Abstract
Growing evidence suggests that perfluorinated compounds (PFCs) contribute to reproductive toxicity, with perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS) being the most extensively studied. These chemicals are known to lower testosterone levels and compromise the integrity of the blood-testis barrier. However, the specific mechanisms of their reproductive toxicity remain largely unknown due to research limitations. In this study, we utilized network pharmacology to pinpoint the core genes and signaling pathways implicated in the reproductive toxicity caused by PFOA and PFOS. Molecular docking was employed to validate the interactions between these compounds and their targets. Key targets identified include CCL2, CXCR4, RPS27A, RPL5, PSMA7, and PSMC1, which are crucial in mediating reproductive toxicity. These genes are primarily involved in the chemokine signaling pathway, viral protein interactions with cytokines and cytokine receptors, and ribosomal functions. This study underscores the effectiveness of combining network toxicology and molecular docking to analyze the toxicity and molecular mechanisms of mixed environmental pollutants.
Keywords: Perfluorooctanoic acid, Perfluorooctane sulfonate, Network pharmacology, Molecular docking, Reproductive toxicity
Graphical abstract

Highlights
-
•
Network toxicology and rapid molecular docking are effective for assessing the toxicity and potential molecular mechanisms of perfluorinated compounds.
-
•
The reproductive toxicity of PFOA and PFOS was analyzed using these techniques.
-
•
Using network toxicology and molecular docking for toxicity assessment can address cost and ethical concerns related to animal use.
1. Introduction
Perfluorinated compounds (PFCs) comprise more than 3000 individual compounds and generally contain a charged functional moiety, such as a sulfonate or carboxylate [1]. A wide range of industrial and household products have been pplied to PFCs for their good physicochemical properties, including textiles, surfactants, furniture, food packaging and lubricants. Despite these advantages, PFCs pose serious long-term risks to the environment and human health [2]. Their toxic effects likely involve multiple targets and pathways, disrupting protein and gene networks rather than just altering individual proteins or genes [3]. Additionally, since consumers often ingest residues of various active substances through their diet, it is important to assess the potential combined effects of these substances [4]. Therefore, innovative and systematic methods are needed to evaluate the health risks of the increasing number of PFCs effectively.
Perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS) are PFCs widely utilized in various industries [5]. Their persistence in the environment and accumulation in the food chain have sparked significant concern [6,7]. PFOA and PFOS have been found in the tissues and plasma of numerous animal species across almost all continents, including polar bears, fish, and birds in North America [8]. Based on epidemiological studies, some scholars have developed health-based guidance values for PFOA and PFOS in the Chinese population, which are 1.52 and 1.56 ng/kg/day, respectively [9]. Despite new alternatives, similar environmental problems continue to arise. The elimination half-life of PFOA in humans is around four years, and for PFOS, it is approximately five years [10,11]. These compounds enter the body mainly through skin contact and ingestion, accumulating in various organs and causing toxicity [12]. Mammalian studies show that rodents exposed to PFOA and PFOS experience weight loss and lipid metabolism issues [13].
The impact of environmental pollutants on reproduction is a significant concern due to its effects on human development. Studies show that PFCs can damage testicular function and lower sperm count [14]. Based on epidemiological studies, some scholars have developed health-based guidance values for PFOA and PFOS in the Chinese population, which are 1.52 and 1.56 ng/kg/day, respectively [15]. Epidemiological research links maternal exposure to PFOA during pregnancy with poor semen quality in offspring [16]. In experiments with white-footed mice, high prenatal doses of PFOS resulted in neonatal death, while lower doses impaired growth and development [17]. For females, PFOA and PFOS may produce reproductive toxicity in the ways of damage to oocytes through oxidative stress and Inhibition of steroid hormone synthesis. Additionally, PFOA and PFOS are linked to reduced testosterone levels and compromised blood-testis barrier integrity in humans [18]. In women, these chemicals are associated with hormone imbalances and higher infertility risks [19]. Despite extensive studies on their reproductive effects in rodents [20,21], the exact mechanisms of PFOA and PFOS toxicity are not well understood.
