Summary
This protocol describes the preparation of a nanoparticle-encapsulated bioink capable of protecting tissue-engineered constructs against bacterial infections while also providing contrast for magnetic resonance (MR) imaging modalities. The report includes details of preparing the methacrylated gelatin-based bioinks and the incorporation of superparamagnetic iron oxide nanoparticles. A detailed protocol is presented for characterizing the bioink, evaluating cell response, and assessing its antibacterial effect. Overall, this article presents a robust approach for the development of antibacterial, MR-visible bioinks suitable for various tissue engineering applications.
For complete details on the use and execution of this protocol, please refer to Theus et al.1
Subject areas: tissue engineering, biotechnology and bioengineering
Graphical abstract
Highlights
-
•
Steps for developing bioinks with bacteriostatic functionality
-
•
Procedure for optimizing scaffold properties at 200 μg/mL concentration
-
•
Protocol for adding nanoparticles to enable noninvasive scaffold imaging
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
This protocol describes the preparation of a nanoparticle-encapsulated bioink capable of protecting tissue-engineered constructs against bacterial infections while also providing contrast for magnetic resonance (MR) imaging modalities. The report includes details of preparing the methacrylated gelatin-based bioinks and the incorporation of superparamagnetic iron oxide nanoparticles. A detailed protocol is presented for characterizing the bioink, evaluating cell response, and assessing its antibacterial effect. Overall, this article presents a robust approach for the development of antibacterial, MR-visible bioinks suitable for various tissue engineering applications.
Before you begin
Cell culture
-
1.Culture of human umbilical vein endothelial cells (HUVECs).
-
a.Warm up VascuLife VEGF Endothelial Medium, Eagle’s Minimum Essential Medium, fetal bovine serum, penicillin/ streptomycin (Pen/Strep) solution, 0.05% trypsin-EDTA solution, phosphate buffered saline (PBS) to 37°C for 30 min before start.
-
b.Culture HUVECs in VascuLife VEGF Endothelial Medium + 1%(v/v) penicillin/streptomycin (Pen/Strep) supplement in T75 ventilated flasks. Change media with fresh warm media every 2–3 days.
-
c.After reaching 90%–95% confluency, wash HUVECs with phosphate buffer saline (PBS) and treat them with 1 mL warm 0.05% trypsin-EDTA for 2–5 min at 37°C for cell detachment.
-
d.Neutralize trypsin-EDTA with 2 mL warm medium, harvest cell suspension, and centrifuge at 300 × g for 5 min at 20–25°C.
-
e.Resuspend the cell pellet with fresh warm medium and mix 10 μL cell suspension with a 10 μL aliquot of trypan blue at a 1:1 ratio for counting the cells.
-
f.After counting, add fresh media and dilute to the desired concentration. Add 5×105 cells in a T75 flask, along with 10 mL fresh media for passaging. Use appropriate aliquots of the cell suspension for subcultures.
-
a.
-
2.Culture of NIH/3T3 fibroblast cell line.
-
a.Culture NIH/3T3 fibroblasts in Eagle’s Minimum Essential Medium supplemented with 10%(v/v) fetal bovine serum (FBS) and 1%(v/v) Pen/Strep supplement in T75 ventilated flasks. Change media with fresh warm media every 2–3 days.
-
b.After reaching 90%–95% confluency, wash NIH/3T3 fibroblasts with PBS and treat them with 2 mL warm trypsin-EDTA for 5–7 min at 37°C for detachment.
-
c.Neutralize trypsin-EDTA with 4 mL warm medium. harvest cell suspension and centrifuge at 300 × g for 5 min at 20–25°C.
-
d.Resuspend the cell pellet with fresh warm medium and mix 10 μL cell suspension with a 10 μL aliquot of trypan blue at a 1:1 ratio for counting the cells.
-
e.Add fresh medium to dilute to desired concentration. Add appropriate aliquots of the cell suspension to new culture vessels for subculture.
-
a.
Media preparation
-
3.Preparation of Luria-Bertani (LB) medium.
-
a.For each 950 mL of deionized (DI) water, add 25 g of premixed LB formulation and mix until dissolved.
-
b.Balance the pH of the solution to ∼7.4–7.6 using 1N NaOH.
-
c.Adjust the final volume of the solution to 1 L.
-
d.Autoclave the solution using liquid cycle at 121.1°C for 30 min.
-
e.Store at 20–25°C.
-
a.
Preparation of general lab items
-
4.Preparation of LB agar for culturing bacteria on plates.
-
a.Add 25 g of premixed LB formulation and 15 g agar for each 1 L of deionized water.
-
b.Mix until fully dissolved.
-
c.Autoclave the solution using liquid cycle at 121.1°C for 30 min.
-
d.After the medium is cooled to a temperature in which it is still liquid but safe enough to handle, pour 50 mL LB agar from bottles into plates.
-
e.Leave plates in biosafety cabinet until agar sets.
-
f.When agar has set, replace the lid, invert the plates, and store in an airtight bag at 4°C.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD31 antibody (dilution: 1:200) | Invitrogen | Cat# ENMA3100 |
| Anti-connexin43 antibody (dilution: 1:200) | Invitrogen | Cat# 71-070-0 |
| Anti F-actin antibody (dilution: 1:200) | Invitrogen | Cat# MA1-80729 |
| DAPI (dilution: 1:1000) | Invitrogen | Cat# R37606 |
| Polyclonal anti-Staphylococcus aureus antibody (dilution: 1:200) | Bio-Rad | Cat# 0300-0084 |
| Alexa Fluor 488 polyclonal antibody (dilution: 1:200) | Invitrogen | Cat# A-11094 |
| Goat anti-rabbit IgG (H + L) cross-adsorbed secondary antibody, Alexa Fluor 555 (dilution: 1:200) | Invitrogen | Cat# A-21428 |
| Bacterial and virus strains | ||
| Staphylococcus aureus (S. aureus) | American Type Cell Culture (ATCC) | Cat# 43300 |
