Abstract
Coronavirus relevancy for human health has surged over the past 20 years as they have a propensity for spillover into humans from animal reservoirs resulting in pandemics such as COVID-19. The diversity within the Coronavirinae subfamily and high infection frequency in animal species worldwide creates a looming threat that calls for research across all genera within the Coronavirinae subfamily. We sought to contribute to the limited structural knowledge within the Gammacoronavirus genera and determined the structure of the viral core replication–transcription complex (RTC) from infectious bronchitis virus using single-particle cryo-electron microscopy. Comparison between our infectious bronchitis virus structure with published RTC structures from other Coronavirinae genera reveals structural differences across genera. Using in vitro biochemical assays, we characterized these differences and revealed their differing involvement in core RTC formation across different genera. Our findings highlight the value of cross-genera Coronavirinae studies, as they show genera-specific features in coronavirus genome replication. A broader knowledge of coronavirus replication will better prepare us for future coronavirus spillovers.
Keywords: viral polymerase, protein complex, RNA virus, structural biology, viral transcription
Coronaviruses belong to the Nidovirales order of positive-sense RNA viruses. Within Nidovirales, this diverse subfamily of viruses is divided into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (1). In 1931, the Gammacoronavirus infectious bronchitis virus (IBV) was the first coronavirus to be discovered (2). Subsequently, additional members of the subfamily have been characterized, including the human seasonal betacoronaviruses HKU1 and OC43 and alphacoronaviruses NL63 and 229E (1). Since 2002, three animal betacoronaviruses have crossed into humans and caused disease outbreaks: SARS-CoV in 2002, MERS-CoV in 2012, and SARS-CoV-2 in 2019 (3, 4, 5). The emergence of SARS-CoV-2, the causative agent of COVID-19, led to a global pandemic that has resulted in large losses of life and significant burdens on both healthcare and the economy. In 2018, a recombinant canine-feline Alphacoronavirus, CCoV-HuPn-2018, was isolated from human patients hospitalized with pneumonia (6). Although CCoV-HuPn-2018 is currently incapable of efficiently infecting humans, it is poised as a preemergent human pathogen (7, 8). The Gammacoronavirus and Deltacoronavirus genera contain numerous avian coronaviruses, and while no avian-to-human spillovers from these genera have been reported, the identification of recent independent infection of Haitian children with porcine deltacoroanvirus along with the consistent threat posed by avian viruses from the close contact between human and avian populations highlights the need for better characterization and monitoring of these animal coronaviruses (9).
The Gammacoronavirus genus is subdivided into three subgenera: Igacovirus, Brangacovirus, and Cegacovirus (10). Igacovirus is currently recognized to have three species including galli, pulli, and anatis (formerly duck coronavirus 2714). Isolates of IBV fall into both galli and pulli species, while anatis is typically found in wild birds (11). Infection of chickens with IBV typically initiates in the respiratory tract, and some strains can additionally infect the reproductive tract and kidneys. Infection of the reproductive tract can lead to a decrease in egg quality, while infection of the kidneys may lead to nephritis and death (12, 13). Respiratory tract infection may also weaken the immune system permitting secondary bacterial pneumonia (14). Having a high prevalence in most parts of the world, IBV has been an immense economic burden on the poultry industry. Despite extensive vaccination campaigns against IBV, the large genetic diversity of the virus arising from mutation and recombination creates difficulties in providing broad protection from IBV infection (15).
Coronavirus genomes encode a large number of structural proteins and nonstructural proteins (nsps) used to replicate viral genomes, assemble new virions, and interact with the infected host cell (16). The 5′ two-thirds of the viral genome encodes the viral nsps responsible for viral RNA replication and transcription. These nsps are the products of polyprotein cleavage and are encoded within two open reading frames: ORF1a and ORF1b, with ORF1b accessed by −1 programmed ribosomal frameshifting at the end of ORF1a to produce either the pp1a or pp1ab polyproteins (17). Across coronavirus genera, ORF1a/b have similar organizations and cleavage products to assemble the necessary machinery for viral RNA synthesis. Within the functionally conserved suite of nsps, nsp12 encodes the RNA-dependent RNA polymerase as well as containing a second active site for a nucleotidyltransferase (18, 19). For viruses of Alphacoronavirus and Betacoronavirus, nsp12 requires the replication factors nsp7 and nsp8 for robust RNA synthesis activity (20, 21). These three nsps form the core polymerase complex that can perform processive RNA synthesis in vitro (1). The majority of the work to characterize coronavirus polymerases has focused on betacoronaviruses leaving members of other genera relatively understudied (22). A prior study of IBV polymerase has indicated an interaction of nsp12 with nsp8 though without a demonstration of polymerase activity or molecular descriptors (23).
Structural studies of coronavirus polymerase complexes have largely focused on complexes from SARS-CoV-2 with limited polymerase structures from SARS-CoV and the Alphacoronavirus porcine epidemic diarrhea virus (PEDV) (21, 22, 24, 25). These structures have revealed similar nsp12 active site architectures and requirements for nsp7 and nsp8 replication factors. Expanding beyond this dataset dominated by structures of Betacoronavirus polymerases affords the opportunity to examine unique features of coronavirus polymerases across the subfamily while also identifying conserved mechanisms between these divergent viruses. Here, we use cryo-electron microscopy (cryo-EM) to solve the structure of the IBV polymerase complex, the first such polymerase structure from the Gammacoronavirus genus. We identified a genus-specific nsp12 loop that in PEDV and IBV contacts a subunit of nsp8. Subsequent biochemical analyses demonstrate the importance of this interaction and point to the potential of genus-specific polymerase assembly pathways. Continued investigation of coronavirus polymerases across this diverse group of viruses will aid in the development of broad-spectrum antiviral therapeutics and inform mechanisms for viral polymerase function.
