Abstract
Asgard archaea are of great interest as the progenitors of Eukaryotes, but little is known about the mobile genetic elements (MGEs) that may shape their ongoing evolution. Here, we describe MGEs that replicate in Atabeyarchaeia, a wetland Asgard archaea lineage represented by two complete genomes. We used soil depth–resolved population metagenomic data sets to track 18 MGEs for which genome structures were defined and precise chromosome integration sites could be identified for confident host linkage. Additionally, we identified a complete 20.67 kbp circular plasmid and two family-level groups of viruses linked to Atabeyarchaeia, via CRISPR spacer targeting. Closely related 40 kbp viruses possess a hypervariable genomic region encoding combinations of specific genes for small cysteine-rich proteins structurally similar to restriction-homing endonucleases. One 10.9 kbp integrative conjugative element (ICE) integrates genomically into the Atabeyarchaeum deiterrae-1 chromosome and has a 2.5 kbp circularizable element integrated within it. The 10.9 kbp ICE encodes an expressed Type IIG restriction-modification system with a sequence specificity matching an active methylation motif identified by Pacific Biosciences (PacBio) high-accuracy long-read (HiFi) metagenomic sequencing. Restriction-modification of Atabeyarchaeia differs from that of another coexisting Asgard archaea, Freyarchaeia, which has few identified MGEs but possesses diverse defense mechanisms, including DISARM and Hachiman, not found in Atabeyarchaeia. Overall, defense systems and methylation mechanisms of Asgard archaea likely modulate their interactions with MGEs, and integration/excision and copy number variation of MGEs in turn enable host genetic versatility.
Asgard archaea, including Loki-, Hermod-, Thor-, Odin-, Baldr-, Freya/Jord-, Sif-, Heimdall-, Atabey-, Asgard-, and Wukongarchaeia, bridge our understanding of the evolution of eukaryotes and prokaryotes (Spang et al. 2015; Seitz et al. 2016; Zaremba-Niedzwiedzka et al. 2017; Imachi et al. 2020; Farag Ibrahim et al. 2021; Sun et al. 2021; Zhang et al. 2021; Tamarit et al. 2022, 2024; Eme et al. 2023; Valentin-Alvarado et al. 2024). Their genomic features, particularly the presence of eukaryotic signature proteins (ESPs), provide insights into the steps leading to eukaryotic cellular complexity. Recent phylogenetic analyses place eukaryotes within Asgard archaea (Spang et al. 2015; Zaremba-Niedzwiedzka et al. 2017), most closely related to Hodarchaeales (Eme et al. 2023). Despite intense interest in their functionality and evolutionary relationships, little has been reported regarding Asgardarchaeota mobile genetic elements (MGEs) that may shape their population diversity, contribute to genome divergence, and facilitate cross-domain horizontal gene transfer (Ghaly et al. 2022). Recent studies identified viruses of Loki-, Odin-, Thor-, and Heimdallarchaeia (Medvedeva et al. 2022; Rambo et al. 2022; Tamarit et al. 2022; Wu et al. 2022), as well as putative transposons carrying cargo genes that replicate within Heimdallarchaeia (Wu et al. 2022), primarily based on CRISPR spacer targeting. However, the limitations of CRISPR-based approaches and the lack of complete genomes in archaea make it challenging to comprehensively identify viruses and other MGEs in this domain. To our knowledge, no plasmids or integrative conjugative elements (ICEs) have been described for Asgard archaea.
Recently, we reported two complete and three near-complete genomes for Atabeyarchaeia, a new group of Asgard archaea, and a complete genome for Freyarchaeia (Valentin-Alvarado et al. 2024). Here, we present a comprehensive analysis of MGEs in Atabeyarchaeia and Freyarchaeia, two groups of Asgard archaea, using a combination of metagenomic, metatranscriptomic, and epigenetic approaches. Leveraging complete metagenome-assembled genomes and read-based population analyses, we track subtle strain variations of integrated MGEs over a soil depth profile. This approach allows us to establish the host, precisely define MGE insertion sites, and determine MGE lengths.
Our study aims to expand the repertoire of known MGEs in Asgard archaea and provide new insights into their potential roles in archaeal evolution and ecology. Additionally, we investigate MGE integration and excision events in natural populations of Atabeyarchaeia and genomically define groups of circular viruses and unclassified MGEs in Asgard archaea. We also describe genomically encoded defense systems of both Atabeyarchaeia and Freyarchaeia, confirming their expression using metatranscriptomic data, and report methylation patterns that distinguish these archaea, including the identification of transcriptionally active MGE-encoded methylases.
This study demonstrates the power of combining complete metagenome-assembled genomes, population-level analyses, and multiomic approaches to uncover the diversity and functionality of MGEs in uncultured microorganisms from complex environmental samples.
Results
Complete Atabeyarchaeia genomes contain integrated genetic mobile elements
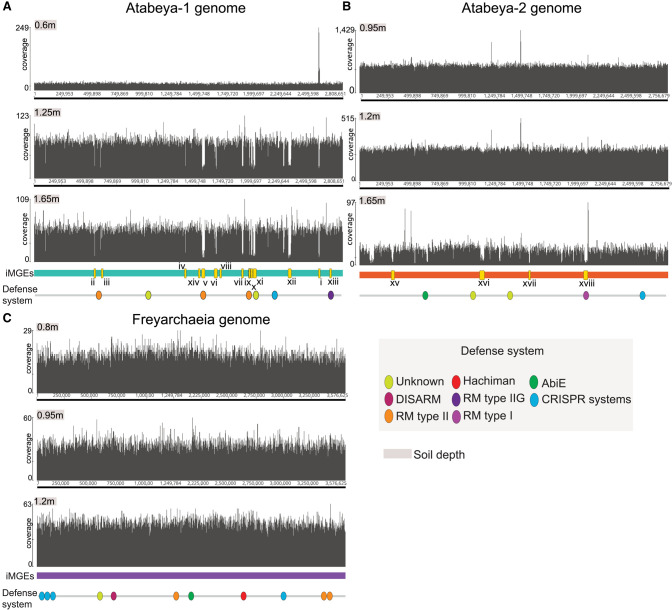
Short- and long-read metagenomic data sets were generated from wetland soil sampled at a single local site (for details, see Methods). We mapped reads from 28 samples collected from soil depths of 60 to 175 cm to the Atabeyarchaeum deiterrae group 1 (GCA_037308085.1), A. deiterrae group 2 (GCA_037310415.1), and Freyarchaeum deiterrae (GCA_037305845.1) genomes previously assembled from this environment and used mapped read details to uncover evidence for integrated, excised, and coexisting circularized MGEs (Fig. 1). The absence of MGEs in some cells can lead to lower-than-average coverage over the integrated region, whereas higher read depth of coverage is likely owing to coexisting extrachromosomal versions of the MGE. The sequences identified occur only once in the genome and do not occur in other genomes from the same sample. By manual inspection of sequencing depth and read alignment discrepancies, we identified 14 chromosomally integrated MGEs in the A. deiterrae group 1 (Atabeya-1) genome (Fig. 2A) and four in the A. deiterrae group 2 (Atabeya-2) genome (Fig. 2B), ranging from 1.3 to 40 kbp in length. No integrated elements were identified in the F. deiterrae genome using this approach (Fig. 2C). Comparison of the sizes of these Atabeyarchaeia MGEs to other known archaeal MGEs reveals that they fall within the typical size range for archaeal plasmids and viruses (Supplemental Fig. S1). The identified MGEs range from 1.3 to 40 kbp, which is consistent with the size distribution observed in other archaeal lineages, including Euryarchaeota and Asgardarchaeota.
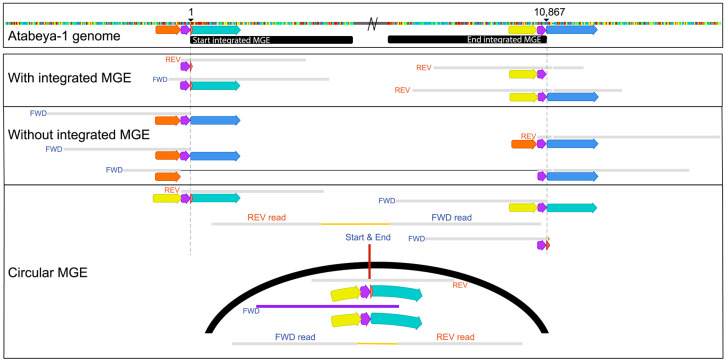
Figure 1.
Read mapping to the reference genome provides evidence for integration and excision, illustrated for the case of one mobile genetic element (MGE). The central region of the integrated sequence of Yucahu-i (between black bars) has been deleted to focus on details of reads mapped to the start and end of the region. Read sequences that match the genome sequences are shown as gray bars; small vertical colored bars adjacent to read portions in agreement with the reference indicate bases that disagree with the reference. The yellow, purple, blue, green, and orange arrows indicate distinct nucleotide sequences (same color, same sequence). In the panel demonstrating that some cells that lack the integrated MGE, one read has been split (the black line links the two parts of a single read) to illustrate agreement with the flanking sequence at both ends of the integrated region. Note that the sequence designated by the purple arrow occurs twice when the MGE is integrated. The vertical colored bars indicate single-nucleotide polymorphisms (SNPs) relative to the reference genome sequence and thus do not represent the sequence itself.
Figure 2.
Chromosomally integrated MGEs and defense systems in soil Asgard Archaea genomes. Each panel (A–C) depicts the coverage across each complete genome, as determined by mapping to metagenome reads derived from three different soil depth profiles. Regions exhibiting low coverage suggest strain variations associated with specific soil depths and may indicate the presence of integrated MGEs in only a subset of cells. Notably, the Freyarchaeia genome exhibits even coverage using reads from all sampling depths, with no discernible integrated MGEs identified. Some of the low-coverage regions are not labeled as potential MGEs; these regions are strain variants with sequences so divergent that read mapping is precluded. Oval symbols indicate predicted defense systems.
