Abstract
Whether DNA methylation changes follow human physical trauma is uncertain. We aimed to investigate if severe trauma was associated with DNA methylation changes. In a prospective, observational, clinical study, we included severely injured adults and adults undergoing elective surgery (controls). Blood was obtained from trauma patients (n = 60) immediately- and 30-45 days post-trauma, and from surgical patients (n = 57) pre-, post-, and 30-45 days post-surgery. Epigenome-wide DNA methylation profiling was performed and analyzed for significant differentially methylated CpGs and -regions (DMRs) within and between groups. Within the trauma group we identified 10,126 significant differentially methylated CpGs and 1169 DMRs. No significant differential methylation was found in the surgical group. In the trauma group, differentially methylated sites were enriched in genes and pathways involved in blood coagulation and inflammatory response. Severe trauma was associated with profound alterations in the DNA methylome of circulating leucocytes, and differential methylation was located in trauma-relevant genes.
Subject terms: Trauma, DNA methylation
Introduction
Traumatic injury is a major cause of death and disability worldwide1. Exposure to severe physical trauma often causes lesions to vital organs and significant blood loss. Alongside the anatomical injuries, the immune system is rapidly and systemically activated as a response to damaged tissue and haemorrhage2. This immune response has the overall purpose of modulating pathogen- and damage-associated molecular patterns (PAMPs and DAMPs) and restoring cells and tissue by initiating repair mechanisms3. The immunologic response is initiated within hours after trauma and has been found to massively activate gene expression related to the innate immune system in circulating leucocytes, while genes involved in the adaptive immune system are suppressed4. This adds to the understanding that the immune response not only acts protectively, but may also have harmful effects resulting in an increased susceptibility to infection and sepsis, which are well-known consequences of trauma and account for a large part of the late (days to weeks after the trauma) mortality5,6.
In addition to the immense activation of the immune system caused by traumatic injury, there is evidence that also genes regulating epigenetic mechanisms are altered in circulating leucocytes following trauma7,8. However, it is yet uncertain whether epigenetic marks also change due to severe trauma in humans. Epigenetics is becoming an increasingly important player in our understanding of disease development and response, and there is increasing evidence that DNA methylation plays a role in the development of mental disease following psychological trauma9–11. However, within the field of physical trauma, DNA methylation has been much less investigated. A few studies using trauma-relevant animal models have suggested that alterations in the DNA methylome occur quickly after trauma and in genetic positions relevant to trauma response12,13. Some pre-clinical models have suggested an association between neurotrauma and global DNA methylation changes in neuronal cells14,15, however, this research field faces the challenges of translating the results into human studies due to the difficulties of obtaining relevant tissue.
Although DNA methylation is highly dynamic at some loci, it has also been found to be rather constant over time at other loci16. Alterations in DNA methylation patterns after trauma have not been studied in detail in humans, but it is well-known that severe physical trauma causes long-term disability for many victims, and it is plausible that changes in the DNA methylation profile can be involved both in the acute response following trauma as well as in the development of long-term outcomes after trauma. Hence, we hypothesized that DNA methylation changes would occur rapidly after exposure to severe physical trauma, that some of the changes in the DNA methylation profile would be stable and persist after one month, and that the post-traumatic DNA methylation profile would be associated with prolonged stay in the intensive care unit.
Our primary objective was to investigate if severe physical trauma was associated with acute alterations in the DNA methylation profile of peripheral leucocytes, and whether the DNA methylation alterations persisted after one month.
Results
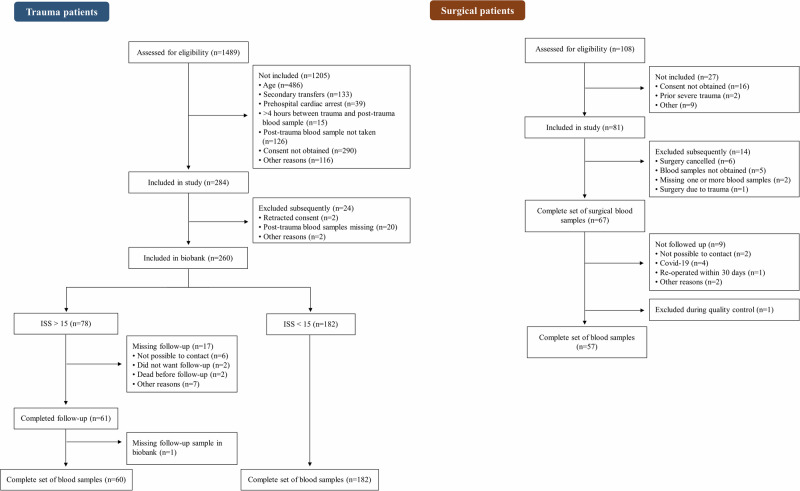
Between June 2019 and April 2021, 1489 trauma patients admitted to the Trauma Center at Rigshospitalet (RH), Copenhagen, Denmark were screened for eligibility (Fig. 1). Of these, 260 (17%) were included in our biobank; 182 (70%) had an injury severity score (ISS) < 15, and 78 (30%) had an ISS > 15 of whom 60 had a complete set of blood samples (post-trauma and 30d-trauma) (Fig. 1). Surgical patients were included between September 2019 and January 2021. During this period, 108 patients were screened, 81 (75%) were included, and 60 patients were followed up after 30–45 days. For two surgical patients, one or more blood samples were missing and the group hence consisted of 58 surgical patients with a complete set of blood samples (pre-surgery, post-surgery, 30d-surgery). One additional surgical patient (three samples) was subsequently excluded, as the sex of the patient registered in the data collection sheet did not match the sex predicted in the quality control of the DNA methylation profile (Fig. 1).
Fig. 1. Flowchart of the screening and inclusion of trauma and surgical patients.
The flowchart displays the different steps from assessment for eligibility to final inclusion for the trauma patients and surgical patients, respectively.
Of the 60 severely traumatized patients, the median age was 43.5 [IQR: 29.75;53] years, and 65% were men (Table 1). All but two patients had sustained blunt trauma, and the median ISS was 18 [IQR: 17;22]. In the group of 57 surgical patients, the median age was 48 [IQR: 35;55] years, and 33% were men. The surgical procedures included spinal fusion (56%), lower extremity surgery (THA/TKA) (25%), and Ganz osteotomy (18%). The median time from trauma or the end of surgery to collection of the post-trauma/post-surgery blood sample was 77 [IQR: 61;101] minutes and 8 [IQR: 5;17] minutes, respectively. Follow-up occurred a median of 38 [IQR: 32;43] days and 38 [IQR: 33;39] days after trauma and surgery, respectively (Table 1).
Table 1.
