Abstract
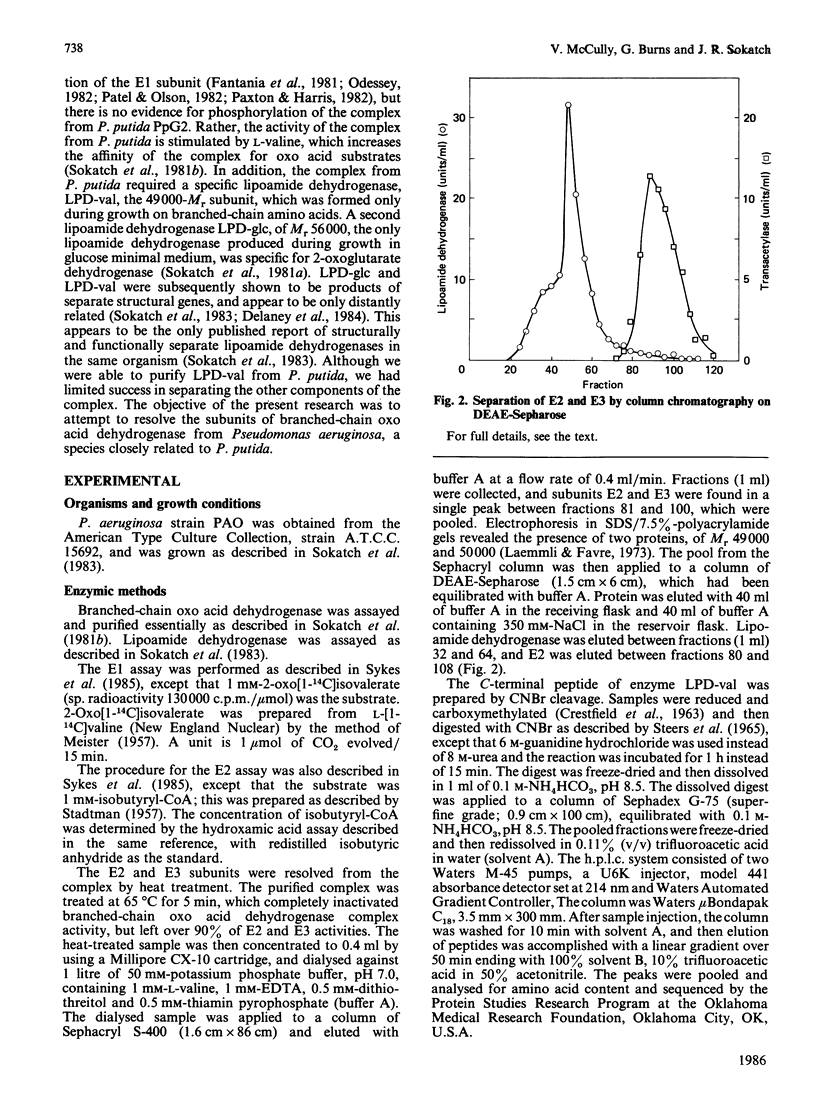
Branched-chain oxo acid dehydrogenase was purified from Pseudomonas aeruginosa strain PAO with the objective of resolving the complex into its subunits. The purified complex consisted of four proteins, of Mr 36,000, 42,000, 49,000 and 50,000. The complex was resolved by heat treatment into the 49,000 and 50,000-Mr proteins, which were separated by chromatography on DEAE-Sepharose. The 49,000-Mr protein was identified as the E2 subunit by its ability to catalyse transacylation with a variety of substrates, with dihydrolipoamide as the acceptor. P. aeruginosa, like P. putida, produces two lipoamide dehydrogenases. One, the 50,000-Mr protein, was identified as the specific E3 subunit of branched-chain oxo acid dehydrogenase and had many properties in common with the lipoamide dehydrogenase LPD-val of P. putida. The second lipoamide dehydrogenase had Mr 54,000 and corresponded to the lipoamide dehydrogenase LPD-glc of P. putida. Fragments of C-terminal CNBr peptides of LPD-val from P. putida and P. aeruginosa corresponded closely, with only two amino acid differences over 31 amino acids. A corresponding fragment at the C-terminal end of lipoamide dehydrogenase from Escherichia coli also showed extensive homology. All three peptides had a common segment of eight amino acids, with the sequence TIHAHPTL. This homology was not evident in any other flavoproteins in the Dayhoff data base which suggests that this sequence might be characteristic of lipoamide dehydrogenase.