| Chemicals, peptides, and recombinant proteins | ||
| Gelatin from porcine skin, Bloom 300 | Sigma-Aldrich | Cat: G2500-1KG |
| Methacrylic anhydride | MilliporeSigma | Cas: 760-93-0 |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | MilliporeSigma | Cas: 85073-19-4 |
| Superparamagnetic iron oxide nanoparticles (SPIONs) | Micromod Partikeltechnologie GmbH | 45-00-252 |
| Antibiotic-Antimycotic | Thermo Fisher Scientific | 15240062 |
| VascuLife VEGF Endothelial Medium Complete Kit | Lifeline Cell Technology | Cat# LL-0003 |
| Eagle’s minimum essential medium | ATCC | Cat# 30-2003 |
| Fetal bovine serum (FBS) | ATCC | Cat# 30-2021 |
| Penicillin-Streptomycin solution 100x | ATCC | Cat# 30-2300 |
| Phosphate-buffered saline (PBS), 1X without calcium and magnesium | Corning | Cat # MT21040CM |
| Trypsin-EDTA (0.05%), phenol red | Gibco | Cat# 25300054 |
| Trypan blue | Bio-Rad | Cat# 1450021 |
| AlamarBlue cell viability reagent | Bio-Rad | Cat# BUF012B |
| Paraformaldehyde (PFA) 4% | Thermo Fisher Scientific | J61899.AP |
| Luria-Bertani (LB) medium | Fisher Scientific | MP113002036 |
| Sodium hydroxide (NaOH) | Sigma-Aldrich | 221465-500G |
| Agarose | Sigma-Aldrich | Cat# A9539-100G |
| 10% neutral buffered formalin | VWR International, Inc. | Cat# 89370-094 |
| Triton X-100 | IBI Scientific | Cat# IB07100 |
| Blocker bovine serum albumin (BSA) | VWR International | Cat# PI37535 |
| Antifade mounting medium | Fisher Scientific | Cat# D1306 |
| Experimental models: Cell lines | ||
| Human umbilical vein endothelial cells (HUVECs) | Lifeline Cell Technology | Cat# FC-0044 |
| NIH/3T3 fibroblasts | ATCC | Cat# CRL-1658 |
| Other | ||
| 500 mL Erlenmeyer flask | VWR | 75804-644 |
| Crystalizing dish | VWR | 470346-184 |
| Magnetic heater/stirrer | Sigma-Aldrich | Z671762-1EA |
| SnakeSkin dialysis tubing (dialysis tubing, 10K MWCO, 35 mm I.D.) | Thermo Fisher Scientific | 88245 |
| Freeze dryer −84°C | Labconco | N/A |
| Bright-field microscope | ||
| BioAssemblyBot 400 | Advanced Solutions | N/A |
| BioAssemblyBot 10 mL cartridges | Advanced Solutions | ASLS-0000133 |
| Mach-1 micromechanical system | Biomomentum | N/A |
| Spherical indenter (D = 0.5 mm) | Biomomentum | Cat# MA034 |
| Motion controller, ESP301, 3-Axis, GPIB, USB, RS232 | Newport | Cat #ESP301-3G |
| Kimwipes delicate task wipers | Kimberly-Clark | Cat# 34155 |
| T75 ventilated flask | VWR | 10062-860 |
| 6-well cell culture plate | Fisher Scientific | 07-200-80 |
| 10-cm Petri dish | VWR | 10416-320 |
| 96-well plate | VWR | 10861-696 |
| Centrifugal tube-15 mL (conical tube) | Greiner | Cat# 188261 |
| Centrifugal tube-50 mL (conical tube) | Greiner | Cat# 210261 |
| Biosafety (laminar flow) cabinet | N/A | N/A |
| Incubator with humidified atmosphere, at 37°C and 5% CO2 | N/A | N/A |
| Water bath with controlled temperature | N/A | N/A |
| Centrifuge | N/A | N/A |
| Confocal microscope | Olympus | Fv1000 |
| Microplate reader | BioTek | Cat# 18531 |
| Vibrating microtome (vibratome) | Leica | VT1200 |
| Tissue culture plate | Sigma-Aldrich | CLS430196 |
| Glass slides | VWR | 48311–703 |
| Glass coverslips | VWR | 10118–642 |
| Nail polish | N/A | N/A |
Step-by-step method details
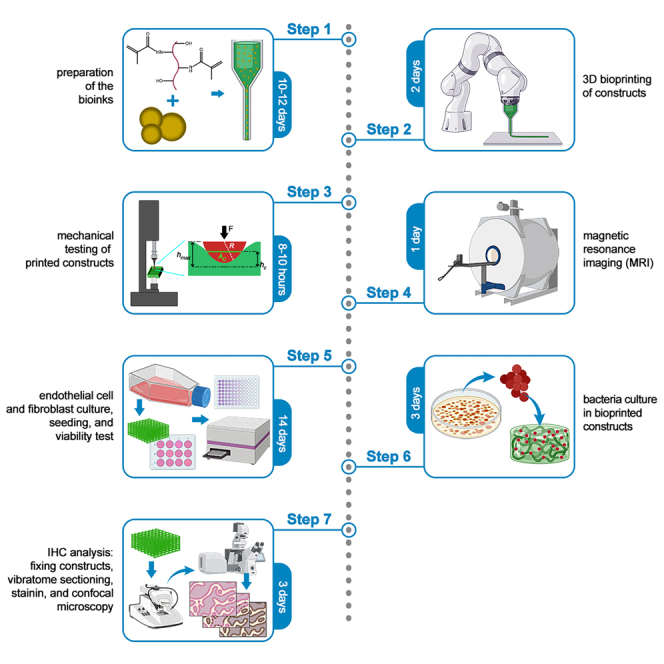
Preparation of the bioinks
Timing: 10–12 days
This section focuses on development and preparation of the SPION encapsulated GelMA bioinks, including synthesis of GelMA, reconstitution of lyophilized GelMA and incorporation of SPIONs within it.
-
1.Synthesis of methacrylated gelatin (GelMA).
-
a.Fill a 2-L crystalizing dish with deionized water and warm up to 50°C as a water bath.
-
b.Warm up 100 mL PBS in a 500 mL Erlenmeyer flask to 50°C.
-
c.Add 10 g of gelatin from porcine skin to the PBS.
-
d.Stir using a magnetic stirrer at 500 RPM for 1 h at 50°C.
 CRITICAL: After 1 h the gelatin should be completely dissolved, resulting in a transparent solution with no visible clumps. If dissolution is not complete, continue stirring for another hour or until gelatin is fully dissolved.
CRITICAL: After 1 h the gelatin should be completely dissolved, resulting in a transparent solution with no visible clumps. If dissolution is not complete, continue stirring for another hour or until gelatin is fully dissolved. -
e.Add an additional 150 mL PBS to the gelatin solution and stir for another 30 min.
-
f.Move the items under a chemical fume hood, add 8 mL methacrylic anhydride (MA) in a dropwise manner to the gelatin solution.
-
g.Allow the reaction to go on under stirring at 50°C for 3 h.
-
h.After the reaction, transfer the product to 50 mL conical tubes and centrifuge at 300 × g for 4 min at 20–25°C.
-
i.Transfer the supernatant to an Erlenmeyer flask and add 150 mL fresh PBS.
-
j.Dialyze the product against DI water for 5 days using dialysis membrane with a molecular weight cutoff of 10 kDa at 50°C. Change the water three times per day.
-
k.Transfer the dialyzed product to 50 mL conical tubes, freeze at −80°C for 10–15 h, and freeze dry for 4 days to obtain the final product.
-
l.Store the final product in −20°C freezer until further use.
 CRITICAL: Due to expansion of water upon freezing, the conical tubes must not be completely filled to avoid cracks and breaking.
CRITICAL: Due to expansion of water upon freezing, the conical tubes must not be completely filled to avoid cracks and breaking.
-
a.
-
2.Reconstitution of GelMA.
-
a.Remove the synthesized GelMA from freezer, weigh, and calculate the amount of PBS required to prepare a 10%(w/v) solution.