Demographics, injury and surgery characteristics, in-hospital characteristics, and blood sampling data for the trauma- and surgical population, respectively
| Trauma patients n = 60 | Surgical patients n = 57 | |
|---|---|---|
| Age, median [IQR] (years) | 43.5 [29.75;53] | 48 [35;55] |
| Male sex, n (%) | 39 (65) | 19 (33) |
| Mechanism of trauma, n (%) | NA | |
| Blunt | 58 (97) | NA |
| Motor vehicle accident | 23 (38) | NA |
| Bicycle accident | 10 (17) | NA |
| Pedestrian struck | 8 (13) | NA |
| Fall from height | 6 (10) | NA |
| Other | 11 (18) | NA |
| Penetrating | 2 (3) | NA |
| Stab | 1 (2) | NA |
| Gunshot | 1 (2) | NA |
| Traumatic Brain Injury | 22 (37) | NA |
| Injury Severity Score, median [IQR] | 18 [17;22] | NA |
| Trauma surgery during hospitalization, n (%) | 44 (73) | NA |
| Orthopaedic surgery, n (%) | 19 (32) | NA |
| Ear-nose-throat surgery, n (%) | 6 (10) | NA |
| Spine surgery, n (%) | 5 (8) | NA |
| Neurosurgery, n (%) | 5 (8) | NA |
| Exploratory laparotomy, n (%) | 3 (5) | NA |
| Other, n (%) | 6 (10) | NA |
| SBP on arrival in TC, median [IQR] (mmHg) (n = 57) | 130 [116–139] | NA |
| Lactate in TC (arterial blood gas), median [IQR] (mmol/L) (n = 37) | 1.7 [1.2–2.9] | NA |
| Lactate in TC (venous blood gas), median [IQR] (mmol/L) (n = 20) | 2.0 [1.6–2.7] | NA |
| Surgical procedure, n (%) | ||
| Spinal fusion | NA | 32 (56) |
| Ganz osteotomy | NA | 10 (18) |
| TKA / revision of TKA | NA | 8 (14) |
| THA / revision of THA | NA | 6 (11) |
| Other | NA | 1 (2) |
| Surgical duration, median [IQR] (minutes) | NA | 126 [100;175] |
| Transfusion of PRBC during surgery, n (patients) (%) | NA | 3 (5) |
| Time from | ||
| pre-trauma blood sampling to trauma, median [IQR] (years) (n = 5) | 2.6 [2.3;4.1] | NA |
| pre-surgery blood sampling to surgical incision, median [IQR] (mins) | NA | 37 [31;49] |
| trauma/end of surgery to blood sampling, median [IQR] (mins) | 77 [61;101] | 8 [5;17] |
| trauma/surgery to follow-up blood sampling, median [IQR] (days) | 38 [32;43] | 38 (33;39) |
| Received in-hospital blood products, n (%) [ml] | 28 (47) | 5 (9) |
| PRBC, n (patients) (%) | 20 (33) | 5 (9) |
| Amount PRBC products, median [IQR] (ml) | 1300 [245;2331] | 490 [245;545] |
| FFP, n (patients) (%) | 23 (38) | 4 (7) |
| Amount FFP products, median [IQR] (ml) | 1080 [905;1712] | 750 [750;750] |
| Platelets, n (patients) (%) | 14 (23) | 0 |
| Amount platelet products, median [IQR] (ml) | 478 [360;718] | NA |
| Sepsis, n (%) | 3 (5) | 1 (2) |
| ICU admission, n (%) | 48 (80) | 3 (5) |
| ICU length of stay, median [IQR] (hours) | 60 [26;100] | 14 [10;22] |
| ICU length of stay > 48 h, n (%) | 27 (45) | 0 (0) |
| Hospital length of stay, median [IQR] (days) | 9 [6;16] | 3 [3;5] |
| In-hospital mortality, n (%) | 1 (2) | 0 (0) |
| Discharge location, n (%) | ||
| Home | 16 (27) | 53 (93) |
| Rehabilitation facility | 2 (3) | 0 (0) |
| Another hospital | 41 (68) | 4 (7) |
IQR interquartile range, NA not applicable, SBP systolic blood pressure, TC trauma center, TKA total knee arthroplasty, THA total hip arthroplasty, PRBC packed red blood cells, ICU intensive care unit, ml millilitres, FFP fresh frozen plasma, mins minutes.
We obtained pre-trauma biological samples for six of the included trauma patients. Of these, five samples were peripheral blood, and one sample was a blood clot. Following DNA extraction from the pre-trauma samples, one sample was excluded due to low DNA quality. Hence, five (8.3%) trauma patients had pre-trauma DNA available for analysis, and these samples had been obtained a median of 2.6 years (range: 17 days to 4.8 years) prior to the trauma admission.
Dynamic changes in the methylome associated with trauma
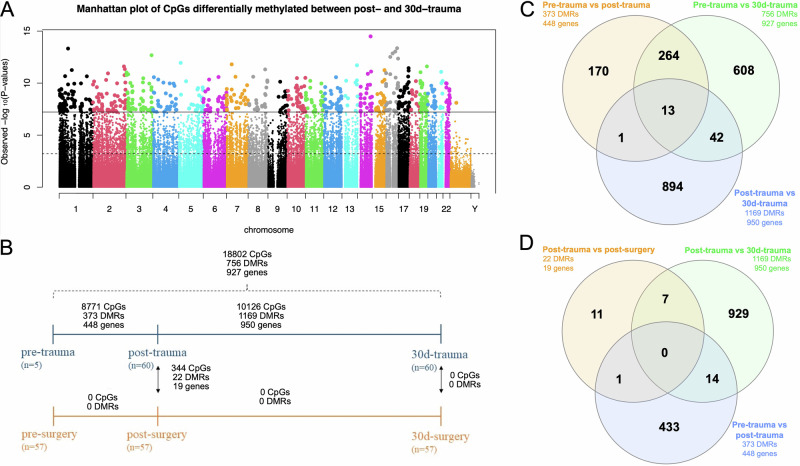
Within the trauma group, analysis of pre-trauma vs. post-trauma DNA methylation profiles (analysis 1 in Fig. 2) revealed 8771 differentially methylated CpGs (with a genomic inflation factor (lambda) of 1.78) and 373 differentially methylated regions (DMRs) (Harmonic Mean False Discovery Rate [HMFDR] < 0.05). Comparing post-trauma to 30d-trauma DNA methylation profiles (analysis 2 in Fig. 2) resulted in 10,126 CpGs (lambda = 1.67) and 1169 DMRs that were differently methylated (HMFDR < 0.05) between the two time points. Assessing pre-trauma vs. 30d-trauma DNA methylation profiles (analysis 3 in Fig. 2) returned 18,802 differentially methylated CpGs (lambda = 1.96) and 756 DMRs (HMFDR < 0.05) (Table 2). The 20 most significant DMRs from analysis 2 and the nearby genes that most likely are regulated by the DMRs are presented in Table 3 and analysis 1 is presented in Supplementary Table 1. These data suggest that both the initial phase as wells as recovery within 30 days from severe trauma result in profound changes in the blood methylome.
