Abstract
Background
Primary Sjögren's syndrome (pSS) stands as a chronic autoimmune disease characterized by an elusive pathogenesis. The synergy of single-cell RNA sequencing and Mendelian randomization (MR) analysis provides an opportunity to comprehensively unravel the contributory role of monocytes/macrophages in the intricate pathogenesis of pSS.
Methods
Differentially expressed genes (DEGs) of various types of immune cells were analyzed after annotating single-cell RNA sequencing (scRNA-seq) data. MR analysis of expression quantitative trait loci (eQTL) and protein quantitative trait loci (pQTL) was conducted to search for key pathogenic genes and proteins. Cellular localization of pathogenic genes was performed based on scRNA-seq data. Variations in signaling pathways between immune cells were further analyzed.
Results
A total of 1434 significant DEGs were identified. Among these, 60 genes exhibited strong relevance to the occurrence of pSS, of which 32 genes differentially expressed in monocytes/macrophages. CTSS was found to be a significant risk protein with a p-value of 0.001 and an odds ratio of 1.384 (1.147–1.669), showing pronounced expression in monocytes/macrophages. Furthermore, monocytes/macrophages displayed heightened expression levels of MXD1, AMPD2, TNFSF10, FTL, UBXN11, CSF3R, and LILRA5. The analysis of intercellular signaling revealed increased signal intensity in both incoming and outgoing signals in monocytes/macrophages. The signaling interactions between monocytes/macrophages, B cells, and T cells exhibited varying degrees of deviation.
Conclusions
This study highlights the significant involvement of monocytes/macrophages in the pathogenesis of pSS, as evidenced by MR analysis and scRNA-seq analysis. This suggests monocytes/macrophages as a focal point for pathogenesis research and potential therapeutic targeting in pSS.
Keywords: Primary Sjögren's syndrome, Monocytes and macrophages, Mendelian randomization, Single-cell sequencing, Pathogenesis
1. Introduction
Primary Sjögren's syndrome (pSS) is a chronic autoimmune disorder characterized by the dysfunction and inflammation of exocrine glands, particularly the salivary and lacrimal glands. It is recognized as one of the most prevalent autoimmune rheumatic diseases. Population-based studies reported an annual incidence of pSS ranging from 3.90 to 11.77 per 100,000 inhabitants, with higher rates observed in Asia [1]. While pSS can manifest at any age, it mainly emerges in individuals aged 40 and above, with a prevalence approximately ten times higher in females than males. Remarkably, around 13 % of pSS patients experience potentially severe complications affecting non-glandular organ systems, such as interstitial lung disease, myelitis, thrombocytopenia, and lymphoma, which may contribute to an elevated mortality risk [2].
The therapeutic management pSS has undergone limited substantive changes in recent decades. The availability of disease-modifying treatment options, particularly for glandular manifestations, remains scarce [3]. A prominent contributing factor could be the incomplete understanding of its pathogenesis. The pathological features of pSS can be classified as either germinal-centre-like structures around epithelial tissues of parenchymal organs or deposition of immunocomplexes caused by excessive activation of B cells [4]. Thus, the activation of B cells stands out as a pivotal element in the pathogenesis of pSS. However, pSS should not be perceived as a solely B-cell-driven disorder. An array of compelling evidence indicated the participation of other immune cell types, encompassing monocytes/macrophages, diverse subsets of T cells, natural killer (NK) cells [5,6]. Various cells intricately intertwine to form an immune response network, contributing to the pathogenesis of pSS.
Previous research tended to center around predetermined cell types and lacked a comprehensive outlook on the disruption of immune network balance. In this study, Mendelian randomization (MR) analyses of expression quantitative trait loci (eQTL) and protein quantitative trait loci (pQTL) were employed to elucidate the key pathogenic genes and proteins of pSS, and the expression patterns of these genes across immune cells were delineated utilizing single-cell RNA sequencing (scRNA-seq) data. Integrated with the analyses of alterations in intercellular signals among immune cells, the study explored the possible crucial pathogenic cells in the progression of pSS. Leveraging a novel analytical approach, this research offered a fresh perspective on the pathogenesis of pSS, thereby contributing to the identification of new therapeutic targets.
2. Methods
2.1. Analysis of single-cell RNA sequencing
The scRNA-seq data of peripheral blood mononuclear cells (PBMCs) from 5 patients with pSS and 5 healthy controls (HC) were retrieved from GEO database under accession GSE157278 (https://www.ncbi.nlm.nih.gov/geo/) [7]. Quality control, analysis, and exploration of the scRNA-seq data were performed using the R package “Seurat” (https://github.com/satijalab/seurat) [8]. The specific process was as follows: 1) The red blood cells were first removed. 2) The unqualified cells were screened according to cell features (nFeature_RNA >200, nCount_RNA <30000 and percent.mt < 30). 3) Data normalization was executed using the package "SCTransform", which provided functions for batch correction and data correction [9]. Although a relatively clear separation was observed between pSS patients and HC, there was no significant batch effect within the pSS patients. Similarly, there was no significant separation within the HC group (Additional file 1 Fig. S1). Therefore, the separation between pSS and HC might tend to be due to biological effects. Clusters of cells were determined through a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm via the "FindClusters" function, employing a resolution of 0.6. The identification of differentially expressed genes was conducted using the "FindAllMarkers" function, with a logFC threshold of 0.25 and min pct of 0.25.
2.2. Cell annotation and differentially expressed genes (DEGs) based on cell types
Cell annotation primarily relied upon published literature and the CellMarker database (http://bio-bigdata.hrbmu.edu.cn/CellMarker/). T cells were identified by expression of CD3D and CD3E. NK cells were characterized by the expression of NKG7, GNLY and CD247. The recognition of B cells were based on CD79A, CD79B, CD37. Monocytes and macrophages exhibited expression characteristics of CD14, CD68, S100A12, LYZ. The recognition genes of NKT cells were NKTR. Dendritic cells (DCs) were distinguished by CST3 and CD1C, while plasmacytoid dendritic cells were identified by LILRA4 and JCHAIN. Mast cell were recognized by GATA2, CPA3 [10]. After cell annotation, differentially expressed genes were analyzed for each cell type with the function “FindAllMarkers”. Genes with adjusted P value < 0.05 and an average log2FC > 0.3 were considered significant DEGs and proceeded to the next stage of analysis.
2.3. Mendelian randomization analysis of eQTLs and pQTLs
The eQTLs utilized in the MR analysis were selected based on significant DEGs and were derived from eQTLGen database (https://eqtlgen.org). The pQTLs for the MR analysis were selected based on significant eQTLs and were sourced from deCODE database (https://www.decode.com/) [11]. The genome-wide association (GWAS) data of pSS was acquired from IEU OpenGWAS project (https://gwas.mrcieu.ac.uk/) and the original data came from the FinnGen study [12]. The criteria for the selection of instrumental variables (IVs) consisted of the following: (1)The IV was associated with the risk factor. (2) The IV was not associated with confounders. (3) The IV influenced outcome only through the risk factor.
In cases where only one single nucleotide polymorphism (SNP) was available for constructing the instrumental variable, the result was determined using the Wald ratio. In scenarios involving multiple SNPs, the primary result was obtained from the inverse-variance weighted regression model (IVW). The selection of IV was based on the threshold of p < 5x10-8e and r = 0.001 [13]. Cochran's Q statistic were performed to assess the level of heterogeneity of each SNP. Detection of horizontal pleiotropy in causal relationships inferred from the MR analysis was conducted using the MR-PRESSO test and MR Egger test. A P value < 0.05 for the main result was considered statistically significant. Leave-one-out analysis was executed to exhibit the individual effects of each SNP on the overall results and demonstrate the robustness of the results. R package “TwoSampleMR” (https://mrcieu.github.io/TwoSampleMR/) and “MR-PRESSO” (https://github.com/rondolab/MR-PRESSO) were applied for MR analysis [14,15]. The scatter plot visualized the casual effect of pQTL on pSS risk. The forest plot depicted the impact of each SNP in the pQTL analysis, along with the overall effect.
2.4. Cell-cell interaction analysis
Cell-cell interaction analysis was conducted by R package “Cellchat” (https://github.com/sqjin/CellChat) [16]. The integrated ligand-receptor interaction database, ChatDB, amalgamates information regarding signaling molecule interactions from the KEGG Pathway database as well as experimental investigations.
Deconvolution of the complex intercellular communications in signaling network using centrality measures from network were used to determine major signaling sources networks across datasets.
