Abstract
Background:
Initial insights into oncology clinical trial outcomes are often gleaned manually from conference abstracts. We aimed to develop an automated system to extract safety and efficacy information from study abstracts with high precision and fine granularity, transforming them into computable data for timely clinical decision-making.
Methods:
We collected clinical trial abstracts from key conferences and PubMed (2012–2023). The SEETrials system was developed with three modules: preprocessing, prompt engineering with knowledge ingestion, and postprocessing. We evaluated the system’s performance qualitatively and quantitatively and assessed its generalizability across different cancer types— multiple myeloma (MM), breast, lung, lymphoma, and leukemia. Furthermore, the efficacy and safety of innovative therapies, including CAR-T, bispecific antibodies, and antibody-drug conjugates (ADC), in MM were analyzed across a large scale of clinical trial studies.
Results:
SEETrials achieved high precision (0.964), recall (sensitivity) (0.988), and F1 score (0.974) across 70 data elements present in the MM trial studies Generalizability tests on four additional cancers yielded precision, recall, and F1 scores within the 0.979–0.992 range. Variation in the distribution of safety and efficacy-related entities was observed across diverse therapies, with certain adverse events more common in specific treatments. Comparative performance analysis using overall response rate (ORR) and complete response (CR) highlighted differences among therapies: CAR-T (ORR: 88 %, 95 % CI: 84–92 %; CR: 95 %, 95 % CI: 53–66 %), bispecific antibodies (ORR: 64 %, 95 % CI: 55–73 %; CR: 27 %, 95 % CI: 16–37 %), and ADC (ORR: 51 %, 95 % CI: 37–65 %; CR: 26 %, 95 % CI: 1–51 %). Notable study heterogeneity was identified (>75 % I2 heterogeneity index scores) across several outcome entities analyzed within therapy subgroups.
Conclusion:
SEETrials demonstrated highly accurate data extraction and versatility across different therapeutics and various cancer domains. Its automated processing of large datasets facilitates nuanced data comparisons, promoting the swift and effective dissemination of clinical insights.
Keywords: Oncology clinical trial, Automated safety and efficacy extraction, Large scale analysis, Conference abstracts, GPT-4, Large language models
1. Introduction
Multiple myeloma (MM) remains an incurable malignancy, marked by elevated rates of relapse and therapy resistance despite significant advancements in treatment options and survival rates over the last 15 years [1]. Furthermore, the introduction of numerous new therapies has led to uncertainty in therapy selection and sequencing, compounded by limited consensus guidelines and recent systematic literature reviews [2, 3]. Effective clinical decision-making is supported by a robust evidence base, typically sourced from clinical trials [4]. The prompt dissemination of clinical trial findings, including both risks and benefits, is fundamental for informed patient care [5] and crucial in refining patient recruitment strategies for subsequent trial phases—particularly in urgent medical fields like oncology [6].
Conference abstracts often serve as crucial sources of initial findings, unveiling clinical trial results and providing insights into the efficacy and safety of investigational treatments. Yet, the initial disclosures via conference abstracts present retrieval and integration challenges, with over half of study outcomes and nearly a third of randomized trial results from these abstracts not advancing to full publication [7–9]. In addition, publication bias disproportionately excludes “not positive, positive but not statistically significant, or negative” results [10,11] despite their critical influence on clinical decision-making. Recognizing these issues, extracting data from both journal articles and conference abstracts is essential for thorough evidence synthesis and for facilitating robust comparisons. Nevertheless, the extraction of clinical outcomes for large-scale analysis can be formidable when relying solely on manual methods.
In the following chapters, we will review related work for automatic data extraction, outline the main contributions of our study, describe the methods for data collection, SEETrials system development, evaluation, and data analysis, and discuss results.
2. Literature review
Advancements in Natural Language Processing (NLP) have notably enhanced medical research by enabling the automatic extraction of data from unstructured texts [12–14]. Recently, large language models (LLMs) have demonstrated exceptional proficiency in contextual understanding and textual data processing [15–17]. Various LLMs, including generative pre-trained transformer (GPT) models, have been examined for information extraction from radiology clinical documents including tumor size, type, and status of invasion or metastasis [18–21], as well as for extracting social determinants of health [22], cognitive exam dates and scores [23], substance use disorder severity [24], and rare disease symptoms and signs [25]. In the clinical trial domain, LLMs show promise in trial information retrieval [26], criteria text generation [27], and clinical trial eligibility criteria analysis [28]. Several studies have explored the capability of LLMs in the automated extraction of treatment safety and efficacy outcome information [29–32] as summarized in Table 1. For instance, Tang et al. [29] and Kartchner et al. [32] extracted the study design, disease characteristics, intervention, comparator, and outcomes. Wang et al. [30] extracted treatment efficacy outcomes as part of an automated systematic literature review system, which included query expansion, article screening, data extraction, and data analysis modules. Although these studies demonstrated the potential of automatic data extraction and evidence generation in complicated clinical trial studies, the model performances, especially in extracting outcome data elements, need improvement. Gartlehner et al. [31] showed a high accuracy of 96.3 % with an F1 score of 0.98 in extracting trial study design and treatment outcome data elements, but this was tested in only 10 studies as a proof-of-concept. Moreover, the application of LLMs to extract comprehensive clinical trial study design and treatment outcomes from conference abstracts, especially with tables, remains largely unexplored.
Table 1.
Summary of studies utilizing LLMs for automated data extraction of treatment safety and efficacy outcomes.
| Author (Year) | Extracted Data Elements | Input Data Format | LLM Models | Results |
|---|---|---|---|---|
| Tang et al. (2024) | Study details, intervention, comparator, and treatment outcomes | Text, 100 abstracts of articles | GPT-3.5 and GPT-4 | The average accuracy of GPT-4 ranged from 0.688 to 0.964 Low performance in intervention (0.688) |
| Kartchner et al. (2023) | Study details, intervention, disease characteristics | Text, Articles. | ChatGPT and GPT-JT | ChatGPT (Accuracy: 0.141–0.956 for study details, interventions, disease characteristics data elements) outperformed GPT-JT in most data fields. The average Precision, Recall, and F1 score were 0.496, 0.431, 0.441, respectively Accuracy for some adverse event data elements was low (e. g., neutropenia, thrombocytopenia, adverse event grade) |
| Wang et al. (2024) | Treatment outcomes (7 efficacy related outcomes) | Text Table, 25 full articles | GPT-4 and Sonnet | Established a customized TrialMind model. The accuracy of the TrialMind model ranges from 0.65 to 0.84 in outcome extraction, indicating that extracting study outcome elements is challenging |
| Gartlehner et al. (2024) | Study details and treatment outcomes | Text, 10 articles | Claude 2 | An accuracy of 96.3 % with an F1 score of 0.98 across 10 study reports |
| SEETrials | Study details and treatment outcomes | Text & Table, 215 conference and article abstracts | GPT-4 | Established a customized SEETrials model. The overall precision, recall, and F1 scores were 0.958, 0.944, 0.951, respectively, across 70 data elements in MM studies. Generalizability tests across other cancers yielded precision, recall, and F1 scores within the range of 0.966–0.986 |
In this study, we present “SEETrials,” an LLM-based pipeline specifically designed to automatically extract detailed safety and efficacy outcome information. The main contributions of our study are summarized below.
Our SEETrials system automatically extracts detailed safety and efficacy outcome information with fine granularity and high accuracy across all data elements through prompt engineering, converting it into a computable format to facilitate downstream data analysis. This includes handling data extraction from tables, a critical capability that not only applies to conference abstracts but can also be lever-aged for extracting data from multiple tables within full-text articles.
By focusing on MM clinical trials and concentrating on three recent classes of interventions including chimeric antigen receptor T cell (CAR-T) therapy, bispecific antibodies (BsAbs) therapy, and antibody-drug conjugates (ADC) studies, we demonstrate the practical utility of our approach in enhancing clinical evidence generation for decision-making.
Our system shows its applicability across various cancer trial studies including both solid and blood cancers.
3. Materials and methods
3.1. Data
For the model development, evaluation, and data analysis, we collected a total of 245 MM clinical trial abstracts (2012–2023) from various drug groups, focusing on five different therapies. The sources included the American Society of Clinical Oncology (ASCO: https://ascopubs.org/jco/meeting), American Society of Hematology conferences (ASH: https://ashpublications.org/blood/issue/142/Supplement%201), and PubMed (https://pubmed.ncbi.nlm.nih.gov/) websites. The selection was based on keywords (“clinical trial” AND “each therapy”).