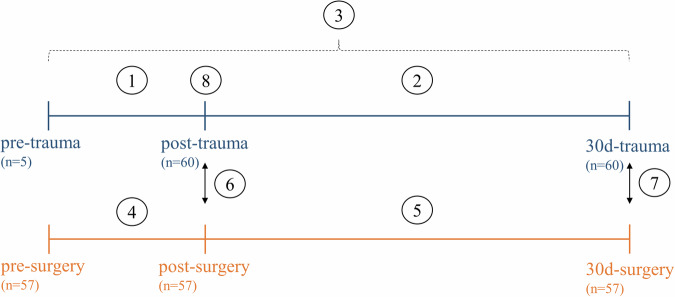
Fig. 2. Overview of the sampling time points and the sample groups to be compared for differential DNA methylation analyses.
Two patient groups (trauma and surgery) were included, and blood sampling was performed at three different time points for each group. As such, the following samples were available for analysis: pre-trauma, post-trauma, 30d-trauma and pre-surgery, post-surgery, 30d-surgery. The following eight analyses were performed: (1) pre-trauma vs. post-trauma, (2) post-trauma vs. 30d-trauma, (3) pre-trauma vs. 30d-trauma, (4) pre-surgery vs. 30d-surgery, (5) post-surgery vs. 30d-surgery, (6) post-trauma vs. post-surgery, (7) 30d-trauma vs. 30d-surgery, (8) uncomplicated vs. complicated recovery within the trauma group.
Table 2.
Number of significant CpGs and Differentially Methylated Regions (DMRs) at FDR < 0.05 for the eight different analyses of trauma patients and elective surgical patients
| Analysis | n | CpGs at FDR < 0.05 | DMRs at HMFDR < 0.05 | Lambda | |
|---|---|---|---|---|---|
| 1 | pre-trauma vs. post-traumaa | 5 | 8771 | 373 | 1.78 |
| 2 | post-trauma vs. 30d-traumaa | 60 | 10126 | 1169 | 1.67 |
| 3 | pre-trauma vs. 30d-traumaa | 5 | 18802 | 756 | 1.96 |
| 4 | pre-surgery vs. post-surgerya | 57 | 0 | 0 | 1.22 |
| 5 | post-surgery vs. 30d-surgerya | 57 | 0 | 0 | 1.15 |
| 6 | post-trauma vs. post-surgery | 117 | 344 | 22 | 1.19 |
| 7 | 30d-trauma vs. 30d-surgery | 117 | 0 | 0 | 1.22 |
| 8 | uncomplicated vs. complicated recovery, post-trauma/30d-traumaa | 60 | 0/0 | 0/0 | 0.98/0.96 |
apaired analysis.
FDR false discovery rate, HMFDR harmonic mean false discovery rate.
Table 3.
List of genes related to the top 20 differentially methylated regions (DMRs) in the analysis of the DNA methylation profile of patients post-trauma vs. 30d-trauma (analysis 2)
| Gene ID(s) | Gene name(s) | hg19 genomic coordinates | No. of CpGs | Percent mean methylation difference | q-value |
|---|---|---|---|---|---|
| DUS2 | Dihydrouridine Synthase 2 | chr16: 68112968-68113598 | 2 | 2.2 | 1.14 × 10−7 |
| VKORC1 | Vitamin K Epoxide Reductase Complex Subunit 1 | chr16:31105982-31107784 | 16 | 0.8 | 1.25 × 10−6 |
| JAK1 | Janus Kinase 1 | chr1:65348569-65348727 | 2 | 3.9 | 1.48 × 10−6 |
| MVD | Mevalonate Diphosphate Decarboxylase | chr16:88720046-88721033 | 4 | 2.3 | 1.59 × 10−6 |
| BLM | BLM RecQ Like Helicase | chr15:91292460-91293212 | 5 | -2.5 | 1.79 × 10−6 |
| MTSS1 | MTSS I-BAR Domain Containing 1 | chr8:125698322-125698676 | 2 | 2.0 | 2.04 × 10−6 |
|
LPP LPP-AS1 |
LIM Domain Containing Preferred Translocation Partner In Lipoma LPP Antisense RNA 1 |
chr3:188285801-188287006 | 9 | 4.6 | 2.08 × 10−6 |
|
ITPR1 EGOT |
Inositol 1,4,5-Trisphosphate Receptor Type 1 Eosinophil Granule Ontogeny Transcript |
chr3:4790361-4791251 | 4 | 2.4 | 2.97 × 10−6 |
|
KCTD7 RABGEF1 |
Potassium Channel Tetramerization Domain Containing 7 RAB Guanine Nucleotide Exchange Factor 1 |
chr7:66255551-66256111 | 4 | 1.5 | 5.61 × 10−6 |
| RAB11FIP1 | RAB11 Family Interacting Protein 1 | chr8:37731969-37732224 | 3 | 1.8 | 6.06 × 10−6 |
| APMAP | Adipocyte Plasma Membrane Associated Protein | chr20:24968599-24968849 | 2 | 3.0 | 6.90 × 10−6 |
| KMT2C | Lysine Methyltransferase 2C | chr7:151840170-151841181 | 3 | -1.3 | 7.14 × 10−6 |
| CCDC88C | Coiled-Coil Domain Containing 88C | chr14:91864095-91864737 | 2 | 0.3 | 8.34 × 10−6 |
| SIK3 | SIK Family Kinase 3 | chr11:116942829-116943299 | 2 | -2.0 | 8.34 × 10−6 |
| USP36 | Ubiquitin Specific Peptidase 36 | chr17: 76789167-76789804 | 2 | 1.4 | 9.06 × 10−6 |
| DGKD | Diacylglycerol Kinase Delta | chr2:234288623-234289457 | 6 | 1.2 | 9.22 × 10−6 |
| AXIN1 | Axin1 | chr16:392436-392723 | 2 | 2.4 | 9.89 × 10−6 |
| CALHM1 | Calcium Homoeostasis Modulator 1 | chr10:105217980-105219759 | 11 | 3.7 | 1.11 × 10−5 |
| LINC00426 | Long Intergenic Non-Protein Coding RNA 426 | chr13:30947309-30948716 | 6 | 1.2 | 1.29 × 10−5 |
| NMUR1 | Neuromedin U Recepter 1 | chr2:232393064-232393816 | 4 | 2.5 | 1.33 × 10−5 |
The overlap of genes with significant DMRs of analyses 1, 2, and 3 revealed that 13 genes (AEBP2, RP11-297D21.4, UBE2K, ZBTB12P1, SSBP3, TRAPPC9, MEF2A, ATP8B2, ERGIC1, PPP1CA, KCNQ1, KCNQ1OT1, ASAP1) were affected by DMRs at all time points (pre-, post-, and 30d-trauma; Fig. 3C). Most genes (277) with DMRs changing as a consequence of trauma were found to be shared between analysis 1 and 3 (Fig. 3C), indicating that the pre-trauma patterns were substantially different from post- and 30d-patterns.
Fig. 3. Overview of the location and number of differentially methylated CpGs and DMRs in the different analyses as well as the overlap of related genes between the different analyses.
