Abstract
Background:
Although regional wall motion abnormality (RWMA) detection is foundational to transthoracic echocardiography, current methods are prone to interobserver variability. We aimed to develop a deep learning (DL) model for RWMA assessment and compare it to expert and novice readers.
Methods:
We used 15,746 transthoracic echocardiography studies—including 25,529 apical videos—which were split into training, validation, and test datasets. A convolutional neural network was trained and validated using apical 2-, 3-, and 4-chamber videos to predict the presence of RWMA in 7 regions defined by coronary perfusion territories, using the ground truth derived from clinical transthoracic echocardiography reports. Within the test cohort, DL model accuracy was compared to 6 expert and 3 novice readers using F1 score evaluation, with the ground truth of RWMA defined by expert readers. Significance between the DL model and novices was assessed using the permutation test.
Results:
Within the test cohort, the DL model accurately identified any RWMA with an area under the curve of 0.96 (0.92–0.98). The mean F1 scores of the experts and the DL model were numerically similar for 6 of 7 regions: anterior (86 vs 84), anterolateral (80 vs 74), inferolateral (83 vs 87), inferoseptal (86 vs 86), apical (88 vs 87), inferior (79 vs 81), and any RWMA (90 vs 94), respectively, while in the anteroseptal region, the F1 score of the DL model was lower than the experts (75 vs 89). Using F1 scores, the DL model outperformed both novices 1 (P = .002) and 2 (P = .02) for the detection of any RWMA.
Conclusions:
Deep learning provides accurate detection of RWMA, which was comparable to experts and outperformed a majority of novices. Deep learning may improve the efficiency of RWMA assessment and serve as a teaching tool for novices.
Keywords: Deep learning, Machine learning, Echocardiography, Coronary artery disease, Ventricular function
INTRODUCTION
The assessment of regional wall motion abnormalities (RWMAs) is paramount for the echocardiographic evaluation of ischemic heart disease. Accurate identification of RWMAs is key to the identification of acute and chronic myocardial infarction, as well as the differentiation of ischemic from nonischemic causes of cardiomyopathy. Currently, the assessment of RWMA relies on qualitative interpretation of the multiple echocardiographic views. However, conceptually this is one of the most difficult skills to learn in echocardiography. Additionally, even for readers who attain expertise, visual RWMA assessment remains prone to interobserver variability.1,2 For example, in a prior study by Hoffman et al.,1 the interobserver agreement for the detection of RWMA using noncontrast two-dimensional (2D) echocardiography was only 37%. Moreover, much of the available evidence supporting qualitative methods for RWMA detection is derived from expert readers at academic medical centers.3,4 The accuracy of RWMA assessment appears to be even worse in novice readers.5,6 There is therefore room for improvement in the current paradigm for RWMA assessment.
One potential method by which to augment reader assessment of RWMA is through artificial intelligence (AI), which has shown the potential to improve the automation and diagnostic accuracy of several tasks in echocardiography. Deep learning (DL) is an AI method in which models are trained directly on echocardiographic images using neural networks to detect a finding or condition.7 The factual data used for training are commonly referred to as the ground truth, which is used to train the AI model, such that its detection comes as close as possible to the ground truth. The power of AI in echocardiography has been demonstrated in a variety of disease states, including hypertrophic cardiomyopathy, cardiac amyloidosis, and valvular heart disease.8–10 Previous studies have also shown promising capabilities for AI to detect RWMAs.11,12 However, further studies are needed to prospectively validate AI-based RWMA assessment using current American Society of Echocardiography (ASE) wall segmentation.13 Additionally, it is unknown which types of readers (i.e., experts and/or novices) are likely to derive the most benefit from AI-assisted RWMA assessment.
The aim of this study was to utilize a large database of 2D echocardiograms to (1) train and validate a novel AI model to detect RWMAs in accordance with ASE segmentation guidelines and (2) compare its accuracy to that of both expert and novice readers.
METHODS
Training, Validation, and Test Cohort Selection and Image Analysis
We identified 15,746 consecutive transthoracic echocardiograms—composed of 25,529 apical 2-, 3-, and 4-chamber Digital Imaging and Communications in Medicine (DICOM) videos—performed at the University of Chicago between 2007 and 2020. The dataset was composed of all patients imaged within this time period who possessed adequate-quality 2-, 3-, and/or 4-chamber videos, including those with RWMAs due to both ischemic and nonischemic causes, as well as patients without RWMA. These studies were randomly assigned to model training (n = 14,072) and validation (n = 1,563) cohorts with no patient overlap between groups. The training and validation cohorts were utilized for model development and fine-tuning using the presence of RWMAs from the clinical transthoracic echocardiography report as the ground truth. A separate test dataset containing 111 studies—including 40 normal and 71 with RWMA—was used to perform the reader study to compare the performance of the AI model to both experts and novice readers, using the consensus determination of the presence of RWMAs from the expert readers as the ground truth. In order to adequately assess the model across all regions, the test cohort was designed to include a similar proportion of RWMAs in each region. All of the readers who formulated the clinical echocardiography reports were level III readers, a majority of whom had >10 years of reading experience. This study was approved by the Institutional Review Board with a waiver of informed consent.
All studies were performed using Philips ultrasound imaging equipment using standard acquisition techniques. For each study, regional wall motion was assessed by an expert reader in accordance with ASE guidelines and standards. We divided the myocardium into 7 regions defined based on the American Heart Association (AHA) 17-segment model, while also conforming to the boundaries of the coronary territories outlined in the ASE guidelines: anteroseptal, anterior, anterolateral, inferolateral, inferior, inferoseptal, and apical (Figure 1).13 Coding of an RWMA in any AHA segment within this region was considered positive, irrespective of the number of AHA segments involved. For the purposes of this study, those with global hypokinesis were considered to have an RWMA only if there were regional differences in wall motion. The overall methodology for model training, validation, and testing is depicted in the Central Illustration.
Figure 1.