The therapies covered in this study are 1) chimeric antigen receptor T cell (CAR-T) therapy 2) Bispecific Antibodies (BsAbs) therapy 3) Antibody-drug conjugates (ADC) therapy 4) Others including Cereblon E3 ligase modulator therapy (CELMoD), Histone deacetylase inhibitor (HDACi) and immune checkpoint inhibitors (ICI). The breakdown of abstract numbers for each therapy and phase is presented in Supplement Table 1.
Of these, 93 abstracts concerning CAR-T, BsAbs, and ADC therapies were earmarked for in-depth quantitative comparison analysis. Exclusion criteria included studies without reported outcomes or those superseding initial results with expanded trial cohorts. A manual review was conducted to validate the extracted data for quantitative analysis.
An additional 115 abstracts across four other cancers—breast, lung, lymphoma, and leukemia—were collected to assess the system’s generalizability. This included acute lymphocytic leukemia (7), acute myelocytic leukemia (6), chronic lymphocytic leukemia (7), and chronic myeloid leukemia (5).
3.2. SEETrials
We developed SEETrials using GPT-4 to extract clinical trial details from annual conference presentations and published journal abstracts. The system’s architecture is delineated in Fig. 1, with each step detailed below. In summary, SEETrials is comprised of three modules, preprocessing, prompt engineering, and post-processing.
Fig. 1.
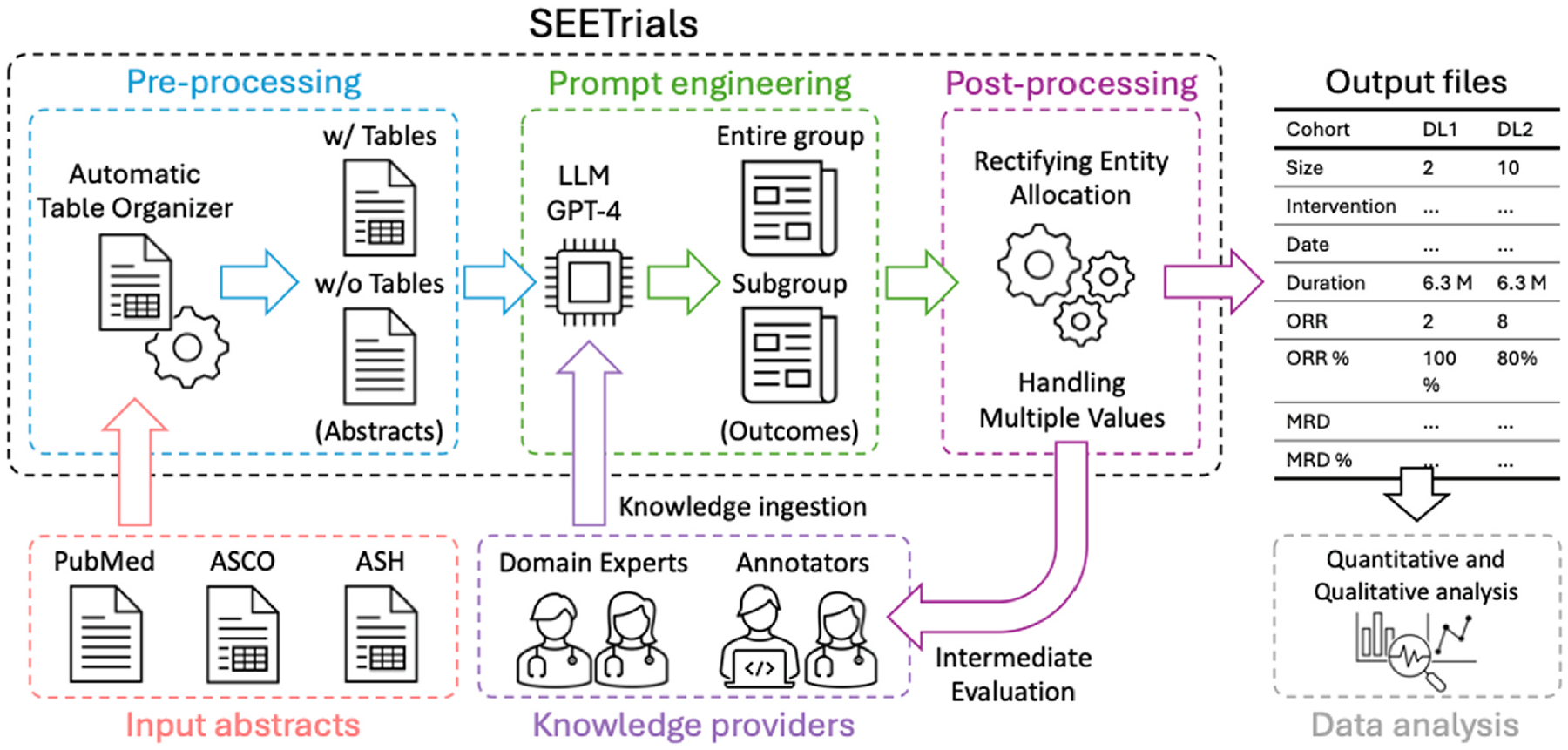
SEETrials system overview.
SEETrials is an autonomous system designed to extract critical details from clinical trial studies presented at annual conferences and published journal abstracts. Utilizing the capabilities of GPT-4, the system is delineated in subsequent sections, with a schematic overview of its components.
ASCO, American Society of Clinical Oncology; ASH, American Society of Hematology; LLM, Large language model; GPT, Generative Pre-trained Transformer; ORR, overall response rate; MRD, minimum residual disease.
3.2.1. Pre-processing
This initial module is responsible for the systematic collection of abstracts from the ASCO and ASH conferences, as well as the PubMed database. Notably, annual conferences like ASCO and ASH permitted the inclusion of table-format data within abstracts. Within this stage, any tables present in the conference abstracts are meticulously converted to a text format to facilitate further analysis.
3.2.1.1. Abstract collection.
PubMed enabled direct saving in text format. Abstracts from ASCO and ASH websites were initially captured in a Word file and then converted to a text file (UTF-8) to facilitate the loading of a substantial number of abstracts for automated data extraction.
3.2.1.2. Automatic table organizer.
To address the formatting challenges posed by documents containing tables (e.g., misaligned or unorganized tables), we introduced an “Automatic Table Organizer” step. This step realigns the columns and rows to ensure the tables are properly formatted.
3.2.1.3. Preparation of two abstract sets for one abstract with table.
After running the Automatic Table Organizer, we prepare two sets of abstracts for input into our GPT system: one set includes the organized tables and the other excludes the tables. This dual input enhances the accuracy of data extraction from both main text and tables.
3.2.2. Prompt engineering
The module focuses on creating tailored prompts that enable the comprehensive and precise extraction and consolidation of data. These prompts are designed to guide the LLM in effectively identifying and merging relevant trial outcomes.
3.2.2.1. Knowledge ingestion.
Prompts in specific areas often benefit from incorporating background knowledge within the prompt. To enhance the LLM’s analytical capabilities, we integrated background knowledge from oncology clinical trials such as definitions and examples of safety and efficacy outcomes into the prompt. Subject experts in this study encompassed various tasks: 1) identifying types of treatment efficacy and safety entities in cancer clinical trials, 2) formulating descriptions for the entire cohort, sub-cohorts, and each arm for different interventions or dosages specified in the prompts, 3) ensuring the clinical accuracy of the prompts, 4) providing feedback for prompt adjustments/calibration, and 5) manually reviewing system output to establish the gold standard for system validation.
3.2.2.2. Cohort/study group identification.
Clinical trial studies present various outcomes from different study groups, including the entire group, specific subgroups (e.g., those with specific biomarkers or risk factors), and individual cohorts. To enhance the accuracy of outcome extraction for each group, subgroup, and cohort, we employed two separate GPT prompts due to the token limit of the GPT-4 API model (8,192): one prompt for “Individual cohorts and entire group”. Another prompt for “ Individual cohorts and subgroup”
3.2.2.3. Combining process with two GPT outcomes.
To merge outputs from the “Individual cohorts and entire group” and “ Individual cohorts and subgroup”, we created an additional prompt. This prompt was designed to combine the extracted information and eliminate any overlaps, ensuring a comprehensive final output.
3.2.3. Post-processing
In the final post-processing module, the extracted outcome data are refined and structured to suit the requirements of subsequent data analysis tasks. This module addresses two key formatting challenges.
3.2.3.1. Handling undesired entity allocations.
Correct any undesired allocations of entities and their corresponding values in the outputs.
