Abstract
Gastrointestinal stromal tumor (GISTs) are clinically heterogenous exhibiting varying degrees of disease aggressiveness in individual patients. We comprehensively describe the genomic and transcriptomic landscape of a cohort of 117 GISTs including 31 low-risk, 18 intermediate-risk, 29 high-risk, 34 metastatic and 5 neoadjuvant GISTs from 105 patients. GISTs have notably low tumor mutation burden but widespread copy number variations. Aggressive GISTs harbor remarkably more genomic aberrations than low-/intermediate-risk GISTs. Complex genomic alterations, chromothripsis and kataegis, occur selectively in aggressive GISTs. Despite the paucity of mutations, recurrent inactivating YLPM1 mutations are identified (10.3%, 7 of 68 patients), enriched in high-risk/metastatic GIST and functional study further demonstrates YLPM1 inactivation promotes GIST proliferation, growth and oxidative phosphorylation. Spatially and temporally separated GISTs from individual patients demonstrate complex tumor heterogeneity in metastatic GISTs. Finally, four prominent subtypes are proposed with different genomic features, expression profiles, immune characteristics, clinical characteristics and subtype-specific treatment strategies. This large-scale analysis depicts the landscape and provides further insights into GIST pathogenesis and precise treatment.
Subject terms: Sarcoma, Cancer genomics, Gastric cancer, Gastric cancer
Gastrointestinal stromal tumours (GISTs) are clinically heterogeneous, with varying degrees of aggressiveness. Here, the authors describe the genomic and transcriptomic landscape of 117 GISTs from 105 patients; they find four molecular subtypes as well as recurrent inactivating YLPM1 mutations in high-risk/metastatic GIST.
Introduction
Gastrointestinal stromal tumor (GIST) is the most common sarcoma, usually originating in the stomach or the small intestine. Most GISTs are initiated by activating mutations in the KIT (75–80%) or PDGFRA (5–10%)1–4. GISTs are clinically heterogenous exhibiting varying degrees of disease aggressiveness in individual patients, with some low-risk lesions remaining stable for years whereas others progress rapidly to widespread metastatic disease4,5. Deciphering the molecular changes contribute to the development of aggressive GIST may shine light on GIST biology and therapeutic strategies. More recently, using cytogenetic approaches and whole-exome sequencing (WES) in a small cohort of patients with GIST, we and others have reported recurrent somatic alterations of DEPDC5 (17.5%)6, DMD (66%)7, MAX (32%)8, SETD2 (11.2% in high-risk GISTs)9 and SDH (9.0%)10 in GISTs. The Cancer Genome Atlas (TCGA) genetic analysis of sarcoma has provided a detailed genomic characterization of other 6 common adult sarcomas11. However, systematic genome- and transcriptome-wide investigation of GIST is lacking.
Approval of KIT tyrosine kinase inhibitors (TKIs) - imatinib (first-line), sunitinib (second-line), regorafenib (third-line) and ripretinib (fourth-line) - and PDGFRA TKI - avapritinib (first-line) - improved the survival of advanced GISTs4,12,13. Inhibition of KIT/PDGFRA with TKIs is the only established systemic therapeutic strategy for GIST. Evolution of TKI resistance mutations is inexorable in the advanced setting, leading to poor patient outcomes3,4. There has been a consensus on the subtyping of GIST based on different driver genes, but few studies focused on the classification combined with other omics data in GISTs.
In this work, by integrative analysis of whole-genome sequencing (WGS), WES and whole-transcriptome sequencing (WTS), we obtain a comprehensive genomic and transcriptomic landscape of GISTs and propose four prominent subtypes with different genomic features, expression profiles, immune characteristics, clinical characteristics and subtype-specific treatment strategies. Moreover, a recurrently mutated gene, YLPM1, in GISTs was discovered and functionally validated.
Results
Overview of cohort characteristics
This study was composed of 117 GIST samples (113 frozen GISTs and 4 GIST cell lines) and 68 matched non-cancerous, normal samples from 105 patients. The 117 GIST samples include 31 low-risk, 18 intermediate-risk, 29 high-risk and 34 metastatic according to the well-established, widely used modified NIH clinicopathological criteria7,9,14. Note that the remaining 5 GISTs with preoperative neoadjuvant TKI therapy cannot be classified as pretreatment impacts mitotic count. A pathology review of specimens by two independent pathologists revealed that the tumor cellularity of tumor tissues surpassed 75% (Fig. S1a). Clinical and pathological features were summarized in Supplementary Data 1.
Integrated multiplatform analysis was performed (Fig. S1b). WES was performed on 59 GISTs and 49 matched normal samples from 49 patients with a median depth of 131.99× (130.46× normal, 133.27× tumor), and, on average, 96.96% of target bases had >30 reads (range, 77.60–99.48%). WGS was performed on 19 GISTs and 19 matched normal samples with a median depth of 54.09× (44.01× normal, 64.17× tumor), and 91.54% of target bases had >30 reads (range, 78.30–97%) (Fig. S1c; Supplementary Data 2). No sample was mixed as revealed by pair-wise comparisons of all Binary Sequence Alignment/Map (BAM) files using BAM-matcher15 (Fig. S1d). WTS was performed on 116 samples (107 frozen GISTs, 4 GIST cell lines and 5 matched normal samples). The average number of paired-end reads for transcriptome sequencing was 201.43 million (range, 158.11–205.71 million) (Supplementary Data 2).
Aggressive GISTs harbor more genomic aberrations
A total of 1729 coding single nucleotide variations (SNVs) and small insertions/deletions (indels) involving 1282 genes were identified (Supplementary Data 3, Fig. S1e and S1f). The median coding mutation rate is 0.67 (range, 0.15–1.70) per megabase (Mb) (Fig. 1a), which is comparable to low-mutation-rate cancers such as chromophobe renal cell carcinoma and chronic lymphocytic leukemia16 (Fig. S2a). In addition, GISTs have the lowest tumor mutation burdens when compared to the other 6 types of sarcomas reported by TCGA (average 1.06 per Mb)11 (Fig. S2b).
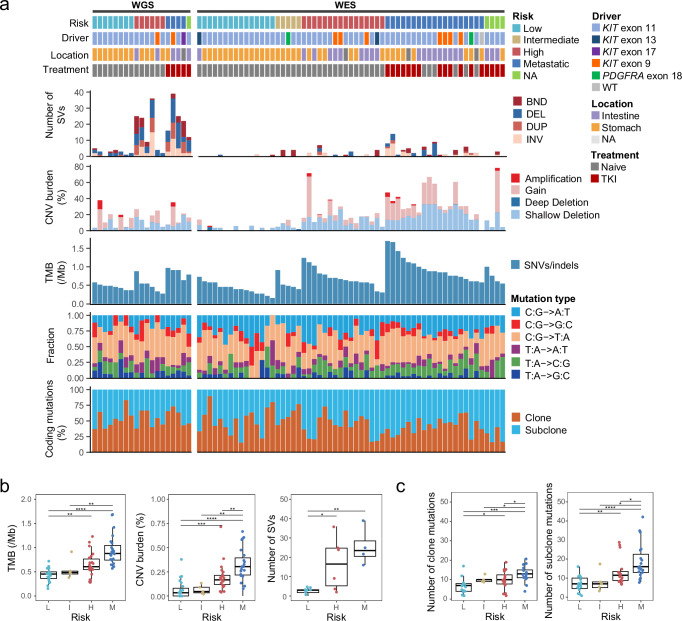
Fig. 1. Molecular landscape of the GIST cohort.
a Each column represents an individual tumor (n = 78). Patients with paired tumor and normal samples are separated into two groups: WGS (n = 19) and WES (n = 59). The top panel shows information about the clinical risk stratification, mutated exon of KIT or PDGFRA, primary tumor sites, and TKI treatment information. Each subsequent panel displays a specific molecular profile. NA, not available. Samples are ranked according to the risk stratification followed by the TMB. In the top panel, TKI indicates TKI treatment prior to surgery (tumor collection), and the GIST samples show pathologically progression (resistance) on TKIs before samples were collected. DEL, deletion; BND, translocation; INV, inversion; DUP, duplication. b Boxplots showing that the alternation burdens increase with risk stratification. Left, TMB of coding mutations, L (n = 23), I (n = 5), H (n = 22), and M (n = 23). Center, the percentage of CNV segments in autosomal genome region, L (n = 23), I (n = 5), H (n = 22), and M (n = 23). Right, the number of SVs in 19 WGS tumors, L (n = 8), H (n = 6), and M (n = 4). c Number of clone (left) and subclone (right) mutations in coding region among different risk stratification. L (n = 15), I (n = 5), H (n = 16), and M (n = 19). The P values in (b, c) are calculated using the two-sided wilcoxon rank-sum test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). L denotes low-risk, I denotes intermediate-risk, H denotes high-risk, M denotes metastatic. The low, centerline, and upper of boxplot represent the first quartile, the median, and the third quartile of data, respectively. The whiskers extend to the largest and smallest values within 1.5 times the interquartile range (IQR). Source data are provided as a Source Data file.
In contrast to the sparse of SNVs and indels, we observed a higher burden of copy number variations (CNVs) in the GIST genomes. In total, 2126 copy number gain segments and 6246 copy number loss segments were identified across the whole cohort, corresponding to an average of 19% (range, 0–78%) of the genome (Fig. 1a). In addition, we detected 336 somatic structural variations (SVs), with the number of SV events per sample ranging from 0 to 39 (Fig. 1a).
As the level of risk increased, the total number (or fraction) of somatic coding mutations, CNVs and SVs of the tumor increased as well (Figs. 1b, 1c and S3a), indicating that an increasing number of alterations were accumulated during GIST progression. Increased genomic complexity (TMB, CNV burden) is positively associated with tumor size and mitotic count (Fig. S3c). Moreover, clonal analysis showed that both clonal and subclonal coding mutations were increased in primary ‘high-risk’ GISTs (GISTs having histological criteria predictive of metastasis) and metastatic GISTs (Fig. 1c), while subclonal coding mutations were more accumulated than clonal coding mutations in high-risk/metastatic GISTs (Fig. S3b), showing that aggressive GISTs was more heterogeneous.
Recurrently mutated genes
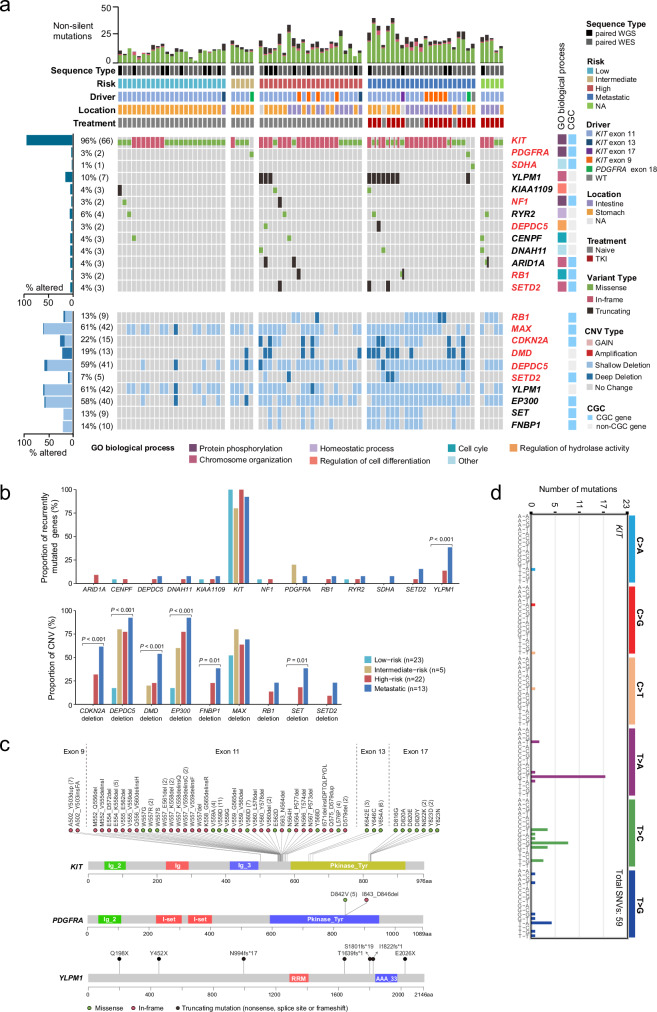
Functional alterations in reported GIST driver genes, including KIT1, PDGFRA2, NF117, SDHA18, DMD7, RB119, CDKN2A19 and SETD29, were successfully identified in our cohort (Fig. 2a). However, KIT (q-value < 0.001) and YLPM1 (q-value < 0.001) were predicted as significantly mutated genes by MutSigC2V20 (Supplementary Data 4). The KIT mutations were mainly located in exons 11, 9, 13 and 17 (Fig. 2a, c). A primary exon 9 mutation (A502_Y503insFA) of KIT was identified and showed a gain-of-function property (Fig. S4a) and was sensitive to both the first-line and the second-line TKIs (imatinib and sunitinib) (Fig. S4b). In line with previous studies2, KIT and PDGFRA mutations were mutually exclusive (Fig. 2a). Of note, YLPM1 (YLP motif-containing protein 1) was the most frequently mutated gene (7 of 68 patients) after KIT in our cohort (Fig. 2a). All of the YLPM1 mutations were protein-truncating mutations throughout its length, indicating the potential role of a tumor suppressor (Fig. 2c). A missense mutation (R585Q, NM_004168) in SDHA predicted to be deleterious was found in a KIT/PDGFRA wild type GIST (Fig. 2a). All mutations in SETD2 were inactivating mutations, occurring predominantly in high-risk/metastatic GISTs as previously reported9 (Fig. 2a and S5). For our recently discovered driver gene DEPDC56, a missense mutation predicted to be damaging was identified in a high-risk GIST (Fig. 2a and S5).
