Abstract
The transcription factor (TF) cone-rod homeobox (CRX) is essential for the differentiation and maintenance of photoreceptor cell identity. Several human CRX variants cause degenerative retinopathies, but most are variants of uncertain significance. We performed a deep mutational scan (DMS) of nearly all possible single amino acid substitutions in CRX using a cell-based transcriptional reporter assay, curating a high-confidence list of nearly 2000 variants with altered transcriptional activity. In the structured homeodomain, activity scores closely aligned to a predicted structure and demonstrated position-specific constraints on amino acid substitution. In contrast, the intrinsically disordered transcriptional effector domain displayed a qualitatively different pattern of substitution effects, following compositional constraints without specific residue position requirements in the peptide chain. These compositional constraints were consistent with the acidic exposure model of transcriptional activation. We evaluated the performance of the DMS assay as a clinical variant classification tool using gold-standard classified human variants from ClinVar, identifying pathogenic variants with high specificity and moderate sensitivity. That this performance could be achieved using a synthetic reporter assay in a foreign cell type, even for a highly cell type-specific TF like CRX, suggests that this approach shows promise for DMS of other TFs that function in cell types that are not easily accessible. Together, the results of the CRX DMS identify molecular features of the CRX effector domain and demonstrate utility for integration into the clinical variant classification pipeline.
Cone-rod homeobox, encoded by CRX, is a photoreceptor-specific transcription factor (TF) essential to both the terminal differentiation of photoreceptors and the maintenance of rod and cone structure and function in adulthood (Swaroop et al. 2010). Over a dozen human sequence variants in CRX have been characterized in the pathogenesis of degenerative retinopathies with a wide range of ages of onset and severity, including retinitis pigmentosa (RP), a “rod-centric” disease with loss of peripheral vision; cone-rod dystrophy (CoRD), a “cone-centric” disease with loss of central visual field acuity progressing to peripheral vision loss; and Leber congenital amaurosis (LCA), an early onset retinal disease causing severe vision loss and blindness (Freund et al. 1997, 1998; Jacobson et al. 1998; Sohocki et al. 1998; Swaroop et al. 1999; Perrault et al. 2003; Nichols et al. 2010; Huang et al. 2012). In ClinVar, the NCBI catalog of human gene variants, ∼150 missense CRX variants have been reported, but more than 80% of these are variants of uncertain significance (VUS), with no clinical or functional evidence of their pathogenicity or lack thereof. With the ever-increasing pace of clinical genome sequencing, CRX VUS will continue to be identified in patients with degenerative retinopathies, and diagnosis of these patients would benefit from better functional characterization of CRX variants.
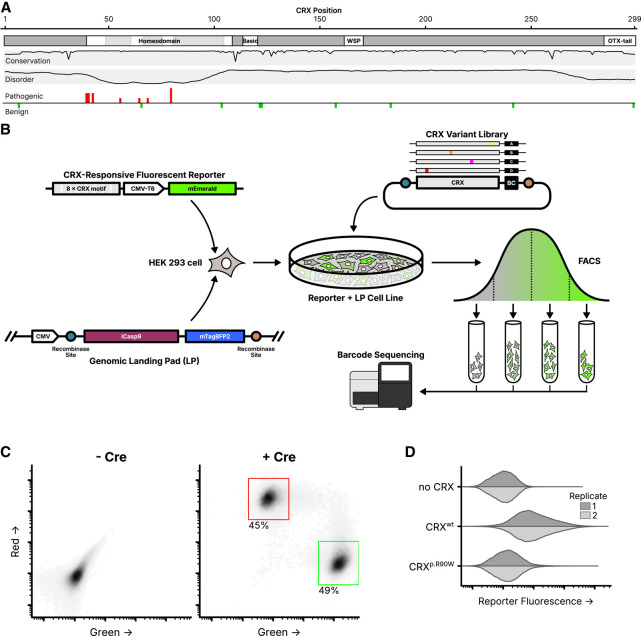
As a TF, CRX primarily functions by regulating the expression of target photoreceptor genes such as the rod and cone opsins (RHO, OPN1SW, and OPN1MW) (Peng and Chen 2005; Corbo et al. 2010). Pathogenic sequence variants in CRX cause profound alterations in the gene expression profiles of photoreceptors that correlate with structural and functional deficits (Tran and Chen 2014; Tran et al. 2014). The p.E80A variant has been observed in patients with severe dominant CoRD (Freund et al. 1997; Sohocki et al. 1998; Chen et al. 2002), while the nearby p.K88N variant has been observed to cause dominant LCA (Nichols et al. 2010). The p.R115Q variant has been reported in patients with RP (Sohocki et al. 2001), while the p.R90W variant has been shown to cause LCA when homozygous, but only mild late-onset CoRD when heterozygous (Swaroop et al. 1999; Chen et al. 2002). Pathogenic missense variants have been identified in both the DNA-binding domain and transcriptional effector domain (Fig. 1A).
Figure 1.
Experimental overview of the CRX deep mutational scan. (A) Known CRX domains, sequence conservation, predicted disorder, and reported ClinVar missense variants. Per-residue conservation was computed using sequences from the UniProt UniRef50 cluster derived from human CRX. Disorder predicted with Metapredict (Emenecker et al. 2021). Missense pathogenic variants (“Pathogenic” and/or “Likely pathogenic”) and benign variants (“Benign” and/or “Likely Benign”) from ClinVar (accessed June 2024); height of bar proportional to number of variants at each position (max = 3). (B) Schematic of the CRX deep mutational scan. A clonal cell line carrying a CRX-responsive fluorescent reporter and genomic landing pad (LP) was generated, and a library of CRX variants was integrated into the LP so that each cell expresses a single variant. Following fluorescence-activated cell sorting (FACS), sequencing was used to determine relative variant barcode abundances in each fluorescence bin, allowing for the calculation of a reporter activity score. (C) LP cells integrated with a 1:1 ratio of plasmids carrying mEmerald (green; arbitrary units) or mCherry2 (red; arbitrary units), with or without a plasmid expressing Cre recombinase (60,000 cells plotted per condition, points shaded by density, percent of cells falling within the indicated gates shown). (D) Reporter activation (green fluorescence; arbitrary units) was measured in Reporter + LP cells with the indicated CRX variants integrated. Two independent biological replicate experiments per sample (distributions plotted from 40,000 cells).
We applied deep mutational scanning, which uses libraries of gene variants combined with pooled high-throughput assays, to measure the functional consequences of all missense CRX variants in a single experiment. Fundamentally, a deep mutational scan (DMS) is the combination of a DNA library of many sequence variants, a method for introducing those variants into a model system in a manner such that the effects of each variant can be individually measured, and a functional assay capable of quantifying the activity of each variant. We developed a cell line carrying a synthetic fluorescent CRX transcriptional reporter and a genomic landing pad (LP) system for controlled single-copy expression of variants. We integrated a plasmid library encoding all single amino acid substitution CRX variants into this cell line so that each individual cell carried a single CRX variant expressed from the LP, sorted cells into bins based on reporter fluorescence, and sequenced variant enrichment across bins to calculate a quantitative activity score for each variant.
We analyzed the DMS variant activity scores in the context of the CRX DNA-binding domain structure, the residue composition of the intrinsically disordered transcriptional effector domain, and for the purposes of clinical variant classification. This work provides both a clinical resource for the classification of CRX variant effects as well as insight into the rules governing the composition and structure of TFs.
Results
To systematically measure the effects of all variants in CRX, we developed a clonal HEK 293-derivative cell line carrying two genomic integrations: a synthetic CRX transcriptional activity reporter, and the aforementioned LP system. The transcriptional reporter contains eight consecutive repeats of a strong consensus CRX-binding motif upstream of a CMV-T6 minimal promoter and the fluorescent protein mEmerald (Loew et al. 2010). The LP contains a strong constitutive cytomegalovirus (CMV) promoter with asymmetric lox sites downstream flanking drug and fluorescent selection markers (Fig. 1B). Upon the addition of a plasmid with matching lox sites and Cre recombinase, the LP can undergo recombinase-mediated cassette exchange (RMCE), allowing for the precise insertion of a CRX variant at the defined genomic LP locus, under the control of the CMV promoter. HEK 293 cells were used due to their experimental tractability, and a history of prior successful reporter assays for CRX (Chen et al. 2002; Peng et al. 2005; Tran et al. 2014). The endogenous CRX locus is repressed and unexpressed in HEK 293 cells.
The use of a genomic LP achieves precise expression of the integrated gene cassette, allowing for the expression of a single CRX variant in each cell. To validate the LP cell line, we cotransfected a 1:1 mixture of two plasmids, carrying either the mEmerald or mCherry2 fluorescent proteins with flanking compatible lox sites. Without the addition of a plasmid expressing Cre recombinase, the fluorescent proteins were not expressed, showing that RMCE is required for protein production (as the plasmids themselves lack a promoter). With the addition of Cre recombinase, cells could be observed with tightly controlled green or red fluorescence but not the expression of both colors, demonstrating that constructs could be successfully expressed upon RMCE and that the cell line only harbored a single LP (Fig. 1C).
