Abstract
After the perception of vegetation proximity by phytochrome photoreceptors, shade-avoider plants initiate a set of responses known as the shade avoidance syndrome (SAS). Shade perception by the phytochrome B (phyB) photoreceptor unleashes the PHYTOCHROME INTERACTING FACTORs and initiates SAS responses. In Arabidopsis (Arabidopsis thaliana) seedlings, shade perception involves rapid and massive changes in gene expression, increases auxin production, and promotes hypocotyl elongation. Other components, such as phyA and ELONGATED HYPOCOTYL 5, also participate in the shade regulation of the hypocotyl elongation response by repressing it. However, why and how so many regulators with either positive or negative activities modulate the same response remains unclear. Our physiological, genetic, cellular, and transcriptomic analyses showed that (i) these components are organized into 2 main branches or modules and (ii) the connection between them is dynamic and changes with the time of shade exposure. We propose a model for the regulation of shade-induced hypocotyl elongation in which the temporal and spatial functional importance of the various SAS regulators analyzed here helps to explain the coexistence of differentiated regulatory branches with overlapping activities.
The regulatory network of shade-induced hypocotyl elongation has 2 main branches that act at different times and within different cells along the hypocotyl axis.
Introduction
When plants grow in high density, the close proximity of vegetation might obstruct sunlight and pose a threat for plant survival. Plants have adopted contrasting avoidance or tolerance strategies to deal with vegetation proximity or shade (Martinez-Garcia and Rodriguez-Concepcion 2023). Specifically, when shade-avoider (sun-loving) plants face this scenario, they display a set of responses known as the shade avoidance syndrome (SAS). Some of the SAS responses acclimate photosynthesis to eventual light shortage caused by the presence of neighboring plants; others focus on redirecting growth to escape from shade by promoting either stem elongation and/or apical dominance (reduced branching) or flowering to produce seeds (Casal 2012; Roig-Villanova and Martínez-Garcia 2016; Morelli et al. 2021). At the seedling stage, hypocotyl elongation is likely the best characterized and most conspicuous SAS response in the shade-avoider plant Arabidopsis (Arabidopsis thaliana) (Casal 2012; Martínez-Garcia et al. 2014) and the focus of this work.
Plants detect neighbor vegetation as changes in the red (R) to far-red light (FR) ratio (R:FR). Plants absorb R and reflect mainly FR from sunlight. Under low planting density, the intensity of incoming sunlight during the day changes but the R:FR (>1.2) remains relatively constant (Smith 1982). By contrast, when neighboring plants are close enough, they can sense plant proximity by detecting the reflected FR from other plants that combines with sunlight and results in a moderate decrease in the R:FR (R:FR 0.5 to 0.3) without reducing light intensity. When neighboring vegetation is denser forming a plant canopy, photosynthetic pigments of the upper leaves act as selective filters that preferentially absorb and deplete blue and R from sunlight but transmit part of green and most FR (Casal 2012). Plant canopy shade presents a drastic reduction of R compared to the FR that results in a very low R:FR ratio (R:FR < 0.06) and a low light intensity in the photosynthetic active radiation region (Martínez-Garcia et al. 2014; Pierik and de Wit 2014; Fiorucci and Fankhauser 2017). The reduced R:FR occurring under both proximity and canopy shade acts as a reliable signal indicative of the nearby presence of vegetation that is perceived by the phytochrome photoreceptors (Fig. 1).
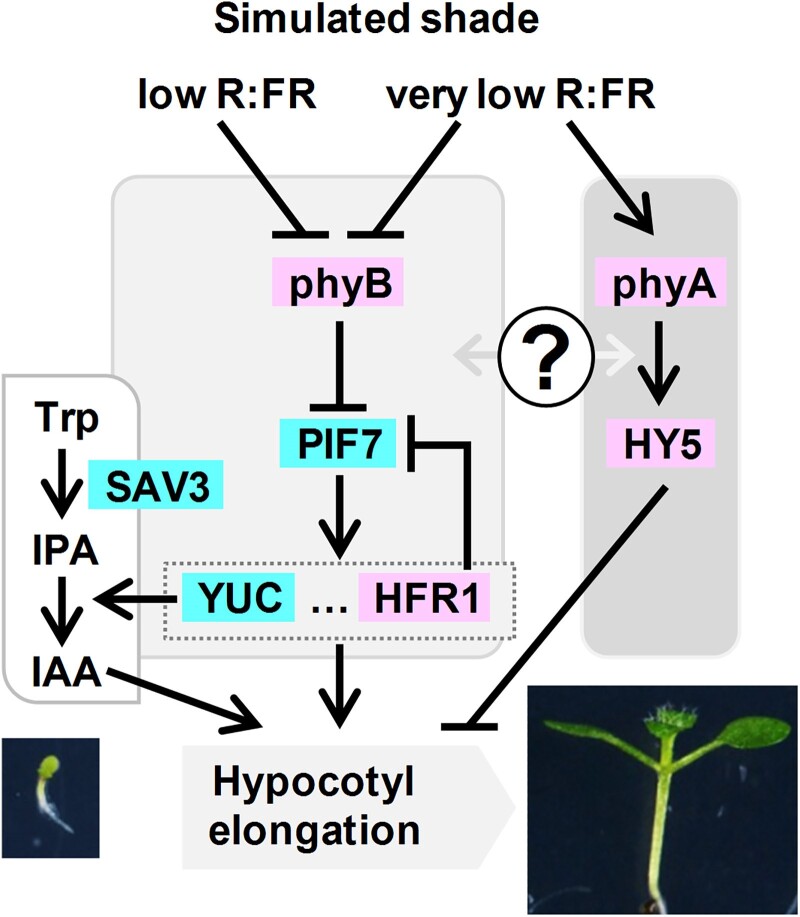
Figure 1.
Simplified model that depicts the genetic components analyzed in this work involved in plant neighbor detection. Color indicates the positive (blue) or negative (pink) contribution to the shade-induced hypocotyl elongation. Aspect of representative seedlings just before (W-grown 2-d-old, bottom left) and after the shade treatment (shade-grown 7-d-old, bottom right) is shown. Arrows indicate positive and bars represent negative regulatory relationships. Question mark indicates an unknown regulatory relationship between the connected components.
As molecular switches, phytochromes exist in 2 photoconvertible isoforms (an inactive R-absorbing Pr form and an active FR-absorbing Pfr form) that are present in an equilibrium that depends on the prevailing R:FR. Under high R:FR (low vegetation density), most phytochromes are in the active Pfr forms and SAS is suppressed, whereas under low R:FR (high vegetation density), the photoequilibrium moves toward the inactive Pr form and SAS is induced. From the 5 phytochromes characterized in A. thaliana (phytochrome A [phyA] to phyE), phyA and phyB have the main roles in controlling SAS responses. Genetic and physiological analyses indicate that photostable phyB is the major phytochrome controlling the SAS (Casal 2012; Martínez-Garcia et al. 2014). Additional genetic analyses also showed that phyA, the only photolabile phytochrome, has an antagonistic role over phyB in the SAS control, particularly under very low R:FR mimicking plant canopy shade. Under low R:FR (proximity shade), wild-type and phyA mutant seedlings present a similar hypocotyl elongation whereas phyB mutants display longer hypocotyls. In contrast, under very low R:FR (canopy shade), wild-type and phyB seedlings elongate less than when grown under low R:FR and phyA seedlings present an exaggerated hypocotyl length. This indicates that phyB is deactivated by both proximity (low R:FR) and canopy (very low R:FR) shade whereas phyA activity is induced only by very low R:FR (Fig. 1) (Yanovsky et al. 1995; Martínez-Garcia et al. 2014; Molina-Contreras et al. 2019). It has been shown that under very low R:FR conditions, phyA protein tends to accumulate (Martínez-Garcia et al. 2014; Yang et al. 2018; Molina-Contreras et al. 2019). For clarity, we will use the term simulated shade to refer to any treatment, including proximity, canopy, or other similar conditions, that lowers the R:FR but has not been specifically defined as such (Roig-Villanova and Martinez-Garcia 2022).
SAS implementation is regulated by, at least, the interaction of active phyB with PHYTOCHROME INTERACTING FACTORS (PIFs), a family of basic-helix-loop-helix (bHLH) transcription factors. When interacting with active phyB, PIFs are phosphorylated. Phosphorylation triggers the degradation of PIF1, PIF3, PIF4, and PIF5 (known as the PIF quartet [PIFQ]) via the 26S proteasome. By contrast, PIF7 phosphorylation has little effect on its stability but it inhibits its DNA-binding activity (Li et al. 2012) and promotes cytoplasm retention, reducing its nuclear import (Huang et al. 2018). Either case, under high R:FR, PIF transcriptional activity is inhibited by the active form of phyB, whereas deactivation of phyB under low R:FR results in PIF accumulation in the nucleus and/or promotion of their DNA-binding activity. This initiates a transcriptional cascade that leads to the expression of dozens of PHYTOCHOME RAPIDLY REGULATED (PAR) genes, several of which encode transcription factors from various families (e.g. bHLH, HD-Zip, and BBX) having positive, negative, or even complex roles in implementing the hypocotyl elongation response (Sessa et al. 2005; Roig-Villanova et al. 2006, 2007; Sorin et al. 2009; Cifuentes-Esquivel et al. 2013; Ciolfi et al. 2013; Gangappa et al. 2013; Kohnen et al. 2016; Gallemi et al. 2017; Gommers et al. 2017; Buti et al. 2020).
PIF7, together with a minor contribution of PIF4 and PIF5, has a major and positive role in promoting the shade-induced hypocotyl elongation. pif7 and the pif4 pif5 pif7 (from now on pif457) showed an attenuated and almost null hypocotyl elongation in response to simulated shade (Li et al. 2012; de Wit et al. 2015). From the various PAR genes, induction of YUCCAs (YUCs) contributes to auxin production together with SHADE AVOIDANCE 3 (SAV3, also known as TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1/WEAK ETHYLENE INSENSITIVE 8 [TAA1/WEI8]) in the 2-step indole-3-acetic acid (IAA) pathway from tryptophan (Trp). Indeed, the Trp aminotransferase encoded by SAV3 catalyzes the conversion from Trp to indole-3-pyruvic acid (IPA), and the flavin monoxigenase encoded by YUC genes catalyzes the IPA oxidative decarboxylation to IAA (Fig. 1) (Brumos et al. 2014; Zheng et al. 2016). The single sav3 and multiple mutants in YUC genes (yuc2 yuc5 yuc8 yuc9 and yuc3 yuc5 yuc7 yuc8 yuc9) had attenuated shade-induced hypocotyl elongation (Li et al. 2012; Kohnen et al. 2016). Together, these results highlight the importance of SAV3- and YUC-mediated production of IAA in this SAS response.
Another PAR gene with a well-known negative role in the shade-induced hypocotyl elongation is LONG HYPOCOTYL IN FAR-RED 1 (HFR1) that encodes a transcriptional cofactor of the bHLH family structurally related to PIFs but lacks the phyB- and DNA-binding ability (Galstyan et al. 2011; Hornitschek et al. 2012). HFR1 heterodimerizes and inhibits the activity of all 4 PIFQ members (PIF1, PIF3, PIF4, and PIF5) (Fairchild et al. 2000; Hornitschek et al. 2009; Shi et al. 2013) and PIF7 (Zhang et al. 2019; Buti et al. 2020; Paulisic et al. 2021). hfr1 hypocotyls display an opposed phenotype to that of pif7 or pif457 seedlings; i.e. they are longer than wild-type ones under simulated shade (Roig-Villanova et al. 2007; Ciolfi et al. 2013; de Wit et al. 2016). Another PIF antagonist is ELONGATED HYPOCOTYL 5 (HY5), known to encode a transcription factor of the basic domain-leucine zipper (bZIP) family. HY5 expression is phyA-dependent, and it is not rapidly or strongly induced in response to certain shade condition (Ciolfi et al. 2013). Hypocotyls of the hy5 mutant seedlings elongate more than the wild-type ones under low R:FR (Sellaro et al. 2011; Bou-Torrent et al. 2015; van Gelderen et al. 2018; Ortiz-Alcaide et al. 2019). Therefore, HY5 acts as a negative SAS regulator. CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) is an E3 ubiquitin ligase that interacts with and modulates the abundance of several SAS regulatory components, including HY5 and HFR1. COP1 accumulates in the nucleus under shade, counteracting the negative impact of HY5 and HFR1 accumulation and modulating therefore this response (Pacín et al. 2013, 2016). Nuclear-pore complex components, chloroplast-derived signals, and epigenetic components also prevent an excessive response to shade, providing additional levels of regulation of this response (Gallemi et al. 2016; Ortiz-Alcaide et al. 2019; Martínez-Garcia and Moreno-Romero 2020).
