Graphical abstract
Keywords: PR-AgNPs; Cell proliferation; Apoptosis; HCT-15, p53; Caspase-3; HSP-70
Abstract
Background
In recent years, biosynthesized nanoparticles has shown a promise as alternative avenue for improving the effectiveness of conventional chemotherapy. Despite, there is a significant gap in existing literature concerning the comprehensive study of biogenic silver nanoparticles derived from terrestrial fern species and their potential effects on cancer cells. This study is aiming to investigate effects of biogenic silver nanoparticles synthesized using aqueous extract of bracken fern Pteridium revolutum on inhibiting cell proliferation and inducing apoptosis in HCT-15 cells.
Methods
Biogenic silver nanoparticles synthesized using aqueous extract of Pteridum revolutum followed by their characterization (UV–Visible spectroscopy, TEM, XRD and FTIR). The impact on cell proliferation of HCT-15 cells was assessed by MTT assay while induction of apoptosis was demonstrated via DNA fragmentation, caspase-3 assay, cell cycle arrest, FITC V- Annexin assay and evaluation of expression of apoptotic genes using real time PCR and western blotting techniques.
Results
Results of UV–Vis spectrum of colloidal solution of CW-AgNPs showed surface plasmon resonance peak at 430 nm. TEM and XRD results confirmed synthesis of spherical shaped, 20–40 nm sized nanoparticles. The results elucidate cytotoxic effect of PR-AgNPs against HCT-15 cells in time and dose dependent manner with IC50 observed at 5.79 ± 0.58 µg /mL after 24 h of exposure. Furthermore, PR-AgNPs induce significant alterations in cellular morphology, elevate DNA DNA fragmentation and enhance expression of p53 and caspase-3 in HCT10 cells.
Conclusion
The findings from this study address the noteworthy antiproliferative effects of PR-AgNPs in cancer cells primarily mediated through activation of intrinsic apoptosis pathway by inducing p53 and caspase-3 genes.
1. Background
In recent years, nanomedicine has revolutionized biomedical science, paving the way for innovative therapeutic strategies and the creation of new pharmaceuticals. Extensive research has highlighted the immense potential of metallic nanoparticles across various sectors, including industrial, healthcare, and biomedicine sectors.1, 2 Particularly, silver and gold nanoparticles have been extensively studied for their synthesis methods and diverse applications.3, 4 Development of cancer is characterized by uncontrolled cell proliferation and tumor formation, has traditionally been treated with conventional chemotherapy, which often results in unwanted toxicities and drug resistance due to non-specific targeting. However, nanotechnology has revolutionized cancer therapeutics by enhancing the efficiency and effectiveness of both conventional and novel chemotherapy drugs.5, 6 Nanoparticles have emerged as promising alternative anticancer agents owing to their unique attributes such as non-toxicity, natural antioxidant properties, biocompatibility, multi-targeting capabilities and their ability to modulate biological processes involved in anticancer efficacy.7, 8 Among these, silver nanoparticles have garnered considerable attention for their biogenic synthesis and biomedical applications, showing anticancer activity in various cancer models, both in vitro and in vivo.9, 10, 11 The phytosynthesized silver nanoparticles using extracts different parts of various medicinal plants have gained considerable attention for their anticancer properties. Various studies have demonstrated that AgNPs synthesized from plant extracts exhibit significant cytotoxicity against a wide range of cancer cells.12, 13, 14 The biogenically synthesized silver nanoparticles demonstrate anticancer effects by modulating cellular processes, including cell cycle, signaling pathways, and apoptosis induction through the mechanisms such as oxidative DNA damage, mitochondrial degradation, and cellular shrinkage.15, 16
Although information regarding silver nanoparticles synthesized from various sources and their applications is available in literature, the studies on their effects on cancer cell growth, cytotoxicity, and gene expression at very low concentrations of biogenic AgNPs synthesized using fern species are limited. Further efforts are required to evaluate their efficacy against cancer cells. Therefore, this research aims to explore the potential anticancer properties of silver nanoparticles synthesized from wild terrestrial fern Pteridium revolutum, (Blume) Nakai (Family: Dennstaedtiaceae) also known as bracken fern. Various secondary metabolites, such as tannins, triterpenoids, alkaloids, flavonoids, phytosterols, and phenols, present in different fern species,17, 18 which are known to enhance their antioxidant potential.19, 20, 21 Although some research has been done on using other terrestrial ferns to synthesize metallic nanoparticles and their applications,22, 23, 24 there is little knowledge about creating silver or other nanoparticles from P. revolutum. This gap highlights the need for more studies to explore their biological potential.
The mechanism of action underlying the inhibition of cancer cell proliferation in response to biogenic nanoparticles synthesized using P. revolutum is not well understood. The current research is concentrated on the biosynthesis of silver nanoparticles using the aqueous leaf extract of P. revolutum (PR-AgNPs) and assessing their impact on modulating cell proliferation and apoptosis pathways in the colon carcinoma (HCT-15) cell line. Apoptosis pathway is being investigated to understand the expression of various genes, including p53, and caspase-3.25, 26 Similarly, the role of heat shock proteins in protecting the cells from oxidative stress and inhibiting cell proliferation in response to nanoparticle exposure remains unexplored. In this study cell proliferation inhibition, indicative of metabolic disturbances affecting cell viability, was assessed via MTT assay. The real-time PCR (RT-PCR) was used to measure the mRNA levels of p53, caspase-3, and HSP-70 genes to study changes in genes related to the apoptosis pathway. Protein levels of selected apoptosis markers were checked with western blotting. Additionally, we investigated the potential mechanism of apoptotic cell death using the caspase-3 assay, cell cycle arrest assay, FITC-Annexin V assay, and DNA fragmentation assay.
2. Methods
2.1. Biosynthesis, purification and characterization of silver nanoparticles
Biosynthesis, purification and characterization of silver nanoparticles were conducted through series of steps. Initially, silver nanoparticles were biosynthesized by reacting 10 mL of aqueous extract of bracken fern P. revolutum with 90 mL of 1 mM silver nitrate (AgNO3) solution. The reaction mixture was then incubated at 80 °C in the dark with continuous agitation, and monitored for the change in color of reaction mixture. Subsequently, the PR-AgNPs nanoparticle mixture was centrifuged at 15,000 rpm for 10 min, followed by multiple washes with distilled water to remove unwanted contaminants. Reduction of silver nitrate by the plant extract was monitored for up to 3 h at 30-minute intervals during the formation of biogenic PR-AgNPs. The initial characterization of biogenic silver nanoparticles was carried out using UV–Visible spectroscopy, where the bioreduction of silver ions to nanoparticles was confirmed by scanning the spectrum of the reaction mixture within the range of 300 to 800 nm. Further characterization of the biosynthesized PR-AgNPs was performed using Transmission Electron Microscopy (TEM), X-ray diffraction (XRD), and Fourier Transform Infrared Spectroscopy (FTIR), dynamic light scattering (DLS) and Zeta potential analysis at the Sophisticated Analytical Instrument Facility located at the Sophisticated Test and Instrumentation Centre, Cochin University, Kerala.
2.2. In vitro evaluation of cell viability and cell proliferation
Cell Line & Cell Culture: The inhibitory effects of PR-AgNPs were investigated using the HCT-15 cell line. The HCT-15 cell line obtained from the cell repository of National Centre for Cell Sciences (NCCS), Pune, India. These cells were cultured in T-25 culture flasks with RPMI-1640 medium supplemented with 10 % heat-inactivated fetal bovine serum (FBS) and Penicillin (100 U/mL)-Streptomycin (100 μg/mL). Culture conditions were maintained at 5 % CO2, 95 % humidity, and a temperature of 37 °C. The primary fibroblasts cells obtained from breast tissue were cultured in DMEM/F12 media supplemented with containing FBS (10 %), Pen-strep100 (U/mL), EGF (10 ng/mL), insulin (5 µg/mL), Pitutary extract (50 ng/mL), hydrocortisone (0.5 µg/mL).
2.3. Cell viability/cell proliferation assay (MTT assay)
The MTT colorimetric assay was employed to evaluate the inhibitory effect of biogenic PR-AgNPs on the proliferation of HCT-15 cells by the method as prescribed earlier by Al-Qasmi.27 Initially, 1x104 cells/well were seeded in a 96-well plate with complete RPMI-1640 medium containing 10 % FBS and incubated at 37 °C with 5 % CO2 for 24 h for recovery. Subsequently, the cells were treated with varying concentrations (ranging from 2.5 to 15 µg/mL) of PR-AgNPs in culture medium and further incubated for 24 h at 37 °C with 5 % CO2. After treatment, the cells were washed with Hanks Balanced Salt Solution (HBSS) medium. Then, 10 µl of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) solution at a concentration of 5 mg/ml was added to each well and incubated at 37 °C with 5 % CO2 for 4 h. Following incubation, 200 μL of DMSO was added to dissolve the formazan crystals, and the absorbance of the developed purple color was measured at 560 nm wavelength using a UV–Vis 1800 spectrophotometer (Shimadzu). The percentage inhibition of growth for treated and untreated cells was determined where the inhibition of cell proliferation induced by PR-AgNPs was expressed as the percentage growth inhibition using the formula: Percentage (%) inhibition = 100- (A560 nm of treated cells / A560 nm of control cells) × 100 %. The experiments were conducted in triplicates to ensure the reproducibility and reliability of the results.
