Abstract
A variety of methods are available to analyze protein–DNA interactions in vivo. Two of the most prominent of these methods are chromatin immunoprecipitation (ChIP) and in vivo footprinting. Both of these procedures have specific limitations. For example, the ChIP assay fails to document where exactly a protein binds in vivo. The precipitation of a specific segment of DNA with antibodies directed against DNA-binding proteins does not necessarily indicate that the protein directly interacts with a sequence in the precipitate but could rather reflect protein–protein interactions. Furthermore, the results of in vivo footprinting studies are inconclusive if a DNA sequence is analyzed that is bound by a specific protein in only a certain fraction of cells. Finally, in vivo footprinting does not indicate which protein is bound at a specific site. We have developed a new procedure that combines the ChIP assay and DMS footprinting techniques. Using this method we show here that antibodies specific for USF1 and NF-E2 precipitate the murine β-globin promoter in MEL cells. DMS footprinting analysis of the DNA precipitated with NF-E2 antibodies revealed a protection over a partial NF-E2-binding site in the β-globin downstream promoter region. We believe that this novel method will generally benefit investigators interested in analyzing protein–DNA interactions in vivo.
INTRODUCTION
Transcription in eukaryotes is a complex process involving the binding of proteins to the promoter, recruitment of transcription complexes, initiation, elongation and termination (1). This process is dynamic and controlled at each step by proteins that bind to the DNA, to the RNA polymerase holocomplex, or both. Although knowledge about transcription complex formation in vitro is extensive, the mechanisms leading to transcription in the context of higher order chromatin in vivo are not understood in detail. A number of techniques are available to study the interaction of proteins with specific segments of DNA in vivo; each of these techniques has distinct limitations (2–6). Chromatin immunoprecipitation (ChIP) is a powerful method to analyze the interaction of proteins with specific chromatin regions in vivo (2,3). Combined with PCR, the ChIP assay is very sensitive in detecting proteins that crosslink to a specific region in chromatin, but it fails to provide information regarding the sequence-specific interaction of these proteins with the DNA. Another method commonly used to examine the interaction of proteins and DNA is in vivo footprinting (4–6). The problem with in vivo footprinting techniques is that they fail to provide clear results if a protein is bound in a sequence-specific manner in only a certain fraction of cells.
We have combined the two techniques and developed a novel method for analyzing protein–DNA interactions in vivo. This new technique is similar in concept to an in vitro protocol developed by Gallarda et al. (7). These authors used immunoprecipitation to select for specific proteins and subsequent footprinting to analyze whether the selected proteins interact with a specific DNA sequence. The technique we have developed here is aimed at analyzing sequence-specific interactions of particular proteins in vivo. As one example, we have analyzed the interaction of proteins with the mouse β-globin downstream promoter region in unsynchronized MEL cells. We show that chromatin fragments precipitated with antibodies against USF1 or NF-E2 reveal strikingly different footprint patterns on the globin genes. This result indicates that this novel method will allow investigators to select specific templates for in vivo footprinting.