This study aimed to uncover the core genes and signaling pathways involved in PFOA- and PFOS-induced reproductive toxicity using network pharmacology. We used molecular docking to validate interactions between these compounds and their targets, shedding light on their toxic mechanisms. This innovative approach provides a rapid method for assessing the reproductive toxicity of PFCs and offers new insights for treating diseases linked to these toxic substances.
2. Methods
2.1. Collection of potential targets for PFOA and PFOS
Limiting the analysis to "Homo sapiens," we identified potential targets for PFOA and PFOS from the ChEMBL database (https://www.ebi.ac.uk/chembl/) [22]. Obtaining the SMILE nodes of PFOA and PFOS from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [23], we then inputted these nodes into the Swiss Target Prediction database (http://www.swisstargetprediction.ch/) to gather overloaded targets [24]. Integrating and deduplicating the Uniprot IDs of all targets from the ChEMBL and Swiss Target Prediction databases, we standardized the potential targets using the Uniprot database (https://www.uniprot.org/id-mapping/) [25]. This process facilitated the construction of a potential target library for PFOA and PFOS.
2.2. Screening for reproductive toxicity targets
We searched for related targets using “reproductive toxicity” as the key word in the GeneCards (https://www.genecards.org/) and OMIM (https://www.omim.org/) databases [26], selecting genes with scores above the median to ensure strong correlation. Standardizing all targets using the Uniprot database specified "Homo sapiens" as the species for target and gene name conversion. Next, we identified common targets between PFOA/PFOS and reproductive toxicity by intersecting their respective target sets. Using the Venny 2.1.0 online platform (https://bioinfogp.cnb.csic.es/tools/venny/), we created a Venn diagram to visualize the overlap.
2.3. Construction of protein-protein interaction (PPI) network
Using the PPI network, we visualized the co-localization, neighborhood, and co-expression of interactions among potential target genes and predicted genes. Genes representing potential targets of PFOA/PFOS-induced reproductive toxicity were analyzed in the String database (https://string-db.org) to gain deeper insights into the toxic mechanisms of PFOA and PFOS. To ensure PPI accuracy and quantity, a confidence score exceeding 0.4 was required, with species limited to "Homo sapiens" [27]. The resulting PPI network was analyzed using Cytoscape software (version 3.10.1) for topology assessment, and core targets were identified based on their degree value ranking. This visualization method allowed for easy identification of key targets, as node size and color depth reflected numerical changes.
2.4. Analysis of target function and pathway enrichment
We utilized the DAVID online tool for Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to identify functional annotations and pathway enrichment related to potential genes. Following this, we performed visual analysis on the Bioinformatics online platform (https://www.bioinformatics.com.cn/) to interpret and present the results of the GO and KEGG analyses.
2.5. Verification of interaction by molecular docking
A molecular docking approach was utilized to forecast the binding affinity between PFOA/PFOS and their identified core targets. The crystal structures of these core proteins were acquired from the RCSB Protein Data Bank (http://www.rcsb.org). The core target protein underwent modifications including dehydration, hydrogenation, and charge balancing using AutoDockTools1.5.7 software, followed by the docking experiment.
3. Results
3.1. Identification of targets of PFOA/PFOS-induced reproductive toxicity
244 potential target proteins of PFOA and PFOS were sourced from the ChEMBL and Swiss Target Prediction databases, integrated through Uniprot. Additionally, 3403 targets highly relevant to reproductive toxicity were screened from GeneCards and OMIM. Utilizing the Venny 2.1.0 online platform, 82 common targets of PFOA/PFOS and reproductive toxicity were identified, detailed in Fig. 1. These 82 targets may play a significant role in PFOA/PFOS-induced reproductive toxicity.
Fig. 1.
Venn diagram of potential targets of PFOA/PFOS and reproductive toxicity.
3.2. PPI network construction and core target screening
Eighty-two potential targets related to reproductive toxicity from PFOA and PFOS were analyzed using the String database, generating a PPI network with 436 edges and an average node degree of 10.6. This network's structure allowed for an in-depth examination of how PFOA and PFOS induce reproductive toxicity. The results are shown in Fig. 2, where node size and color indicated different aspects of node importance.
Fig. 2.
PPI network of the potential action targets of PFOA and PFOS induced reproductive toxicity. The darker colors, larger fonts, and larger shapes of genes indicate higher betweenness, closeness, and degree values.
