Abstract
Introduction:
The discovery of gene editing techniques has opened a new era within the field of biology and enabled scientists to manipulate nucleic acid molecules. CRISPR-Cas9 genome engineering has revolutionized this achievement by successful targeting the DNA molecule and editing its sequence. Since genomic changes are the basis of the birth and growth of many tumors, CRISPR-Cas9 method has been successfully applied to identify and manipulate the genes which are involved in initiating and driving some neoplastic processes.
Methods:
By review of the existing literature on application of CRISPR-Cas9 in cancer, different databases, such as PubMed and Google Scholar, we started data collection for "CRISPR-Cas9", "Genome Editing", "Cancer", "Solid tumors", "Hematologic malignancy" "Immunotherapy", "Diagnosis", "Drug resistance" phrases. Clinicaltrials.gov, a resource that provides access to information on clinical trials, was also searched in this review.
Results:
We have defined the basics of this technology and then mentioned some clinical and preclinical studies using this technology in the treatment of a variety of solid tumors as well as hematologic neoplasms. Finally, we described the progress made by this technology in boosting immune-mediated cell therapy in oncology, such as CAR-T cells, CAR-NK cells, and CAR-M cells.
Conclusion:
CRISPR-Cas9 system revolutionized the therapeutic strategies in some solid malignant tumors and leukemia through targeting the key genes involved in the pathogenesis of these cancers.
Keywords: CRISPR-Cas9, Gene editing, Cancer, Solid tumors, Hematologic malignancy, Immunotherapy, Diagnosis, Drug resistance
Introduction
DNA repair by deletion or replacement of the sequences involved in diseases and generation of nonpathogenic genes are the main areas of genetic manipulation in gene therapy. These therapeutic approaches are promising strategies for treating many non-curable human diseases, such as some hemoglobinopathies, acquired immunodeficiency syndrome (AIDS), diabetes, metabolic and neurodegenerative diseases, and cancer.1-3 The precise manipulation of the target regions, including specific gene activation or inactivation, nucleotide sequence replacement, gene deletion, and rearrangement, usually enables appropriate nucleases to be embedded in the editing platforms.3 At the first attempts, few products purported to be impressive in the treatment of patients with head and neck carcinomas and certain brain tumors.These products used a cut-and-paste technique to eliminate defects and insert the correct gene in the right place on the chromosome.3 However, these initial gene therapy tools integrated a copy of exogenous DNA into the genome with undesirable effects, including abnormal protein expression and insertion mutations. In recent years, to avoid these drawbacks, programmable nucleases have been widely used in gene editing platforms.3 This review briefly describes the basics of the most successful gene editing technologies, such as clustered regularly interspersed short palindromic repeats (CRISPR) −Cas9, their applications, and achievements in some prevalent cancer treatments. CRISPR-Cas9-based genome editing has generated massive interest in applied biology and medicine due to the easy targeting of Cas9, high effectiveness as a site-specific nuclease, and the potential for highly complex changes.
Different kinds of genome editing tools
Effective genome editing requires four main kinds of DNA-binding proteins: zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), Meganucleases, and Cas9 nuclease (Table 1).4 The first programmable nucleases that alter the stem cell genome were ZFNs. 4 ZFNs contain two domains: a zinc finger domain that recognizes DNA targets and a nuclease domain, which is originated from the FokI restriction enzyme. The DNA-binding domain can bind to target sites which are about 18 bp in length or longer and allows each ZFN motif to recognize 3-6 bp. The second domain cleaves ZFN target sites, which are often rich in guanines and consist of 5′-GNN-3′.5 As ZFN heterodimers can be quickly packaged as 1 kb/monomer, they can be carried by adenoviral vectors. The limited targeting density and high off-target effects of ZFNs often hinder their usefulness due to resulting cytotoxicity.6
Table 1. Comparison of the four major classes of DNA-binding proteins .
| ZFN | TALEN | Cas9 | Meganuclease | Ref | |
| Specificity | Low | Medium | High | High | 9 |
| Targeting constraints | Difficult to target non-G-rich sequences | 5ʹ targeted base must be a T for each TALEN monomer | Targeted sequence must precede a PAM | Targeting novel sequences often results in low efficiency | 9 |
| Ease of engineering | Difficult | Moderate | Easy | Difficult | 9,10 |
| Cleavage efficiency | Low efficiency | Efficient | Efficient Highly | Low efficiency | 9 |
| Ease of multiplexing | Low | Low | High | Low | 10 |
Another programmable nuclease is TALEN. TALENs consist of a FokI nuclease domain at the carboxyl terminus and a DNA-binding domain at the amino-terminal region. The first domain terminates almost 12–20 bps nearby the spacer sequence.5 The DNA-binding domains of TALEN contained nearly identical amino acid replicates of 34 residues in length, varying only at positions 12 and 13. This hypervariable di-residue area makes the specific nature of the TALEN editing tool. TALENs can modify any desired DNA sequence, and in spite of their enormous size, they can be delivered using various tools, namely codon-divergent repeat variable di-residues (RVDs), adenoviral vectors, and mRNA- or protein-based gene transfer procedures.5 However, the primary obstacles faced by TALEN approaches include the need for intricate molecular cloning for every new DNA target and the limited effectiveness in screening genomes within targeted cells.7
Meganucleases are the other members of the endonuclease family that specifically recognize and cleave large segments of DNA (12-40 nucleotides). Although they have large recognition sites and are less cytotoxic than ZFNs, the number of naturally occurring meganucleases is limited and insufficient to support all the potential loci of interest.8 In addition, the expensive and time-consuming process of developing sequence-specific enzymes for all potential sequences makes meganuclease less favorable.9
By making double-strand breaks (DSBs) at particular genome sites, the mentioned gene editing tools tackled the major obstacle of genetic engineering. However, some severe limitations, such as the complex and time-consuming design of the constructs, remained significant challenges.
CRISPR-Cas9
The Cas9 nuclease combined with a short guide RNA, discerns the DNA target using the Watson-Crick base pairing model. The guide RNA sequence (corresponds to the phage sequences which is a natural mechanism of antiviral defense) can be simply replaced with the desired sequence and retarget Cas9. Multiplexed CRISPR-Cas9 gene targeting can now be controlled with a short guide RNA in place of several bulky proteins (Supplementary movie).11
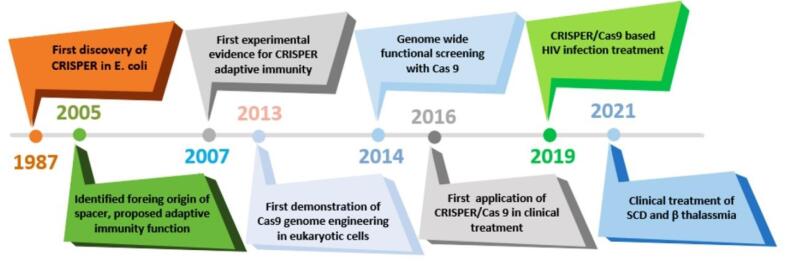
The history of CRISPR started with the study of the isozyme transformation of alkaline phosphatase by the intestinal alkaline phosphatase (IAP) enzyme in Escherichia coli in 1987.11 Nakata et al detected 29 replicates of nucleotides downstream of the IAP gene, which, unlike most replication elements, were separated by interspaced sequences.12 Afterwards, Mojica et al categorized repetitive interspaced sites as an unparalleled family of clustered repetitive elements that exist in more than 40% of sequenced bacteria and 90% of archaea. Jansen and Mojica used CRISPR to describe bacterial sequences containing an interspaced repeat array.12 The importance of these sequences became apparent when the researchers realized the similarity of the phage genome sequence with the sequences between CRISPR loci. It was also discovered that the CRISPR locus is a part of adaptive immunity targeting foreign viruses.12 Fig. 1 shows the timeline of the historical events surrounding the discovery and identification of the CRISPR–Cas9 system.
Fig. 1.
A chronological overview of key milestones in the development of CRISPR-Cas9 genome editing technology.
The CRISPR-Cas9 system functions as an acquired immune system against invading viruses and plasmids in bacteria and archaea.13 When a foreign DNA fragment enters the cells, it is cleaved by Cas nuclease enzymes, and a part called a spacer is placed in the locus of the crisper gene between two repetitive sequences (adaptation step).14 In the next step, known as the expression step, the spacer sequences are used as a template to generate short CRISPR RNA sequences (crRNA). In addition, after transcription of a part of the CRISPR gene, CrRNA-activating-trans (tracrRNA) is produced, then two sequences are joined. This complex is a guide sequence in the next step (interference) and guides the Cas protein to the foreign DNA. Once the Cas binds to the invading DNA, its HNH nuclease domain mediates the cleavage of the complementary crRNA strand, and the Like-RuvC1 nuclease domain cleaves the opposite strand. Since the discovery of the CRISPR system, 45 families of Cas proteins have been described with different roles, including crRNA synthesis, uptake and insertion of new spacer sequences, and invasive DNA cleavage.
The Cas9 protein is responsible for the cleavage of DNA, and the guide RNA (sgRNA) is comprised of crRNA and tracrRNA.15 The Cas9 protein is derived from Streptococcus pyogenes and is responsible for destroying two DNA strands and is delivered to the target site by sgRNA.15 This two-membered protein consists of target recognition and the nuclease lobe. The nuclease lobe also contains RuvC and HNH domains; both domains generate blunt-ended double-stranded DNA (DSB); RuvC (the retroviral integrase protein) cleaves non-target double-stranded DNA (DSB), whereas HNH cleaves targeted DNA strands. siRNA effectively identifies specific sites and directs Cas9 to the target region. The high specificity, efficiency, and accuracy of sgRNA are related to the 10-12 initial nucleotides at the end of '3, located upstream of the proto-spacer adjacent motif (PAM), and direct the Cas9 protein to the desired site.16,17 Eventually, the assembly of tracrRNA, crRNA precursor (pre-crRNA), and Cas9 forms a complex in which tracrRNA is responsible for activating RNase III to accelerate the maturation of pre-crRNA. The tracrRNA with a hairpin structure is transcribed from a repetitive region.17
CRISPR-Cas9 delivery
CRISPR-Cas9 gene editing faces challenges in delivering its components. Viral vectors have been used for decades in genetic engineering, with recombinant retroviral vectors receiving much attention due to their low off-target effects, high accuracy, and specificity for targeting the cell genome. Lentiviruses have been selected as a suitable delivery option due to the large size of Cas9 (4.1 KB).18 However, several main problems, including the possibility of mutations and immunogenicity, have limited the use of adenoviral and lentiviral vectors. Plasmid DNA is another vehicle for delivery of CRISPR components, which has been used in in vitro and in vivo studies. However, problems such as the high persistence of CRISPR RNPs inside the cell, consequent rise in off-target effects, and probability of mutagenicity limit their optimal use.19 To overcome these challenges, researchers have introduced non-viral methods due to their advantages, including high packing ability, fewer side effects, high accuracy, and spatiotemporal specificity for targeting the genome. Several compounds, such as graphene oxide, liposomes, gold nanoparticles, zeolitic imidazole, DNA nanoclews, and cationic polymers, have been associated with promising results in delivering CRISPR-Cas9 components.20 However, the large size of Cas9, the high negative charge of sgRNAs, the difficulty of protecting Cas9 and sgRNAs from degradation, and the Cas9-loaded non-viral particles process are also crucial concerns for non-viral carriers. Researchers have developed modified lipid nanoparticles that can deliver RNPs to modify gene expression and significantly reduce serum levels in mice. These results indicate the generalizable approach of these nanoparticles for editing different regions of the genome, biological modeling, and therapeutic interventions.21
Application of CRISPR-Cas9 in treatment of malignancies
Non-lethal genetic alterations are trigger points in cancers. This initial damage may be caused by environmental exposure (viral, chemical, toxic endogenous molecules, radiation, and unknown factors) or may be inherited in the germ lines of victims.22 At the beginning of cancer gene editing, TALENs and ZFNs acted as a powerful class of tools for cancer therapy but their complicated design and time-consuming experimental processes hindered their clinical usage.18 RNA interference (RNAi) was another successful cancer treatment, but it was accompanied by unwanted gene insertion, low specificity, transfection, and a low immune response.23 Genome editing by CRISPR-Cas9 has recently elucidated to be successfully applied in the correction of genes involved in initiation or driving in carcinogenesis.CRISPR-Cas9 has been a significant priority for cancer treatment due to its simplicity and high accuracy.24
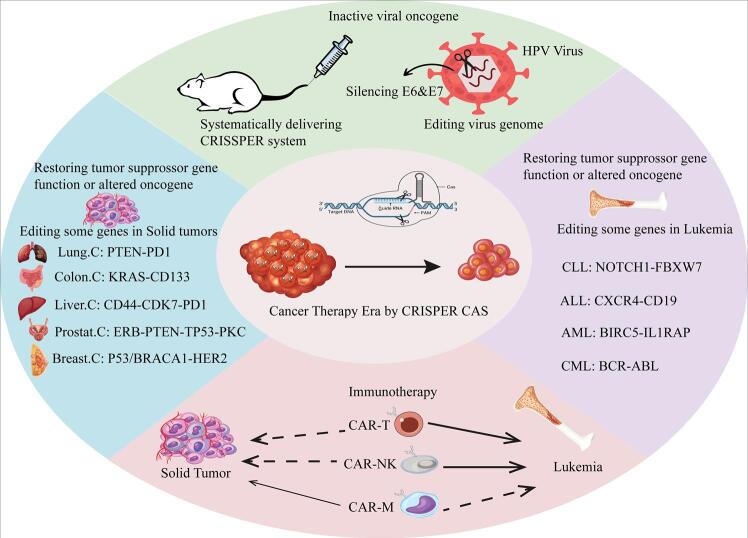
There are two main strategies for genetic editing in tumor cells: inactivation of active viral oncogenes and repairing altered oncogenes or silenced tumor suppressor genes.2 As the CRISPR-Cas9 editing tool originates from the bacterial adaptive immune system, it has an inherent benefit in protecting against viral infections. This editing system can directly remove or inactivate viral infections by manipulating the virus genome and may offer new opportunities to prevent and treat virus-associated malignancies.2 Several viruses have been associated with cancer development, including hepatitis B virus (HBV) and hepatitis C virus (HCV) in hepatocellular carcinoma (HCC), Epstein-Barr virus (EBV) in nasopharyngeal carcinoma, human papillomavirus (HPV) in uterine cervix squamous cell carcinoma, and human Herpesvirus-8 disease (HHV8) in Kaposi sarcoma. Eliminating or inactivating these carcinogenic viruses can intercept or reverse the process of tumor formation.2 The studies targeting the genomic structure of viruses are limited but show promising results. Zhen et al transferred Cas9 and sgRNAs into HPV-16-positive cervical cancer cells (SiHa), targeting HPV16 E6 or E7 and observed inhibition of the proliferation of cancer cells in a mouse xenograft model.25 Another study utilized the CRISPR-Cas9 system to inactivate E6 or E7 and induce senescence in HPV18 immortalized HeLa cells.26 They reported that E6 or E7-knocked out HeLa cells showed senescence characteristics (such as large surface area and high expression of β-galactosidase) and increased expression of p53/p21 and pRb/p21 genes. In addition, Jubair et al systemically delivered the CRISPR-Cas9 system to an animal model of cervical cancer with high expression of the E6 and E7 proteins. They observed tumor omission and increased survival in the treated group compared to untreated animals.27 Regarding EBV and cancer, a study investigated the role of latent membrane protein 1 (LMP1) in nasopharyngeal carcinoma (NPC) CNE-2 cell growth and the effects of LMP1-knockout on EBV infection and CNE-2 cell growth using the CRISPR-Cas9 system. The researchers found that LMP1 overexpression promoted CNE-2 cell growth compared to LMP2A overexpression. LMP1 knockout significantly hindered EBV proliferation in CNE-2 cells and markedly inhibited LMP1-mediated promotion of cell growth. The knockout of either LMP1 or LMP2A blocked the activation of eukaryotic translation initiation factor 4E (eIF4E) induced by EBV infection or LMP1/LMP2A overexpression. The study concluded that LMP1-mediated promotion of NPC cell growth can be effectively blocked by CRISPR-Cas9-mediated LMP1 knockout, and precise LMP1 knockout might be a promising method for targeted inhibition of EBV infection and NPC cell growth.28
These types of examinations aiming to abolish the viral oncogenes are limited and this era is open to be studied vastly in combatting virus-related malignancies.