2.4. Cell morphology
HCT-15 cells were seeded at a density of 1x106 cells/well in a 6-well plate with RPMI-1640 medium without FBS. Subsequently, the cells were treated with various concentrations of PR-AgNPs, ranging from 2.5 to 15.0 µg/mL. After a 24-hour exposure to the AgNPs, alterations in cell morphology were examined using a phase-contrast microscope (Primovert, Carl Zeiss).
2.5. Study of apoptosis
2.5.1. Caspase- 3 assay
The control and PR-AgNP (IC50: 5.79 µg/mL) treated cells were harvested and washed with ice-cold PBS. Caspase-3 activity in HCT-15 cells was assessed using a caspase-3 assay kit (Sigma Aldrich USA) following the manufacturer's protocol. Cell lysis was performed using 100 μL of lysis buffer (composed of 50 mM HEPES (pH 7.4), 5 mM CHAPS, and 5 mM DTT) for 30 min at 4 °C, followed by centrifugation at 12,000 X g for 20 min to collect protein extracts. Equal volumes (10 μl) of protein extracts were combined with assay buffer (containing 20 mM HEPES (pH 7.4), 0.1 % CHAPS, 10 mM DTT, and 1 mM EDTA), and incubated with the caspase-3 substrate (acetyl-Asp-Glu-Val-Asp p-nitroanilide (Ac-DEVD-pNA)) and caspase-3 inhibitor (Ac-DEVD-CHO) for 4 h. Absorbance was then measured at 405 nm using a double-beam UV–Vis spectrophotometer. Comparative analyses were performed with non-induced cells and in the presence of a caspase-3 inhibitor. The assay for caspase-3 was repeated in triplicates to ensure the repeatability and consistency of the results.
2.6. DNA fragmentation assay
To assess apoptosis, a DNA fragmentation assay was conducted. Approximately 1x105 cells were seeded in 6-well plates and treated with PR-AgNPs (IC50: 5.79 µg/mL), alongside untreated control cells, and then incubated at 37 °C in a 5 % CO2 environment for 24 and 48 h. Subsequently, the cells were harvested and lysed in 0.3 mL of cell lysis buffer containing 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.2 % Triton X-100, and 0.5 % sodium dodecyl sulfate (SDS). Following incubation of the cell lysate with 0.5 mg/mL of RNase A at 37 °C for 1 h and 0.2 mg/mL of proteinase K at 55 °C for 1 h, DNA was precipitated by adding 1/10th volume of 5 M sodium chloride and an equal volume of isopropanol at −20 °C for 16 h, followed by centrifugation at 12,000 x g for 30 min at 4 °C. The resulting DNA pellet was washed with 70 % ice-cold ethanol, air-dried, and then resuspended in T10E1 buffer (pH 8.0). The DNA samples were analyzed using a 1.5 % (w/v) low EEO agarose gel containing 1 μg/mL ethidium bromide, visualized, and photographed using a gel documentation system (BioRad Laboratories, USA). The experimental procedure for DNA fragmentation was repeated three times for the confirmation of the results.
2.7. Annexin-V-FITC/ PI assay
In vitro ability of PR-AgNPs to induce apoptotic cell death in HCT-15 cells was studied by the FITC-Annexin-V assay as described earlier by Vafaei and coworkers.28 A total of 1 × 105 HCT-15 cells were seeded in a 6-well plate and treated with PR-AgNPs at 5.79 µg/mL concentration, alongside untreated control cells. After 24 h of incubation, the cells were collected and rinsed with ice-cold phosphate-buffered saline (PBS). Subsequently, apoptosis was assessed using the FITC-Annexin V/Dead Cell Apoptosis Kit (Invitrogen). The cells were suspended in 1X annexin-binding buffer and then stained with FITC-Annexin V and propidium iodide (PI) at room temperature for 15 min. The stained cells were analyzed using Attune NxT flow cytometer at 488 nm dual wavelength with the Ex/ Em (499/521). Detection of the green fluorescence of Annexin V-FITC through the FITC channel (FL1 or BL1) detection.
2.8. Cell cycle arrest assay
A total of 1 × 105 HCT-15 cells were exposed to PR-AgNPs at concentrations of 5.79 µg/mL. Following a 24-hour incubation period, the cells were detached using trypsinization and rinsed with 1X PBS. Subsequently, the cell pellet was briefly vortexed with 70 % ethanol for cell fixation at 4 °C for 30 min. After fixation, the cells were washed with 1X PBS to eliminate ethanol residues. Next, the cells were treated with RNase A and then stained with propidium iodide (PI) for 30 min. The fluorescence intensity was assessed and recorded using Attune™ NxT Flow Cytometer, blue/red; Make: Thermo Fisher Scientific) in FL-2 or BL2 channels with the Ex/ Em 499/521.
2.9. Western blot analysis
To investigate mechanistic apoptosis induction by PR-AgNPs, western blot analysis was conducted on HCT-15 cells. Cells were seeded at 1 x 105 in 6-well plates with RPMI-1640 medium supplemented with 10 % FBS and grown for 24 h hours at 37 °C in a 5 % CO2, 95 % humidity. Subsequently, the cells were treated with PR-AgNPs (IC50: 5.79 µg/mL) in RPMI-1640 for 24 h. After treatment, cells were harvested, lysed, and centrifuged at 12000Xg for 30 min. Protein concentration was determined, and equal amounts of protein (50 µg/lane) were resolved on 10 % SDS-PAGE. Proteins were transferred to a nitrocellulose membrane, blocked with Tris buffer saline with Tween-20 (TBST) containing 5 % casein for 1 h, and incubated with primary antibodies (anti-p53, anti-caspase-3, anti-HSP70, and anti-Actin) for 1 h. After washing with TBST, the membrane was incubated with secondary antibodies (horseradish peroxidase-conjugated goat anti-rabbit IgG and rabbit anti-mouse IgG) for 1 h and immunoreactive bands were detected using chemiluminescence Western BrightTm ECL detection system (Advansta). After standardization of the western blotting protocol, final confirmation of the results was checked by repeating the experiment thrice three times to ensure the consistency of the results.
2.10. Real time PCR analysis for apoptotic gene expression
The expression changes of p53, caspase-3, and HSP-70 mRNA were analyzed by real-time PCR using cDNA synthesized from RNA extracted from control and PR-AgNPs-treated HCT-15 cells. RNA was extracted from control and PR-AgNPs (IC50: 5.79 µg /mL) treated HCT-15 cells using Trizol reagent, and cDNA was synthesized from 5 µg of total RNA using a cDNA synthesis kit. Real-time PCR was performed with primers (Table 1) SYBR green PCR Master mix, and cycling conditions included an initial denaturation at 95 °C for 10 min followed by 40 cycles of amplification at (95 °C for 15 sec, 55 °C for 60 sec. Ct values were extracted for each gene, and ΔΔCt values were determined to compare gene expression between samples. The results of the experiments were confirmed by repeating the assay procedures three times.
Table 1.
List of Primer sequences of apoptotic genes used for expression studies by RT-PCR in response to PR-AgNPs treatment.
| Sr.No. | Gene | Oligonucleotide Sequence | Tm(0C) | Amplicon size (bp) |
|---|---|---|---|---|
| 1 | P53 | FP 5′-ACTAAGCGAGCACTGCCCAA-3′RP 5′-ATGGCGGGAGGTAGACTGAC-3′ | 5456 | 175 |
| 2 | Caspase3 | FP 5′-GTG GCA TTG AGA CAG ACA GTG G-3′RP 5′-GCCAAG AAT AAT AAC CAG GTG C-3′ | 5754 | 110 |
| 3 | HSP70 | FP 5′-ACC ACT TCG TGG AGG AGT TCA AGA −3′ RP 5′-ACG TGT AGA AGT CGA TGC CCT CAA −3′ |
5757 | 171 |
| 4 | Actin | FP 5′-TCT GGC ACC ACA CCT TCT ACA ATG-3′ RP 5′- AGC ACA GCC TGG ATA GCA ACG −3′ |
5756 | 200 |
2.11. Statistical analysis
Statistical analyses were conducted on all assays performed in triplicate, and the results are presented as mean ± SEM of three independent experiments. The IC50 values with 95 % confidence intervals are reported as Mean ± SEM of three independent experiments. Student's t-test was employed to determine the significance of the difference between the means of gene and protein expression levels in control and PR-AgNPs treated samples, with ***p < 0.005 considered statistically significant. All statistical analyses were conducted using SPSS (IBM Version 20.0) software.