3. Results
3.1. Analysis and identification of peripheral blood mononuclear cells
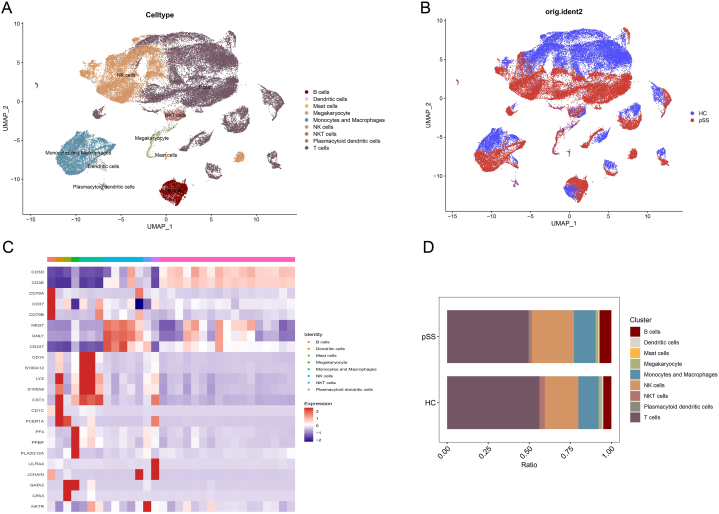
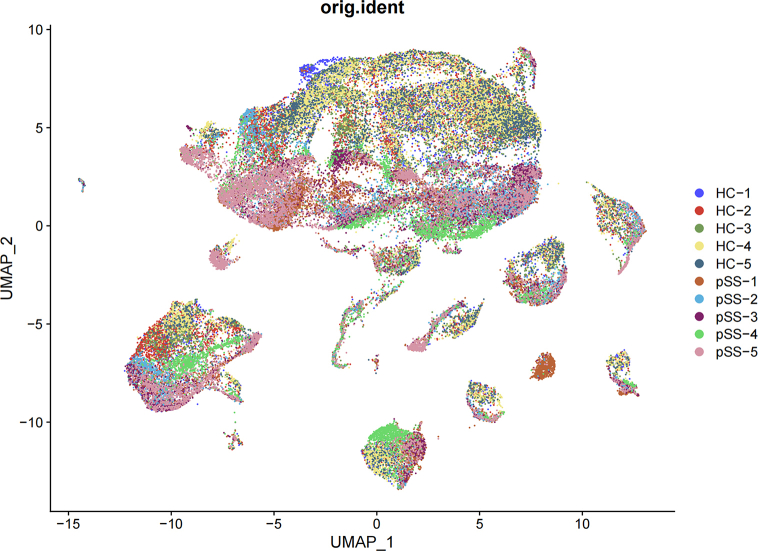
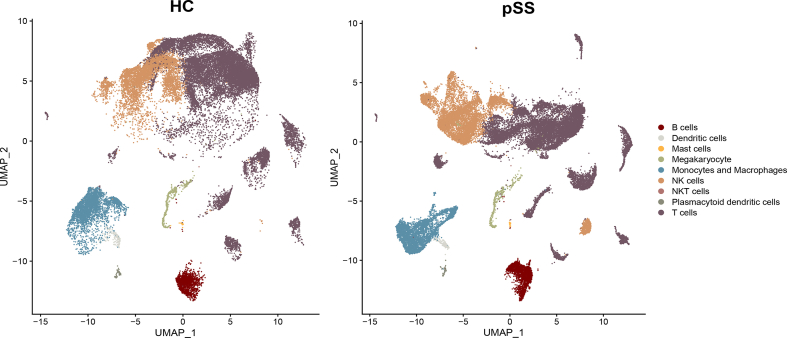
Substandard cells were selectively excluded after quality control, resulting in the analysis of 28289 cells from five healthy controls and 31086 cells from five patients with pSS. A total of 30 clusters were identified, mainly divided into B cells, dendritic cells, mast cells, megakaryocytes, monocytes/macrophages, NK cells, plasmacytoid dendritic cells and T cells. The aggregation of distinct cell types was depicted in Fig. 1A using umap, while Fig. 1B and Additional file 2 Fig. S2 showcased the categorization of these cells based on whether they originated from healthy controls or pSS patients. The expression of characteristic genes (based on the average logFC) in each cluster were performed in Fig. 1C, in which the phenotype of each cell type can be clearly seen. Fig. 1D illustrated the relative proportions of diverse cell types in patients with pSS as compared to HC.
Fig. 1.
Clustering and annotation of single cells (A) Cell type annotation for different cell clusters (B) Cell clustering of pSS patients and HC (C) Expression of characteristic molecules in different cell types (D) Cell ratio of pSS patients and HC.
3.2. Differentially expressed genes (DEGs) based on cell types
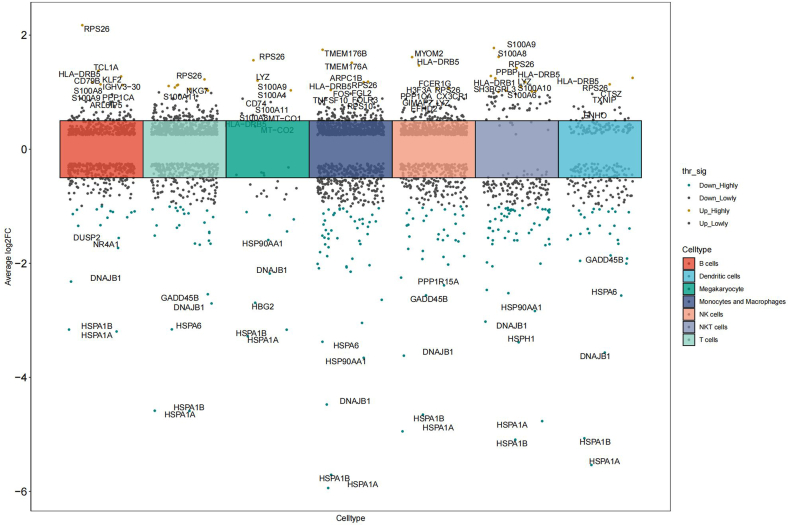
Genes with adjusted P value < 0.05 and average log2FC > 0.3 were designated as significant DEGs. A total of 785 significant DEGs were ascertained in monocytes/macrophages, compared to 473 in B cells, 254 in dendritic cells, 34 in megakaryocytes, 577 in NK cells, 231 in NKT cells, and 610 in T cells. Among the 785 genes exhibiting significant differential expression in monocytes/macrophages, 440 were up-regulated and 345 were down-regulated. More detailed information about the significant DEGs of each cell type could be found in the Additional file 3 Table S1. Mast cells and plasmacytoid dendritic cells were precluded in this analysis due to the insufficient cell count. Fig. 2 visualized the significant DEGs across each cell type, accompanied by labels for the top five up-regulated and down-regulated genes.
Fig. 2.
Differentially expressed genes in different immune cells.
3.3. Mendelian randomization analysis of eQTLs
There were 1434 significant DEGs after the elimination of duplicates, of which 1318 eQTLs without repetitions were matched. These eQTLs were taken as exposure, with pSS serving as the outcome, and subsequent MR analysis was performed separately on each eQTL. Wald ratio was adopted as main result for single SNP, IVW was adopted as the main result. The main result P value < 0.05 was considered significant. These outcomes underwent further scrutiny to assess the fulfillment of the three hypotheses. Two eQTLs associated with genes NELL2 and KLRC3 were subjected to recalibration, involving the removal of IVs that did not meet the criteria of hypothesis three. After verification by MR-PRESSO and MR-EGGER, it is found that eQTL related to CSF3R performed significant heterogeneity. Therefore, the main source of heterogeneity, rs3917922, was removed. The re-calculated outcome remained statistically significant, while heterogeneity was successfully mitigated. A total of 60 genes were ultimately identified to be significant related to the occurrence of pSS. Among these genes, 25 exhibited odds ratios below 1, indicating a protective effect, while 35 displayed odds ratios above 1, signifying risk factors. Of these 60 genes, 32 were significantly differentially expressed in monocytes/macrophages, suggesting the profound involvement of monocytes/macrophages in the pathogenesis of pSS.
However, whether genes were risk or protective does not precisely coincide with their up-regulated or down-regulated states. On one hand, of the 35 risk genes, 23 exhibited up-regulation, while the remaining 12 displayed down-regulation. On the other hand, of the 25 protective genes, 14 were down-regulated and 11 up-regulated. This outcome highlights that while up-regulated risk-associated genes and down-regulated protective genes constitute the predominant trend, instances of reverse patterns also emerge, thus emphasizing the intricate and multifaceted roles played by these genes in the pathogenesis of pSS.
