Abstract
It has long been presumed impossible to measure telomeres in vertebrate DNA by PCR amplification with oligonucleotide primers designed to hybridize to the TTAGGG and CCCTAA repeats, because only primer dimer-derived products are expected. Here we present a primer pair that eliminates this problem, allowing simple and rapid measurement of telomeres in a closed tube, fluorescence-based assay. This assay will facilitate investigations of the biology of telomeres and the roles they play in the molecular pathophysiology of diseases and aging.
INTRODUCTION
The traditional method of measuring telomere length in samples of total human genomic DNA determines a mean terminal restriction fragment (TRF) length (1). The method requires large amounts of DNA (0.5–5 µg/individual) and time (3–5 days). Furthermore, the relative mean TRF lengths of individuals can vary by as much as 5% depending on the particular restriction enzymes used, suggesting the existence of subtelomeric restriction site polymorphisms and/or subtelomeric length polymorphisms that may confound the identification of primary factors accounting for inter-individual variation in the mean length of the true telomeric repeat sequence. More recently, methods have been developed that allow multiple samples to be compared for their relative content of just the telomeric hexamer repeat itself (2–5). However, none of these methods is as simple and as amenable to rapid high throughput processing of large numbers of samples as the method presented below.
Our strategy for determining relative telomere lengths by quantitative PCR was to measure, for each DNA sample, the factor by which the sample differed from a reference DNA sample in its ratio of telomere repeat copy number to single copy gene copy number. This ratio should be proportional to the average telomere length. The quantity of telomere repeats in each experimental sample was measured as the level of dilution of an arbitrarily chosen reference DNA sample that would make the experimental and reference samples equivalent with regard to the number of cycles of PCR needed to generate a given amount of telomere PCR product during the exponential phase of PCR amplification. Similarly, the relative quantity of the single copy gene in each experimental sample was expressed as the level of dilution of the reference DNA sample needed to match it to the experimental sample with regard to the number of cycles of PCR needed to generate a given amount of single copy gene PCR product during the exponential phase of the PCR. For each experimental sample the ratio of these dilution factors is the relative telomere to single copy gene (T/S) ratio. Thus T/S = 1 when the unknown DNA is identical to the reference DNA in its ratio of telomere repeat copy number to single copy gene copy number. The reference DNA sample (to which all of the experimental samples in a given study are compared) can be from a single individual or it can be a pooled sample from multiple individuals. The T/S ratio of one individual relative to the T/S ratio of another should correspond to the relative telomere lengths of their DNA.
MATERIALS AND METHODS
Research subjects
Genomic DNA was extracted directly from blood samples by standard procedures. The samples used to compare quantitative PCR versus Southern blot approaches to telomere measurement were donated by 95 individuals (47 females and 48 males, age range 5–94 years) from Utah families that are part of the Centre pour les Etudes du Polymorphisme Humaine (CEPH) collection used world wide to build the human genetic linkage map (6). Purified DNA samples were diluted in 96-well microtiter source plates to ∼1.75 ng/µl in 10 mM Tris–HCl, 0.1 mM EDTA, pH 7.5 (TE–4, final volume 300 µl/well), heated to 95°C for 5 min in a thermal cycler, quick chilled by transfer to an ice/water bath for 5 min, centrifuged briefly at 730 g, sealed with adhesive aluminum foil and stored at 4°C until the time of assay.
Quantitative PCR
Real time kinetic quantitative PCR determines, for each sample well, the Ct, i.e. the fractional cycle number at which the well’s accumulating fluorescence crosses a set threshold that is several standard deviations above baseline fluorescence (7). A plot of Ct versus log(amount of input target DNA) is linear, allowing simple relative quantitation of unknowns by comparison to a standard curve derived from amplification, in the same plate, of serial dilutions of a reference DNA sample. For this study, telomere (T) PCRs and single copy gene (S) PCRs were always performed in separate 96-well plates. Repeated measures of the T/S ratio in the same DNA sample gave the lowest variability when the sample well position (i.e. row and column coordinates) for T PCR on the first plate matched its well position for S PCR on the second plate.