In the PPI network, the darker the color and the larger the node area, which indicate the node is more important. Six core targets crucial for PFOA/PFOS-induced reproductive toxicity were identified through topological analysis based on their degree values. These targets include Ribosomal protein S27a (RPS27A), C-X-C chemokine receptor type 4 (CXCR4), C-C motif chemokine 2 (CCL2), Ribosomal Protein L5 (RPL5), Proteasome 26S subunit ATPase 1 (PSMC1), and proteasome subunit alpha type-7 (PSMA7) (Table 1). They are expressed across various tissues and organs, playing critical roles in regulating transcription, protein modification, and inflammatory responses.
Table 1.
Core targets screened from PPI network.
| Gene | Degree | Closeness centrality | Betweenness centrality | Topological coefficient |
|---|---|---|---|---|
| RPS27A | 28 | 0.506666 | 0.129092 | 0.293126 |
| CXCR4 | 25 | 0.550724 | 0.162562 | 0.199384 |
| CCL2 | 24 | 0.493506 | 0.080804 | 0.258986 |
| RPL5 | 22 | 0.434285 | 0.039878 | 0.390442 |
| PSMC1 | 22 | 0.431818 | 0.011701 | 0.449631 |
| PSMA7 | 21 | 0.493506 | 0.073648 | 0.320987 |
3.3. Functional and pathway enrichment analysis of core targets
In this study, 82 key targets were analyzed for GO enrichment using the David online tool, revealing 133 biological processes (BP), 43 cellular components (CC), and 44 molecular functions (MF) in GO entries. Fig. 3 presents the top 10 entries of −lgP values for BP, CC, and MF. Biological processes related to reproductive toxicity induced by PFOA and PFOS primarily involve mRNA stability regulation and calcium-mediated signaling. The analysis of cellular components showed significant enrichment of genes related to the secretory granule lumen and nucleoplasm. Molecular function analysis revealed close association with virus receptor activity and metalloaminopeptidase activity.
Fig. 3.
GO enrichment analysis of the core genes of PFOA and PFOS induced reproductive toxicity.
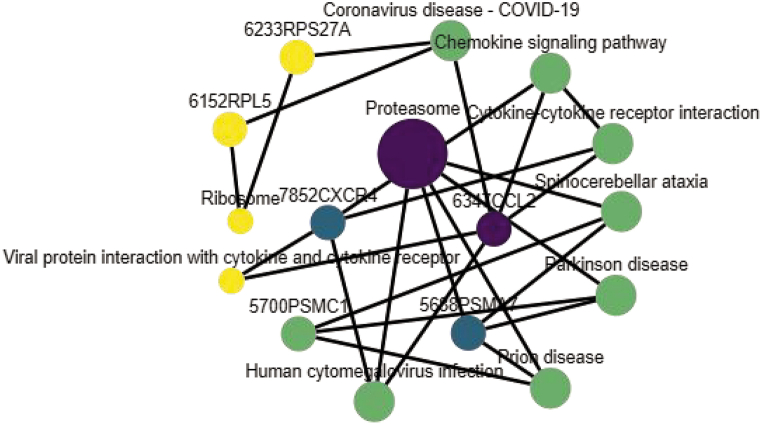
Additionally, 82 potential targets underwent KEGG analysis using the David database to identify their involvement in specific signaling pathways. This analysis yielded 30 KEGG pathway enrichment analyses, with the top 20 items shown in the bubble plot of Fig. 4 based on the −lgP value. Key targets were primarily enriched in pathways such as the chemokine signaling pathway, ribosome, viral protein interaction with cytokine and cytokine receptor, and proteasome. The proteasome pathway is particularly significant as the main degradation pathway for misfolded proteins and those undergoing proteolysis during protein synthesis. Its functions include cell cycle control, apoptosis, and inflammation, all relevant to reproductive toxicity. Continuous observation of the entire life cycle of animals is crucial for understanding reproductive toxicity, highlighting the importance of proteasomes in this process. A network relationship diagram of KEGG pathways and core targets was created using the top 10 KEGG enrichment pathways and 6 core targets, illustrating the relationship between each pathway and genes (Fig. 5).
Fig. 4.