3. Results
3.1. Biosynthesis and characterization of PR-AgNPs
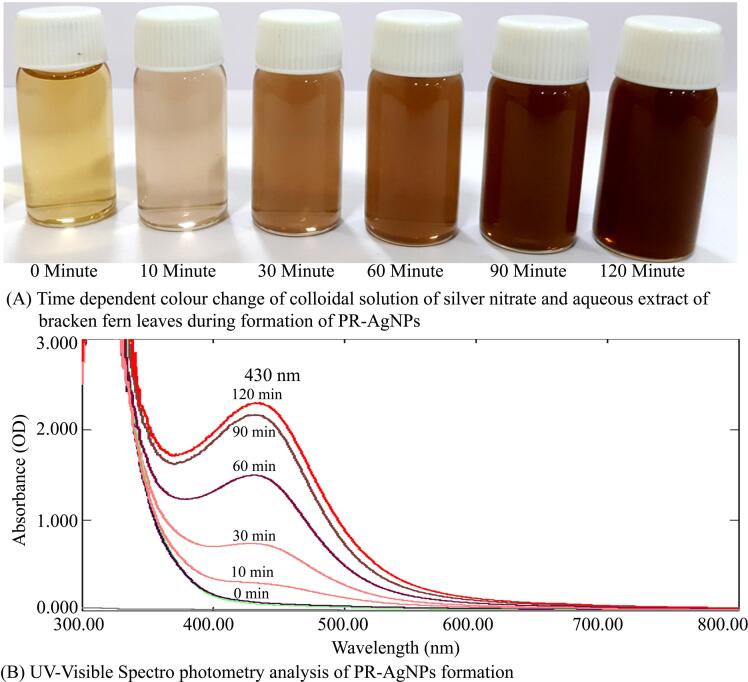
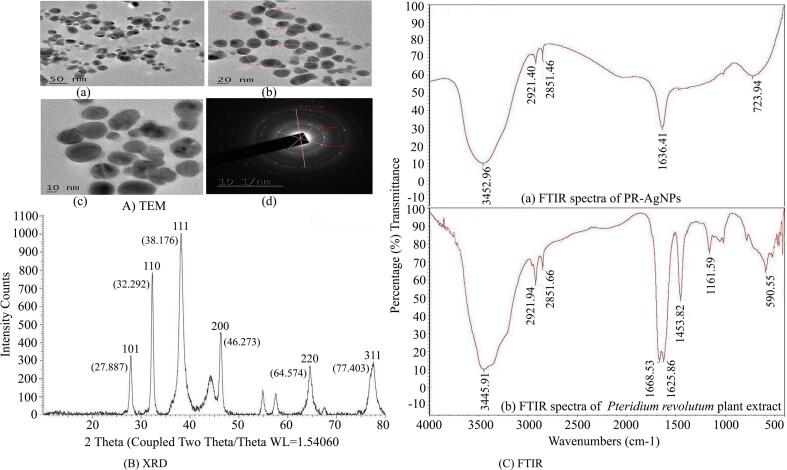
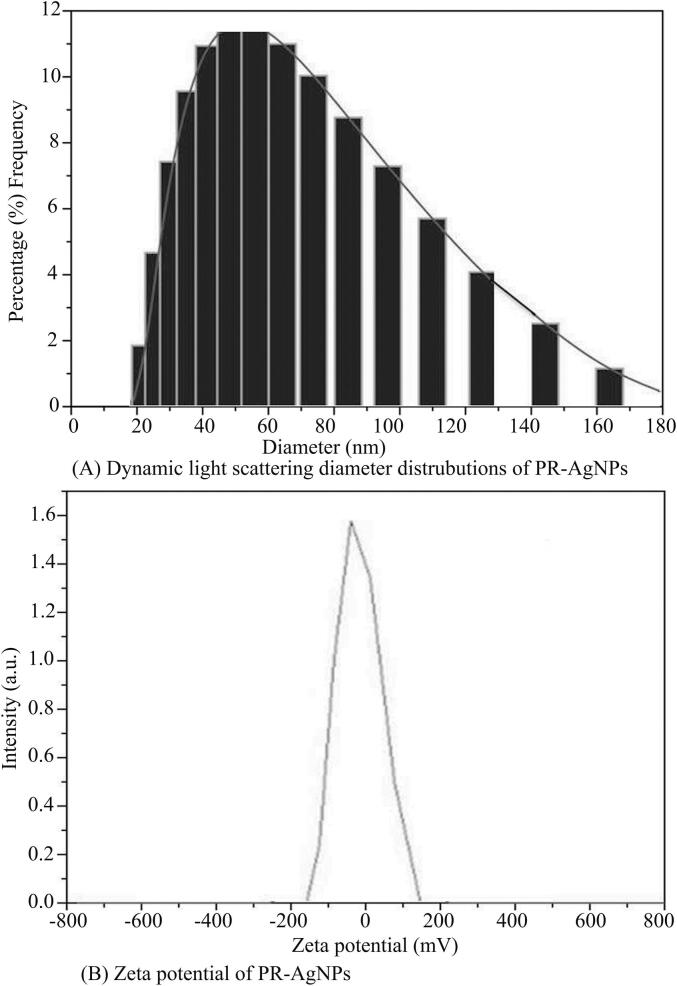
The alteration of silver nitrate solution color, from colorless to a brown, was observed after incubation at 80 °C in the absence of light, as depicted in Fig. 1A. This transformation, prompted by the aqueous extract of P. revolutum, confirmed the conversion of metallic silver to silver nanoparticles. It was supported by the evidences from literature that the active secondary metabolites including phenols, alkanes, ketones, alcohols, alkaloids, terpenoids and flavonoids present in plants can act as capping agents and help in bioreduction of silver ions into nanosilver during the formation of silver nanoparticles. Additionally, UV–visible spectroscopy was utilized to characterize the formation of PR-AgNPs by measuring absorption spectra within the 300–800 nm range. The reduction of silver ions into PR-AgNPs over time was tracked through UV–Visible spectra taken at 30-minute intervals. Notably, the presence of an absorption peak at 430 nm indicated the synthesis of silver nanoparticles of varying sizes, as illustrated in Fig. 1B. The results indicated that the bioreduction of AgNO3 commenced within 30 min of incubation, with complete formation of silver nanoparticles observed after 120 min. Further characterization of the morphology, size, and shape of the biosynthesized PR-AgNPs was conducted using transmission electron microscopy (TEM), revealing spherical particles within the 20–40 nm size range (Fig. 2A). Structural analysis of the biogenic PR-AgNPs was performed using XRD, which revealed characteristic peaks at 2θ values of 38.176°, 46.373°, 64.574°, and 77.403° corresponding to 111, 200, 220, and 311 lattice planes of silver, confirming the crystalline structure of the biogenic PR-AgNPs (Fig. 2B). The FTIR spectra of both P. revolutum and PR-AgNPs were analyzed within the range of 500–4000 cm-1 to identify the functional groups responsible for the bioreduction of silver ions into silver nanoparticles. The aqueous extract of bracken fern exhibited two major peaks at 3445.91 cm-1 and 1668.53 cm-1, while the PR-AgNPs displayed distinct peaks at 3452.96 cm-1 and 1636.41 cm-1. The peaks at 3445.91 cm-1 and 3452.96 cm-1 corresponded to the stretching vibration of the O–H group of alcohol/phenol, while the peaks at 1668.53 cm-1 and 1636.41 cm-1 corresponded to the stretching vibrations of carbonyl (C = C) groups of amides/alkynes during the formation of biogenic PR-AgNPs (Fig. 2C). The results of FTIR indicated the presence of O–H, C = C and CH groups which confirmed the presence of hydroxyl and carboxyl groups of flavonoids of plant extract. This indicated the role of phytoconstituents present in P. revolutum in bioreduction of metallic sliver to silver nanoparticles. Further, size distribution of PR-AgNPs was additionally assessed and verified using dynamic light scattering (DLS) analysis, revealing the presence of smaller nanoparticles (20–40 nm) as well as medium-sized ones (50–70 nm) (Fig. 3A). Similarly, the zeta potential measurement was used to evaluate the surface charge and stability of the colloidal PR-AuNPs solution. The biosynthesized AgNPs exhibited a zeta potential of −13.8 mV, indicating that the nanoparticles carried a negative charge (Fig. 3B). The relatively low zeta potential of PR-AgNPs may be attributed to the polar functional groups of organic compounds found in the aqueous extract of P. revolutum. The biosynthesized PR-AgNPs remained stable for over three months, showing no signs of color change or aggregation and absorption peak at 430 nm. This stability is likely due to the steric effects created by the phytoconstituents adsorbed onto the nanoparticle surface.
