Cervical cancer disproportionally affects Native American women. Sociodemographic and behavioral factors might contribute to this disparity via alteration of vaginal microbiota. Here, we show the association between these factors and vaginal dysbiosis and immune activation, which can be implicated in high-risk HPV infection among Native American and other racial/ethnic populations.
Abstract
Vaginal dysbiosis is implicated in persistent human papillomavirus (HPV) infection and cervical cancer. Yet, there is a paucity of data on the vaginal microbiome in Native American communities. Here, we aimed to elucidate the relationships between microbiome, HPV, sociodemographic, and behavioral risk factors to better understand an increased cervical cancer risk in Native American women. In this pilot study, we recruited 31 participants (16 Native American and 15 non-Native women) in Northern Arizona and examined vaginal microbiota composition, HPV status, and immune mediators. We also assessed individuals’ sociodemographic information and physical, mental, sexual, and reproductive health. Overall, microbiota profiles were dominated by common Lactobacillus species (associated with vaginal health) or a mixture of bacterial vaginosis–associated bacteria. Only 44% of Native women exhibited Lactobacillus dominance, compared with 58% of non-Native women. Women with vaginal dysbiosis also had elevated vaginal pH and were more frequently infected with high-risk HPV. Furthermore, we observed associations of multiple people in a household, lower level of education, and high parity with vaginal dysbiosis and abundance of specific bacterial species. Finally, women with dysbiotic microbiota presented with elevated vaginal levels of proinflammatory cytokines. Altogether, these findings indicate an interplay between HPV, vaginal microbiota, and host defense, which may play a role in the cervical cancer disparity among Native American women. Future longitudinal studies are needed to determine the mechanistic role of vaginal microbiota in HPV persistence in the context of social determinants of health toward the long-term goal of reducing health disparities between non-Hispanic White and Native American populations.
Prevention Relevance: Cervical cancer disproportionally affects Native American women. Sociodemographic and behavioral factors might contribute to this disparity via alteration of vaginal microbiota. Here, we show the association between these factors and vaginal dysbiosis and immune activation, which can be implicated in high-risk HPV infection among Native American and other racial/ethnic populations.
Introduction
Human papillomavirus (HPV) infection and cervical cancer disproportionately impact Native American women relative to non-Hispanic White (NHW) women (1). According to Indian Health Service data from 1999 to 2009, Native American women had approximately a two-fold higher incidence and associated mortality rate than NHW women (2). In Arizona between 2016 and 2020, Hispanic and American Indian/Alaska Native (AI/AN) women had the highest age-adjusted rates of cervical cancer, with 7.8 and 7.1 cases per 100,000 women, respectively, compared with 5.6 in NHW women (3). The associated mortality was also 1.5 times higher in Hispanic women compared with NHW, whereas data on AI/AN were suppressed because the number of cases was too small (3). This cervical cancer disparity is primarily attributed to a lack of screening, unequal access to healthcare, and quality of care. Yet, biologic factors within the local microenvironment, such as vaginal microbiome, might contribute to HPV infection, HPV persistence, and cervical carcinogenesis in Native American women.
Vaginal microbiome is a collection of microorganisms residing in the vagina (referred to as the vaginal microbiota) with “theater” of their activity (such as a spectrum of molecules produced by microorganisms and coexisting hosts structured by the surrounding environment; ref. 4). In most reproductive-age women, the lower reproductive tract (vagina and cervix) is colonized by one or few Lactobacillus species (including Lactobacillus crispatus, Lactobacillus iners, Lactobacillus gasseri/paragasseri, and Lactobacillus jensenii/mulieris), which protect the host from invading pathogens, by lowering vaginal pH through lactic acid production and the secretion of antimicrobial compounds (5). However, when vaginal dysbiosis occurs, protective lactobacilli are depleted and replaced by a diverse consortium of anaerobic bacteria, such as Gardnerella, Fannyhessea, Prevotella, Megasphaera, and Sneathia (5). This dramatic shift in the microbiota composition may lead to bacterial vaginosis (BV) and manifest with symptoms, such as vaginal discharge, malodor, and elevated pH; yet, some women with Lactobacillus-depleted microbiota are asymptomatic (6).
Notably, vaginal dysbiosis has been implicated in an increased risk of acquisition of sexually transmitted infections (STI), including HPV (7). Growing evidence also suggests the causal link between dysbiosis and HPV persistence and development of cervical dysplasia (8–10). Intriguingly, women representing racial/ethnic minorities have higher rates of BV or vaginal dysbiosis (11), which consequently may increase their risk of cervical cancer. For example, in the United States, Black and Hispanic women have significantly higher rates of BV or vaginal dysbiosis (5, 11, 12). These differences in the microbiota composition among racial/ethnic groups are multifactorial and have been attributed to individuals’ behavior, lifestyle, diet, socioeconomics, environment, genetic ancestry, and immune interactions (10).
However, our knowledge of the vaginal microbiota composition and HPV infection patterns in Native American communities is very limited (1). In fact, most prior microbiome studies do not include Native American women. In this pilot study, we elucidate the relationships between the vaginal microbiota, HPV, immune markers, and sociodemographic and lifestyle factors in a cohort of women from Northern Arizona to better understand an increased cervical cancer risk in Native American communities.
Materials and Methods
Ethics statement
This prospective observational study was approved by the Institutional Review Board at the University of Arizona (reference no. 1510171298). The study design was respectfully discussed with The Partnership for Native American Cancer Prevention (NACP) Outreach Core, the Community Advisory Board of the Native Americans for Community Action (NACA) Family Health Center, and the University of Arizona Tribal Consultation for guidance on cultural relevance and appropriateness. All patients provided written informed consent to participate, and the study was conducted in accordance with federal guidelines and regulations and the Declaration of Helsinki.
Study participants
Participants were recruited during their visit to the NACA Family Health Center (a non-Indian Health Service clinic) in Flagstaff, AZ, USA, between December 2020 and April 2022. The NACA clinic provides a variety of health and human services (including physicals, immunizations, disease screening, STI testing, women’s health, and other medical services) to urban Native Americans, low-income and other underserved families, and individuals who experience health and socioeconomic disparities. This clinic provides services to Native and non-Native people in Northern Arizona regardless of their insurance and healthcare coverage. The NACA's mission is to provide preventive wellness strategies, empower, and advocate for Native people and others in need to create a healthy community based on Harmony, Respect, and Indigenous values. Recruitment strategies included flyers describing the study posted at the clinic, on the website as well as social media, postcards given to patients during their visits, and word of mouth from NACA staff to eligible participants. Thirty-one participants were enrolled and contributed to the study. We included individuals of any race or ethnicity aged 18 to 55 years who were premenopausal. Postmenopausal women were excluded because of the known impact of menopause on the vaginal microbiota composition (13). Data on tribal affiliation were not collected. Exclusion criteria included the following: being pregnant, currently menstruating, or postmenopausal; currently on antibiotics, antifungals, antivirals, or topical steroids; having a current vaginal infection (including BV), vulvar infection, urinary tract infection, or STI or within the previous 3 weeks; and having sexual intercourse within 48 hours. Demographic, socioeconomic, medical history, and sexual/reproductive health data were collected from culturally tailored, self-reported surveys (14). In addition, participants were administered the Perceived Stress Scale (PSS10) survey, developed by Cohen and colleagues (15), and a Patient Reported Outcome Measurement System survey, National Institutes of Health Toolbox Global Health v1.2, which includes measurements of physical health, mental health, general health and ability to carry out social activities. T-scores were calculated to express final scores for the Global Health and PSS10 survey responses.