The mechanisms that connect SAS components have been established in a few cases: (i) HFR1 inhibits PIF activity; (ii) HY5 appears to be mainly associated with phyA action (Ciolfi et al. 2013; Zhang et al. 2018), although it has not been explored; and (iii) other transcription factors, including the growth-promoting PIFs, would be mostly linked to the phyB-dependent pathway (Casal 2012; Roig-Villanova and Martínez-Garcia 2016) (Fig. 1). The antagonistic phyA/HY5/HFR1 and phyB/PIF/SAV3 activities likely provide young seedlings with the capacity to rapidly elongate when impeding competition is nearby and also to attenuate excessive growth when growing under a canopy. However, several key aspects of the genetic architecture of the SAS regulatory network remain unclear. These include features regarding whether these pathways or components operate concurrently on the same cell type and, if they do, how they are connected (Fig. 1). To address these issues, we have carried out (i) genetic analyses, to establish if different SAS components work in the same or different regulatory branches or modules of the network; (ii) temporal analyses, to learn when the different components analyzed act in controlling the shade-induced hypocotyl elongation; and (iii) spatial analyses, to identify the cells targeted along the hypocotyl axis epidermis by each SAS regulator. Besides, we explored molecular connections between HY5 and PIFs, 2 antagonistic SAS components. Our findings indicated that these components are grouped in, at least, 2 main modules or branches that act at different times and impact the elongation of distinct cells along the hypocotyl axis. We also show that HY5 acts as a node, although its functional relationship with PIF457 changes with the time of shade exposure.
Results
The SAS regulatory network is organized in at least 2 genetically differentiated modules or branches
We first prepared a series of genetic crosses focusing on a few mutants in negative (phyA, phyB, HY5, and HFR1) or positive (SAV3, PIF4, PIF5, and PIF7) SAS components (Fig. 1). These mutants result in strong shade-related hypocotyl phenotypes. From these components, only PIF4, PIF5, and PIF7 (PIF457) show some redundancy in controlling the shade-induced hypocotyl elongation (Li et al. 2012; Hersch et al. 2014).
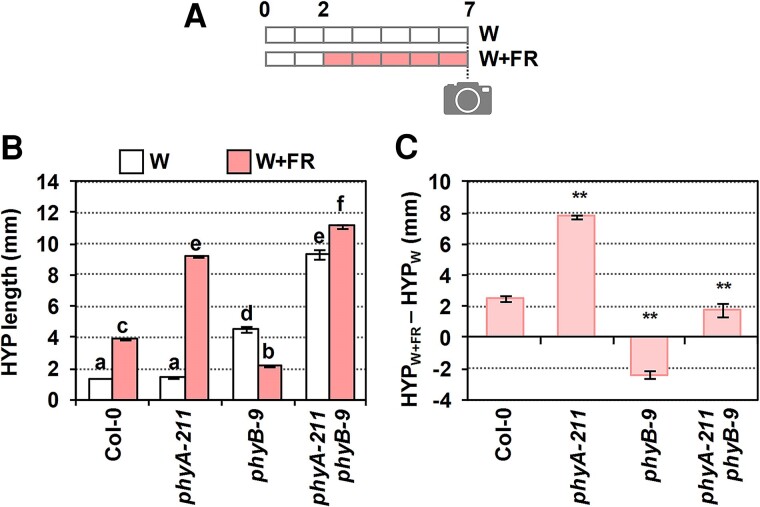
The hypocotyl length of the single and double phyA and phyB mutant seedlings in response to simulated shade was first analyzed (Fig. 2). In continuous white light (W) (that simulates sunlight of high R:FR), the length of Col-0 and phyA hypocotyls was similar, whereas that of phyB hypocotyls was longer and those of phyA phyB double mutant seedlings were the longest, as expected. In continuous W + FR (very low R:FR), phyA hypocotyls were longer and phyB hypocotyls shorter, respectively, than the wild type (Fig. 2, A and B). The antagonistic activity of phyA and phyB under simulated shade indicates that the W + FR conditions employed in these experiments mimic canopy shade, in contrast with those proximity shade conditions in which phyA action is negligible (Martínez-Garcia et al. 2014). Importantly, the phyA phyB hypocotyl length in W + FR was even longer than in W (Fig. 2B), in agreement with the conclusion that other phytochromes regulate the shade-induced hypocotyl elongation (Devlin et al. 2003). To better visualize the effect of simulated shade in controlling hypocotyl elongation, the difference in hypocotyl length in W + FR and W (HYPW + FR − HYPW) was calculated (Fig. 2C). This representation showed that phyA phyB double mutant hypocotyls had an intermediate shade-induced elongation response compared to those of phyA and phyB single mutants (Fig. 2C), suggesting that the effect of the 2 phytochromes is additive. This is interpreted as indicative that these 2 phytochromes act likely independently of one another in controlling the shade-induced hypocotyl length. In the following set of experiments, the HYPW + FR − HYPW is shown when comparing the different mutants (raw data are included as Supplementary data).
Figure 2.
Genetic interaction of phyA and phyB in the shade-induced hypocotyl elongation. A) Cartoon showing the design of the experiment. Seedlings were germinated and grown in W (R:FR > 1.5) for 2 d, and then they either maintained in W or transferred to simulated shade (W + FR, R:FR, 0.02) for 5 more days. On Day 7, pictures were taken and hypocotyl (HYP) length was measured. B) HYP length of Col-0, phyA-211, phyB-9, and phyA-211 phyB-9 double mutant seedlings after growing in W (HYPW) or W + FR (HYPW + FR). Values are means and error bars are Se of 3 independent replicates. C) Elongation response (HYPW + FR − HYPW) of lines shown in B). Values of HYPW and HYPW + FR (shown in B) were used to calculate HYPW + FR − HYPW. Se was propagated accordingly. In B), different letters denote significant differences (2-way ANOVA with the Tukey test, P < 0.05) among means. In C), asterisks indicate significant differences (based on the 2-way ANOVA) between the mutant and wild-type genotypes in response to simulated shade (**P < 0.01).
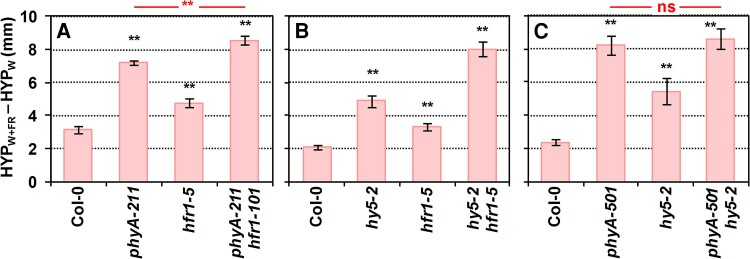
We next produced double mutants deficient in other negative regulators (phyA hfr1, hy5 hfr1, and phyA hy5) and analyzed their shade-induced hypocotyl elongation response (Fig. 3; Supplementary Fig. S1) (no phyB mutant was included in these crosses as its phenotype was observed more clearly in W). Seedlings of phyA hfr1 and hy5 hfr1 double mutants elongated more than the single mutants (Fig. 3, A and B), suggesting that they worked additively, in agreement with previous information (Kim et al. 2002; Ciolfi et al. 2013). By contrast, phyA hy5 seedlings elongated as much as the phyA single mutant (Fig. 3C), indicating that phyA was epistatic over HY5. During deetiolation under monochromatic FR, hundreds of phyA-associated genes that are phyA regulated have been identified as putative direct targets of phyA. These direct targets are likely to be cotargeted by phyA in association with many known light-related transcription factors, such as HY5 (Chen et al. 2014). It is therefore expected that several other DNA-binding or light-related transcription factors act downstream phyA. Following this model, epistasis reflects the upstream activity of phyA over HY5 under our very low R:FR conditions, and it also involves the additional action of other factors (e.g. HYH). Together, our genetic analyses suggested that (i) phyA and HY5 act in the same branch of the SAS regulatory network and that (ii) HFR1 acts independently of phyA and HY5 in controlling this response.
Figure 3.
Genetic interaction of SAS-negative regulators in the shade-induced hypocotyl elongation. Difference of hypocotyl length (HYP) in simulated shade, W + FR (HYPW + FR) and W (HYPW) of Col-0, A) phyA-211, hfr1-5, phyA-211 hfr1-101, B) hy5-2, hfr1-5, hy5-2 hfr1-1, C) phyA-501, hy5-2, and phyA-501 hy5-2. Seedlings were grown as in Fig. 2A. Means and Se of 3 independent replicates were used to calculate the shown values of HYPW + FR − HYPW and to propagate the Se. Error bars are the propagated Se. Black asterisks indicate significant differences (based on the 2-way ANOVA) between the mutant and wild-type genotypes in response to simulated shade (**P < 0.01). In A) and C), red asterisks indicate significant differences (based on the 2-way ANOVA) between the double mutants and phyA single genotypes in response to simulated shade (**P < 0.01; ns, not significant).
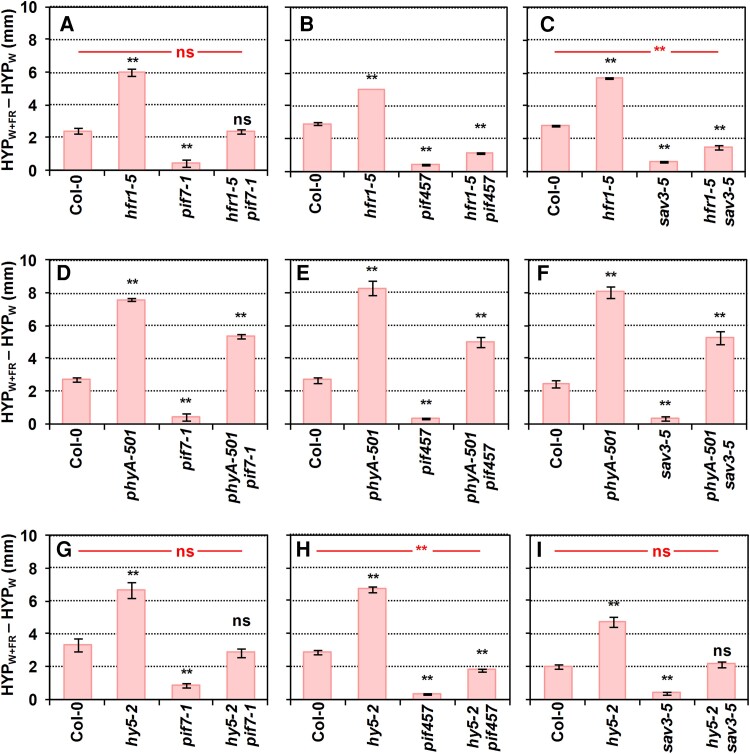
We also generated multiple mutants of positive and negative SAS regulators (Fig. 4; Supplementary Fig. S2). As before, phyB mutants were excluded from these crosses. Phenotypic analyses showed that hfr1 pif7 length was intermediate between the single mutants (Fig. 4A; Supplementary Fig. S2A) consistent with HFR1 interacting with and inhibiting PIF7 activity (Fiorucci and Fankhauser 2017; Buti et al. 2020; Paulisic et al. 2021). The shade-induced elongation of hypocotyls of an hfr1 pif457 quadruple mutant line was reduced compared to hfr1 pif7. This is consistent with the reported role of HFR1 as a transcriptional cofactor by heterodimerizing and inhibiting the transcriptional activity of PIF4 and PIF5 (Hornitschek et al. 2009; Galstyan et al. 2011). However, the elongation response of the hfr1 pif457 mutant was still higher than that of pif457 seedlings (Fig. 4B; Supplementary Fig. S2B), reflecting the minor contribution of additional factors. Mutant hfr1 plants were also crossed with sav3, known to have a role in the shade-induced auxin biosynthesis (Tao et al. 2008). Phenotypic analyses (Fig. 4C; Supplementary Fig. S2C) showed that the phenotype of hfr1 sav3 mutants was significantly longer than in sav3 seedlings and virtually as short as hfr1 pif457, in agreement with the described role of PIF457 in promoting the SAV3-dependent IAA biosynthesis (Hornitschek et al. 2012; Li et al. 2012). These results not only pointed to a very minor contribution of other PIFs but also to a SAV3-independent auxin mechanism in these shade conditions.
Figure 4.
