ABSTRACT
Objective:
Schiff bases are versatile chemical compounds extensively used in various applications, including as catalysts, polymer stabilizers, pigments, dyes, and building blocks for organic synthesis. In addition, they exhibit a wide range of biological activities, such as antifungal, antibacterial, antiviral, antiproliferative, anti-inflammatory, and antipyretic effects.
Methods:
A novel Schiff base ligand (HL) was synthesized by condensing isatin with 2,6-diaminopyridine and isoleucine, followed by the preparation of transition metal complexes. The ligand and complexes were characterized using techniques such as elemental analysis, IR, 1H-NMR, UV-vis spectroscopy, mass spectrometry, and thermal analysis. Antimicrobial, antiproliferative activities, and structural investigations through X-ray diffraction and scanning electron microscopy were also evaluated.
Results:
The complexes were identified as [Cr(L)Cl(H2O)]Cl·2H2O, [Fe(L)Cl2], [M(L)]Cl·nH2O, and [M(L)(H2O)2]Cl, where M represents Mn(II), Cu(II), Cd(II), Co(II), Zn(II), and Ni(II). Thermogravimetric analysis showed initial water loss, followed by decomposition of anionic compounds and ligands. The ligand forms a uninegative-tetradentate bond with the metal ions, and all complexes, except Fe(III), exhibit electrolytic behavior. Most complexes displayed tetrahedral geometry, while Ni(II), Co(II), and Zn(II) had octahedral geometry. The metal complexes showed enhanced antibacterial, antifungal, and antiproliferative activity against MCF-7 breast cancer cells compared to the free ligand. Molecular docking studies indicated inhibitory potential against receptors 1GS4, 2HQ6, 3DJD, and 5JPE.
Conclusion:
These newly synthesized ligands and complexes show promise as therapeutic agents against infections and cancer, though further studies are needed to understand their mechanisms.
Keywords: Biological activity, isoleucine Schiff base, metal complexes, molecular docking, Schiff bases, spectrum investigations
Introduction
Transition metal complexes find broad applications in fields such as photochemistry, biological systems, material synthesis, and catalysis, due to their remarkable diversity in chemical, optical, and magnetic properties.[1-3] Schiff bases, a prominent class of compounds, are extensively utilized as chemical catalysts, polymer stabilizers, pigments, dyes, and as precursors in organic synthesis. Beyond their functional applications, Schiff bases exhibit a wide range of biological activities, including antifungal, antibacterial, antimalarial, antiproliferative, anti-inflammatory, antiviral, and antipyretic effects.[4-6] These versatile compounds feature imine (or azomethine) groups, which are essential for their bioactivity and are commonly found in both naturally occurring and synthetic materials.[7] The biological efficacy of Schiff bases has been shown to be closely linked to the presence of the imine group, making them important candidates for therapeutic and pharmaceutical development.[8]
Isoleucine is a well-known amino acid recognized for its role in enhancing muscle tissue healing and repair, promoting coagulation at injury sites, and increasing endurance. Its primary role in the body involves supporting energy production and aiding recovery after intense physical activity, making it an essential nutrient for athletes and bodybuilders.[9]
The objective of the present study was to synthesize and characterize a novel Schiff base ligand through the condensation of isatin with the donor amino acid isoleucine and 2, 6-aminopyridine. The resulting metal complexes were characterized using elemental analysis, thermal analysis, molar conductivity, and infrared spectroscopy to determine their structure. The cobalt (Co[II]) and nickel (Ni[II]) complexes were found to be amorphous, while the complexes of isoleucine ligand with Co(II), Ni(II), iron (Fe[III]), manganese (Mn[II]), copper (Cu[II]), zinc (Zn[II]), and cadmium (Cd[II]) were found to be crystalline. Scanning electron microscopy (SEM) images confirmed the nanostructured nature of these compounds. The antibacterial and anticancer efficacy of the synthesized ligands and their M(II)/(III) complexes were assessed, revealing significant bioactivity. In addition, molecular docking studies were performed to evaluate the potential biological interactions of the synthesized ligand with key receptors, including 1GS4, 2HQ6, 3DJD, and 5JPE, suggesting promising biological properties.
Methods
Materials and reagents
The highest purity and analytical reagent grade were used for each compound. Isatin, isoleucine, 2,6-diaminopyridine, CrCl3·6H2O, MnCl2·2H2O, NiCl2·6H2O, FeCl3·6H2O, and CoCl2·6H2O were purchased from Acros, Strem Chemicals, and Sigma-Aldrich, respectively. The organic solvents, including diethyl ether, dimethylformamide (DMF), and ethyl alcohol, were of spectroscopic grade from BDH. Deionized water was routinely used for all preparations. Other chemicals, such as sodium bicarbonate, isotonic trypan blue, dimethyl sulfoxide (DMSO), RPMI-1640 media, penicillin, streptomycin, trypsin, acetic acid, sulforhodamine B (SRB), and trichloroacetic acid (TCA), were supplied by Sigma Chemical Co., St. Louis, MO, USA.
Human tumor cell lines (breast cells) obtained from the American Type Culture Collection were cryopreserved in liquid nitrogen at −180°C. These cell lines were maintained through serial sub-culturing at the National Cancer Institute in Cairo, Egypt.
Solutions
Stock solutions of metal complexes (1 × 10-3 mol/L) were prepared by dissolving an accurately weighed amount of the ligand and its metal complexes in DMF. Conductivity measurements for each metal complex were carried out using these stock solutions. For UV-visible spectrophotometric analysis, solutions of the Schiff base ligand and its metal complexes (1 × 10-4 mol/L) were prepared by diluting the original stock solutions.
Reagents for anticancer experiments
A fresh stock solution of the Schiff base ligand (1 × 10-3 mol/L, 0.003514 g/L) was prepared in 95% ethanol. DMSO from Sigma Chemical Co., St. Louis, MO, USA, was used for cryopreserving cells, while RPMI-1640 medium (Sigma Chemical Co., St. Louis, MO, USA) was used for cell growth and maintenance. The preparation of the medium involved weighing 10.4 g of RPMI-1640 powder, adding 2 g of sodium bicarbonate, and diluting it to a final volume of 1 L, followed by thorough mixing to ensure complete dissolution. The medium was then sterilized using a Millipore bacterial filter (0.22 μm).
The RPMI-1640 medium (Sigma Chemical Co., St. Louis, MO, USA) was prepared using sodium bicarbonate. For cell viability counting, a 0.05% isotonic Trypan blue solution in normal saline was obtained from Sigma Chemical Co., St. Louis, MO, USA. In addition, the RPMI-1640 medium was supplemented with 10% fetal bovine serum (heat-inactivated at 56°C for 30 min), 100 units/mL penicillin, and 2 mg/mL streptomycin, all provided by Sigma Chemical Co. Trypsin (0.025%, w/v) from the same supplier was used for cell harvesting. To dissolve the unbound SRB dye, 1% (v/v) acetic acid (Sigma Chemical Co., St. Louis, MO, USA) was used. SRB, a protein-binding dye, was dissolved in 1% acetic acid, and a 50% TCA stock solution was prepared for protein precipitation by adding 50 μL of the stock to 200 μL of RPMI-1640 medium per well to achieve a final concentration of 10%. The solvents used for other procedures included 100% isopropanol and 70% ethanol.
Instruments
Microanalyses for carbon, hydrogen, and nitrogen were conducted at Cairo University’s Microanalytical Center using a CHNS-932 (LECO) Vario Elemental Analyzer. For metal content determination, solid complexes were dissolved in concentrated HNO3 followed by dilution with deionized water. The metal content was analyzed at the Egyptian Petroleum Research Institute using inductively coupled plasma atomic emission spectrometry. Fourier-transform infrared spectroscopy (FT-IR) spectra (4000–400 400 cm−1) were recorded using a Perkin-Elmer 1650 spectrometer with KBr pellets.
Electronic spectra of the samples were obtained at room temperature in ethanol using a Shimadzu 3101PC spectrophotometer. Proton nuclear magnetic resonance (^1H-NMR) spectra were recorded on a 300 MHz Varian-Oxford Mercury at room temperature, with TMS as an internal standard. Mass spectra were acquired at 70 eV using an EI method on an MS-5988 GS-MS Hewlett-Packard instrument at the Microanalytical Center, National Center for Research, Egypt.
Molar conductivities of 1 × 10−3 mol/L solutions of the solid complexes in DMF were measured using a Jenway 4010 conductivity meter. Thermogravimetric analysis and differential thermogravimetric analysis (TGA and DTG) of the solid complexes were performed from room temperature to 1000°C using a Shimadzu TG-50H thermal analyzer.
X-ray powder diffraction (XRD) experiments were carried out with a PANalytical X’pert PRO diffractometer equipped with a secondary monochromator and Cu radiation (λ = 1.542 Å) at 45 kV, 35 mA, with a scanning speed of 0.02°/s. The relative intensities (I/I0) and corresponding spacings (d, Å) of the diffraction peaks were determined between 20° and 60°. The diffraction patterns and relative intensities were compared with the International Centre for Diffraction Data files to confirm the crystal structure.
Preparation of Schiff base ligand
To synthesize the Schiff base ligand (HL), isoleucine (30.49 mmol, 4 g) was added dropwise to L1 (30.94 mmol, 7.26 g) in ethanol. The mixture was then refluxed and stirred for approximately 2 h at a temperature between 100°C and 150°C, resulting in the formation of a dark purple solid. As shown in Scheme 1, the solid was filtered, recrystallized, washed with ethanol, and dried under vacuum.
Scheme 1.
Schiff base ligand preparation (HL1)
HL1
(cyclopenta-2,4-dien-1-yl)(cyclopenta-2,4-dien-1-yl)(1-((8-aminonaphthalen-1-yl)imino)ethyl) (HL) was obtained as a dark purple solid. FT-IR (v, cm−1): (OH) 3434 br, (C=O) 1705 w, azomethine (C=N) 1619 sh, (COO−) asymmetric 1456 s, (COO−) symmetric 1360 w. The yield was 80,%, with a melting point of 180°C. The calculated percentages for C, H, and N in C19H21N5O2 were 64.88%, 5.98%, and 19.92%, respectively. The experimental values were C 64.74%, H 5.98%, and N 19.92%. ¹H-NMR (300 MHz, DMSO–d6, ppm) revealed the following peaks: 4.20 (d, 1H, NH), 6.74–7.24 (m, 8H, aromatic ring), 8.33 (s, 2H, NH2), 10.30 (s, 1H, OH group). UV-Vis absorption maxima were observed at 215 nm (π-π* transition of the aromatic ring), 248 nm (n-π* transition of the azomethine group), and 339 nm (n-π* transition of the azomethine group).