These 18 integrated MGEs in Atabeyarchaeia genomes were classified based on their genomic content, size, and phylogenetic analysis of proteins associated to MGEs (for details, see Methods). The classification resulted in five insertion sequence-like transposons (ISs), three putative ICEs (7.9–12 kbp in length) carrying integration machinery and cargo genes, two defense islands, six provirus, and two elements that could not be definitively classified owing to their unique gene contents (Supplemental Table S1). To date, the only Asgard nonviral integrated MGEs reported are Heimdallarchaeia “aloposons” (Wu et al. 2022), which are transposons that carry cargo genes. These previously reported MGEs do not display any similarity at the nucleotide level with those found in Atabeyarchaeia. However, some Atabeyarchaeia integrated MGEs and these aloposons encode partition proteins (ParB-like) that are distantly related (Supplemental Fig. S2A). The Atabeyarchaeia tyrosine-like integrases are most closely related to those found in genomes of Njordarchaeales, Bathyarchaeia, and Aenigmarchaeota, which share a similar ecological distribution in terrestrial wetlands and also occur in deep ocean sediments (Supplemental Figs. S2B, S3; Seitz et al. 2019).
Ten of the integrated MGEs coexist in circularized forms with their integrated versions in the same metagenomic samples (e.g., Fig. 1). One of these of particular interest is Atabeya-1 MGE-i (Yucahu-i, in homage to the son of the Taíno goddess Atabey, reflecting our previous designation of the host archaeon as Atabeyarchaeia), for which the coexisting circularized version in 60 cm deep soil is four times more abundant than the integrated version (Fig. 3A). Some of the reads span the genome, which indicates that a subset of Atabeya-1 genomes lack or have excised this integrated element (Figs. 1, 3B), enabling us to determine the exact length of the Yucahu-i to be 10,867 bp. The MGE is inserted following an AATTAACTTAT sequence that is also present at the end of the integrated Yucahu-i and occurs within the excised, circularized version. This region likely represents the attachment (att) site, a unique location within the genome. The low GC content (9%) compared with the genome-wide average (∼50%) suggests that the DNA in this area may exhibit increased susceptibility to cleavage during processes such as excision or integration.
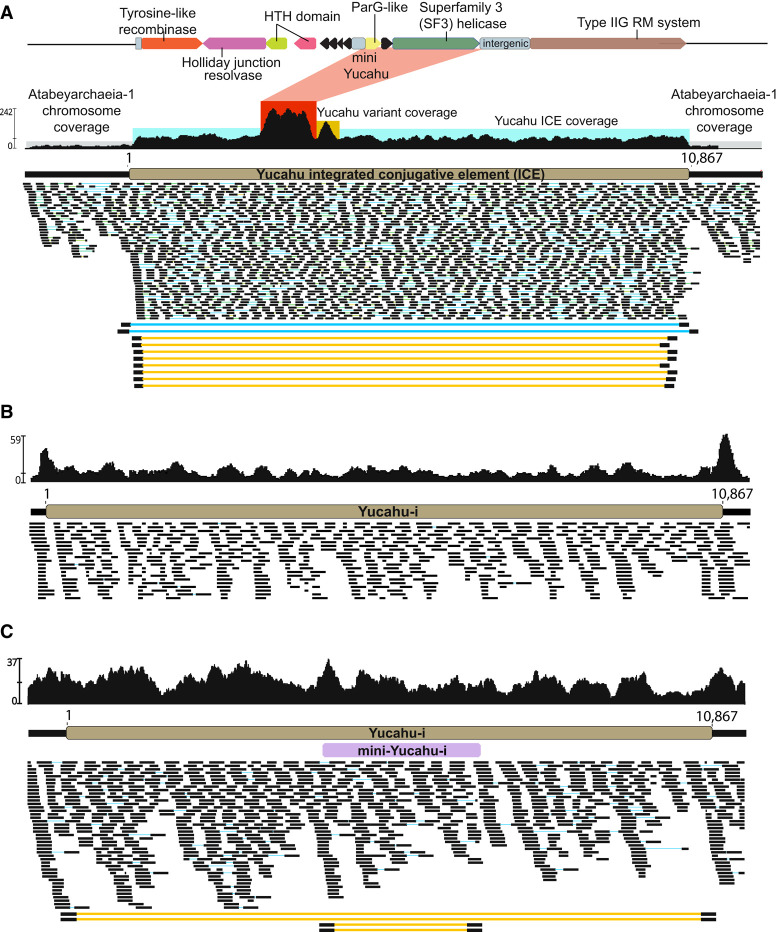
Figure 3.
Integration and excision of Yucahu. (A) For the 60 cm sample, elevated coverage and paired reads indicate that Yucahu-i is integrated into the genome, excised from some genomes (blue lines), and coexists in circularized form (yellow lines). The red box indicates elevated coverage from a related gene from another genome. (B) For the 165 cm sample, low coverage over the MGE and read sequence discrepancies indicate that most cells in this sample lack the MGE. (C) For the 70 cm sample, coverage and paired read information indicate that Yucahu-i is integrated into essentially all cells. The circularized Yucahu-i is present but rare. Paired reads pointing out internal to the MGE indicate that a 2644 bp element has integrated into the plasmid and coexists in circularized form.
The Yucahu-i element includes 11 open reading frames (ORFs) (Fig. 3A). Some of the gene products could be functionally annotated using protein homology and in silico structural prediction. The first gene encodes a tyrosine recombinase/integrase that likely recognizes and cuts at the AATTAACTTAT motif in the genome and in the circularized version (resulting in Yucahu-i linearization) and may be involved in integration of the linear sequence. The subsequent gene is a Holliday junction resolvase, which likely acts in conjunction with the integrase. We are uncertain if a host integration factor is required, but it is possible that two of the following genes predicted to encode DNA-binding proteins, based on their HTH-domains, may have this function. Yucahu-i also encodes a superfamily 3 (SF3) helicase that may unwind the DNA and initiate plasmid replication (Guo and Huang 2010), and a novel Type IIG restriction-modification (IIG RM) protein fusion that combines endonuclease and methyltransferase (MTase) activities. Phylogenetic analyses suggest the IIG RM sequence shares its most recent common ancestor with sequences found in DPANN archaeal genomes (>60% amino acid identity). Basal to this clade are many sequences from bacteria, which supports the inference that the origin of the sequences in question is likely archaeal, potentially acquired via horizontal gene transfer (Supplemental Fig. S4). Metatranscriptomic data indicate that the IIG RM gene is transcribed (Supplemental Table S2). Based on predicted protein functions and presence of the excised, circular (copy ratio up to 4×) MGE, Yucahu-i is likely an ICE (Wozniak and Waldor 2010).
We investigated how frequently Yucahu-i was integrated in, or coexisted in circular form with, the Atabeya-1 genome by systematically analyzing reads from the 20 soil metagenomes that contained this archaeon (Supplemental Fig. S5; Supplemental Table S3). In 11 samples, Yucahu-i is integrated into essentially all Atabeya-1 cells; however, the data indicate substantial variation in presence/absence of the integrated version and in the copy number of the circularized version (Fig. 3A; Supplemental Fig. S5; Supplemental Table S3). A few reads from the 70 cm deep soil revealed evidence for the circularization of a 2644 bp element that is integrated within Yucahu-i. We refer to this as mini-Yucahu-I (Fig. 3C). Its presence highlights the genetic plasticity of the plasmid. The mini-Yucahu-i carries a putative ParG, a hypothetical protein, and Holliday junction ATP-dependent DNA helicase RuvB. Interestingly, the identical 11 bp Yucahu-i putative attachment motif is also present adjacent to, and within, a 7848 bp integrated element in the Atabeya-2 genome (iMGE-xvi). However, the genomes share no detectable similarity; the percentage of identity of the tyrosine integrases are <25%; and they are phylogenetically unrelated (Supplemental Fig. S6).
Viruses and plasmids targeted by CRISPR systems
To explore exogenous MGEs of Atabeyarchaeia and Freyarchaeia, we mined CRISPR spacers from their genomes and matched them to unbinned metagenomic scaffolds from the same wetland soil. More than 30 putative MGE scaffolds are confidently targeted by CRISPR spacers and thus are predicted to have once replicated within Atabeyarchaeia (Supplemental Table S4). We manually curated them and obtained one complete 20.8 kbp circular plasmid genome; two circular, complete 40.1 kbp genomes for a pair of closely related viruses; and a circular complete 26.7 kbp genome for an unclassified MGE.
The 20.8 kbp plasmid has 24 ORFs, primarily encoding hypothetical proteins (Supplemental Fig. S7; Supplemental Tables S5, S6). It also encodes plasmid proteins such as protein repressor ribbon–helix–helix protein from the CopG family (Gomis-Rüth et al. 1998), usually present in bacterial conjugative plasmids. Other predicted proteins are implicated in autonomous replication, such as a DNA primase-helicase, mini-topoisomerase VI, and a tyrosine integrase, as well as other genes, involved in nucleic acid processing. Seven of these proteins contain transmembrane domains, suggesting the presence of a putative conjugative system or a secretion-like system (Supplemental Fig. S7). A protein with a Glu–Glu motif was annotated as an integral membrane CAAX-like protease self-immunity, based on structural modeling and phylogeny (Supplemental Fig. S8). It encodes a NTPase with similar function to ParB, a protein typically associated with plasmid chromosome partitioning during replication. Phylogenetic analysis places this protein within a clade that contains MGEs recently discovered in Heimdallarchaeia (Supplemental Fig. S2A). Interestingly, this clade also contains ParB-like proteins from publicly available draft genomes of Lokiarchaeia and Thorarchaeia, along with other archaeal genomes (e.g., Sulfolobales), suggesting that this plasmid lineage is widespread in other Asgard archaea. Also included are sequences from Streptomyces plasmids. Therefore, these plasmid partitioning genes may have undergone inter-domain horizontal gene transfer.
We identified a circular 40,094 bp novel virus predicted to infect Atabeyarchaeia-2 based on CRISPR spacer matches. A search against the IMG/VR v4 (Camargo et al. 2023) database revealed no matches for these spacers to any known viral sequence, highlighting the novelty of both the virus and the host. Genomic analysis of the virus with geNomad (Camargo et al. 2024) classified this group of viruses within the class Caudoviricetes (realm Duplodnaviria) (Supplemental Table S7). We named this virus “Opia” after a mythical creature associated with the Taíno goddess Atabey. Interestingly, we found this virus integrated into the end of a 2.58 Mbp PacBio-derived genome fragment from a related Atabeya-2 strain (Atabeya-2′), confirming its host association and a putatively temperate replication cycle.