To demonstrate the sensitivity of the synthetic CRX transcriptional reporter, we separately integrated three different constructs into the genomic LP: a transcriptionally inactive control construct (the miRFP670 fluorescent protein), wild-type CRX, or the known hypomorphic p.R90W CRX variant (Fig. 1D). HEK 293 cells do not natively express their endogenous copy of CRX, minimizing background protein expression that could interfere with assay measurements (Supplemental Fig. S1). As expected, wild-type CRX alone was capable of activating the reporter construct and driving expression of the fluorescent reporter protein, while the p.R90W variant protein failed to activate the reporter above background fluorescence as compared to the inactive control.
A cell-based CRX reporter system quantitatively measures variant activity
We cloned a library of all possible single amino acid substitution CRX variants, and associated each variant with a unique random barcode sequence using Pacific Biosciences (PacBio) long-read sequencing (Supplemental Fig. S2A). This library was integrated into the clonal cell line carrying the CRX reporter and genomic LP by RMCE. The fluorescence of each cell is proportional to the level of reporter activation by the CRX variant expressed in that cell. We sorted the cells into four bins based on fluorescence, extracted genomic DNA, and sequenced variant barcodes. The relative abundance of each barcode in each bin, and thus the relative abundance of each variant in each bin, were used to calculate per-variant activity scores (Fig. 1B; Supplemental Fig. S2B–D). After filtering and quality control for low representation or read abundance, we were able to calculate activity scores for 5285 variants out of 5662 designed variants.
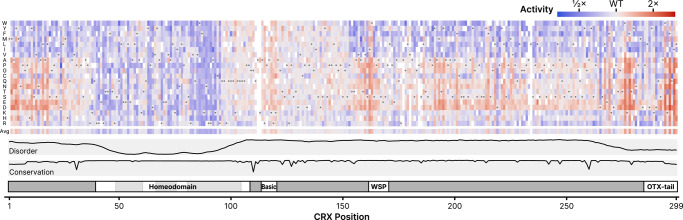
The computed variant activity scores for each variant show patterns of sensitivity to substitutions that correspond with different structural domains of CRX (Fig. 2). Residues in the homeodomain are particularly sensitive to substitution, as seen in the average variant effect track. In the central region of CRX (residues ∼95–150), apart from the basic domain, residues are largely insensitive to substitution. In the remainder of the disordered regions (including at the N-terminus), substitutions for specific classes of amino acids (such as negatively charged residues or aromatics) show specific increases or decreases in activity. In the C-terminus, substitutions of a number of residues tend to substantially affect CRX transcriptional activity, with substitutions of specific residues systematically increasing or decreasing activity. Below, we discuss each of these trends in more detail.
Figure 2.
DMS activity scores for all measured single amino acid CRX substitutions. Activity scores were normalized to wild-type CRX; the wild-type amino acid at each position is indicated by the gray circle in each column. The average row shows the mean activity score for all substitutions at each position. Disorder, domains, and conservation are shown as in Figure 1. Empty boxes indicate variants not measured in the DMS assay, due to drop-out during the library cloning or variant measurement steps. An interactive version of this figure is available in Supplemental Interactive S1.
Activity scores can be used to classify known pathogenic CRX variants with high specificity
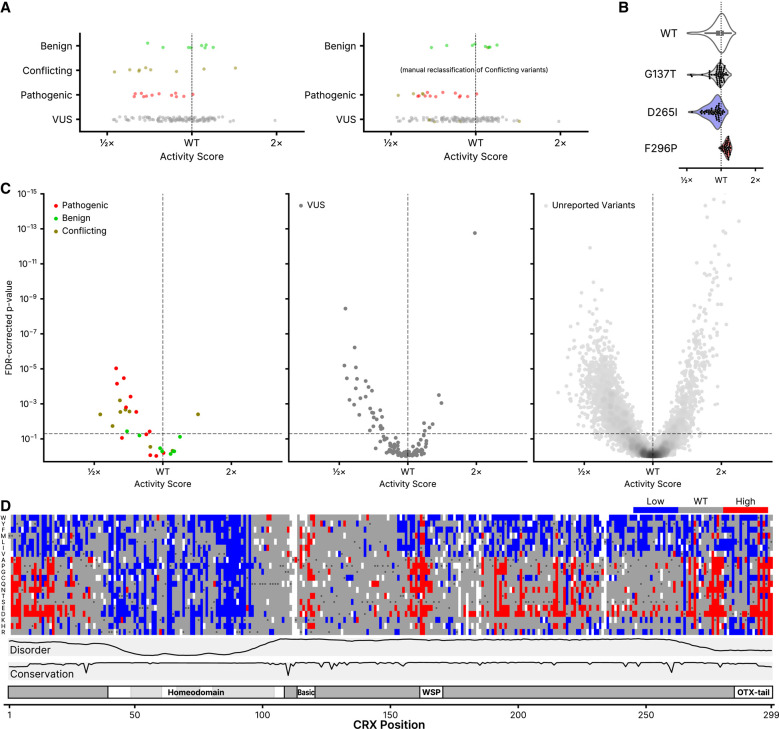
Only 21 missense CRX variants have been unambiguously classified in ClinVar (Fig. 1A), but we can use the activity scores for these variants to benchmark the classification performance of the DMS assay. Although the variant classifiers in clinical use typically aggregate multiple evidence sources (including evolutionary conservation, modeling, and functional data), looking at the classification performance of this single functional assay can give insight into its performance and utility for incorporation as a clinical evidence source. The DMS activity scores for ClinVar variants are shown in Figure 3A. A number of the CRX variants in ClinVar have a “conflicting” classification, meaning that submitting clinical laboratories disagree on the pathogenicity of the variant—in Figure 3A, we also plot scores with a manual reclassification of these variants, taking the modal class for each variant. Regardless of reclassification, although the scores for the benign and pathogenic variants show some clustering, not all variants can be clearly separated by activity score alone. To assign a statistical confidence to each variant, we took advantage of the design of the DMS assay—specifically, that each variant is marked by multiple sequence barcodes. The distributions of barcode-level activity scores for several selected variants are shown in Figure 3B. We leveraged the fact that in the CRX variant library, wild-type CRX is barcoded thousands of times. We used a two-sample Kolmogorov–Smirnov (K–S) test to compare the shapes of each variant's barcode-level activity distribution to that of wild-type CRX. Combining the DMS activity scores with the FDR-corrected K–S P-value, we were able to set cutoffs for variants with significantly low or high activity (Fig. 3C).
Figure 3.
Classification of CRX variants. (A) DMS activity scores for variants reported to ClinVar in each of the indicated classes (Pathogenic includes “Pathogenic” and/or “Likely pathogenic”; Benign includes “Benign” and/or “Likely Benign”). On the right, “Conflicting” variants are shown reclassified based on the modal reported ClinVar classification. (B) Barcode-level activity measurements for wild-type CRX and the indicated representative wild-type-like (p.G137T), low-activity (p.D265I), and high-activity (p.F296P) variants. Each black dot represents a unique barcoded construct; not shown for wild-type CRX due to it being barcoded thousands of times. (C) Volcano plots showing classifications for the indicated ClinVar variants (left and middle panels) or all other variants not yet reported in ClinVar (right panel). FDR-corrected P-values were computed from a two-sample K–S test comparing each variant's barcode-level measurements to those of wild-type CRX, as visualized in (B). The horizontal line corresponds to an FDR-corrected P-value of 0.05; the vertical line corresponds to a normalized activity score of 1 (wild-type). For visualization purposes, the y-axis is clipped to 10−15; 13 variants are hidden with activity scores greater than wild-type and P-values up to 10−26. (D) Quantized DMS activity scores for each variant, coloring low- and high-activity variants using the significance cutoffs shown as dotted lines in (C). Disorder, domains, and conservation are shown as in Figure 1.
Using these criteria, we found that the DMS assay is a high-specificity pathogenicity classifier (i.e., the type I error rate is low): seven of eight benign variants have DMS activity scores that are not significantly different from wild-type CRX, while for the 12 pathogenic variants, 7 show significantly reduced activity scores. If we include data from the manually reclassified “conflicting” ClinVar variants, we can revise this performance to 8/9 benign variants and 11/16 pathogenic variants for which the DMS activity measurements alone are concordant with ClinVar. Using the K–S test significance criteria, we generated a list of high-suspicion activity-altering variants (Fig. 3D). When combined with other clinical data sources, this list of variants identifies substitutions with the highest predicted impact on CRX activity, and thus greatest likelihood for pathogenicity. Using the OddsPath framework (Tavtigian et al. 2018) for benchmarking functional assay data for variant classification, this corresponds to an OddsPath score of 4.67 (6.19 if including the manually curated classifications of conflicting variants), meeting the criterion for the PS3_Moderate evidence level in the ACMG variant classification framework (Brnich et al. 2020). Because of the small number of reported CRX variants in ClinVar, PS3_Moderate is the maximum achievable evidence level even for an assay with perfect variant discrimination.
