Figure 1. Study overview.
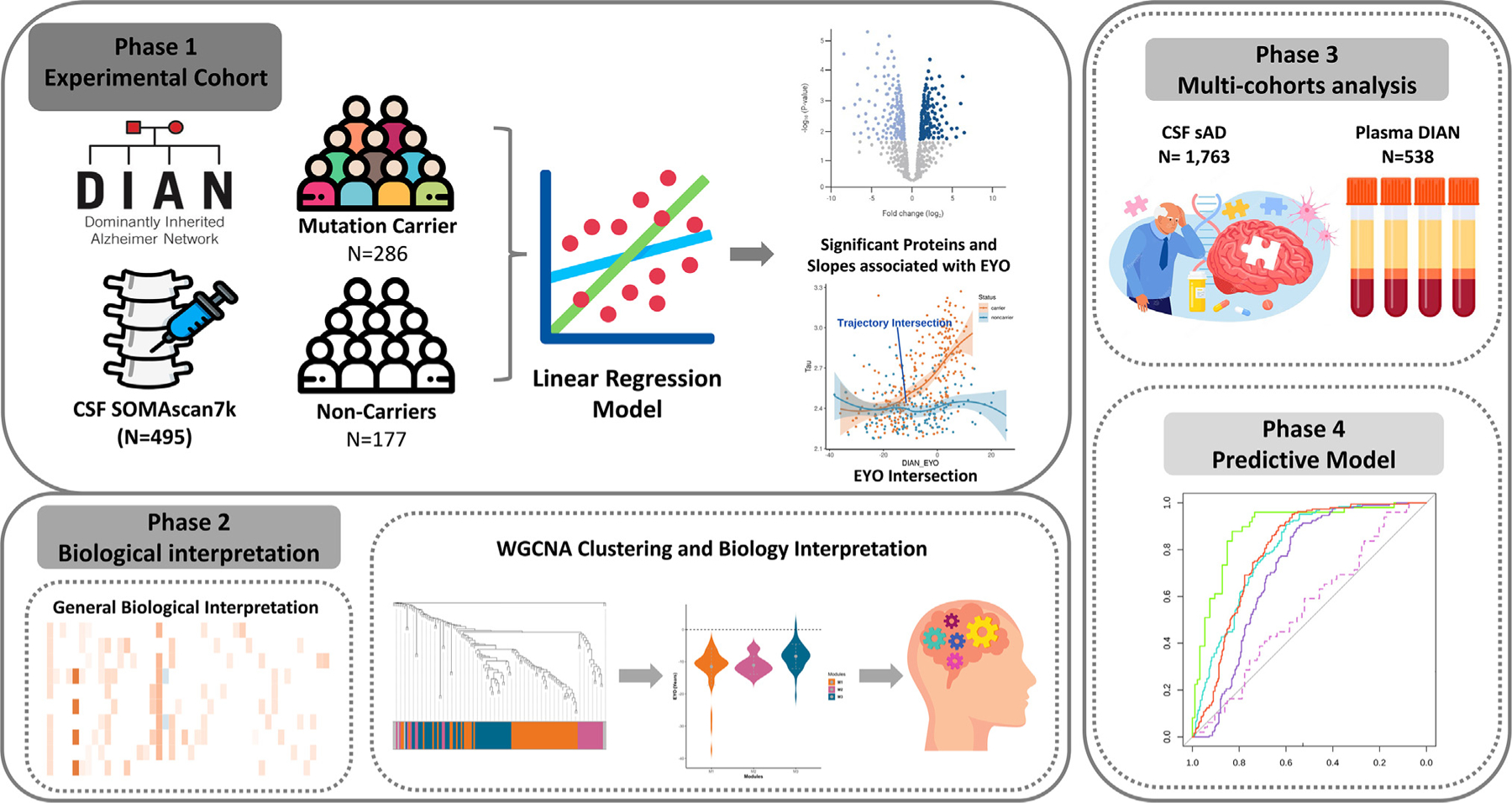
A total of 6,163 proteins were measured in CSF sample from 286 mutation carriers (MCs) and 177 non-carriers (NCs). Differential pseudo-trajectory analyses were performed between MCs and NCs. Trajectory intersections were calculated for significant pseudo-trajectory proteins. Biological functions were identified by protein co-expression network analysis and pathway enrichment. A total of 1,763 sAD CSF samples and 538 DIAN plasma samples were analyzed to validate the approach and contextualize the findings. Several publicly available external proteomic datasets were used to validate our findings as well. Finally, the LASSO model was used to select significant trajectory proteins and to create predictive models for ADAD.
