Abstract
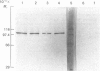
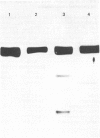
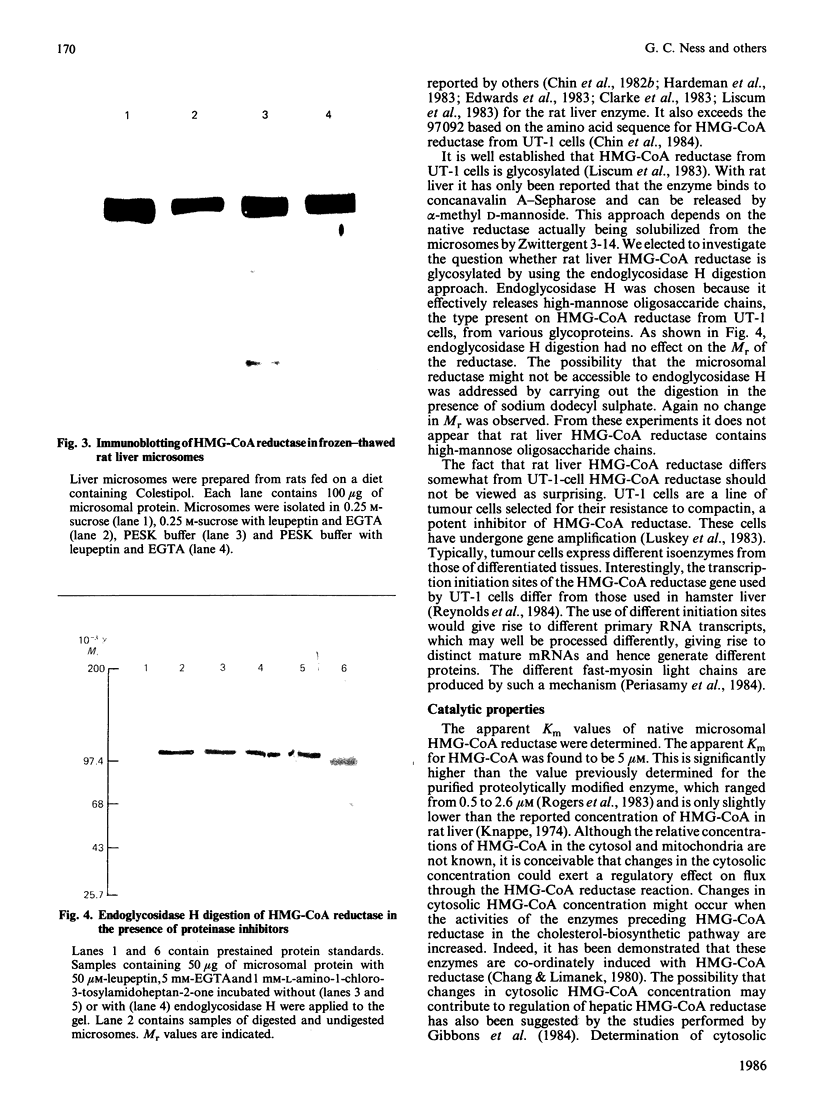
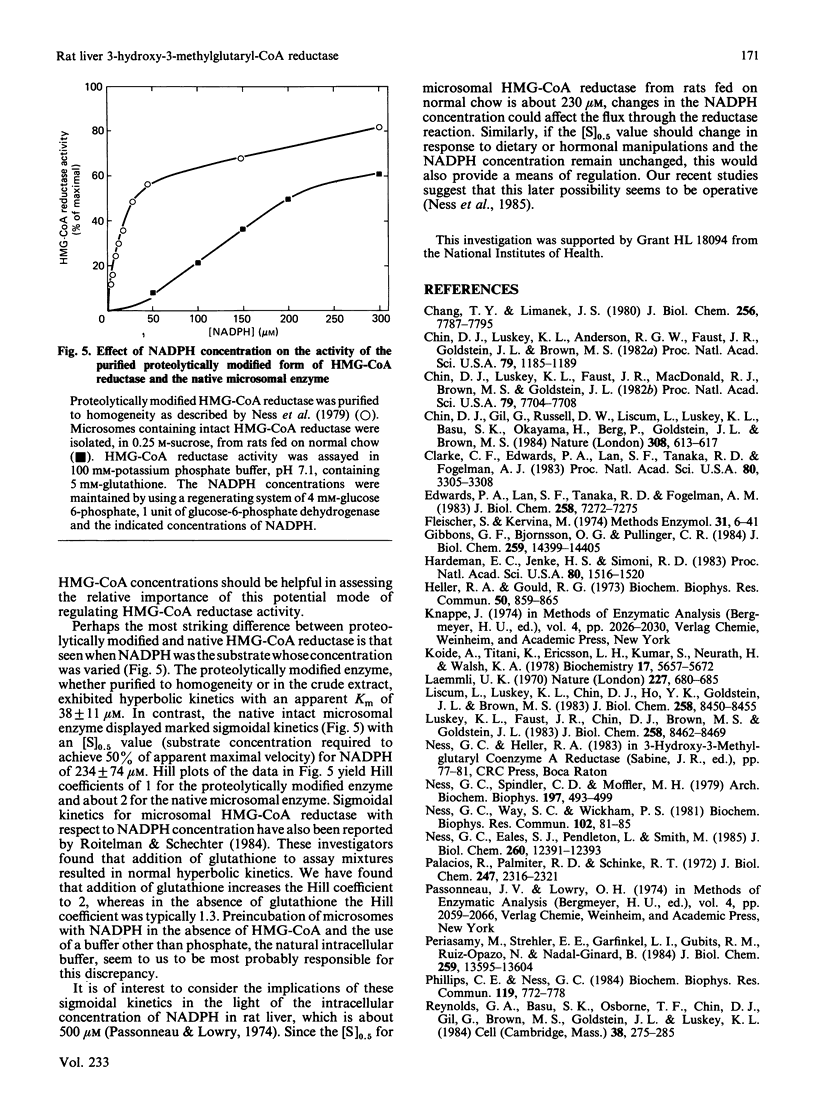
A procedure for the preparation of rat liver microsomal fractions essentially devoid of contaminating lysosomes is described. When this preparation was examined by immunoblotting with a rabbit antiserum to rat 3-hydroxy-3-methylglutaryl-CoA reductase, a single band corresponding to an Mr of 100000 was observed. No evidence was found for glycosylation of rat liver-3-hydroxy-3-methylglutaryl-CoA reductase. Native rat liver microsomal 3-hydroxy-3-methylglutaryl-CoA reductase differs from the purified proteolytically modified species in that it displays allosteric kinetics towards NADPH.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang T. Y., Limanek J. S. Regulation of cytosolic acetoacetyl coenzyme A thiolase, 3-hydroxy-3-methylglutaryl coenzyme A synthase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and mevalonate kinase by low density lipoprotein and by 25-hydroxycholesterol in Chinese hamster ovary cells. J Biol Chem. 1980 Aug 25;255(16):7787–7795. [PubMed] [Google Scholar]
- Chin D. J., Gil G., Russell D. W., Liscum L., Luskey K. L., Basu S. K., Okayama H., Berg P., Goldstein J. L., Brown M. S. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase, a glycoprotein of endoplasmic reticulum. Nature. 1984 Apr 12;308(5960):613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- Chin D. J., Luskey K. L., Anderson R. G., Faust J. R., Goldstein J. L., Brown M. S. Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold increase in 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1185–1189. doi: 10.1073/pnas.79.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D. J., Luskey K. L., Faust J. R., MacDonald R. J., Brown M. S., Goldstein J. L. Molecular cloning of 3-hydroxy-3-methylglutaryl coenzyme a reductase and evidence for regulation of its mRNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7704–7708. doi: 10.1073/pnas.79.24.