Fig. 1.
Biosynthesis and characterization of silver nanoparticles using Pteridium revolutum leaf extract. (A) Time dependent color change of colloidal solution of silver nitrate and aqueous extract of Pteridium revolutum. (B) The absorption spectrum of colloidal solution of AgNO3 and aqueous extract of P. revolutum at different time intervals during the formation of PR-AgNPs. The surface plasmon resonance of biogenic PR-AgNPs exhibited a strong peak at 430 nm.
Fig. 2.
Characterization of PR-AgNP nanoparticless: (A) TEM micrograph of PR-AgNPs. (B) XRD pattern of PR-AgNPs (C) FTIR spectra of (a) PR-AgNPs and (b) aqueous extract of Pteridium revolutum.
Fig. 3.
(A) Dynamic light scattering image for measuring size distribution of PR-AgNPs. (B) Zeta potential pf PR-AgNPs.
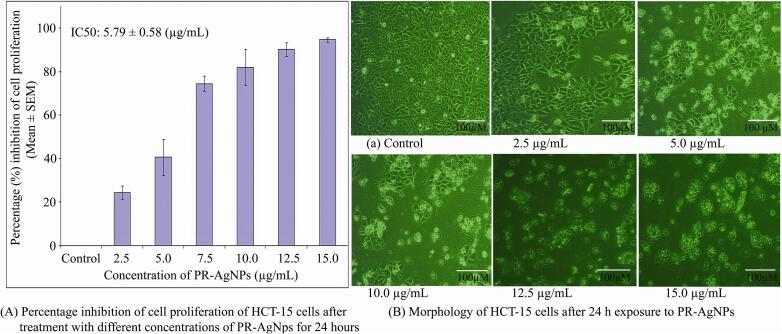
3.2. Biogenic PR-AgNPs inhibit cell proliferation in colon cancer HCT-15 cells
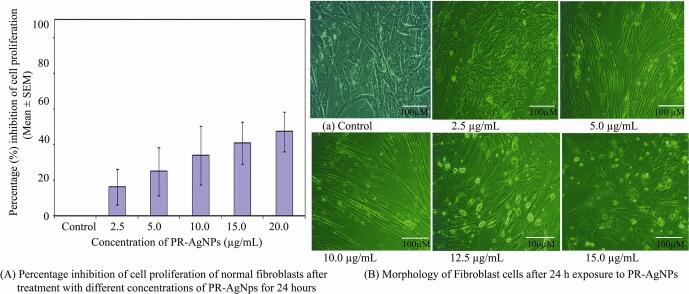
In vitro studies were conducted to evaluate the inhibitory effects of biogenic PR-AgNPs on the proliferation of colon cancer (HCT-15) cells. The findings revealed significant cytotoxicity against HCT-15 cells in a dose-dependent manner (2.5 – 15.0 µg/mL), with cell viability decreasing as the concentration of PR-AgNPs increased (Fig. 4A). The results of the MTT assay clearly demonstrated that biogenic PR-AgNPs effectively suppress cell proliferation in HCT-15 cells after 24 h of exposure. The fifty percent inhibitory concentration (IC50) and IC90 of PR-AgNPs necessary to inhibit cell growth in HCT-15 cells after 24 h were 5.79 ± 0.58 µg/mL and 13.82 ± 1.38 µg/mL, respectively. Complete inhibition of cell growth in HCT-15 cells was achieved at a concentration of 15 µg/mL of PR-AgNPs. The cytomorphology of HCT-15 cells exhibited noticeable morphological abnormalities, including a shrunken appearance, upon treatment with PR-AgNPs at concentrations ranging from 10 to 15 µg/mL (Fig. 4B), contrasting with the healthy appearance of untreated control cells. Biosynthesized PR-AgNPs prompted intracellular suicide in HCT-15 cells, resulting in alterations in cell morphology characterized by compromised cell membrane integrity, cellular shrinkage, cytoplasm condensation, growth arrest, and ultimately, apoptotic cell death. The normal fibroblast cells derived from adjacent breast cancer tissue were exposed to PR-AgNPs in order to demonstrate the cytotoxicity results which indicated comparatively low toxicity on fibroblast cells. When we exposed fibroblast cells to 2.5–––20 μg/mL PR-AgNPs, we observed that even higher concentration i.e, 15 μg/mL inhibits less than 40.70 ± 11.83 percentage cell growth in fibroblast cells after 24 h exposure. (Supplementary Fig. 1).
Fig. 4.
Representative histogram showing in vitro dose dependent cytotoxicity of biogenic PR-AgNPs on HCT-15 cells. (A) Inhibition of cell proliferation at different concentrations (2.5, 5, 7.5,10, 12.5 and 15 µg/ml) of PR-AgNPs was measured by MTT assay. The results represent the means of three independent experiments, and error bars represent the standard error of the mean. (B) Cell morphology of HCT-15 cells treated with PR-AgNPs. All images are taken at 20X magnification with Carl Zeiss phase contrast microscope, Scale bars, 100 µm.
3.3. Biogenic PR-AgNPs induced apoptosis in HCT-15 cells through cell cycle arrest, caspase-3 activation and DNA damage
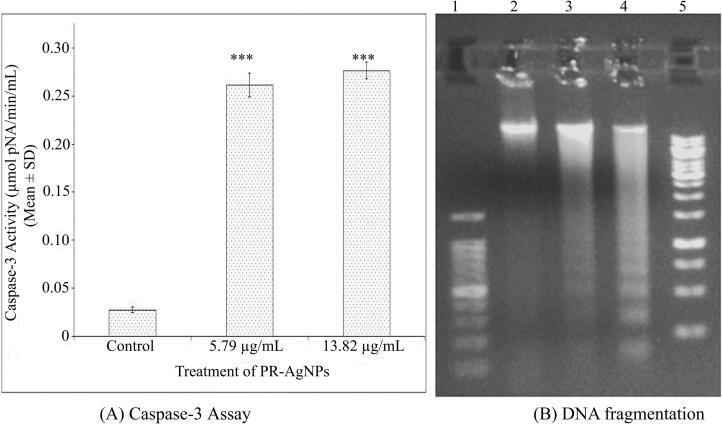
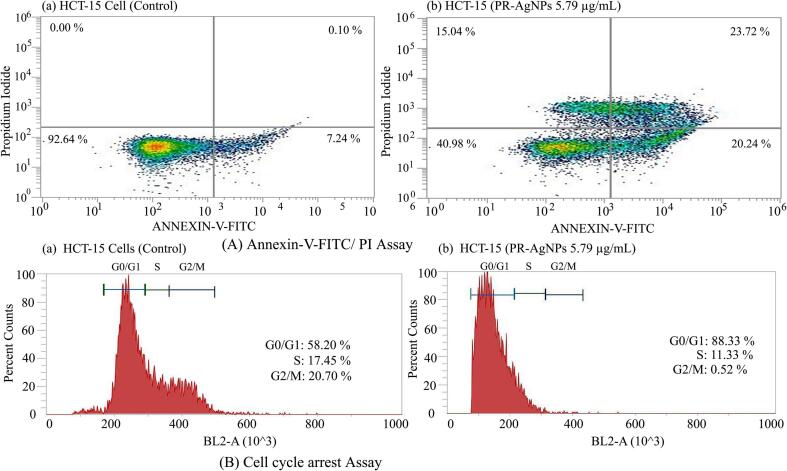
The induction of apoptosis in HCT-15 cells upon exposure to PR-AgNPs was demonstrated through caspase-3 assay. The findings revealed activation of caspase-3 activity as a contributing factor to apoptotic cell death following 24-hour exposure to PR-AgNPs. Specifically, when HCT-15 cells were treated with 5.79 ± 0.58 µg/mL and 13.82 ± 1.38 µg/mL of PR-AgNPs, a significant increase in caspase-3 activity was observed in treated cells (p < 0.005) compared to untreated control cells, as illustrated in Fig. 5A. Moreover, the induction of apoptosis in colon cancer cells was further investigated through the analysis of DNA fragmentation patterns in cells exposed to PR-AgNPs. DNA damage within cells in response to stressors is a distinctive hallmark of the apoptosis process, or programmed cell death. Upon treatment of HCT-15 cells with PR-AgNPs, apoptotic cells displayed characteristic extensive double-strand breaks in cellular DNA, resulting in a ladder-like appearance (Fig. 5B, Lane 3–4), whereas DNA from untreated control cells did not exhibit significant DNA fragmentation (Fig. 5B, Lane 2) when examined through agarose gel electrophoresis. Thus, the extensive DNA damage observed in PR-AgNPs exposed cells provides evidence of apoptosis induction in HCT-15 cells. Furthermore, flow cytometry analysis alongwith Annexin-V-FITC/PI dual staining validated the presence of apoptotic cells. HCT-15 cells treated with 5.79 µg/mL of PR-AgNPs exhibited a concentration-dependent increase in both early and late apoptotic cells compared to untreated controls (Fig. 6A). The flow cytometry results revealed that after 24 h of PR-AgNPs treatment, HCT-15 cells experienced both early and late apoptosis, with 20.24 % of cells undergoing early apoptosis and 23.72 % undergoing late apoptosis compared to untreated control cells. These findings suggest that PR-AgNPs induce cell death in colon cancer cells primarily through early apoptosis. Moreover, cell cycle analysis results indicated apoptotic cell death in HCT-15 cells following PR-AgNPs exposure was associated with cell cycle arrest. Specifically, HCT-15 cells treated with a concentration of 5.79 µg/mL of PR-AgNPs for 24 h and stained with PI showed cell cycle arrest at the G2/M phase (Fig. 6B).