Preparing of metal complexes
A heated solution (60°C) of the appropriate metal chloride (1.1383 mmol) was combined with a hot solution of the HL ligand (0.4 g/L, 1.1383 mmol) in 25 mL of ethanol to form the metal complexes. The mixture was stirred under reflux for 1.5 h, resulting in the precipitation of the complexes. The precipitates were filtered, washed with hot ethanol and acetone, and then dried in a vacuum desiccator over anhydrous calcium chloride.
[Cr(III)(L)Cl(H2O)]Cl·2H2O
Brown solid, m.p. >300°C, 75% yield. Analysis for C19H27Cl2CrN5O5 :(%): Calc. C, 43.16; H, 5.11; N, 13.25; Cr, 9.84. Λm (Ω−1mol−1cm2) = 70. FT-IR (v, cm−1): v(C=O) 1711w, azomethine ν(C=N) 1645s, ν (COO−)asym 1462s, ν(COO−)sym1320w, ν(H2O stretch) 980w, 875m, ν(M–O) 568w, ν(M–O of H2O) 520w, ν(M–N) 477w.
[Mn(II)(L)(H2O)2]Cl.3H2O
Brown solid, m.p. >300°C, 77% yield. Analysis for C19H27Cl2CrN5O5 (%): Calc. C, 42.88; H, 5.83; N, 13.16; Mn, 10.33. Found: C, 42.56; H, 5.56; N, 12.94; Mn, 10.14. Λm = 76. FT-IR: ν(C=O) 1707m, azomethine ν(C=N) 1620s, ν(COO−)asym 1464s, ν(COO−)sym 1310w, ν(H2O stretch) 922w, 875w, ν(M–O) 594w, ν(M–O of H2O) 540w, ν(M–N) 493w.
[Fe(III)(L)Cl2]·2H2O
Reddish-brown solid, m.p. >300°C, 76% yield. Analysis for C19H25Cl2FeN5O4 (%): Calc. C, 44.34; H, 4.86; N, 13.61; Fe, 10.86. Found: C, 44.12; H, 4.34; N, 13.44; Fe, 10.65. Λm = 31. FT-IR: ν(C=O) 1707m, azomethine ν(C=N) 1616s, ν(COO−)asym 1458s, ν(COO−)sym 1324w, ν(M–O) 544m, ν(M–N) 477w.
[Co(II)(L)]Cl·2H2O
Brown solid, m.p. >300°C, 79% yield. Anal. for C19H25ClCoN5O4 (%): Calc. C, 47.32; H, 4.77; N, 14.53; Co, 12.23. Found: C, 47.28; H, 4.33; N, 14.39; Co, 12.07. Λm = 118. FT-IR: ν(C=O) 1705w, azomethine ν(C=N) 1641s, ν(COO−)asym 1463s, ν(COO−)sym 1330w, ν(M–O) 588w, ν(M–N) 486w.
[Ni(II)(L)]Cl·4H2O
Brown solid, m.p. >300°C, 75% yield. Analysis for C19H29ClNiN5O6 (%): Calc. C, 43.72; H, 5.56; N, 13.42; Ni, 11.25. Found: C, 43.28; H, 5.35; N, 13.16; Ni, 11.09. FT-IR: ν(C=O) van., azomethine ν(C=N) 1625s, ν(COO−)asym 1461s, ν(COO−)sym 1310w, ν(M–O) 591w, ν(M–N) 486w.
[Cu(II)(L)(H2O)2]Cl·H2O
Brown solid, m.p. 205°C, 72% yield. Analysis for C19H27ClCuN5O5 (%): Calc. C, 45.20; H, 5.35; N, 13.88; Cu, 12.60. Found: C, 45.07; H, 5.17; N, 13.82; Cu, 12.07. Λm = 61. FT-IR: ν(C=O) 1720m, azomethine ν(C=N) 1618s, ν(COO−) asym 1458s, ν(COO−)sym 1310w, ν(H2O stretch) 920w, 876w, ν(M–O) 597w, ν(M–O of H2O) 520w, ν(M–N) 477w.
[Zn(II)(L)]Cl·2H2O
Brown solid, m.p. >300°C, 74% yield. Analysis for C19H25ClZnN5O4 (%): Calc. C, 46.69; H, 5.12; N, 14.34; Zn, 13.39. Found: C, 46.47; H, 5.09; N, 14.28; Zn, 13.22. FT-IR: ν(C=O) 1705w, azomethine ν(C=N) 1621s, ν(COO−)asym 1460s, ν(COO−)sym 1324w, ν(M–O) 582w, ν(M–N) 499w.
[Cd(II)(L)(H2O)2]Cl·H2O
Brown solid, m.p. >300°C, 73% yield. Analysis for C19H27ClCdN5O5 (%): Calc. C, 41.21; H, 4.88; N, 12.65; Cd, 20.32. Found: C, 41.17; H, 4.21; N, 12.65; Cd, 20.24. FT-IR: ν(C=O) 1712m, azomethine ν(C=N) 1621sh, ν(COO−)asym 1464s, ν(COO−)sym 1320w, ν(H2O stretch) 920w, 876m, ν(M–O) 596w, ν(M–O of H2O) 520w, ν(M–N) 486w.
Spectrophotometric investigations
The absorption spectra of the free ligand and its metal complexes (1 × 10−4 mol/L) were recorded across the 200–700-nm wavelength range.
Biological activities
Anti-microbiological effect
A 5-mm filter paper disk was placed in 250-mL flasks containing 20 mL of the tested solution at a concentration of 100 mg/mL. The flasks were autoclaved for 20 min at 121°C. Using the agar diffusion method, two fungal strains (Candida albicans and Aspergillus fumigatus) and four bacterial strains (Gram-positive Bacillus subtilis and Staphylococcus aureus, Gram-negative Salmonella spp. and Escherichia coli) were inoculated on LB agar plates. A saturated disk with the tested solution was placed in the center of each plate. Four chemical samples were placed at evenly spaced points, 2 cm from the center. DMSO was used as the reference solvent.
The plates were incubated at 25°C for 48 h to observe the formation of inhibitory or clear zones around each disk. The antibacterial activity was determined by subtracting the diameter of the inhibition zone obtained with DMF (control). Amikacin and ketoconazole were used as reference compounds for antibacterial and antifungal activity, respectively. Each experiment was performed in triplicate, and the average results are reported.
Anticancer activities
The cytotoxicity of the compounds was evaluated using the method described by Skehan et al.[10,11] A total of 104 cells per well were plated in a 96-well plate and incubated for 24 h to allow cell adhesion. The cell monolayers in triplicate wells were treated with different concentrations of the test compounds (0, 5, 12.5, 25, 50, and 100 μg/mL) for each dose. The treated cells were then incubated at 37°C with 5% CO2 for 48 h. After the incubation period, the cells were fixed, washed, and stained with SRB dye. Excess dye was removed with acetic acid, and the remaining bound dye was solubilized in Tris-ethylenediaminetetraacetic acid buffer. The optical density (O.D.) of each well was measured at 564 nm using an enzyme-linked immunosorbent assay microplate reader. The mean values for each drug concentration were calculated after automatically subtracting the mean background absorbance. The cell survival curve was plotted by correlating the drug concentration with the surviving cell fraction.
Cell survival estimation
The percentage of surviving cells was calculated using the formula: O.D. (treated cells)/O.D. (untreated cells) = survival fraction (control cells). The IC50 values represent the concentration of thymoquinone required to inhibit cell growth by 50%. Each experiment was performed in triplicate for every cell line.
Molecular docking
Molecular docking studies were conducted using the MOE 2008 software. The crystal structures of human colon cancer antigen 10 (PDB ID: 2HQ6), A. fumigatus fructosamine oxidase (PDB ID: 3DJD), and mutant human androgen receptors (ARccr) from androgen-independent prostate cancer (PDB ID: 1GS4) were used to predict potential binding modes and their lowest binding energies. In addition, serine/threonine protein phosphatase (PPZ1) from C. albicans (PDB ID: 5JPE) was included for comparison in an interactive molecular graphics program.[6] The receptor and Schiff base ligand structures were input in PDB format, and the compound structures were generated using the Gaussian03 program. The receptor crystal structures (1GS4, 2HQ6, 3DJD, and 5JPE) were obtained from the Protein Data Bank (http://www.rcsb.org/pdb). Water molecules, co-crystallized ligands, counterions (Cl−), and unsupported elements (e.g., K, Na, and Hg) were removed from the docking experiments, while the amino acid chains were preserved.[12]
Results
Schiff base ligand characterization
A novel Schiff base ligand was synthesized by reacting isoleucine with L1 (isatin and 2,6-diaminopyridine) in a 1:1 ratio. Spectral and elemental analyses were carried out to determine its composition. The resulting ligand was a solid, dark purple compound, stable at room temperature, and soluble in common organic solvents like DMSO and DMF. The calculated values were in strong agreement with the experimental data.
The FTIR spectrum of the Schiff base ligand (HL) confirmed its formation, showing a strong band at 1619 cm−1 attributed to ν(C=N), along with two distinct bands at 1456 and 1360 cm−1 corresponding to the asymmetric (COO−) and symmetric (COO−1) stretching vibrations, respectively [13,14] The unbound ligand displayed a broad band at 3434 cm−1 due to O-H stretching and a band at 1705 cm−1 from the carbonyl (C=O) group, further supporting the successful synthesis of the Schiff base.[15,16]
Metal complex characterization
Elemental analysis
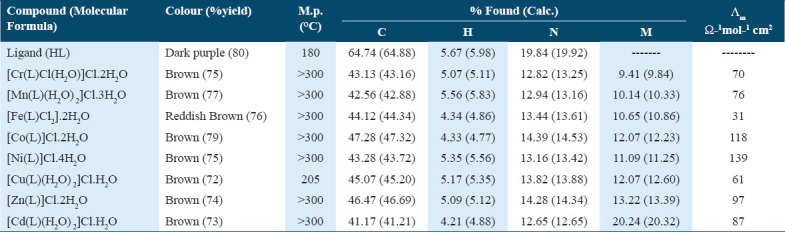
Metal complexes with an ML-type composition were synthesized by reacting the Schiff base in hot ethanol with an ethanolic solution of the corresponding metal chloride in a 1:1 ratio. The experimental and theoretical elemental analyses of the metal complexes showed good agreement. The results of the elemental analysis (C, H, N, and M), along with the molecular formulas and melting points, are presented in Table 1.