Fig. 2. Landscape of genomic alterations in 78 GISTs.
a Integrated plots of clinical and genomic alterations in 78 GISTs from 68 patients. The middle panel shows selected mutated genes and variant types. Known driver genes of GIST and recurrently mutated genes detected in at least 3 patients are included. The mutation frequency of each gene is shown as a bar plot on the left with the number of affected cases labeled in parentheses. The corresponding GO biological process for each gene are shown as colored blocks on the right. Blue annotations on the right indicate whether the genes are in the Cancer Gene Census (CGC) list. The bottom panel shows selected focal CNV genes detected by GISTIC2.0. The copy number: CN = 0 indicates a Deep Deletion, CN = 1 indicates a Shallow Deletion, CN = 3 indicates a Gain and ≥4 indicates an Amplification. The red gene symbols indicate known GIST drivers. b Bar plots illustrating relative proportion of recurrently mutated genes (top) and copy number alterations (bottom) by different risk. c Lollipop plots showing the distribution of all non-silent mutations in KIT, PDGFRA, and YLPM1. The scale bars represent the length (amino acids) of the protein sequence and the protein domains of the gene are indicated by colors. The number in parentheses denotes the number of patients. d 96-mutation spectrum of KIT mutations in GISTs. A total of 59 SNVs are identified in 46 GISTs. The distributions of KIT mutations are different from the overall SNV distributions, showing T > C and T > A bias. Source data are provided as a Source Data file.
In addition to these verified GIST driver genes, we identified 5 recurrently mutated genes (≥3 cases), including RYR2 (6%), ARID1A (4%), KIAA1109 (4%), CENPF (4%) and DNAH11 (4%). ARID1A have been listed in the Cancer Gene Census (CGC v90)21. All of the ARID1A mutations were truncated (frameshift mutations or nonsense mutations), showing the potential tumor suppressor roles (Figs. S5 and 2a). These data show that GISTs, exhibiting substantial heterogeneity at the mutational level, are possibly initiated by KIT/PDGFRA mutations and then driven by a diverse spectrum of less frequently mutated “gene hills”, which may be different for each patient. Gene Ontology analysis was performed to show that these mutated genes were enriched in 7 biological processes: protein phosphorylation (KIT, PDGFRA, and NF1), chromosome organization (YLPM1, ARID1A, and SETD2), homeostatic process (RYR2), regulation of cell differentiation (KIAA1109), regulation of hydrolase activities (DEPDC5) and cell cycle (RB1, and CENPF), thus indicating that abnormalities of these biological processes involve in GIST pathogenesis (Fig. 2a).
YLPM1 Inactivation in GISTs
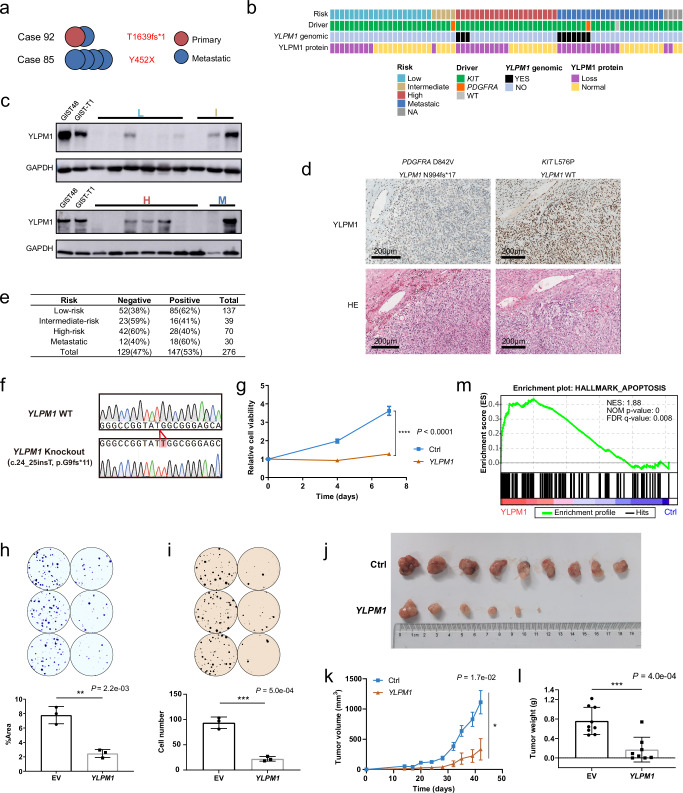
Somatic homozygous YLPM1 mutations (SNVs and indels) were confirmed by Sanger sequencing in the WES/WGS cohort (Fig. S6a), and a homozygous deletion of YLPM1 was also validated by genomic quantitative PCR, of which the deletion region (exon 1–4) could be inferred by IGV (Fig. S6b and S6c). YLPM1 mutations are more common in gastric GISTs. YLPM1 inactivating mutations, when present within a primary tumor, were perpetuated in subsequent metastatic GISTs (case 92, Fig. 3a) and, when present in any metastatic lesions, were also detected in other metastases from the same patient (case 85, Fig. 3a). YLPM1 copy number variations (CNVs) were detected in 42 of 68 (61%) patients (Fig. 2a), including shallow deletions in 40 of 68 (59%) patients and deep deletions in 2 of 68 (3%) patients. Heterozygous deletion of chromosome 14q is one of the most frequent genomic events in GISTs, as reported previously22. Human YLPM1 locates in 14q24. Heterozygous deletion of chromosome 14q likely counts for the frequent shallow deletions. Homozygous YLPM1 mutations and deletions were identified in 9 of 68 (13%) patients (Fig. 2a, S7a and S7b). Genomic alteration of YLPM1 is correlated with telomere length (Fig. S8), in line with previous studies that YLPM1 is involved in telomere maintenance23. Genomic YLPM1 aberrations were observed only infrequently in 253 non-GIST sarcomas (3%) and in 10,953 Pan-Cancers (3%) in the TCGA Pan-Cancer Atlas program24 (Fig. S7a and S7b), which is significantly lower than the frequency in GISTs (P < 0.0001), showing that YLPM1 inactivation selectively occurs in GISTs.
Fig. 3. High frequency of YLPM1 inactivation in GISTs and YLPM1 inactivation promotes tumor growth and proliferation in vitro and in vivo.
a Multiple tumors from the same patients share the same YLPM1 mutation. b Summary of YLPM1 genomic and protein aberrations in 73 GISTs. c YLPM1 protein inactivation is demonstrated by immunoblotting of GIST biopsies. YLPM1 wild-type GIST48 and GIST-T1 cell lines are used as positive controls. YLPM1 inactivation is defined by relative expression level (YLPM1/GAPDH) < 0.3, normalized to GIST-T1. All panels represent data from 3 times independent experiments. d Hematoxylin and eosin stains (bottom) and YLPM1 immunohistochemistry (IHC) stains (top): case 94 with wild-type YLPM1 shows retained YLPM1 expression; case 104 with YLPM1 frameshift mutation shows a loss of YLPM1 expression. e Summary of YLPM1 expression assessed by IHC in tissue microarrays validation cohort. f–m Restoration of YLPM1 suppresses tumor growth and proliferation in YLPM1-inactivated GISTs. f Sanger sequencing shows that GIST-T1YLPM1 KO isogenic cells are successfully established. g Lentivirus-mediated YLPM1 restoration reduces the viability of GIST-T1YLPM1 KO cells, as assessed by CellTiter-Glo viability assay. Data are presented as mean values ± s.d. n = 3. h Crystal violet staining assays show that restoration of YLPM1 suppresses cell proliferation. Representative plates (top) and mean percentage area (bottom) are shown. Data are presented as mean values ± s.d. n = 3. i Restoration of YLPM1 inhibits anchorage-independent growth. Representative plates (top) and mean colony numbers (bottom) are shown. Data are presented as mean values ± s.d. n = 3. j–l Restoration of YLPM1 suppresses the growth of GIST-T1YLPM1 KO xenografts in nude mice. Photo images (j) (n = 9 mice for Ctrl, n = 8 mice for YLPM1 restoration, note that no tumor growth in 2 mice), growth curves (k), and tumor weight (l) of transplanted tumors are shown. Error bars are the mean ± s.e.m.. m GSEA reveals that genes involved in Hallmark apoptosis gene set are upregulated in GIST-T1YLPM1 KO group. NES, normalized enrichment score. NOM P-value, Nominal P-value. All the P values are calculated using the two-sided Student’s t test. Source data are provided as a Source Data file.
We further assessed the inactivation frequency of YLPM1 at protein level by immunoblotting in 73 GISTs from 64 patients. YLPM1 protein loss was demonstrated in 31 of 64 (48%) patients, irrespective of whether they had KIT or PDGFRA mutations (Fig. 3b, Supplementary Data 5). 32% (10 of 31) of the patients with loss of YLPM1 expression were classified as low-risk or intermediate-risk (Fig. 3b, c). Then, we performed immunohistochemistry to validate the frequency to which YLPM1 protein expression was lost. YLPM1 expression was negative in 47% (129/276) of GISTs on tissue microarray validation cohort, including 75 low or intermediate risk (Fig. 3e), showing that YLPM1 inactivation could be an early event in GIST pathogenesis. Genomic alteration of YLPM1 is correlated with protein expression (Fisher’s exact text, P = 0.0021). Considering that the inactivation frequency of YLPM1 at the protein level is higher than that at the genomic level, we tested whether the promoter hypermethylation leads to YLPM1 inactivation. WTS data and DNA methylation studies indicated that dysregulation of DNA methylation was not common in the regulation of YLPM1 expression in GISTs (Fig. S9, Supplementary Data 6). Similar high frequency of DMD protein loss versus relatively low frequency of DMD genomic changes are also found in GIST7, showing non-genomic inactivation mechanisms in GIST. Whether non-genomic mechanisms, such as post-transcriptional modifications, lead to YLPM1 protein loss in the low-risk GISTs merits further investigation.
The biological function of YLPM1 was investigated using various GIST models. We established YLPM1-knockout (KO) cells from GIST-T1 cells that retained YLPM1 expression using a CRISPR/Cas9 system (Fig. S10a). YLPM1 knockout facilitated the cell growth and proliferation in both short-term (Fig. S10b) and long-term (Fig. S10c) assays. YLPM1 knockout also promoted the three-dimensional anchorage-independent growth (Fig. S10d). In contrast, re-expression of YLPM1 in YLPM1-KO isogenic GIST-T1 cells (GIST-T1YLPM1 KO) reduced the number of viable cells (Fig. 3f, g) and proliferative properties (Fig. 3h, i). To determine whether the inhibition of cell proliferation was manifested in vivo, we generated 4 xenografts (GIST-T1 with or without YLPM1 KO, GIST-T1YLPM1 KO with or without YLPM1 restoration) in nude mice. In vivo experiments showed that YLPM1 KO promoted tumor growth to a certain extent, while YLPM1 restoration markedly attenuated tumor growth although the tumors contained oncogenic KIT mutation (Fig. S10e and S10f, Fig. 3j–l, Fig. S10g). Collectively, these results demonstrate that YLPM1 inactivation promotes GIST proliferation and growth. An increasing body of evidence demonstrates that some cancers are heavily reliant on oxidative phosphorylation (OXPHOS) to promote tumorigenesis25,26, and many recent studies have revealed that OXPHOS inhibition is effective in targeting these cancers27. OXPHOS is the metabolic pathway by which cells use enzymes to oxidize nutrients (glucose, fat and protein), thereby releasing energy, which is used to reform ATP. The mitochondrial oxygen consumption rate (OCR) is a measurement of oxygen utilization in cells and is an indicator of mitochondrial function. In addition to its role in cell growth and proliferation, we show that YLPM1 knockout also increases OCR and ATP productions, promoting oxidative phosphorylation in GIST (Fig. S10g, h).
To determine the global gene expression patterns regulated by YLPM1 inactivation in GIST, we performed RNA-seq on paired cell lines (GIST-T1 with or without YLPM1 KO, GIST-T1YLPM1 KO with or without YLPM1 restoration). Gene set enrichment analysis (GSEA) revealed that cell cycle-related genes, including mitotic spindle genes, were upregulated in YLPM1 KO GIST-T1 cells (Fig. S10i), while apoptosis-related genes were positively enriched in YLPM1-restored GIST-T1YLPM1 KO cells (Fig. 3m). Immune gene sets related to interferon response (HALLMARK INTERFERON-ALPHA RESPONSE and HALLMARK INTERFERON-GAMMA RESPONSE) were downregulated in the KO group (Fig. S10i). These results suggested that YLPM1 may play roles in the regulation of immunity in GISTs. Given the critical roles of the KIT pathways in GIST tumorgenesis, we further explored whether YLPM1 regulates oncogenic KIT pathways. YLPM1 did not regulate the KIT pathways (Fig. S11a) and did not modulate the sensitivity of GIST to KIT inhibitors (Fig. S11b).
Mutational signatures
To gain insight into the etiology and the mutational processes that contribute to GISTs, we attempted to decipher the mutational signatures in somatic mutation catalogs. The mutational spectrum of 96 mutation classes between WGS and WES showed high correlation when only coding regions were included, which reflected the consistency of different sequencing platforms and sub-cohorts (Fig. S12a and S12b). The correlation dropped significantly when including non-coding mutations, C > A transversion mutations decreased, whereas T > C and T > A mutations increased (Fig. S12a). These findings indicated distinct mutational processes in coding and non-coding regions. Interestingly, the mutation spectrum (mainly T > A, T > C) of KIT, which composes 3.7% of all coding SNVs, is completely different from the distribution (C > T, C > A) of overall mutation catalogue (Fig. 2d). The T > A mutations of KIT were predominantly enriched in the GpTpT context, which may be associated with a specific mutagenesis process. Due to the low mutation burden in GISTs, stable de novo signature discovery was only available in genome-wide SNVs of 19 WGS datasets. Two signatures most similar to COSMIC signature 5 and signature 8 were extracted by BayesNMF28 (Fig. S12c–e). Signature 5 shows clock-like properties, exhibiting transcriptional strand bias for T > C substitutions at the ApTpN context29. Signature 8, found in breast cancer and medulloblastoma, exhibits weak strand bias for C > A substitutions29. Regrettably, both of these two signatures are flat signatures with an unknown etiology29.