A Manhattan plot of CpGs differentially methylated between post-trauma and 30d-trauma samples. The solid horizontal line indicates the cutoff for Holm (step-down Bonferroni) significance, and the dashed lines indicate FDR significance. B Number of significant differentially methylated CpGs and DMRs and the related genes for the included analyses. C Venn diagram of overlapping genes with DMRs identified (FDR < 0.05) in analyses 1, 2, and 3. D Venn diagram of overlapping genes with DMRs identified (FDR < 0.05) in analyses 1, 2, and 6.
Dynamic changes in the methylome associated with surgery
No significant (FDR < 0.05) differentially methylated CpGs and hence no DMRs were identified when the pre-surgery was compared with the post-surgery DNA methylation profiles (analysis 4 in Fig. 2). This pattern was also seen when post-surgery was compared to 30d-surgery DNA methylation profiles (analysis 5 in Fig. 2) where no significant CpGs nor any DMRs were identified (Table 2). These data suggest that no systematic sustained changes in the blood methylome persist after the selected types of elective surgery, which contrasts with what we observed within the trauma group.
Comparing post-trauma with post-surgery
We identified 344 single CpGs with significant differential methylation and 22 DMRs between post-trauma and post-surgery DNA methylation profiles (analysis 6 in Fig. 2) (Table 2). We found no significant CpGs nor any DMRs when comparing the 30d-trauma sample set to the 30d-surgery sample set (analysis 7 in Fig. 2) (Table 2). These data suggest that some differences exist in the blood methylome shortly after severe trauma, but not after 30 days when the trauma cohort was compared to the surgical cohort. When comparing the 19 genes associated with the 22 DMRs identified we found that only 1 and 7 genes overlapped with analysis 1 (pre- vs post-trauma) and 2 (post-vs 30d-trauma), respectively (Fig. 3D). This suggests that the profound changes observed in the paired analysis of trauma patients are not consistent enough to be significant at group-by-group comparison.
Comparing uncomplicated with complicated recovery
We identified no significant CpGs nor any significant DMRs when comparing the post-trauma DNA methylation profiles of trauma patients with an uncomplicated recovery ( ≤ 48 h in the ICU) and trauma patients with a complicated recovery ( > 48 h in the ICU) (analysis 8 in Fig. 2). The analysis was done both at post- and 30-day trauma time points and our data hence do not provide evidence that the methylome of post-trauma circulating leucocytes could serve as a predictive biomarker for trauma recovery using this analytical approach.
Enrichment analyses
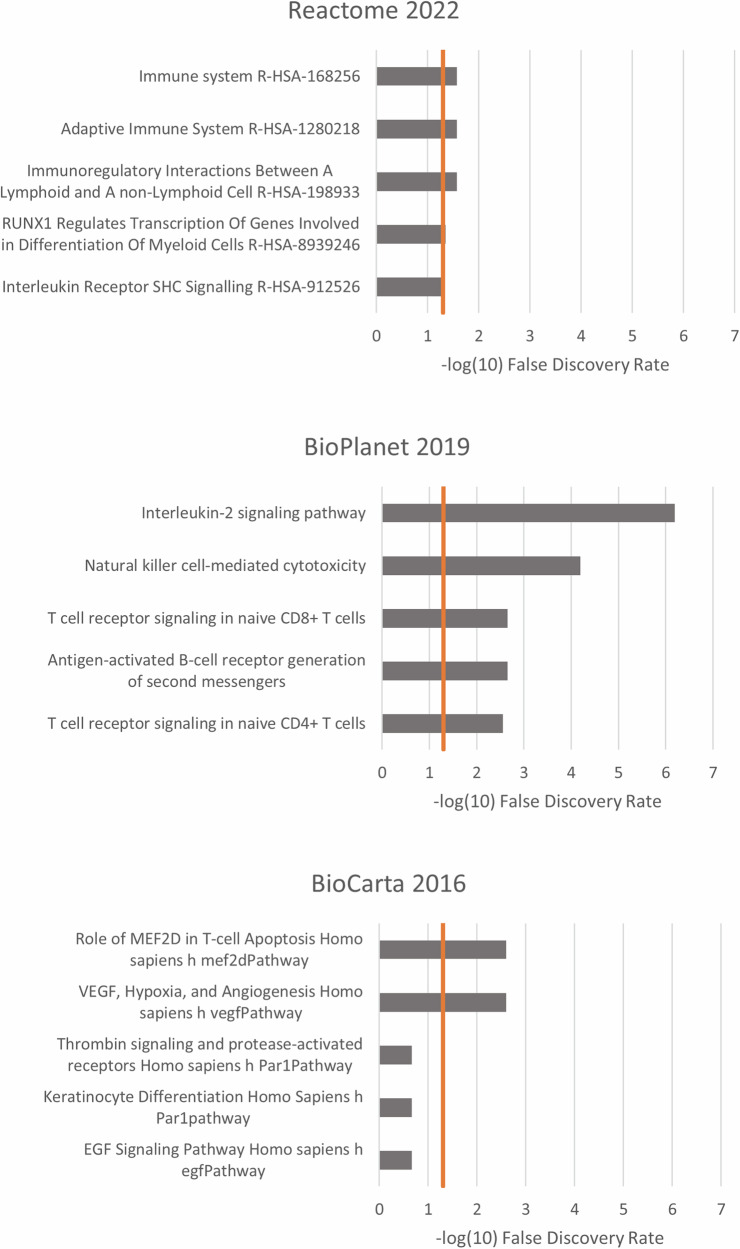
Enrichment analyses of the genes related to recovery (DMRs in the post-trauma vs. 30d-trauma analysis, analysis 2) resulted in several significantly associated pathways within all three included pathway databases (Fig. 4). In general, the most affected pathways were related to the immune system including pathways in both the innate and adaptive immune system.
Fig. 4. Pathway analyses; post-trauma vs. 30d-trauma.
The 5 highest represented pathways of the genes related to the significant DMRs in the post-trauma vs. 30d-trauma analysis from the pathway databases Reactome 2022, BioPlanet 2019, and BioCarta 2016 (through EnrichR). The x-axis is the -log10 False Detection Rate (FDR). The vertical line is the FDR cut-off of 0.05 (−log10(0.05) = 1.3).
Discussion
We found that severe physical trauma was associated with profound alterations in the DNA methylation profile in human leucocytes. Furthermore, the locations of the DNA methylation changes were associated with trauma-relevant genes and biological pathways. We found no association between the post-trauma DNA methylation profile and complicated recovery.
The present study should be considered as a preliminary study. The main purpose was to identify whether severe physical trauma in humans was associated with changes in the DNA methylome of circulating leucocytes. Overall, our results suggest that the exposure to severe trauma is associated with substantial changes in the DNA methylome that potentially are clinically relevant. Our findings are in line with a transcriptomics study of severe, blunt trauma patients, which found that more than 80% of the cellular functions and pathways in leucocytes were maximally altered within twelve hours after trauma4. We identified DNA methylation alterations between samples taken shortly after trauma and 30–45 days later, but we cannot be certain when in this time interval the alterations occurred. We assume that a large proportion of the DNA methylation changes occurred rather quickly after trauma as an acute response to tissue injury and haemorrhage, but more studies are needed to further specify the timing and trend of the DNA methylation response to trauma.