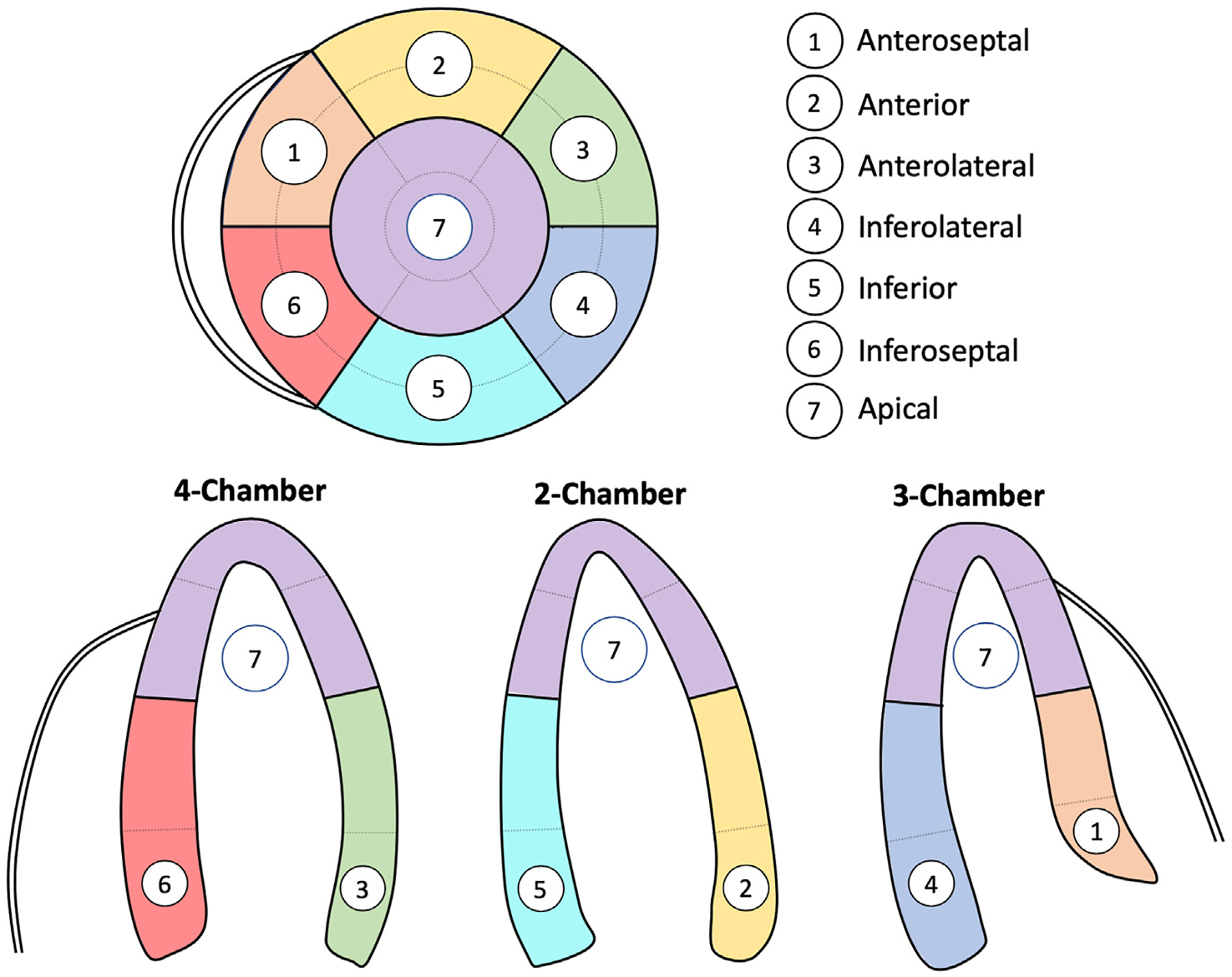
Definitions of each of the 7 ASE regions that were used for model training and validation.
Central Illustration.
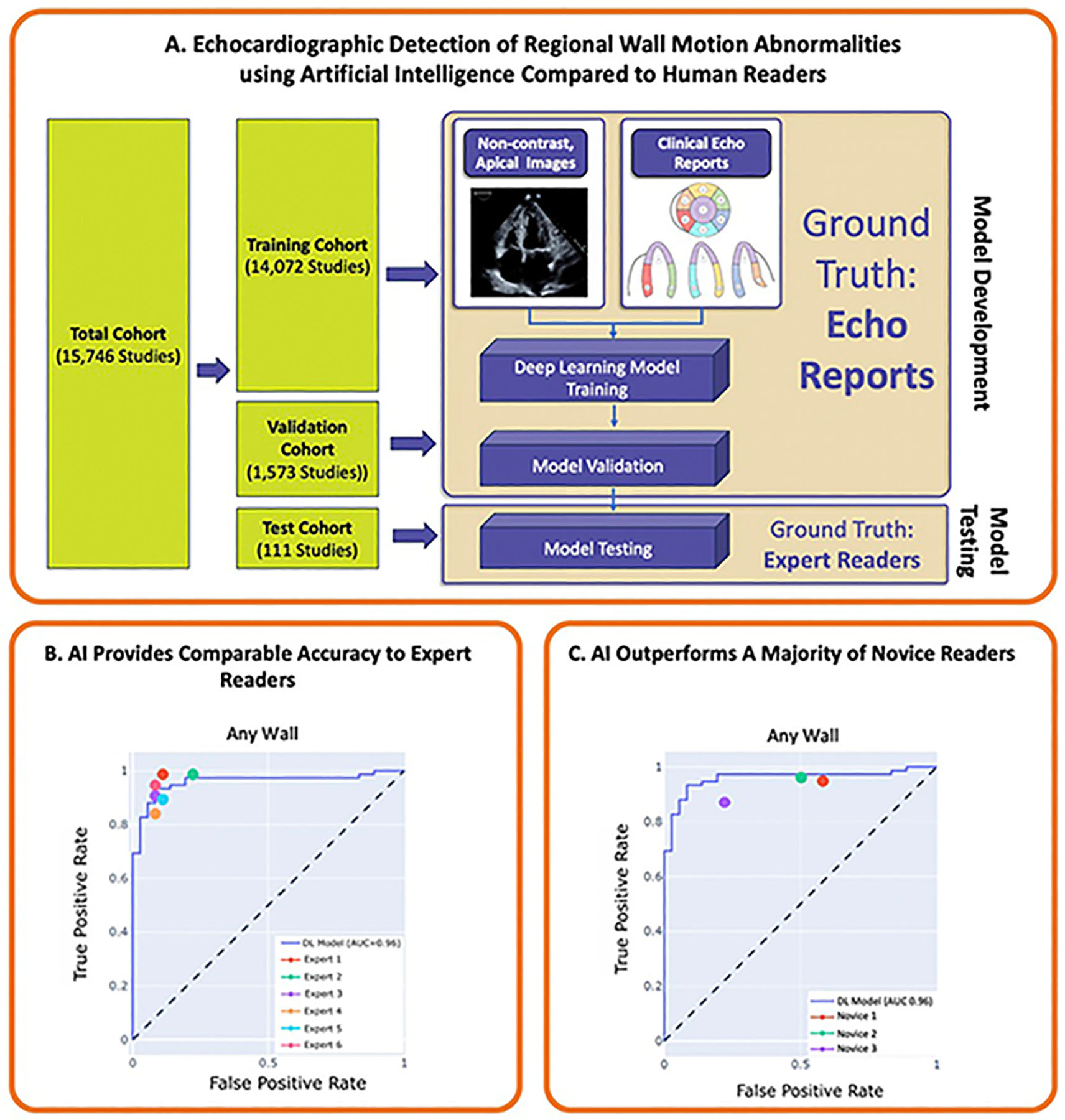
Illustration of key findings of the study. The AI model was trained and validated using apical 2-, 3-, and 4-chamber images to predict the ground truth of RWMA from the clinical echocardiography report. The model was subsequently tested using a reader study format. Here, the AI model demonstrated comparable accuracy to the ground truth of expert readers and outperformed a majority of novice readers for RWMA detection.