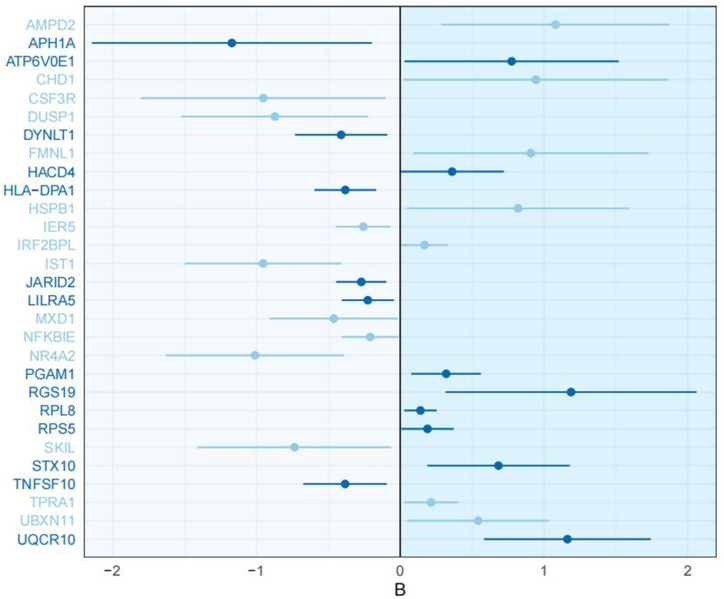
Among the 35 risk genes, 10 were up-regulated in monocytes/macrophages, while 8 were down-regulated. Simultaneously, among the 25 protective genes, 9 were down-regulated in monocytes/macrophages, while 5 were up-regulated. This indicated that the involvement of monocytes/macrophages in the pathogenesis of pSS was also complicated. Detailed information of MR analysis were performed in Table 1. Fig. 3 illustrated the causal effect of pathogenic genes in monocytes/macrophages.
Table 1.
Mendelian randomization analysis of significant eQTLs.
| gene | method | b | se | OR | P-value | MR-PRESSO | Cochran's Q | MR-Egger |
|---|---|---|---|---|---|---|---|---|
| JARID2 | IVW | −0.271 | 0.089 | 0.763 | 0.002 | 0.606 | 0.406 | 0.609 |
| IL32 | IVW | 0.203 | 0.084 | 1.225 | 0.015 | 0.520 | 0.318 | 0.566 |
| SEC62 | Wald ratio | −0.547 | 0.235 | 0.579 | 0.020 | / | / | / |
| MXD1 | IVW | −0.462 | 0.227 | 0.630 | 0.042 | / | 0.673 | 0.855 |
| MEF2C | IVW | 0.213 | 0.096 | 1.237 | 0.027 | 0.588 | 0.553 | 0.392 |
| RPS5 | Wald ratio | 0.190 | 0.093 | 1.209 | 0.040 | / | / | / |
| FTL | Wald ratio | 0.894 | 0.452 | 2.444 | 0.048 | / | / | / |
| DYNLL1 | Wald ratio | 1.129 | 0.488 | 3.092 | 0.021 | / | / | / |
| DHRS7 | IVW | 0.254 | 0.086 | 1.289 | 0.003 | 0.646 | 0.395 | 0.672 |
| CHMP4B | IVW | −0.162 | 0.077 | 0.850 | 0.036 | 0.893 | 0.833 | 0.677 |
| UBE2I | IVW | −0.264 | 0.098 | 0.768 | 0.007 | / | 0.655 | / |
| STX10 | IVW | 0.683 | 0.252 | 1.981 | 0.007 | 0.245 | 0.114 | 0.859 |
| TBCB | IVW | −0.231 | 0.099 | 0.794 | 0.020 | / | 0.817 | / |
| HSPB1 | Wald ratio | 0.820 | 0.395 | 2.270 | 0.038 | / | / | / |
| PTGDS | IVW | 0.310 | 0.130 | 1.363 | 0.017 | / | 0.529 | 0.510 |
| BACH2 | IVW | −0.453 | 0.160 | 0.636 | 0.005 | 0.758 | 0.619 | 0.918 |
| ATP6V0E1 | Wald ratio | 0.775 | 0.380 | 2.171 | 0.041 | / | / | / |
| AMPD2 | Wald ratio | 1.081 | 0.405 | 2.946 | 0.008 | / | / | / |
| UAP1 | Wald ratio | 0.333 | 0.163 | 1.395 | 0.041 | / | / | / |
| APH1A | Wald ratio | −1.171 | 0.497 | 0.310 | 0.018 | / | / | / |
| CSF3R | IVW | −0.952 | 0.434 | 0.386 | 0.022 | 0.051 | 0.060 | 0.140 |
| IRF2BPL | IVW | 0.169 | 0.084 | 1.184 | 0.043 | 0.501 | 0.591 | 0.311 |
| DUSP1 | Wald ratio | −0.872 | 0.331 | 0.418 | 0.009 | / | / | / |
| TNFSF10 | IVW | −0.383 | 0.148 | 0.682 | 0.010 | 0.523 | 0.308 | 0.878 |
| ZBTB1 | Wald ratio | 1.721 | 0.513 | 5.592 | 0.001 | / | / | / |
| SDF2L1 | IVW | 0.677 | 0.265 | 1.967 | 0.011 | / | 0.632 | / |
| SKIL | IVW | −0.735 | 0.343 | 0.479 | 0.032 | / | 0.672 | / |
| RPS3A | IVW | 0.568 | 0.269 | 1.765 | 0.035 | / | 0.756 | / |
| NFKBIE | IVW | −0.208 | 0.101 | 0.812 | 0.039 | 0.534 | 0.560 | 0.291 |
| DYNLT1 | IVW | −0.411 | 0.164 | 0.663 | 0.012 | / | 0.841 | / |
| NR4A2 | Wald ratio | −1.011 | 0.316 | 0.364 | 0.001 | / | / | / |
| CHD1 | Wald ratio | 0.944 | 0.470 | 2.570 | 0.045 | / | / | / |
| AZIN1 | IVW | 0.196 | 0.094 | 1.216 | 0.037 | 0.820 | 0.947 | 0.615 |
| UBXN11 | IVW | 0.542 | 0.250 | 1.719 | 0.030 | / | 0.451 | / |
| SPON2 | IVW | 0.303 | 0.112 | 1.354 | 0.007 | 0.516 | 0.534 | 0.699 |
| G6PD | Wald ratio | 1.314 | 0.492 | 3.720 | 0.008 | / | / | / |
| ITGB2 | IVW | 0.153 | 0.064 | 1.166 | 0.016 | 0.703 | 0.774 | 0.785 |
| RPL8 | IVW | 0.141 | 0.058 | 1.151 | 0.014 | / | 0.343 | / |
| CYB561A3 | Wald ratio | 1.019 | 0.443 | 2.769 | 0.021 | / | / | / |
| IER5 | IVW | −0.257 | 0.097 | 0.774 | 0.008 | / | 0.643 | 0.779 |
| CTSS | IVW | 0.203 | 0.074 | 1.225 | 0.006 | 0.798 | 0.775 | 0.270 |
| KRTCAP2 | Wald ratio | −1.211 | 0.412 | 0.298 | 0.003 | / | / | / |
| TPRA1 | Wald ratio | 0.214 | 0.095 | 1.239 | 0.025 | / | / | / |
| UQCRQ | Wald ratio | 0.882 | 0.445 | 2.416 | 0.047 | / | / | / |
| CITED2 | IVW | −0.267 | 0.129 | 0.766 | 0.039 | / | 0.715 | 0.612 |
| S1PR1 | Wald ratio | −0.847 | 0.286 | 0.429 | 0.003 | / | / | / |
| PGAM1 | IVW | 0.320 | 0.123 | 1.376 | 0.010 | / | / | / |
| RGS19 | Wald ratio | 1.188 | 0.445 | 3.281 | 0.008 | / | / | / |
| NOP10 | IVW | 0.137 | 0.069 | 1.147 | 0.047 | 0.402 | 0.427 | 0.865 |
| IST1 | Wald ratio | −0.954 | 0.278 | 0.385 | 0.001 | / | / | / |
| UQCR10 | Wald ratio | 1.163 | 0.296 | 3.201 | 0.000 | / | / | / |
| NELL2 | IVW | −0.178 | 0.090 | 0.837 | 0.011 | 0.648 | 0.841 | 0.109 |
| FMNL1 | Wald ratio | 0.908 | 0.417 | 2.480 | 0.029 | / | / | / |
| LILRA5 | IVW | −0.225 | 0.092 | 0.798 | 0.014 | 0.677 | 0.322 | 0.590 |
| SEMA4D | IVW | −0.205 | 0.080 | 0.814 | 0.010 | 0.752 | 0.704 | 0.512 |
| HACD4 | IVW | 0.361 | 0.184 | 1.434 | 0.050 | 0.331 | 0.135 | 0.186 |
| HLA-C | IVW | 0.337 | 0.114 | 1.401 | 0.021 | 0.171 | 0.002 | 0.343 |
| KLRC3 | IVW | 0.154 | 0.062 | 1.167 | 0.013 | 0.961 | 0.967 | 0.923 |
| HLA-DPA1 | IVW | −0.382 | 0.110 | 0.683 | 0.000 | 0.433 | 0.086 | 0.922 |
| HLA-DQA1 | Wald ratio | −0.532 | 0.096 | 0.587 | 0.000 | / | / | / |
Fig. 3.
