Abstract
Hemipteran insects transmit viruses when infesting plants, during which vectors activate salicylic acid (SA)-regulated antiviral defenses. How vector-borne plant viruses circumvent these antiviral defenses is largely unexplored. During co-infections of begomoviruses and betasatellites in plants, betasatellite-encoded βC1 proteins interfere with SA signaling and reduce the activation of antiviral resistance. βC1 inhibits SA-induced degradation of NbNPR3 (Nicotiana benthamiana nonexpressor of pathogenesis-related genes 3), a negative regulator of SA signaling. βC1 does not bind directly to NbNPR3, but regulates NbNPR3 degradation via heat shock protein 90s (NbHSP90s). NbHSP90s bind to both NbNPR3 and βC1 and suppress SA signaling. This viral success strategy appears to be conserved as it is also documented for viral proteins encoded by two aphid-borne viruses. Our findings reveal an exquisite mechanism that facilitates the persistence of vector-borne plant viruses and provide important insights into the intricacies of the virus life cycle.
Subject terms: Plant immunity, Virus-host interactions, Microbe
Amid virus inoculation by hemipteran insects, vector infestation induces salicylic acid (SA)-mediated plant antiviral defense. Here it is found that the vector-borne plant virus circumvents this antiviral defense via disrupting the SA signaling pathway.
Introduction
As a group of obligate parasites, plant viral pathogens pose considerable threats to crop production worldwide. Efficient inter-plant spread is vital for virus persistence and disease outbreaks in the field. To achieve productive inter-plant dissemination, the majority of plant viruses rely on arthropod vectors, mostly hemipteran insects such as whiteflies, aphids and leafhoppers1,2. Insect vectors ingest viruses from infected plants and then transmit the viruses to uninfected plants through feeding1–3. Intimate interactions occur amongst the insect vector, virus, and host plant during this virus acquisition and transmission cycle. The compatibility among the three kinds of organisms determines the efficiency of virus transmission by insect vectors and in planta virus infection post inoculation3. For example, during feeding-mediated virus inoculation, the preference of an insect vector for host plants and plant resistance to viruses dictate the chance of virus inoculation by a vector and virus propagation in planta, respectively. The intricate dynamics of interactions among vectors, viruses, and plants play a pivotal role in determining the spread of viruses and, consequently, the occurrence of viral disease epidemics within agricultural ecosystems.
Insect colonization and feeding on host plants induce an array of changes in plant physiology and biochemistry. Of these changes, plant hormone biosynthetic and signaling pathways are among the most significant, as they directly control the expression of a plethora of downstream genes associated with defenses against pathogens and pests4. Activation of the jasmonate (JA)-signaling pathway is a hallmark of plant responses to insect herbivores with chewing mouthparts4. In contrast, hemipteran insects with piercing-sucking mouthparts, such as whiteflies and aphids, often induce the accumulation of salicylic acid (SA) and the expression of SA-regulated genes5–9. Notably, the SA-signaling pathway is also important in virus-plant interactions, as SA regulates plant antiviral defenses4,7,10. Therefore, concomitant with a vector delivering a virus to initiate in planta infection, hemipteran infestation activates the SA-signaling pathway and this inevitably impacts the success of these viruses7. While vector-mediated transmission is indispensable in the life cycle of many plant viruses, whether and how this important group of pathogens adapt to insect infestation-associated defenses remain enigmatic.
Viral proteins are often versatile, capable of modulating a repertoire of plant biological processes such as morphogenesis, reproduction, and immunity. As major regulators of plant defenses, hormonal pathways are often targeted by viral proteins to suppress plant immunity4,11–18. For example, during the infections of begomoviruses and their betasatellites, the betasatellite-encoded βC1 promotes the success of their whitefly vectors by downregulating JA biosynthesis and signal transduction11,12. Surprisingly, although vector-mediated transmission is indispensable for the life cycle of many plant viruses, whether and how these viruses interact with vector infestation-induced plant defenses remain unexplored. For plant viruses whose insect vectors induce SA accumulation in plants, elucidating the strategies deployed to manage the SA-signaling pathways in these tripartite interactions will provide critical insights into the molecular mechanisms that promote viral persistence.
In this work, we demonstrate that the viral vector Bemisia tabaci (whitefly) induces SA accumulation, which deters begomovirus infection of host plants. However, when begomoviruses and their associated betasatellite co-infect a host, the betasatellite-encoded βC1 dampens the antiviral resistance triggered by whitefly-induced SA. We show that this strategy is active in two begomovirus-betasatellite complex-host plant interactions. In addition, aphid-borne viruses that do not associate with satellites also recruit virus-encoded proteins to deploy this virus ‘survival’ strategy. Mechanistically, viral proteins slow SA-induced degradation of Nicotiana benthamiana NPR3 (a SA receptor and negative regulator of SA-regulated response) by interacting with heat shock protein 90s (NbHSP90s). Our findings unravel an important aspect of the insect vector-virus-plant interactions that facilitates the infection of vector-borne plant viruses.
Results
Begomoviral betasatellites mitigate vector-induced SA accumulation to dampen host plant resistance to begomoviruses
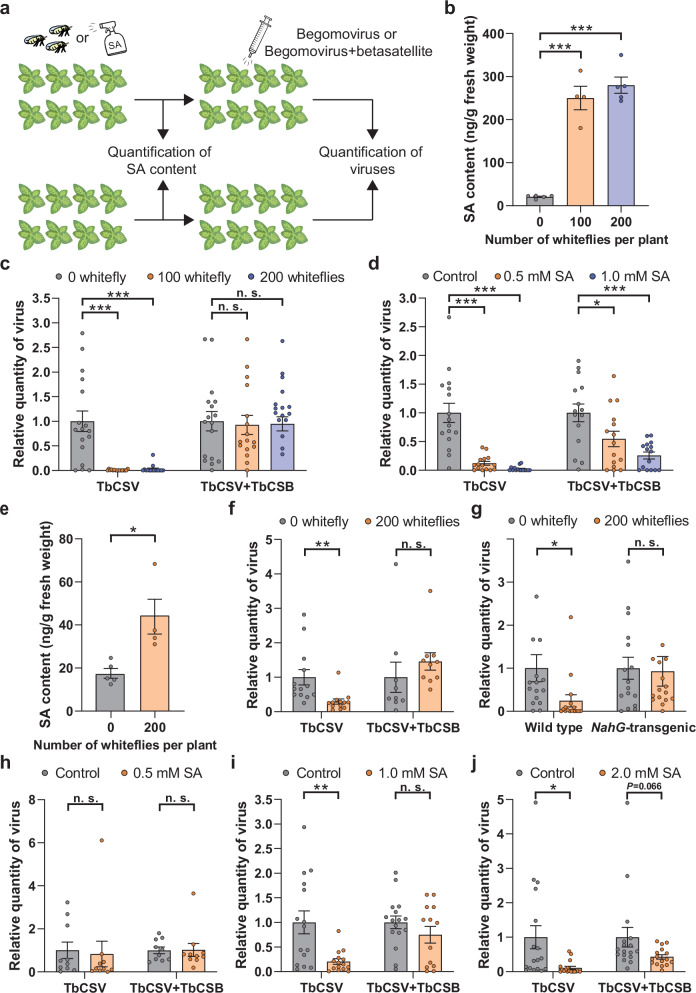
We investigated the role of hemipteran vector-activated defenses in host plant antiviral resistance. The quadripartite interactions of the Bemisia tabaci (whitefly), the host plant tobacco (Nicotiana tabacum), the tobacco curly shoot virus (TbCSV) (genus Begomovirus, family Geminiviridae), and its associated betasatellite (TbCSB) were investigated19. Plants were subjected to whitefly infestation for 48 h or served as non-infested controls. Some of the plants were used for the analysis of phytohormones, and the other plants were agro-inoculated with TbCSV or TbCSV+TbCSB after the removal of whitefly adults. TbSCV was quantified at 10 days post inoculation, when whitefly nymphs were feeding and the viruses have replicated extensively, moved systematically, and viral symptoms have fully developed (Fig. 1a). Whitefly feeding and egg deposition for 48 h significantly increased the level of endogenous SA and JA-Ile, but not JA in tobacco (Fig. 1b, Supplementary Fig. 1a, b). To further validate the accumulation of SA, we repeated the whitefly infestation experiment and profiled SA and the major SA conjugate (SA O-β-D-glucoside, SAG) in whitefly-infested and non-infested tobacco plants20. Whitefly infestation significantly increased the content of SA and SAG (Supplementary Fig. 1c, d). In TbCSV-challenged plants, antiviral resistance was detected in response to whitefly infestation, as reflected by lower virus levels (Fig. 1c). However, when whitefly-infested plants were inoculated with TbCSV+TbCSB, resistance was diminished as TbCSV levels were similar to non-infested plants (Fig. 1c). The enhanced plant resistance to the TbCSV was also reflected in the decreased occurrence of viral symptoms (Supplementary Fig. 2a–f).
Fig. 1. Whitefly infestation-induced SA accumulation increases plant resistance to TbCSV, but not to the TbCSV + TbCSB complex.
a Schematic representation of experimental design. b SA content in tobacco plants upon whitefly infestation (48 h). c Quantity of TbCSV in tobacco plants that were first infested by whiteflies (48 h) and then inoculated with TbCSV or TbCSV+TbCSB. d Quantity of TbCSV in tobacco plants that were first sprayed with ethanol (control) or SA and then inoculated with TbCSV or TbCSV+TbCSB. e SA content in Nicotiana benthamiana plants upon whitefly infestation (48 h). f Quantity of TbCSV in N. benthamiana plants that were first infested by whiteflies (48 h) and then inoculated with TbCSV or TbCSV+TbCSB. g Quantity of TbCSV in wild type and NahG-transgenic N. benthamiana plants that were first infested by whiteflies and then inoculated with TbCSV. h–j Quantity of TbCSV in N. benthamiana plants that were first sprayed with ethanol (control) or SA (0.5, 1 or 2 mM) and then inoculated with TbCSV or TbCSV+TbCSB. N = 4–5 samples (2–3 plants per sample) for (b, e), 15–16 plants for (c, d), 9–13 plants for (f), 16 plants for (g), 10 plants for (h), 13–16 plants for (i), 16–17 plants for (j). Data are mean ± SEM. n. s. stands for no significant difference, *P < 0.05, **P < 0.01, and ***P < 0.001 (one-way ANOVA for (b–d); two-sided Student’s t test for e–j). Source data and the exact P-values are provided in Source Data file.
To further document the role of SA in whitefly-induced resistance to TbCSV, tobacco plants were first sprayed with SA and subsequently inoculated with TbCSV or TbCSV+TbCSB (Fig. 1a). Foliar SA spray dramatically increased the total content of SA in and on the surface of tobacco leaves, but did not significantly affect the content of JA and JA-Ile in the leaves (Supplementary Fig. 3a–c). It should be noted that in SA-sprayed plants, many SA molecules may stay on the surface of leaves. Relative to the control, 1.0 mM SA treatment substantially reduced TbCSV levels to ~2% of that in controls (Fig. 1d). While 1.0 mM SA treatment also reduced TbCSV quantity in TbCSV+TbCSB-inoculated plants, the reduction was less pronounced with TbCSV levels reaching only 26% of control levels (Fig. 1d). The SA-induced increases in plant resistance to TbCSV or TbCSV+TbCSB were correlated with viral symptoms in inoculated plants (Supplementary Fig. 2a–e, g). Similar results were obtained when 0.5 mM SA was sprayed (Fig. 1d, Supplementary Fig. 2g).
Similar to tobacco, whitefly infestation significantly induced SA accumulation, but did not significantly change JA and JA-Ile levels in the model plant N. benthamiana (Fig. 1e, Supplementary Fig. 1e, f). In addition, whiteflies significantly increased plant resistance to TbCSV, but not to TbCSV+TbCSB (Fig. 1f). Furthermore, whitefly-induced antiviral resistance was abolished in NahG-transgenic N. benthamiana plants (Fig. 1g). NahG encodes a salicylate hydroxylase and NahG-transgenic plants do not accumulate SA21. The role of TbCSB in regulating SA-controlled antiviral resistance was also supported by exogenous SA treatments. While 0.5 mM SA did not significantly affect plant resistance to either TbCSV or TbCSV+TbCSB (Fig. 1h), 1.0 and 2.0 mM SA provoked a strong resistance to TbCSV, but not to TbCSV+TbCSB (Fig. 1i, j).
