Abstract
In a recent publication in Cell, Xie et al.1 report a sensitive and scalable method for the detection and characterization of native glycoRNAs and identify acp3U, an abundant modified nucleoside discovered 50 years ago in tRNAPhe, as one of the primary attachment sites for N-glycans.
The 2021 discovery by Flynn et al., illustrating that RNA can be glycosylated, offered yet another twist in the now long and meandering pathway of the resurgence of this incredible biopolymer to the peak of cellular biology.2 Besides a handful of “exotic” glycosylated queuosine nucleosides, identified in tRNAs decades earlier,3 the separation between nucleic acid research and glycobiology seemed to be almost absolute up to that point. The groundbreaking discovery that sialylated and fucosylated glycans were conjugated to noncoding RNAs to form glycoRNAs, which were found to be displayed on cell surfaces, unraveled this notion. Flynn’s exploitation of novel azide-containing precursors to hijack the biosynthetic pathways and facilitate bioorthogonal labeling of sialic acid yielded then the first glimpse into cell-surface glycoRNAs but left numerous questions open: (1) which RNAs are subjected to glycosylation and how/where are they synthesized; (2) which nucleosides serve as the attachment site(s); and (3) what are the biological roles of such hybrid biopolymers?
In a recent study in Cell, Xie et al. report RNA-optimized periodate oxidation and aldehyde labeling (rPAL), a refined and more sensitive approach for the enrichment, isolation, and characterization of glycoRNAs. Importantly, they establish the nature of the hitherto unknown linkage, connecting RNA to N-glycans.1 Capitalizing on the distinct reactivity patterns of 1,2-diols in sialic acids, periodate oxidation to an aldehyde, followed by a stable oxime formation through a reaction with an aminooxy-containing reagent or a solid support, has led to the labeling of glycoRNAs. By coupling this approach to highly sensitive mass spectrometry analyses, 3-(3-amino-3-carboxypropyl) uridine (acp3U) was proposed as one of the glycan-linkage nucleosidic anchors (Figure 1).
Figure 1. Deciphering the molecular structure of glycoRNA.
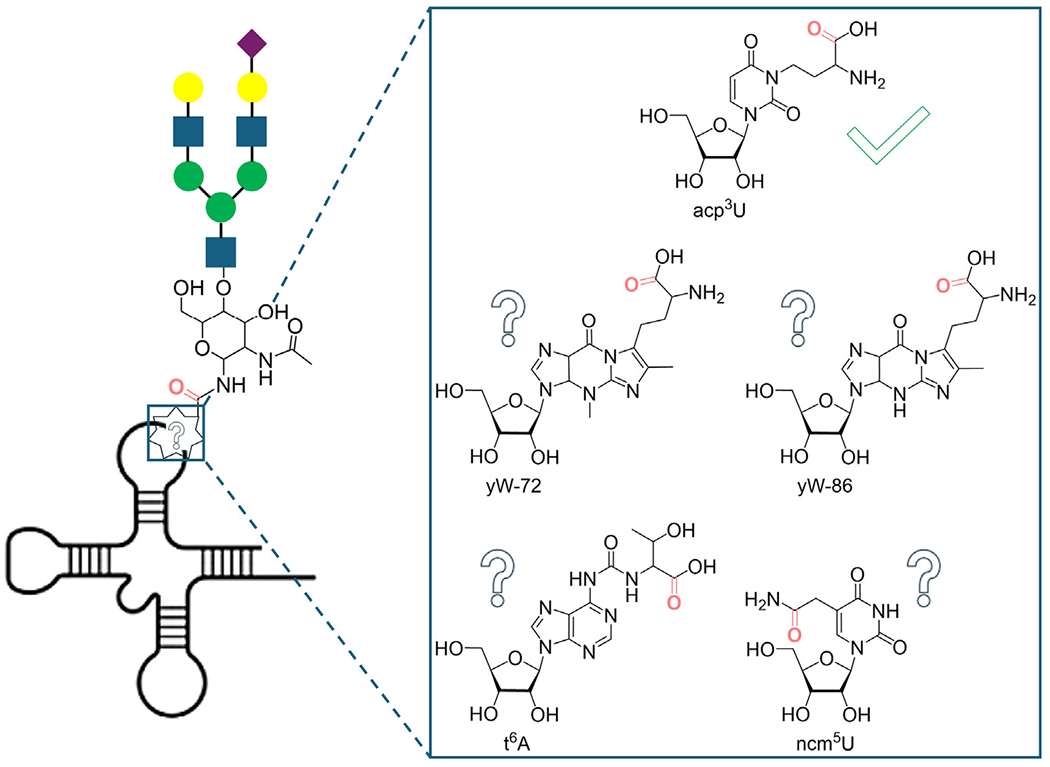
Cartoon representation of a generic sialicglycoRNA (left). An amino-acid-containing nucleoside, acp3U, a verified N-glycan anchoring nucleoside, is shown next to other carboxylic-acid- and carboxamide-containing nucleosides, which may also serve as attachment sites for oligosaccharides (right).