Artificial Intelligence Model Training and Validation
After accounting for the 111 studies that were utilized for the test dataset, the remaining 15,635 studies were randomly assigned 90%/10% into model training and validation sets. Apical 2-, 3-, and 4-chamber non-contrast-enhanced videos were manually annotated from the complete echocardiographic examination. Using these videos as input, we developed a three-dimensional convolutional neural network to predict the ground truth of presence of RWMAs (Figure 2A). For model training and validation, the ground truth for RWMAs was defined from the clinical echocardiography report.
Figure 2.
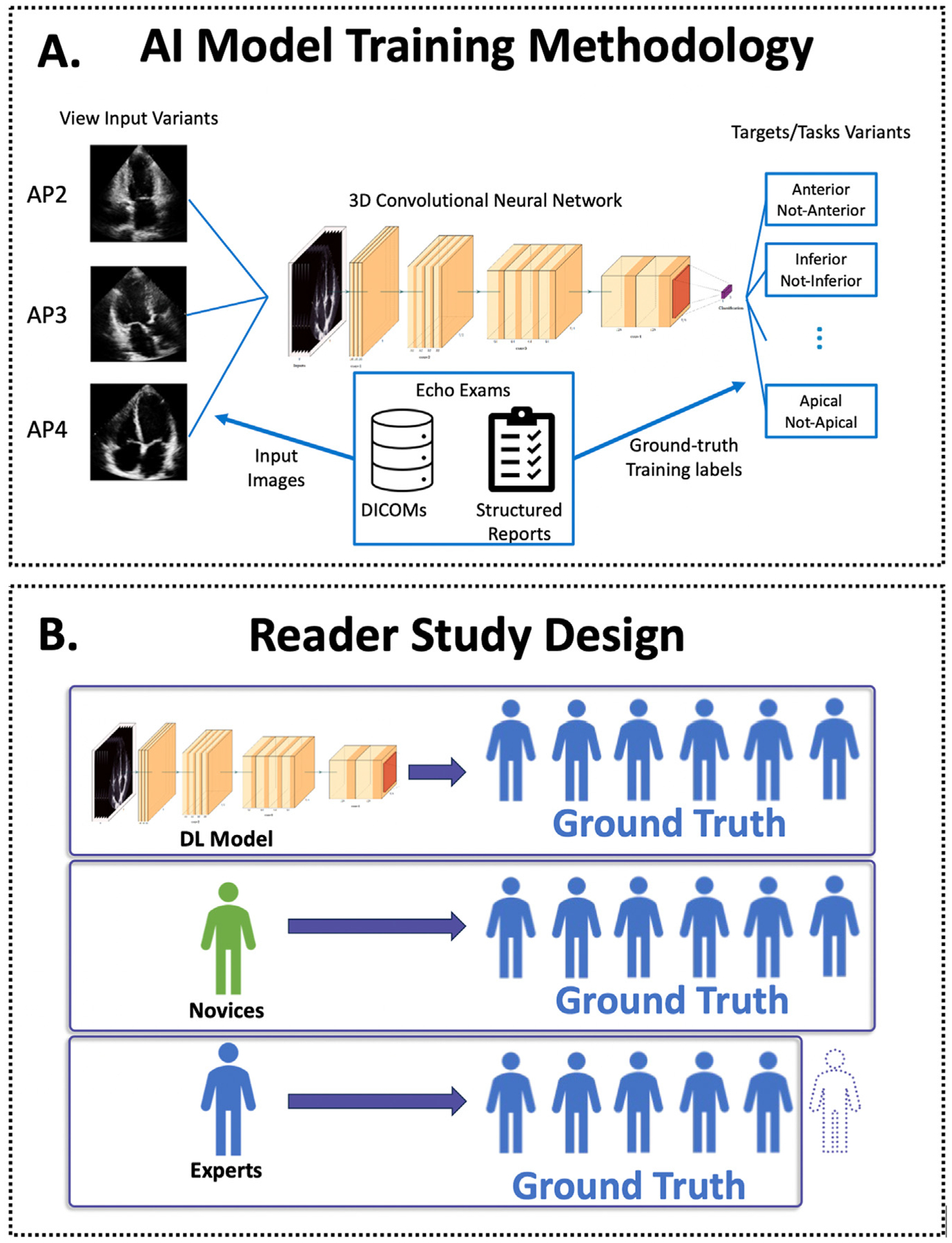
(A) Flowsheet depicting the workflow for training the AI model; apical images were input into the 10-layer convolutional neural network to predict the ground truth of RWMAs derived from the clinical echocardiogram reports. (B) Model testing was performed via a reader study in which the accuracy of the AI model was compared to expert and novice readers. Artificial intelligence and novice accuracy were assessed using a ground truth of RWMA derived from a majority of the 6 experts. Experts were assessed using a rotating panel of other experts such that an expert was not included in their own ground truth.
The convolutional neural network works by analyzing imaging features derived from raw echocardiographic videos—in this case apical 2-, 3-, and 4-chamber views—in order to predict the presence and location of RWMAs as defined from the clinical reports. More specifically, the network processes ultrasound image sequences by convolving both over the space and time dimension of the input. The model itself used the full scan-converted B-mode images, without annotations. No spatial cropping was used. The clips were split by cardiac cycle, using the electrocardiogram, that is, only using complete cardiac cycles. The images were resampled to a matrix size of 120 × 120 using B-spline interpolation. The temporal axis was not resampled. We did consider multiple cycles per view, if available. The model processes each cardiac cycle from each view independently. The available predictive values from each view and each cycle are then averaged to generate an aggregate predicted probability for RWMAs. We trained 1 model for each of the 7 wall sectors.13 In the case of the left ventricular apex, the probabilities from all available apical 2-, 3-, and 4-chamber cardiac cycles were aggregated. The model is an end-to-end DL model that directly learns from the images, that is, there is no special treatment for a specific region like the apex.