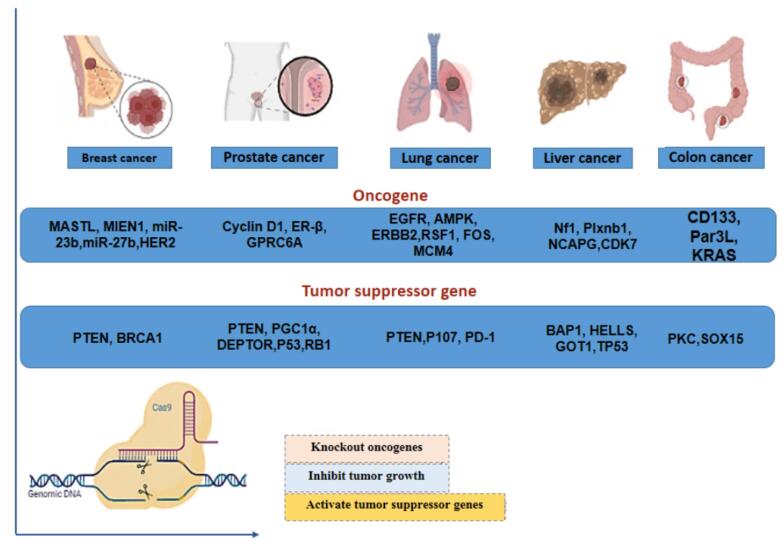
Tumor cells may acquire several types of genetic alterations affecting proto-oncogenes, tumor-suppressing genes, DNA repair genes, or genes involved in apoptosis and cell survival. These alterations include point mutations, deletions, amplifications, and non-random chromosomal abnormalities such as translocations, deletions, and inversions. Different cancers usually exhibit distinct DNA fingerprints related to one (or more) of the mentioned genetic alterations. CRISPR-Cas9 technique can disrupt the activity of oncogenes and restore tumor suppressor gene activity (Fig. 2).29 Some of these DNA alterations are the main genetic drivers of carcinogenesis in certain tumors, which can be regarded as hallmarks for molecular diagnosis and hotcakes in targeted tumor therapy.
Fig. 2.
The utilization of the CRISPR-Cas9 tool shows promise in treating various solid tumors by effectively targeting oncogenes and tumor-suppressor genes in cancer therapies, including those for breast, lung, liver, prostate, and colon cancers. Cas9 has extensively improved or suppressed the activity of different oncogenes and reduced tumor growth.
In the following sections, we will notify some interesting reports on gene editing made by CRISPR-Cas9 on some of the major organs’ cancers.
Cancers targeted by CRISPR-Cas9 in preclinical and clinical practice
Breast cancer
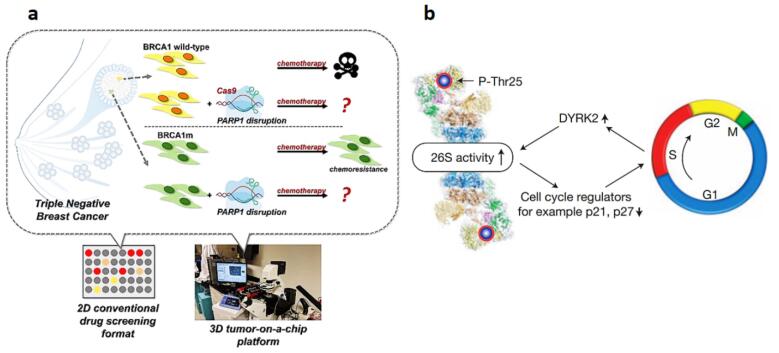
Today, breast cancer is the second leading cause of cancer-related deaths among females, with heterogeneous molecular characteristics, subtypes, and diverse clinical consequences.30 According to the expression of estrogen receptors (ERs), the breast epithelium is classified into four subgroups: luminal A, luminal B, Her2-enriched, and triple-negative breast cancer.31 CRISPR-Cas9 has recently gained much attention for genetic manipulation targeting HER-2, MIEN1, and MASTL oncogenes as well as PTEN and BRCA1 tumor suppressor genes. Concerning the role of mir-23b and mir-27b, Hannafon et al knocked out these genes using CRISPR-Cas9 in the MCF-7 cell line, showing a potent oncogenic role for mir-23b and mir27b in breast cancer. These data were compatible with the in vivo results, showing a reduction in cancer progression in xenograft nude mice following miR-23b and miR-27b knockout.32 HER-2 was directly targeted via the CRISPR-Cas9 editing tool in Wang and colleagues’ experiment. They demonstrated the efficient role of this technique in inhibiting the growth of BT-474, SKBR-3, and MCF-7 cell lines.33 Mintz et al used 2D and 3D tumor-chip models to evaluate the synergistic effect of various chemotherapies and CRISPR-Cas9 in BRCA1 wild type and BRCA1 mutant (BRCA1m) cell lines, namely as MDA-MB-231 and MDA-MB-436, respectively (Fig. 3A).34 BRCA1m displayed higher sensitivity to three indicative chemotherapy medicines, including docetaxel, gemcitabine, and doxorubicin in comparison to the wild-type form. Similarly, delivery of sgRNA to HCC1806 and MCF10A cells leads to considerable inactivation of both alleles of APOBEC3G by inhibiting cell proliferation and blocking the G1/S transition in breast cancer.35 In another survey, the role of the Migration and Invasion Enhancer (MIEN1) gene in the development and spread of breast cancer was confirmed by gene deletion.36 Likewise, Pulver et al reported that genome editing through lentiviral administration of the CRISPR-Cas9 targeting tumor suppressor genes in a mouse model significantly increased breast cancer resistance in triple-negative breast cancer.37
Fig. 3.
(a) Tumor-on-chip models to examine the combined therapeutic effects of different types of chemotherapy treatments and CRISPR-Cas9 in two breast cancer cell lines. 34 (b) A model of the 26S proteasome's reversible phosphorylation. Cell cycle-dependent Rpt3-T25 phosphorylation controlled by DYRK2 facilitates the degradation of crucial proteins like p21 and p27, thereby promoting cell cycle progression 40 (Reprinted with permission under the Copyright Clearance Center).
Dai et al identified tumor regulatory functions for essential mTOR and Hippo pathways components in triple-negative breast cancer (TNBC). They showed the therapeutic relevance of their findings using in vitro drug-synergy matrix models and in vivo patient-derived xenografts. Moreover, they found that pharmacological inhibition of mTORC1/2 and oncoprotein YAP efficiently reduces tumorigenesis in TNBC, and torin1-mediated mTORC1/2 inhibition promotes macropinocytosis, further facilitating verteporfin uptake and enhancing its pro-apoptotic effects in cancer cells. Their study underscored the power and robustness of in vivo CRISPR genome-wide screening in identifying clinically relevant and innovative therapeutic modalities in cancer.38 Inhibition of MASTL kinase effectively condensed human mammary tumor cell line growth, suggesting that MASTL interruption is a privileged choice for breast cancer treatment39 and Guo et al could interrupt the carcinogenesis of the proteasome-addicted human breast tumor cells in vivo by knocking out dual-specificity tyrosine-regulated kinase 2 (DYRK2) using the CRISPR-Cas9 technology (Fig. 3B).40
Considering the role of tumor suppressor genes in breast cancer progression, Annunziata et al reported a novel approach to validate candidate tumor suppressors that may be involved in invasive lobular breast carcinoma (ILC) in vivo. ILC is known for the elimination of E-cadherin, the cell-cell adhesion molecule. To model ILC, lentiviral vectors encoding either Cre recombinase, the CRISPR/Cas9 system, or both were utilized to inject female mice with conditional alleles of the Cdh1 gene. By employing CRISPR-Cas9-mediated somatic gene editing, the PTEN gene was specifically disrupted in ILC-initiating cells.41
To assess the usefulness of targeted somatic manipulation of missense mutations in breast cancer, a knock-in mouse with the Cre-conditional expression of a cytidine base editor was created. To evaluate the impact of defined allelic variations on mammary carcinogenesis, a designed sgRNA-encoding vector was applied to introduce specific point mutations. This model was successfully applied in triple-negative breast cancer (TNBC) simulations.42 Table 2 delineates the studies with CRISPR-Cas9 system that had efficient results in antitumor therapy by targeting oncogenes and tumor suppressor genes in breast cancer.
Table 2. Studies using CRISPR-Cas9 in the treatment of solid tumor cell line/animal model .
| Cell line/animal model | Gene | Assay setting | Vector | CRISPR effect | |
|---|---|---|---|---|---|
| Breast Cancer | MCF-7 | miR-23b and miR-27b | In vitro | lentiviral vectors | Educed tumor growth in xenograft nude mice |
| BT-474, SKBR-3, MCF-7 | HER2 | In vitro | Plasmid | Inhibits cell proliferation | |
| MDA-MB-231, MDA-MB-436 | PARP1 | In vitro | Plasmid | Knockout PARP1 inhibitors | |
| MCF10A, HCC1806 | APOBEC3G | In vitro | pX459 Plasmid | Knockout of both alleles of APOBEC3 | |
| MDA-MB-231 | MIEN1 | In vitro | PX458 and MLM3636 plasmids | Disruption of the MIEN1 gene | |
| K14Cre; p53F/F; Cas9 mouse model | BRCA1 or BRCA2 | In vivo | lentiviral vectors | Somatic gene disruption of tumor suppressor genes | |
| Athymic nude mice | MASTL kinase | In vivo | pLKO.1 lentiviral plasmid | MASTL knockdown | |
| Athymic Nude-Foxn1nu mice | DYRK2 | In vivo | Retroviruses | Knockout of DYRK2 | |
| pSECC-sgPten in Cdh1F/F | Cdh1 | In vivo | lentiviral vectors | ILC development | |
| WB1P-Cas9 mice | BRCA1 and p53 | In vivo | Lenti-sgRNA-Myc lentiviral vectors | Point mutations with high efficiency in one or multiple endogenous genes | |
| Liver Cancer | Huh7 and Hep38 | CDK7 | In vitro | lentiviral vectors | CDK7 knockout |
| C3A | CD44 | In vitro | Lentivirus plasmid | Knockout of CD44 | |
| Huh7, Hep3B and PLC BALB/c mice |
LncBRM | In vitro/In vivo | lenti Cas9-EGFPvector | Knockout of the IncBRM | |
| Hep3B, HepG2, SNU387, SNU398, SNU449, HuH7 and PLC/PRF/5 BALB/c nude mice | ERK2 (MAPK1) | In vitro/In vivo | lentiviral vectors | Sensitizes several liver cancer cell lines to sorafenib | |
| Hep3B, HepG2, and Huh7 p53flox/flox mice | Nf1, Plxnb1, Flrt2, and B9d1 | In vitro/In vivo | pX330 vector/ lentiV2 | Inactivation of Plxnb1, Nf1, Flrt2, and B9d1 | |
| HEK293 and BEL7402 nude mice | survivin | In vitro/In vivo | Lactose-derived branched cationic biopolymer (LBP) | Downregulate of survivin oncogene | |
| Huh7,HepG2, Hep3B, PCL/PLF5, and HEK293T orthotopic Huh7 mice | kinesin spindle protein (KSP) | In vitro/In vivo | E1E2-pseudotyped lentiviral vectors | Powerful KSP gene disruption | |
| SMMC-7721 BALB/c nu/nu mice | HIF-1α | In vitro/In vivo | pLenti-Cas9-sgRNA719/720/721-egfp vectors | HIF-1α knockout | |
| T-cells | PD-1 | Clinical trial | - | Recruiting | |
| Lung Cancer | A549, NCI-H460 | PTEN | In vitro | Plasmid | Knockout of both alleles of PTEN |
| NCI-H1975, NCI-H1650 | EGFR | In vitro | lentiviral vectors | Knockout of EGFR | |
| 634T, 821T4 and 807LN/Genetically engineered mouse model | AMPK | In vitro/In vivo | lentiviral vectors | AMPK deletion in KrasG12D lung tumors | |
| Trp53flox/flox; Rb1flox/flox;Rosa26LSLLuciferase/LSL-Luciferase mice | p107 | In vivo | Adenoviral vectors | Knockout of genes | |
| Prostate Cancer | PC-3, LNCap, DU145, 22Rv1/Athymic nude mice | GPRC6A | In vitro/In vivo | lentiviral vector | Blocking of GPRC6A |
| PC-3 cells | TP53 | In vitro | Plasmid | Repair the TP53 414delC gene region | |
| 2924V cell line/ | PTEN | In vitro | Plasmid | Inactivation of PTEN | |
| ERβcrispr−/− mouse | ERβ | In vivo | - | Delete the ERβ gene | |
| Human PC cells, PC3 and DU145, & the murine PC cell line C57BL/6 male TRAMP mouse | IL30 | In vitro/In vivo | lentiviral vector | IL30 deletion | |
| LNCap, PC-3 nude mice | PLK1 | In vitro/In vivo | A10-liposome- CRISPR/Cas9 chimeras | Silencing PLK1 gene |
Liver cancer
Hepatocellular carcinoma (HCC) is the one of the most common malignancies leading to death in both men and women.18 Some of the most significant mutations predisposing to HCC are activation of Plxnb1, NCAPG, CDK7, CD44, Nf1 oncogenes,18,43 and silencing of BAP1, HELLS, and P53 suppressor genes.44-46 Preliminary reports on the application of the CRISPR-Cas9 system to silence some of the genes involved in liver carcinogenesis are described in Table 2.