The experimental approach is an adaptation of a robust method previously developed for the characterization of sialoglycoproteins and glycolipids.4 This chemistry was coupled to sequential window acquisition of all theoretical mass spectra (SWATH-MS), a liquid chromatography-tandem mass spectrometry (LC-MS/MS) technique, which benefited from a search engine enabling the analysis of modified RNA nucleosides.5 rPAL offers a more direct approach to labeling sialoglycoRNAs in comparison to the previously reported one, which relied on N-azidoacetylmannosamine-tetraacylated (Ac4ManNAz), a modified metabolic precursor for sialic acid, where an azide moiety ultimately serves as a click handle.2 A comparison of the two methods revealed at least a 25-fold higher signal for the rPAL method over the metabolite-based strategy.
This effective enrichment process, applied to HEK293 and K562 cells, coupled with RNA digestion and specific enzymatic cleavages, released the nucleoside(s) from the glycans and ultimately facilitated the characterization of the linkage between the RNA and the glycan moieties. SWATH-MS analysis revealed a total of 34 unique nucleosides, including the native A, C, G, and U, with distinct nucleoside compositions, depending on the cell line and the approach used for their release. Focusing on 7-aminocarboxy-propylwyosine (yW-72), 7-aminocarboxypropyl-demethylwyosine (yW-86), and acp3U, all carboxylate-containing modified nucleosides detected (Figure 1), revealed the latter as the most abundant across all enzymatic release approaches and cell lines. As a highly conserved modified uridine, acp3U is found in bacterial and mammalian tRNAs.6,7 It has been shown to confer thermal stability on tRNA and be of significance to cell physiology.8
To corroborate the identification of acp3U as an RNA-glycan linking nucleoside, two additional experiments were conducted. The use of an amidase, known to efficiently hydrolyze the amide linkage of N-glycans in heavy (H218O) water, had shown the expected mass increase for the MS signals. Additionally, an enzymatic release approach that included endoglycosidases showed the presence of acp3U-GlcNAc, supporting this modified nucleoside as the modification site.
To further probe the role of acp3U, the rPAL protocol was conducted in two DTWD2 knockout cell lines deprived of an enzyme responsible for installing acp3U in tRNAs.8 A 10% decrease in rPAL signal intensity and a gel shift were seen. Similar observations were made in a direct amidase digestion of glycoRNAs, hydrolyzing the amide bond between the RNA and the glycans. These results point to acp3U as a central glycan anchoring site. A biosynthetic pathway for the generation of cell-surface glycoRNAs was therefore proposed, including an amidation of acp3U, N-glycosylation by oligosaccharide transferase, and then trafficking to the cell surface accompanied by N-glycan modifications. Careful inspection of the experimental observations hint, nonetheless, at the possibility that in the absence of acp3U, the cells likely utilize other nucleosides to anchor shorter and more diverse glycoRNAs.