-
b.Dissolve 0.5%(w/v) lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) in the PBS by vortexing at 20–25°C for 2–3 min.
-
c.Move the PBS containing the LAP to a conical tube and immerse the lyophilized GelMA sponge in the solution.
-
d.Store the mixture at 37°C for the GelMA to fully dissolve.
-
e.Mix the superparamagnetic iron oxide nanoparticles (SPIONs) (commercially purchased) with the GelMA solution at a concentration of 100 μg/mL, 200 μg/mL, or 500 μg/mL.
-
f.Store the prepared bioink at 4°C for up to a week.
-
a.
3D bioprinting of the constructs
Timing: 2 days
This section uses the bioinks prepared in the previous section to perform 3D bioprinting, starting with the design, the process of printing, assessment of fidelity and printing of constructs for testing.
-
3.3D bioprinting.
-
a.Design the required 3D geometry using Autodesk Fusion 360 CAD (or other relevant) software and export the design as Standard Triangle Language (STL) file format.Note: A suggested standard design for further analysis would include a cube of 12 × 12 × 4 mm3, with a lattice infill and pore sizes of 1 mm.
-
b.Incubate the bioink at a 37°C water bath for 10–15 min for the GelMA to become liquid.
-
c.Transfer the bioink into the 10 mL cartridges for BioAssemblyBot (BAB) 3D bioprinter and insert the piston.
-
d.Arrange the 3D designed model and any replicates on the BAB stage in associated TSIM software and ensure that they do fit on the print stage.
-
e.Add the model to the TSIM library on the BAB using a flash drive or through network connection to the device.
-
f.Make sure that the BAB bioprinter is connected to the pressurized air source (cylinder or direct line) and turn it on.
-
g.Turn on the HEPA filter and fans.
-
h.Load the bioink cartridge into a printer “hand” (printhead) and load the hand.
-
i.Run nozzle size calibration.
-
j.Manually home the hand to the printing surface and save the coordinates.
-
k.Home the BAB from the bioprinter menu.
-
l.Set the pneumatic pressure.
-
m.Start the bioprinting session.Note: Optimal printing parameters for this bioink and design were identified to be a printing speed of 5 mm/s, pressure of 20 psi, temperature of 27°C, and layer height of 0.2 mm. Parameters can be optimized following characterization of prints and may be different at different times/locations.
-
n.Store the printed constructs in PBS supplemented with 2%(w/v) antibiotic-antimycotic at 4°C.
-
a.
-
4.Characterization of bioprints.
-
a.Optimization of bioprinting parameters can be done by measuring printing fidelity. To start, design and print a two-layer 10 × 10 mm grid structure with a line distance of 2 mm, following the steps in the protocol section 3.
-
b.Image the constructs using bright-field microscopy at 1.5X and 4X magnifications.
-
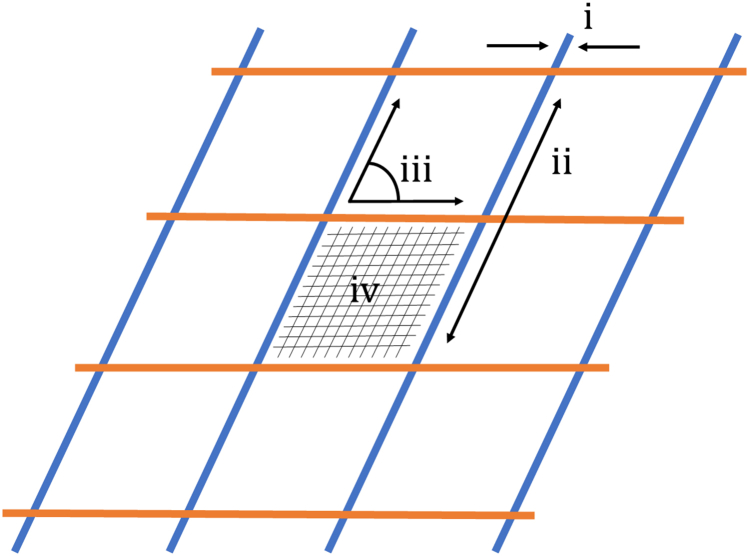
c.Transfer images to FIJI ImageJ software and measure the following parameters, also shown schematically in Figure 1.
-
i.Strand diameter: the ratio of printed (extruded) strand diameter to that in the CAD model.
-
ii.Strand length: the ratio of printed strand length to that in the CAD model.
-
iii.Strand angle: the ratio of the angle between printed strands compared to that in the CAD model.
-
iv.Inter-strand area: the ratio of the actual area constrained by bioprinted strands compared to that in the CAD model.
-
i.
-
d.Tune the bioprinting parameters (described above) to achieve fidelity ratios approaching a value of 1.0 for all different geometric parameters.
-
a.
Figure 1.
Schematic representation of fidelity measurements with measurement i being the line width measurement, ii the line length, iii the strand angle and iv the area enclosed by printed strands
Mechanical testing of bioprinted constructs
Timing: 8–10 h
This section focuses on assessing the stiffness of constructs bioprinted using different concentrations of SPIONs.
-
5.Microindentation of the bioprinted scaffolds.
-
a.Bioprint n = 5 samples with no SPIONs (control) and n = 5 samples for each different concentration of SPIONs (100, 200, and 500 μg/mL) following steps 2 and 3 of this protocol.
-
b.Using a spatula, place the sample on the stage of the Mach-1 micromechanical system.
-
c.Carefully remove excess water from the surface of the sample using Kimwipe tissue paper to get an accurate reading.
-
d.Move the stage using the movement screws to bring the point of interest right under the probe for indentation.
 CRITICAL: Avoid indenting on top of any non-uniformity (e.g., bubble or a hollow channel).
CRITICAL: Avoid indenting on top of any non-uniformity (e.g., bubble or a hollow channel). -
e.Bring the probe close to the surface using the “down” function in the manual controls section of the software.
 CRITICAL: Be careful to not actually touch the sample surface and only bring the probe as close as possible.
CRITICAL: Be careful to not actually touch the sample surface and only bring the probe as close as possible. -
f.Run the “Find Contact” sequence in the Mach-1 motion software to enable the probe to find contact with the surface. The find contact sequence has two functions within it.
-
i.Zero load which can be added from the function menu in the software.
-
ii.Find contact which can also be added from the function menu. This is the main function to find contact with the surface of the sample, which would be the starting point of indentation.
-
i.
-
g.After finding contact, load the indentation sequence. Before execution, save the data in a text file. The indentation sequence consists of three functions.
-
i.Move relative which make the probe to indent on the surface up to a depth of 100 μm at a speed of 2 μm/s.
-
ii.Once the 100 μm depth is reached the probe waits at that position for 1 min due to the wait function of the sequence.
-
iii.Once the wait is over, the probe moves back up to the zero-position using a second move relative function.
-
i.
-
h.After finishing the program, manually move the probe up using the “up” function in the manual controls section of the software.
-
i.Move the stage to the next point of interest for the next microindentation.
