Abstract
Unlike mild DNA damage exposure, DNA damage repair (DDR) is reported to be ineffective in full-grown mammalian oocytes exposed to moderate or severe DNA damage. The underlying mechanisms of this weakened DDR are unknown. Here, we show that moderate DNA damage in full-grown oocytes leads to aneuploidy. Our data reveal that DNA-damaged oocytes have an altered, closed, chromatin state, and suggest that the failure to repair damaged DNA could be due to the inability of DDR proteins to access damaged loci. Our data also demonstrate that, unlike somatic cells, mouse and porcine oocytes fail to activate autophagy in response to DNA double-strand break-inducing treatment, which we suggest may be the cause of the altered chromatin conformation and inefficient DDR. Importantly, autophagy activity is further reduced in maternally aged oocytes (which harbor severe DNA damage), and its induction is correlated with reduced DNA damage in maternally aged oocytes. Our findings provide evidence that reduced autophagy activation contributes to weakened DDR in oocytes, especially in those from aged females, offering new possibilities to improve assisted reproductive therapy in women with compromised oocyte quality.
Subject terms: Meiosis, Chromosome segregation, Double-strand DNA breaks, Macroautophagy
DNA damage repair machinery is not robust in full-grown mammalian oocytes. Here, the authors demonstrate that mammalian oocytes fail to activate autophagy in response to DNA DSBs, correlating with an altered chromatin structure, improper localization of DNA damage repair proteins and increased DNA damage.
Introduction
Infertility is a highly prevalent global issue. In the US, at least 10% of women within reproductive age experience infertility1–3. Furthermore, female fertility starts to decline in the early thirties, and this decline accelerates after the age of 354. A major cause of this decline is reduced oocyte quality. Female gametes (oocytes) are produced through two rounds of meiotic divisions following a single round of DNA replication. For poorly understood reasons, oocyte meiosis is notoriously prone to errors leading to a high rate of aneuploidy, the leading genetic cause of miscarriage and congenital abnormalities (e.g., Down’s syndrome). Indeed, the incidence of aneuploidy is at least 10times higher in oocytes than in spermatozoa (male gametes), and frequently occurs in meiosis I as opposed to meiosis II5. This high rate of aneuploidy in oocytes increases exponentially with the advancement of age6. Therefore, understanding the molecular mechanisms regulating oocyte meiosis is essential to unveiling how these mechanisms are further perturbed in oocytes from females of advanced reproductive age.
DNA damage is a major problem leading to premature aging, genome instability, mutagenesis and cancer in somatic cells7–13. DNA double-strand breaks (DSBs) are considered the most lethal form of DNA damage which occurs constantly in almost all cell types, due to assaults by endogenous and exogenous environmental agents. As a result, cells have evolved a DNA damage response that involves a complex network of signals responsible for the activation of specific machineries mediating DNA damage sensing, cell cycle regulation, DNA damage repair (DDR) and apoptosis14,15. In somatic cells, following DNA damage, the cell activates a DNA damage checkpoint, an essential step to allow the required time for DDR by inducing cell cycle arrest. If the DNA damage is not repaired, apoptosis will be triggered to induce a cell death16–18. During early fetal development, mammalian oocytes enter meiosis followed by meiotic recombination. Meiotic recombination requires the formation of programmed DNA DSBs. At that stage, oocytes respond efficiently to this programmed DNA damage and repair these programmed DNA breaks by homologous recombination19. Shortly after birth, oocytes undergo an arrest at prophase I of meiosis I which is accompanied by oocyte growth (i.e. increase in oocyte volume, growing oocytes). Quiescent growing oocytes can efficiently repair DNA damage in apoptosis-inhibited mice by activating RAD51-mediated DDR machinery20. After puberty and prior to ovulation, arrested full-grown oocytes resume meiosis I. In contrast to somatic cells and growing oocytes, full-grown mammalian oocytes can progress through meiosis despite having DNA damage21. Surprisingly, studies in mouse and human oocytes suggest that the DNA damage response is further weakened in full-grown oocytes of reproductively aged females, compared to young adult females22–25. Why the DNA damage response (i.e., to exogenous DNA damage) is not robust in full-grown mammalian oocytes, especially in those from females with advanced reproductive age, remains an unresolved question, representing a significant gap in our knowledge of why oocyte meiosis I is notoriously prone to meiotic errors.
Autophagy is a cellular quality control mechanism which plays an important role in maintaining cellular homeostasis by degrading and recycling unnecessary cytoplasmic proteins and organelles in response to diverse stress conditions, such as nutrient deprivation, infection and genotoxic stress26,27. Autophagy includes three major forms: microautophagy, chaperon-mediated autophagy and macroautophagy28,29. Macroautophagy is the most prevalent form of autophagy. In macroautophagy, hereafter referred to as autophagy, a double-membrane vesicle known as an autophagosome forms which will target and isolate damaged cytoplasmic components such as protein aggregates and organelles from the rest of the cell. The autophagosome will then fuse with a lysosome to form an autolysosome to accomplish degradation of the content by lysosomal enzymes30,31. In somatic cells, autophagy is activated in response to DNA damage and plays an important role in regulating several cellular functions including DDR32–36. Emerging evidence indicate that autophagy inhibition in DNA-damaged cells results in QSTM1/p62 upregulation, E3 ligase RNF168s activity inhibition and H2A ubiquitination reduction35. Following DNA damage, histone ubiquitination is critical for altering chromatin structure37, an important step for DDR38–43. We and others showed that treating oocytes with MG-132 (a proteasomal inhibitor) or rapamycin, known to stimulate autophagy, improved the developmental competence of mammalian oocytes44–47 and decreased DNA damage48. While these studies do not directly examine autophagy or its status following exogenous DNA damage, their findings imply the possible participation of autophagy in mitigating DNA damage. Therefore, the role of autophagy in the response of full-grown mammalian oocytes to exogenous DNA damage remains unknown.
Here we demonstrate that full-grown oocytes behave differently than somatic cells in response to DNA damage. We found that full-grown oocytes fail to activate autophagy in response to exogenous DSB inducers. The inability of full-grown oocytes to activate autophagy correlated with (1) altered chromatin architecture, (2) the failure of DDR protein, RAD51, to localize to DNA damaged sites, (3) their inefficiency to repair damaged DNA, and (4) increased aneuploidy incidence. Importantly, induction of autophagy in DNA-damaged oocytes rescued altered chromatin architecture, increased RAD51 localization to the DNA, decreased DNA DSBs, and reduced aneuploidy incidence. Our results suggest that failure to activate autophagy in response to DNA DSBs contributes to inefficient DDR function in full-grown oocytes.
Results
DNA-damaged oocytes progress through meiosis I leading to aneuploidy
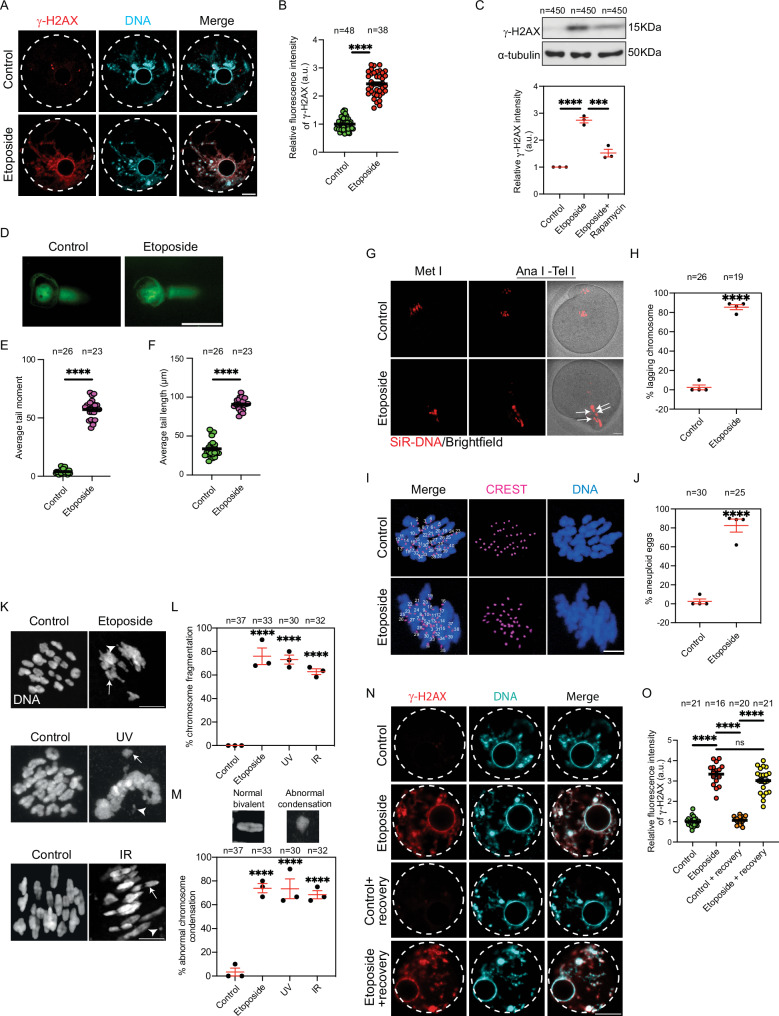
To examine the response of full-grown prophase I-arrested (germinal vesicle, GV) oocytes to exogenous DNA damage, we treated full-grown GV oocytes with etoposide, a topoisomerase II inhibitor, that is widely used as a DSB inducer in somatic cells49,50 and mammalian oocytes21,25,51,52. Upon inducing DSBs, histone H2AX is rapidly phosphorylated on Ser139 (γH2AX) by Ataxia Telangiectasia Mutated (ATM) kinase to trigger signaling for DDR53–55. Once DNA is repaired, γH2AX decreases56. Therefore, γH2AX is recognized as a sensitive molecular marker for DSBs in somatic cells and oocytes57–60. Similar to previous reports21,25,61, treating full-grown GV oocytes (collected from sexually mature mice and incubated with milrinone, a phosphodiesterase inhibitor, to prevent meiotic resumption) with etoposide (50 μg/ml for 3 h51) induced DSBs as evidenced by the significant increase of γH2AX fluorescence pixel intensity (Fig. 1A, B and Supplementary Fig. 1A) and γH2AX levels by Western blot analysis (Fig. 1C) in etoposide-treated oocytes, relative to DMSO-treated controls. To confirm this finding, we employed the alkaline comet assay, an effective method to detect DNA fragmentation/damage in eukaryotic cells. Alkaline comet assay measures DNA tail moment and length (See Material and Methods for more detail), which correlate with DNA damage62. Indeed, etoposide-treated oocytes exhibited a significant increase in both DNA tail length and moment, compared to DMSO-treated controls (Fig. 1D–F). However, caspase 3, an apoptosis marker, did not change significantly in etoposide-treated oocytes, compared to control oocytes (Supplementary Fig. 1B). Induction of DSBs in full-grown GV oocytes does not completely prevent meiotic resumption and progression when the oocytes are allowed to mature in vitro25,61. Similar to previous reports51, approximately 40% of DNA-damaged oocytes (induced by 50 µg/ml etoposide), which resumed meiosis and underwent nuclear envelope breakdown (NEBD), were able to extrude the first polar body (PB) and reached metaphase II stage when matured in milrinone-free medium (Supplementary Fig. 1C). However, there was a significant delay in the timing of PB extrusion (PBE) in etoposide-treated oocytes when compared to control oocytes (Supplementary Fig. 1D). Using time-lapse confocal imaging, we observed lagging chromosomes during anaphase I/telophase I in ~80% of DNA-damaged oocytes which were able to extrude the PB (Fig. 1G,H; Supplementary Movies 1 and 2), a phenotype that highly correlates with aneuploidy63,64. Using in situ chromosome counting technique, we found that ~80% of etoposide-treated oocytes that extruded the PB were aneuploid, compared to ~3% aneuploidy in controls (Fig. 1I,J). Thus, the exposure of full-grown GV oocytes to DNA damage leads to the development of aneuploid gametes. Because most chromosome segregation errors during meiosis I arise from mistakes occurring during metaphase I65, we examined metaphase I stage in DNA-damaged oocytes in detail. Consistent with previous reports52,61, we observed higher rates of chromosome misalignment (Supplementary Fig. 1E, F) and chromosome fragmentation (Fig. 1K, L) in etoposide-treated oocytes when compared to control oocytes. Interestingly, we found that almost all chromosome fragments in etoposide-treated oocytes contain kinetochores (Supplementary Fig. 1G_c,d). Kinetochores are large protein complex that link spindle microtubules to centromeric DNA of chromosomes. Therefore, we asked whether chromosome fragmentation in etoposide-treated oocytes is induced by microtubule-induced tension on chromosomes. Treatment of DNA-damaged oocytes with nocodazole, a microtubule depolymerizing drug, decreased chromosome fragmentation (Supplementary Fig. 1G, H), indicating that microtubule-induced tension on chromosomes leads to chromosome fragmentation at metaphase I in DNA-damaged oocytes. These results are consistent with previous observations that microtubules can establish stable attachments with kinetochores in DNA-damaged oocytes66. During meiosis I, homologous chromosomes must be bioriented and correctly attached (at kinetochores) to opposite spindle poles, necessary for accurate chromosome segregation during anaphase I and telophase I. The presence of separated kinetochores in DNA-damaged oocytes (Supplementary Fig. 1G_c,d) may explain why homologous chromosomes do not properly segregate during anaphase/telophase I, leading to aneuploidy. Importantly, we found that chromosome architecture is altered in DNA-damaged oocytes. In contrast to control oocytes in which chromosomes have a normal bivalent morphology, etoposide-treated oocytes had significantly higher rates of abnormal chromosome condensation (i.e., loss of normal bivalent morphology) (Fig. 1K, M). These chromosome condensation defects and chromosome fragmentation are not specific to etoposide treatment because similar phenotypes were observed when DSBs were induced by a brief exposure to ultraviolet (UV) B radiation (302 nm, 30 s) or ionizing radiation (1 Gy) (Fig. 1K–M). These results demonstrate that the exposure of full-grown oocytes to DSBs at the GV stage alters chromosome architecture at metaphase I and increases their risk of developing aneuploid gametes.