MATERIALS AND METHODS
ChIP and dimethylsulfate (DMS) treatment
ChIP was performed essentially as described by Forsberg et al. (8) with the following modifications. Cultures of 4 × 108 MEL cells were grown in RPMI with 10% fetal bovine serum and 1% ABAM (antibiotic–antimycotic; Gibco BRL) in 5% CO2 at 37°C. Crosslinking was induced by adding 1% (v/v) formaldehyde and incubation for 10 min at room temperature on a shaker. After stopping the crosslinking reaction by adding 0.125 M glycine and incubation for 5 min (with shaking at room temperature), the cells were pelleted at 2000 r.p.m. The cells were then washed twice in 25 ml ice-cold phosphate-buffered saline (PBS) including protease inhibitors. Nuclei were isolated by resuspending the cell pellet in 1 ml ice-cold swelling buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40 and protease inhibitors), split into two aliquots and incubated on ice for 10 min. Chromatin was fragmented by subjecting the nuclei to restriction enzyme digestion with 200 U PstI for 4 h at 37°C and 100 U PstI for an additional 16 h at 37°C. The nuclei were then incubated with 200 U RNase cocktail (Ambion) and an additional 100 U aliquot of PstI for 2 h at 37°C. Nuclei were pelleted at 4°C for 5 min at 5000 r.p.m. and lysed in 1 ml lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris–HCl pH 8.1) on ice for 20 min. Completion of restriction enzyme digestion was verified by electrophoresis on a 2% agarose gel followed by Southern blotting using a radioactive probe hybridizing to the human β-globin gene (data not shown). The lysate was combined and transferred to a 15 ml conical tube and diluted with 9 ml dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.1, 167 mM NaCl and protease inhibitors). An aliquot of 500 µl protein A–Sepharose beads (Pharmacia) was added to the diluted nuclear lysate and incubated for 2 h at 4°C while rotating. The beads were pelleted for 10 min at 2000 r.p.m. and the supernatant was divided into three aliquots. An aliquot of 25 µl of the appropriate antibody (USF1 or NF-E2; Santa Cruz Biotechnology) or no antibody was added to the aliquoted supernatant and incubated at 4°C overnight while rotating. Protein A–Sepharose beads were washed twice in blocking buffer [3% BSA, 0.05% sodium azide, and protease inhibitor in 1× TE (10 mM Tris–HCl pH 8.1, 1 mM EDTA)]. The chromatin was then immunoprecipitated with 600 µl blocked protein A–Sepharose for 2 h at 4°C on a rotator. The immunoprecipitates were pelleted at 13 000 r.p.m. for 30 s and 1 ml of the no antibody supernatant was saved and labeled as ‘input’. Half of the input chromatin was ethanol precipitated and resuspended in two aliquots of 20 µl ddH2O and 800 µl DMS buffer (50 mM sodium cacodylate, 1 mM EDTA) and the other half was saved for the ChIP/PCR analysis. The supernatants of the samples precipitated with USF1 and NF-E2 antibodies were discarded and the pellets were washed by rotating at 4°C for 5 min with 1 ml each of low salt wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1 and 150 mM NaCl), high salt wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1 and 500 mM NaCl), LiCl wash (0.25 M LiCl, 1% NP-40, 1% sodium desoxycholate, 1 mM EDTA and 10 mM Tris–HCl pH 8.1) and twice with 1× TE. Of the immunoprecipitates, 80% was resuspended in 800 µl DMS buffer and 20% was left in TE buffer for the ChIP/PCR analysis (see Fig. 1C).
Figure 1.
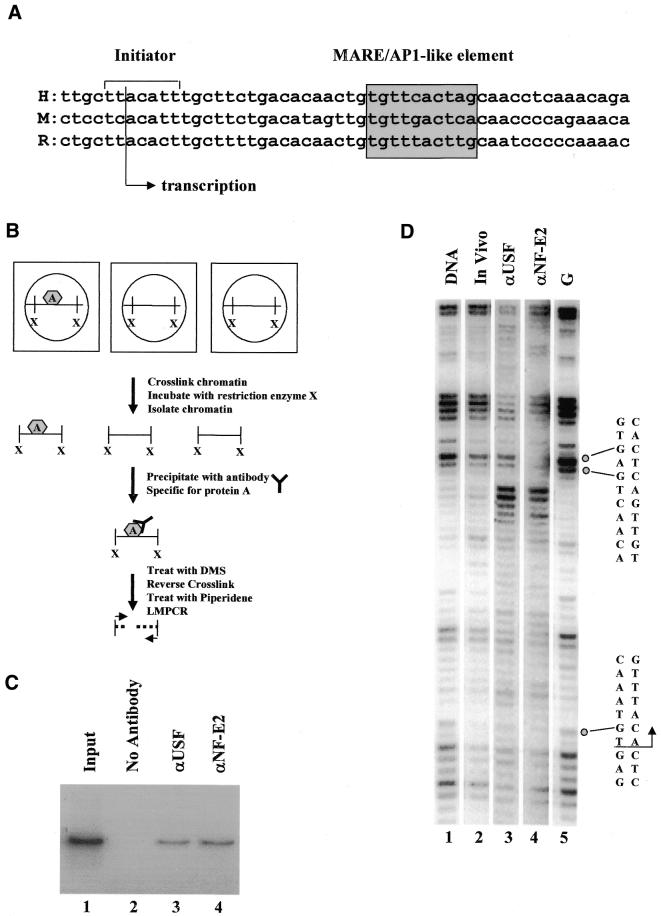