-
j.Once at least three points are measured on one sample, move the probe to the highest possible point to avoid any damage, and place the next sample on the stage. Repeat the measurements for at least three samples in each group.
-
k.Once all the samples are tested, remove the probe from the machine carefully and wipe again with DI water and Kimwipe. Keep the probe in a protective case.
-
l.Calculate the elastic modulus () using the following formula:Where:
-
i.( = reduced elastic modulus).
-
ii.= 1 (constant in this experiment).
-
iii..
-
iv..
-
v.= peak unloading distance.
-
vi.= peak unloading force.
-
vii.(constant for spherical probe).
-
i.
-
a.
MR imaging
Timing: 1 day
This section describes the protocol to demonstrate the MR visibility of constructs by incorporating different concentrations of SPIONs, preparing the samples and performing the MR imaging.
-
6.Perform the following tasks.
-
a.Prepare GelMA bioinks containing 100 μg/mL, 200 μg/mL, and 500 μg/mL of SPIONS following the step 2 of this protocol.
-
b.Bioprint the SPION-encapsulated constructs following the step 3 of this protocol.
-
c.Prepare a 2%(w/v) solution of agarose in DI water by adding the agarose powder to the water.
-
i.Microwave at 1000 W for 2 min.
-
ii.Store the gel at 20–25°C until further use.
 CRITICAL: Make sure the container is not sealed, otherwise the pressure might break the container.
CRITICAL: Make sure the container is not sealed, otherwise the pressure might break the container.
-
i.
-
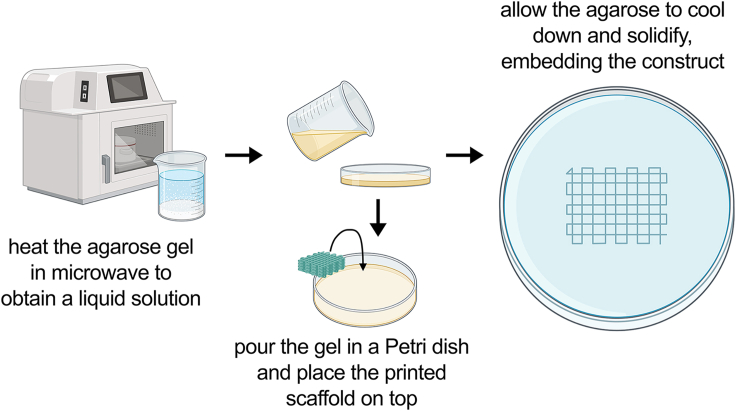
d.As shown schematically in Figure 2, to fix the bioprinted constructs, heat the agarose gel in a microwave at 1000 W for 30 s to 1 min.
-
i.Pour the gel in a petri dish and place the constructs on top, allowing them to sink slowly into the gel.
-
ii.Let the gel cool at 20–25°C for 20–30 min.
-
i.
-
e.Image the fixed constructs using an MR imaging/spectroscopy system (e.g., 9.4T/20 cm Bruker animal MR imaging system), obtaining T2∗ weighted images.
-
a.
Figure 2.
Schematical representation of the steps for embedding the printed constructs in agarose
Endothelial cell (EC) and NIH3T3 culture, viability, and growth
Timing: 14 days
This section describes the culture of endothelial cells and NIH3T3, seeding them on the bioprinted constructs and evaluating their viability and proliferation on the bioprinted constructs.
-
7.HUVEC and NIH/3T3 cell culture.
-
a.Prior to cell seeding, maintain HUVECs in T75 flasks with VascuLife VEGF endothelial medium supplemented with 1%(v/v) Pen/Strep until confluency.
-
b.Maintain NIH/3T3 fibroblasts in 10 cm petri dishes with NIH/3T3 fibroblast culture medium (Eagle’s Minimum Essential Medium) + 1%(v/v) Pen/Strep until confluency.
-
c.For 2D cultures, seed HUVECs or NIH/3T3 fibroblasts at 1×104 cells/cm2 in a 6-well plate with 2 mL cell solution per well.
-
i.Change media every other day and culture for the desired time (e.g., 7 days). Make sure to avoid cell confluency within the wells.
-
i.
-
d.For 3D cultures, seed the HUVECs or NIH/3T3 fibroblasts at 2×106 cells per construct.
-
i.Culture 3D bioprinted constructs in 6-well plates and change media every other day for the desired time (e.g., 14 days).
-
i.
-
a.
-
8.Cell viability and growth measurements.
-
a.Prepare fresh AlamarBlue solution by adding AlamarBlue reagent to culture media at 1:10 volumetric ratio and mixing to get a uniform solution.
-
b.Aspirate culture media in the 6-well plates and add equal volume (enough to fully cover the cellular constructs) of AlamarBlue solution to each well.
-
c.Incubate the cell culture plates in a humidified incubator at 37°C with 5% CO2 for 4 h while protected from light.
-
d.Gently shake the plates for 2 min every 1 h to distribute the AlamarBlue reagents uniformly in the cultures.
-
e.After 4 h incubation, take 100 μL supernatant per well, transfer to a 96-well plate in triplicates. Make sure to avoid bubbles (can burst the bubbles using a clean pipette tip).
-
f.Read the fluorescence absorbance at 550 and 600 nm using a microplate reader.
-
g.Once the AlamarBlue assay is done, aspirate the AlamarBlue solution and change to fresh culture media.
-
h.Perform the AlamarBlue assay on serial time points during culture (e.g., days 3 and 7 of 2D cultures, and days 3, 7, 10, and 14 of 3D cultures).
-
a.
Bacterial culture
Timing: 3 days
Given the antibacterial nature of SPIONs, this section describes culture of Staphylococcus aureus, infecting bioprinted constructs with them and assessing the antibacterial effects of the SPIONs when incorporated in the bioink.
-
9.Plating and culturing Staphylococcus aureus (S. aureus) from frozen stock.
-
a.Transfer frozen aliquot of S. aureus onto an LB agar plate using the loop, streaking across the plate from left to right and top to bottom.
-
b.Invert the plates and incubate for 10–15 h at 37°C.
-
a.
-
10.Inoculating liquid S. aureus culture.
-
a.Transfer 10 mL of LB medium to 15 mL conical tube.
-
b.If selection for an antibiotic resistance marker in your bacterial strain is required, add the antibiotic in the aliquoted 10 mL of LB medium to get a final concentration of 1X.
-
c.Transfer 2 mL of the LB antibiotic (for S. aureus 100 μg/mL ampicillin can be used) media into each culture tube.
-
d.Using a sterile p200 pipette tip, select a single colony from the LB agar plate and place toothpick into the medium.
-
e.Incubate at 37°C for 24 h if on a shaker or incubate for 48 h if no shaking is done.
-
a.
-
11.Infect bioprinted constructs with S. aureus.
-
a.After the bioprinted constructs have been cellularized and grown in static culture for 3 days, aspirate media completely from each well without disturbing the construct.
-
b.Count and pipette 108 colony-forming units of bacteria from the inoculated liquid culture directly onto each bioprinted cellular construct.