To determine if these principles are recapitulated in another whitefly-begomovirus-plant interaction, we examined the impact of whitefly infestation and SA treatments on tomato resistance to the tomato leaf curl China virus (ToLCCNV) (genus Begomovirus, family Geminiviridae) and its associated betasatellite (ToLCCNB)22. Endogenous SA levels in tomato leaves significantly increased upon whitefly feeding, while JA and JA-Ile levels were unchanged (Supplementary Fig. 4a–c). Whitefly and the 2.0 and 4.0 mM SA treatments significantly increased tomato resistance to ToLCCNV, but not to ToLCCNV+ToLCCNB (Supplementary Fig. 4d–f). As N. benthamiana is also a host for ToLCCNV, we further investigated these interactions using NahG plants or SA treatments. Similar to tomato, whitefly-induced resistance to ToLCCNV was observed in wild type plants, but not in NahG plants (Supplementary Fig. 4g). In addition, exogenous SA significantly increased plant resistance to ToLCCNV, but not to ToLCCNV+ToLCCNB (Supplementary Fig. 4h, i). Collectively, these data indicate that whiteflies increased plant resistance to begomoviruses by inducing SA accumulation and this SA-regulated antiviral resistance was abolished when a betasatellite was co-inoculated with its partner begomovirus.
Betasatellite-encoded βC1 mitigates the negative effects of SA treatment on begomovirus infection by interfering with SA signaling
The ability of betasatellites to abate the SA-regulated resistance against begomoviruses could be due to catabolism of SA or interference with a critical step in SA signaling. In both tobacco and N. benthamiana plants, TbCSV infection did not significantly alter SA content and coinfection of TbCSV and TbCSB slightly enhanced SA levels (Supplementary Fig. 5a, c). Similar results were obtained for ToLCCNV and ToLCCNV+ToLCCNB infections of tomato plants (Supplementary Fig. 5b). To further examine the modulation of plant SA-signaling pathway by begomoviruses and betasatellites, we analyzed the transcript level of two SA-sentinel genes (PR1a and PR2) in control and virus-infected N. benthamiana. When compared to control, both TbCSV and TbCSV+TbCSB significantly increased the transcript level of PR1a and PR2 (Supplementary Fig. 5d, e), suggesting the activation of SA-signaling pathway. These results show that the begomovirus-betasatellite complexes did not reduce SA levels and suggest that betasatellites may directly or indirectly interfere with SA signaling.
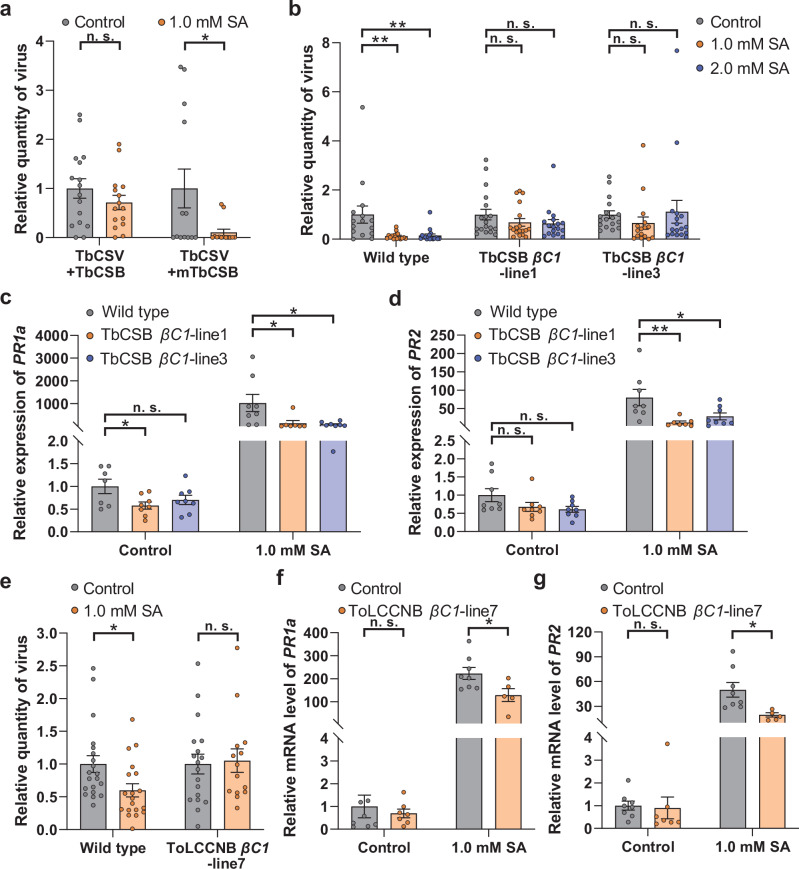
Betasatellites encode βC1, an essential pathogenicity determinant in many begomovirus-betasatellite complexes23,24. To determine if βC1 disabled the SA-regulated resistance against begomoviruses, we compared TbCSV infection after SA treatment when this virus was paired with a wild type or mutant TbCSB (mTbCSB) that is unable to express βC125. We found that a functional βC1 is critical to subdue the SA-regulated resistance against begomovirus in N. benthamiana (Fig. 2a). To further examine the role of βC1 as a suppressor of SA-regulated antiviral immunity, transgenic N. benthamiana plants that ectopically express TbCSB βC1 were constructed and verified by RT-PCR (Supplementary Fig. 6a). Two independent 35S:βC1 transgenic lines displayed typical βC1-induced symptoms, such as leaf curl and curly shoots (Supplementary Fig. 6c). In the wild type plants, exogenous SA significantly increased plant resistance to TbCSV as evidenced by decreased TbCSV levels (Fig. 2b). Whereas, in both βC1-transgenic lines, SA treatment did not induce resistance to TbCSV (Fig. 2b). To assess the activity of the SA-signaling pathway in these interactions, the transcript level of PR1a and PR2 was monitored by qPCR. After SA treatment, PR1a and PR2 transcripts were less abundant in the βC1-transgenic plant lines than in the control (Fig. 2c, d).
Fig. 2. βC1 mitigates the deleterious effects of SA treatment on begomoviruses infection and interferes with SA signaling.
a Quantity of TbCSV in N. benthamiana plants that were first sprayed with ethanol (control) or SA and then inoculated with TbCSV+TbCSB or TbCSV+mutant TbCSB (mTbCSB). b Quantity of TbCSV in wild type and transgenic N. benthamiana plants that were first sprayed with ethanol (control) or SA and then inoculated with TbCSV; c, d Relative mRNA level of PR1a (c) and PR2 (d) in wild type and TbCSB βC1-transgenic N. benthamiana plants that were sprayed with ethanol (control) or SA. e Quantity of ToLCCNV in wild type and transgenic N. benthamiana plants that were first sprayed with ethanol (control) or SA and then inoculated with ToLCCNV. f, g Relative mRNA level of PR1a (f) and PR2 (g) in wild type and ToLCCNB βC1-transgenic N. benthamiana plants that were sprayed with ethanol (control) or SA. N = 13–16 plant for (a), 16–18 plants for (b), 7–8 samples (2-3 plants per sample) for (c, d), 14–20 plants for e, 5-8 samples (2-3 plants per sample) for e and f. Data are mean ± SEM. n. s. stands for no significant difference, *P < 0.05, and **P < 0.01 (one-way ANOVA for b-d; two-sided Student’s t test for a, e–g). Source data and the exact P values are provided in Source Data file.
To evaluate if the ToLCCNB βC1 influenced SA signaling and antiviral immunity in a manner similar to TbCSB βC1, transgenic N. benthamiana plants overexpressing ToLCCNB βC1 were constructed and verified by RT-PCR (Supplementary Fig. 6b). ToLCCNB βC1-transgenic plants displayed mild upward leaf curl (Supplementary Fig. 6d). Similar to TbCSB βC1-transgenic plants, the SA-induced resistance against ToLCCNV seen in wild type plants was abolished in the ToLCCNB βC1-transgenic plants (Fig. 2e). Moreover, ToLCCNB βC1 significantly downregulated the transcript level of PR1a and PR2 upon SA treatment but not in the control plants (Fig. 2f, g). Taken together, these data demonstrate that betasatellite-encoded βC1 mitigates the negative effects of SA treatment on begomovirus infection and interferes with the SA-signaling pathway.
N. benthamiana HSP90s are bona fide targets of TbCSB βC1
To explore how βC1 interferes with the activation of the SA-signaling pathway, proteins that interact with TbCSB βC1 were identified. GST and GST-βC1 proteins were expressed in Escherichia coli and purified. GST or GST-βC1 were incubated with total N. benthamiana protein extracts. Pull-down of GST-βC1 and its interactors identified a 100-kDa protein band, which was subjected to mass spectrometry analysis (Supplementary Fig. 7a). Intriguingly, six N. benthamiana heat shock protein 90 (HSP90) proteins were identified (Supplementary Table 1). Using hidden Markov model search, we identified the entire complement of NbHSP90s in the N. benthamiana genome (www.nbenth.com)26. We inferred their evolutionary origins based on a phylogenic tree using HSP90s from A. thaliana as a reference (Supplementary Fig. 7b). We selected the mass spectrometry-identified NbHSP90-2, NbHSP90-10 and NbHSP90-12 for further study, as they had diverged sequences (evidenced by belonging to different clades) and sequence lengths (Supplementary Fig. 7b, Supplementary Table 1).
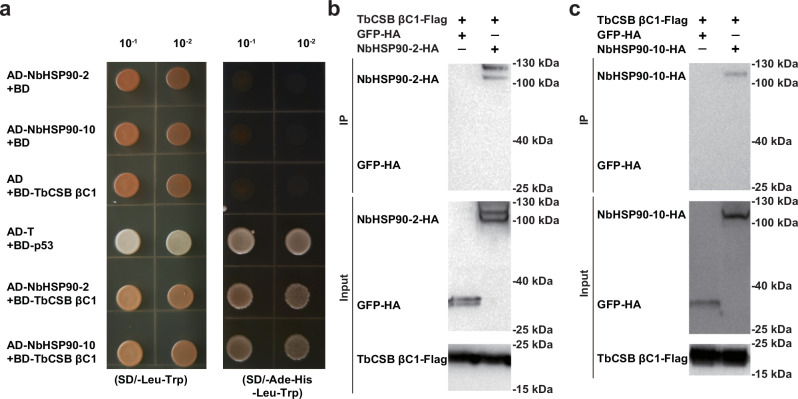
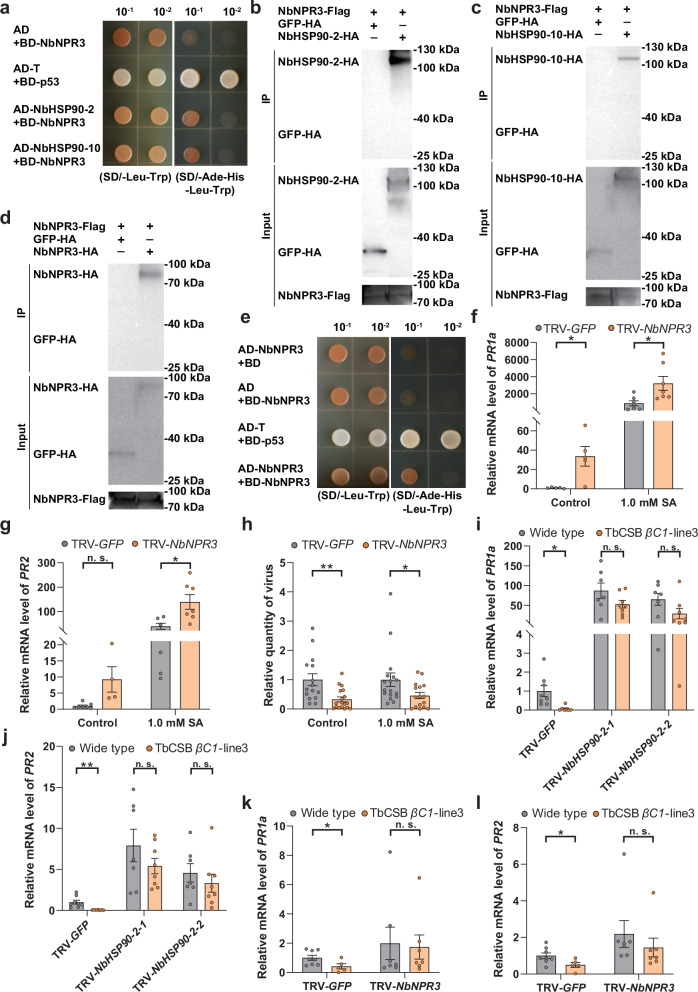
To verify the interaction between TbCSB βC1 and NbHSP90 proteins, bimolecular fluorescence complementation (BiFC) assays were performed. The TbCSB βC1 was fused with C-terminal domain of YFP (TbCSB βC1-cYFP) and NbHSP90s was fused to the N-terminal domain of YFP (HSP90-nYFP). Transient co-expression of the two classes of proteins in N. benthamiana demonstrated that YFP fluorescence was detected in all three TbCSB βC1-NbHSP90 combinations, but not in negative controls (Supplementary Fig. 8). Yeast two-hybrid assays were also performed to examine in vitro protein-protein interactions. Full-length sequences of TbCSB βC1 and NbHSP90-2 (or NbHSP90-10) were fused to the GAL4 DNA-binding and activation domains, respectively. Yeast transformants harboring BD-T + AD-p53 (positive control), BD-TbCSB βC1 + AD-NbHSP90-2 or BD-TbCSB βC1 + AD-NbHSP90-10 readily grew on selection medium (Fig. 3a). No growth of yeast transformants harboring BD + AD-NbHSP90-2, BD + AD-NbHSP90-10 or BD-βC1 + AD was found (Fig. 3a). These data indicate that TbCSB βC1 associates with NbHSP90 proteins.