The decision to conduct re-sampling of all included patients 30–45 days post trauma or surgery was based on the above-mentioned transcriptomics study. Here they included various blood sampling intervals post trauma with the final sampling occurring 28 days after trauma revealing significant alterations in gene expression at this time point. Due to limitations preventing us from including multiple sampling times in our preliminary study, we found it most relevant to include a late re-sampling time. In addition, the inclusion of a 30-day survival follow-up aligns with the Utstein Uniform reporting of Trauma data17 thereby ensuring the 30- day follow-up is both reasonable and comparable to other trauma outcomes.
Three different analyses (1, 2, and 3) were performed for the trauma group. Although little overlap in genes related to the significant DMRs was found between the three analyses, 13 genes were found differentially methylated in all analyses (Fig. 3C). This included MEF2A, which is a transcription factor that plays a role in skeletal- and cardiac muscle development18, and reduced activity hereof has been associated with coronary artery disease19. Similarly, PP1 is important in the regulation of cardiac function, hence dysregulation has been associated with cardiac dysfunction including cardiac arrhythmias and heart failure20,21. Whether the altered methylation pattern in relation to trauma could have biological/pathological consequences remains to be investigated further, although we have recently demonstrated that twins exposed to trauma have a significantly increased risk of immune-mediated disease several years after trauma compared to their co-twins22.
We found a considerable number of significant DNA methylation alterations by comparing the DNA methylation profiles of blood samples taken within four hours after severe trauma and samples taken 30–45 days after the trauma. Analyzing the location of the methylation changes, we found these to be present in several genes relevant to traumatic injury and systemic responses. These genes included VKORC1, which is involved in the activation of vitamin K and thus an important factor in the synthesis of coagulation factors23; JAK1, which has a key role in mediating the inflammatory response24; and KMT2C, which is involved in histone methylation and hence regulation of gene expression25. Subsequently, we performed pathway analyses of the significant DMRs and found several statistically significant pathways across the three included pathway databases (Fig. 4). The highest represented pathways were related to vascular endothelial growth factor (VEGF) and different components of the immune system. VEGF is a signal protein with well-established functions in endothelial cell development and permeability, and seems to elicit altered expression following lung injury and haemorrhagic shock26. The numerous significant pathways related to genes involved in the immune system fits well with the established understanding that both the innate and adaptive immune system are involved in the trauma response2,27. In particular, several T-cell signalling pathways were identified in the enrichment analyses, which is in line with the pathway analyses of the transcriptomics study by Xiao et al. finding several T-cell signalling pathways to be downregulated following severe blunt trauma4. It is not surprising that the activated pathways involve those related with the immune system, but in the context of this study it indicates that the identified DNA methylation changes after trauma contribute to the activation of these specific pathways.
In analysis 1 and 3, we had the unique possibility of including pre-trauma DNA samples from five trauma patients. Although this only constituted a minor proportion of the trauma group and the uncertainty of the analyses was consequently higher (higher lambda), it was noteworthy that a large number of significant CpGs and DMRs was identified in both analyses including the pre-trauma samples (analyses 1 and 3). Still, it must be kept in mind that the five pre-trauma samples were obtained at varying time points prior to the patients’ trauma (range: 17 days to 4.8 years), allowing for other exposures than trauma to affect the DNA methylation profile. However, the results of the analyses including the pre-trauma samples still support the findings from the other analyses that severe trauma was associated with alterations in the DNA methylation profile.
To our surprise we did not find significant differentially methylated CpGs or DMRs within the surgical group. Some possible reasons why we did not detect an association between major surgery and DNA methylation changes in peripheral leucocytes are that the different surgical procedures were too heterogenous to provide a homogeneous DNA methylation response. Furthermore, it is possible that elective surgery during anaesthesia causes fewer profound changes as numerous physiological variables are kept within the normal range, and thus rarely reached extremes as for severely injured patients. Lastly, the surgical patients’ underlying medical condition leading to surgery may involve a persistent or chronic inflammation which could potentially mask a DNA methylation signal.
The between-group analyses revealed differential methylation between post-trauma and post-surgery DNA methylation profiles. Unexpectedly, we did not find any differentially methylated CpGs nor DMRs comparing 30d-trauma to 30d-surgery DNA methylation profiles. A possible explanation could be that the groups had returned to a “normal” state this long after the trauma/surgery, but inadequate statistical power in the unpaired analysis could also be important. The issue of low statistical power may also be true for the comparison of trauma patients with an uncomplicated and complicated recovery. This too was an unpaired analysis with only 33 and 27 patients in the complicated and uncomplicated recovery group, respectively. Indeed paired analysis has previously been shown to provide a statistical power to studies of DNA methylation28,29.
DNA methylation alterations have been investigated in a few trauma-relevant animal models. One study compared DNA methylation profiling in lung tissue in rats exposed to haemorrhage and subsequent saline resuscitation with sham rats12. That study also used the Reactome database for pathway analyses and, like in our study, found the highest represented pathways to include VEGF interactions and interleukin signalling. Probably the most studied trauma-relevant animal model is different traumatic brain injury (TBI) models, though TBI is traditionally considered a special subset of trauma. A study of rats exposed to blast-overpressure TBI found significant DNA methylation alterations in neurons and glia eight months after exposure, suggesting that post-traumatic changes in the DNA methylome are long lasting at least following specific types of injury in animals30.
The strengths of this study lie in the inclusion of a rather large and unselected trauma population with few exclusion criteria. Furthermore, we included severe trauma patients despite the heterogeneity of specific anatomical injuries. This increases the generalizability of the findings, and it may be speculated that the detected DNA methylation alterations are associated with a general host response to trauma and not to specific anatomical injuries. The post-trauma blood samples were drawn before obtaining patient consent, which allowed post-trauma samples to be taken rather quickly after trauma and with little variation within the group.