Two master mixes of PCR reagents were prepared, one with the T primer pair, the other with the S primer pair. Thirty microliters of T master mix was added to each sample well and standard curve well of the first plate and 30 µl of S master mix was added to each sample well and standard curve well of the second plate. For each individual in whom the T/S ratio was assayed, three identical 20 µl aliquots of the DNA sample (35 ng/aliquot) were added to plate 1 and another three aliquots were added to the same well positions in plate 2. For each standard curve, one reference DNA sample was diluted serially in TE–4 by ∼1.68-fold per dilution to produce five concentrations of DNA ranging from 0.63 to 5 ng/µl, which were then distributed in 20 µl aliquots to the standard curve wells on each plate. The plates were then sealed with a transparent adhesive cover, centrifuged briefly at 730 g and stored at 4°C in the dark until the PCR was performed (0–3 days later).
The composition of T and S PCRs were identical except for the oligonucleotide primers. The final concentrations of reagents in the PCR were 150 nM 6-ROX and 0.2× Sybr Green I (Molecular Probes), 15 mM Tris–HCl pH 8.0, 50 mM KCl, 2 mM MgCl2, 0.2 mM each dNTP, 5 mM DTT, 1% DMSO and 1.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems). The final telomere primer concentrations were: tel 1, 270 nM; tel 2, 900 nM. The final 36B4 (single copy gene) primer concentrations were: 36B4u, 300 nM; 36B4d, 500 nM. The primer sequences (written 5′→3′) were: tel 1, GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT; tel 2, TCCCGACTATCCCTATCCCTATCCCTATCCCTATCC-CTA; 36B4u, CAGCAAGTGGGAAGGTGTAATCC; 36B4d, CCCATTCTATCATCAACGGGTACAA. [The 36B4 gene, which encodes acidic ribosomal phosphoprotein PO, is located on chromosome 12 (8).]
All PCRs were performed on the Prism 7700 Sequence Detection System (Applied Biosytems, Foster City, CA), a thermal cycler equipped to excite and read emissions from fluorescent molecules during each cycle of the PCR. The thermal cycling profile for both amplicons began with a 95°C incubation for 10 min to activate the AmpliTaq Gold DNA polymerase. For telomere PCR, there followed 18 cycles of 95°C for 15 s, 54°C for 2 min. For 36B4 PCR, there followed 30 cycles of 95°C for 15 s, 58°C for 1 min. ABI’s SDS v.1.7 software was then used to generate the standard curve for each plate and to determine the dilution factors of standards corresponding to the T and S amounts in each sample.
Relative T/S ratios will reflect relative length differences in telomeric DNA only if the number of copies of S per cell that are effectively PCR-amplified is the same in all individuals being studied. To test whether PCR with the 36B4 primers met this requirement, we determined, by quantitative PCR, the relative ratio of 36B4 gene copies to β-globin gene copies in the experimental DNAs versus the reference DNA. The 36B4 PCR was as described above; the conditions for β-globin PCR were exactly as for 36B4, except that β-globin primers (HBG1, GCTTCTGACACAACTGTGTTCACTAGC; and HBG2, CACCAACTTCATCCACGTTCACC; both at a final concentration of 400 nM) were used instead of 36B4 primers. The relative ratio (36B4/β-globin) for all experimental DNAs in comparison with the reference DNA was ~1.0 (average = 1.0; range = 0.97 – 1.08), as expected, indicating that equal copy numbers of the 36B4 gene per cell were amplified in all DNA samples (data not shown).
