Abstract
Recently, methane has been considered a next-generation carbon feedstock due to its abundance and it is main component of shale gas and biogas. Methylomonas sp. DH-1 has been evaluated as a promising industrial bio-catalyst candidate. Succinate is considered one of the top building block chemicals in the agricultural, food, and pharmaceutical industries. In this study, succinate production by Methylomonas sp. DH-1 was improved by combining adaptive laboratory evolution (ALE) technology with genetic engineering in the chromosome of Methylomonas sp. DH-1, such as deletion of bypass pathway genes (succinate dehydrogenase and succinate semialdehyde dehydrogenase) or overexpression of genes related with succinate production (citrate synthase, pyruvate carboxylase and phosphoenolpyruvate carboxylase). Through ALE, the maximum consumption rate of substrate gases (methane and oxygen) and the duration maintaining high substrate gas consumption rates was enhanced compared to those of the parental strain. Based on the improved methane consumption, cell growth (OD600) increased more than twice, and the succinate titer increased by ~ 48% from 218 to 323 mg/L. To prevent unwanted succinate consumption, the succinate semialdehyde dehydrogenase gene was deleted from the genome. The first enzyme of TCA cycle (citrate synthase) was overexpressed. Pyruvate carboxylase and phosphoenolpyruvate carboxylase, which produce oxaloacetate, a substrate for citrate synthase, were also overproduced by a newly identified strong promoter. The new strong promoter was screened from RNA sequencing data. When these modifications were combined in one strain, the maximum titer (702 mg/L) was successfully improved by more than three times. This study demonstrates that successful enhancement of succinic acid production can be achieved in methanotrophs through additional genetic engineering following adaptive laboratory evolution.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-024-02557-0.
Keywords: Methane, Methanotroph, Succinate, Gas fermentation
Introduction
Methane is the second most widely distributed greenhouse gas (GHG) after carbon dioxide. It accounts for about 14 percent of global annual GHG emission. The mean life span of methane in atmosphere was estimated as 9–12 years which was shorter than that of carbon dioxide. However, methane has 25 times the global warming potential (GWP) of carbon dioxide because it can trap more heat than carbon dioxide [1]. In fact, over 60% of global methane emission comes from human activities [2, 3]. It is important to point out that the effects of global warming can be significantly reduced if we can reduce anthropogenic methane emissions [4]. Furthermore, methane is the significant component in shale gas and biogas. And it is considered a next-generation carbon feedstock because of its abundance and reduced carbon [4–8].
Methanotrophs are well-known for their ability to utilize methane gas at mild conditions while much harsher conditions are required for chemical methane conversion [9, 10]. Recently, many studies have been conducted to convert methane, a potent greenhouse gas, into high-value compounds using methanotrophs [11]. Genetically engineered methanotrophic bacteria produce many potential chemical products, such as succinic acid, lactic acid, carotenoids, and 1, 4-butanediol (1, 4-BDO) [12, 13]. Methylomicrobium buryatense 5G and Methylomicrobium alcaliphilum 20Z, showing fast growth rates, have been found to produce lactic acid, C-4 carboxylic acid, and 2, 3-butanediol (2,3-BDO) [14–17].
Methylomonas sp. DH-1, a type I methanotroph isolated from brewery waste sludge, has been considered a good platform strain for methane bioconversion because of its expeditious growth rate and highly efficient bioconversion of methane to methanol and propane to acetone [18, 19]. Its complete genome sequence was reported in 2018 [20]. It was revealed that, both the ribulose monophosphate (RuMP) and the complete serine cycle existed in Methylomonas sp. DH-1 genome. The RuMP cycle has the role of fixing and rearranging formaldehyde made from methane. The serine cycle which can also play a role for C1 carbon fixation, includes two carboxylases. Phosphoenolpyruvate (PEP) carboxylase converts PEP to oxaloacetate with addition of CO2. And pyruvate carboxylase converts pyruvate to oxaloacetate with addition of CO2 [20]. The presence of phosphoenolpyruvate carboxylase (ppc) and pyruvate carboxylase (pc) and the complete TCA cycle in its genome provide advantages to produce TCA cycle-related chemicals, such as succinate and 1,4-butanediol (1,4-BDO) [20].