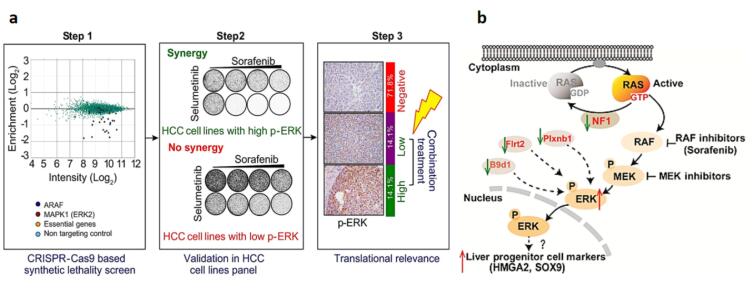
Feng et al conducted a genome-wide CRISPR-Cas9-based knockout screen and revealed a relationship between the anti-tumor effects of metformin and the levels of DOCK1. They established patient-derived HCC organoids to characterize the role of DOCK1 in determining metformin sensitivity. They also found that combining metformin with DOCK1 inhibition may provide a promising personalized therapeutic strategy for metformin-resistant HCC patients.47 Wang et al gave an idea of lentiviral delivery of sgRNA into Huh7 and Hep38 liver cell lines and described CDK7, a cyclin-dependent kinase, as a hit target in hepatic cell growth.48 Additionally, Huh7 and Hep38 growth rates were noticeably reduced by CRISPR-Cas9-mediated CDK7, indicating that this gene may be a good target for HCC treatment. Successful knockout of the CD44 gene in HCC, a transcriptional regulatory gene, was achieved by lentiviral delivery of sgRNA and Cas9 to C3A-iCSCs liver cancer stem cells (C3A-induced cancer stem cells). The results of this research indicate the function of CD44 in the poor differentiation of cancer stem cells, high malignant degree of tumor cells, and tumor growth.49 Zhu et al knocked out the IncBRM gene in two liver cancer cell lines, Huh7 and Hep3B. They noted that knocking out the IncBRM gene affected the development of the serial sphere while not affecting the expression of nearby genes, suggesting that IncBRM has a trans role.50 In addition, another experiment used the CRISPR-Cas9 system to reverse the limited benefits of sorafenib as the recommended treatment for people with advanced HCC. They suggested that inhibition of ERK2 (MAPK1) causes a variety of HCC cell lines to be susceptible to sorafenib. Thus, it can be assumed that higher levels of ERK (more than 30% of all liver tumors) leading to a better response to this polytherapy, and inhibiting different kinases, such as ERK and MEK, represent a promising therapeutic tool in HCC treatment (Fig. 4A).51 Song et al used CRISPR-Cas9 and discovered that the genes Plxnb1, B9d1, Flrt 2, and Nf1 inhibit liver tumors.52 When sgRNA and Cas9 were delivered to p53-/-; Myc; Cas9 hepatocytes, the NF1 gene was successfully knocked out, and subsequent liver cancer inhibition in a mouse model was achieved (Fig. 4B). An in vivo experiment used a lactose-derived branched cationic biopolymer (LBP) with the superior properties of gene transfection, biocompatibility, compacting, degradability, and HCC-targeting for the delivery of CRISPR-Cas9 to an orthotopic mouse model of HCC. Targeting survivin oncogenes exhibited excellent anti-cancer activities as well as effective gene editing performances.53 Besides, systemic administration of E1E2-pseudotyped lentiviral vectors could deliver Cas9 and sgRNA-specific for kinesin spindle protein (KSP) to Huh7 tumors in orthotopic Huh7 mice. This particular delivery system effectively disrupted the KSP gene, thereby potently suppressing the growth of HCC.54 Due to the essential role of hypoxia-inducible factor 1-α (HIF-1α) in the pathogenesis, invasiveness, and angiogenesis of HCC, it was knocked out in SMMC-7721 cells (liver cancer cells). The HIF-1α knocked out tumor cells considerably and showed low invasiveness and migration. In addition, this strategy suppressed the engraft growth of SMMC-7721 tumors and significantly extended the survival of mice bearing HCC with a decreased CD31 expression level (a tumor angiogenesis marker) and enhanced apoptosis in the tumor cells.55 A clinical trial [NCT04417764] started to knockout PD-1 receptors using the CRISPR-Cas9 system in autologous T lymphocytes taken from individuals with advanced HCC combined with trans-catheter arterial chemoembolization.56
Fig. 4.
(a) An in vitro and in vivo experiment employed CRISPR-Cas9 to assess the combined inhibitory effects of selumetinib and sorafenib in the treatment of liver tumors.51 In HCC cell lines with elevated p-ERK, Selumetinib augments the response to sorafenib, and inhibiting ERK2 enhances the sensitivity to sorafenib in HCC. Synthetic inhibition of ERK kinase results in a synthetic lethal effect. In cancers with high p-ERK levels, combination therapy is highly likely to yield benefits (Reprinted with permission under the Copyright Clearance Center). (b) Diagram showing how the loss of potential tumor suppressor genes causes the MAPK pathway to be activated and causes the liver progenitor cell markers HMGA2/SOX9 to be induced in liver cancer activity (Reprinted with permission under the Copyright Clearance Center).
Lung cancer
Many genes have been linked to the evolution and progression of different histological types of lung cancers, but there are limited studies using molecular manipulation of these genes by CRISPR-Cas9.57,58 Perumal et al utilized plasmid delivery (non-viral) of CRISPR-Cas9 to A549 and NCI-H460 (Slug/Snail non-small lung cancer cells) and successfully knocked out the PTEN gene. They disclosed that knocking out the PTEN gene was associated with high rates of cancer cell growth, metastasis, and invasion. Wild-type PTEN suppresses the phosphorylation of β-catenin while enhancing GSK-3β and AKT pathways.59 Another study showed that the presence of T > G, A > G, and C > G point mutations can be treated using CRISPR-Cas9 to target specific parts of the genome in L858R mutant lung cancer cells.60 Eichner et al removed the murine AMPK genes (α1 and α1) and thereby the size of murine lung tumors (KrasG12D) was significantly reduced.61 In another survey, CRISPR-Cas9 helped to knockout p107 and related p130 genes (tumor suppressor genes) in a mouse model, resulting in the development and progression of small cell lung cancer (SCLC).62 They also showed the possibility of employing CRISPR-Cas9 technology to stimulate the deletion of tumor suppressor genes in animal models of SCLC. In 2020, an in vivo epigenetic CRISPR screen in a KRAS-mutant lung adenocarcinoma (LUAD) model identified Asf1a as an immunotherapeutic target. Li et al constructed an epigenetic-focused sgRNA library and performed an in vivo CRISPR screen in the KrasG12D/P53−/− (KP) LUAD model. They found that loss of the histone chaperone Asf1a in tumor cells sensitizes tumors to anti-PD-1 treatment. Mechanistic studies revealed that tumor cell-intrinsic Asf1a deficiency induced immunogenic macrophage differentiation in the tumor microenvironment by upregulating GM-CSF expression and potentiated T-cell activation in combination with anti-PD-1.63 The outcomes of a phase I clinical trial in individuals with advanced non-small cell lung cancer [NCT02793856] revealed that infusion of PD-1-edited T cells modified by transfection of Cas9 and sgRNA to T cells by electroporation increased the median progression-free survival with no serious adverse event.64 The effects of the CRISPR-Cas9 editing technology on different lung tumor cell lines are illustrated in Table 2.
Colon cancer
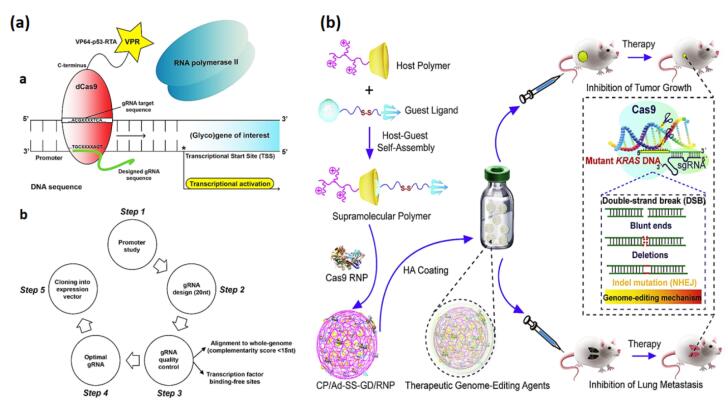
Colorectal cancer (CRC) cells exhibit increased fucosylation of the Lewisx antigen, attributed to the abnormal expression of pertinent fucosyltransferases like fucosyltransferase 4 (FUT4) and fucosyltransferase 9 (FUT9).65 In this regard, Blanas et al activated the expression of fucosyltransferases (Fut4 and Fut9) at the transcriptional level in a mouse colon cancer cell (MC38) via the CRISPR-d Cas9-VPR system, which resulted in the expression of functional Lewisx antigen on the cell surface (Fig. 5A). In both MC38-FUT4 and MC38-FUT9 cells, Lewisx was mostly transported by N-linked glycans and had a significant effect on MC38-glycovariants' core-fucosylation, sialylation, antennarity, and N-glycan subtypes. Data from this study demonstrated the CRISPR-dCas9-VPR system's use for amplifying the expression of glycosyltransferase as a propitious technique in glycobiology and oncology experiments.65 Similarly, lentiviral delivery of the CRISPR-Cas9 system to colon cancer cells (LoVo) caused CD133 knockout and blocked tumorigenic features, cell migration, and metastasis.66 Restoration of a loss-of-function mutation of Protein Kinase C (PKC) through CRISPR-Cas9 interrupted anchorage-independent proliferation in a patient-derived colon cancer cell line and decreased tumor proliferation in a xenograft model.67 In a different experiment, Cas9/sgRNA lentiviral delivery into the CaCO-2 cell line was able to remove the Par3L protein in colon cancer cells by suppressing cell growth, apoptosis induction, and activation of the cascade-3 pathway.68 Due to the sensitivity of Par3L proteins to antitumor chemotherapies, the inactivation of Par3L by inhibiting AMPK signaling transduction can be a prospective target for treating CRC cell proliferation. Michels et al designed a platform for pooled CRISPR-Cas9 screening in human colon organoids to identify tumor suppressors. The researchers in this project developed optimal conditions and strict guide RNA requirements for screening in 3D organoids. They demonstrated that this technology permits unbiased detection of genes that confer positive selection in vitro and after xenotransplantation.69 In an in vitro and in vivo experiment, delivery of hyaluronic acid (HA)-associated CP/Ad-SS-GD/RNP nanocomplexes to a CRC mouse model efficiently suppressed the growth and invasion of tumor cells by specifically focusing on the KRAS mutant gene using CRISPR-Cas9 (Fig. 5B).21 Additionally, the effective transport of Cas9 RNP through CP/Ad-SS-GD/RNP nanocomplexes to 293T cells and CRC cells leads to significant genome-editing capability in vitro. This particular delivery system is a supramolecular polymer to facilitate effective controlled delivery of Cas9 RNP. This system is formulated through the complexation of disulfide-bridged biguanidyl adamantine (Ad-SS-GD) with β-cyclodextrin-conjugated low-molecular-weight polyethyleneimine (CP) via supramolecular assembly, resulting in the formation of CP/Ad-SS-GD.
Fig. 5.
(a) A model illustrating the CRISPR-dCas9-VPR system (a) and an overview (b) the CRISPR-dCas9-VPR technology's five-step experimental design, specifically applied to transcription.65 (b) The administration of Hyaluronic acid-associated CP/Ad-SS-GD/RNP nano-complexes in a colorectal tumor mouse model (Reprinted with permission under the Copyright Clearance Center).