Widespread copy number variations in GISTs
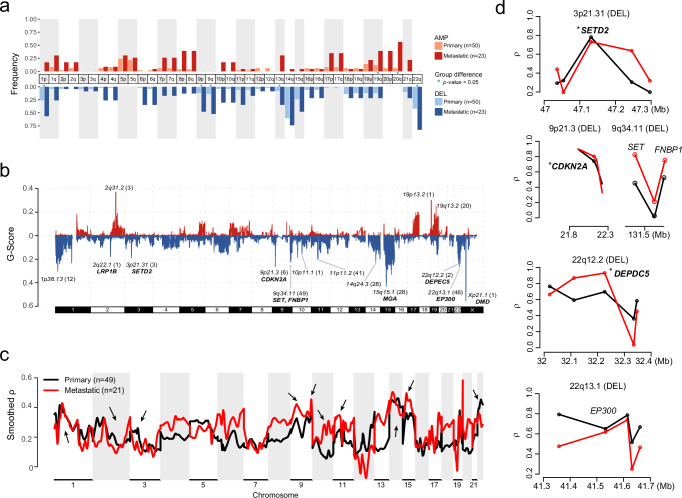
To identify the CNV signatures30 in GISTs, we extracted the CNV signatures with copy number, segment size, and the heterozygosity status of CNVs from GISTs and identified 8 CNV signatures, among which COSMIC_CN1 and COSMIC_CN9 were detected in more than 50% of GISTs (Fig. Supplementary Data 7, S13a, b). COSMIC_CN1 was characterized by heterozygous segments with a total copy number (TCN) of 2 and sizes exceeding 40 Mb, whereas COSMIC_CN9 was identified as a signature of chromosomal instability on a diploid background and exhibited elevated levels in metastatic GIST (Fig. S13a, c). We next sought to delineate the recurrent CNV events using the Genome Identification of Significant Targets in Cancer (GISTIC) 2.0 algorithm. At the arm-level CNVs, deletions in 22q (57%), 1p (39%), 9q (26%), 9p (23%) and 13q (20%), and amplifications in 19q (21%) and 20q (22%) were more frequently occurred in metastatic GISTs (Supplementary Data 8, Fig. 4a), as reported previously3,4,31.
Fig. 4. Genomic imbalances.
a Chromosome arm-level CNV frequencies in different risk stratification of 73 GISTs. Dark red, red and light red represent the amplification (AMP) frequencies in primary (low-risk, intermediate-risk and high-risk, n = 50) and metastatic GISTs (n = 23), respectively. Dark blue, blue and light blue represent the deletion (DEL) frequencies in low or intermediate-risk, high-risk and metastatic GISTs, respectively. Arms with significant group differences are denoted by green asterisks (both the GISTIC q value and chi-square test P value < 0.05). b Focal-level copy number gains and losses across chromosomes 1-22 and X detected by GISTIC 2.0, with the G-score labeled on the vertical axis. Selected cancer-associated genes are labeled in the significant peak regions. c, d Associations between quantitative measurements of CNV and gene expression in different risk groups using MVisAGe R-package. Black represent primary GISTs (n = 49) and red represent metastatic GISTs (n = 21). c Genome-wide plot of smoothed gene-level Pearson correlation coefficients (smoothed ρ values) across chromosomes 1–22. Arrows indicate focal-CNV peaks from GISTIC. d Unsmoothed ρ values and selected genes are plotted based on genomic positions in selected regions from focal-CNV peaks. The asterisks indicate known drivers in GIST. Source data are provided as a Source Data file.
At the focal CNVs, a total of 15 peak regions (involving 211 genes) were detected, clearly with more deletions than amplifications (12 versus 3) (Fig. 4b, Supplementary Data 9, a). The recurrent deletions included several known tumor suppressor genes (CDKN2A, DEPDC5, and DMD), which were solely identified in the narrow peaks with fewer than 10 genes and occurred predominantly in high-risk or metastatic GISTs, as reported6,7,9,19 (Figs. 2a, b and 4b). Other cancer gene in focal CNV peaks included SET and FNBP1 (9q34.11 deletion) were also enriched in the high-risk or metastatic GISTs.
As copy number gains and losses are often accompanied by corresponding changes in gene expression, we further inferred the Pearson correlation coefficients (ρ value) between CNVs and mRNA expression (CN/GE) by MVisAGe32. Many of the genes within broad and focal regions showed a higher correlation in metastatic GISTs than that in primary GISTs (Fig. 4c). Several well-known GIST drivers were observed to have high CN/GE correlations (Fig. 4d), such as CDKN2A, DEPDC5 and SETD2 (ρ = 0.73, 0.8 and 0.74, respectively). We further performed rank-based GSEA using hallmark pathways derived from the Molecular Signature Database (MSigDB) to identify pathways for high CN/GE correlation genes (ρ > 0.535, Supplementary Data 9,b). Many of these genes were implicated in MYC Targets, oxidative phosphorylation and DNA repair related pathways (Fig. S14a). We also found 86 genes with high CN/GE correlation (ρ > 0.535, Supplementary Data 9, b) were occurred in the focal peaks (Fig. S14b). Moreover, both FNBP1, EP300 and their surrounding genes showed modest CN/GE correlations (ρ = 0.64 and ρ = 0.63, respectively) (Fig. 4d). Expectedly, GISTs with DMD deletions showed significantly lower expression than those with normal copy number status (Fig. S14c).
Chromothripsis and kataegis events occur selectively in aggressive GISTs
A total of 336 structural variants (SVs) were identified (median of 12 SVs per sample, range 0–39) (Supplementary Data 10, a) using Manta33, including 7418 genes. The most frequently affected genes were DMD, LRP1B and ACOT7 (each in 4 cases), and the breakpoints of DMD deletions were successfully verified by Sanger sequencing (Supplementary Data 10,b, 13e, Fig. S15a).
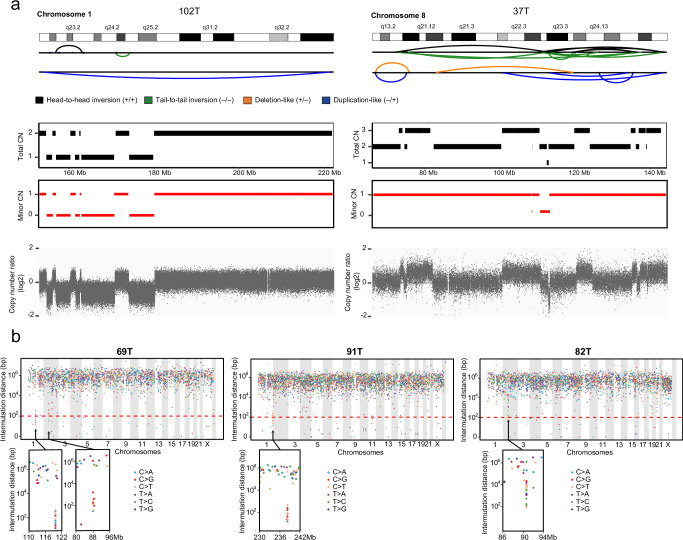
We next scanned the genome in WGS data of 19 GISTs to search for two complex aberrations that have rarely been reported in GISTs: 1) chromothripsis, in which large numbers of broken fragments occur simultaneously, clustered on one or a few chromosomes, randomly stitching back together the resulting pieces, leading to continuously copy number oscillations between 2 or sometimes 3 states34; 2) kataegis, a pattern of hypermutations identified in a short distance, with mutations biased toward a single DNA strand, co-localized with rearrangement29,35. 10.5% (2 of 19) of GISTs were identified as having chromothripsis regions (Fig. 5a, S15b and Supplementary Data 11), including one TKI-naïve, gastric GIST and one imatinib-treated GIST. We also identified 4 kataegis events in 3 GISTs (Fig. 5b and S15e), including 2 intestinal GISTs (1 TKI-naïve, and 1 TKI-treated) and 1 imatinib-treated, gastric GIST. These suggested that the occurrence of chromothripsis and kataegis in GIST showed no preference for anatomical sites and also independent of TKI therapy. Among these 3 GISTs with kataegis regions, 69T and 91T had predominant C > T or C > G mutations in a TpC context, which is a typical feature that is probably caused by APOBEC activity29. Notably, all these 5 GISTs with chromothripsis or kataegis were aggressive GISTs (45% versus 0%, high-risk/metastatic versus low or intermediate-risk, P < 0.0001), potentially indicating the late occurrence of chromothripsis and kataegis events in GIST progression.
Fig. 5. Complex genomic aberrations.
a SV and CNV profiles for two aggressive cases with CN oscillation features of chromothripsis. (Left) Evidence of chromothripsis on chromosome 1 in a metastatic GIST with CN oscillations between 2 CN levels and LOH. (Right) Evidence of chromothripsis on chromosome 8 in a high-risk GIST with CN oscillations that span 3 CN levels. The chromosome location and the SV calls in the chromothripsis region are shown on the top panel. The subsequent panel displays the total CN (black rectangle), minor CN (red rectangle) and the total copy number log-ratio for SNPs (gray dots) within the affected region. CN, copy number. b Rainfall plots showing the inter-mutation distance versus the genomic position for 3 GISTs with localized hypermutations. The horizontal axis shows mutations ordered by chromosome loci (from the first mutated position on chromosome 1 to the last mutated position on chromosome Y), and the vertical axis represents the inter-mutation distance. The lower section shows the localized hypermutation loci in detail. Source data are provided as a Source Data file.
Chromothripsis and kataegis have been associated with telomere crisis, which is triggered by the continuous shortening of telomeres, leading to genome instability35. To achieve replicative immortality, most tumors maintain their telomere length through reactivation of TERT expression, while approximately 10-15% tumors employ recombination-dependent alternative lengthening of telomere (ALT) pathway to maintain telomere length, which is prevalent in tumors of mesenchymal origin36. Due to a lack of knowledge of telomere in GISTs, the most common mesenchymal tumors of the gastrointestinal tract, we first used TelomereHunter37 to estimate telomere content in 19 WGS GISTs, then examined the variation of genes related to telomeres maintenance. Consistent with other cancers38, GISTs have a lower telomere content than the matched normal samples, presumably reflecting the greater replicative drive and consequent telomere attrition in cancer cells (Fig. S15c). Truncating mutations in ATRX and DAXX, which have been correlated with longer telomere length mediated by ALT pathway, were absent in our cohort. TERT genomic aberrations (amplification, promoter mutations and SV) in these GISTs were also not detected. Of note, TERT expression in the 5 GISTs with chromothripsis or kataegis was indeed higher than those without these events (Fig. S15d), although no significant difference in their telomere content between these two groups. These results suggested that the chromothripsis and kataegis were likely the cause of the TERT upregulation and GISTs might rely on different telomere stabilization mechanisms39.
Clonal evolution
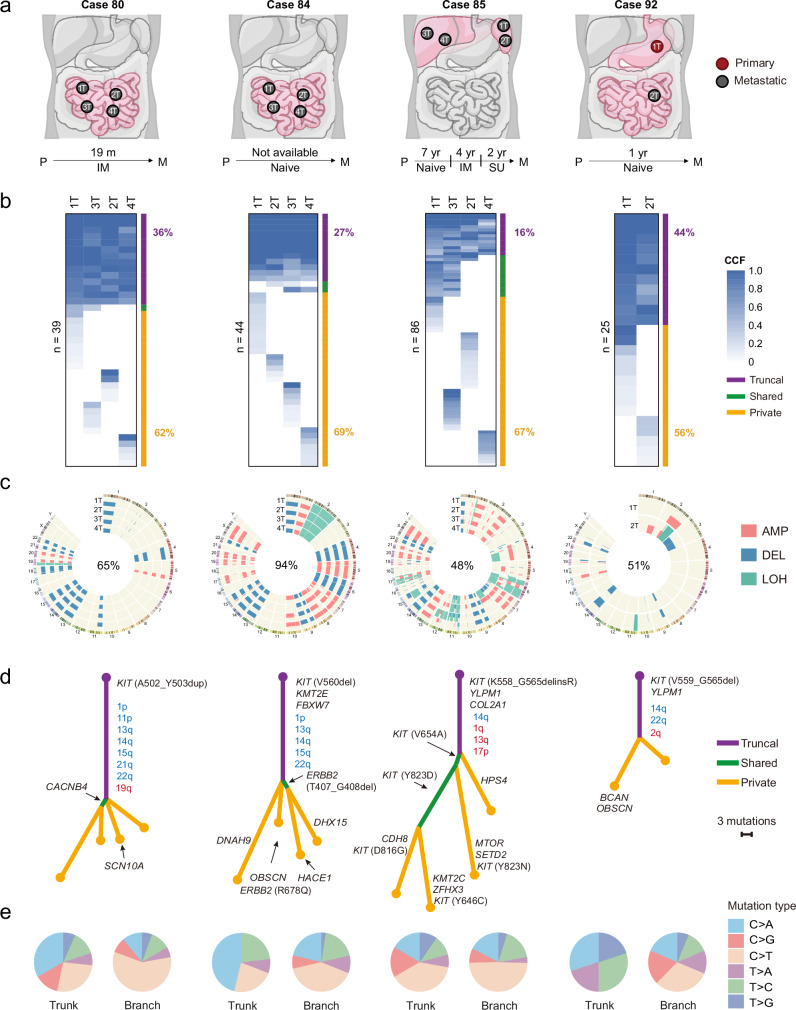
To understand the subclonal structure of GISTs and discriminate early and late driving events more accurately, we performed clonal evolution analysis on 4 metastatic cases with multiple lesions. Cases 80, 84 and 85 had 4 geographically separate lesions and case 92 had longitudinal lesions in the natural history of the disease (Fig. 6a). 2 of these 4 cases (cases 84 and 92) did not receive any TKI treatment, whereas case 80 was treated with imatinib for 19 months and case 85 received long-term treatment with imatinib and sunitinib successively after recurrence (Fig. 6a). Lesions from each case shared fewer mutations (36%, 27%, 16%, and 44% for cases 80, 84, 85 and 92, respectively) (Fig. 6b), whereas more mutations (62%, 69%, 67%, and 56%, respectively) were private to a single lesion (Fig. 6b), implying a polyclonal origin of the individual lesions in metastatic patients. Lesions from each case shared fewer mutations, but more CNV events (65%, 94%, 48%, and 51% for cases 80, 84, 85 and 92, respectively) (Fig. 6c). Interestingly, we found that the proportion of mutations and CNVs shared among lesions in case 85 were the lowest (Fig. 6b, c), and the calculated tumor heterogeneity was the highest (Fig. S16a), which was consistent with the notion that long-term TKI treatment induced more private aberrations in tumors.