The study has several limitations. Firstly, pre-trauma blood samples from all included trauma patients would be desirable, however, as traumatic exposure is unpredictable, this was not possible. Instead, we included a group of patients undergoing elective surgery of the spine or lower extremity. The surgical group differed from the trauma group in the sex distribution, as the trauma group constituted 65% males, whereas the surgical group only included 33% males. We did, however, adjust for sex in our statistical analyses. The surgical- and trauma groups also differed in terms of injury type. Patients within the surgical group sustained similar trauma under controlled conditions, whereas the injuries within the trauma group were heterogeneous and included all anatomical regions. However, our focus was not on the specific anatomical injuries, but rather the potential immune reaction to major injury regardless of injury type. In hindsight, the two groups may not be as comparable in terms of immune response as we had expected, which is a limitation. Secondly, we used whole blood as our model tissue with the rationale being that whole blood is readily available and imposes minimal risk for the patient when obtained. Another reason was that circulating leucocytes in whole blood play an important role in the rapid and massive immune response to major trauma and have the ability to swiftly travel to the site(s) of injury. It is, however, very likely that investigating other trauma-relevant tissues (brain, liver, lung, kidney, muscle, etc.) would reveal other epigenetic marks. Thirdly, we included only patients having sustained severe trauma (ISS > 15) and our results cannot necessarily be extrapolated to trauma patients with minor trauma. Fourthly, there is a potential selection bias in the patients followed up after 30–45 days. Within the trauma group, patients who were lost to follow-up included patients with active psychiatric disease, patients without a home and/or phone, and patients who died before day 30. Lastly, we have excluded trauma patients who had received packed red blood cells (PRBC) before the first blood sample was obtained and limited inclusion of surgical procedures to those with an expected low risk of blood transfusion. We have not been able to control the transfusion of blood products during subsequent hospital admission. As such, almost half of the trauma patients, but only 9% of the surgical patients received any blood products during the hospital stay. Blood products are, however, leucocyte-reduced and almost free of DNA.
As changes in DNA methylation may be long-lasting or even permanent, it may be speculated that potential DNA methylation alterations following traumatic exposure could have prolonged epigenetic effects with implications for the future health of the host. As such, DNA methylation profiles could be relevant as future biomarkers and pharmacological targets.
In conclusion, severe physical trauma was associated with profound changes in the DNA methylation patterns in human circulating leucocytes and several differentially methylated regions were located in trauma-relevant genes and pathways.
Methods
Ethics, approvals, and study registration
The study protocol was approved by The Committees on Health Research Ethics for the Capital Region of Denmark (approval number: H-19010072), hence, the study complied with all relevant ethical regulations including the Declaration of Helsinki. The handling of data complied with the rules and regulations set forward by the data responsible institution in the Capital Region of Denmark (approval numbers: VD-2019-161 / P-2021-126). The study was registered on ClinicalTrials.gov (registration number: NCT03974048). Informed consent for participation was obtained as described below.
Study design
Study setup and reporting were conducted in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines31. This was a prospective, observational epigenome-wide association study (EWAS) investigating a possible association between severe traumatic injury and alterations in the whole blood DNA methylation profile. Ideally, the study would have been set up to compare the pre-trauma DNA methylation profile with the post-trauma DNA methylation profile of a cohort of patients exposed to severe traumatic injury. However, as traumatic injury is acute and unpredictable, we assumed it would only be possible to obtain pre-trauma samples from a small proportion of trauma patients. Therefore, a cohort of patients admitted for elective surgery was included as a control group. The surgical procedure was considered a “controlled” form of traumatic injury, and for this group we were able to obtain both pre-surgery and post-surgery samples. Pre-trauma blood/DNA samples were obtained from the Danish National Biobank (DNB), when inclusion of all patients was complete.
Study population and informed consent
Trauma patients: We included trauma patients 18–65 years of age who were admitted to the Trauma Center at RH following a trauma team activation from June 2019 to April 2021. Exclusion criteria were secondary transfers, prehospital cardiac arrest, more than four hours between the time of trauma and the first study related blood sample, and transfusion with PRBC before the first study related blood sample was obtained. Prehospital transfusion with freeze-dried or liquid plasma or in-hospital transfusion with fresh frozen plasma were not exclusion criteria. Trauma patients were included regardless of the severity of the trauma. After the initial examination and treatment phase (tertiary survey) all patients had an ISS determined by review of their medical records performed by a certified Abbreviated Injury Scale (AIS) coding specialist usually within seven days32. Patients with ISS > 15 (considered severe trauma) were included for DNA methylation profiling and compared with the surgical control group. Blood samples from trauma patients with ISS < 15 were stored, but not handled further in this study. It is not possible to obtain an ISS of 15.
All trauma patients were considered temporarily unable to provide informed consent at the time of inclusion in the study. Informed consent was sought from the patient or his/her next-of-kin as soon as possible after the acute clinical examination and treatment phase.
Surgical patients: In the control group, we included patients 18–65 years of age who were scheduled for major, elective orthopaedic or spine surgery at either RH or RH/Glostrup Hospital between September 2019 and January 2021. Surgical procedures included Ganz osteotomy, lower extremity surgery (total hip arthroplasty [THA], total knee arthroplasty [TKA]), or spinal fusion with an expected duration of at least 60 min and anticipated minor to moderate blood loss. We excluded patients who had surgery due to fractures or cancer and patients who had previously been exposed to severe trauma (assessed by asking the patient about prior traumatic exposure). Patients who were re-operated within 30 days after the surgery were excluded from the follow-up blood sample.
All surgical patients provided informed consent before inclusion in the study.
Study procedures
Trauma patients: All trauma patients admitted to the Trauma Center at RH who were initially considered eligible for inclusion had a study blood sample (“post-trauma” blood sample, see below) taken in the trauma bay by a laboratory technician. Hereafter, a member of the research staff assessed the medical record in relation to the in- and exclusion criteria and sought informed consent if the patient was still eligible for inclusion. Post-trauma blood samples from patients deemed ineligible or where informed consent was not obtained, were destroyed.
Pre-trauma blood samples: Upon completion of trauma patient inclusion, we queried the DNB for previously collected blood- or DNA samples from the included trauma patients with ISS > 15. The DNB contains millions of biological samples from the Danish population, and these samples were either excess material from other research biobanks or national screenings. We used the included trauma patients’ unique Central Person Register (CPR) number (which is assigned to all Danish citizens) to identify biological samples containing DNA, which had been collected for another purpose before the patients’ trauma date.
Surgical patients: The surgical patients had a peripheral venipuncture (“pre-surgery” blood sample) performed immediately prior to incision. Again, immediately after surgery and no later than four hours after wound closure another peripheral venipuncture (“post-surgery” blood sample) was performed.
Follow-up at 30–45 days: We contacted trauma patients with an ISS > 15 and the surgical patients in whom both the pre-surgery and post-surgery blood sample had been obtained. After consent, a follow-up peripheral venipuncture was performed 30–45 days after trauma/surgery (“30d-trauma” and “30d-surgery” blood sample, respectively). Thus, all included trauma patients had a set of two or three blood samples ((pre-trauma), post-trauma, 30d-trauma), and all included surgical patients had a set of three blood samples (pre-surgery, post-surgery, 30d-surgery).
Data collection: Demographic, trauma- and surgery-related, and in-hospital data were obtained by the research staff from the patients’ electronic medical records.
Outcome measures
Outcome measures included differential DNA methylation of either individual CpG sites or regions in the genome of whole blood DNA. We compared findings within the trauma group (analysis 1, 2, and 3; paired analyses), within the surgical group (analyses 4 and 5; paired analyses), and between the two study groups (analyses 6 and 7; unpaired analyses) (Fig. 2). In addition, the DNA methylation profiles of the post-trauma blood samples were compared for trauma patients with an uncomplicated vs. complicated recovery (analysis 8; unpaired). A complicated recovery was defined as a stay exceeding 48 h in the intensive care unit (ICU) during the trauma-related hospital admission.
