Abstract
Background
Gastric cancer (GC) is a deadly malignancy with an ever-increasing incidence worldwide. The cellular communication network (CCN) family serves as matricellular proteins and exerts their various functions via regulating cell proliferation and differentiation. This study aimed to perform an integrated analysis of CCNs to predict the prognosis in GC.
Methods
The microarray datasets were obtained from Gene Expression Omnibus database to identify the differentially expressed genes between GC and non-tumor tissues. Functional enrichment and genetic alteration analysis revealed the biological functions and alteration status associated with CCNs. We analyzed the mRNA and protein expressions of CCN family in GC patients. Furthermore, the prognostic value of distinct CCN family members were analyzed using the Kaplan–Meier plotter database. Finally, the human gastric cancer cell lines were used for in vitro experiments to further validate the role of WISP1.
Results
26 genes were firstly identified to be significantly highly expressed in gastric tumor tissues. CCN family genes were identified to predict the prognosis in GC. Among the six CCNs, WISP1 is upregulated in GC tissues and its highly expression is associated with poor survival in GC patients. Moreover, a significant correlation is found between the expression of WISP1 and the pathological stage of patients with GC. Additionally, in vitro experiments demonstrated that WISP1 promotes the proliferation and invasive potential of GC cells, suggesting it may be a potential therapeutic target for GC.
Conclusions
A comprehensive bioinformatic analysis of CCN genes provides new insights into the potential roles of this family in GC. Importantly, WISP1 may be a good prognostic predictor and a potential therapeutic target for GC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-024-01459-2.
Keywords: Gastric cancer, CCN family, WISP1, Prognosis, Biomarker
Introduction
Gastric cancer (GC) is one of the most common malignant tumors and imposes a substantial health burden globally. Early diagnosis and multidisciplinary therapeutic strategies are pivotal to improve the prognosis of patients with GC. The heterogeneity in GC is a considerable obstacle, underscoring the need for precision treatment strategies [1, 2]. Molecular studies on cancers have yielded a large number of effective information for exploring new therapy. Advances against GC have lagged compared with other tumors; therefore, more research studies should be performed to explore reliable prognostic biomarkers and novel therapeutic targets to prolong the survival of patients with GC.
As a reliable technique used for years, gene chip could quickly identify differentially expressed genes and effectively provide slice information in the open databases [3]. Consequently, science researchers could detect a variety of valuable data to continue new investigations based on these data. It is particular note that diverse bioinformatics methods could help us better identify significant genes related to tumor prognosis and further explore the underlying mechanisms. Numerous studies have already identified differentially expressed genes in GC tumorigenesis via bioinformatics analysis. However, the prognostic value of these genes has not been acceptable in clinic.
Here, we firstly selected three gene chips from Gene Expression Omnibus (GEO) and obtained the commonly up-expressed genes in these datasets. Among these genes, we focused on the cellular communication network (CCN) family genes, which serve as matricellular proteins and have extensive extracellular binding partners. CCN family genes exert their various functions via regulating cell proliferation and differentiation [4]. It consists of six unique members, including cysteine-rich 61 (CYR61)/CCN1, connective tissue growth factor (CTGF)/CCN2, nephroblastoma overexpressed (NOV)/CCN3, and WNT1 inducible signaling pathway protein genes (WISP1-3, also known as CCN4-6).
Accumulating evidence revealed that CCN family members have been associated with the growth of various tumors. Previous published data also suggested that CCN family proteins have contributed to the carcinogenesis of GC [5–7]. More importantly, matricellular proteins are still crucial for the interaction between tumor and its microenvironment [8]. However, little is known regarding the prognostic value of distinct CCN family members in GC. Therefore, we further investigated the message RNA (mRNA) expression and clinical significance of CCN family genes in GC based on several databases. Through an integrative bioinformatic analysis, we found WISP1 as the most interest of GC prognosis assessment. Moreover, the expression of WISP1 in GC cells were inhibited via siRNA, the cell proliferation and migration were in return attenuated. Our study indicated that WISP1 might have potential value in prognosis and treatment of GC.
Materials and methods
Microarray profile and gene screening
We analyzed the gene expression profile of GSE19826, GSE54129 and GSE79973 which were all from GPL570 Platform. Differentially expressed genes (DEGs) between tumor tissues and adjacent normal tissues was firstly identified by the ‘limma’ R package. DEGs between GC samples and normal gastric samples were also assessed with GEO2R tools. |logFC| > 2 and adjust P value < 0.05 were selected as the thresholds [9]. We next utilized the Venn software to identify the commonly DEGs in these datasets.
Functional enrichment analysis
We utilized the Database for Annotation, Visualization and Integrated Discovery (DAVID) to visualize the DEGs enrichment of biological processes and pathways. Metascape is another effective tool to provide a comprehensive gene list annotation and analysis resource [10]. We also used this database to perform pathway enrichment analysis of CCN family genes.
Genetic alteration analysis
We used the cBioPortal website and obtained the alteration status of CCN family genes in patients with GC from the dataset “Stomach Adenocarcinoma (TCGA, PanCan 2018)”. The data was downloaded and plotted by GraphPad Prism v7.00 (GraphPad Software Inc., USA).
Gene expression analysis
We used Gene Expression Profiling Interactive Analysis (GEPIA) to compare the data of RNA sequencing expression between tumor samples and normal controls, as well as different pathological stages. And we evaluated the expression of CCN family genes in GC from several datasets in ONCOMINE. The significance thresholds include p 0.05, a fold change of 2, and a gene rank in the top 10%. We obtained the fragments per kilobase of exon per million fragments mapped (FPKM) values and the protein expression of each CCN family gene in patients with GC from the Pathology Atlas part. The data was downloaded and plotted by GraphPad Prism v7.00 (GraphPad Software Inc., USA).