-
c.Let the bacteria culture onto the constructs for 2–3 h in an incubator at 37°C with 5% CO2 and 95% humidity.
-
d.After 3 h, fill the well until the construct is fully submerged with endothelial medium (without antibiotic supplements), and store back in the incubator.
-
e.After 24 h, remove the well plate from incubator for analysis.
-
a.
CRITICAL: Perform all steps described in this section in a sterilized biosafety cabinet.
Immunohistochemistry (IHC) analysis
Timing: 3 days
This step outlines the steps required to perform immunohistochemistry analysis, enabling visualization of cellular structures within the bioprinted constructs.
-
12.Sample fixation.
-
a.Remove cell culture media and wash the samples with PBS (2X, 3 min). Ensure complete removal of PBS.
-
b.Add enough volume of 10% formalin solution (4% formaldehyde) to each well to fully cover the constructs.
-
c.Incubate for 30 min at 20–25°C.
-
d.Remove fixation media and wash with PBS.
-
a.
-
13.Embedding constructs in agarose.
-
a.Prepare a 4% agarose solution by weighing out the appropriate amount of agarose powder and dissolving it in sterile water Heat the solution until the agarose is completely dissolved, then allow it to cool down to 40–50°C.
-
b.Place the fixed constructs into suitable dishes. Pour the molten agarose solution over the samples, ensuring complete coverage. Avoid trapping air bubbles in the agarose.
 CRITICAL: Bubbles could cause artefacts in MR imaging and also damage the tissue sections when slicing.
CRITICAL: Bubbles could cause artefacts in MR imaging and also damage the tissue sections when slicing. -
c.Allow the agarose to solidify at 20–25°C or by placing the molds on ice for faster solidification.
-
d.Once the agarose has solidified, carefully remove the molds. Trim the excess agarose around the embedded constructs using a sharp blade or scalpel, leaving only the desired area of interest embedded in agarose.
-
a.
-
14.Sectioning.
-
a.Ensure that the vibratome is clean and properly calibrated. Prepare the cutting chamber by filling it with deionized water. Add ice to the outer container.
-
b.Carefully mount the construct onto the cutting stage of the vibratome. Ensure that it is securely attached and positioned properly for slicing.
-
c.Set the desired slicing parameters on the vibratome, a slice thickness of 200 μm, cutting speed of 1.5 mm/s, and an amplitude of 1.5 mm.
-
d.Start the vibratome to initiate the slicing process. Monitor the slicing process closely to ensure that the sections are being cut accurately and consistently. Adjust the cutting parameters if necessary to optimize the slicing quality.
-
e.As the vibratome slices through the sample, collect the resulting sections sequentially in a multiwell plate filled with PBS. Handle the sections carefully to prevent damage or distortion.
-
f.Store the samples at 4°C for a long-term period.
-
a.
-
15.Permeabilization and blocking (duration: 1–2 h).
-
a.Prepare 0.5%(v/v) Triton X-100 in PBS and add a volume necessary to cover the construct for 15–20 min at 20–25°C.
-
b.Wash constructs with PBS three times, with each wash taking 10 min.
-
c.Prepare the blocking solution by dissolving 1%(w/v) bovine serum albumin (BSA) in PBS and add enough amount to cover the constructs for 60 min at 20–25°C.
-
d.Wash fixed samples three times with PBS with each wash taking 5 min at 20–25°C.
-
a.
-
16.Primary staining (duration: 10–15 h).
-
a.Remove PBS from the samples. Do not let the constructs dry out.
-
b.Dilute the primary antibody with a 1/200 ratio in PBS and add to each sample. Incubate the samples for 10–15 h at 4°C. Primary antibodies for this protocol are:
-
i.Anti CD31.
-
ii.Anti-connexin43.
-
iii.Anti F-actin.
-
iv.DAPI.
-
v.Polyclonal anti-Staphylococcus aureus.
-
i.
-
a.
-
17.Secondary staining (duration: 1–2 h).
-
a.Wash samples for 5 min with PBS (3 times). Remove PBS after each wash.
-
b.Prepare a 1/1000 ratio of secondary antibodies corresponding to primary species in PBS and incubate for 1–2 h at 20–25°C while covered in foil.
-
c.Wash samples for 5 min with PBS (3 times); keep them covered in foil while washing.
-
a.
-
18.Mounting samples on glass slides (duration: 30 min).
-
a.Add a drop of mounting medium onto the glass slide (about 40 μL).
-
b.Place the sections on the glass slide and add a drop of mounting medium on top of the sample.
-
c.Carefully place a glass coverslip onto the construct. Try to avoid the formation of bubbles.
-
d.Seal the coverslip edges with the nail polish.
-
e.Store the slides covered at 4°C until imaging. Protect them from light.
-
a.
-
19.Confocal imaging.
-
a.Turn on the confocal laser scanning microscope.
-
b.Carefully place the prepared glass slide with the mounted samples onto the microscope stage. Ensure that the sample is positioned correctly under the objective lens and secured in place.
-
c.Use the microscope’s eyepiece or camera system to locate the region of interest on the sample. Use the coarse and fine focus knobs to bring the sample into initial focus.
-
d.Choose the appropriate imaging parameters, including the excitation wavelength and emission wavelength range based on the fluorophores used for staining and the desired imaging objectives.
-
e.Set the imaging parameters, such as the scan speed, laser intensity, and laser gain, based on the visualized images. Adjust the z-axis settings when conducting 3D imaging or z-stack acquisition.
-
f.Initiate the image acquisition process using the software interface of the confocal microscope (e.g., Leica). Capture images of the samples at the desired locations and optical sections. Ensure that the exposure time and image averaging settings are optimized to obtain high-quality images with sufficient signal-to-noise ratio and minimal autofluorescence.
-
g.Perform sequential scanning to avoid spectral overlap between channels. Acquire images for each fluorophore separately or set up multichannel imaging for simultaneous detection of multiple fluorophores.
-
h.Process the acquired images using FIJI ImageJ to enhance contrast, remove background noise, and adjust brightness and contrast levels if necessary. Perform image stitching and 3D reconstruction for z-stack images.
-
a.
Expected outcomes
The field of additive biomanufacturing and in particular, 3D bioprinting, has found extensive and rapidly growing applications in tissue engineering and regenerative medicine, both in the basic science research and clinical applications. This is due to the unique capability of these technologies to create versatile and highly complex physiological architectures in a tunable, high-throughput, and reproducible manner.2,3 The choice of biomaterial(s) used as bioink plays a pivotal role in the success of tissue bioprinting strategies. Among various biomaterials used for this purpose, methacrylated gelatin (GelMA) is one of the most widely used hydrogels, due to its biocompatibility, adaptability with several bioprinting strategies, its tunable structure, and its availability.4,5,6 While research using bioprinting to engineer tissue analogues has been progressing rapidly, translation of this success into clinical setting still suffers from significant challenges. One of these hurdles is bacterial infection in the bioprinted constructs used as implants in the in vivo applications.1,7 This protocol describes a novel approach, via incorporating superparamagnetic iron oxide nanoparticles (SPIONs) in the hydrogel-based bioinks. As robust antibacterial agents, the embedded SPIONs confer significant antibacterial properties to the bioprinted scaffolds. These nanoparticles are also known as contrast agents for magnetic resonance imaging (MRI), facilitating further in vivo tracking of the tissue engineered constructs.