Determination of mean TRF length
Mean TRF lengths were determined as described by Slagboom et al. (9) with minor modifications. Approximately 0.5 µg of purified whole blood DNA was digested to completion with HaeIII restriction endonuclease. Digested samples were then mixed with DNA size standards (phage λ DNA digested with HindIII and a 1 kb ladder ranging from 1 to 10 kb), electrophoresed through agarose gels and blotted to a nylon membrane. Blots were hybridized with a 32P-end-labeled oligonucleotide, (TTAGGG)7, washed to remove non-specifically bound probe, exposed to a phosphor plate for 1–5 days and scanned with a PhosphorImager (Molecular Dynamics). Blots were then stripped of the telomere probe, hybridized with a mixture of two radiolabeled oligos, one for the 1 kb ladder, the other specific for the 23 kb λ HindIII fragment, washed, exposed to a phosphor plate and scanned. The size standard images and telomere smear images were then superimposed to locate the positions of the size intervals within the telomere smears. Mean TRF length was then calculated (10) as mean TRF length = Σ(ODi)/Σ(ODi/Li), where ODi is total radioactivity above background in interval i and Li is the average length of i in base pairs. This entire procedure was performed twice, i.e. the two mean TRF length values determined on each individual were obtained from two independent experiments.
RESULTS
Primer design
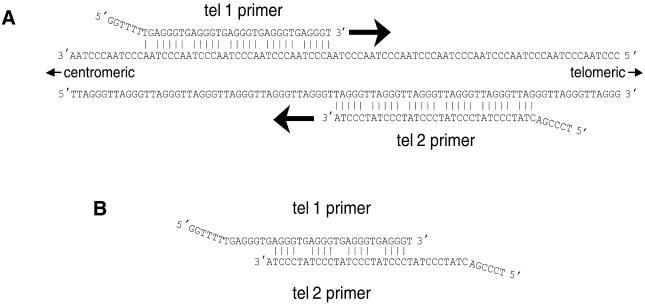
Figure 1 shows why the oligonucleotide primer pair designed for this study should specifically amplify telomeric hexamer repeats without generating primer dimer-derived products. Each primer is designed to allow DNA polymerase to extend from its 3′-end when it is hybridized to telomere hexamer repeats but not when it is hybridized to the other primer. Note also that the last six bases on the 5′-end of each primer cannot base pair with the telomere sequence when the rest of the primer is optimally hybridized. The complements of these 5′-sequences are generated at the 3′-ends of all products that are completed in each cycle of the PCR, thereby blocking those 3′-ends from initiating DNA synthesis in the middle of telomere amplification products in subsequent cycles.
Figure 1.
Annealing of primers tel 1 and tel 2 to genomic DNA and to each other in the first round of PCR. (A) Annealing of primers to genomic DNA. The tel 1 primer can hybridize to any available partially complementary 31 bp stretch along the strand of telomeric DNA oriented 5′→3′ toward the centromere. The tel 2 primer can hybridize to any partially complementary 33 bp stretch along the strand oriented 5′→3′ toward the end of the chromosome. In both of these primer–template hybridizations, every sixth base is mismatched, however, the last five bases at the 3′-end of the primers are perfectly matched to complementary bases in the template. Addition of bases by DNA polymerase begins at the 3′-ends of the annealed primers and proceeds in the direction of the large arrows. (B) Annealing of primers to each other. The strongest possible hybridizations of the primers to each other involve a repeated pattern of six bases containing four consecutive paired bases followed by two mismatched bases, an example of which is shown here. Note that the 3′-terminal base of each primer cannot form a stable base pair with the base opposite it, thereby blocking addition of bases by DNA polymerase.