Survival analysis
We analyzed the association between the expressions of CCN family genes and survival of patients with GC via Kaplan Meier-plotter. Moreover, we assessed the prognostic values of CCN family genes in patients with different clinicopathological characteristics. The specific Affymetrix identity card (ID) of each CCN family gene selected in the Kaplan–Meier Plotter analysis were exhibited in Table S1. The survival data was downloaded and plotted by GraphPad Prism v7.00 (GraphPad Software Inc., USA).
Cell lines, reagents and antibodies
Human gastric cancer cell lines Hgc-27 and SNU-1 were were cultured in RPMI 1640 medium (GIBCO BRL, Paisley, UK) supplemented with 10% fetal bovine serum (Invitrogen Life Technologies), and incubated in a humidified incubator at 37 °C supplemented with 5% carbon dioxide. Lipofectamine™ RNAiMAX was obtained from Invitrogen Corporation (Carlsbad, CA, USA). Human WISP1 short interfering RNA (siRNA) was purchased from Ribobio (Guangzhou, China). Antibodies against GAPDH and WISP1 were purchased from Abcam (Cambridge, MA, USA).
Western blot analysis
Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in lysis buffer (containing RIPA, phosphatase inhibitor and protease inhibitor) for 15 min on ice. Cell debris was removed by centrifugation at 12,000×g for 15 min at 4 °C. Protein lysates were analyzed by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. Protein bands of interest were revealed by blotting with the respective antibodies.
Flow cytometry
Cell apoptosis was analyzed by an Annexin-V/Propidium Iodide according to the manufacturer’s protocol. Cells were collected and analyzed on Beckman flow cytometer.
Colony formation assay
Cells were seeded at a low density (400 cells per well) in six-well plates, and cultured at 37 °C for 7 days in medium. At the end of incubation, the cells were fixed with formalin for 15 min, stained with crystal violet for 15 min, and the numbers of colonies were counted using AlphaImager HP system (ProteinSimpre, CA, USA).
Scratch test
The cells in each group were seeded in six-well plates and transfected when the cells reached 50% confluence. After transfection for 48 h, culture solution was replaced and a vertical scratch was made in the cell culture surface using a 100 µL pipette tip. The cells were rinsed with PBS thrice and the culture medium was changed. Images were acquired and the scratch distance was measured at 0 h and 24 h.
Detection of key regulated factor
We analyzed the data in the Transcription Regulatory Relationships Unraveled Sentence-based Text mining (TRRUST) database and obtained information on which transcription factors involved in the regulation of CCN family genes.
Statistical analysis
Data are presented as mean ± standard deviation. Comparisons between groups were performed using the Student’s t test on statistical product service solutions (SPSS) software (version21.0). A p value less than 0.05 was considered statistically significant.
Results
Identification of differentially expressed genes in GC
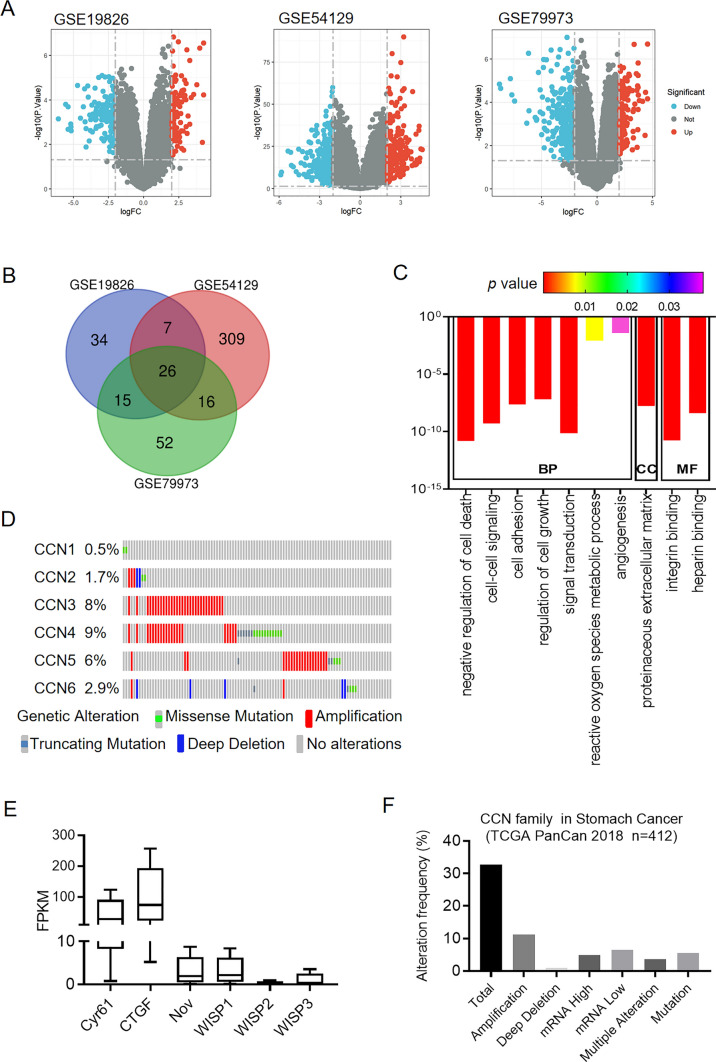
The flow chart of this study is shown in Fig. 1. We explored gene expression information of GC from GEO database and obtained the genomic profiles of GSE19826, GSE54129 and GSE79973. Volcano plots were generated to visualize the expression patterns of these differentially expressed genes in these databases (Fig. 2A). Venn diagram software was next used to detect the commonly up-regulated genes in these datasets (Fig. 2B). In total, 26 genes were identified to be significantly highly expressed in gastric tumor tissues compared with normal controls (Table S2). Interestingly, these highly expressed genes except retinoic acid receptor responder 1 (RARRES1) was significantly associated with poor overall survival in patients with GC (Figs. S1 and 2). Of these genes, we pay attention to WISP1 which belongs to CCN family and plays crucial roles in many physiological and pathological processes.
Fig. 1.
