angew Chem. Int. Ed. Engl. 58, 15646–15649 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Wu J., Shaidani S., Theodossiou S. K., Hartzell E. J., Kaplan D. L., Localized, on-demand, sustained drug delivery from biopolymer-based materials. Expert Opin. Drug Deliv. 19, 1317–1335 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi W., Nensel A. K., Droho S., Fattah M. A., Mokashi-Punekar S., Swygart D. I., Burton S. T., Schwartz G. W., Lavine J. A., Gianneschi N. C., Thrombospondin-1 proteomimetic polymers exhibit anti-angiogenic activity in a neovascular age-related macular degeneration mouse model. Sci. Adv. 9, eadi8534 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson M. P., Randolph L. M., James C. R., Davalos A. N., Hahn M. E., Gianneschi N. C., Labelling polymers and micellar nanoparticles via initiation, propagation and termination with ROMP. Polym. Chem. 5, 1954–1964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Instrumentation
Figs. S1 to S35
Tables S1 to S6