Prostate cancer
The second-leading cause of cancer-related mortality in male is prostate cancer. Point mutations, structural rearrangements, single nucleotide polymorphisms (SNPs), and copy number variations in specific genes have all been implicated in the development of prostate cancer.18 GPRC6A, P53, PTEN, cyclin D1, and ER-β play significant roles in the pathogenesis of prostate cancer.18,70 The role of CRISPR-Cas9 as a therapeutic approach for prostate cancer has been proven in several studies due to its great precision and limited off-target effects. The GPRC6A receptor was noticeably inactivated after introduction of CRISPR-Cas9 through lentiviral delivery into the human xenograft mouse model (PC-3), and the mitigation of prostate cancer cells was diminished.71 Recently, Batir et al72 proved that lentiviral delivery of sgRNA to a human prostate tumor cell line (PC3) resulted in the successful repair of 26% of the 414delC mutation in TP53. Takao et al. knocked out the PTEN gene in mouse prostate tumor cells. This action activates cyclin D1 and phosphorylates the RAC-alpha serine/threonine kinase. According to their findings, prostate tumor cells lacking PTEN trigger some genes involved in cancer cell survival.73 Fenner et al also determined the ER-β gene as a prostate tumor suppressor gene in mice with prostate cancer by CRISPR-Cas9.74 An in vivo study in a murine prostate cancer model investigated the loss of functions of two FOX transcription factors: FOXA1 and FOXP1. The result showed that deficiency of Foxp1 increased proliferation in combination with loss of Pten, while loss of Foxa1 induced epithelial plasticity in prostate cancer. The researchers also found that Foxp1 is a repressor for the androgen-regulated target Tmprss2, and there is a negative correlation between FOXP1 and TMPRSS2 expression in a human prostate cancer dataset.75 IL-30 stimulates the proliferation, invasion, and migration of prostate cancer cells. These effects are associated with STAT1/STAT3 phosphorylation and upregulation of BCL2 and NFKB1 and some chemokines. Depletion of the IL-30 gene by the CRISPR-Cas led to upregulation of reduction of tumor suppressor genes like SOCS3 and downregulation of STAT3, NFKB1 BCL2, NFKB1, CXCL5, STAT3, IGF1, and CXCL5 in syngeneic and xenograft PC models.76 Polo-like kinase 1 (PLK1), involved in prostate cancer pathogenesis, was remarkably inhibited by an aptamer-liposome-CRISPR/Cas9 chimera delivery system and caused tumor necrosis in treated mice compared to the control group.77
Leukemia
Although chemotherapy and transplantation have improved cure rates of patients diagnosed with leukemia, there are still many patients showing relapse due to regrowth of minimal residual disease (MRD) or developing drug resistance.78 Some specific and well-known genetic events are linked to most leukemia and lymphoma, rendering them appropriate targets for gene editing.79 In the following paragraphs, we briefly notice the studies that directly address manipulating the main genes involved in the pathogenesis and aggressiveness of leukemia. Most of the achievements in treating acute (and less commonly chronic) leukemia have mainly been yielded by the discovery of CAR technology, which will be mentioned in the CAR-cell section. Table 3 summarizes some of the therapeutic benefits of the CRISPR-Cas9 system in several types of leukemia.
Table 3. Therapeutic effects of CRISPR-Cas9 on various types of leukemia .
| Cell line/animal model | Gene | Assay setting | Vector | CRISPR effect | |
| ALL | LM-TK−, TE671, HEK293T, CHO-K1, CHO lec1, and CHO 6B2 | CASD1 | In vitro | Plasmid | CASD1 knockout and reduction of 9-O-acetylation |
| p210 BCR-ABL1T315I mutation mouse model | BCR-ABL1 fusion gene | In vitro/In vivo | Lentiviral vector | Disrupts the BCR-ABL1 fusion gene | |
| NSG mice | RIP1 | In vivo | Plasmid | Disruption of RIP1 | |
| 9-15C/ non-irradiated NOD/SCID/IL-2rγnull (NSG) mice | CXCR4 | In vitro/In vivo | Plasmid | Deletion of CXCR4 gene | |
| T cells | CD52 and TRAC | Clinical trial | Lentiviral vector | Recruiting | |
| T cells | CD19 | Clinical trial | - | Recruiting | |
| AML | HL-60 and KG-1 | BIRC5 | In vitro | Lentiviral vector | Suppression of BIRC5 |
| BALB/cJ, NOD.Cg-Prkdcscid Il2rgtm1Wjl, NSG | IL1RAP | In vivo | lipidoid nanoparticle | IL1RAP knockout | |
| CLL | MEC-1 | Notch1 | In vitro | Plasmid | Emulate NOTCH1 mutations |
| HG3 | FBXW7 | In vitro | Plasmid | FBXW7 mutations increase in NOTCH1 target gene expression | |
| CML | K562/ e NOD/SCID mice | BCR-ABL gene | In vitro/In vivo | PEG-PLGA nanoparticles | Knocked out the BCR-ABL fusion |
| K562, K562/G01, U937, HL60 and CML stem/progenitor cells/ NOD/SCID mice | BCR-ABL gene | In vitro/In vivo | Plasmid | Disrupt BCR-ABL gene |
Acute lymphoblastic leukemia
Studies have shown that 9-O-acetylation of sialic acids (Sias) in ALL is crucial for lymphoblast medication resistance.79,80 Baumann et al showed the crucial role of CASD1 (a sialate O-acetyltransferase) in the process of 9-O-acetylated sialoglycans synthesis by knocking out this gene.81 T315I mutation is resistant to all approved tyrosine kinase inhibitor. Tan et al82 reported that in vivo delivery of CRISPR-Cas9 efficiently postpones the rapid progression of Ph+ ALL (a subgroup of ALL with c-kit expression) with this mutation. McComb et al showed that the SMAC (second mitochondrial-derived caspase-activators) gene reactivates apoptosis of tumor cells mediated by RIP1. Knocking out this gene in refractory and relapsed ALL xenograft models represses RIP1 expression, impairing cell death.83 To confirm the crucial role of CXCR4 in the pathogenesis of ALL, the deletion of CXCR4 in a B-ALL cell line was carried out, leading to a reduction in the in vivo aggressiveness of ALL.84 During the patient recruitment phase of a clinical trial [NCT04557436], allogeneic-engineered human T-cells (also referred to as TT52CAR19+TCRαβ-) were employed in the treatment of CD19+ B cell leukemia. The lentiviral vector carrying CRISPR constructions was used for editing the CD52 and TRAC loci and transiently supplied Cas9. These constructions were applied to transduce the cells for the expression of an anti-CD19 chimeric antigen receptor (CAR19). Through T cell-mediated cytotoxicity, CD19+ leukemia and normal CD19+ B cells were eliminated in response to recognition by TT52CAR19 T-cells.85 The other trial [NCT04154709] has been designed for the evaluation of the safety and feasibility of CTA101 (a CD19/CD22 dual-targeted CAR-T cell product developed using CRISPR-Cas9 technology) for treating relapsed or refractory CD19+ B-ALL.86
Acute myelocytic leukemia (AML)
Targeting suppression of survivin gene expression in AML, Narimani et al knocked out BIRC5 in AML cell lines, namely HL-60 and KG-1, through site-specific cleavage of its locus. Additionally, inhibition of BIRC5 caused cell viability to decrease and caused necrosis and cell death in the mentioned cell lines.87 Moreover, HO et al introduced leukemia stem cells (LSCs) as a critical player in the pathogenesis and relapse of AML and removed them successfully as a therapy model. They utilized a reducible lipidoid-encapsulated Cas9/single guide RNA (sgRNA) ribonucleoprotein [lipidoid nanoparticle (LNP)–Cas9 RNP] to target the interleukin-1 receptor accessory protein (IL1RAP) and eliminate leukemic stem cells (LSCs). Loading of LNP-Cas9 RNP and the chemokine CXCL12α onto mesenchymal stem cell membrane–coated nanofibril (MSCM-NF) was also done to mimic the environment of bone marrow and increase the capacity of LSC targeting. The in vitro data revealed that LSCs could migrate to MSCM-NF scaffolds, and LNP-Cas9 RNP-mediated gene editing reduced LSCs' capacity to form colonies.88
Chronic lymphoblastic leukemia
Chronic lymphoblastic leukemia (CLL) is the most prevalent form of adult chronic leukemia characterized by a progressive increase in clonal mature-appearing CD5+ B cells in the bone marrow, lymph nodes, and peripheral blood cells. The most relevant mutated genes in CLL are Notch signaling genes, TP53, and ATM.89,90 Arruga et al used CRISPR-Cas9 technology to create several NOTCH1 variants. They demonstrated the role of NOTCH1 signaling in tumor growth and chemotaxis through the downregulation of the tumor suppressor DUSP22 and suggested the contribution of NOTCH1 mutations to the pathogenesis and prognosis in CLL.91 CRISPR-Cas9 technology was used to knockout FBXW7, another mutated gene in NOTCH1 signaling. Following truncation of this gene, the activity of the intracellular domain of NOTCH1, c-MYC protein, and HIF-1α was increased in the HG3 cell line. It has also been reported that CLL patients with dysregulation of FBXW7 could be functionally related to NOTCH1 mutations.92
Chronic myeloblastic leukemia
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder primarily identified by the reciprocal translocation between chromosomes 9 and 22 (BCR-ABL fusion).93 CRISPR-Cas9 technology has received more attention as an exquisite therapeutic protocol for CML and as in vitro model development.94,95 For this purpose, Liu et al used an encapsulated CRISPR-Cas9 plasmid with poly (ethylene glycol)-b-poly (lactic acid-co-glycolic acid) (PEG-PLGA)-based cationic lipid-assisted polymeric nanoparticles (CLANs) to directly target the overhanging BCR-ABL gene fusion region in a mouse model. They effectively knocked out BCR-ABL fusion following the intravenous injection of CLANs carrying pCas9/gBCR-ABL in mice CML models. They observed an improvement in the survival rate and proposed the CRISPR-Cas9 system utilization combined with nanocarriers as an efficient therapy for CML.96 In another study, BCR-ABL fusion was stopped using FokI nuclease (RFN) delivery through the CRISPR-Cas9 editing system. This demonstrated that RFNs effectively impaired BCR-ABL and inhibited cell proliferation in CML lines (K562, K562/G01, U937, and HL60) and CML stem/progenitor cells. In an additional survey, RFNs remarkably suppressed the leukemogenesis capacity of CML cells in a xenograft model.97 Table 3 details the therapeutic benefits of the CRISPR-Cas9 system in several types of leukemia.
CAR-T cells and CRISPR
Currently, novel and promising avenues for immunotherapy have been created by chimeric antigen receptors (CAR). This fusion protein comprises a selected single-chain variable fragment (ScFV) from a specific monoclonal antibody and one or more intracellular T-cell receptor–derived signaling domains.98 Modification of autologous or allograft T-cells to express CAR (CAR-T cells) leads to an efficient and selective recognition of tumor-associated antigens (TAA).99 By making a universal "off-the-shelf" cellular product or altering immune cells to overcome resistance in hematological or solid malignancies, CRISPR-Cas9-based genome editing provides the possibility of more effective immunotherapy.100 The CRISPR-Cas9 system unquestionably helps address the unmet needs of CAR-T cell-based therapy for malignancies despite multiple roadblocks regarding the safety, effectiveness, and scalability.101 Template-free and non-DSB genome editing procedures, multiplex disruption of inhibitory molecules and signaling pathways, targeted gene knock-in, reducing the frequency of cytokine release syndrome (CRS) and GVHD, and manufacturing universal CAR T-cell products using normal donor leukocytes or iPSCs are some examples of such advancements of CRISPR technique.102,103
Several preclinical studies have utilized the CRISPR technique, which has dramatically improved the function of CAR-T cells.104,105 Eyquem et al designed a dual strategy of CD19 CAR knock-in and TCR knockout, which reduced the likelihood of GVHD and simultaneously prevented CAR signaling by optimizing CAR internalization and re-expression kinetics.106 Another study used CRISPR-Cas9 technology to simultaneously disrupt PD-1, endogenous T-cell receptor (TRAC), and beta-2 microglobulin (B2M) genes. The results indicated the long-term survival of glioma mouse models following intracerebral injection of this universal CAR-T cell product.107
CRISPR-Cas9 application for generating feasible, effective, safe, and universal CAR-T cells has already entered the clinical phase (So far, 18 trials have been registered at clinicaltrials.gov). Most of these clinical studies are related to hematologic neoplasms with the aim of targeting CD19 markers (Table 4).