The AI model was trained in a supervised manner, where the ground truth of RWMAs was obtained from finding codes extracted from structured reports by expert readers that were created during routine exams. A standard cross-entropy loss function and the Adam optimization algorithm were used for model training. Hyperparameters such as confidence threshold, learning rate, and model size were selected using the validation set. To reduce overfitting and improve generalization, we used data augmentation techniques such as random brightness, contrast, and shifting augmentation. The output of the convolutional neural network is a per-region confidence of RWMA detection between 0 and 1. We set a threshold confidence level of 0.4 to define the presence or absence of RWMAs based on the validation set receiver operator curve (ROC) performance with the goal of having a balanced trade-off between true- and false-positive prediction rates.
Comparison of AI Model With Expert and Novice Readers
Using the distinct 111 patient test dataset, we performed a reader study to assess the accuracy of the AI model in comparison to 6 expert and 3 novice readers. Expert readers were defined as board-certified echocardiographers with level III competency in echocardiography.14 Novice readers were defined as fellows with ≥3 months dedicated training in echocardiography who had not yet passed their echocardiography board examinations. Although only apical views were used by the AI model, experts and nonexpert readers were provided with all standard views, including parasternal and contrast-enhanced images when available. For the reader study, expert and nonexpert readers were blinded to the prediction of the AI model.
The definitions of the ground truth for each group for the reader study are depicted in Figure 2B. The AI model and novice performance were assessed using the ground truth for RWMAs formed by a 6-way majority vote of all experts, that is, at least 4 of the 6 experts. Experts’ performance was assessed using a ground truth for RWMAs comprising a rotating majority vote of at least 3 of the remaining 5 experts, such that the expert being evaluated was never part of the ground truth that they were evaluated against. The performance of the AI model, expert, and novice readers were compared in the test dataset using F1 scores, defined as harmonic mean of precision and recall, as well as ROC analysis. The F1 score is a commonly used metric for evaluating the performance of DL models, balancing the model’s ability to detect RWMAs and minimizing the misclassification of an RWMA when it is not present.15 A ROC analysis was used to determine the optimal threshold for detecting RWMAs, while balancing sensitivity and specificity, and to evaluate the overall diagnostic accuracy of the DL model in the different regions.
We tested for significance of the differences in the F1 scores between the DL algorithm and novices using the permutation test. P values < .05 were considered significant. A permutation test is a statistical procedure that helps assess whether an observed difference or relationship between groups in a dataset is likely a real effect or if it could have happened due to random chance.16 Statistical analysis and metric calculation were performed using the Python packages scipy and scikit-learn. The convolutional neural networks were trained with PyTorch.
RESULTS
Model Training and Validation
Clinical and echocardiographic characteristics for the training and validation cohorts are listed in Table 1. Within the 25,529 training and validation images, abnormal regional wall motion was noted within the structured echocardiography reports in 10,722 (42%) of cases, with apical abnormalities being the most common. The prevalence of abnormal wall motion in the inferior, anterolateral, inferolateral, inferoseptal, anterior, anteroseptal, and apical regions was 22%, 13%, 18%, 17%, 18%, 9%, and 23%, respectively.
Table 1.
Characteristics of the training, validation, and test cohorts
| Parameter | Training and validation cohort (n = 15,635 patients, n = 25,529 DICOM) |
Test cohort (n = 111 studies) |
|---|---|---|
| Clinical data | ||
| Age, years, mean (IQR) | 70.7 (60–83) | 77.4 (69–90) |
| Gender, male, % | 48 | 51 |
| Race, % | ||
| White | 51 | 53 |
| Black | 31 | 27 |
| Other | 18 | 20 |
| Height, cm, mean (IQR) | 170 (163–178) | 170 (163–178) |
| Weight, kg, mean (IQR) | 79 (67–93) | 81 (69–97) |
| Systolic blood pressure, mm Hg | 130 ± 23 | 126 ± 25 |
| Diastolic blood pressure, mm Hg | 72 ± 15 | 68 ± 12 |
| Echocardiographic data, cm | ||
| IVSd | 1.01 | 1.04 |
| PWd | 1.00 | 1.04 |
| LVIDd | 5.06 | 5.04 |
| EF (Biplane method of disks), % | 52.22 | 39.6 |
| Wall motion abnormality, n (%) | ||
| Inferior | 5,705 (22) | 48 (43) |
| Inferolateral | 4,681 (18) | 46 (41) |
| Anterolateral | 3,352 (13) | 43 (39) |
| Anterior | 4,495 (18) | 46 (41) |
| Anteroseptal | 2,508 (10) | 34 (31) |
| Inferoseptal | 4,221 (17) | 42 (38) |
| Apical | 5,745 (23) | 52 (47) |
| Any RWMA | 10,722 (42) | 66 (59) |
A subset of measurements is available for the test cohort. EF, Left ventricular ejection fraction; IQR, interquartile range; IVSd, interventricular septal thickness; LVIDd, left ventricular end-diastolic diameter; PWd, posterior wall thickness.
Comparison of AI Model With Expert and Novice Readers
Clinical and echocardiographic characteristics for test cohort are also listed in Table 1. In the test cohort, an RWMA was present in 66 studies (59% of cases). The prevalence of inferior, anterolateral, inferolateral, inferoseptal, anterior, anteroseptal, and apical wall motion abnormalities was 43%, 39%, 42%, 38%, 42%, 31%, and 47%, respectively. The area under the curve of the AI model for detection of any RWMA in the test cohort was 0.96 (0.92–0.98).
The F1 scores of the AI model and the average of the 6 experts within the test dataset are displayed in Table 2. The mean F1 scores of the experts and the DL model were similar for 6 of 7 regions: anterior (86 vs 84), anterolateral (80 vs 74), inferolateral (83 vs 87), inferoseptal (86 vs 86), apical (88 vs 87), inferior (79 vs 81), and any RWMA (90 vs 94), respectively, while in the anteroseptal region the F1 score of the DL model was lower than the experts (75 vs 89). Similarly, the F1 score (94 vs 90), recall (92 vs 92), and accuracy (92 vs 87) were similar for the DL model versus the experts, respectively, for any RWMA. Receiver operator curve analysis demonstrated similar performance for the AI model and the 6 expert readers for detection of any RWMA, while individual differences were observed in specific myocardial areas, being most pronounced in the anterolateral and anteroseptal regions (Figure 3).
Table 2.