Blood sampling and circulating leucocyte DNA methylation profiling
Blood sampling was performed by trained laboratory technicians by peripheral venipuncture in the antecubital fossa using the SAFETY Blood Collection Set + Holder (Greiner Bio-One, Kremsmünster, Austria). A total volume of 21 millilitres (9 + 9 + 3 mililiters) was drawn in VACUETTE® EDTA coated tubes (Greiner Bio-One, Kremsmünster, Austria). All blood samples were taken to the biobank within five hours upon obtainment and stored at 4 degrees Celsius until further handling. Samples delivered to the biobank on weekdays (Monday-Friday) from 8 AM to 3 PM were handled on the same day, and samples delivered at other times were handled on the first coming weekday. Samples were separated into full blood samples and plasma samples. Full blood samples were pipetted into 850 µL matrix tubes (Thermo Fisher Scientific, Waltham, Massachusetts, USA), and samples for plasma obtainment were centrifuged at 1800 Relative Centrifugal Force (RFG) at 4 degrees Celsius for 10 minutes and stored in 850 µL matrix tubes (Thermo Fisher Scientific, Waltham, Massachusetts, USA). All samples were subsequently stored at -80 degrees Celsius.
Only samples from patients with a complete set of blood samples were further processed. DNA was extracted from whole blood using the Chemagen technology (ChemagicStar, Perkin Elmer) following the instructions given by the manufacturer. In brief, DNA was isolated with magnetic beads after addition of lysis buffer and proteases to the samples, and finally eluated by adding an elution buffer.
The extracted DNA was bisulfite treated to convert unmethylated cytosines to uracil. Then, the DNA was hybridized to Infinium MethylationEPIC BeadChips (Illumina, San Diego, CA, USA) and further underwent single base extension, fluorescent staining, signal amplification, and scanning using the iScan System (all procedures were performed by Eurofins Genomics, Galten, Denmark). Finally, the intensities of the probes were extracted and analysed using the GenomeStudio Software V2001.1 (Illumina, San Diego, CA, USA). Each MethylationEPIC BeadChip held eight samples, and samples were randomly distributed on the BeadChips to minimize systematic bias. Each BeadChip interrogated 863,904 CpG sites distributed throughout the genome33.
Data analysis
IDAT files were extracted from the GenomeStudio Software V2001.1 (Illumina, San Diego, CA, USA) for each sample and imported into Rstudio version 4.1.0 (2021-05-08) – “Camp Pontanezen” using the minfi package (version 1.38.0)34,35. The level of methylation at each CpG site was measured as a beta(β)-value ranging from 0 (no methylation) to 1 (complete methylation). The ShinyMethyl package (version 1.28.0) was used to assess the quality of the data36. Individual samples that did not pass quality control in order to be sufficiently analysed were excluded (trauma patients: n = 0, surgical patients, n = 1) from further analysis.
Samples passing quality control (trauma patients: n = 60, surgical patients, n = 57) were normalized using a Subset-quantile Within Array Normalization (SWAN)35 method to reduce the technical variability within and between arrays. CpGs and single nucleotide extensions containing single nucleotide polymorphisms (SNPs) were dropped. To control for batch effects, including changes in cell type composition, we calculated surrogate variables using the SmartSVA package (version 0.1.3)37 and included the surrogate variables as covariates in subsequent analyses.
For each sample group comparison (see paragraph on outcome measures and Fig. 2) differential methylation at single CpG sites were determined using the CpGAssoc package (version 2.6.0)38 after inclusion of the surrogate variables as covariates. To correct for multiple testing a False Discovery Rate (FDR) using the Benjamini-Hochberg method was applied resulting in a q-value (adjusted p-value). A q-value below 0.05 was considered statistically significant. All analyses were, in addition to surrogate variables, adjusted for age and sex. Paired analyses within the trauma group were adjusted for ISS, and paired analyses within the surgical group were adjusted for type of surgical procedure.
DMRs were identified and ranked using the DMRcate package (version 2.6.0)39. We used a bandwidth of 1000 nucleotides (lambda = 1000), a scaling factor of 2 (C = 2), and corrected the results for multiple testing using the Benjamini-Hochberg method as recommended by the authors of the DMRcate package39. Annotation was done according to hg19.
Descriptive statistics were reported with median and interquartile range (IQR) and frequencies and percentages for continuous and categorical variables, respectively.
Sample size
As there was limited knowledge on DNA methylation changes following severe trauma, we were not able to properly estimate the effect size of trauma on the DNA methylation profile. Based on studies of non-trauma populations that have found effect sizes of 0–25% difference in genome-wide DNA methylation levels between cases and controls40–42, we assumed that a 15% change in the mean methylation difference between our comparison groups was relevant to detect following exposure to severe trauma. A simulation study set up to estimate power and adequate sample size for EWAS43 found that to detect a 15% mean methylation difference between two groups with 80% power and a genome-wide significance level of 1 × 10−6, 54 patients were needed in each group. Thus, we needed to include 54 trauma patients and 54 surgical patients to obtain 80% power at a genome-wide significance level of 1 × 10−6. To account for potential DNA samples of low quality we decided to continue inclusion of patients until we had a full set of blood samples from 60 trauma patients and 60 surgical patients.
Enrichment analyses
To identify perturbated pathways or biological functions, we performed enrichment analyses of the genes related to the significant DMRs. We used EnrichR44 and included the following pathway databases: Reactome 202245, BioPlanet 201946, and BioCarta 2016.
Supplementary information
Acknowledgements
The study was financially supported by the Research Fund of Rigshospitalet, University of Copenhagen. These materials have received financial support from The Danish Victims Fund (grant number 19-610-00046). The execution, content, and results of the materials are the sole responsibility of the authors. The analysis and viewpoints that have been made evident from the materials belong to the authors and do not necessarily reflect the views of The Council of The Danish Victims Fund. The study has been financially supported by Merchant L. F. Foght’s Fund (Danish: Grosserer L. F. Foghts Fond). The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript. The study has been financially supported by the Danish Society of Anaesthesiology and Intensive Care Medicine’s Fund. The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author contributions
Conceptualization, T.O.E, K.A., L.S.R., M.S., J.S.; Methodology, T.O.E, K.A., E.S., A.W.P., T.N., L.S.R., M.S., J.S.; Investigation, T.O.E, L.E., T.A., M.L.L., A.C., A.H.J., N.B., F.D., M.V.; Data Curation, T.O.E, K.A.; Writing – Original Draft, T.O.E.; Writing – Review & Editing, T.O.E, K.A., L.E., T.A., M.L.L., A.C., A.H.J., N.B., F.D., M.V., E.S., A.W.P., T.N., L.S.R., M.S., J.S.; Visualization, T.O.E, K.A.; Supervision, K.A., L.S.R., M.S., J.S.; Project Administration, T.O.E, J.S.; Funding Acquisition, T.O.E., L.S.R., M.S., J.S.