Fig. 6. Delineation of the metastatic evolution of GISTs.
a Diagram of metastatic foci (P = primary GIST; M=metastatic GIST), time to recurrence (m=months, yr=years) and TKI target therapy (IM = imatinib, SU = sunitinib, Naïve = no TKI therapy). b Heatmaps indicate the cancer cell fraction (CCF) of non-silent mutations in each lesion from 4 patients. The number of mutations is labeled on the left. The percentage of truncal (purple) and private (yellow) mutations is labeled on the right. c Identified regions of CNV and copy-neutral (CN-LOH) in each lesion from 4 patients. The percentage of truncal CNVs is labeled at the center of the Circos plot. d Phylogenetic trees of 4 patients based on all non-silent mutations. Branch and trunk lengths are proportional to their number mutations. Selected cancer-associated genes and truncal CNV arms are indicated with arrows. For non-silent mutations: purple=mutations present in all samples; green=mutations shared by partial samples, yellow=private mutations. For CNVs: truncal arm-level copy number deletion events are labeled in blue, and truncal arm-level copy number amplification events are labeled in red. e Relative contribution of 6 base mutations in the trunks (left circles) and branches (right circles). Trunk equals to truncal SNVs in (b), branch equals to shared and private SNVs in (b). Source data are provided as a Source Data file.
Next, we constructed phylogenetic trees for these tumors using all non-silent mutations, and mapped shared arm-level CNVs onto the trunks. Based on this analysis, we classified these aberrations into 2 categories: (i) predominant truncal aberrations, including KIT primary mutations, 14q loss, 22q loss, 1p loss, and YLPM1 inactivating mutations. These aberrations occur on all trunks and are likely to be necessary for initiating tumor proliferation and early development of GISTs; (ii) subclonal aberrations, which make up the majority of aberrations and always occur on branches, indicating that they are important for tumor adaption during GIST progression (Fig. 6d and S16b). Notably, after TKI treatment, KIT primary mutations that are sensitive to imatinib still locate on the trunk of the case 85, and many different KIT secondary mutations occur on the branches. Among these mutations, V654A and Y823D are clonal mutations during parallel evolution, while D816G, Y646C and Y823N are specific to single lesions, reflecting tumor heterogeneity caused by TKI therapy (Fig. 6d). As expected, KIT primary mutations in GISTs were clonal based on the cancer cell fraction score40 (Fig. S16c), further demonstrating the role of KIT as an early driver. Apart from KIT, several cancer genes with functional mutations may also play roles in branched evolution, such as ERBB2 (R678Q), SETD2 (E119X) and KMT2C (K2797fs) (Fig. 6d). In addition, the relative contribution of 6 base mutations between the trunks and branches showed significant differences, indicating that different mutation process involved in the GIST progression (Fig. 6e).
Molecular subtypes of GISTs
Molecular subtypes provide guidance for cancer prognosis and precision therapy41. We presented a transcriptome-based molecular classification in GISTs. Principle component analysis was used to exclude outlier samples, no tumor samples were excluded at this step except for 5 normal samples as well as 4 cell lines (Fig. S17a). Then we performed consensus clustering by resampling randomly selected certain dataset from the remaining 107 fresh frozen tumors. According to the consensus clustering matrix (Fig. S17b) and the “elbow” point in the relative change in area under the consensus distribution function plot, we identified 5 as the optimal number of subtypes (Fig. S17c) using unsupervised k-means clustering based on the top 1500 most variable genes, measured by median absolute deviation (Fig. S17d). Cluster 5 with only one KIT/PDGFRA wild type GIST was discarded in the subsequent analysis. Silhouette analysis also confirmed that 4 clusters were stable (Fig. S17e). Moreover, the reproducibility of our clustering results was externally validated by microarray expression profiles of the Japanese42 and Complexity Index in Sarcomas (CINSARC)43 cohorts (Fig. S18).
To determine the expression pattern in the 4 mRNA subtypes, we performed differential expression analysis and totally identified 520 differentially expressed genes. Unsupervised clustering of these 520 genes demonstrated the intrinsic heterogeneity among the 4 mRNA subtypes (C1-C4) (Fig. 7a). The expression levels of several driver genes were distinct among different subtypes. KIT was highly expressed in C1-C3 and lowest in C4, whereas PDGFRA was highest in C4 (Fig. S19a). To further illustrate the differences, we performed rank-based GSEA using hallmark pathways derived from the Molecular Signature Database (MSigDB) to identify pathways differentially over-represented in each of the 4 subtypes. The expression of immune-related interferon response-associated genes (INTERFERON-ALPHA RESPONSE, INTERFERON-GAMMA RESPONSE, etc) was upregulated in C1, and the expression of cell cycle-related genes (G2M CHECKPOINT, E2F TARGETS, etc) was downregulated in C1, while C3 was just the opposite (Supplementary Data 12). Furthermore, the expression of two metabolism-related gene sets, OXIDATIVE PHOSPHORYLATION and GLYCOLYSIS, was upregulated and downregulated in C2, respectively (Supplementary Data 12). Positive enrichments were not detected in C4, but the gene expression associated with CHOLESTEROL HOMEOSTASIS, E2F TARGETS and ANDROGEN RESPONSE was significantly downregulated (Supplementary Data 12).
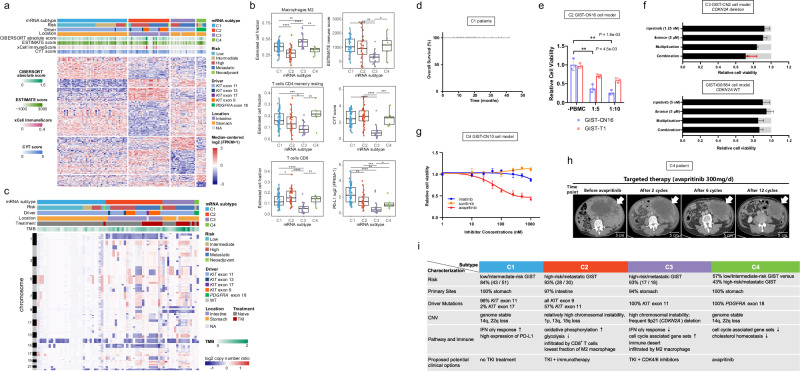
Fig. 7. Molecular subtypes of GISTs.
a Consensus clustering results of GISTs (n = 106) based on the RNA expression. Heatmap shows 520 differentially expressed genes among 4 subtypes. The number of tumors for C1, C2, C3, and C4 subtype is 51, 30, 18, and 7, respectively. Clinical risk stratification, driver mutations, location, immune scores from CIBERSORT, ESTIMATE and xCell and cytolytic (CYT) score are shown. b Boxplots showing the estimated cell fractions, immune score, CYT score and PD-L1 expression among 4 subtypes. The P values are calculated using the two-sided wilcoxon rank-sum test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). The low, centerline, and upper of boxplot represent the first quartile, the median, and the third quartile of data, respectively. The whiskers extend to the largest and smallest values within 1.5 times IQR. c Copy-number-based clustering results. Heatmap shows log2 copy-number ratio across the genome. d Overall survival of C1 patients (n = 29). e T cell-mediated tumor cell killing assay in C2 subtype cells. After 3 days of incubation of GIST-CN16 or GIST-T1 cells with PBMC, CTG viability assay was performed. Data are presented as mean values ± s.d. n = 3. The P values are calculated using the two-sided Student’s t test. f Cell viability assay reveals the synergistic effect of KIT inhibitor and CDK4/6 inhibitor. Top, GIST-CN2 primary cells established from C3 subtype GIST (94T, CDKN2A deletion). Bottom, GIST430/654 cell line (CDKN2A WT). Light gray bars indicate control values. “Multiplication” indicates expected effect of combined treatment if single-treatment effects are multiplied; red arrow indicates actual effect of combination. Data are presented as mean values ± s.d. n = 3. g GIST-CN10 primary cells established from PDGFRA D842V-mutant, C4 subtype show response to avapritinib, but resistance to imatinib or sunitinib. Data are presented as mean values ± s.d. n = 3. h Clinical data with avapritinib confirm evidence of activity in patient with metastatic C4 subtype GIST. Avapritinib induces rapid radiographic clinical response. i Highlights of the genomic features, expression profiles, immune characteristics and potential treatment strategies for the GIST subtypes. Source data are provided as a Source Data file.
Immunotherapy has become a clinically validated treatment for many difficult-to-treat cancers and has great potential for development44. Preclinical studies have reported that imatinib in combination with immunotherapy can improve the antitumor activity of targeted agents in GIST mouse models45,46, but limited efficacy observed with immunotherapy in unselected GIST patients47,48. Therefore, we performed CIBERSORT, ESTIMATE and several immunotherapy related index49 to identify which subtype might benefit from immunotherapy. The CIBERSORT analysis revealed that M2 macrophages (average 0.37, 0.10-0.70) were the most infiltrated immune cells, followed by CD4+ memory resting T cells (average 0.15, 0-0.38) and CD8+ T cells (average 0.13, 0.01-0.38), and the remaining 19 types of immune cells made up a very low proportion (Fig. S20a). When it comes to specific molecular subtypes, immune suppressive cells (M2 macrophages) showed the lowest infiltration in C2 and the highest in C3. In contrast, CD8+ T cells were most enriched in C2 and the lowest in C3 (Fig. 7b). Immunohistochemistry staining also confirmed that the infiltration of CD8+ T cells in C3 was the lowest (Fig. S20b), which was in line with the results from the mRNA expression analysis. In addition, the amount of CD4+ memory resting T cells in C3 was also the lowest (Fig. 7b). The results of ESTIMATE were consistent with CIBERSORT: C3 showed the lowest immune infiltration, indicating that C3 was an immune desert subtype (Fig. 7b). Moreover, the expression levels of immune checkpoint genes (PD-1 and PD-L1) and immunoinhibitory molecules in C3 were significantly downregulated compared with the other subtypes (Fig. 7b, Fig. S20c and S20d). These findings supported that C3 was featured with an immune desert phenotype, which was unlikely benefit from immunotherapy. Although the immune score of C1, C2 and C4 showed no difference, the fraction of M2 macrophages was the lowest in C2, while CYT score, which represents the cytolytic activity of the immune infiltrates50, and the CD8+ T cells fraction of C2 were higher than that of C1 and C4 (Fig. 7b). Interestingly, the synergistic effects of imatinib combined with immunotherapy were all occurred in KIT V559del transgenic mice with spontaneous small intestinal GISTs45,46, indicating that C2 (small intestinal GISTs predominantly) is most likely benefit from TKI plus immunotherapy.
Since genomic alterations in high-risk or metastatic GISTs were significantly higher than that in low or intermediate-risk GISTs and most of the GISTs in C2 and C3 mRNA subtypes were high-risk or metastatic GISTs (Figs. 7a, 6c), we then compared the TMB and CNV burden among 4 mRNA subtypes. GISTs in C1 and C4 subtypes were more stable, carrying lower mutational burden. In contrast, both the C2 and C3 subtypes displayed higher mutational burden (Fig. S19d). To investigate the enrichment of CNV signatures in our mRNA subtypes, we performed correlation analyses between signature intensity and molecular subtypes. Specifically, we observed that COSMIC_CN9, which was strongly associated with metastatic GIST, was enriched in the C2 and C3 subtypes (Fig. S13c). Conversely, C1 subtype exhibited a high level of COSMIC_CN1 signature (Fig. S13c). Notably, COSMIC_CN1 demonstrated an opposing pattern to COSMIC_CN9, consistent with normal diploid characteristics (Fig. S13b). We also explored the associations between mRNA subtypes and the arm level CNVs and identified a high frequency of 9p deletions in the C3 subtype (Fig. 7c, S19b), which influenced CDKN2A, a crucial gene involved in cell cycle regulation and was associated with responses to CDK4/6 inhibitors51. Moreover, a significant frequent of 1p and 15q deletions were exhibited in C2 subtype. Next, we employed GSEA on expression data to detect differentiated genomic regions for each mRNA subtype with collection C1 (positional gene sets) in MSigDB. We identified the expression of genes in multiple genomic regions were enriched in specific mRNA subtype. In particular, genes annotated as 1p34 and 15q15 were significantly downregulated in C2, while genes annotated as 9p21 were significantly downregulated in C3 (Fig. S19c), supporting that CNV occurring at these genomic regions were associated with gene expression.