Comparison of F1, recall, and accuracy between AI model and the expert readers
| Wall sector | F1 score | Recall | Accuracy | |||
|---|---|---|---|---|---|---|
| AI model | Experts | AI model | Experts | AI model | Experts | |
| Anterior | 84 (76, 91) | 86 (79–93) | 88 (80, 96) | 84 (76–93) | 85 (78, 92) | 88 (82–93) |
| Anterolateral | 74 (66, 84) | 80 (72–87) | 78 (68, 91) | 78 (67–86) | 78 (70, 85) | 82 (75–88) |
| Inferolateral | 87 (80, 92) | 83 (75–90) | 88 (81, 95) | 82 (73–90) | 87 (82, 92) | 85 (78–90) |
| Inferoseptal | 86 (80, 92) | 86 (78–92) | 86 (77, 94) | 86 (76–94) | 88 (82, 93) | 85 (81–93) |
| Anteroseptal | 75 (64, 83) | 89 (83–95) | 66 (52, 78) | 89 (81–96) | 81 (72, 87) | 90 (84–95) |
| Apical | 87 (80, 94) | 88 (81–94) | 98 (93, 100) | 88 (81–94) | 87 (81, 94) | 89 (84–94) |
| Inferior | 81 (72, 88) | 79 (70–87) | 91 (82, 98) | 79 (67–89) | 82 (75, 87) | 81 (74–89) |
| Any Wall | 94 (90, 96) | 90 (84–94) | 92 (86, 97) | 92 (87–97) | 92 (87, 96) | 87 (81–93) |
Values are stated as median and 95% CI.
Figure 3.
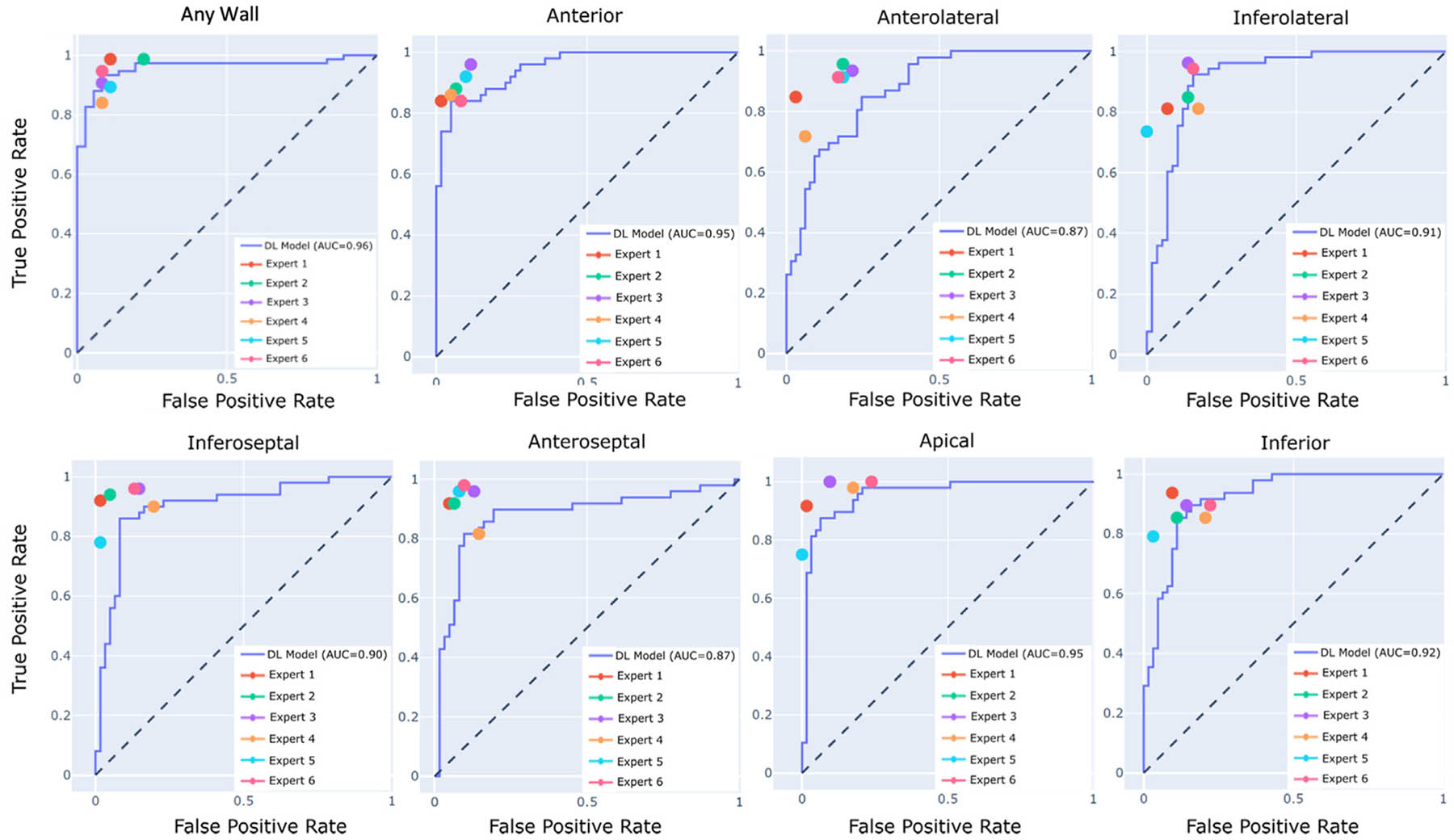
Receiver operator curve analysis depicting accuracy of the AI model within the test cohort on a per-region basis. Expert accuracies represented by the 6 color dots are compared against the AI model represented by the ROC curves.
The F1 scores of the AI model and the 3 novice readers are shown in Table 3. As compared with the AI model, both novices 1 and 2 performed significantly worse for the detection of any RWMA, as compared to the AI model (P = .002 and .02, respectively). The AI model performed significantly better than novice 2 for the detection of apical wall motion abnormalities (P = .006). Additionally, the AI model outperformed novice 2 for the detection of RWMAs in the inferolateral, inferoseptal, and inferior regions (P = .03, .02, and .05, respectively). There were no significant differences in F1 scores between novice 3 and the AI model. With respect to recall scores, the DL model performed significantly worse than novice 1 and novice 3 in the anteroseptal region. The ROC analysis showed similar or better performance of the AI model compared with all 3 novice readers with the exception of the anterolateral and anteroseptal region (Figure 4).
Table 3.