Table 4. Clinical trials based on the application of CRISPR-Cas9 strategy for improving the efficacy of CAR-T cell .
| Rank | Clinical Trial identifier | Status | Clinical trial phase | Disease | CAR Antigen | Target Locus of CRISPR | NK source | Number enrolled | Interventions | Dosage | Starting date | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NCT03166878 | Unknown | Phase 1/2 | CD19+ leukemia and lymphoma | CD19 | Endogenous TCR and B2M genes | Allogeneic | 80 | Universal CRISPR-Cas9 Gene-Editing CAR-T Cells Targeting CD19 | Unknown | June 2017 | China |
| 2 | NCT03398967 | Unknown | Phase 1/2 | B-cell malignancies | CD19, and CD20 or CD22 | Unknown | Allogenic | 80 | Universal Dual Specificity CD19 and CD20 or CD22 CAR-T Cells | Unknown | January 2, 2018 | China |
| 3 | NCT03545815 | Unknown | Phase 1 | Solid Tumors | Mesothelin | PD-1 and TCR gene-knocked out | Unknown | 10 | anti-mesothelin CAR-T cells | Unknown | March 19, 2018 | China |
| 4 | NCT03747965 | Unknown | Phase 1 | Solid Tumors | Mesothelin | PD-1 gene-knocked out | Unknown | 10 | Mesothelin-directed CAR-T cells | Unknown | November 2018 | China |
| 5 | NCT05812326 | Completed | Phase 1/2 | MUC1-positive breast cancer | MUC1 | PD-1 | Autologous | 15 | AJMUC1- PD-1 gene knockout anti-MUC1 CAR-T cells | 3×105/kg, 1×106/kg 3×106/kg | May 17, 2019 | China |
| 6 | NCT04035434 | Recruiting | Phase 1 | B-cell malignancies | CD19 | Unknown | Allogenic | 143 | CD19-directed CAR-T-cell (CTX110) | Unknown | July 22, 2019 | Unknown |
| 7 | NCT04037566 | Recruiting | Phase 1 | CD19+ leukemia or lymphoma | CD19 | Endogenous HPK1 | Autologous | 40 | CD19-specific CAR-T cells with edited endogenous HPK1 (XYF19 CAR-T cells) | Unknown | August 2019 | China |
| 8 | NCT04438083 | Active, not recruiting | Phase 1 | Renal Cell Carcinoma | CD70 | Unknown | Allogeneic | 107 | CRISPR-Cas9-Engineered T Cells (CTX130) | Unknown | June 16, 2020 | USA |
| 9 | NCT04502446 | Recruiting | Phase 1 | T or B cell malignancies | CD70 | Unknown | Allogeneic | 45 | Anti-CD70 CRISPR-Cas9-Engineered T Cell | Unknown | July 31, 2020 | USA |
| 10 | NCT04557436 | Active, not recruiting | Phase 1 | B-ALL | CD19 | CD52 and TRAC loci | Allogenic | 10 | CRISPR-CAR Genome Edited T Cells (PBLTT52CAR19) | Unknown | August 12, 2020 | UK |
| 11 | NCT04767308 | Not yet recruiting | Early phase 1 | CD5+ Hematopoietic Malignancies | CD5 | Unknown | Autologous | 18 | Anti-CD5 CAR-T Cells | 1×106/kg 2×106/kg 3×106/kg |
March 2021 | Unknown |
| 12 | NCT04637763 | Recruiting | Phase 1 | B cell non-Hodgkin lymphoma | CD19 | Unknown | Allogeneic | 72 | CRISPR-edited allogeneic CAR-T cell therapy targeting CD19 | Unknown | May 26, 2021 | USA |
| 13 | NCT04976218 | Recruiting | Phase 1 | EGFR positive solid tumors | EGFR | TGF-β receptor Ⅱ | Unknown | 30 | TGFβR-KO CAR-EGFR T Cells | 1-2×105/kg 1×106/kg 1×107/kg |
March 15, 2022 | China |
| 14 | NCT05397184 | Recruiting | Phase 1 | T Cell Malignancies | CD7 | Unknown | Allogeneic | 10 | Cryopreserved BE CAR7 T cells | Unknown | April 19, 2022 | UK |
| 15 | NCT05037669 | Withdrawn | Phase 1 | CD19+ Leukemia and Lymphoma | CD19 | Endogenous TCR, HLA-class I and HLA-class II | allogeneic | 0 | CRISPR-edited T Cells (PACE) Gene Edited to Eliminate Endogenous TCR, HLA-class I and HLA-class II and Engineered to Express Anti-CD19 CAR | Unknown | July 2022 | Unknown |
| 16 | NCT05722418 | Recruiting |
Phase 1 |
Multiple myeloma | BCMA | Unknown | allogeneic | 50 | CRISPR-Edited Allogeneic Anti-BCMA CAR-T Cell | Unknown | February 6, 2023 | USA |
| 17 | NCT05643742 | Recruiting | Phase 1/2 | B-cell malignancies | CD19 | Unknown | allogeneic | 120 | Anti-CD19 Allogeneic CRISPR-Cas9-Engineered T Cells (CTX112) | Unknown | March 10, 2023 | USA |
| 18 | NCT05795595 | Recruiting |
Phase 1/2 |
Solid tumors | CD70 | Unknown | allogeneic | 250 | Anti-CD70 Allogeneic CRISPR-Cas9-Engineered T Cells (CTX131) i | Unknown | April 2023 | USA |
A part of the data from the phase 1 clinical trial [NCT04557436] was published in 2022. In this experiment, the CRISPR-Cas9 technology was applied to improve the efficacy of CAR19 T cells in such a way that T-cell receptor α-chain CD52 antigen was disrupted in CAR19 T cells (TT52CAR19 T cells) through lentiviral vector carrying Cas9 and sgRNA. Following the lymphodepletion regimen, six children with relapsed/refractory CD19-positive B-ALL received a single dose of CAR T-cells (spanning 0.8×106 to 2.0×106/kg). Although no immediate toxicity was seen, grade II cytokine release syndrome, skin GVHD, and transient grade IV neurotoxicity were observed in some patients. Generally, safety, feasibility, and therapeutic application of CRISPR-engineered immunotherapy have been confirmed.108 Additionally, during a clinical trial performed by Guo et al, TCR and B2M double-removed CAR-T cells were created by lentiviral and electroporation delivery of the CRISPR-Cas9 editing tool. In this study, the lymphodepletion regimen was followed by allogeneic CAR T-cell administrationin two patients with R/R diffuse large B-cell lymphoma at a dose of 1.5×106/kg and 1.21×106/kg cells for each subject. Despite disease progression, both patients showed expansion and biological acting without GVHD.109 It is charming to point to the results of phase I clinical trial [NCT04035434], which examined the effect of AAV-mediated anti-CD19 CAR-T cells and CRISPR-Cas9-disrupted TRAC and B2m genes. Although long-term outcomes are still pending, interim findings in NHL patients showed a 38% remission rate with no GVHD. Only one case of severe neurotoxicity related to viral reactivation has been reported.110 A similar approach was used to test the effect of CRISPR-Cas9 mediated-anti-CD70 CAR incorporation into the TRAC locus with a second edit TRAC, B2m, and CD70 in 15 T-cell lymphoma volunteers. The preliminary results of this trial showed complete remission (29%), no detectable GVHD, and a tolerable toxicity profile.111
Finally, regarding improvements in accuracy and efficiency, as well as the possibility of multiplex targeting of genes, antigens, and checkpoint molecules,112 it is hoped that positive data from these clinical studies will revolutionize the treatment of a larger variety of malignancies and ameliorate their outcomes.
CAR-NK cell and CRISPR
Natural killer (NK) cells are fundamental components of the innate immune system developing in the bone marrow (BM) and secondary lymphoid tissues.113 They play antiviral and antitumor roles in multiple ways, like releasing lysing granules (perforin and granzyme), cytokine secretion, and antibody-dependent cell cytotoxicity (ADCC).114 NK cells have the innate ability to identify and eliminate transformed cells without prior sensitization, independent of the MHC class antigen presentation. Furthermore, NK cells can distinguish abnormal cells from normal cells through KIR receptors and ligand mismatch, resulting in targeted cytotoxicity and reduced off-target complications.115,116
Given their specified biological characteristics, strong inherent ability to combat cancers, and favorable history of safety in medical practice, NK cells have attracted considerable interest as a promising alternative framework for CAR engineering in the last few years. It is possible to obtain pure populations of NK cells from both autologous and allogeneic sources, such as peripheral blood, umbilical cord blood, different kinds of stem cells (induced pluripotent stem cells and hematopoietic stem cells), and NK cell lines such as NK-92.117 Nevertheless, genetic engineering of NK cells faces some limitations, such as a high rate of cell apoptosis, low efficiency of transduction, and loss of transgene expression after expansion.118 Toward cancer immunotherapy research, CRISPR has proven to be effective in the construction of CAR and TCR structures and the identification of novel immune checkpoint genes.113 Regarding the application of gene editing of NK cells, CRISPR has been utilized for optimizing NK cell metabolism, targeting checkpoint molecules, enhancing antibody therapies, and producing of CAR-NK cells.119
Immunotherapy using CAR-NK cells has the advantage of being performed in HLA-unmatched individuals. However, the injected NK cells are recognized and destroyed by the immune system of the recipient limiting their lifespan in the body and reducing the chance of success in repeated injections. Hoerster et al. utilized CRISPR-Cas9 to suppress HLA class I molecule expression by deleting the B2M gene in NK cells, and then genetically modifying the NK cells to express a single-chain HLA-E molecule. Their study showed that it is possible to access non-HLA-matched primary human NK cells as an “off-the-shelf” product with a high level of safety.120 In a one-step CRISPR delivery through a retroviral particle (RP) system, Jo et al successfully knocked out the TIGIT gene and simultaneously generated anti-epidermal growth factor receptor (EGFR)-CAR NK cells. They generated large amounts of gene-edited CAR-NK cells that have great potential to integrate site-specific CAR transgenes.121 Another targeted gene integration into NK cells was achieved through an innovative electroporation delivery system of Cas9/RNP and ssAAV6 or scAAV6 gene combinations.118 By leveraging the precise targeting capability of CRISPR-Cas9 for the selective introduction of genetic materials into primary NK cells, Kararoudi et al generated numerous high-purity CD33-targeting CAR NK cells with significant antileukemic activity. Cas9 ribonucleoprotein complexes in combination with single-stranded (ss) or self-complementary (sc) adeno-associated virus (AAV)-mediated gene delivery have been used for site-directed gene editing of NK cells.122 Their theory was confirmed in a subsequent study, and the RNP/scAAV6 combination was introduced as a promising approach for clinical application, owing to the stable transduction of NK cells.123 In this direction, the cytokine-inducible Src homology 2-containing (CIS) protein, a critical negative regulator of interleukin 15 (IL-15) signaling, was targeted using CRISPR. Coalescence of this strategy and CAR-NK cells indicated enhanced efficacy of natural killer (NK) cells against tumors in comparison to each modification in a lymphoma mouse model.124 An in vitro study tested the effect of disrupting three distinct inhibitory checkpoints (CBLB, NKG2A, and TIGIT) on the functionality of NK cells using the CRISPR-Cas9 technique in combination with CD276-CAR or CD19-CAR NK-92 cells in U-937 or U-937 CD19/tag AML cell lines. It was concluded that the cytotoxicity of triple-knockout CD276-CAR-NK-92 cells markedly became greater when targeting different AML cell lines.125
Thus, the CRISPR-Cas9 technique has received much attention because of its ability to increase the antitumor capabilities of NK cells. This can be achieved by inserting CARs into NK cells as well as by permanently disabling or integrating specific genes related to NK cell exhaustion, activation, and tolerance functions.126
CAR-M cells and CRISPR
Macrophages (M) are long-lived phagocytic cells that reside in nearly all organs and are vital components of the innate immune system.Macrophages exhibit two functional phenotypes. Classically activated macrophages (M1 type) mainly produce NO and lysosomes, enhance their ability to kill microorganisms, and act as pro-inflammatory cells. The alternatively activated macrophages (M2 type) are principally involved in the repair process.127 A significant portion of the macrophage population within the tumor microenvironment is composed of the M2 phenotype, probably to restore the destructive effects of tumor cells.128 In addition, there is a unique subpopulation of macrophages in solid tumors known as tumor-associated macrophages (TAMs). These TAMs facilitate metastasis, promote invasion and angiogenesis, and augment immunosuppression.129 Due to the infiltration of solid tissues, genetically manipulated macrophages (CAR-M) could be more effective than CAR-T or CAR-NK cells in eliminating solid tumors. However, the pivotal role of macrophages in combating tumor cells depends on their pro-inflammatory state.130
Although the macrophages have been used to combat cancer for many years, early clinical trials showed that transplanting purified macrophages had little antitumor effects, suggesting that additional pro-inflammatory signals are required to battle against tumors.131,132 Opposing signals impede macrophages from destroying targeted tumor cells.133 Cancer cells use some mechanisms (such as self-labeling) to avoid being destroyed by macrophages. One of these significant tools is the "don't eat me" signal between CD47 and the macrophage membrane receptor SIRPα. Exploring these positive and negative signals has attracted scientific attention in CAR-M therapy by manipulating specific gene(s) to augment the ability of macrophages to bind to tumor cells (via antigen identification) and activate their activity in tumor lysis by activating certain enzymes.134,135
One era of the CAR-M studies is to augment immune cell infiltration into tumors via the extracellular matrix (ECM) by producing CAR-147 macrophages. CD147 plays a crucial role in ECM remodeling via the expression of matrix metalloproteinases (MMPs).136 CAR-iPSC-induced macrophages (CAR-iMACs) containing CD86 and FcRγ domains result in M2 macrophages. However, the addition of IFN-γ can polarize the macrophages to M1 to enhance the phagocytic capacity of CAR-iMACs against solid tumors.137
Although the efficacy and safety profile of CAR-M cells have been studied in laboratory animals, their effects in humans are still unknown, and the use of viral transfection in CAR gene transfer may result in unpredictable effects on treatment.138 Zhang et al applied CRISPR-Cas9 technology to insert a third-generation anti-GD2 CAR (14G2a-CD28-OX40-CD3ζ) into the AAVS1 locus of H9 human pluripotent stem cells (hPSCs) for the development of antigen-dependent antitumor macrophages. Their findings indicated that the antitumor activity of CAR-Ms was not affected by the M1/M2 phenotype induced by LPS, IFNγ, and IL-4. Additionally, when hPSC-derived macrophages were exposed to tumor cells, CAR-M expression led to their polarization towards the M1 state. A complementary in vivo study showed that CAR-Ms produced from the arterial hemogenic endothelium (HE) have much greater antitumor activity than wild-type macrophages.139
Focusing on the ACOD1 gene (as a potent regulator of the pro-inflammatory state), human ACOD1 knockout macrophages were developed using an induced pluripotent stem cell-derived CAR macrophage (CAR-iMAC) platform. The engineered iMACs presented a more robust and enduring polarization towards the pro-inflammatory state. Moreover, elevated production of reactive oxygen species (ROS) as well as enhanced phagocytic and cytotoxic activities targeting cancer cells in vitro were observed. Finally, by transplantation into ovarian and pancreatic cancer mouse models, ACOD1-depleted CAR iMACs showed an improved capacity to suppress tumor cells, leading to a longer lifespan in the treated mice.140 Three clinical trials were approved by the FDA to assess the efficacy of CAR-M-based approaches in solid tumors until the end of June 2023. One of these trials encompasses a cohort investigation aimed at ascertaining the antitumor efficacy of fresh CAR-macrophages in organoids derived from patients with breast cancer [NCT05007379]. The other trial used CAR-macrophages for the management of solid tumors, overexpressing HER2 [NCT04660929].In the third trial, a phase 1 study of intra-peritoneal MCY-M11 (Mesothelin-targeting CAR) in peritoneal mesothelioma and ovarian cancer treatment was designed [NCT03608618].