Biospecimen collection and processing
Participants provided two physician- or self-collected vaginal swabs based on participant preference. The first swab was collected using an ESwab collection system with liquid Amies medium (Cat. No. 480C, COPAN Diagnostics). The second swab was collected using FLOQSwabs nylon-flocked swab (Cat. No. 502CS01, COPAN Diagnostics) and used to measure vaginal pH by Hydrion pH test paper 4.5 to 7.0 (Cat. No. 334, Micro Essential Laboratory) followed by placing the swab in a tube containing 1 mL of sterile 0.9% saline solution (Cat. No. S5815, Teknova). Following the collection, vaginal specimens were immediately placed on ice and frozen at −20°C within 1 hour. For long-term storage, specimens were shipped on dry ice and transferred to −80°C. The first vaginal swabs were used for DNA extraction and subsequent HPV genotyping and 16S rRNA sequencing analyses. Swabs were thawed on ice, and total DNA was isolated using the DNeasy PowerSoil Pro Kit (Qiagen, RRID:SCR_008539) following the manufacturer’s instructions. DNA samples were aliquoted and stored at −80°C for further analysis. The second vaginal swabs were used for the quantification of soluble proteins. Swabs were thawed on ice and vigorously shaken in saline solution. Samples were clarified by centrifugation (700× g for 10 minutes at 4°C) and aliquoted to avoid freeze/thaw cycles and stored at −80°C for further analysis.
HPV genotyping
Detection of 14 high-risk HPV (hrHPV) genotypes, HPV16, HPV18, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51, HPV52, HPV56, HPV58, HPV59, HPV66, HPV68, and one emerging possibly high-risk genotype, HPV53, was performed by isothermal amplification with real-time fluorescence detection on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, RRID:SCR_020239) using purified vaginal DNA samples and the AmpFire HPV High Risk Genotyping Assay (Cat. No. GHPVF100, Atila Biosystems) following the manufacturer’s instructions.
Microbiome analysis
Microbiome analysis was performed using DNA extracted from vaginal swabs and 16S ribosomal RNA (rRNA) gene sequencing. The hypervariable region 4 (V4) of the 16S rRNA gene was amplified by PCR using 515F and 806R primers with Golay barcode tags on the forward primer (16). Amplicons were purified from agarose gel and quantified using the Quant-iT dsDNA High Sensitivity Assay (Cat. No. Q33120, Invitrogen, RRID:SCR_008452). Amplicons were pooled in equimolar concentrations, followed by quality verification with the Bioanalyzer DNA 1000 chip (Agilent Technologies, RRID:SCR_013575). The pool was combined with 1% PhiX control and sequenced on the MiSeq platform using the 600-cycle MiSeq Reagent Kit v3 (Illumina, RRID:SCR_016379). Bioinformatic analyses were performed using QIIME 2 (RRID:SCR_021258; ref. 17). Following demultiplexing, amplicon quality filtering, denoising and chimera removal, and amplicon sequence variant (ASV) definition were performed using DADA2 (q2-dada2 plugin) with no trimming or truncation of reads required because of high sequence quality (18). ASVs were taxonomically classified using q2-feature-classifier (19) using the 202 release of Genome Taxonomy Database (20) and the vaginal microenvironment-weighted classifier (21, 22) based on the STIRRUPS database (23), which increases the frequency at which species level identification can be obtained from 16S sequencing data (Supplementary Table S1). Any ASVs that were not classified as bacterial phylum or that were annotated as mitochondria were removed from the data as likely nonmicrobial features. Microbial differential abundance and microbiome composition analyses were performed using R (RRID:SCR_001905). For the microbial differential abundance analysis, microbiome tables from QIIME 2 were loaded with QIIME2R (https://github.com/jbisanz/qiime2R/blob/master/inst/CITATION). ASV counts were combined with taxonomy identification and filtered to species level for downstream analyses. The R package Analysis of Compositions of Microbiomes with Bias Correction (ANCOM-BC) package version 2.6.0 (24) was used to determine differentially abundant taxa between the groups based on age, body mass index (BMI), educational level, household size, marital status, self-reported race, vaginal pH, number of pregnancies, hrHPV status, perceived stress scores, general health scores, physical health t-scores, mental health t-scores, and ability to carry out social scores. The formula was based on the categorical or continuous groupings with structural zeroes included. P values were corrected for multiple comparisons using the FDR method (q value < 0.05 was considered significant). The output data was visualized with Prism 9.0 (GraphPad, RRID:SCR_002798). For the microbiome composition analysis, microbiome tables from QIIME 2 were converted with QIIME2R to relative abundances. Taxa were then filtered to remove those with less than 0.1% relative abundance within a sample and those with less than 5% prevalence across the samples. A hierarchical clustering of taxa at the species level was performed using ClustVis (RRID:SCR_017133; ref. 25) and based on Euclidean distance and Ward linkage. A heatmap was used to visualize the relative abundance data and patient-related annotations, including self-reported racial groups, household size, education level, number of pregnancies, HPV status, Lactobacillus dominance, bacterial species predominance, and vaginal pH. In addition, the correlation between vaginal microbiota species was calculated using Spearman’s rank correlation analysis.
Quantification of soluble proteins
Levels of 42 proteins (EGF, eotaxin/CCL11, fibroblast growth factor 2 (FGF2), Flt3L, fractalkine/CX3CL1, G-CSF, granulocyte–macrophage colony-stimulating factor (GM-CSF), GROα/CXCL1, IFNα2, IFNγ, IL1α, IL1β, IL1Ra, IL6, IL8/CXCL8, IL9, IL10, IL12 (p40), IL12 (p70), IL13, IL15, IL17A, IL17E/IL25, IL18, IL22, IL27, IP10/CXCL10, M-CSF, MCP1/CCL2, MCP3/CCL7, MDC/CCL22, MIG/CXCL9, MIP1α/CCL3, MIP1β/CCL4, platelet-derived growth factor (PDGF)–AA, PDGF-AB/BB, RANTES/CCL5, sCD40L, TGF-α, TNFα, TNFβ, and VEGF-A) were measured in vaginal swab samples using the Milliplex MAP Human Cytokine Chemokine Panel A Magnetic Bead Immunoassay (Cat. No. HCYTA60K, Millipore, RRID:SCR_008983) in accordance with the manufacturer’s protocols. Data were collected with a Bio-Plex 200 instrument (Bio-Rad, RRID:SCR_018026) and analyzed using Bio-Plex Manager 6.0 software (Bio-Rad, RRID:SCR_014330). A five-parameter logistic regression curve fit was used to determine the concentration. All samples were assayed in duplicate. The concentration values below the detection limit were substituted with 0.5 of the minimum detectable concentration provided in the manufacturer’s instructions. Logarithmic transformation was applied to normalize the data.
Statistical analyses
Statistical differences between continuous variables were determined using a two-sample independent t test, whereas differences between categorical variables were determined using Fisher’s exact test or χ2 test. P values less than 0.05 were considered significant. Statistical analyses were performed using Prism 10 software (GraphPad, RRID:SCR_002798).
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The 16S rRNA gene sequences were not deposited in the NCBI database due to limited consent, the presence of possible human reads within the dataset, and Indigenous data sovereignty. Instead, we included a table with ASV counts and corresponding taxonomic annotations in the supplementary materials (Supplementary Table S1).
Results
Study population
In this pilot, cross-sectional study, a total of 31 participants were recruited and enrolled at the Native Americans for Community Action (NACA) clinic in Flagstaff, AZ. Table 1 shows patients’ demographics, socioeconomics, and other characteristics. The mean age of participants was 29.1 years old (ranging from 18 to 46). About race or ethnicity, participants predominantly identified themselves as Native American (or AI/AN; 51.6%), White (41.9%), or Hispanic/Latina (16.1%). The majority of participants shared that they spoke English as a first language (90.3%) and were heterosexual (80.6%). About socioeconomics, many of the participants identified that they were single (45.2%) or living with a partner (35.5%), had some college education (32.3%) or had a bachelor’s degree (25.8%), were working full-time (71.0%), part-time (12.9%), or keeping house/raising children full-time (16.1%), living with at least two other people in a household (64.5%), and earning less than $75,000 per household (61.3%). Most participants identified that they did not use antibiotics within the last 3 months (80.6%) and did not smoke (80.6%). The data showed that the majority of women were sexually active in the last year (77.4%). Many participants did not use any contraception (35.5%). Participants who used contraceptives most frequently reported the use of condoms (32.3%) or birth control pills (19.4%). About parity, the majority of participants were nulliparous (64.5%) followed by individuals who were primiparous (19.4%). About prophylactic vaccination and screening, most women received at least one dose of the HPV vaccine (51.6%) and had a Pap smear within the last 3 years (64.5%).
Table 1.