Comparison of F1 scores, recall, and accuracy between the AI model and novice readers
| Wall sector | F1 score comparison | |||
|---|---|---|---|---|
| AI model | Novice 1 | Novice 2 | Novice 3 | |
| Anterior | 84 (76, 91) | 86 (79, 93) | 76 (65, 85) | 80 (70, 88) |
| Anterolateral | 74 (66, 84) | 74 (64, 82) | 69 (59, 78) | 83 (75, 90) |
| Inferolateral | 87 (80, 92) | 81 (74, 88) | 76 (67, 84)* | 86 (78, 91) |
| Inferoseptal | 86 (80, 92) | 83 (75, 88) | 74 (65, 81)* | 87 (80, 93) |
| Anteroseptal | 75 (64, 83) | 83 (73, 89) | 77 (70, 86) | 83 (74, 90) |
| Apical | 87 (80, 94) | 75 (66, 84)* | 80 (73, 87) | 87 (80, 93) |
| Inferior | 81 (72, 88) | 75 (65, 83) | 71 (62, 80)* | 86 (80, 91) |
| Any wall | 94 (90, 96) | 85 (80, 91)* | 88 (82, 92)* | 88 (82, 93) |
| Recall score comparison | ||||
| Anterior | 88 (80, 96) | 90 (82, 98) | 70 (59, 82)* | 79 (64, 88) |
| Anterolateral | 78 (68, 91) | 85 (77, 94) | 74 (60, 84) | 85 (77, 94) |
| Inferolateral | 88 (81, 95) | 84 (74, 93) | 74 (63, 84) | 86 (77, 94) |
| Inferoseptal | 86 (77, 94) | 96 (91, 100) | 84 (74, 94) | 90 (80, 97) |
| Anteroseptal | 66 (52, 78) | 96 (90, 100)* | 85 (75, 93) | 90 (82, 98)* |
| Apical | 98 (93, 100) | 96 (89, 100) | 89 (78, 97) | 96 (91, 100) |
| Inferior | 91 (82, 98) | 83 (73, 92) | 83 (71, 92) | 88 (78, 96) |
| Any wall | 92 (86, 97) | 95 (91, 99) | 96 (92, 100) | 87 (81, 94) |
| Accuracy comparison | ||||
| Anterior | 85 (78, 92) | 87 (81, 93) | 81 (73, 87) | 82 (75, 88) |
| Anterolateral | 78 (70, 85) | 76 (68, 83) | 72 (66, 80)* | 86 (78, 92) |
| Inferolateral | 87 (82, 92) | 82 (75, 88) | 78 (72, 86) | 86 (80, 92) |
| Inferoseptal | 88 (82, 93) | 82 (75, 88) | 73 (66, 80)* | 88 (82, 94) |
| Anteroseptal | 81 (72, 87) | 82 (74, 89) | 78 (71, 86) | 84 (76, 91) |
| Apical | 87 (81, 94) | 73 (65, 82)* | 80 (72, 86) | 87 (81, 93) |
| Inferior | 82 (75, 87) | 76 (68, 85) | 72 (64, 80) | 88 (82, 93) |
| Any wall | 92 (87, 96) | 78 (70, 85)* | 82 (75, 87)* | 84 (77, 90) |
Values are stated as median and 95% CI.
P < .05 between AI model and novice.
Figure 4.
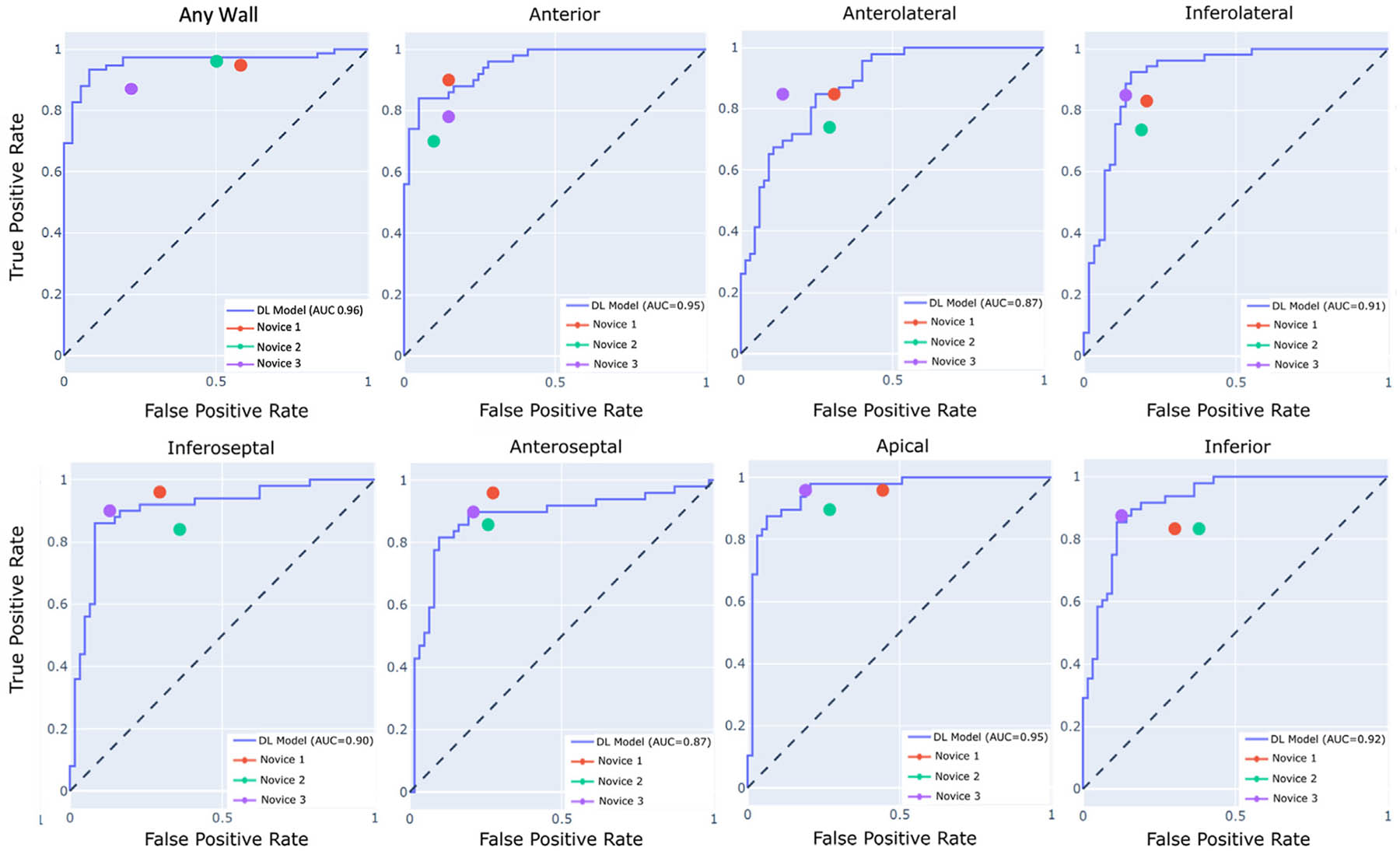
Receiver operator curve analysis depicting accuracy of the AI model within the test cohort on a per-region basis. Novice accuracies represented by the 3 color dots are compared against the AI model represented by the ROC curves.