Population-level allele frequencies are available for some CRX variants in databases such as gnomAD (https://gnomad.broadinstitute.org) and All of Us (https://databrowser.researchallofus.org). The pleiotropic clinical phenotypes of CRX-associated disease, which can present in dominant and recessive contexts with varying severity and onset, limit the utility of a strict variant frequency threshold. Nevertheless, visualization of our DMS activity scores versus variant allele frequencies shows that variants with high allele frequencies in the general population tend to have wild-type-like activity in our assay (Supplemental Fig. S3A,B). This is particularly true for variants with multiple homozygous individuals reported. Variants with significantly reduced or increased activity in the DMS assay tend to have lower allele frequencies, and only a single homozygous individual is reported for a single high-activity variant.
Variants can affect protein abundance, but abundance alone does not explain changes in activity
We selected a group of low- and high-activity variants to individually validate in one-at-a-time integration experiments (Supplemental Fig. S4A). For these variants, we also performed western blots to assess their effects on CRX protein abundance, and evaluated the correlation between DMS activity scores and protein abundance (Supplemental Fig. S4B). For each variant, we performed two replicate blots with each of two antibodies (CRX A-9 and CRX B-11), for a total of four replicate blots (Supplemental Fig. S4C). We assessed blot intensity relative to a β-tubulin control for each sample, and normalized abundance within each blot to that of wild-type CRX. In general, we did not observe a correlation between protein abundance and DMS activity for the tested variants. Some variants, such as the p.N281C low-activity variant, showed reduced abundance compared to wild-type CRX, while two wild-type-like variants (p.A59Y and p.Q84M) showed increased abundance. Further characterization would be required to systematically probe the relationship of protein abundance and activity for CRX, but this analysis at least suggests that abundance does not explain the observed effects on activity for all measured variants.
Variant activity scores in the homeodomain closely align with a predicted structural model
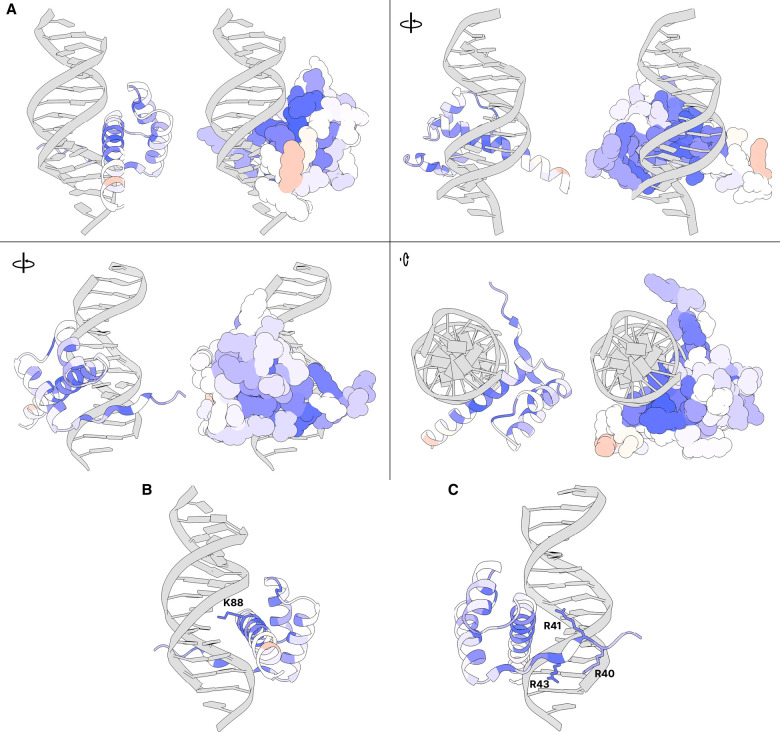
In general, variants in the homeodomain show significantly reduced activity scores compared to nonhomeodomain variants (Brunner–Munzel test P = 2.59 × 10−114). Even within the homeodomain, however, some positions are more sensitive to substitution than others. To benchmark our assay results and evaluate how closely they reflect known physical constraints of CRX-DNA binding, we mapped the average per-residue DMS activity scores onto a structure of the CRX homeodomain in complex with DNA (Fig. 4A; Supplemental Movie S1). Because an experimentally determined structure of CRX is not available, we aligned the AlphaFold2-predicted CRX structure to crystal structures of other homeodomains in the paired family, which is highly structurally conserved. Notably, DMS activity scores are highly concordant with the structural model. Amino acid residues in the major groove are particularly sensitive to substitution, even when compared to residues in the same α-helix that do not lie in the major groove. Furthermore, residues on the DNA-facing sides of the two α-helices not in the major groove are more sensitive to substitution than residues on the outward-facing sides of the same α-helices.
Figure 4.
Average DMS activity scores superimposed on a predicted structure of the CRX homeodomain in complex with DNA. (A) Various views of residues 38–104 of an AlphaFold predicted structure of CRX aligned to a crystal structure of Drosophila paired in complex with DNA (PDB 1FJL) (Wilson et al. 1995). For each view, a cartoon ribbon model is shown on the left and a space-filling atomic model is on the right. Supplemental Movie S1 animates a 360° rotation of this structure. (B) Close-up of residues in the major groove with K88 highlighted (side chain shown). (C) Close-up of minor groove-contacting residues with arginine residues highlighted (side chains shown). In all panels, residues are colored by the average DMS activity score, as shown in the “Average” track in Figure 2.
CRX is a K50-type homeodomain TF, which refers to the lysine (K) residue at the 50th position of the homeodomain sequence which is crucial for determining DNA-binding specificity (Noyes et al. 2008). In CRX, residue 88 is the K50 position, and substitutions at this position do indeed have particularly large effects on activity (Figs. 2, 4B). It is also known that arginine residues at the N-terminus of the homeodomain interact with the minor groove to further control motif specificity (Noyes et al. 2008). These residues are particularly sensitive to substitution as predicted, even when compared to immediately adjacent non-DNA-contacting residues (Fig. 4C). Taken together, the strong concordance of the DMS activity scores with the structural model in general and at known crucial residues provides support that the DMS assay is accurately measuring the ability of CRX variants to bind and activate the transcriptional reporter, and provides mechanistic and structural insight into substitution sensitive and nonsensitive amino acid residues. Furthermore, these results lend confidence to the biological relevance of the synthetic HEK 293-based transcriptional reporter used in these experiments.
Patterns of substitution activity in the transcriptional effector domain support an acidic exposure model
In addition to its use in classifying clinical CRX variants, the DMS assay reveals molecular features of the disordered CRX transcriptional effector domain. Throughout the effector domain, substitutions that add a negative charge (aspartic acid or glutamic acid) tend to increase activity, while substitutions that remove existing negatively charged residues (particularly D24, D219, D265, D271, D284, D287, and D290) tend to reduce activity. Conversely, substitutions that add aromatic residues (phenylalanine, tryptophan, and tyrosine) or aliphatic hydrophobic residues with larger side chains (methionine, leucine, isoleucine, and valine) tend to decrease activity, while substitutions that remove existing residues in these classes tend to increase activity, especially when replaced with polar uncharged or negatively charged residues (Fig. 2).
Further underscoring the composition-sensitivity of the effector domain, the effects of residue substitutions can be used to cluster amino acids by the chemical properties of their side chains. We performed unsupervised hierarchical clustering by substituted residue on positions in the disordered region of the transcriptional effector domain (residues 2–38 and 153–264) (Fig. 5A). Substituted amino acids largely cluster by their biochemical properties, supporting the idea that the disordered transcriptional effector domain is primarily sensitive to overall amino acid composition, rather than the identity of particular residues at certain positions.
Figure 5.