Table 1.
Analytical and physical data of Schiff base ligand (HL) and its metal complexes
Molecular conductivity evaluations
Molar conductance values for the metal complexes in DMF at 25°C and 1 × 10−3 mol/L concentration are shown in Table 1. The results indicate that the Co(II), Cr(III), Cu(II), Cd(II), Ni(II), Zn(II), and Mn(II) complexes are ionic and electrolytic, with conductance values ranging from 61 to 139 Ω−1mol−1cm². The Fe(III) complex, however, displayed a lower molar conductance of 31 Ω−1mol−1cm², suggesting weak ionic (non-electrolytic) behavior, with the anion located within the inner coordination sphere.[17]
IR spectral studies
The IR spectra of the free ligand (HL) and its metal complexes were examined to identify key bands in the 4000–400 cm−1 range, as shown in Table 2. The spectra were compared to determine which coordination sites were involved in chelation. Coordination to the metal ions through the azomethine nitrogen was evidenced by a slight shift in the azomethine (νC=N) stretching frequency, moving to 1616–1645 cm−1 after complexation.[18]
Table 2.
IR spectra (4000–400 cm−1) of ligand (HL2) and its metal complexes
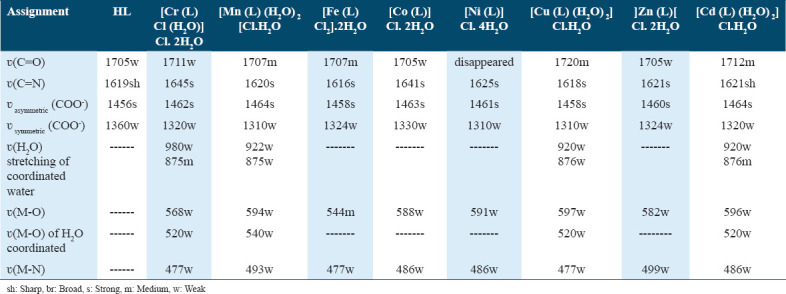
The presence of water molecules in the free ligand was confirmed by a broad band at 3434 cm−1, corresponding to the O-H stretching vibration. This band persisted in the complexes, appearing between 3339 and 3430 cm−1. Interference from NH2 vibrations, often associated with O-H, complicates the distinction of these bands, which can be further analyzed through ¹H-NMR. The free ligand’s asym(COO−1) and sym(COO−1) stretching vibrations were observed at 1456 and 1360 cm−1, respectively.[15,16]
For the metal complexes, a shift in the asym(COO−1) and sym(COO−1) bands to 1458–1464 cm−1 and 1310 –1330 cm−1, respectively, indicated coordination through the carboxylate O atom. New bands in the 544–597 cm−1 (M-O) and 477–499 cm−1 (M-N) ranges further supported the bonding of oxygen and nitrogen to the central metal ions.[19,20] The presence of coordinated water molecules in the Cr(III), Cu(II), Mn(II), and Cd(II) complexes was confirmed by weak bands at 920–980 cm−1 (rocking) and 875–876 cm−1 (wagging) for the ν(H2O) vibration.[21,22] Weak stretching vibrations of coordinated water were observed at 520–540 cm−1.[23]
The tetradentate ligand contributed two azomethine nitrogen atoms, one carboxylate oxygen, and one amino nitrogen to the complex. In octahedral complexes, two additional coordination sites were occupied by either two chloride ions or a combination of water and chloride molecules.
1H-NMR spectral analyses
Comparing the proton signal positions of the Zn(II) and Cd(II) complexes to those of the ligand indicates that all signals remained within the expected range, with only slight shifts attributed to ligand-metal ion interactions.[19] The proton signal at 4.20 ppm (d, 1H, NH) in the [Zn(L)]Cl·2H2O and [Cd(L)(H2O)2]Cl·H2O complexes corresponds to the NH of isatin. This signal, however, shifted to 4.13–4.00 ppm in these complexes.
The multiplet observed in the range of 6.74–7.24 ppm (m, 8H, Ar-H) was found at 6.49–7.26 ppm and 6.90–7.08 ppm in the [Zn(L)]Cl·2H2O and [Cd(L)(H2O)2]Cl·H2O complexes, respectively, indicating the presence of aromatic protons. In addition, the signal at 8.33 ppm, attributed to NH2 protons, shifted to 8.20 ppm in both the [Zn(L)]Cl·2H2O and [Cd(L)(H2O)2]Cl·H2O complexes.[24,25]
Mass spectrometric analyses
The mass spectrum of the ligand HL1 exhibited a molecular ion peak at (m/z) 352.22 amu, as shown in Figure 1. This result is consistent with the predicted formula, which indicates that the ligand moiety has an atomic mass of 351 amu, identified as C19H21N5O2. The spectrum of the Cu(II) complex revealed a mass peak at m/z 501.25 amu, aligning with the complex’s formula weight of 504 amu. Furthermore, the identification of the Schiff base ligand peak at 353.49 amu in the mass spectra of the complex provides evidence of its successful synthesis.
Figure 1.
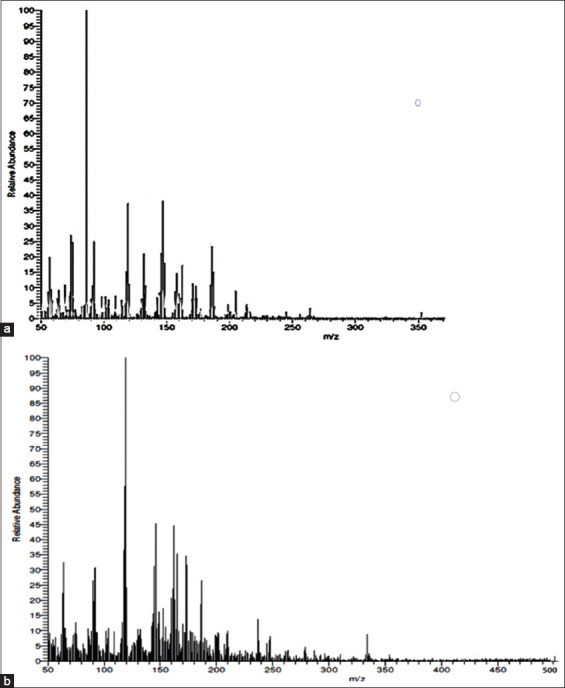
Mass spectra of (a) Schiff base ligand (HL) and (b) [Cu(l)(H2O)2]Cl.H2O
Examination of UV-visible absorption
The electronic spectra of the Schiff base ligand and its metal complexes were recorded in DMF solvent over a wavelength range of 200–700 nm. In the UV-visible spectra of the Schiff base ligand, two absorption bands were observed at 215 and 248 nm. These absorption bands are likely attributed to the π-π* and conjugation transitions of the aromatic rings. In addition, the absorption band at 339 nm may result from the n-π* transition of the azomethine or amino group. However, this band was absent in all the metal complexes, indicating that the azomethine nitrogen was coordinated to the metal ions. Each metal complex exhibited absorption spectra in the range of 216–236 nm, containing bands associated with the π-π* transition.
Elemental spectra
The diffused reflectance spectra of the Cr(III) complex in DMF revealed three distinct bands at 19,920 cm−1(1), 26,246 cm−1 (2), and 28,901 cm−1 (3). These bands correspond to the transitions 4A2g(F)4T2g(P), 4A2g(F)4T2g(F), and 4A2g(F)4T2g(F), respectively, indicating the octahedral geometry of the Cr(III) complex.[26]
In the case of the Mn(II) complex, the diffused reflectance spectra displayed two bands at 27,322 cm−1and 21,834 cm−1, attributed to the transitions 4T1g6A1g and 4T2g(G)6A1g, respectively, confirming the presence of an octahedral structure.[27]
For the Fe(III) chelate, a band at 20,161 cm−1 in the diffused reflectance spectra was linked to the 6A1g4T2g(G) transition, further supporting the octahedral geometry of the complex.[28,29] In addition, a band at 26,178 cm−1 was observed, likely resulting from charge transfer from the ligand to the metal. The diffused reflectance spectra of the Co(II) complex prominently displayed the 4A24T1 (F) transition, indicative of a tetrahedral environment around the Co(II) ion.[30] In the Ni(II) complex, a broad band centered at 16,666 cm−1 was identified as the 3T13T1 (P) transition, confirming the tetrahedral geometry of the Ni(II) ion.[31]
The electronic spectra of six-coordinate Cu(II) complexes (2D free ions) are expected to split into B1g, A1g, B2g, and Eg levels. Although few examples of such bands have been reported, three spin-allowed transitions were predicted in the visible and near-infrared regions.[32,33] These bands, in increasing energy order, were associated with the transitions 2B1g2A1g (dx2-y2 dz2), 2B1g2B2g (dx2-y2 dxy), and 2B1g 2Eg (dx2-y2 dxz, dyz), revealing d-d transition bands at 16,556, 24,154, and 29,411 cm−1, respectively. The order of these energy levels is influenced by the ligand field strength and the Jahn-Teller effect, which can cause deformation of the octahedral geometry into a tetragonal configuration.[33,34]
Both the Zn(II) and Cd(II) complexes exhibited diamagnetism. Empirical formulas indicate that the Cd(II) complex adopts an octahedral geometry, while the Zn(II) complex has a tetrahedral structure.
X-ray diffraction spectroscopy of powder
Conducting an XRD analysis in the range of 0° to 260° for the ligand was essential to understand the lattice dynamics of the Schiff base ligand and its metal complexes. The XRD results raised questions about the assertion that each solid represented a unique compound with a well-defined structure. The findings indicated that the complexes of Cr(III), Fe(III), Mn(II), Cu(II), Zn(II), and Cd(II) were crystalline, while the complexes of Co(II) and Ni(II) were amorphous. The average crystallite size (n) can be determined from the XRD pattern using the Debye-Scherrer equation.
ξ =Kλ/1/2cos Θ
The equation incorporates the X-ray wavelength (1.542475 Å), a constant KKK (typically assumed to be 0.95 for organic compounds[35,36]), and the full width at half maximum (FWHM) of the reference diffraction peak in radians. The dislocation density of a crystal is defined as the number of dislocation lines per unit area. There is a linear relationship between the average particle diameter and its corresponding value, as demonstrated in references.[37]
δ = 1/ξ2
The calculated ξ values for the HL, Cr(III), Mn(II), Fe(III), Cu(II), Zn(II), and Cd(II) complexes are 95.29, 69.85, 41.60, 79.87, 43.80, 41.82, and 41.80 nm, respectively. The ξ values were found to be 1 × 10−3 nm² for the ligand (HL) and Fe(III), 2 × 10−3 and 6 × 10−3 nm² for Cr(III) and Mn(II), and 5 × 10−3 nm² for the Cu(II), Zn(II), and Cd(II) complexes. These results indicate that all of these complexes are nanosized.