Agarose gel electrophoresis of the telomere PCR product
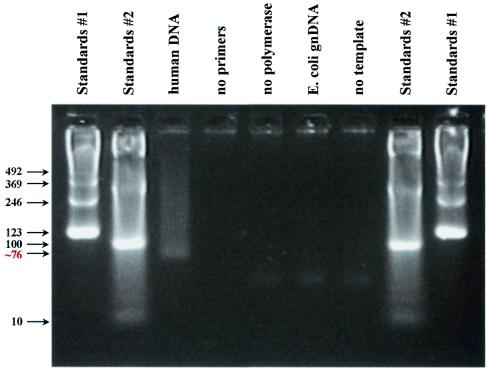
In the presence of 35 ng human DNA, telomere PCR product was detectable by Sybr Green I staining in the ABI Prism 7700 Sequence Detection System beginning around nine cycles of PCR. After 22 cycles of telomere PCR, the product was visualized by agarose gel electrophoresis, ethidium bromide staining and UV transillumination as a smear beginning with greatest intensity at the smallest predicted possible size, 76 bp (the sum of the lengths of the two primers), and progressively fading to background at ∼500 bp (Fig. 2). Note that products of the same length as the telomeres are not expected with this assay; rather, primarily the shortest possible product (76 bp) is expected, at a copy number proportional to the number of sites available for primer binding in cycle 1 of the PCR (and, therefore, proportional to total telomere length). When the genomic DNA template was omitted from the telomere PCR and when Escherichia coli DNA (a genome without telomeres) was substituted for human DNA, no product was detectable out to 22 cycles of PCR (Fig. 2).
Figure 2.
Agarose gel electrophoresis following PCR with the tel 1 and tel 2 primers. PCR was as described in Materials and Methods, except that 22 cycles of PCR were performed instead of 18 cycles. Eight microliters of reaction product were then loaded into each lane of a 4% (Nusieve 3:1) agarose gel and electrophoresis carried out at ∼2.5 V/cm in 45 mM Tris–borate, 1 mM EDTA with ethidium bromide present at a concentration of 0.5 µg/ml. The gel was then transilluminated with UV light and digital photography was performed with the Stratagene EagleEye System. Lanes 1, 2, 8 and 9, size standards. For lane 3 the template for PCR was 35 ng of total human genomic DNA. The remaining samples differed from the lane 3 sample as follows: lane 4, no primers; lane 5, no polymerase; lane 6, E.coli genomic DNA as template instead of human genomic DNA; lane 7, no template. The bottom of the smear in lane 3 was estimated to contain a 76 bp product, based on a comparison of its mobility with the mobilities of the size standards in a plot of log(number of base pairs) versus distance migrated from the origin.
Any autosomal single copy gene can serve as the basis for normalizing the telomere quantitative PCR signal. We chose the 36B4 gene, because it has already been validated for gene dosage studies (8). In the presence of 35 ng of human DNA, 30 cycles of 36B4 PCR yielded only the expected 74 bp product. This single copy gene PCR yielded no product when genomic DNA was omitted from the PCR and when E.coli DNA was substituted for human DNA.
Reproducibility of T/S ratio measurements
To test the reproducibility of the T/S ratio from well to well spatially across the 96-well array, 35 ng of a single individual’s DNA sample was added to all 96 wells of each of two plates and the PCRs carried out as above. Since the amount of the PCR product approximately doubles in each cycle of the PCR, the T/S ratio is approximately [2Ct(telomeres)/2Ct(36B4)]–1 = 2–ΔCt. The average ΔCt was –9.05, i.e. the single copy gene PCR needs about nine more cycles of PCR than the telomere PCR in order to produce as much fluorescent signal as the telomere PCR; the standard deviation of ΔCt was 1.48%. The relative T/S ratio (T/S of one sample relative to the T/S of another sample) is ![]() . Using this formula, a relative T/S ratio for each of the 96 wells was determined, as the T/S of the well relative to the mean T/S for all 96 wells. The standard deviation of this relative T/S ratio (reflecting the well-to-well variation in T/S across the 96-well plate) was 9.4%.
. Using this formula, a relative T/S ratio for each of the 96 wells was determined, as the T/S of the well relative to the mean T/S for all 96 wells. The standard deviation of this relative T/S ratio (reflecting the well-to-well variation in T/S across the 96-well plate) was 9.4%.