Fig. 5.
(A) Representative histogram showing illustration of caspase-3 activity in HCT-15 cells. Error bars denote the standard error of the mean (SEM) derived from three independent experiments (n = 3). Significant differences in caspase-3 activity (µmolpNA/min/ml) between control and PR-AgNPs exposed samples were assessed using Student’s t-test, with the significance levels marked as (***p < 0.005). (B) The agarose gel images presented are representative illustrations of DNA fragmentation observed in HCT-15 cells treated with PR-AgNPs. Lane 1 exhibits 100 bp DNA marker, while Lane 2 displays DNA from control cells. Subsequently, Lane 3 corresponds to DNA from cells treated with 5.79 µg/ml of PR-AgNPs, and Lane 4 represents the DNA from cells treated with 13.82 µg/ml of PR-AgNPs. Lane 5 indicates the 1Kb DNA marker.
Fig. 6.
(A) Confirmation of apoptosis by Annexin-V-FITC/PI staining assay. (a) Control HCT-15 cells (b) HCT-15 cells treated with IC50: 5.79 µg/ml PR-AgNPs for 24 h. The cells were stained with Annexin-V-FITC and propidium iodide and the percentage of apoptotic cell population was analyzed by flow cytometry. (B) Cell cycle arrest assay where (a) control cells and (b) HCT-15 cells were treated with 5.79 µg/ml PR-AgNPs for 24 h and the cell cycle distribution was analyzed by flow cytometry.
3.4. Biogenic PR-AgNPs altered expression of p53, caspase-3 genes in HCT-15 cells
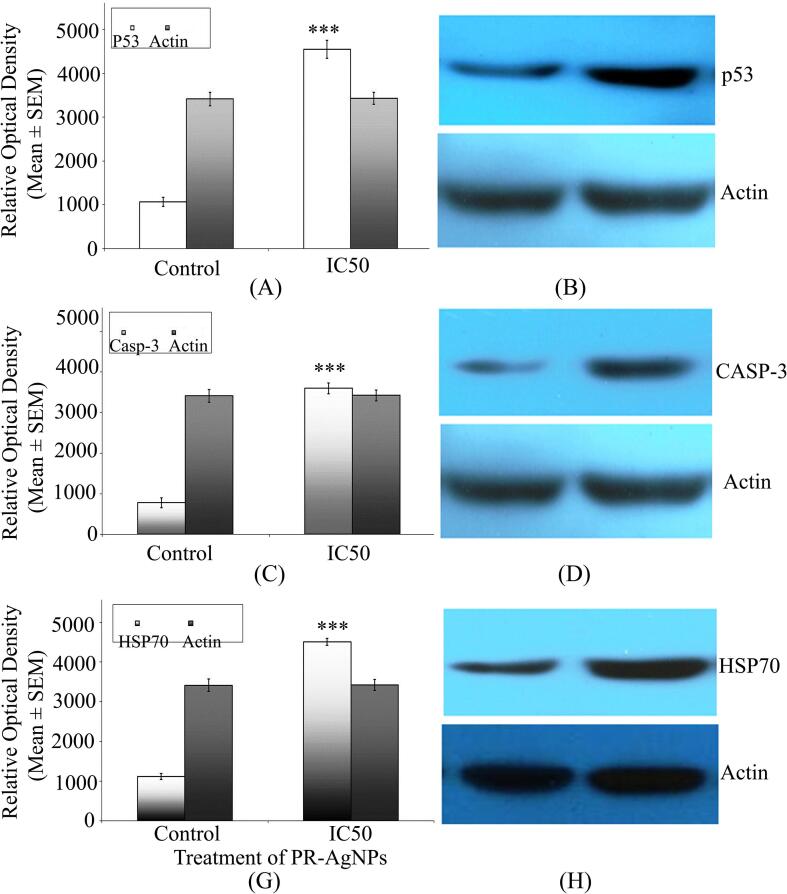
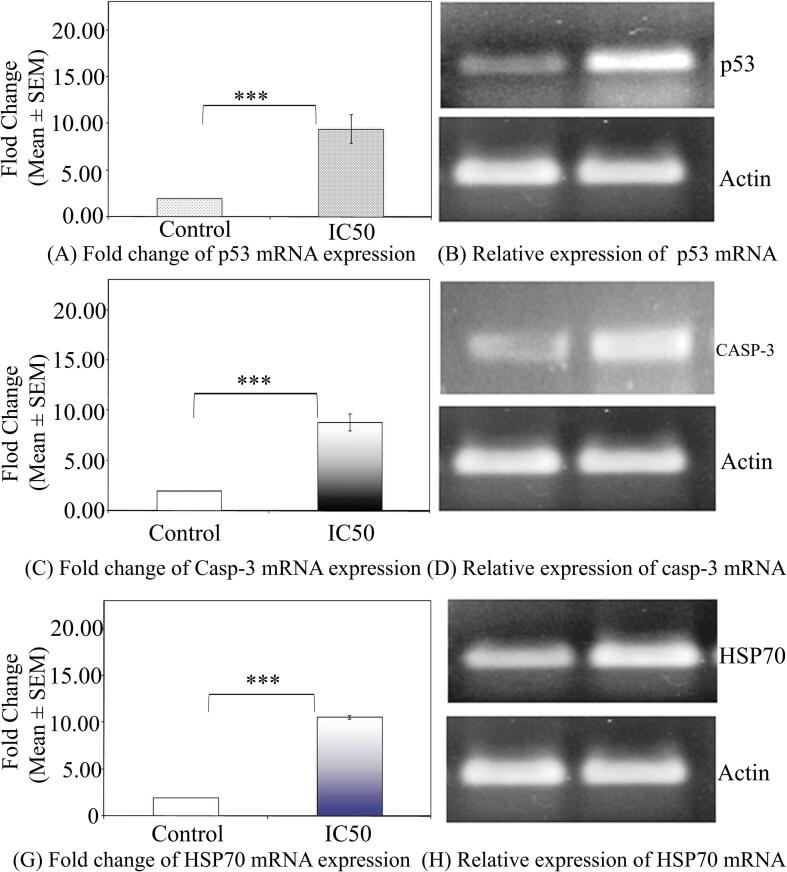
In this research, we investigated the protein-level expression of apoptotic pathway genes such as p53, and caspase-3 in HCT-15 cells following exposure to biogenic PR-AgNPs using western blotting. The results demonstrate a notable increase in the levels of p53 proteins in HCT-15 cells after 24-hour exposure to the IC50 dose of biogenic PR-AgNPs. Furthermore, the presence of PR-AgNPs led to an elevation in caspase-3 enzyme levels, signifying the activation of the apoptosis pathway in HCT-15 cells via the caspase-3 pathway. Significant differences were observed in the protein expression levels of p53, and caspase-3 in HCT-15 cells treated with PR-AgNPs for 24 h compared to untreated controls (p < 0.005) (Fig. 7A-D). Actin served as a loading control, showing consistent intensity bands across all samples, indicating uniform protein concentration (Fig. 7B, D, F). The expression pattern of apoptosis pathway genes at the mRNA level was observed in the results of real-time PCR. Relative expression levels of each control and treated group were determined after normalizing cycle threshold (Ct) values of p53, and caspase-3 genes to the mRNA levels of the housekeeping actin gene. The real-time PCR results were presented as fold changes in mRNA expression levels, indicating a statistically significant increase of 3–4 fold in p53, and caspase-3 mRNA expression levels in PR-AgNPs treated HCT-15 cells (Fig. 8A-B). The mRNA expression levels of apoptotic genes were significantly up-regulated (p < 0.005) in PR-AgNPs treated HCT-15 cells compared to untreated cells. Furthermore, the results demonstrated a consistent correlation between caspase-3 mRNA expression and p53 mRNA expression in response to biogenic AgNPs treatment (Fig. 8A-D).
Fig. 7.