Results
IBV shares replication factor requirements for RNA binding and synthesis with Alphacoronavirus and Betacoronavirus
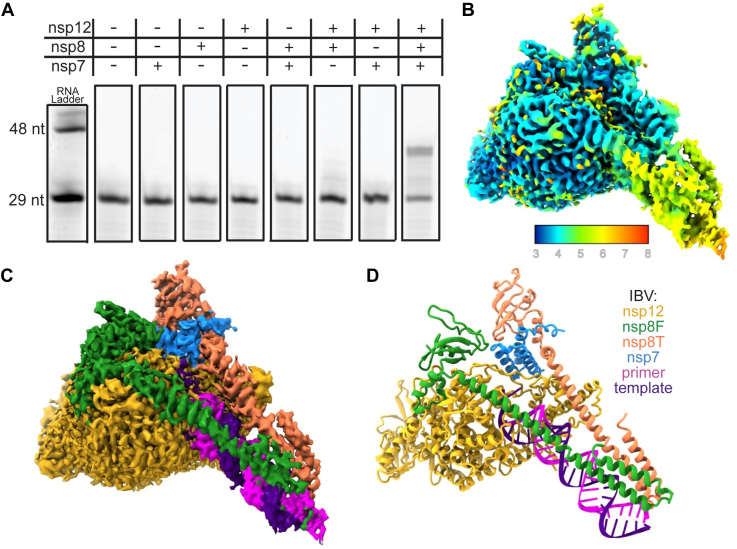
Recombinantly expressed and purified IBV nsp7, nsp8, and nsp12 (Fig. S1) combined with a fluorescently labeled RNA primer/template pair altered the mobility of the RNA on native-PAGE in a manner demonstrating that both nsp7 and nsp8 are required for RNA binding to nsp12 (Fig. S2). Adding nucleotides to this complex similarly demonstrated the IBV nsp12 requirement for both nsp7 and nsp8 for promoting RNA synthesis activity by primer extension assay (Figs. 1A and S3). The results of these assays indicate similar requirements for nsp7 and nsp8 for RNA binding and polymerase activity across the coronavirus subfamily (20, 21, 25).
Figure 1.
Structure of an active IBV polymerase complex.A, extension of a short (29 nt) RNA primer to the length of the template RNA (38 nt) in the presence of the IBV polymerase complex. B and C, cryo-EM reconstruction of the IBV polymerase complex colored by local resolution (B) or chain (C). D, atomic model of the IBV polymerase complex built using the cryo-EM reconstruction. IBV, infectious bronchitis virus; nsp, nonstructural protein.
Structure of the IBV polymerase complex
Single-particle cryo-EM was used to solve the structure of the IBV nsp7-nsp8-nsp12–RNA complex. Our cryo-EM reconstruction has a resolution of 3.5 Å (Figs. 1, B and C, S4, and S5, Table S1). Clearly visible in the map are densities for all components, including nsp12, nsp7, two copies of nsp8, and the RNA substrate with a complex stoichiometry of 1:2:1 for nsp7:nsp8:nsp12 which is consistent with other coronavirus polymerase structures (Fig. 1D) (21, 24, 25). Similar to Alphacoronavirus and Betacoronavirus polymerases, one protomer of nsp8 binds the nsp12 fingers domain (nsp8F) while a second protomer binds to nsp12 as a nsp7-nsp8 heterodimer adjacent to the nsp12 thumb domain (nsp8T). The identification of nsp8F is congruent with a previous biochemical study of IBV nsp12 demonstrating an interaction of nsp8 with nsp12 residues 1 to 400 which encompasses nearly the entirety of our observed nsp8F binding site on nsp12 (23). As previously observed in coronavirus polymerase structures bound to duplex RNAs, the N-terminal extensions of each nsp8 form long helices to contact upstream double-stranded RNA extending from the polymerase active site (21, 25). The IBV polymerase and nucleotidyltransferase active sites are well resolved and well conserved both in sequence and structure among SARS-CoV, SARS-CoV-2, and PEDV polymerases (SARS-CoV: 6NUR, SARS-CoV-2: 7KRP, PEDV: 8URB), suggesting the broad applicability of antiviral drugs targeting these sites (26, 27).
Structurally observed insertions and deletions in the Gammacoronavirus polymerase complex
Despite the high sequence and structural homology of coronavirus polymerases, we identified insertions and deletions in nsp8 and nsp12 that result in unique conformations within the IBV polymerase complex structure. Many of these regions are distal to known active sites and protein–protein interfaces, and their influence on the viral polymerase remains a topic for future study.
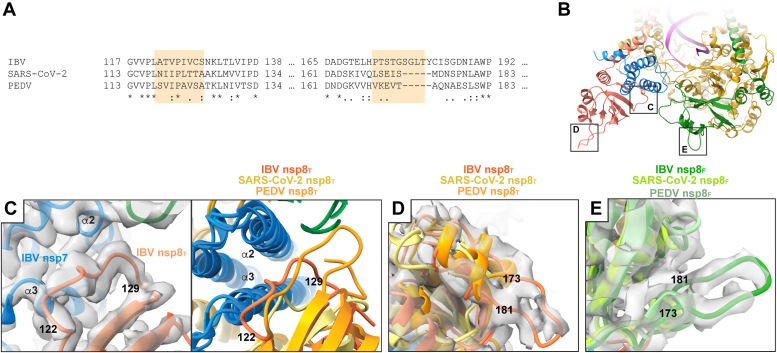
IBV nsp8 loop 173 to 181 contains an insertion not observed in other genera of coronaviruses (Figs. 2A and S6). This loop sequence is well conserved among Igacovirus nsp8s while Brangocovirus nsp8s contain an additional three amino acid insertion and the Cegacovirus nsp8s have a nine amino acid deletion (Fig. S7). Betacoronavirus nsp8s possess shorter loops in this nsp8 region with Embecovirus members, such as murine hepatitis virus, having nsp8 loops nine amino acids shorter than IBV. Similarly, Alphacoronavirus and Deltacoronavirus nsp8s have loops five and 13 amino acids shorter than IBV, respectively. This IBV nsp8 region lacks secondary structure and forms an extended loop from the nsp8 C-terminal head domains, while in SARS-CoV, SARS-CoV-2, and PEDV, this loop forms a pair of short helices (Fig. 2). This loop is clearly visible in the IBV reconstructed density in both nsp8F and nsp8T though the density is weaker at the distal end of the loop particularly for nsp8T. Additional examination of IBV nsp8T reveals an 18° rotation in the conformation of the nsp8 head domain relative to nsp7 when compared to corresponding domains from SARS-CoV-2 (24). This rotated nsp8T head domain is similar to the orientation of nsp8T for PEDV (20). IBV nsp8 122 to 129 is highly conserved among Gammacoronavirus though poorly conserved across Coronavirinae and in nsp8T makes contacts with nsp7. IBV and PEDV nsp8T regions 122 to 129 (IBV) make extensive contacts to nsp7 α2, while in the SARS-CoV and SARS-CoV-2 equivalent nsp8T regions, the interactions are more restricted to nsp7 α3 (Figs. 2, S6 and S7) (21, 24, 25).