The surface morphology of the ligand and metal complexes was assessed using SEM analysis [Figure 2]. The SEM micrographs revealed significant differences between the free ligand and the Co(II) chelate, attributed to the coordination of metal ions at the ligand’s donor sites.[38] The micrographs of the complex indicated that this coordination altered the surface morphology of the metal complex.
Figure 2.
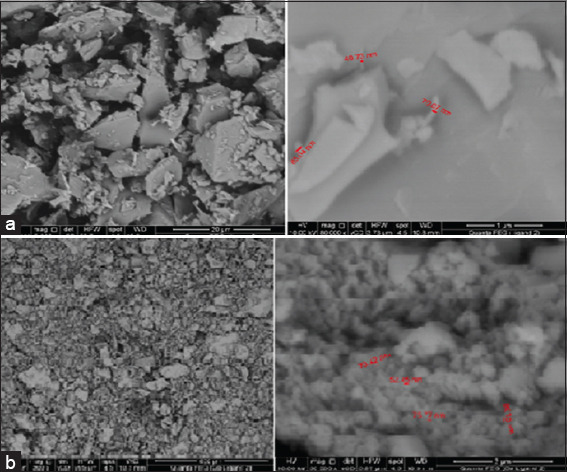
Scanning electron microscopy for (a) Schiff base ligand (HL) and (b) Co(l)[Cl.2H2O
The image of the Schiff base ligand HL2 displayed tubular rods that varied in size but maintained a consistent shape. In contrast, the SEM micrograph of the Co(L)[Cl.2H2O] complex revealed irregularly fractured shapes resembling ice rocks, with randomly distributed particles resting atop these structures. The average particle size of the [Co(L)[Cl.2H2O] complex was measured at 71.94 nm, while the HL2 ligand had an average particle size of 78 nm. Based on the average grain size determined through SEM analysis, it was concluded that the compound is polycrystalline with nanosized grains.[39]
Thermal analysis investigations (TGA and DTG)
The thermal properties of the ligand and its complexes were characterized using TGA and DTG over a temperature range from room temperature to 1000°C. These methods allow for the assessment of water molecules present either within or outside the inner coordination sphere of the central metal ion, as well as provide insights into the general mechanism of thermal decomposition for these chelates. The temperature ranges and corresponding mass loss percentages are detailed in Table 3 of the supplementary data.
Table 3.
Thermoanalytical Results (TGA and DTG) of the (HL) Ligand and Its metal complexes
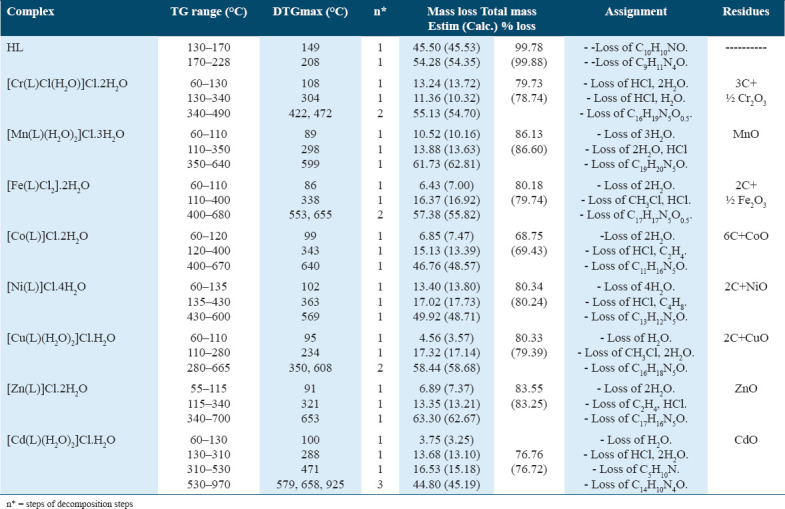
The TGA data for the HL2 Schiff base ligand (C19H21N5O2) revealed two distinct stages of decomposition. The first stage occurred between 130 and 170°C, with a maximum temperature peak at 149°C, resulting in a mass loss of 45.50% (estimated mass loss = 45.53%). The ligand component (C9H11N4O) underwent complete degradation in the final stage, occurring between 170 and 228°C, which resulted in a mass loss of 54.28% (calculated mass loss = 54.35%). The DTG curve indicated a total calculated mass loss of 99.88%, with a maximum peak temperature of 208°C, confirming a weight loss of 99.78%.
The thermogravimetric (TGA) curve for the [Cr(L)Cl(H2O)]Cl·2H2O chelate exhibited four weight loss events. In the first stage, occurring between 60 and 130°C with a peak at 108°C, two water molecules and one hydrochloride molecule were lost, resulting in an estimated mass loss of 13.24% (calculated mass loss = 13.72%). The second stage took place between 130 and 340°C, peaking at 304°C, leading to the loss of one coordinated water molecule and one hydrochloride molecule, with a mass loss of 11.36% (calculated mass loss = 10.32%).
The final two stages occurred between 340 and 490°C, with peaks at 422°C and 472°C. During this period, the C16H19N5O0.5 fragment was lost, resulting in an estimated mass loss of 55.13% (calculated mass loss = 54.70%). Overall, the total mass loss was 78.74% (or 79.73% of the total weight).
The [Mn(L)(H2O)2]Cl·3H2O molecule displayed three stages of degradation. The first stage, occurring between 60 and 110°C with a peak at 89°C, resulted in a weight loss of 10.52% (estimated mass loss = 10.16%) due to the loss of three water molecules. The second stage, between 110 and 350°C, involved the loss of two coordinated water molecules and one hydrochloride molecule, resulting in a mass loss of 13.88% (calculated mass loss = 13.63%).
The final stage reached a peak temperature of 599°C, resulting in the loss of a C19H20N5O fragment with an estimated mass loss of 61.73% (calculated mass loss = 52.81%). The total mass loss was 86.13% (or 86.60% based on calculated values), leaving MnO as the residue.
For the [Fe(L)Cl2]·2H2O complex, four degradation phases were observed between 60 and 680°C. In the first stage, two water molecules were lost between 60 and 110°C, with a peak at 86°C and a mass loss of 6.43% (estimated mass loss = 7.00%). The second stage, peaking at 338°C and occurring between 110 and 400°C, resulted in the loss of one hydrochloride and one CH3Cl molecule, corresponding to a mass loss of 16.37% (calculated mass loss = 16.92%).
During the final two stages, a C17H17N5O molecule was lost between 400 and 680°C, with peaks at 553°C and 655°C, resulting in a mass loss of 57.38% (estimated mass loss = 55.82%). The residue at the end of the thermogram was Fe2O3, contaminated with carbon, with a total estimated mass loss of 80.18% (calculated mass loss = 79.74%).
The [Co(L)]Cl·2H2O chelate displayed three phases of thermal degradation between 60 and 670°C. The first stage, occurring between 60 and 120°C with a peak at 99°C, resulted in an estimated mass loss of 6.85% (calculated mass loss = 7.47%) due to the release of two hydrated water molecules. In the second stage, between 120 and 400°C with a maximum peak at 343°C, the complex lost one hydrochloride molecule and one C2H4 gas, resulting in an estimated mass loss of 15.13% (calculated mass loss = 13.39%).
During the final stage of disintegration, which occurred between 400 and 670°C with a peak temperature of 640°C, a C11H16N5O fragment was lost, resulting in a calculated mass loss of 48.57% and an estimated mass loss of 46.76%. At the conclusion of the thermogram, CoO, contaminated with carbon, was identified, with a total estimated mass loss of 68.75% (calculated mass loss = 69.43%).
The thermogravimetric (TGA) curve for the [Ni(L)]Cl·4H2O complex displayed three weight loss events. The first stage of decomposition occurred between 60 and 135°C, peaking at 102°C, during which four water molecules were lost, resulting in an estimated mass loss of 13.40% (calculated mass loss = 13.80%). In the second stage, between 135 and 430°C with a peak at 363°C, one hydrochloride molecule and a C4H8 fragment were lost, leading to an estimated mass loss of 17.02% (calculated mass loss = 17.73%).
The final stage involved the loss of a C13H12N5O fragment, resulting in NiO contaminated with two carbon atoms as residues. This stage, occurring between 430 and 600°C with a peak temperature of 569°C, had an estimated mass loss of 49.92% (calculated mass loss = 48.71%). Overall, there was an 80.34% reduction in body weight (calculated mass loss = 80.24%).
The TG curve for the [Zn(L)]Cl·2H2O complex peaked at 91°C in the 55–115°C range, corresponding to a mass loss of 6.89% (estimated weight loss = 7.37%) due to the loss of two hydrated water molecules. The second phase, occurring between 115 and 340°C and peaking at 321°C, resulted in a mass loss of 13.35% (estimated mass loss = 13.21%) from the loss of a hydrochloride molecule and C2H4 gas.
In the penultimate stage, between 340 and 700°C, with a peak at 653°C, the C17H16N5O fragment was lost, resulting in a mass loss of 63.30% (calculated = 62.67%). The thermogram concluded with the identification of ZnO, which had lost 83.55% of its total weight (calculated mass loss = 83.25%).
For the [Cd(L)(H2O)2]Cl·H2O chelate, six breakdown phases were observed between 60 and 970°C. The first breakdown stage, occurring between 60 and 130°C with a maximum temperature of 100°C, had an estimated mass loss of 3.75% (calculated mass loss = 3.25%) due to the release of a hydrated water molecule. In the second stage, between 130 and 310°C, with a peak at 288°C, the complex lost two coordinated water molecules and one hydrochloride molecule, resulting in an estimated mass loss of 13.68% (calculated mass loss = 13.10%).
The third stage, occurring between 310 and 530°C, with a peak temperature of 471°C, involved the loss of a C5H10N fragment, yielding an estimated mass loss of 16.53% (calculated mass loss = 15.18%). The fourth, fifth, and sixth stages occurred between 530 and 970°C, with three temperature peaks at 579, 658, and 925°C, corresponding to the complete decomposition of the ligand C14H10N4O and an estimated mass loss of 44.80% (calculated mass loss = 45.19%). The thermogram concluded with the identification of CdO, which had a total estimated mass loss of 76.76% (calculated mass loss = 76.72%).