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. F., Edwards P. A., Lan S. F., Tanaka R. D., Fogelman A. M. Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase mRNA levels in rat liver. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3305–3308. doi: 10.1073/pnas.80.11.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Tanaka R. D., Fogelman A. M. Mevalonolactone inhibits the rate of synthesis and enhances the rate of degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in rat hepatocytes. J Biol Chem. 1983 Jun 25;258(12):7272–7275. [PubMed] [Google Scholar]
- Fleischer S., Kervina M. Subcellular fractionation of rat liver. Methods Enzymol. 1974;31:6–41. doi: 10.1016/0076-6879(74)31005-1. [DOI] [PubMed] [Google Scholar]
- Gibbons G. F., Björnsson O. G., Pullinger C. R. Evidence that changes in hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase activity are required partly to maintain a constant rate of sterol synthesis. J Biol Chem. 1984 Dec 10;259(23):14399–14405. [PubMed] [Google Scholar]
- Hardeman E. C., Jenke H. S., Simoni R. D. Overproduction of a Mr 92,000 protomer of 3-hydroxy-3-methylglutaryl-coenzyme A reductase in compactin-resistant C100 cells. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1516–1520. doi: 10.1073/pnas.80.6.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R. A., Gould R. G. Solubilization and partial purification of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochem Biophys Res Commun. 1973 Feb 5;50(3):859–865. doi: 10.1016/0006-291x(73)91324-7. [DOI] [PubMed] [Google Scholar]
- Koide A., Titani K., Ericsson L. H., Kumar S., Neurath H., Walsh K. A. Sequence of the amino-terminal 349 residues of rabbit muscle glycogen phosphorylase including the sites of covalent and allosteric control. Biochemistry. 1978 Dec 26;17(26):5657–5672. doi: 10.1021/bi00619a012. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and its mRNA in rat liver as studied with a monoclonal antibody and a cDNA probe. J Biol Chem. 1983 Jul 10;258(13):8450–8455. [PubMed] [Google Scholar]