Fig. 1. DNA-damaged oocytes progress through meiosis I resulting in aneuploidy.
A Full-grown germinal vesicle (GV) oocytes were incubated in milrinone-containing CZB medium supplemented with DMSO (control) or 50 μg/ml etoposide for 3 h followed by immunostaining with γH2AX antibody. B Quantification of γH2AX fluorescence intensity. C Control and etoposide-treated GV oocytes were assessed by Western blot analysis. Quantification of γH2AX (lower panel). D Control and etoposide-treated GV oocytes were examined for DNA damage by alkaline comet assay. Scale bars represent 100 μm. E Quantification of DNA tail moment. F Quantification of DNA tail length. G Control and etoposide-treated GV oocytes were in vitro matured in CZB medium containing SiR-DNA. Shown are representative time-lapse images. The white arrow represents lagging chromosomes. H Quantification of lagging chromosome percentage. I Control and etoposide-treated GV oocytes were in vitro matured for metaphase II followed by in situ chromosome counting to assess aneuploidy. J Quantification of aneuploidy percentage. K Etoposide- ultraviolet B (UV)- or ionizing radiation (IR)- treated GV oocytes were assessed for DNA morphology at metaphase I. White arrows represent abnormal chromosome condensation and white arrow heads represent a fragmented chromosome. L Quantification of chromosome fragmentation percentage. M Quantification of abnormal chromosome condensation percentage. N Full-grown GV oocytes were incubated in milrinone-containing CZB medium supplemented with DMSO (control) or etoposide for 3 h. A subset of control and etoposide-treated oocytes were fixed after 3 h, whereas the remaining oocytes were released from etoposide and cultured in milrinone-containing CZB medium for 16 h (recovery) prior to fixation. The oocytes were immunostained with γH2AX antibody. O Quantification of γH2AX fluorescence intensity. DNA was labeled with DAPI. Scale bars represent 10 μm. The white dashed circle represents the nucleus. Data are expressed as mean ± SEM. Each dot in plot graphs represents an oocyte except C, H, J, L and M represents the average of an experimental replicate. Two-tailed Student-t-test (B, E, F, H and J), one-way ANOVA (C, L, M and O) were used to analyze the data. Values with asterisks are significantly different, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The total number of analyzed oocytes is specified above each graph/column. All experiments were replicated three times except G, H, I and J (four replicates). Source data are provided as a Source Data file.
Somatic cells can efficiently repair most DNA damage if they are given longer time for recovery67. We asked whether providing DNA-damaged oocytes with a longer recovery duration would allow for efficient DDR. Following a 3 h etoposide treatment (50 μg/ml), GV oocytes were cultured in etoposide-free medium containing milrinone (to prevent meiotic resumption and to keep all oocytes arrested at the GV stage) for an additional 16 h followed by assessing γH2AX levels. Interestingly, full-grown GV oocytes were not able to fully repair DSBs as γH2AX did not decrease significantly after the extended incubation in etoposide-free medium (Fig. 1N,O). This finding is consistent with a previous observation that, at 6 h post-etoposide washout (50 or 100 μg/ml etoposide treatment for 3 h), the levels of γH2AX did not return to the baseline levels and were approximately 7-10-fold higher than those in the control group68. Given that oocytes can fully repair mild DNA damage (induced by a short incubation, 15 min, with a lower concentration, 25 μg/ml, of etoposide) over a period of 10 h52, our findings suggest that full-grown GV oocytes have a limited capacity to repair moderate levels of DNA damage (induced by 50 μg/ml etoposide treatment for 3 h in this study), and at that stage, one or more of DDR mechanisms are not fully functional.
Mammalian oocytes do not activate autophagy in response to DSBs
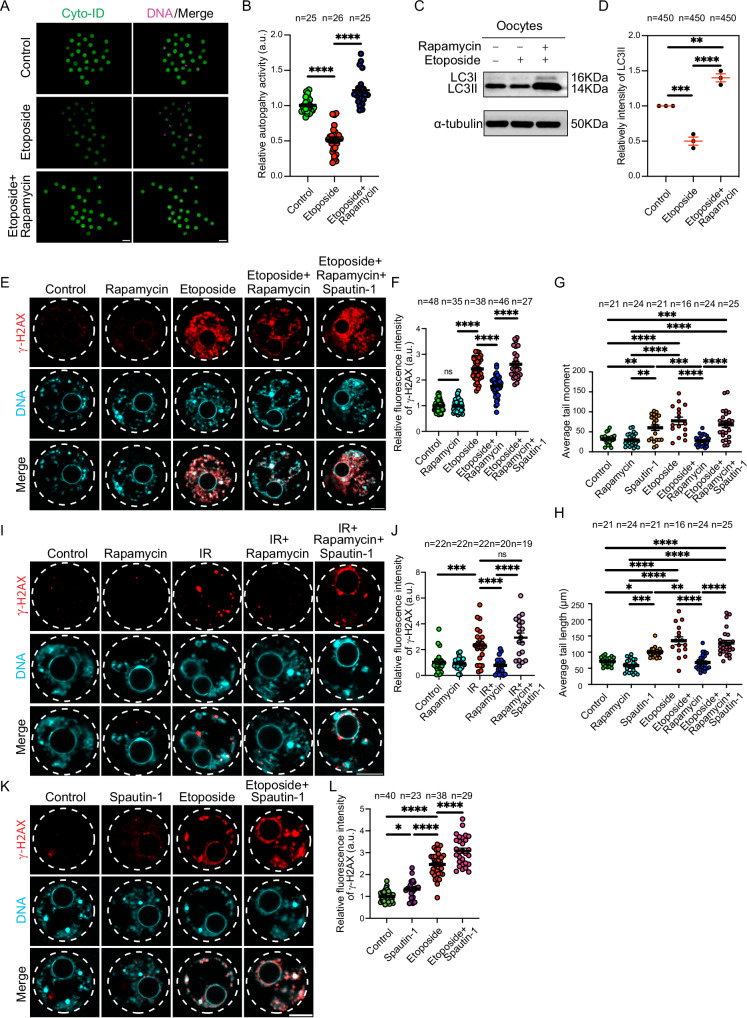
In somatic cells, autophagy is activated in response to DNA damage and plays a significant role in DDR32–36. Treating mammalian oocytes (in the absence of exogenous DNA damage) with rapamycin, an autophagy inducer, is accompanied by decreased γH2AX levels48. However, it is unknown whether rapamycin can rescue exogenous DNA damage, or whether it acts through autophagy induction. Therefore, the involvement of autophagy in the DNA damage response of mammalian oocytes is largely unknown. Given the limited capacity of full-grown oocytes to repair moderate levels of DSBs, we asked whether the autophagy-mediated DDR mechanism is dysfunctional in oocytes. To this end, full-grown GV oocytes were collected and cultured with etoposide (to induce DSBs) and milrinone (to maintain the oocytes arrested at GV stage) before assessing autophagy activity. Strikingly, in contrast to somatic cells in which autophagy is stimulated in response to DNA damage32–34,36, autophagy activity (assessed by an autophagy Cyto-ID detection assay) failed to be activated and was significantly decreased in live DNA-damaged oocytes, compared to controls (Fig. 2A, B). When autophagy is activated, the cytosolic form of the pro-autophagic, microtubule-associated protein 1 light chain 3 (LC3-I) becomes conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to autophagosomal membranes69. Because LC3-II is specifically correlated with autophagosomes and autolysosomes, it is a reliable marker of autophagic activity in both somatic cells and oocytes70–72. Consistent with Cyto-ID assay results, LC3-II levels, determined by Western blot densitometry, were significantly reduced in etoposide-treated oocytes when compared to controls (Fig. 2C, D). This contrasts with granulosa cells (somatic cells), in which LC3-II was increased following etoposide treatment (Supplementary Fig. 2B). We then asked whether the failure of the oocyte to upregulate autophagy in response to DSBs is conserved in mammalian oocytes. Immature porcine cumulus oocyte complexes (COCs) were collected and cultured in medium containing milrinone (to prevent meiotic resumption) and etoposide (to induce DSBs) followed by assessing autophagy activity. Similar to mouse oocytes, porcine oocytes exhibited a lack of autophagy activation when treated with etoposide (Supplementary Fig. 2C, D). This contrasts with somatic cumulus cells, in which autophagy activity increased following etoposide treatment (Supplementary Fig. 2C, E). Thus, autophagy activation in response to exogenously induced DSBs in mammalian oocytes is not functional, unveiling a significant difference between somatic cells and full-grown oocytes, and raising the question of whether inefficient autophagy activation is the cause, at least in part, of deficient DDR in mammalian oocytes.
Fig. 2. Reduced autophagy is the likely cause of weakened DNA damage repair in oocytes.
A Full-grown germinal vesicle (GV) oocytes were incubated in milrinone-containing CZB medium supplemented with DMSO (control), etoposide or etoposide + rapamycin for 3 h followed by autophagy activity measurements by using Cyto-ID detection kit. Representative images are shown. Scale bar represents 100 μm. B Quantification of autophagy activity in A. C Full-grown GV oocytes (150 oocytes per group) were incubated in milrinone-containing CZB medium supplemented with the indicated treatments for 3 h followed by Western blot analysis with LC3 and α-tubulin antibodies. Representative images are shown. D Quantification of LC3II in C. Each dot in the plot graph represents the average of an experimental replicate. (E) Full-grown GV oocytes were incubated in milrinone-containing CZB medium supplemented with the indicated treatments for 3 h. Oocytes were fixed and immunostained with γH2AX antibody. Representative images are shown. Scale bar represents 10 μm. F Quantification of γH2AX fluorescence intensity in E. G, H Full-grown GV oocytes were incubated in milrinone-containing CZB medium supplemented with the indicated treatments for 3 h followed by alkaline comet assay to assess DNA tail moment (G) and DNA tail length (H), quantified from Fig. 5A. (I) Full-grown GV oocytes were exposed to ionizing radiation (IR, 1 Gy) and incubated with the indicated treatments in milrinone-containing CZB medium for 3 h followed by immunostaining with γH2AX antibody. Representative images are shown. Scale bar represents 10 μm. J Quantification of γH2AX fluorescence intensity in (I). K Full-grown GV oocytes were incubated in milrinone-containing CZB medium supplemented with the indicated treatments for 3 h. Oocytes were fixed and immunostained with γH2AX antibody. Representative images are shown. Scale bar represents 10 μm. L Quantification of γH2AX fluorescence intensity in (K). DNA was stained with DAPI. Data are expressed as mean ± SEM. Two-tailed Student-t-test (B), One-way ANOVA was used to analyze the data. Values with asterisks differ significantly, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above each graph. Source data are provided as a Source Data file.