Structural analysis
The complex architectures of Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), Zn(II), and Cd(II) are illustrated in the Figure 3. These structures are derived from a variety of physicochemical and spectroscopic data that have been previously presented and discussed.[40]
Figure 3.
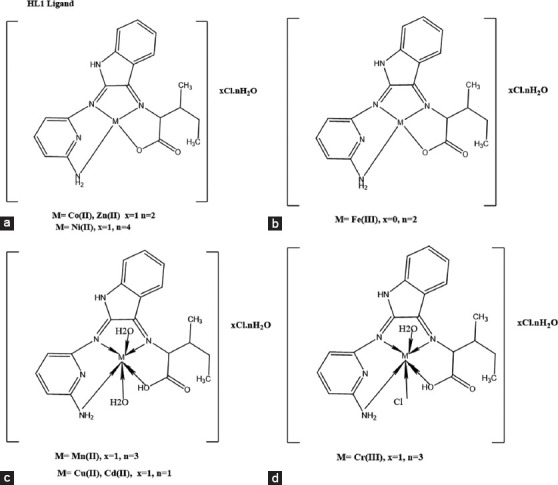
(a-d) Structure of metal complexes of Schiff base ligand (HL)
Antimicrobial activity
The size of the inhibition zone is influenced by factors such as the concentration of the antibacterial agent, diffusion rate, incubation conditions, and the culture medium. The behavior of the complexes under investigation was explained through chelation theory. Chelation reduces the polarity of the metal atom because the donor groups partially share the metal’s positive charge, causing electron delocalization throughout the chelate ring. This increases the lipophilicity of the metal atom, facilitating its penetration through the lipid layers of cell membranes.
The antifungal and antibacterial activities of the Schiff base ligand (HL2) and its metal complexes were tested in vitro against two fungal species (A. fumigatus and C. albicans), two Gram-positive bacteria (S. aureus and B. subtilis), and two Gram-negative bacteria (Salmonella species and E. coli). The results of these antimicrobial tests are shown in Table 4 and Figures 4 and 5. For comparison, DMSO was used as a negative control, while amikacin and ketoconazole served as positive standards for antibacterial and antifungal tests, respectively.
Table 4.
Biological activity of Schiff base ligand (HL) and its metal complexes with Gram-positive bacteria and Gram-negative bacteria and two fungi species
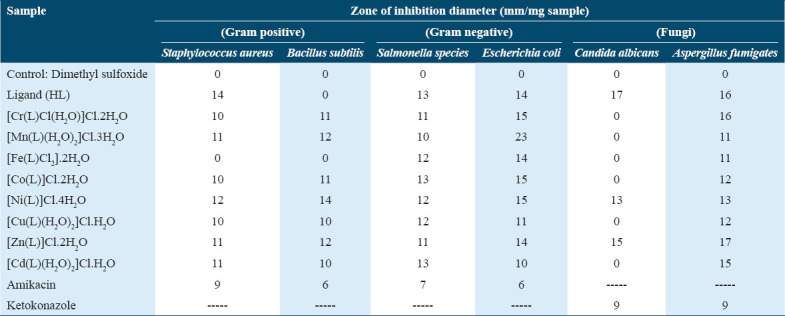
Figure 4.
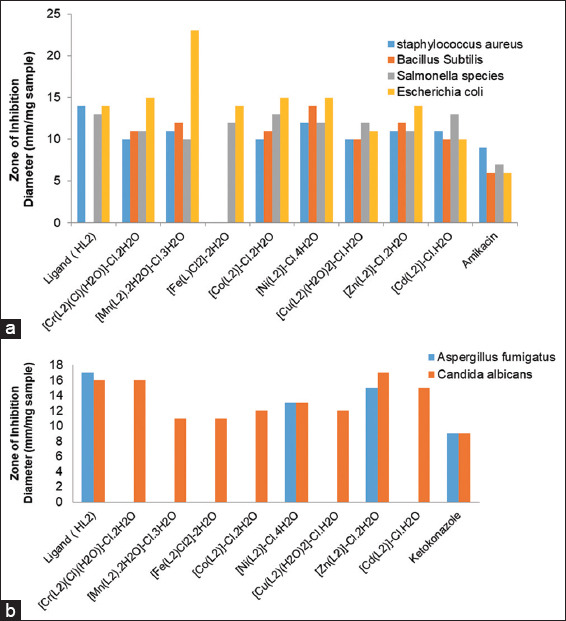
Biological activity of Schiff base ligand (HL) and its metal complexes with (a) Gram-positive and Gram-negative bacteria and (b) two fungi species
Figure 5.
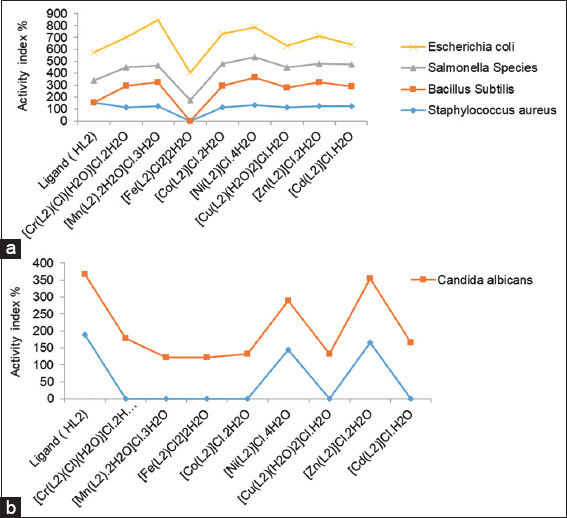
Activity index value of Schiff base ligand (HL) and its metal complexes against (a) four bacterial species (b) two fungi species
The results indicated that while the Fe(III) complex exhibited no activity, the ligand was more biologically active than the complexes against S. aureus, a Gram-positive bacterium. When B. subtilis, another Gram-positive bacterium, was used, all complexes demonstrated better biological activity than the free ligand (HL), except for the Fe(III) complex, which showed no antibacterial activity [Figure 4].
Regarding the Gram-negative Salmonella species, the Cr(III), Mn(II), Fe(III), Ni(II), Cu(II), and Zn(II) complexes had lower biological activity than the free HL2 ligand. However, the Cd(II) and Co(II) complexes exhibited similar activity to the free ligand.
For E. coli, another Gram-negative bacterium, the Cu(II) and Cd(II) complexes showed higher biological activity compared to the free HL2 ligand. While the Cr(III), Mn(II), Co(II), and Ni(II) complexes demonstrated less activity than the free ligand, they were still more active than the free ligand. The Mn(II) complex displayed the strongest antibacterial activity against E. coli. Both the Fe(III) and Zn(II) complexes exhibited activity comparable to the free HL ligand.
In antifungal studies involving A. fumigatus, the Zn(II) and Ni(II) complexes showed lower activity compared to the free HL ligand. In contrast, the Cr(III), Mn(II), Fe(III), Co(II), Cu(II), and Cd(II) complexes were active.[41,42] The biological activity of the synthesized Schiff base ligand and its metal complexes was further validated by calculating the activity index using the following equation:
Activity index (A) = (Inhibition zone of standard drug (mm)/Inhibition zone of compound (mm) × 100
As illustrated in Figure 6.
Figure 6.
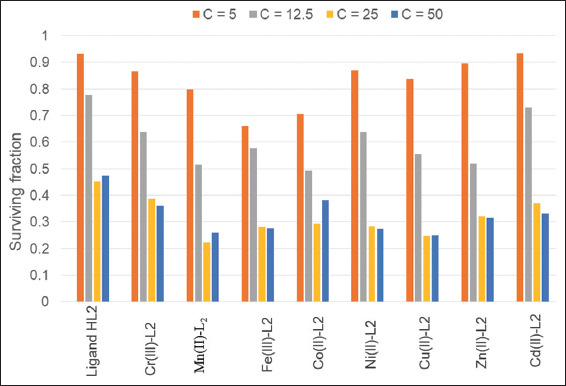
Anticancer/antiproliferative activity of Schiff base ligand (HL) and its metal complexes
Based on these results, the Mn(II) and Ni(II) complexes showed the highest activity indexes among all the compounds. For the fungus C. albicans, the Zn(II) complex had the highest activity index, while the free ligand showed the highest activity index against A. fumigatus [Figure 5].
Anticancer characteristics
Anticancer properties
The anticancer effects of the ligand and its complexes were tested against the human breast cancer cell line MCF-7 at concentrations ranging from 100 to 150 μg/mL. These results revealed that all metal complexes were highly effective against breast cancer cells, with inhibition ratios ranging from 74% to 86%. Figure 6 demonstrates the activity pattern across different drug concentrations [Table 5].
Table 5.
Antiproliferative/anticancer activity of Schiff base ligand (HL) and its metal complexes
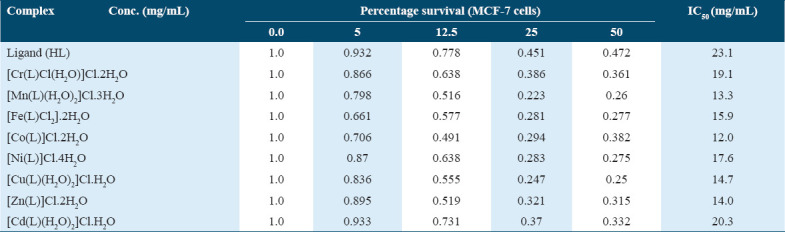
Cell growth inhibition resulted in a 70% reduction in cell proliferation. Neither the ligand nor any of the metal complexes affected the MCF-7 cell line. The IC50 values of the metal complexes ranged from 12 to 20.3 μg/mL, indicating the concentration required to inhibit cell growth by 50% compared to untreated control cells. The IC50 values of the compounds under investigation ranged from 12 to 23.1 μg/mL. The free ligand exhibited an IC50 value of 23.1 μg/mL, while the Co(II) complex showed a lower value of 12.0 μg/mL.
The Zn(II) and Mn(II) complexes displayed strong anticancer activity with IC50 values of 14.0 μg/mL and 13.3 μg/mL, respectively. The Cu(II), Fe(III), Ni(II), Cr(III), and Cd(II) complexes had IC50 values of 14.7, 15.9, 17.6, 19.1, and 20.3 μg/mL, respectively.
Molecular docking studies of the ligand and its metal complexes
Molecular docking is a key technique in computer-aided drug design, allowing scientists to model molecular recognition processes between a drug and its target protein. The primary goal of molecular docking is to determine the optimal binding structure between a protein and a drug that minimizes the system’s overall energy. This approach is commonly used to identify the most favorable mode of interaction between a small molecule and its target protein.