To test the reproducibility of T/S quantitation at the same well position in pairs of T and S PCR plates over time, five pairs of T/S PCRs on the same DNA sample were carried out at each of three different well positions. Wells with a serially diluted reference DNA were included on each plate so that relative quantities of T and S could be determined from standard curves. The standard deviation for the T/S ratio for the five paired PCRs done at each well position was determined. The average of the standard deviations at the three well positions was 6.9%.
Correlation between mean TRF lengths and relative T/S ratios
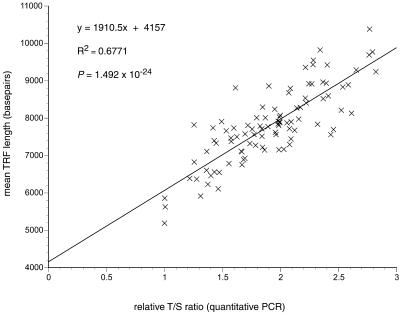
Relative T/S ratios in DNA samples from 95 individuals (47 females and 48 males, age range 5–94 years; 6) were measured by this assay by the standard curve method (see standard curves, Fig. 3) and compared with relative mean TRF lengths in the same samples (Fig. 4) measured by traditional Southern blot methods (9). Figure 5 shows the strong correlation between these two very different approaches to measuring telomeres: by linear regression analysis the correlation coefficient, r2, for the relationship of T/S ratio to TRF length was 0.677 and the P value was 1.4915 × 10–24.
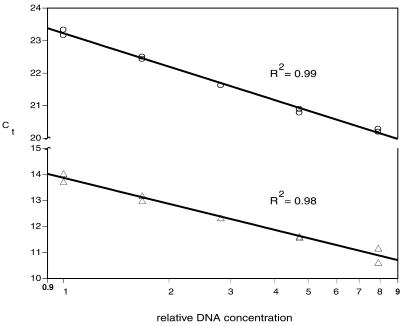
Figure 3.
Standard curves used to measure the relative T/S ratio. Five DNA concentrations over an 8-fold range were generated by serial dilution (dilution factor ∼1.68) and aliquoted to microtiter plate wells; the final amounts per well ranged from 12.64 to 100 ng, with the middle quantity approximately matching that of the samples being assayed. The Ct of a DNA sample is the fractional number of PCR cycles to which the sample must be subjected in order to accumulate enough product to cross a set threshold of magnitude of fluorescent signal. Any individual or pooled human DNA sample may be used to create the standard curves, as long as the Ct of each assayed sample falls within the range of Ct values of the standard curves. Circles, single copy gene 36B4; triangles, telomere.
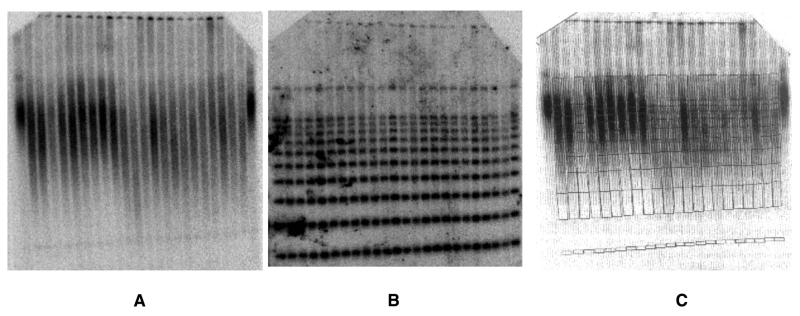
Figure 4.