The causal effect of genes significantly related to the occurrence of pSS in monocytes/macrophages (dark blue: up-regulated genes in monocytes/macrophages, light blue: genes down-regulated in monocytes/macrophages).
3.4. Mendelian randomization analysis of pQTLs
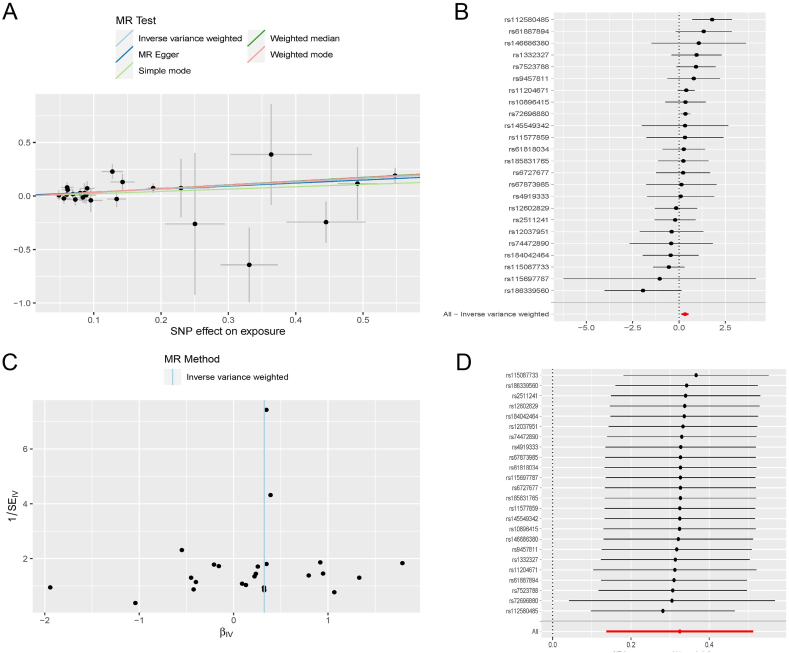
Sixty genes related to the occurrence of pSS were previously been identified. The pQTLs related to these genes were further validated by MR analysis to explore whether the proteins expressed by these genes were key pathogenic proteins. A total of 31 pQTLS were matched in the DECODE database. Among these proteins, CTSS emerged as a significant risk factor for the development of pSS, with a P-value of 0.001 and an OR of 1.384(1.147–1.669) according to the result of IVW. More detailed information, along with outcomes from four other methodologies, was presented in Table 2. Twenty-four SNPs that fit the hypothesis were selected as IV during the analysis, all possessing F-values exceeding 10. Detailed information were performed in the Additional file 3 Table S2. The P-value of MR-PRESSO was 0.48 and the P-value of MR-egger intercept test was 0.70, indicating no heterogeneity. The P-value of Cochran's Q test was 0.37, indicating no pleiotropy. Fig. 4A performed the casual effect of CTSS on pSS risk. Fig. 4B showed the effect of each SNP and also the total effect.Through the leave-one-out analysis, it was ascertained that no single SNP exerted undue influence on the overall outcome, thus confirming the robustness of the result. The funnel plot manifested that the two sides were basically symmetrical, indicating a lack of significant bias on either side.
Table 2.
Mendelian randomization analysis of CTSS.
| Method | b | se | pval | or |
|---|---|---|---|---|
| MR Egger | 0.283 | 0.145 | 0.064 | 1.327(0.999–1.764) |
| Weighted median | 0.347 | 0.121 | 0.004a | 1.415(1.116–1.796) |
| Inverse variance weighted | 0.325 | 0.096 | 0.001a | 1.384(1.147–1.669) |
| Simple mode | 0.215 | 0.273 | 0.438 | 1.240(1.147–1.669) |
| Weighted mode | 0.341 | 0.115 | 0.007a | 1.407(1.123–1.763) |
P < 0.05.
Fig. 4.
Mendelian randomization analysis of CTSS (A) Scatterplot of the effect sizes (B) Forrest plot of the effect size of each SNP (C) Funnel plot (D) Leave-one-out analysis.
3.5. The expression of key genes in immune cells
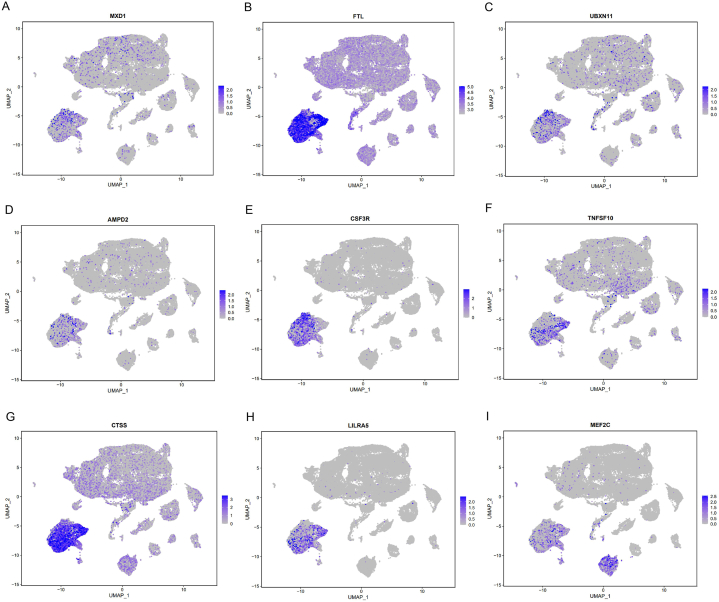
The expression of 60 key genes related to the occurrence of pSS was shown in Fig. 5 and Additional file 4 Fig. S3. MXD1, AMPD2, TNFSF10, FTL, UBXN11, CTSS, CSF3R and LILRA5 exhibited heightened expression levels within monocytes/macrophages compared to other cell types (Fig. 5A–H). Notably, the expression of CTSS, CSF3R and LILRA5 in monocytes/macrophages was relatively specific, with CTSS having been previously identified as a key pathogenic protein in the earlier analysis. Moreover, MEF2C displayed elevated expression in both monocytes/macrophages and B cells (Fig. 5I). The distinct expression pattern of these pivotal genes within monocytes/macrophages further accentuated the potential pivotal role of monocytes/macrophages in the complicated pathogenesis of pSS.
Fig. 5.
The expression of key genes in different cell types (A) MXD1 (B) FTL (C) UBXN11 (D) AMPD2 (E) CSF3R (F) TNFSF10 (G) CTSS (H) LILRA5 (I) MEF2C.