Genetic interaction of pairs of SAS-negative and -positive regulators in the shade-induced hypocotyl elongation. Difference of hypocotyl length (HYP) in simulated shade, W + FR (HYPW + FR) and W (HYPW) of Col-0 (A) hfr1-5, pif7-1, hfr1-5 pif7-1, B) hfr1-5, pif457, hfr1-5 pif457, C) hfr1-5, sav3-5, hfr1-5 sav3-5, D) phyA-501, pif7-1, phyA-501 pif7-1, E) phyA-501, pif457, phyA-501 pif457, F) phyA-501, sav3-5, phyA-501 sav3-5, G) hy5-2, pif7-1, hy5-2 pif7-1, H) hy5-2, pif457, hy5-2 pif457, and I) hy5-2, sav3-5, and hy5-2 sav3-5. Seedlings were grown as in Fig. 2A. Means and Se of 3 independent replicates were used to calculate the shown values of HYPW + FR − HYPW and to propagate the Se. Error bars are the propagated Se. Asterisks indicate significant differences (based on the 2-way ANOVA) between the mutant and wild-type genotypes in response to simulated shade (**P < 0.01; ns, not significant).
In contrast with the previous crosses, phyA pif7, phyA pif457, and phyA sav3 shade-induced hypocotyl elongation was similar between them and closer in length to that of phyA than to pif7, pif457, or sav3 (Fig. 4, D to F; Supplementary Fig. S2, D to F). Together, these results suggested that the elongation repression imposed by phyA under simulated shade is mostly independent on PIF457 or the rapid shade-induced and SAV3-dependent auxin biosynthesis. Finally, hypocotyls of hy5 pif7, hy5 pif457, and hy5 sav3 seedlings showed an intermediate elongation compared to the parental pif7, pif457, sav3, and hy5 lines (Fig. 4, G to I; Supplementary Fig. S2, G to I). Together, these results suggest an additive activity for these regulators in the control of this shade-induced elongation response.
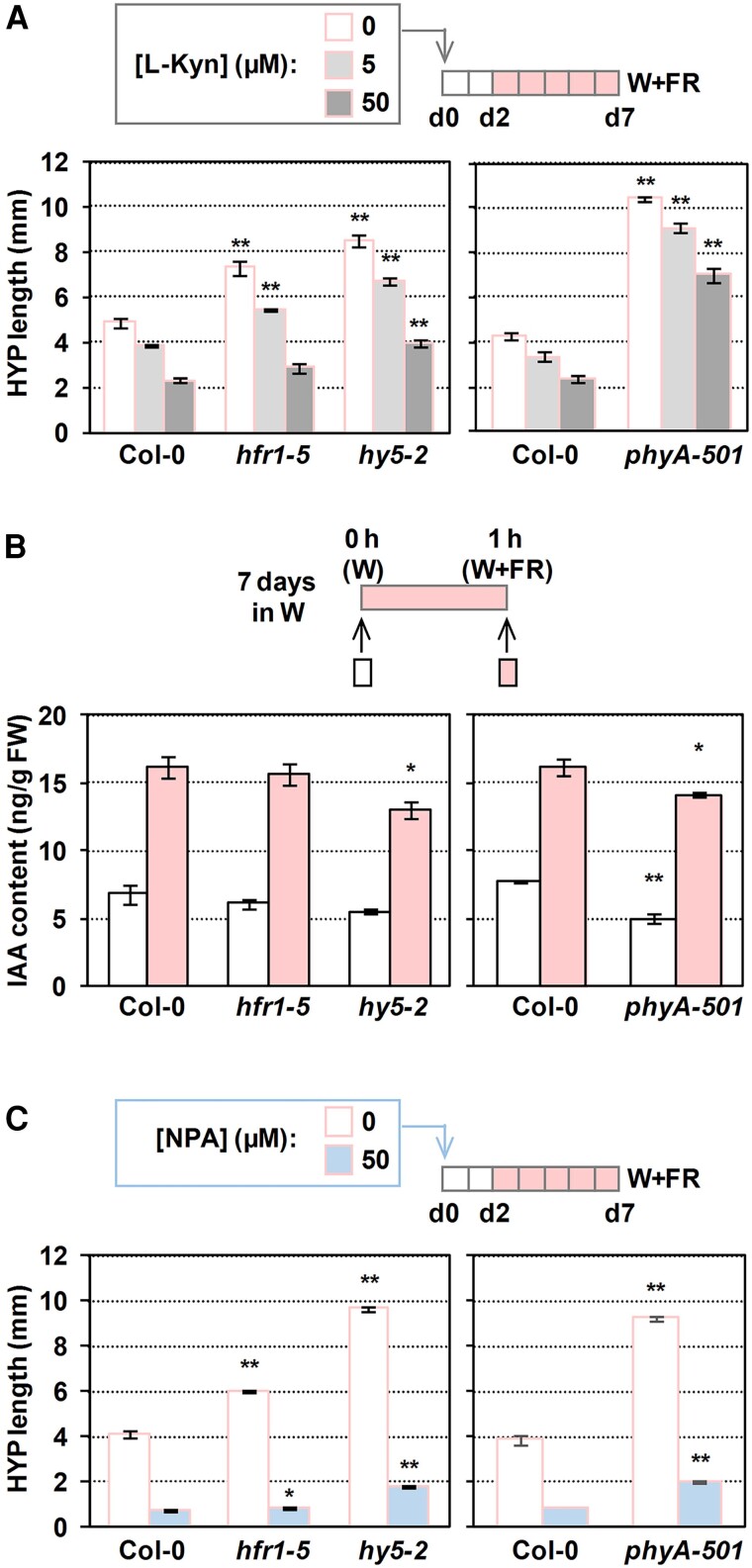
The variations in SAV3 requirements between the 2 proposed branches led us to explore the contribution of auxin synthesis and transport in these modules. The cotyledons of A. thaliana and Brassica rapa seedlings perceive shade and trigger local IAA synthesis in the cotyledons themselves (Fig. 1). Then, IAA is transported from cotyledons to the hypocotyl, where cellular elongation occurs (Procko et al. 2014). Treatments of wild-type seedlings with the auxin biosynthesis inhibitor L-kynurenine (L-kyn) that effectively and specifically targets Trp aminotransferases such as SAV3 (He et al. 2011) or with the auxin transport inhibitor naphthylphthalamic acid (NPA) abolish the shade-induced hypocotyl elongation response in wild-type seedlings (Fig. 5) (Sorin et al. 2009; Keuskamp et al. 2010; Hersch et al. 2014; Zheng et al. 2016). In our W + FR conditions, the extra elongation of hfr1 compared to wild-type hypocotyls was completely abolished by the highest doses of L-kyn applied (Fig. 5A). Auxin quantification indicated that IAA levels in Col-0 and hfr1 seedlings increased to similar values after 1 h of W + FR treatment (Fig. 5B), in contrast with published information (Hersch et al. 2014). Nonetheless, this result suggests that HFR1 does not have a strong and measurable impact on the IAA levels in our growth/shade conditions, at least at the time of shade treatment analyzed. The extra hypocotyl elongation of phyA and hy5 seedlings, which was less affected by L-kyn than the wild type (Fig. 5A), and the attenuated IAA levels after 1 h of W + FR in both hy5 and phyA (Fig. 5B) suggested that shade-induced elongation in these 2 mutants was not fully dependent on auxin levels. As IAA levels seem to be under negative feedback control, as in several auxin-signaling mutants (Suzuki et al. 2015; Takato et al. 2017), these results are consistent with phyA and hy5 having an altered auxin responsiveness (Cluis et al. 2004; Yang et al. 2018).
Figure 5.
Contribution of auxin synthesis and polar transport in the shade-induced hypocotyl response of phyA, hy5, and hfr1 seedlings. A) Effect of L-kyn on the shade-induced hypocotyl (HYP) length of Col-0, hfr1-5, hy5-2, and phyA-501 seedlings. B) IAA content in Col-0, hfr1-5, hy5-2, and phyA-501 seedlings grown in W for 7 d and then treated for 1 h with W + FR (R:FR = 0.02). Whole seedlings were collected to measure IAA levels. Data are means and error bars are Se of 4 biological replicates. FW, fresh weight. C) Effect of NPA on the shade-induced HYP length of Col-0, hfr1-5, hy5-2, and phyA-501 seedlings. In A) and C), inhibitors were applied in the media, seedlings were grown in W for 2 d and then transferred to W + FR for 5 d, and values are means and error bars are Se of 3 independent replicates. Asterisks indicate significant differences (Student's t-test) relative to the wild type growing under the same light treatment (ns, not significant, *P < 0.05, **P < 0.01).
The extra elongation of hfr1 mutant seedlings is almost abolished by the application of the auxin transport inhibitor NPA (Fig. 5C), which indicates that the action of PIF-HFR1 in modulating the shade-induced hypocotyl elongation is mostly dependent on auxin produced somewhere else and transported to the hypocotyl, as proposed (Hersch et al. 2014). By contrast, the resistance to the inhibitory effect of NPA of phyA and hy5 (Fig. 5C) suggested that these 2 factors share similar mechanisms to repress shade-induced elongation and, in contrast with HFR1, are less dependent on auxin transport to promote elongation.
Together, these results are consistent with a network architecture in which these components are likely organized in at least 2 separate branches or modules, one involving the activity of PIF457, HFR1, and SAV3 to promote the shade-induced hypocotyl elongation and a second one requiring the activity of PHYA and HY5 to repress it.
SAS regulatory components act in different moments during the shade-induced hypocotyl elongation
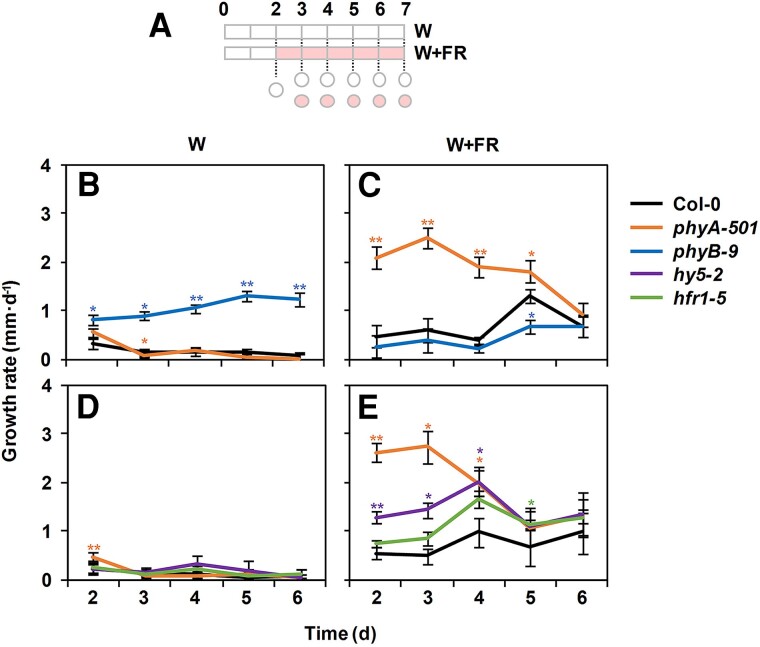
We next studied when the different components act upon exposure of young seedlings to W + FR. Growth rates were first determined in wild-type (Col-0) seedlings grown under W and W + FR. To do so, hypocotyl length was measured daily from Days 2 to 7 in different groups of seedlings, and the variations in the daily growth rate were estimated for each genotype and light treatment (Fig. 6A). Under W, Col-0 growth rate remained low but constant along the period analyzed (Fig. 6, B and D), whereas under W + FR, it went up from Day 5 onwards (Fig. 6, C and E). As an additional control, the growth rate of the phyB mutant hypocotyls was also estimated. Importantly, under W, phyB growth rate increased with the age of the seedlings (Fig. 6B). Under W + FR, phyB growth rate mimicked that of Col-0 but the peak at Day 5 was attenuated in the mutant (Fig. 6C), consistent with its reduced elongation compared to Col-0 (Fig. 2) (Martínez-Garcia et al. 2014; Molina-Contreras et al. 2019). Under W, the growth rate of phyA, hy5, and hfr1 hypocotyls was constant along time and similar to that of Col-0 (Fig. 6, B and D). Under W + FR, phyA and, to a lower extent, hy5 growth rate was much higher than that of Col-0 hypocotyls on Days 2 to 4 but progressively dropped to values closer to those of Col-0 (Fig. 6, C and E). By contrast, hfr1 followed a pattern of growth rate similar to Col-0 and, if anything, it elongated slightly faster than Col-0 in the second half of the period of time analyzed (Fig. 6E). To visualize the repressor activity of the different regulators, the growth rate of the wild type was subtracted to that of each mutant grown in those conditions where the phenotype is more obvious: W for phyB and W + FR for hfr1, hy5, and phyA. This representation confirmed our previous conclusions (Supplementary Fig. S3). In summary, although the temporal activity of the regulators overlapped, phyA represses hypocotyl elongation more strongly at the beginning of seedling development (from Days 2 to 4), HY5 at the beginning but shows a peak in the middle of this period (Day 4) and HFR1 (and phyB) appeared as more active at the second half of the period analyzed (from Days 5 to 7). These results provided a framework that separates the temporal action of the participating components.