KEGG enrichment analysis of the core genes of PFOA and PFOS induced reproductive toxicity.
Fig. 5.
Network relationship diagram of KEGG pathway and core genes.
3.4. Molecular docking verification
Molecular docking analysis examined how PFOA/PFOS interact with six core targets (CCL2, CXCR4, RPS27A, RPL5, PSMA7, and PSMC1) identified in the PPI network. And the scoring equation included the sum of several effects such as electrostatic interaction, van der Waals’ force, hydrogen bonding contributions and intermolecular conflicts. Studies have shown that stable complexes are formed when the free binding energy is negative. A lower binding energy suggests a higher probability of ligand receptor binding. The results indicated strong binding between PFOA/PFOS and all six core target proteins, with binding energies below 0 (Fig. 6). For example, among them, the binding affinity of PFOA to CXCR4 was the strongest, with a value of −13.16 kcal/mol. PFOA formed 4 hydrogen bonds with ASP-122 and ASP-120 in the docking pocket of CXCR4. These core targets may play a critical role in the molecular mechanism of reproductive toxicity induced by PFOA and PFOS.
Fig. 6.
Molecular docking analysis of PFOA/PFOS and the core genes. (A) The results of ligand-target binding energy values. (B) PFOA-CCL2, (C) PFOA-CXCR4, (D) PFOA-RPS27A, (E) PFOA-RPL5, (F) PFOA-PSMA7, (G) PFOA-PSMC1, (H) PFOS-CCL2, (I) PFOS-CXCR4, (J) PFOS-RPS27A, (K) PFOS-RPL5, (L) PFOS-PSMA7, (M) PFOS-PSMC1.
4. Discussion
PFOA and PFOS, widely used in consumer and industrial products, have raised concerns due to their persistence in the environment and harmful effects on humans and animals [28]. Evidence increasingly connects PFC exposure to reproductive toxicity, including reduced testosterone levels akin to those seen in infertile men [29]. However, current research on PFOA and PFOS is constrained, focusing on their toxicological effects rather than fully understanding their reproductive toxicity mechanisms. Using network pharmacology, our study identified six core genes—CCL2, CXCR4, RPS27A, RPL5, PSMA7, and PSMC1—as potential targets of PFOA/PFOS-induced reproductive toxicity, confirmed by molecular docking.
In this study, we analyzed the GO molecular functions and KEGG pathway of the reproductive toxicity-related targets and validated them by molecular docking techniques. Enrichment analysis revealed several important pathways, including chemokine signaling pathway, ribosome, viral protein interaction with cytokine and cytokine receptor, and proteasome. Regarding the chemokine pathway, the core targets we obtained, CCL2 and CXCR4, also belong to chemokines. And for the ribosome pathway, the core targets RPS27A and RPL5 are also associated with ribosomes. Moreover, the research indicates that core genes related to PFOA and PFOS-induced reproductive toxicity are also closely associated with chemokines and protein synthesis mediated by different molecules, such as core targets RPS27A and RPL5.
Ribosomes, essential for protein synthesis, contain RPS27A, a subunit of ribosomal protein 40S. RPS27A plays key roles in ribosome formation, protein synthesis regulation [30], transcriptional control, and protein modifications [31], contributing to sperm vitality and potentially impacting reproduction. RPL5, involved in various cancers, is linked to ribosome defects associated with cancer development [32]. In many tumors, RPL5 heterozygote inactivation rates are high, with evidence suggesting an anti-tumor effect in breast cancer, which is influenced by reproductive factors [33].
CXCR4 and CCL2, core targets within the chemokine class, guide cell movement by binding to and activating cell surface receptors, initiating signal transduction toward the chemokine's gradient [34]. Initial studies on the chemokine stromal derived factor 1 (CXCL12) were proposed to be enhanced in several diseases including those which affect the female reproductive tract, While its well-known receptor was CXCR4. These include endometriosis, Asherman's syndrome, endometrial cancers, and ovarian cancers. Being stimulated by the chemokine CXCL12, the CXCR4/CXCR7 cascade is involved in tumor proliferation, migration, and metastasis [35]. This migration is crucial for many biological processes, with CXCR4 recruiting immune cells and contributing to inflammation, while CCL2 stimulates tumor cell proliferation [36], impacting breast development and cancer progression [37].