3.2.3.2. Handling multiple values in a single column.
In some cases, the value’s columns contained more than two values, like “9 patients (28 %). We try to ensure that each cell within the value’s column contains a singular value, optimizing the presentation for computational analysis.
3.3. Evaluation
The evaluation took place using two approaches: Quantitative and qualitative evaluation. For qualitative evaluation, predominantly appearing error types were evaluated. For Quantitative evaluation, precision, recall, and F1 scores as well as accuracy of SEETrials are reported for each abstract. Accuracy (Accuracy = TP + TN/TP + TN + FP + FN) measures the proportion of correct predictions, both True positive and True Negative. Precision ((Precision = TP/(TP + FP)) is calculated as the ratio of correctly predicted positive entities to the total predicted positive entities. Recall, also known as sensitivity, is calculated as the ratio of correctly predicted positive entities to all actual positive entities (Recall = TP/(TP + FN)). The F1-score is the harmonic mean of precision and recall and is calculated using the formula: F1-score = 2 × ((Precision × Recall)/(Precision + Recall)). In these equations, TP stands for true positives, FP stands for false positives, and FN stands for false negatives.
3.4. Data analysis
In the descriptive data analysis, we extensively scrutinized the distribution of entities and provided a quantitative overview, encompassing both the absolute number and the percentage of abstracts where each entity is mentioned out of the total abstracts analyzed for each therapy. This analysis was performed in conjunction with the categorization of entities based on different trial phases.
We also conducted the quantitative statistical analysis using R software (version 4.0.0), employing the meta, metasens, and metafor packages. A random-effects model was employed, weighting studies by the inverse variance method to favor large population studies. Logit transformations normalized the proportions of overall response rates (ORR), complete response (CR), Neutropenia (≥Grade 3), and Cytokine Release Syndrome (CRS) for effect size pooling. Subgroup analysis was performed for each therapy to investigate treatment effects comprehensively. Statistical heterogeneity among studies was assessed using the I2 inconsistency index and Cochran Q test. Effect sizes were reported with 95 % confidence to address variability. Two-sided P values with a significance level of P < 0.05 determined statistical relevance.
4. Results
4.1. Evaluation of the SEETrials
Our SEETrials system demonstrated high precision, recall (sensitivity), and F1 scores, achieving overall metrics of 0.964, 0.988, and 0.974, respectively, across all trial phases. The overall accuracy was 0.952. Supplement Table 2 displays the average performance scores for both the overall dataset and each phase of the MM studies. Comprehensive details, including the performance scores for individual abstracts, as well as true positive, false positive, and false negative numbers, are provided in Supplement Table 3.
To further validate SEETrials across a broader range of cancer types, we implemented our system on clinical trial reports from four other randomly chosen cancer types, including both solid cancers (lung and breast) and hematologic cancers (lymphoma and leukemia). The average precision, recall, F1 scores, and accuracy were 0. 979, 0.992, 0.985, 0.971(breast cancer), 0.988, 0.984, 0.985, 0.972(lung cancer), and 0.983, 0.988, 0.985, 0.971(leukemia/lymphoma), respectively. Supplement Table 4 presents the average performance scores for each cancer type, while Supplement Table 5 provides comprehensive details, including the performance scores for individual abstracts, along with the numbers of true positive, false positive, and false negative.
4.2. Qualitative error analysis
We conducted a qualitative error analysis on all documents with errors (68 from MM, 28 from Leukemia/Lymphoma, and 35 from Lung/Breast cancer studies). We classified errors into three categories, including extraction failures, cohort recognition errors, and inaccurate extractions, and quantified. Extraction failure was the most common error across all cancer studies, occurring in 55.9 % (n = 38) of MM, 60.7 % (n = 17) of Leukemia/Lymphoma, and 65.7 % (n = 23) of Lung/Breast cancer studies. Cohort recognition errors were found in 26.5 % (n = 18), 25 % (n = 7), and 14.3 % (n = 5), respectively. Inaccurate extraction occurred in 17.6 % (n = 12), 14.3 % (n = 4), and 20.2 % (n = 7) of studies, respectively (Supplement Table 6). Notably, no instances of hallucination were observed during the data extraction process. Supplement Table 7 provides illustrative examples of cohort recognition errors. Clinical trial study abstracts often contain diverse clinical information for multiple cohorts/arms, including dose-escalation, dose-expansion, recommended phase 2 dosage (RP2D) groups, entire groups, and subgroups. The system occasionally struggled to allocate values correctly due to the lack of explicit descriptions in the text. Additional errors included extracting the author’s affiliation as the study location in PubMed abstracts, cohort size discrepancies, and occasional extraction of only percentage values without other relevant information. Improvements are needed for consistent value extraction and allocation.
4.3. Analyzing efficacy and safety data from abstracts
We extracted data from 130 abstracts related to CAR-T, 63 to BsAbs, 38 to ADC, 6 to CELMoD, 4 to HDACi, and 4 to ICI therapies. Among these, there were 75, 47, 46, and 16 abstracts corresponding to phases 1, 1/2, 2, and 3. 61 abstracts did not have phase information. The distribution of clinical trial phases in each therapy is detailed in Supplement Table 1. The complete list of extracted data elements from all studies is detailed in Supplement Fig. 1.
Fig. 2 visually summarizes frequently appearing efficacy and safety-related entity percentages across the three therapies, CAR-T, BsAbs, and ADC. We present a comprehensive breakdown of trial numbers and the percentages of each efficacy-related and safety-related entity out of all mentioned entities (Figs. 3 and 4, respectively) categorized by clinical trial phases (phases 1, 1/2, 2, and 3) for both efficacy and safety-related entities.
Fig. 2.
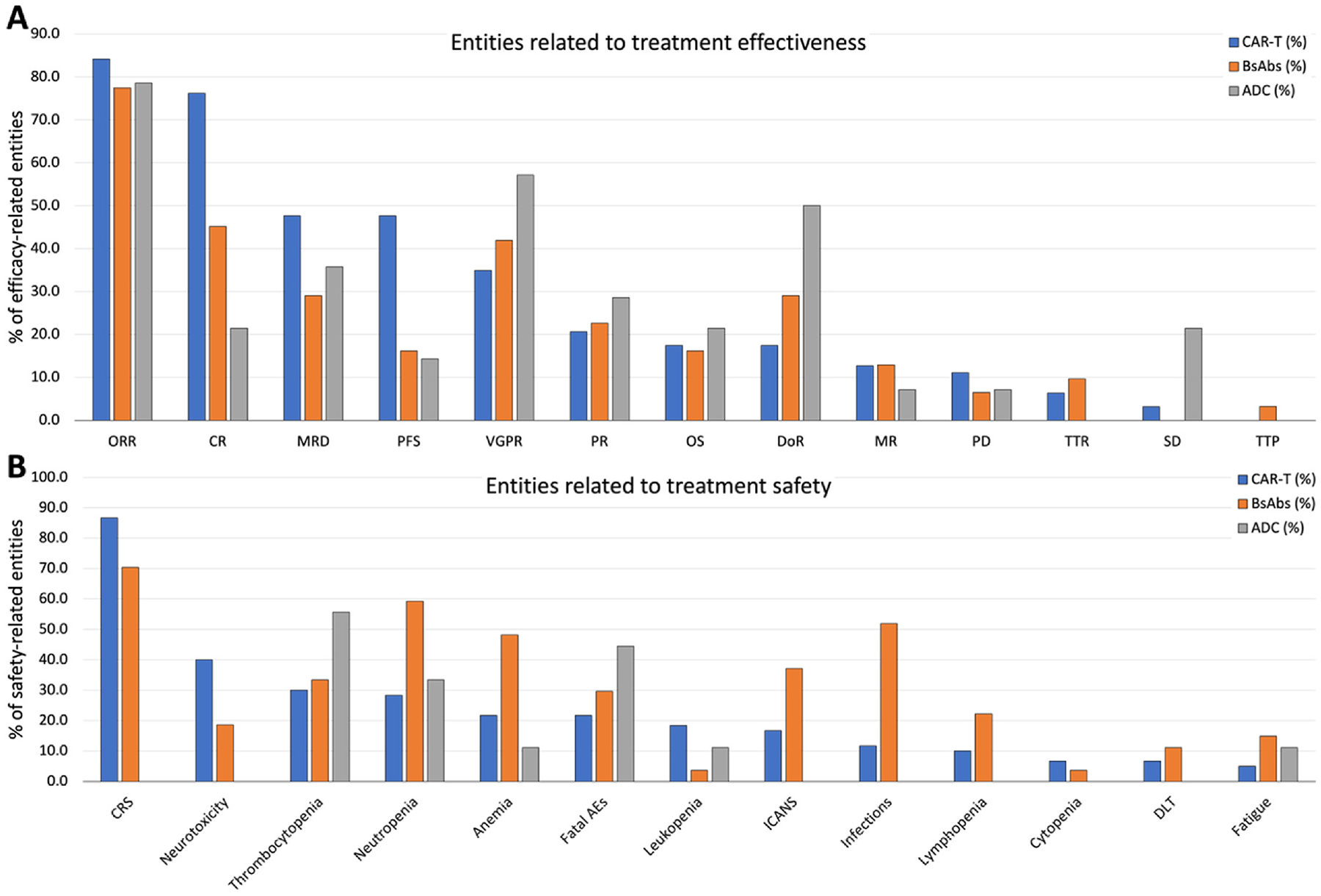
The comparative landscape of efficacy and safety entities across CAR-T, BsAbs, and ADC therapies. This visual summary illustrates the percentages of 11 efficacy and 13 safety-related entities across CAR-T cell therapy, BsAbs, and ADC therapies, providing a comprehensive overview of their comparative clinical profiles. A) Entities related to treatment effectiveness. B) Entities related to treatment safety.
