Abstract
A method is described for fitting the velocities obtained from progress curves to a steady-state rate equation. It is based on the method of Markus & Plesser [(1981) in Kinetic Data Analysis: Design and Analysis of Enzyme and Kinetic Data (Edrenyi, ed.), pp. 317-339, Plenum Press, New York]. The obstacle of needing good initial estimates of kinetic parameters is removed by using the parameters provided graphically by a minor modification of the method of Yun & Suelter [(1977) Biochim, Biophys. Acta 480, 1-13]. This progress-curved-based method allows the same discrimination among rival models as do the initial-velocity-based methods, with a great saving of experimental time. The BASIC and FORTRAN 77 programs are deposited as Supplementary Publication SUP 50132 (17 pages) at the British Library (Lending Division), Boston Spa, Wetherby, West Yorkshire LS23 7BQ, U.K., from whom copies can be obtained on the terms indicated in Biochem. J. (1986) 233, 5-6.
Full text
PDF

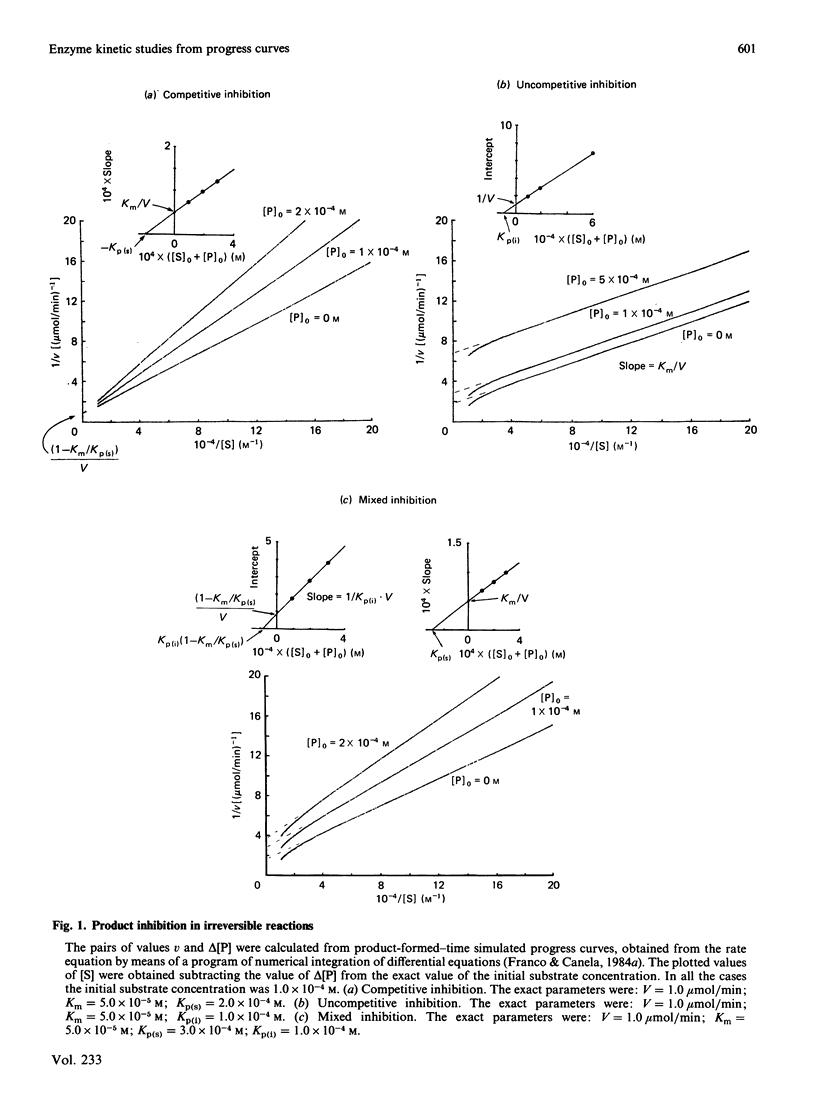
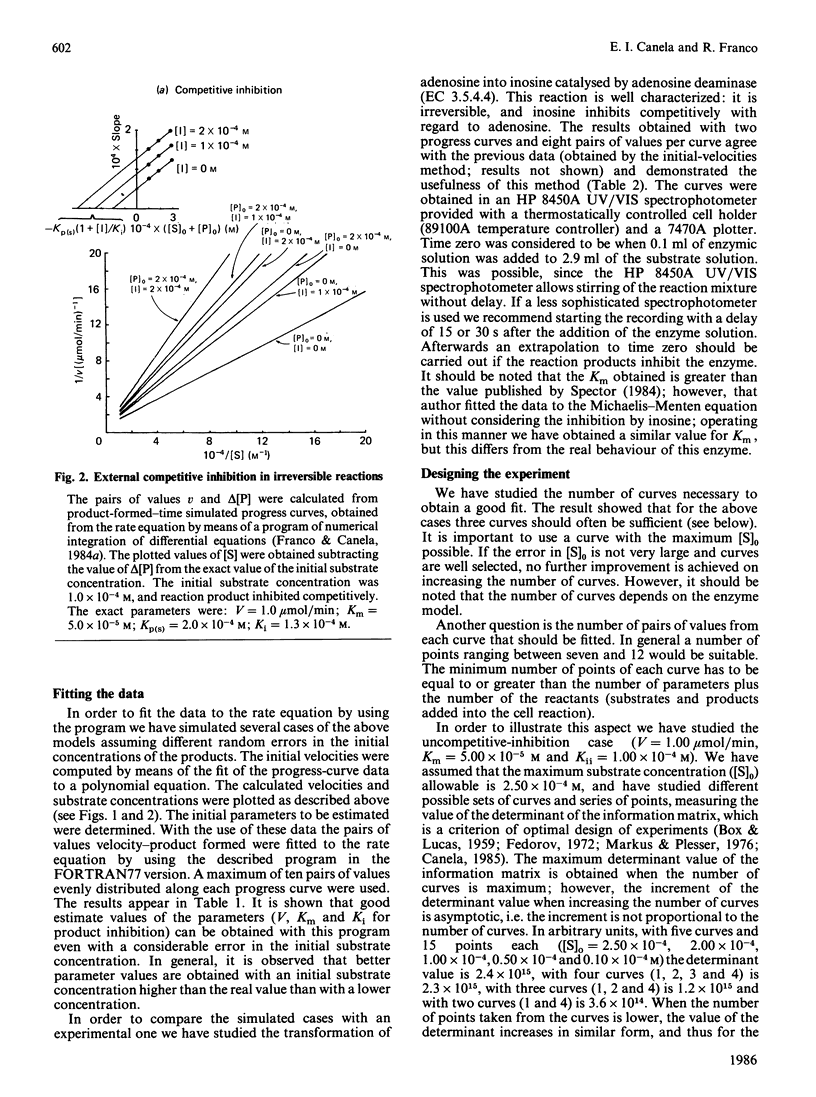
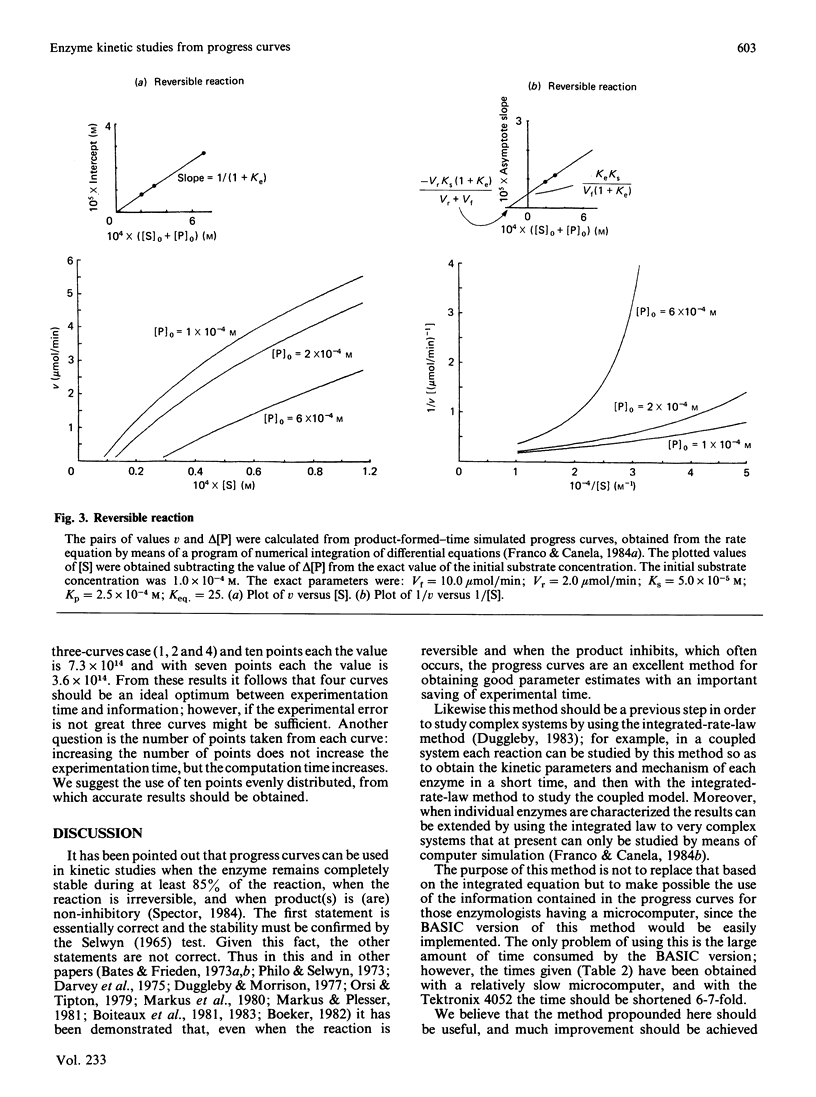
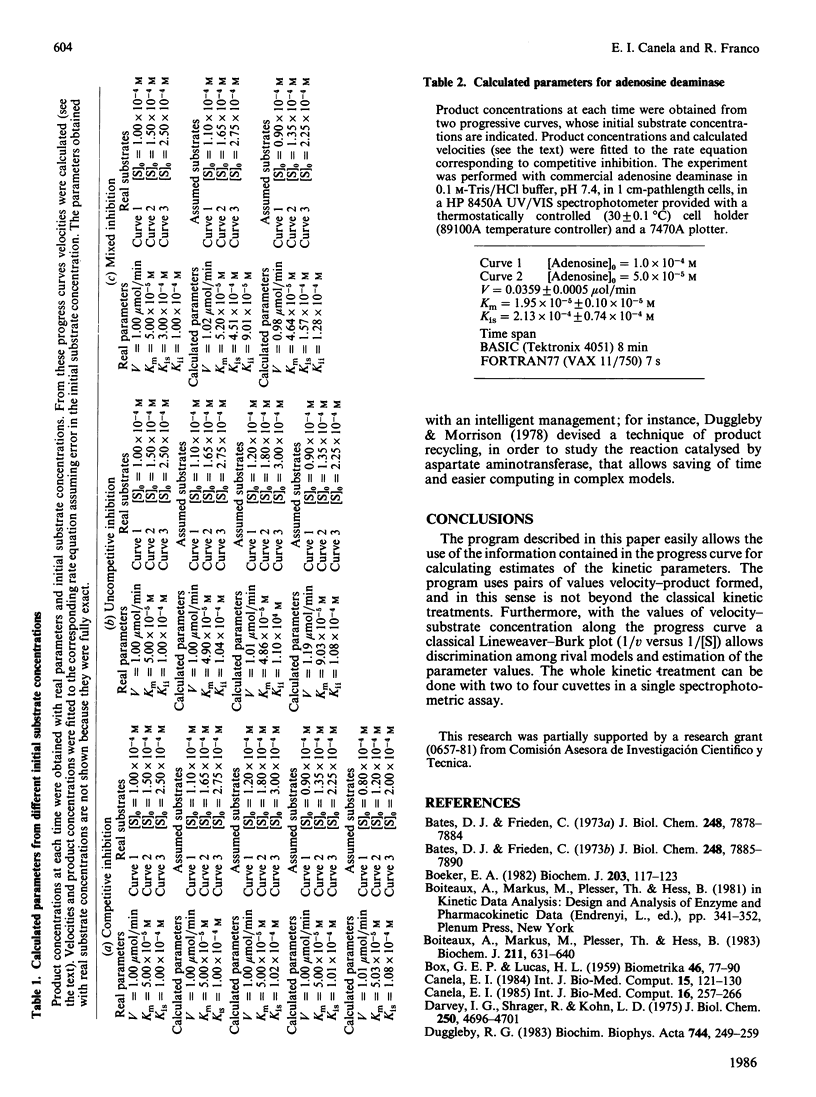

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bates D. J., Frieden C. Full time course studies on the oxidation of reduced coenzyme by bovine liver glutamate dehydrogenase. Use of computer simulation to obtain rate and dissociation constants. J Biol Chem. 1973 Nov 25;248(22):7885–7890. [PubMed] [Google Scholar]