Reduced autophagy contributes to a weakened DNA damage response in oocytes
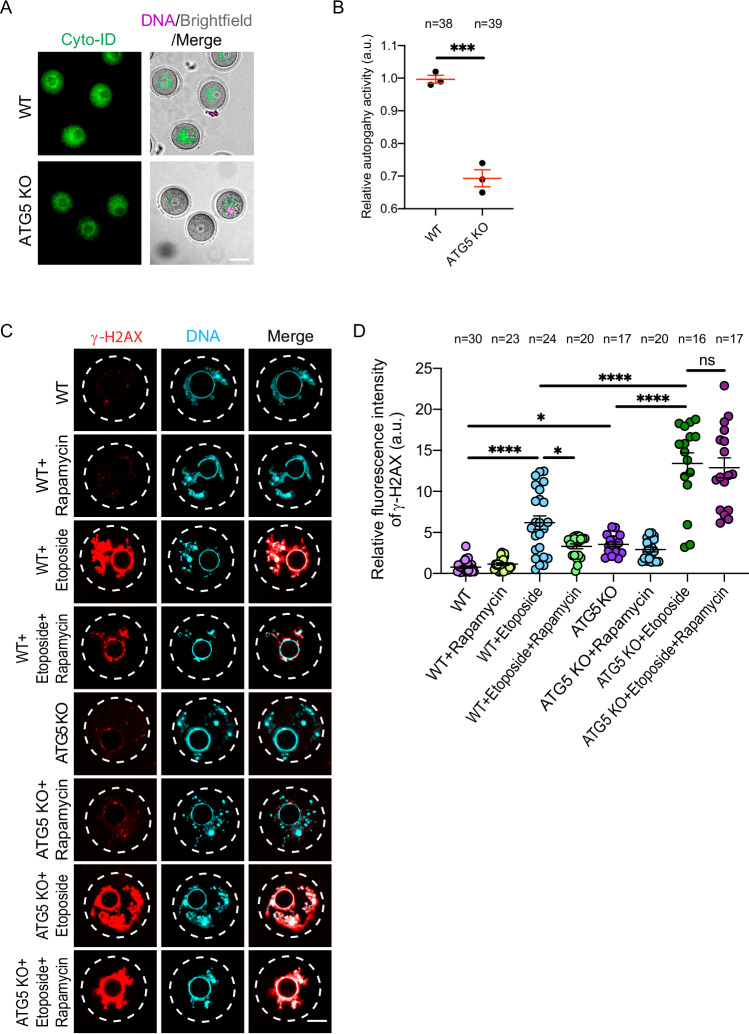
Whether autophagy induction can rescue exogenous DNA damage is unknown. We treated full-grown GV oocytes with etoposide alone or etoposide and rapamycin, a well-established autophagy inducer in both somatic cells and oocytes44,73,74, followed by assessing the DNA DSB status. Expectedly, treating mouse oocytes with rapamycin stimulated autophagy as evidenced by the significant increase of autophagic activity in live oocytes, compared to control oocytes (Supplementary Fig. 2A) and its ability to rescue decreased autophagy in etoposide-treated oocytes (Fig. 2A–D). Although we did not observe a reduction in γH2AX levels in control (non-etoposide-treated) oocytes treated with rapamycin, rapamycin treatment decreased etoposide-induced DSBs as evidenced by decreased γH2AX levels, compared to the etoposide-only treated group (Fig. 2E, F). Again, autophagy induction by rapamycin significantly decreased DNA damage in etoposide-treated oocytes as evidenced by decreased DNA tail moment and length (Fig. 2G, H). To further confirm our conclusion, we induced DNA DSBs using two different methods: UV and ionizing radiation. Compared to controls, a brief exposure of the oocytes to ionizing radiation (1 Gy) significantly increased γH2AX levels (Fig. 2I, J). Similarly, a brief exposure of the oocytes to UV (302 nm for 30 s) significantly increased cyclobutane pyrimidine dimer (CPD) (Supplementary Fig. 2F, G), the dominant form of DNA lesions in UV-exposed cells75,76 and γH2AX levels (Supplementary Fig. 2H, I), compared to controls. Importantly, rapamycin treatment significantly reduced DSBs in oocytes exposed to ionizing radiation (Fig. 2I, J) or UV (Supplementary Fig. 2H, I). Rapamycin induces autophagy by inhibiting mTOR73 which is involved in a variety of cellular functions in addition to autophagy induction77. To confirm that rapamycin rescues DSBs through autophagy induction specifically, we inhibited autophagy using an mTOR-independent pathway. Spautin-1 is a potent small molecule inhibitor that effectively suppresses autophagy by targeting ubiquitin-specific peptidases, USP10 and USP13, leading to the degradation of Beclin-1, a crucial protein necessary for autophagy initiation78. In the following experiments, the co-treatment of oocytes with spautin-1 and rapamycin was used as an additional control group to establish whether rapamycin effects specifically result from autophagy induction, but not through other mTOR-dependent pathways. Treating mouse oocytes with spautin-1 efficiently decreased autophagy (Supplementary Fig. 2A) and increased the level of DSBs when compared to control oocytes (Fig. 2K, L), suggesting that full-grown oocyte contains certain level of autophagy necessary to respond to minimal DNA damage induced by endogenous DNA assaults. Inhibition of autophagy with spautin-1 further increased γH2AX levels in etoposide-treated oocytes (Fig. 2K, L). Autophagy inhibition using spautin-1, which inhibits autophagy via mTOR-independent pathway, abolished the rescue effect of autophagy induction by rapamycin in DNA-damaged oocytes (Fig. 2E, F; Fig. 2I, J and Supplementary Fig. 2H, I). Proteasomal inhibitor, MG-132, is also known to induce autophagy indirectly by inhibiting the proteosome degradation system, leading ultimately to increased LC3-I to LC3-II conversion79,80. Consistent with our observations, MG-132 supplementation significantly decreased γH2AX levels in etoposide-treated oocytes (Supplementary Fig. 3A, B). Autophagy inhibition using spautin-1 abolished the effect of MG-132 on reducing DSBs (Supplementary Fig. 3A, B), suggesting that MG-132 decreases DSBs through autophagy activation. It is unlikely that autophagy induction (using rapamycin or MG-132) reduced exogenously induced DSBs by affecting cell cycle progression because the aforementioned experiments were performed while the full-grown oocytes were arrested at the GV stage. To further confirm our findings, we generated mice deficient of Autophagy-Related Gene 5 (ATG5) specifically in oocytes (Atg5−/− oocytes) by crossing ATG5 floxed allele mice81 with Gdf9-Cre mice. Expectedly, Atg5−/− oocytes exhibited lower levels of autophagy activity than wildtype (WT) oocytes (Fig. 3A,B). Atg5−/− oocytes exhibited higher levels of DSBs as evidenced by the significant increase of γH2AX fluorescence pixel intensity when compared to WT oocytes (Fig. 3C, D). Moreover, Atg5−/− oocytes were sensitive to a low level of DNA damage. Compared to WT oocytes, Atg5−/− oocytes showed significantly higher levels of γH2AX following the exposure to a low concentration (10 μg/ml) of etoposide treatment (Fig. 3C, D). Importantly, rapamycin treatment significantly decreased etoposide-induced DSBs in WT oocytes, but not in Atg5−/− oocytes (Fig. 3C, D). Taken together, these results indicate that rapamycin decreases DSBs by inducing autophagy and that reduced autophagy contributes to the increased sensitivity of full-grown oocytes to DSBs.
Fig. 3. ATG5-depleted oocytes are sensitive to a low level of DNA damage and exhibit elevated levels of DNA double-strand breaks.
A Full-grown wildtype (WT) or ATG5-depleted (ATG5 KO) germinal vesicle (GV) oocytes were assessed for autophagy activity by using Cyto-ID detection kit. Representative images are shown. Scale bar represents 50 μm. B Quantification of autophagy activity in A. C Full-grown WT or ATG5 KO GV oocytes were incubated in milrinone-containing CZB medium supplemented with the indicated treatments for 3 h followed by fixation and immunostaining with γH2AX antibody. Etoposide was used at a low concentration (10 μg/ml). Representative images are shown. DNA was stained with DAPI. Scale bar represents 10 μm. D Quantification of γH2AX fluorescence intensity in C. Data are expressed as mean ± SEM. One-way ANOVA was used to analyze the data. Values with asterisks differ significantly, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above each graph. Source data are provided as a Source Data file.
Autophagy has both DNA damage protection and repair roles in oocytes by promoting RAD51 recruitment to the DNA
To understand how autophagy regulates the response of mouse oocytes to DSBs, we first investigated whether autophagy protects against DSBs, repairs DSBs, or both. To answer this question, two experiments were conducted. To first investigate whether autophagy protects against DSBs, we stimulated autophagy by treating the oocytes with rapamycin for 2 h followed by exposing the oocytes to etoposide, for an additional 1 h, in a rapamycin-free milrinone-containing medium. Our results revealed that inducing autophagy before DNA damage significantly decreased γH2AX intensity when compared to non-rapamycin-pretreated oocytes or rapamycin+spautin-1-pretreated oocytes (Supplementary Fig. 4A–C), indicating that high levels of autophagy can protect against DSBs in mouse oocytes. Then to investigate whether autophagy decreased DSBs by stimulating DDR, we treated the oocytes with etoposide for 1 h followed by culturing the oocytes in etoposide-free medium with or without rapamycin for an additional 2 h. Again, our results revealed that increasing autophagy after etoposide treatment significantly decreased γH2AX fluorescence pixel intensity when compared to non-rapamycin-treated oocytes (Supplementary Fig. 4D–F). Taken together, our results indicate that autophagy plays a role in both the protection and repair mechanisms against DSBs in mouse oocytes.
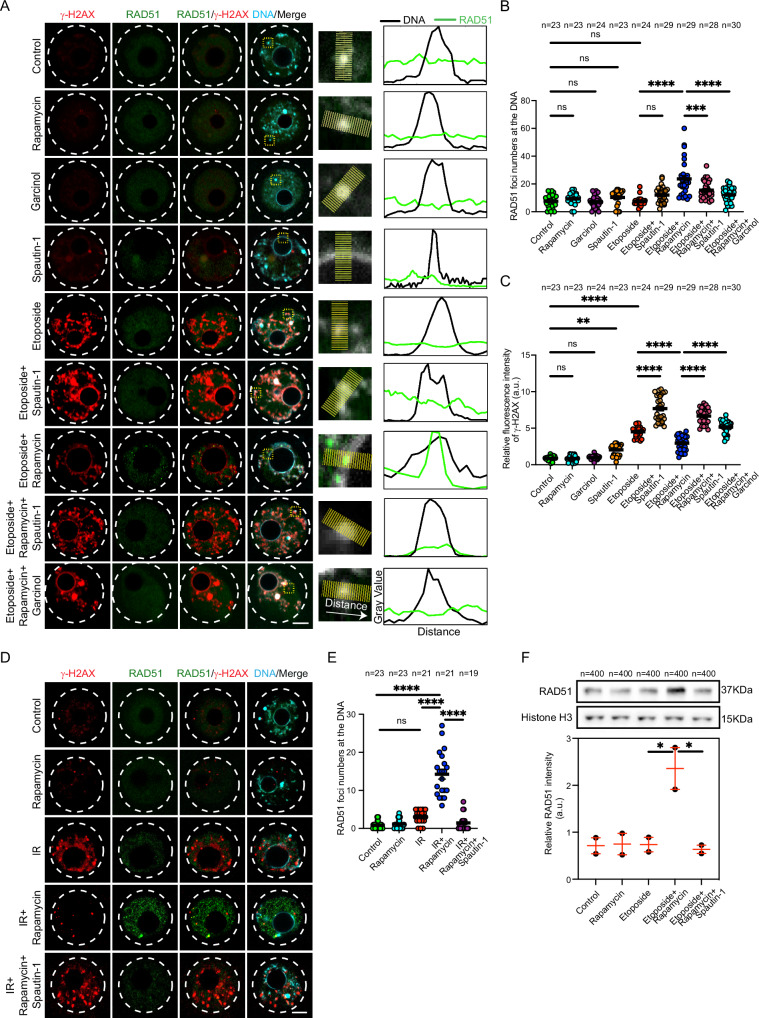
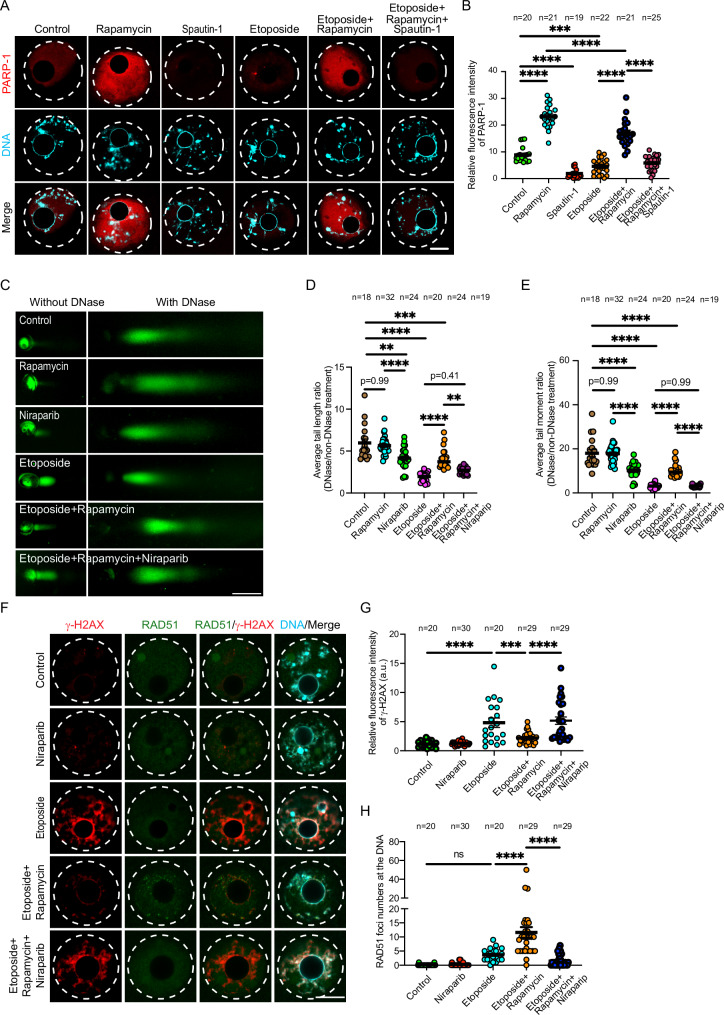
To understand the molecular mechanism of autophagy-mediated DDR, we screened several DNA damage checkpoint and repair proteins in DNA-damaged oocytes incubated with or without rapamycin. Upon DNA DSBs, ATM kinase is activated, a necessary step for DDR initiation. When damaged DNA is repaired, ATM is inactivated82,83. Levels of phospho-ATM (p-ATM) were elevated in etoposide-treated oocytes (Supplementary Fig. 4G, H), suggesting that DNA damage sensing mechanism is functional in oocytes. As expected, treatment with rapamycin, but not with rapamycin+spautin-1, significantly decreased p-ATM levels in etoposide-treated oocytes (Supplementary Fig. 4G, H), further confirming the rescue effect of autophagy induction on DSBs. To investigate the efficiency of DDR mechanism, we assessed the localization of RAD51 in mouse oocytes. RAD51 is an essential protein for DDR by homologous recombination84. In somatic cells, RAD51 expression is activated following DNA damage and plays a critical role in DDR by its recruitment to damaged-DNA foci85,86. To our surprise, even in the presence of elevated DSBs, the number of RAD51 foci (i.e., RAD51 aggregates as a result of their localization to DNA) at the damaged DNA did not vary significantly in etoposide-treated oocytes (Fig. 4A, B) or oocytes exposed to ionizing radiation (Fig. 4D, E) when compared to those in control oocytes. Strikingly, autophagy induction with rapamycin treatment, but not with rapamycin+spautin-1, significantly increased RAD51 localization to the DNA in DNA-damaged oocytes induced by etoposide (Fig. 4A, B), ionizing radiation (Fig. 4D, E) or UV exposure (Supplementary Fig. 2H, J). To further confirm RAD51 localization to the DNA, we isolated DNA-protein complex and assessed RAD51 levels in the extracted chromatin of DNA-damaged oocytes treated with or without rapamycin. RAD51 protein levels did not vary significantly between control and etoposide-treated oocytes. Autophagy induction with rapamycin treatment, but not with rapamycin+spautin-1, significantly increased RAD51 levels in the extracted chromatin of etoposide-treated oocytes (Fig. 4F). Consistent with our observations, inducing autophagy using MG-132 treatment, but not with MG-132+spautin-1, significantly increased RAD51 foci numbers in etoposide-treated oocytes (Supplementary Fig. 3A, C). To further investigate the efficiency of DDR mechanism, we assessed the localization of Ku80, a critical protein for non-homologous end-joining repair pathway, in mouse oocytes68. Consistent with RAD51, even in the presence of elevated DSBs, the numbers of Ku80 foci at the DNA did not increase significantly in etoposide-treated oocytes when compared to controls (Supplementary Fig. 5A, B). Importantly, autophagy induction by rapamycin, but not by rapamycin+spautin-1, increased the numbers of Ku80 foci in DNA-damaged oocytes (Supplementary Fig. 5A, B), albeit lower than its effect on RAD51 localization to the DNA. Together, these results show that autophagy promotes the recruitment of the DDR proteins RAD51 and Ku80 to damaged DNA through an uncharacterized mechanism.