Determination of mean terminal restriction fragment lengths in human DNA samples spiked with DNA size standards. The size standards, a 1 kb ladder ranging from 1 to 10 kb and HindIII-digested phage λ DNA, were added to each individual’s DNA sample after complete restriction enzyme digestion of the human genomic DNA and heat inactivation of the restriction enzyme. (A) Southern blot hybridized with a 32P-end-labeled (TTAGGG)7 oligonucleotide probe. (B) The same blot after stripping the telomere repeat probe and rehybridizing with a mixture of two radiolabeled oligonucleotides, one that binds all components of the 1 kb ladder, the other specific for the 23.13 kb λ HindIII fragment. (C) A grid image of the standard ladders of (B), generated using ImageQuaNT software tools (Molecular Dynamics), was superimposed on the image of the telomere smears of (A) to locate the positions of the size intervals within the smears.
Figure 5.
Correlation of relative T/S ratios determined by quantitative PCR and mean TRF lengths determined by Southern blot analysis in DNA samples prepared directly from blood draws on 95 individuals. All relative T/S ratios plotted here have values ≥1.0, because the initial T/S ratios determined using the standard curves have all been normalized to the lowest T/S ratio (0.69) observed among the samples. The equation for the linear regression line best fitting the data is shown.
For the triplicate measurements of relative T/S ratios in these 95 samples, the proportion of the total variation attributable to experimental error (1 – r2, where r2 is the correlation coefficient) was 13.1%, and for the duplicate measurements of mean TRF length it was 3.4%. The coefficient of variation (average standard deviation) for the relative T/S ratios was 5.8%. Assuming a normal distribution for relative T/S ratios in repeated measurements of the same sample, samples differing in average telomere length by as little as 11.4% (1.96 × SD) should be distinguishable by this method at the 95% confidence level.
DISCUSSION
The correlation between relative T/S ratios measured by quantitative PCR and relative TRF lengths measured by the traditional Southern blot approach in these 95 genomic DNA samples (Fig. 5) strongly supports the conclusion that the new PCR method does indeed measure relative telomere lengths.
Individuals that have the same mean length of terminal hexamer repeat array may nevertheless have very different mean TRF lengths, due to restriction site polymorphisms and/or length polymorphisms in the subtelomeric regions that determine the length of the subtelomeric portion of the TRFs. Early studies showed that the mean length of this subtelomeric portion of the TRFs in unrelated individuals ranged from 2.5 to 6 kb (reviewed in 11, p. 958). In our dataset we estimate the average value for the mean length of the subtelomeric portion of the TRFs to be ∼4.2 kb, based on the value of the y-intercept in Figure 5 (where T/S goes to 0); furthermore, we estimate the mean length of this non-telomeric DNA in the TRFs to vary by up to 2 kb between individuals, based on the range of mean TRF lengths we observed in individuals sharing nearly the same T/S ratio (see Fig. 5). Similarly, in comparing TRF length measurements with telomeric DNA measurements by quantitative fluorescence in situ hybridization (Q-FISH) with a fluorescein-labeled peptide nucleic acid (CCCTAA)3 probe, Hultdin et al. (12) estimated the mean length of the subtelomeric portion of the TRFs to be 3.2 kb, and they observed at least a 2 kb variation in the mean TRF lengths of individuals sharing nearly the same level of Q-FISH telomere signal (12) (Fig. 4).
Furthermore, we have found that the value measured for the difference in mean TRF length between two DNA samples can be greatly affected by the choice of restriction enzyme(s) used to release the terminal restriction fragments; for example, a 590 bp mean TRF length difference between two siblings following complete digestion with a combination of RsaI + HinfI decreased to only a 170 bp difference when HaeIII was used instead (R. M. Cawthon, unpublished observations). Therefore, investigators aiming to identify primary factors accounting for inter-individual variation in the mean length of the true telomeric repeat sequence may do well to avoid measuring TRF lengths and focus instead on methods that determine the relative quantities of the telomeric hexamer repeats per se.