- Bates D. J., Frieden C. Treatment of enzyme kinetic data. 3. The use of the full time course of a reaction, as examined by computer simulation, in defining enzyme mechanisms. J Biol Chem. 1973 Nov 25;248(22):7878–7884. [PubMed] [Google Scholar]
- Boeker E. A. Initial rates. A new plot. Biochem J. 1982 Apr 1;203(1):117–123. doi: 10.1042/bj2030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux A., Markus M., Plesser T., Hess B., Malcovati M. Analysis of progress curves. Interaction of pyruvate kinase from Escherichia coli with fructose 1,6-bisphosphate and calcium ions. Biochem J. 1983 Jun 1;211(3):631–640. doi: 10.1042/bj2110631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela E. I. A free derivative program for non-linear regression analysis of enzyme kinetics to be used on small computers. Int J Biomed Comput. 1984 Mar-Apr;15(2):121–130. doi: 10.1016/0020-7101(84)90024-2. [DOI] [PubMed] [Google Scholar]
- Canela E. I. A microcomputer method for designing optimal experiments for estimating enzyme kinetic parameters. Int J Biomed Comput. 1985 May;16(3-4):257–266. doi: 10.1016/0020-7101(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Darvey I. G., Shrager R., Kohn L. D. Integrated steady state rate equations and the determination of individual rate constants. J Biol Chem. 1975 Jun 25;250(12):4696–4701. [PubMed] [Google Scholar]