CAR-T, chimeric antigen receptor T cell; BsAbs, Bispecific antibody; ADC, antibody-drug conjugate; ORR, overall response rate; CR, complete response; VGPR, very good partial response; PR, partial response; PFS, progression-free survival; MRD, minimum residual disease; OS, overall survivor; DoR, duration of response; SD, stable disease; PD, progressive disease; TTR, time to response; MR, minimal response; TTP, time to progress; TTTD, time to treatment discontinuation; TTTF, time to treatment failure; TTNT, time to next treatment; DCR, disease control rate; CRS, cytokine release syndrome; Aes, adverse events; ICANS, immune effector cell associated neurotoxicity syndrome.
Fig. 3.
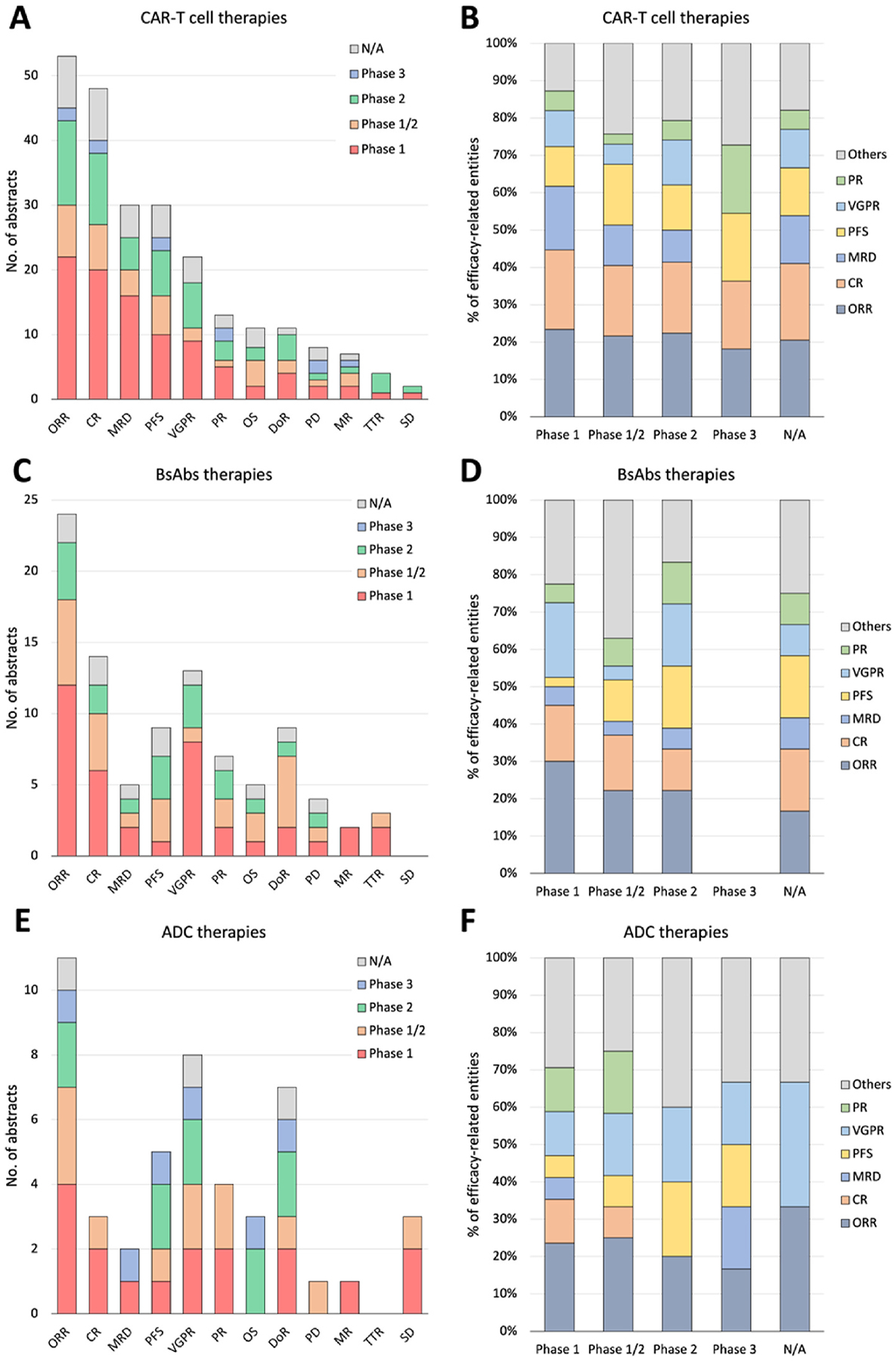
A detailed breakdown of abstract numbers with each efficacy-related entity (A, C, E) and percentages of each entity out of all mentioned entities (B, D, F) is presented, categorizing clinical trials into phases 1, 1/2, 2, and 3. A and B: CAR-T cell therapies. C and D: BsAbs therapies. E and F: ADC therapies.
CAR-T, chimeric antigen receptor T cell; BsAbs, Bispecific antibody; ADC, antibody-drug conjugate; ORR, overall response rate; CR, complete response; (VG)PR, (very good) partial response; PFS, progression-free survival; MRD, minimum residual disease; OS, overall survivor; DoR, duration of response; SD, stable disease; PD, progressive disease; TTR, time to response; MR, minimal response.
Fig. 4.