DISCUSSION
In our study, we developed a novel AI model aimed to detect RWMAs, which was subsequently validated by comparing its accuracy to that of both expert and novice readers. This model demonstrated excellent accuracy, which was equivalent to that of experts and outperformed a majority of the novice readers (Central Illustration).
In our study, the AI model demonstrated comparably high accuracy in the test dataset, supporting the generalizability of our results. Although the model was reasonably accurate on a regional basis, it was most accurate for the detection of any RWMA. Because echocardiography is the frontline assessment tool in patients with known or suspected ischemic heart disease, the detection of any RWMA is therefore likely to be most clinically relevant, as this identifies patients who may warrant further diagnostic evaluation. The model performed best for detection of anterior and apical RWMA. Conversely, the model performed worst for the detection of RWMAs in the anterolateral and anteroseptal regions. One potential explanation for this is that these latter regions are more prone to endocardial dropout—particularly on non-contrast-enhanced images.17 This may have disproportionately affected the accuracy of the AI model relative to the readers who had access to contrast-enhanced images. Further studies are needed to assess whether the inclusion of parasternal views and/or contrast-enhanced views—which may provide improved segmental visualization—can further improve the accuracy of DL for RWMA assessment.
Among AI techniques, DL is uniquely suited to visual tasks, such as RWMA assessment, due to its ability to rapidly analyze large amounts of spatial and temporal imaging data, automatically selecting important features without the need for manual selection or quantification.18,19 One potential drawback of our model was the need for manual annotation of views for the convolutional neural network, which may slightly increase performance time. The integration of existing technologies for automated view identification will likely further improve model efficiency in the future.20,21
Our study expands on a growing body of literature supporting the accuracy of AI for the echocardiographic detection of RWMAs. Previously, Huang et al.11 had developed an AI model that demonstrated a high degree of accuracy for the detection of RWMAs in both internal and external validation. Similarly, using a smaller 300 patient cohort, Kusunose et al.12 developed an AI model based on coronary distributions, which demonstrated comparable accuracy to that of echocardiographers. One advantage of our study was the division of wall segments into 7 myocardial regions, which conformed to the boundaries of the coronary perfusion territories as defined in the current ASE guidelines.13 This is important as the compliance of the model with current reporting standards helps to ensure that the AI output is easily interpretable and clinically actionable. Further studies are needed to assess whether expansion of DL-based RWMA assessment to a 17-segment model provides incremental value.
In addition to demonstrating high accuracy of the detection of RWMAs in the test cohort, the AI model provided comparable accuracy to that of expert readers. Our findings parallel those of Kusonose et al.12 An advantage of AI methods over conventional visual analysis is that they can be performed in seconds, providing rapid and accurate information. Artificial intelligence–based RWMA assessment therefore has the potential to augment expert reads by rapidly highlighting areas of concern for RWMAs, improving the ease and efficiency of interpretation. Artificial intelligence assistance may be particularly helpful as a user support tool given the rising utilization of transthoracic echocardiography, which may increase workload, potentially at the risk of accuracy.22,23 Another potential benefit of AI for RWMA assessment is in improving the consistency of echocardiography reporting. As in prior studies, we observed substantial interobserver variability with respect to RWMA detection, even among expert readers,1,2 as reflected by the data reported in Figure 3. Further studies are needed to determine whether the integration of the AI methodology into clinical interpretation can reduce interobserver variability with respect to RWMA assessment.
Lastly, our AI model outperformed a majority of novice readers for RWMA assessment. In their study, Kusunose et al. compared the performance of their AI model to resident physicians. Our study differs in that we utilized novice echocardiographers who—unlike resident physicians—have received formal training in echocardiography. Our study suggests the potential for AI to augment the accuracy of novices for RWMA detection even with such training. Given this, it is perhaps possible that AI could potentially allow for distribution of expert-level RWMA analysis to geographic regions where such expertise is not otherwise available. Artificial intelligence models could also serve as an educational tool to improve novices’ ability to assess regional wall motion. It should be noted that we did not assess whether the use of AI augmented novice performance; further studies may help to better elucidate whether access to AI model predictions would provide direct benefit to novice readers.
Limitations
One limitation of our study is that we used images obtained from a single ultrasound vendor. However, as the model uses DICOM images as input, it is unlikely that these results would not be generalizable to other vendors. Extended follow-up studies should aim to validate our findings using multicenter, multivendor data. Additionally, although no images were excluded, some studies were partially incomplete and did not contain all 3 apical views, resulting in a discordance between the 15,746 echo studies and 25,529 images included; however, we chose not to exclude these studies in order to maximize the size of our dataset for model development. Additionally, rather than randomly select patients for the test dataset, we chose to include a similar number of RWMAs from each myocardial region in the test dataset. Accordingly, age, left ventricular ejection fraction, and prevalence of RWMA differed between the training/validation and test datasets. We felt this was important because random sampling would be likely to overrepresent more common RWMAs, such as apical and inferior territories, thus skewing the performance of the model in the reader study. Additionally, using a testing pool with many healthy patients—which are easy to classify—could have also shifted the results in favor of the model.
We only assessed readers within a single tertiary academic echocardiography laboratory. Further studies are needed to determine whether AI could augment RWMA assessment at centers where such a high level of expertise is not available. Another potential limitation of this study is the use of expert wall motion interpretation on resting 2D echocardiogram as the ground truth rather than invasive coronary angiography or noninvasive stress testing. One reason for this choice was that RWMA may be caused by a variety of conditions, including ischemia, infarction, and even nonischemic etiologies, such as cardiac sarcoidoisis or takotsubo cardiomyopathy. Even high-grade stenosis without myocardial infarction may not manifest in RWMAs under resting conditions; similarly, occult prior myocardial infarction may manifest in an RWMA in the absence of epicardial stenosis.24 Further studies may be useful to determine whether AI is similarly effective at detecting stress-induced RWMA.
Moreover, the use of the clinical reports for the classification of RWMAs during training and validation may have affected the development of the DL model. Presumably readers generating those reports would have access to clinical data that might influence the interpretation of the exam, as readers may be more likely to assign an RWMA when they have knowledge of a history of coronary disease or prior infarction. Our model also did not utilize parasternal or contrast-enhanced images. However, this is likely only to the disadvantage of the model, as readers were provided with all views including parasternal and contrast-enhanced images when present. Further studies are needed to determine whether the inclusion of parasternal and/or contrast-enhanced images can further improve the DL-based assessment of RWMA.