Figure 2.
Sequence divergent features in coronavirus nsp8.A, a multiple sequence alignment of IBV, SARS-CoV-2, and PEDV nsp8 highlights sequence and structurally divergent regions. Highlighted regions correspond to the numbered IBV nsp8 loops in C–E. B, an overview of the IBV replication-transcription complex indicates structurally divergent nsp8 regions shown in greater detail in C–E. C, a focused view of IBV nsp8T 122 to 129 shows conformational heterogeneity across genera [PEDV (8URB) and SARS-CoV-2 (6XEZ)]. Reconstructed IBV RTC density is shown to support the observed conformation. D, a focused view of the nsp8T 173 to 181 loop shows an extended loop conformation (IBV RTC reconstructed density) with superimposed PEDV and SARS-CoV-2 nsp8T adopting alternate conformations. E, a focused view of nsp8F 173 to 191 showing an extended loop conformation (IBV reconstructed density) with superposed PEDV and SARS-CoV-2 nsp8F adopting alternate conformations. IBV, infectious bronchitis virus; nsp, nonstructural protein; PEDV, porcine epidemic diarrhea virus; RTC, replication–transcription complex.
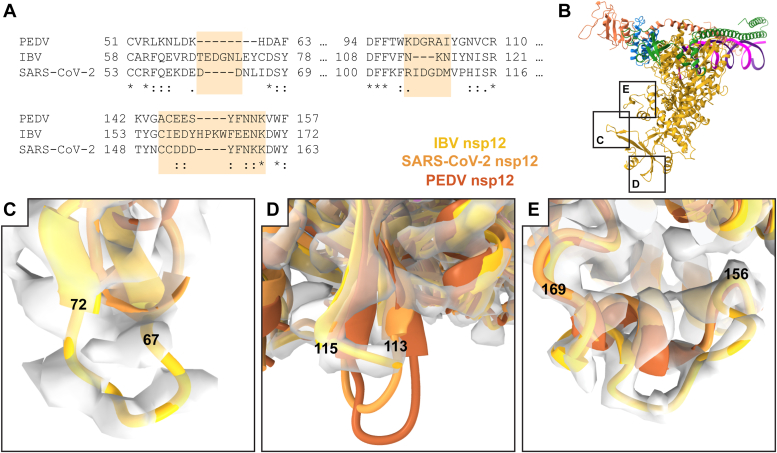
In addition to nsp8, there are several insertions and deletions in IBV nsp12 particularly in the N-terminal nucleotidyltransferase domain (19). There is a large insertion in IBV nsp12 loop 67 to 72 when compared to Alphacoronavirus and Betacoronavirus nsp12s (Fig. S8). This loop sequence is well conserved in Gammacoronavirus galli and pulli with some length polymorphisms in other Gammacoronavirus nsp12s (Fig. S9). In Alphacoronavirus and Betacoronavirus nsp12, this loop is four and eight amino acids shorter, respectively, and one to two amino acids longer in Deltacoronavirus. This loop is positioned on the opposite side of the nucleotidyltransferase domain from the enzyme active site and is distal to known protein-binding sites for nsp7, nsp8, nsp9, and nsp13 (Fig. 3) (24, 28, 29).
Figure 3.
Sequence divergent features in coronavirus nsp12.A, a multiple sequence alignment of IBV, SARS-CoV-2, and PEDV nsp12 highlights sequence and structurally divergent regions in the N-terminal region of nsp12. B, an overview of the IBV replication-transcription complex indicates the locations of structurally divergent nsp12 regions shown in greater detail in C–E. C, an insertion in IBV nsp12 67 to 72 presents as an extended loop. Reconstructed IBV RTC density is shown with superimposed PEDV (8URB) and SARS-CoV-2 (6XEZ) structures. D, IBV nsp12 has a shortened loop in the 113 to 115 region (IBV reconstructed density) as compared to SARS-CoV-2 and PEDV nsp12. E, an insertion in IBV nsp12 156 to 169 (IBV reconstructed density) presents as an altered conformation compared to SARS-CoV-2 and PEDV. IBV, infectious bronchitis virus; nsp, nonstructural protein; PEDV, porcine epidemic diarrhea virus; RTC, replication–transcription complex.
There is a shortened loop in IBV nsp12 113 to 115 that is conserved in all Gammacoronavirus nsp12s except for Cegacovirus nsp12s which are one amino acid shorter, similar to Deltacoronavirus nsp12s (Fig. S8 and S9). In contrast, Alphacoronavirus and Betacoronavirus nsp12s are four amino acids longer in this region (Fig. S8). This nsp12 loop lies within the nucleotidyltransferase domain but again is distant from the enzyme active site and known protein-binding sites (Fig. 3).