Figure 6.
Effect of phyA, phyB, HFR1, and HY5 mutations on the hypocotyl growth rate under W and simulated shade. A) Cartoon of the experiment design. Seeds were germinated and grown for 2 d under W and then either kept under W or transferred to simulated shade (W + FR, R:FR = 0.02) for 5 more days. Circles indicate the days on which hypocotyl lengths were measured to estimate the daily growth rate. Growth rate of Col-0, phyA-501, and phyB-9 hypocotyls grown in W B) or W + FR C). Growth rate of Col-0, phyA-501, hfr1-5, and hy5-2 hypocotyls grown in W D) or W + FR E). In B) to E), growth rate values were estimated as the difference of average hypocotyl length after 2 consecutive days. Values are means and error bars are Se of 3 independent biological replicates.
SAS regulatory components target overlapping but different regions along the hypocotyl axis
In A. thaliana, hypocotyl elongation is a result of cell elongation (not cell division). Among the different tissues of this organ, the epidermis is of particular importance in mediating auxin-induced growth in the hypocotyl (Procko et al. 2016). In W-grown hypocotyls, the pattern of epidermal cell length takes place in all cells over the entire growth period (from 1 to 9 d after germination), although the area of fastest growth moves acropetally, from the base (Cells 2 to 4) on Days 1 to 2 to the middle (Cells 10 to 12) of the hypocotyl on Days 7 to 9 (Gendreau et al. 1997). In dark-grown seedlings, growth also initiates in the hypocotyl basal cells but, in this case, cells that elongated fastest move up much more rapidly and only a few cells upwards: from Cell 1 at 36 to 48 h to Cells 3 to 4 at 72 h from germination (Gendreau et al. 1997). As there is not much information about how A. thaliana hypocotyls elongate in response to simulated shade at the cell level, we first established the pattern of epidermal cell length in wild-type (Col-0) hypocotyls grown under W and W + FR. Using confocal microscopy, the length of several files of epidermal cells along the hypocotyl longitudinal axis per treatment was measured (Supplementary Fig. S4). Cell length in W-grown hypocotyls was similar along the hypocotyl (Supplementary Fig. S5). By contrast, W + FR treatment enhanced the elongation of all cells compared to W treatment, although the pattern of epidermal cell length was not uniformly distributed, with cells located in the lower half of the hypocotyl elongating the most (Supplementary Fig. S5). A similar conclusion was reached when representing the difference in length between cells grown in W + FR and W in each position, with Cells 7 to 8 being the ones that grew the most, becoming about 170 to 250 µm longer than cells in the same position of W-grown hypocotyls (Fig. 7, Col-0 panels). These results indicate that the shade-induced hypocotyl elongation of wild-type seedlings resulted from a bell-shaped non-symmetrical (skewed) elongation pattern of epidermal cells peaking around Cells 7 to 8 from the base. A skewed distribution of cell elongation in the hypocotyl has also been observed in dark-, W-, or low blue-grown hypocotyls of A. thaliana (Gendreau et al. 1997; Keuskamp et al. 2010), shade-exposed B. rapa hypocotyls (Procko et al. 2014), and end-of-day-FR-treated cowpea epicotyls (Martinez-Garcia and Garcia-Martinez 1992).
Figure 7.
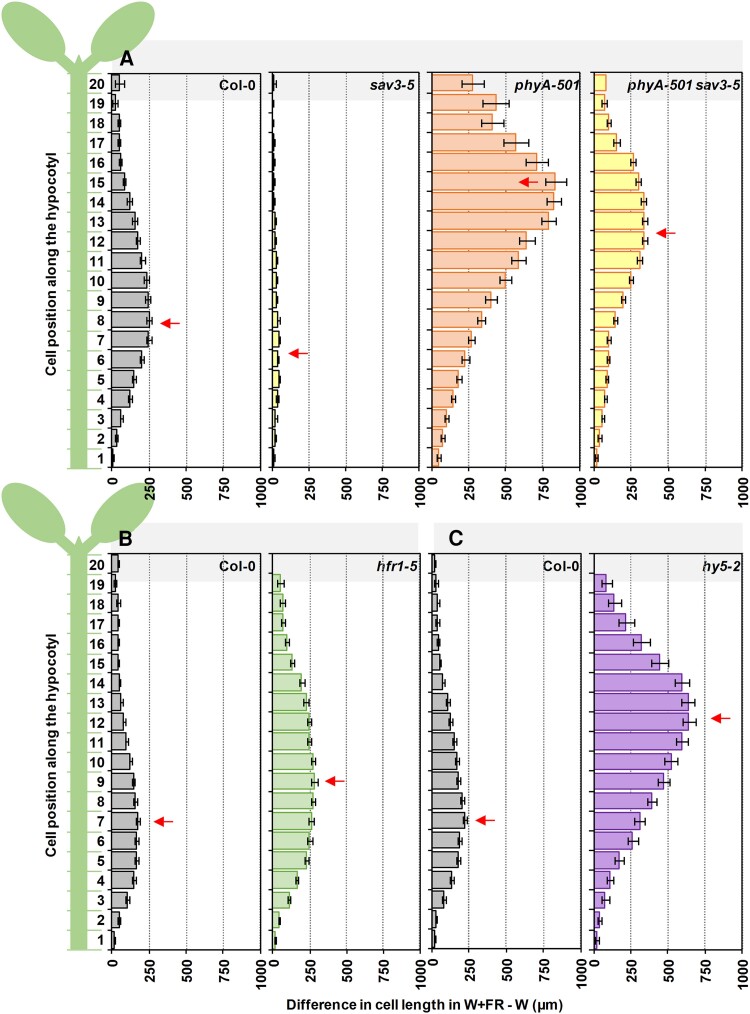
Distribution of the epidermal cell length from base to top induced by simulated shade in hypocotyls of wild-type or SAS mutant seedlings. Schematic representation of the 20 cells composing a cell row of the epidermis along the longitudinal axis of an A. thaliana hypocotyl (left). Difference in length in simulated shade (W + FR) and W for each of the 20 epidermal cells in hypocotyls of Col-0, A) sav3-5, phyA-501, phyA-501 sav3-5, B) hfr1-5, and C) hy5-2. Seedlings were grown as in Fig. 2A. Arrows point to cell with a highest difference in length. Values have been estimated after mean lengths of at least 15 cells of 2 cell rows per hypocotyl from 7 different hypocotyls per genotype and growth condition. Error bars represent Se.
Independently of the primary site of action of the studied SAS regulators (e.g. cotyledons or hypocotyls), their activities converge on the elongation of hypocotyls. To establish whether the convergence affected the same or different hypocotyl cells, we next analyzed the shade-induced cell length in hypocotyls of seedlings deficient in specific SAS regulators (Supplementary Fig. S5; Fig. 7). The hyporesponsive sav3 hypocotyls showed a similar pattern of cell elongation as wild type but strongly attenuated and slightly shifted to lower cells (elongation peak in Cells 5 to 7 that elongated ∼40 µm more than the same cells in W-grown hypocotyls) (Fig. 7A). In the shade-hyperresponsive hfr1 seedlings, the peak of cell length was widened, with Cell 9 showing the maximum of elongation (∼280 µm longer than Cell 9 in W-grown hypocotyls) (Fig. 7B). In the case of hy5 and phyA, also hyperresponsive to shade, cell length was strongly enhanced and the elongation peak moved to the upper half of the hypocotyl (Cell 12 in hy5 that elongated ∼640 µm more than the same cell in W-grown hypocotyls; Cell 15 in phyA that elongated ∼830 µm more than the same cell in W-grown hypocotyls) (Fig. 7, A and C).
As the peak cell number was associated with the difference in hypocotyl length in W + FR and W (HYPW + FR − HYPW) (Supplementary Fig. S6), we wondered if the redistribution of cell growth was a consequence of the enhanced hypocotyl shade-induced elongation shown by these genotypes. To check this possibility, we analyzed the cell length in phyA sav3 hypocotyls, whose shade-induced hypocotyl elongation was similar to that of hfr1 and lower than hy5 hypocotyls (Fig. 6). The peak of cell elongation in phyA sav3 seedlings (Cell 13) was closer to that of hy5 (Cell 12) and phyA (Cell 15). In addition, the most responsive cell in phyA sav3 seedlings elongated more (Cell 13, ∼340 µm) than in hfr1 (Cell 9, ∼280 µm). These results reinforced the conclusion that phyA acted by repressing the elongation of a group of cells located in the upper half of the hypocotyl (Fig. 7A). In this case, peak cell number did not associate with the HYPW + FR − HYPW (Supplementary Fig. S6).
Altogether, these analyses indicate that (i) the hypocotyl cells more responsive to simulated shade are located in the lower half of the wild-type hypocotyls (centered in Cells 7 to 8), (ii) deficiency in SAS-negative regulators keeps the pattern of epidermal cell length but affects the peak cell number, and (iii) although the target cells of the various SAS-negative regulators overlap, the peak cell number due to loss of HY5 and PHYA function is strongly shifted toward the upper half of the hypocotyl. These results are consistent with phyA and HY5 activities repressing the cells of the upper part of the hypocotyl whereas HFR1 more clearly repressed the elongation of cells located in the lower half of the hypocotyl, providing a spatial framework that separates the action of the participating components.
PIF457 and HY5 modulate the expression of shared shade-regulated genes
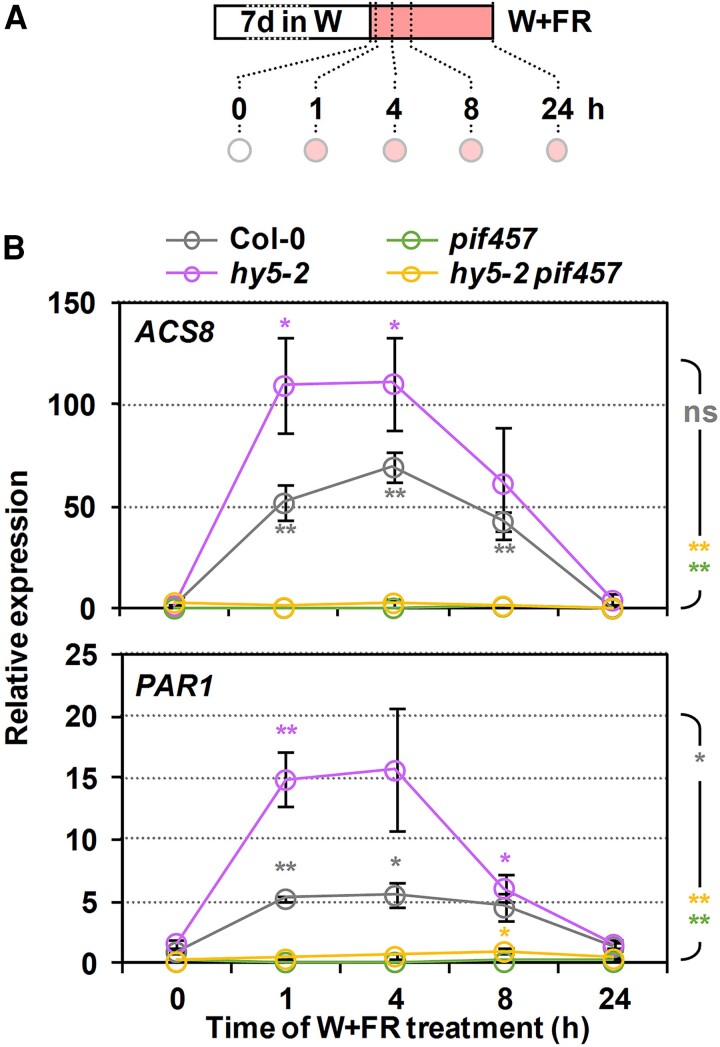
Despite the temporal differences observed between phyA, HY5, and PIFs/HFR1/SAV3, their activities overlap and eventually converge in controlling hypocotyl elongation. Hence, we aimed to further investigate possible convergence points between these 2 groups of regulators. Evidence in other photomorphogenic or temperature-regulated responses showing that HY5 directly interacts with PIF1/PIF3 proteins (Chen et al. 2013) and HY5 and PIF activities converge at a shared cis-regulatory element (Toledo-Ortiz et al. 2014; Gangappa and Kumar 2017; Zhang et al. 2017) led us to explore shade-induced changes of PIF457 and HY5 in the expression of shared targets genes. We focused on 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE 8 (ACS8) and PAR1, identified as both potentially putative HY5 binding targets (Lee et al. 2007) and direct PIFs targets (Khanna et al. 2007; Gallego-Bartolome et al. 2011; Yang et al. 2018). The expression of both genes was significantly promoted in Col-0 after 1 to 8 h of W + FR treatment (compared to the beginning of the treatment) and decreased after 24 h of the shade treatment (Fig. 8). In hy5, ACS8, and PAR1, expression was also induced after 1 to 8 h. By contrast, in pif457 and hy5 pif457, the expression of ACS8 and PAR1 remained virtually unaffected by the W + FR treatment (Fig. 8B). These results suggest that PIF457 activates whereas HY5 represses ACS8 and PAR1 expression. Importantly, HY5 activity depends on PIF457 transcriptional activation. These expression analyses were carried out in 7-d-old seedlings. These older seedlings elongated mildly to W + FR treatments (Supplementary Fig. S7), although the profile of response was consistent to what was observed in younger seedlings (Fig. 4H) suggesting that the same genetic components and molecular mechanisms are still functional.