Participant demographics and characteristics.
| Characteristic | N = 31 |
|---|---|
| Age [mean (SD)] | 29.1 (6.6) |
| Range | 18–46 |
| Racea [n (%)] | |
| American Indian or Alaska Native | 16 (51.6) |
| White/Caucasian | 13 (41.9) |
| Asian | 2 (6.5) |
| Mixed or multiracial | 1 (3.2) |
| Refused to answer | 1 (3.2) |
| Missing | 1 (3.2) |
| Ethnicity [n (%)] | |
| Hispanic/Latina | 5 (16.1) |
| Non-Hispanic | 26 (83.9) |
| First language [n (%)] | |
| English | 28 (90.3) |
| Non-English | 3 (9.7) |
| Sexual orientation [n (%)] | |
| Heterosexual | 25 (80.6) |
| Bisexual | 5 (16.1) |
| Pansexual | 1 (3.2) |
| Body mass index [mean (SD)] | 28.0 (7.4) |
| Marital status [n (%)] | |
| Single | 14 (45.2) |
| Living with partner | 11 (35.5) |
| Married | 3 (9.7) |
| Divorced | 2 (6.5) |
| Missing | 1 (3.2) |
| Completed education [n (%)] | |
| Less than high school | 2 (6.5) |
| High school diploma, GED | 4 (12.9) |
| Some college credit/no degree | 10 (32.3) |
| Associate degree/technical school | 3 (9.7) |
| Bachelor’s degree | 8 (25.8) |
| Master’s or doctoral degree | 4 (12.9) |
| Daily activitya [n (%)] | |
| Work full-time | 22 (71.0) |
| Work part-time | 4 (12.9) |
| Unemployed or laid-off | 2 (6.5) |
| Keeping house or raising children full-time | 5 (16.1) |
| Student | 2 (6.5) |
| Disabled | 1 (3.2) |
| Household income [n (%)] | |
| <$10,000 | 3 (9.7) |
| $10,000–$24,999 | 3 (9.7) |
| $25,000–$49,999 | 5 (16.1) |
| $50,000–$74,999 | 8 (25.8) |
| $75,000–$99,999 | 4 (12.9) |
| ≥$100,000 | 3 (9.7) |
| Refuse to answer | 5 (16.1) |
| Household size [including self; n (%)] | |
| 1 | 2 (6.5) |
| 2 | 9 (29.0) |
| 3 | 10 (32.3) |
| 4 | 5 (16.1) |
| 5 | 3 (9.7) |
| 6+ | 2 (6.5) |
| Antibiotics use [in the last 3 months; n (%)] | |
| Yes | 6 (19.4) |
| No | 25 (80.6) |
| History of smoking [at least 100 cigarettes in lifetime; n (%)] | |
| Yes | 6 (19.4) |
| No | 25 (80.6) |
| Number of sexual partners in lifetime [n (%)] | |
| 0 | 1 (3.2) |
| 1–2 | 11 (35.5) |
| 3–5 | 4 (12.9) |
| 6–7 | 1 (3.2) |
| 8+ | 12 (38.7) |
| Missing | 2 (6.5) |
| Number of sexual partners in last year [n (%)] | |
| 0 | 3 (9.7) |
| 1–2 | 23 (74.2) |
| 3–5 | 1 (3.2) |
| Missing | 4 (12.9) |
| Contraception usea [n (%)] | |
| Condoms | 10 (32.3) |
| Birth control pills | 6 (19.4) |
| Intrauterine device | 4 (12.9) |
| Contraception injection | 3 (9.7) |
| Contraception implant | 1 (3.2) |
| None | 11 (35.5) |
| Number of pregnancies [n (%)] | |
| 0 | 20 (64.5) |
| 1 | 6 (19.4) |
| 2 | 1 (3.2) |
| 3 | 1 (3.2) |
| 5+ | 3 (9.7) |
| Vaccinated against HPVb [n (%)] | |
| Yes | 16 (51.6) |
| No | 10 (32.3) |
| Do not know | 5 (16.1) |
| Most recent Pap smear [n (%)] | |
| Within the last year | 13 (41.9) |
| More than 1 year ago, but within the last 3 years | 7 (22.6%) |
| More than 3 years ago, but within the last 5 years | 8 (25.8) |
| Never had a Pap smear | 2 (6.5) |
| Missing | 1 (3.2) |
Abbreviations: GED, general education development; SD, standard deviation.
Not mutually exclusive.
Received at least one dose.
HPV status
First, we determined the distribution of high-risk HPV genotypes in our cohort. High-risk HPV genotypes were detected in 22.6% of participants, including five Native American women and two NHW women. Four participants (12.9%) were infected with a single high-risk genotype and three participants (9.7%) were infected with two different high-risk genotypes. Detected HPV genotypes included HPV39, HPV45, HPV52, HPV53, HPV58, and HPV59. Notably, the current nonavalent HPV vaccine does not target half of the genotypes detected in this cohort (such as HPV39, HPV53, and HPV59; Table 1).
Vaginal microbiome composition
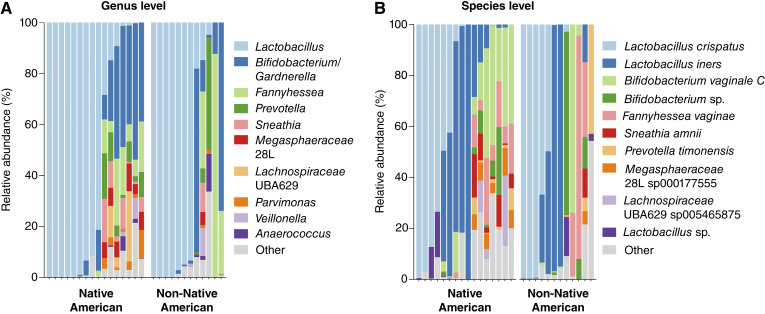
To examine the vaginal microbiota communities, we utilized the 16S rRNA gene sequencing technique. We were able to amplify DNA of sufficient quantity and quality (at least 4,000 reads) for microbiome analysis from 28 out of 31 vaginal swab samples. We calculated the relative abundance of bacterial taxa on the genus (Fig. 1A) and species (Fig. 1B; Supplementary Fig. S1) levels and compared taxonomic compositions between Native American women and women of other races (referred to as non-Native American). Composition bar plots were used to visualize the vaginal microbial profiles of each participant. Overall, vaginal microbiota profiles were similar between Native American and non-Native American women. In some women, vaginal microbiota was dominated by health-associated Lactobacillus species (mainly Lactobacillus crispatus or Lactobacillus iners), whereas in other women vaginal microbiota were depleted of health-associated Lactobacillus species and consisted of a mixture of common BV-associated bacteria (Gardnerella, Fannyhessea, Prevotella, Sneathia, and Megasphaera). When analyzed across all participants, BV-associated species positively correlated with each other, whereas L. crispatus but not L. iners negatively correlated with BV-associated bacteria (Supplementary Fig. S2). In addition, we also observed that pathobionts and opportunistic pathogens correlated with each other; however, these bacterial species were not highly abundant in our cohort.
Figure 1.
Vaginal microbiota composition of Native American women in our study. Bar plots show the relative abundance of taxa assigned to the A, genus or B, species level grouped based on race. Vaginal microbiota profiles were similar between Native American and non-Native American women from Northern Arizona. The most prevalent Lactobacillus species included Lactobacillus crispatus and Lactobacillus iners. Dysbiotic Lactobacillus-depleted profiles consisted of typical communities of anaerobic bacteria associated with bacterial vaginosis (BV), including Gardnerella vaginalis [assigned as Bifidobacterium vaginale in the Genome Taxonomy Database (GTDB)], Fannyhessea vaginae (formerly known as Atopobium vaginae), Sneathia vaginalis (formerly known as Sneathia amnii), Prevotella timonensis, Megasphaera lornae (assigned as 28L sp00017755 in GTDB), and Clostridiales genomosp. BVAB1 (assigned as UBA629 sp005465875 in GTDB).