IBV nsp12 contains a four amino acid insertion in loop 156 to 169 compared to Alphacoronavirus, Betacoronavirus, and Deltacoronavirus nsp12s (Fig. S8). In Gammacoronavirus, the nsp12 loop 156 to 169 (IBV) is conserved across G. galli and pulli species, while Gammacoronavirus anatis and Brangacovirus contain an additional 10 amino acid insertion with more divergent sequences (Fig. S9). Cegacovirus nsp12s have shorter loops of similar length to the other coronavirus genera (Figs. S8 and S9). Structurally, this region of nsp12 appears to be conserved in both Alphacoronavirus and Betacoronavirus nsp12s which is unsurprising given nsp12s′ moderate sequence conservation between these two genera. The insertion in IBV nsp12 loop 156 to 169 results in a shortened helical region and more extended loop region that extends outward from the polymerase (Fig. 3). While loop sequence insertions at this position have so far only been noted for coronaviruses infecting avian species, the lack of insertions here among avian viruses of Deltacoronavirus and the poor representation of Cegacovirus sequences that infect mammals in databases warns against identifying this loop as a potential host species determinant.
Sequence and structural comparison of the IBV nsp12 loop 264 to 278
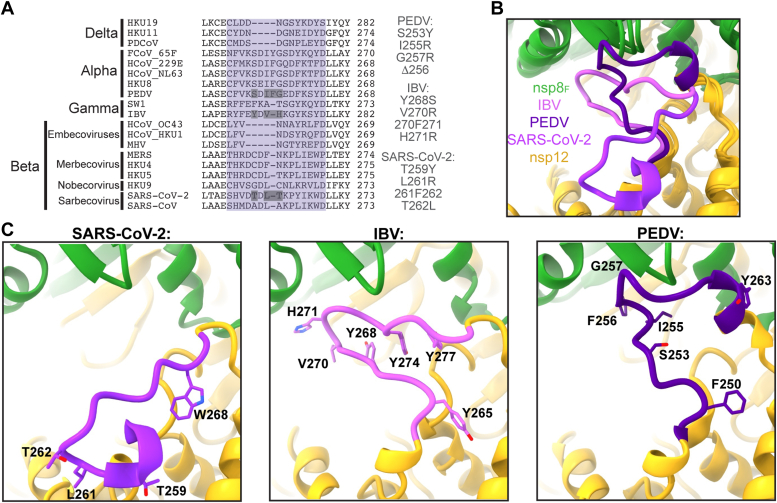
In the IBV nsp12 interface domain, amino acids 264 to 278 form a large loop that is in a dramatically different conformation when compared to the homologous nsp12 region in both Alphacoronavirus and Betacoronavirus (Fig. 4B) (21, 25). This loop extends from the interface domain to contact the head domain of nsp8F. We define this nsp12 loop as being flanked by well-conserved residues L279 and L280 that form the hydrophobic pocket for nsp8F region 103 to 129 and nsp12 E263 that forms a well-conserved salt bridge with K294 (IBV nsp12 numbering). The corresponding region in PEDV nsp12 (249–264) also extends to contact the nsp8F head domain but adopts a different conformation to use distinct regions on both nsp12 and nsp8F to form the protein–protein interaction (21). In contrast, the corresponding loop in Betacoronavirus SARS-CoV and SARS-CoV-2 nsp12s extends away from the core complex not forming interactions with known replication factors (24, 25).
Figure 4.
Altered sequence and structure of nsp12–nsp8Finteractions.A, multiple sequence alignment of nsp12 residues 260 to 282 (IBV) across the coronavirus subfamily. The loop region of interest is highlighted in purple, and residues that were mutated in this study are listed to the right and have gray boxes around them. B, superimposition of IBV, PEDV (8URB), and SARS-CoV-2 (6YYT) highlighting the altered conformations of the nsp12 loop (IBV residues 264–278). C, individual snapshots of the nsp12 loop (IBV residues 264–278) from IBV (left), PEDV (center), and SARS-CoV-2 (right). IBV, infectious bronchitis virus; nsp, nonstructural protein; PEDV, porcine epidemic diarrhea virus; RTC, replication–transcription complex.
Examining sequence alignments for this region of nsp12, the Betacoronavirus subgenera Nobecovirus, Sarbecovirus, and Merbecovirus have loop lengths similar to Gammacoronavirus nsp12 while Embecovirus nsp12s have loops that are four amino acids shorter (Fig. 4A). While there are no structural data to provide insight into this nsp12 region for Deltacoronavirus polymerases, sequence alignment indicates a shortening of this loop by three amino acids relative to IBV. Hence, we hypothesize that the nsp12 loop does not contact nsp8F in Deltacoronavirus nsp12s. In contrast Alphacoronavirus nsp12s have a one amino insertion, adding a Phe at position 256 (PEDV numbering). PEDV nsp12 F256 lies at the apex of the nsp12 loop and packs into the hydrophobic surface between nsp12 and nsp8F. To test the role of F256 in PEDV complex assembly, we produced a recombinant PEDV nsp12 with F256 deleted (PEDV nsp12 ΔF256) which resulted in nearly a complete loss of polymerase activity using our aforementioned in vitro RNA primer extension assay (Fig. 5). Complementary to this PEDV nsp12 deletion, we inserted a Phe into the corresponding nsp12 positions of IBV (270F271) and SARS-CoV-2 (261F262). Neither of these insertions diminished the ability of the mutant polymerases to bind RNA or extend primers (Figs. 4, 5, S10 and S11). In IBV nsp12, while this loop contacts nsp8F, the 270F271 insertion would be expected to be surface exposed owing to different use of this nsp12 region to contact the nsp8F head domain.
Figure 5.