Figure 8.
Effect of hy5 and pif457 mutations on the shade regulation of the expression of ACS8 and PAR1. A) Cartoon of the experiment design. Seeds were grown for 7 d under W and then transferred to simulated shade (W + FR, R:FR = 0.02) for the indicated time before harvesting samples (circles). B) Relative expression of ACS8 (top) and PAR1 (bottom) in Col-0, hy5-2, pif457, and hy5-2 pif457 at the indicated times of simulated shade treatment. Values are means and error bars are Se of 3 independent biological replicates. Expression is presented relative to the Col-0 genotype at 0 h. Asterisks around the symbols indicate significant differences (Student's t-test) relative to the same genotype at 0 h. Asterisk at the right indicate significant differences between the different mutants and the wild type in response to simulated shade (2-way ANOVA); ns, not significant, *P < 0.05, **P < 0.01.
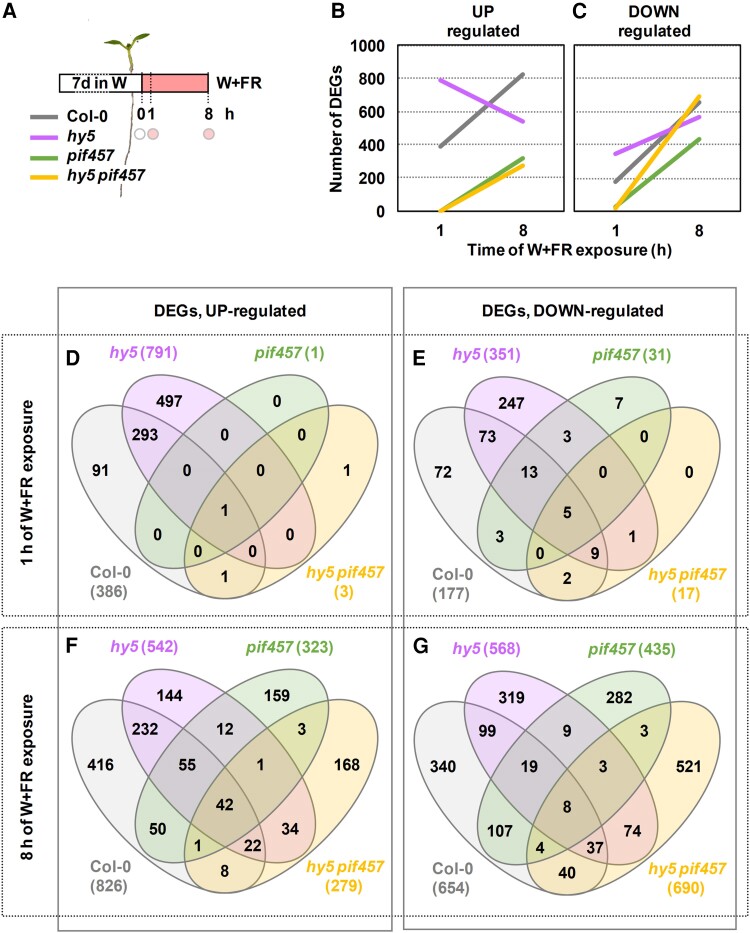
To expand our understanding of the role and interaction of HY5 and PIF457 activities, we carried out RNA sequencing (RNA-seq) of the time points 0, 1, and 8 h after shade exposure of the 4 genotypes (Col-0, hy5, pif457, and hy5 pif457) (Fig. 9A). We identified differentially expressed genes (DEGs) upregulated (fold change [FC] ≥ 1.5, P < 0.05) and downregulated (FC ≤ 0.667, P < 0.05) after 1 and 8 h of shade treatment compared to 0 h for each genotype analyzed (Supplementary Tables S1 to S4). After 1 h of W + FR, 386 and 791 DEGs were induced and 177 and 351 were repressed in wild-type and hy5 seedlings, respectively. Importantly, only 1 and 3 DEGs were induced and 31 and 17 were repressed in pif457 and hy5 pif457 seedlings, respectively (Fig. 9, B to E). From these early shade-modulated DEGs, 294 upregulated genes were shared between hy5 (out of 791 genes, 37.2%) and Col-0 (out of 386 genes, 76.2%) (Fig. 9D) and 100 downregulated genes were shared between hy5 (out of 351, 28.5%) and Col-0 (out of 674, 14.8%) (Fig. 9E). As 748 DEGs (497 upregulated and 251 downregulated) appeared only in hy5 but not in Col-0, we concluded that HY5 has a dual role as both activating and repressing rapid shade-modulated gene expression. The vast majority of these DEGs did not change in pif457 and hy5 pif457 (Fig. 9, D and E), indicating that PIF457 is basically required for all the changes in gene expression that take place after 1 h of simulated shade exposure. An important but weaker impact of PIF457 on gene expression was detected after 3 h of shade exposure (Ince et al. 2022).
Figure 9.
Effect of hy5 and pif457 mutations on the shade-regulated transcriptome. A) RNA-seq was performed with RNA extracted from Col-0, hy5-2, pif457, and hy5-2 pif457 seedlings at the indicated times (circles) of simulated shade treatment. Seedlings were grown as in Fig. 8A. Three independent biological replicates were used for each genotype and treatment. Evolution of the number of upregulated B) and downregulated C) DEGs in response to 1 and 8 h of W + FR in Col-0, hy5-2, pif457, and hy5-2 pif457 seedlings grown as indicated in A). Venn diagrams showing the overlap of upregulated D, F) and downregulated E, G) DEGs after 1 D, E) and 8 h F, G) of W + FR treatment between Col-0, hy5-2, pif457, and hy5-2 pif457 seedlings.
After 8 h of W + FR, 826 and 542 DEGs were induced and 654 and 568 were repressed in Col-0 and hy5 seedlings, respectively. After this time of W + FR exposure, a substantial number of DEGs were detected in pif457 (323 upregulated and 435 downregulated) and hy5 pif457 (279 upregulated and 690 downregulated) seedlings (Fig. 9, B to G). Venn diagrams indicated that, from the total number of DEGs identified in all genotypes (1,347 upregulated and 1,865 downregulated), a large fraction appeared as upregulated (65.9%: 416 in Col-0, 144 in hy5, 159 in pif457, and 168 in hy5 pif457) or downregulated (78.4%: 340 in Col-0, 319 in hy5, 282 in pif457, and 521 in hy5 pif457) only in 1 genotype, whereas the rest appeared in at least 2 genotypes (Fig. 9, F and G). Based on the highest significance and the enrichment fold of overlapping genes, we concluded that the set of DEGs of hy5 pif457 (upregulated and downregulated) is closer to this in hy5 than pif457. The number of misregulated DEGs in hy5 pif457 is lowest when compared to hy5 and highest when compared to pif457 (Supplementary Fig. S8). These results indicate that, after 8 h of simulated shade, (i) the expression of a substantial amount of DEGs does not require PIF457 activity and (ii) the DEG identity is closer to hy5 than pif457, in contrast to what happens at 1 h.
Regarding the functional prediction, the DEGs belonged to similar GO term categories in all genotypes (except in pif457 and hy5 pif457 after 1 h of W + FR, in which no GO term enrichment was found because of the massive drop in DEG number) (Supplementary Table S5). Importantly, no obvious and specific processes were differentially affected by HY5 (at 1 h) or HY5 and PIF457 at later times that could easily explain the differences in growth detected among the genotypes (Fig. 5H).
Together, we concluded that (i) PIF457 and HY5 have a strong impact in the early shade-regulated changes in gene expression, although (ii) the leading role of PIF457 at this early time of shade exposure dissipates after longer periods (8 h) of treatment.
Discussion
In the A. thaliana shade-induced hypocotyl elongation, the function of phyB and its effect on the PIF457-HFR1 and auxin biosynthesis via SAV3/YUCs in the cotyledons are well established (Tao et al. 2008; Li et al. 2012; Ciolfi et al. 2013; Kohnen et al. 2016; Fiorucci and Fankhauser 2017; Paulisic et al. 2021) (Fig. 1). The observed genetic interactions between sav3/pif7/pif457 and hfr1 (Fig. 4, A to C) and the pharmacological applications of L-kyn and NPA on hfr1 seedlings (Fig. 5A) are consistent with this scenario. The genetic analyses with SAS-negative and SAS-positive regulators (Figs. 2 to 4), the enhanced resistance to L-kyn and NPA shown by phyA and hy5 seedlings in response to shade, and the changes in the rapid shade-induced IAA production (relative to Col-0) (Fig. 5) supported that phyA and HY5 act in a different branch than PIF457-HFR1 and SAV3/YUCs.
The strong shade-induced elongation of phyA pif457 and phyA sav3 hypocotyls (Fig. 4, D to I) indicated that these mutants, therefore, might elongate either using IAA generated from a PIF457- and SAV3-independent biosynthesis pathway or without the need of de novo synthesis of IAA. Indeed, IAA can be produced from IAA-conjugated with amino acids molecules in the hypocotyl, IAA that is able to elicit the shade-induced hypocotyl elongation independently of the SAV3-mediated IAA biosynthesis in cotyledons (Zheng et al. 2016). However, this increase in IAA potentially produced in the hypocotyl by this alternative pathway does not seem to be high enough for being detected when quantifying IAA in whole phyA or hy5 seedlings (Fig. 5B).
HY5 is involved in the phyA-mediated gene repression in prolonged low R:FR (Ciolfi et al. 2013), and both phyA and HY5 act very early in the seedling development (Days 2 to 5) (Fig. 6). The phyA-HY5 early suppression seems fundamental for seedling establishment and survival soon after germination in deep shade environments (Yanovsky et al. 1995). Because deep canopy conditions are usually accompanied by reductions in the light intensity, in these early stages of the seedling development, the mechanisms of elongation are very dependent on changes of auxin sensitivity (Hersch et al. 2014) that can be modulated directly by phyA action on the stability of the auxin signaling repressors Aux/IAA (Yang et al. 2018) and by HY5 on the promotion of the expression of negative regulators of auxin signaling (Cluis et al. 2004). The PIF457-HFR1 module appears to be working in early and late seedling development (Fig. 6) and even in other organs and stages of development, such as petiole length and lamina size in leaves (de Wit et al. 2015), responses that appear to be more dependent on changes of auxin levels. Therefore, our results provide a temporal framework with different dependence on auxin sensitivity and levels that support the functional separation of these 2 signaling modules.