3.6. Overall analysis of intercellular signaling
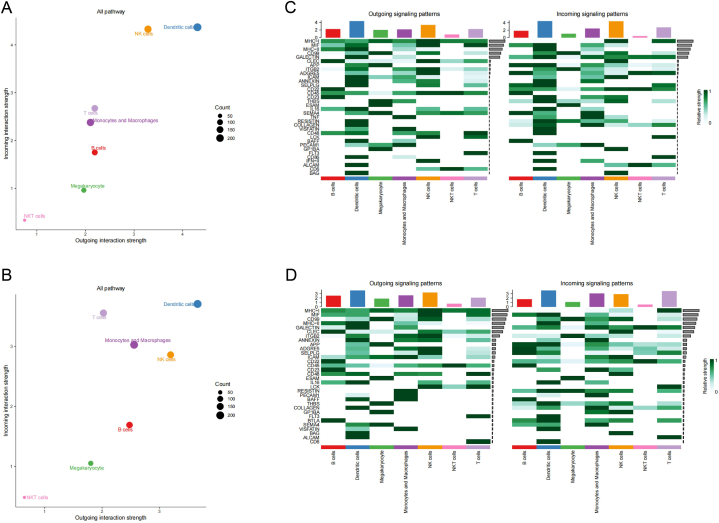
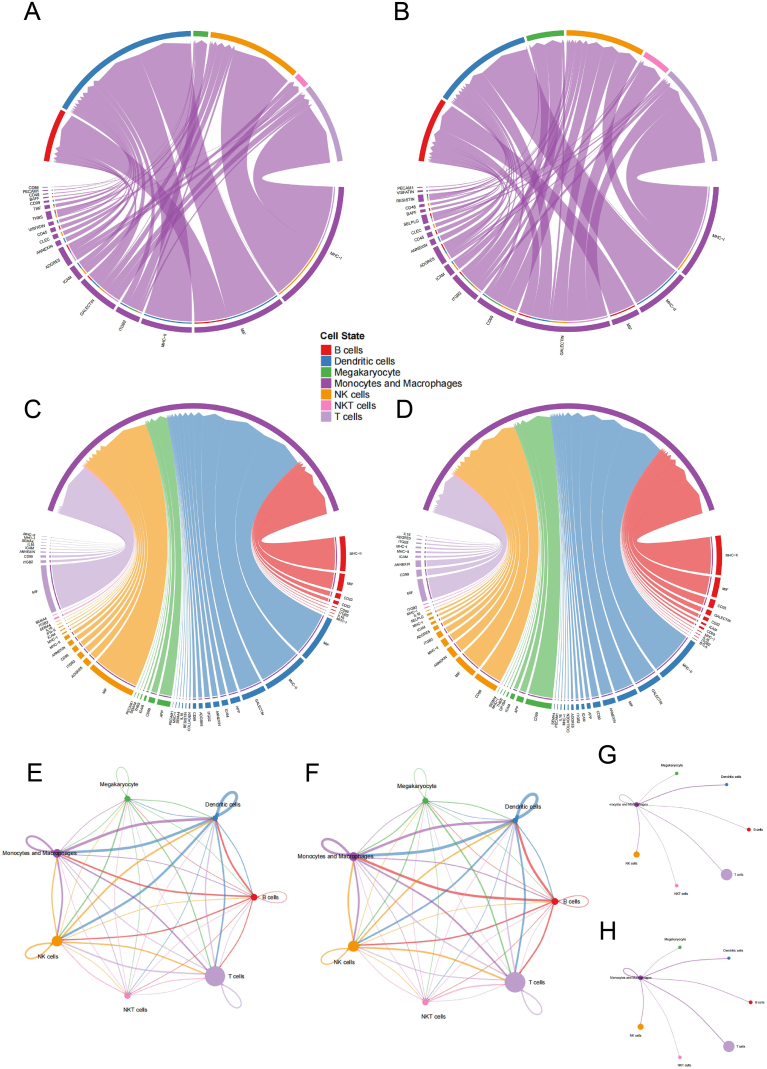
The total incoming and outgoing signal strengths for each cell type were presented in Fig. 6A and B. The findings revealed noticeable signal alterations in monocytes/macrophages, T cells, B cells, and NK cells. Among them, monocytes/macrophages exhibited apparent enhancement in both incoming signal and outgoing signals. T cells displayed enhanced incoming signal intensity, while B cells demonstrated an increased outgoing signal. In contrast, NK cells exhibited a decrease in incoming signal intensity alongside an elevated outgoing signal.
Fig. 6.
Global intercellular signaling between HC and pSS patients (A) Overall incoming and outgoing signal strength in HC (B) Strength of intercellular signaling pathway in HC (C) Overall incoming and outgoing signal strength in pSS patients (D) Strength of intercellular signaling pathway in pSS patients.
Fig. 6C and D depicted the major intercellular signal pathways in HC and pSS patients. The relative ranking of “CD99”, "ADGRE5″, "CD48″, “LCK”, “PCAM1” and “BAFF” in the signaling pathway increased, while the ranking of “MHC-II”, “APP”, “ICAM”, “THBS”, “SEMA4”, “COLLAGEN” and “VISFATIN” decreased. "TNF," "CD86," and "IFN-II", observed in HC, were absent in pSS patients due to inadequate signal strength, while "BLTA" was exclusively observed in pSS patients.
3.7. Monocytes/macrophages-associated intercellular signaling pathways
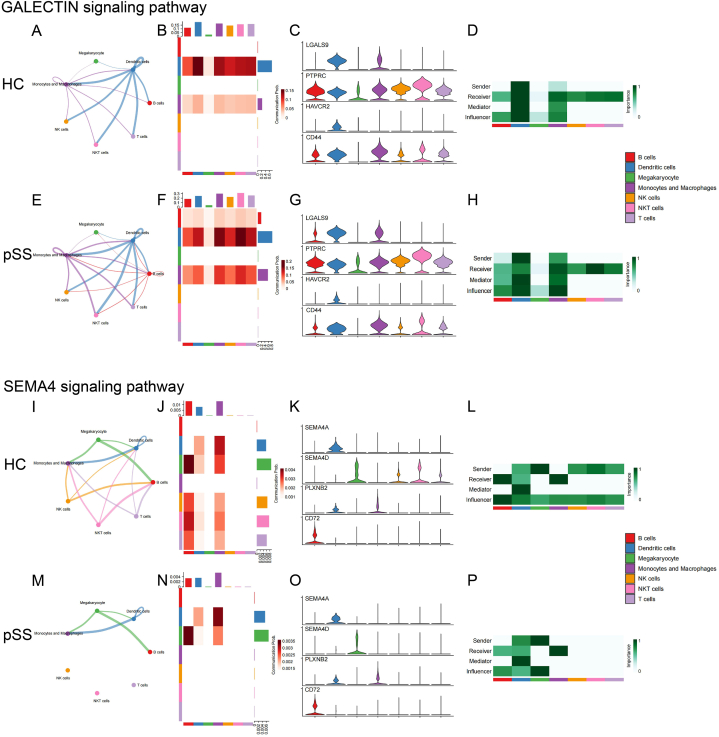
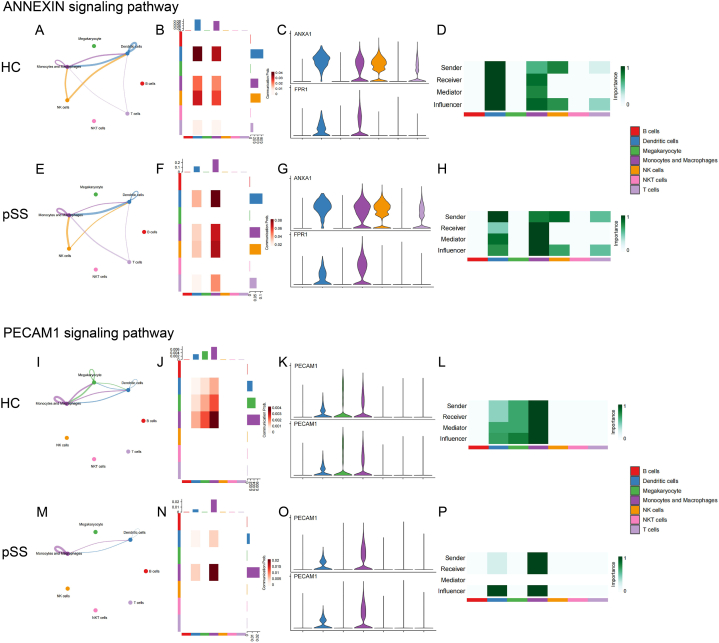
The main outgoing signal pathways of monocytes/macrophages were “MHC-I”, “ADGRE5”, “ICAM”, “RESISTIN”, “BAFF” and “VISFATIN”, while the main incoming signal pathways of monocytes/macrophages were “GALECTIN”, “COLLAGEN” and “SEMA4”. Meanwhile, “ANNEXIN” and “PECAM1” exhibited both strong signal inputs and outputs. Fig. 7 displayed the monocytes/macrophages-associated intercellular signaling pathways between HC and pSS patients.
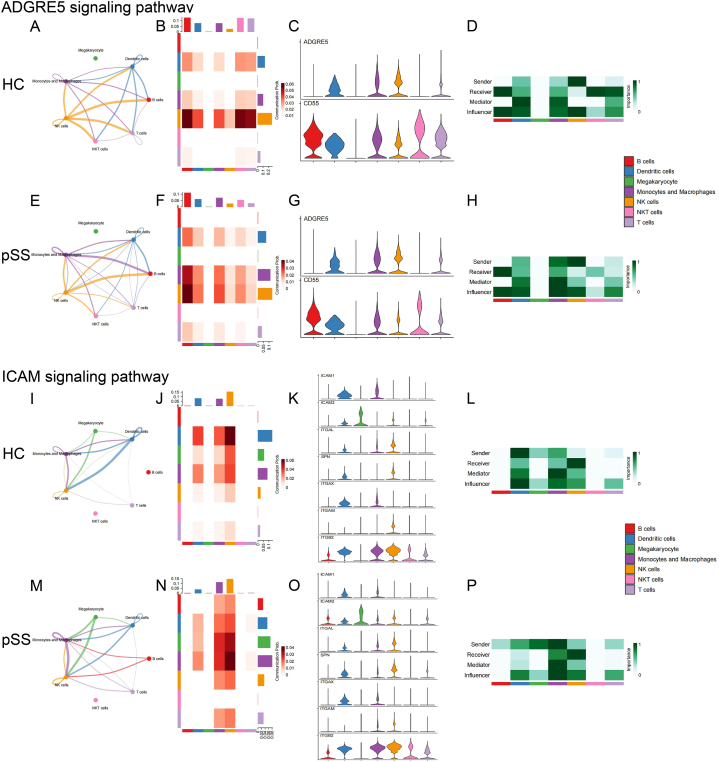
Figure 8-1.