Fig. 3. TbCSB βC1 interacts with N. benthamiana HSP90s (NbHSP90s).
a Interactions between TbCSB βC1 and NbHSP90s in the yeast two-hybrid assay. Yeast AH109 transformed with the indicated plasmid combinations were spotted with 10-fold serial dilutions on synthetic dextrose media SD/-Leu-Trp and SD/-Ade-His-Leu-Trp. b Interaction between TbCSB βC1 and NbHSP90-2 in the co-IP assay. TbCSB βC1-Flag and NbHSP90-2-HA were co-expressed in N. benthamiana leaves, and TbCSB βC1-Flag+GFP-HA served as a negative control. TbCSB βC1-Flag interacting proteins were immunoprecipitated with anti-Flag beads. GFP-HA and NbHSP90-HA proteins were detected with anti-HA antibodies and TbCSB βC1-Flag with anti-Flag antibodies in input samples and after co-IP. c Interaction between TbCSB βC1 and NbHSP90-10 in co-IP assay. TbCSB βC1-Flag and NbHSP90-10-HA were co-expressed, and TbCSB βC1-Flag+GFP-HA served as a negative control. Source data are provided in Source Data file.
Co-immunoprecipitation experiments corroborated TbCSB βC1-NbHSP90s interactions. TbCSB βC1-Flag was co-expressed with NbHSP90-2-HA, NbHSP90-10-HA or GFP-HA. TbCSB βC1-Flag and its interactors were isolated using the anti-Flag beads. NbHSP90-2-HA and NbHSP90-10-HA were specifically detected in the samples with TbCSB βC1-Flag (Fig. 3b, c). To identify the domain of HSP90 proteins that interacted with TbCSB βC1, N-terminal domain (ND), middle domain (MD) and C-terminal domain (CD) of NbHSP90-2 and NbHSP90-10 were tested for their interactions with TbCSB βC1 using BiFC. All three domains of NbHSP90-2 and NbHSP90-10 interacted with TbCSB βC1 (Supplementary Fig. 9). Collectively, these results suggest that N. benthamiana HSP90s are bona fide targets of TbCSB βC1.
NbHSP90s negatively regulate the SA-signaling pathway and plant resistance to TbCSV + TbCSB complex
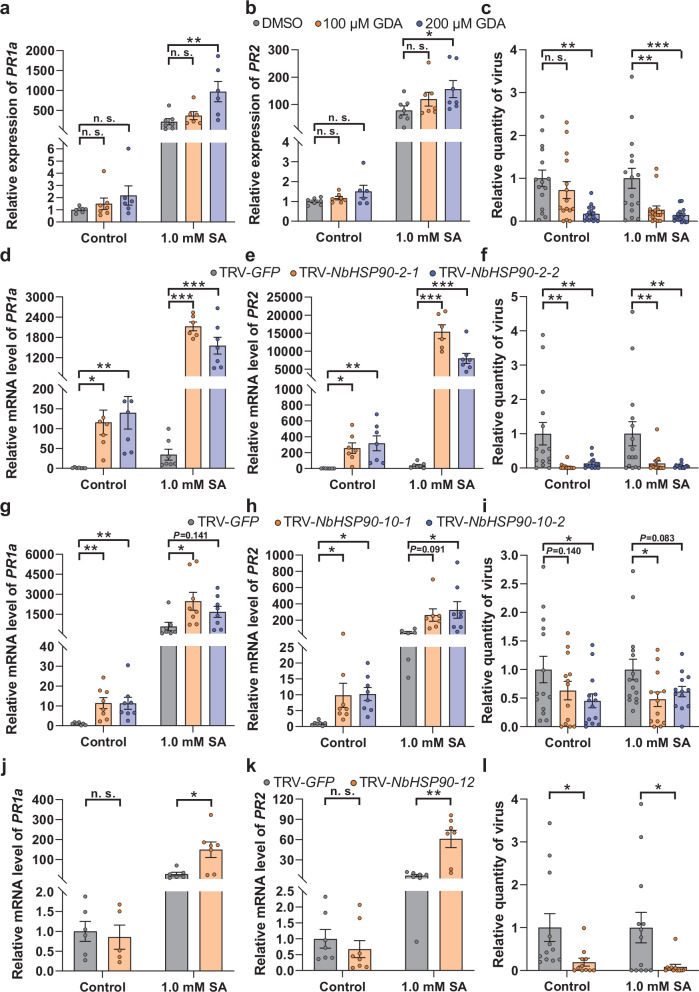
To explore the roles of NbHSP90s in the regulation of SA-signaling pathway and antiviral defenses, we used geldanamycin (GDA), a specific inhibitor of HSP90s, to inactivate HSP90s prior to SA treatment of N. benthamiana. GDA treatments caused an increase in PR1a and PR2 transcript level in SA-treated samples but not in the control (Fig. 4a, b). To confirm the suppression of SA-signaling pathway by HSP90s, virus-induced gene silencing (VIGS) was used to downregulate NbHSP90-2, NbHSP90-10 and NbHSP90-12 (Supplementary Figs. 10a–c). Diminished levels of NbHSP90-2 were associated with significantly higher levels of PR1a and PR2 transcripts in both SA and control treatments (Fig. 4d, e). Similar results with NbHSP90-10 silencing were observed. There was a significant increase in PR1a and PR2 transcripts in both SA and control treatments in all but two samples; these samples displayed similar strong trends (Fig. 4g, h). Unlike NbHSP90-2 and −10, silencing of NbHSP90-12 increased PR1a and PR2 mRNA levels after SA treatment but not in the control (Fig. 4j, k), suggesting NbHSP90-12 may have a slightly different role relative to NbHSP90-2 and −10 in regulating SA-signaling pathway.
Fig. 4. NbHSP90s negatively regulates SA signaling and plant resistance against TbCSV + TbCSB complex in N. benthamiana plants.
a, b Relative mRNA level of PR1a (a) and PR2 (b) in N. benthamiana plants that were first treated with geldanamycin (GDA) or DMSO and then sprayed with ethanol (control) or SA. c Quantity of TbCSV in N. benthamiana plants that were first sprayed with ethanol (control) or SA and then inoculated with TbCSV+TbCSB+GDA or TbCSV+TbCSB+DMSO. d, e Relative mRNA level of PR1a (d) and PR2 (e) in N. benthamiana plants that were first inoculated with TRV-GFP or TRV-NbHSP90-2 and then sprayed with ethanol (control) or SA. f Quantity of TbCSV in N. benthamiana plants that were first sprayed with ethanol (control) or SA and then inoculated with TbCSV+TbCSB+TRV-GFP or TbCSV+TbCSB+TRV-NbHSP90-2. g, h Relative mRNA level of PR1a (g) and PR2 (h) in N. benthamiana plants that were first inoculated with TRV-GFP or TRV-NbHSP90-10 and then sprayed with ethanol (control) or SA. i Quantity of TbCSV in N. benthamiana plants that were first sprayed with ethanol (control) or SA and then inoculated with TbCSV+TbCSB+TRV-GFP or TbCSV+TbCSB+TRV-NbHSP90-10. j, k Relative mRNA level of PR1a (j) and PR2 (k) in N. benthamiana plants that were first inoculated with TRV-GFP or TRV-NbHSP90-12 and then sprayed with ethanol (control) or SA. l Quantity of TbCSV in N. benthamiana plants that were first inoculated with TbCSV+TbCSB+TRV-GFP or TbCSV+TbCSB+TRV-NbHSP90-12 and then sprayed with ethanol (control) or SA. N = 6–7 samples (2-3 plants per sample) for (a, d, e), 5–7 samples (2–3 plants per sample) for (b, j), 15–16 plants for (c), 14–15 plants for (f), 7-8 samples (2-3 plants per sample) for (g, k), 6–8 samples (2–3 plants per sample) for (h), 12-15 plants for (i), 11–13 plants for l. Data are mean ± SEM. n. s. stands for no significant difference, *P < 0.05, **P < 0.01, and ***P < 0.001 (one-way ANOVA for (a–i); two-sided Student’s t test for j–l). Source data and the exact P values are provided in Source Data file.
We then explored the roles of NbHSP90s in plant resistance against the begomovirus-betasatellite complex. GDA treatments and silencing of NbHSP90-2, NbHSP90-10 and NbHSP90-12 significantly enhanced plant resistance to TbCSV+TbCSB as evidenced by decreased TbCSV quantity (Fig. 4c, f, i, l). Collectively, these data suggest that NbHSP90s are negative regulators of SA signaling and plant resistance against begomovirus-betasatellite complex.
NbHSP90s interact with NbNPR3, a negative regulator of SA signaling
HSP90s often interact with each other and exist as oligomers27. BiFC assays were used to assess the interactions among the three NbHSP90s. BiFC interaction signals were detected in all pair-wise combinations of NbHSP90s, but not in the negative controls, indicating the assembly of homo- and hetero-NbHSP90 complexes (Supplementary Fig. 11a–c). Co-immunoprecipitation assays corroborated the interaction between NbHSP90-2 and NbHSP90-10, NbHSP90-2 and NbHSP90-2, and NbHSP90-10 and NbHSP90-10 (Supplementary Fig. 11d–f).
HSP90s often regulate biological processes by stabilizing client proteins27. In plants, receptors of auxin- and jasmonate-signaling pathways are HSP90 clients28,29. Given the roles of NbHSP90s and the known role of Arabidopsis’s SA receptor NPR proteins in the regulation of SA-signaling pathway30,31, we first cloned an NbNPR gene and then examined the interaction between NbHSP90s and the NbNPR. The cloned NbNPR was named as NbNPR3, as its deduced protein sequence contained a canonical BTB/POZ domain, ankyrin repeats and a NPR1-like C terminus domain (Supplementary Fig. 12a)32, and phylogenetic analysis revealed that the NbNPR was most closely related to AtNPR3 relative to other AtNPR proteins (Supplementary Fig. 12b). While NbNPR3 diverged significantly from AtNPR3 in protein sequence, NbNPR3 has an EAR-like motif in its C-terminus (VDLNEVP), similar to AtNPR3’s EAR motif (VDLNETP) (Supplementary Fig. 12c)31.