PSMA7, a core subunit of the 20S proteasome's α subunit, regulates intracellular protein degradation via the ubiquitin proteasome system. Reduced PSMA7 and acylaminoacyl peptide hydrolase (APEH) expression decreases proteasome activity, impairing protein degradation and inhibiting high-grade serous ovarian cancer growth [38]. In contrast, the 26S proteasome subunit ATPase (PSMC) family, including PSMC1-6, forms a heterohexamer loop and is part of the 19S regulatory particles. Silencing PSMC genes can impede prostate cancer development and metastasis by affecting processes such as promotion, apoptosis, and migration [39].
Exposure of developing oocytes to PFCs may disrupt embryonic development and reproductive outcomes. Environmental endocrine disruptors have been linked to adverse effects on the female reproductive system, potentially increasing the risk of tumors in hormone-sensitive organs such as the breast, ovaries, and endometrium [40]. The majority of the six core targets identified through PPI network analysis are linked to tumor development. KEGG analysis has identified three significant cancer-related pathways: the chemokine signaling pathway, ribosome pathway, and viral protein interaction with cytokine and cytokine receptor pathway. These results suggest a strong association between PFOA/PFOS-induced reproductive toxicity and cancer, particularly in hormone-sensitive organs.
The combination of network toxicology and molecular docking utilizes advances in bioinformatics, genomics, and big data to explore the mechanisms of mixed environmental pollutants. This methodology enhances the efficiency of ecotoxicological research. However, current investigations, including our own, are constrained by the reliance on computational techniques to understand the molecular mechanisms behind the reproductive toxicity of PFOA and PFOS. These mechanisms require further validation through pharmacological and clinical studies. However, research based on network pharmacology and molecular docking technology only qualitatively predicts the toxic targets of ingredients, and clear physiological effects still need to be verified through animal experiments or even clinical trials. Given these inherent limitations, we believe that the combined molecular docking method of network pharmacology is only suitable to evaluate certain pollutants that directly interact with proteins expressed by certain genes.
5. Conclusions
In this study, a novel approach combining network pharmacology and molecular docking was used to explore the targets and pathways involved in PFOA/PFOS-induced reproductive toxicity. Core targets such as CCL2, CXCR4, RPS27A, RPL5, PSMA7, and PSMC1 are implicated in mediating reproductive toxicity, primarily through pathways like chemokine signaling, viral protein interaction with cytokines, and ribosomal pathways. Molecular docking further supported the association between PFOA/PFOS and these targets. This study represents a significant step forward in analyzing the toxicity and molecular mechanisms of potential mixed environmental pollutants using network toxicology and molecular docking.
CRediT authorship contribution statement
Liang Chen: Writing – original draft, Funding acquisition, Formal analysis, Conceptualization. Shanshan Liang: Investigation, Formal analysis, Conceptualization. Jiaxin Li: Investigation. Qian Li: Conceptualization. Qingwen Sun: Writing – review & editing, Funding acquisition.
Data availability statement
Data will be made available on request.
Funding statement
The work was supported by the National Natural Science Foundation of China (82160803), and High level Innovative Talents Project of Guizhou Province (Guizhou Science and Technology Cooperation Platform Talents-GCC [2023] 077).