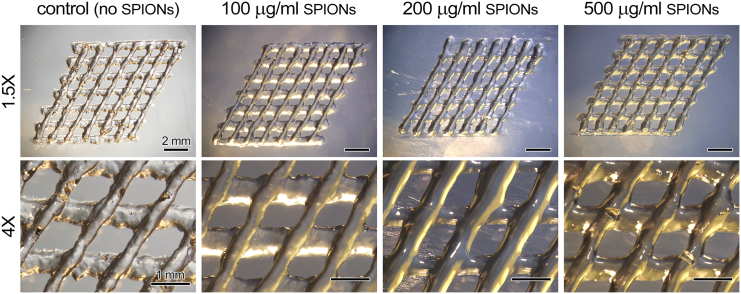
The protocol starts with synthesis of GelMA, which takes place through a one-pot reaction, substituting the amine groups on the gelatin backbone with methacrylate groups. The reported protocol results in a substitution degree ranging from 80 to 90%. Following this step, SPIONs are mixed with the liquid GelMA to create the antibacterial MR-visible bioinks. Incorporation of micro and nano particles in bioinks has been reported before as rheology modifiers, affecting printability of the bioinks in the extrusion process depending on the concentration of the particles.8 Concentrations mentioned in this protocol expect to maintain the printability of GelMA without any negative impact, as demonstrated in Figure 3.
Figure 3.
Printability of gelatin methacrylate (GelMA) based bioink, incorporating different concentrations of superparamagnetic iron oxide nanoparticles (SPIONs)
Figure adapted from Theus et al.1
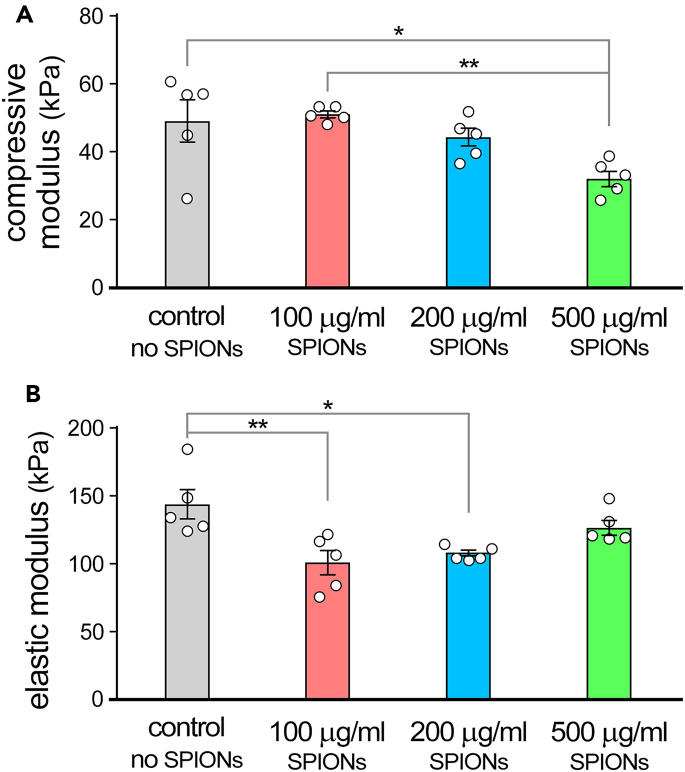
Besides printability, mechanical properties of the bioink play a pivotal role in success of the tissue engineering strategies by modulating the cell response.9 Incorporation of micro and nano particles can affect the organization of polymer chains within the bioink and crosslinking dynamics, hence, modulate mechanical properties such as compressive or elastic modulus.1,8 Consequently, a reduction in both compressive and elastic modulus of the bioprinted constructs can be expected following encapsulation of SPIONs, as shown in Figure 4.
Figure 4.
Mechanical characterization of 3D bioprinted scaffolds
Mechanical properties of 3D bioprinted constructs with different concentrations of superparamagnetic iron oxide nanoparticles (SPIONs), (A) compressive modulus and (B) elastic modulus
Figure adapted from Theus et al.1
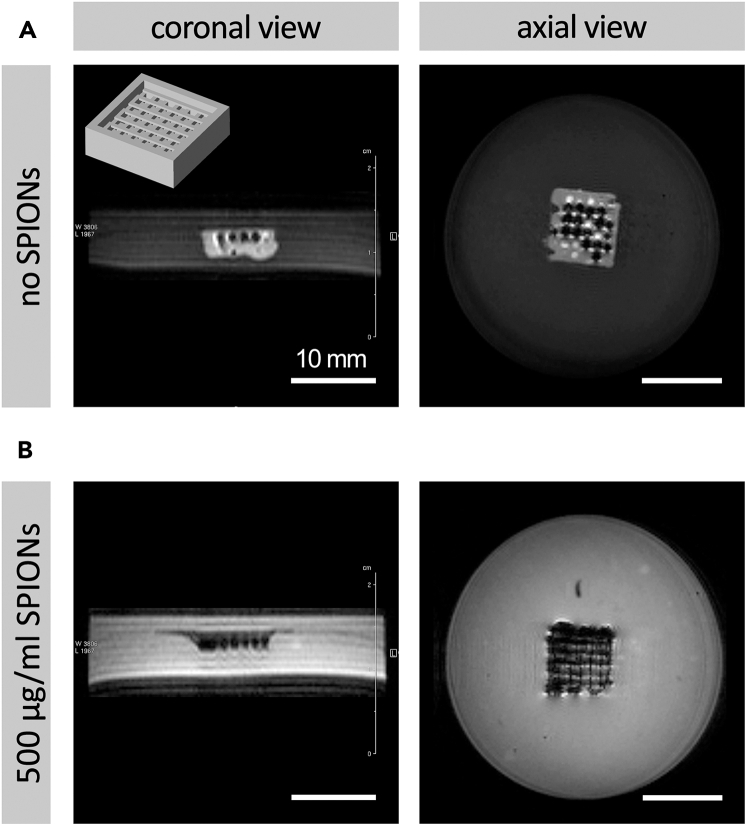
One of the major advantages offered by incorporation of SPIONs in bioinks is providing contrast for MR imaging. SPIONs have been used as contrast agents in a variety of applications, such as drug delivery via tissue engineered scaffolds.10,11 Encapsulation of these FDA-approved particles within hydrogels can therefore enable, for the first time, formulating novel bioink formulations with significant imaging contrast, making the printed scaffolds visible in MR imaging strategies. The resulting imaging contrast will be related to the concentration of encapsulated SPIONs. Some MR images demonstrating this effect are presented in Figure 5.
Figure 5.