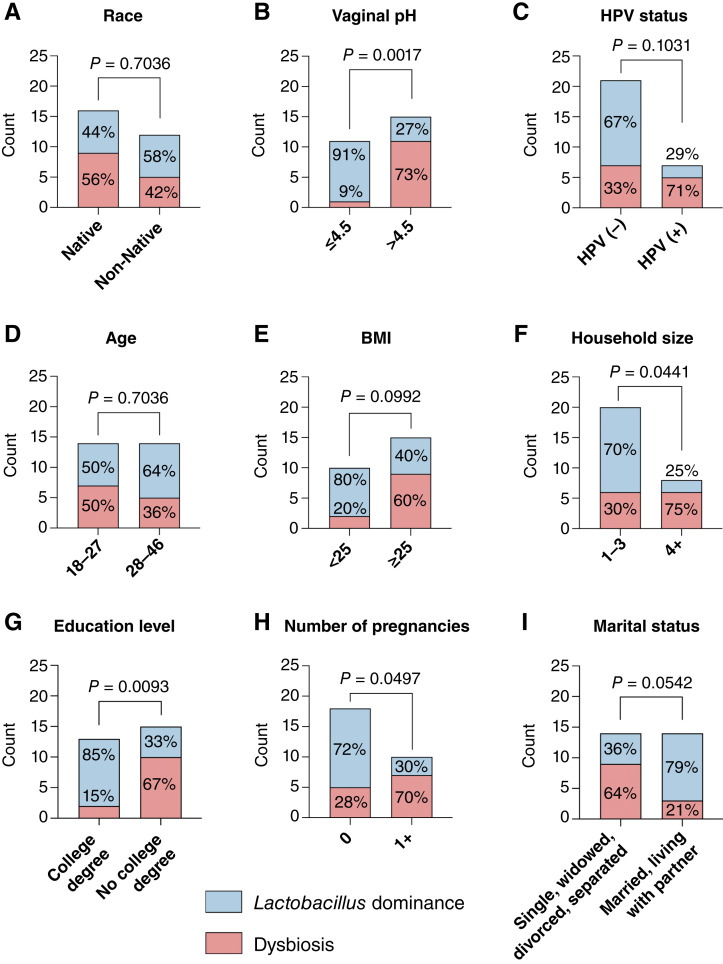
Next, we compared the relative abundance of health-associated Lactobacillus species between Native American and non-Native American women, and we did not observe any significant differences (P = 0.9363) between the groups (Supplementary Fig. S3). We also analyzed Lactobacillus abundance in the context of vaginal pH and high-risk HPV status. Women with normal vaginal pH (<4.5) had significantly (P = 0.0094) higher levels of Lactobacillus spp. when compared with women with abnormal vaginal pH (≥4.5; Supplementary Fig. S3). Similarly, HPV-negative women also tended (P = 0.0682) to have higher levels of Lactobacillus spp. compared with HPV-positive women (Supplementary Fig. S3). Once we categorized microbial profiles based on the Lactobacillus dominance (defined as ≥80% relative abundance of Lactobacillus; ref. 26), we also found no significant differences between the racial groups (Fig. 2A). Only 44% of Native American women exhibited Lactobacillus dominance compared with 58% of non-Native women. Vaginal pH was significantly (P = 0.0017) associated with Lactobacillus dominance with 91% of women with normal pH having Lactobacillus-dominant microbiota (Fig. 2B). In contrast, high-risk HPV infection was associated with Lactobacillus-depleted microbiota, although this association did not reach significance (P = 0.1031). Women infected with high-risk HPV also tended to have microbiome depleted of Lactobacillus (Fig. 2C). Seventy-one percent of HPV-positive women exhibit non-Lactobacillus-dominant microbiota, whereas only 33% of HPV-negative women had vaginal dysbiosis. Overall, these data show a strong association of vaginal dysbiosis with abnormal vaginal pH regardless of race/ethnicity, which might contribute to HPV infection.
Figure 2.
Associations of Lactobacillus dominance with race, vaginal pH, HPV status, and socioeconomic and lifestyle factors. Stacked bar plots show the number of participants with Lactobacillus-dominant microbiota or dysbiotic Lactobacillus-depleted microbiota among the groups based on A, race, B, vaginal pH, C, HPV status, D, age, E, BMI, F, size of household, G, education level, H, number of pregnancies, and I, marital status. P values were calculated using Fisher’s exact test. Vaginal microbiota profiles were dominated by Lactobacillus species in 44% of Native American women, which was similar to levels observed in non-Native American participants (58%). Lactobacillus dominance was highly associated with low vaginal pH (≤4.5). HPV-positive women also tended to have lower Lactobacillus abundance compared with HPV-negative women. Vaginal microbiota profiles were also more frequently dominated by Lactobacillus species in participants with BMI < 25, living in smaller households (1–3 people), having a college (bachelor’s, master’s, and doctorate) or associate/technical degree, who were nulliparous, married or living with a partner. No difference was observed between the younger and older participants (dichotomized based on the median age).
Impact of sociodemographic and lifestyle factors on vaginal microbiome
Because the vaginal microbiota composition can be impacted by multiple factors, we investigated the relationship between Lactobacillus dominance and other self-reported sociodemographic and behavioral data. There was no significant association between participants’ age and Lactobacillus dominance (P = 0.7036; Fig. 2D). However, participants with BMI equal to or greater than 25 tended to have more frequently Lactobacillus-depleted microbiota compared with participants with BMI less than 25 (P = 0.0992; Fig. 2E). In addition, we observed significant associations between the vaginal microbiota composition and participants’ household size (Fig. 2F), an education level (Fig. 2G) and the number of pregnancies (Fig. 2H). Women living in smaller households (three or fewer people including study participant; P = 0.0441), women with a higher level of education (with college or associate/technical degree; P = 0.0093), as well as nulliparous women (P = 0.0497) exhibited more often Lactobacillus dominance. Individuals who were married or living with a partner also tended (P = 0.0542) to have more frequently Lactobacillus-dominant microbiota compared with women who were single, divorced/separated, or widowed (Fig. 2I). We did not observe any significant associations of Lactobacillus dominance with employment status, household income, antibiotic use, history of smoking, number of sexual partners, contraception use, or HPV vaccination status (Supplementary Table S2). In addition, we examined associations of the vaginal microbiota composition with self-reported measurements of physical health, mental health, general health, carrying out social activities, and perceived stress. There were no significant associations of Lactobacillus dominance with any of these health measurements (Supplementary Table S3). Furthermore, neither HPV status (Supplementary Table S4) nor race (Supplementary Table S5) related to higher health T-scores, except for Native American women reporting better general health compared with non-Native Americans (P = 0.0013).