(A) The histogram provided is a representation of the relative levels of p53, caspase-3, and HSP70 proteins derived from HCT-15 cells, as resolved in western blot analysis and quantitated through densitometric analysis. Error bars signify the standard error of the mean (SE) across three independent experiments (n = 3). Significant differences, determined via Student’s t-test (***) with a threshold of p < 0.005, indicate a notable increase in samples treated with PR-AgNPs (5.79 µg/ml) collected at 24 h post-exposure compared to the control group. The results represent the means of three independent experiments, and error bars represent the standard error of the mean. (B) A representative western blot is provided to illustrate the expression of p53, caspase-3, and HSP70 proteins from HCT-15 cells treated with the IC50 concentration (5.79 µg/ml) of PR-AgNPs at 24 h (N = 3). Lane-1 depicts the control, while lane-2 shows cell protein extracts from samples exposed to PR-AgNPs for 24 h.
Fig. 8.
(A) Representative histogram showing the relative levels of p53, caspase-3, and HSP70 cDNA in HCT-15 cells. Significant differences in the levels of p53, caspase-3, and HSP70 (***p < 0.005) were observed, indicating a significant increase in samples obtained from the 24-hour post-exposure period compared to the control. The results represent the means of three independent experiments, and error bars represent the standard error of the mean. (B) Representative amplicons of p53, caspase-3, HSP70, and Actin genes from HCT-15 cells exposed to the IC50 concentration (5.79 µg/ml) of PR-AgNPs are depicted. Lane 1 represents the control, while lane 2 shows mRNA levels from PR-AgNPs exposed samples at 24 h exposure time.
3.5. Biogenic PR-AgNPs altered expression of HSP-70 mRNA and protein in HCT-15 cells
Heat shock protein 70 (HSP-70) acts as a molecular chaperone, playing a vital role in maintaining cellular homeostasis by binding to and stabilizing other proteins to prevent denaturation or aggregation under stressful conditions. Our investigation involved assessing the expression levels of both HSP-70 protein and mRNA using western blotting and real-time PCR, respectively. It was evidenced from our results of western blot analysis showed significant up-regulation of HSP-70 in PR-AgNPs treated HCT-15 cells (p < 0.005) over the unexposed control cells (Fig. 7 G-H). Additionally, a statistically significant increase in the expression of HSP-70 mRNA was observed in HCT-15 cells treated with biogenic AgNPs (p < 0.005) compared to the control cells (Fig. 8G). Results of both western blot analysis and real time PCR revealed significant difference of 3–5 fold change of HSP70 protein as well as mRNA expression in HCT-15 cells after exposure with PR-AgNPs suggesting the role of HSP-70 in mitigation of AgNP induced stress.
4. Discussion
Plant extract mediated bio-synthesis of nanoparticles is an advanced and forthcoming approach in the field of nanoscience research with an intention to develop therapeutic regimen in management of malignant ailments. Terrestrial pteridophytes, are group of lower plants enriched with wide range of phytoconstituents including alkaloids, flavonoids and polyphenols and are known for their bio-potential activities. Considering the biomedical potential of terrestrial ferns, we explored the potential of aqueous extract of P. revolutum for synthesizing silver nanoparticles and evaluated their anti-proliferative and apoptotic effects on cancer cells. Several studies demonstrated biosynthesis metal nanoparticles using bio-components of plant extracts,29, 30, 31, 32 but there remained insufficient knowledge about use of lower cryptogams in biosynthesis of silver nanoparticles.22, 33, 34 This study marks the initial endeavor to comprehend the biosynthesis of silver nanoparticles utilizing bracken fern, along with exploring the potential mechanism behind the inhibition of cell proliferation and induction of apoptosis by biogenic PR-AgNPs. The reduction of silver ions to nanoparticles was identified by the alteration in color of the reaction mixture from colorless to reddish-brown (observed at 120 min) and confirmed through UV–Visible spectroscopy within the range of 300–800 nm. The surface plasmon resonance, with a peak maximum at 430 nm resembling silver nanoparticles, was observed, consistent with findings from previous studies.35, 36 The FTIR analysis elucidated involvement of probable functional chemical groups (O–H, C = C) of phytoconstituents in bioreduction and stabilization of PR-AgNPs. The FTIR peaks of PR-AgNPs at 3452.96 cm−1 and 1636.41 cm−1 (O–H group of alcohol and C = C group of alkynes) were similar to 3445.91 cm−1, 1668.53 cm−1 peaks obtained for aqueous extract of bracken fern. Thus, in line with prior research, the results of FTIR analysis provided strong evidence for the involvement of active phytochemical components in reducing metallic silver ions to silver nanoparticles.37, 38 Likewise, the confirmation of PR-AgNPs by TEM certified 20–40 nm sized spherical silver nanoparticles and XRD peaks of 38.176°, 46.373°, 64.574° and 77.403° indicated the crystalline nature of biosynthesized PR-AgNPs.
Previously, numerous studies have demonstrated the anticancer potential of biogenic nanoparticles against various cancer cell lines, including human breast cancer,39 cervical adenocarcinoma,35 hepatocellular carcinoma,11, 24 and lung cancer.40 However, insubstantial information is available regarding biosynthesis of nanoparticles using lower ferns and their role in cytotoxicity.22, 24 Furthermore, understanding of the mechanisms underlying the inhibition of cell proliferation process through the alteration of cellular pathways remains unclear. In this context, we aimed to understand the mechanism of action of PR-AgNPs in inhibiting cell proliferation and activating the apoptosis pathway in HCT-15 cells. The biosynthesized PR-AgNPs exerted strong anti-proliferative effect on colon carcinoma HCT-15 cells with maximum cell growth inhibition (96.27 %) at 15 µg/mL and IC50 of 5.79 ± 0.58 µg/mL. The distinct changes in cell morphology, characterized by cell shrinkage, contraction, and loss of cell membrane integrity, were observed in HCT-15 cells upon exposure to concentrations of PR-AgNPs at 5 µg/mL and above. For instance, these results were similar to those reported in previous studies where biosynthesized AgNPs had cytotoxic effects against breast cancer MCF-7 and MDA-MB-231 and HCT15 cells.35, 39 This could be because of nineteen different types of secondary plant metabolites present in Pteridium sp.40
Furthermore, PR-AgNPs exhibited minimal cytotoxicity against normal fibroblast cells with cell viability remaining above (%) at the concentration of 20ug/ml tested.
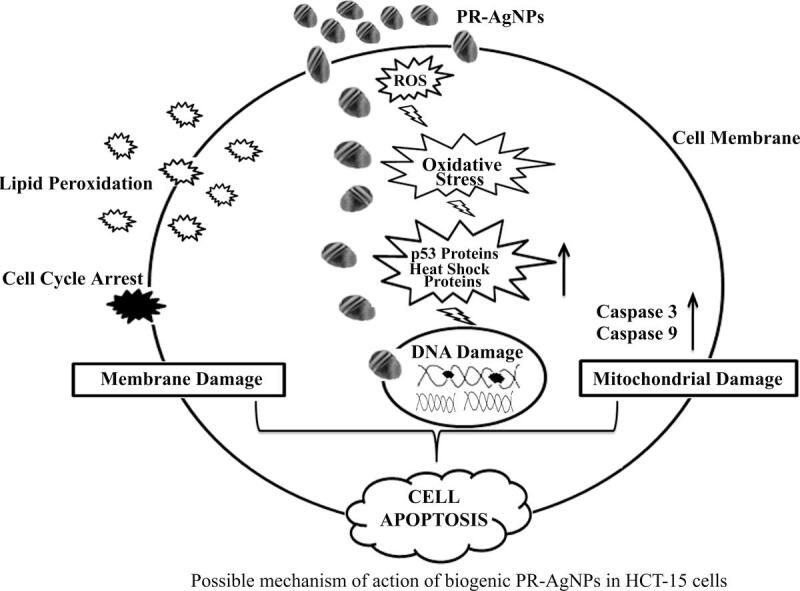
This selective toxicity also suggests that the biogenic AgNPs may selectively target the cancer cells with higher metabolic activity and increased sensitivity of cancer cells towards oxidative stress.41 This selectivity is a crucial factor in reducing the adverse side effects on healthy cells which is a significant challenge in conventional chemotherapy. Decreased cell proliferation and increased macromolecular damage including nuclear fragmentation exhibit cellular apoptosis. Upon treatment of HCT-15 cells with biogenic PR-AgNPs, extensive DNA damage in cellular DNA, resembling a ladder pattern, were observed, indicating the infiltration of AgNPs into the cell nucleus. Similar findings of DNA damage in various cancer cells upon exposure to silver nanoparticles have been reported in previous studies.35, 42 Consistently, our results demonstrate DNA fragmentation in HCT-15 cells following exposure to PR-AgNPs. In the present study, the results confirmed significant increase in caspase-3 expression in HCT-15 cells exposed to PR-AgNPs, indicating the induction of apoptosis. The distinctive changes observed in biological functions, such as inhibition of cell proliferation and induction of apoptosis, as crucial mechanisms governing cell survival or cell death in response to unfavorable physiological conditions. The caspase-3 and p53 mediated pathway is known for its role in cell cycle arrest through activation of target genes concerned with apoptosis.43, 44 On account of this, when HCT-15 cells were exposed to PR-AgNPs for 24 h, a notable increase in the expression of p53, and caspase-3 was observed, indicating the nanoparticles involvement in triggering apoptosis. Previous studies demonstrated that the AgNPs induce apoptosis through intrinsic mitochondrial pathway which correlated with our findings by activation of caspase-3.45 The flow cytometry analysis using Annexin V/PI revealed significant increase in apoptotic cells in response to PR-AgNPs treatment. For instance, 5.79 µg/mL concentration of PR-AgNPs caused 20.24 % early apoptosis as well as 23.72 % late apoptosis as compared to 7.24 % early apoptosis and 0.10 % late apoptosis in untreated control cells. These results are consistent with previous findings showing biogenic AgNPs induce apoptosis in cancer cell lines.46 Furthermore, cell cycle analysis revealed that PR-AgNPs induced G2/M phase arrest in HCT-15 cells preventing the cells from further proliferating. Similar findings were reported prostate cancer cells, where AgNPs caused cell cycle arrest at G2/M phase highlighting the mechanism for inhibition of cell proliferation by biogenic AgNPs.47 Heat shock proteins are pivotal in safeguarding cells during instances of toxicity. Among them, HSP70 stands out as a widely researched stress biomarker, known for its resilience against various stressors like temperature fluctuations, radiation exposure, drug interactions, and nanoparticle exposure.48, 49 Our results indicate a noticeable increase, approximately 3–4 folds, in the expression of HSP-70 in HCT-15 cells treated with PR-AgNPs compared to untreated cells. This suggests that HSP70 could be a stress biomarker in response to silver nanoparticles. However, more research is needed to see how it responds to other types of nanoparticles.