The Opia virus genome encodes structural proteins typical of archaeal tailed viruses, including capsid-like, phage head morphogenesis, portal, and tail-like proteins, as well as a large terminase subunit (KEGG: K06909). Phylogenetic analysis of capsid and terminase proteins groups Opia with other Asgard viruses, notably Nidhogg virus (Fig. 4A,B). The genome also contains genes for nucleic acid processing, including a Mu-like prophage protein, putative transposase, tyrosine recombinase-like enzyme, site-specific DNA-MTase, and a ParB-like NTPase.
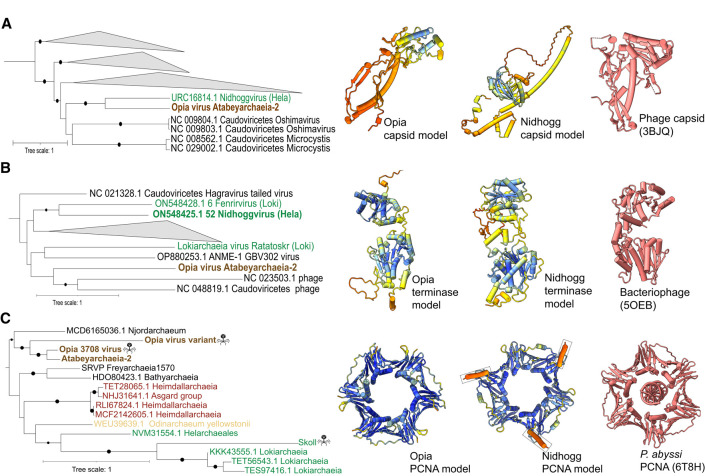
Figure 4.
Evolutionary relationships and structural conservation of Opia virus hallmark proteins. (A–C) Maximum-likelihood phylogenetic trees of capsid (A), terminase large subunit (B), and proliferating cell nuclear antigen (PCNA; C). Trees are midpoint-rooted; circles indicate bootstrap support >70%. Insets show structural models: Opia virus (infecting Atabeyarchaeia, this study) and Nidhogg virus (infecting Helarchaeales) colored by AlphaFold3 confidence (blue to red, high to low), with best FoldSeek match in coral. Reference structures: bacteriophage capsid (PDB: 3BJQ) for A, large terminase from thermophilic bacteriophage D6E (PDB: 5OEB) for B, and P. abyssi PCNA (PDB: 6T8H) for C. Scale bars represent one substitution per site. Opia virus variants cluster with other Asgardarchaeota MGEs, highlighting their evolutionary relationships within this archaeal phylum.
Structural models of Opia virus hallmark proteins, generated using AlphaFold 3, reveal conservation with known viral and archaeal structures (Fig. 4). The capsid and terminase models show similarity to Nidhogg virus and reference bacteriophage structures, suggesting potentially functional conservation across diverse viral lineages. The PCNA-like protein model closely resembles that of Nidhogg virus and Pyrococcus abyssi PCNA, indicating a conserved role in DNA replication.
A DNA polymerase sliding clamp subunit (PCNA-like protein) was identified (Fig. 4C), which may promote viral DNA synthesis and manipulate host pathways. Phylogenetic analysis places the Opia PCNA-like sequence within an Asgard archaea clade, with its closest homolog in Njordarchaeales (MCD6165036.1) from the Auka vent field (Speth et al. 2022), suggesting that viruses related to Opia virus integrate into other Asgard genomes. Similar proteins are found in the Sköll viral genome infecting Lokiarchaeia and other archaeal viruses (Raymann et al. 2014; Mizuno et al. 2019; Medvedeva et al. 2022; Rambo et al. 2022; Tamarit et al. 2022).
At the whole-proteome level, the Opia virus clusters with other Atabeyarchaeia MGEs and has similarity to the Ratatoskr, Nidhogg, Skoll, and Fenrir viruses (Rambo et al. 2022), known to infect various Asgard archaea (Fig. 5A). Analysis of genome sizes among reported Asgard viruses, including Opia virus from this study, reveals a diverse range of genomic lengths (Fig. 5B; Supplemental Table S8). The Opia virus, with its 40,094 bp genome, falls within the mid-range of Asgard viral genome sizes, which span from ∼15 kb to >100 kb. This variation in genome size likely reflects the diverse replication strategies and host interactions among Asgard-infecting viruses, with Opia representing an intermediate complexity within this spectrum.
Figure 5.
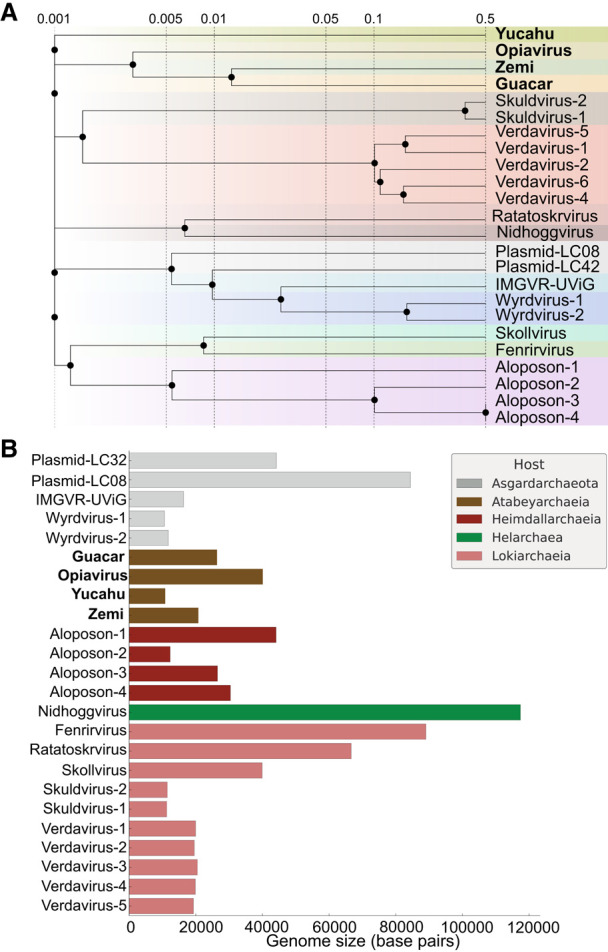
Comparative genomics of Asgardarchaeota MGEs. (A) Whole-proteome similarity network of MGEs from Atabeyarchaeia and other Asgardarchaeota (Medvedeva et al. 2022; Rambo et al. 2022; Wu et al. 2022). Nodes represent individual MGEs, with edge thickness indicating similarity strength. Yucahu, Opia virus, Zemi, and Guacar form a distinct cluster, separate from other Asgard MGEs. Analysis based on NCBI database entries. (B) Genome size distribution of reported Asgard viruses/MGEs, including those from this study.
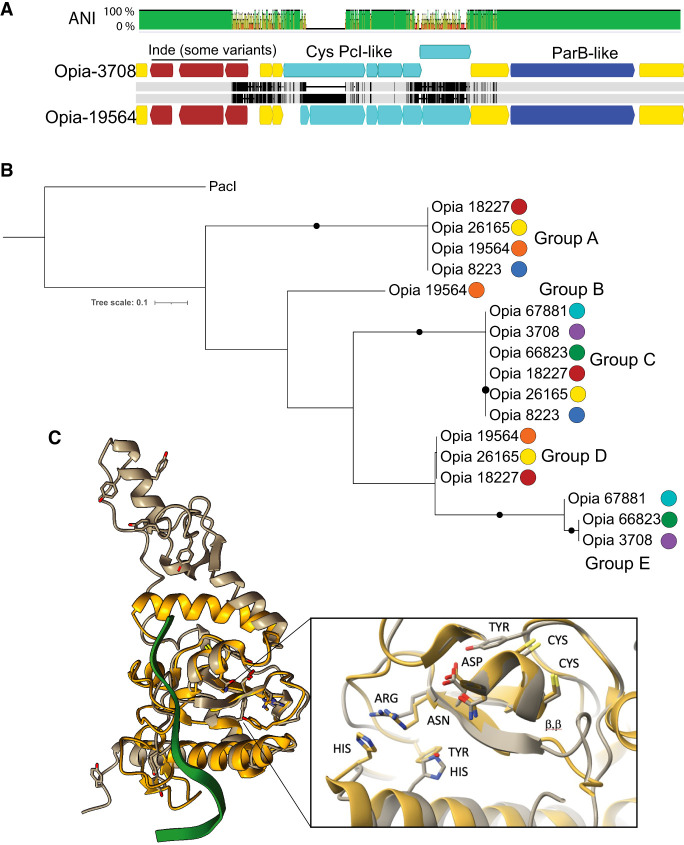
We identified at least seven distinct Opia virus variant genotypes from different samples. The sequences align near-perfectly over >85% of the genomes (Fig. 6A). All Opia variants (and their identifiable fragments) are exactly targeted by one CRISPR spacer present in loci of both Atabeya-2 and Atabeya-2′ (two identical sequential spacers in Atabeya-2′), despite the presence of the Opia provirus in the Atabeya-2′ genome. The provirus was only partially recovered, so it is impossible to say whether the integrated version differs in the targeted region. A hotspot in the Opia virus genomes encodes a series of genes that are distinctly different in some variants. Included are up to six small cysteine-rich proteins, many with predicted double Zn-binding domains (Fig. 6A). The genes for specific cysteine-rich proteins occur in different combinations from different genotypes (e.g., one has sequence types A, B, D; another has A, C, D; and another has C, E) (Fig. 6B). In addition, a three-gene block (one of which has sequence variants) and adjacent intergenic sequences are variably present/absent. Finally, different versions of ParB-like partition proteins occur in the variable region, and some lack a C-terminal endonuclease domain (Fig. 6A).
Figure 6.
Sequence variation in the cysteine-rich proteins of Opia viruses and comparison to Pacl, a rare-cutting HNH restriction endonuclease. (A) The aligned ∼5.6 kbp variable region of two Opia viral genotypes. Light gray bars indicate perfect nucleotide identity, and thin vertical black lines indicate SNPs. The three genes labeled in brown are present in both Opia-3708 (top) and Opia-19564 (bottom) but are absent in some other genotypes. Light blue genes are cysteine-rich (For 3708L, 10, five, nine, five, 10; for Opia-19564, three, 14, five, nine, four, 10 cysteines per protein). Dark blue genes encode ParB-like proteins that are very divergent between some genotypes (Opia-3708 vs. Opia-66823, 67881). Before the three-gene indel and after the ParB-like gene, the ∼35 kb regions of all genomes are essentially identical. (B) Phylogenetic tree including the 17 larger cysteine-rich proteins from the variable regions of seven Opia genomes (numbers represent genome names) (for context, see Supplemental Material). Proteins with identical sequences occur in five different combinations across the genotypes. (C) Comparison of the active site region of PacI (with nine cysteines) and the structure of Opia-19564 protein 16 (with 10 cysteines; silver). Active site residues of PDB 3m7k (gold) are displayed based on figure 1 of Shen et al. (2010). The critical ββα-metal motifs are well aligned. Tyrosine and histidine residues exist in proximity to the active site, although their locations differ somewhat, possibly owing to fold inaccuracy.