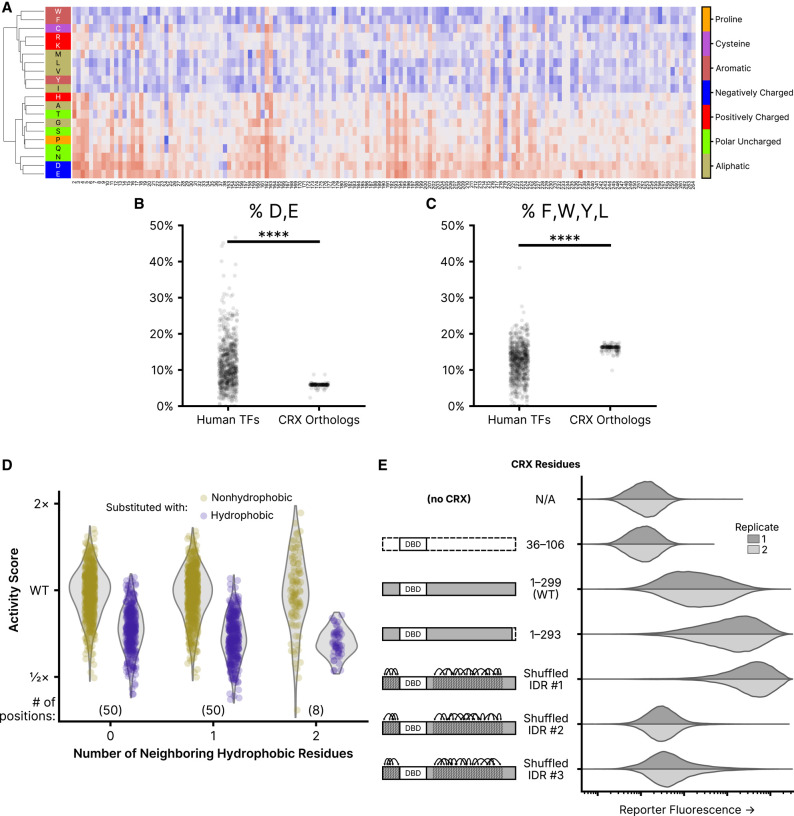
Residue class preferences in the CRX transcriptional effector domain. (A) Unsupervised hierarchical clustering (UPGMA method) of per-position activity scores for residues in the disordered region of the transcriptional effector domain (residues 2–38 and 153–264). Residues colored by class. Substitution activity scores are colored as in Figure 2. (B) Abundance of aspartic acid (D) and glutamic acid (E) residues in disordered regions of all human TFs and CRX orthologs. (C) The abundance of phenylalanine (F), tryptophan (W), tyrosine (Y), and leucine (L) residues in disordered regions of all human TFs and CRX orthologs. For (B) and (C), significance was tested by a two-sided Mann–Whitney U test; P < 1 × 10−39. (D) Comparison of the effects of substituting nonhydrophobic positions in wild-type CRX with hydrophobic (F, W, Y, M, I, L, or V) or nonhydrophobic amino acids, separated by the number of neighboring hydrophobic residues. The analysis is limited to positions in the disordered transcriptional effector domain; the number of positions in each neighbor group is shown in parentheses. (E) Reporter activation (green fluorescence; arbitrary units) was measured in Reporter + LP cells with the indicated CRX variants integrated. Two independent biological replicate experiments per sample (distributions plotted from 50,000 cells).
The balance of effects between substitutions affecting negatively charged and aromatic residues in the transcriptional effector domain is notable in light of the acidic exposure model of transcriptional activation domains (Staller et al. 2022). The acidic exposure model proposes that strong transcriptional activators are facilitated by a balance of acidic and aromatic residues, in which acidic residues permit the solubilization of hydrophobic motifs. Even among TF effector domains, the CRX transcriptional effector domain is particularly low in negative charge and high in aromatic residue content (Fig. 5B,C). This is consistent with the observed substitution effects of residues in these classes: substitutions of negatively charged residues to amino acids with uncharged or positively charged side chains uniformly decrease CRX transcriptional activity while substitutions of uncharged or positively charged residues to negatively charged amino acids increase CRX transcriptional activity, and the reverse is true for aromatic residues.
To examine the role of local sequence context in determining the functional effect of hydrophobic substitutions, we compared the activity scores of hydrophobic substitutions (hydrophobic residues defined as F, W, Y, M, L, I, or V) to all other substitutions at nonhydrophobic residues in the transcriptional effector domains (residues 2–38 and 153–264), stratifying positions according to the number of immediately adjacent hydrophobic residues (Fig. 5D). We fit an interaction model between the class of substituted residue and the number of neighboring hydrophobic residues, finding that the deleterious effect of hydrophobic substitutions increased with the local density of hydrophobic residues (likelihood ratio test P = 2.37 × 10−4). Though subtle, the context dependence of hydrophobic substitution effects indicates that the effector domain function is sensitive to residue spacing as well as composition.
To further test whether the CRX transcriptional effector domain exhibits any residue position requirements or is primarily sensitive to overall amino acid composition, we tested three CRX variants in which we shuffled the position of the amino acids in the disordered region. To shuffle the amino acids, we broke the disordered regions into five residue windows and randomized the order of the residues within each window. This ensures global sequence chemistry is preserved while local sequence order is altered. Two of the three shuffled variants showed reduced transcriptional activity compared to wild-type CRX, but the third showed the strongest reporter activation of any CRX variant tested (Fig. 5E). From this result, it is clear that the effector domain is not entirely position-insensitive, as three arbitrary amino acid rearrangements demonstrated substantially different effects on transcriptional activity. Nevertheless, there is clearly a qualitative difference in position sensitivity between the ordered and disordered regions of CRX.
We also tested a variant truncating the six most C-terminal residues of the OTX-tail motif. Substitutions of any of these amino acids (WKFQIL), which include several aromatic and leucine residues, largely increased transcriptional activity in the DMS assay, particularly when replaced with aliphatic or negatively charged side chains. Consistently, the 1–293 truncation variant activates the CRX transcriptional reporter more strongly than wild-type CRX, although not as strongly as the strongest shuffled variant. This result suggests that these residues of the OTX-tail motif may play a repressive role in the CRX transcriptional effector domain.
Discussion
We conducted a systematic characterization of variant effects in the photoreceptor TF CRX. Using a synthetic fluorescent reporter, we measured the activity of nearly all possible missense variants in CRX, and assigned each a functional activity score. Substitutions showed unique patterns of variant activity in different functional domains of CRX. Residues in the DNA-binding homeodomain were particularly sensitive to any substitution, and per-residue average activity scores closely aligned to a predicted structure of the CRX homeodomain in complex with DNA. In the intrinsically disordered transcriptional effector domain, variant effects showed a strong association with biochemical amino acid class rather than position, and substitution effects were consistent with the acidic exposure model of transcriptional activation domains.
From the observed patterns of variant effect, CRX can be broken into three main regions. The DNA-binding domain, roughly spanning residues 40–95, is highly sensitive to substitution, and many substitutions result in reduced reporter activity, particularly for DNA-contacting or folding-essential residues. The central intrinsically disordered region of CRX, by contrast, spanning residues 95–155, is largely insensitive to substitution, with the exception of some residues in the basic domain that tend to result in increased activity upon substitution. Lastly, the C- and N-terminal disordered regions show strong sensitivity to amino acid class, with both increases and decreases in activity depending on substitution. While disordered regions lack a fixed 3D structure and are, therefore, often assumed to be relatively insensitive to mutations, our work clearly reveals chemically interpretable patterns in mutational sensitivity.
As discussed, the acidic exposure model of activation domains suggests that strong transcriptional activators require a balance of acidic residues with aromatic and leucine residues (Staller et al. 2022). Compared to other TF-disordered domains, CRX has a high proportion of aromatic and leucine residues in the disordered transcriptional effector domain and a low proportion of acidic residues (Fig. 5B,C). This may explain why substitutions increasing negative charge or decreasing aromaticity result in increased activation—the CRX transcriptional effector may have a residue composition that is not the same as that of an “ideal” strong activator. We also observed that the local density of hydrophobic residues in the transcriptional effector domain affected variant activity, with substitutions of hydrophobic amino acids in sites with a higher number of neighboring hydrophobic residues tending to show stronger reductions in transcriptional activity. This effect appears consistent with functional constraints on the relative spacing of aliphatic and aromatic residues observed in other hydrophobic-enriched IDRs (Martin et al. 2020, Holehouse et al. 2021, Jonas et al. 2023), indicating that the introduction of hydrophobic patches in the CRX transcriptional effector domain may impair solubility or accessibility of residues required for activation. CRX and other homeodomain TFs have been shown to be capable of acting as both activators and repressors at different target sequences (White et al. 2016; DelRosso et al. 2023; Shepherdson et al. 2024). Further characterization is required, but balancing activation and repression in the transcriptional effector domain may necessitate the “intermediate” activation strength residue composition observed for CRX. If so, the pathogenicity of high-activity variants could be further compounded by impaired repression at particular target cis-regulatory elements.
The CRX transcriptional effector domain also has a high proportion of proline and serine residues. If any of these serine residues were to undergo phosphorylation, they would acquire a negative charge that could potentially contribute to the residue balance requirements of the acidic exposure model. Proline-directed protein kinases, including members of the MAPK family, have been shown to target NRL, a key TF expressed in rods and a close partner of CRX (Swain et al. 2007). Furthermore, other protein kinases, including PKC family members, have been reported to be essential for rod cell development in mice (Pinzon-Guzman et al. 2011). Whether these or other protein kinase enzymes interact with CRX and what the impact may be of any possible serine phosphorylation on the CRX transcriptional effector domain will require further study.