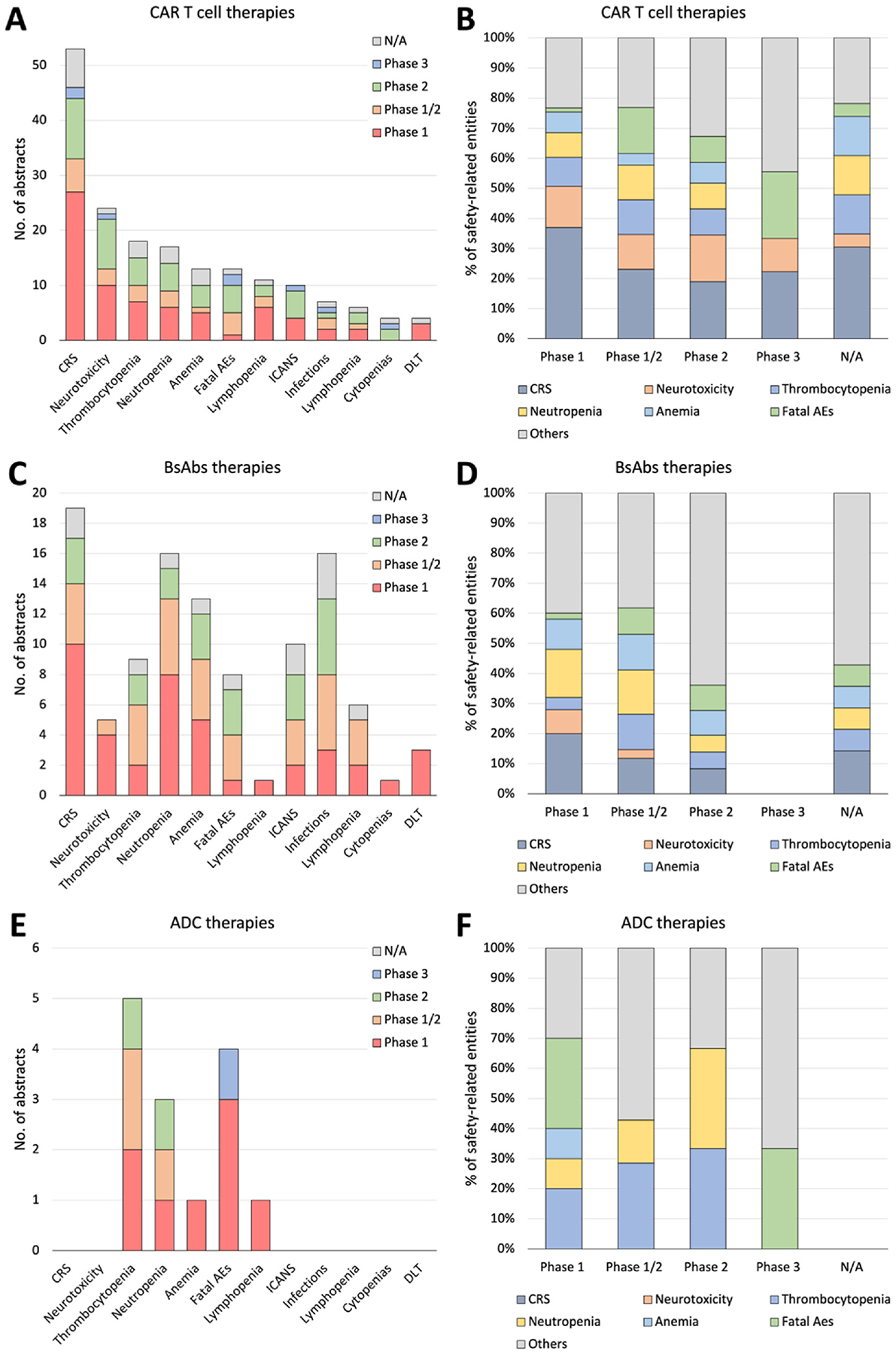
A detailed breakdown of abstract numbers with each safety-related entity (A, C, E) and percentages of each entity out of all mentioned entities (B, D, F) is presented, categorizing clinical trials into phases 1, 1/2, 2, and 3. A and B: CAR-T cell therapies. C and D: BsAbs therapies. E and F: ADC therapies.
CAR-T, chimeric antigen receptor T cell; BsAbs, Bispecific antibody; ADC, antibody-drug conjugate; CRS, cytokine release syndrome; Fatal AEs, fatal adverse events; ICANS, immune effector cell associated neurotoxicity syndrome; DLT, Dose Limiting Toxicity.
The ORR emerged as the most frequently reported entity in clinical trial abstracts, constituting 84.1 % in CAR-T, 77.4 % in BsAbs, and 78.6 % in ADC studies. CR was mentioned in 76.2 % of CAR-T and 45.2 % of BsAbs abstracts. Very good partial responses (VGPR) were mentioned in 57.1 % of ADC and 41.9 % of BsAbs abstracts (Supplement Table 8). Supplement Table 9 provides the percentage of abstracts mentioning each safety-related entity, categorized by phases and therapies. For ORR, 41.5 %, 50.0 %, and 36.4 % were from phase 1 studies, and 15.1 %, 25.0 %, and 27.3 % were from phase 1/2 studies for CAR-T, BsAbs, and ADC, respectively. Similarly, for VGPR, 40.9 %, 61.5 %, and 25.0 % were from phase 1 studies, and 31.8 %, 23.1 %, and 25 % were from phase 2 studies for CAR-T, BsAbs, and ADC, respectively.
In safety-related entity analysis, cytokine release syndrome (CRS) was prominently mentioned in CAR-T (86.7 % of abstracts) and BsAbs (70.4 % of abstracts) studies but not in ADC studies (0 %). Infection was frequently cited in BsAbs (51.9 %) compared to CAR-T (11.7 %) and ADC (0 %) abstracts. Conversely, thrombocytopenia was more commonly mentioned in ADC (55.6 %), compared to CAR-T (30 %) and BsAbs (33.3 %). Notably, Fatal adverse events (AEs) or Severe AEs more prevalently appeared in ADC (44.4 % and 44.4 %, respectively) abstracts compared to CAR-T (21.7 % and 3.3 %) and BsAbs (29.6 % and 7.4 %) (Supplement Table 10). When assessing the percentage of abstracts mentioning each safety-related entity by phases and therapies (Supplement Table 11), our data revealed that most safety-related entities such as CRS, neurotoxicity, and thrombocytopenia were mentioned more frequently in phase 1 studies compared to phase 1/2 or phase 2 studies, except for fatal AEs in CAR-T (7.7 %, 30.8 %, and 38.5 %, respectively) and BsAbs (12.5 %, 37.5 %, and 37.5 %, respectively).
For the comparative analysis of treatment outcome values among CAR-T, BsAbs, and ADC therapies, we included a total of 93 distinct studies identified using NCT IDs. For studies lacking NCT IDs, we manually examined the authors, their affiliations, and study titles. Of these, 87 were included in the efficacy comparison and 74 were included in the safety comparison. Supplement Table 12 provides a detailed breakdown of the trial numbers by therapies and phases. The degrees of study heterogeneity for ORR (I2 = 87 %, 97 %, and 96 %, respectively) (Supplement Fig. 2A) and CR (I2 = 90 %, 91 %, and 94 %, respectively) (Fig. 4B) were high (over 75 %) in all three therapies even after categorized by phases for ORR (Phase 1: I2 = 81 %, 76 %, and 85 %; Phase 2: I2 = 88 %, 97 %, and 95 %, respectively) (Fig. 5A and B) and for CR (Phase 1: I2 = 87 %, 39 %, and 96 %; Phase 2: I2 = 91 %, 96 %, and NA, respectively) (Fig. 5C and D) except phase 1 BsAbs studies (I2 = 39 %). The 95 % CIs of estimated ORR and CR were 84–92 % and 53–66 % (CAR-T), 55–73 % and 16–37 % (BsAbs), and 37–65 % and 1–51 % (ADC). We conducted a similar analysis on CRS (including studies mentioning all grades but excluding studies only mentioning grade ≥ 3) and Neutropenia grade ≥ 3 for each therapy. The 95 % CIs of estimated CRS and grade ≥ 3 neutropenia were 64–86 % and 65–93 % (CAR-T), 51–74 % and 38–55 % (BsAbs), and N/A and 2–29 % (ADC). The results were visualized in Supplement Figure 3 (overall) and Fig. 6(for phases 1 and 2) (see Fig. 6).
Fig. 5.