Analysis of protein–DNA interactions in the murine β-globin downstream promoter region by a combination of chromatin immunoprecipitation and DMS footprinting. (A) Sequence alignment of the adult β-globin downstream promoter region. Shown are three sequences of the β-globin downstream promoter region from human (H), mouse (M) and rabbit (R) (13). The shaded box highlights the position of the MARE/Ap1-like sequence and the arrow points to the transcription start site. (B) Diagram outlining the experimental procedure for footprinting ChIP-selected templates (see Materials and Methods for details). (C) ChIP experiment showing the interaction of USF1 and NF-E2 with the murine βmaj-globin promoter in MEL cells. MEL cells were grown under standard conditions and crosslinking was induced with formaldehyde. After fragmentation, the chromatin was precipitated with no antibodies (lane 2), antibodies against USF1 (αUSF, lane 3) or antibodies against the p45 subunit of NF-E2 (αNF-E2, lane 4). Lane 1 shows the PCR result of the input, which serves as a positive control. (D) LMPCR footprint analysis of non-selected or ChIP-selected chromatin. MEL cells were grown under standard conditions and treated with formaldehyde. Isolated nuclei from these cells were incubated with PstI and the fragmented chromatin was precipitated with USF or NF-E2 antibodies. The precipitates were incubated with DMS for 90 s and piperidine, and analyzed by LMPCR (lane 3, USF-selected chromatin; lane 4, NF-E2-selected chromatin). In vivo footprinting was also performed on MEL cells that were not treated with formaldehyde (lane 2, treated with DMS for 90 s; lane 1 shows the DMS/piperidine cleavage pattern in in vitro treated MEL genomic DNA). Lane 5 shows the G-ladder of the β-globin downstream promoter region. Indicated on the right are the positions of the initiator and MARE-like sequences, as well as the start site and direction of transcription. Open circles indicate protected G residues in NF-E2-selected chromatin.
DMS treatment of the immunoprecipitated chromatin was performed using the Maxam and Gilbert guanine-specific sequencing reaction with 0.1% DMS for 15, 45 or 90 s at room temperature (9). The reaction was stopped by adding 50 µl DMS stop buffer (1.5 M sodium acetate pH 7.0, 1 M 2-mercaptoethanol), followed by two ethanol precipitations in a dry ice bath. The DMS-treated and non-DMS-treated chromatin was then eluted from the beads by incubating twice with 250 µl elution buffer (1% SDS, 0.1 M NaHCO3), shaking at 1000 g for 15 min at 65°C, each time saving the supernatant. An aliquot of 200 mM NaCl was added to the eluates and crosslinking was reversed by incubation at 65°C for 5 h. Proteins were digested with 40 µg/ml proteinase K in 10 mM EDTA and 40 mM Tris pH 6.5 for 1 h at 37°C. Immunoprecipitated DNA was purified using a Qiagen kit and eluted with 180 µl ddH2O. To cleave the DMS-treated DNA, 20 µl piperidine were added and incubated at 95°C for 30 min. The DNA was washed twice by adding 1 ml ddH2O, dried in a Speed Vac and resuspended in 50 µl 1× TE. Of the DMS-treated immunoprecipitated DNA, 10% was used for ligation-mediated PCR (LMPCR)-assisted in vivo footprinting. An aliquot of the precipitated DNA was also analyzed by PCR using primers specific for the murine β-globin downstream promoter region (forward primer, 5′-GACAAACATTATTCAGAGGGAGTACCC; reverse primer, 5′-AGGTGCACCATGATGTCTGTTTCTGG) using a protocol previously published by Forsberg et al. (8).