- Luskey K. L., Faust J. R., Chin D. J., Brown M. S., Goldstein J. L. Amplification of the gene for 3-hydroxy-3-methylglutaryl coenzyme A reductase, but not for the 53-kDa protein, in UT-1 cells. J Biol Chem. 1983 Jul 10;258(13):8462–8469. [PubMed] [Google Scholar]
- Ness G. C., Eales S. J., Pendleton L. C., Smith M. Activation of rat liver microsomal 3-hydroxy-3-methylglutaryl coenzyme A reductase by NADPH. Effects of dietary treatments. J Biol Chem. 1985 Oct 15;260(23):12391–12393. [PubMed] [Google Scholar]
- Ness G. C., Spindler C. D., Moffler M. H. Purification of 3-hydroxy-3-methylglutaryl coenzyme A reductase from rat liver. Arch Biochem Biophys. 1979 Oct 15;197(2):493–499. doi: 10.1016/0003-9861(79)90272-8. [DOI] [PubMed] [Google Scholar]
- Ness G. C., Way S. C., Wickham P. S. Proteinase involvement in the solubilization of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochem Biophys Res Commun. 1981 Sep 16;102(1):81–85. doi: 10.1016/0006-291x(81)91491-1. [DOI] [PubMed] [Google Scholar]
- Palacios R., Palmiter R. D., Schimke R. T. Identification and isolation of ovalbumin-synthesizing polysomes. I. Specific binding of 125 I-anti-ovalbumin to polysomes. J Biol Chem. 1972 Apr 25;247(8):2316–2321. [PubMed] [Google Scholar]
- Periasamy M., Strehler E. E., Garfinkel L. I., Gubits R. M., Ruiz-Opazo N., Nadal-Ginard B. Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J Biol Chem. 1984 Nov 10;259(21):13595–13604. [PubMed] [Google Scholar]
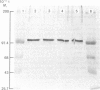
- Phillips C. E., Ness G. C. Topography of rat liver microsomal 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochem Biophys Res Commun. 1984 Mar 15;119(2):772–778. doi: 10.1016/s0006-291x(84)80317-4. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Roitelman J., Shechter I. Regulation of rat liver 3-hydroxy-3-methylglutaryl coenzyme A reductase. Evidence for thiol-dependent allosteric modulation of enzyme activity. J Biol Chem. 1984 Jan 25;259(2):870–877. [PubMed] [Google Scholar]
- Trouet A. Isolation of modified liver lysosomes. Methods Enzymol. 1974;31:323–329. doi: 10.1016/0076-6879(74)31034-8. [DOI] [PubMed] [Google Scholar]