The methods published to date for the relative quantitation of telomeric DNA per se in cell-free DNA preparations all normalize the telomere signal to another cellular DNA signal of high copy number, either centromeric alphoid DNA (3,4) or Alu repetitive DNA (5). However, the level of inter-individual variation in the copy number of centromeric alphoid DNA and Alu DNA sequences per cell is not known; if such variation is significant, then differences between individuals in relative telomere lengths measured by these methods may be quite inaccurate. By normalizing the quantity of telomere repeats to the quantity of a single copy gene, the PCR method of telomere measurement presented here avoids this problem.
In conclusion, we have demonstrated measurement of relative average telomere lengths by quantitative PCR in a closed tube, fluorescence-based assay, using a carefully designed pair of oligonucleotide primers. In this assay the telomere signal is normalized to the signal from a single copy gene to generate a T/S ratio. The resolution of the assay (detecting differences >∼11.4%) should be adequate for many genetic and epidemiological studies, since T/S values vary ∼2.5 fold among age- and sex-matched individuals (data not shown). The assay is simple, rapid and readily scalable to achieve a high throughput of samples. It should prove useful in the investigation of the biology of telomeres and the roles they play in the molecular pathophysiology of multiple diseases and aging.
Acknowledgments
ACKNOWLEDGEMENTS
I thank Mark Leppert for access to the Utah CEPH DNA samples, Richard Kerber and Ken Smith for guidance with statistical analysis and Robert Weiss, Jeff Stevens, Shannon Odelberg, Hilary Coon and Elizabeth O’Brien for helpful discussions. This work was supported by the Allied Signal Award for Research on Aging and by NIH grants K01 AG00767, R29CA69421 and AG13478.
REFERENCES
- 1.Allshire R.C., Dempster,M. and Hastie,N.D. (1989) Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res., 17, 4611–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lansdorp P.M., Verwoerd,N.P., van de Rijke,F.M., Dragowska,V., Little,M.T., Dirks,R.W., Raap,A.K. and Tanke,H.J. (1996) Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet., 5, 685–691. [DOI] [PubMed] [Google Scholar]
- 3.Bryant J.E., Hutchings,K.G., Moyzis,R.K. and Griffith,J.K. (1997) Measurement of telomeric DNA content in human tissues. Biotechniques, 23, 476–478, 480, 482, passim. [DOI] [PubMed] [Google Scholar]
- 4.Norwood D. and Dimitrov,D.S. (1998) Sensitive method for measuring telomere lengths by quantifying telomeric DNA content of whole cells. Biotechniques, 25, 1040–1045. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura Y., Hirose,M., Matsuo,H., Tsuyama,N., Kamisango,K. and Ide,T. (1999) Simple, rapid, quantitative and sensitive detection of telomere repeats in cell lysate by a hybridization protection assay. Clin. Chem., 45, 1718–1724. [PubMed] [Google Scholar]
- 6.White R., Leppert,M., Bishop,D.T., Barker,D., Berkowitz,J., Brown,C., Callahan,P., Holm,T. and Jerominski,L. (1985) Construction of linkage maps with DNA markers for human chromosomes. Nature, 313, 101–105. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 8.Boulay J.L., Reuter,J., Ritschard,R., Terracciano,L., Herrmann,R. and Rochlitz,C. (1999) Gene dosage by quantitative real-time PCR. Biotechniques, 27, 228–230, 232. [DOI] [PubMed] [Google Scholar]
- 9.Slagboom P.E., Droog,S. and Boomsma,D.I. (1994) Genetic determination of telomere size in humans: a twin study of three age groups. Am. J. Hum. Genet., 55, 876–882. [PMC free article] [PubMed] [Google Scholar]
- 10.Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- 11.Levy M.Z., Allsopp,R.C., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Telomere end-replication problem and cell aging. J. Mol. Biol., 225, 951–960. [DOI] [PubMed] [Google Scholar]
- 12.Hultdin M., Gronlund,E., Norrback,K., Eriksson-Lindstrom,E., Just,T. and Roos,G. (1998) Telomere analysis by fluorescence in situ hybridization and flow cytometry. Nucleic Acids Res., 26, 3651–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]