Differentially abundant vaginal taxa
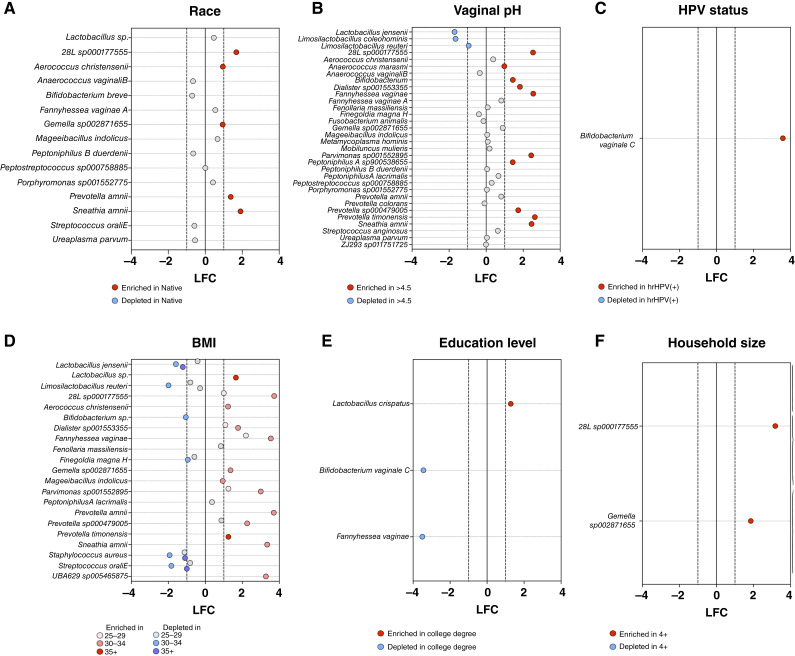
To identify microbial features that were differential between sociodemographic groups and to understand the role of the vaginal microbiome in risk factors associated with HPV infection, we applied ANCOM-BC (24). Participants who identified as Native American had profiles with significantly enriched in Gemella sp002871655 (q < 0.0001) and Megasphaeraceae 28L sp000177555 (q < 0.0001) as well as Aerococcus christensenii (q < 0.0001), Prevotella amnii (q < 0.0001), and most enriched in Sneathia amnii (q < 0.0001; Fig. 3A). These signatures may be related to health disparities observed in HPV infection and cervical cancer. Investigation of vaginal pH, in which elevated pH is associated with vaginal dysbiosis, revealed that eight vaginal species were statistically significantly depleted in abnormal vaginal pH (>4.5), and 24 species were enriched (Fig. 3B). Participants with abnormal pH had significantly enriched [log fold change (LFC) of 1] levels of Prevotella timonensis (q < 0.00001), Fannyhessea vaginae (q < 0.00001), Megasphaereaceae 28L sp000177555 (q < 0.00001), Sneathia amnii (q < 0.00001), Parvimonas sp001552895 (q < 0.00001), Dialister sp001553355 (q < 0.00001), Prevotella sp000479005 (q < 0.00001), unclassified Bifidobacterium species (q < 0.00001), Peptoniphilus A sp900538655 (q < 0.00001), and Anaerococcus marasmi (q < 0.00001) compared with those with normal vaginal pH (4.5; Fig. 3B). However, profiles with an abnormal pH had significant depletion (LFC of 1) of Limosilactobacillus reuteri (q < 0.0001), Limosilactobacillus coleohominis (q < 0.0001), and Lactobacillus jensenii (q < 0.0001; Fig. 3B). Although a small sample size in this pilot cohort, one taxon was significantly enriched in participants who were hrHPV-positive, Bifidobacterium vaginale C (q < 0.0001; Fig. 3C), also known as Gardnerella vaginalis. Participants with a high BMI of 35+ had enrichment in Prevotella timonensis (q < 0.0001) and an unclassified Lactobacillus species (q < 0.0001) compared with profiles of participants with a BMI of <25. Vaginal profiles of 35+ were also depleted in Streptococcus oralis E (q < 0.0001), an unclassified Bifidobacterium species (q < 0.0001), Staphylococcus aureus (q < 0.0001) and Lactobacillus jensenii (q < 0.0001; Fig. 3D). Interestingly, we observed that the vaginal microbiota of participants with a college degree were highly enriched in Lactobacillus cripatus (q < 0.0001) and depleted in Bifidobacterium vaginale C (q < 0.0001) and Fannyhessea vaginae (q < 0.0001; Fig. 3E). Other dysbiotic bacteria were also enriched in vaginal profiles from participants who reported households with four or more individuals, including Gemella sp002871655 (q < 0.0001) and Megasphaeraceae 28L sp000177555 (q < 0.0001; Fig. 3F). No significant taxa were observed for categorical groupings of marital status and the number of pregnancies. Additional analyses on continuous variables of age, ability to carry out social scores, general health scores, mental health t-scores, physical health t-scores, and perceived stress t-scores also observed no significant differences.
Figure 3.
Microbial features associated with sociodemographic factors, vaginal pH, and HPV status. Differentially abundant taxa between the groups based on A, race, B, vaginal pH, C, HPV status, D, BMI, E, education level, and F, household size were identified at the species level using ANCOM-BC. P values were corrected for multiple comparisons using the FDR method; taxa with q < 0.05 are depicted. Red and blue dots indicate taxa with LFC > 1 and LFC < −1, respectively. Numerous dysbiotic vaginal species are enriched in participants with abnormal vaginal pH, lower education level, who live in larger households, identify themselves as Native American, have higher BMI, and are hrHPV-positive, whereas Lactobacillus and Limosilactobacillus species were enriched in women with normal pH, higher education level and lower BMI.
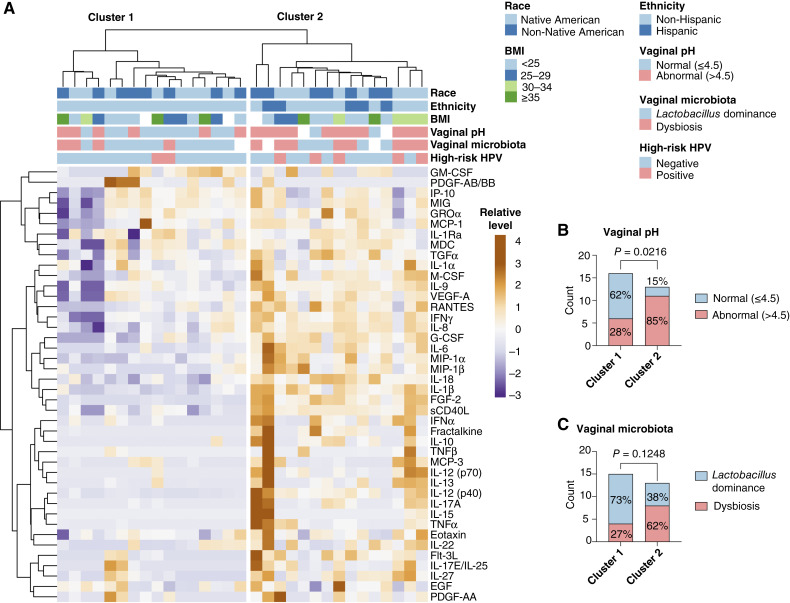
Vaginal immune mediator profiles
To characterize vaginal immune mediator profiles, we quantified concentrations of 42 cytokines, chemokines, and growth factors in vaginal swab samples. Unsupervised hierarchical clustering analysis was utilized to visualize relative levels of immune markers and compare the protein profiles among participants (Fig. 4A). The generated heatmap and dendrogram revealed two distinct clusters: cluster 1 with relatively low levels of immune mediators and cluster 2 with elevated levels of immune mediators. To further characterize these clusters, patient-related demographic metadata (race, ethnicity, and BMI), as well as vaginal pH, vaginal microbiota composition, and high-risk HPV status data, were also plotted on the heatmap. The distribution of demographic metadata did not significantly (P > 0.05) differ between the clusters. Yet, cluster 2 had a significantly (P = 0.0216) higher rate of patients with abnormal vaginal pH (85%; Fig. 4B) as well tended (P = 0.1248) to have more patients with dysbiotic Lactobacillus-depleted microbiota (65%; Fig. 4C) in contrast to cluster 1 with only 28% and 27% of participants, respectively, exhibiting these negative cervicovaginal microenvironment features.
Figure 4.
Vaginal profiles of cytokines, chemokines, and growth factors. A, heatmap reflects relative levels of proteins in vaginal swab samples. Unsupervised hierarchical clustering was used to assess the similarity between protein profiles. Two main clusters based on Euclidean distance and Ward linkage were observed. B, Stacked bar plots show the distribution of patients with normal and abnormal pH or C,Lactobacillus-dominant and dysbiotic Lactobacillus-depleted microbiota between the clusters. P values were calculated using the Fisher exact test. Unsupervised hierarchical clustering analysis indicates that immune marker levels associate with vaginal pH and Lactobacillus dominance.