Fermentative succinate production is an attractive option to replace fossil oil-based production of bulk chemicals [21–24]. A wide variety of bacteria, such as Pasteurellaceae species, Escherichia coli, and Corynebacterium glutamicum can form succinic acid during anaerobic fermentative metabolism [25]. In the presence of bicarbonate, C. glutamicum does not grow under anaerobic conditions but efficiently converts glucose to succinate, lactate, and acetate [26]. Additionally, non-recombinant organisms have been isolated and studied, especially anaerobic rumen bacteria such as Anaerobiospirillum succiniciproducens, Actinobacillus succinogenes, and Mannheimia succiniciproducens [27]. In a previous study, genetically engineered Methylomonas sp. DH-1 (DS-GL) was shown to produce succinate at a titer value of 195 mg/L at 127 h cultivation time in 5L scale bioreactor. Its maximum OD600 was 6.3 after 82 h incubation [28]. The genome of the DS-GL strain was modified to have glyoxylate shunt genes with a disrupted succinate dehydrogenase gene (sdhB). With the DS-GL strain, the oxidative TCA cycle and glyoxylate pathways, which operate under aerobic conditions, were proven to be suitable for succinate production in Methylomonas sp. DH-1 [28].
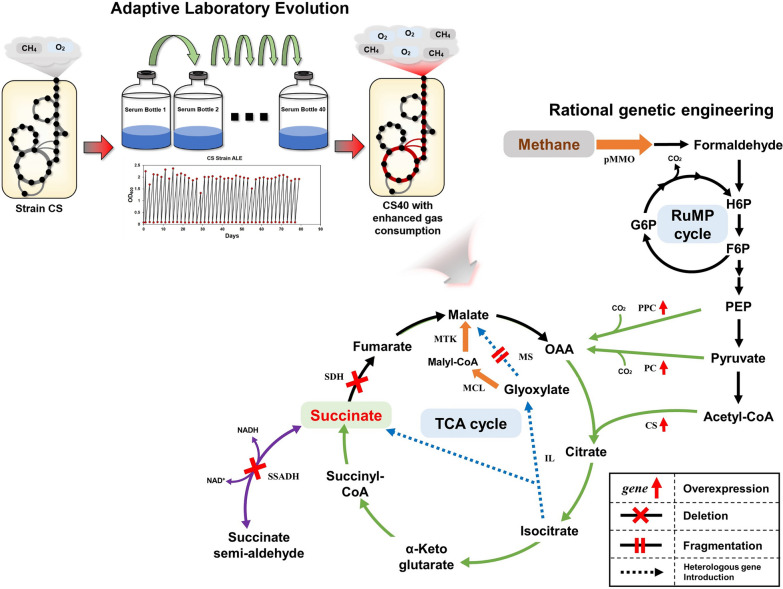
In this study, the strain (CS: Cre-lox plus Succinate production) containing sdhB deletion and glyoxylate shunt genes was constructed again based on the strain (DH-Cre) with a genome-integrated Cre-lox system [29]. The potential for increasing succinic acid production through adaptive laboratory evolution (ALE) and additional genetic engineering was tested with this strain (Fig. 1). In addition, the genetic mutations that occurred through ALE were analyzed, and based on RNA sequencing data, a novel strong promoter applicable to methanotrophs was proposed.
Fig. 1.