Furthermore, patient-derived cell models mimicking the molecular subtypes were established for study on potential therapeutic strategies. C1 subtype patients show favorable prognosis after complete surgical resection alone even without any TKI treatment (Fig. 7d). GIST primary cell culture (GIST-CN16) was established from C2 subtype GIST (Fig. S21). We have evaluated the C2 subtype with a T cell-mediated tumor cell killing assay, which is the fundamental principle of immune checkpoint inhibitor therapy and a valuable tool for immuno-oncology discovery projects. We further co-cultured GIST-CN16 with human peripheral blood mononuclear cells (PBMC), which is more sensitive to the killing of immune cells than C3 subtype cells (GIST-T1) (Fig. 7e). Next, we analyzed the data of patients with GIST from a randomized Phase 2 immunotherapy trial by Singh et al recently47. Among the 6 patients with long term benefit >6 months, 5 had tumors that originated in the small intestine, while the origin of the 6th patient’s tumor was unknown. C3 subtype harbors frequent 9p deletions, which influenced CDKN2A. GIST primary cell culture (GIST-CN2) was established from C3 subtype GIST (94 T, CDKN2A deletion). Strong synergistic effects of ripretinib (KIT inhibitor) and ibrance (CDK4/6 inhibitor) were observed in this C3-derived GIST model (Fig. 7f). C4 subtype contains PDGFRA mutations. GIST-CN10 primary cells established from PDGFRA D842V-mutant, C4 GIST show response to avapritinib, but resistance to imatinib or sunitinib (Fig. 7g). In addition, clinical data with avapritinib confirm evidence of activity in C4 patients with metastatic GISTs driven by PDGFRA mutation (Fig. 7h), representing a validation of previous drug-therapy strategy on PDGFRA–mutant GIST52.
Combined with genomic variations, expression profiles, immune characteristics, and clinical information, we summarized the key features for the 4 mRNA subtypes and proposed hypothesis regarding treatment strategy (Fig. 7i): C1 (genome stable subtype), mainly consists of low or intermediate-risk, gastric GISTs with mostly KIT exon 11 mutations, favorable prognosis after complete surgical resection alone (Fig. 7d); C2 (CD8+ inflamed subtype), mainly consists of the high-risk/metastatic, intestinal GISTs, might ultimately have a potential response to TKI in combination with immunotherapy (Fig. 7e); C3 (immune desert subtype), almost high-risk/metastatic, gastric GISTs with predominantly KIT exon 11 mutations, may not benefit from immunotherapy, but are potential candidates for treatment with CDK4/6 inhibitors and TKIs (Fig. 7f); C4 (PDGFRA-driven subtype), all PDGFRA-mutated GISTs, treatment with PDGFRA inhibitor avapritinib should be considered (Fig. 7g, h).
Discussion
Although GIST is the most common sarcoma, it was not included in TCGA sarcoma project11. One remarkable difference between GISTs and most other sarcomas is the fact that the varying degrees of disease aggressiveness has been well characterized in GISTs11,53. Despite containing similar oncogenic KIT/PDGFRA mutations, most GISTs manifest different clinical behaviors, which vary widely from clinically small tumors to locally invasive, distant metastatic tumors with high mitotic activity. Additional genomic events may influence the variable behavior of these tumors, and advanced tumors are likely to harbor more genomic alterations associated with highly aggressive behavior. This study provides a blueprint of the sequential genetic alterations responsible for clinical progression from low-risk to advanced, lethal GIST. GISTs, even highly aggressive GISTs harbor remarkably few coding mutations, having one of the lowest somatic coding mutation rates observed in a human cancer thus far (Fig. S2). As GIST progresses, the total number of somatic coding mutations, CNVs and SVs increases, indicating that increasing number of alterations accumulate during clonal expansion in GIST progression. For example, copy number alterations in CDKN2A, DEPDC5, RB1 and DMD are more frequent in aggressive GISTs than in low or intermediate-risk GISTs (P < 0.001, Fig. 2a, b). Similarly, chromothripsis and kataegis predominantly occur in aggressive GISTs, demonstrating that the two massive genomic rearrangement events play a major role in shaping the architecture of aggressive GIST genomes as major processes that drive GIST genome evolution. Our comprehensive genomic landscape profiling has validated previous findings with reasonably large cohort, such as that GISTs have low level of TMB, a higher level of CNVs and that these increase with tumor progression54–56.
Most studies only evaluated one metastatic lesion per GIST patient, thus largely underestimated the tumor heterogeneity. Statistical evaluation of the variant allele frequency for coexisting mutations and mutation analyses in spatially and temporally separated GISTs from individual patients demonstrated substantially complex tumor heterogeneity in metastatic GISTs. This may have clinical implications, as the failure of TKI therapies is related to tumor heterogeneity and the constant, adaptive evolution of GISTs in the context of TKI treatment response and resistance.
This is a multi-omics study on a large cohort of KIT/PDGFRA-mutate GISTs demonstrating distinct molecular subtypes. An oncogenic KIT mutation was identified in our GIST cohort, manifesting large cohort. Another major finding is that molecular subtypes are sensitive to different therapeutic strategies. The dramatic success of immune checkpoint blockade therapies in a variety of difficult-to-treat cancers has nearly standardized immunomodulation as an approach for cancer treatment44. In view of the limited efficacy observed with immunotherapy in unselected advanced GIST47,48, there has been a research focus on potential patients through molecular subtyping. The enrichment of CD8+ T cells and T-cell mediated killing assay suggests that patients with the C2 (CD8+ inflamed subtype, intestinal GISTs) might ultimately have a potential response to immunotherapy. The GISTs with the C3 (immune desert subtype) showed frequent CDNK2A aberrations. Because CDKN2A has been associated with responses to CDK4/6 inhibitors51 and strong synergistic effects of KIT inhibitor and CDK4/6 inhibitor were observed in C3-derived GIST model, our study provides potential combination strategy for clinical translation to treat patients with C3 subtype GISTs. This study brings information because CDK4 inhibitor alone has limited efficacy in patients with CDKN2A–deleted GISTs57. There is a high overlap between established risk classification and the C1-C4 classifications. C1 subtype basically describes low-risk and intermediate-risk GIST, and C4 subtype contains PDGFRA mutations. C1 and C4 represent a validation of good prognosis on patients with low-risk or intermediate-risk GIST and a validation of previous drug-therapy strategy on PDGFRA-mutant GIST. Our hypothesis regarding subtype-specific treatment strategies were mainly based on analyses of genomic and transcriptomic data and experimental study; prospectively well-designed clinical trials should be added before we translate our results into clinical practice.
One of the most fundamental traits of cancer cells involves their abilities to sustain proliferation58. Despite the low somatic coding mutation rates, GISTs harbor frequent YLPM1 mutations. Our findings of recurrent genomic alterations, together with functional data, highlight the YLPM1 as a potential GIST-specific tumor suppressor, the inactivation of which contributes to GIST sustaining proliferative signaling. Many questions remain unanswered. YLPM1 is broadly expressed across tissues59. Why are YLPM1-inactivated mutations so frequent in GIST? Nonetheless, it indicates that the YLPM1 plays a more prominent role in GIST pathogenesis. More than 40% of GISTs have YLPM1 inactivation. YLPM1 restoration in YLPM1-inactivated GIST suppresses tumor growth in nude mice (Fig. 3j–l). In addition to its role in cell proliferation, YLPM1 inactivation also promotes oxidative phosphorylation in GIST (Fig. S10g, h). Therefore, molecular interventions targeting YLPM1 deficiency may have therapeutic potential for GISTs.
In summary, our integrative analysis of GIST multi-omics is a valuable tool that provides a complementary and more comprehensive understanding of GIST pathogenesis and offers an opportunity to expedite translation of basic research to more-precise treatment in the clinic.
Methods
Ethical statement
This research complies with all relevant ethical regulations. All samples were collected with institutional review board approval by Shanghai Jiaotong University School of Medicine, Renji Hospital Ethics Committee, with the approved ID 2018-029. All animal experiments were conducted in accordance with the protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the Shanghai Institute of Nutrition and Health, Chinese Academy of Science (with approved ID SIBS-2017-WYX-1).
Specimens and pathological evaluation
113 de-identified tumor specimens and 68 matched normal samples (5 peripheral blood samples and 63 non-cancerous tissues) were collected from 101 GIST patients surgically dissected at Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine. In addition, 4 GIST cell lines were also included in the study. All samples were immediately frozen in liquid nitrogen and stored at −80 °C. Thin slices of snap-frozen, optimal cutting temperature embedded tissue blocks were sent for hematoxylin-eosin (HE) and CD117 staining. After independent review by two professionals (Dr. Yanying Shen and Dr. Daoqiang Tang) with expertise in GIST diagnosis, DNA and RNA were extracted, and all samples were characterized by the presence of KIT or PDGFRA mutation. Risk stratification was evaluated according to the well-established modified NIH clinicopathological criteria. Detailed clinicopathological data are available in Supplementary Data 1. All samples were collected with institutional review board approval (with approved ID 2018-029). Informed written consent was obtained from all human participants. In addition to approval by the local IRB, this study has been reviewed by and is compliant with the Chinese Ministry of Science and Technology for the Review and Approval of Human Genetic Resources.
Cell lines
HEK 293T was obtained from the American Type Culture Collection (ATCC # ACS-4500, RRID: CVCL_4V93) and used for functional studies. The following five GIST cell lines were subjected to WES and/or transcriptome sequencing. GIST-T1 (case 90T, KIT exon 11: V560_Y578del mutation) was generously provided by Dr. Takahiro Taguchi60. The remaining 4 cell lines were developed and kindly provided by Dr. Jonathan Fletcher laboratory at Brigham and Women’s Hospital as previously reported61,62. GIST882 (case 101T, KIT exon 13: K642E mutation) was established from a TKI-naive, metastatic GIST6,61. GIST430/654 (case 89T, KIT exon 11: V560_L576del plus exon 13: V654A) and GIST48 (case 98T, KIT exon 11: V560D plus exon 17: D820A) were established from GIST that had progressed, after initial clinical response, during imatinib therapy62. GISTNS72A (case 105T) is a KIT/PDGFRA wild type GIST cell line harbored a NF1 frameshift mutation (exon 28: A1240fs*8).
HEK 293T and GIST882 were maintained in RPMI 1640 medium (HyClone #SH30027.01) and the remaining cell lines were cultured with IMDM medium (HyClone #SH30228.01). Both two types of medium were supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific #10099141) and 1% penicillin/streptomycin (Thermo Fisher Scientific #15140122). All these cells were cultured at 37 °C in a 5% CO2 humidified atmosphere. All cell lines were routinely tested for microbial contamination (including mycoplasma) and identities have been authenticated by Sanger sequencing.
Primary cell culture
Tumor tissues were obtained from surgical resection of the GIST patient tumor samples. GIST-CN2 primary culture was established from a TKI resistant, metastatic GIST patient (case 94T, male, KIT exon 11: L576P plus exon 13: V654A). GIST-CN10 primary culture was established from a metastatic GIST (male) with PDGFRA D842V mutation. GIST-CN16 primary culture was established from a TKI resistant, metastatic GIST patient (female, KIT exon 9: A502_Y503dup plus exon 17: N822K). Tumor tissues were collected in serum-free IMDM medium and cut into small fragments (5 mm3) with a sterile scalpel or scissors. Add collagenase type I (Gibco # 17100-017) to 50-200 U/mL with 3 mM CaCl2 and incubate at 37 °C for 6 h. Disperse cells by passing through a cell strainer. Wash dispersed cells several times by centrifugation in PBS. Seed cells into culture dish containing IMDM media. After then, the cells were cultured in IMDM containing 10% fetal bovine serum and penicillin-streptomycin mixed solution.
DNA and RNA isolation
High molecular weight genomic DNA from frozen tissues or whole blood samples was isolated using the QIAamp DNA Mini Kit (Qiagen; #51306, Germany) according to the manufacturer’s protocol. RNA was isolated using a TRIzol chloroform method.
Whole exome/genome sequencing
1 μg genomic DNA was randomly fragmented by Covaris to an average size of 250–300 bp (WES) or 200-400 bp (WGS). Fragmented DNAs were tested by agarose gel electrophoresis and purified with the AxyPrep Mag PCR clean up kit. The selected fragments were subjected to end-repair, 3’ adenylated, adapter ligation, PCR amplifying and the PCR products were recovered using the AxyPrep Mag PCR clean up Kit. For WES, libraries were constructed using an Agilent SureSelect Human All Exon V6 kit, followed by 2 × 150-bp paired-end sequencing on the Illumina HiSeq X10 platform. For WGS, the double-stranded PCR products were heat denatured and circularized by the splint oligo sequence. Single-strand circle DNA (ssCir DNA) was formatted as the final library and qualified by an Agilent Technologies 2100 bioanalyzer and ABI StepOnePlus Real-Time PCR System. The qualified libraries were sent for 2 × 100-bp paired-end sequencing on the BGISEQ-500 platform (BGI-Wuhan, China).
Bioinformatics analysis of DNA sequencing data
The raw sequencing data was processed with the following steps by SOAPnuke (https://github.com/BGI-flexlab/SOAPnuke) (1) reads containing sequencing adapters were removed; (2) reads having more than 50% bases with a base quality <5 were removed; (3) reads with an unknown base (‘N’ base) ratio more than 10% were removed. The Sentieon Genomics pipeline (https://www.sentieon.com/) was used to align the paired-end reads to the UCSC hg19 human reference genome and subsequent preprocessing, including PCR duplication marking, base quality score recalibration and local realignment of the aligned reads. BAM-matcher was used to rapidly determine whether the two bam files represented samples from the same biological source15.
Variant calling
To achieve a better trade-off between sensitivity and specificity, six callers were used to identify somatically acquired single-nucleotide variants (SNVs) and small insertions or deletions (indels) in paired samples: MuSE (v1.0, default parameters)63, Strelka2 (v2.99, default parameters)64, MuTect2 (GATK v4.1.0.0, default parameters)65, SomaticSniper (v1.0.5.0, -q 1 -Q 40 -L -G)66 and Lancet (v1.07, default parameters)67 for single nucleotide variants (SNVs), while Strelka2 (v2.99, default parameters), MuTect2 (GATK v4.1.0.0, default parameters), Lancet (v1.07, default parameters) and SvABA (v0.2.1, default parameters)68 were used for small indels. Mutations detected by two or more algorithms were retained. The mean value of variant allele frequency from several algorithms was calculated as the final variant allele frequency (VAF). To extract high-confidence somatic variants, the following SNVs and indels were eliminated: (i) the variants reported in dbSNP (v150) but not in COSMIC (v88); (ii) the variants reported in 1000 Genome Project April 2015 release and The Exome Aggregation Consortium (ExAC) database release 0.3 with a frequency of > 0.01; and (iii) the variants with a VAF < 0.05.