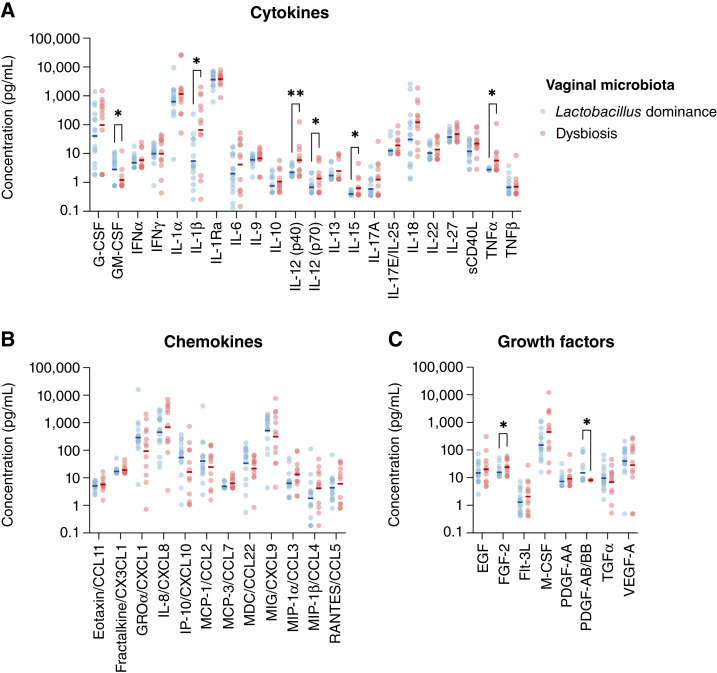
Subsequently, we compared individual concentrations of proteins between women with Lactobacillus-dominant microbiota and women with dysbiotic Lactobacillus-depleted microbiota. Four proinflammatory cytokines, IL1β, IL12 (p70), IL15, and TNFα, and one regulatory cytokine, IL12 (p40), were significantly (P < 0.05) increased in women with vaginal dysbiosis compared with women with Lactobacillus dominance (Fig. 5A). No chemokines were significantly altered between the groups (Fig. 5B). One growth factor, FGF2, was also increased in women with dysbiosis (Fig. 5C). Furthermore, women with Lactobacillus dominance had increased levels of two immune markers, GM-CSF and PDGF-AB/BB, when compared with women with dysbiosis (Fig. 5A and C).
Figure 5.
Vaginal levels of immune markers in the context of Lactobacillus dominance. Scatter plots show the concentration of A, cytokines, B, chemokines, and C, growth factors in vaginal swab samples in women with Lactobacillus-dominant or dysbiotic Lactobacillus-depleted microbiota. P values were calculated using two sample independent t tests (*, P < 0.05; **, P < 0.01). Women with dysbiotic Lactobacillus-depleted microbiota exhibited increased vaginal levels of IL1β, IL12 (p40), IL12 (p70), IL15, TNFɑ, and FGF2, when compared with women with Lactobacillus-dominant microbiota. In addition, GM-CSF and PDGF-AB/BB were significantly decreased in women with dysbiosis.
Discussion
Emerging evidence strongly suggests that dysbiosis or polymorphic microbiomes are implicated in the development of cancer (10, 27). In the lower female reproductive tract, dysbiosis occurs when protective lactobacilli are replaced by a polymicrobial consortium of anaerobic bacteria (6). In cervical cancer, these dysbiotic bacterial communities have been linked to HPV acquisition and persistence, as well as the development and progression of cervical neoplasia (8, 9). Indeed, numerous cross-sectional studies have shown that women infected with HPV or with cervical neoplasia more frequently harbor Lactobacillus-depleted microbiota (26, 28–32). In addition, only a few longitudinal studies have demonstrated that women with vaginal dysbiosis have higher odds of HPV persistence and disease progression compared with women with Lactobacillus dominance (33–36).
Because racial/ethnic minorities have higher rates of dysbiosis (likely because of socioeconomic, behavioral/lifestyle factors at individual, relational, community, and societal levels; ref. 37), the vaginal microbiota might be an important driver of adverse health outcomes and contribute to cervical cancer health disparities in these populations (10). In our previous study, we demonstrated that Hispanic/Latina women from the Phoenix (AZ) metropolitan area had frequently Lactobacillus-depleted microbiota, which was strongly associated with HPV status, severity of cervical disease, and elevated cervicovaginal levels of proinflammatory cytokines (26). Yet, the relationship between HPV and vaginal microbiota in Native American women is still unrecognized (1).
In this pilot study, we recruited Native and non-Native women of reproductive age. The microbiome analysis revealed that the majority of participants in our cohort (Native and non-Native) exhibited higher rates of vaginal dysbiosis (66% and 42%, respectively) compared with previously reported NHW cohorts (up to 90%; refs. 5, 12). This is in accordance with a small study of 70 Native American women from the Northwestern Plains (38), which reported vaginal dysbiosis in 60% of their participants. About predominant Lactobacillus species, women in our cohort were frequently colonized by L. crispatus or L. iners, which are the most common species of vaginal lactobacilli and also found in the Northwestern Plains cohort (38). Intriguingly, these species might not equally benefit the host: L. crispatus has been associated with optimal health outcomes, whereas the role of L. iners in health and disease remains controversial (5). For example, a recent meta-analysis revealed that L. crispatus correlated with decreased detection of hrHPV and cervical dysplasia, whereas L. iners did not (9).
Our study also revealed that the interplay between HPV, vaginal microbiota composition, and immune activation may play a role in Native American women, similar to our findings in Hispanic women (26). In this cohort of women, individuals infected with hrHPV were more likely to harbor Lactobacillus-depleted microbiota. Women with dysbiosis had elevated vaginal pH and increased cervicovaginal levels of key cytokines (e.g., IL1β and TNFα), which have been previously reported to be elevated in women with BV or vaginal dysbiosis (6). These findings further suggest that the host-microbe interactions in the cervicovaginal microenvironment might result in chronic inflammation, which, in consequence, can promote cervical carcinogenesis. The exact mechanisms by which chronic inflammation contributes to HPV-mediated carcinogenesis are still unclear; thus further investigations are required to identify specific microbial signatures or/and communities contributing to an inflammatory state in the cervicovaginal microenvironment.
Because the vaginal microbiome is a dynamic ecosystem that can be disturbed by multiple factors, we investigated associations of the vaginal microbiota composition with collected patient-related metadata within the entire cohort. Noteworthily, we found that regardless of race/ethnicity, women with higher education levels were more likely colonized by Lactobacillus species (particularly with L. crispatus). This aligns with other studies investigating the vaginal microbiota in pregnant African American women (39) and nonpregnant premenopausal Brazilian (40) or Finnish women (41). The Brazilian study also identified other factors associated with Lactobacillus-depleted microbiota: smoking and a higher number of sexual partners (40). We did not observe these associations in our cohort; however, we might not have adequate sample size to detect these effects with only a few participants who were smokers or had more than two sexual partners within the last year. In addition, we found that women who were married or cohabiting with a partner had higher odds of Lactobacillus dominance in accordance with a previous study of women undergoing cervical cancer screening in Helsinki, Finland (41). On the other hand, living in larger households and history of pregnancy were associated with higher rates of vaginal dysbiosis. It is well established that the vaginal microbiota is frequently disturbed postpartum (42, 43) and multiple reports showed the influence of parity on the vaginal microbiome (44–46). For example, three independent studies of predominantly African American pregnant women (44), pregnant Finnish women (45), and nonpregnant Belgian women (46) showed significant associations of high parity with vaginal dysbiosis. This phenomenon could be explained by a cumulative effect of multiple pregnancies and increased microbiota diversity after live birth but warrants further investigation (44). The same reports also found the effect of obesity on the vaginal microbiota (44, 46). We observed a similar trend with BMI in our cohort. Other studies in nonpregnant populations also consistently show an association between obesity with a lower likelihood of Lactobacillus dominance and an increased risk of BV (47). It is postulated that hormone and immune system alterations in obese individuals may create an environment favoring dysbiotic bacteria, yet the mechanistic link between obesity and vaginal dysbiosis is still incompletely understood (47). Intriguingly, high BMI has also been linked to decreased detection of precancerous cervical lesions, and, consequently, increased risk of cervical cancer development (48). Finally, we did not detect any meaningful impact of self-reported measurements of general, physical, or mental health (including perceived stress) on the vaginal microbiota composition. This conflicts with a previous report on Northwestern Plains Native American women showing an association of psychosocial stress tied to historic loss-associated trauma with increased odds of vaginal dysbiosis (38). However, the use of different survey instruments to measure stress/trauma might explain these conflicting results. Because there is growing evidence of the role of psychosocial stress in modulating immune function and microbiota composition, additional larger translational studies with comprehensive evaluation of various stress/trauma measurements are required to further validate these associations in Native American women.