Yeast two-hybrid and co-IP assays revealed NbHSP90-2 and NbHSP90-10 specifically interacted with NbNPR3 (Fig. 5a–c), and NbNPR3 formed homo-oligomers (Fig. 5d, e). To explore the role of NbNPR3 in SA signaling, NbNPR3 was downregulated using VIGS and ectopically expressed in a transgenic 35S:NPR3 N. benthamiana line. NbNPR3 silencing significantly upregulated the transcript level of PR1a and PR2 (Fig. 5f, g). In addition, increased plant resistance against TbCSV-TbCSB complex was seen when NbNPR3 was silenced, as viral quantity in both SA-treated and control plants decreased significantly (Fig. 5h). In contrast, the 35S:NPR3 N. benthamiana line exhibited no significant differences in either SA signaling or plant resistance against the TbCSV-TbCSB complex relative to control (Supplementary Fig. 13a–d). These data suggest that the quantities of NbNPR3 in planta are sufficient to exert its suppression of SA-signaling. Collectively, these data suggest that NbHSP90s interact with NbNPR3, a negative regulator of SA-signaling and plant resistance against begomoviruses.
Fig. 5. NbHSP90s interact with NbNPR3, a negative regulator of SA signaling and both NbHSP90s and NbNPR3 are required for the suppression of SA signaling by TbCSB βC1.
a Interactions between NbHSP90s and NbNPR3 in the yeast two-hybrid assay. b, c Interaction between NbNPR3 and NbHSP90-2 (b) or NbHSP90-10 (c) in the co-IP assay. NbNPR3-Flag and NbHSP90s-HA were co-expressed in plants, and NbNPR3-Flag+GFP-HA served as a negative control. d, e Interactions between NbNPR3 and NbNPR3 in the co-IP (d) and yeast two-hybrid (e) assay. f, g Relative mRNA level of PR1a (f) and PR2 (g) in N. benthamiana plants that were first inoculated with TRV-GFP or TRV-NbNPR3 and then sprayed with ethanol (control) or SA. h Quantity of TbCSV in N. benthamiana plants that were first inoculated with TbCSV+TbCSB+TRV-GFP or TbCSV+TbCSB+TRV-NbNPR3 and then sprayed with ethanol (control) or SA. i, j Relative mRNA level of PR1a (i) and PR2 (j) in wild type and TbCSB βC1-transgenic N. benthamiana plants that were first inoculated with TRV-GFP or TRV-NbHSP90-2 and then sprayed with SA. k, l Relative mRNA level of PR1a (k) and PR2 (l) in wild type and TbCSB βC1-transgenic N. benthamiana plants that were first inoculated with TRV-GFP or TRV-NbNPR3 and then sprayed with SA. N = 5–7 samples (2–3 plants per sample) for (f), 4–7 samples (2–3 plants per sample) for (g), 15–18 plants for (h), 7–8 samples (2–3 plants per sample) for (i, j), 5–8 samples (2–3 plants per sample) for (k, l). Data are mean ± SEM. n. s. stands for no significant difference, *P < 0.05, and **P < 0.01 (two-sided Student’s t test for f–l). Source data and the exact P-values are provided in Source Data file.
Both NbHSP90s and NbNPR3 are required for the suppression of SA signaling by βC1
To explore the roles of NbHSP90s and NbNPR3 in the suppression of SA signaling by TbCSB βC1, we used VIGS to silence NbHSP90s or NbNPR3 in wild type and βC1-transgenic N. benthamiana plants. VIGS silencing of GFP served as a negative control (Supplementary Fig. 10e, f). Upon SA treatment, PR1a and PR2 transcript levels were suppressed in the TRV-GFP-treated βC1-transgenic lines when compared to TRV-GFP-treated wild type plants (Fig. 5i–l). When NbHSP90-2 was silenced, PR1a and PR2 RNAs were abundant and the differences between wild type and βC1-transgenic plants were abolished (Fig. 5i, j). Similarly, after SA treatment of NbNPR3-silenced wild type and βC1-transgenic plants, PR1a and PR2 transcript levels were similar (Fig. 5k, l). Taken together, these data indicate that both NbHSP90s and NbNPR3 were required for the suppression of SA signaling by TbCSB βC1.
SA-induced NbNPR3 degradation is suppressed by TbCSB βC1 in a NbHSP90s-dependent manner
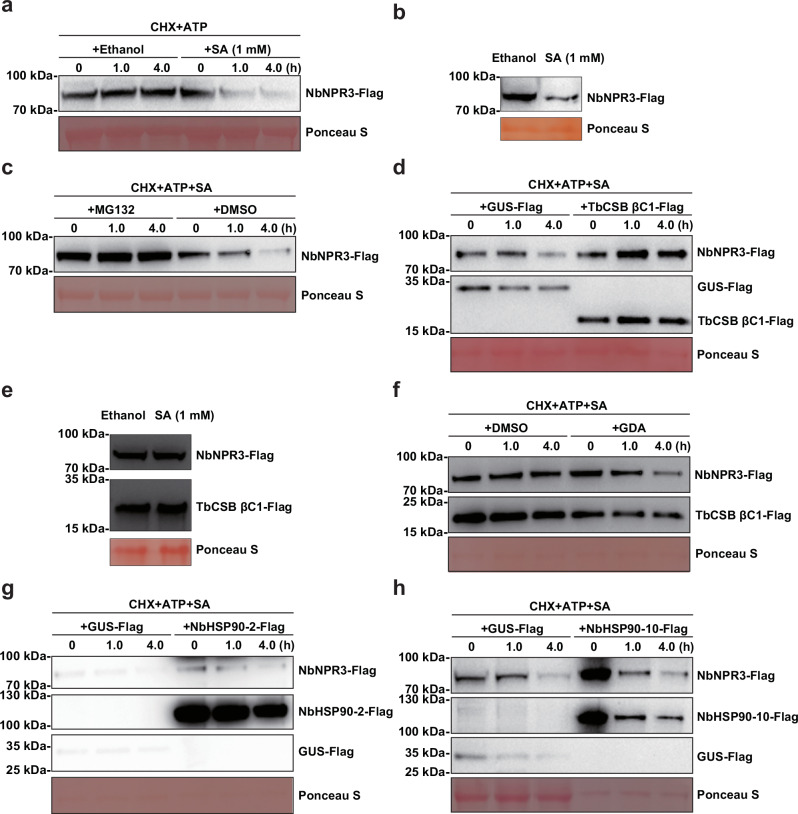
Yeast-two hybrid assays indicated that there was no direct interaction between TbCSB βC1 and NbNPR3 (Supplementary Fig. 14), suggesting TbCSB βC1 may modulate NbNPR3 through NbHSP90s. As SA modulates the stability of the Arabidopsis NPR3 and HSP90s modulate the stability of their client proteins28,29,33, we tested the effects of SA on NbNPR3 stability in two assays. In the semi-in vivo degradation assay NbNPR3-Flag was transiently expressed in N. benthamiana and protein extracts were used to assess NbNPR3 protein stability in the presence of cycloheximide (CHX) and adenosine triphosphate (ATP). CHX was used to stop protein translation, and ATP provided energy for the protein degradation machinery. In the in vivo degradation assay, NbNPR3-Flag was transiently expressed in N. benthamiana leaves, and SA was sprayed on these leaves. In both semi-in vivo and in vivo degradation assay, SA treatment provoked the degradation of NbNPR3 (Fig. 6a, b). The addition of proteasome inhibitor MG132 significantly inhibited the SA-induced degradation of NbNPR3 in the semi-in vivo degradation assay (Fig. 6c). To test the effect of TbCSB βC1-Flag on NbNPR3 stability, NbNPR3-Flag was co-expressed with TbCSB βC1-Flag and then subjected to two degradation assays. Both assays revealed that TbCSB βC1-Flag interfered with SA-induced NbNPR3 degradation (Fig. 6d, e). Notably, when NbHSP90s were inhibited by the addition of GDA in the semi-in vivo degradation assay, even though TbCSB βC1 was present NbNPR3 levels declined relative to the DMSO control indicating that in the presence of SA, NbHSP90s were needed for the suppression of NbNPR3 turnover by TbCSB βC1 (Fig. 6f). To test the roles of NbHSP90s in modulating NbNPR3 turnover, we co-expressed NbNPR3-Flag with NbHSP90-2-Flag (or NbHSP90-10-Flag) or GUS-Flag (control), and then conducted the semi-in vivo degradation assay. The presence of NbHSP90-2-Flag or NbHSP90-10-Flag in the protein extracts did not suppress SA-induced degradation of NbNPR3; this may be due to the turnover of the two NbHSP90s in the presence of SA, or the in planta quantities of NbHSP90s are sufficient for their modulation of NbNPR3 degradation (Fig. 6g, h).
Fig. 6. SA-induced NbNPR3 degradation is interrupted by TbCSB βC1 in a NbHSP90s- dependent manner.
a Effect of SA treatment on the degradation of NbNPR3-Flag in semi-in vivo assays. NbNPR3-Flag was expressed in N. benthamiana leaves, proteins extracted, and cycloheximide (CHX) (final concentration 100 µM) and adenosine triphosphate (ATP) (final concentration 20 mM) added. Next, SA or ethanol (control) was added and samples were subjected to incubation. NbNPR3-Flag protein was detected with anti-Flag antibodies. b Effect of SA on the degradation of NbNPR3-Flag in the in vivo assays. NbNPR3-Flag was expressed in N. benthamiana leaves. Leaves were sprayed with SA or ethanol (control) and five hours later collected to determine NbNPR3 levels. c Effect of MG132 on the SA-induced degradation of NbNPR3-Flag in semi-in vivo assays. DMSO was added as control. d Effect of TbCSB βC1-Flag on the SA-induced degradation of NbNPR3-Flag in semi-in vivo assay. GUS-Flag was used as control. NbNPR3-Flag was co-expressed with TbCSB βC1-Flag or GUS-Flag (control). Protein extracts were mixed with CHX, ATP and SA, and then subjected to protein degradation. e Effect of TbCSB βC1-Flag on the SA-induced degradation of NbNPR3-Flag in the in vivo degradation assay. NbNPR3-Flag was co-expressed with TbCSB βC1-Flag and treated with ethanol (control) or SA. Levels of NbNPR3-Flag and TbCSB βC1-Flag were determined five hours later. f Effect of GDA on the suppression of SA-induced degradation of NbNPR3-Flag by TbCSB βC1-Flag in the semi-in vivo degradation assay. DMSO was added as control. g, h Effect of NbHSP90-2-Flag (g) and NbHSP90-10-Flag (h) on the SA-induced degradation of NbNPR3-Flag in the semi-in vivo degradation assay. Ponceau S staining was performed to determine the amount of protein load. Source data are provided in Source Data file.
HSP90s stabilize client proteins by direct binding28,29. While yeast two-hybrid assays indicated that TbCSB βC1 did not interact with NbNPR3 directly (Supplementary Fig. 14), TbCSB βC1 may promote NPR3 stability by facilitating the interaction between NbHSP90s and NbNPR3. To test this hypothesis, GST and GST-βC1 were expressed in E. coli and purified (Supplementary Fig. 15e). NbNPR3-Flag and NbHSP90-2-HA were co-expressed in N. benthamiana and proteins extracted (Supplementary Fig. 15f, g). The NbNPR3-Flag+NbHSP90-2-HA extract was mixed with various amounts of GST or GST-βC1 and NbNPR3-flag interacting proteins were isolated using anti-Flag beads. The amount of NbHSP90-2-HA associated with NbNPR3-Flag was similar in all GST and GST-βC1 treatments (Supplementary Fig. 15a–d), suggesting that in vitro TbCSB βC1 does not enhance NbNPR3-NbHSP90s interactions. Collectively, these data suggest that SA-induced degradation of NbNPR3 is proteasome-dependent and TbCSB βC1 suppressed NbNPR3 degradation by interacting with NbHSP90s.