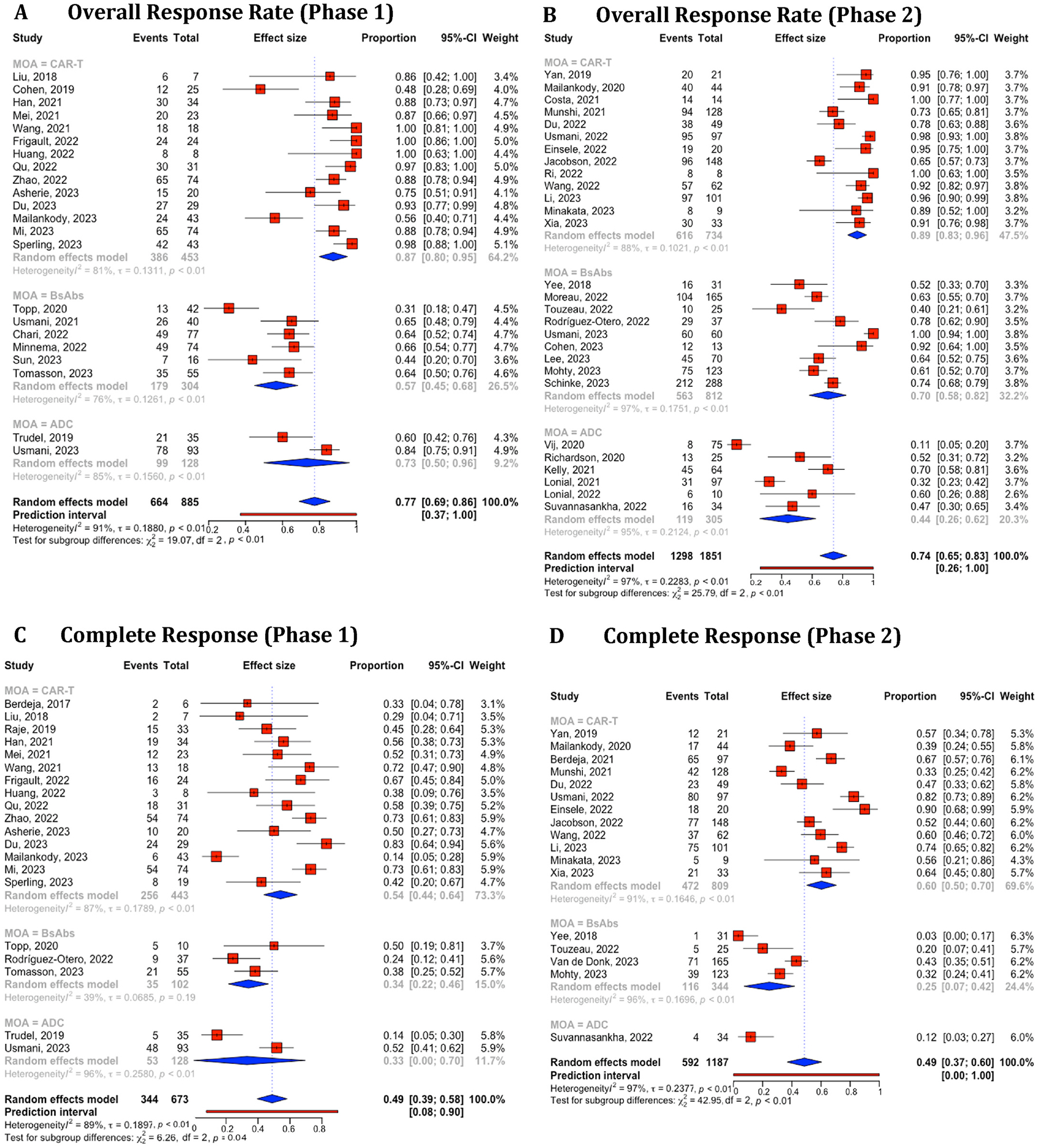
Combined and subgroup analysis of overall response rate and complete response based on therapies categorized in phase 1 and 2 trials. A. Overall response rates in phase 1 trial. B. Overall response rates in phase 2 trial. C. Complete response in phase 1 trial. D. Complete response in phase 2 trial. Horizontal lines through the squares indicate 95 % Confidence Intervals (CIs). The diamond symbol aggregates these estimates, presenting the pooled mean effect size and its 95 % CI. CAR-T, chimeric antigen receptor T cell; BsAbs, Bispecific antibody; ADC, antibody-drug conjugate.
Fig. 6.
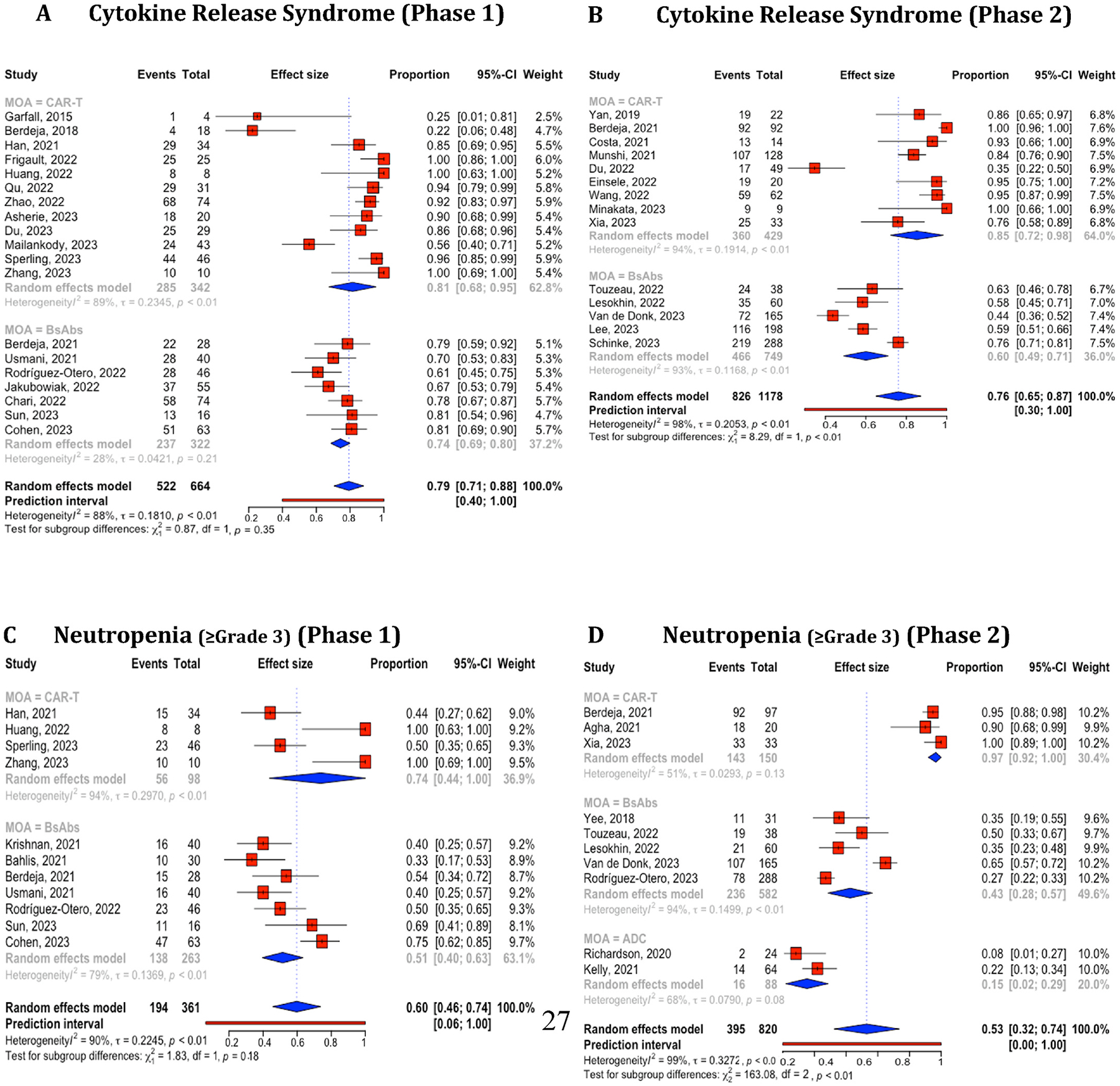
Combined and subgroup analysis of cytokine release syndrome and neutropenia (≥Gr3) based on therapies categorized in phase 1 and 2 trials. A. Cytokine release syndrome in phase 1 trial. B. Cytokine release syndrome in phase 2 trial. C. Neutropenia (≥Gr3) in phase 1 trial. D. Neutropenia (≥Gr3) in phase 2 trial. Horizontal lines through the squares indicate 95 % Confidence Intervals (CIs). The diamond symbol aggregates these estimates, presenting the pooled mean effect size and its 95 % CI.CAR-T, chimeric antigen receptor T cell; BsAbs, Bispecific antibody; ADC, antibody-drug conjugate.
5. Discussion
Our knowledge-driven LLM system, SEETrials, demonstrated the capabilities of GPT-4 in extracting and processing clinical trial data. The success of SEETrials in automatically identifying intervention outcomes and their associations with specific cohorts is underscored by robust performance metrics and speaks to the potential of LLMs to revolutionize data extraction from condensed and complex medical texts.
The system’s proficiency in handling data from annual conference abstracts is particularly noteworthy. These abstracts are often the first public report of a clinical trial’s findings and can influence clinical practice ahead of peer-reviewed publications. SEETrials’ capacity to process this information rapidly and reliably is invaluable, ensuring that the latest clinical insights are accessible to healthcare professionals and researchers without delay. During the validation phase, we observed that manually abstracting a conference proceeding takes approximately 30 min per abstract. For a large conference like ASCO, which features around 7000 online abstracts—including approximately 1500 related to hematologic malignancies and over 500 focused on plasma cell dyscrasias—summarizing the plasma cell dyscrasia-related abstracts alone would require around 250 h of manual effort. In contrast, SEETrials processes the same volume of information in just 3–6 min per abstract, depending on the presence of a table, resulting in more than 80%-time savings. On average, conducting a systematic literature review takes 17 months, with a range from 12 to 24 months [33], and the data extraction step, which is one of the most time-consuming parts of the process, could benefit significantly from automation. SEETrials also showcased the generalizability of a prompt initially tailored for specific cancer (i.e., multiple myeloma) to effectively extract trial information from diverse cancer types (breast cancer, lung cancer, and leukemia/lymphoma) with consistently high-performance scores across various disease clinical trial studies (F1 scores within the 0.979–0.992 range). This enables uniform analysis of clinical trial outcomes across diseases. To our knowledge, this study pioneers exploring GPT-powered models for comprehensive clinical trial outcomes extraction from diverse abstract types, in addition to our previously established system “AutoCriteria”—a generalizable clinical trial eligibility criteria extraction system powered by a large language model [28].