The high-specificity classification we observed for clinical variants, with a low false positive rate (type I error) but moderate false negative rate (type II error), is reasonable given the design of the reporter assay. In the genome, CRX binds numerous cis-regulatory elements with varied and nonconsensus-binding motif sequences (Corbo et al. 2010). The synthetic CRX transcriptional reporter used in this study, which contains only high-affinity consensus motif sequences, may be insensitive to subtle variants that alter DNA binding at nonideal motifs. Furthermore, it is completely insensitive to variants that cause a gain of novel binding motifs. For instance, the p.E80A CRX variant, a known pathogenic variant in humans, displays wild-type-like activity in the DMS assay. It has recently been shown that while the p.E80A CRX variant prefers the same consensus motif as wild-type CRX, it is more tolerant of base mismatches in the motif, an effect to which the DMS reporter may not be sensitive (Zheng et al. 2023). Of note, the E80 site has two other characterized substitutions in ClinVar: p.E80G and p.E80K. In our assay, only the p.E80K variant showed significantly reduced activity compared to wild-type CRX. Glycine and alanine are more similar biochemically to each other than either is to lysine, and it is not unreasonable that our synthetic CRX reporter is sensitive to some homeodomain perturbations and not others. We note that the DMS classifications are concordant with ClinVar for all other colocated pathogenic variants (no benign variants are colocated).
Furthermore, the LP + reporter cell line is a synthetic transcriptional environment, lacking many of the partner TFs typically expressed in photoreceptors. In the retina, CRX is known to cobind at cis-regulatory elements with a number of TFs, including NRL, OTX2, RAX, RORB, and others (Friedman et al. 2021). How the presence or absence of these factors influences the functional consequences of CRX variants remains to be seen. However, the fact that ectopic expression of CRX variants and the addition of a single transcriptional reporter construct to a nonphotoreceptor cell type is enough to produce data so consistent with existing structural data, variant classifications, and activation domain models is a testament to the power of the reductionist approach used in this DMS assay. When combined with evolutionary sequence conservation data, population variant allele frequencies, structural and biochemical models, and other forms of evidence in the ACMG variant classification framework, the DMS activity scores presented here have the potential to aid in the future classification of previously uncharacterized clinical CRX variants.
The approach used in this study, with a synthetic transcriptional reporter and nonnative cell type, may be broadly applicable to other TFs with expression limited to specific cell types that are difficult to obtain or culture. Compared to the growth and survival assays that are more typical of deep mutational scans in mammalian cells (Maes et al. 2023), transcriptional reporters have the benefit of being a direct readout of TF protein activity. This may help to increase their sensitivity to the effects of protein variants, even in a nonphysiologic context.
That a CRX variant with shuffled residues in the intrinsically disordered transcriptional effector domain was capable of activating the reporter more strongly than wild-type CRX further supports that CRX did not evolve to encode the strongest possible activation domain, consistent with the discussion above of a potential balance of activation and repression. Furthermore, the varied effects on transcriptional activity observed across the three shuffled variants suggest that the precise residue order has an effect on activation strength, even if specific positions are generally less individually sensitive to substitution than those in the ordered and folded homeodomain. This conclusion is in line with analyses of other DNA-binding proteins (Sanborn et al. 2021; Langstein-Skora et al. 2022; Jonas et al. 2023; Mindel et al. 2023).
Considerable effort has been undertaken in recent years to develop computational predictors of variant effect, and several methods, including PolyPhen-2 (Adzhubei et al. 2010), CADD (Schubach et al. 2024), AlphaMissense (Cheng et al. 2023), ESM1b (Brandes et al. 2023), and EVE (Frazer et al. 2021) are either in clinical use or under consideration for that purpose. We present the CRX variant classifications for the DMS and these methods in Supplemental Figure S5, along with a comparison of their classifications for specifically the variants present in ClinVar. We note, however, that it is common for computational classifiers to use ClinVar variants as part of their training and/or validation data, which may cause overfitting. For instance, PolyPhen-2, AlphaMissense, EVE, and ESM1b all show strong concordance with ClinVar despite considerable differences in their classifications of non-ClinVar variants. Additionally, all five computational classifiers shown in Supplemental Figure S5 call the p.L299F variant as pathogenic. This variant was initially classified as “Likely pathogenic” on first ClinVar submission in 2014 but, subsequent to the training/development of these computational classifiers, was recently reclassified to “Benign” based on additional clinical reports. In the DMS assay, p.L299F displays wild-type-like activity. Ultimately, we are enthusiastic about the prospects of computational variant classification to address the challenges posed by the large number of VUS in the genome, but we believe that integration of these methods with functional assay results is the most productive path forward in the near term.
The variant activity scores in the intrinsically disordered transcriptional effector domain are particularly relevant in light of recent developments of machine learning-based computational variant effect predictors trained on protein sequence homology and structural data. The AlphaMissense, ESM1b, and EVE models predict variant pathogenicity from, primarily, evolutionary protein sequence conservation data. This is of particular relevance to proteins with intrinsically disordered regions, however, because disordered proteins are not subject to the same evolutionary sequence constraints as structured proteins (Holehouse and Kragelund 2024). Visualization of variant classifications derived from the DMS activity scores compared to classifications from AlphaMissense, ESM1b, and EVE show concordance in the structured homeodomain, but there is less agreement in the disordered transcriptional effector domain (Supplemental Fig. S5). Future machine learning-based variant effect predictors would likely benefit from the incorporation of functional mutational scanning measurements in their training data, particularly for proteins with intrinsically disordered regions like CRX. New machine learning-based methods trained on both sequence conservation and functional mutational scanning data may eventually permit the use of these classifiers for direct clinical variant classification as a sole evidence source, a practice which the ACMG discourages for the current generation of computational variant classifiers.
The limitations of this work primarily relate to the intrinsic limitations of reporter assays in nonprimary cell types. As discussed, the synthetic transcriptional reporter developed in this study cannot capture the full breadth of CRX-bound cis-regulatory elements in the genome, and may be insensitive to certain variants that subtly alter DNA-binding specificity. This assay also primarily measures variant effects on activation, and would not be sensitive to a hypothetical variant that specifically disrupts the repressive functions of CRX without affecting activation, if such a variant were possible. Furthermore, while extremely tractable, the HEK 293-derived cell line used for these experiments does not reflect the unique transcriptional environment of photoreceptors. For instance, the substitution-tolerant central region of CRX could conceivably be necessary for protein–protein interactions with partner TFs not expressed in HEK 293 cells. It is noteworthy that our DMS assay identified a number of missense variants that appear to increase transcriptional activity relative to wild-type CRX. As can be seen in Figure 1A, all of the pathogenic missense CRX variants reported in ClinVar fall in or around the homeodomain, and tend to decrease transcriptional activity. High-activity pathogenic variants may exist but have not yet been reported clinically (possibly as a result of subtle clinical phenotypes), or it may be that these variants increase transcriptional activity but do cause clinical retinal disease. Alternatively, the change in activity we measure may not occur in photoreceptors (e.g., due to the transcriptional regulatory environment or the presence of cofactors). Ultimately, the small number of pathogenic variants reported in ClinVar for CRX is limiting, but we anticipate that the ongoing expansion of clinical genome sequencing and further molecular characterization studies will improve our understanding of the CRX activation domain in the future. Nevertheless, the observed concordance of DMS activity scores with homeodomain structural models, the consistency of variant substitutions with the acidic exposure model of transcriptional activation domains, and the clinical variant classification performance all lend support to the validity of the DMS measurements.
Methods
For a more detailed description of cell line generation and library cloning, please see the Supplemental Methods. For a list of primers and plasmids used in this study, please see Supplemental Tables S1 and S2, respectively.
Generation of the landing pad cell line
All cell lines were cultured in 90% DMEM (Gibco 11965092), 10% heat-inactivated fetal bovine serum (Gibco 16140089) supplemented with Penicillin–Streptomycin (Gibco 15140122). Transfections were performed with Lipofectamine 3000 and P3000 reagent (Invitrogen L3000015) with Opti-MEM (Gibco 31985062). All flow cytometry was performed on a Cytoflex S flow cytometer (Beckman-Coulter, V4-B2-Y4-R3 model).
In experiments where constructs were integrated into the LP, cells were transfected with a mixture of the indicated constructs and Addgene 11916, a plasmid expressing Cre recombinase. Drug selection against the iCasp9 inducible caspase present in the naive LP was performed using 5 nM AP1903/Rimiducid (MedChemExpress HY-16046) (Straathof et al. 2005). For positive drug selection of integrants, 1 μg/mL puromycin (Sigma-Aldrich P8833) was used. Following integration, cells were continually grown in media supplemented with 5 nM AP1903 and 1 μg/mL puromycin.
To generate the LP cell line, HEK 293 cells were cotransfected with 400 ng pJLS83, a plasmid carrying the LP sequence, and 400 ng Addgene 105927, a plasmid expressing Cas9 and a sgRNA targeting the human Rosa26 safe harbor locus, and individual clones were sorted using a Cytoflex SRT (Beckman-Coulter, V5-B2-Y5-R3 model) and expanded.