We predicted the structures of the largest cysteine-rich protein from Opia-3708 (gene 46) and two Opia-19564 proteins (genes 16, 18). All three represent different protein sequence clusters, but they share core structural components, including two sets of four cysteines, and an alpha helix in proximity to paired antiparallel beta strands (i.e., ɑ, ββ,-metal; Fig. 6C). HHpred predicts the Opia-3708 protein (197 aa) to be related to an HNH endonuclease, and the best match for the three-dimensional structure (PDB 3M7K) is the Rare-Cutting HNH Restriction Endonuclease PacI, a homing endonuclease that is one of the smallest restriction endonucleases known (142 aa) (Shen et al. 2010). Zinc bound by the four cysteines is required for the DNA cleavage by PacI endonucleases. The H, DR, and CxxCN catalytic residues of 3M7K HNH endonuclease are generally conserved in the Opia proteins (e.g., Opia-19564_16) (Fig. 6C). However, the expected tyrosine residue precedes rather than follows the DR motif, and its placement in the predicted structure is offset from that in 3M7K. The histidine active site residue is also slightly differently positioned (Fig. 6C). These discrepancies may be attributed to uncertainties in protein folding. However, the positioning of histidine in the location typically occupied by tyrosine suggests its potential involvement in DNA cleavage, as occurs in other HNH endonucleases. Elsewhere, the predicted structures have large regions of positively charged surface, likely involved in DNA binding. These findings suggest that the Opia proteins share characteristics with PacI restriction endonucleases, yet they may represent a novel class of enzymes, likely with homing endonuclease function (Fig. 6B). The biochemically characterized PacI homodimer has a target recognition sequence of 5′-TTAATTAA-3′ and cleaves between the internal thymine residues. Pacl endonucleases rely on the absence of the recognition site elsewhere in the host genome. We could not determine the recognition sequence for the Opia Pacl-like homing restriction endonuclease, but apparently it was possible for different combinations of six variants to insert in the same region of a series of Opia genotypes.
We further reconstructed a circular, complete 26,349 bp genome (MGE-9917) for another circularized element that is targeted by three CRISPR spacers from Atabeya-1. This element, named “Guacar” after the twin son of Atabey, could not be definitively classified as either a virus or plasmid based on its predicted protein functions. All Guacar genes are encoded on the same strand. The genome features a CT-rich intergenic tandem repeat region with six, seven, eight, or nine units of 8 bp in length (variants identified using mapped reads). MGE-9917 encodes at least 14 proteins with transmembrane domains, two of which are 1402 and 1202 amino acids in length and lack related sequences in the NCBI database (Supplemental Tables S5, S6). The genome contains a protein that combines an N-terminal ParB-like nuclease domain with a C-terminal tRNA G10 N-methylase Trm11. Additionally, Guacar-9917 includes genes for a tyrosine recombinase, a transposase, and a Type IV methyl-directed restriction enzyme featuring an HNH motif. A family of related elements occurs in virtually all of the deep soil samples. One version differs owing to the presence of a transposase that is related to those found in the Opia viruses.
Defense systems and epigenetic regulations in Atabeyarchaeia and Freyarchaeia
We used DefenseFinder (Tesson et al. 2022) and PADLOC (Payne et al. 2022) to identify 10 defense systems in Freyarchaeia, five in Atabeyarchaeia-1, and six in Atabeyarchaeia-2. Freyarchaeia harbored at least four different defense system classes, including type I-B, III-A, and III-D CRISPR-Cas systems; IIG RM systems; the Hachiman antiphage defense system; and the antiphage system, defense island system associated with restriction-modification (DISARM) (Supplemental Fig. S9A; Supplemental Tables S9, S10). The identification of DISARM in Freyarchaeia adds to the growing list of defense systems recently reported in Asgard archaea (Leão et al. 2024). The DISARM system comprises drmABC, a MTase (drmMI, N6 adenine-specific MTase or drmMII, C5 cytosine-specific DNA MTase), and drmD or drmE (Ofir et al. 2018). The Freyarchaeia system includes drmA, drmB, drmC, drmMII, drmE, and drmD (helicase similar to the RNA polymerase [RNAP]-associated SWI2/SNF2 protein), classifying this system as a DISARM class II (DISARM-II). Interestingly, drmD is a homolog typically found in DISARM class I. The DISARM methylase modifies host CCWGG motifs to distinguish its own DNA from foreign DNA. A specific conformation of the DrmAB complex (trigger loop) inhibits the complex to prevent an autoimmune response (Bravo et al. 2022). DrmA is responsible for DNA targeting in DISARM through multiple nonspecific interactions with the DNA backbone (Bravo et al. 2022). By not requiring a specific sequence for DNA binding, DrmA distinguishes this defense system from other common restriction-modification systems, endowing DISARM with a broad spectrum of action against viruses (Tesson et al. 2022). Phylogenetic analysis of the helicase DrmA places the gene within the Euryarchaeota and Chloroflexota, suggesting that this system has been laterally transferred (Supplemental Fig. S9B). The Hachiman system is encoded by hamA, a DNA endonuclease (DUF1837), and hamB, a ski2-family helicase (pfam00271) bearing relation to archaeal Hel308 (Doron et al. 2018; Tuck et al. 2024). Recently, a model for Hachiman-mediated defense was reported (Tuck et al. 2024). Helicase HamB senses DNA structures from DNA damage (or structurally related replication intermediates), unleashes further DNA degradation through HamA endonuclease activity (Tuck et al. 2024), and confers antiphage immunity through abortive infection. Whether Hachiman-mediated immunity performs a similar role in archaea and synergizes with other immune systems remain unknown. Potentially, an ATP-dependent endonuclease is an overcoming lysogenization defect (OLD) upstream of the hamAB locus, providing synergistic protection against invading MGEs.
We used DNA polymerase kinetics from Pacific Biosciences (PacBio) metagenomic sequencing data and the Restriction Enzyme database (REBASE) to illuminate the DNA methylation patterns and methylases in the genomes of Freyarchaeia, Atabeya-1, and Atabeya-2. In the genome of Atabeya-1, we identified 13 methylation sequence motifs, of which seven were directly linked to a specific methylase gene. Similarly, Atabeya-2 has 11 methylation motifs, five of which could be linked to a methylase. Freyarchaeia has only five detectable methylation motifs (Supplemental Table S11A,B). Interestingly, one of those motifs is CCWGG, which has been characterized as a motif targeted by DISARM class II.
Atabeya-1 and Atabeya-2 both have 4-methylcytosine (m4C) and 6-methyladenosine (m6A) methylation. Atabeya-1 Yucahu-i MGE encodes a IIG RM system that targets an m6A methylation motif and is the only candidate enzyme that could methylate the Atabeya-1 genome. The Yucahu-i system is analogous to the MmeI family, which typically recognizes a 6–7 bp motif with adenine as the penultimate base (Morgan et al. 2009). The motif GYATGAG (m6A) was methylated at 66% of sites within the Atabeya-1 genome and could represent the active methylation motif of this RM system. In contrast, Freyarchaeia has five m4C motifs but no m6A methylation motifs.
To validate these findings, we analyzed previously sequenced Oxford Nanopore soil data (Schoelmerich et al. 2024). Both technologies support the m6A motifs. Although there was no Nanopore model for calling m4C, overextended versions of the m4C motifs were also detectable as m6A motifs (i.e., GCGm4C was detected as GCGCm6A) (Supplemental Table S11B,C). Additionally, two potential 5mC motifs were called: GATm5C in Freyarchaeia-1 and RG5mCWGCY in Atabeya-1.
Integrated mobile-like regions encode ESPs
We identified two small GTPases in a region of the Atabeya-1 genome that appears to be enriched in genes often associated with MGEs. This region is absent in some Atabeya-1 strains. The classification of these proteins as GTPases is supported by sequence and structural homology, as well as structural predictions in comparison to reference eukaryotic sequences (Supplemental Text). Based on phylogenetic analysis, these proteins cluster with Arf GTPases from other Asgardarchaeota and eukaryotes (Supplemental Fig. S10).
Discussion
Here, we describe chromosomally integrated and coexisting MGEs that replicate in Atabeyarchaeia by leveraging extensive sequencing of a series of soil samples in which soil depth and biogeochemical conditions select for different strain variant populations. By examining read mappings across multiple samples from the same environment, we established a larger repertoire of MGEs than could be found from the analysis of any single metagenome. Integrated elements and coexisting circularized MGEs range from 2.5 to 40 kb in length; all complete MGEs were circular, and at least some replicate bidirectionally. We could not confidently identify MGEs by changes in read mapping abundances in the Freyarchaeia genome. This might indicate stable integration into the Freyarchaeia genome across the entire population studied (thus excision sites and coexisting versions of circular elements were not detected by our methods). Assuming that Freyarchaeia do carry MGEs that excise, the best approach to finding them would be to analyze populations from a larger set of samples. The presence of coexisting integrated and free, circularized MGEs, likely mostly plasmids, as well as variation in copy number of circularized elements and in the fraction of cells with integrated elements, suggests regular movement of MGEs into and out of the Atabeyarchaeia chromosomes. Insertion/excision and variation in copy number may enable Atabeyarchaeia to respond to changes in their environment. For example, MGEs may behave synergistically, and increase in MGE copy number (thus gene content) serves as a response to increased pressure from other MGEs (Krupovic et al. 2019). The tiny mini-Yucahu indicates another layer of genomic variability, as just this portion of the host MGE can excise.
Interestingly, the attachment motifs for Yucahu-i plasmid-like Atabeya-1 MGE and for a circular, unclassified and essentially unrelated MGE linked to Atabeya-2 are exactly the same, implying that the very different integrases of each (25% aa ID) recognize and cut at the same motif. Protein sequence divergence may enable host chromosome specificity, yet the active site apparently evolved to target the same motif.