Mutant nsp12 primer extensions.A, point mutations and insertions of nsp12 from SARS-CoV-2 (left), IBV (center), and PEDV (right) were tested for their ability to form the core-RTC and extend RNA in vitro. Uncropped gels are presented in Fig. S11. B, for each coronavirus, primer extension results are presented as % activity of the polymerase complex with respect to wildtype nsp12. Error bars indicate standard deviation of the technical replicates (3–12). Using an unpaired t test comparing each mutant to wildtype, “∗” denotes p < 0.05, “∗∗” denotes p < 0.001, and “∗∗∗” denotes p < 0.0001. Primer extensions were carried out in 10 mM Tris-Cl (pH8), 2 mM MgCl2, 1 mM DTT, and 10 mM NaCl (SARS-CoV-2 and PEDV) or 100 mM K-Glu (IBV). Reagents were combined at concentrations of nsp12—500 nM, nsp7—1.5 μM, nsp8—1.5 μM, and RNA—250 nM. After the addition of NTPs (500 μM), extensions ran for 1 min at room temperature (SARS-CoV-2) or 30 °C (PEDV), or 30 min at 30 °C (IBV). C, point mutations and insertions of nsp12 from SARS-CoV-2 (left), IBV (center), and PEDV (right) were further tested for their ability to form the core-RTC in vitro using an electrophoretic mobility shift assay. Complexes were assembled in buffer containing 10 mM Tris-Cl (pH 8), 2 mM MgCl2, 1 mM DTT, and 10 mM NaCl (SARS-CoV-2 and PEDV) or 10 mM potassium glutamate (IBV). Proteins were combined at nsp12—1 μM, nsp7—3 μM, and nsp8—3 μM with RNA at 250 nM. Complexes were assembled at room temperature before being analyzed via native-PAGE. Uncropped raw images of gels are presented in Fig. S10. EMSA, electrophoretic mobility shift assay; IBV, infectious bronchitis virus; nsp, nonstructural protein; PEDV, porcine epidemic diarrhea virus; RTC, replication–transcription complex.
Examining the conformation of the IBV nsp12 264 to 278 loop, Tyr residues at 268, 274, and 277 undergo aromatic stacking and likely stabilize this unique loop conformation. To test the importance of the tyrosine stacking in maintaining the IBV nsp12 264 to 278 loop conformation, we created an IBV nsp12 with a Y268S mutation. IBV nsp12 Y268S showed a more than 50% reduction in polymerase activity as well as a strong defect in RNA binding (Figs. 4, 5, S10 and S11). These defects in polymerase activities likely are caused by a failure to properly assemble IBV nsp8F on nsp12 Y268S and point to the importance of this IBV nsp12 loop region for polymerase complex binding to RNA. Complementary mutations in PEDV nsp12 (S253Y) or SARS-CoV-2 nsp12 (T259Y) did not have major effects on viral polymerase activities supporting the observed distinct conformations and interactions of these nsp12 loop regions (Figs. 4, 5, S10 and S11).
The presence of an aromatic residue at nsp12 277 (IBV numbering, SARS-CoV-2 W268, PEDV Y263) is well conserved across the coronavirus subfamily; however, these aromatic residues are placed in very different contexts within this loop region. In PEDV, nsp12-Y263 is surface exposed, while in IBV, nsp12-Y277 participates in the tyrosine stacking interaction that stabilizes this unique loop conformation. In contrast, SARS-CoV and SARS-CoV-2 nsp12-W268 is oriented into a hydrophobic pocket on nsp12 which may drive the diversion of this loop into the observed outward directed conformation. Structural comparison of the SARS-CoV-2 nsp12-W268 hydrophobic pocket shows that the homologous pocket is occupied by IBV nsp12-Y265 or PEDV-F250 (Fig. 4). The functional constraint of needing to insert an aromatic residue into this hydrophobic pocket may be driving loop conformational differences and presentation of this nsp12 loop to nsp8F in IBV and PEDV polymerase complexes.
To further examine specific interactions at the IBV nsp12 264 to 278 and PEDV nsp12 249 to 264 loop apexes with their respective nsp8F head domains, we generated nsp12 mutants for key residues in the protein interfaces and complementary mutations in other coronavirus polymerases (Figs. 4, 5, S10 and S11). In addition to PEDV nsp12 F256, I255 contributes to the buried hydrophobic surface between nsp12 and the nsp8F head domain. A PEDV nsp12-I255R mutation reduced primer extension activity by 50% and prevented strong RNA binding to the PEDV polymerase complex. Homologous mutations to IBV nsp12 (V271R) and SARS-CoV-2 nsp12 (L261R) did not have large effects on either polymerase complex. For IBV nsp12, H271 resides at the apex of the 264 to 278 loop to contact the nsp8F head domain. IBV nsp12-H271R had a 50% reduction in polymerase activity and a significant loss in RNA-binding activity. Mutations to homologous positions in PEDV nsp12 (G257R) or SARS-CoV-2 nsp12 (T262L) had no effect on either polymerase primer extension or RNA binding activity. These targeted mutations highlight the distinct interactions of each genera’s nsp12 loop with nsp8F and that disrupting these interactions has genus-specific negative impacts on polymerase activity.
Discussion
Here, we have presented the first structure of a Gammacoronavirus polymerase complex showing the IBV RNA polymerase bound to its essential replication factors and RNA. Structural comparisons highlight a loop in IBV nsp12 (residues 264–278) that is in an alternate conformation than previous polymerase complex structures from PEDV and SARS-CoV-2. Mutagenesis of key residues in this protein region among these three polymerase complexes were a detriment to IBV and PEDV polymerase RNA-binding and primer extension activities. The inability of these polymerases to bind RNA is likely a result of defects in the nsp12s′ ability to assemble properly with replication factor nsp8F. It has been previously shown that replication factors nsp7 and nsp8 are essential for nsp12 RNA binding and that disruptions to the nsp8F head domain–nsp12 interaction resulted in polymerases incapable of extending primers or binding RNA despite not fully blocking nsp8F subunit binding to the complex (20, 21). We identify this nsp12 loop as a genus-specific structural feature that is functionally important for the proper assembly of Alphacoronavirus and Gammacoronavirus polymerases. Work on SARS-CoV-2 polymerase complexes has identified the binding of nsp8F as a rate-limiting step in polymerase assembly (30). The observed altered interactions of IBV and PEDV nsp12 with nsp8F may indicate the existence of alternate polymerase assembly pathways across diverse viruses. Our observed structural differences among viral nsps with functional consequences for polymerase activity highlight the need to consider alternate assembly and functional pathways across diverse coronavirus genera more broadly. In addition, the altered interactions and potential assembly pathways could provide a barrier for cross-genera RTC assemblies during co-infection. Interestingly, the IBV nsp12 264 to 278 and PEDV nsp12 249 to 264 loops neighbor SARS-CoV-2 nsp12 P323. During the early months of the COVID-19 pandemic, variant strains carrying a nsp12 P323L mutation along with a D614G mutation in the viral spike rapidly rose to prominence (31). The impacts of the SARS-CoV-2 nsp12 P323L mutation on polymerase activity remain unclear, but the spatial proximity of this mutation to the observed altered loop conformations in IBV and PEDV polymerases creates the possibility that this region of the nsp12 polymerase has a role in modulating the activity and assembly of the coronavirus polymerase complex across viral evolution.