Linker LMPCR
LMPCR was essentially performed as described by Hornstra and Yang (5) with the following modifications. Approximately 2 µg DMS-treated genomic DNA or 10% immunoprecipitated DNA was annealed to 0.6 pmol gene-specific primer (MPA 1) by denaturing at 96°C for 10 min followed by annealing at 47°C for 30 min in a 15 µl solution of 1× Vent buffer (10 mM Tris–HCl pH 8.9 and 40 mM NaCl). Primer extension was performed by adding a 15 µl solution of 10 mM Tris–HCl pH 8.9, 40 mM NaCl, 0.5 mM dNTPs and 2 U Vent polymerase (New England Biolabs) and incubating at 53°C for 1 min, 55°C for 1 min, 57°C for 1 min, 60°C for 1 min, 62°C for 1 min, 66°C for 1 min, 68°C for 3 min and 76°C for 3 min. A 20 µl dilution solution (110 mM Tris–HCl pH 7.5, 18 mM MgCl2, 50 mM DTT and 0.0125% BSA) was then added to the extension reaction. Blunt-end ligation was performed by adding a 25 µl solution of 100 pmol asymmetric double-stranded linker, 10 mM MgCl2, 20 mM dithiothreitol, 3 mM ATP, 0.005% BSA and 4.5 U T4 ligase (Ambion) to the reaction and incubating at 17°C overnight. The ligation products were purified by standard phenol/chloroform extractions and ethanol precipitation including 10 µg/µl tRNA. The ligated DNA was resuspended in 20 µl ddH2O and added to 80 µl PCR mix [10 mM Tris–HCl pH 8.9, 40 mM NaCl, 3 mM MgCl2, 0.25 mM dNTPs, 20 pmol gene-specific PCR primer (MPA 3), 20 pmol linker primer 2 and 0.5 U Taq polymerase; Gibco BRL]. This PCR mixture was initially denatured at 95°C for 5 min and then subjected to 20 cycles of PCR under the following conditions: 95°C for 20 s, 65°C for 1 min, 72°C for 1 min with an increase of 5 s/cycle and an additional 5 cycles of 95°C for 20 s, 65°C for 1 min, 72°C for 2 min 30 s, followed by a final extension at 72°C for 15 min. The PCR products were purified by phenol/chloroform extraction and ethanol precipitation and resuspended in 30 µl ddH2O. Initially, 3 µl of the PCR products were size-fractionated on a 0.4 mm thick 5% polyacrylamide gel made with 8 µg/ml ammonium persulfate, then electrotransfered for 30 min to a nylon membrane (Hybond N+; Amersham).
Radiolabeled probes were synthesized using the Prime-a-Probe kit (Ambion) from a gel-purified PCR product (PCR primers: β-maj FWD, 5′-GACAAACATTATTCAGAGGGAGTACCC-3′; MPB 1, 5′-TCTGTCTCCAAGCACCCAA-3′) containing the region of interest. To radiolabel the probe, ∼150 ng template DNA was mixed with 0.3 µg gene-specific primer (MPA 3), used in the PCR step of the LMPCR protocol, and brought up to 8 µl in ddH2O. This primer plus template mixture was denatured at 95°C for 10 min and immediately placed on dry ice to snap freeze. Then 5 µl of 5× Decaprime buffer containing dATP, dGTP and dTTP (Ambion) was incubated with 10 µl [α-32P]dCTP (3000 Ci/mmol) and 1 U/µl Klenow fragment at 37°C for 30 min. The reaction was quenched on ice and stopped by adding 35 µl formamide loading dye. The probe was denatured at 95°C for 10 min and purified on a 5% denaturing poylacrylamide gel. After exposing the gel to film (Type 57; Polaroid) the probe was cut out of the gel and crushed and soaked in 4 ml hybridization buffer (250 mM Na2PO4 pH 7.2, 7% SDS, 1% BSA). The probe was hybridized to the blots at 65°C overnight. The blots were washed three times at 65°C in washing solution (20 mM Na2PO4 pH 7.2, 1% SDS) and visualized by autoradiography. The following primers were used for LMPCR: MPA 1, 5′-ATGTCCAGGGAGAAATATCG-3′; MPA 3, 5′-TGAAGGGCCAATCTGCTCACACAGG-3′.