To screen outlines with the integration of the LP construct on multiple alleles, clones were cotransfected with 600 ng Addgene 11916, a plasmid expressing Cre recombinase, and a 1:1 mixture of 200 ng pJLS119 and 200 ng pJLS120, plasmids carrying the mEmerald and mCherry2 fluorescent proteins. Cells were measured by flow cytometry after AP1903 + puromycin purification of LP-integrated cells, and any clones yielding cells exhibiting both green and red fluorescence were discarded. The final validated clonal LP cell line was frozen at low passage numbers, in CryoStor CS10 freezing medium (Biolife Solutions 210102).
Generation of the CRX reporter cell line
A synthetic CRX reporter was synthesized carrying eight repeats of a strong consensus CRX-binding motif, CTAATCCC, each padded with a 2 bp spacer motif (AG) (White et al. 2016). This reporter sequence was cloned upstream of the CMV-T6 minimal promoter (Loew et al. 2010) and the mEmerald fluorescent protein to produce pJLS96.
To produce lentivirus carrying the reporter, HEK 293T cells were transfected with a mixture of 8 μg Addgene 12260, 1 μg Addgene 12259, 1 μg pJLS96, and 40 µL PEI in 500 µL Opti-MEM, and the virus was concentrated with Lenti-X Concentrator (TaKaRa 631231) following the manufacturer's recommended protocol. To create a “dead” reporter control, a second batch of lentivirus was produced using pJLS97, a variant of pJLS96 in which the CRX motifs were replaced with a variant known to abrogate CRX binding (CTACTCCC) (White et al. 2016).
LP cells were separately transduced with the pJLS96 and pJLS97 lentiviruses. After 72 h, the bulk populations of transduced cells were transfected with 400 ng pJLS38, a plasmid expressing human CRX. Three days posttransfection, individual pJLS96-transduced cells were sorted to generate single clonal cell lines, gating for positive blue channel fluorescence to select for the presence of the LP construct and for positive green fluorescence, relative to pJLS38-transfected cells transduced with the pJLS97 “dead” reporter construct.
Selected clones were cotransfected with 400 ng Addgene 11916 and 400 ng of either pJLS99, pJLS100, or pJLS101, plasmids carrying wild-type CRX, the p.R90W hypomorphic CRX variant, or miRFP670 (a transcriptionally inactive control protein), respectively. Following the successful isolation of integrated cells, cells were screened by flow cytometry to maximize the dynamic range of mEmerald fluorescence between the pJLS99- and pJLS100-transfected cells. The final validated clonal LP + reporter cell line was frozen at low passage numbers, in CryoStor CS10 freezing medium.
Cloning the CRX variant library
A DNA library comprising all possible human CRX single residue substitution variants was ordered as a combinatorial variant library from Twist Bioscience, with the synthesis of a designed codon for each variant. The library construct was ligated between the AflII (NEB R0520) and NheI (NEB R3131) sites in pJLS84v2, a plasmid carrying recombinase sites matching the LP, using T4 Ligase (NEB M0202).
The ligation product was purified using a Monarch PCR and DNA Cleanup Kit (NEB T1030) following the manufacturer's recommended protocol. The purified ligation product was transformed into 10-beta Electrocompetent Escherichia coli (NEB C3020) using a Bio-Rad GenePulser Xcell Electroporation System with PC Module (Bio-Rad 1652662) with the following conditions: 2000 V, 200 Ω, and 25 μF.
Following electroporation, transformation products were pooled. 1:1000 and 1:10,000 dilutions of the outgrowth were plated on LB plates carrying 50 μg/mL kanamycin (LB + Kan50, Sigma-Aldrich K1377) and incubated. One thousand seven hundred microliters of the outgrowth was inoculated into 200 mL LB + Kan50 and incubated until an OD600 of 3.0 was reached. Based on colony counts from the dilution plates, the 200 mL culture was inoculated with ∼5.4 million colony-equivalents. Plasmid DNA was purified using a GenElute HP Plasmid Maxiprep Kit (Sigma-Aldrich NA0310) following the manufacturer's recommended protocol and concentrated using an Eppendorf Vacufuge plus, yielding the “step one” CRX DMS library.
To add barcodes to this library, a short random barcoding oligo (“variant barcode,” “vBC”) was ordered as an Ultramer DNA Oligo from Integrated DNA Technologies (pJLS84v2 + BC1). One microgram of the step one plasmid library was linearized with XhoI (NEB R0146). The digest product was run on a 1% agarose gel and purified using a Monarch DNA Gel Extraction Kit (NEB T1020) following the manufacturer's recommended protocol. The linearized plasmid library and barcoding oligo were assembled with NEBuilder HiFi DNA Assembly Master Mix (NEB E2621) at a 1:5 molar ratio, targeting ∼200 fmol total DNA in the assembly reaction. The assembly reaction was purified using a Monarch PCR and DNA Cleanup Kit.
The purified, assembled, barcoded library was transformed following the same protocol as the step one DMS library. The 200 mL culture was inoculated with 40 μL of outgrowth and grown to an OD600 of 2.0 (∼13 h). Based on colony counts from the dilution plates, the 200 mL culture was inoculated with ∼85,000 colony-equivalents. Plasmid DNA was purified and concentrated using the same procedure as for the step one DMS library, yielding the “step two” CRX DMS library.
To add the puromycin N-acetyltransferase (PAC) cassette to the step two library, a fragment carrying an internal ribosome entry site (IRES) and PAC coding sequence was amplified from pJLS98 using primers JClPr88 and JClPr89. One microgram of the step two plasmid library was linearized with SpeI-HF (NEB R3133) and purified following the same protocol as the step two library. The IRES-PAC cassette and linearized step two library were assembled and purified following the same protocol used to generate the step two library. The purified, assembled, PAC-containing library was transformed following the same protocol used to generate the step one DMS library. The 200 mL culture was inoculated with 1850 μL of outgrowth and grown to an OD600 of 3.5 (∼11 h). Plasmid DNA was purified and concentrated using the same procedure as for the step one DMS library, yielding the “step three” CRX DMS library.
A second short random barcoding oligo (“random barcode,” “rBC”) was ordered as an Ultramer DNA Oligo from Integrated DNA Technologies (pJLS84v2 + BC2). One microgram of the step three plasmid library was linearized with XhoI and purified following the same protocol as the step two library. The barcoding oligo and linearized step three library were assembled and purified following the same protocol used to generate the step two library.
The purified, assembled, barcoded library was transformed following the same protocol used to generate the step one DMS library. The 200 mL culture was inoculated with 1850 μL of outgrowth and grown to an OD600 of 3.0 (∼10 h). Plasmid DNA was purified and concentrated using the same procedure as for the step one DMS library, yielding the final CRX DMS library.
Associating variants with barcodes
To associate CRX variants with barcodes, the step three CRX DMS library was sequenced using a PacBio Revio long-read sequencer. Briefly, 4 μg of the barcoded plasmid library was linearized by digestion with NruI-HF (NEB R3192) and purified using a Monarch PCR and DNA Cleanup Kit. The purified linearized library was used to prepare a PacBio sequencing library using an SMRTbell prep kit 3.0 (PacBio 102-141-700), following the manufacturer's recommended protocol (PacBio Protocol 102-166-600 REV02) with the following modifications: DNA shearing was skipped and the 20 μL of the purified linearized library was used directly as input for Repair and A-tailing after the addition of 27 μL of Low TE buffer; the final cleanup with SMRTbell cleanup beads was not performed (no size selection). The prepared library was sequenced on a single Revio SMRT cell.
Reads were aligned to a synthetic reference sequence comprising the expected library plasmid structure with wild-type CRX and “N” nucleotides in place of the expected barcode location using minimap2 (Li 2018) (https://github.com/lh3/minimap2, v2.24, with parameters -A2 -B4 -O12 -E2 –end-bonus = 13 –secondary = no –cs = long). The resulting PAF file was parsed with a custom Python script (see Supplemental Code, “call_variants.py”) to generate a barcode-to-variant map. From 9.5 million total HiFi reads, 63.8% contained a valid barcode and a single missense CRX variant or wild-type sequence. The remaining reads represent a mixture of sequencing errors, low-quality reads, or valid reads of constructs that failed barcoding, acquired indels during cloning, or contained more than a single CRX missense variant. Of 75,946 observed barcodes on full-length plasmid reads, 86.8% mapped to a single CRX variant, while 7.3% mapped to wild-type CRX. Of the remaining barcodes, 4.9% mapped to CRX constructs with more than a single missense variant, while only ∼1% of barcodes could not be unambiguously assigned—i.e., were observed to co-occur with different variants in different reads. All barcodes not uniquely mapping to a single CRX missense variant or wild-type CRX were discarded.