Experimental procedures
DNA constructs
All IBV, PEDV, and SARS-CoV-2 nsp gene sequences were codon optimized (Genscript). Sequences for G. galli IBV/M41/Y28 proteins originate from the GenBank sequence QWC71293.1. SARS-CoV-2 protein sequences originate from the GenBank sequence UHD90671.1. PEDV protein sequences originate from the GenBank sequence AKJ21892.1. IBV nsp7 and PEDV nsp7 were cloned into the pET46 vector with C-terminal tobacco etch virus (TEV) protease cleavage site and hexahistidine tag. IBV nsp8 was cloned into pET45 vector with an N-terminal hexahistidine tag and TEV protease cleavage site. IBV nsp12, PEDV nsp8, and SARS-CoV-2 nsp7 and nsp8 were cloned into pET46 vectors with N-terminal hexahistidine tags and TEV protease cleavage sites. SARS-CoV-2 and PEDV nsp12 were cloned into pFastBac vectors with C-terminal TEV cleavage site and Strep II tags. Mutant nsp12 vectors were made by performing site-directed mutagenesis on the wildtype nsp12 vectors. The sequences of all open reading frames in plasmids were confirmed using Sanger sequencing.
Protein expression
Nsp7 and nsp8 were expressed in Rosetta 2pLysS Escherichia coli (E. coli) cells (Novagen). Cultures were grown at 37 °C until they reached an A600 of 0.6 to 0.8 where they were induced with isopropyl β-D-1-thiogalactopyranoside at a final concentration of 500 μM and incubated overnight at 16 °C. Bacterial cells were pelleted and resuspended in wash buffer [10 mM Tris-Cl, 300 mM sodium chloride, 30 mM imidazole, 2 mM dithiothreitol (DTT), pH 8]. Cells were then lysed using a microfluidizer (Microfluidics), and lysate was cleared using centrifugation and filtration. Lysate supernatant was used to batch bind to Ni-NTA beads (Qiagen) for 30 min before loading onto a gravity column. Beads were washed with wash buffer, and then protein was eluted from beads using elution buffer (10 mM Tris-Cl, 300 mM sodium chloride, 300 mM imidazole, and 2 mM DTT, pH 8). Eluted proteins were buffer exchanged by dialysis (10 mM Tris-Cl, 300 mM sodium chloride, and 2 mM DTT, pH 8) while cleaving off the tag with TEV protease (1% w/w) at 4 °C overnight. Proteins were passed back over a Ni-NTA column, collecting the flowthrough containing the cleaved protein sample. Protein was concentrated, then loaded onto a Superdex 200 10/300 Increase GL column (Cytiva) for size exclusion (25 mM Tris-Cl, 300 mM sodium chloride, and 2 mM DTT, pH 8). Protein peak fractions were pooled and concentrated, then aliquoted and flash-frozen with liquid nitrogen. Proteins were stored at −80 °C until use. See also Fig. S1.
IBV nsp12 was expressed and purified using the same protocol as above but with alternate buffers. Ni-NTA wash buffer contained 25 mM sodium-Hepes, 300 mM sodium chloride, 30 mM imidazole, 1 mM magnesium chloride, and 2 mM DTT, pH 7.5. Ni-NTA elution buffer contained 10 mM sodium-Hepes, 300 mM sodium chloride, 300 mM imidazole, 1 mM magnesium chloride, and 2 mM DTT, pH 7.5. Dialysis buffer contained 10 mM sodium-Hepes, 300 mM sodium chloride, 1 mM magnesium chloride, and 2 mM DTT, pH 7.5. Size-exclusion buffer contained 25 mM sodium-Hepes, 300 mM sodium chloride, 100 μM magnesium chloride, and 2 mM tris(2-carboxyethyl)phosphine, pH 7.5. See also Fig. S1.
PEDV and SARS-CoV-2 nsp12 pFastBac vectors were transformed into DH10Bac E. coli to generate recombinant Bacmid plasmids. Bacmid plasmids were transfected into Sf9 cells to produce baculovirus stocks that were then amplified twice before being used to infect Sf21 cells. After 2 days of incubation at 27 °C, infected Sf21 cells were pelleted, resuspended in wash buffer (25 mM sodium-Hepes, 300 mM sodium chloride, 1 mM magnesium chloride, and 5 mM DTT, pH 7.4) with an added 143 uL of BioLock (IBA Lifesciences) and lysed with a microfluidizer. Lysed cells were cleared via centrifugation and filtration. Lysates were bound to streptactin beads (IBA Lifesciences) in batch for 30 min then loaded onto a gravity column. Beads were washed with wash buffer, and proteins eluted with elution buffer (strep wash buffer with additional 2.5 mM desthiobiotin). Proteins were then concentrated and further purified via SEC on a Superdex 200 10/300 Increase GL column with SEC buffer (25 mM Hepes, 300 mM sodium chloride, 100 μM magnesium chloride, and 2 mM tris(2-carboxyethyl)phosphine, pH 7.4). Protein peak fractions were pooled, concentrated, aliquoted, and then flash-frozen with liquid nitrogen. Aliquoted samples were stored at −80 °C until use. See also Fig. S1.