The intercellular signal strength, the intensity of the signals sent and received by each cell type, the expression of receptors and ligands on the cell surface, and the role of each cell type in the signaling network of the main outgoing signal pathways in monocytes/macrophages between HC and pSS patients (A)-(H) ADGRE5 signaling pathway (I)-(P) ICAM signaling pathway.
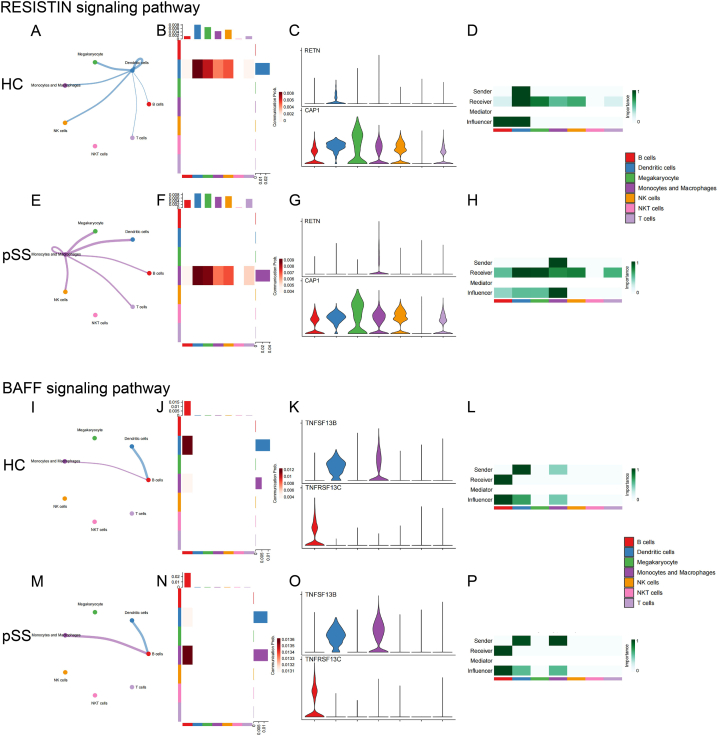
Figure 8-2.
The intercellular signal strength, the intensity of the signals sent and received by each cell type, the expression of receptors and ligands on the cell surface, and the role of each cell type in the signaling network of the main outgoing signal pathways in monocytes/macrophages between HC and pSS patients (A)-(H) RESISTIN signaling pathway (I)-(P) BAFF signaling pathway.
Fig. 9.
The intercellular signal strength, the intensity of the signals sent and received by each cell type, the expression of receptors and ligands on the cell surface, and the role of each cell type in the signaling network of the main incoming signal pathways in monocytes/macrophages between HC and pSS patients (A)–(H) GALECTIN signaling pathway (I)–(P) SEMA4 signaling pathway.
Fig. 10.
The intercellular signal strength, the intensity of the signals sent and received by each cell type, the expression of receptors and ligands on the cell surface, and the role of each cell type in the signaling network of the signaling pathways with high incoming and outgoing signals in monocytes/macrophages between HC and pSS patients (A)–(H) ANNEXIN signaling pathway (I)–(P) PECAM1 signaling pathway.
Fig. 7.
Monocytes/macrophages-associated intercellular signaling pathways (A) Pathways send from monocytes/macrophages in HC (B) Pathways send from monocytes/macrophages in pSS patients (C) Pathways send to monocytes/macrophages in HC (D) Pathways send to monocytes/macrophages in pSS patients (E) Signal interaction between immune cells in HC (F) Signal interaction between immune cells in patients with pSS (G) Signal intensity that monocytes/macrophages send to other immune cells in HC (H) Signal intensity that monocytes/macrophages send to other immune cells in patients with pSS.
3.8. Changes in monocytes/macrophages-related intercellular signaling pathways
This section provided a detailed analysis of pathways that have undergone noteworthy and significant alterations. According to the signal pattern, these pathways could be divided into three types. The first were the pathways that mainly displayed the outgoing signals, including “ADGRE5”, “ICAM”, “RESISTIN” and “BAFF”. The second were the pathways characterized by incoming signals, such as “GALECTIN” and “SEMA4”. The last were the pathways with both high outgoing and high incoming signals, such as “ANNEXIN” and “PECAM1”.
The results showed enhanced communication between monocytes/macrophages and other immune cells in pSS patients, including increased intercellular interaction and enhanced signaling strength. In pSS patients, monocytes/macrophages sent more signals to T cells (“RESISTIN” and “GALECTIN”), and received more signals from T cells (“ADGRE5”, “ICAM” and “ANNEXIN”). Similarly, signals sent to B cells (“ADGRE5”, “RESISTIN”, “BAFF” and “GALECTIN”) and received from B cells (“ICAM”) were both enhanced. Likewise, the intercellular interaction between monocytes/macrophages and NK cells also increased (“ADGRE5”, “RESISTIN”, “GALECTIN” and “ANNEXIN”). In addition, several pathways showed enhanced signaling to monocytes/macrophages themselves (“ADGRE5”, “RESISTIN”, “GALECTIN”, “ANNEXIN” and “PECAM1”).
Table 3 summarized the specific changes of these signaling pathways in three aspects: intercellular interaction, signal strength, and the roles in the signal networks. Descriptions such as “New” or “Absent” or “Heightened” or “Attenuated” in the table were based on the comparison of pSS patients with healthy controls. Table 3 also provided links to figures that depicted the changes for each signaling pathway.
Table 3.
Changes of monocytes/macrophages-related intercellular signaling pathways.
| Pathways | Intercellular interaction | Signal strength | Roles in signal networks | Figure | |
|---|---|---|---|---|---|
| High Outgoing Signaling | |||||
| ADGRE5 | New: Mono/Mac signaling to NK cells and receiving signals from T cells |
Heightened: Outgoing signaling toward B cells; Self-signaling |
Intensified: Senders Influencers |
Figs. 8–1 A-H | |
| ICAM | New: Mono/Mac signaling to T cells and receiving signals from B cells |
Heightened: Incoming signaling from B cells and T cells |
Intensified: Senders Receivers Influencers |
Figs. 8–1 I-P | |
| RESISTIN | New: Mono/Mac signaling to DCs, B cells, T cells and NK cells; Mono/Mac self-signaling Absent: Mono/Mac receiving signals from DCs |
Heightened: Outgoing signaling toward DCs, B cells, T cells and NK cells; Self-signaling |
Intensified: Senders Receivers Influencers |
Figs. 8–2 A-H | |
| BAFF | / | Heightened: Outgoing signaling toward B cells |
Intensified: Senders |
Figs. 8–2 I-P | |
| High Incoming Signaling | |||||
| GALECTIN | / | Heightened: Outgoing signaling toward DCs, B cells, T cells, NK cells and NKT cells; Self-signaling |
Intensified: Senders Receivers |
Fig. 9A–H | |
| SEMA4 | Absent: Mono/Mac receiving signals from T cells, NK cells and NKT cells |
Attenuated: Incoming signaling from T cells, NK cells and NKT cells |
Weakened: Senders |
Fig. 9I–P | |
| High incoming and outgoing signals | |||||
| ANNEXIN | / | Heightened: Incoming signaling from DCs, T cells and NK cells; Self-signaling |
Intensified: Senders Receivers Mediators Influencers |
Fig. 10A–H | |
| PECAM1 | / | Heightened: Self-signaling |
Weakened: Mediators | Fig. 10I–P | |
4. Discussion
This study provided a comprehensive perspective that highlighted the critical role of monocytes/macrophages in the pathogenesis of pSS, a facet previously not well-addressed. The results furnished a twofold substantiation of this notion, encompassing both the expression of pathogenic genes and proteins within monocytes/macrophages, as well as the signal fluctuations of monocytes/macrophages within the immune cell network.
Macrophages display remarkable vigor in the realm of innate immunity while they also contribute to the initiation of adaptive immunity by recruiting various immune cells, including lymphocytes. Macrophages has been classified as M1 macrophages, which promote inflammation or as alternatively M2 macrophages, which mitigate inflammation and facilitate tissue repair [17].
The pathological progression of pSS may, in part, be attributed to tissue-resident macrophages. Analyses revealed a conspicuous abundance of M1 macrophage infiltration in pSS patients compared to normal human salivary gland tissue [18]. A more specific analysis showed that the presence of macrophages in the labial salivary glands in pSS patients was positively associated with biopsy focus score, parenchymal-organ involvement, lymphoma and cryoglobulinemia [19]. The process of salivary glands damage can be partially explained by the secretion of IL-18, nitric oxide and metalloproteases by macrophages [[20], [21], [22]]. Beyond the salivary glands, macrophages can exert similar effects on other epithelial tissues such as the ocular epithelium [23]. In addition, several studies have suggested that both tissue-resident and circulating macrophages are involved in the development of lymphoma in pSS patients [19,20,24], which also indicating an essential interplay between macrophages and lymphocytes in pSS.