Measuring variant activity
To conduct the DMS, LP + reporter cells were transfected with 2 μg of the final CRX DMS library, as well as 8 μg Addgene 11916, 50 μL Lipofectamine 3000, and 50 μL P3000, in 3 mL Opti-MEM. Transfected cells underwent drug selection to isolate successful integration events, first with 5 nM AP1903 and then with 5 nM AP1903 and 1 μg/mL puromycin combined. Cells were sorted into four bins based on reporter fluorescence on a Sony SY3200 fluorescence-activated cell sorter, recovering between 500,000 and 3,000,000 cells per bin. Sort distributions and gating strategies for each replicate are shown in Supplemental Data S1. Sorted fractions were plated in a fresh flask and harvested 3–6 days postsort.
Genomic DNA was extracted from each fraction using a Monarch Genomic DNA Purification Kit (NEB T3010). Barcode sequences were amplified from gDNA using a two-step protocol: gDNA was first amplified with primers JLSPr141–144 and JLSPr165–168 + 171 + 172 using Q5 High-Fidelity DNA Polymerase (NEB M0491) for 20 cycles with an annealing temperature of 65°C, and then with primers IDT10_i7_NN and IDT10_i5_NN, where NN is replaced with the unique indexing barcode ID, for sample multiplexing (10 cycles, 65°C annealing temperature). Prepared fraction amplicon libraries were sequenced on an Illumina NovaSeq X Plus instrument in a series of shared 10B flow cells, targeting 100 million reads per replicate.
Reads were cleaned with fastp (Chen et al. 2018) (https://github.com/OpenGene/fastp, v0.23, with default parameters). Barcodes were extracted, counted, and analyzed using custom scripts (see Supplemental Code, “extract_bcs.sh,” “count_bcs.py,” and “analysis.ipynb”). vBCs were mapped to variants using the results of the PacBio long-read sequencing of the barcoded plasmid library. vBCs failing to match an expected barcode from the PacBio sequencing were error-corrected up to a Hamming distance of 1, if and only if they could be unambiguously mapped to an expected variant. All reads with nonmapping or ambiguous vBCs were discarded. vBC–rBC pairs with fewer than three reads were discarded. vBCs with fewer than 10 unique rBCs in any of the four bins were discarded. Raw activity scores were computed by summing the number of rBCs per vBC in each of the four bins divided by the sum of the number of rBCs across all four bins, weighted by the mean fluorescence intensity of each bin. Raw activity scores for each variant were divided by the wild-type CRX activity score, yielding the normalized DMS activity score used throughout this paper (Supplemental Table S3).
Visualizing activity scores on a predicted CRX structural model
To visualize mean per-variant DMS activity scores on a structural model of CRX in complex with DNA, we first downloaded a crystal structure of the Drosophila paired homeodomain in complex with a DNA target (Protein Data Bank [PDB; https://www.rcsb.org] 1FJL), as well as the AlphaFold2-predicted structure for CRX (corresponding to UniProt O43186). Using UCSF ChimeraX (v1.7, https://www.cgl.ucsf.edu/chimerax/), we aligned the CRX-predicted structure to the paired homeodomain crystal structure with the “matchmaker” tool (Meng et al. 2006). After alignment, we hid the Drosophila paired structure, leaving the CRX-predicted structure oriented with just the DNA target crystal structure from PDB 1FJL.
Western blotting for CRX abundance
Cells were harvested by removal of media and addition of RIPA buffer (Cell Signaling Technology 9806, diluted to 1× in deionized water) supplemented with 1× Halt Protease Inhibitor Cocktail (Thermo Scientific 78430) and incubated on ice for 30 min. The cell lysate was centrifuged at 21,000g for 10 min and the supernatant was mixed with Blue Loading Buffer (Cell Signaling Technology 7722) per the manufacturer's recommended ratio. The combined supernatant and loading buffer were boiled for 8 min at 97°C and cooled to room temperature. Samples were run on 4%–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad 4561096) in Tris/Glycine/SDS buffer (Bio-Rad 1610732, diluted to 1× in deionized water). Transfers were performed using a Bio-Rad Trans-Blot Turbo instrument, RTA Mini 0.2 µm PVDF Transfer Kit (Bio-Rad 1704272), and Trans-Blot Turbo Transfer Buffer (Bio-Rad 10026938). CRX (A-9) (Santa Cruz Biotechnology sc-377138), CRX (B-11) (Santa Cruz Biotechnology sc-377207), and β-Tubulin Rabbit Ab (Cell Signaling Technology 2146S) were used, along with Anti-rabbit IgG, HRP-linked (Cell Signaling Technology 7074S) and Anti-mouse IgG, HRP-linked (Cell Signaling Technology 7076S). TBST (EZ BioResearch S-1012, diluted to 1× in deionized water) and EveryBlot Blocking Buffer (Bio-Rad 12010020) were used for washing and blocking, respectively. To estimate abundance from blot band intensity, the “Quantity Tools” feature of Image Lab (Bio-Rad 12012931) was applied to raw blot images (SCN format) from a ChemiDoc XRS + System (Bio-Rad 1708265), using the “Relative” quantitation feature (relative to the β-tubulin band within each lane). After relative quantitation, band abundances were normalized to the wild-type CRX band present on each blot. These β-tubulin- and wild-type CRX-normalized abundances were used for further analysis and plotting.
Analysis of transcriptional effector domain residue composition
The composition of transcriptional effector domains from CRX orthologs was compared to that of disordered regions found in annotated human TFs. CRX ortholog sequences were obtained from the UniProt UniRef50 cluster containing human CRX. Activating and bifunctional human TFs were curated by Soto et al. (2022). Disordered regions were predicted from these sequences using metapredict v2.6 (Emenecker et al. 2021). We compared the fraction of D, E, and F, W, Y, and L residues in the predicted C-terminal IDRs of the CRX orthologs to the distribution observed for all IDRs predicted in the set of human TFs, evaluating a difference in the mean fraction by Mann–Whitney U test.
Local sequence effects of hydrophobic substitutions
Activity scores for substitutions occurring at nonhydrophobic residue positions in the transcriptional effector domain were fit to an interaction model between the class of substitution (hydrophobic or nonhydrophobic) and the number of immediately adjacent hydrophobic residues (0, 1, or 2). The following model was fit with ordinary least squares using the Python package statsmodels v0.14.0.
To determine the significance of the interaction term, a likelihood ratio test was performed against a reduced model excluding the interaction.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE262060. Source code used for the analyses presented in this work is available at GitHub (https://github.com/barakcohenlab/crx-dms-manuscript) and as Supplemental Code.
Supplemental Material
Acknowledgments
We are grateful to Jessica Hoisington-Lopez and Maria-Lynn Crosby at the Washington University Center for Genome Sciences’ DNA Sequencing Innovation Lab as well as the staff at the Washington University Genome Technology Access Center for their assistance with the generation of high-throughput sequencing data sets. We are also grateful to the staff of the Alvin J. Siteman Cancer Center Flow Cytometry Core for their assistance with fluorescence-activated cell sorting. This work was supported by the National Institutes of Health: National Institute of General Medical Sciences R01GM092910, National Human Genome Research Institute R21HG012146, and National Eye Institute R01EY027784 to B.A.C.; National Cancer Institute 1DP2CA290639 to A.S.H.; National Institute of General Medical Sciences R01GM121755 to M.A.W.; and National Eye Institute F30EY033640 to J.L.S.