Perception of the R:FR by phyB in the control of the hypocotyl elongation occurs mainly in the cotyledons, where PIF457-HFR1 promotes elongation by inducing IAA production (Tanaka et al. 2002; Keuskamp et al. 2010; Procko et al. 2014; Kohnen et al. 2016). Newly synthesized IAA is then transported to the adjacent hypocotyl where cell elongation is promoted. As before, differences were observed between the 2 signaling modules: the activity of the PIF457-HFR1 module, that it is SAV3-dependent, affects the elongation of cells in the middle-lower half of the hypocotyl that is spatially distinct from that of phyA-HY5, that mainly represses cell elongation in the upper half (Fig. 7). It seems, therefore, that the strong repression imposed by phyA-HY5 in the upper half of the wild-type hypocotyls takes place at the beginning of seedling development. This temporal and spatial separation of the PIF457-HFR1 (together with SAV3) and phyA-HY5 regulatory activities is consistent with an acropetal gradient of hypocotyl growth (from the base to the top) in response to simulated shade, as it was observed in both dark- and W-grown seedlings (Gendreau et al. 1997).
Additional levels of regulation refer to (i) when the different SAS components and modules act relative to the beginning of the simulated shade exposure and (ii) their level of molecular interaction. Our expression analyses indicate that PIF457 is essential to modulate gene expression immediately after shade exposure. In clear contrast, HY5 suppressed the number of DEGs after 1 h although its activity was strongly dependent on PIF457 at this early time after shade exposure (Figs. 8 and 9, D and E), which suggests a connection of the 2 mentioned branches at these initial stages after shade exposure. Previously, it has been demonstrated physical interaction between HY5 and some PIFs and HFR1 and convergence of their transcriptional activities in non-shade-related processes (Chen et al. 2013; Jang et al. 2013; Toledo-Ortiz et al. 2014; Zhang et al. 2017). Thus, PIF457-HFR1 and HY5 could be key players connecting the 2 regulatory modules rapidly after shade exposure. However, the crosstalk between PIF457-HFR1 and HY5 activities seems dynamic and changes with time. After 8 h of shade exposure, the transcriptome was clearly affected by shade even in the absence of PIF457 (Fig. 9, B, C, F, and G), reflecting that a large percentage of the expression changes caused after shade perception by phyB happens bypassing PIF457 activity. Hence, expression of these DEGs depends either on other PIFs (e.g. PIF1 and PIF3) or on the effect exerted by unknown but non-PIF regulators whose transcriptional activity is also connected to the reduction in phyB activity caused by shade. In addition, after 8 h of shade exposure, the transcriptome divergence between the various genotypes, even between pif457 and hy5 pif457 (Fig. 9, F and G), points to a change in the molecular relationship of PIFs and HY5 that appear in this moment to act independently from each other. What sustains this dynamic relationship is unknown, although it might involve changes in the accessibility of these regulators to the same target promoters with time triggered by shade perception (e.g. caused by the increase in the abundance of transcriptional regulators–cofactors that can affect their DNA-binding abilities), a shade-induced divergence of their spatial pattern of expression that impedes PIFs and HY5 to be expressed in the same cells, and/or by epigenetic processes that alter chromatin compaction, also known to influence the accessibility and binding of transcription factors to regulatory elements in the DNA (Martínez-Garcia and Moreno-Romero 2020).
In the SAS regulation, PIFs are usually presented as positive regulators by promoting the expression of genes involved in hypocotyl elongation. Our RNA-seq analyses support that they also have an important function in the repression of gene expression, as it has been previously described for some PIFs in shade-induced processes related with metabolic or architectural responses (hence, not related with cell elongation) (Toledo-Ortiz et al. 2010; Xie et al. 2017; Jia et al. 2020). Similarly, although HY5 acts mainly inducing gene expression (Burko et al. 2020), it has an important role in the shade repression of genes that, after just 1 h of shade exposure, requires PIF457 (Fig. 9, D and E).
Our findings propose a model for the regulation of shade-induced hypocotyl elongation that incorporates the temporal and spatial functional importance of the various SAS regulators analyzed in here. These components are grouped in 2 main modules or branches: (i) a well-defined pathway in which PIF457-HFR1 participates, it is highly dependent on auxin produced via SAV3 and YUCs mostly in the cotyledons, acts along all seedling development (from Days 2 to 7 from germination), and targets cells in the middle-lower region of the hypocotyl; and (ii) a less well-characterized pathway with phyA and HY5 as main components, that is less dependent on SAV3-related auxin biosynthesis and polar transport, it has an important role in the early seedlings development (Days 2 to 5 after germination) and targets cells in the upper region of the hypocotyl. In these processes, PIF457 transcriptional activity is fundamental at 1 h of W + FR and its importance dissipates at later times (8 h). By contrast, the importance of HY5 regulatory role increases at longer times of shade exposure, when its expression is also reported to enhance (Ciolfi et al. 2013), likely because of the delayed accumulation of phyA. Importantly, the molecular interaction between these transcriptional regulators is dynamic and moves from epistasis, soon after shade exposure, to additivity, at later hours (based on both transcriptomic and hypocotyl elongation experiments). Because of the reported interaction of HY5 with some PIFs and HFR1, it might act connecting both branches that, therefore, are crosstalking along the seedling development.
Materials and methods
Plant material and growth conditions
All the A. thaliana plant material used was in the Col-0 background. Mutants used in this study were described before: phyA-501 (Martínez-Garcia et al. 2014), hy5-2 (Ortiz-Alcaide et al. 2019), hfr1-5 (Roig-Villanova et al. 2007), pif7-1 (Li et al. 2012), and sav3-5, also known as wei8-4/tir2-3 (Stepanova et al. 2008). The multiple mutants pif457 (pif4-101 pif5-3 pif7-1) (de Wit et al. 2015), phyA-211 hfr1-101 (Duek et al. 2004), and phyA-211 phyB-9 (Strasser et al. 2010) used in this study were described elsewhere. To produce seeds of the various A. thaliana genotypes, plants were grown in the greenhouse under long day photoperiod (16-h light, 8-h dark).
Fluence rates were measured with a Spectrosense2 meter associated with a 4-channel sensor (Skye Instruments Ltd., www.skyeinstruments.com), which measures PAR (400 to 700 nm) and 10-nm windows in the blue (464 to 473 nm), R (664 to 674 nm), and FR (725 to 735 nm) regions. Light spectra were generated using a Flame Model Spectrometer with Sony Detector (FLAME-S; Ocean Optics).
Pharmacological treatments
When indicated, the medium was supplemented with different concentrations of L-kyn (Sigma-Aldrich) or NPA (Duchefa). L-kyn was dissolved at 50 mm in DMSO. NPA was dissolved at 5 mm in DMSO. Stock solutions were kept at −20 °C until use.
Genetic crosses and genotyping
Mutants were crossed to generate the following multiple mutants: phyA hy5 (phyA-501 hy5-2), phyA pif7 (phyA-501 pif7-1), phyA hfr1 (phyA-501 hfr1-5), phyA sav3 (phyA-501 sav3-5), hy5 pif7 (hy5-2 pif7-1), hy5 hfr1 (hy5-2 hfr1-5), hy5 sav3 (hy5-2 sav3-5), hfr1 pif7 (hfr1-5 pif7-1), hfr1 pif457 (hfr1-5 pif457), hy5 pif457 (hy5-2 pif457), and phyA pif457 (phyA-501 pif457). After crosses, seedlings in the segregating F2 generation were preselected searching by the predicted phenotypes, if any. In any case, the genetic identity of the plants was established by genotyping the preselected plants by PCR using specific primers (Supplementary Table S6).
Measurements of hypocotyl length
For hypocotyl growth assays, seeds were sterilized and sown in solid agar plates without sucrose (GM–; 0.215% [w/v] MS salts plus vitamins, 0.025% [w/v] MES pH 5.80) (Roig-Villanova et al. 2019). After 3 to 6 d of stratification, plates were incubated in growth chambers at 22 °C under continuous W provided by 4 cool-white vertical fluorescence tubes (F36W/840, Sylvania) for 2 d (PAR of 20 to 25 µmol·m−2·s−1, R:FR > 2.5). After that time, plates were either maintained in W or transferred to simulated shade (W + FR) for 5 d. Simulated shade was generated by enriching W with supplementary FR (peak at 725 nm) provided by 4 horizontal LED lamps (Philips GreenPower LED module FR) (PAR of 20 to 25 µmol·m−2·s−1, R:FR of about 0.02). Details of the resulting light spectra are shown as Supplementary Fig. S9 (Molina-Contreras et al. 2019). At Day 7, seedlings were flattened down on the petri dishes and pictures of them were taken. Each biological replicate corresponded to ∼25 seedlings per treatment and genotype. Experiments were done with at least 3 biological replicates. Hypocotyl measurements were carried out by using the National Institutes of Health (NHS) ImageJ software (Bethesda, MD, USA; http://rsb.info.nih.gov/). Hypocotyl measurements from the different biological replicates were averaged.
Hypocotyl measurements for the temporal analyses
Seedlings were grown for up to 7 d either in W or W + FR, as described in the previous section. In these experiments (Fig. 6; Supplementary Fig. S3), hypocotyl length measurements were made daily from pictures taken from plants of different ages, from Day 2 until Day 7 after germination (6 time points). By subtracting the hypocotyl length of 2 consecutive days, the growth rate (mm·day−1) from Days 2 to 6 was calculated for each genotype and light treatment (W and W + FR). Each biological replicate corresponded to ∼25 seedlings per treatment, genotype, and time point. Experiments were done with 3 biological replicates. Hypocotyl measurements from the different biological replicates were averaged. These averaged data were used to calculate the growth rate.
Hormone analyses
About 50 seedlings per biological replica of the different genotypes and treatments (that ranged from 80 to 120 mg) were immediately frozen in liquid nitrogen. Hormone extraction and analysis were performed as described (Simura et al. 2018) with a few modifications. Briefly, around 100 mg of fresh material was extracted in 1 mL of 50% (v/v) acetonitrile prepared with ultrapure water, adding 2.5 ng of [2H5]IAA as internal standard in a ball mill (MillMix 20, Domel) for 10 min at 17 rps, followed by 5 min of sonication. After sonication, the samples were centrifuged at 4,000 × g at 4 °C for 10 min. Finally, supernatants were filtered through SPE columns (OASIS HLB 30 mg 1 cc, Waters), recovering the eluent. Finally, 0.5 mL of 30% (v/v) acetonitrile prepared in ultrapure water was added to the SPE columns and the eluent was recovered joint to the previous ones.
Chromatographic separations were performed on a reverse-phase C18 column (50 × 2.1 mm, 1.6-µm particle size, Luna-Omega, Phenomenex) using a acetonitrile:water (both supplemented with 0.1% [v/v] formic acid) gradient at a flow rate of 300 µL/min. IAA was detected with a triple quadrupole mass spectrometer connected online to the output of the column though an orthogonal Z-spray electrospray ion source (Xevo TQ-S). Finally, IAA content was quantified by interpolation in a standard curve prepared with commercial IAA (Sigma) using the MassLynx v4.2 software.
Cell length measurements along the hypocotyl axis for spatial analysis
For the cell length measurements, about 100 seedlings were grown either in W or W + FR, as previously described. On Day 7, hypocotyls were measured and the mean value for each group (genotype and treatment) was calculated. About 15 individuals with a hypocotyl length of the estimated averaged value ± 5% were selected. Cotyledons and roots were removed from these seedlings and the remaining hypocotyls were fixed and stained with Calcofluor White (Sigma-Aldrich) to visualize cell walls. Briefly, hypocotyls were submerged in a 1× PBS solution (137 mm NaCl [8.06 g/L], 2.7 mm KCl [0.22 g/L], 10 mm Na2HPO4 [1.15 g/L], 18 mm KH2HPO4 [0.20 g/L]) with 4% (w/v) paraformaldehyde for 60 min at room temperature. Then, hypocotyls were washed twice for 1 min in 1× PBS and cleared after transferring them to ClearSee solution (10% [w/v] xylitol (Sigma), 15% [w/v] sodium deoxycholate [Sigma], 25% [w/v] urea [Sigma]). The clearing was carried out for at least 1 wk at room temperature. Before taking images, hypocotyls were stained with 100 µg·mL−1 Calcofluor in ClearSee solution for 120 min and washed twice with ClearSee solution for 2 d (Kurihara et al. 2015).
Images of fixed and stained plant material were taken by using confocal microscopy (Zeiss LSM 780). Calcofluor White stained samples were imaged with 405-nm excitation Argon laser and detected at 425 to 475 nm (Kamiya et al. 2015). In most of the images, gain and laser intensity were adjusted to remove background noise. Cell growth measurements were carried out using the NHS ImageJ software on the obtained pictures. At least 15 cells of 2 cell files per hypocotyl from 7 seedlings were measured for each genotype and growth condition. Values were averaged for each of the about 20 cells that constitute a cell file (from bottom to top) along the hypocotyl longitudinal axis.