The novel cluster of genomically similar Opia viruses of Atabeya-2 and Atabeya-2′ encode sequential cysteine-rich proteins inferred to have nuclease activity owing to their distant homology with PacI. A set of two or three of five protein types occurs in the seven genomes, but the set present is generally different, except in the case of yellow and orange genotypes (Fig. 5B), which both have group A, B, and D protein types. These involve combinatorial patterns (Fig. 5B) that may have arisen via recent recombination events in which these putative endonucleases may have played a role. The multiple variants might form heterodimers rather than the normal homodimers expected for Pacl, possibly extending target recognition. The results suggest the importance of diverse nuclease activity for these viruses.
The Opia virus proteomes exhibit similarities (e.g., capsid, tube and tail proteins) to those of tailed viruses, which commonly replicate in bacterial and archaeal hosts from hypersaline environments (Senčilo and Roine 2014). They are quite distinct from those of eukaryotic viruses, supporting the suggestion that, despite the evolutionary relationship between Asgard archaea and eukaryotes, their viruses display no obvious evolutionary relationships (Medvedeva et al. 2022; Rambo et al. 2022; Tamarit et al. 2022).
Genome context (dominated by MGE-associated genes) and apparent excision of the region encoding the GTPases from some strain genotypes supports the inference that some Atabeyarchaeia MGEs encode ESPs. ARF GTPases are involved in membrane trafficking in eukaryotes and have been previously described as ESPs in Asgard archaea (Spang et al. 2015; Eme et al. 2023). If it is established that this is an active MGE that can excise, the presence of these GTPases provides the first indication that increase in cellular complexity could be associated with the transfer of ESPs via MGEs (Supplemental Figs. S10–S12; Supplemental Tables S5, S11).
Our results suggest several examples of genes integrated into Atabeyarchaeia genomes or in their coexisting MGEs that have nearest homologs in the bacterial domain (e.g., ParB, DrmA, and IIG RM enzymes). These findings are consistent with recent work on cross-domain gene transfer via movement of integrons associated with diverse MGEs (Ghaly et al. 2022) and extend earlier work inferring the acquisition of archaeal genes by bacteria (e.g., Hug et al. 2013). As we discover new Asgard archaea from genome-resolved metagenomes, we can expect to find further parallels between bacterial and archaeal immune systems. These associations could have implications for the evolution of eukaryotic immune systems (Wein and Sorek 2022).
IIG RM systems, to our knowledge, have not previously been associated with archaeal MGEs. The observation that the only methylase seemingly able to methylate at the host genome's well-represented m6A motif is carried by Yucahu-i, as well as that it is transcriptionally active, suggests that the Yucahu-i Type IIG plays a significant role in host genome epigenetic modification. This MGE-encoded system may protect its host Atabeyarchaeia against infection by other MGEs. This behavior aligns with the emerging perspective that defense systems themselves can serve as MGEs (Rocha and Bikard 2022; Wu et al. 2022).
To our knowledge, these are the first metagenome-derived Asgard archaeal complete genomes for which methylation patterns have been reported. PacBio sequences corresponding to these complete, manually curated genomes (Valentin-Alvarado et al. 2024) were used to infer the methylation motifs and to determine the fraction of sites that were methylated, as well as the methylation patterns of their newly reported MGEs. Using REBASE, which features all biochemically characterized methylases, it was possible to link methylated sites with likely methylases encoded on both genomes and MGEs. Relatively little is known about genome methylation in archaea, especially in Asgard archaea (Anton and Roberts 2021). These genomes and their methylation motif data presented here provide a starting point for detailed biochemical studies to expand the known inventory of archaeal methylases. The higher number of methylation motifs in Atabeyarchaeia compared with the Freyarchaeia genome could be an evolutionary response to the larger inventory of MGEs associated with Atabeyarchaeia.
We leveraged the read diversity inherent to population genomic data, long-read sequencing, methylation pattern analysis, comparative genomics, and functional and structural prediction to explore integrated and coexisting MGEs of one group of Asgard archaea. These analyses brought to light an extensive landscape of MGEs that associate with Atabeyarchaeia, including viruses, plasmids, and as-yet-unclassified entities (Fig. 7). The excision, insertion, and changes in copy number of these MGEs may enable adaptation to changing conditions and have contributed evolution, possibly to the acquisition and spread of genes linked to the origin of cellular complexity. The availability of MGEs that could be adapted for delivery of genome editing tools in a community context (Rubin et al. 2022) may pave the way for genetic manipulation of these archaea.
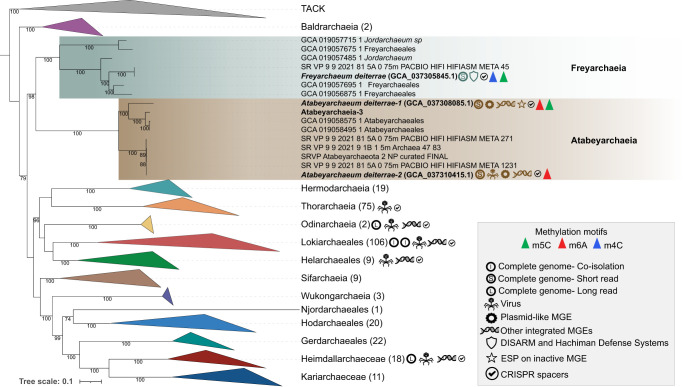
Figure 7.
Phylogenetic placement of Asgard archaeal genomes and distribution of associated MGEs. The phylogeny shows the evolutionary relationships among different Asgard archaeal lineages, with symbols indicating the presence and types of published MGEs in various candidate groups. Descriptions of the phylogenomic analyses have been previously described in detail (Valentin-Alvarado et al. 2024). Our work on Atabeyarchaeia and Freyarchaeia is highlighted, demonstrating the novel MGEs identified in this study and the presence of two novel defense systems in Freyarchaeia. This figure provides a comprehensive overview of the current state of knowledge regarding MGEs across the Asgardarchaeota phylum.
Our study opens up avenues for future research. The first relates to comparative genomics of MGEs across the Asgard archaea and beyond, including investigations across diverse environments and temporal studies. Using enrichments of Asgard archaea or, if they become available, pure cultures, experimental studies could test the adaptive significance of specific MGEs. With enrichments, genetic manipulation methods designed for community editing may be used to inactivate prophages or eliminate the function of specific genes (e.g., methylases). Methylases are of particular interest, given that novel methylation patterns and restriction-modification systems were identified in Atabeyarchaeia and Freyarchaeia and are likely involved in epigenetic regulation and host–MGE interactions. Specifically, the unassigned methylase genes for unassigned motifs reported here per genome, are immediate targets for biochemical studies via heterologous expression. Of particular interest would be the functional characterization of MGE-encoded genes, including methylases, potentially linked to the development of multicellularity.
Methods
Sample acquisition, nucleic acid extraction, and sequencing
Metagenomic data were generated from deep wetland soil in Lake County, California (Al-Shayeb et al. 2022; Valentin-Alvarado et al. 2024). Briefly, we collected soil cores from a seasonally flooded wetland (SRVP) in Lake County, California, in October 2018, October 2019, November 2020, and October 2021 (38°41′39″N 122°31′36″W 571 m). Samples were frozen in the field using dry ice and kept at −80 C until extraction. The Qiagen PowerSoil max DNA extraction kit was used to extract DNA from 5–10 g of soil, and the Qiagen AllPrep DNA/RNA extraction kit was used to extract RNA from 2 g of soil. Samples were sequenced by the QB3 sequencing facility at the University of California, Berkeley on a NovaSeq 6000. Read lengths for the 2018 DNA samples and the RNA samples were 2 × 150 bp and 2 × 250 bp for the 2019–2021 DNA samples. A sequencing depth of 10 Gb was targeted for each of 2018, 2020, and 2021 samples and 20 Gbp for each of the 2019 samples. PacBio sequencing was obtained from a subset of deep soil samples from 2021 via the University of Maryland sequencing facility. Samples from September 9, 2021, from 140 cm and 75 cm were sequenced using a Sequel II to generate PacBio HiFi reads. Reads were quality trimmed using BBDuk (bbduk.sh minavgquality = 20 qtrim = rl trimq = 20) (Bushnell 2014) and assembled with hifiasm-meta (Feng et al. 2022).
Discovery of integrated genetic elements using closed complete genomes
Our manual approach to identifying integrated MGEs was based on three criteria:
Anomalous coverage: We evaluated regions with unusually low or high coverage compared with the host chromosome. Coverage changes occurring in localized regions often indicate MGE boundaries. We used the following rule to determine candidate integrated genetic elements: If a region's coverage was significantly different from the overall host chromosome (e.g., 3× higher or lower), we delineated it as a candidate MGE (see section Coverage Calculation of Integrated MGEs and Host-Chromosome).
Short reads supporting excision: We examined reads that indicated potential excision events. This included paired reads with large separations that mapped to locations before and after the integration boundaries, or partially discrepant reads in which the discrepant portion matched the sequence on the opposite side of the integration region (see Fig. 1).
Gene annotations: We analyzed the functional annotations of genes within candidate regions (see section Functional Annotation of Candidate MGEs). Although these often included functions associated with plasmids or viruses, we also frequently observed an unusually high proportion of hypothetical proteins in these elements.
To implement this strategy, metagenome reads were aligned to the reference genome sequences and coverage anomalies and read discrepancies identified. Read coverage was employed to determine the relative abundance and distribution of each MGE across the different samples. Additionally, in-depth examination of reads sequence discrepancies allowed for the identification of integration sites and mini elements within other integrated elements. To further characterize the MGEs, we compared genomes lacking the MGE with those containing them. This combination of approaches enabled us to accurately determine the length of the MGE and pinpoint the specific sites of integration within the plasmid.
Coverage calculation of integrated MGEs and host-chromosome
We aligned metagenome reads to reference genomes using the BBMap's short-read aligner. Using minimum identity threshold of 0.95, ambiguously mapping reads were discarded. The coverage values of the MGEs and total genome coverage values were calculated from the alignment/map (BAM) files. The positions of integrated elements were defined. For the genome coverage calculation, the entire genome was divided into two sections: the region before the start of the larger integrated element and the region after the end of the larger integrated element. This approach enabled systematic, efficient calculation of the coverage of integrated elements within genomes.