Given the significance of these findings, we conducted molecular docking studies to explore the binding modes and energies of the prepared Schiff base ligand with various receptors. Specifically, we examined the interactions with the crystal structures of the deglycation enzyme fructosamine oxidase from A. fumigatus (PDB ID: 5JPE) and a mutant human androgen receptor (ARccr), derived from androgen-independent prostate cancer (PDB ID: 1GS4).
The docking results showed binding energies for the receptors 1GS4, 2HQ6, 3DJD, and 5JPE at −7.8, −3.0, −5.5, and −4.6 kcal/mol, respectively. These results suggest that this theoretical data could play an important role in guiding future research on the design and application of ligands for the treatment of prostate and colon cancer. The detailed molecular docking data are presented in Table 6, with visual representations shown in Figure 7.
Table 6.
Energy values obtained in docking calculations of Schiff base ligand (HL2) with receptors of mutant human androgen (ARccr) derived from an androgen-independent prostate cancer (PDB ID: 1GS4), crystal structure of Colon Cancer Antigen 10 from Homo sapiens (PDB ID: 2HQ6), crystal structure of deglycating enzyme fructosamine oxidase from Aspergillus fumigatus (PDB ID: 3DJD) and yeast-specific serine/threonine protein phosphatase (PPZ1) of Candida albicans (PDB ID: 5JPE)
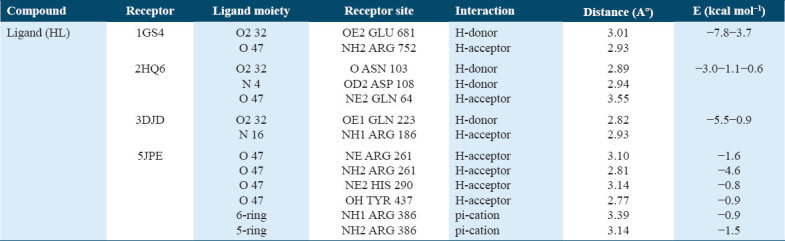
Figure 7.
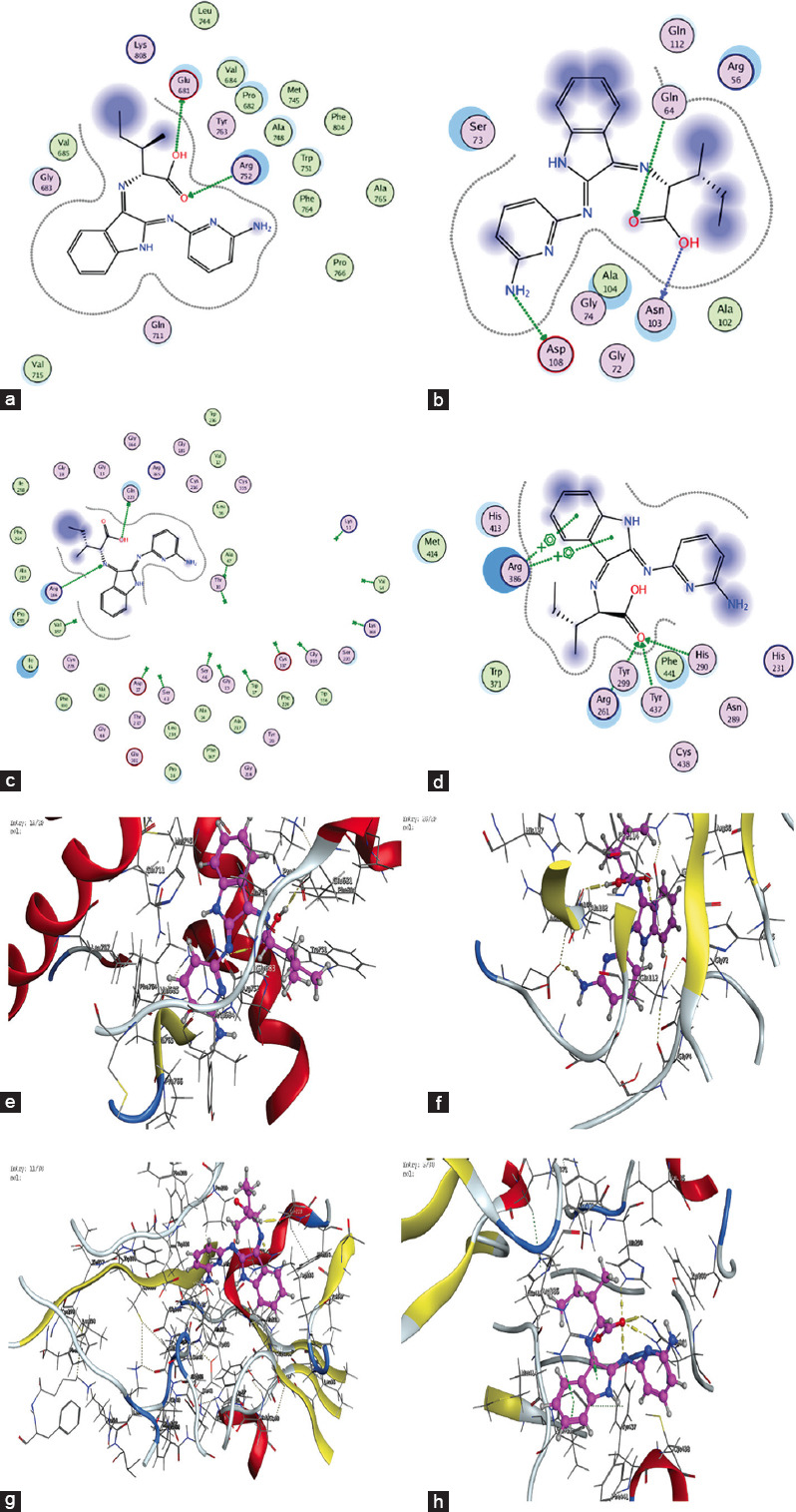
2D and 3D Interaction Plots of the Ligand (HL) with Various Receptors (a) 1GS4, (b) 2HQ6, (c) 3DJD, (d) 5JPE – 2D interaction plots (e) 1GS4, (f) 2HQ6, (g) 3DJD, and (h) 5JPE – 3D interaction plots
Discussion
The synthesis and characterization of Schiff base ligands and their metal complexes remain a focal point in coordination chemistry, with numerous recent studies highlighting their significance in diverse applications such as antimicrobial, anticancer, and catalytic activities.[3] In this study, the Schiff base ligand (HL) was synthesized and confirmed through various spectroscopic techniques, including FT-IR, ¹H-NMR, mass spectrometry, and UV-Visible spectroscopy. The formation of the Schiff base was evidenced by the characteristic ν(C=N) stretching band at 1619 cm−1, which shifted on coordination to the metal ions, indicating successful chelation. This shift in frequency is consistent with recent studies on Schiff base-metal complexes, where coordination through azomethine nitrogen is well documented.[35,36]
The metal complexes were further characterized by XRD, revealing that most complexes (Cr[III], Mn[II], Fe[III], Cu[II], Zn[II], and Cd[II]) had a crystalline structure, while the Co(II) and Ni(II) complexes were amorphous. The crystalline nature of the metal complexes suggests a well-defined lattice, which is beneficial for stability and reactivity.[43] Amorphous complexes, such as Co(II) and Ni(II), may exhibit different physical properties, such as solubility and bioavailability, which can impact their applications in biological systems.[44]
The molecular conductivity results showed that most metal complexes were ionic and electrolytic, except for Fe(III), which exhibited non-electrolytic behavior.[38,45] This observation aligns with studies indicating that the nature of the metal ion and its coordination environment significantly influences the electrical properties of the complex.[45,46] The presence of coordinated water molecules in some complexes, such as Cr(III) and Cd(II), further supports the proposed octahedral geometries, a common structural feature in Schiff base-metal complexes.
The antimicrobial activity of Schiff base ligands and their metal complexes has been widely explored, with many recent studies showing enhanced activity upon complexation.[41,47] In this study, the Schiff base ligand exhibited moderate antibacterial and antifungal activity, particularly against S. aureus and A. fumigatus. However, most metal complexes demonstrated significantly improved antimicrobial properties, with Mn(II) and Ni(II) complexes showing the highest antibacterial activity.
The enhanced biological activity of the metal complexes can be attributed to chelation, which increases the lipophilicity of the metal center, facilitating its passage through the lipid membranes of bacterial and fungal cells.[42,48] This is consistent with the chelation theory, which postulates that complexation reduces the polarity of the metal ion by sharing its positive charge with the donor atoms of the ligand.[49] The resulting increase in lipophilicity enhances the penetration of the complex into the microbial cell, leading to improved biological activity.
The Cr(III), Mn(II), and Cu(II) complexes also displayed notable activity against Gram-negative bacteria, such as Salmonella species and E. coli, which are typically more resistant to antimicrobial agents. This broad-spectrum activity of the metal complexes makes them promising candidates for further development as antimicrobial agents.[50,51] Interestingly, the Fe(III) complex showed little to no activity across the tested microorganisms, a finding that could be linked to the lower bioavailability and stability of Fe(III) in biological environments.
The Schiff base ligand and its metal complexes were evaluated for their anticancer potential against the human breast cancer cell line MCF-7. The results revealed that all the metal complexes exhibited strong anticancer activity, with inhibition ratios ranging from 74% to 86%. The Co(II) complex showed the greatest efficacy, with an IC50 value of 12.0 μg/mL, making it a promising candidate for anticancer therapy. The anticancer activity of Schiff base-metal complexes has been widely studied, with many reports indicating that metal complexation enhances the cytotoxic effects of the ligand.[52,53] The increased activity of the metal complexes in this study can be attributed to the ability of metal ions to interact with cellular biomolecules, such as DNA and proteins, leading to cell cycle arrest and apoptosis.[54] The IC50 values of the Zn(II), Mn(II), Cu(II), and Ni(II) complexes were also within the range of highly active anticancer agents, indicating that these complexes can induce significant cytotoxic effects at relatively low concentrations.