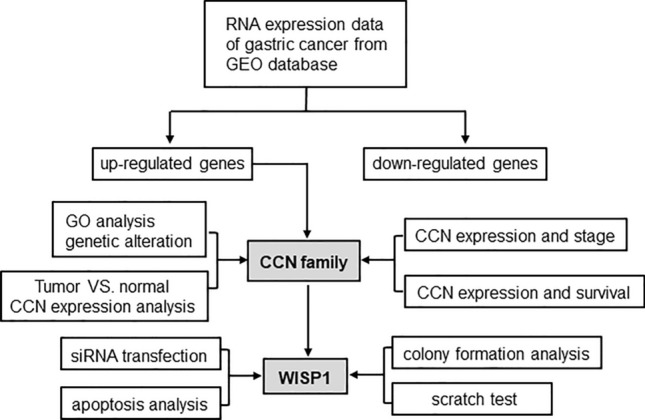
Flow chart of data collection, analysis and validation
Fig. 2.
Gene ontology enrichment analysis and the genetic alterations of the CCN family genes. A Volcano plots of the differentially expressed genes in the GSE19826, GSE54129 and GSE79973 databases. B Venn diagram to show the commonly up-regulated genes. C Significantly enriched gene ontology terms of CCN family. D The genetic alteration profiles of the six CCN family genes in STAD from cBioportal. E Genetic alteration frequency of CCN family genes in STAD using cBioPortal. F Relative expression differences of CCN family genes in STAD from HAP database
Functional enrichment analysis of CCN family genes in GC
To better understand the functions of CCN family, we performed the functional enrichment analysis using DAVID 6.8. As shown in Fig. 2C, among all the significant enriched functions in the biological process (BP) category, negative regulation of cell death, regulation of cell growth, signal transduction, reactive oxygen species metabolic process, and angiogenesis were involved in the tumorigenesis and progression of GC. Cell–cell signaling and cell adhesion were associated with tumor microenvironment. The proteinaceous extracellular matrix was the only significant enriched item in the cell component (CC) category. In the molecular function (MF) category, the CCN family members were mainly involved in integrin binding and heparin binding activities. Table 1 showed the specific genes involved in these biological activities. Therefore, the function of CCN family is regulating cell–cell and cell-matrix interaction and in turn modulating cell growth.
Table 1.
Gene ontology analysis of CCN family genes
| Category | Term count | Genes | |
|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0060548~negative regulation of cell death | 5 | NOV, WISP2, WISP1, CTGF, CYR61 |
| GOTERM_BP_DIRECT | GO:0007267~cell–cell signaling | 5 | NOV, WISP2, WISP1, CTGF, CYR61 |
| GOTERM_BP_DIRECT | GO:0007155~cell adhesion | 5 | NOV, WISP2, WISP1, CTGF, CYR61 |
| GOTERM_BP_DIRECT | GO:0001558~regulation of cell growth | 4 | WISP2, WISP1, CTGF, CYR61 |
| GOTERM_BP_DIRECT | GO:0007165~signal transduction | 5 | NOV, WISP2, WISP1, CTGF, CYR61 |
| GOTERM_BP_DIRECT | GO:0072593~reactive oxygen species metabolic process | 2 | CTGF, CYR61 |
| GOTERM_BP_DIRECT | GO:0001525~angiogenesis | 2 | NOV, CTGF |
| GOTERM_CC_DIRECT | GO:0005578~proteinaceous extracellular matrix | 5 | NOV, WISP2, WISP1, CTGF, CYR61 |
| GOTERM_MF_DIRECT | GO:0005178~integrin binding | 5 | NOV, WISP2, WISP1, CTGF, CYR61 |
| GOTERM_MF_DIRECT | GO:0008201~heparin binding | 5 | NOV, WISP2, WISP1, CTGF, CYR61 |
We also performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses and found no significant pathway was enriched in the CCN family. This limited information may be attributable to insufficient studies on CCN family. We next conducted the functional enrichment analysis with Metascape database. The functions of CCN family genes were mainly enriched in naba extracellular matrix glycoproteins pathway (Table S3).
The genetic alterations of CCN family members in patients with GC
We next explored the genetic alterations of CCN family members in 412 patient samples with stomach adenocarcinoma via cBioportal database. Stomach adenocarcinoma (STAD) is the most common histological type (~ 95%) of GC [7]. In detail, CCN1, CCN2, CCN3, CCN4, CCN5, and CCN6 were altered in 0.5, 1.7, 8, 9, 6, and 2.9% of these samples (Fig. 2D). WISP1 (CCN3) was frequently altered CCN family gene with an alteration rate of 8%. More than 30% of GC patients showed genetic aberrations of CCN family genes and the most common alteration was mRNA deregulation including mRNA high and mRNA low (Fig. 2E). Additionally, we compared the mRNA level among CCN family members in STAD via Human Protein Atlas (HPA) database, the mRNA level of WISP2 was quite low (Fig. 2F).
The expressions of CCN family members in patients with GC
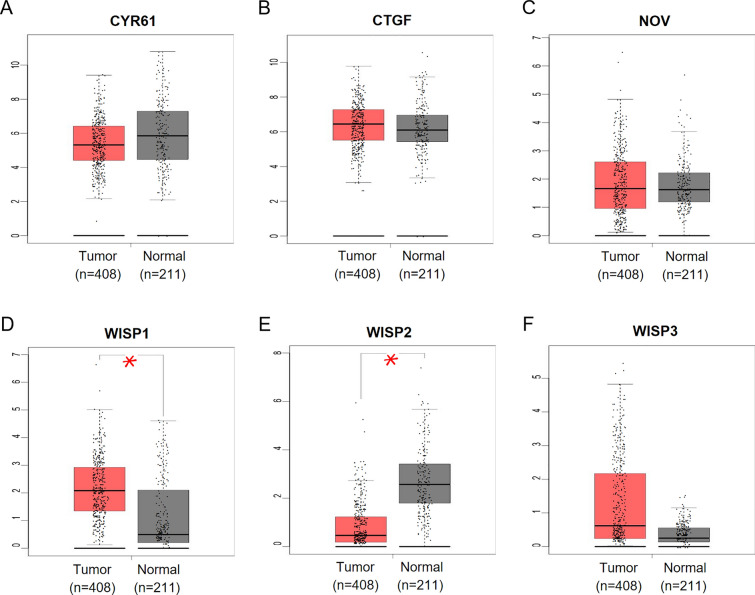
We explored the expression levels of CCN family genes in STAD and normal tissues with GEPIA. Based on the data form GEPIA, only the expression of WISP1 in GC tissues was significantly elevated while the expression of WISP2 were significantly reduced (Fig. 3). The expressions of CTGF, NOV and WISP3 were slightly increased in tumor tissues without a significant p-value.