5. Conclusion
It is evident from the results obtained from our study that biosynthesized PR-AgNPs inhibit cell proliferation in colon cancer cells by triggering the intrinsic apoptosis pathway. This mechanism is mediated through the activation of genes such as p53 and caspase-3, as confirmed by both real-time PCR and immunoblotting analysis.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Availability of data and material: All data generated and analyzed in this study are included in this manuscript.
Funding: Intramural funding.
CRediT authorship contribution statement
Kailas D. Datkhile: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Project administration, Investigation, Funding acquisition, Data curation, Conceptualization. Pratik P. Durgawale: Writing – review & editing, Writing – original draft, Validation, Resources, Methodology, Investigation, Formal analysis, Data curation. Nilam J. Jagdale: Writing – original draft, Validation, Software, Resources, Methodology, Investigation. Ashwini L. More: Resources, Methodology, Investigation. Satish R. Patil: Writing – review & editing, Validation, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Kailas D. Datkhile reports financial support was provided by Krishna Vishwa Vidyapeeth (Deemed to be University). Kailas D. Datkhile reports a relationship with Krishna Vishwa Vidyapeeth (Deemed to be University) that includes: employment. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors gratefully acknowledge all the facilities and financial support provided by the Krishna Institute of Medical Sciences ‘Deemed to be University’ Karad, India for experimental work. Authors are thankful to Mr. Santosh Jadhav for technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgeb.2024.100428.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
(A) Representative histogram showing in vitro dose dependent cytotoxicity of PR-AgNPs on normal fibroblast cells with 2.5, 5.0, 10.0, 12.5, 15.0 µg/mL derived from normal breast tissue. The results represent the means of three independent experiments and error bars represent the standard error of mean. (B) Cell morphology of fibroblasts treated with PR-AgNPs. All images are taken at 20X magnification with Carl Zeiss phase contrast microscope, Scale bars, 100 µm.
References
- 1.Rathore B., Sunwoo K., Jangili P., et al. Nanomaterial designing strategies related to cell lysosome and their biomedical applications: a review. Biomaterials. 2019;211:25–47. doi: 10.1016/j.biomaterials.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Yaqoob S.B., Adnan R., Rameez Khan R.M., Rashid M. Gold, silver, and palladium nanoparticles: a chemical tool for biomedical applications. Front Chem. 2020;8:376. doi: 10.3389/fchem.2020.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naidu K.B., Govender P., Adam J.K. Biomedical applications and toxicity of nanosilver: a review. Med Technol SA. 2015;29:13–19. [Google Scholar]
- 4.Menon S., Rajesh Kumar S., Venkat Kumar S. A review on biogenic synthesis of gold nanoparticles, characterization, and its applications. Resource-Efficient. Technol. 2017;3(4):516–527. [Google Scholar]
- 5.Patra J.K., Das G., Fraceto L.F., et al. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnol. 2018;16:71. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Y., Gu Z., Yao C., Luo D., Yang D. Nucleic acid–based functional nanomaterials as advanced cancer therapeutics. Small. 2019;15:1900172. doi: 10.1002/smll.201900172. [DOI] [PubMed] [Google Scholar]
- 7.Cordani M., Somoza A. Targeting autophagy using metallic nanoparticles: A promising strategy for cancer treatment. Cell Mol Life Sci. 2019;76:1215–1242. doi: 10.1007/s00018-018-2973-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y., Zhou Y., Liu L., et al. Nanoparticle- based drug delivery in cancer therapy and Its role in overcoming drug resistance. Front Mol Biosci. 2020;7:193. doi: 10.3389/fmolb.2020.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes H.I.O., Martins C.S.M., Prior J.A.V. Silver nanoparticles as carriers of anticancer drugs for efficient target treatment of cancer cells. Nanomaterials (basel) 2021;11(4):964. doi: 10.3390/nano11040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain N., Jain P., Rajput D., Patil U.K. Green synthesized plant-based silver nanoparticles: therapeutic prospective for anticancer and antiviral activity. Micro and Nano Syst Lett. 2021;9(1):5. [Google Scholar]
- 11.Al-Hhedhairy A.A., Wahab R. Silver nanoparticles: an instantaneous solution for anticancer activity against human liver (HepG2) and breast (MCF-7) cancer cells. Metals. 2022;12(1):148. [Google Scholar]
- 12.Noorbazargan H., Amintehrani S., Dolatabadi A., et al. Anti-cancer & anti-metastasis properties of bioorganic-capped silver nanoparticles fabricated from Juniperus chinensis extract against lung cancer cells. AMB Expr. 2021;11:61. doi: 10.1186/s13568-021-01216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirzaie A., Badmasti F., Dibah H., et al. Phyto-fabrication of silver nanoparticles using Typha azerbaijanensis aerial part and root extracts. Iran J Public Health. 2022;51(5):1097–1106. doi: 10.18502/ijph.v51i5.9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asadipour E., Asgari M., Mousavi P., Piri-Gharaghie T., Ghajari G., Mirzaie A. Nano-biotechnology and challenges of drug delivery system in cancer treatment pathway: review article. Chem Biodiver. 2023;20(6):e202201072. doi: 10.1002/cbdv.202201072. [DOI] [PubMed] [Google Scholar]
- 15.Foldbjerg R., Dang A.D., Autrup H. Cytotoxicity and genotoxicity of silver nanoparticles in the human lung cancer cell line, A549. Arch Toxicol. 2011;85:743–750. doi: 10.1007/s00204-010-0545-5. [DOI] [PubMed] [Google Scholar]
- 16.Buttacavoli M., Albanese N.N., Di G., et al. Anticancer activity of biogenerated silver nanoparticles: Anintegrated proteomic investigation. Oncotarget. 2018;9:9685–9705. doi: 10.18632/oncotarget.23859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao H., Chai T.T., Wang X., et al. Phytochemicals from fern species: potential for medicine applications. Phytochem Rev. 2017;16(3):379–440. doi: 10.1007/s11101-016-9488-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baskaran X.R., Geo Vigila A.V., Zhang S.Z., Feng S.X., Liao W.B. A review of the use of pteridophytes for treating human ailments. J Zhejiang Univ Sci B. 2018;19(2):85–119. doi: 10.1631/jzus.B1600344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarial R., Singh L., Thakur S., Zularisam A.W., Sakinah M., Kanwar S.S. Evaluation of antilipolytic, antioxidant and antibacterial activities of selected ferns. J Appl Pharm Sci. 2017;7(6):150–156. [Google Scholar]
- 20.Halder K., Chakroborty S. An account of antioxidant potential in pteridophytes: a biochemical perspective. Intl J Bioinformatics Biological Sci. 2018;6(1):15–24. [Google Scholar]