Viral proteins encoded by aphid-borne plant viruses target NbHSP90s and NbNPR3 for the suppression of SA signaling
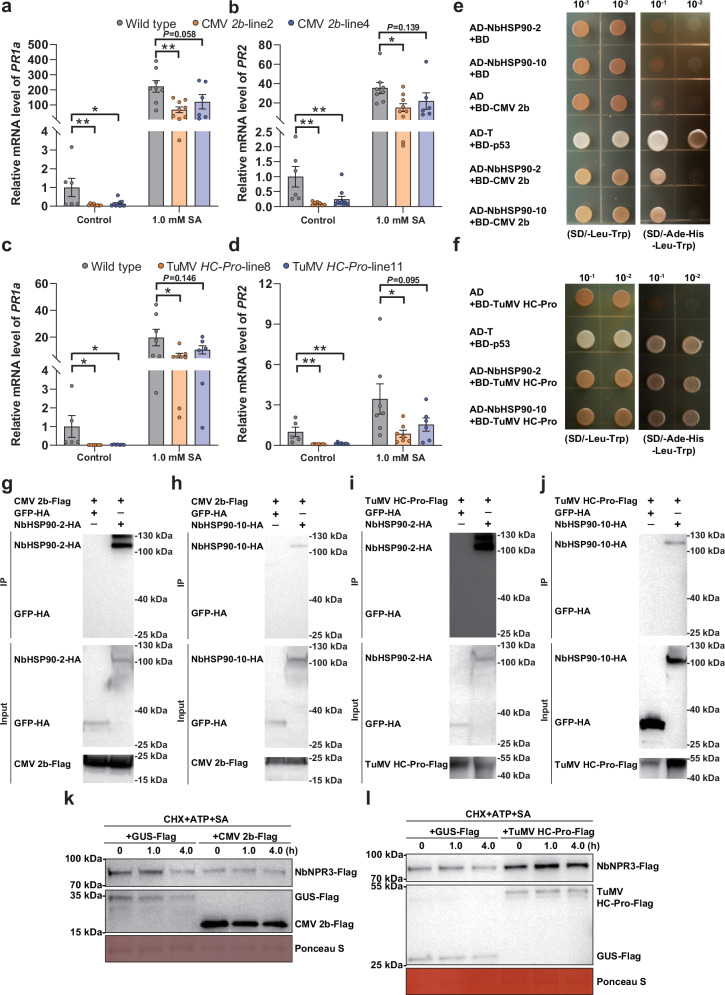
We have shown that begomovirus-betasatellite complexes, which are transmitted by whiteflies, interfere with SA signaling to promote virus success. To determine if viral suppression of SA signaling is a strategy utilized by other vector-borne plant viruses, we investigated the modulation of the SA-signaling pathway by two aphid-borne plant viruses, namely cucumber mosaic virus (CMV) (genus Cucumovirus, family Bromoviridae) and turnip mosaic virus (TuMV) (genus Potyvirus, family Potyviridae). Aphid-borne plant viruses were chosen for experimentation because they are widespread and of substantial economic significance in agriculture1,3,34. CMV and TuMV encode the 2b and HC-Pro proteins, respectively, both of which were reported to interfere with SA signaling13,15. To verify whether these viral proteins modulate the same steps in SA-dependent plant immunity, CMV 2b and TuMV HC-Pro transgenic N. benthamiana plants were constructed and verified through RT-PCR and phenotyping (Supplementary Fig. 16a–d). CMV 2b and TuMV HC-Pro transgenic plants displayed severe downward leaf curl and mild upward leaf curl, respectively (Fig. S16b, d). When compared to the wild type plants, CMV 2b-transgenic plants expressed the SA-response genes PR1a and PR2 at significantly lower levels (Fig. 7a, b). Similar patterns were documented in the TuMV HC-Pro-transgenic plants (Fig. 7c, d).
Fig. 7. Viral proteins encoded by aphid-borne plant viruses target NbHSP90s and NbNPR3 for the suppression of SA signaling.
a, b Relative mRNA level of PR1a (a) and PR2 (b) in wild type and CMV 2b-transgenic N. benthamiana plants. c, d Relative mRNA level of PR1a (c) and PR2 (d) in wild type and TuMV HC-Pro-transgenic N. benthamiana plants. e, f Interactions between CMV 2b (e) or TuMV HC-Pro (f) with NbHSP90s in a yeast two-hybrid assay. Yeast strain AH109 transformed with the indicated plasmid combinations was spotted with 10-fold serial dilutions on synthetic dextrose media SD/-Leu-Trp and SD/-Ade-His-Leu-Trp. g, h Interaction between CMV 2b and NbHSP90-2 (g) or NbHSP90-10 (h) in the co-IP assay. CMV 2b-Flag and NbHSP90-2-HA were co-expressed and CMV 2b-Flag+GFP-HA served as a negative control. i, j Interaction between TuMV HC-Pro and NbHSP90-2 (i) or or NbHSP90-10 (j) in the co-IP assay. k, l Effect of CMV 2b (k) and TuMV HC-Pro (l) on the SA-induced degradation of NbNPR3-Flag in the semi-in vivo assays. NbNPR3-Flag was co-expressed with CMV 2b-Flag (or TuMV HC-Pro) or GUS-Flag (control). Protein extracts were mixed with CHX, ATP and SA and then subjected to semi-in vivo protein degradation. NbNPR3-Flag protein was analyzed with anti-Flag antibodies at different time points. Ponceau S staining was performed to determine the amount of protein load. N = 6–10 samples (2–3 plants per sample) for (a, b), 5–7 samples (2–3 plants per sample) for (c, d). Data are mean ± SEM. n. s. stands for no significant difference, *P < 0.05, and **P < 0.01 (one-way ANOVA for a–d). Source data and the exact P values are provided in Source Data file.
Yeast two-hybrid assays revealed that both CMV 2b and TuMV HC-Pro interact with NbHSP90-2 and NbHSP90-10 (Fig. 7e, f). In addition, co-IP assays revealed that NbHSP90-2-HA and NbHSP90-10-HA were specifically immunoprecipitated by CMV 2b-Flag (Fig. 7g, h) and TuMV HC-Pro-Flag (Fig. 7i–j). Finally, using the semi-in vivo degradation assay, SA-induced degradation of NbNPR3 was inhibited by CMV 2b and TuMV HC-Pro (Fig. 7k, l). These results suggest that aphid-borne plant viruses use a mechanism similar to that of βC1 to interfere with SA signaling via targeting NbHSP90s and NbNPR3.
Discussion
Vectors contribute directly to the spread and epidemics of vector-borne plant viruses1–3. To facilitate efficient virus spread, vector-borne plant viruses significantly manipulate host plants and vectors in various ways. For example, some viruses suppress plant defenses against insects to promote vector population growth, which is beneficial to virus transmission and spread3,11,12. Additionally, some vector-borne viruses manipulate the sensory behavior of insect vectors influencing their choice of host plants to promote virus transmission3,35. Inherent in the vector-virus-plant interactions is the challenge that hemipteran vectors trigger plant defenses that actively deter the success of the viruses that they transmit. Here, we provide first insights into this plant virus-vector conundrum and the virus innovations to resolve this challenge. We show that when confronted with stresses associated with vector infestation-mediated inoculation, viruses promote and sustain in planta infection by deploying viral proteins to counter these defenses. In the long-term evolution, once a viral protein acquired this function, the virus encoding this protein variant will outcompete other viral individuals as it promotes virus infection in plants, while keeping the essential insect feeding-mediated inoculation processes intact. This appears to be an evolutionarily conserved virus-success strategy that has evolved multiple times, as it was observed in two distantly-related whitefly-borne viruses and two aphid-borne viruses.
We used the tripartite whitefly-begomovirus-plant interactions to elucidate the molecular mechanisms that promote virus success on whitefly-infested plants (Fig. 8). Two begomoviruses (TbCSV and ToLCCNV) and their associated betasatellites (TbCSB and ToLCCNB, respectively) were studied using multiple host plant species. Our agro-inoculation studies showed that these begomoviruses or begomovirus-betasatellite complexes caused a marginal increase in SA levels and significant upregulation of the expression of SA-sentinel genes. Upon whitefly feeding and infestation, plant gene expression is reprogrammed leading to a dramatic rise in SA levels and activation of SA-signaling pathway6,7. We show that whitefly-induced activation of SA-regulated defenses resulted in an amplification of plant resistance against begomoviruses (Fig. 8). To short circuit these vector-activated antiviral defenses, TbCSV and ToLCCNV pair with their cognate betasatellites to actively suppress the SA-signaling pathway.
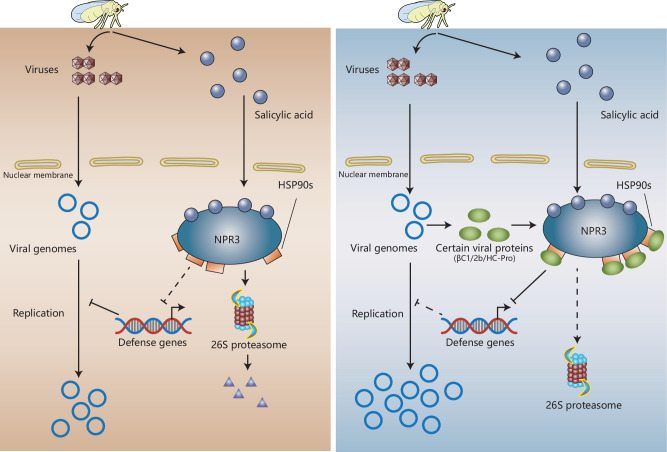
Fig. 8. A working model for the dynamics of SA-signaling pathway induction and suppression during insect vector-virus-host plant interactions.
Infestation by insect vectors induces SA accumulation. Concomitantly, viruliferous vectors secrete viruses into plants. SA promotes the degradation NPR3-HSP90s complex via 26S proteasomes. Degradation of the negative regulator NPR3 allows for the activation of defense gene expression, which inhibits virus replication providing the host plant immunity to the viral invader (left panel). Viral proteins encoded by vector-borne plant viruses (2b or HC-Pro) or their associated betasatellites (βC1) antagonize the SA-induced antiviral immunity (right panel). These proteins bind to HSP90s and antagonize the SA-induced degradation of NPR3 through yet unknown mechanisms. The persistence of NPR3 inhibits the expression of SA-regulated genes and associated antiviral resistance, thereby promoting virus replication.
Currently, SA is known to induce resistance to viral replication, cell-to-cell movement, and systemic movement in plants36,37. Yet the defense genes that mediated the antiviral function of SA, particularly the downstream genes that directly target viruses are poorly characterized36. PR genes, including PR1a and PR2, do not seem to contribute to plant antiviral defenses36. However, PR1a and PR2 transcript levels are a reliable and widely-used indicator of the activation status of the SA-signaling pathway36–38. Here we used two complementary assays with PR1a and PR2 transcripts serving sentinels for SA-signaling pathway activity and in planta virus infection as the indicator of SA-regulated plant antiviral defenses. The two pronged approach provided the opportunity to test the role of viral proteins and plant factors, including HSP90 and NPR3, and to explore the prime question of whether and how viral proteins modulate SA-controlled plant antiviral defenses.
Here, we show that the βC1 proteins encoded by the betasatellites TbCSB and ToLCCNB have a previously unrecognized role in the vector-virus-plant tripartite interactions. While whitefly infestation significantly induces SA accumulation in plants, βC1 proteins actively obstruct SA signaling and the deployment of SA-regulated defenses. Roles of betasatellites in virus pathogenesis and disease symptom development are well known19,22–24.βC1 is also a known suppressor of RNA silencing (an antiviral defense) and the JA-signaling pathway11,12,23. By disrupting JA biosynthesis and signaling, the tomato yellow leaf curl China betasatellite-encoded βC1 promotes the success of its whitefly vector on virus-infected plants11,12. Here, we show that the rises of SA that are concomitant with whitefly feeding and infestation are also counteracted by betasatellite-encoded βC1 proteins. By muting SA signaling, βC1 proteins antagonize the vector-induced resistance to begomoviruses.