CTSS, a cysteine endopeptidase belonging to the papain family, is notably abundant within lysosomes. CTSS is highly expressed primarily in antigen-presenting cells (APCs) and is involved in major histocompatibility complex class II (MHC-II) mediated immune responses, implying that inhibition of CTSS could be a promising approach in attenuating autoantigen-specific CD4+T cells responses [25]. Overexpression of CTSS in lacrimal gland was considered to be associated with the ocular manifestations of pSS and therapeutic interventions targeting CTSS have demonstrated efficacy in non-clinical studies of pSS [[26], [27], [28]]. Our findings indicated the significance of CTSS as a key risk gene in pSS, while the relatively specific expression of CTSS in monocytes/macrophages suggested that monocytes/macrophages portrayed exceptional significance in the MHC-II mediated immune response in pSS.
The results of intercellular signals suggested an intensification of interactions between monocytes/macrophages and other immune cells, especially B cells and T cells, in pSS patients when compared to HC.
The obvious changes in the communication between monocytes/macrophages and B cells in pSS patients are noteworthy, given the widely acknowledged role of B cells as effector cells in pSS pathogenesis [29]. The BAFF signaling pathway, chiefly composed of BAFF and BAFFR, stands as a pivotal signal governing the survival and maturation of B cells [30]. Elevated BAFF levels have been reported in pSS patients, correlating with heightened lymphocytic infiltration that may partly contribute to ectopic germinal center formation [31]. The encouraging outcomes from BAFF receptor blockade treatment trials hold promise as a potential breakthrough in the current therapeutic landscape, albeit requiring further clinical data for validation [32]. The intensified BAFF signaling pathway between monocytes/macrophages and B cells observed in the study could imply an increased capacity of monocytes/macrophages to stimulate B cells. Contrastingly, the enhanced GALECTIN signaling pathway suggested an inhibition of B cell triggered by monocytes/macrophages, as galectin-9, binding with CD45, suppressed calcium signaling via a Lyn-CD22-SHP-1 dependent mechanism, ultimately dampening B cell activation [33]. Another notable observation was that B cells expressed more ICAM2 and monocytes/macrophages expressed more ITGAM in pSS patients, culminating in the reinforcement of ICAM2-(ITGAM + ITGB2) and ICAM2-(ITGAL + ITGB2) pathways, thereby facilitating cell adhesion [34,35]. The observed increased ADGRE5-CD55 signaling also supported enhanced adhesion to supporting cells [36].
Similarly, enhanced cell adhesion was observed between monocytes/macrophages and T cells in pSS patients, mainly evident through increased ADGRE-CD55 and ICAM signaling. In addition, a noteworthy alteration was the reduction in SEMA4D expression, not only observed in T cells but also in NK cells. It functioned in T cell priming [37] and was able to drive CD8+ T-Cell lesional trafficking in oral lichen planus via CXCL9/CXCL10 up-regulations [38].
Another crucial aspect is the self-regulation of monocytes/macrophages. While RETN was primarily expressed on DCs in HC, scRNA-seq data revealed a remarkably shift, with demonstrating monocytes/macrophages as the main source of RETN expression in pSS patients. Meanwhile, the up-regulation of resistin was observed during monocyte/macrophage differentiation, shedding light on resistin's role in monocyte/macrophage function [39,40]. Monocytes/macrophages also exhibited a significant enhancement of cell-adhesion-related pathways, including not only the previously discussed ADGRE5 and ICAM signaling pathways, but also ANXA1-FPR1 signaling, given that FPR1 participated in a signal relay which regulated the infiltration of phagocytes to inflammatory sites [41]. Meanwhile, enhanced PECAM1 signaling could also promote macrophage migration, as it was reported to be essential for leukocyte transendothelial migration under most inflammatory conditions [42]. However, PECAM1 could act as a negative feedback regulator of inflammation, as evidenced by its ability to inhibit LPS-induced inflammation through TLR4 [43].
The complicated interactions among immune cells, involving a myriad of pathways, presented a holistic tableau in which B cells and T cells might have been stimulated by monocytes/macrophages, and the adhesion and aggregation might have been enhanced between them. The alterations in the signaling network of monocytes/macrophages suggested that they were not only direct effectors but also adept communicators among immune cells, potentially fueling the enduring state of chronic inflammation observed in pSS.
The main limitation of this study stems from the absence of experimental evidence, partly attributed to the study's overarching focus on the overall immune cell landscape, while subsequent experiments that delve deeper into specific molecular components would be beneficial. Another limitation is the number of participants qualified for the study, which is due to the impracticality of large-scale application of scRNA sequencing. In addition, problems such as horizontal pleiotropy and weak IVs in MR analysis may lead to unrobust results. Although we used some methods to eliminate unqualified IVs, such as MR-PRESSO test and F-value, to reduce the risk when performing MR Analysis, there is still a risk of potential bias. Batch effects are another issue that deserves attention. Despite efforts to reduce the impact of the batch effect, latent risks may also exist.
In summary, the study underscored the pivotal contribution of monocytes/macrophages to pSS pathogenesis, substantiated by both Mendelian randomization analysis and comprehensive cell-to-cell interaction assessment. However, the absence of direct evidence definitively establishing macrophage function in pSS pathogenesis remains a non-negligible gap. In light of this, monocytes/macrophages are anticipated to be a focal point for pathogenesis research and potential therapeutic targeting in pSS.
CRediT authorship contribution statement
Yimei Ding: Writing – original draft, Methodology, Formal analysis, Data curation. Xue Luan: Writing – review & editing, Supervision. Jiaqi Hou: Writing – original draft, Validation, Funding acquisition.
Data availability statement
The scRNA-seq data used in this study can be obtained from GEO database under the accession number GSE157278. The GWAS data used in this study can be obtained from the openGWAS database with the GWAS ID finn-b-M13_SJOGREN.
Ethics approval
Review and/or approval by an ethics committee was not needed for this study because all data in this study were derived from public databases. Appropriate ethics approval has been provided at the publication of original articles. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (Award Number: 82104719).
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Jiaqi Hou reports financial support was provided by National Natural Science Foundation of China. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We want to acknowledge the participants and investigators of the FinnGen and deCODE study. We also appreciate the data shared by Xiaoping Hong et al.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e39130.
Abbreviations
- pSS
primary Sjögren's syndrome
- MR
Mendelian randomization
- DEG
differentially expressed gene
- eQTL:
expression quantitative trait loci
- pQTL:
protein quantitative trait loci
- NK
natural killer
- scRNA-seq
single-cell RNA sequencing
- PBMC
peripheral blood mononuclear cell
- HC
healthy controls
- DC
dendritic cell
- GWAS
genome-wide association
- IV
instrumental variable
- IVW
inverse-variance weighted
- SNP
nucleotide polymorphism
- OR
odds ratio
- APC
antigen-presenting cell
- MHC-II
histocompatibility complex class II
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Additional file 3: Table S1. Detailed information about the significant DEGs of each cell type. Table S2. Detailed information about the instrumental variables of CTSS.
Additional file 1: Fig. S1. Cell clustering of each sample.
Additional file 2: Fig. S2. Cell clustering of pSS and HC.
Additional file 4: Fig. S3. The expression of 51 key genes related to the occurrence of pSS in immune cells.