Full text
PDF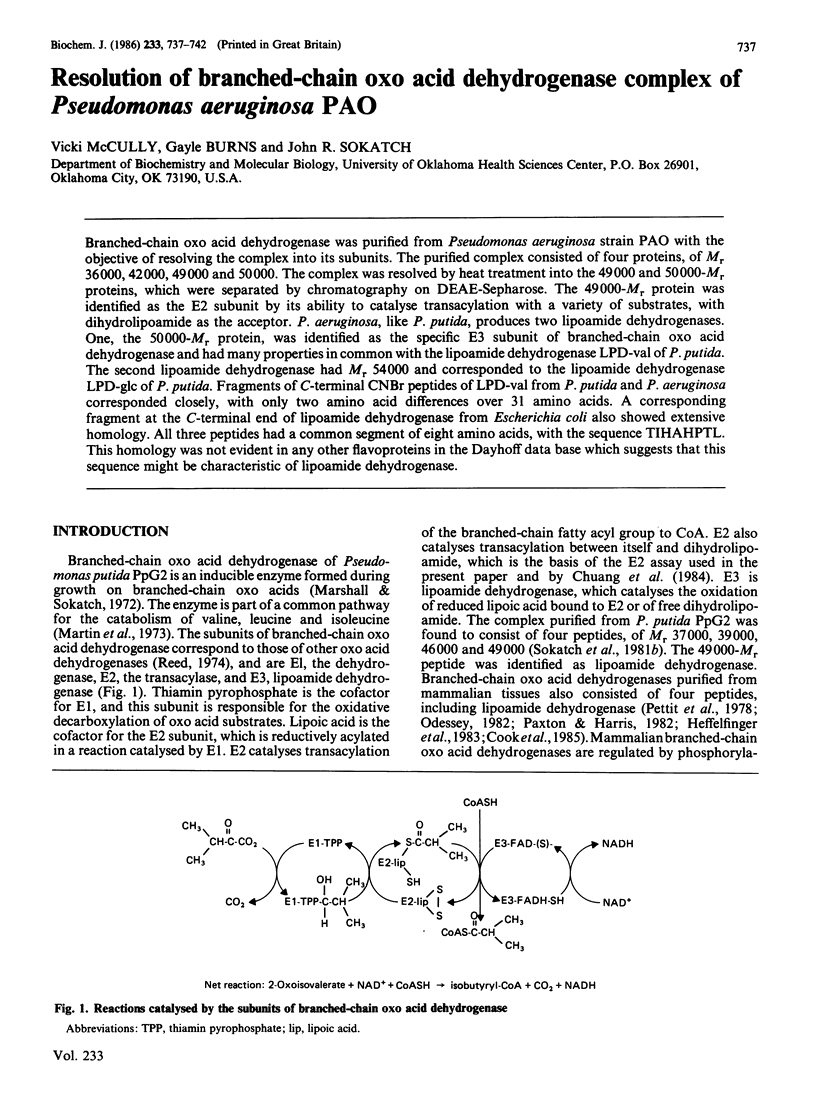




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CRESTFIELD A. M., MOORE S., STEIN W. H. The preparation and enzymatic hydrolysis of reduced and S-carboxymethylated proteins. J Biol Chem. 1963 Feb;238:622–627. [PubMed] [Google Scholar]
- Chuang D. T., Hu C. C., Ku L. S., Niu W. L., Myers D. E., Cox R. P. Catalytic and structural properties of the dihydrolipoyl transacylase component of bovine branched-chain alpha-keto acid dehydrogenase. J Biol Chem. 1984 Jul 25;259(14):9277–9284. [PubMed] [Google Scholar]
- Cook K. G., Bradford A. P., Yeaman S. J. Resolution and reconstitution of bovine kidney branched-chain 2-oxo acid dehydrogenase complex. Biochem J. 1985 Feb 1;225(3):731–735. doi: 10.1042/bj2250731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. Relating proteins by amino acid composition. Methods Enzymol. 1983;91:60–75. doi: 10.1016/s0076-6879(83)91011-x. [DOI] [PubMed] [Google Scholar]
- Darlison M. G., Spencer M. E., Guest J. R. Nucleotide sequence of the sucA gene encoding the 2-oxoglutarate dehydrogenase of Escherichia coli K12. Eur J Biochem. 1984 Jun 1;141(2):351–359. doi: 10.1111/j.1432-1033.1984.tb08199.x. [DOI] [PubMed] [Google Scholar]
- Delaney R., Burns G., Sokatch J. R. Relationship of lipoamide dehydrogenases from Pseudomonas putida to other FAD-linked dehydrogenases. FEBS Lett. 1984 Mar 26;168(2):265–270. doi: 10.1016/0014-5793(84)80259-8. [DOI] [PubMed] [Google Scholar]
- Fatania H. R., Lau K. S., Randle P. J. Inactivation of purified ox kidney branched-chain 2-oxoacid dehydrogenase complex by phosphorylation. FEBS Lett. 1981 Sep 28;132(2):285–288. doi: 10.1016/0014-5793(81)81180-5. [DOI] [PubMed] [Google Scholar]
- Fox B., Walsh C. T. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982 Mar 10;257(5):2498–2503. [PubMed] [Google Scholar]
- HAGER L. P., KORNBERG H. L. On the mechanism of alpha-oxoglutarate oxidation in Escherichia coli. Biochem J. 1961 Jan;78:194–198. doi: 10.1042/bj0780194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffelfinger S. C., Sewell E. T., Danner D. J. Identification of specific subunits of highly purified bovine liver branched-chain ketoacid dehydrogenase. Biochemistry. 1983 Nov 22;22(24):5519–5522. doi: 10.1021/bi00293a011. [DOI] [PubMed] [Google Scholar]
- Henderson C. E., Perham R. N., Finch J. T. Structure and symmetry of B. stearothermophilus pyruvate dehydrogenase multienzyme complex and implications for eucaryote evolution. Cell. 1979 May;17(1):85–93. doi: 10.1016/0092-8674(79)90297-6. [DOI] [PubMed] [Google Scholar]
- Kaneda T. Fatty acids of the genus Bacillus: an example of branched-chain preference. Bacteriol Rev. 1977 Jun;41(2):391–418. doi: 10.1128/br.41.2.391-418.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Lowe P. N., Hodgson J. A., Perham R. N. Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched-chain 2-oxo acids in Bacillus subtilis. Biochem J. 1983 Oct 1;215(1):133–140. doi: 10.1042/bj2150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall V. D., Sokatch J. R. Regulation of valine catabolism in Pseudomonas putida. J Bacteriol. 1972 Jun;110(3):1073–1081. doi: 10.1128/jb.110.3.1073-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. R., Marshall V. D., Sokatch J. R., Unger L. Common enzymes of branched-chain amino acid catabolism in Pseudomonas putida. J Bacteriol. 1973 Jul;115(1):198–204. doi: 10.1128/jb.115.1.198-204.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey V., Williams C. H., Jr On the reaction mechanism of yeast glutathione reductase. J Biol Chem. 1965 Nov;240(11):4470–4480. [PubMed] [Google Scholar]