Fig. 4. Autophagy promotes RAD51 recruitment to damaged DNA.
A Full-grown germinal vesicle (GV) oocytes were incubated in milrinone-containing CZB medium supplemented with the indicated treatments for 3 h. Oocytes were fixed and immunostained with RAD51 and γH2AX antibodies. Right panels represent zoomed examples of fluorescence intensity of RAD51 at the DNA using the plot profiles function in Image J. B Quantifications of the number of RAD51 foci localized at the DNA. C Quantifications of γH2AX fluorescence intensity. D Control and DNA-damaged oocytes (exposed to ionizing radiation, IR) were incubated in milrinone-containing CZB medium supplemented with the indicated treatments for 3 h. The oocytes were fixed at GV stage and immunostained with RAD51 and γH2AX antibodies. E Quantifications of the number of RAD51 foci localized at the DNA in (D). Shown are representative images. DNA was stained with DAPI. Scale bars represent 10 μm. F Full-grown GV oocytes were incubated with the indicated treatments for 3 h followed by chromatin extraction and Western blot analysis with RAD51 and Histone H3 antibodies. Quantification of RAD51 protein level (lower panel). Each dot in the plot graph represents the average of an experimental replicate. Data are expressed as mean ± SEM. One-way ANOVA was used to analyze the data. Values with asterisks differ significantly, ∗p < 0.05, ∗∗p < 0. 01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The total number of analyzed oocytes from three independent replicates (4 F, two replicates) is specified above each graph. Source data are provided as a Source Data file.
Autophagy regulates chromatin remodeling in DNA-damaged oocytes
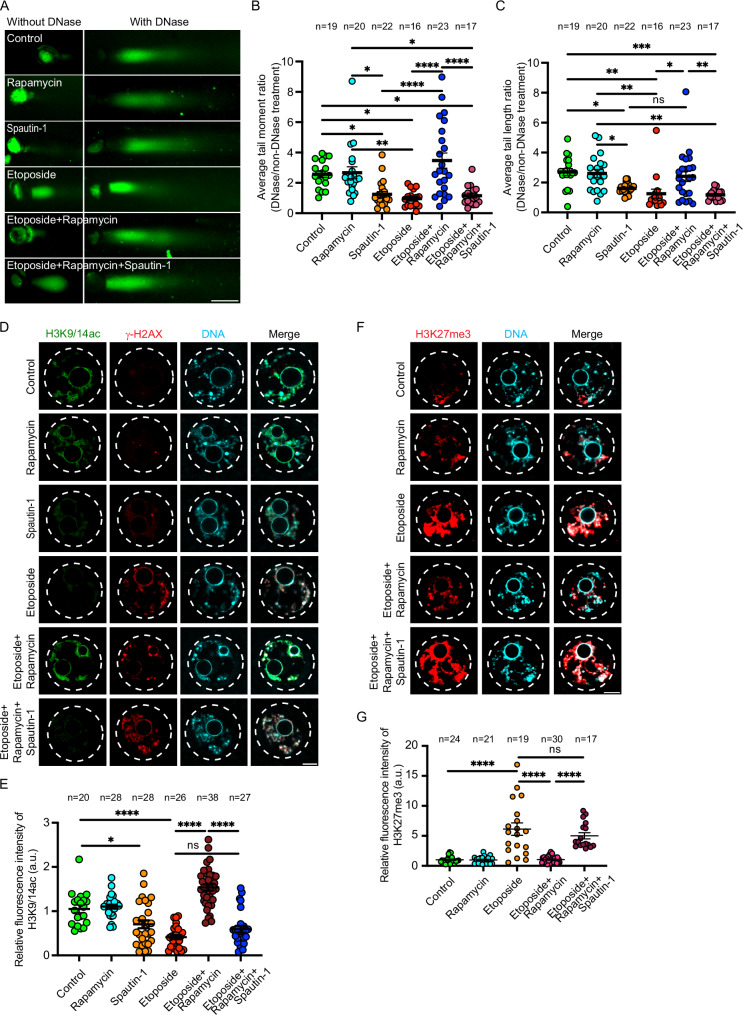
Chromatin remodeling from a condensed closed chromatin to an open chromatin structure is required during DDR, to render damaged DNA accessible to DDR proteins38–43. In somatic cells, autophagy-deficiency decreases RNF168-mediated histone H2A ubiquitination, leading to a more closed chromatin structure35. Considering the abnormal chromosome condensation phenotype observed in DNA-damaged oocytes, we hypothesized that the reduced RAD51 localization to DNA-damaged sites is due to steric hindrance by closed chromatin structure, and consequently, autophagy induction enables RAD51 localization to DNA-damaged sites by causing the chromatin to open. To assess chromatin conformation in DNA-damaged oocytes, we employed the DNase sensitivity assay, an accurate and commonly used method to study chromatin states in mammalian cells87–92.This assay is premised on the ability of DNase I enzyme to cut open DNA/chromatin faster than closed DNA/chromatin. Therefore, when the chromatin is open, the ratio between DNA fragmentation in DNase treated vs. non-DNase treated (DNase/non-DNase ratio) is higher than that of closed chromatin. DNA-damaged oocytes (by etoposide-treatment and irradiation) exhibited a significant decrease in tail moment DNase/non-DNase ratio and tail length DNase/non-DNase ratio when compared to control oocytes (Fig. 5A–C and Supplementary Fig. 6A–D), indicating that the chromatin is relatively closed in DNA-damaged oocytes. We also found that reducing autophagy levels by spautin-1 treatment resulted in a closed chromatin structure, as shown by decreased tail moment DNase/non-DNase ratio and tail length DNase/non-DNase ratio, compared to controls (Fig. 5A–C). The induction of autophagy by rapamycin, but not by rapamycin+spautin-1, in DNA-damaged oocytes caused the chromatin to open, as shown by the significant increase in tail moment DNase/non-DNase ratio and DNA tail length DNase/non-DNase ratio, compared to DNA-damaged oocytes (Fig. 5A–C and Supplementary Fig. 6A–D), indicating that autophagy modulates chromatin conformation in mouse oocytes. To confirm our results, we examined the levels of H3K9/K14 acetylation, a marker of open chromatin93–95, and H3K27me3, a marker preferentially associates with closed heterochromatin96–98, in DNA-damaged oocytes. Consistent with DNase sensitivity assay results, induction of DSBs by etoposide treatment or ionizing radiation resulted in a more closed chromatin state in DNA-damaged oocytes as shown by decreased H3K9/K14 acetylation levels (Fig. 5D,E and Supplementary Fig. 6E, F) and increased H3K27me3 levels (Fig. 5F, G). Importantly, autophagy induction by rapamycin treatment, but not by rapamycin+spautin-1 treatment, increased open chromatin status in DNA-damaged oocytes as shown by the significant increase of H3K9/K14 acetylation levels (Fig. 5D,E and Supplementary Fig. 6E, F) and the decrease of H3K27me3 levels (Fig. 5F, G). Thus, autophagy promotes chromatin remodeling in DNA-damaged oocytes. Histone acetylation is a major regulator of chromatin state and is largely regulated by histone deacetylases (HDACs) and histone acetyl transferases (HATs). Therefore, HAT inhibitors have been shown to induce a closed chromatin structure, whereas HDAC inhibitors have been shown to induce an open chromatin status99–102. We found that, following DSB induction, the chromatin is relatively closed with reduced RAD51 localization to the DNA. To determine whether open chromatin conformation is required for RAD51 to access DNA-damaged sites, we treated DNA-damaged oocytes with an HDAC inhibitor, trichostatin A, to promote open chromatin100. Treating the oocytes with trichostatin A increased RAD51 localization to the DNA in etoposide-treated oocytes (Supplementary Fig. 7A, B), indicating that, similar to somatic cells38–43, chromatin remodeling to an open chromatin structure is required for DDR protein localization to the DNA. A known phenomenon of trichostatin A is the phosphorylation of H2A by ATM even in the absence of actual DNA damage103. Consistent with this finding, we did not observe a reduction in γH2AX intensity in oocytes treated with both etoposide and trichostatin A (Supplementary Fig. 7A, C), suggesting that increased RAD51 localization to the DNA is due to the trichostatin A-induced open chromatin structure. To confirm that the open chromatin conformation caused by autophagy induction is the cause, at least in part, of increased RAD51 localization to the damaged DNA, we asked whether increasing closed chromatin state abolishes the increased RAD51 localization to the DNA in rapamycin-treated DNA-damaged oocytes. Importantly, treating the oocytes with garcinol, a HAT inhibitor that has been shown to increase closed chromatin structure in somatic and cancer cells102,104,105, decreased RAD51 localization to the DNA and increased γH2AX levels in rapamycin-treated DNA-damaged oocytes (Fig. 4A–C). Taken together, our findings indicate that in DNA-damaged oocytes, autophagy increases RAD51 localization to the DNA at least partially by promoting an open chromatin structure.
Fig. 5. Autophagy regulates chromatin remodeling in DNA-damaged oocytes.
A Full-grown germinal vesicle (GV) oocytes were incubated in milrinone-containing CZB medium supplemented with indicated treatments for 3 h. The DNase-treated and non-DNase-treated oocytes were subjected to the alkaline comet assay. Shown are representative images. Scale bar represents 100 μm. B Quantification of average DNA tail moment ratio (DNase/non-DNase treatment) in (A). C Quantification of average DNA tail length ratio (DNase/non-DNase treatment) in (A). D Full-grown GV oocytes were incubated in milrinone-containing CZB medium and exposed to the indicated treatments. The oocytes were fixed and immunostained with H3K9/14ac and γH2AX antibodies. Scale bar represents 10 μm. E Quantification of H3K9/14ac fluorescence intensity in (D). Shown are representative images. F Full-grown GV oocytes were incubated in milrinone-containing CZB medium and exposed to the indicated treatments. The oocytes were fixed and immunostained with H3K27me3 antibody. G Quantification of H3K27me3 fluorescence intensity in F. Shown are representative images. DNA was stained with DAPI. Scale bar represents 10 μm. Data are expressed as mean ± SEM. One-way ANOVA was used to analyze the data. Values with asterisks differ significantly, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above each graph. Source data are provided as a Source Data file.