- 21.Wang X., Ding G., Liu B., Wang Q. Flavonoids and antioxidant activity of rare and endangered fern: Isoetes sinensis. PLoS One. 2020;15(5):e0232185. doi: 10.1371/journal.pone.0232185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baskaran X., Geo Vigila A.V., Parimelazhagan T., Muralidhara-Rao D., Zhang S. Biosynthesis, characterization, and evaluation of bioactivities of leaf extract-mediated biocompatible silver nanoparticles from an early tracheophyte, Pteris tripartita Sw. Int J Nanomedicine. 2016;11:5789–5806. doi: 10.2147/IJN.S108208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Femi-Adepoju A.G., Dada A.O., Otun K.O., Adepoju A.O., Fatoba O.P. Green synthesis of silver nanoparticles using terrestrial fern (Gleichenia Pectinata (Willd.) C. Presl.): characterization and antimicrobial studies. Heliyon. 2019;5(4):e01543. doi: 10.1016/j.heliyon.2019.e01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das G., Patra J.K., Shin H.S. Biosynthesis, and potential effect of fern mediated biocompatible silver nanoparticles by cytotoxicity, antidiabetic, antioxidant and antibacterial, studies. Mater Sci Eng C Mater Biol Appl. 2020;114 doi: 10.1016/j.msec.2020.111011. [DOI] [PubMed] [Google Scholar]
- 25.Karthik S., Sankar R., Varunkumar K., Ravikumar V. Romidepsin induces cell cycle arrest, apoptosis, histone hyperacetylation and reduces matrix metalloproteinases 2 and 9 expression in bortezomib sensitized non-small cell lung cancer cells. Biomed Pharmacother. 2014;68(3):327–334. doi: 10.1016/j.biopha.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Kuppusamy P., Ichwan S.J., Al-Zikri P.N., et al. In Vitro anticancer activity of Au, Ag nanoparticles synthesized using Commelina nudiflora L. aqueous extract against HCT-116 colon cancer cells. Biol Trace Elem Res. 2016;173(2):297–305. doi: 10.1007/s12011-016-0666-7. [DOI] [PubMed] [Google Scholar]
- 27.Al-Qasmi N. Sustainable and efficacy approach of green synthesized cobalt oxide (Co3O4) nanoparticles and evaluation of their cytotoxicity activity on cancerous cells. Molecules. 2022;27:8163. doi: 10.3390/molecules27238163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vafaei S., Sadat Shandiz S.A., Piravar Z. Zinc-phosphate nanoparticles as a novel anticancer agent: an in vitro evaluation of their ability to induce apoptosis. Biol Trace Elem Res. 2020;198:109–117. doi: 10.1007/s12011-020-02054-6. [DOI] [PubMed] [Google Scholar]
- 29.Khandel P., Yadav R.K., Soni D.K., Kanwar L., Shahi S.K. Biogenesis of metal nanoparticles and their pharmacological applications: present status and application prospects. J Nanostructure Chem. 2018;8(3):217–254. [Google Scholar]
- 30.Datkhile K.D., Durgawale P.P., Patil M.N., Jagdale N.J., Deshmukh V.N. Biosynthesis, characterization and evaluation of biological properties of biogenic gold nanoparticles synthesized using Nothapodytes foetida leaf extract. Nanosci Nanotec Asia. 2021;11(1):84–96. [Google Scholar]
- 31.Lekha D.C., Shanmugam R., Madhuri K., et al. Review on silver nanoparticle synthesis method, antibacterial activity, drug delivery vehicles, and toxicity pathways: recent advances and future aspects. J Nanomaterials Article ID. 2021;4401829:11 pages. [Google Scholar]
- 32.Arif R., Uddin R. A review on recent developments in the biosynthesis of silver nanoparticles and its biomedical applications. Med Devices Sens. 2021;4(1):e10158. [Google Scholar]
- 33.Chatterjee A., Khatua S., Acharya K., Sarkar J. A green approach for the synthesis of antimicrobial biosurfactant silver nanoparticles by using fern. Digest J Nanomater Biostruct. 2019;14(2):479–490. [Google Scholar]
- 34.Gupta P.K., Ranganath K.V.S., Dubey N.K., Mishra L. Green synthesis, characterization and biological activity of synthesized ruthenium nanoparticles using fishtail fern, sago palm, rosy periwinkle and holy basil. Current Sci. 2019;17(8):1308–1317. [Google Scholar]
- 35.Datkhile K.D., Patil S.R., Durgawale P.P., et al. Biogenic silver nanoparticles synthesized using Mexican poppy plant inhibits cell growth in cancer cells through activation of intrinsic apoptosis pathway. Nano Biomed Eng. 2020;12(3):241–252. [Google Scholar]
- 36.Datkhile K.D., Durgawale P.P., Patil S.R. Biogenic synthesis of silver nanoparticles using Lasiosiphon eriocephalus (Decne): In vitro assessment of their antioxidant, antimicrobial and cytotoxic activities. Pharm Nanotechnol. 2023;11(2):180–193. doi: 10.2174/2211738511666221207153116. [DOI] [PubMed] [Google Scholar]
- 37.Hembram K.C., Kumar R., Kandha L., Parhi P.K., Kundu C.N., Bindhani B.K. Therapeutic prospective of plant-induced silver nanoparticles: application as antimicrobial and anticancer agent. Artif Cells Nanomed Biotechnol. 2018;46(sup3):S38–S51. doi: 10.1080/21691401.2018.1489262. [DOI] [PubMed] [Google Scholar]
- 38.Ranoszek-Soliwoda K., Tomaszewska E., Małek K., Celichowski G., Orlowski Krzyzowska M., Grobelny J. The synthesis of monodisperse silver nanoparticles with plant extracts. Colloids Surf B Biointerfaces. 2019;177:19–24. doi: 10.1016/j.colsurfb.2019.01.037. [DOI] [PubMed] [Google Scholar]
- 39.Gurunathan S., Han J.W., Eppakayala V., Jeyaraj M., Kim J.H. Cytotoxicity of biologically synthesized silver nanoparticles in MDA-MB-231 human breast cancer cells. Biomed Res Int. 2013;2013 doi: 10.1155/2013/535796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panneerselvam C., Murugan K., Roni M., et al. Fern-synthesized nanoparticles in the fight against malaria: LC/MS analysis of Pteridium aquilinum leaf extract and biosynthesis of silver nanoparticles with high mosquitocidal and antiplasmodial activity. Parasitol Res. 2016;115(3):997–1013. doi: 10.1007/s00436-015-4828-x. [DOI] [PubMed] [Google Scholar]
- 41.Lokina S., Narayanan K.B., Roopan S.M., Rajasekar S. Selective cytotoxicity of silver nanoparticles towards cancer cells: Advances and perspectives. Colloids Surf B Biointerfaces. 2020;193 [Google Scholar]
- 42.Gurunathan S., Qasim M., Park C., Yoo H., Kim J.H., Hong K. Cytotoxic potential and molecular pathway analysis of silver nanoparticles in human colon cancer cells HCT116. Int J Mol Sci. 2018;19(8):2269. doi: 10.3390/ijms19082269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanipandian N., Li D., Kannan S. Induction of intrinsic apoptotic signaling pathway in A549 lung cancer cells using silver nanoparticles from Gossypium hirsutum and evaluation of in vivo toxicity. Biotechnol Rep (amst) 2019;23:e00339. doi: 10.1016/j.btre.2019.e00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumari R., Saini A.K., Kumar A., Saini R.V. Apoptosis induction in lung and prostate cancer cells through silver nanoparticles synthesized from Pinus roxburghii bioactive fraction. J Biol Inorg Chem. 2020;25(1):23–37. doi: 10.1007/s00775-019-01729-3. [DOI] [PubMed] [Google Scholar]
- 45.Asharani P.V., Hande M.P., Valiyaveettil S. Antiproliferative activity of silver nanoparticles. Biomaterials. 2011;32(29):6985–6993. [Google Scholar]
- 46.Kim J., Lee J., Lee S., Oh J. Synthesis of silver nanoparticles using Diospyros lotus extract and its cytotoxic effects in human breast cancer cells. J Mol Struct. 2019;1179:239–243. [Google Scholar]
- 47.Sharma V.K., Yngard R.A., Lin Y. Silver nanoparticles: Green synthesis and their antimicrobial activities. Adv Coll Interf Sci. 2018;145(1):83–96. doi: 10.1016/j.cis.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Chae Y.J., Pham C.H., Lee J., Bae E., Yi J., Gu M.B. Evaluation of the toxic impact of silver nanoparticles on Japanese medaka (Oryzias latipes) Aquat Toxicol. 2009;94(4):320–327. doi: 10.1016/j.aquatox.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 49.Ahamed M., Posgai R., Gorey T.J., Nielsen M., Hussain S.M., Rowe J.J. Silver nanoparticles induced heat shock protein 70, oxidative stress and apoptosis in Drosophila melanogaster. Toxicol Appl Pharmacol. 2010;242(3):263–269. doi: 10.1016/j.taap.2009.10.016. [DOI] [PubMed] [Google Scholar]