Functional annotation of candidate MGEs
We functionally annotated putative MGEs using multiple databases and tools. Protein families and domains were identified using TIGRFAMs 15.0 and Pfam v31.0 (both downloaded September 2018; Haft et al. 2013; Paladin et al. 2021). Carbohydrate-active enzymes were annotated using CAZymes (dbCAN-HMMdb-V7, dbCAN v2, downloaded September 2018; Drula et al. 2022). Peptidases were identified using MEROPS (November 2018) (Mistry et al. 2007). Hydrogenases were annotated using HydDB (November 2018) (Søndergaard et al. 2016). Clusters of orthologous groups (COGs) were assigned using the NCBI COG database (https://www.ncbi.nlm.nih.gov/research/cog; November 2018). Membrane transporters were identified using TransportDB (http://www.membranetransport.org/transportDB2/index.html; November 2018; Galperin et al. 2021). We employed InterProScan (Jones et al. 2014; v5.50–84.0) for comprehensive protein annotation. All annotations were performed using the pipeline described by Dombrowski et al. (2020) with default parameters. Detailed results are provided in Supplemental Table S6.
For MGE-specific annotations, we utilized several manually curated databases: Nucleo-Cytoplasmic Virus Orthologous Groups (NCVOGs) (Yutin et al. 2009), prokaryotic Virus Orthologous Groups (pVOGs) (Grazziotin et al. 2017), Prokaryotic Virus Remote Homologous Groups (PHROGs) (Terzian et al. 2021), Phage Artificial Neural Networks (PhANNs) (Cantu et al. 2020), Virus Orthologous Groups (VOGs) (Bao et al. 2004), and Giant virus metagenome-assembled genomes (GVMAG) (Schulz et al. 2020; Rambo et al. 2022). Viral and plasmid protein hallmarks were classified using geNomad v. 2.16 (Camargo et al. 2024). Given the challenges in annotating archaeal MGEs owing to limited reference databases and validated features, we conducted manual inspections supplemented with phylogenetic and gene synteny analyses for these sequences. Detailed results are provided in Supplemental Table S5.
Identification and genome curation of Atabeyarchaeia-associated exogenous MGEs
We used metagenomic data sets to search for candidate MGEs associated with Atabeyarchaeia and Freyarchaeia. Screening was based on taxonomic profile, GC contents, and CRISPR-based targeting. All the candidate contigs were manually curated using Geneious Prime-2023.1.2 (https://www.geneious.com). The manually curated genomes were de novo reconstructed from high-quality Illumina metagenomic data. Manual genome curation methods generally follow the method of Chen et al. (2020). Long-read PacBio data were used to verify and expand the sequence data set. Replichores of complete genomes were predicted according to the GC skew, and cumulative GC skew was calculated by the iRep package gc_skew.py (https://github.com/christophertbrown/iRep). We classified complete MGE genomes as viruses if they contained viral structural genes. Genomes lacking viral structural genes were categorized as plasmids or other MGEs (such as transposons or conjugative elements) based on length, the presence of transposases (classified using ISFinder) (Siguier et al. 2006), genomic composition, and the presence of circularized and excised forms. Genomes not fitting these categories were left unclassified. We further validated our viral and/or plasmid candidates using geNomad v. 2.16 (Supplemental Table S7; Camargo et al. 2024).
CRISPR-Cas systems and classification of soil Asgard-associated viruses
CRISPR-Cas systems in Atabeyarchaeia and Freyarchaeia genomes were identified using CRISPRCasTyper v1.8.0 (Russel et al. 2020). Spacers were extracted from reads by mapping reads to the corresponding CRISPR arrays via BBMap (https://sourceforge.net/projects/bbmap/). Recruited spacers were matched against all assembled scaffolds with one or fewer mismatch using Bowtie v1.3.1 (Langmead et al. 2009). Scaffolds that are targeted by CRISPR spacers and not affiliated with microbial genomes were curated manually to completion. The phylogenetic classification was predicted based on genome-wide similarities using ViPTree whole-proteome-based similarity of MGE from Atabeyarchaeia and other Asgardarchaeota (Supplemental Table S8).
Comparative analysis of archaeal MGEs sizes
To contextualize the sizes of the identified MGEs in Atabeyarchaeia, we compiled a comprehensive data set of archaeal MGEs from various sources. We downloaded all complete reference plasmids from the NCBI Plasmid Browser (https://www.ncbi.nlm.nih.gov/genome/browse#!/plasmids/) and incorporated MGEs previously published (Al-Shayeb et al. 2022; Medvedeva et al. 2022; Rambo et al. 2022; Wu et al. 2022). The data set included various types of MGEs such as plasmids, viruses, iMGEs, mini-Borgs, Aloposons, and unclassified MGEs from diverse archaeal hosts including ANME-1, Asgard archaea, DPANN, Euryarchaeota, and Thermoproteota. We filtered the data set to include only MGEs with total genome sizes ≤ 100,000 bp to focus on elements comparable in size to those found in Atabeyarchaeia. The resulting data were visualized using a scatter plot, with MGE types represented by different shapes and host lineages distinguished by colors, allowing for a comprehensive comparison of MGE sizes across archaeal taxa (Supplemental Fig. S1).
Methylation analysis via REBASE and single-molecule, real-time
Methylation patterns within the genomes of Atabeyarchaeia and Freyarchaeia were investigated by mapping PacBio circular-consensus reads metagenomic reads to each of the three curated circular reference genomes for Atabeyarchaeia-1, Atabeyarchaeia-2, and Freyarchaeia using minimap2. The resulting BAM files were then processed and analyzed to identify methylation patterns using the ipdSummary and motifMaker commands in the single-molecule, real-time (SMRT) link analysis software package (v11.0, PacBio). To annotate MTase activities and restriction enzyme sites, the sequenced genomes and identified methylated motifs were compared against REBASE (Roberts et al. 2015; Blow et al. 2016). This comparison enabled the annotation of methylation sites and the determination of specific motifs associated with MTase activity and restriction-modification systems within these archaeal genomes.
Methylation motif validation with nanopore sequencing
Reads were base-called by dorado using the 4 kHz v.4.0.1 5 mC and 6 mA Rerio models (https://github.com/nanoporetech/rerio). Reads were aligned to the genomes with minimap2 (Li 2018). To exclude partial mappings and reads from other organisms, alignments with <80% alignment or >5% SNPs were excluded. Modifications were aggregated with “modkit pileup,” and motifs were called with “modkit find-motifs” (https://github.com/nanoporetech/modkit). Motifs were refined by inspecting the methylation distribution of related or off-by-one motifs to see if methylation was increased or decreased by more specific or more generic motifs.
Identification and network analysis of archaeal phage integrases
Phage integrase sequences were extracted from a custom database of NCBI archaeal genomes and archaeal-associated MGEs using HMMER (v3.3.2; Finn et al. 2011). The Pfam-established E-value cutoff for the Phage_integrase family (PF00589) was used as the threshold for sequence identification. The resulting integrase sequences were then analyzed using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST; Gerlt et al. 2015) to generate a sequence similarity network. The network was visualized and further analyzed using Cytoscape (v3.9.1; Shannon et al. 2003).
Phylogenetic analyses of proteins present in MGEs
Hallmark and poorly characterized proteins
For the terminase, PCNA, viral capsid, CAAX, and Type IIG RM fusion protein, we compiled the top 25 to 50 best matches for candidate proteins from the NCBI database, ggKbase, and UniProt (E-value cutoff of 1 × 10−5, 70% query coverage). The sequences were aligned using MAFFT (v7.310) (Katoh and Standley 2013) with the parameters ‐‐localpair ‐‐maxiterate 1000 ‐‐reorder. Following alignment, each was trimmed using TrimAl, applying a gap threshold of 0.7. The final alignments underwent manual inspection using Geneious (v11.0.20.1 + 1; https://www.geneious.com; see above). Maximum likelihood trees were inferred using IQ-TREE v1.6.12 (v1.4), (Nguyen et al. 2015), employing the auto option for model selection and a bootstrap value of 1000 and identifying the best-fit model for constructing the final trees. The details of all models used are included in the description of each figure. Trees were visualized using iTOL (Letunic and Bork 2021). All hallmark MGEs protein alignments and trees have been provided in the Supplemental Material for further reference.
GTPases
A curated set of proteins for the eukaryotic Arf family described by Vargová et al. (2021) was used to search in both soil-associated Atabeyarchaeia and Freyarchaeia genomes and MGEs. A nonredundant subset of the references and Asgardarchaeota hits with >25% protein identity were aligned and trimmed with MAFFT auto (v7.310) (Katoh and Standley 2013) and TrimAl -gt 0.5 (v1.4.rev15) (Capella-Gutiérrez et al. 2009). An initial phylogeny was produced with IQ-TREE (v.1.6.1) (Nguyen et al. 2015), and the LG + R9 model was chosen according to BIC.
By blasting the MGE GTPases against the NCBI database (June 19, 2024), we added sequences with percentage identity ≥35% with >71% query coverage. Our expanded reference set includes 10 Asgardarchaeota (one Baldrarchaeia [previously misidentified as Odindarchaeia], six Lokiarchaeia, and three Heimdallarchaeia) and two grassland soil-associated Deferrimicrobium sp. sequences. We also added one eukaryotic sequence from Hericium alpestre collected from forest deadwood that had high query coverage (70%–71%) and percentage identity (33.33%–34.92%) to both putative MGE GTPases. Sequences were aligned with MAFFT (v7.490) (Katoh and Standley 2013), trimmed with TrimAl -gt 0.5 (v1.4.rev22) (Capella-Gutiérrez et al. 2009), and used to create a maximum likelihood tree with IQ-TREE (v1.6.12) (Nguyen et al. 2015). LG + R7 was the best-fit model chosen according to BIC (Supplemental Fig. S10). The phylogeny was visualized with iTOL (Letunic and Bork 2021).
Hallmark viral proteins structural analysis
Protein structures for Opia virus hallmark proteins (capsid, terminase large subunit, and PCNA) were predicted using AlphaFold 3 (Abramson et al. 2024). The resulting models were compared to known structures using Foldseek (van Kempen et al. 2024) to identify structural homologs. Reference structures from the Protein Data Bank (PDB; https://www.rcsb.org) were used for comparison: bacteriophage capsid (PDB: 3BJQ), thermophilic bacteriophage D6E large terminase (PDB: 5OEB), and P. abyssi PCNA (PDB: 6T8H). Structural alignments and visualizations were performed using UCSF Chimera X v1.8 (Meng et al. 2023). Confidence scores from AlphaFold 3 predictions were mapped onto the structures using a blue-to-red color gradient, representing high to low confidence, respectively.