PARP1 activation is a downstream pathway of autophagy in mouse oocytes
In somatic cells, Poly [ADP-ribose] Polymerase 1 (PARP-1) is activated in response to DNA damage, a necessary step to promote open chromatin status and to facilitate DDR protein recruitment106,107. Indeed, treating granulosa (somatic) cells with etoposide increased PARP-1, compared to DMSO-treated cells (Supplementary Fig. 2B). Again, in contrast to somatic cells, the oocytes failed to stimulate PARP-1 (Fig. 6A, B and Supplementary Fig. 8A) in response to DNA damage, a phenomenon similar to the autophagy activation failure in DNA-damaged oocytes. Autophagy is necessary for the maintenance of NAD(H) pool, which is necessary for PARP-1 activation and function108. Moreover, autophagy inhibition downregulates PARP-1 expression in hepatoma cells exposed to radiation-induced damage109. To test whether the lack of PARP-1 activation in DNA-damaged oocytes is related to autophagy reduction, we assessed PARP-1 after autophagy induction in DNA-damaged oocytes. Interestingly, autophagy induction increased PARP-1 levels in DNA-damaged oocytes compared to those in etoposide-treated oocytes, as shown by Western blot (Supplementary Fig. 8A) and immunocytochemistry (Fig. 6A, B) analyses. We then examined whether autophagy induction rescued DDR in DNA-damaged oocytes by stimulating PARP-1. We utilized niraparib, a highly selective FDA-approved inhibitor of PARP-1 and PARP-2110. Inhibition of PARP-1 by using niraparib eliminated autophagy-mediated open chromatin configuration in DNA-damaged oocytes (Fig. 6C–E). Importantly, PARP-1 inhibition abolished the rescue effect of autophagy on DDR in DNA-damaged oocytes (rapamycin+etoposide), as evidenced by increased γH2AX levels (Fig. 6F, G), increased both DNA tail length and moment (Supplementary Fig. 8B, C) and decreased RAD51 localization to the DNA (Fig. 6F, H). Taken together, these data indicate that autophagy induction promotes DDR protein localization by inducing open chromatin structure, at least in part through PARP-1 activation.
Fig. 6. Autophagy-induced PARP-1 activation regulates chromatin remodeling in DNA-damaged oocytes.
A, B Full-grown germinal vesicle (GV) oocytes were incubated with the indicated treatments for 3 h in milrinone-containing CZB medium followed by immunostaining with PARP-1 antibody. B Quantification of PARP-1 fluorescence intensity in (A). C Full-grown GV oocytes were cultured in milrinone-containing CZB medium and exposed to the indicated treatments. The DNase-treated and non-DNase-treated oocytes were subjected to the alkaline comet assay. Representative images are shown. Scale bars represent 100 μm. D Quantification of average DNA tail length ratio (DNase/non-DNase treatment). E Quantification of average DNA tail moment ratio (DNase/non-DNase treatment). F Full-grown GV oocytes were incubated in milrinone-containing CZB medium supplemented with the indicated treatments for 3 h. Oocytes were fixed and immunostained with γH2AX and RAD51 antibodies. G Quantification of γH2AX fluorescence intensity. H Quantification of RAD51 foci numbers at the DNA. DNA was stained with DAPI. Representative images are shown. Scale bars represent 10 μm. Data are expressed as mean ± SEM. One-way ANOVA was used to analyze the data. Values with asterisks differ significantly, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above each graph. Source data are provided as a Source Data file.
Reduced autophagy is the likely cause of weakened DNA damage response in oocytes from reproductively aged mice
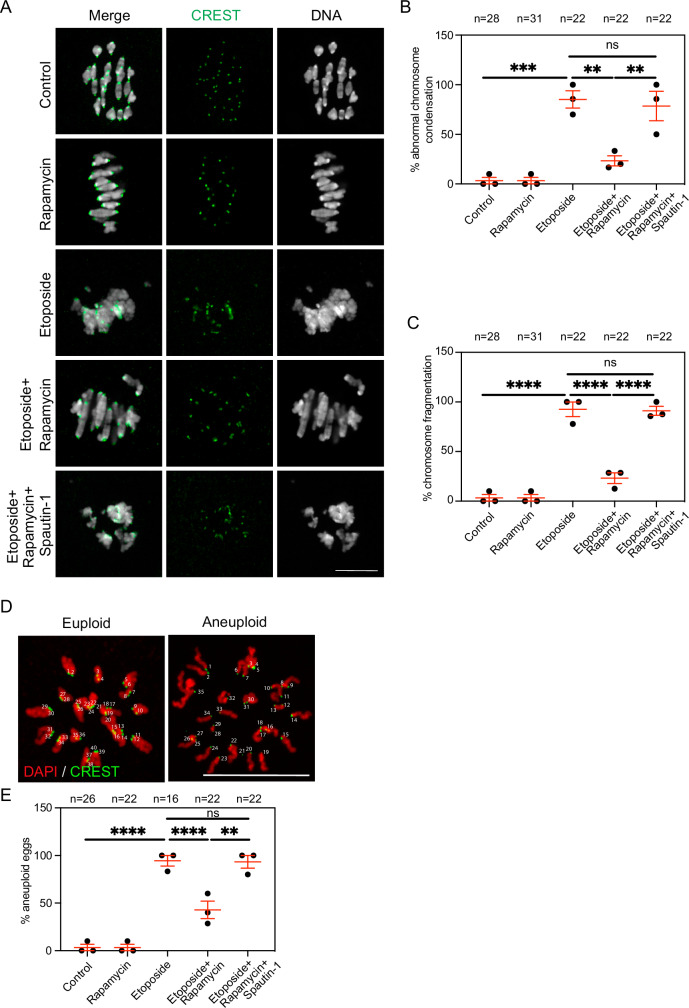
The main consequences of DSB induction in mouse oocytes are decreased PBE, delayed meiotic progression, altered chromosome morphology, increased chromosome fragmentation, and the development of aneuploid gametes. We found that autophagy induction increased the percentage of PBE (Supplementary Fig. 1C), accelerated the meiotic progression (Supplementary Fig. 1D), decreased the incidence of abnormal chromosome condensation (Fig. 7A, B), decreased chromosome fragmentation (Fig. 7A, C) and, decreased the incidence of aneuploidy (Fig. 7D, E) in DNA-damaged oocytes. Thus, autophagy induction may provide a promising approach to overcome the consequences of exposing full-grown mammalian oocytes to exogenously induced DSBs. Although the DDR is not robust in full-grown mammalian oocytes, including humans and mice, it is further weakened in oocytes from females of advanced reproductive age due to poorly understood reasons21,24,25. Thus, oocytes from aged females represent a model of elevated DNA damage without chemical or radiation treatment. Autophagy reduction is a hallmark of aging as it decreases gradually with the advancement of age, leading to the development of a wide variety of disorders including neurodegenerative and metabolic diseases, and cancer111. This study shows that reduced autophagy correlates with increased DNA damage in full-grown GV oocytes. Thus, we further hypothesize that reduced autophagy contributes to the weakened DDR in oocytes from aged mice. To this end, we assessed autophagic activity in GV oocytes collected from aged mice (12-14-month-old). Oocytes collected from young mice (6-8-week-old) were used as controls (excluding the effect of early meiotic recombination in response to programmed DNA breaks). Consistent with our hypothesis, the autophagic activity was significantly decreased in oocytes collected from aged vs. young mice (Supplementary Fig. 9A, B), raising the question of whether autophagy induction can rescue increased DSBs in aged mouse oocytes. In line with previous reports24,61, oocytes from aged mice exhibited higher levels of DSBs (increased γH2AX intensity) compared to those in young mouse oocytes (Supplementary Fig. 9C, D). Importantly, autophagy induction in oocytes from aged mice significantly reduced γH2AX intensity to a level comparable to that of young mouse oocytes (Supplementary Fig. 9C, D). Expectedly, oocytes from aged mice showed higher rates of aneuploidy when compared to those from young mice (Supplementary Fig. 9E, F). We found that autophagy induction by rapamycin, but not by rapamycin+spautin-1, decreased the incidence of aneuploidy in oocytes from aged mice (Supplementary Fig. 9E, F). Taken together, our results show that reduced autophagy correlates with increased DNA damage in oocytes from aged mice.
Fig. 7. Autophagy induction rescues the consequences of DNA damage in mouse oocytes.
A Full-grown germinal vesicle (GV) oocytes were incubated with the indicated treatments for 3 h in milrinone-containing CZB medium followed by washing and in vitro maturation for 7 h (metaphase I) in milrinone-free CZB medium. Metaphase I oocytes were fixed and immunostained with CREST antibody to label kinetochores and stained with DAPI to label DNA. Representative images are shown. B Quantification of abnormal chromosome condensation phenotype in A. C Quantification of chromosome fragmentation phenotype in (A). D Full-grown GV oocytes were incubated with the indicated treatments for 3 h in milrinone-containing CZB medium followed by washing and in vitro maturation for 14 h (metaphase II). Oocytes were treated with monastrol for 2 h, fixed and immunostained with CREST antibody to label kinetochores. Oocytes were scored either as euploid (containing 40 kinetochores) or aneuploid (containing ± 40 kinetochores). Representative images are shown. E Quantification of aneuploidy percentage in D. Scale bars represent 10 μm. DNA was stained with DAPI. Data are expressed as mean ± SEM. Each dot in the plot graph represents the average of an experimental replicate. One-way ANOVA was used to analyze the data. Values with asterisks differ significantly, ∗∗p <0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. The total number of analyzed oocytes (from three independent replicates) is specified above each graph. Source data are provided as a Source Data file.
Discussion
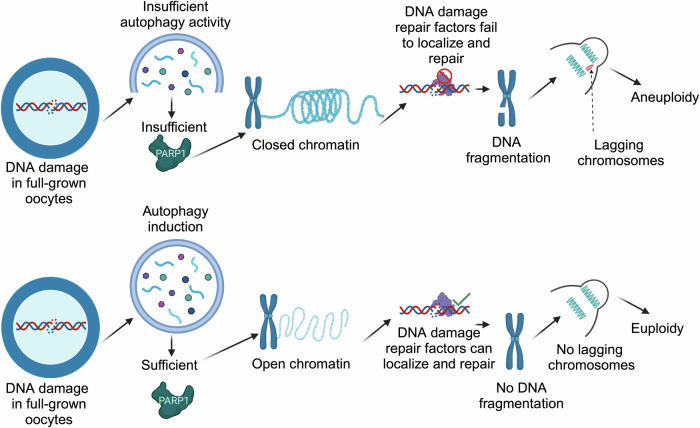
For largely unknown reasons, full-grown GV oocytes have a limited capacity to repair damaged DNA. We find that, unlike somatic cells, full-grown oocytes do not efficiently activate autophagy in response to moderate levels of DSBs. We show that this diminished autophagy activation in full-grown oocytes contributes to reduced PARP-1 activation, altered chromatin conformation and the failure of RAD51 recruitment to the DNA in response to moderate DNA damage levels. Induction of autophagy rescues altered chromatin conformation, increased RAD51 localization to the DNA, increased DDR efficiency, decreased chromosome fragmentation and decreased the incidence of aneuploidy in DNA-damaged oocytes (Fig. 8). We also show that autophagy is reduced in oocytes from reproductively aged mice, which is the cause, at least partially, of the increased severity of DNA damage in maternally aged oocytes.
Fig. 8. Schematic model summarizing the impact of DNA double-stand breaks on oocyte meiosis I and the role of autophagy in DNA damage repair.
Following the induction of DNA double-stand breaks in fully grown prophase I-arrested oocytes, autophagy activity does not increase, leading to insufficient PARP-1 levels, altered chromatin conformation, reduced localization of DNA damage repair factors to the DNA and an increased incidence of aneuploidy. Created in BioRender. Londono, A. (2023) BioRender.com/j60r980.