Herein, we also identified several microbial features associated with sociodemographic factors. Intriguingly, the vaginal microbiota of Native American women were highly enriched in Sneathia vaginalis (formerly known as S. amnii). In our previous study of Hispanic and non-Hispanic women, we found that this bacterium was associated with all stages of cervical carcinogenesis (HPV infection, dysplasia, and invasive carcinoma; ref. 26), as well as Hispanic ethnicity and abnormal vaginal pH (26). In addition, our systematic review of microbiome studies in Hispanic/Latina populations in North and South America revealed Sneathia to be consistently increased in women across cervical carcinogenesis (49). Though we did not observe a significant increase in Sneathia in HPV-positive individuals in this pilot study (likely because of the small sample size), Sneathia was more abundant in women who identified as Native American and had abnormal vaginal pH and higher BMI. Overall, Sneathia might be a microbial marker associated with HPV-related carcinogenesis across populations disproportionally affected by cervical cancer and requires further in vitro studies using human organotypic cell culture models (e.g., three-dimensional cervical epithelial cell model based on a rotating wall vessel bioreactor technology; ref. 50) to evaluate mechanisms by which this bacterium impacts HPV infection and the progression of disease.
Although this pilot study begins to advance our knowledge of HPV and the vaginal microbiome in Native American women, it is important to acknowledge its limitations, including a relatively small sample size (because of the COVID19 pandemic) and the cross-sectional design, thus we may not have had the power to detect effects of some factors (e.g., smoking or sexual activity) on vaginal dysbiosis in our cohort and we cannot generalize our findings. It is also important to note that patients recruited in our study were recruited in urban settings in Northern Arizona. More studies are needed across urban and rural populations, as well as geographic regions, to account for differences in experiencing adverse social determinants of health, gaps in the access and quality of healthcare, and other socioeconomic and/or cultural differences between these racial/ethnic groups and populations (1). Despite these limitations, we were able to identify associations of the vaginal microbiota with HPV and immune markers, as well as sociodemographic and lifestyle/behavioral factors, which provides a more comprehensive understanding of individual risk factors related to increased cervical cancer risk. In the future, longitudinal studies—which include Native American women—are required to move from association to causation. Investigation of social determinants of health contributing to geographic and racial/ethnic differences in the cervical cancer burden is needed to adequately address this health disparity. In addition, features of structural racism (a system of policies that support an unfair advantage for some people and unfair treatment of others based on their race/ethnicity) should be investigated in the context of cervical cancer. Finally, an improved understanding of the mechanistic role of the vaginal microbiota in HPV-related carcinogenesis is still needed to identify potential intervention strategies to increase health equity in Native American populations.
Supplementary Material
Supplementary Figure S1: Vaginal microbiota profiles cluster based on Lactobacillus dominance and not clinical or sociodemographic factors.
Supplementary Figure S2: BV-associated bacteria positively correlate with each other and negatively correlate with Lactobacillus species, including L. crispatus and L. jensenii.
Supplementary Figure S3: Associations of Lactobacillus abundance with race, vaginal pH, and HPV status.
Supplementary Table S1: 16S rRNA sequencing data.
Supplementary Table S2: Differences in demographic, socioeconomic and behavioral/life style factors between participants with Lactobacillus-dominant microbiota and non-Lactobacillus-dominant microbiota.
Supplementary Table S3: Differences in global health and perceived stress measurements between participants with Lactobacillus-dominant microbiota and non-Lactobacillus-dominant microbiota.
Supplementary Table S4: Differences in global health and perceived stress measurements between hrHPV-positive and hrHPV-negative participants.
Supplementary Table S5: Differences in global health and perceived stress measurements between Native and non-Native participants.
Acknowledgments
We would like to thank the patients who enrolled in the study and the NACA clinic staff for their kind assistance in patient recruitment, sample, and clinical data collection and Elisa Martinez for her clinical research coordination, regulatory assistance and support in training clinic staff in this study. We thank Carol Goldtooth from the NACP Outreach Core and the NACP Community Advisory Board members: Miguel Flores Jr, LuAnn Leonard, and Shannon Williams for their critical review of the manuscript. We respectfully acknowledge the University of Arizona and Northern Arizona University are on the land and territories of Indigenous peoples. Today, Arizona is home to 22 federally recognized tribes, with Tucson being home to the O’odham and the Yaqui, and with Phoenix being home to the Ak-Chin Indian Community, Fort McDowell Yavapai Nation, Gila River Indian Community, and Salt River Pima-Maricopa Indian Community. Committed to diversity and inclusion, the University strives to build sustainable relationships with sovereign Native Nations and Indigenous communities through education offerings, partnerships, and community service. Northern Arizona University sits at the base of the San Francisco Peaks, on homelands sacred to Native Americans throughout the region. We honor their past, present, and future generations, who have lived here for millennia and will forever call this place home. This study was funded by NIH National Cancer Institute Awards for the Partnership of Native American Cancer Prevention: U54CA143924 (M.M. Herbst-Kralovetz) and U54CA143925 (N.R. Lee, J.G. Caporaso) and by NIH National Cancer Institute Informatics Technology for Cancer Research Award 1U24CA248454-01 (J.G. Caporaso). In addition, P. Łaniewski and N.R. Jimenez were supported by the Guiding U54 Investigator Development to Sustainability (GUIDeS) program under the U54CA143924 award.
Footnotes
Note: Supplementary data for this article are available at Cancer Prevention Research Online (http://cancerprevres.aacrjournals.org/).
Authors’ Disclosures
P. Łaniewski reports grants from the National Institutes of Health (NIH), National Cancer Institute (NCI) during the conduct of the study. D.J. Roe reports grants from the NIH NCI during the conduct of the study. M.M. Herbst-Kralovetz reports grants from the NIH NCI during the conduct of the study, as well as personal fees from Freya Biosciences outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
P. Łaniewski: Data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft. T.R. Joe: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. N.R. Jimenez: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. T.L. Eddie: Resources, writing–review and editing. S.J. Bordeaux: Resources, writing–review and editing. V. Quiroz: Resources, writing–review and editing. D.J. Peace: Resources, writing–review and editing. H. Cui: Formal analysis, methodology, writing–review and editing. D.J. Roe: Resources, formal analysis, methodology, writing–review and editing. J.G. Caporaso: Resources, formal analysis, supervision, funding acquisition, validation, investigation, writing–review and editing. N.R. Lee: Resources, formal analysis, supervision, funding acquisition, validation, investigation, writing–review and editing. M.M. Herbst-Kralovetz: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, project administration, writing–review and editing.