Structural analyses and structural phylogeny of the ESP
The protein sequences of small GTPases found were analyzed along with eukaryotic small GTPases identified as sequence homologues (Supplemental Figs. S10–S12). These sequences were submitted for structural modeling using ColabFold v1.5.240 (Mirdita et al. 2022). Multisequence alignments were performed using the MMseqs2 (Steinegger and Söding 2017) mode and the AlphaFold2_ptm models. Two recycling steps were employed to improve model prediction. The structural models were used as queries to search for structural homologues in the RCSB Protein Data Bank using FoldSeek easy-search feature, with a cutoff of >15% identity and <1 × 10−5 E-value.
Protein structures identified by FoldSeek were integrated with the models generated in ColabFold (V1.5.240; Mirdita et al. 2022). A multistructural alignment (MSTA) of these structures was carried out using the default parameters of mTM-align (Supplemental Fig. S12; Dong et al. 2018). The resulting MSTA was further analyzed using IQ-TREE v2.0.3 (model LG + I + G4 chosen according to BIC), yielding the dendrogram in Supplemental Figure S11A. The pairwise matrix obtained from the mTM-align process (Supplemental Table S12) was utilized to select proteins suitable for 3D reconstruction of their alignments (Supplemental Fig. S11B). This was done using the Needleman–Wunsch algorithm and the BLOSUM-62 matrix within the ChimeraX software (Meng et al. 2023)
Data access
The metagenomic data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA1050611. The Asgard archaea genomes can be accessed via ggKbase (https://ggkbase.berkeley.edu/SRVP_asgard/organisms). The complete viral genome sequences generated in this study have been submitted to the NCBI BioSample database (https://www.ncbi.nlm.nih.gov/biosample/) under accession numbers SAMN43308743, SAMN43308744, and SAMN43308745. The methylation data and candidate MTases identified in this study are available via REBASE (http://rebase.neb.com/rebase/private/pacbio_Banfield10.html). All the read mapping files, protein sequence alignments, and phylogenetic tree and modeled structures are available via Zenodo (https://zenodo.org/records/12617226).
Supplemental Material
Acknowledgments
This research was funded by the Bill & Melinda Gates Foundation (grant number INV-037174 to J.F.B.), a University of California Dissertation-Year Fellowship (to L.E.V.A.), a Stengl-Wyer Graduate Fellowship and University of Texas Graduate Continuing Fellowship (to K.E.A.), and the Innovative Genomics Institute at UC Berkeley. This work was also supported by the Moore-Simons Project on the Origin of the Eukaryotic Cell, Simons Foundation 73592LPI (https://doi.org/10.46714/735925LPI to B.J.B.) and Simons Foundation early career award 687165 (to B.J.B.). The findings and conclusions contained within are those of the authors and do not necessarily reflect positions or policies of the Bill & Melinda Gates Foundation. D.F.S. is an Investigator of the Howard Hughes Medical Institute, and the work was supported by the U. S. Department of Energy under Award number DE-SC0016240 to D.F.S. We thank Dr. Basem Al-Shayeb and Dr. Marie C. Schoelmerich for their contribution to the field work and generation of sequence data sets; Dr. Jacob West-Roberts, Shufei Lei, and Jordan Hoff for bioinformatics support; Dr. Mart Krupovic and Dr. Gavin Knott for helpful discussion; and Dr. Lin LinXing Chen for critical reading over the manuscript.
Author contributions: L.E.V.A. and J.F.B. designed the study. L.E.V.A., J.F.B., A.C.C., and L.-D.S. collected samples and extracted nucleic acids. L.E.V.A., R.S., A.C.C., and J.F.B. generated metagenomic data. L.E.V.A. and J.F.B. performed genome binning, and J.F.B., L.E.V.A., and L.-D.S. conducted genome curation. L.E.V.A. and J.F.B. analyzed the MGE genomes. L.E.V.A., K.E.A., and P.L. analyzed the functionally annotated MGEs. A.C.C. and L.E.V.A. performed correlation analyses. L.E.V.A. conducted phylogenetic analyses of hallmark MGE proteins. L.E.V.A., A.C.C., and J.F.B. analyzed MGE integration and excision events. L.E.V.A. and L.-D.S. analyzed whole viral proteomes. B.A.A. provided guidance on transposase and MGE classification. K.E.A., P.L., V.D.A., and B.J.B. analyzed eukaryotic signature proteins. A.C.C., L.E.V.A., and R.J.R. performed methylation analyses. R.S. contributed to bioinformatics analyses. B.J.B. and D.F.S. provided feedback on study design, methodology, and financial support. L.E.V.A. and J.F.B. wrote the manuscript, with contributions from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279480.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
J.F.B. is a cofounder of Metagenomi. D.F.S. is a cofounder and scientific advisory board member of Scribe Therapeutics. R.J.R. works for New England Biolabs, a company that sells research reagents including restriction enzymes and DNA MTases to the scientific community. L.L. is an employee of Oxford Nanopore Technologies and is a stock or stock option holder of Oxford Nanopore Technologies. The other authors declare that they have no competing interests.
References
- Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, et al. 2024. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630: 493–500. 10.1038/s41586-024-07487-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shayeb B, Schoelmerich MC, West-Roberts J, Valentin-Alvarado LE, Sachdeva R, Mullen S, Crits-Christoph A, Wilkins MJ, Williams KH, Doudna JA, et al. 2022. Borgs are giant genetic elements with potential to expand metabolic capacity. Nature 610: 731–736. 10.1038/s41586-022-05256-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton BP, Roberts RJ. 2021. Beyond restriction modification: epigenomic roles of DNA methylation in prokaryotes. Annu Rev Microbiol 75: 129–149. 10.1146/annurev-micro-040521-035040 [DOI] [PubMed] [Google Scholar]
- Bao Y, Federhen S, Leipe D, Pham V, Resenchuk S, Rozanov M, Tatusov R, Tatusova T. 2004. National center for biotechnology information viral genomes project. J Virol 78: 7291–7298. 10.1128/JVI.78.14.7291-7298.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Clark TA, Daum CG, Deutschbauer AM, Fomenkov A, Fries R, Froula J, Kang DD, Malmstrom RR, Morgan RD, et al. 2016. The epigenomic landscape of prokaryotes. PLoS Genet 12: e1005854. 10.1371/journal.pgen.1005854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo JPK, Aparicio-Maldonado C, Nobrega FL, Brouns SJJ, Taylor DW. 2022. Structural basis for broad anti-phage immunity by DISARM. Nat Commun 13: 2987. 10.1038/s41467-022-30673-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA. [Google Scholar]
- Camargo AP, Nayfach S, Chen I-MA, Palaniappan K, Ratner A, Chu K, Ritter SJ, Reddy TBK, Mukherjee S, Schulz F, et al. 2023. IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res 51: D733–D743. 10.1093/nar/gkac1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo AP, Roux S, Schulz F, Babinski M, Xu Y, Hu B, Chain PSG, Nayfach S, Kyrpides NC. 2024. Identification of mobile genetic elements with geNomad. Nat Biotechnol 42: 1303–1312. 10.1038/s41587-023-01953-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu VA, Salamon P, Seguritan V, Redfield J, Salamon D, Edwards RA, Segall AM. 2020. PhANNs, a fast and accurate tool and web server to classify phage structural proteins. PLoS Comput Biol 16: e1007845. 10.1371/journal.pcbi.1007845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-X, Anantharaman K, Shaiber A, Eren AM, Banfield JF. 2020. Accurate and complete genomes from metagenomes. Genome Res 30: 315–333. 10.1101/gr.258640.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N, Williams TA, Sun J, Woodcroft BJ, Lee J-H, Minh BQ, Rinke C, Spang A. 2020. Undinarchaeota illuminate DPANN phylogeny and the impact of gene transfer on archaeal evolution. Nat Commun 11: 3939. 10.1038/s41467-020-17408-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Peng Z, Zhang Y, Yang J. 2018. mTM-align: an algorithm for fast and accurate multiple protein structure alignment. Bioinformatics 34: 1719–1725. 10.1093/bioinformatics/btx828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359: eaar4120. 10.1126/science.aar4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drula E, Garron M-L, Dogan S, Lombard V, Henrissat B, Terrapon N. 2022. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res 50: D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L, Tamarit D, Caceres EF, Stairs CW, De Anda V, Schön ME, Seitz KW, Dombrowski N, Lewis WH, Homa F, et al. 2023. Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes. Nature 618: 992–999. 10.1038/s41586-023-06186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag Ibrahim F, Zhao R, Biddle Jennifer F. 2021. “Sifarchaeota,” a novel Asgard phylum from Costa Rican sediment capable of polysaccharide degradation and anaerobic methylotrophy. Appl Environ Microbiol 87: e02584–20. 10.1128/AEM.02584-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Cheng H, Portik D, Li H. 2022. Metagenome assembly of high-fidelity long reads with hifiasm-meta. Nat Methods 19: 671–674. 10.1038/s41592-022-01478-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. 2021. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res 49: D274–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, Whalen KL. 2015. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim Biophys Acta 1854: 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaly TM, Tetu SG, Penesyan A, Qi Q, Rajabal V, Gillings MR. 2022. Discovery of integrons in Archaea: platforms for cross-domain gene transfer. Sci Adv 8: eabq6376. 10.1126/sciadv.abq6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Solá M, Acebo P, Párraga A, Guasch A, Eritja R, González A, Espinosa M, del Solar G, Coll M. 1998. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J 17: 7404–7415. 10.1093/emboj/17.24.7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazziotin AL, Koonin EV, Kristensen DM. 2017. Prokaryotic Virus Orthologous Groups (pVOGs): a resource for comparative genomics and protein family annotation. Nucleic Acids Res 45: D491–D498. 10.1093/nar/gkw975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Huang L. 2010. A superfamily 3 DNA helicase encoded by plasmid pSSVi from the hyperthermophilic archaeon Sulfolobus solfataricus unwinds DNA as a higher-order oligomer and interacts with host primase. J Bacteriol 192: 1853–1864. 10.1128/JB.01300-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41: D387–D395. 