From the available evidence, we can infer that the clinical translation of CAR-M cells necessitates a reliable source of expansion that can be obtained through the use of PBMCs, HSCs, or iPSCs with the assurance of safety and efficacy.141 The iPSC origin may be an attractive source because of its ability to reproduce indefinitely and transform into almost any differentiated cell type. It is worth noticing that most CAR-M gene transfers have been via viral transfection that may lead to the occurrence of insertion mutations and have unforeseeable implications for therapeutic applications. Utilization of the safe and effective CRISPR-Cas9 technique in gene editing facilitates the refinement of M cell-based treatments, particularly by producing a universal "off-the-shelf" cellular product and redirecting effector cells to overcome resistance in various neoplasms, spanning from hematological to solid tumors.142
Challenges of CRISPR-Cas9
Despite being effective in the treatment of different types of cancers, such as any genetic manipulation, the CRISPR/Cas9 system has some limitations. CRISPR/Cas9 systems suffer from the main limitation of having potential off-target effects. Therefore, the progress of CRISPR-Cas9-based cancer therapeutic strategies has been impeded due to their probability of causing cancer. Alternatively, the delivery systems, including viral and bacteriophage-derived vectors, have the potential to result in gene and cell toxicity (discussed in the CRISPR-Cas9 delivery section).143,144 These drawbacks can be hindered by the high precision of the 5′ end sequence of sgRNA.145 The modification of Cas9 activity is beneficial to decrease off-target effects, as the higher the Cas9 activity, the greater the number of off-targets due to non-specific cleavage. Generally, on-target effects can be provided by some parameters such as gRNA design (decrease the length of gRNAs to 20 bases), Cas9 structure (changing the polarity of Cas9’s domains and usage protein form of Cas9), gRNA/Cas9 ratio, and target site originality.145,146
Ethical concern
Due to promising advantages, such as easy handling, high accuracy, and affordability, CRISPR-Cas9 technology made a revolution in the genetic engineering era. Even though it has jumped rapidly into different fields of human diseases, including cancer therapy, several unsolved questions regarding its clinical applications and bioethical issues remain. Most concerns are related to its ability to genetically modify human germ line cells and embryos, which may create undesirable genetic alterations in the treated patient or influence the fate of future reproduction. Although scientists agree with the utilization of CRISPR-Cas9 for providing models of human diseases and comprehending the cellular and molecular processes of disease, its use for eugenics or enhancement purposes should be forbidden. Therefore, the scientific community and bioethical, social, legal, and governmental parties should be comprehensively informed of the advantages and disadvantages of this technology in clinical applications.
Conclusion
Tumor cells undergo various genetic alterations affecting proto-oncogenes, tumor-suppressing genes, DNA repair genes, and genes involved in apoptosis and cell survival. CRISPR-Cas9 technology has revolutionized the detection and treatment of genetic disorders as well as cancers. This technique showed efficient and precise editing/replacing of individual genes and reconstruction of targets in different types of cancer cells and significantly suppressed the activity of different oncogenes, leading to reduced tumor growth. The use of the CRISPR-Cas9 tool exhibited promises in the treatment of different solid tumors by effectively targeting oncogenes and tumor-suppressor genes in cancer therapies, including those associated with breast, lung, liver, prostate, and colon cancers. The application of CRISPR-Cas9 for generating feasible, effective, and safe CAR-T cells, CAR-NK cells, and CAR-M cells has already progressed to the clinical phase. While over a dozen of in vivo studies have been conducted to treat solid and hematologic cancers by using this technique, transition from the laboratory to the bedside still require further clinical trials.
Review Highlights
What is the current knowledge?
√ Advancements in conducting clinical trials for CRISPR-based cancer therapies.
√ Therapeutic potentials of CRISPR-Cas9 system in treating solid tumors and leukemia.
What is new here?
√ CRISPR/Cas9 revealed the mechanisms of oncogenes and tumor suppressor genes involved in cancer pathogenesis.
√ Optimistic use of CRISPR- Cas9 in production of effective CAR-M, CAR-NK, CAR-T cells in cancer therapy.
Acknowledgements
The authors thank Nooshin Naeimi for the language editing of the manuscript.
Competing Interest
The authors declare no conflict of interest.
Ethical Statement
Authors declare no ethical issues.
Funding Statement
This work was supported by NIMAD Institute, Grant No. 971251.
References
- 1.Kaufmann KB, Büning H, Galy A, Schambach A, Grez M. Gene therapy on the move. EMBO Mol Med. 2013;5:1642–61. doi: 10.1002/emmm.201202287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao-Jie L, Hui-Ying X, Zun-Ping K, Jin-Lian C, Li-Juan J. CRISPR-Cas9: a new and promising player in gene therapy. J Med Genet. 2015;52:289–96. doi: 10.1136/jmedgenet-2014-102968. [DOI] [PubMed] [Google Scholar]
- 3.Wirth T, Parker N, Ylä-Herttuala S. History of gene therapy. Gene. 2013;525:162–9. doi: 10.1016/j.gene.2013.03.137. [DOI] [PubMed] [Google Scholar]
- 4.Chandrasegaran S, Carroll D. Origins of Programmable Nucleases for Genome Engineering. J Mol Biol. 2016;428:963–89. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J, Bayarsaikhan D, Bayarsaikhan G, Kim J-S, Schwarzbach E, Lee B. Recent advances in genome editing of stem cells for drug discovery and therapeutic application. Pharmacology & Therapeutics. 2020;209:107501. doi: 10.1016/j.pharmthera.2020.107501. [DOI] [PubMed] [Google Scholar]
- 6. Song M, Kim Y-H, Kim J-S, Kim H. Chapter Five - Genome Engineering in Human Cells. In: Doudna JA, EJ Sontheimer, editors. Methods in Enzymology. Academic Press; 2014. p. 93-118.
- 7.Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5:1. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaslavskiy M, Bertonati C, Duchateau P, Duclert A, Silva GH. Efficient design of meganucleases using a machine learning approach. BMC Bioinformatics. 2014;15:191. doi: 10.1186/1471-2105-15-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdallah NA, Prakash CS, McHughen AG. Genome editing for crop improvement: Challenges and opportunities. GM Crops Food. 2015;6:183–205. doi: 10.1080/21645698.2015.1129937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, Bayarsaikhan D, Bayarsaikhan G, Kim JS, Schwarzbach E, Lee B. Recent advances in genome editing of stem cells for drug discovery and therapeutic application. PharmacolTher. 2020;209:107501. doi: 10.1016/j.pharmthera.2020.107501. [DOI] [PubMed] [Google Scholar]
- 11.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 14.Ishino Y, Krupovic M, Forterre P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J Bacteriol. 2018;200:e00580–17. doi: 10.1128/jb.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asmamaw M, Zawdie B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biologics. 2021;15:353–61. doi: 10.2147/btt.S326422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Dong H, Cui Y, Cong L, Zhang D. Application of different types of CRISPR/Cas-based systems in bacteria. Microbial Cell Factories. 2020;19:172. doi: 10.1186/s12934-020-01431-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazafa A, Mumtaz M, Farooq MF, Bilal S, Chaudhry SN, Firdous M, et al. CRISPR/Cas9: A powerful genome editing technique for the treatment of cancer cells with present challenges and future directions. Life Sci. 2020;263:118525. doi: 10.1016/j.lfs.2020.118525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Liu L, Lang J, Cheng K, Wang Y, Li X, et al. A CRISPR-Cas13a system for efficient and specific therapeutic targeting of mutant KRAS for pancreatic cancer treatment. Cancer Lett. 2018;431:171–81. doi: 10.1016/j.canlet.2018.05.042. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Chen Y, Xin H, Wan T, Ping Y. Near-infrared optogenetic engineering of photothermal nanoCRISPR for programmable genome editing. Proc Natl Acad Sci U S A. 2020;117:2395–405. doi: 10.1073/pnas.1912220117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan T, Chen Y, Pan Q, Xu X, Kang Y, Gao X, et al. Genome editing of mutant KRAS through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy. J Control Release. 2020;322:236–47. doi: 10.1016/j.jconrel.2020.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Coppedè F. Genes and the Environment in Cancer: Focus on Environmentally Induced DNA Methylation Changes. Cancers (Basel) 2023;15:1019. doi: 10.3390/cancers15041019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian Z, Liang G, Cui K, Liang Y, Wang Q, Lv S, et al. Insight Into the Prospects for RNAi Therapy of Cancer. Front Pharmacol. 2021;12:644718. doi: 10.3389/fphar.2021.644718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Lage M, Puig-Serra P, Menendez P, Torres-Ruiz R, Rodriguez-Perales S. CRISPR/Cas9 for Cancer Therapy: Hopes and Challenges. Biomedicines. 2018;6:105. doi: 10.3390/biomedicines6040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. BiochemBiophys Res Commun. 2014;450:1422–6. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Inturi R, Jemth P. CRISPR/Cas9-based inactivation of human papillomavirus oncogenes E6 or E7 induces senescence in cervical cancer cells. Virology. 2021;562:92–102. doi: 10.1016/j.virol.2021.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Jubair L, Fallaha S, McMillan NAJ. Systemic Delivery of CRISPR/Cas9 Targeting HPV Oncogenes Is Effective at Eliminating Established Tumors. Mol Ther. 2019;27:2091–9. doi: 10.1016/j.ymthe.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huo H, Hu G. CRISPR/Cas9-mediated LMP1 knockout inhibits Epstein-Barr virus infection and nasopharyngeal carcinoma cell growth. Infect Agent Cancer. 2019;14:30. doi: 10.1186/s13027-019-0246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White MK, Khalili K. CRISPR/Cas9 and cancer targets: future possibilities and present challenges. Oncotarget. 2016;7:12305–17. doi: 10.18632/oncotarget.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Łukasiewicz S, Czeczelewski M, Forma A, Baj J, Sitarz R, Stanisławek A. Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies-An Updated Review. Cancers (Basel) 2021;13:4287. doi: 10.3390/cancers13174287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park S, Koo JS, Kim MS, Park HS, Lee JS, Lee JS, et al. Characteristics and outcomes according to molecular subtypes of breast cancer as classified by a panel of four biomarkers using immunohistochemistry. Breast. 2012;21:50–7. doi: 10.1016/j.breast.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Hannafon BN, Cai A, Calloway CL, Xu Y-F, Zhang R, Fung K-M, et al. miR-23b and miR-27b are oncogenic microRNAs in breast cancer: evidence from a CRISPR/Cas9 deletion study. BMC Cancer. 2019;19:642. doi: 10.1186/s12885-019-5839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Sun W. CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett. 2017;385:137–43. doi: 10.1016/j.canlet.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 34.Mintz RL, Lao YH, Chi CW, He S, Li M, Quek CH, et al. CRISPR/Cas9-mediated mutagenesis to validate the synergy between PARP1 inhibition and chemotherapy in BRCA1-mutated breast cancer cells. BioengTransl Med. 2020;5:e10152. doi: 10.1002/btm2.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendes de Almeida R BS, Clara Ribeiro A, Mascarenhas P, Bekman E, Barahona I. Inactivation of APOBEC3G gene in breast cancer cells using the CRISPR/Cas9 systemAnn Med. 2019 May. 28;51(Suppl 1):40. doi: 10.1080/07853890.2018.1561848. [DOI] [Google Scholar]
- 36.Van Treuren T, Vishwanatha JK. CRISPR deletion of MIEN1 in breast cancer cells. Plos One. 2018;13:e0204976. doi: 10.1371/journal.pone.0204976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pulver E dSA, Bouwman P, Annunziato S, Jonkers J. PO-333 somatic engineering of mammary gland epithelial cells using CRISPR/Cas9 for rapid testing of breast cancer susceptibility genes in mouse models. ESMO Open. 2018;3:A151–A2. doi: 10.1101/gad.279190.116. [DOI] [Google Scholar]
- 38.Dai M, Yan G, Wang N, Daliah G, Edick AM, Poulet S, et al. In vivo genome-wide CRISPR screen reveals breast cancer vulnerabilities and synergistic mTOR/Hippo targeted combination therapy. Nat Commun. 2021;12:3055. doi: 10.1038/s41467-021-23316-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Álvarez-Fernández M, Sanz-Flores M, Sanz-Castillo B, Salazar-Roa M, Partida D, Zapatero-Solana E, et al. Therapeutic relevance of the PP2A-B55 inhibitory kinase MASTL/Greatwall in breast cancer. Cell Death Differ. 2018;25:828–40. doi: 10.1038/s41418-017-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo X, Wang X, Wang Z, Banerjee S, Yang J, Huang L, et al. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat Cell Biol. 2016;18:202–12. doi: 10.1038/ncb3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Annunziato S, Kas SM, Nethe M, Yücel H, Del Bravo J, Pritchard C, et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 2016;30:1470–80. doi: 10.1101/gad.279190.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Annunziato S, Lutz C, Henneman L, Bhin J, Wong K, Siteur B, et al. In situ CRISPR-Cas9 base editing for the development of genetically engineered mouse models of breast cancer. Embo J. 2020;39:e102169. doi: 10.15252/embj.2019102169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Gao B, Tan PY, Handoko YA, Sekar K, Deivasigamani A, et al. Genome-wide CRISPR knockout screens identify NCAPG as an essential oncogene for hepatocellular carcinoma tumor growth. Faseb J. 2019;33:8759–70. doi: 10.1096/fj.201802213RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Qi X, Zeng Z, Wang L, Wang J, Zhang T, et al. CRISPR/Cas9-mediated p53 and Pten dual mutation accelerates hepatocarcinogenesis in adult hepatitis B virus transgenic mice. Sci Rep. 2017;7:2796. doi: 10.1038/s41598-017-03070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuller S, Sieker J, Riemenschneider P, Köhler B, Drucker E, Weiler SME, et al. HELLS Is Negatively Regulated by Wild-Type P53 in Liver Cancer by a Mechanism Involving P21 and FOXM1. Cancers (Basel) 2022;14:459. doi: 10.3390/cancers14020459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Artegiani B vVL, Lindeboom RGH, Seinstra D, Heo I, Tapia P, et al. Probing the Tumor Suppressor Function of BAP1 in CRISPR-Engineered Human Liver Organoids. Cell Stem Cell 2019; 24: 927-43.e6. 10.1016/j.stem.2019.04.017. [DOI] [PubMed]
- 47.Feng J, Lu H, Ma W, Tian W, Lu Z, Yang H, et al. Genome-wide CRISPR screen identifies synthetic lethality between DOCK1 inhibition and metformin in liver cancer. Protein Cell. 2022;13:825–41. doi: 10.1007/s13238-022-00906-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C, Jin H, Gao D, Wang L, Evers B, Xue Z, et al. A CRISPR screen identifies CDK7 as a therapeutic target in hepatocellular carcinoma. Cell Res. 2018;28:690–2. doi: 10.1038/s41422-018-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han S, Guo J, Liu Y, Zhang Z, He Q, Li P, et al. Knock out CD44 in reprogrammed liver cancer cell C3A increases CSCs stemness and promotes differentiation. Oncotarget. 2015;6:44452–65. doi: 10.18632/oncotarget.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nature Communications. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C JH, Gao D, Evers B, Jin G-Z, Xue Z, et al. Phospho-ERK is a response biomarker to a combination of sorafenib and MEK inhibition in liver cancer. J Hepatol. 2018;69:1057–65. doi: 10.1101/252452. [DOI] [PubMed] [Google Scholar]
- 52. Song CQ, Li Y, Mou H, Moore J, Park A, Pomyen Y, et al. Genome-Wide CRISPR Screen Identifies Regulators of Mitogen-Activated Protein Kinase as Suppressors of Liver Tumors in Mice. Gastroenterology 2017; 152: 1161-73.e1. 10.1053/j.gastro.2016.12.002. [DOI] [PMC free article] [PubMed]
- 53.Qi Y, Liu Y, Yu B, Hu Y, Zhang N, Zheng Y, et al. A Lactose-Derived CRISPR/Cas9 Delivery System for Efficient Genome Editing In Vivo to Treat Orthotopic Hepatocellular Carcinoma. Adv Sci (Weinh) 2020;7:2001424. doi: 10.1002/advs.202001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee S, Kim Y-Y, Ahn HJ. Systemic delivery of CRISPR/Cas9 to hepatic tumors for cancer treatment using altered tropism of lentiviral vector. Biomaterials 2021; 272: 120793. 10.1016/j.biomaterials.2021.120793. [DOI] [PubMed]
- 55.Liu Q, Fan D, Adah D, Wu Z, Liu R, Yan QT, et al. CRISPR/Cas9-mediated hypoxia inducible factor-1α knockout enhances the antitumor effect of transarterial embolization in hepatocellular carcinoma. Oncol Rep. 2018;40:2547–57. doi: 10.3892/or.2018.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. TACE Combined With PD-1 Knockout Engineered T Cell in Advanced Hepatocellular Carcinoma, Identifier NCT04417764. U.S. National Library of Medicine, 2019. https://clinicaltrials.gov/study/NCT04417764. Accessed 2024-01-28.