- Duggleby R. G. Determination of the kinetic properties of enzymes catalysing coupled reaction sequences. Biochim Biophys Acta. 1983 May 18;744(3):249–259. doi: 10.1016/0167-4838(83)90197-8. [DOI] [PubMed] [Google Scholar]
- Duggleby R. G., Morrison J. F. Progress curve analysis in enzyme kinetics: model discrimination and parameter estimation. Biochim Biophys Acta. 1978 Oct 12;526(2):398–409. doi: 10.1016/0005-2744(78)90131-6. [DOI] [PubMed] [Google Scholar]
- Duggleby R. G., Morrison J. F. The analysis of progress curves for enzyme-catalysed reactions by non-linear regression. Biochim Biophys Acta. 1977 Apr 12;481(2):297–312. doi: 10.1016/0005-2744(77)90264-9. [DOI] [PubMed] [Google Scholar]
- Franco R., Canela E. I. A program for the numerical integration of enzyme kinetic equations using small computers. Int J Biomed Comput. 1984 Nov-Dec;15(6):419–432. doi: 10.1016/0020-7101(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Franco R., Canela E. I. Computer simulation of purine metabolism. Eur J Biochem. 1984 Oct 15;144(2):305–315. doi: 10.1111/j.1432-1033.1984.tb08465.x. [DOI] [PubMed] [Google Scholar]