Imaging properties of 3D bioprinted constructs
Demonstration of magnetic resonance (MR) imaging contrast provided by superparamagnetic iron oxide nanoparticles (SPIONs) within 3D bioprinted constructs, (A) MRI of bioprinted constructs with no SPIONS, and (B) MRI of bioprinted constructs containing 500 μg/mL SPIONs
Figure adapted from Theus et al.1
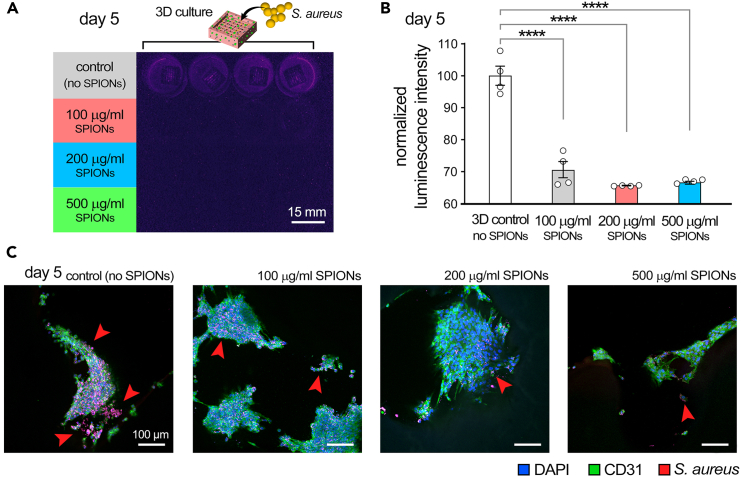
Finally, one main rationale for incorporation of SPIONs in the bioprinting workflow is to prevent infection. Nanoparticles such as SPIONs have shown significant efficiency in fighting infections, including antibiotic resistant ones, caused by their ability to bind to bacteria membrane, pass through the membrane, and cause oxidative stress.12,13,14 This protocol presents culture of S. aureus, one of the most common causes of hospital acquired and wound infections.15,16 It is expected that the incorporation of SPIONS greatly reduces the population (growth) of S. aureus cultured in the 3D bioprinted scaffolds, as shown in Figure 6. This is while the adverse impact of SPIONs on viability of HUVECs and fibroblasts is expected to be minimal,17 as demonstrated in Figure 7.
Figure 6.
The impact of superparamagnetic iron oxide nanoparticles (SPIONs), incorporated within 3D bioprinted constructs, on S. aureus population and growth in the 3D cultures
(A) luminescence imaging, (B) quantification based on luminescence intensity, and (C) immunostained slices demonstrating colocalization of bacterial colonies with endothelial cells (ECs) with green showing ECs, red showing S. Aureus, and blue signaling cell nucleus
Figure adapted from Theus et al.1
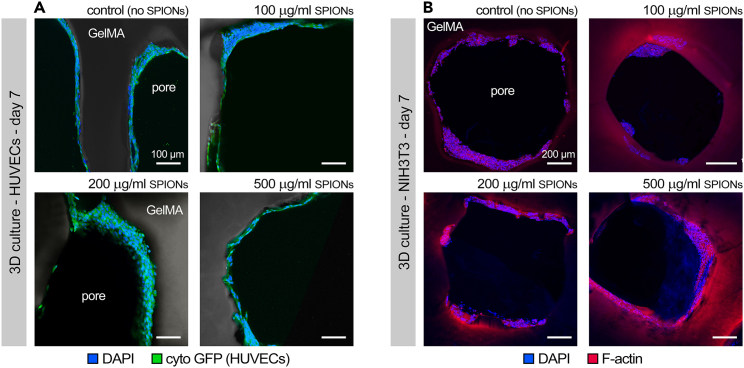
Figure 7.
Immunohistochemical analysis of cellular bioprints
Immunohistochemistry images of human umbilical vein endothelial cells (HUVECs, A) and fibroblasts (B) after 7 days of culture, demonstrating formation of multiple layers of cells and absence of major toxicity by the superparamagnetic iron oxide nanoparticles (SPIONs).
Overall, this protocol presents a facile approach for development of antibacterial, MR-visible bioinks capable of incorporation in a wide range of bioprinting strategies. Conferring this unique set of functionalities to engineered scaffolding devices could significantly increase their potential for clinical translation into a wide variety of biomedical applications.
Limitations
While this protocol provides a direct route for development of antibacterial bioinks, certain limitations are associated with it. First, a deeper characterization of the SPIONs, including their chemical composition, surface groups (i.e., the protein corona effect), or the particle size would offer a deeper understanding of their mechanism of action, potentially enhancing the antibacterial efficacy and interaction with cells. Furthermore, the cytocompatibility assay suggested in this protocol is limited to two cell types and a 14-day evaluation time. A more in depth cytocompatibility analysis incorporating stem cells and in vivo experiments, for extended time periods, would potentially shed further light on the impact of SPIONs on cells and enable further optimization of the nanoparticles. Finally, this protocol limits the study of antibacterial effects to certain species. While generally SPIONs are known to be effective against different strains of bacteria, studying the impact of these nanoparticles on various bacteria strains in a bioprinting context could further demonstrate the benefits and applicability of this approach.
Troubleshooting
Problem 1
Preparation of the bioinks, step 1: GelMA is synthesized with a lower degree of substitution, resulting in weaker gels.
Potential solution
Several approaches could be used to address this issue; first, increasing the synthesis duration could enhance efficiency. In case further modification is required, modulation of buffer pH to 8–9 can be used to increase the synthesis efficiency.
Problem 2
Preparation of the bioinks, step 2: Aggregation of SPIONs while mixing with GelMA.
Potential solution
Especially in higher concentrations, there is a possibility of SPION aggregation affecting uniformity and printability of the bioinks. Maintaining the SPION concentration at lower levels would generally be recommended, as that would also ensure adequate cytocompatibility of the particles for cell culture assays. In the case of particle aggregation, slow addition of SPIONs is suggested, while GelMA is being stirred (more robustly) at 37°C.
Problem 3
3D bioprinting of constructs, step 3: SPION precipitation during printing.
Potential solution
Perform printing parameter optimizations without SPIONs; next, mix SPIONs in the bioink; increase the printing pressure to allow increasing nozzle movement speed. A more rapid printing process would reduce the chance for SPIONs to sediment within the deposited ink. Alternatively, optimize printing parameters for a lower temperature of GelMA; after mixing with SPIONs rapidly cool down GelMA to a temperature of approximately 15°C and print at this temperature. In this case, the physical gelation of GelMA can prevent precipitation in the bioink.
Problem 4
Mechanical testing of bioprinted constructs, step 6: Bioprinted hydrogel constructs dry out during microindentation.
Potential solution
Keep samples in PBS until measurement; dry the surface of each construct right before measuring the stiffness.
Problem 5
Mechanical testing of bioprinted constructs, step 6: Microindentation device not finding contact point on the surface of bioprinted constructs.
Potential solution
This issue might occur with hydrogels, mainly due to surface water or nonuniformities on the surface of construct. The first solution is to ensure the sample surface is properly dried with tissue paper. Alternatively, increase the contact depth in the find contact sequence.