Author contributions: J.L.S., D.M.G., J.L, Z.S., S.P.P: investigation; J.L.S.: writing—original draft; J.L.S., D.M.G., J.L., Z.S., S.P.P., A.S.H., M.A.W., B.A.C.: writing—review and editing; A.S.H., M.A.W., B.A.C.: supervision.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.279415.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
B.A.C. is on the scientific advisory board of Patch Biosciences. The other authors declare no competing interests.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N, Goldman G, Wang CH, Ye CJ, Ntranos V. 2023. Genome-wide prediction of disease variant effects with a deep protein language model. Nat Genet 55: 1512–1522. 10.1038/s41588-023-01465-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brnich SE, Abou Tayoun AN, Couch FJ, Cutting GR, Greenblatt MS, Heinen CD, Kanavy DM, Luo X, McNulty SM, Starita LM, et al. 2020. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med 12: 3. 10.1186/s13073-019-0690-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang Q-L, Xu S, Liu I, Li LY, Wang Y, Zack DJ. 2002. Functional analysis of cone–rod homeobox (CRX) mutations associated with retinal dystrophy. Hum Mol Genet 11: 873–884. 10.1093/hmg/11.8.873 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Novati G, Pan J, Bycroft C, Žemgulytė A, Applebaum T, Pritzel A, Wong LH, Zielinski M, Sargeant T, et al. 2023. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science (New York, NY) 381: eadg7492. 10.1126/science.adg7492 [DOI] [PubMed] [Google Scholar]
- Corbo JC, Lawrence KA, Karlstetter M, Myers CA, Abdelaziz M, Dirkes W, Weigelt K, Seifert M, Benes V, Fritsche LG, et al. 2010. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res 20: 1512–1525. 10.1101/gr.109405.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelRosso N, Tycko J, Suzuki P, Andrews C, Aradhana N, Mukund A, Liongson I, Ludwig C, Spees K, Fordyce P, et al. 2023. Large-scale mapping and mutagenesis of human transcriptional effector domains. Nature 616: 365–372. 10.1038/s41586-023-05906-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenecker RJ, Griffith D, Holehouse AS. 2021. Metapredict: a fast, accurate, and easy-to-use predictor of consensus disorder and structure. Biophys J 120: 4312–4319. 10.1016/j.bpj.2021.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer J, Notin P, Dias M, Gomez A, Min JK, Brock K, Gal Y, Marks DS. 2021. Disease variant prediction with deep generative models of evolutionary data. Nature 599: 91–95. 10.1038/s41586-021-04043-8 [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick J-AS, Duncan A, et al. 1997. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91: 543–553. 10.1016/S0092-8674(00)80440-7 [DOI] [PubMed] [Google Scholar]
- Freund CL, Wang Q-L, Chen S, Muskat BL, Wiles CD, Sheffield VC, Jacobson SG, Mclnnes RR, Zack DJ, Stone EM. 1998. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet 18: 311–312. 10.1038/ng0498-311 [DOI] [PubMed] [Google Scholar]
- Friedman RZ, Granas DM, Myers CA, Corbo JC, Cohen BA, White MA. 2021. Information content differentiates enhancers from silencers in mouse photoreceptors. eLife 10: e67403. 10.7554/eLife.67403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse AS, Kragelund BB. 2024. The molecular basis for cellular function of intrinsically disordered protein regions. Nat Rev Mol Cell Biol 25: 187–211. 10.1038/s41580-023-00673-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holehouse AS, Ginell GM, Griffith D, Böke E. 2021. Clustering of aromatic residues in prion-like domains can tune the formation, state, and organization of biomolecular condensates. Biochemistry 60: 3566–3581. 10.1021/acs.biochem.1c00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Xiao X, Li S, Jia X, Wang P, Guo X, Zhang Q. 2012. CRX variants in cone–rod dystrophy and mutation overview. Biochem Biophys Res Commun 426: 498–503. 10.1016/j.bbrc.2012.08.110 [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Huang Y, Hanna DB, Freund CL, Affatigato LM, Carr RE, Zack DJ, Stone EM, McInnes RR. 1998. Retinal degenerations with truncation mutations in the cone-rod homeobox (CRX) gene. Invest Ophthalmol Vis Sci 39: 2417–2426. [PubMed] [Google Scholar]
- Jonas F, Carmi M, Krupkin B, Steinberger J, Brodsky S, Jana T, Barkai N. 2023. The molecular grammar of protein disorder guiding genome-binding locations. Nucleic Acids Res 51: 4831–4844. 10.1093/nar/gkad184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langstein-Skora I, Schmid A, Emenecker RJ, Richardson MOG, Götz MJ, Payer SK, Korber P, Holehouse AS. 2022. Sequence- and chemical specificity define the functional landscape of intrinsically disordered regions. bioRxiv 10.1101/2022.02.10.480018 [DOI]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew R, Heinz N, Hampf M, Bujard H, Gossen M. 2010. Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol 10: 81. 10.1186/1472-6750-10-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes S, Deploey N, Peelman F, Eyckerman S. 2023. Deep mutational scanning of proteins in mammalian cells. Cell Rep Methods 3: 100641. 10.1016/j.crmeth.2023.100641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T. 2020. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science (New York, NY) 367: 694–699. 10.1126/science.aaw8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. 2006. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics 7: 339. 10.1186/1471-2105-7-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindel V, Brodsky S, Cohen A, Manadre W, Jonas F, Carmi M, Barkai N. 2023. Intrinsically disordered regions of the Msn2 transcription factor encode multiple functions using interwoven sequence grammars. Nucleic Acids Res 52: 2260–2272. 10.1093/nar/gkad1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols LL, Alur RP, Boobalan E, Sergeev YV, Caruso RC, Stone EM, Swaroop A, Johnson MA, Brooks BP. 2010. Two novel CRX mutant proteins causing autosomal dominant Leber congenital amaurosis interact differently with NRL. Hum Mutat 31: E1472–E1483. 10.1002/humu.21268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–1289. 10.1016/j.cell.2008.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G-H, Chen S. 2005. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci 22: 575–586. 10.1017/S0952523805225063 [DOI] [PubMed] [Google Scholar]
- Peng G-H, Ahmad O, Ahmad F, Liu J, Chen S. 2005. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet 14: 747–764. 10.1093/hmg/ddi070 [DOI] [PubMed] [Google Scholar]
- Perrault I, Hanein S, Gerber S, Barbet F, Dufier J-L, Munnich A, Rozet J-M, Kaplan J. 2003. Evidence of autosomal dominant Leber congenital amaurosis (LCA) underlain by a CRX heterozygous null allele. J Med Genet 40: e90. 10.1136/jmg.40.7.e90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzon-Guzman C, Zhang SS-M, Barnstable CJ. 2011. Specific protein kinase C isoforms are required for rod photoreceptor differentiation. J Neurosci 31: 18606–18617. 10.1523/JNEUROSCI.2578-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Yeh BT, Feigerle JT, Hao CV, Townshend RJ, Lieberman Aiden E, Dror RO, Kornberg RD. 2021. Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to Mediator. eLife 10: e68068. 10.7554/eLife.68068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubach M, Maass T, Nazaretyan L, Röner S, Kircher M. 2024. CADD v1.7: using protein language models, regulatory CNNs and other nucleotide-level scores to improve genome-wide variant predictions. Nucleic Acids Res 52: D1143–D1154. 10.1093/nar/gkad989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherdson JL, Friedman RZ, Zheng Y, Sun C, Oh IY, Granas DM, Cohen BA, Chen S, White MA. 2024. Pathogenic variants in CRX have distinct cis-regulatory effects on enhancers and silencers in photoreceptors. Genome Res 34: 243–255. 10.1101/gr.278133.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, McInnes RR, Daiger SP. 1998. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet 63: 1307–1315. 10.1086/302101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohocki MM, Daiger SP, Bowne SJ, Rodriquez JA, Northrup H, Heckenlively JR, Birch DG, Mintz-Hittner H, Ruiz RS, Lewis RA, et al. 2001. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat 17: 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto LF, Li Z, Santoso CS, Berenson A, Ho I, Shen VX, Yuan S, Fuxman Bass JI. 2022. Compendium of human transcription factor effector domains. Mol Cell 82: 514–526. 10.1016/j.molcel.2021.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller MV, Ramirez E, Kotha SR, Holehouse AS, Pappu RV, Cohen BA. 2022. Directed mutational scanning reveals a balance between acidic and hydrophobic residues in strong human activation domains. Cell Syst 13: 334–345.e5. 10.1016/j.cels.2022.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straathof KC, Pulè MA, Yotnda P, Dotti G, Vanin EF, Brenner MK, Heslop HE, Spencer DM, Rooney CM. 2005. An inducible caspase 9 safety switch for T-cell therapy. Blood 105: 4247–4254. 10.1182/blood-2004-11-4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain P, Kumar S, Patel D, Richong S, Oberoi P, Ghosh M, Swaroop A. 2007. Mutations associated with retinopathies alter mitogen-activated protein kinase-induced phosphorylation of neural retina leucine-zipper. Mol Vis 13: 1114–1120. [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Wang Q-L, Wu W, Cook J, Coats C, Xu S, Chen S, Zack DJ, Sieving PA. 1999. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum Mol Genet 8: 299–305. 10.1093/hmg/8.2.299 [DOI] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. 2010. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci 11: 563–576. 10.1038/nrn2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Greenblatt MS, Harrison SM, Nussbaum RL, Prabhu SA, Boucher KM, Biesecker LG. 2018. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet Med 20: 1054–1060. 10.1038/gim.2017.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Chen S. 2014. Mechanisms of blindness: animal models provide insight into distinct CRX-associated retinopathies. Dev Dyn 243: 1153–1166. 10.1002/dvdy.24151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Zhang A, Zhang X, Huecker JB, Hennig AK, Chen S. 2014. Mechanistically distinct mouse models for CRX-associated retinopathy. PLoS Genet 10: e1004111. 10.1371/journal.pgen.1004111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Kwasnieski JC, Myers CA, Shen SQ, Corbo JC, Cohen BA. 2016. A simple grammar defines activating and repressing cis-regulatory elements in photoreceptors. Cell Rep 17: 1247–1254. 10.1016/j.celrep.2016.09.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS, Guenther B, Desplan C, Kuriyan J. 1995. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell 82: 709–719. 10.1016/0092-8674(95)90468-9 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Sun C, Zhang X, Ruzycki P, Chen S. 2023. Missense mutations in CRX homeodomain cause dominant retinopathies through two distinct mechanisms. eLife 12: RP87147. 10.7554/eLife.87147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.