- Markus M., Plesser T., Boiteux A., Hess B., Malcovati M. Analysis of progress curves. Rate law of pyruvate kinase type I from Escherichia coli. Biochem J. 1980 Sep 1;189(3):421–433. doi: 10.1042/bj1890421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus M., Plesser T. Design and analysis of progress curves in enzyme kinetics. Biochem Soc Trans. 1976;4(2):361–364. doi: 10.1042/bst0040361. [DOI] [PubMed] [Google Scholar]
- Orsi B. A., Tipton K. F. Kinetic analysis of progress curves. Methods Enzymol. 1979;63:159–183. doi: 10.1016/0076-6879(79)63010-0. [DOI] [PubMed] [Google Scholar]
- Philo R. D., Selwyn M. J. Use of progress curves to investigate product inhibition in enzyme-catalysed reactions. Application to the soluble mitochondrial adenosine triphosphatase. Biochem J. 1973 Nov;135(3):525–530. doi: 10.1042/bj1350525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn M. J. A simple test for inactivation of an enzyme during assay. Biochim Biophys Acta. 1965 Jul 29;105(1):193–195. doi: 10.1016/s0926-6593(65)80190-4. [DOI] [PubMed] [Google Scholar]
- Spector T. Progress curve analysis of adenosine deaminase-catalyzed reactions. Anal Biochem. 1984 Apr;138(1):242–245. doi: 10.1016/0003-2697(84)90796-6. [DOI] [PubMed] [Google Scholar]
- Yun S. L., Suelter C. H. A simple method for calculating Km and V from a single enzyme reaction progress curve. Biochim Biophys Acta. 1977 Jan 11;480(1):1–13. doi: 10.1016/0005-2744(77)90315-1. [DOI] [PubMed] [Google Scholar]