Problem 6
Endothelial cell (EC) and NIH3T3 culture, viability, and growth, step 8: AlamarBlue readings at later time points are not reliable / reproducible.
Potential solution
While cellular samples used for the AlamarBlue assay can be washed and reused at later time points, residual, colored reagents can cause inaccuracy in the colorimetric measurements at later time points. Aside from washing the samples completely, an alternative approach can be to use separate samples for each time point.
Problem 7
Immunohistochemistry (IHC) analysis, step 19: IHC images not showing clear signal.
Potential solution
In case IHC signal is not clear, the first solution is to increase the concentration of primary antibodies used. Alternatively, the incubation time for secondary antibody staining can be increased to obtain a stronger signal. In case excessive autofluorescence or artefacts from GelMA disturbs the clear IHC imaging, cutting tissue sections at smaller thicknesses or using the tissue clearing methods could be tested.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Vahid Serpooshan (vahid.serpooshan@bme.gatech.edu).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to and will be answered by the technical contact, Dr. Mehdi Salar Amoli (mehdi.salar.amoli@gatech.edu).
Materials availability
All reagents used are commercially available.
Data and code availability
No datasets or codes were reported in this protocol.
Acknowledgments
This research was funded by the National Institutes of Health (NIH) grant number R01 HL131017 and the National Science Foundation (NSF) CAREER award number 2044657 (to V.S.).
Author contributions
M.S.A.: writing – original draft; L.J.: writing – original draft; S.R.: writing – original draft; Y.S.: writing – original draft; M.S.: writing – original draft; L.N., B.H., M.L.T., C.N.L., and M.M.: writing – review and editing and data acquisition; H.B.-H. and V.S.: writing – review and editing, conceptualization, and funding acquisition.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Mehdi Salar Amoli, Email: mehdi.salar.amoli@gatech.edu.
Vahid Serpooshan, Email: vahid.serpooshan@bme.gatech.edu.
References
- 1.Theus A.S., Ning L., Kabboul G., Hwang B., Tomov M.L., LaRock C.N., Bauser-Heaton H., Mahmoudi M., Serpooshan V. 3D bioprinting of nanoparticle-laden hydrogel scaffolds with enhanced antibacterial and imaging properties. iScience. 2022;25 doi: 10.1016/j.isci.2022.104947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serpooshan V., Guvendiren M. Editorial for the Special Issue on 3D Printing for Tissue Engineering and Regenerative Medicine. Micromachines. 2020;11 doi: 10.3390/mi11040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang Y., Miller K., Guan J., Kiratitanaporn W., Tang M., Chen S. 3D bioprinting of complex tissues in vitro: state-of-the-art and future perspectives. Arch. Toxicol. 2022;96:691–710. doi: 10.1007/s00204-021-03212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mobaraki M., Ghaffari M., Yazdanpanah A., Luo Y., Mills D.K. Bioinks and bioprinting: A focused review. Bioprinting. 2020;18 doi: 10.1016/j.bprint.2020.e00080. [DOI] [Google Scholar]
- 5.Ning L., Mehta R., Cao C., Theus A., Tomov M., Zhu N., Weeks E.R., Bauser-Heaton H., Serpooshan V. Embedded 3D Bioprinting of Gelatin Methacryloyl-Based Constructs with Highly Tunable Structural Fidelity. ACS Appl. Mater. Interfaces. 2020;12:44563–44577. doi: 10.1021/acsami.0c15078. [DOI] [PubMed] [Google Scholar]
- 6.Yin P., Su W., Li T., Wang L., Pan J., Wu X., Shao Y., Chen H., Lin L., Yang Y., et al. A modular hydrogel bioink containing microsphere-embedded chondrocytes for 3D-printed multiscale composite scaffolds for cartilage repair. iScience. 2023;26 doi: 10.1016/j.isci.2023.107349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rastin H., Ramezanpour M., Hassan K., Mazinani A., Tung T.T., Vreugde S., Losic D. 3D bioprinting of a cell-laden antibacterial polysaccharide hydrogel composite. Carbohydr. Polym. 2021;264 doi: 10.1016/j.carbpol.2021.117989. [DOI] [PubMed] [Google Scholar]
- 8.Galliger Z., Vogt C.D., Helms H.R., Panoskaltsis-Mortari A. Extracellular Matrix Microparticles Improve GelMA Bioink Resolution for 3D Bioprinting at Ambient Temperature. Macromol. Mater. Eng. 2022;307:2200196. doi: 10.1002/mame.202200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi B., Xu Q., Liu W. An overview of substrate stiffness guided cellular response and its applications in tissue regeneration. Bioact. Mater. 2022;15:82–102. doi: 10.1016/j.bioactmat.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulińska-Litewka J., Łazarczyk A., Hałubiec P., Szafrański O., Karnas K., Karewicz A. Superparamagnetic Iron Oxide Nanoparticles-Current and Prospective Medical Applications. Materials. 2019;12:617. doi: 10.3390/ma12040617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen M.P., Thuy V.T.T., Kim D. Integration of iron oxide nanoparticles and polyaspartamide biopolymer for MRI image contrast enhancement and an efficient drug-delivery system in cancer therapy. Nanotechnology. 2020;31 doi: 10.1088/1361-6528/ab8f49. [DOI] [PubMed] [Google Scholar]
- 12.Gholami A., Mohammadi F., Ghasemi Y., Omidifar N., Ebrahiminezhad A. Antibacterial activity of SPIONs versus ferrous and ferric ions under aerobic and anaerobic conditions: a preliminary mechanism study. IET Nanobiotechnol. 2020;14:155–160. doi: 10.1049/iet-nbt.2019.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geilich B.M., Gelfat I., Sridhar S., van de Ven A.L., Webster T.J. Superparamagnetic iron oxide-encapsulating polymersome nanocarriers for biofilm eradication. Biomaterials. 2017;119:78–85. doi: 10.1016/j.biomaterials.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Taylor E.N., Kummer K.M., Durmus N.G., Leuba K., Tarquinio K.M., Webster T.J. Superparamagnetic Iron Oxide Nanoparticles (SPION) for the Treatment of Antibiotic-Resistant Biofilms. Small. 2012;8:3016–3027. doi: 10.1002/smll.201200575. [DOI] [PubMed] [Google Scholar]
- 15.Mahmoudi M., Serpooshan V. Silver-Coated Engineered Magnetic Nanoparticles Are Promising for the Success in the Fight against Antibacterial Resistance Threat. ACS Nano. 2012;6:2656–2664. doi: 10.1021/nn300042m. [DOI] [PubMed] [Google Scholar]
- 16.Tong S.Y.C., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G., Jr. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/cmr.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei H., Hu Y., Wang J., Gao X., Qian X., Tang M. Superparamagnetic Iron Oxide Nanoparticles: Cytotoxicity, Metabolism, and Cellular Behavior in Biomedicine Applications. Int. J. Nanomedicine. 2021;16:6097–6113. doi: 10.2147/ijn.S321984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets or codes were reported in this protocol.