Lastly, the use of DL for RWMA assessment does represent a “black box.” Because the neural network works by processing raw images, we are unable to determine which aspects of the images are most important for RWMA assessment.
CONCLUSION
This study indicates that the use of this new AI algorithm provides highly accurate detection of RWMAs. The automated DL-based assessment of RWMAs has the potential to improve the efficiency of all readers and may serve as a teaching tool for novice readers.
HIGHLIGHTS.
Currently, assessment of regional wall motion is prone to interobserver variability.
We developed a DL model for regional wall motion assessment.
The model demonstrated excellent accuracy, equivalent to that of expert readers.
The model outperformed the majority of the novice readers.
It may prove useful, as it rapidly highlights areas of concern.
ACKNOWLEDGMENT
We thank Dr. Akhil Narang for his help with this study.
CONFLICTS OF INTEREST
In addition to employment relationships between several authors (N.T.G., L.O., N.P., P.E., A.S., S.W., D.P., I.W.S., A.M.C., T.S.) and Philips Healthcare, R.M.L. received research grants from this company. The remaining authors have nothing to disclose.
Abbreviations
- 2D
Two-dimensional
- AHA
American Heart Association
- AI
Artificial intelligence
- ASE
American Society of Echocardiography
- DICOM
Digital Imaging and Communications in Medicine
- DL
Deep learning
- ROC
Receiver operator curve
- RWMA
Regional wall motion abnormality
REFERENCES
- 1.Hoffmann R, Borges AC, Kasprzak JD, et al. Analysis of myocardial perfusion or myocardial function for detection of regional myocardial abnormalities. An echocardiographic multicenter comparison study using myocardial contrast echocardiography and 2D echocardiography. Eur J Echocardiogr 2007;8:438–48. [DOI] [PubMed] [Google Scholar]
- 2.Schnaack SD, Siegmund P, Spes CH, et al. Transpulmonary contrast echocardiography: effects on delineation of endocardial border, assessment of wall motion and interobserver variability in stress echocardiograms of limited image quality. Coron Artery Dis 2000;11:549–54. [DOI] [PubMed] [Google Scholar]
- 3.Medina R, Panidis IP, Morganroth J, et al. The value of echocardiographic regional wall motion abnormalities in detecting coronary artery disease in patients with or without a dilated left ventricle. Am Heart J 1985;109: 799–803. [DOI] [PubMed] [Google Scholar]
- 4.Hundley WG, Kizilbash AM, Afridi I, et al. Effect of contrast enhancement on transthoracic echocardiographic assessment of left ventricular regional wall motion. Am J Cardiol 1999;84:1365–8, A1368–1369. [DOI] [PubMed] [Google Scholar]
- 5.Fathi R, Cain P, Nakatani S, et al. Effect of tissue Doppler on the accuracy of novice and expert interpreters of dobutamine echocardiography. Am J Cardiol 2001;88:400–5. [DOI] [PubMed] [Google Scholar]
- 6.Picano E, Lattanzi F, Orlandini A, et al. Stress echocardiography and the human factor: the importance of being expert. J Am Coll Cardiol 1991; 17:666–9. [DOI] [PubMed] [Google Scholar]
- 7.Johnson KW, Torres Soto J, Glicksberg BS, et al. Artificial intelligence in Cardiology. J Am Coll Cardiol 2018;71:2668–79. [DOI] [PubMed] [Google Scholar]
- 8.Elias P, Poterucha TJ, Rajaram V, et al. Deep learning electrocardiographic analysis for detection of left-sided valvular heart disease. J Am Coll Cardiol 2022;80:613–26. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Gajjala S, Agrawal P, et al. Fully automated echocardiogram interpretation in clinical practice. Circulation 2018;138:1623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goto S, Mahara K, Beussink-Nelson L, et al. Artificial intelligence-enabled fully automated detection of cardiac amyloidosis using electrocardiograms and echocardiograms. Nat Commun 2021;12:2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang MS, Wang CS, Chiang JH, et al. Automated recognition of regional wall motion abnormalities through deep neural network interpretation of transthoracic echocardiography. Circulation 2020;142:1510–20. [DOI] [PubMed] [Google Scholar]
- 12.Kusunose K, Abe T, Haga A, et al. A deep learning approach for assessment of regional wall motion abnormality rom echocardiographic images. JACC Cardiovasc Imaging 2020;13:374–81. [DOI] [PubMed] [Google Scholar]
- 13.Lang RM, Badano LP, Mor-Avi V, et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr 2015;28:1–39.e14. [DOI] [PubMed] [Google Scholar]
- 14.Wiegers SE, Ryan T, Arrighi JA, et al. 2019 ACC/AHA/ASE advanced training statement on echocardiography (revision of the 2003 ACC/AHA clinical competence statement on echocardiography): a Report of the ACC Competency Management Committee. Catheter Cardiovasc Interv 2019;94:481–505. [DOI] [PubMed] [Google Scholar]
- 15.Hicks SA, Strumke I, Thambawita V, et al. On evaluation metrics for medical applications of artificial intelligence. Sci Rep 2022;12:5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst MD. Permutation methods: a basis for exact inference. Stat Sci 2004; 19:676–85. 610. [Google Scholar]
- 17.Mathias W Jr., Arruda AL, Andrade JL, et al. Endocardial border delineation during dobutamine infusion using contrast echocardiography. Echocardiography 2002;19:109–14. [DOI] [PubMed] [Google Scholar]
- 18.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 19.Sermesant M, Delingette H, Cochet H, et al. Applications of artificial intelligence in cardiovascular imaging. Nat Rev Cardiol 2021;18:600–9. [DOI] [PubMed] [Google Scholar]
- 20.Howard JP, Tan J, Shun-Shin MJ, et al. Improving ultrasound video classification: an evaluation of novel deep learning methods in echocardiography. J Med Artif Intell 2020;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madani A, Arnaout R, Mofrad M, et al. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit Med 2018;1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thachil R, Hanson D. Artificial intelligence in echocardiography: a disruptive technology for democratizing and standardizing health. J Am Soc Echocardiogr 2022;35:A14–6. [DOI] [PubMed] [Google Scholar]
- 23.Blecker S, Bhatia RS, You JJ, et al. Temporal trends in the utilization of echocardiography in Ontario, 2001 to 2009. JACC Cardiovasc Imaging 2013; 6:515–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Angelis G, De Luca A, Merlo M, et al. Prevalence and prognostic significance of ischemic late gadolinium enhancement pattern in nonischemic dilated cardiomyopathy. Am Heart J 2022;246:117–24. [DOI] [PubMed] [Google Scholar]