Recent studies have shown that Schiff base ligands, when complexed with transition metals, can trigger the generation of reactive oxygen species (ROS), which play a crucial role in inducing apoptosis in cancer cells.[55,56] The Co(II) and Zn(II) complexes in particular may facilitate ROS production, leading to oxidative stress and subsequent cancer cell death. The involvement of metal ions in stabilizing the ligand structure and facilitating its interaction with cancer cell membranes further enhances the anticancer potential of these complexes.[57]
Molecular docking studies were performed to investigate the binding interactions between the Schiff base ligand and various target proteins, including the fructosamine oxidase from A. fumigatus (PDB ID: 5JPE) and the mutant androgen receptor from prostate cancer (PDB ID: 1GS4). The docking results revealed favorable binding energies, with the Schiff base ligand exhibiting strong interactions with the 1GS4 receptor (−7.8 kcal/mol) and moderate interactions with 2HQ6, 3DJD, and 5JPE receptors. Molecular docking is a powerful tool for predicting the binding affinity of ligands to target proteins, and the results of this study suggest that the Schiff base ligand and its metal complexes could serve as potential therapeutic agents for the treatment of prostate and colon cancers.[58,59] The strong binding interaction with the androgen receptor (1GS4) is particularly significant, as this receptor plays a key role in the progression of androgen-independent prostate cancer.[60] The moderate binding energies observed for the other receptors indicate that the Schiff base ligand may also have potential as a multi-target therapeutic agent.
Conclusion
The tetradentate ligand L1 (isatin with 2,6-diaminopyridine) condenses to form the Schiff base HL2, which coordinates to Cr(III), Mn(II), Fe(III), Cu(II), Ni(II), Co(II), Zn(II), and Cd(II) ions through two azomethine nitrogens, one amino nitrogen, and one deprotonated carboxylic oxygen. The resulting ligand is uninegative and tetradentate. The complexes of Cr(III), Mn(II), Cu(II), Fe(III), and Cd(II) exhibit octahedral geometry, while those of Ni(II), Co(II), and Zn(II) do not.
All complexes, except for the non-electrolytic Fe(III) complex, are ionic electrolytes, with typical ML-type compositions such as [Cr(L)Cl(H2O)] and variations for other metals. Antimicrobial tests indicated that Fe(III) complexes had the lowest activity indices, while Mn(II) and Ni(II) complexes demonstrated the highest. Although all metal complexes exhibited anticancer activities with the ligand, the Co(II) complex had a higher IC50 value of 12 μg/mL. Molecular docking studies revealed that the receptors 1GS4, 2HQ6, 3DJD, and 5JPE had the lowest binding energies, measured at 7.8, 3.0, −5.5, and −4.6 kcal/mol, respectively.
Author’s Contributions
WD, BSM, GGM, RHT, and AF designed the study concept, methods, and performed data analysis. WD, BSM, and GGM performed experimentation, WD and AF wrote the original manuscript. WD, BSM, and GGM were responsible for supervision and resources. WD, BSM, and GGM critically evaluated the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The project was funded by the Deanship of Scientific Research at Jouf University (Grant No. DSR2020-02-2592).
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at Jouf University for funding this work through Research Grant No - DSR2020-02-2592.
References
- 1.Wen J, Wang F, Zhang X. Asymmetric hydrogenation catalyzed by first-row transition metal complexes. Chem Soc Rev. 2021;50:3211–37. doi: 10.1039/d0cs00082e. [DOI] [PubMed] [Google Scholar]
- 2.Li G, Zhu D, Wang X, Su Z, Bryce MR. Dinuclear metal complexes:Multifunctional properties and applications. Chem Soc Rev. 2020;49:765–838. doi: 10.1039/c8cs00660a. [DOI] [PubMed] [Google Scholar]
- 3.Boulechfar C, Ferkous H, Delimi A, Djedouani A, Abdesalem K, Boublia A, et al. Schiff bases and their metal Complexes:A review on the history, synthesis, and applications. Inorg Chem Commun. 2023;150:110451. [Google Scholar]
- 4.Karad RP, Singare AS, Korde NS. Biological activities of metal complexes of Schiff bases-a review. Int J Adv Res Trends Eng Technol. 2022;9:921–59. [Google Scholar]
- 5.Dhedan RM, Alsahib SA, Ali RA. A brief review on schiff base, synthesis, and their antimicrobial activities. Russian J Bioorgan Chem. 2023;49:S31–52. [Google Scholar]
- 6.Al-Hawarin JI, Abu-Yamin AA, Abu-Saleh AA, Saraireh IA, Almatarneh MH, Hasan M, et al. Synthesis, characterization, and DFT calculations of a new sulfamethoxazole Schiff base and its metal complexes. Materials (Basel) 2023;16:5160. doi: 10.3390/ma16145160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almehmadi YA, McGeehan J, Guzman NJ, Christensen KE, Yamazaki K, Dixon DJ. Iridium-catalysed synthesis of C, N, N-cyclic azomethine imines enables entry to unexplored nitrogen-rich 3D chemical space. Nat Synth. 2024;3:1168–75. [Google Scholar]
- 8.Bryndal I, Stolarczyk M, Mikołajczyk A, Krupińska M, Pyra A, Mączyński M, et al. Pyrimidine Schiff bases:Synthesis, structural characterization and recent studies on biological activities. Int J Mol Sci. 2024;25:2076. doi: 10.3390/ijms25042076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ely IA, Phillips BE, Smith K, Wilkinson DJ, Piasecki M, Breen L, et al. A focus on leucine in the nutritional regulation of human skeletal muscle metabolism in ageing, exercise and unloading states. Clin Nutr. 2023;42:1849–65. doi: 10.1016/j.clnu.2023.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Brandão LF, Alcantara GB, Matos MF, Bogo D, Freitas DS, Oyama NM, et al. Cytotoxic evaluation of phenolic compounds from lichens against melanoma cells. Chem Pharm Bull (Tokyo) 2013;61:176–83. doi: 10.1248/cpb.c12-00739. [DOI] [PubMed] [Google Scholar]
- 11.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 12.Balakrishnan C, Subha L, Neelakantan MA, Mariappan SS. Synthesis, spectroscopy, X-ray crystallography, DFT calculations, DNA binding and molecular docking of a propargyl arms containing Schiff base. Spectrochim Acta A Mol Biomol Spectrosc. 2015;150:671–81. doi: 10.1016/j.saa.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Ibrahim S, Gavisiddegowda P, Deepakumari HN, Kollur SP, Naik N. Newly synthesized benzothiazole derived ligand and its Co (III) and Ru (III) complexes as biological potent molecules:Chemical preparation, structure, antimicrobial, in vitro and in vivo cytotoxicity studies. Biointerface Res Appl Chem. 2022;12:7817–44. [Google Scholar]
- 14.Bhardwaj V. Design, Synthesis and Applications of Novel Supramolecular Assemblies. India: Maharaja Sayajirao University of Baroda; 2022. [Google Scholar]
- 15.Sharma N, Prakash R, Chaturvedi K. Spectroscopic and antimicrobial studies of mixed ligand complexes of transition metal (II) ions with nitro quinoline and dibenzoyl methane. Sci Rev Chem Commun. 2012;2:108–14. [Google Scholar]
- 16.Chaudhary N. In vitro antibacterial studies of some transition metal complexes of Schiff base derived from 2-aminophenol and furan-2-carbaldehyde. Arch Appl Sci Res. 2013;5:227–31. [Google Scholar]
- 17.Mohamed GG, Omar MM, Moustafa BS, AbdEl-Halim HF, Farag N. Spectroscopic investigation, thermal, molecular structure, antimicrobial and anticancer activity with modelling studies of some metal complexes derived from isatin Schiff base ligand. Inorg Chem Commun. 2022;141:109606. [Google Scholar]
- 18.Abdelrahman MS, Omar FM, Saleh AA, El-Ghamry MA. Synthesis, molecular modeling, and docking studies of a new pyridazinone-acid hydrazone ligand, and its nano metal complexes. Spectroscopy, thermal analysis, electrical properties, DNA cleavage, antitumor, and antimicrobial activities. J Mol Struct. 2022;1251:131947. [Google Scholar]
- 19.Kumar N, Kaushal R, Awasthi P. Non-covalent binding studies of transition metal complexes with DNA:A review. J Mol Struct. 2023;1288:135751. [Google Scholar]
- 20.Zülfikaroğlu A, Ataol CY, Çelikoğlu E, Çelikoğlu U, İdil O. New Cu (II), Co (III) and Ni (II) metal complexes based on ONO donor tridentate hydrazone:Synthesis, structural characterization, and investigation of some biological properties. J Mol Struct. 2020;1199:127012. [Google Scholar]
- 21.El-Sonbati A, Diab MA, Morgan SM, Abou-Dobara MI, El-Ghettany AA. Synthesis, characterization, theoretical and molecular docking studies of mixed-ligand complexes of Cu (II), Ni (II), Co (II), Mn (II), Cr (III), UO2 (II) and Cd (II) J Mol Struct. 2020;1200:127065. [Google Scholar]
- 22.Kargar H, Aghaei-Meybodi F, Behjatmanesh-Ardakani R, Elahifard MR, Torabi V, Fallah-Mehrjardi M, et al. Synthesis, crystal structure, theoretical calculation, spectroscopic and antibacterial activity studies of copper (II) complexes bearing bidentate Schiff base ligands derived from 4-aminoantipyrine:Influence of substitutions on antibacterial activity. J Mol Struct. 2021;1230:129908. [Google Scholar]
- 23.Hamed AA, Sharaby CM, Amine MF, Ammar YA, El-Sherif AA. In vitro anticancer and antibacterial assessment of novel metal complexes of 1, 3-di-[P-Tolyl]-2, 4-di-[9H-Purin-6-Yl]-2, 4-dichlorocyclodiphosph (V) Azane. Egypt J Chem. 2022;65:843–57. [Google Scholar]
- 24.Ali OA, Elangovan N, Mahmoud SF, El-Gendey MS, Elbasheer HZ, El-Bahy SM, et al. Synthesis, characterization, vibrational analysis and computational studies of a new Schiff base from pentafluoro benzaldehyde and sulfanilamide. J Mol Struct. 2022;1265:133445. [Google Scholar]