RNA extraction and gene expression analyses
Seven-day-old seedlings grown in W or W + FR were harvested (about 35 mg per sample) and frozen in liquid nitrogen. RNA was extracted using commercial kits (Maxwell RSC Plant RNA kits; www.promega.com) and quantified using NanoDropTM 8000 spectrophotometer (Thermo Fisher Scientific). Two micrograms of total RNA were retrotranscribed to cDNA in a final volume of 20 µL by using the Transcriptor First Strand cDNA Synthesis Kit (Roche, www.roche.com) or the NZY First-Strand cDNA Synthesis Kit, separate oligos (NZYtech) following the manufacturer's instructions. Subsequently, cDNA was diluted 10-fold and stored at −20 °C for further analysis.
Relative mRNA abundance was determined via reverse transcription quantitative PCR (RT-qPCR) in a final volume of 10 µL made up of 0.3 µm of both forward and reverse primers, 5 µL of the LightCycler 480 SYBR Green I Master Mix (Roche), and 2 µL of 10-fold diluted cDNA (Molina-Contreras et al. 2019). The RT-qPCR was carried out in LightCycler 480 Real-Time PCR system (Roche). The analysis was performed with 3 independent biological replicates (∼30 seedlings per biological replicate) for each condition and 3 technical replicates for each biological replicate. ELONGATION FACTOR 1α (EF1α) was used as endogenous reference genes to normalize the expression of the genes of interest. Primers used for the RT-qPCR analyses are provided in Supplementary Table S7.
Statistical analyses
These analyses were carried out using the Real Statistics Resource Pack, an Excel add-in that extends Excel's standard statistics capabilities. For the statistical analyses, we compared 3 values corresponding to 3 replicates in the case of relative expression and hypocotyl length.
RNA-seq: processing, analyses, and data availability
Total RNA for sequencing was obtained as in the expression analyses by RT-qPCR. The total RNA samples were quantified using the Qubit RNA BR Assay Kit (Thermo Fisher Scientific), and the RNA integrity was assessed with the Agilent RNA 6000 Pico Bioanalyzer 2100 Assay (Agilent).
The RNA-seq libraries were prepared using the KAPA Stranded mRNA-Seq Illumina Platforms Kit (Roche), following the manufacturer's recommendations, starting with 500 ng of total RNA. The size and quality of the libraries were evaluated using a High Sensitivity DNA Bioanalyzer assay (Agilent). The libraries were sequenced on the HiSeq 4000 (Illumina) with a read length of 2 × 51 bp using the HiSeq 4000 SBS Kit (Illumina), following the manufacturer's protocol for dual indexing. Image analysis, base calling, and quality scoring of the run were performed using the manufacturer's software Real Time Analysis (RTA 2.7.7). RNA-seq data have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE268032.
RNA-seq reads were mapped against A. thaliana reference genome (TAIR10) from Ensembl Plants, using STAR aligner version 2.5.3a (Dobin et al. 2013) and ENCODE parameters. Annotated genes were quantified with RSEM v1.3.0 (Li and Dewey 2011), using Ensembl annotation release 47. Differential expression analysis was performed with limma v3.4.2 R package. Counts were normalized with TMM (Robinson and Oshlack 2010) and transformed with the “voom” function. The linear model was fitted with the voom-transformed counts and contrasts were extracted. Genes were considered differentially expressed (DEG) with an adjusted P < 0.05. From these, we selected those whose 0.667 ≥ FC ≥ 1.5.
GO enrichment
The list of DEGs (Supplementary Tables S1 to S4) was used to identify the enrichment in GO terms using the agriGO online analyses tool.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ACS8 (At4g37770), EF1α (At5g60390), HFR1 (At1g02340), HY5 (At5g11260), PAR1 (At2g42870), PHYA (At1g09570), PHYB (At2g18790), PIF4 (At2G43010), PIF5 (At3g59060), PIF7 (At5g61270), and SAV3 (At1g70560).
Supplementary Material
Acknowledgments
We are grateful to Christian Fankhauser (University of Lausanne, Switzerland) for providing pif457 and phyA-211 hfr1-101 seeds, to Pablo Cerdán (Fundación Instituto Leloir, Argentina) for providing phyA-211 phyB-9 seeds, to Javier Brumós (IBMCP) for comments on the manuscript, and to Javier Forment (IBMCP) for helping in GEO data submission.
Contributor Information
Pedro Pastor-Andreu, Centre for Research in Agricultural Genomics (CRAG), CSIC-IRTA-UAB-UB, Barcelona 08193, Spain.
Jordi Moreno-Romero, Centre for Research in Agricultural Genomics (CRAG), CSIC-IRTA-UAB-UB, Barcelona 08193, Spain; Institute for Plant Molecular and Cell Biology (IBMCP), CSIC-UPV, València 46022, Spain; Departament de Bioquimica I Biologia Molecular, Universitat Autònoma de Barcelona, Barcelona 08193, Spain.
Mikel Urdin-Bravo, Institute for Plant Molecular and Cell Biology (IBMCP), CSIC-UPV, València 46022, Spain.
Julia Palau-Rodriguez, Institute for Plant Molecular and Cell Biology (IBMCP), CSIC-UPV, València 46022, Spain.
Sandi Paulisic, Centre for Research in Agricultural Genomics (CRAG), CSIC-IRTA-UAB-UB, Barcelona 08193, Spain.
Elizabeth Kastanaki, Group of Plant Vascular Development, Swiss Federal Institute of Technology (ETH) Zurich, Zurich CH-8092, Switzerland.
Vicente Vives-Peris, Departament de Biologia, Bioquimica I Ciències Naturals, Universitat Jaume I, Castelló de la Plana 12071, Spain.
Aurelio Gomez-Cadenas, Departament de Biologia, Bioquimica I Ciències Naturals, Universitat Jaume I, Castelló de la Plana 12071, Spain.
Anna Esteve-Codina, Functional Genomics Team, Centro Nacional de Análisis Genómico (CNAG), Universitat de Barcelona, Barcelona 08028, Spain.
Beatriz Martín-Mur, Functional Genomics Team, Centro Nacional de Análisis Genómico (CNAG), Universitat de Barcelona, Barcelona 08028, Spain.
Antía Rodríguez-Villalón, Group of Plant Vascular Development, Swiss Federal Institute of Technology (ETH) Zurich, Zurich CH-8092, Switzerland.
Jaume F Martínez-García, Centre for Research in Agricultural Genomics (CRAG), CSIC-IRTA-UAB-UB, Barcelona 08193, Spain; Institute for Plant Molecular and Cell Biology (IBMCP), CSIC-UPV, València 46022, Spain.
Author contributions
J.F.M.-G. designed the research; P.P.-A., J.M.-R., J.P.-R., S.P., E.K., V.V.-P., A.G.-C., and A.R.-V. performed the research and analyzed their respective data; A.E.-C. and B.M.-M. performed and analyzed RNA-seq data; J.F.M.-G., P.P.-A., and J.M.-R. wrote the paper with revisions, contributions, and/or comments from all other authors.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. Genetic interaction of SAS negative regulators in the shade-induced hypocotyl elongation.
Supplementary Figure S2. Genetic interaction of pairs of SAS-negative and -positive regulators in the shade-induced hypocotyl elongation.
Supplementary Figure S3. Effect of phyA, phyB, HFR1, and HY5 mutations on the hypocotyl growth rate under W and simulated shade.
Supplementary Figure S4. Representative confocal images of Col-0 hypocotyls grown under W or simulated shade to measure epidermal cell length.
Supplementary Figure S5. Distribution of the epidermal cell length from base to top induced by simulated shade in hypocotyls of wild-type or SAS mutant seedlings.
Supplementary Figure S6. Correlation between peak of cell number and difference in length in simulated shade and W-grown hypocotyls.
Supplementary Figure S7. Effect of hy5 and pif457 mutations on the shade-induced hypocotyl elongation of older seedlings.
Supplementary Figure S8. Overlap of upregulated and downregulated gene groups between Col-0, hy5, pif457, and hy5 pif457 after 8 h of shade treatment.
Supplementary Figure S9. Light spectra of the treatments given in this study.
Supplementary Table S1. Bioset of upregulated DEGs in Col-0 seedlings in response to 1 to 8 h of W + FR.
Supplementary Table S2. Bioset of upregulated DEGs in hy5 seedlings in response to 1 to 8 h of W + FR.
Supplementary Table S3. Bioset of upregulated DEGs in pif457 seedlings in response to 1 to 8 h of W + FR.
Supplementary Table S4. Bioset of upregulated DEGs in hy5 pif457 seedlings in response to 1 to 8 h of W + FR.
Supplementary Table S5. Functional enrichment groups based on GO term analyses.
Supplementary Table S6. Primers used for genotyping the different lines.
Supplementary Table S7. Primers used for gene expression analyses.
Funding
P.P.-A. was funded by a short-term EMBO fellowship that covered his stay at the ETH, Zurich. J.M.-R. has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Slodowska-Curie (H2020-MSCA-IF-2017) grant agreement 797473. J.P.-R. is supported by a predoctoral fellowship form Agencia Estatal de Investigación (PRE2021-099195). M.U.-B. is supported by a predoctoral fellowship from the Spanish Ministerio de Universidades (FPU20/05486). Our research is supported by grants BIO2017-85316-R from MCIN/AEI/10.13039/501100011033, “ERDF A way of making Europe”, PID2020-115782GB-I00 from MCIN/AEI/10.13039/501100011033, PROMETEU/2021/065 from Generalitat Valenciana, and the EC-H2020-PRIMA UToPIQ project (PCI2021-121941) funded by Agencia Estatal de Investigación. We also acknowledge the support of the CERCA Programme/Generalitat de Catalunya.