Fig. 3.
Gene expression of CCN family genes in STAD. Comparison of the mRNA expression of CYR61 (A), CTGF (B), NOV (C), WISP1 (D), WISP2 (E), and WISP3 (F) in gastric tumor tissue (left plot n = 408) and normal tissue (right plot n = 211) from GEPIA database
In addition, we analyzed the transcriptional levels of CCN family members in GC and normal gastric tissues using ONCOMINE database. The transcription levels of CYR61, CTGF, NOV, WISP1, and WISP3 in different types of GC tissues were significantly elevated compared with normal gastric tissues in these datasets shown in Table 2. Three datasets including Cui dataset [11], Derrico dataset [12] and Wang dataset [13], suggested that the mRNA expressions of CTGF, WISP1 and WISP3 in GC were prominently more than 1.5-fold higher than in normal gastric controls. The transcriptional level of WISP2 in GC tissues exhibited no significant alteration in all the datasets [11–14]. The possible explanation for the different results in these databases might be the different samples included and detection methods selected. We considered excluding several CCN family members such as WISP2 from further analysis, however, we preferred to identify the potential of the whole CCN family as effective prognostic biomarker in GC.
Table 2.
The elevated mRNA levels of CCN family members in different types of GC (ONCOMINE)
| Gene | Type | Fold change | p-value | Normal cases | Tumor cases | Dataset |
|---|---|---|---|---|---|---|
| CYR61 | Mixed type vs. normal | 2.283 | 3.51e−4 | 28 | 8 | Chen |
| Diffuse type vs. normal | 1.351 | 0.041 | 29 | 13 | Chen | |
| Intestinal type vs. normal | 1.259 | 0.044 | 29 | 66 | Chen | |
| CTGF | Mixed type vs. normal | 1.849 | 0.01 | 29 | 8 | Chen |
| Diffuse type vs. normal | 1.638 | 0.005 | 29 | 13 | Chen | |
| Mixed type vs. normal | 2.326 | 4.24e−4 | 31 | 4 | DErrico | |
| Gastric cancer vs. normal | 1.677 | 0.03 | 15 | 12 | Wang | |
| NOV | Diffuse type vs. normal | 1.189 | 0.034 | 20 | 13 | Chen |
| WISP1 | Gastric cancer vs. normal | 2.117 | 1.59e−4 | 80 | 80 | Cui |
| Mixed type vs. normal | 6.069 | 1.07e−6 | 31 | 4 | DErrico | |
| Diffuse type vs. normal | 3.452 | 0.001 | 31 | 6 | Derrico | |
| Intestinal type vs. normal | 4.941 | 3.03e−9 | 31 | 26 | DErrico | |
| Gastric cancer vs. normal | 2.948 | 0.01 | 15 | 12 | Wang | |
| WISP3 | Gastric cancer vs. normal | 3.206 | 0.003 | 80 | 80 | Cui |
| Intestinal type vs. normal | 2.360 | 0.011 | 31 | 26 | DErrico | |
| Gastric cancer vs. normal | 5.970 | 0.003 | 15 | 12 | Wang |
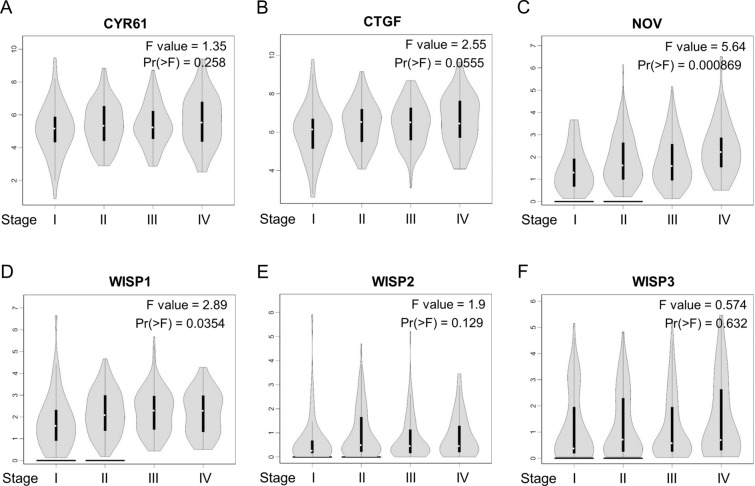
We then assessed the alteration of CCN family expressions during the gastric tumor progression, finding a significant relationship between the expression of NOV and WISP1 and pathological stage (Fig. 4). With the progression of the gastric tumor, the expression of NOV and WISP1 increased, suggesting that NOV and WISP1 might play a pivotal role in the promotion and progression of GC. In addition, by analyzing the protein expressions of CCN family form HPA database, we found that only WISP1 expression was increased in gastric tumor tissues (Table S4). This result mainly due to insufficient sample size and ineffectiveness of antibodies.
Fig. 4.