RNA substrate preparation
RNA primers with 5′ fluorescein tags (6-FAM) were annealed to longer template RNA substrates. Formation of duplex RNAs was carried out in RNA annealing buffer (2.5 mM potassium chloride, 2.5 mM Hepes, and 0.5 mM magnesium chloride, pH 7.4), with a primer:template ratio of 1:1.2. After mixing, samples were heated at 95 °C for 5 min, then slowly cooled until reaching room temperature. Annealed substrates were used immediately or stored at −20 °C.
RNA primer for in vitro assays
5ʹ – CAUUCUCCUAAGAAGCUAUUAAAAUCACA– 3ʹ
RNA template for in vitro assays
5ʹ – AAAAAGGGUUGUGAUUUUAAUAGCUUCUUAGGAGAAUG– 3ʹ
RNA primer for structure determination
5ʹ – CAUUCUCCUAAGAAGCUAUUAAAAUCACAGAUU– 3ʹ
RNA template for structure determination
5ʹ – CAGUGUCAUGGAAAAACAGAAAAAUCUGUGAUUUUAAUAGCUUCUUAGGAGAAUG– 3ʹ
Primer extension
Primer extension assays were carried out in 20 μl volumes with final buffer concentrations of 10 mM Tris-Cl pH 8, 2 mM magnesium chloride, 1 mM DTT, and either 10 mM sodium chloride (PEDV and SARS-CoV-2) or 100 mM potassium glutamate (IBV). Nsp7 and nsp8 replication proteins and nsp12 were combined at final concentrations of 1.5 μM and 500 nM, respectively. After combining, proteins were incubated together at room temperature for 15 min followed by the addition of duplex RNA substrate to 250 nM. After another 15-min incubation at room temperature, 500 μM of each ribonucleotide was added to begin the extension. Reaction conditions were specific to each viral polymerase complex: 1 min at room temperature for SARS-CoV-2, 1 min at 30 °C for PEDV, and 30 min at 30 °C for IBV. Reactions were then quenched by adding two volumes of denaturing RNA gel loading buffer (95% formamide (v/v), 2 mM EDTA, and 0.75 mM bromophenol blue). Quenched reactions were heated at 95 °C for 15 min, then loaded on a denaturing urea-PAGE gel (8 M urea, 15% polyacrylamide) and run in TBE running buffer (89 mM Tris-Cl, 89 mM boric acid, and 2 mM EDTA, pH 8.3). Gels were imaged using a GE Typhoon FLA 9000 scanner, using FAM tag excitation at 470 nm and measuring emission at 530 nm. The bands were analyzed using ImageJ (32).
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays were carried out in 20 μl reaction volumes in buffer conditions of 10 mM Tris-Cl pH 8, 2 mM magnesium chloride, 1 mM DTT, and either 10 mM sodium chloride (PEDV and SARS-CoV-2) or 10 mM potassium glutamate (IBV). Proteins were combined at final concentrations of nsp7 (3 μM), nsp8 (3 μM), and nsp12 (1 μM). Viral proteins were first mixed and allowed to incubate at room temperature for 15 min. RNA substrate was then added (250 nM final conc.), and the reaction incubated for an additional 15 min at room temperature. Finally, 10× nondenaturing gel loading buffer (10 mM Tris-Cl, 1 mM EDTA, 50% (v/v) glycerol, and 0.75 mM bromophenol blue) was added to the reaction. Samples were run on a 4.5% nondenaturing PAGE gel with IBV and SARS-CoV-2 run on TBE native-PAGE gels, while PEDV was run on clear native-PAGE gels (33). Gels were scanned using a Typhoon imager scanning for FAM fluorescence. Bands were quantitated using ImageJ (32).
Specimen preparation for cryoEM
IBV complexes were initially assembled at a total protein concentration of 2 mg/ml in cryoEM freezing buffer (10 mM Tris-Cl pH 8, 100 mM potassium glutamate, 2 mM MgCl2, and 1 mM DTT). Proteins and RNA were mixed at a ratio of 2:3:1:1.2 nsp7:nsp8:nsp12:RNA. To assemble the complexes, proteins were diluted in freezing buffer, then combined and incubated at 25 °C for 15 min before RNA was added and incubated for another 15 min at 25 °C. After assembly, complexes were concentrated to 4 mg/ml total protein using ultrafiltration with a 100 kDa molecular weight cutoff. Samples were stored on ice prior to grid freezing.
Samples were frozen on UltraAuFoil R1.2/1.3 300 mesh grids (Quantifoil) using a Vitrobot Mark IV (ThermoFisher Scientific). Grids were freshly glow discharged using a GloQube Plus (Quorom) for 20 s with a current of 20 mA in an air atmosphere, creating a negative surface charge. Immediately prior to blotting, 0.5 μl of 42 mM 3-([3-cholamidopropyl] dimethylammonio)-2-hydroxy-1-propanesulfonate was added to 3 μl of sample. Three microliters of sample with 3-([3-cholamidopropyl] dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO, 6 mM final) was spotted onto grids before double-sided blotting and vitrification in liquid ethane. Vitrobot chamber conditions were set to 100% humidity and 4 °C.
CryoEM data collection, processing, and model building
EPU (ThermoFisher Scientific) was used for data collection on a Talos Arctica 200 keV transmission electron microscope (ThemoFisher Scientific). Movies were collected using a K3 direct electron detector (Gatan) in CDS mode. A GIF quantum energy filter was used with a slit width of 20 eV. Data were collected with no stage tilt at a magnification of 79000× with a pixel size of 1.064 Å and a defocus range of −0.5 to −2.0 μm with a step size of 0.5 μm. Total dose per movie was 60 e-/Å2.
Data were processed using cryoSPARC v4.3.0 (34). After patch motion correction and CTF estimation, 2,633,255 particles were picked using blob picker and extracted at a box size of 256 pixels. Particles were subjected to multiple rounds of 2D classification before three ab initio models were generated. Particles were classified by heterogeneous refinement using the three ab initio models as initial models. Output maps and classified particle stacks from heterogenous classification were used as inputs for nonuniform refinement. Particles from the class that resembled a polymerase complex were further classified by producing two ab initio models that were then used for heterogeneous refinement. The final reconstruction was produced using nonuniform refinement with 179,183 particles (Figs. S4, S5, Table S1).