Exposure of somatic cells to mild DNA damage is sufficient to induce a cell cycle arrest (by activating the G2/M checkpoint), providing sufficient time for repairing DNA damage. If DNA damage is not repaired, the cell will activate the p53-dependent apoptotic pathway. For poorly understood reasons, the DNA damage response is not robust, and both p53- and p63-mediated apoptosis are not functional in full-grown GV oocytes112. Only exposure to severe, but not mild or moderate, DNA damage can prevent meiotic resumption in full-grown oocytes21, increasing the risk of developing aneuploid gametes. Hence, full-grown oocytes employ an alternative mechanism to hinder meiotic progression following DNA damage by activating another surveillance mechanism, the spindle assembly checkpoint (SAC), leading to a cell cycle arrest at metaphase I stage52,61. This is surprising because the SAC is known to be weak in oocytes, compared to somatic cells113–115. Therefore, even though the SAC is activated in response to DNA damage, a considerable percentage of mouse oocytes can progress through meiosis I and reach metaphase II stage. Indeed, similar to a previous report51, we found that, approximately 40% of etoposide-treated oocytes that underwent NEBD were able to extrude the PB, likely because the DNA damage was below the SAC activation threshold. Human oocytes can similarly progress through meiosis I and reach metaphase II while harboring damaged DNA25, suggesting that not only DNA damage-induced cell cycle arrest mechanism is weak, but the DDR mechanism is also inefficient. We found that autophagy promotes chromatin remodeling and DDR function in in GV-arrested oocytes, independently of affecting the cell cycle. Additionally, when DNA-damaged oocytes were allowed to mature in the presence of rapamycin, there was no delay in the timing of NEBD or PBE. Thus, reduced autophagy contributes to the weakened DDR mechanism in DNA-damaged oocytes, but not to the inefficient initiation of cell cycle arrest. However, future research is essential to determine whether excessive autophagy induction affects other cytoplasmic events in mammalian oocytes.
The full-grown oocyte exhibits relatively low levels of autophagy under normal physiological conditions, which increases only after fertilization72. This basal level of autophagy activity is likely required to protect the oocyte against mild endogenous DNA assaults. Indeed, autophagy inhibition by spautin-1 increased DSBs in oocytes and further exacerbated the severity of DNA damage in etoposide-treated oocytes. In somatic cells, autophagy is activated in response to various stressors including exogenous DNA damage. Similar to somatic cells, full-grown oocyte can activate autophagy in response to several stressors such as cryopreservation and heat stress116,117. However, we found that, in response to moderate DNA damage induction, full-grown oocytes failed to activate autophagy, and this basal autophagy level is even decreased. We speculate that upon DSB induction, the basal level of autophagic activity reaches its capacity during the initial DDR response. Failure to further activate autophagy in DNA-damaged full-grown oocytes contributes to their limited capacity to repair DNA damage. It would be an interesting future direction to determine underlying the molecular mechanisms that might be responsible for this differential autophagic response to environmental stressors and the failure to activate autophagy in response to DNA damage in full-grown GV oocytes.
Similar to somatic cells, nongrowing and growing prophase I oocytes (from prepubertal mice) can efficiently repair severe DNA damage in apoptosis-inhibited mice by activating ATM, phosphorylating histone H2AX, and translocating RAD51 to DNA-damaged sites20. This contrasts with full-grown oocytes that can repair mild DNA damage, but not moderate DNA damage (this study). Our data reveal that although full-grown oocytes (from sexually mature mice) can activate ATM and phosphorylate histone H2AX in response to moderate DNA damage, they are inefficient in recruiting RAD51 to the DNA and repairing DSBs, even after being arrested at the GV stage for 16 h. It is interesting that oocytes can efficiently repair moderate DNA damage at earlier developmental stage (nongrowing and growing), but not at later stages (fully grown), suggestive of a growth-related decline in DDR machinery. Indeed, nongrowing and growing oocytes are characterized by decondensed euchromatin, whereas full-grown oocytes are characterized by condensed heterochromatin118,119. Analyzing the autophagy activity within the oocyte throughout its development and growth may provide insights into the underlying mechanisms behind the changes in chromatin structure associated with oocyte growth and the decline in DDR in full-grown oocytes.
Aging-associated chromatin structure changes are more complex than previously thought. First, these changes are not universal. For example, in aged mice, a reduction in heterochromatin (closed chromatin) was observed in excitatory neurons, but not in inhibitory neurons, heart, bone marrow or skeletal muscles120. Intriguingly, aging correlates with an increased formation of localized heterochromatin at specific genomic loci, known as heterochromatin redistribution phenomenon121,122. Given that DNA-damaged sites are random and unpredictable, developing and optimizing techniques to assess chromatin structural changes specifically at age-associated DNA-damaged sites is crucial to determine the involvement of chromatin structural changes in the weakened DDR observed in maternally aged oocytes.
Although DNA damage is a fundamental problem for life and its underlying mechanism has been thoroughly investigated in somatic cells, its consequences on oocyte meiosis are still poorly understood. Our data provide evidence that DSBs lead to aneuploidy in oocytes. We reveal that inefficient autophagy activation contributes to weakened DNA damage response in oocytes, particularly in those from females of advanced reproductive age. We also show that autophagy induction decreases DSBs and aneuploidy in maternally aged oocytes. This is particularly important because female gametes, especially those from women with advanced reproductive age, are notoriously prone to increased aneuploidy. At least two major causes of aneuploidy in maternally aged oocytes have been demonstrated: loss of centromere cohesion and defective kinetochore-microtubule attachments123–125. Although much work has aimed to improve oocyte quality of women with maternally advanced age, success has been limited, suggesting the involvement of additional unknown contributing factors. Our data shed new light on autophagy deficiency as another contributing factor to increased aneuploidy in mammalian oocytes, including those from females with advanced maternal age.
Methods
Ethics
All experiments were performed and complies with all relevant ethical regulations in accordance with the Animal Care and Use guidelines of the University of Missouri (Animal Care Quality Assurance Reference Number, 39341).
Mouse strains
Sexually mature CF1 female mice (Mus musculus, Envigo, Indianapolis, IN, USA) and Atg5flox/flox Gdf9-Cre female mice were used in this study. Mice lacking ATG5 specifically in oocytes were generated by crossing Atg5 floxed allele mice81 (B6.129s-Atg5<tmlMvob, RBRC02975, RIKEN BioResource Research Center, Japan) with Gdf9-Cre (Tg(Gdf9-icre)5092Coo/J, Jackson Laboratories)126. Control wildtype (WT) mice are from the same genetic background but lack the Cre recombinase transgene. Mice were housed at 21 °C and 55% humidity in 12 h/12 h light/dark cycle and ad libitum access to food and water. Unless otherwise specified, oocytes were collected from 6-8-week-old mice. Aged oocytes were collected from 12 to 14-month-old mice.
Oocyte collection and in vitro maturation
Fully grown, GV-intact oocytes were collected from CF1 mice previously primed (44–48 h before collection) with pregnant mare serum gonadotropin (PMSG, Lee BioSolutions, Maryland Heights, MO, USA #493-10-10), as previously described127–129. The COCs were denuded mechanically by pipetting and cultured in bicarbonate-free minimal essential medium (MEM) supplemented with 3 mg/ml polyvinylpyrrolidone (PVP) and 25 mM HEPES (pH 7.3) under mineral oil (MilliporeSigma, St. Louis, MO, USA #P2307, #H3784 and #M8410). The GV oocytes were then transferred to Chatot, Ziomek, and Bavister (CZB) medium and cultured at 37˚C with 5% CO2 in humidified air for either 3 (arrested at GV stage), 7 (metaphase I), or 16 h (metaphase II).
Porcine ovaries were collected from a local slaughterhouse and transported to the University of Missouri within 2 h. The COCs were aspirated from 3-8 mm follicles by using an 18-gauge needle attached to a 10 mL syringe. Oocytes with homogenous cytoplasm and surrounded by more than 3 layers of cumulus cells were selected. The COCs were cultured in milrinone-containing chemically defined maturation medium, TCM 199 (Invitrogen, Grand Island, NY), supplemented with 10% bovine calf serum, 1 mM L-glutamine, 0.2 mM sodium pyruvate, 20 μg/mL follicle stimulating hormone [FSH; Folltropin-V, Vetoquinol, TX, USA], 2 μg/mL β-estradiol and 10 μg/mL gentamycin at 38.5 °C and 5% CO2 in humidified air. After culture, cumulus cells were removed by vortexing for 3 min in the presence of 0.1% hyaluronidase.
To prevent meiotic resumption, milrinone, a phosphor diesterase inhibitor, was added to the medium (2.5 μM, MilliporeSigma #M4659)130. Etoposide (MilliporeSigma #E1383), rapamycin (MilliporeSigma #553211), MG-132 (MilliporeSigma #474790), spautin-1 (Selleckchem, Houston, TX, USA #S7888), niraparib (Selleckchem #S7625), nocodazole (MilliporeSigma #M1404), monastrol (MilliporeSigma #M8515), garcinol (Santa Cruz Biotechnology, Dallas, TX, USA #78824-30-3) and trichostatin A (Selleckchem #S1045) were dissolved in DMSO and added to CZB culture medium at a final concentration of 50 µg/ml, 100 nM, 10 µM, 20 µM, 10 µM, 5 µM, 100 µM, 10 µM and 500 nM, respectively. DMSO was added to CZB culture medium as a vehicle control at 0.05% concentration. SiR-DNA was added to the maturation medium at a final concentration of 500 nM to label the DNA during time-lapse confocal live imaging.
Immunocytochemistry and confocal microscopy
Oocytes were fixed for 20 min at room temperature in a freshly prepared 2% paraformaldehyde solution (MilliporeSigma #P6148) dissolved in phosphate buffer saline (PBS). After fixation, oocytes were permeabilized using 0.1% Triton X-100 in PBS solution for 20 min. Oocytes were then incubated in blocking solution (PBS containing 0.3% BSA and 0.01% Tween-20) for an additional 20 min. The oocytes were then incubated in primary antibody for 1 h at room temperature prior to 3 successive washes in blocking solution (8 min each). Oocytes were then incubated in secondary antibody for another 1 h at room temperature followed by washing in blocking solution (3 times, 8 min each). The oocytes were mounted on slides using Vectashield containing 4′,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI; Vector Laboratories, Burlingame, CA, USA) to label the DNA, under a coverslip with gentle compression. Omission of primary antibodies and the use of isotype-specific immunoglobulins served as negative controls. Fluorescence signals were observed under a 63× oil objective using Leica Stellaris 5 confocal microscope. Oocytes were captured using 1, 3 or 5 μm Z-intervals.
All oocytes in the same experiment were imaged and processed simultaneously. The laser power for imaging all groups was adjusted to a level where the signal intensity is just below the saturation point in the group showing the highest fluorescence intensity. Fluorescence intensity was quantified using NIH Image J software (National Institute of Health, Bethesda, MD, USA) with same processing parameters.
Antibodies: The following primary antibodies were used: conjugated α-tubulin-AlexaFluor 488 (Life Technologies #322588), CREST autoimmune serum (Antibodies Incorporated, Davis, CA, USA #15-234), anti-RAD51 (MilliporeSigma, St. Louis, MO, USA #ABE257), anti-p-ATM (Santa Cruz Biotechnology, Dallas, TX, USA #sc47739), anti-γH2AX (Abcam, Boston, MA, USA #ab11174), anti-α-Tubulin antibody (MilliporeSigma # T6074), anti-LC3A/B antibody (Cell Signaling #4148), anti-γH2AX (MilliporeSigma #05-636), anti-Ku80 (NSJ Bioreagents, San Diego, CA, USA #R31953), anti-Acetyl-Histone H3 K9/14 (Cell Signaling #9677), anti-Histone H3K27me3 (Active Motif, Carlsbad, CA, USA #39155). The CPD assay was carried out using the OxiSelect Cellular UV-Induced DNA Damage CPD Staining Kit (Cell Biolabs # STA-327, San Diego, CA, USA).
Time-lapse confocal microscopy
Oocytes were transferred to milrinone-free CZB medium. Brightfield and SiR-DNA image acquisition was started following NEBD using Leica Stellaris 5 confocal microscope equipped with a microenvironmental chamber to maintain the oocytes at controlled CO2 (5%) and temperature (37 °C) in a humidified air. Images of single oocytes were captured at 5 µm Z-intervals every 40 min. Images were processed using NIH Image J software.
In situ chromosome counting
Metaphase II stage oocytes (14 h) were transferred to CZB medium supplemented with 100 μM monastrol, a cell-permeable Eg5 kinesin inhibitor to induce monopolar spindle formation with subsequent chromosome dispersion, for additional 2 h131–133. Oocytes were then fixed in a freshly prepared 2% paraformaldehyde solution followed by immunostaining using CREST autoimmune serum antibody (to label kinetochores). Oocytes were imaged under a 63× oil objective using Leica Stellaris 5 confocal microscope at 0.7-μm Z-intervals to capture all kinetochores. The total number of kinetochores were acquired after analyzing all confocal sections using NIH Image J software.
Chromatin extraction
Cells (400 oocytes) were cross-linked by a final concentration of 1% formaldehyde solution for 10 min on a rocking platform (50-100 rpm) and terminated by 0.125 M glycine. Chromatin extraction was carried out using the Chromatin Extraction Kit (Abcam #ab117152, USA). To enhance chromatin extraction, the cells were sonicated twice (20 s each). The sample was allowed to cool on ice for 30 s between sonication pulses. Anti-RAD51 (Abcam #ab176458) and Anti-Histone H3 (Abcam #ab1791) antibodies were used for western blot analysis.