Correlation between different expressed CCN family genes and the pathological stage of patients with STAD. A CYR61, B CTGF, C NOV, D WISP1, E WISP2, F WISP3. *p < 0.05
The prognostic value of CCN family genes in patients with GC
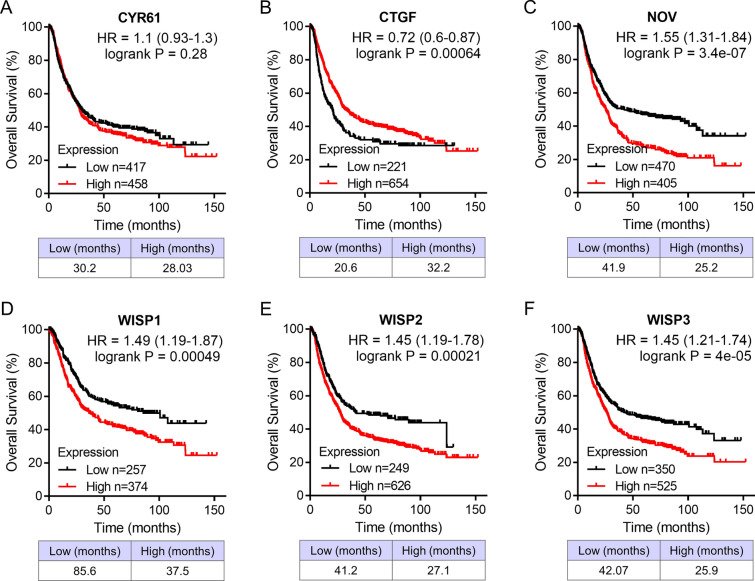
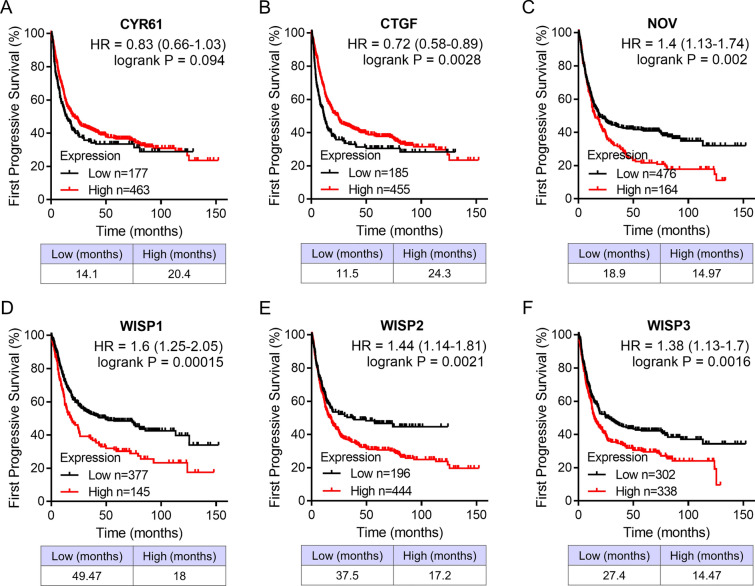
We analyzed the association between the mRNA expressions of CCN family genes and clinical outcome in patients with GC. The patients had been divided into high and low expression groups in the Kaplan–Meier Plotter database. Patients with high expressed CTGF was significantly related to better overall survival, whereas patients with high transcriptional levels of NOV, WISP1, WISP2, and WISP3 were remarkably associated with shorter overall survival (Fig. 5; CYR61, HR = 1.1, 95% CI 0.93–1.3, p = 0.28; CTGF, HR = 0.72, 95% CI 0.6–0.87, p = 0.00064; NOV, HR = 1.55, 95% CI 1.31–1.84, p = 3.4e−07; WISP1, HR = 1.49, 95% CI 1.19–1.87, p = 0.00049; WISP2, HR = 1.45, 95% CI 1.19–1.78, p = 0.00021; WISP3, HR = 1.45, 95% CI 1.21–1.74, p = 4e−05). The prognostic value of differentially expressed CCN family genes in the first progressive survival was also assessed. Similar with the result in overall survival, high expressed CTGF and low expressed NOV, WISP1, WISP2, and WISP3 were significantly associated with longer first progressive survival (Fig. 6; CYR61, HR = 0.83, 95% CI 0.66–1.03, p = 0.094; CTGF, HR = 0.72, 95% CI 0.58–0.89, p = 0.0028; NOV, HR = 1.4, 95% CI 1.31–1.74, p = 0.002; WISP1, HR = 1.6, 95% CI 1.25–2.05, p = 0.00015; WISP2, HR = 1.44, 95% CI 1.14–1.81, p = 0.0021; WISP3, HR = 1.38, 95% CI 1.13–1.7, p = 0.0016).
Fig. 5.
Overall survival analysis of CCN family genes in patients with GC. Survival curves of CYR61 (A), CTGF (B), NOV (C), WISP1 (D), WISP2 (E), and WISP3 (F) analyzed via the Kaplan–Meier plotter database
Fig. 6.
Survival analysis of CCN family genes in patients with diffuse GC. First progressive survival curves of CYR61 (A), CTGF (B), NOV (C), WISP1 (D), WISP2 (E), and WISP3 (F) analyzed via the Kaplan–Meier plotter database
According to the Lauren classification, GC is separated into diffuse and intestinal types histologically [15]. Therefore, we investigated the prognostic value of CCN family genes in these two cancer types. Subgroup analyses indicated that higher expression levels of all the CCN family members were correlated with poorer overall survival (OS) in patients with intestinal type (Fig. S3; CYR61, HR = 2.25, 95% CI 1.63–3.11, p = 4e−07; CTGF, HR = 1.53, 95% CI 1.11–2.1, p = 0.0084; NOV, HR = 3.17, 95% CI 2.05–4.9, p = 4.7e−08; WISP1, HR = 1.65, 95% CI 1.14–2.4, p = 0.0077; WISP2, HR = 2.01, 95% CI 1.46–2.77, p = 1.3e−05; WISP3, HR = 2.05, 95% CI 1.45–2.9, p = 3.7e−05). And elevated mRNA expressions of CTGF, NOV and WISP2 were associated with poorer OS in patients with diffuse type (Fig. S4; CYR61, HR = 1.3, 95% CI 0.9–1.89 p = 0.16; CTGF, HR = 1.45, 95% CI 1.03–2.04, p = 0.031; NOV, HR = 2.05, 95% CI 1.41–2.97, p = 0.00011; WISP1, HR = 1.39, 95% CI 0.98–1.96, p = 0.064; WISP2, HR = 1.66, 95% CI 1.1–2.51, p = 0.015; WISP3, HR = 0.78, 95% CI 0.55–1.09, p = 0.14).