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Liang Chen reports financial support, article publishing charges, equipment, drugs, or supplies, and writing assistance were provided by National Natural Science Foundation of China. Qingwen Sun reports financial support, article publishing charges, and equipment, drugs, or supplies were provided by High level Innovative Talents Project of Guizhou Province. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Cai Y., Wang Q., Zhou B., Yuan R., Wang F., Chen Z., Chen H. A review of responses of terrestrial organisms to perfluorinated compounds. Sci. Total Environ. 2021;793 doi: 10.1016/j.scitotenv.2021.148565. [DOI] [PubMed] [Google Scholar]
- 2.Wu J.Y., Hua Z.L., Gu L., Li X.Q., Gao C., Liu Y.Y. Perfluorinated compounds (PFCs) in regional industrial rivers: interactions between pollution flux and eukaryotic community phylosymbiosis. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111876. [DOI] [PubMed] [Google Scholar]
- 3.Liu X., Zhu Y., Liu T., Xue Q., Tian F., Yuan Y., Zhao C. Exploring toxicity of perfluorinated compounds through complex network and pathway modeling. J. Biomol. Struct. Dyn. 2020;38:2604–2612. doi: 10.1080/07391102.2019.1637281. [DOI] [PubMed] [Google Scholar]
- 4.Heise T., Schmidt F., Knebel C., Rieke S., Haider W., Geburek I., Niemann L., Marx-Stoelting P. Hepatotoxic combination effects of three azole fungicides in a broad dose range. Arch. Toxicol. 2018;92:859–872. doi: 10.1007/s00204-017-2087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassan H.F., Bou Ghanem H., Abi Kharma J., Abiad M.G., Elaridi J., Bassil M. Perfluorooctanoic acid and perfluorooctane sulfonate in human milk: first survey from Lebanon. Int. J. Environ. Res. Public Health. 2023;20:821. doi: 10.3390/ijerph20010821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierozan P., Cattani D., Karlsson O. Perfluorooctane sulfonate (PFOS) and perfluorooctanoic acid (PFOA) induce epigenetic alterations and promote human breast cell carcinogenesis in vitro. Arch. Toxicol. 2020;94:3893–3906. doi: 10.1007/s00204-020-02848-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson S.L., Zeng X., Guan W., Sundaram R., Mendola P., Putnick D.L., Waterland R.A., Gunasekara C.J., Kannan K., Gao C., Bell E.M., Yeung E.H. Perfluorooctanoic acid (PFOA) or perfluorooctane sulfonate (PFOS) and DNA methylation in newborn dried blood spots in the Upstate KIDS cohort. Environ. Res. 2021;194 doi: 10.1016/j.envres.2020.110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jantzen C.E., Annunziato K.A., Bugel S.M., Cooper K.R. PFOS, PFNA, and PFOA sub-lethal exposure to embryonic zebrafish have different toxicity profiles in terms of morphometrics, behavior and gene expression. Aquat. Toxicol. 2016;175:160–170. doi: 10.1016/j.aquatox.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Luo Z.M., Huang Y., Wang J.W., Ouyang G. Recent trends in degradation strategies of PFOA/PFOS substitutes. Chemosphere. 2023;315 doi: 10.1016/j.chemosphere.2022.137653. [DOI] [PubMed] [Google Scholar]
- 10.Boesen S.A.H., Long M., Wielsøe M., Mustieles V., Fernandez M.F., Bonefeld-Jørgensen E.C. Exposure to Perflouroalkyl acids and foetal and maternal thyroid status: a review. Environ. Health. 2020;19:107. doi: 10.1186/s12940-020-00647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratier A., Casas M., Grazuleviciene R., Slama R., Småstuen Haug L., Thomsen C., Vafeiadi M., Wright J., Zeman F.A., Vrijheid M., Brochot C. Estimating the dynamic early life exposure to PFOA and PFOS of the HELIX children: emerging profiles via prenatal exposure, breastfeeding, and diet. Environ. Int. 2024;186 doi: 10.1016/j.envint.2024.108621. [DOI] [PubMed] [Google Scholar]
- 12.Liang L., Pan Y., Bin L., Liu Y., Huang W., Li R., Lai K.P. Immunotoxicity mechanisms of perfluorinated compounds PFOA and PFOS. Chemosphere. 2022;291 doi: 10.1016/j.chemosphere.2021.132892. [DOI] [PubMed] [Google Scholar]