Western blotting
Oocytes or granulosa cells (after COC denudation) were lysed in 1% SDS, 1% β-mercaptoethanol, 20% glycerol and 50 mM Tris–HCl (pH 6.8). Lysed oocytes were heated at 95 °C for 10 min. Proteins were then separated by electrophoresis using SDS-PAGE (10% SDS polyacrylamide precast gel) and transferred to nitrocellulose membranes using Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA, USA). Membranes were blocked in 2% skim milk in Tris-buffered saline with 0.1% Tween 20 (TBS-T) for 1 h at room temperature and incubated overnight at 4 °C with primary antibodies diluted in a blocking solution. After washing in TBS-T (3 times for 10 min each), the membranes were incubated in secondary antibodies (anti-Rabbit IgG-HRP, ThermoFisher Scientific, Waltham, MA, USA #A16104 and anti-mouse IgG-HRP, Bio-Rad #1706516) diluted in blocking solution for 1 h at room temperature followed by 3 times washing in TBS-T solution. Protein signals were detected using the Clarity Max Western ECL Substrate (Bio-Rad #1705062S) following the manufacturer’s protocol. NIH Image J software was used to do protein quantification.
Antibodies: anti-PARP1 (Cell Signaling #9532), anti-γH2AX (Abcam #ab11174), anti-α-tubulin antibody (MilliporeSigma #T6074) and LC3A/B antibody (Cell Signaling #4148).
Detection of autophagic activity
Autophagy was assessed using Cyto-ID autophagy detection kit (Enzo Life Sciences, Farmingdale, NY, USA #ENZ-51031) following the manufacturer’s protocol with some modifications. Briefly, oocytes were incubated with the reaction mix (1 μL in 500 μL CZB medium) at 37˚C with 5% CO2 in humidified air for 15 min. DNA was stained by adding Hoechst 33342 (25 μg/mL) to the same medium for an additional 5-7 min. Oocytes were then rinsed in PVP-PBS before imaging live oocytes using Leica DMI8 fluorescence microscope. The images were analyzed using NIH Image J software.
Detection of intracellular caspase 3 activity
Caspase 3 (DEVD)2 detection kit assay (Immunochemistry Technologies, Bloomington, MN, USA #935) was used to detect caspase 3 activity following the manufacturer’s protocol. Metaphase I oocytes were incubated with the reaction mix (1 μl of DMSO-diluted stock solution in 250 μl CZB culture medium) for 30 min in a humidified atmosphere of 5% CO2 at 37 °C. The omission of caspase 3 substrate served as a negative control. Stained oocytes were washed in PVP-PBS before imaging using Leica DMI8 fluorescence microscope. Images within the same experiment were captured and analyzed under the same parameters using NIH image J software.
Alkaline comet assay/ DNase sensitivity assay
Full-grown GV oocytes were permeabilized in 0.1% Triton X-100 in PBS for 10 min followed by transfer to 50 µl (5 Units) DNase I enzyme (ThermoFisher Scientific #18047019) or DNase I reaction buffer (non-DNase-treated). The LMAgarose (Trevigen, Gaithersburg, MD, USA #4250-050-02) was preheated in a tube in boiling water for 30 min before cooling down to 37 °C. Oocytes treated with or without DNase I were mixed with LMAgarose on comet assay slides (Trevigen #4250-050-03). The slides were incubated at 4 °C in the dark with high humidity for 30 min followed by incubation in lysis solution (Trevigen #4250-050-01) overnight at 4 °C. The slides were then immersed in a freshly prepared alkaline unwinding solution (pH>13, 200 mM NaOH, 1 mM EDTA in dH2O) for 1 h at 4 °C in the dark prior to submerging in 4 °C alkaline electrophoresis solution (pH > 13, 200 mM NaOH, 1 mM EDTA in dH2O). Electrophoresis was run at 21 V for 30 min using the Comet Assay ES unit. The slides were then immersed twice in dH2O for 5 min each followed by another wash in 70% ethanol for 5 min. Slides were dried at 37 °C for 15 min. SYBR-Gold (ThermoFisher Scientific, #S11494) was placed on dried agarose for 30 min at room temperature before complete drying at 37 °C. Samples were imaged using Leica DMI8 fluorescence microscope. Tail moment was calculated based on the following equation: tail moment = tail length × (fluorescence intensity of the DNA in the tail part /total fluorescence intensity of the DNA in both the head and tail). The images were analyzed using NIH Image J software.
Quantification and statistical analysis
One-way ANOVA, Two-tailed Student t-test and chi-square contingency test were used for the evaluation of experimental statistical significance using GraphPad Prism software (GraphPad v9, San Diego, CA, USA). ANOVA was followed by the Tukey post hoc test. Each experiment was replicated at least three times except Fig. 4F and Supplementary Fig. 2D, E (two replicates). The data were expressed as means ± SEM. p-value less than 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank all Balboula lab members, Luhui Zhang and Jason Rizo for their valuable help and support. The authors thank Zhiyuan Chen for his technical expertise and valuable help. The authors also thank Wen-Xing Ding for sharing Atg5flox/flox mice from his laboratory colony. This research was supported by a research grant from the American Society for Reproductive Medicine, the NIH (R35 GM142537), the NIH (R01 HD113358) and the National Institute of Food and Agriculture, grant number 2022-67015-36301 to A.Z.B. P.S. was funded by USDA National Institute of Food and Agriculture, Agriculture and Food Research Initiative Competitive grant number 2020-67015-31017 and seed funding from the College of Agriculture, Food and Natural Resources, University of Missouri.
Author contributions
A.Z.B. conceived experiments; A.Z.B., F.S. and N.N.A.; designed experiments; F.S., N.N.A. and D.L-V. performed and analyzed experiments; F.S. and A.Z.B. wrote the manuscript; A.Z.B. secured funding. C.A.S., H.Q., M.S.O., Y.A., M.T. R.M.R., A.M.K., P.S., A.L.P. provided technical expertise and contributed to study design, data analysis and interpretation. All authors revised, edited and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
No sequence or proteomic data has been generated in this study. All the data can be found in either the main text or the supplementary materials. Source data are provided with this paper. All raw image data are available from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fei Sun, Nourhan Nashat Ali.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53559-w.
References
- 1.Sun, H. et al. Global, regional, and national prevalence and disability-adjusted life-years for infertility in 195 countries and territories, 1990–2017: results from a global burden of disease study, 2017. Aging (Albany NY)11, 10952–10952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerrits, T. et al. Infertility in the Global South: Raising awareness and generating insights for policy and practice. Facts Views Vis. ObGyn9, 39–39 (2017). [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease, C. & Prevention. (2019).
- 4.American College of, O. & Gynecologists. (2020).
- 5.Hassold, T., Hall, H. & Hunt, P. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet.16, R203–R208 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Gruhn, J. R. et al. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science365, 1466–1469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin, L. J. DNA damage and repair: relevance to mechanisms of neurodegeneration. J. Neuropathol. Exp. Neurol.67, 377–387 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl, T. Instability and decay of the primary structure of DNA. Nature362, 709–715 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann, W. K. & Paules, R. S. DNA damage and cell cycle checkpoints. FASEB J.10, 238–247 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Burgoyne, P. S., Mahadevaiah, S. K. & Turner, J. M. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet.10, 207–216 (2009). [DOI] [PubMed] [Google Scholar]
- 12.De Bont, R. & van Larebeke, N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis19, 169–185 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Negrini, S., Gorgoulis, V. G. & Halazonetis, T. D. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol.11, 220–228 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Kastan, M. B. DNA damage responses: mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial Award Lecture. Mol. Cancer Res.6, 517–524 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pailas, A., Niaka, K., Zorzompokou, C. & Marangos, P. The DNA Damage Response in Fully Grown Mammalian Oocytes. Cells11. 10.3390/cells11050798 (2022). [DOI] [PMC free article] [PubMed]
- 17.Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature461, 1071–1078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindahl, T. & Barnes, D. E. Repair of Endogenous DNA Damage. Cold Spring Harb. Symposia Quant. Biol.65, 127–134 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Hunter, N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harbor Perspectives in Biology, a016618. 10.1101/cshperspect.a016618 (2015). [DOI] [PMC free article] [PubMed]
- 20.Stringer, J. M., Winship, A., Zerafa, N., Wakefield, M. & Hutt, K. Oocytes can efficiently repair DNA double-strand breaks to restore genetic integrity and protect offspring health. Proc. Natl Acad. Sci. USA117, 11513–11522 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marangos, P. & Carroll, J. Oocytes progress beyond prophase in the presence of DNA damage. Curr. Biol.22, 989–994 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Oktay, K., Turan, V., Titus, S., Stobezki, R. & Liu, L. BRCA mutations, DNA repair deficiency, and ovarian aging. Biol. Reprod.93, 67–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horta, F., Catt, S., Ramachandran, P., Vollenhoven, B. & Temple-Smith, P. Female ageing affects the DNA repair capacity of oocytes in IVF using a controlled model of sperm DNA damage in mice. Hum. Reprod.35, 529–544 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Titus, S. et al. Impairment of BRCA1-related DNA double-strand break repair leads to ovarian aging in mice and humans. Sci. Transl. Med.5, 172ra121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remillard-Labrosse, G. et al. Human oocytes harboring damaged DNA can complete meiosis I. Fertil. Steril.113, 1080–1089.e1082 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Kundu, M. & Thompson, C. B. Autophagy: basic principles and relevance to disease. Annu Rev. Pathol.3, 427–455 (2008). [DOI] [PubMed] [Google Scholar]
- 27.He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet.43, 67–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klionsky, D. J. & Emr, S. D. Autophagy as a regulated pathway of cellular degradation. Science290, 1717–1721 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filomeni, G., De Zio, D. & Cecconi, F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ.22, 377–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gozuacik, D. & Kimchi, A. Autophagy and cell death. Curr. Top. Dev. Biol.78, 217–245 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Kroemer, G., Marino, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell40, 280–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, Y., Quach, C. & Liang, C. Autophagy modulator plays a part in UV protection. Autophagy12, 1677–1678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomes, L. R., Menck, C. F. M. & Leandro, G. S. Autophagy Roles in the Modulation of DNA Repair Pathways. Int J Mol Sci18. 10.3390/ijms18112351 (2017). [DOI] [PMC free article] [PubMed]
- 34.Gillespie, D. A. & Ryan, K. M. Autophagy is critically required for DNA repair by homologous recombination. Mol. Cell Oncol.3, e1030538 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Y. et al. Autophagy Regulates Chromatin Ubiquitination in DNA Damage Response through Elimination of SQSTM1/p62. Mol. Cell63, 34–48 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Eliopoulos, A. G., Havaki, S. & Gorgoulis, V. G. DNA Damage Response and Autophagy: A Meaningful Partnership. Front Genet7, 204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fierz, B. et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol.7, 113–119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czaja, W., Mao, P. & Smerdon, M. J. Chromatin remodelling complex RSC promotes base excision repair in chromatin of Saccharomyces cerevisiae. DNA Repair (Amst.)16, 35–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dion, V. & Gasser, S. M. Chromatin movement in the maintenance of genome stability. Cell152, 1355–1364 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Citterio, E. et al. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell Biol.20, 7643–7653 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ataian, Y. & Krebs, J. E. Five repair pathways in one context: chromatin modification during DNA repair. Biochem Cell Biol.84, 490–504 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Bao, Y. Chromatin response to DNA double-strand break damage. Epigenomics3, 307–321 (2011). [DOI] [PubMed] [Google Scholar]
- 43.House, N. C., Koch, M. R. & Freudenreich, C. H. Chromatin modifications and DNA repair: beyond double-strand breaks. Front Genet5, 296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, J. et al. Effect of autophagy induction and cathepsin B inhibition on developmental competence of poor quality bovine oocytes. J. Reprod. Dev.66, 83–91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, Q. et al. Generation of fertile cloned rats by regulating oocyte activation. Science302, 1179 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Sutovsky, P. & Prather, R. S. Nuclear remodeling after SCNT: a contractor’s nightmare. Trends Biotechnol.22, 205–208 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Whitworth, K. M. et al. Method of oocyte activation affects cloning efficiency in pigs. Mol. Reprod. Dev.76, 490–500 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Yang, Q. et al. Rapamycin improves the quality and developmental competence of mice oocytes by promoting DNA damage repair during in vitro maturation. Reproductive Biology and Endocrinology20, 10.1186/s12958-022-00943-0 (2022). [DOI] [PMC free article] [PubMed]
- 49.Alvarez-Quilon, A. et al. ATM specifically mediates repair of double-strand breaks with blocked DNA ends. Nat. Commun.5, 3347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamamori-Adachi, M. et al. DNA damage response induced by Etoposide promotes steroidogenesis via GADD45A in cultured adrenal cells. Sci. Rep.8, 9636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leem, J., Bai, G. Y., Kim, J. S. & Oh, J. S. Melatonin protects mouse oocytes from DNA damage by enhancing nonhomologous end-joining repair. J. Pineal Res. 67, e12603 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Collins, J. K., Lane, S. I. R., Merriman, J. A. & Jones, K. T. DNA damage induces a meiotic arrest in mouse oocytes mediated by the spindle assembly checkpoint. Nat. Commun.6, 8553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bassing, C. H. et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl Acad. Sci. USA99, 8173–8178 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burma, S., Chen, B. P., Murphy, M., Kurimasa, A. & Chen, D. J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem.276, 42462–42467 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Stiff, T. et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res.64, 2390–2396 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Chowdhury, D. et al. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell20, 801–809 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Mah, L. J., El-Osta, A. & Karagiannis, T. C. γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia24, 679–686 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Sharma, A., Singh, K. & Almasan, A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol.920, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Ma, J. Y. et al. The effects of DNA double-strand breaks on mouse oocyte meiotic maturation. Cell Cycle12, 1233–1241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin, F. et al. Different fates of oocytes with DNA double-strand breaks in vitro and in vivo. Cell Cycle13, 2674–2680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marangos, P. et al. DNA damage-induced metaphase I arrest is mediated by the spindle assembly checkpoint and maternal age. Nat. Commun.6, 8706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olive, P. L. & Banath, J. P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc.1, 23–29 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Ganem, N. J. & Pellman, D. Linking abnormal mitosis to the acquisition of DNA damage. J. Cell Biol.199, 871–881 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mihajlovic, A. I., Haverfield, J. & FitzHarris, G. Distinct classes of lagging chromosome underpin age-related oocyte aneuploidy in mouse. Dev. Cell56, 2273–2283 e2273 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Antonarakis, S. E., Lyle, R., Dermitzakis, E. T., Reymond, A. & Deutsch, S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat. Rev. Genet.5, 725–738 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Lane, S. I. R. et al. DNA damage induces a kinetochore-based ATM/ATR-independent SAC arrest unique to the first meiotic division in mouse oocytes. Development144, 3475–3486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, Y., Liu, Z., Selby, C. P. & Sancar, A. Long-term, genome-wide kinetic analysis of the effect of the circadian clock and transcription on the repair of cisplatin-DNA adducts in the mouse liver. J. Biol. Chem.294, 11960–11968 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ratnayaka-Gamage, N. D., Alesi, L. R., Zerafa, N., Stringer, J. M. & Hutt, K. J. Xrcc5/KU80 is not required for the survival or activation of prophase-arrested oocytes in primordial follicles. Front Endocrinol. (Lausanne)14, 1268009 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanida, I., Ueno, T. & Kominami, E. LC3 and autophagy. Methods Mol. Biol.445, 77–88 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Kabeya, Y. et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci.117, 2805–2812 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Lee, S. H., Hiradate, Y., Hoshino, Y., Tanemura, K. & Sato, E. Quantitative analysis in LC3-II protein in vitro maturation of porcine oocyte. Zygote22, 404–410 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Tsukamoto, S., Kuma, A. & Mizushima, N. The role of autophagy during the oocyte-to-embryo transition. Autophagy4, 1076–1078 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Sarkar, S., Ravikumar, B., Floto, R. A. & Rubinsztein, D. C. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ.16, 46–56 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Noda, T. & Ohsumi, Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem.273, 3963–3966 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Mitchell, D. L. The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem. Photobiol.48, 51–57 (1988). [DOI] [PubMed] [Google Scholar]
- 76.Pfeifer, G. P. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem. Photobiol.65, 270–283 (1997). [DOI] [PubMed] [Google Scholar]
- 77.Castellanos, M., Gubern, C. & Kadar, E. mTOR: Exploring a new potential therapeutic target for stroke. Molecules to Medicine with mTOR: Translating Critical Pathways into Novel Therapeutic Strategies, 105–122. 10.1016/B978-0-12-802733-2.00012-8 (2016).