References
- 1. Morales CG, Jimenez NR, Herbst-Kralovetz MM, Lee NR. Novel vaccine strategies and factors to consider in addressing health disparities of HPV infection and cervical cancer development among Native American women. Med Sci (Basel) 2022;10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. White MC, Espey DK, Swan J, Wiggins CL, Eheman C, Kaur JS. Disparities in cancer mortality and incidence among American Indians and Alaska Natives in the United States. Am J Public Health 2014;104(Suppl 3):S377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. United States Cancer Statistics: Data Visualizations. Atlanta (GA): Centers for Disease Control and Prevention. [cited 2024 Aug 16]. Available from:https://gis.cdc.gov/Cancer/USCS/#/AtAGlance/ [Google Scholar]
- 4. Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome 2020;8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011;108(Suppl 1):4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muzny CA, Łaniewski P, Schwebke JR, Herbst-Kralovetz MM. Host–vaginal microbiota interactions in the pathogenesis of bacterial vaginosis. Curr Opin Infect Dis 2020;33:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tamarelle J, Thiébaut ACM, de Barbeyrac B, Bébéar C, Ravel J, Delarocque-Astagneau E. The vaginal microbiota and its association with human papillomavirus, Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium infections: a systematic review and meta-analysis. Clin Microbiol Infect 2019;25:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Norenhag J, Du J, Olovsson M, Verstraelen H, Engstrand L, Brusselaers N. The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis. BJOG 2020;127:171–80. [DOI] [PubMed] [Google Scholar]
- 9. Wang H, Ma Y, Li R, Chen X, Wan L, Zhao W. Associations of cervicovaginal lactobacilli with high-risk human papillomavirus infection, cervical intraepithelial neoplasia, and cancer: a systematic review and meta-analysis. J Infect Dis 2019;220:1243–54. [DOI] [PubMed] [Google Scholar]
- 10. Łaniewski P, Ilhan ZE, Herbst-Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol 2020;17:232–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peebles K, Velloza J, Balkus JE, McClelland RS, Barnabas RV. High global burden and costs of bacterial vaginosis: a systematic review and meta-analysis. Sex Transm Dis 2019;46:304–11. [DOI] [PubMed] [Google Scholar]
- 12. Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, et al. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology (Reading) 2014;160:2272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Łaniewski P, Herbst-Kralovetz MM. Connecting microbiome and menopause for healthy ageing. Nat Microbiol 2022;7:354–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee NR, Winer RL, Cherne S, Noonan CJ, Nelson L, Gonzales AA, et al. Human Papillomavirus prevalence among American Indian women of the Great Plains. J Infect Dis 2019;219:908–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav 1983;24:385–96. [PubMed] [Google Scholar]
- 16. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 2018;6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, Hugenholtz P. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res 2022;50:D785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bokulich NA, Łaniewski P, Adamov A, Chase DM, Caporaso JG, Herbst-Kralovetz MM. Multi-omics data integration reveals metabolome as the top predictor of the cervicovaginal microenvironment. PLoS Comput Biol 2022;18:e1009876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaehler BD, Bokulich NA, McDonald D, Knight R, Caporaso JG, Huttley GA. Species abundance information improves sequence taxonomy classification accuracy. Nat Commun 2019;10:4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fettweis JM, Serrano MG, Sheth NU, Mayer CM, Glascock AL, Brooks JP, et al. Species-level classification of the vaginal microbiome. BMC Genomics 2012;13(Suppl 8):S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat Commun 2020;11:3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 2015;43:W566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Łaniewski P, Barnes D, Goulder A, Cui H, Roe DJ, Chase DM, et al. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep 2018;8:7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lythgoe MP, Mullish BH, Frampton AE, Krell J. Polymorphic microbes: a new emerging hallmark of cancer. Trends Microbiol 2022;30:1131–4. [DOI] [PubMed] [Google Scholar]
- 28. Audirac-Chalifour A, Torres-Poveda K, Bahena-Román M, Téllez-Sosa J, Martínez-Barnetche J, Cortina-Ceballos B, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS One 2016;11:e0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Godoy-Vitorino F, Romaguera J, Zhao C, Vargas-Robles D, Ortiz-Morales G, Vázquez-Sánchez F, et al. Cervicovaginal fungi and bacteria associated with cervical intraepithelial neoplasia and high-risk human papillomavirus infections in a Hispanic population. Front Microbiol 2018;9:2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee JE, Lee S, Lee H, Song Y-M, Lee K, Han MJ, et al. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 2013;8:e63514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oh HY, Kim B-S, Seo S-S, Kong J-S, Lee J-K, Park S-Y, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect 2015;21:674.e1–9. [DOI] [PubMed] [Google Scholar]
- 32. Mitra A, MacIntyre DA, Lee YS, Smith A, Marchesi JR, Lehne B, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep 2015;5:16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brotman RM, Shardell MD, Gajer P, Tracy JK, Zenilman JM, Ravel J, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis 2014;210:1723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Di Paola M, Sani C, Clemente AM, Iossa A, Perissi E, Castronovo G, et al. Characterization of cervico-vaginal microbiota in women developing persistent high-risk Human Papillomavirus infection. Sci Rep 2017;7:10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Usyk M, Zolnik CP, Castle PE, Porras C, Herrero R, Gradissimo A, et al. Cervicovaginal microbiome and natural history of HPV in a longitudinal study. PLoS Pathog 2020;16:e1008376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mitra A, MacIntyre DA, Ntritsos G, Smith A, Tsilidis KK, Marchesi JR, et al. The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat Commun 2020;11:1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewis FMT, Bernstein KT, Aral SO. Vaginal microbiome and its relationship to behavior, sexual health, and sexually transmitted diseases. Obstet Gynecol 2017;129:643–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Borgogna J-LC, Anastario M, Firemoon P, Rink E, Ricker A, Ravel J, et al. Vaginal microbiota of American Indian women and associations with measures of psychosocial stress. PLoS One 2021;16:e0260813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dixon M, Dunlop AL, Corwin EJ, Kramer MR. Joint effects of individual socioeconomic status and residential neighborhood context on vaginal microbiome composition. Front Public Health 2023;11:1029741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marconi C, El-Zein M, Ravel J, Ma B, Lima MD, Carvalho NS, et al. Characterization of the vaginal microbiome in women of reproductive age from 5 regions in Brazil. Sex Transm Dis 2020;47:562–9. [DOI] [PubMed] [Google Scholar]
- 41. Virtanen S, Rantsi T, Virtanen A, Kervinen K, Nieminen P, Kalliala I, et al. Vaginal microbiota composition correlates between Pap smear microscopy and next generation sequencing and associates to socioeconomic status. Sci Rep 2019;9:7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. MacIntyre DA, Chandiramani M, Lee YS, Kindinger L, Smith A, Angelopoulos N, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep 2015;5:8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nunn KL, Witkin SS, Schneider GM, Boester A, Nasioudis D, Minis E, et al. Changes in the vaginal microbiome during the pregnancy to postpartum transition. Reprod Sci 2021;28:1996–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Romero R, Theis KR, Gomez-Lopez N, Winters AD, Panzer JJ, Lin H, et al. The vaginal microbiota of pregnant women varies with gestational age, maternal age, and parity. Microbiol Spectr 2023;11:e0342922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kervinen K, Holster T, Saqib S, Virtanen S, Stefanovic V, Rahkonen L, et al. Parity and gestational age are associated with vaginal microbiota composition in term and late term pregnancies. EBioMedicine 2022;81:104107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lebeer S, Ahannach S, Gehrmann T, Wittouck S, Eilers T, Oerlemans E, et al. A citizen-science-enabled catalogue of the vaginal microbiome and associated factors. Nat Microbiol 2023;8:2183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garg A, Ellis LB, Love RL, Grewal K, Bowden S, Bennett PR, et al. Vaginal microbiome in obesity and its impact on reproduction. Best Pract Res Clin Obstet Gynaecol 2023;90:102365. [DOI] [PubMed] [Google Scholar]
- 48. Clarke MA, Fetterman B, Cheung LC, Wentzensen N, Gage JC, Katki HA, et al. Epidemiologic evidence that excess body weight increases risk of cervical cancer by decreased detection of precancer. J Clin Oncol 2018;36:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mancilla V, Jimenez NR, Bishop NS, Flores M, Herbst-Kralovetz MM. The vaginal microbiota, human papillomavirus infection, and cervical carcinogenesis: a systematic review in the latina population. J Epidemiol Glob Health 2024;14:480–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Łaniewski P, Herbst-Kralovetz MM. Bacterial vaginosis and health-associated bacteria modulate the immunometabolic landscape in 3D model of human cervix. NPJ Biofilms Microbiomes 2021;7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Vaginal microbiota profiles cluster based on Lactobacillus dominance and not clinical or sociodemographic factors.
Supplementary Figure S2: BV-associated bacteria positively correlate with each other and negatively correlate with Lactobacillus species, including L. crispatus and L. jensenii.
Supplementary Figure S3: Associations of Lactobacillus abundance with race, vaginal pH, and HPV status.
Supplementary Table S1: 16S rRNA sequencing data.
Supplementary Table S2: Differences in demographic, socioeconomic and behavioral/life style factors between participants with Lactobacillus-dominant microbiota and non-Lactobacillus-dominant microbiota.
Supplementary Table S3: Differences in global health and perceived stress measurements between participants with Lactobacillus-dominant microbiota and non-Lactobacillus-dominant microbiota.
Supplementary Table S4: Differences in global health and perceived stress measurements between hrHPV-positive and hrHPV-negative participants.
Supplementary Table S5: Differences in global health and perceived stress measurements between Native and non-Native participants.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The 16S rRNA gene sequences were not deposited in the NCBI database due to limited consent, the presence of possible human reads within the dataset, and Indigenous data sovereignty. Instead, we included a table with ASV counts and corresponding taxonomic annotations in the supplementary materials (Supplementary Table S1).