RESULTS AND DISCUSSION
The developmental stage-specific expression of the human β-globin gene is regulated primarily by transcription factors that interact with gene-proximal cis DNA elements (10). We have shown that USF1 and USF2 interact with a downstream promoter element of the human β-globin gene in vitro (K. Leach, unpublished results). In the same study we have shown that using the ChIP assay these proteins are crosslinked to the β-globin gene in erythroid cells in vivo. In addition, data published by Sawado et al. (11) as well as our own experiments revealed that NF-E2 can be crosslinked to the β-globin gene in vivo. The crosslinking of NF-E2 to the globin promoter is somewhat surprising as there is no consensus DNA-binding site in the globin gene. However, a sequence located downstream of the transcription start site reveals partial homology to the NF-E2 consensus element, also referred to as the MARE (maf recognition element, Fig. 1A; 12–14). The NF-E2 consensus sequence is 5′-TGCTGASTCAY-3′ (S = G or C; Y = T or C; 14). The sequence in the mouse β-globin downstream promoter region differs in one position (the third) from the consensus sequence. Conventional in vivo footprinting did not reveal significant protection of the NF-E2-binding site in MEL cells (Fig. 1D, lanes 1 and 2). To analyze the possibility that NF-E2 interacts with the β-globin downstream element in vivo in only a fraction of erythroid cells, we have developed a novel method that combines the ChIP assay with DMS footprinting.
The procedure (outlined in Fig. 1B) involves the crosslinking of proteins and DNA using formaldehyde (1%), followed by digestion of permeabilized nuclei with restriction enzymes, precipitation with specific antibodies, treatment and cleavage of precipitated chromatin restriction fragments with DMS and piperidine and analysis of the cleavage pattern by LMPCR (for details see Materials and Methods). Figure 1C shows that antibodies against USF1 and NF-E2 precipitate crosslinked chromatin fragments containing the murine adult β-globin gene, as expected from previous data (K. Leach, unpublished results; 11). The subsequent DMS footprint of the precipitated chromatin fragments revealed interesting characteristics of the precipitated fragments (Fig. 1D). First, the overall footprint pattern of fragments selected with USF antibodies is different from those precipitated with NF-E2 antibodies (compare Fig. 1D, lanes 3 and 4). This is particularly obvious over the MARE-like sequence, which shows clear protection only in templates selected by NF-E2 precipitation. The difference in the cleavage pattern between the templates selected by USF or NF-E2 antibodies could reflect the possibility that the proteins interact with the globin gene at different stages of the transcription cycle. Alternatively, the results could reflect the possibility that low occupancy of these sites would strongly reduce the probability of selecting templates which have both proteins bound simultaneously. Second, the overall footprint pattern of fragments precipitated with USF antibodies is similar to the pattern found in unselected templates, whereas the footprint pattern for NF-E2-selected fragments is different. This result suggests that only a small fraction of cells may have NF-E2 bound at the β-globin gene.
We are currently in the process of examining the interaction of NF-E2 with the non-consensus MARE-binding site in the downstream promoter region in detail using a combination of in vitro and in vivo methods. The potential sequence-specific interaction of NF-E2 with the β-globin downstream promoter region is interesting in light of the fact that Johnson et al. (15) previously showed that RNA polymerase II is recruited to both the LCR and to the adult β-globin gene and that the transfer of the polymerase from the LCR to the globin gene depends on the presence of NF-E2 (p45). In this respect it is possible that NF-E2 is part of the recruitment process that mediates the interaction of RNA polymerase II with the β-globin promoter.
It is important to note that formaldehyde crosslinking does not appear to change the cleavage pattern in the DNA region analyzed in these experiments; the overall pattern is similar between untreated cells (Fig. 1D, lanes 1 and 2) and crosslinked cells (lanes 3 and 4). In addition, we found that incubation of genomic DNA with 1% formaldehyde (with subsequent reversal of the crosslink) does not change the overall DMS cleavage pattern in the DNA region that we have analyzed by LMPCR (data not shown).