The evolutionary gain-of-function of βC1 as a suppressor of SA signaling may be attributed to viral adaptation to whitefly-mediated transmission. The betasatellites TbCSB and ToLCCNB are not essential for TbCSV and ToLCCNV infections, respectively, but these betasatellites intensify virus infection symptoms19,22. The widespread association of begomoviruses and their cognate betasatellites suggests that there are potent selection pressures that drive the persistence of betasatellites and conservation of their βC1 proteins24. Previously-identified selection pressures mainly include plant antiviral defenses such as RNA interference and autophagy23. Additionally, the need of begomoviruses to reprogram plant development and systematically move represent additional selection pressures23,24. Notably, these selection pressures result from in planta infection of begomoviruses. Here we identified a novel selection pressure that promotes the persistence of betasatellites and βC1 proteins, namely SA accumulation in whitefly-infested plants. This selection pressure is associated with insect vector-mediated transmission, a process of prime importance in the life cycle of begomoviruses.
Mechanistically, βC1 proteins suppress SA signaling by prolonging the lifespan of a negative regulator of SA signaling - NbNPR3 (Fig. 8). βC1 proteins exert their effects on NbNPR3 protein turnover indirectly, as βC1 and NbNPR3 are not binding partners. βC1 exerts its control by interacting with NbHSP90s, which are molecular chaperones that bind to NbNPR3. NbNPR3 degradation is mediated by the 26S proteasome and this degradation is blocked by βC1 in a NbHSP90s-dependent manner. By preventing NbNPR3 turnover, SA signaling is suppressed, along with its associated antiviral defenses (Fig. 8).
HSP90s are a group of highly conserved molecular chaperones that participate in the activation, stabilization, and maturation of client proteins, thereby modulating biological processes27. Plant HSP90s regulate auxin- and JA-signaling pathways by stabilizing auxin and JA receptors, respectively28,29. Here, we demonstrate that a putative SA receptor (NbNPR3) is a HSP90 client. NPR proteins are key regulators of the SA-signaling pathway in A. thaliana, and AtNPR1, AtNPR3 and AtNPR4 are proposed SA receptors30,31,33,39,40. While AtNPR1 positively regulates SA signaling, AtNPR3 and AtNPR4 are negative regulators that function as adaptors of cullin3-based E3 ligase to mediate the degradation of the positive regulators AtNPR1 and AtEDS130,31,39–41. Here, we identified NbNPR3, an ortholog of AtNPR3, as a negative regulator of SA signaling in N. benthamiana.
Intriguingly, we show that NbNPR3 regulation in N. benthamiana may be distinct from that in Arabidopsis. In both plants, the proteasome regulates NPR3 protein levels. In N. benthamiana, NbNPR3 degradation is induced by SA. In contrast, SA promotes AtNPR3 stability so as to sustain the turnover of AtNPR1, thereby ensuring the full induction of SA-induced immunity31. Here the response of NbNPR3 level to SA is similar to that of phosphorylated AtNPR1 monomers. AtNPR1 oligomer and monomer levels rise in response to SA, but subsequent phosphorylation of AtNPR1 monomers promotes their turnover in the nucleus and full induction of immunity42. Our discovery of the role of NbHSP90s in regulating NPR3 turnover adds an important dimension to this regulatory pathway and our findings highlight the divergence of NPR3 protein regulation in different plant species. Further investigations on the roles of HSP90 in regulating the function of NPR proteins in the model plant Arabidopsis and beyond will uncover more regulatory mechanisms in the SA-signaling pathway.
At present, the mechanism of βC1 action is not known. However, we know that βC1 does not directly bind to NbNPR3 nor does it enhance NbHSP90 binding to its client NbNPR3. All three HSP90 domains (i.e., ND, MD and CD) that are important for HSP90 function27 interact with TbCSB βC1, indicating TbCSB βC1 may manipulate the function of one or multiple NbHSP90 domains. Therefore, it is possible that βC1 modulates HSP90 folding activity to execute its regulatory role on NPR3 stability or, potentially, other (yet to be discovered) HSP90 functions that are important in regulating NPR3 stability. When βC1 is present, NbHSP90 may mask or expose NbNPR3 residues critical for turnover. It is possible that sites in NbNPR3 that are critical for interaction with E3 ubiquitin ligases and ubiquitination may be hidden to prevent polyubiquitination and targeting to the proteasome. Alternatively, polyubiquitylated sites may be exposed, allowing the deubiquitylation machinery to access to NbNPR3. Both scenarios would extend the longevity of NbNPR3 longevity, thereby promoting virus success. A third scenario involves the interaction of NbHSP90 with certain E3 ubiquitin ligases, leading to the ubiquitination of NbHSP90 and subsequent proteasomal degradation of the NbHSP90-NbNPR3 complex. The binding of βC1 to NbHSP90 may interfere with the recruitment of E3 ubiquitin ligases, thereby inhibiting the ubiquitination of NbHSP90 and the degradation of the NbHSP90-NbNPR3 complex. Future studies could focus on the interplay between NPR3 and HSP90s, which dictates the proteasomal degradation of NPR3, and its modulation by viral proteins in N. benthamiana and other plant species.
Together with previous reports14,17,18, our data suggest that several vector-borne plant viruses encode suppressors of the SA-signaling pathway. We show that in addition to betasatellite-encoded βC1, HSP90 interacts with other viral proteins that are known to disrupt SA signaling in plants including TuMV HC-Pro and CMV 2b13,15. Furthermore, SA-induced NPR3 degradation is reduced in the presence of these viral proteins. Therefore, there appears to be a functional convergence of viral proteins from three distantly-related families (Geminiviridae, Bromoviridae and Potyviridae), suggesting that diminishing SA signaling is important in the evolution of hemipteran vector-virus-plant interactions. The circumventing of host defenses is of broad significance due to the requirement of vectors for the transmission of these plant viruses. In theory, the molecular maneuvers facilitated by two betasatellite-encoded βC1 proteins, HC-Pro of TuMV, and 2b of CMV may be conserved among other plant viruses vectored by the Hemiptera, whose infestations induce plant antiviral defenses such as SA-signaling pathway. For viruses vectored by the other arthropods, the investigation of vector infestation-induced changes in virus-host interactions and viral mitigation strategies may further reveal the intricacies of the virus life cycle.
Here we show that begomovirus-betasatellite complexes deploy a previously unreported defense-evading strategy using HSP90 and NPR3 to counteract the consequences of vector infestation and vector-mediated virus inoculation. Suppressing the vector-induced SA signaling appears to be evolutionarily conserved, as we also document it in two aphid-transmitted viruses. The detailed molecular mechanisms of the viral protein-HSP90-NPR3 regulatory unit are being elucidated and will provide critical insights into the complexities of the vector-virus-host plant triad interactions.
Methods
Plants
Tobacco (Nicotiana tabacum cv. NC89), tomato (Solanum lycopersicum cv. Moneymaker), and N. benthamiana plants were grown in growth chambers at 26 ± 2 °C, 60–80% relative humidity with 14/10 h light/dark cycles (light intensity, 200 μmol m−2 s−1). Cotton (Gossypium hirsutum cv. Zhe-Mian 1793) plants were grown in insect-proof greenhouses under natural lighting at 25 ± 3 °C and used for whitefly rearing. Plant seeds were sown in soil and plants were transplanted approximately two weeks later. Three-four weeks later, tobacco plants were at 3–4 true-leaf stage, tomato plants were at 3-4 true-leaf stage, and N. benthamiana plants were at 5-6 true-leaf stage. They were used for experimentation unless otherwise specified. Cotton plants were grown for 8–9 weeks to 9–11 true-leaf stage and then used for whitefly rearing. Transgenic N. benthamiana H2B-RFP plants expressing red fluorescent protein fused to the C-terminus of histone 2B were provided by Dr. Xueping Zhou (Institute of Biotechnology, Zhejiang University). NahG-transgenic N. benthamiana plants were provided by Dr. Xinzhong Cai (Institute of Biotechnology, Zhejiang University). TbCSB βC1 (GenBank accession code: AJ421484.1), ToLCCNB βC1 (GenBank accession code: AJ704612), TuMV HC-Pro (GenBank accession code: AB194797.1), CMV 2b (GenBank accession code: NC_002035.1), or NbNPR3 (GenBank accession code: OR725689) sequences were cloned into the pBWA(V)HS-3xFlag vector and transgenic N. benthamiana plants constitutively expressing these Flag-tagged proteins were then generated using agrobacterium-mediated transformation by Biorun Co., Ltd. (China). To verify transgenic events, reverse transcription-polymerase chain reaction (RT-PCR) was conducted to detect the transcripts of TbCSB βC1, ToLCCNB βC1, TuMV HC-Pro, or CMV 2b. For NbNPR3, transgenic events were verified by quantifying NbNPR3 mRNAs in plants with qPCR.
Viruses and agro-inoculation
Infectious clones of TbCSV isolate Y35 (GenBank accession code: AJ420318) and its associated betasatellite TbCSB (GenBank accession code: AJ421484), and ToLCCNV isolate G18 (GenBank accession code: AJ558119) and its associated betasatellite ToLCCNB (GenBank accession code: AJ704612) were provided by Dr. Xueping Zhou (Institute of Biotechnology, Zhejiang University). Mutant TbCSB (mTbCSB) that was unable to express βC1 was previously described in ref. 25. Agrobacteria containing infectious clones of TbCSV or TbCSB (or mTbCSB) were cultured separately until OD600 reached 1.5–2.0, cells were pelleted and re-suspended in resuspension buffer (10 mM MgCl2, 10 mM MES, and 200 µM acetosyringone). TbCSV was agro-inoculated into plants either alone or in a 1:1 ratio with TbCSB (or mTbCSB), and the final OD600 value of agrobacteria containing infectious clones of TbCSV was kept constant between TbCSV and TbCSV+TbCSB (or mTbCSB) agrobacteria solutions. ToLCCNV and ToLCCNV+ToLCCNB agrobacteria solutions were prepared following the same protocol. One-mL syringes were used to introduce the agrobacteria into leaves of tobacco, tomato, and N. benthamiana plants. Unless specified otherwise, plants were sampled for virus detection at 10 days post inoculation.
Whitefly rearing and infestation
Bemisia tabaci MEAM1 (mtCOI GenBank accession code: KM821540) was reared on cotton plants. For experiments using plants that were infested with whiteflies, adult whiteflies were released into insect-proof cages with tobacco, tomato or N. benthamiana plants in plant growth chambers. The number of plants in the cages varied from 7 to 19 and 100 or 200 whiteflies per plant were released into each cage. Non-infested plants were used as control. Forty-eight hours later, whiteflies were removed and plants were sampled for the analysis of hormone content or subjected to virus inoculation.
Extraction of plant DNA and RNA and quantitative PCR (qPCR) analysis
For the analysis of virus quantity and plant gene transcript level, the first apical fully-expanded leaf from treated or control plants was harvested and frozen in −80 °C until use. For the quantification of viral DNAs, DNAs were extracted from leaves using the Plant Genomic DNA Kit (Tiangen, China). For the analysis of gene transcripts, RNAs were isolated using TRIzol (Invitrogen, USA) and cDNAs were synthesized using an Evo M-MLV RT Kit with gDNA Clean for qPCR (Accurate Biology, China). qPCR analysis was performed using SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology, China) and CFX96 Real-Time PCR Detection System (Bio-Rad, USA). Primers are listed in Supplementary Data 1.