References
- Bou-Torrent J, Toledo-Ortiz G, Ortiz-Alcaide M, Cifuentes-Esquivel N, Halliday KJ, Martinez-Garcia JF, Rodriguez-Concepcion M. Regulation of carotenoid biosynthesis by shade relies on specific subsets of antagonistic transcription factors and cofactors. Plant Physiol. 2015:169(3):1584–1594. 10.1104/pp.15.00552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumos J, Alonso JM, Stepanova AN. Genetic aspects of auxin biosynthesis and its regulation. Physiol Plant. 2014:151(1):3–12. 10.1111/ppl.12098 [DOI] [PubMed] [Google Scholar]
- Burko Y, Seluzicki A, Zander M, Pedmale UV, Ecker JR, Chory J. Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell. 2020:32(4):967–983. 10.1105/tpc.19.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buti S, Pantazopoulou CK, van Gelderen K, Hoogers V, Reinen E, Pierik R. A gas-and-brake mechanism of bHLH proteins modulates shade avoidance. Plant Physiol. 2020:184(4):2137–2153. 10.1104/pp.20.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Shade avoidance. Arabidopsis Book. 2012:10:e0157. 10.1199/tab.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Li B, Li G, Charron JB, Dai M, Shi X, Deng XW. Arabidopsis phytochrome a directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell. 2014:26(5):1949–1966. 10.1105/tpc.114.123950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R. Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell. 2013:25(5):1657–1673. 10.1105/tpc.112.104869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Esquivel N, Bou-Torrent J, Galstyan A, Gallemi M, Sessa G, Salla Martret M, Roig-Villanova I, Ruberti I, Martinez-Garcia JF. The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 2013:75(6):989–1002. 10.1111/tpj.12264 [DOI] [PubMed] [Google Scholar]
- Ciolfi A, Sessa G, Sassi M, Possenti M, Salvucci S, Carabelli M, Morelli G, Ruberti I. Dynamics of the shade-avoidance response in Arabidopsis. Plant Physiol. 2013:163(1):331–353. 10.1104/pp.113.221549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004:38(2):332–347. 10.1111/j.1365-313X.2004.02052.x [DOI] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 2003:133(4):1617–1629. 10.1104/pp.103.034397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M, Keuskamp DH, Bongers FJ, Hornitschek P, Gommers CMM, Reinen E, Martinez-Ceron C, Fankhauser C, Pierik R. Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr Biol. 2016:26(24):3320–3326. 10.1016/j.cub.2016.10.031 [DOI] [PubMed] [Google Scholar]
- de Wit M, Ljung K, Fankhauser C. Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 2015:208(1):198–209. 10.1111/nph.13449 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013:29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C. The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol. 2004:14(24):2296–2301. 10.1016/j.cub.2004.12.026 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000:14(8):2377–2391. www.genesdev.org/cgi/doi/10.1101/gad.82800 [PMC free article] [PubMed] [Google Scholar]
- Fiorucci AS, Fankhauser C. Plant strategies for enhancing access to sunlight. Curr Biol. 2017:27(17):R931–R940. 10.1016/j.cub.2017.05.085 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolome J, Arana MV, Vandenbussche F, Žádníková P, Minguet EG, Guardiola V, Van Der Straeten D, Benkova E, Alabadí D, Blázquez MA. Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 2011:67(4):622–634. 10.1111/j.1365-313X.2011.04621.x [DOI] [PubMed] [Google Scholar]
- Gallemi M, Galstyan A, Paulisic S, Then C, Ferrandez-Ayela A, Lorenzo-Orts L, Roig-Villanova I, Wang X, Micol JL, Ponce MR, et al. DRACULA2 is a dynamic nucleoporin with a role in regulating the shade avoidance syndrome in Arabidopsis. Development. 2016:143(9):1623–1631. 10.1242/dev.130211 [DOI] [PubMed] [Google Scholar]
- Gallemi M, Molina-Contreras MJ, Paulisic S, Salla-Martret M, Sorin C, Godoy M, Franco-Zorrilla JM, Solano R, Martinez-Garcia JF. A non-DNA-binding activity for the ATHB4 transcription factor in the control of vegetation proximity. New Phytol. 2017:216(3):798–813. 10.1111/nph.14727 [DOI] [PubMed] [Google Scholar]
- Galstyan A, Cifuentes-Esquivel N, Bou-Torrent J, Martinez-Garcia JF. The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 2011:66(2):258–267. 10.1111/j.1365-313X.2011.04485.x [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell. 2013:25(4):1243–1257. 10.1105/tpc.113.109751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Kumar SV. DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 2017:18(2):344–351. 10.1016/j.celrep.2016.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997:114(1):295–305. 10.1104/pp.114.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers CM, Keuskamp DH, Buti S, van Veen H, Koevoets IT, Reinen E, Voesenek LA, Pierik R. Molecular profiles of contrasting shade response strategies in wild plants: differential control of immunity and shoot elongation. Plant Cell. 2017:29(2):331–344. 10.1105/tpc.16.00790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011:23(11):3944–3960. 10.1105/tpc.111.089029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc Natl Acad Sci U S A. 2014:111(17):6515–6520. 10.1073/pnas.1320355111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012:71(5):699–711. 10.1111/j.1365-313X.2012.05033.x [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009:28(24):3893–3902. 10.1038/emboj.2009.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang Q, Jiang Y, Yang C, Wang Q, Li L. Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. Elife. 2018:7:e31636. 10.7554/eLife.31636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince YÇ, Krahmer J, Fiorucci AS, Trevisan M, Galvão VC, Wigger L, Pradervand S, Fouillen L, Van Delft P, Genva M, et al. A combination of plasma membrane sterol biosynthesis and autophagy is required for shade-induced hypocotyl elongation. Nat Commun. 2022:13(1):5659. 10.1038/s41467-022-33384-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Chua NH. Three transcription factors, HFR1, LAF1 and HY5, regulate largely independent signaling pathways downstream of phytochrome A. Plant Cell Physiol. 2013:54(6):907–916. 10.1093/pcp/pct042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Kong X, Hu K, Cao M, Liu J, Ma C, Guo S, Yuan X, Zhao S, Robert HS et al. PIFs coordinate shade avoidance by inhibiting auxin repressor ARF18 and metabolic regulator QQS. New Phytol. 2020:228(2):609–621. 10.1111/nph.16732 [DOI] [PubMed] [Google Scholar]
- Kamiya T, Borghi M, Wang P, Danku JMC, Kalmbach L, Hosmani PS, Naseer S, Fujiwara T, Geldner N, Salt DE. The MYB36 transcription factor orchestrates Casparian strip formation. Proc Natl Acad Sci U S A. 2015:112(33):10533–10538. 10.1073/pnas.1507691112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci U S A. 2010:107(52):22740–22744. 10.1073/pnas.1013457108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell. 2007:19(12):3915–3929. 10.1105/tpc.107.051508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Woo JC, Song PS, Soh MS. HFR1, a phytochrome A-signalling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J. 2002:30(6):711–719. 10.1046/j.1365-313X.2002.01326.x [DOI] [PubMed] [Google Scholar]
- Kohnen MV, Schmid-Siegert E, Trevisan M, Petrolati LA, Senechal F, Muller-Moule P, Maloof J, Xenarios I, Fankhauser C. Neighbor detection induces organ-specific transcriptomes. Revealing patterns underlying hypocotyl-specific growth. Plant Cell. 2016:28(12):2889–2904. 10.1105/tpc.16.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D, Mizuta Y, Sato Y, Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015:142(23):4168–4179. 10.1242/dev.127613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007:19(3):731–749. 10.1105/tpc.106.047688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011:12(1):323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012:26(8):785–790. 10.1101/gad.187849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Moreno-Romero J. Shedding light on the chromatin changes that modulate shade responses. Physiol Plant. 2020:169(3):407–417. 10.1111/ppl.13101 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Rodriguez-Concepcion M. Molecular mechanisms of shade tolerance in plants. New Phytol. 2023:239(4):1190–1202. 10.1111/nph.19047 [DOI] [PubMed] [Google Scholar]
- Martínez-Garcia JF, Gallemi M, Molina-Contreras MJ, Llorente B, Bevilaqua MR, Quail PH. The shade avoidance syndrome in Arabidopsis: the antagonistic role of phytochrome A and B differentiates vegetation proximity and canopy shade. PLoS One. 2014:9(10):e109275. 10.1371/journal.pone.0109275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García JF, García-Martínez JL. Interaction of gibberellins and phytochrome in the control of cowpea epicotyl elongation. Physiol Plant. 1992:86(2):236–244. 10.1034/j.1399-3054.1992.860208.x [DOI] [Google Scholar]
- Molina-Contreras MJ, Paulišić S, Then C, Moreno-Romero J, Pastor-Andreu P, Morelli L, Roig-Villanova I, Jenkins H, Hallab A, Gan X, et al. Photoreceptor activity contributes to contrasting responses to shade in Cardamine and Arabidopsis seedlings. Plant Cell. 2019:31(11):2649–2663. 10.1105/tpc.19.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli L, Paulišić S, Qin W, Iglesias-Sanchez A, Roig-Villanova I, Florez-Sarasa I, Rodriguez-Concepcion M, Martinez-Garcia JF. Light signals generated by vegetation shade facilitate acclimation to low light in shade-avoider plants. Plant Physiol. 2021:186(4):2137–2151. 10.1093/plphys/kiab206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Alcaide M, Llamas E, Gomez-Cadenas A, Nagatani A, Martinez-Garcia JF, Rodriguez-Concepcion M. Chloroplasts modulate elongation responses to canopy shade by retrograde pathways involving HY5 and abscisic acid. Plant Cell. 2019:31(2):384–398. 10.1105/tpc.18.00617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacín M, Legris M, Casal JJ. COP1 re-accumulates in the nucleus under shade. Plant J. 2013:75(4):631–641. 10.1111/tpj.12226 [DOI] [PubMed] [Google Scholar]
- Pacín M, Semmoloni M, Legris M, Finlayson SA, Casal JJ. Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol. 2016:211(3):967–979. 10.1111/nph.13965 [DOI] [PubMed] [Google Scholar]
- Paulisic S, Qin W, Arora Veraszto H, Then C, Alary B, Nogue F, Tsiantis M, Hothorn M, Martinez-Garcia JF. Adjustment of the PIF7-HFR1 transcriptional module activity controls plant shade adaptation. EMBO J. 2021:40(1):e104273. 10.15252/embj.2019104273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, de Wit M. Shade avoidance: phytochrome signalling and other aboveground neighbour detection cues. J Exp Bot. 2014:65(11):2815–2824. 10.1093/jxb/ert389 [DOI] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J. The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 2016:30(13):1529–1541. 10.1101/gad.283234.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Crenshaw CM, Ljung K, Noel JP, Chory J. Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 2014:165(3):1285–1301. 10.1104/pp.114.241844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010:11(3):R25. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, Martínez-García JF. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007:26(22):4756–4767. 10.1038/sj.emboj.7601890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martínez-García JF. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 2006:141(1):85–96. 10.1104/pp.105.076331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Martinez-Garcia JF. Molecular regulation of plant responses to shade. In: Lüttge U, Cánovas FM, Risueño MC, Leuschner C, Pretzsch H, editors. Progress in botany. 84. Cham: Springer; 2022. p. 221–240. [Google Scholar]
- Roig-Villanova I, Martínez-García JF. Plant responses to vegetation proximity: a whole life avoiding shade. Front Plant Sci. 2016:7:236. 10.3389/fpls.2016.00236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Yanovsky MJ, Casal JJ. Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J. 2011:68(5):919–928. 10.1111/j.1365-313X.2011.04745.x [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005:19(23):2811–2815. 10.1101/gad.364005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell. 2013:25(10):3770–3784. 10.1105/tpc.113.117424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimura J, Antoniadi I, Široká J, Tarkowská D, Strnad M, Ljung K, Novák O. Plant hormonomics: multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 2018:177(2):476–489. 10.1104/pp.18.00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. Light quality, photoperception, and plant strategy. Ann Rev Plant Physiol. 1982:33(1):481–518. 10.1146/annurev.pp.33.060182.002405 [DOI] [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martinez-Garcia JF. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 2009:59(2):266–277. 10.1111/j.1365-313X.2009.03866.x [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie D-Y, Doležal K, Schlereth A, Jürgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008:133(1):177–191. 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD. Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci U S A. 2010:107(10):4776–4781. 10.1073/pnas.0910446107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Yamazaki C, Mitsui M, Kakei Y, Mitani Y, Nakamura A, Ishii T, Soeno K, Shimada Y. Transcriptional feedback regulation of YUCCA genes in response to auxin levels in Arabidopsis. Plant Cell Rep. 2015:34(8):1343–1352. 10.1007/s00299-015-1791-z [DOI] [PubMed] [Google Scholar]
- Takato S, Kakei Y, Mitsui M, Ishida Y, Suzuki M, Yamazaki C, Hayashi K-I, Ishii T, Nakamura A, Soeno K, et al. Auxin signaling through SCFTIR1/AFBs mediates feedback regulation of IAA biosynthesis. Biosci Biotechnol Biochem. 2017:81(7):1320–1326. 10.1080/09168451.2017.1313694 [DOI] [PubMed] [Google Scholar]
- Tanaka S-I, Nakamura S, Mochizuki N, Nagatani A. Phytochrome in cotyledons regulates the expression of genes in the hypocotyl through auxin-dependent and -independent pathways. Plant Cell Physiol. 2002:43(10):1171–1181. 10.1093/pcp/pcf133 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer J-L, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008:133(1):164–176. 10.1016/j.cell.2008.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Rodriguez-Concepcion M . Direct regulation of phytoene synthase gene expression carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci USA. 2010:107(25):11626–11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014:10(6):e1004416. 10.1371/journal.pgen.1004416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Paalman R, Keuskamp D, Hayes S, Pierik R. Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell. 2018:30(1):101–116. 10.1105/tpc.17.00771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu Y, Wang H, Ma X, Wang B, Wu G, Wang H. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat Commun. 2017:8(1):348. 10.1038/s41467-017-00404-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xie F, Jiang Y, Li Z, Huang X, Li L. Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev Cell. 2018:44(1):29–41.e4. 10.1016/j.devcel.2017.11.017 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Casal JJ, Whitelam GC. Phytochrome A, phytochrome B and HY4 are invovled in hypocotyl growth responses to natural radiation in Arabidopsis: weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ. 1995:18(7):788–794. 10.1111/j.1365-3040.1995.tb00582.x [DOI] [Google Scholar]
- Zhang X, Huai J, Shang F, Xu G, Tang W, Jing Y, Lin R. A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 2017:174(4):2487–2500. 10.1104/pp.17.00418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li C, Zhou Y, Wang X, Li H, Feng Z, Chen H, Qin G, Jin D, Terzaghi W, et al. TANDEM ZINC-FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell. 2018:30(4):835–852. 10.1105/tpc.17.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Yang C, Jiang Y, Li L. A PIF7-CONSTANS-centered molecular regulatory network underlying shade-accelerated flowering. Mol Plant. 2019:12(12):1587–1597. 10.1016/j.molp.2019.09.007 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Guo Y, Novák O, Chen W, Ljung K, Noel JP, Chory J. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants. 2016:2(4):16025. 10.1038/nplants.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.