Data availability
According to Danish law, it is not allowed for us to share individual patient data. As such, we are not able to make the data available to the public. However, it can be possible to share de-identified patient data with individual, named researchers following a specific IRB approval.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-024-00438-4.
References
- 1.Geneva, W. H. O. Global Health Estimates 2020: Deaths by Cause, Age, Sex, by Country and by Region, 2000-2019.https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates (2020).
- 2.Lord, J. M. et al. The systemic immune response to trauma: An overview of pathophysiology and treatment. Lancet384, 1455–1465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billiar, T. R. & Vodovotz, Y. Time for trauma immunology. PLoS Med14, e1002342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao, W. et al. A genomic storm in critically injured humans. J. Exp. Med.208, 2581–2590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunst, M. et al. Changing epidemiology of trauma deaths leads to a bimodal distribution. Bayl. Univ. Med. Cent. Proc.23, 349–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobrino, J. & Shafi, S. Timing and causes of death after injuries. Bayl. Univ. Med. Cent. Proc.26, 120–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sillesen, M., Li, Y. & Alam, H. B. Transfusion strategies are associated with epigenetic changes following blunt trauma. Shock50, 24–30 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Sillesen, M. et al. Histone deactylase gene expression profiles are associated with outcomes in blunt trauma patients. J. Trauma Acute Care Surg.80, 26–33 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Rutten, B. P. F. et al. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol. Psychiatry23, 1145–1156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith, A. K. et al. Epigenome-wide meta-analysis of PTSD across 10 military and civilian cohorts identifies methylation changes in AHRR. Nat. Commun.11, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng, H. et al. Childhood trauma, DNA methylation of stress-related genes, and depression: findings from two monozygotic twin studies. Psychosom. Med.80, 599–608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonde, A. et al. Hemorrhage and saline resuscitation are associated with epigenetic and proteomic reprogramming in the rat lung. Injury52, 2095–2103 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Sun, H. et al. DNA hydroxymethylation mediated traumatic spinal injury by influencing cell death–related gene expression. J. Cell. Biochem.119, 9295–9302 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Wong, V. S. & Langley, B. Epigenetic changes following traumatic brain injury and their implications for outcome, recovery and therapy. Neurosci. Lett.625, 26–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagalakshmi, B., Sagarkar, S. & Sakharkar, A. J. Epigenetic mechanisms of traumatic brain injuries. Prog. Mol. Biol. Transl. Sci.157, 263–298 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Feinberg, A. P. The key role of epigenetics in human disease prevention and mitigation. N. Engl. J. Med.378, 1323–1334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringdal, K. G. et al. The Utstein template for uniform reporting of data following major trauma: A joint revision by SCANTEM, TARN, DGU-TR and RITG. Scand. J. Trauma. Resusc. Emerg. Med.16, 1–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pon, J. R. & Marra, M. A. MEF2 transcription factors: developmental regulators and emerging cancer genes. Oncotarget7, 2297–2312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, L., Fan, C., Topol, S. E., Topol, E. J. & Wang, Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Sci.302, 1578–1581 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer-Roxlau, S. et al. Differential regulation of protein phosphatase 1 (PP1) isoforms in human heart failure and atrial fibrillation. Basic Res. Cardiol. 112, (2017). [DOI] [PubMed]
- 21.Heijman, J., Dewenter, M., El-Armouche, A. & Dobrev, D. Function and regulation of serine/threonine phosphatases in the healthy and diseased heart. J. Mol. Cell. Cardiol.64, 90–98 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Eskesen, T. O. et al. Association of Trauma With Long-Term Risk of Death and Immune-Mediated or Cancer Disease in Same-Sex Twins. JAMA Surg. 10.1001/jamasurg.2023.1560 (2023). [DOI] [PMC free article] [PubMed]
- 23.Oldenburg, J., Watzka, M., Rost, S. & Müller, C. R. VKORC1: molecular target of coumarins. J. Thromb. Haemost.5, 1–6 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Spinelli, F. R., Colbert, R. A. & Gadina, M. JAK1: number one in the family; number one in inflammation? Rheumatol. (U. Kingd.)60, II3–II10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Froimchuk, E., Jang, Y. & Ge, K. Histone H3 lysine 4 methyltransferase KMT2D. Gene627, 337–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loftus, T. J. et al. Effects of trauma, hemorrhagic shock, and chronic stress on lung vascular endothelial growth factor. J. Surg. Res.210, 15–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber-Lang, M., Lambris, J. D. & Ward, P. A. Innate immune responses to trauma review-article. Nat. Immunol.19, 327–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, H. H. et al. Influence of genetic background and tissue types on global DNA methylation patterns. PLoS One5, (2010). [DOI] [PMC free article] [PubMed]
- 29.Almstrup, K. et al. Pubertal development in healthy children is mirrored by DNA methylation patterns in peripheral blood. Sci. Rep.6, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haghighi, F. et al. Neuronal DNA methylation profiling of blast-related traumatic brain injury. J. Neurotrauma32, 1200–1209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Elm, E. et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J. Clin. Epidemiol.61, 344–349 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Gennarelli, T & Wodzin, E. E. Abbreviated Injury Scale (c) 2005 Update 2008. (Association for the Advancement of Automotive Medicine, 2016).
- 33.Wu, M. C. & Kuan, P. F. A guide to illumina beadchip data analysis. Methods in Molecular Biology vol. 1708 (2018). [DOI] [PubMed]
- 34.Aryee, M. J. et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics30, 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maksimovic, J., Gordon, L. & Oshlack, A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol.13, 1–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fortin, J. P., Fertig, E. & Hansen, K. shinyMethyl: Interactive quality control of Illumina 450k DNA methylation arrays in R. F1000Research3, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen, J. et al. Fast and robust adjustment of cell mixtures in epigenome-wide association studies with SmartSVA. BMC Genomics18, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barfield, R. T., Kilaru, V., Smith, A. K. & Conneely, K. N. CpGassoc: An R function for analysis of DNA methylation microarray data. Bioinformatics28, 1280–1281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters, T. J. et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin8, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakyan, V. K. et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet7, 1–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Javierre, B. M. et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res20, 170–179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeilinger, S. et al. Tobacco Smoking Leads to Extensive Genome-Wide Changes in DNA Methylation. PLoS One8, (2013). [DOI] [PMC free article] [PubMed]
- 43.Tsai, P. C. & Bell, J. T. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int. J. Epidemiol.44, 1429–1441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr. Protoc.1, 139–148 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gillespie, M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res50, D687–D692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang, R. et al. The NCATS BioPlanet – an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front. Pharmacol.10, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
According to Danish law, it is not allowed for us to share individual patient data. As such, we are not able to make the data available to the public. However, it can be possible to share de-identified patient data with individual, named researchers following a specific IRB approval.