Inhibition of WISP1 dampens proliferation and migration of GC cells
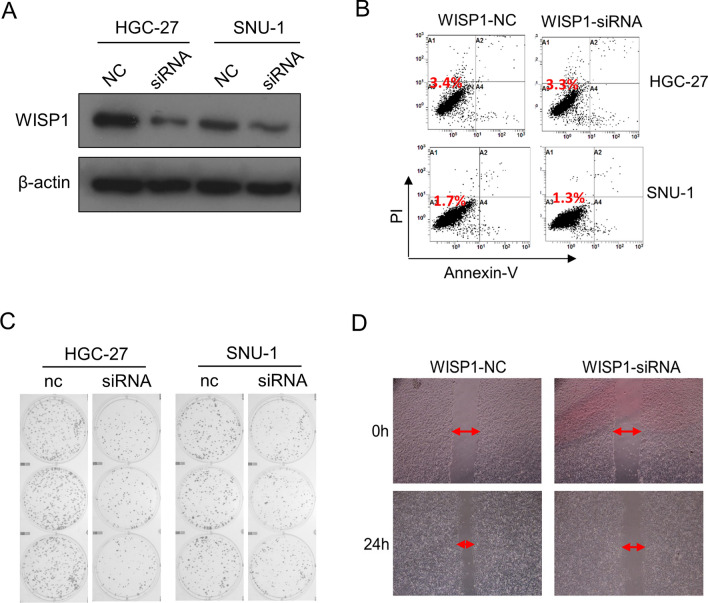
Our data suggested that targeting WISP1 could be a promising strategy for the treatment of GC. Therefore, we preliminarily investigated the effects of WISP1 expression level on the proliferation and migration of GC cells. We used RNA interference technology to downregulate WISP1 expression. After cellular transfection of WISP1-siRNA and WISP1-negative control (NC), WSIP1 expression in each cell lines were detected using western blot. As shown in Fig. 7A, WISP1 expression was significantly lower in the WISP-siRNA group compared with the control group. Decreased WISP1 exhibited no affection on cell activity (Fig. 7B). To determine the effect of WISP1 on the proliferation and migration of GC cells, colony formation and scratch test were performed. The activity of colony formation in the WISP1-siRNA group was significantly attenuated compared with the WISP1-NC groups (Fig. 7C). In addition, the cell scratch distance in the WISP1-siRNA group was significantly larger compared with the WISP1-NC groups after 24 h (Fig. 7D).
Fig. 7.
Inhibition of WISP1 dampens proliferation and migration of GC cells. A siRNA lowers WISP1 expression in HGC-27 and SNU-1 cells. B apoptosis of GC cells after transfection for 48 h. C Colony formation of GC cells after transfection. D Scratch test results in SNU-1 cells after transfection. The experiment was repeated 3 times
Key transcription factors of CCN family genes in GC
We further evaluated possible transcription factor targets of CCN family using TRRUST database. Unfortunately, the most valuable member WISP1 was not searched in TRRUST. CYR61, CTGF, NOV, and WISP2 were included in this database. We found that two transcription factors including Wilms tumor 1 (WT1) and signal transducer and activator of transcription 3 (STAT3) were involved in the regulation of CCN family genes (Table 3). Specifically, WT1 was the key transcription factor for CTGF and NOV. STAT3 was the key transcription factor for CYR61 and CTGF.
Table 3.
Key regulated factor of CCN family members in GC (TRRUST)
| Key TF | Description | Regulated gene | p-value | FDR |
|---|---|---|---|---|
| WT1 | Wilms tumor 1 | CTGF, NOV | 0.000133 | 0.000267 |
| STAT3 | Signal transducer and activator of transcription 3 (acute-phase response factor) | CYR61, CTGF | 0.000827 | 0.000827 |
Discussion
It is crucial to explore regulators driving GC tumor development and biomarkers predicting postoperative patients with GC. With the emergence of bioinformatics technology, several studies have utilized TCGA or GEO datasets to find numerous biomarkers related to prognosis and survival in GC [16, 17]. We started studies related to differentially expressed genes in GC, linking the findings to potential therapeutic opportunities. We thoroughly explored three profile datasets (GSE19826, GSE54129 and GSE79973) and found a total of 26 commonly up-regulated genes in GC compared with normal controls. Among all these genes, we further in-depth observed the CCN family genes which have been well known as major players in fundamental biological processes [18].
We focused on the function of CCN family genes using Gene Ontology (GO) enrichment analysis and KEGG pathway enrichment analysis. The functions of these genes are mainly involved in the cell–cell and cell–stroma interactions, and cell growth. It has been reported that the multiple functionalities of CCN proteins are result of interacting with various cytokines, extracellular matrices, and cell membrane proteins in microenvironment [19]. Therefore, CCN proteins can participate in specific extracellular physiological and pathological activities in the tumor microenvironment. The tumor microenvironment is different depending on the tissue types and differentiation stages, thus the diverse biological actions of CCN family are complicated. Previous study has reported that the wnt/β-catenin signaling pathway was regulated by WISP2 in GC cells [20] and WISP3 depletion induced suppression of wnt/β-catenin signaling and its downstream genes in GC [21]. However, no significantly pathway was associated with CCN family in our KEGG pathway enrichment analysis.
We next explored the genetic alterations, mRNA expression and prognostic value of CCN family in GC. Most CCN family genes except WISP2 was highly expressed in different types of GC respectively. The inconsistent results between GEPIA and ONCOMINE could be related to the different criteria for patient/sample inclusion and grouping, and the methods for data analysis. However, WISP1 was the only CCN family gene highly expressed in GC tissues. WISP1 reflected high expression in GC samples compared with normal controls in both RNA and protein levels. Moreover, we found that the expression of WISP1 was elevated during the tumor progression. We still explored the prognostic value of each CCN family member in patients with CG as well as different histological types of GC. Patients with low expression of WISP1 were significantly related to better survival. Despite several CCN family genes exert potential prognostic value in GC, we considered WISP1 was the most promising one based on our and other researchers’ data [6, 22].