This method for glycoRNA labeling, along with the discovery of acp3U as an RNA-linking component, represents an important step forward in this new and evolving field of glycoRNA biology. This approach possesses, however, certain caveats: (1) it is specific to sialylated glycoRNAs; (2) the periodate oxidation can also oxidize the 2’,3’ vicinal diol at the 3’ terminus of RNA, albeit to a lower extent; and (3) the release-dependent distinct findings suggest a certain protocol bias. For instance, the carboxylic-acid-bearing modified nucleoside, N6-threonylcarbamoyladenosine (t6A) (Figure 1), was found in much higher abundance than acp3U in one cleavage method but was not detected at all in another. An additional intriguing future pathway would be to search for amide-modified nucleosides, such as 5-carbamoylmethyluridine(ncm5U) (Figure 1), as potential glycan anchoring sites, as well as O-linked conjugates.9
The important advances reported in this contribution can help address open questions in this nascent field, particularly the biological role(s) of glycoRNAs and their abundance in other domains of life. Further into the future, correlations between the structure and frequency of glycoRNAs and certain metabolic pathways and/or pathologies may be drawn. With additional venues for the study of glycoRNAs, such as imaging approaches,10 we should be expecting exciting years ahead, full of discoveries in this emerging glycoRNA world.
ACKNOWLEDGMENTS
Research in Y.T.’s laboratory is supported by the NIGMS of the National Institutes of Health under award number R35 GM139407.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Xie Y, Chai P, Till NA, Hemberger H, Lebedenko CG, Porat J, Watkins CP, Caldwell RM, George BM, Perr J, et al. (2024). The modified RNA base acp3U is an attachment site for N-glycans in glycoRNA. Cell 187, 5228–5237.e7. 10.1016/j.cell.2024.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, George BM, Majzoub K, Villalta PW, Carette JE, and Bertozzi CR (2021). Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184, 3109–3124.e22. 10.1016/j.cell.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasai H, Nakanishi K, Macfarlane RD, Torgerson DF, Ohashi Z, McCloskey JA, Gross HJ, and Nishimura S (1976). Letter: The structure of Q* nucleoside isolated from rabbit liver transfer ribonucleic acid. J. Am. Chem. Soc 98, 5044–5046. 10.1021/ja00432a071. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Li XJ, Martin DB, and Aebersold R (2003). Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol 21, 660–666. 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 5.. Xie Y, De Luna Vitorino FN, Chen Y, Lempiäinen JK, Zhao C, Steinbock RT, Lin Z, Liu X, Zahn E, Garcia AL, et al. (2023). SWAMNA: a comprehensive platform for analysis of nucleic acid modifications. Chem. Commun 59, 12499–12502. 10.1039/d3cc04402e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman S, Li HJ, Nakanishi K, and Van Lear G (1974). 3-(3-amino-3-carboxy-n-propyl)uridine. The structure of the nucleoside in Escherichia coli transfer ribonucleic acid that reacts with phenoxyacetoxysuccinimide. Biochemistry 13, 2932–2937. 10.1021/bi00711a024. [DOI] [PubMed] [Google Scholar]
- 7.Ohashi Z, Maeda M, McCloskey JA, and Nishimura S (1974). 3-(3-Amino-3-carboxypropyl)uridine: a novel modified nucleoside isolated from Escherichia coli phenylalanine transfer ribonucleic acid. Biochemistry 13, 2620–2625. 10.1021/bi00709a023. [DOI] [PubMed] [Google Scholar]
- 8.Takakura M, Ishiguro K, Akichika S, Miyauchi K, and Suzuki T (2019). Biogenesis and functions of aminocarboxypropyluridine in tRNA. Nat. Commun 10, 5542. 10.1038/s41467-019-13525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCown PJ, Ruszkowska A, Kunkler CN, Breger K, Hulewicz JP, Wang MC, Springer NA, and Brown JA (2020). Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA 11, e1595. 10.1002/wrna.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Guo W, Mou Q, Shao X, Lyu M, Garcia V, Kong L, Lewis W, Ward C, Yang Z, et al. (2024). Spatial imaging of glycoRNA in single cells with ARPLA. Nat. Biotechnol 42, 608–616. 10.1038/s41587-023-01801-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