Quantification of phytohormones
Quantification of phytohormones was conducted with two methods. For the first method, SA was quantified alone or with JA and JA-Ile. The second and third apical leaves were harvested from plants and ground to a fine powder in liquid nitrogen using a mortar and pestle. The powder (0.15 g) was transferred into a 1.5 mL centrifuge tube and mixed with 1 mL of ethyl acetate containing 10 ng of D4-SA, D6-JA and D6-JA-Ile (Quality Control Chemicals, USA). Samples were vortexed for 15 min and centrifuged. Supernatants were collected and then evaporated using a vacuum concentrator at 30 °C. The dry residues were re-suspended in 110 μL of MeOH: H2O (1:1, v/v) and mixed by vortexing for 15 min. After centrifugation, 100 μL of the supernatants were collected and analyzed with a high-performance liquid chromatography-tandem mass spectrometry system (TripleTOF 5600 + , SCIEX, USA). The contents of SA, JA and JA-Ile were calculated as normalized to D4-SA, D6-JA and D6-JA-Ile, respectively. For the second method, SA and SAG were quantified in the whitefly-infested and non-infested plant samples by RuiYuan (Nanjing, China) Co., Ltd. The second and third apical leaves were harvested and powdered using liquid nitrogen. SA and SAG were extracted using acetonitrile containing 8 ng of D4-SA (Sigma, USA), and resuspended in methanol. SA and SAG were analyzed with QSight 420 ultra-performance liquid chromatography-tandem mass spectrometry system (PerkinElmer, USA). The contents of SA and SAG were calculated using a standard curve made from a series of solutions of SA (or SAG) standard chemicals (Sigma, USA) with D4-SA (Sigma, USA).
Salicylic acid and geldanamycin treatments
Salicylic acid (SA, Sigma-Aldrich, USA) was dissolved in ethanol to make a 2 M stock solution. SA was diluted to 4.0, 2.0, 1.0 or 0.5 mM in 0.2% ethanol. Ethanol (0.2%) was used as control. A hand sprayer was used to apply 0.5 mL of SA or ethanol solution per plant. Unless specified otherwise, plants were sprayed once per day for three consecutive days. For each plant species, 1.0 mM SA solutions were first tested in preliminary trails and then SA concentrations for experimentation were determined based on the results obtained. Geldanamycin (GDA, Selleck, USA) was dissolved in DMSO to make a 100 mM stock solution, and diluted to 100 or 200 μM working solution. DMSO was used as control. To analyze the effect of GDA on TbCSV infection, TbCSV+TbCSB (cultured and re-suspended in a 1:1 ratio) was mixed with 100 or 200 μM GDA solution or DMSO, and co-inoculated into N. benthamiana plants. To analyze the effects of GDA on the transcript level of SA-regulated genes, GDA (100 or 200 μM) or DMSO were infiltrated into N. benthamiana leaves. The next day, plants were sprayed with SA or ethanol solutions for three consecutive days. Twenty-four hours after the last spray, leaves were harvested and subjected to gene transcript analysis as described above.
GST Pull-down and mass spectrometry assay
The coding region of TBCSB βC1 was cloned into the pGEX-6p-1 vector to express a glutathione S-transferase (GST)-βC1 fusion protein. Primers are listed in Supplementary Data 1. The recombinant plasmids were transferred into E. coli strain BL21 for protein expression. Cells were induced with 0.2 mM IPTG for 12 h at 16 °C, lysed by ultrasonication and then centrifuged. Supernatants were collected and mixed with glutathione agarose beads (GE Healthcare, USA) at 4 °C for 2 h. The beads were loaded into 6-mL affinity chromatography columns (Sangon, China) and washed with 5 mL 1xPBS for five times. After verification by SDS-PAGE and Coomassie blue staining, the bead-bound GST-βC1 and GST proteins were mixed with total N. benthamiana proteins. N. benthamiana proteins were extracted from 2.0 g leaves from 5-6 true-leaf stage plants using a native extraction buffer (50 mM Tris-HCl, pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM dithiothreitol, and 1 mM PMSF). After incubation with leaf proteins for 4 h at 4 °C in a revolver, the beads were washed with 1 mL 1xPBS for five times and analyzed with SDS-PAGE and Coomassie blue staining. The protein bands that were present in GST-βC1 treatment but not in GST treatment were collected as one sample, and analyzed by mass spectrometry (LTQ Orbitrap Elite, Thermo Fisher, USA) using default parameters. Mass spectrometry results were analyzed with Proteome Discoverer (version 2.0) and the captured peptides were annotated using the software SEQUEST with default parameters and the N. benthamiana protein database (www.nbenth.com) was used as the reference database26.
Sequence analysis of NbHSP90s and NbNPR3
To obtain the entire complement of HSP90s (Pfam entry PF00183) in N. benthamiana plants, we searched the N. benthamiana protein database (www.nbenth.com) using hidden Markov models. The hmmsearch program in HMMER (version 3.4) was used, and the E-value was e-5. In total, 18 sequences were obtained, among which 12 sequences with the length over 698 amino acids were subjected to phylogenic analysis with A. thaliana HSP90 proteins43. For NbNPR3, the deduced amino acid sequence of the obtained DNA sequence was used to predict functional domains in NCBI. CDD v3.20-59693 PSSMs database was used and expect value threshold was 0.01. Next, phylogenic relationships between NbNPR3 and A. thaliana NPR proteins44 were determined. MEGA6 and the incorporated Neighbor-Joining (NJ) algorithm were used45. The reliability of the phylogenetic analysis was examined by percentages obtained through 1000 bootstrap iterations of the datasets.
Bimolecular fluorescence complementation (BiFC)
The full-length coding regions of TBCSB βC1, TuMV HC-Pro, CMV 2b, NbHSP90-2, NbHSP90-10, and NbHSP90-12 were cloned into p2YC vector, and the full-length coding regions of NbHSP90-2, NbHSP90-10, and NbHSP90-12 were cloned into p2YN vector. p2YC and p2YN contain sequences encoding cYFP (159-238 of the YFP protein) and nYFP (1-158 of the YFP protein), respectively (provided by Dr. Xueping Zhou, Zhejiang University). GenBank accession codes are OR727908 for NbHSP90-2, OR727909 for NbHSP90-10, and OR727910 for NbHSP90-12. Additionally, the N-terminal, middle and C-terminal domains of NbHSP90-2 and NbHSP90-10, namely NbHSP90-2 ND (1-221 aa), NbHSP90-10 ND (1-312 aa), NbHSP90-2 MD (245-591 aa), NbHSP90-10 MD (328-676 aa), NbHSP90-2 CD (592-699 aa), and NbHSP90-10 CD (677-793 aa) were cloned into p2YN vector. All primers are listed in Supplementary Data 1. These recombinant plasmids were transferred into Agrobacterium tumefaciens EHA105. Transgenic N. benthamiana H2B-RFP plants were co-infiltrated with agrobacteria containing recombinant p2YC vectors and recombinant p2YN vectors in equal ratios. Fluorescence was examined using a confocal microscope (Zeiss LSM800, Germany) at two days post inoculation (RFP excitation, 590 nm; RFP detection, 582-650 nm; YFP excitation, 524 nm; YFP detection, 450-585 nm).
Yeast two-hybrid
The full-length coding regions of TBCSB βC1, TuMV HC-Pro, and CMV 2b were cloned into the pGBKT7 vector, and NbHSP90-2, NbHSP90-10 and NbNPR3 were cloned into the pGADT7 vector. All primers are listed in Supplementary Data 1. Recombinant pGBKT7 and pGADT7 plasmids (50 ng/µL) were co-transformed into Saccharomyces cerevisiae AH109 Chemically Competent Cells (Weidi Biology, China) as per the manufacturer’s protocol. All transformants were plated on SD/-Leu-Trp dropout selective medium, and then transferred to SD/-Ade-His-Leu-Trp selective medium to verify interactions.
Co-immunoprecipitation (co-IP)
The coding sequences of TBCSB βC1, TuMV HC-Pro, CMV 2b, NbHSP90-2, NbHSP90-10, NbHSP90-12, and NbNPR3 were cloned into pBWA(V)HS-3xFlag vectors. The sequences of NbHSP90-2, NbHSP90-10, NbHSP90-12, and NbNPR3 were cloned into pCAMBIA1300-3xHA vectors, and pCAMBIA1300-GFP−3xHA was used as control. GenBank accession code is OR725689 for NbNPR3. Primers are listed in Supplementary Data 1. These recombinant plasmids were mobilized into A. tumefaciens EHA105 by electro-transformation. N. benthamiana leaves were co-infiltrated with agrobacteria containing a recombinant pBWA(V)HS-3xFlag vector and agrobacteria containing recombinant pCAMBIA1300-3xHA vectors (1:1 ratio). At two days post inoculation, proteins were extracted from 0.5 g of the leaves with 1 mL IP buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10% glycerol, 0.1% Tween 20, 1 mM β-mercaptoethanol, 2.5 mM imidazole, 1 mM DTT, and 1 mM PMSF). Soluble proteins were immunoprecipitated with 20 μL anti-Flag M2 Magnetic Beads (Sigma-Aldrich, USA) at 4 °C for 2 h, washed six times using IP buffer and proteins were subjected to SDS-PAGE and western blot analysis. Flag-tagged proteins and HA-tagged proteins were detected with Anti-Flag Tag Mouse Monoclonal Antibody (EarthOx, E022060-01) and Anti-HA Tag Mouse Monoclonal Antibody (EarthOx, E022010-01), respectively.
To explore the effects of TBCSB βC1 on the interaction between NbHSP90-2 and NbNPR3, GST and GST-TBCSB βC1 were expressed in E. coli cells, purified using glutathione agarose beads (GE Healthcare, USA) and eluted using 20 mM glutathione. Proteins were verified by SDS-PAGE and Coomassie blue staining. NbHSP90-2-HA was co-expressed with NbNPR3-Flag in N. benthamiana for two days. Proteins were extracted, and verified with SDS-PAGE and western blot analysis. The NbHSP90-2-HA+NbNPR3-Flag extract was mixed with various amount of GST or GST-TBCSB βC1, respectively, and then co-immunoprecipitated using anti-Flag M2 Magnetic Beads (Sigma-Aldrich, USA) as described above.
Virus-induced gene silencing (VIGS)
For VIGS of NbNPR3, NbHSP90-2, NbHSP90-10, and NbHSP90-12, around 300 bp of the coding sequences were cloned into pTRV-RNA2 for virus-induced gene silencing46 using primers listed in Supplementary Data 1. Recombinant plasmids were introduced into A. tumefaciens EHA105 by electro-transformation. Agrobacteria containing recombinant pTRV2 or pTRV1 were cultured and re-suspended to an OD600 of 0.2. Equal amounts of agrobacteria cells containing recombinant pTRV2 and pTRV1 were mixed and inoculated into N. benthamiana plants using 1 mL syringes. pTRV1 + pTRV2-PDS served as the positive control and pTRV1 + pTRV2-GFP as the negative control. One week later, plants were sampled for gene-silencing efficiency, treated with SA or ethanol for three consecutive days and assessed for gene transcripts using qPCR. To explore the effects of gene silencing on virus infection, TbCSV+TbCSB agrobacteria solutions were mixed with pTRV1 + pTRV2 agrobacteria solutions (1:1, v/v), and then used for agro-inoculation. At 7 days post inoculation, plants were sprayed with SA or ethanol solutions for three consecutive days. Three days post the last spray, plants were sampled for virus detection.
Semi-in vivo and in vivo degradation of NbNPR3
Semi-in vivo degradation of NbNPR3 was analyzed following the protocol described in Shen et al.47 with minor modifications. pBWA(V)HS-3xFlag vector expressing Flag-tagged NbNPR3, TBCSB βC1, TuMV HC-Pro, CMV 2b, NbHSP90-2, and NbHSP90-10 were used, and pBWA(V)HS-GUS−3xFlag served as a control. All the recombinant plasmids were mobilized into A. tumefaciens EHA105. Agrobacteria cells containing pBWA(V)HS-NbNPR3−3xFlag plasmid was inoculated into N. benthamiana leaves; 24 h later, proteins were extracted with native extraction buffer as described above. Plant extracts were mixed with cycloheximide (CHX) (final concentration: 100 μM) and adenosine triphosphate (ATP) (final concentration: 20 mM) and used for SA or MG132 treatments. For SA treatments, SA was added to the protein extract to a final concentration of 1 mM; the control sample was mixed with an equal volume of ethanol. For the proteasome inhibitor treatment, MG132 was added to a final concentration of 100 μM; an equal volume of DMSO was used for the control. The samples were agitated in an Eppendorf Thermomixer at 25 °C and 100 μL aliquots were collected at 0, 1 and 4 h. Samples were subjected to SDS-PAGE and western blot analysis using Anti-Flag Tag Mouse Monoclonal Antibody (EarthOx, E022060-01). The blots were stained with Ponceau S to estimate protein loading, and the most obvious bands were presented.