Previous studies revealed that WISP1 was upregulated in multiple cancers including breast cancer [23], non-small cell lung cancer [24], colon cancer [25], esophageal squamous cell carcinoma [26], endometrial endometrioid adenocarcinoma [27], prostate cancer [28], and gastric cancer [29]. In addition, it reported that scirrhous gastric carcinoma cells with WISP1 genetic alteration enhanced the invasiveness of co-cultured gastric carcinoma cells [30]. Another research study revealed an oncogene role of WISP1 in GC, due to inhibiting WISP1 leads to suppression of cyclin D1 and epithelial–mesenchymal transition (EMT) progression and further decreases in cell proliferation, migration and invasion in GC cells [31]. In addition, forced expression of WISP1 in fibroblast cells enhanced the invasive abilities of co-cultured GC cells [30]. These data suggested the great potential of WISP1 as a good prognostic biomarker in GC and targeting WISP1 could be a promising strategy for the treatment of GC.
CYR61, CTGF, and WISP1 have been generally involved in cell proliferation and tumor growth, in contrast, NOV, WISP2 and WISP3 have exerted inhibition activity on cell proliferation and tumor growth [32, 33]. Recently, accumulating evidence has demonstrated that WISP1-3 exhibited different biological functions in a broad spectrum of human cancers. Although we considered WISP1 was the most promising CCN family gene in the prognosis of GC, we could not underestimate the prognostic value and important roles of the other CCN family members in GC. For instance, NOV is still a promising prognostic biomarker and therapeutic target, and deserve more detailed investigation [34].
Our in vitro experiments provided evidence that the inhibition of WISP1 could suppress proliferation and migration of GC cells. The development of treatment strategies to down-regulation of WISP1 could provide a promising therapeutic target treating patients with GC. The specific mechanisms of WISP1 including its signal pathway and influencing factors are still remain unclear. We further explored the transcription factor of the CCN family and found that WT1 and STAT3 might be key transcription factors in the regulation of CCN family genes. The regulation between STAT3 and CCN family members is still unknown. Importantly, our present data possibly provide future research directions.
Limitations
Limitations do exist in our present study. The expression and prognostic value of CCN family genes at the protein level, rather than the mRNA level, are really needed to be further investigated. And more functional experiments are urgently needed to validate these present conclusions. Another challenge is to illustrate the specific role of WISP1 in the etiology of GC. In the future study, we may combine gene expression level of WISP1 and cell biological behavior analysis to further explore the pathogenesis of GC.
Conclusion
In this study, we firstly identified different expression genes between GC and normal gastric tissues via analyzing public microarray datasets. Based on the important functions of CCN family, we aim to find effective biomarkers from CCN family to predict the prognosis of patients with GC. Further comprehensive analysis indicated that WISP1 might be a potential prognostic biomarker and drug therapeutic target for GC. The molecular mechanism regulating the malignant biological behaviour of tumour cells deserves further investigation. In addition, it is reasonable to develop novel targeted strategies against WISP1 for GC.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to the contributors of data to GEO, DAVID 6.8, Metascape, cBioPortal, GEPIA, OMCOMINE, HPA, the Kaplan–Meier plotter, and TRRUST.
Author contributions
Chen HT, Liu YJ and Yang MQ conceived and designed the experiments; Chen HT, Zhang XM, Zhang Z and Yang SR analyzed the data; Zhang Z, Li GQ and Li X contributed to the data curation; Chen HT wrote-original draft preparation; Liu YJ and Yang MQ participated in the writing-review and editing.
Funding
This work was supported by Shenzhen Science and Technology Innovation Commission Project (JCYJ20220531093612027) and Shenzhen City San Ming project (SZSM202211036).
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Huanting Chen, Xiaomin Zhang and Zhe Zhang contributed equally to this article.
Contributor Information
Yajie Liu, Email: liuyajie_pkuszh@163.com.
Mengqi Yang, Email: yangmq_sysu@163.com.
References
- 1.Micucci M, Xiang BZ, Ting CM, Kwan HY, Mari M, Retini M, Burattini S, Osman R, Okeke UJ, Abdullah FO, et al. Matching traditional Chinese medicine and western medicine-based research: advanced nutraceutical development for proactive gastric cancer prevention. World J Gastrointest Oncol. 2024;16(9):3798–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seo SB, Lim J, Kim K, Maeng I, Rho HW, Son HY, Kim E, Jang E, Kang T, Jung J, et al. Nucleic acid amplification circuit-based hydrogel (NACH) assay for one-step detection of metastatic gastric cancer-derived exosomal miRNA. Adv Sci. 2024;2024:e2407621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kosuge M, Ito J, Hamada M. Landscape of evolutionary arms races between transposable elements and KRAB-ZFP family. Sci Rep. 2024;14(1):23358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ascencao K, Lheimeur B, Szabo C. Regulation of CyR61 expression and release by 3-mercaptopyruvate sulfurtransferase in colon cancer cells. Redox Biol. 2022;56:102466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng TY, Wu MS, Hua KT, Kuo ML, Lin MT. Cyr61/CTGF/Nov family proteins in gastric carcinogenesis. World J Gastroenterol. 2014;20(7):1694–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y, Li W, Yang Y, Li Y, Zhao Y. WISP1 indicates poor prognosis and regulates cell proliferation and apoptosis in gastric cancer via targeting AKT/mTOR signaling pathway. Am J Transl Res. 2020;12(11):7297–311. [PMC free article] [PubMed] [Google Scholar]