10.1093/nar/gks1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, Williams KH, Tringe SG, Banfield JF. 2013. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1: 22. 10.1186/2049-2618-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, et al. 2020. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577: 519–525. 10.1038/s41586-019-1916-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Makarova KS, Wolf YI, Medvedeva S, Prangishvili D, Forterre P, Koonin EV. 2019. Integrated mobile genetic elements in Thaumarchaeota. Environ Microbiol 21: 2056–2078. 10.1111/1462-2920.14564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leão P, Little ME, Appler KE, Sahaya D, Aguilar-Pine E, Currie K, Finkelstein IJ, De Anda V, Baker BJ. 2024. Asgard archaea defense systems and their roles in the origin of eukaryotic immunity. Nat Commun 15: 6386. 10.1038/s41467-024-50195-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49: W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedeva S, Sun J, Yutin N, Koonin EV, Nunoura T, Rinke C, Krupovic M. 2022. Three families of Asgard archaeal viruses identified in metagenome-assembled genomes. Nat Microbiol 7: 962–973. 10.1038/s41564-022-01144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng EC, Goddard TD, Pettersen EF, Couch GS, Pearson ZJ, Morris JH, Ferrin TE. 2023. UCSF ChimeraX: tools for structure building and analysis. Protein Sci 32: e4792. 10.1002/pro.4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2022. ColabFold: making protein folding accessible to all. Nat Methods 19: 679–682. 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Bateman A, Finn RD. 2007. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics 8: 298. 10.1186/1471-2105-8-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno CM, Prajapati B, Lucas-Staat S, Sime-Ngando T, Forterre P, Bamford DH, Prangishvili D, Krupovic M, Oksanen HM. 2019. Novel haloarchaeal viruses from Lake Retba infecting Haloferax and Halorubrum species. Environ Microbiol 21: 2129–2147. 10.1111/1462-2920.14604 [DOI] [PubMed] [Google Scholar]
- Moreno-Cinos C, Goossens K, Salado IG, Van Der Veken P, De Winter H, Augustyns K. 2019. ClpP protease, a promising antimicrobial target. Int J Mol Sci 20: 2232. 10.3390/ijms20092232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RD, Dwinell EA, Bhatia TK, Lang EM, Luyten YA. 2009. The MmeI family: type II restriction-modification enzymes that employ single-strand modification for host protection. Nucleic Acids Res 37: 5208–5221. 10.1093/nar/gkp534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir G, Melamed S, Sberro H, Mukamel Z, Silverman S, Yaakov G, Doron S, Sorek R. 2018. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol 3: 90–98. 10.1038/s41564-017-0051-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladin L, Raj S, Richardson LJ, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49: D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne LJ, Meaden S, Mestre MR, Palmer C, Toro N, Fineran PC, Jackson SA. 2022. PADLOC: a web server for the identification of antiviral defence systems in microbial genomes. Nucleic Acids Res 50: W541–W550. 10.1093/nar/gkac400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo IM, Langwig MV, Leão P, De Anda V, Baker BJ. 2022. Genomes of six viruses that infect Asgard archaea from deep-sea sediments. Nat Microbiol 7: 953–961. 10.1038/s41564-022-01150-8 [DOI] [PubMed] [Google Scholar]
- Raymann K, Forterre P, Brochier-Armanet C, Gribaldo S. 2014. Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in archaea. Genome Biol Evol 6: 192–212. 10.1093/gbe/evu004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. 2015. REBASE: a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43: D298–D299. 10.1093/nar/gku1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, Bikard D. 2022. Microbial defenses against mobile genetic elements and viruses: Who defends whom from what? PLoS Biol 20: e3001514. 10.1371/journal.pbio.3001514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BE, Diamond S, Cress BF, Crits-Christoph A, Lou YC, Borges AL, Shivram H, He C, Xu M, Zhou Z, et al. 2022. Species- and site-specific genome editing in complex bacterial communities. Nat Microbiol 7: 34–47. 10.1038/s41564-021-01014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel J, Pinilla-Redondo R, Mayo-Muñoz D, Shah SA, Sørensen SJ. 2020. CRISPRCasTyper: automated identification, annotation, and classification of CRISPR-Cas loci. CRISPR J 3: 462–469. 10.1089/crispr.2020.0059 [DOI] [PubMed] [Google Scholar]
- Schoelmerich MC, Ly L, West-Roberts J, Shi L-D, Shen C, Malvankar NS, Taib N, Gribaldo S, Woodcroft BJ, Schadt CW, et al. 2024. Borg extrachromosomal elements of methane-oxidizing archaea have conserved and expressed genetic repertoires. Nat Commun 15: 5414. 10.1038/s41467-024-49548-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F, Roux S, Paez-Espino D, Jungbluth S, Walsh DA, Denef VJ, McMahon KD, Konstantinidis KT, Eloe-Fadrosh EA, Kyrpides NC, et al. 2020. Giant virus diversity and host interactions through global metagenomics. Nature 578: 432–436. 10.1038/s41586-020-1957-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz KW, Lazar CS, Hinrichs K-U, Teske AP, Baker BJ. 2016. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J 10: 1696–1705. 10.1038/ismej.2015.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz KW, Dombrowski N, Eme L, Spang A, Lombard J, Sieber JR, Teske AP, Ettema TJG, Baker BJ. 2019. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun 10: 1822. 10.1038/s41467-019-09364-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senčilo A, Roine E. 2014. A Glimpse of the genomic diversity of haloarchaeal tailed viruses. Front Microbiol 5: 84. 10.3389/fmicb.2014.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen BW, Heiter DF, Chan S-H, Wang H, Xu S-Y, Morgan RD, Wilson GG, Stoddard BL. 2010. Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI. Structure 18: 734–743. 10.1016/j.str.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34: D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søndergaard D, Pedersen CNS, Greening C. 2016. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep 6: 34212. 10.1038/srep34212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173–179. 10.1038/nature14447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth DR, Yu FB, Connon SA, Lim S, Magyar JS, Peña-Salinas ME, Quake SR, Orphan VJ. 2022. Microbial communities of Auka hydrothermal sediments shed light on vent biogeography and the evolutionary history of thermophily. ISME J 16: 1750–1764. 10.1038/s41396-022-01222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinegger M, Söding J. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35: 1026–1028. 10.1038/nbt.3988 [DOI] [PubMed] [Google Scholar]
- Sun J, Evans PN, Gagen EJ, Woodcroft BJ, Hedlund BP, Woyke T, Hugenholtz P, Rinke C. 2021. Recoding of stop codons expands the metabolic potential of two novel Asgardarchaeota lineages. ISME Commun 1: 30. 10.1038/s43705-021-00032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarit D, Caceres EF, Krupovic M, Nijland R, Eme L, Robinson NP, Ettema TJG. 2022. A closed Candidatus Odinarchaeum chromosome exposes Asgard archaeal viruses. Nat Microbiol 7: 948–952. 10.1038/s41564-022-01122-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarit D, Köstlbacher S, Appler KE, Panagiotou K, De Anda V, Rinke C, Baker BJ, Ettema TJG. 2024. Description of Asgardarchaeum abyssi gen. nov. spec. nov., a novel species within the class Asgardarchaeia and phylum Asgardarchaeota in accordance with the SeqCode. Syst Appl Microbiol 47: 126525. 10.1016/j.syapm.2024.126525 [DOI] [PubMed] [Google Scholar]
- Terzian P, Olo Ndela E, Galiez C, Lossouarn J, Pérez Bucio RE, Mom R, Toussaint A, Petit M-A, Enault F. 2021. PHROG: families of prokaryotic virus proteins clustered using remote homology. NAR Genom Bioinform 3: lqab067. 10.1093/nargab/lqab067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson F, Hervé A, Mordret E, Touchon M, d'Humières C, Cury J, Bernheim A. 2022. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat Commun 13: 2561. 10.1038/s41467-022-30269-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck OT, Adler BA, Armbruster EG, Lahiri A, Hu JJ, Zhou J, Pogliano J, Doudna JA. 2024. Hachiman is a genome integrity sensor. bioRxiv 10.1101/2024.02.29.582594 [DOI] [PubMed]
- Valentin-Alvarado LE, Appler KE, De Anda V, Schoelmerich MC, West-Roberts J, Kivenson V, Crits-Christoph A, Ly L, Sachdeva R, Greening C, et al. 2024. Asgard archaea modulate potential methanogenesis substrates in wetland soil. Nat Commun 15: 6384. 10.1038/s41467-024-49872-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kempen M, Kim SS, Tumescheit C, Mirdita M, Lee J, Gilchrist CLM, Söding J, Steinegger M. 2024. Fast and accurate protein structure search with Foldseek. Nat Biotechnol 42: 243–246. 10.1038/s41587-023-01773-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargová R, Wideman JG, Derelle R, Klimeš V, Kahn RA, Dacks JB, Eliáš M. 2021. A eukaryote-wide perspective on the diversity and evolution of the ARF GTPase protein family. Genome Biol Evol 13: evab157. 10.1093/gbe/evab157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein T, Sorek R. 2022. Bacterial origins of human cell-autonomous innate immune mechanisms. Nat Rev Immunol 22: 629–638. 10.1038/s41577-022-00705-4 [DOI] [PubMed] [Google Scholar]
- Wozniak RAF, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8: 552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- Wu F, Speth DR, Philosof A, Crémière A, Narayanan A, Barco RA, Connon SA, Amend JP, Antoshechkin IA, Orphan VJ. 2022. Unique mobile elements and scalable gene flow at the prokaryote-eukaryote boundary revealed by circularized Asgard archaea genomes. Nat Microbiol 7: 200–212. 10.1038/s41564-021-01039-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6: 223. 10.1186/1743-422X-6-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541: 353–358. 10.1038/nature21031 [DOI] [PubMed] [Google Scholar]
- Zhang J-W, Dong H-P, Hou L-J, Liu Y, Ou Y-F, Zheng Y-L, Han P, Liang X, Yin G-Y, Wu D-M, et al. 2021. Newly discovered Asgard archaea Hermodarchaeota potentially degrade alkanes and aromatics via alkyl/benzyl-succinate synthase and benzoyl-CoA pathway. ISME J 15: 1826–1843. 10.1038/s41396-020-00890-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.