All SNVs and indels were subsequently annotated by ANNOVAR (v20170716)69.
Variant validation by Sanger sequencing
To validate variants identified by WES and WGS, we selected 403 SNVs/Indels (292 randomly selected variants according to different VAF levels, 106 KIT variants and 6 PDGFRA variants) and recurrent SVs of LRP1B, DMD and CDKN2A and designed specific PCR primers by NCBI Primer-BLAST. PCR reactions were performed on a ProFlex 96-Well PCR System (Applied Biosystems) using VazymeTM 2×Taq Master Mix (Vazyme, #P112-02), followed by direct sequencing on an ABI 3730 DNA Sequence Analyzer. The Chromatograms of paired tumor and normal samples were aligned by SnapGene 4.2.4. A somatic variant was considered validated if the mutant peaks were only existed in tumor samples. Finally, Sanger sequencing successfully confirmed 336 variants out of 351 selected SNVs/indels with variant allele frequencies ranging from 5% to 100%, indicating a high accuracy (95.72%) of our variant calling pipeline (Supplementary Data 13d). Fourteen variants were not verified due to low sensitivity of Sanger sequencing (limitation for detected allele frequency less than 20%). These results show that our sequencing and mutation calling pipeline is robust.
Mutational signature analysis
To identify mutational signature in the WGS data of 19 GISTs, we first generated the mutational catalogue (96 trinucleotide mutation contexts) using SigProfilerMatrixGenerator (v1.1)70 and performed the BayesNMF algorithm28 to extract de novo signatures. We ran the Bayesian NMF 200 times with the hyperparameter for the inverse gamma prior setting to 10 (a = 10) and the iterations were terminated when the tolerance for convergence was <10−7. All independent runs in our data set extracted 2 signatures. These 2 signatures were then compared to the known mutational processes from the COSMIC v2 signature database by calculating the cosine similarity as following71:
| 1 |
where K is the number of mutation types (K = 96), Ak and Bk are the k-th components of signature A and signature B.
Copy number variation analysis
Copy number variations were estimated by FACETS (v0.5.11)72. Snp-pileup (-q15 -Q20 -P100 -r25, 0) was run to create an input for each paired sample which calculated the reference and variant read counts of common SNPs. A critical value (cval) of 150 was used to run FACETS with the recommended parameters. Then, the broad and focal CNVs were identified by GISTIC2.0 (v2.0.22)73 with the following parameters: amplification threshold = 0.1; deletion threshold = 0.1; arm level peel off = 1; join segment size = 8; gene GISTIC = 1; confidence level = 0.99; broad length cutoff = 0.8; and remove the X-chromosome = 0. We also removed regions corresponding to germline copy-number alterations generated from TCGA when performing GISTIC analysis. Chromosome arms were labeled as ‘altered’ in each sample if the log2 copy ratio > 0.1 or < −0.1. Arm-level CNV differences between different cluster or risk groups were calculated by chi-squared test for amplifications and deletions, respectively. Copy numbers of DMD were estimated manually from WES and WGS data using the plots of read depths in both tumor and normal samples.
Loss of heterozygosity (LOH) calls were determined in paired samples using FACETS (v0.5.11, cval = 150)72. Only regions of autosomal chromosomes detected by FACETS that had a minor allele copy number equal to zero and major copy number greater than zero were considered to have undergone LOH.
The CNV matrix (48 contexts) was generated using SigProfilerMatrixGenerator70 from 78 GISTs. The extraction of CNV signatures was analyzed using SigProfilerExtractor74 with parameters set to 1000 iterations, a minimum number of 2 signatures and a maximum number of 30 signatures. The best number of CNV signatures selected was 6. These de novo signatures of CNV were subsequently decomposed into COSMIC (v3.3) signatures and/or novel signatures.
Structural variation analysis
Structural variations (SVs) were detected using Manta (v1.5.0)33 (https://github.com/Genomon-Project/GenomonSV) with default parameters. Firstly, SVs that did not have a “PASS” filter status were discarded. Then, the following filters were used to obtain high-confidence SVs:
-
(i)
for WGS samples, SVs with either the altered spanning-paired-read counts or the altered split-read counts of tumor samples equal to 0;
-
(ii)
for WES samples, SVs that meet any of the following filters were excluded: either the altered spanning-paired-read counts or the altered split-read counts of normal samples more than 0; the ratio of altered spanning-paired-read counts to refer spanning-paired-read counts less than or equal to 0.1; the ratio of altered split-read counts to refer split-read counts less than or equal to 0.1.
Inference of chromothripsis and kataegis
To identify and visualize chromothripsis-like patterns in the cancer genomes, the copy number (CN) and SV data were used as input for ShatterSeek (v0.4) (https://github.com/parklab/ShatterSeek) with default parameters. We applied the same criteria as previous studies to define a positive call34. To avoid missing chromothripsis due to few SVs in GIST, we manually inspected the number of switches between copy-number states (2 and 3 status) for each chromosome. An open-source tool named SeqKat (v0.0.8)75 (https://github.com/cran/SeqKat) was used to predict kataegis regions from WGS samples with default parameters (minimum hypermutation score cutoff = 5, maximum inter-mutation distance cutoff = 3.2 and minimum SNV count cutoff = 4) base on SNVs. Both the hypermutation score and an APOBEC mediated kataegic score along with the start and end position of each detected event were determined by SeqKat. Then KataegisPortal (https://github.com/MeichunCai/KataegisPortal) was used to visualize the kataegis events. The occurrence of kataegis was also inferred by SigProfilerClusters (v.1.0.1) with default parameters76.
Clonal analysis
ABSOLUTE (v1.0.6)77 was used to estimate the purity and ploidy of paired samples. The clonal status of somatic mutations in coding regions was determined by assessing the cancer cell fraction (CCF)40. The CCF of each mutation was estimated by the VAF, tumor purity (p), local copy number of the tumor sample (CNtumor) and normal sample (CNnormal) using the following formula:
| 2 |
Specifically, the local copy number of the normal sample (CNnormal) was assumed to be 2 and the sex chromosomes were excluded from this analysis. For each mutation, alternative reads a, and total depth N could be estimated using a binomial distribution P(CCF) = binom (a|N, VAF(CCF)). Then, CCF values can be calculated over a uniform grid of 100 CCF values (0.01, 1) and subsequently normalized to obtain a posterior distribution. Mutations were defined as clone events if the 95% confidence interval overlapped 1; otherwise, the mutations were determined to be subclone events40.
Tumor heterogeneity analysis
The tumor heterogeneity levels were estimated based on SNVs and indels profiling for each of the four patients with multi-lesion sequencing. For each pair of lesions from a patient, the pairwise tumor heterogeneity level was calculated as the proportion of private mutations in total mutations of this paired lesions. Finally, we defined the tumor heterogeneity index for each patient as the mean of these pairwise tumor heterogeneity levels for all possible pairs of lesions from a patient.
Phylogenetic tree analysis
For 4 patients with multi-regions, sequences within 20 bp upstream and downstream of the somatic non-synonymous mutations (SNVs and indels) were extracted to construct phylogenetic tree of each patient based on maximum likelihood by MEGA software (version 6.06)78. All the phylogenetic trees were further optimized and the potential driver genes were labeled manually using Adobe Illustrator.
Transcriptome sequencing and data analysis
Approximately 1 μg of total RNA was used for library construction. Oligo(dT)-attached magnetic beads were used to purified mRNA. Purified mRNA was fragmented into small pieces with fragment buffer at appropriate temperature. Then First-strand cDNA was generated using random hexamer-primed reverse transcription, followed by a second-strand cDNA synthesis. Subsequently, A-Tailing Mix and RNA Index Adapters were added by incubation for end repair. The cDNA fragments obtained from the previous step were amplified by PCR, and the products were purified by Ampure XP Beads, and then dissolved in EB solution. The products were validated on an Agilent Technologies 2100 bioanalyzer for quality control. The double stranded PCR products from the previous step were heated denatured and circularized by the splint oligo sequence to obtain the final library. Single strand circle DNA (ssCir DNA) was formatted as the final library. The final library was amplified to generate DNA nanoballs (DNBs) that had more than 300 copies of one molecular,
DNBs were loaded into the patterned nanoarray and 100 bp paired-end reads were generated on the BGISEQ-500 platform (BGI-Shenzhen, China).
In the raw RNA sequencing data, rRNA reads were first removed and then reads were discarded by SOAPnuke (https://github.com/BGI-flexlab/SOAPnuke) for 1) reads containing sequencing adapters; 2) low-quality reads (>50% bases with quality <5); 3) reads with more than 10% unknown bases. The clean reads were mapped to the UCSC human transcriptome (hg19) by Bowtie279 with parameters “-q --phred64 --sensitive --dpad 0 --gbar 99999999 --mp 1,1 --np 1 --score-min L, 0, −0.1 -I 1 -X 1000 --no-mixed --no-discordant -p 1 -k 200”. The fragments Per Kilobase of exon model per Million mapped fragments (FPKM) value of all genes and isoforms were estimated by RSEM (v1.2.3) with default parameters.
For YLPM1 knockout/restored and control samples, RNA sequencing were performed on a BGI-500/MGISEQ-2000 sequencer (BGI-Wuhan, China). All reads that passed quality metrics were mapped to the UCSC hg19 human genome.
Fusion gene discovery and validation
Since the normal counterparts for GIST (interstitial cells of Cajal, ICC) are not available, RNA-based fusion detection is prone to false positives80. Fusion genes and their respective fusion points were detected by following algorithms: FusionCatcher (v.1.10)81, Arriba (v1.1.0) (https://github.com/suhrig/arriba), STAR-Fuison (v1.6.0)82 and SOAPfuse (v1.18)83. Fusion genes called from the 116 tumor samples were compared to those from 5 adjacent normal tissues to remove germline fusion alterations. Then, we only retained fusion events detected by two or more algorithms. In addition, genes annotated as probably false positive by FusionCatcher were also excluded. The resulting fusion genes were further filtered (Supplementary Data 14).
Correlations of mRNA expression and copy number
Both the expression and quantitative DNA copy number data were filtered by restricting to common samples as well as genes in 22 autosomal chromosomes whose copy number values and gene expression values had non-zero variance. For each gene, the gene-level DNA copy number values were produced by the CNTools (v0.9.5) (http://bioconductor.org/packages/3.12/bioc/html/CNTools.html) which yielded gene expression and quantitative DNA copy number values. Gene-level Pearson correlation coefficients were computed and plotted using MVisAGe R package for the log-transformed FPKM values and quantitative DNA copy number values. Smoothed Pearson correlation coefficients were plotted to identify chromosomal regions for which DNA copy number was most highly correlated with gene expression. The smoothing parameter was used to create plots of smoothed Pearson correlation coefficients over larger genomic regions and unsmoothing parameter was used for focal regions based on manual review32.
mRNA expression cluster analysis
The molecular subtypes were obtained by unsupervised hierarchical clustering of mRNA expression for 107 tumor samples. We first calculated the median absolute deviation (MAD) for each gene across all tumors to determine the optimal number of genes for clustering. Then we tested the clustering results by choosing genes based on the MAD from the top 5% to the top 50% and finally chose the MAD top 1500 genes to perform clustering. Expression values were log2-transformed by log2(FPKM + 1) for clustering using ConsensusClusterPlus (v1.42.0) on R (v3.4) with 1000 permutations84. Options included maxK = 10, pItem = 0.8, pFeature = 1, clusterAlg = “hc,” distance = “pearson”, and a seed value of 1262118388.71279.
The optimal number of clusters was determined from the cumulative density function (CDF), which plots the corresponding empirical cumulative distribution, defined over a range between 0 and 1, and from a calculation of the proportion increase in the area under the CDF curve. The preferred clustering result was determined when any further increase in cluster number (k) did not lead to a corresponding remarkable increase in the CDF area. We also used silhouette width to identify samples that most closely represent of these molecular subtypes by the R package ‘cluster’85. The differentially expressed genes (one subtype vs any other subtypes) were detected by DEseq2 method (Love et al., 2014) (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) with threshold of the log2 fold changes >=1 and the adjust-P-value < 0.05. A total of 520 genes in top MAD 1500 were selected.
Additionally, the expression data from two independent cohorts (Japanese cohort and CINSARC cohort) were downloaded to validate these molecular subtypes42,43. The expression of genes with multiple probes in this dataset was determined from the median value of these probes. Since the expression profiles from validated cohorts were generated by different microarray, we used ComBat (http://www.bu.edu/jlab/wp-assets/ComBat) to remove the batch effects and tested the clustering results by choosing genes based on the SD from the top 1000 to the top 7000. Finally, we chose the SD top 1100 genes to perform clustering with the same scheme.
Cytolytic activity and immune cell signatures
The total immune components for each sample were analyzed using the ESTIMATE (v1.0.13) based on the gene expression data86. Cytolytic activity (CYT) was calculated as the geometric mean normalized expression of two key cytolytic effectors, granzyme A (GZMA) and perforin (PRF1)50. Then, we used CIBERSORT (v1.0.5) to quantify the fractions of infiltrating immune cells with 1000 permutations49 and an externally validated leukocyte gene signature matrix (LM22). The LM22 signature was generated from 547 genes to distinguish 22 cell phenotypes, including 7 T cell types, B cells, macrophages, monocytes, and NK cells49. The immune infiltration score was also estimated using the online platform of Tumor Immune Estimation Resource 2.0 (TIMER2.0, http://timer.cistrome.org/)87, which integrated multiple algorithms of immune infiltration estimations including TIMER, xCell, QUANTISEQ, MCP-counter, EPIC, CIBERSORT-ABS, and CIBERSORT49.