- 13.Wang L., Wang Y., Liang Y., Li J., Liu Y., Zhang J., Zhang A., Fu J., Jiang G. PFOS induced lipid metabolism disturbances in BALB/c mice through inhibition of low density lipoproteins excretion. Sci. Rep. 2014;4:4582. doi: 10.1038/srep04582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi W., Zhang Z., Li M., Dong H., Li J. Reproductive toxicity of PFOA, PFOS and their substitutes: a review based on epidemiological and toxicological evidence. Environ. Res. 2024;250 doi: 10.1016/j.envres.2024.118485. [DOI] [PubMed] [Google Scholar]
- 15.Wan H.T., Lai K.P., Wong C.K.C. Comparative analysis of PFOS and PFOA toxicity on sertoli cells. Environ. Sci. Technol. 2020;54:3465–3475. doi: 10.1021/acs.est.0c00201. [DOI] [PubMed] [Google Scholar]
- 16.Vested A., Ramlau-Hansen C.H., Olsen S.F., Bonde J.P., Kristensen S.L., Halldorsson T.I., Becher G., Haug L.S., Ernst E.H., Toft G. Associations of in utero exposure to perfluorinated alkyl acids with human semen quality and reproductive hormones in adult men. Environ. Health Perspect. 2013;121:453–458. doi: 10.1289/ehp.1205118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narizzano A.M., Lent E.M., Hanson J.M., East A.G., Bohannon M.E., Quinn M.J., Jr. Reproductive and developmental toxicity of perfluorooctane sulfonate (PFOS) in the white-footed mouse (Peromyscus leucopus) Reprod. Toxicol. 2022;113:120–127. doi: 10.1016/j.reprotox.2022.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Cheng X., Wei Y., Zhang Z., Wang F., He J., Wang R., Xu Y., Keerman M., Zhang S., Zhang Y., Bi J., Yao J., He M. Plasma PFOA and PFOS levels, DNA methylation, and blood lipid levels: a Pilot study. Environ. Sci. Technol. 2022;56:17039–17051. doi: 10.1021/acs.est.2c04107. [DOI] [PubMed] [Google Scholar]
- 19.Heffernan A.L., Cunningham T.K., Drage D.S., Aylward L.L., Thompson K., Vijayasarathy S., Mueller J.F., Atkin S.L., Sathyapalan T. Perfluorinated alkyl acids in the serum and follicular fluid of UK women with and without polycystic ovarian syndrome undergoing fertility treatment and associations with hormonal and metabolic parameters. Int. J. Hyg Environ. Health. 2018;221:1068–1075. doi: 10.1016/j.ijheh.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Conley J.M., Lambright C.S., Evans N., Medlock-Kakaley E., Dixon A., Hill D., McCord J., Strynar M.J., Ford J., Gray L.E., Jr. Cumulative maternal and neonatal effects of combined exposure to a mixture of perfluorooctanoic acid (PFOA) and perfluorooctane sulfonic acid (PFOS) during pregnancy in the Sprague-Dawley rat. Environ. Int. 2022;170 doi: 10.1016/j.envint.2022.107631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxwell D.L., Oluwayiose O.A., Houle E., Roth K., Nowak K., Sawant S., Paskavitz A.L., Liu W., Gurdziel K., Petriello M.C., Richard Pilsner J. Mixtures of per- and polyfluoroalkyl substances (PFAS) alter sperm methylation and long-term reprogramming of offspring liver and fat transcriptome. Environ. Int. 2024;186 doi: 10.1016/j.envint.2024.108577. [DOI] [PubMed] [Google Scholar]
- 22.Huang S. A novel strategy for the study on molecular mechanism of prostate injury induced by 4,4'-sulfonyldiphenol based on network toxicology analysis. Journal of applied toxicology : JAT. 2024;44:28–40. doi: 10.1002/jat.4506. [DOI] [PubMed] [Google Scholar]
- 23.Guo B., Zhao C., Zhang C., Xiao Y., Yan G., Liu L., Pan H. Elucidation of the anti-inflammatory mechanism of Er Miao San by integrative approach of network pharmacology and experimental verification. Pharmacol. Res. 2022;175 doi: 10.1016/j.phrs.2021.106000. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y., Liu X., Li G. Integrated bioinformatics and network pharmacology to identify the therapeutic target and molecular mechanisms of Huangqin decoction on ulcerative Colitis. Sci. Rep. 2022;12:159. doi: 10.1038/s41598-021-03980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X., Zhang M., Song Y., Sun B., Lin L., Song X., Li C. Integrated network pharmacology to investigate the mechanism of Salvia miltiorrhiza Bunge in the treatment of myocardial infarction. J. Cell Mol. Med. 2023;27:3514–3525. doi: 10.1111/jcmm.17932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Q., Liu C., Wang X., Rong K., Zhu M., Duan L., Zheng P., Mi Y. Exploring the mechanism of curcumin in the treatment of colon cancer based on network pharmacology and molecular docking. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F., Gu Y., Yan Y., Wang G. Based on network pharmacology and molecular docking to predict the mechanism of TMDZ capsule in the treatment of IS. Medicine. 2023;102 doi: 10.1097/MD.0000000000034424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Z., Wen Y., Wang B., Deng S., Zhang F., Fu Z., Yuan Y., Zhang D. Toxic effects of per- and polyfluoroalkyl substances on sperm: epidemiological and experimental evidence. Front. Endocrinol. 2023;14 doi: 10.3389/fendo.2023.1114463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarapore P., Ouyang B. Perfluoroalkyl chemicals and Male reproductive health: Do PFOA and PFOS Increase risk for Male infertility? Int. J. Environ. Res. Public. Health. 2021;18:3794. doi: 10.3390/ijerph18073794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montellese C., van den Heuvel J., Ashiono C., Dörner K., Melnik A., Jonas S., Zemp I., Picotti P., Gillet L.C., Kutay U. USP16 counteracts mono-ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit. Elife. 2020;9 doi: 10.7554/eLife.54435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J., Zhao H., Chen L., Liu M. Multifaceted functions of RPS27a: an unconventional ribosomal protein. J. Cell. Physiol. 2023;238:485–497. doi: 10.1002/jcp.30941. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H., Liu J., Dang Q., Wang X., Chen J., Lin X., Yang N., Du J., Shi H., Liu Y., Han J. Ribosomal protein RPL5 regulates colon cancer cell proliferation and migration through MAPK/ERK signaling pathway. BMC Mol. Cell. Biol. 2022;23:48. doi: 10.1186/s12860-022-00448-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fancello L., Kampen K.R., Hofman I.J., Verbeeck J., De Keersmaecker K. The ribosomal protein gene RPL5 is a haploinsufficient tumor suppressor in multiple cancer types. Oncotarget. 2017;8:14462–14478. doi: 10.18632/oncotarget.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozga A.J., Chow M.T., Luster A.D. Chemokines and the immune response to cancer. Immunity. 2021;54:859–874. doi: 10.1016/j.immuni.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin R., Ren W., Ya G., Wang B., He J., Ren S., Jiang L., Zhao S. Role of chemokines in the crosstalk between tumor and tumor-associated macrophages. Clin. Exp. Med. 2023;23:1359–1373. doi: 10.1007/s10238-022-00888-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu M., Wang Y., Xia R., Wei Y., Wei X. Role of the CCL2-CCR2 signalling axis in cancer: mechanisms and therapeutic targeting. Cell Prolif. 2021;54 doi: 10.1111/cpr.13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller A.K., Köhler U.A., Trzebanski S., Vinik Y., Raj H.M., Girault J.A., Ben-Chetrit N., Maraver A., Jung S., Lev S. Mouse modeling dissecting macrophage-breast cancer communication uncovered roles of PYK2 in macrophage recruitment and breast tumorigenesis. Adv. Sci. 2022;9 doi: 10.1002/advs.202105696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tangri A., Lighty K., Loganathan J., Mesmar F., Podicheti R., Zhang C., Iwanicki M., Drapkin R., Nakshatri H., Mitra S. Deubiquitinase UCHL1 Maintains protein Homeostasis through the PSMA7-APEH-proteasome Axis in high-grade serous ovarian Carcinoma. Mol. Cancer Res. 2021;19:1168–1181. doi: 10.1158/1541-7786.MCR-20-0883. [DOI] [PubMed] [Google Scholar]
- 39.Chen Q., Fu L., Hu J., Guo G., Xie A. Silencing of PSMC2 inhibits development and metastasis of prostate cancer through regulating proliferation, apoptosis and migration. Cancer Cell Int. 2021;21:235. doi: 10.1186/s12935-021-01934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czarnywojtek A., Jaz K., Ochmańska A., Zgorzalewicz-Stachowiak M., Czarnocka B., Sawicka-Gutaj N., Ziółkowska P., Krela-Kaźmierczak I., Gut P., Florek E., Ruchała M. The effect of endocrine disruptors on the reproductive system - current knowledge. ur Rev. Med. Pharmacol. Sci. 2021;25:4930–4940. doi: 10.26355/eurrev_202108_26450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.