References
- 1.Qin B., Wang J., Yang Z., et al. Epidemiology of primary Sjögren's syndrome: a systematic review and meta-analysis. Ann. Rheum. Dis. 2015;74(11):1983–1989. doi: 10.1136/annrheumdis-2014-205375. [DOI] [PubMed] [Google Scholar]
- 2.Flores-Chávez A., Kostov B., Solans R., et al. Severe, life-threatening phenotype of primary Sjögren's syndrome: clinical characterisation and outcomes in 1580 patients (GEAS-SS Registry) Clin. Exp. Rheumatol. 2018;36(3):121–129. Suppl 112. [PubMed] [Google Scholar]
- 3.Ramos-Casals M., Brito-Zerón P., Bombardieri S., et al. EULAR recommendations for the management of Sjögren's syndrome with topical and systemic therapies. Ann. Rheum. Dis. 2020;79(1):3–18. doi: 10.1136/annrheumdis-2019-216114. [DOI] [PubMed] [Google Scholar]
- 4.Mavragani C.P. Mechanisms and new strategies for primary Sjögren's syndrome. Annu. Rev. Med. 2017;68:331–343. doi: 10.1146/annurev-med-043015-123313. [DOI] [PubMed] [Google Scholar]
- 5.Chivasso C., Sarrand J., Perret J., et al. The involvement of innate and adaptive immunity in the initiation and perpetuation of Sjögren's syndrome. Int. J. Mol. Sci. 2021;22(2) doi: 10.3390/ijms22020658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nocturne G., Mariette X. Advances in understanding the pathogenesis of primary Sjögren's syndrome. Nat. Rev. Rheumatol. 2013;9(9):544–556. doi: 10.1038/nrrheum.2013.110. [DOI] [PubMed] [Google Scholar]
- 7.Hong X., Meng S., Tang D., et al. Single-cell RNA sequencing reveals the expansion of cytotoxic CD4(+) T lymphocytes and a landscape of immune cells in primary Sjögren's syndrome. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.594658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satija R., Farrell J.A., Gennert D., et al. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20(1):296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha D., Kumar A., Kumar H., et al. dropClust: efficient clustering of ultra-large scRNA-seq data. Nucleic Acids Res. 2018;46(6) doi: 10.1093/nar/gky007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferkingstad E., Sulem P., Atlason B.A., et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021;53(12):1712–1721. doi: 10.1038/s41588-021-00978-w. [DOI] [PubMed] [Google Scholar]
- 12.Kurki M.I., Karjalainen J., Palta P., et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–518. doi: 10.1038/s41586-022-05473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi X., Wei T., Hu Y., et al. The associations between plasma soluble Trem1 and neurological diseases: a Mendelian randomization study. J. Neuroinflammation. 2022;19(1):218. doi: 10.1186/s12974-022-02582-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemani G., Zheng J., Elsworth B., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verbanck M., Chen C.Y., Neale B., et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin S., Guerrero-Juarez C.F., Zhang L., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morell M., Varela N., Marañón C. Myeloid populations in systemic autoimmune diseases. Clin. Rev. Allergy Immunol. 2017;53(2):198–218. doi: 10.1007/s12016-017-8606-7. [DOI] [PubMed] [Google Scholar]
- 18.Gong H., Qiu X., Li P., et al. Immune infiltration analysis reveals immune cell signatures in salivary gland tissue of primary Sjögren's syndrome. Front. Med. 2023;10 doi: 10.3389/fmed.2023.1033232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christodoulou M.I., Kapsogeorgou E.K., Moutsopoulos H.M. Characteristics of the minor salivary gland infiltrates in Sjögren's syndrome. J. Autoimmun. 2010;34(4):400–407. doi: 10.1016/j.jaut.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Manoussakis M.N., Boiu S., Korkolopoulou P., et al. Rates of infiltration by macrophages and dendritic cells and expression of interleukin-18 and interleukin-12 in the chronic inflammatory lesions of Sjögren's syndrome: correlation with certain features of immune hyperactivity and factors associated with high risk of lymphoma development. Arthritis Rheum. 2007;56(12):3977–3988. doi: 10.1002/art.23073. [DOI] [PubMed] [Google Scholar]
- 21.Konttinen Y.T., Platts L.A., Tuominen S., et al. Role of nitric oxide in Sjögren's syndrome. Arthritis Rheum. 1997;40(5):875–883. doi: 10.1002/art.1780400515. [DOI] [PubMed] [Google Scholar]
- 22.Gliozzi M., Greenwell-Wild T., Jin W., et al. A link between interferon and augmented plasmin generation in exocrine gland damage in Sjögren's syndrome. J. Autoimmun. 2013;40:122–133. doi: 10.1016/j.jaut.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alam J., de Paiva C.S., Pflugfelder S.C. Desiccation induced conjunctival monocyte recruitment and activation - implications for keratoconjunctivitis. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.701415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciccia F., Guggino G., Rizzo A., et al. Interleukin (IL)-22 receptor 1 is over-expressed in primary Sjogren's syndrome and Sjögren-associated non-Hodgkin lymphomas and is regulated by IL-18. Clin. Exp. Immunol. 2015;181(2):219–229. doi: 10.1111/cei.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi G.P., Villadangos J.A., Dranoff G., et al. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 1999;10(2):197–206. doi: 10.1016/s1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 26.Klinngam W., Fu R., Janga S.R., et al. Cathepsin S alters the expression of pro-inflammatory cytokines and MMP-9, partially through protease-activated receptor-2, in human corneal epithelial cells. Int. J. Mol. Sci. 2018;19(11) doi: 10.3390/ijms19113530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klinngam W., Janga S.R., Lee C., et al. Inhibition of cathepsin S reduces lacrimal gland inflammation and increases tear flow in a mouse model of Sjögren's syndrome. Sci. Rep. 2019;9(1):9559. doi: 10.1038/s41598-019-45966-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamm-Alvarez S.F., Janga S.R., Edman M.C., et al. Tear cathepsin S as a candidate biomarker for Sjögren's syndrome. Arthritis Rheumatol. 2014;66(7):1872–1881. doi: 10.1002/art.38633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nocturne G., Mariette X. B cells in the pathogenesis of primary Sjögren syndrome. Nat. Rev. Rheumatol. 2018;14(3):133–145. doi: 10.1038/nrrheum.2018.1. [DOI] [PubMed] [Google Scholar]
- 30.Smulski C.R., Eibel H. BAFF and BAFF-receptor in B cell selection and survival. Front. Immunol. 2018;9:2285. doi: 10.3389/fimmu.2018.02285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding J., Zhang W., Haskett S., et al. BAFF overexpression increases lymphocytic infiltration in Sjögren's target tissue, but only inefficiently promotes ectopic B-cell differentiation. Clin Immunol. 2016;169:69–79. doi: 10.1016/j.clim.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Dörner T., Posch M.G., Li Y., et al. Treatment of primary Sjögren's syndrome with ianalumab (VAY736) targeting B cells by BAFF receptor blockade coupled with enhanced, antibody-dependent cellular cytotoxicity. Ann. Rheum. Dis. 2019;78(5):641–647. doi: 10.1136/annrheumdis-2018-214720. [DOI] [PubMed] [Google Scholar]
- 33.Giovannone N., Liang J., Antonopoulos A., et al. Galectin-9 suppresses B cell receptor signaling and is regulated by I-branching of N-glycans. Nat. Commun. 2018;9(1):3287. doi: 10.1038/s41467-018-05770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winograd-Katz S.E., Fässler R., Geiger B., et al. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 2014;15(4):273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 35.Swaim C.D., Scott A.F., Canadeo L.A., et al. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol Cell. 2017;68(3):581–590. doi: 10.1016/j.molcel.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunn H.A., Orlandi C., Martemyanov K.A. Beyond the ligand: extracellular and transcellular G protein-coupled receptor complexes in physiology and pharmacology. Pharmacol. Rev. 2019;71(4):503–519. doi: 10.1124/pr.119.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maleki K.T., Cornillet M., Björkström N.K. Soluble SEMA4D/CD100: a novel immunoregulator in infectious and inflammatory diseases. Clin Immunol. 2016;163:52–59. doi: 10.1016/j.clim.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Ke Y., Dang E., Shen S., et al. Semaphorin4D drives CD8(+) T-cell lesional trafficking in oral lichen planus via CXCL9/CXCL10 upregulations in oral keratinocytes. J. Invest. Dermatol. 2017;137(11):2396–2406. doi: 10.1016/j.jid.2017.07.818. [DOI] [PubMed] [Google Scholar]
- 39.Patel L., Buckels A.C., Kinghorn I.J., et al. Resistin is expressed in human macrophages and directly regulated by PPAR gamma activators. Biochem. Biophys. Res. Commun. 2003;300(2):472–476. doi: 10.1016/s0006-291x(02)02841-3. [DOI] [PubMed] [Google Scholar]
- 40.Savage D.B., Sewter C.P., Klenk E.S., et al. Resistin/Fizz3 expression in relation to obesity and peroxisome proliferator-activated receptor-gamma action in humans. Diabetes. 2001;50(10):2199–2202. doi: 10.2337/diabetes.50.10.2199. [DOI] [PubMed] [Google Scholar]
- 41.Liu M., Zhao J., Chen K., et al. G protein-coupled receptor FPR1 as a pharmacologic target in inflammation and human glioblastoma. Int Immunopharmacol. 2012;14(3):283–288. doi: 10.1016/j.intimp.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dasgupta B., Dufour E., Mamdouh Z., et al. A novel and critical role for tyrosine 663 in platelet endothelial cell adhesion molecule-1 trafficking and transendothelial migration. J. Immunol. 2009;182(8):5041–5051. doi: 10.4049/jimmunol.0803192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rui Y., Liu X., Li N., et al. PECAM-1 ligation negatively regulates TLR4 signaling in macrophages. J. Immunol. 2007;179(11):7344–7351. doi: 10.4049/jimmunol.179.11.7344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 3: Table S1. Detailed information about the significant DEGs of each cell type. Table S2. Detailed information about the instrumental variables of CTSS.
Data Availability Statement
The scRNA-seq data used in this study can be obtained from GEO database under the accession number GSE157278. The GWAS data used in this study can be obtained from the openGWAS database with the GWAS ID finn-b-M13_SJOGREN.