- 7.An M, Mehta A, Min BH, Heo YJ, Wright SJ, Parikh M, Bi L, Lee H, Kim TJ, Lee SY, et al. Early immune remodeling steers clinical response to first-line chemoimmunotherapy in advanced gastric cancer. Cancer Discov. 2024;14(5):766–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Liu X, Wang X, Lu J, Tian Y, Liu Q, Xue J. Matricellular proteins: potential biomarkers in head and neck cancer. J Cell Commun Signal. 2024;18(2):e12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis S, Meltzer PS. GEOquery: a bridge between the Gene expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–7. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo J, Ni Z, Zhang M, Kong X, Hoffman LL, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39(4):1197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45(3):461–9. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29(1):77–83. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14(8):3208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–64. [DOI] [PubMed] [Google Scholar]
- 16.Zang X, Jiang J, Gu J, Chen Y, Wang M, Zhang Y, Fu M, Shi H, Cai H, Qian H, et al. Circular RNA EIF4G3 suppresses gastric cancer progression through inhibition of beta-catenin by promoting delta-catenin ubiquitin degradation and upregulating SIK1. Mol Cancer. 2022;21(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y, Luo K, Liu M, Chen L, Gao X, Zhang L, Li X, Zhang H. Comprehensive analysis reveals that P4HA3 is a prognostic and diagnostic gastric cancer biomarker that can predict immunotherapy efficacy. Sci Rep. 2024;14(1):22959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perbal A, Perbal B. The CCN family of proteins: a 25th anniversary picture. J cell Commun Signal. 2016;10(3):177–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takigawa M. The CCN proteins: an overview. Methods Mol Biol. 2017;1489:1–8. [DOI] [PubMed] [Google Scholar]
- 20.Gong Z, Mu Y, Chen J, Chu H, Lian P, Wang C, Wang J, Jiang L. Expression and significance of cyclophilin J in primary gastric adenocarcinoma. Anticancer Res. 2017;37(8):4475–81. [DOI] [PubMed] [Google Scholar]
- 21.Fang F, Zhao WY, Li RK, Yang XM, Li J, Ao JP, Jiang SH, Kong FZ, Tu L, Zhuang C, et al. Silencing of WISP3 suppresses gastric cancer cell proliferation and metastasis and inhibits Wnt/beta-catenin signaling. Int J Clin Exp Pathol. 2014;7(10):6447–61. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang LH, Wang Y, Fan QQ, Liu YK, Li LH, Qi XW, Mao Y, Hua D. Up-regulated Wnt1-inducible signaling pathway protein 1 correlates with poor prognosis and drug resistance by reducing DNA repair in gastric cancer. World J Gastroenterol. 2019;25(38):5814–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirza Z, Ansari MS, Iqbal MS, Ahmad N, Alganmi N, Banjar H, Al-Qahtani MH, Karim S. Identification of novel diagnostic and prognostic gene signature biomarkers for breast cancer using artificial intelligence and machine learning assisted transcriptomics analysis. Cancers. 2023;15(12):3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soon LL, Yie TA, Shvarts A, Levine AJ, Su F, Tchou-Wong KM. Overexpression of WISP-1 down-regulated motility and invasion of lung cancer cells through inhibition of rac activation. J Biol Chem. 2003;278(13):11465–70. [DOI] [PubMed] [Google Scholar]
- 25.Desnoyers L, Arnott D, Pennica D. WISP-1 binds to decorin and biglycan. J Biol Chem. 2001;276(50):47599–607. [DOI] [PubMed] [Google Scholar]
- 26.Nagai Y, Watanabe M, Ishikawa S, Karashima R, Kurashige J, Iwagami S, Iwatsuki M, Baba Y, Imamura Y, Hayashi N, et al. Clinical significance of wnt-induced secreted protein-1 (WISP-1/CCN4) in esophageal squamous cell carcinoma. Anticancer Res. 2011;31(3):991–7. [PubMed] [Google Scholar]
- 27.Tang Q, Jiang X, Li H, Lin Z, Zhou X, Luo X, Liu L, Chen G. Expression and prognostic value of WISP-1 in patients with endometrial endometrioid adenocarcinoma. J Obstet Gynaecol Res. 2011;37(6):606–12. [DOI] [PubMed] [Google Scholar]
- 28.Tsui KH, Liu CL, Yeh HL, Liu MK, Li CH, Chen WH, Jiang KC, Li HR, Thuy Dung PV, Hsiao M, et al. WNT1-inducible signaling pathway protein 1 activation through C-X-C motif chemokine ligand 5/C-X-C chemokine receptor type 2/leukemia inhibitory factor/leukemia inhibitory factor receptor signaling promotes immunosuppression and neuroendocrine differentiation in prostate cancer. iScience. 2024;27(8):110562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Song Y, Ye M, Hu X, Wang ZP, Zhu X. The emerging role of WISP proteins in tumorigenesis and cancer therapy. J Transl Med. 2019;17(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka S, Sugimachi K, Saeki H, Kinoshita J, Ohga T, Shimada M, Maehara Y, Sugimachi K. A novel variant of WISP1 lacking a Von Willebrand type C module overexpressed in scirrhous gastric carcinoma. Oncogene. 2001;20(39):5525–32. [DOI] [PubMed] [Google Scholar]
- 31.Jia S, Qu T, Feng M, Ji K, Li Z, Jiang W, Ji J. Association of Wnt1-inducible signaling pathway protein-1 with the proliferation, migration and invasion in gastric cancer cells. Tumour Biol. 2017;39(6):1010428317699755. [DOI] [PubMed] [Google Scholar]
- 32.Jun JI, Lau LF. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat Rev Drug Discov. 2011;10(12):945–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie D, Yin D, Wang HJ, Liu GT, Elashoff R, Black K, Koeffler HP. Levels of expression of CYR61 and CTGF are prognostic for tumor progression and survival of individuals with gliomas. Clin Cancer Res. 2004;10(6):2072–81. [DOI] [PubMed] [Google Scholar]
- 34.Luan Y, Zhang H, Ma K, Liu Y, Lu H, Chen X, Liu Y, Zhang Z. CCN3/NOV regulates proliferation and neuronal differentiation in mouse hippocampal neural stem cells via the activation of the Notch/PTEN/AKT pathway. Int J Mol Sci. 2023;24(12):10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.