Assessment of telomere content
The telomere content was determined using TelomereHunter37 with default parameters on DNA bam files from 19 paired WGS-sequenced patients and 49 paired WES-sequenced patients. The sequencing reads of four most common telomeric repeat types (TTAGGG, TCAGGG, TGAGGG, and TTGGGG) were quantified, and the log2 ratio of telomere content between tumor and normal samples was calculated. We also performed telomere length (TL) analysis on 19 paired WGS-sequenced patients using Telseq88.
Establishment of isogenic GIST-T1 YLPM1 knockout subline
Short guide RNAs (sgRNAs) targeting human YLPM1 was ligated into pSpCas9(BB)−2A-Puro vector (PX459 from Addgene, #62988, RRID: Addgene_62988). Thermo Fisher’s Neon transfection system (MPK5000) was used to transfer the sgRNA-pSpCas9 plasmid into GIST-T1 cells, and then GIST-T1 YLPM1 knockout subline was established with single-cell-cloning techniques. Specific steps are as follows: (a) GIST-T1 cells with 90-100% confluency were prepared into single cell suspension by trypsinization and counted; (b) 1.5×10^6 cells were washed twice with 500 μL PBS; (c) Cells were resuspended with suspension buffer (Thermo, #MPK1096), then 10 μl of sgRNA-pSpCas9 plasmid (total 1ug) was added; (d) 100 μL of the cell suspension in step (c) were taken for electroporation transfection under the pulse condition of 1500 mV, 20 ms, and the transfected cells were plated evenly into a 6 cm dish containing IMDM medium; (e) After the cells were confluent, the cells were re-plated into a 96-well plate using the infinite dilution method. Cells were passaged again after confluency and DNA was isolated to confirm the oncogenic KIT mutation.
YLPM1 restoration
Lentivirus preparations were produced by co-transfecting lentiviral YLPM1 construct, and helper virus packaging plasmid pCMVΔ8.9 and pHCMV-VSV-G into 293 T cells using Lipofectamine 3000 (Invitrogen, #L3000015). Lentivirus was harvested at 24, 36, 48 and 60 h post-transfection and stored at −80 °C. GIST cell transductions were carried out overnight with 8 μg/ml polybrene (Sigma; #107689), and transduced cells were selected with 2 μg/ml puromycin (Millipore; #540411).
CRISPR knockout
Short guide RNAs (sgRNAs) targeting human YLPM1 were designed on the basis of the Optimized CRISPR Design web tool (http://crispr.mit.edu/) and cloned into lentiCRISPRv2 vector (Addgene, plasmid #52961, RRID: Addgene 52961) individually. The YLPM1 sgRNA sequences were shown in Supplementary Data 13,c. GIST-T1 cells were infected for 16 hours in the supernatant containing 8 μg/ml polybrene, and then treated with 2 μg/ml puromycin one day after infection. Transformant pools were confirmed by genomic Sanger sequencing.
Gene set enrichment analysis (GSEA)
To discriminate major biologic characteristics and processes in defined four mRNA subtypes, GSEA (v4.0.1, RRID:SCR 003199) was applied separately to each mRNA subtype (one subtype vs. the rest) in the Molecular Signature Database (MSigDB v7.0, Hallmark) downloaded from the GSEA website (http://www.broadinstitute.org/gsea/). We also applied GSEA on mRNA expression data in MSigDB positional gene sets (MSigDB v7.0, collection C1) to evaluate the effect of CNVs and gene expression among four mRNA subtypes. Significantly enriched gene sets were defined based on an FDR < 0.25 and absolute normalized enrichment score (NES) > 1.0.
For YLPM1 knockout/restored and control samples, the differentially expressed genes were subjected to gene set enrichment analysis (GSEA). GSEA were carried out using the pre-ranked mode using log2 fold-change values with default settings.
Real-time quantitative RT-PCR and quantitative PCR
To verify the RNA-seq results after knocking out YLPM1 in GIST-T1, the RNA was reverse-transcribed into cDNA using the TransScript First-Strand cDNA Synthesis SuperMix (TransGen; #AT301); indicated primers were listed in Supplementary Data 13,a. Genomic DNA of 56 T and 56 N isolated as previously mentioned were used to verify YLPM1 homozygous deletion. For quantitative of the deletion range of YLPM1, YLPM1 exons 1/4/5 and the last exon of nearby FCF1 gene were amplified and GAPDH was used as reference. qPCR was performed for target-gene-expression analysis or exon detection of YLPM1 using the Vazyme™ ChamQ Universal SYBR qPCR Master Mix (Vazyme; #Q711-02); indicated primers were listed in Supplementary Data 13,b. Amplification accuracy was verified by melting curve analysis. Relative mRNA expression was normalized to GAPDH expression as an internal amplification control. Reactions were carried out on a CFX ConnectTM Real-time PCR Detection System (Bio‐Rad Laboratories, Inc). The CT values (the cycle at which the change in fluorescence for the SYBR dye passes a significance threshold) were used for data normalization. ΔCT values (CTCtrl-CTYLPM1) of triplicate samples were used to calculate copy number changes relative to control DNA using Microsoft Excel. Bar graphs show mean ± s.d. of three technical replicates for each sample.
Cell viability and imatinib sensitivity assays
Viability studies were performed using the CellTiter-Glo luminescent assay (Promega, Madison, WI, G7572). Cells were plated at 6000 (restore) or 10,000 (knockout) cells per well in a 96-well flat-bottomed plate. For imatinib sensitivity assays, cells were treated with 5 gradient concentrations of imatinib. Luminescence was measured using a BioTek Gen5TM Microplate Readers (BioTek, Winooski, VT, #H1210-018), 72‐96 hours after drug treatment according to the manufacturer’s instructions.
Western blotting
Whole cell lysates from cell lines were prepared using lysis buffer (1% NP-40, 50 mM Tris-HCl pH 8.0, 100 mM sodium fluoride, 30 mM sodium pyrophosphate, 2 mM sodium molybdate, 5 mM EDTA, 2 mM sodium orthovanadate) containing protease inhibitors (10 μg/mL aprotinin, 10 μg/mL leupeptin and 1 mM phenylmethylsulfonyl fluoride). The lysates were then rocked overnight at 4oC and cleared by centrifugation at 18,000 g for 30 min at 4oC. The lysate protein concentrations were determined using a Quick Start™ Bradford 1×Dye Reagent (Bio-Rad, #5000205). Electrophoresis and western blotting were performed using standard techniques. The following primary antibodies were incubated at 4 °C overnight: PCNA (Santa Cruz, #sc-56, 1:500, RRID:AB_628110), p-KITY721 (Cell Signaling Technologies, #3391, 1:1000, RRID:AB_2131153), KIT (Agilent, #R7145, 1:1000, RRID:AB_2131465), p-MAPKThr202/Tyr204 (Cell Signaling Technology, #9101, 1:1000, RRID:AB_331646), MAPK (Cell Signaling Technology, #9102, 1:1000, RRID:AB_330744), p-AKTSer473 (Cell Signaling Technology, #9271, 1:1000, RRID:AB_329825), AKT (Cell Signaling Technology, #9272, 1:1000, RRID:AB_329827) and GAPDH (Sigma, #G8795, 1:1000, RRID:AB_1078991), YLPM1(Novus Biologicals, #NBP2-22326, 1:2000). Bands were detected using HRP-labeled secondary antibodies and the hybridization signals were detected by chemiluminescence (Immobilon Western, Millipore Corporation, MA) and captured using an Amersham Imager 600 imagers (GE Healthcare; #29083461). Relative protein quantification was performed with Image Quant TL 8.1 (GE Healthcare, RRID:SCR_018374) software.
Immunohistochemistry
Freshly collected tumors were fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 5-μm sections. To further confirm the YLPM1 inactivation frequency in GISTs, 2 tissue microarrays (TMAs) including 278 GIST samples were constructed by Suzhou Xinxin Biotechnology Co., Ltd (Xinxin Biotechnology Co, Suzhou, China)89. Paraffin-embedded GIST tissue blocks were stained with hematoxylin-eosin to confirm the diagnoses, and then marked at fixed points with most typical histological characteristics under a microscope. Three-micron-thick sections were cut from the recipient blocks and transferred to glass slides with an adhesive tape transfer system for ultraviolet cross linkage. Immunohistochemical staining was conducted with the BenchMark XT automated slide-staining system (Roche, Basel, Switzerland). Antigen retrieval and primary antibody incubation conditions were set at 95 °C for 30 minutes and at 37 °C for 30 minutes, respectively. We used primary antibodies against CD117 (ready for use; Maixin Bio Co., Ltd., Fuzhou, China), SDHB (ready for use; Maixin Bio Co., Ltd., Fuzhou, China), YLPM1 (1:400; Novus Biologicals, #NBP2-22326) and CD8 (ready for use; Maixin Bio Co., Ltd., Fuzhou, China). Slides were scanned and photographed with a Motic VM Digital Slide System (Motic China Group Co., Ltd). The staining intensity and percentage of positive cells were recorded by two pathologists and a consensus score was obtained for each slide. The proportion of CD8 positive cells was quantitatively evaluated using software for digital bioimage analysis (QuPath) (RRID: SCR_018257)90.
Soft agar assay and colony formation assay
Six-well plates were first layered with 0.6% bottom Noble agar (BD Difco ™, #214220) containing RPMI1640 medium (Hyclone, #SH30197.03) with 10% FBS (Gibco, #10270-106), 1% L-Glutamine (Gibco, #25030-081) and 1% penicillin/streptomycin. GIST-T1 cells (10,000 cells per well) were transduced with control or YLPM1 lentivirus and seeded in 0.35% top agar. Cells were allowed to grow for 4 weeks and then stained with 1 ml of 1 mg/ml methyl thiazol tetrazolium (MTT, Sigma #M5655) for 3 hours. Colonies were counted by ImageJ software (National Institutes of Health, USA). All the assays were performed in triplicate wells, with the entire study replicated three times.
Colony formation assays were conducted by seeding GIST-T1 or YLPM1-KO isogenic GIST-T1 cells (500 cells per well) transduced with the Ctrl, YLPM1 sgRNA or full length YLPM1 lentivirus into six-well plates and allowed to grow for 3 weeks. Then, the cells were fixed with 4% paraformaldehyde for 10 minutes and stained with crystal violet solution (Shanghai Sangon Biotechnology Co, # E607309-0100) for 15 mins. After rinse with distilled water, the colony images were obtained using a scanner (Microtek, TMA 1600III) and counted by ImageJ software (National Institutes of Health, USA). All assays were carried out in in triplicate wells, with the entire study replicated three times.
Xenograft tumor model
The animal experiments were approved by Institutional Animal Care and Use Committee of the Shanghai Institute of Nutrition and Health, Chinese Academy of Science (with.approved ID SIBS-2017-WYX-1). All the mice were fed with standard laboratory diet and maintained in a pathogen-free environment (20–26 °C, 40–70% humidity) on a 12-h light/12-h dark cycle with food and water supplied throughout the experiment period. GIST cells (2×106) in PBS/Matrigel mixture were injected subcutaneously into 6-week-old male BALB/c nude mice. The resulting tumors were measured every three days. Tumor volumes were calculated using the formula: tumor volume = length× width× width/2. Once the largest tumor diameter reached the maximal tumor diameter allowed under our institutional protocol, all mice were killed and tumors were collected, weighed and photographed. The maximal tumor diameter allowed by Institutional Animal Care and Use Committee is 2.0 cm.
Bisulfite genomic sequencing
We performed bisulfite conversion on 0.5 μg of genomic DNA from each sample using the ZYMO EZ DNA Methylation Gold Kit (Zymo Research, #D5005) according to the manufacturer’s instructions. The CpG islands associated with the YLPM1 locus were predicted with UCSC Genome Browser. A 382 bp fragment containing 48 CpG sites was amplified from bisulfite-treated DNA using the YLPM1-Bi-F3 primer 5′-GGAAGATGGTAATTACGAGTCGTT-3′ and YLPM1-Bi-R3 primer 5′- GAACCCCGACGAAACCTCAAA-3′. The PCR conditions were as follows: 95 °C denaturation for 5 min on initial cycle, followed by 40 cycles of 10 s denaturation at 95°C and 30 s annealing at 48 °C. PCR products were subcloned into the pMD19-T vector (Takara, #6031), and 10 clones from each PCR reaction were sequenced to determine the methylated status of cytosines at the 48 CpG sites.
The oxygen consumption rate (OCR) assay
A day before the assay, GIST-T1 cells were seeded at 20000 cells per well in a seahorse XFe24 cell culture microplate. The cells were allowed to adhere to the plate for 24 h in a 37 °C humidified incubator with 5% CO2. After 24 h incubation, cells were washed with XF-based medium DMEM supplemented with 10 mM glucose, 2 mM glutamine and 1 mM pyruvate (adjusted to pH7.4) and maintained at 37 °C in a non-CO2 incubator for 1 h to allow for pre-equilibration with the XF Assay Medium. Oxygen consumption rate was measured using XFe24 (Seahorse; Agilent) in the presence or absence of the following reagents: 1 μM oligomycin, 1 μM FCCP (MCE) and 0.5 μM Rotenone/Antimycin (MCE). The mitochondrial respiration was analyzed using Agilent Seahorse Wave software. Afterward, cells were stained with DAPI (1:1000), and washed twice with PBS for normalization.