- 78.Schott, C. R., Ludwig, L., Mutsaers, A. J., Foster, R. A. & Wood, G. A. The autophagy inhibitor spautin-1, either alone or combined with doxorubicin, decreases cell survival and colony formation in canine appendicular osteosarcoma cells. PLoS ONE13, e0206427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ge, P. F. et al. Inhibition of autophagy induced by proteasome inhibition increases cell death in human SHG-44 glioma cells. Acta Pharm. Sin.30, 1046–1052 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seguin, S. J. et al. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ.21, 1838–1851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hara, T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- 82.Goodarzi, A. A. et al. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J.23, 4451–4461 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tang, M. et al. SIRT7-mediated ATM deacetylation is essential for its deactivation and DNA damage repair. Sci. Adv.5, eaav1118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim, K.-H. et al. The role of Rad51 in safeguarding mitochondrial activity during the meiotic cell cycle in mammalian oocytes. Sci. Rep.6, 34110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tashiro, S., Walter, J., Shinohara, A., Kamada, N. & Cremer, T. Rad51 Accumulation at Sites of DNA Damage and in Postreplicative Chromatin. J. Cell Biol.150, 283–292 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tarsounas, M., Davies, A. A. & West, S. C. RAD51 localization and activation following DNA damage. Philos. Trans. R. Soc. Lond. B Biol. Sci.359, 87–93 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsompana, M. & Buck, M. J. Chromatin accessibility: a window into the genome. Epigenetics Chromatin7, 33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weintraub, H. & Groudine, M. Chromosomal subunits in active genes have an altered conformation. Science193, 848–856 (1976). [DOI] [PubMed] [Google Scholar]
- 89.Jin, W. et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature528, 142–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kwon, H., Imbalzano, A. N., Khavari, P. A., Kingston, R. E. & Green, M. R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature370, 477–481 (1994). [DOI] [PubMed] [Google Scholar]
- 91.Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sheffield, N. C. et al. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res.23, 777–788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu, J., Cheng, K. C. L. & Yuen, K. W. Y. Histone H3K9 and H4 Acetylations and Transcription Facilitate the Initial CENP-AHCP−3 Deposition and De Novo Centromere Establishment in Caenorhabditis elegans Artificial Chromosomes. Epigenetics & Chromatin11. 10.1186/s13072-018-0185-1 (2018). [DOI] [PMC free article] [PubMed]
- 94.Ohzeki, J.-I. et al. Breaking the HAC Barrier: Histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J.31, 2391–2402 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bailey, L. T., Northall, S. J. & Schalch, T. Breakers and amplifiers in chromatin circuitry: acetylation and ubiquitination control the heterochromatin machinery. Curr. Opin. Struct. Biol.71, 156–163 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu, C. et al. Histone H1 facilitates restoration of H3K27me3 during DNA replication by chromatin compaction. Nat. Commun.14, 4081 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siegenfeld, A. P. et al. Polycomb-lamina antagonism partitions heterochromatin at the nuclear periphery. Nat. Commun.13, 4199 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hosogane, M., Funayama, R., Nishida, Y., Nagashima, T. & Nakayama, K. Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet.9, e1003698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qu, K. et al. Chromatin accessibility landscape of cutaneous T cell lymphoma and dynamic response to HDAC inhibitors. Cancer Cell32, 27–41 e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Toth, K. F. et al. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci.117, 4277–4287 (2004). [DOI] [PubMed] [Google Scholar]
- 101.Liu, B. et al. Inhibition of histone deacetylase 1 (HDAC1) and HDAC2 enhances CRISPR/Cas9 genome editing. Nucleic Acids Res.48, 517–532 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Walker, C. J. et al. Nuclear mechanosensing drives chromatin remodelling in persistently activated fibroblasts. Nat. Biomed. Eng.5, 1485–1499 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee, J. S. Activation of ATM-dependent DNA damage signal pathway by a histone deacetylase inhibitor, trichostatin A. Cancer Res Treat.39, 125–130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oike, T. et al. Garcinol, a histone acetyltransferase inhibitor, radiosensitizes cancer cells by inhibiting non-homologous end joining. Int J. Radiat. Oncol. Biol. Phys.84, 815–821 (2012). [DOI] [PubMed] [Google Scholar]
- 105.Balasubramanyam, K. et al. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J. Biol. Chem.279, 33716–33726 (2004). [DOI] [PubMed] [Google Scholar]
- 106.Ray Chaudhuri, A. & Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol.18, 610–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sinha, S., Molla, S. & Kundu, C. N. PARP1-modulated chromatin remodeling is a new target for cancer treatment. Med. Oncol.38, 118 (2021). [DOI] [PubMed] [Google Scholar]
- 108.Kataura, T. et al. Autophagy promotes cell survival by maintaining NAD levels. Dev. Cell57, 2584–2598.e2511 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang, X. et al. Autophagy suppresses radiation damage by activating PARP-1 and attenuating reactive oxygen species in hepatoma cells. Int J. Radiat. Biol.95, 1051–1057 (2019). [DOI] [PubMed] [Google Scholar]
- 110.LaFargue, C. J., Dal Molin, G. Z., Sood, A. K. & Coleman, R. L. Exploring and comparing adverse events between PARP inhibitors. Lancet Oncol.20, e15–e28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barbosa, M. C., Grosso, R. A. & Fader, C. M. Hallmarks of aging: an autophagic perspective. Front Endocrinol. (Lausanne)9, 790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suh, E. K. et al. p63 protects the female germ line during meiotic arrest. Nature444, 624–628 (2006). [DOI] [PubMed] [Google Scholar]
- 113.Gui, L. & Homer, H. Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development139, 1941–1946 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kolano, A., Brunet, S., Silk, A. D., Cleveland, D. W. & Verlhac, M. H. Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc. Natl. Acad. Sci. USA109, E1858–E1867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kyogoku, H. & Kitajima, T. S. Large cytoplasm is linked to the error-prone nature of oocytes. Dev. Cell41, 287–298.e284 (2017). [DOI] [PubMed] [Google Scholar]
- 116.Bang, S., Shin, H., Song, H., Suh, C. S. & Lim, H. J. Autophagic activation in vitrified–warmed mouse oocytes. REPRODUCTION148, 11–19 (2014). [DOI] [PubMed] [Google Scholar]
- 117.Latorraca, L. B. et al. Autophagy is a pro-survival adaptive response to heat shock in bovine cumulus-oocyte complexes. Scientific Reports10, 10.1038/s41598-020-69939-3 (2020). [DOI] [PMC free article] [PubMed]
- 118.Zuccotti, M., Piccinelli, A., Giorgi Rossi, P., Garagna, S. & Redi, C. A. Chromatin organization during mouse oocyte growth. Mol. Reprod. Dev.41, 479–485 (1995). [DOI] [PubMed] [Google Scholar]
- 119.Luciano, A. M. et al. Large-scale chromatin morpho-functional changes during mammalian oocyte growth and differentiation. Eur. J. Histochem56, e37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang, Y. et al. Single-cell epigenome analysis reveals age-associated decay of heterochromatin domains in excitatory neurons in the mouse brain. Cell Res. 32, 1008–1021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sedivy, J. M., Banumathy, G. & Adams, P. D. Aging by epigenetics–a consequence of chromatin damage? Exp. Cell Res. 314, 1909–1917 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tsurumi, A. & Li, W. X. Global heterochromatin loss: a unifying theory of aging? Epigenetics7, 680–688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chiang, T., Duncan, F. E., Schindler, K., Schultz, R. M. & Lampson, M. A. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr. Biol.20, 1522–1528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shomper, M., Lappa, C. & FitzHarris, G. Kinetochore microtubule establishment is defective in oocytes from aged mice. Cell Cycle13, 1171–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Duncan, F. E. et al. Chromosome cohesion decreases in human eggs with advanced maternal age. Aging Cell11, 1121–1124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lan, Z. J., Xu, X. & Cooney, A. J. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod.71, 1469–1474 (2004). [DOI] [PubMed] [Google Scholar]
- 127.Schultz, R. M., Montgomery, R. R. & Belanoff, J. R. Regulation of mouse oocyte meiotic maturation: Implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev. Biol.97, 264–273 (1983). [DOI] [PubMed] [Google Scholar]
- 128.Stein, P. & Schindler, K. Mouse oocyte microinjection, maturation and ploidy assessment. J. Vis. Exp. 10.3791/2851 (2011). [DOI] [PMC free article] [PubMed]
- 129.Kincade, J. N., Hlavacek, A., Akera, T. & Balboula, A. Z. Initial spindle positioning at the oocyte center protects against incorrect kinetochore-microtubule attachment and aneuploidy in mice. Sci. Adv.9, eadd7397 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsafriri, A., Chun, S. Y., Zhang, R., Hsueh, A. J. & Conti, M. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev. Biol.178, 393–402 (1996). [DOI] [PubMed] [Google Scholar]
- 131.Duncan, F. E., Chiang, T., Schultz, R. M. & Lampson, M. A. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol. Reprod.81, 768–776 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Balboula, A. Z. & Schindler, K. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet.10, e1004194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Londono-Vasquez, D., Rodriguez-Lukey, K., Behura, S. K. & Balboula, A. Z. Microtubule organizing centers regulate spindle positioning in mouse oocytes. Dev. Cell57, 197–211.e193 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
No sequence or proteomic data has been generated in this study. All the data can be found in either the main text or the supplementary materials. Source data are provided with this paper. All raw image data are available from the corresponding author upon request. Source data are provided with this paper.