- 25.Baliram TV, Sanjay KP, Arvind MP. Synthesis, spectral characterization and antitubercular study of novel quinoline Schiff base and its metal complexes. Anal Chem Lett. 2021;11:523–38. [Google Scholar]
- 26.El-Ghamry MA, Elzawawi FM, Abdel Aziz AA, Nassir KM, Abu-El-Wafa SM. New Schiff base ligand and its novel Cr (III), Mn (II), Co (II), Ni (II), Cu (II), Zn (II) complexes:Spectral investigation, biological applications, and semiconducting properties. Sci Rep. 2022;12:17942. doi: 10.1038/s41598-022-22713-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Essang SB, Omoregie HO. Synthesis and characterization of mixed ligand Mn (ii), Co (ii), Ni (ii) and Cu (ii) complexes of some hydrazones and nitrogen donor ligands. Lautech J Eng Technol. 2020;14:136–52. [Google Scholar]
- 28.Mahmoud WH, Mohamed GG, El-Dessouky MM. Synthesis, structural characterization, in vitro antimicrobial and anticancer activity studies of ternary metal complexes containing glycine amino acid and the anti-inflammatory drug lornoxicam. J Mol Struct. 2015;1082:12–22. [Google Scholar]
- 29.Gomes RN, Silva ML, Gomes KS, Lago JH, Cerchiaro G. Synthesis, characterization, and cytotoxic effects of new copper complexes using Schiff-base derivatives from natural sources. J Inorg Biochem. 2024;250:112401. doi: 10.1016/j.jinorgbio.2023.112401. [DOI] [PubMed] [Google Scholar]
- 30.Kowalkowska-Zedler D, Dołęga A, Nedelko N, Łyszczek R, Aleshkevych P, Demchenko I, et al. Structural, magnetic and spectral properties of tetrahedral cobalt (II) silanethiolates:A variety of structures and manifestation of field-induced slow magnetic relaxation. Dalton Trans. 2020;49:697–710. doi: 10.1039/c9dt03722e. [DOI] [PubMed] [Google Scholar]
- 31.Siraj I, Ado H. Synthesis and characterization of potentially bioactive sulfamethoxazole isatin Schiff base and its Mn (II), Fe (II) and Ni (II) complexes. ChemSearch J. 2021;12:27–33. [Google Scholar]
- 32.Pierloot K. The CASPT2 method in inorganic electronic spectroscopy:From ionic transition metal to covalent actinide complexes. Mol Phys. 2003;101:2083–94. [Google Scholar]
- 33.Kumar P, SantaLucia DJ, Kaniewska-Laskowska K, Lindeman SV, Ozarowski A, Krzystek J, et al. Probing the magnetic anisotropy of Co (II) complexes featuring redox-active ligands. Inorg Chem. 2020;59:16178–93. doi: 10.1021/acs.inorgchem.0c01812. [DOI] [PubMed] [Google Scholar]
- 34.Wang QL, Zhao B, Liao DZ, Yan SP, Cheng P, Jiang ZH. Synthesis, structure, spectroscopic and magnetic properties of a novel two-dimensional copper (II) complex Na 2 [Cu (pba)]·6H 2 O [pba=1, 3-propylenebis (oxamato) Trans Metal Chem. 2003;28:326–30. [Google Scholar]
- 35.Sinicropi MS, Ceramella J, Iacopetta D, Catalano A, Mariconda A, Rosano C, et al. Metal complexes with Schiff bases:Data collection and recent studies on biological activities. Int J Mol Sci. 2022;23:14840. doi: 10.3390/ijms232314840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakur S, Jaryal A, Bhalla A. Recent advances in biological and medicinal profile of schiff bases and their metal complexes:An updated version (2018–2023) Results Chem. 2024;7:101350. [Google Scholar]
- 37.Alaghaz AN, Zayed MZ, Alharbi SA, Ammar RA, Chinnathambi A. Synthesis, spectroscopic identification, thermal, potentiometric and antibacterial activity studies of 4-amino-5-mercapto-S-triazole Schiff's base complexes. J Mol Struct. 2015;1087:60–7. [Google Scholar]
- 38.AbouEl-Enein SA, Emam S, Wagdy RM, Abouzayed F. Spectral and thermal investigation of novel biologically active (N-(1, 5-dimethyl-3-oxo-2-phenyl-2, 3-dihydro-1H-pyrazol-4-yl)-2-(1, 5-dimethyl-3-oxo-2-phenyl-2, 3-dihydro-1H-pyrazol-4-yl-amino)-2-oxo-cetimidic acid) ligand and its metal complexes. J Mol Struct. 2020;1215:128230. [Google Scholar]
- 39.Kariper IA. A novel method for producing nanostructured CdSe thin film. Surf Rev Lett. 2020;27:1950175. [Google Scholar]
- 40.Kürkçüoğlu GS, Yeşilel OZ, Kekeç S, Şahin O. Synthesis, crystal structure and spectroscopic investigations of heteronuclear Co (III)/Cu (II), Co (III)/Cd (II) and Fe (III)/Cd (II) 3D coordination polymers with 4-(2-aminoethyl) pyridine. J Mol Struct. 2023;1274:134540. [Google Scholar]
- 41.Singh P, Yadav P, Sodhi PK, Tomer A, Mehta SB. Advancement in the synthesis of metal complexes with special emphasis on Schiff base ligands and their important biological aspects. Results Chem. 2024;7:101222. [Google Scholar]
- 42.Turecka K, Chylewska A, Kawiak A, Waleron KF. Antifungal activity and mechanism of action of the Co (III) coordination complexes with diamine chelate ligands against reference and clinical strains of Candida spp. Front Microbiol. 2018;9:1594. doi: 10.3389/fmicb.2018.01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young RJ, Huxley MT, Pardo E, Champness NR, Sumby CJ, Doonan CJ. Isolating reactive metal-based species in Metal-Organic Frameworks-viable strategies and opportunities. Chem Sci. 2020;11:4031–50. doi: 10.1039/d0sc00485e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siddiki AN, Islam S, Begum S, Salam A. Synthesis, spectral characterization, thermal behavior and biological activities study of ternary metal complexes of alanine and 1, 8-diaminonapthalene with Co (III), Ni (II), Cu (II), Zn (II) and Cd (II) Mater Today Proc. 2021;46:6374–81. [Google Scholar]
- 45.Bendia S, Bourzami R, Weiss J, Ouari K. Structural investigation of the catalytic activity of Fe (III) and Mn (III) Schiff base complexes. Polyhedron. 2021;202:115206. [Google Scholar]
- 46.Jevtovic V, Alshamari AK, Milenković D, Marković JD, Marković Z, Dimić D. The effect of metal ions (Fe, Co, Ni, and Cu) on the molecular-structural, protein binding, and cytotoxic properties of metal pyridoxal-thiosemicarbazone complexes. Int J Mol Sci. 2023;24:11910. doi: 10.3390/ijms241511910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain S, Rana M, Sultana R, Mehandi R, Uddin R. Schiff base metal complexes as antimicrobial and anticancer agents. Polycyclic Aromat Compd. 2023;43:6351–406. [Google Scholar]
- 48.Piatek M. Evaluation of the Mode of Action of Novel Metal-Based Antimicrobial Compounds. Ireland: National University of Ireland Maynooth; 2023. [Google Scholar]
- 49.Kislik V. Competetive complexation/solvation theory of solvent extraction. II. Solvent extraction of metals by acidic extractants. Sep Sci Technol. 2002;37:2623–57. [Google Scholar]
- 50.Waters JE, Stevens-Cullinane L, Siebenmann L, Hess J. Recent advances in the development of metal complexes as antibacterial agents with metal-specific modes of action. Curr Opin Microb. 2023;75:102347. doi: 10.1016/j.mib.2023.102347. [DOI] [PubMed] [Google Scholar]
- 51.Millanao AR, Mora AY, Villagra NA, Bucarey SA, Hidalgo AA. Biological effects of quinolones:A family of broad-spectrum antimicrobial agents. Molecules. 2021;26:7153. doi: 10.3390/molecules26237153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matela G. Schiff bases and complexes:A review on anti-cancer activity. Anticancer Agents Med Chem. 2020;20:1908–17. doi: 10.2174/1871520620666200507091207. [DOI] [PubMed] [Google Scholar]
- 53.Khan S, Alhumaydhi FA, Ibrahim MM, Alqahtani A, Alshamrani M, Alruwaili AS, et al. Recent advances and therapeutic journey of Schiff base complexes with selected metals (Pt, Pd, Ag, Au) as potent anticancer agents:A review. Anticancer Agents Med Chem. 2022;22:3086–96. doi: 10.2174/1871520622666220511125600. [DOI] [PubMed] [Google Scholar]
- 54.Aldulmani SA, Hakami O, Abdel-Nasser M, Alaghaz MA. Applied Organometallic Chemistry. United States: Wiley; 2024. Synthesis, antimicrobial, DNA interactions, in vitro cytotoxicity, and cell cycle analysis efficiency of newly nano-sized Cu (II), Ni (II), Co (II), and Zn (II) Thio-Schiff base complexes of 2-((E)-((4, 5-Dimethyl-2-(((E)-4-methylbenzylidene) amino) phenyl) imino) methyl)-benzenethiol; p. e7731. [Google Scholar]
- 55.Li X, Wang Y, Li M, Wang H, Dong X. Metal complexes or chelators with ROS regulation capacity:Promising candidates for cancer treatment. Molecules. 2021;27:148. doi: 10.3390/molecules27010148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mondal SS, Jaiswal N, Bera PS, Tiwari RK, Behera JN, Chanda N, et al. Cu (II) and Co (II/III) complexes of N, O-chelated Schiff base ligands:DNA interaction, protein binding, cytotoxicity, cell death mechanism and reactive oxygen species generation studies. Appl Organ Chem. 2021;35:e6026. [Google Scholar]
- 57.Abdolmaleki S, Aliabadi A, Khaksar S. Riding the metal wave:A review of the latest developments in metal-based anticancer agents. Coord Chem Rev. 2024;501:215579. [Google Scholar]
- 58.Al-Adilee KJ, Jawad SH, Kyhoiesh HA, Hassan HM. Synthesis, characterization, biological applications, and molecular docking studies of some transition metal complexes with azo dye ligand derived from 5-methyl imidazole. J Mol Struct. 2024;1295:136695. [Google Scholar]
- 59.El-Zahed MM, El-Sonbati AZ, Ajadain FM, Diab MA, Abou-Dobara MI. Synthesis, spectroscopic characterization, molecular docking and antimicrobial activity of Cu (II), Co (II), Ni (II), Mn (II) and Cd (II) complexes with a tetradentate ONNO donor Schiff base ligand. Inorg Chem Commun. 2023;152:110710. [Google Scholar]
- 60.Ezeorah CJ, Ekowo LC, Eze SI, Groutso T, Atiga S, Okafor SN, et al. Synthesis, characterization, and in silico studies of 2-[(E)-(2, 5-dimethoxybenzylidene) amino] phenol and 3-[(E)-(2, 5-dimethoxybenzylidene) amino] phenol. J Mol Struct. 2022;1270:133902. [Google Scholar]