To build a starting coordinate model, we used AlphaFold 2 to create the IBV nsp12 RdRP and docked nsp7, nsp8, and duplex RNA from model 6YYT.pdb into our cryoEM reconstruction using ChimeraX (25, 35, 36, 37). Model building and sequence changes were performed in Coot (38). Iterative real-space refinement in Phenix and model building and adjustments using both ISOLDE and Coot were done to generate the final coordinate model (38, 39, 40).
Data availability
The IBV core polymerase electron density map has been deposited in the Electron Microscopy Data Bank (EMDB: 45805), and the complex model has been deposited in the Protein Data Bank (PDB: 9CPO). All other data can be found within the manuscript and supporting information document.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare no conflicts of interest with the contents of this article.
Acknowledgments
Author contribution
R.N.K., T.K.A., and P.J.H. writing–review & editing; R.N.K., T.K.A., and P.J.H. writing–original draft; R.N.K. project administration; R.N.K., T.K.A., and P.J.H. methodology; R.N.K., T.K.A., and P.J.H. investigation; R.N.K. funding acquisition; R.N.K., T.K.A., and P.J.H data curation; R.N.K., T.K.A., and P.J.H conceptualization.
Funding and additional information
This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases AI158463 to R.N.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Craig Cameron
Supporting information
References
- 1.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schalk A., Hawn M. An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 1931;78:413–423. [Google Scholar]
- 3.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M., et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vlasova A.N., Diaz A., Damtie D., Xiu L., Toh T.-H., Lee J.S.-Y., et al. Novel canine coronavirus isolated from a hospitalized patient with pneumonia in east Malaysia. Clin. Infect. Dis. 2022;74:446–454. doi: 10.1093/cid/ciab456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., Chen D., Wang Y., Li X., Qiu Y., Zheng M., et al. Characterization of CCoV-HuPn-2018 spike protein-mediated viral entry. J. Virol. 2023;97 doi: 10.1128/jvi.00601-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tortorici M.A., Walls A.C., Joshi A., Park Y.-J., Eguia R.T., Miranda M.C., et al. Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein. Cell. 2022;185:2279–2291.e17. doi: 10.1016/j.cell.2022.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lednicky J.A., Tagliamonte M.S., White S.K., Elbadry M.A., Alam Md.M., Stephenson C.J., et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature. 2021;600:133–137. doi: 10.1038/s41586-021-04111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchenko V., Danilenko A., Kolosova N., Bragina M., Molchanova M., Bulanovich Y., et al. Diversity of gammacoronaviruses and deltacoronaviruses in wild birds and poultry in Russia. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-23925-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wille M., Holmes E.C. Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses. FEMS Microbiol. Rev. 2020;44:631–644. doi: 10.1093/femsre/fuaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crinion R.A., Ball R.A., Hofstad M.S. Abnormalities in laying chickens following exposure to infectious bronchitis virus at one day old. Avian Dis. 1971;15:42–48. [PubMed] [Google Scholar]
- 13.Winterfield R.W., Hitchner S.B. Etiology of an infectious nephritis-nephrosis syndrome of chickens. Am. J. Vet. Res. 1962;23:1273–1279. [PubMed] [Google Scholar]
- 14.Nair V.L., Swayne D.E., Glisson J.R., McDougald L.R., Nolan L.K., Suarez D.L. 13th ed. Wiley; Somerset: 2013. Diseases of Poultry. [Google Scholar]
- 15.Bali K., Bálint Á., Farsang A., Marton S., Nagy B., Kaszab E., et al. Recombination events shape the genomic evolution of infectious bronchitis virus in europe. Viruses. 2021;13:535. doi: 10.3390/v13040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., et al. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X., Liu Y., Weiss S., Arnold E., Sarafianos S.G., Ding J. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 2003;31:7117–7130. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., et al. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson T.K., Hoferle P.J., Chojnacki K.J., Lee K.W., Coon J.J., Kirchdoerfer R.N. An alphacoronavirus polymerase structure reveals conserved replication factor functions. Nucleic Acids Res. 2024;52:5975–5986. doi: 10.1093/nar/gkae153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malone B., Urakova N., Snijder E.J., Campbell E.A. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan Y.W., Fung T.S., Shen H., Huang M., Liu D.X. Coronavirus infectious bronchitis virus non-structural proteins 8 and 12 form stable complex independent of the non-translated regions of viral RNA and other viral proteins. Virology. 2018;513:75–84. doi: 10.1016/j.virol.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 26.Shannon A., Fattorini V., Sama B., Selisko B., Feracci M., Falcou C., et al. A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase. Nat. Commun. 2022;13:621. doi: 10.1038/s41467-022-28113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park G.J., Osinski A., Hernandez G., Eitson J.L., Majumdar A., Tonelli M., et al. The mechanism of RNA capping by SARS-CoV-2. Nature. 2022;609:793–800. doi: 10.1038/s41586-022-05185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campagnola G., Govindarajan V., Pelletier A., Canard B., Peersen O.B. The SARS-CoV nsp12 polymerase active site is tuned for large-genome replication. J. Virol. 2022;96 doi: 10.1128/jvi.00671-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schagger H., Cramer W.A., Vonjagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 34.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 35.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng E.C., Goddard T.D., Pettersen E.F., Couch G.S., Pearson Z.J., Morris J.H., et al. UCSF ChimeraX : Tools for structure building and analysis. Protein Sci. 2023;32:e4792. doi: 10.1002/pro.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croll T.I. Isolde : a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The IBV core polymerase electron density map has been deposited in the Electron Microscopy Data Bank (EMDB: 45805), and the complex model has been deposited in the Protein Data Bank (PDB: 9CPO). All other data can be found within the manuscript and supporting information document.