T cell-mediated tumor cell killing assay
To analyze T cell-mediated tumor cell killing, human T cells were activated by culturing human PBMC in ImmunoCult-XF T cell expansion medium (10981, Stemcell) with ImmunoCult human CD3/CD28 T cell activator (10971, Stemcell) and IL-2 (10 ng/mL, 78036, Stemcell) for 7 days. Then adhered GIST-CN16 or GIST-T1 cells were co-cultured with activated human T cells at a ratio of 1:5 or 1:10 for 72 h. T cells and cell debris were washed with PBS, and living cells were measured by CellTiter-Glo luminescent assay (Promega, Madison, WI, G7572).
Statistical analysis
Wilcoxon rank-sum test and Kruskal-Wallis test were used to calculate the comparisons between two groups or among multi-groups in continuous variable, respectively. Fisher’s exact and Pearson’s chi-squared test were used for comparison of response rate difference. Correlations between two groups were analyzed by Pearson’s correlation. All statistical analyses were done using standard R packages (R, v3.4.0).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The authors thank Dr. Jonathan Fletcher at Brigham and Women’s Hospital/Harvard Medical School for GIST cell lines, Dr. Takahiro Taguchi for the GIST-T1 cell line. We also thank Zhonghui Weng and Kai Wang from the animal facility of Shanghai Institute of Nutrition and Health for animal care during the coronavirus pandemic. This work was supported by grants from the National Natural Science Foundation of China to Yuexiang Wang (82072974, 82120108020, 81572642), to Hui Cao (82072669) and to Lin Tu (81702303), from the National Key Research and Development Program of China to Yuexiang Wang (2023YFE0117900), from the Innovation Program of Shanghai Science and technology committee to Yuexiang Wang (20Z11900300), from the Guangdong Provincial Key Laboratory of Human Disease Genomics to Kui Wu (2020B1212070028), and from the Postdoctoral Fellowship Program of CPSF to Simin Wang (GZB20240790).
Author contributions
F.-F.X., S-M.W., H.C., K.W., and Y.-X.W. conceived and designed the research. F.-F.X., S.-Z.L., D.-B.L., X.-J.L., X.-X.L., F.-J.J., Y.-Z.P., C.-L.Z., Y.-M.C., Y.-X.L., F.-F.X., H.H., T.H., K.L., X.-F.Z., W.-B.B., X.-N.J., S.-M.W., and Y.-X.W. performed experiments, bioinformatics investigation and analyzed the data. Y.-Y.S., and D.-Q.T. reviewed the histopathologic diagnoses. H.C., Y.-Y.S., M.W., L.T., L.-X.Y., C.Z., W.-Y.Z., and X.-L.M. provided samples and gathered detailed clinical information. D.-B.L., H.C., Y.H., and K.W. provided scientific advice and helpful comments into the project. F.-F.X., S.-Z.L, X.-J.L., X.-X.L., F.-J.J., S.-M.W., and Y.-X.W. prepared figures and tables. F.-F.X., S.-Z.L, D.-B.L, X.-J.L., X.-X.L., F.-J.J., S.-M.W., and Y.-X.W. wrote the manuscript. All authors read and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The sequencing datasets (raw WGS, WES, and WTS data) generated in this study have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA005970) database under accession code HRA005970. The expression data of CINSARC cohort and Japanese cohort was downloaded from the Array Express accession: E-MTAB-373 and the NCBI database under accession GSE136755. The somatic mutation datasets (MAF file) of SARC analyzed in this study were downloaded from the GDC Portal [https://gdc.cancer.gov/about-data/publications/sarc_2017]. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Feifei Xie, Shuzhen Luo, Dongbing Liu, Xiaojing Lu, Ming Wang, Xiaoxiao Liu, Fujian Jia.
Contributor Information
Simin Wang, Email: wangsm@sinh.ac.cn.
Hui Cao, Email: caohuishcn@hotmail.com.
Kui Wu, Email: wukui@genomics.cn.
Yuexiang Wang, Email: yxwang76@sibs.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53821-1.
References
- 1.Hirota, S. et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science279, 577–580 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Heinrich, M. C. et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science299, 708–710 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Corless, C. L., Barnett, C. M. & Heinrich, M. C. Gastrointestinal stromal tumours: origin and molecular oncology. Nat. Rev. Cancer11, 865–878 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Blay, J. Y., Kang, Y. K., Nishida, T. & von Mehren, M. Gastrointestinal stromal tumours. Nat. Rev. Dis. Prim.7, 22 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Abraham, S. C., Krasinskas, A. M., Hofstetter, W. L., Swisher, S. G. & Wu, T. T. Seedling” mesenchymal tumors are common incidental tumors of the esophagogastric junction. Am. J. Surg. Pathol.31, 1629–1635 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Pang, Y. et al. Mutational inactivation of mTORC1 repressor gene DEPDC5 in human gastrointestinal stromal tumors. Proc. Natl Acad. Sci. USA116, 22746–22753 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, Y. et al. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat. Genet.46, 601–606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaefer, I. M. et al. MAX inactivation is an early event in GIST development that regulates p16 and cell proliferation. Nat. Commun.8, 14674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, K. K. et al. SETD2 histone modifier loss in aggressive GI stromal tumours. Gut65, 1960–1972 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Killian, J. K. et al. Recurrent epimutation of SDHC in gastrointestinal stromal tumors. Sci. Transl. Med.6, 268ra177 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell171, 950–965 e28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith, B. D. et al. Ripretinib (DCC-2618) is a switch control kinase inhibitor of a broad spectrum of oncogenic and drug-resistant KIT and PDGFRA variants. Cancer Cell35, 738–751 e9 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Grunewald, S. et al. Resistance to avapritinib in PDGFRA-driven GIST is caused by secondary mutations in the PDGFRA kinase domain. Cancer Discov.11, 108–125 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Joensuu, H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum. Pathol.39, 1411–1419 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Wang, P. P., Parker, W. T., Branford, S. & Schreiber, A. W. BAM-matcher: a tool for rapid NGS sample matching. Bioinformatics32, 2699–2701 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson, J. et al. NF1-associated gastrointestinal stromal tumors have unique clinical, phenotypic, and genotypic characteristics. Am. J. Surg. Pathol.29, 1170–1176 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Italiano, A. et al. SDHA loss of function mutations in a subset of young adult wild-type gastrointestinal stromal tumors. BMC Cancer12, 408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich, M. C. et al. Genomic aberrations in cell cycle genes predict progression of KIT-mutant GISTs. Clin. Sarcoma Res.9, 3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature505, 495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sondka, Z. et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer18, 696–705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Rifai, W., Sarlomo-Rikala, M., Andersson, L. C., Knuutila, S. & Miettinen, M. DNA sequence copy number changes in gastrointestinal stromal tumors: tumor progression and prognostic significance. Cancer Res.60, 3899–3903 (2000). [PubMed] [Google Scholar]
- 23.Armstrong, L. et al. A role for nucleoprotein Zap3 in the reduction of telomerase activity during embryonic stem cell differentiation. Mech. Dev.121, 1509–1522 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Hoadley, K. A. et al. Cell-of-Origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell173, 291–304.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo, Y. et al. PKClambda/iota loss induces autophagy, oxidative phosphorylation, and NRF2 to promote liver cancer progression. Cancer Cell38, 247–262 e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramchandani, D. et al. Copper depletion modulates mitochondrial oxidative phosphorylation to impair triple negative breast cancer metastasis. Nat. Commun.12, 7311 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molina, J. R. et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med.24, 1036–1046 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Kim, J. et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet.48, 600–606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steele, C. D. et al. Signatures of copy number alterations in human cancer. Nature606, 984–991 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Rifai, W., Sarlomo-Rikala, M., Miettinen, M., Knuutila, S. & Andersson, L. C. DNA copy number losses in chromosome 14: an early change in gastrointestinal stromal tumors. Cancer Res.56, 3230–3233 (1996). [PubMed] [Google Scholar]
- 32.Walter, V., Du, Y., Danilova, L., Hayward, M. C. & Hayes, D. N. MVisAGe identifies concordant and discordant genomic alterations of driver genes in squamous tumors. Cancer Res.78, 3375–3385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, X. et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics32, 1220–1222 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Korbel, J. O. & Campbell, P. J. Criteria for inference of chromothripsis in cancer genomes. Cell152, 1226–1236 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Maciejowski, J., Li, Y., Bosco, N., Campbell, P. J. & de Lange, T. Chromothripsis and kataegis induced by telomere crisis. Cell163, 1641–1654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dilley, R. L. & Greenberg, R. A. ALTernative telomere maintenance and cancer. Trends Cancer1, 145–156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feuerbach, L. et al. TelomereHunter - in silico estimation of telomere content and composition from cancer genomes. BMC Bioinform.20, 272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barthel, F. P. et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet.49, 349–357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voronina, N. et al. The landscape of chromothripsis across adult cancer types. Nat. Commun.11, 2320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGranahan, N. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med.7, 283ra54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong, J. et al. Genomic and transcriptomic characterization of natural killer T cell lymphoma. Cancer Cell37, 403–419 e6 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Ohshima, K. et al. Driver gene alterations and activated signaling pathways toward malignant progression of gastrointestinal stromal tumors. Cancer Sci.110, 3821–3833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagarde, P. et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin. Cancer Res.18, 826–838 (2012). [DOI] [PubMed] [Google Scholar]
- 44.de Miguel, M. & Calvo, E. Clinical challenges of immune checkpoint inhibitors. Cancer Cell38, 326–333 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Balachandran, V. P. et al. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat. Med.17, 1094–1100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seifert, A. M. et al. PD-1/PD-L1 blockade enhances T-cell activity and antitumor efficacy of imatinib in gastrointestinal stromal tumors. Clin. Cancer Res.23, 454–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh, A. S. et al. A randomized phase II study of nivolumab monotherapy or nivolumab combined with lpilimumab in patients with advanced gastrointestinal stromal tumors. Clin. Cancer Res.28, 84–94 (2022). [DOI] [PubMed] [Google Scholar]
- 48.Reilley, M. J. et al. Phase I clinical trial of combination imatinib and ipilimumab in patients with advanced malignancies. J. Immunother. Cancer5, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Leary, B., Finn, R. S. & Turner, N. C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol.13, 417–430 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Elie, D. Avapritinib approved for GIST subgroup. Cancer Discov.10, 334 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Taylor, B. S. et al. Advances in sarcoma genomics and new therapeutic targets. Nat. Rev. Cancer11, 541–557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen, B. et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell185, 563–575 e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park, J. et al. Genetic Characterization of Molecular Targets in Korean Patients with Gastrointestinal Stromal Tumors. J. Gastric Cancer20, 29–40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li, P. et al. Genetic alterations in cell cycle regulation-associated genes may promote primary progression of gastrointestinal stromal tumors. Lab Invest.100, 426–437 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Toulmonde, M. et al. Activity and safety of palbociclib in patients with advanced gastrointestinal stromal tumors refractory to imatinib and sunitinib: a biomarker-driven phase II study. Clin. Cancer Res.25, 4611–4615 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom.13, 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taguchi, T. et al. Conventional and molecular cytogenetic characterization of a new human cell line, GIST-T1, established from gastrointestinal stromal tumor. Lab Invest.82, 663–665 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Tuveson, D. A. et al. STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene20, 5054–5058 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Bauer, S., Yu, L. K., Demetri, G. D. & Fletcher, J. A. Heat shock protein 90 inhibition in imatinib-resistant gastrointestinal stromal tumor. Cancer Res.66, 9153–9161 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Fan, Y. et al. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol.17, 178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods15, 591–594 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol.31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larson, D. E. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics28, 311–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narzisi, G. et al. Genome-wide somatic variant calling using localized colored de Bruijn graphs. Commun. Biol.1, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wala, J. A. et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res.28, 581–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, H. & Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc.10, 1556–1566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bergstrom, E. N. et al. SigProfilerMatrixGenerator: a tool for visualizing and exploring patterns of small mutational events. BMC Genomics20, 685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexandrov, L. B., Nik-Zainal, S., Wedge, D. C., Campbell, P. J. & Stratton, M. R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep.3, 246–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res.44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol.12, R41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Islam, S. M. A. et al. Uncovering novel mutational signatures by de novo extraction with SigProfilerExtractor. Cell Genom.2, None (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yousif, F. et al. The origins and consequences of localized and global somatic hypermutation. bioRxiv10.1101/287839 (2018).
- 76.Bergstrom, E. N., Kundu, M., Tbeileh, N. & Alexandrov, L. B. Examining clustered somatic mutations with SigProfilerClusters. Bioinformatics38, 3470–3473 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol.30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol.30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haas, B. J. et al. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol.20, 213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nicorici, D. et al. FusionCatcher—a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv10.1101/011650 (2014).
- 82.Haas, B. J. et al. STAR-Fusion: fast and accurate fusion transcript detection from RNA-seq. bioRxiv10.1101/120295 (2017).
- 83.Jia, W. et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol.14, R12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics26, 1572–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lovmar, L., Ahlford, A., Jonsson, M. & Syvänen, A. C. Silhouette scores for assessment of SNP genotype clusters. BMC Genomics6, 35 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun.4, 2612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res.48, W509–W514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ding, Z. et al. Estimating telomere length from whole genome sequence data. Nucleic Acids Res.42, e75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ni, B. et al. The nerve-tumour regulatory axis GDNF-GFRA1 promotes tumour dormancy, imatinib resistance and local recurrence of gastrointestinal stromal tumours by achieving autophagic flux. Cancer Lett.535, 215639 (2022). [DOI] [PubMed] [Google Scholar]
- 90.Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep.7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The sequencing datasets (raw WGS, WES, and WTS data) generated in this study have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA005970) database under accession code HRA005970. The expression data of CINSARC cohort and Japanese cohort was downloaded from the Array Express accession: E-MTAB-373 and the NCBI database under accession GSE136755. The somatic mutation datasets (MAF file) of SARC analyzed in this study were downloaded from the GDC Portal [https://gdc.cancer.gov/about-data/publications/sarc_2017]. Source data are provided with this paper.