- Odessey R. Purification of rat kidney branched-chain oxo acid dehydrogenase complex with endogenous kinase activity. Biochem J. 1982 Apr 15;204(1):353–356. doi: 10.1042/bj2040353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T. B., Olson M. S. Evidence for the regulation of the branched chain alpha-keto acid dehydrogenase multienzyme complex by a phosphorylation/dephosphorylation mechanism. Biochemistry. 1982 Aug 31;21(18):4259–4265. doi: 10.1021/bi00261a012. [DOI] [PubMed] [Google Scholar]
- Paxton R., Harris R. A. Isolation of rabbit liver branched chain alpha-ketoacid dehydrogenase and regulation by phosphorylation. J Biol Chem. 1982 Dec 10;257(23):14433–14439. [PubMed] [Google Scholar]
- Pettit F. H., Yeaman S. J., Reed L. J. Purification and characterization of branched chain alpha-keto acid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEERS E., Jr, CRAVEN G. R., ANFINSEN C. B., BETHUNE J. L. EVIDENCE FOR NONIDENTICAL CHAINS IN THE BETA-GALACTOSIDASE OF ESCHERICHIA COLI K12. J Biol Chem. 1965 Jun;240:2478–2484. [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Gebrosky J., Sokatch D. J. Isolation of a specific lipoamide dehydrogenase for a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):639–646. doi: 10.1128/jb.148.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Roberts C. M. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):647–652. doi: 10.1128/jb.148.2.647-652.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Sahm J. G., Reyes-Maguire M. Mutations affecting lipoamide dehydrogenases of Pseudomonas putida. J Bacteriol. 1983 Feb;153(2):969–975. doi: 10.1128/jb.153.2.969-975.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M. E., Darlison M. G., Stephens P. E., Duckenfield I. K., Guest J. R. Nucleotide sequence of the sucB gene encoding the dihydrolipoamide succinyltransferase of Escherichia coli K12 and homology with the corresponding acetyltransferase. Eur J Biochem. 1984 Jun 1;141(2):361–374. doi: 10.1111/j.1432-1033.1984.tb08200.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983 Jul 1;133(3):481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Lewis H. M., Darlison M. G., Guest J. R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983 Oct 3;135(3):519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- Sykes P. J., Menard J., McCully V., Sokatch J. R. Conjugative mapping of pyruvate, 2-ketoglutarate, and branched-chain keto acid dehydrogenase genes in Pseudomonas putida mutants. J Bacteriol. 1985 Apr;162(1):203–208. doi: 10.1128/jb.162.1.203-208.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington D. J., Rosemeyer M. A. Glutathione reductase from human erythrocytes. Molecular weight, subunit composition and aggregation properties. Eur J Biochem. 1975 Dec 15;60(2):459–466. doi: 10.1111/j.1432-1033.1975.tb21024.x. [DOI] [PubMed] [Google Scholar]