- 57.Jiang C LX, Zhao Z. Applications of CRISPR/Cas9 Technology in the Treatment of Lung Cancer. Trends Mol Med. 2019;25:1039–49. doi: 10.1016/j.molmed.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Nair J NA, Veerappan S, Sen D. Translatable gene therapy for lung cancer using Crispr CAS9-an exploratory review. Cancer Gene Ther. 2020;27:116–24. doi: 10.1038/s41417-019-0116-8. [DOI] [PubMed] [Google Scholar]
- 59.Perumal E, So Youn K, Sun S, Seung-Hyun J, Suji M, Jieying L, et al. PTEN inactivation induces epithelial-mesenchymal transition and metastasis by intranuclear translocation of β-catenin and snail/slug in non-small cell lung carcinoma cells. Lung Cancer. 2019;130:25–34. doi: 10.1016/j.lungcan.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 60.Cheung AH, Chow C, Zhang J, Zhou Y, Huang T, Ng KC, et al. Specific targeting of point mutations in EGFR L858R-positive lung cancer by CRISPR/Cas9. Lab Invest. 2018;98:968–76. doi: 10.1038/s41374-018-0056-1. [DOI] [PubMed] [Google Scholar]
- 61. Eichner LJ, Brun SN, Herzig S, Young NP, Curtis SD, Shackelford DB, et al. Genetic Analysis Reveals AMPK Is Required to Support Tumor Growth in Murine Kras-Dependent Lung Cancer Models. Cell Metab 2019; 29: 285-302.e7. 10.1016/j.cmet.2018.10.005. [DOI] [PMC free article] [PubMed]
- 62.Ng SR, Rideout WM, 3rd 3rd, Akama-Garren EH, Bhutkar A, Mercer KL, Schenkel JM, et al. CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proc Natl Acad Sci U S A. 2020;117:513–21. doi: 10.1073/pnas.1821893117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li F, Huang Q, Luster TA, Hu H, Zhang H, Ng WL, et al. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung Adenocarcinoma. Cancer Discov. 2020;10:270–87. doi: 10.1158/2159-8290.Cd-19-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Y, Xue J, Deng T, Zhou X, Yu K, Deng L, et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;26:732–40. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 65.Blanas A, Cornelissen LAM, Kotsias M, van der Horst JC, van de Vrugt HJ, Kalay H, et al. Transcriptional activation of fucosyltransferase (FUT) genes using the CRISPR-dCas9-VPR technology reveals potent N-glycome alterations in colorectal cancer cells. Glycobiology. 2019;29:137–50. doi: 10.1093/glycob/cwy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li W, Cho MY, Lee S, Jang M, Park J, Park R. CRISPR-Cas9 mediated CD133 knockout inhibits colon cancer invasion through reduced epithelial-mesenchymal transition. PLoS One. 2019;14:e0220860. doi: 10.1371/journal.pone.0220860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antal CE, Hudson AM, Kang E, Zanca C, Wirth C, Stephenson NL, et al. Cancer-associated protein kinase C mutations reveal kinase's role as tumor suppressor. Cell. 2015;160:489–502. doi: 10.1016/j.cell.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li T, Liu D, Lei X, Jiang Q. Par3L enhances colorectal cancer cell survival by inhibiting Lkb1/AMPK signaling pathway. BiochemBiophys Res Commun. 2017;482:1037–41. doi: 10.1016/j.bbrc.2016.11.154. [DOI] [PubMed] [Google Scholar]
- 69. Michels B MM, Streibl B, Zhan T, Menche C, Abou-El-Ardat K, et al. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung Adenocarcinoma. Cell Stem Cell 2020; 26(5):782-792.e7. 10.1016/j.stem.2020.04.003. [DOI]
- 70.Wallis CJ, Nam RK. Prostate Cancer Genetics: A Review. Ejifcc. 2015;26:79–91. [PMC free article] [PubMed] [Google Scholar]
- 71.Ye R, Pi M, Cox JV, Nishimoto SK, Quarles LD. CRISPR/Cas9 targeting of GPRC6A suppresses prostate cancer tumorigenesis in a human xenograft model. Journal of Experimental & Clinical Cancer Research. 2017;36:90. doi: 10.1186/s13046-017-0561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Batır MB, Şahin E, Çam FS. Evaluation of the CRISPR/Cas9 directed mutant TP53 gene repairing effect in human prostate cancer cell line PC-3. Mol Biol Rep. 2019;46:6471–84. doi: 10.1007/s11033-019-05093-y. [DOI] [PubMed] [Google Scholar]
- 73.Takao A YK, Karnan S, Ota A, Uemura H, De Velasco MA, et al. Generation of PTEN-knockout (-/-) murine prostate cancer cells using the CRISPR/Cas9 system and comprehensive gene expression profiling. Oncol Rep. 2018;40:2455–66. doi: 10.3892/or.2018.6683. [DOI] [PubMed] [Google Scholar]
- 74.Fenner A. CRISPR–Cas9 ERβ deletion reveals roles in prostate. Nature Reviews Urology. 2020;17:192–3. doi: 10.1038/s41585-020-0302-3. [DOI] [PubMed] [Google Scholar]
- 75.Cai H, Agersnap SN, Sjøgren A, Simonsen MK, Blaavand MS, Jensen UV, et al. In Vivo Application of CRISPR/Cas9 Revealed Implication of Foxa1 and Foxp1 in Prostate Cancer Proliferation and Epithelial Plasticity. Cancers (Basel) 2022;14:4381. doi: 10.3390/cancers14184381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sorrentino C, D'Antonio L, Ciummo SL, Fieni C, Landuzzi L, Ruzzi F, et al. CRISPR/Cas9-mediated deletion of Interleukin-30 suppresses IGF1 and CXCL5 and boosts SOCS3 reducing prostate cancer growth and mortality. J Hematol Oncol. 2022;15:145. doi: 10.1186/s13045-022-01357-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhen S, Takahashi Y, Narita S, Yang YC, Li X. Targeted delivery of CRISPR/Cas9 to prostate cancer by modified gRNA using a flexible aptamer-cationic liposome. Oncotarget. 2017;8:9375–87. doi: 10.18632/oncotarget.14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luskin MR, Murakami MA, Manalis SR, Weinstock DM. Targeting minimal residual disease: a path to cure? Nat Rev Cancer. 2018;18:255–63. doi: 10.1038/nrc.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montaño A, Forero-Castro M, Hernández-Rivas JM, García-Tuñón I, Benito R. Targeted genome editing in acute lymphoblastic leukemia: a review. BMC Biotechnol. 2018;18:45. doi: 10.1186/s12896-018-0455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Visser EA, Moons SJ, Timmermans S, de Jong H, Boltje TJ, Büll C. Sialic acid O-acetylation: From biosynthesis to roles in health and disease. J Biol Chem. 2021;297:100906. doi: 10.1016/j.jbc.2021.100906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baumann AM, Bakkers MJ, Buettner FF, Hartmann M, Grove M, Langereis MA, et al. 9-O-Acetylation of sialic acids is catalysed by CASD1 via a covalent acetyl-enzyme intermediate. Nat Commun. 2015;6:7673. doi: 10.1038/ncomms8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan YT, Ye L, Xie F, Wang J, Müschen M, Chen SJ, et al. CRISPR/Cas9-mediated gene deletion efficiently retards the progression of Philadelphia-positive acute lymphoblastic leukemia in a p210 BCR-ABL1(T315I) mutation mouse model. Haematologica. 2020;105:e232–e6. doi: 10.3324/haematol.2019.229013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McComb S, Aguadé-Gorgorió J, Harder L, Marovca B, Cario G, Eckert C, et al. Activation of concurrent apoptosis and necroptosis by SMAC mimetics for the treatment of refractory and relapsed ALL. Sci Transl Med. 2016;8:339ra70. doi: 10.1126/scitranslmed.aad2986. [DOI] [PubMed] [Google Scholar]
- 84.Randhawa S, Cho BS, Ghosh D, Sivina M, Koehrer S, Müschen M, et al. Effects of pharmacological and genetic disruption of CXCR4 chemokine receptor function in B-cell acute lymphoblastic leukaemia. Br J Haematol. 2016;174:425–36. doi: 10.1111/bjh.14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. TT52CAR19 Therapy for B-cell Acute Lymphoblastic Leukaemia (B-ALL) (PBLTT52CAR19), Identifier NCT04557436. U.S. National Library of Medicine, 2016. https://clinicaltrials.gov/study/NCT04557436. Accessed 2024-02-06..
- 86. CTA101 UCAR-T Cell Injection for Treatment of Relapsed or Refractory CD19+ B-cell Acute Lymphoblastic Leukemia, Identifier NCT04154709. U.S. National Library of Medicine, 2019. https://clinicaltrials.gov/study/NCT04154709. Acessed 2024-02-07..