Importantly, our study extends beyond mere data extraction. By conducting a comparative analysis of CAR-T, BsAbs, and ADC therapies specifically within MM clinical trials, we have gleaned insights into the varying safety and efficacy profiles across different treatment modalities and trial phases. The predominance of certain efficacy markers, such as ORR, in phase 1 trials underscores the importance of these early-phase trials in gauging initial treatment impact. Conversely, the emphasis on survival and treatment duration in phase 2 trials highlights the shift towards understanding the long-term benefits and potential risks of therapies as they progress through the clinical trial pipeline. Our findings also align with existing literature, revealing consistent safety profiles in CAR-T, BsAbs, and ADC studies such as higher CRS rates in CAR-T therapy and infection rates in ADCs [34,35]. Understanding these outcomes’ variations across therapies and phases is crucial for contextualizing clinical trial results.
Moreover, our findings highlight a concerning trend: the increase in reports of fatal adverse events in later trial phases, particularly for CAR-T and ADC therapies. It suggests that early-phase studies may not fully capture the adverse event profile, especially for treatments with complex mechanisms of action or those applied to severely ill patient populations. This observation calls for a re-evaluation of safety monitoring protocols, especially as therapies advance beyond the dose-escalation stages. We also observed a higher number of abstracts mentioning fatal or severe AEs entities in ADC studies compared to CAR-T and BsAbs.
We noted significant variability in the effectiveness of the same treatments, even when categorized by phases, for outcomes such as Objective Response Rate (ORR), Complete Response (CR), Cytokine Release Syndrome (CRS), and neutropenia. A possible reason for the exceptionally low ORR observed in some Phase 1 studies may be attributed to combining data from different dosage groups, as indicated in the abstracts.
5.1. Limitations and future works
We recognize several limitations in our study. The exclusive focus on abstracts, while practical, limits the depth of our analysis. Critical nuances contained within full-text articles are often lost in abstracts, which could lead to incomplete or skewed understandings of the safety and efficacy profiles of therapies. Additionally, the omission of details such as prior treatment histories, specific high-risk patient groups, and biomarker statuses from our analysis may have prevented a fully informed assessment of the treatments’ impacts. Expanding the system’s capabilities to include full-text articles—particularly for extracting data from tables, detailed patient histories, and biomarker information—could significantly enhance the dataset and lead to more comprehensive insights.
Clinical trial reporting often contains conflicting information. To address this, we utilized SEETrials to analyze outcomes on a large scale, comparing results between preliminary conference abstracts and subsequent full publications. This approach could be crucial in evaluating the consistency and evolution of reported data [36], providing valuable insights for the medical community and shaping clinical decision-making. Furthermore, integrating SEETrials’ extracted results with expert-curated databases like HemOnc could foster a collaborative approach to oncology data analysis, ensuring that the most relevant and accurate information is available to guide patient care and research [37]. Additionally, SEETrials can be integrated into existing clinical trial workflows by automating the data extraction process from systematic literature reviews to real-time analysis of ongoing clinical trials. In the pre-trial phase, SEETrials can assist with a comprehensive systematic literature review; during trials, it can be used to monitor safety profiles and efficacy trends through conference proceeding analysis; and in the post-trial phase, it can be used to generate evidence for regulatory submissions.
The extracted data can be integrated into a user-friendly interface for question-and-answer purposes, with the added functionality of generative LLMs to deliver up-to-date care information for clinical practice. To ensure interpretability and explainability of the model’s extractions, features can be implemented that allow users to trace data extractions back to specific text or table locations in the original abstracts. The interface can also present summarized results with visualized graphs, offering a comprehensive view of large volumes of data processed through the SEETrials pipeline. This would provide a powerful tool for enhancing the efficiency and accuracy of clinical decision-making and research.
SEETrials’ modular architecture supports flexible scaling. To ensure that SEETrials can handle large-scale deployment, across different deployment scenarios, the system needs to be tested on datasets comprising thousands of clinical trial abstracts from medical conferences. This is essential to verify its scalability while maintaining performance and accuracy.
6. Conclusion
We developed SEETrials, an LLM-based system for automatically extracting detailed safety and efficacy outcomes from oncology clinical trials, including those with complex data structures like tables. SEETrials demonstrated high accuracy (0.964), precision (0.964), recall (0.988), and F1 score (0.974) across 70 data elements in multiple myeloma trial studies, with strong generalizability across other cancer types.
Our analysis of CAR-T, bispecific antibodies, and ADC therapies revealed significant variations in treatment outcomes and identified notable heterogeneity across studies. SEETrials proves to be an efficient, accurate, and broadly applicable tool for enhancing clinical evidence generation and supporting decision-making in oncology research.
Supplementary Material
FuNDING
We acknowledge the contribution of our coauthor JLW, who received the HemOnc U24 grant from National Cancer Institute (NCI), with grant number CA265879.
Conflict of interest statement
KL, HP, SD, LH, NO, FM, JW, and XW are currently employees of IMO Health Inc. JLW reports funding from NCI/NIH, related to the work, funding from AACR and Brown Physicians Incorporated, consulting from Westat, and ownership of HemOnc.org LLC, outside the scope of the work. No other conflict of interest.
Footnotes
CRediT authorship contribution statement
Kyeryoung Lee: Writing – review & editing, Writing – original draft, Data curation, Conceptualization. Hunki Paek: Writing – original draft, Formal analysis, Data curation, Conceptualization. Liang-Chin Huang: Visualization, Data curation. C Beau Hilton: Writing – review & editing. Surabhi Datta: Writing – review & editing, Data curation. Josh Higashi: Software. Nneka Ofoegbu: Data curation. Jingqi Wang: Software. Samuel M. Rubinstein: Writing – review & editing. Andrew J. Cowan: Writing – review & editing. Mary Kwok: Writing – review & editing. Jeremy L. Warner: Writing – review & editing. Hua Xu: Writing – review & editing. Xiaoyan Wang: Writing – review & editing, Conceptualization.
Ethical statement
It is important to acknowledge that leveraging artificial intelligence techniques, including LLMs, in clinical settings to support clinical decision-making carries inherent risks and should be applied with caution. Automated systems can introduce errors, which may affect downstream analyses and potentially lead to incorrect conclusions. Recognizing this, our primary effort in this study has been to enhance performance accuracy and develop a reliable LLM model across all data element extraction, confirming its efficacy in various types of oncology clinical trial studies. Despite significant improvements, we note that an automatic extraction system can occasionally make errors in extracting and correctly allocating the values within clinical trial cohorts. To mitigate these risks, we can further fine-tune the model using domain-specific data from oncology clinical trials to further improve the precision and recall of data extraction. It is also essential to establish a human-in-the-loop system that enhances data quality while benefiting from the increased speed provided by automation.
Finally, we acknowledge that although the ethical and regulatory burden is lighter when handling published journal articles and conference proceedings, data usage and licensing issues, especially for large-scale automated data extraction, must still be considered. Additionally, ensuring transparency in the model’s functionality and accurate extraction and interpretation of information without bias or misinterpretation is a key ethical responsibility.
- this material has not been published in whole or in part elsewhere.
- the manuscript is not currently being considered for publication in another journal.
- all authors have been personally and actively involved in substantive work leading to the manuscript and will hold themselves jointly and individually responsible for its content.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2024.101589.
Data availability
The prompts and codes used in this study are provided in Supplement Figs. 4 and 5.
The source code is also available at https://github.com/applebyboy/SEEtrials.git.