Several methods are available to fragment crosslinked chromatin for ChIP, including sonication, MNase digestion and restriction enzyme digestion (2,3,16,17). We reasoned that sonication or MNase digestion might fragment the chromatin in a way that not all of the resulting templates would be amplifiable by LMPCR and in addition may result in a high background. We therefore chose to fragment the crosslinked chromatin by restriction enzyme digestion (16). The disadvantage in using this method is that long incubation with the restriction enzyme allows endogenous nucleases to attack the chromatin in accessible regions. In some of our experiments, we observed inconsistent LMPCR banding patterns in ChIP-selected templates in the higher molecular weight ranges (data not shown). This problem can be solved by incubating the nuclei for shorter periods of time with higher concentrations of restriction enzyme (K. Vieira, unpublished results; 17). Another issue that has to be considered is the fact that the precipitation of templates using ChIP results in different concentrations of templates. It is thus important to perform a titration of the templates precipitated with different antibodies.
The combination of ChIP and DMS footprinting will offer investigators the possibility to analyze protein–DNA interactions in DNA regions that are bound by protein in only a small fraction of cells. This method will thus allow for the selection of templates that may represent a specific stage in the transcription cycle.
Acknowledgments
ACKNOWLEDGEMENTS
We thank our colleagues Padraic Levings, Kelly Leach and Christof Dame for encouragement and helpful suggestions. We thank Gail Green for expert technical assistance and members of the Yang laboratory (University of Florida), particularly Christine Mione and Sara Rodriguez-Jato, for help with the in vivo footprinting. This work was supported by grants from the NIH (DK 58209 and DK 52356) to J.B.
REFERENCES
- 1.Roeder R.G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci., 21, 327–335. [PubMed] [Google Scholar]
- 2.Solomon M.J., Larsen,P.L. and Varshavsky,A. (1988) Mapping protein–DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell, 53, 937–947. [DOI] [PubMed] [Google Scholar]
- 3.Orlando V. and Paro,R. (1993) Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell, 75, 1187–1198. [DOI] [PubMed] [Google Scholar]
- 4.Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornstra I.K. and Yang,T.P. (1993) In vivo footprinting and genomic sequencing by ligation-mediated PCR. Anal. Biochem., 213, 179–193. [DOI] [PubMed] [Google Scholar]
- 6.Becker P.B., Weih,F. and Schütz,G. (1993) Footprinting of DNA-binding proteins in intact cells. Methods Enzymol. , 218, 568–587. [DOI] [PubMed] [Google Scholar]
- 7.Gallarda J.L., Foley,K.P., Yang,Z.Y. and Engel,J.D. (1989) The beta-globin stage selector element factor is erythroid-specific promoter/enhancer binding protein NF-E4. Genes Dev., 3, 1845–1859. [DOI] [PubMed] [Google Scholar]
- 8.Forsberg E.C., Downs,K.M. and Bresnick,E.H. (2000) Direct interaction of NF-E2 with hypersensitive site 2 of the β-globin locus control region in living cells. Blood, 96, 334–339. [PubMed] [Google Scholar]
- 9.Maxam A.M. and Gilbert,W. (1980) Sequencing end-labled DNA with base-specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 10.Stamatoyannopoulos G. and Nienhuis,A. (1994) Hemoglobin switching. In Stamatoyannopoulos,G., Nienhuis,A., Majerus,P. and Varmus,H. (eds), The Molecular Basis of Blood Diseases. W. B. Saunders, Philadelphia, PA, pp. 107–155.
- 11.Sawado T., Igarashi,K. and Groudine,M. (2001) Activation of β-major globin gene transcription is associated with recruitment of NF-E2 to the β-globin LCR and gene promoter. Proc. Natl Acad. Sci. USA, 98, 10226–10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motohashi H., Shavit,J.A., Igarashi,K., Yamamoto,M. and Engel,J.D. (1997) The world according to maf. Nucleic Acids Res., 25, 2953–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Globin Gene Server Website (2001) Pennsylvania State University. http://globin.cse.psu.edu.
- 14.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Prüß,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson K., Christensen,H., Zhao,B. and Bresnick,E.H. (2001) Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell, 8, 465–471. [DOI] [PubMed] [Google Scholar]
- 16.Boyd K.E. and Farnham,P.J. (1997) Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol., 17, 2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dekker J., Rippe,K., Dekker,M. and Kleckner,N. (2002) Capturing chromosome conformation. Science, 295, 1306–1311. [DOI] [PubMed] [Google Scholar]