To explore the impact of TBCSB βC1, TuMV HC-Pro, CMV 2b, NbHSP90-2, and NbHSP90-10 on SA-induced degradation of NbNPR3 in semi-in vivo degradation assay, NbNPR3-Flag was co-expressed with these proteins in N. benthamiana. NbNPR3-Flag co-expressed with GUS-Flag was used as control. At 24 h post inoculation, proteins were extracted with native extraction buffer and CHX, ATP and SA were added as described above. To explore the effects of GDA on NbNPR3 degradation, TbCSB βC1-Flag was co-expressed with NbNPR3-Flag. Protein extracts were then mixed with CHX, ATP, and SA. One aliquot of the extract was then mixed with GDA (final concentration: 100 μM) and the second aliquot was mixed with DMSO (control). Protein integrity was assessed as described above.
For in vivo degradation assay, agrobacteria cells containing pBWA(V)HS-NbNPR3−3xFlag plasmid was inoculated into N. benthamiana leaves. Twenty-four hours later, 1.0 mM SA or 0.2% ethanol (control) was sprayed. Leaves were harvested at 5 h post SA spray and then subjected to protein extraction and the detection of NbNPR3-Flag. To explore the impact of TbCSB βC1 on SA-induced NbNPR3 degradation, TbCSB βC1-Flag was co-expressed with NbNPR3-Flag in N. benthamiana leaves. Leaves were then treated with SA and then subjected to protein extraction and the detection of TbCSB βC1-Flag and NbNPR3-Flag.
Statistics and reproducibility
For the comparison of percentage of symptomatic plants, Fisher’s exact test of independence was used. qPCR data of virus quantity and gene transcript level were normalized to plant actin using 2-ΔCt method. Comparisons were conducted using two-sided Student’s independent t-test in experiments with only two treatments, and one-way analysis of variance (ANOVA) along with Fisher’s least significant difference (LSD) was used in experiments with more than two treatments. To clearly illustrate the differences, the data of virus quantity and gene transcript level in each of the experiments were normalized to that of control. All data were presented as the mean ± standard errors of mean (mean ± SEM) and differences were considered significant when P < 0.05. The P value is provided for those comparisons wherein data displayed strong trends, but were not statistically different. All statistical analyses were conducted using SPSS Statistics 21.0 and EXCEL. All experiments were repeated at least once with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Myron Zalucki (the University of Queensland, Australia) and Qiaomei Wang (Zhejiang University, China) for their comments on an earlier version of the manuscript. Financial support for this study was provided by the National Key Research and Development Program (2021YFC2600100) (X.W.W. and L.L.P.), the National Natural Science Foundation of China [32202397 (L.L.P.) and 31930092 (S.S.L. and L.L.P.)] and the earmarked fund for China Agriculture Research System (CARS-23-C05) (L.L.P.).
Author contributions
Conceptualization, S.S.L. and L.L.P.; Data curation, J.R.Z., and L.L.P.; Formal analysis, J.R.Z., S.S.L., and L.L.P.; Funding acquisition, X.W.W., S.S.L., and L.L.P.; Investigation, J.R.Z., Y.M.L., D.L., and Y.J.W.; Methodology, J.R.Z., Y.M.L., S.X.Z., and L.L.P.; Project administration, J.R.Z., and L.L.P.; Supervision, S.S.L., and L.L.P.; Validation, L.L.P.; Writing-review & editing, J.R.Z., X.W.W., S.S.L., L.L.W., and L.L.P.; All authors read and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks Zhengqing Fu, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The datasets supporting the conclusions of this article are deposited in Figshare (10.6084/m9.figshare.27273666). The NCBI accession codes are AJ420318 for TbCSV isolate Y35, AJ421484 for TbCSB, AJ558119 for ToLCCNV isolate G18, AJ704612 for ToLCCNB, AJ421484.1 for TbCSB βC1, AJ704612 for ToLCCNB βC1, AB194797.1 for TuMV HC-Pro, NC_002035.1 for CMV 2b, OR727908 for NbHSP90-2, OR727909 for NbHSP90-10, and OR727910 for NbHSP90-12, OR725689 for NbNPR3 and KM821540 for the mtCOI sequence of Bemisia tabaci MEAM1. Protein mass spectrometry raw data is deposited in a ProteomeXchange partner repository under the accession code PXD056381. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Consent for publication
All authors approved the manuscript and gave consent for publication.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jing-Ru Zhang, Yi-Ming Liu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53894-y.
References
- 1.Whitfield, A. E., Falk, B. W. & Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology479-480, 278–289 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wang, X. W. & Blanc, S. Insect transmission of plant single-stranded DNA viruses. Annu. Rev. Entomol.66, 389–405 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Eigenbrode, S. D., Bosquepérez, N. A. & Davis, T. S. Insect-borne plant pathogens and their vectors: Ecology, evolution, and complex interactions. Annu. Rev. Entomol.63, 169–191 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Pieterse, C. M. J., Van der Does, D., Zamioudis, C., Leon-Reyes, A. & Van Wees, S. C. M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol.28, 489–521 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Chaman, M. E., Copaja, S. V. & Argandoña, V. H. Relationships between salicylic acid content, phenyl-alanine ammonia-lyase (PAL) activity, and resistance of barley to aphid infestation. J. Agr. Food Chem.51, 2227–2231 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Zarate, S. I., Kempema, L. A. & Walling, L. L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol.143, 866–875 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, P. et al. Vector and nonvector insect feeding reduces subsequent plant susceptibility to virus transmission. N. Phytol.215, 699–710 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Hodge, S., Bennett, M., Mansfield, J. W. & Powell, G. Aphid-induction of defence-related metabolites in Arabidopsis thaliana is dependent upon density, aphid species and duration of infestation. Arthropod-Plant Inte13, 387–399 (2019). [Google Scholar]
- 9.Liu, Q. P., Li, S. L. & Ding, W. Aphid-induced tobacco resistance against Ralstonia solanacearumis associated with changes in the salicylic acid level and rhizospheric microbial community. Eur. J. Plant Pathol.157, 465–483 (2020). [Google Scholar]
- 10.Chivasa, S., Murphy, A. M., Naylor, M. & Carr, J. P. Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell9, 547–557 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, T. et al. Begomovirus-whitefly mutualism is achieved through repression of plant defenses by a virus pathogenicity factor. Mol. Ecol.21, 1294–1304 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Li, R. et al. Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell26, 4991–5008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji, L. H. & Ding, S. W. The suppressor of transgene RNA silencing encoded by cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Mol. Plant-Microbe Inte.14, 715–724 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Cheng, X. F. et al. Sumoylation of turnip mosaic virus RNA polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell29, 508–525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poque, S. et al. Potyviral gene-silencing suppressor HCPro interacts with salicylic acid (SA)-binding protein 3 to weaken SA-mediated defense responses. Mol. Plant-Microbe Inte.31, 86–100 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Jiang, Z. H. et al. Barley stripe mosaic virus γb protein targets thioredoxin h-type 1 to dampen salicylic acid-mediated defenses. Plant Physiol.189, 1715–1727 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong, Q. et al. Molecular basis of methyl-salicylate-mediated plant airborne defence. Nature622, 139–148 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Zhang, H. H. et al. Different viral effectors suppress hormone mediated antiviral immunity of rice coordinated by OsNPR1. Nat. Commun.14, 3011 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Z. H., Xie, Y. & Zhou, X. P. Tobacco curly shoot virus DNA is not necessary for infection but intensifies symptoms in a host-dependent manner. Phytopathology95, 902–908 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Lee, H. I., Leon, J. & Raskin, I. Biosynthesis and metabolism of salicylic acid. Proc. Natl Acad. Sci. USA.92, 4076–4079 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaffney, T. et al. Requirement of salicylic-acid for the induction of systemic acquired-resistance. Science261, 754–756 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Yang, X. L., Guo, W., Ma, X. Y., An, Q. L. & Zhou, X. P. Molecular characterization of tomato leaf curl China virus, infecting tomato plants in China, and functional analyses of its associated betasatellite. Appl. Environ. Microbiol.77, 3092–3101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, X. L., Guo, W., Li, F. F., Sunter, G. & Zhou, X. P. Geminivirus-associated betasatellites: Exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci.24, 519–529 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Mubin, M. et al. Journey of begomovirus betasatellite molecules: From satellites to indispensable partners. Virus Genes56, 16–26 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Wu, Y. J., Liu, Y. M., Li, H. Y., Liu, S. S. & Pan, L. L. Temporal dynamic of the ratio between monopartite begomoviruses and their associated betasatellites in plants, and its modulation by the viral gene βC1. Viruses15, 954 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranawaka, B. et al. A multi-omic Nicotiana benthamiana resource for fundamental research and biotechnology. Nat. Plants9, 1558–1571 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.di Donato, M. & Geisler, M. HSP90 and co-chaperones: A multitaskers’ view on plant hormone biology. FEBS Lett.593, 1415–1430 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Zhang, X. C., Millet, Y. A., Cheng, Z., Bush, J. & Ausubel, F. M. Jasmonate signalling in Arabidopsis involves SGT1b-HSP70-HSP90 chaperone complexes. Nat. Plants1, 15049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, R. et al. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun.7, 10269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu, Z. Q. et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature486, 228–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding, Y. L. et al. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell173, 1454–1467 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Rochon, A., Boyle, P., Wignes, T., Fobert, P. R. & Després, C. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell18, 3670–3685 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou, Y., Park, S. H. & Chua, N. H. UBP12/UBP13-mediated deubiquitination of salicylic acid receptor NPR3 suppresses plant immunity. Mol. Plant16, 232–244 (2023). [DOI] [PubMed] [Google Scholar]
- 34.Scholthof, K. B. G. et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol.12, 938–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S. F., Guo, H. J., Ge, F. & Sun, Y. C. Apoptotic neurodegeneration in whitefly promotes the spread of TYLCV. eLife9, e56168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy, A. M., Zhou, T. & Carr, J. P. An update on salicylic acid biosynthesis, its induction and potential exploitation by plant viruses. Curr. Opin. Virol.42, 8–17 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Zhao, S. & Li, Y. Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog.17, e1009242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen, J. et al. Reprogramming and remodeling: transcriptional and epigenetic regulation of salicylic acid-mediated plant defense. J. Exp. Bot.71, 5256–5268 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y. L. et al. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J.48, 647–656 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Wu, Y. et al. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep.1, 639–647 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Chang, M. et al. PBS3 protects EDS1 from proteasome-mediated degradation in plant immunity. Mol. Plant12, 678–688 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Spoel, S. H. et al. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell137, 860–872 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishna, P. & Gloor, G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress and. Chaperon6, 238–246 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, G. S., Holub, E. B., Alonso, J. M., Ecker, J. R. & Fobert, P. R. An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J.41, 304–318 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol.30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen, Q. T. et al. Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded βC1. Mol. Plant9, 911–925 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Liu, Y. L., Schiff, M., Marathe, R. & Dinesh-Kumar, S. P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J.30, 415–429 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The datasets supporting the conclusions of this article are deposited in Figshare (10.6084/m9.figshare.27273666). The NCBI accession codes are AJ420318 for TbCSV isolate Y35, AJ421484 for TbCSB, AJ558119 for ToLCCNV isolate G18, AJ704612 for ToLCCNB, AJ421484.1 for TbCSB βC1, AJ704612 for ToLCCNB βC1, AB194797.1 for TuMV HC-Pro, NC_002035.1 for CMV 2b, OR727908 for NbHSP90-2, OR727909 for NbHSP90-10, and OR727910 for NbHSP90-12, OR725689 for NbNPR3 and KM821540 for the mtCOI sequence of Bemisia tabaci MEAM1. Protein mass spectrometry raw data is deposited in a ProteomeXchange partner repository under the accession code PXD056381. Source data are provided with this paper.