- 87.Narimani M, Sharifi M, Hakhamaneshi MS, Roshani D, Kazemi M, Hejazi SH, et al. BIRC5 Gene Disruption via CRISPR/Cas9n Platform Suppress Acute Myelocytic Leukemia Progression. Iran Biomed J. 2019;23:369–78. doi: 10.29252/ibj.23.6.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ho TC, Kim HS, Chen Y, Li Y, LaMere MW, Chen C, et al. Scaffold-mediated CRISPR-Cas9 delivery system for acute myeloid leukemia therapy. Sci Adv. 2021;7:eabg3217. doi: 10.1126/sciadv.abg3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122–30. doi: 10.1182/blood-2015-07-659144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Delgado J, Nadeu F, Colomer D, Campo E. Chronic lymphocytic leukemia: from molecular pathogenesis to novel therapeutic strategies. Haematologica. 2020;105:2205–17. doi: 10.3324/haematol.2019.236000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arruga F, Gizdic B, Bologna C, Cignetto S, Buonincontri R, Serra S, et al. Mutations in NOTCH1 PEST domain orchestrate CCL19-driven homing of chronic lymphocytic leukemia cells by modulating the tumor suppressor gene DUSP22. Leukemia. 2017;31:1882–93. doi: 10.1038/leu.2016.383. [DOI] [PubMed] [Google Scholar]
- 92.Close V, Close W, Kugler SJ, Reichenzeller M, Yosifov DY, Bloehdorn J, et al. FBXW7 mutations reduce binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in CLL. Blood. 2019;133:830–9. doi: 10.1182/blood-2018-09-874529. [DOI] [PubMed] [Google Scholar]
- 93.Jain P, Kantarjian H, Cortes J. Chronic myeloid leukemia: overview of new agents and comparative analysis. Curr Treat Options Oncol. 2013;14:127–43. doi: 10.1007/s11864-013-0234-8. [DOI] [PubMed] [Google Scholar]
- 94. Vuelta E, Ordoñez JL, Sanz DJ, Ballesteros S, Hernández-Rivas JM, Méndez-Sánchez L, et al. CRISPR/Cas9-Directed Gene Trap Constitutes a Selection System for Corrected BCR/ABL Leukemic Cells in CML. Int J Mol Sci 2022; 23. 10.3390/ijms23126386. [DOI] [PMC free article] [PubMed]
- 95.Vuelta E, García-Tuñón I, Hernández-Carabias P, Méndez L, Sánchez-Martín M. Future Approaches for Treating Chronic Myeloid Leukemia: CRISPR Therapy. Biology (Basel) 2021;10:118. doi: 10.3390/biology10020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Y, Zhao G, Xu CF, Luo YL, Lu ZD, Wang J. Systemic delivery of CRISPR/Cas9 with PEG-PLGA nanoparticles for chronic myeloid leukemia targeted therapy. Biomater Sci. 2018;6:1592–603. doi: 10.1039/c8bm00263k. [DOI] [PubMed] [Google Scholar]
- 97.Luo Z, Gao M, Huang N, Wang X, Yang Z, Yang H, et al. Efficient disruption of bcr-abl gene by CRISPR RNA-guided FokI nucleases depresses the oncogenesis of chronic myeloid leukemia cells. Journal of Experimental & Clinical Cancer Research. 2019;38:224. doi: 10.1186/s13046-019-1229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Razeghian E, Nasution MKM, Rahman HS, Gardanova ZR, Abdelbasset WK, Aravindhan S, et al. A deep insight into CRISPR/Cas9 application in CAR-T cell-based tumor immunotherapies. Stem Cell Res Ther. 2021;12:428. doi: 10.1186/s13287-021-02510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang C, Liu J, Zhong JF, Zhang X. Engineering CAR-T cells. Biomarker Research. 2017;5:22. doi: 10.1186/s40364-017-0102-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mazinani M, Rahbarizadeh F. New cell sources for CAR-based immunotherapy. Biomarker Research. 2023;11:49. doi: 10.1186/s40364-023-00482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Salas-Mckee J, Kong W, Gladney WL, Jadlowsky JK, Plesa G, Davis MM, et al. CRISPR/Cas9-based genome editing in the era of CAR T cell immunotherapy. Hum VaccinImmunother. 2019;15:1126–32. doi: 10.1080/21645515.2019.1571893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dimitri A, Herbst F, Fraietta JA. Engineering the next-generation of CAR T-cells with CRISPR-Cas9 gene editing. Molecular Cancer. 2022;21:78. doi: 10.1186/s12943-022-01559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khan A, Sarkar E. CRISPR/Cas9 encouraged CAR-T cell immunotherapy reporting efficient and safe clinical results towards cancer. Cancer Treat Res Commun. 2022;33:100641. doi: 10.1016/j.ctarc.2022.100641. [DOI] [PubMed] [Google Scholar]
- 104. Schmidts A, Marsh LC, Srivastava AA, Bouffard AA, Boroughs AC, Scarfò I, et al. Cell-based artificial APC resistant to lentiviral transduction for efficient generation of CAR-T cells from various cell sources. J Immunother Cancer 2020; 8. 10.1136/jitc-2020-000990. [DOI] [PMC free article] [PubMed]
- 105.Tang N, Cheng C, Zhang X, Qiao M, Li N, Mu W, et al. TGF-β inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI Insight. 2020;5:e133977. doi: 10.1172/jci.insight.133977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJC, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–7. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Choi BD, Yu X, Castano AP, Darr H, Henderson DB, Bouffard AA, et al. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. J Immunother Cancer. 2019;7:304. doi: 10.1186/s40425-019-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ottaviano G, Georgiadis C, Gkazi SA, Syed F, Zhan H, Etuk A, et al. Phase 1 clinical trial of CRISPR-engineered CAR19 universal T cells for treatment of children with refractory B cell leukemia. Sci Transl Med. 2022;14:eabq3010. doi: 10.1126/scitranslmed.abq3010. [DOI] [PubMed] [Google Scholar]
- 109.Guo Y, Tong C, Su L, Zhang W, Jia H, Liu Y, et al. CRISPR/Cas9 genome-edited universal CAR T cells in patients with relapsed/refractory lymphoma. Blood Advances. 2022;6:2695–9. doi: 10.1182/bloodadvances.2021006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McGuirk J BC BM, Ho PJ, Murthy HS, Dickinson MJ, et al. A phase 1 dose escalation and cohort expansion study of the safety and efficacy of allogeneic CRISPR-Cas9–engineered T cells (CTX110) in patients (Pts) with relapsed or refractory (R/R) B-cell. J Clin Oncol. 2021;39:TPS7570. doi: 10.1200/JCO.2021.39.15_suppl.TPS7570. [DOI] [Google Scholar]
- 111. Iyer SP SR HP, Hu B, Zain J, Prica A, et al. The COBALT-LYM study of CTX130: a phase 1 dose escalation study of CD70-targeted allogeneic CRISPR-Cas9–engineered CAR T cells in patients with relapsed/refractory (R/R) T-cell malignancies. THemaSphere 2022; 173:S21..
- 112.Qasim W. Genome-edited allogeneic donor “universal” chimeric antigen receptor T cells. Blood. 2023;141:835–45. doi: 10.1182/blood.2022016204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Elmas E, Saljoughian N, de Souza Fernandes Pereira M, Tullius BP, Sorathia K, Nakkula RJ, et al. CRISPR Gene Editing of Human Primary NK and T Cells for Cancer Immunotherapy. Frontiers in Oncology 2022; 12. 10.3389/fonc.2022.834002. [DOI] [PMC free article] [PubMed]
- 114.Wang F, Lau JKC, Yu J. The role of natural killer cell in gastrointestinal cancer: killer or helper. Oncogene. 2021;40:717–30. doi: 10.1038/s41388-020-01561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17:1025–36. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 116.Vivier E, Ugolini S, Blaise D, Chabannon C, Brossay L. Targeting natural killer cells and natural killer T cells in cancer. Nat Rev Immunol. 2012;12:239–52. doi: 10.1038/nri3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Basar R, Daher M, Rezvani K. Next-generation cell therapies: the emerging role of CAR-NK cells. Blood Adv. 2020;4:5868–76. doi: 10.1182/bloodadvances.2020002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Naeimi Kararoudi M, Likhite S, Elmas E, Yamamoto K, Schwartz M, Sorathia K, et al. Optimization and validation of CAR transduction into human primary NK cells using CRISPR and AAV. Cell Rep Methods. 2022;2:100236. doi: 10.1016/j.crmeth.2022.100236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Naeimi Kararoudi M, Nagai Y, Elmas E, de Souza Fernandes Pereira M, Ali SA, Imus PH, et al. CD38 deletion of human primary NK cells eliminates daratumumab-induced fratricide and boosts their effector activity. Blood. 2020;136:2416–27. doi: 10.1182/blood.2020006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hoerster K, Uhrberg M, Wiek C, Horn PA, Hanenberg H, Heinrichs S. HLA Class I Knockout Converts Allogeneic Primary NK Cells Into Suitable Effectors for “Off-the-Shelf” Immunotherapy. Front Immunol. 2021;11:586168. doi: 10.3389/fimmu.2020.586168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jo DH, Kaczmarek S, Shin O, Wang L, Cowan J, McComb S, et al. Simultaneous engineering of natural killer cells for CAR transgenesis and CRISPR-Cas9 knockout using retroviral particles. Mol Ther Methods Clin Dev. 2023;29:173–84. doi: 10.1016/j.omtm.2023.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Naeimi Kararoudi M, Elmas E, Likhite S, Schwartz M, Sorathia K, Yamamoto K, et al. CD33 Targeting Primary CAR-NK Cells Generated By CRISPR Mediated Gene Insertion Show Enhanced Anti-AML Activity 2020; 136: 3. 10.1182/blood-2020-142494. [DOI]
- 123. Kararoudi MN, Likhite S, Elmas E, Yamamoto K, Schwartz M, Sorathia K, et al. CRISPR-Targeted CAR Gene Insertion Using Cas9/RNP and AAV6 Enhances Anti-AML Activity of Primary NK Cells. bioRxiv 2021; 2021.03.17.435886. 10.1101/2021.03.17.435886. [DOI]
- 124.Daher M, Basar R, Gokdemir E, Baran N, Uprety N, Nunez Cortes AK, et al. Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR-NK cells. Blood. 2021;137:624–36. doi: 10.1182/blood.2020007748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ureña-Bailén G DJ-M, Hou Y, Dirlam A, Roig-Merino A, Schleicher S, et al. Preclinical Evaluation of CRISPR-Edited CAR-NK-92 Cells for Off-the-Shelf Treatment of AML and B-ALL. Int J Mol Sci. 2022;23:12828. doi: 10.3390/ijms232112828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xie G, Dong H, Liang Y, Ham JD, Rizwan R, Chen J. CAR-NK cells: A promising cellular immunotherapy for cancer. EBioMedicine. 2020;59:102975. doi: 10.1016/j.ebiom.2020.102975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sloas C, Gill S, Klichinsky M. Engineered CAR-Macrophages as Adoptive Immunotherapies for Solid Tumors. Front Immunol. 2021;12:783305. doi: 10.3389/fimmu.2021.783305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen Y, Yu Z, Tan X, Jiang H, Xu Z, Fang Y, et al. CAR-macrophage: A new immunotherapy candidate against solid tumors. Biomed Pharmacother. 2021;139:111605. doi: 10.1016/j.biopha.2021.111605. [DOI] [PubMed] [Google Scholar]
- 131.Lin C, Zhang J. Chimeric antigen receptor engineered innate immune cells in cancer immunotherapy. Sci China Life Sci. 2019;62:633–9. doi: 10.1007/s11427-018-9451-0. [DOI] [PubMed] [Google Scholar]
- 132.Alvey C, Discher DE. Engineering macrophages to eat cancer: from "marker of self" CD47 and phagocytosis to differentiation. J Leukoc Biol. 2017;102:31–40. doi: 10.1189/jlb.4RI1216-516R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li X, Liu R, Su X, Pan Y, Han X, Shao C, et al. Harnessing tumor-associated macrophages as aids for cancer immunotherapy. Molecular Cancer. 2019;18:177. doi: 10.1186/s12943-019-1102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Villanueva MT. Macrophages get a CAR. Nature Reviews Immunology. 2020;20:273. doi: 10.1038/s41577-020-0302-9. [DOI] [PubMed] [Google Scholar]
- 135.Huang Z, Zhang Z, Jiang Y, Zhang D, Chen J, Dong L, et al. Targeted delivery of oligonucleotides into tumor-associated macrophages for cancer immunotherapy. J Control Release. 2012;158:286–92. doi: 10.1016/j.jconrel.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 136.Toole BP. Emmprin (CD147), a cell surface regulator of matrix metalloproteinase production and function. Curr Top Dev Biol. 2003;54:371–89. doi: 10.1016/s0070-2153(03)54015-7. [DOI] [PubMed] [Google Scholar]
- 137.Zhang L, Tian L, Dai X, Yu H, Wang J, Lei A, et al. Pluripotent stem cell-derived CAR-macrophage cells with antigen-dependent anti-cancer cell functions. Journal of Hematology & Oncology. 2020;13:153. doi: 10.1186/s13045-020-00983-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang L, Chen Y, Liu X, Li Z, Dai X. The Application of CRISPR/Cas9 Technology for Cancer Immunotherapy: Current Status and Problems. Front Oncol. 2022;11:704999. doi: 10.3389/fonc.2021.704999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang J, Webster S, Duffin B, Bernstein MN, Steill J, Swanson S, et al. Generation of anti-GD2 CAR macrophages from human pluripotent stem cells for cancer immunotherapies. Stem Cell Reports. 2023;18:585–96. doi: 10.1016/j.stemcr.2022.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang X, Su S, Zhu Y, Cheng X, Cheng C, Chen L, et al. Metabolic Reprogramming via targeting ACOD1 promotes polarization and anti-tumor activity of human CAR-iMACs in solid tumors. bioRxiv 2023; 2023.04.20.537647. 10.1101/2023.04.20.537647. [DOI]
- 141.Paasch D, Meyer J, Stamopoulou A, Lenz D, Kuehle J, Kloos D, et al. Ex Vivo Generation of CAR Macrophages from Hematopoietic Stem and Progenitor Cells for Use in Cancer Therapy. Cells. 2022;11:994. doi: 10.3390/cells11060994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 143.Karn V, Sandhya S, Hsu W, Parashar D, Singh HN, Jha NK, et al. CRISPR/Cas9 system in breast cancer therapy: advancement, limitations and future scope. Cancer Cell International. 2022;22:234. doi: 10.1186/s12935-022-02654-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25:249–54. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kimberland ML, Hou W, Alfonso-Pecchio A, Wilson S, Rao Y, Zhang S, et al. Strategies for controlling CRISPR/Cas9 off-target effects and biological variations in mammalian genome editing experiments. J Biotechnol. 2018;284:91–101. doi: 10.1016/j.jbiotec.2018.08.007. [DOI] [PubMed] [Google Scholar]