References
- [1].Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA, et al. Diagnosis and management of multiple myeloma: a review. JAMA 2022;327:464. 10.1001/jama.2022.0003. [DOI] [PubMed] [Google Scholar]
- [2].Van Nieuwenhuijzen N, Frunt R, May AM, Minnema MC. Therapeutic outcome of early-phase clinical trials in multiple myeloma: a meta-analysis. Blood Cancer J 2021;11:44. 10.1038/s41408-021-00441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tanenbaum B, Miett T, Patel SA. The emerging therapeutic landscape of relapsed/refractory multiple myeloma. Ann Hematol 2023;102:1–11. 10.1007/s00277-022-05058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Subbiah V The next generation of evidence-based medicine. Nat Med 2023;29: 49–58. 10.1038/s41591-022-02160-z. [DOI] [PubMed] [Google Scholar]
- [5].Chen EX, Tannock IF. Risks and benefits of phase 1 clinical trials evaluating new anticancer agents: a case for more innovation. JAMA 2004;292:2150. 10.1001/jama.292.17.2150. [DOI] [PubMed] [Google Scholar]
- [6].Weber JS, Levit LA, Adamson PC, Bruinooge S, Burris HA, Carducci MA, et al. American society of clinical oncology policy statement update: the critical role of phase I trials in cancer research and treatment. J Clin Orthod 2015;33:278–84. 10.1200/JCO.2014.58.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wright EC, Kapuria D, Ben-Yakov G, Sharma D, Basu D, Cho MH, et al. Time to publication for randomized clinical trials presented as abstracts at three gastroenterology and hepatology conferences in 2017. Gastro Hep Adv 2023;2: 370–9. 10.1016/j.gastha.2022.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Scherer RW, Meerpohl JJ, Pfeifer N, Schmucker C, Schwarzer G, von Elm E. Full publication of results initially presented in abstracts. Cochrane Database Syst Rev 2018;11:MR000005. 10.1002/14651858.MR000005.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Scherer RW, Ugarte-Gil C, Schmucker C, Meerpohl JJ. Authors report lack of time as main reason for unpublished research presented at biomedical conferences: a systematic review. J Clin Epidemiol 2015;68:803–10. 10.1016/j.jclinepi.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ioannidis JPA. Effect of the statistical significance of results on the time to completion and publication of randomized efficacy trials. JAMA 1998;279:281. 10.1001/jama.279.4.281. [DOI] [PubMed] [Google Scholar]
- [11].Stern JM, Simes RJ. Publication bias: evidence of delayed publication in a cohort study of clinical research projects. BMJ 1997;315:640–5. 10.1136/bmj.315.7109.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, Osmani V. Natural Language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform 2019;7:e12239. 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cook MJ, Yao L, Wang X. Facilitating accurate health provider directories using natural language processing. BMC Med Inform Decis Mak 2019;19:80. 10.1186/s12911-019-0788-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dave AD, Ruano G, Kost J, Wang X. Automated extraction of pain symptoms: a Natural Language approach using electronic health records. Pain Physician 2022; 25:E245–54. [PubMed] [Google Scholar]
- [15].Singhal K, Azizi S, Tu T, Mahdavi SS, Wei J, Chung HW, et al. Large Language models encode clinical knowledge. 10.48550/ARXIV.2212.13138; 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang X, Chen A, PourNejatian N, Shin HC, Smith KE, Parisien C, et al. A large language model for electronic health records. Npj Digit Med 2022;5:194. 10.1038/s41746-022-00742-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Arsenyan V, Shahnazaryan D. Large Language models for biomedical causal graph construction. 10.48550/ARXIV.2301.12473; 2023. [DOI] [Google Scholar]
- [18].Reichenpfader D, Müller H, Denecke K. Large language model-based information extraction from free-text radiology reports: a scoping review protocol. Health Informatics 2023. 10.1101/2023.07.28.23292031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fink MA, Bischoff A, Fink CA, Moll M, Kroschke J, Dulz L, et al. Potential of ChatGPT and GPT-4 for data mining of free-text CT reports on lung cancer. Radiology 2023;308:e231362. 10.1148/radiol.231362. [DOI] [PubMed] [Google Scholar]
- [20].Hasani AM, Singh S, Zahergivar A, Ryan B, Nethala D, Bravomontenegro G, et al. Evaluating the performance of Generative Pre-trained Transformer-4 (GPT-4) in standardizing radiology reports. Eur Radiol 2023. 10.1007/s00330-023-10384-x. [DOI] [PubMed] [Google Scholar]
- [21].Ge J, Li M, Delk MB, Lai JC. A comparison of large language model versus manual chart review for extraction of data elements from the electronic health record. Gastroenterology 2023. 10.1101/2023.08.31.23294924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Guevara M, Chen S, Thomas S, Chaunzwa TL, Franco I, Kann BH, et al. Large language models to identify social determinants of health in electronic health records. NPJ Digit Med 2024;7:6. 10.1038/s41746-023-00970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang H, Jethani N, Jones S, Genes N, Major VJ, Jaffe IS, et al. Evaluating Large Language models in extracting cognitive exam dates and scores. medRxiv 2024. 10.1101/2023.07.10.23292373.2023.07.10.23292373. [DOI] [Google Scholar]
- [24].Mahbub M, Dams GM, Srinivasan S, Rizy C, Danciu I, Trafton J, et al. Leveraging Large Language models to extract information on substance use disorder severity from clinical notes: a zero-shot learning approach. 10.48550/ARXIV.2403.12297; 2024. [DOI] [Google Scholar]
- [25].Shyr C, Hu Y, Bastarache L, Cheng A, Hamid R, Harris P, et al. Identifying and extracting rare diseases and their phenotypes with Large Language models. J Healthc Inform Res 2024;8:438–61. 10.1007/s41666-023-00155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hu Y, Ameer I, Zuo X, Peng X, Zhou Y, Li Z, et al. Zero-shot clinical entity recognition using ChatGPT. 10.48550/ARXIV.2303.16416; 2023. [DOI] [Google Scholar]
- [27].Peikos G, Symeonidis S, Kasela P, Pasi G. Utilizing ChatGPT to enhance clinical trial enrollment. 10.48550/ARXIV.2306.02077; 2023. [DOI] [Google Scholar]
- [28].Datta S, Lee K, Paek H, Manion FJ, Ofoegbu N, Du J, et al. AutoCriteria: a generalizable clinical trial eligibility criteria extraction system powered by large language models. J Am Med Inform Assoc 2023. 10.1093/jamia/ocad218.ocad218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tang Y, Xiao Z, Li X, Zhang Q, Chan EW, Wong IC, et al. Large Language model in medical information extraction from titles and abstracts with prompt engineering strategies: a comparative study of GPT-3.5 and GPT-4. 10.1101/2024.03.20.24304572; 2024. [DOI] [Google Scholar]
- [30].Wang Z, Cao L, Danek B, Zhang Y, Jin Q, Lu Z, et al. Accelerating clinical evidence synthesis with Large Language models. 10.48550/ARXIV.2406.17755; 2024. [DOI] [Google Scholar]
- [31].Gartlehner G, Kahwati L, Hilscher R, Thomas I, Kugley S, Crotty K, et al. Data extraction for evidence synthesis using a large language model: a proof-of-concept study. Res Synth Methods 2024. 10.1002/jrsm.1710. [DOI] [PubMed] [Google Scholar]
- [32].Kartchner D, Ramalingam S, Al-Hussaini I, Kronick O, Mitchell C. Zero-Shot information extraction for clinical meta-analysis using Large Language models. In: The 22nd workshop on biomedical Natural Language Processing and BioNLP shared tasks. Toronto, Canada: Association for Computational Linguistics; 2023. p. 396–405. 10.18653/v1/2023.bionlp-1.37. [DOI] [Google Scholar]
- [33].Borah R, Brown AW, Capers PL, Kaiser KA. Analysis of the time and workers needed to conduct systematic reviews of medical interventions using data from the PROSPERO registry. BMJ Open 2017;7:e012545. 10.1136/bmjopen-2016-012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Swan D, Murphy P, Glavey S, Quinn J. Bispecific antibodies in multiple myeloma: opportunities to enhance efficacy and improve safety. Cancers 2023;15:1819. 10.3390/cancers15061819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Raje N, Anderson K, Einsele H, Efebera Y, Gay F, Hammond SP, et al. Monitoring, prophylaxis, and treatment of infections in patients with MM receiving bispecific antibody therapy: consensus recommendations from an expert panel. Blood Cancer J 2023;13:116. 10.1038/s41408-023-00879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lee K, Paek H, Huang L-C, Datta S, Annan A, Ofoegbu N, et al. Unveiling consistency: a large-scale analysis of conference proceedings and subsequent publications in oncology clinical trials using large language models. J Clin Orthod 2024;42. 10.1200/JCO.2024.42.16_suppl.7568.7568-7568. [DOI] [Google Scholar]
- [37].Warner JL, Dymshyts D, Reich CG, Gurley MJ, Hochheiser H, Moldwin ZH, et al. HemOnc: a new standard vocabulary for chemotherapy regimen representation in the OMOP common data model. J Biomed Inform 2019;96:103239. 10.1016/j.jbi.2019.103239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The prompts and codes used in this study are provided in Supplement Figs. 4 and 5.
The source code is also available at https://github.com/applebyboy/SEEtrials.git.