Schematic diagram showing the changes caused by adaptive laboratory evolution and the genetic engineering strategy. The cell growth and methane/oxygen consumption characteristics of the parental strain (CS strain) were improved by ALE. The succinate production performance of the mutant strain was enhanced by additional genetic engineering to enhance the TCA cycle and remove unnecessary pathways. The genes whose expression increased in CS40 comparing to CS were indicated by orange arrows. The oxidative branch of the TCA cycle and the glyoxylate pathway are indicated by the green and dashed blue lines, respectively. The following enzyme abbreviations were used: pMMO, particle methane monooxygenase; PPC, phosphoenolpyruvate carboxylase; PC, pyruvate carboxylase; CS, citrate synthase; MCL, malyl-CoA lyase; MTK, malate thiokinase; MS, malate synthase; IL, isocitrate lyase; SDH, succinate dehydrogenase; SSADH, succinate semialdehyde dehydrogenase
Materials and methods
Strains and plasmids
Methylomonas sp. DH-1 was kindly provided by Professor Eun Yeol Lee (Department of Chemical Engineering, Kyung Hee University, Republic of Korea) [18]. The mutant strain containing the integrated Cre-lox system was kindly provided by the inventor Professor Yong Jun Choi (School of Environmental Engineering, University of Seoul, Republic of Korea) [29, 30].
All strains and plasmids used in this study are listed in Table 1. Primers used for cloning and genetic engineering are listed in the Supplementary Table 1. To delete succinate-semialdehyde dehydrogenase (ssadh, AYM39_17625), upstream and downstream 1kb DNA regions flanking the ssadh gene were amplified by PCR from the DH-1 genome. The polymerase for amplifying PCR fragments used nPfu-forte (Enzynomics, Korea). The kanamycin resistance cassette was amplified from the pDCKIU vector [29]. Amplified DNA fragments were ligated using Gibson assembly (NEB, USA) and inserted into the pTOP Blunt V2 vector (Enzynomics, Korea) to generate pTOP-ssadh. To construct an overexpression vector for the citrate synthase (gltA, AYM39_19030) gene, the strong promoter, PmxaF (AYM39_15615), was used. The sequences of the PmxaF and gltA genes were assembled as one fragment by overlap extension PCR. The assembled DNA fragment was cloned at the SbfI site of the pTOP-ssadh vector to generate pTOP-ssadh-gltA. For promoter exchange of pyruvate carboxylase (pc, AYM39_19225) and phosphoenolpyruvate carboxylase (ppc, AYM39_12335), 1 kb upstream and downstream sequences around the start codon of the target genes, kanamycin cassette, and 400 bp DNA sequence upstream of AYM39_18825 were amplified and ligated by Gibson assembly as described above (Supplementary Fig. S6). To confirm protein expression from the cold shock protein promoter, DNA fragments of green fluorescence protein (GFP) and promoters of pc, ppc, and AYM39_18825 were joined by overlap extension PCR. The joined fragment was inserted between the SbfI and BglII sites of the pTOP-ssadh vector (Supplementary Fig. S7).
Table 1.
Bacterial strains and plasmids used in this study
| Name | Description | References |
|---|---|---|
| Strains | ||
| E. Coli TOP10 | Cloning host strain; | Invitrogen |
| Methylomonas sp. DH-1 | Wild-type strain, A novel Type I methanotroph isolated from brewery waste sludge | Hur et al. [19] |
| DH-Cre | Methylomonas sp. DH-1 Cre-lox | Kang et al. [29] |
| CS | DH-Cre ΔsdhB-ms-il | This study |
| CS40 | Evolved strain from CS | This study |
| CS401 | CS40 PAYM39_18825-pc | This study |
| CS402 | CS40 PAYM39_18825-ppc | This study |
| CS403 | CS401 PAYM39_18825-ppc | This study |
| CS404 | CS40 Δssadh | This study |
| CS405 | CS403 Δssadh PmxaF-gltA | This study |
| CS40-GFP1 | CS40 Ppc-GFP | This study |
| CS40-GFP2 | CS40 Pppc-GFP | This study |
| CS40-GFP3 | CS40 P18825-GFP | This study |
| Plasmid | ||
| pDCKIU | pUC; Cre-lox system integration | Kang et al. [29] |
| pCM184-F12-sdh-ms-il | pCM184; sdhB deletion, intergration of ms-il from E. coli MG1655 | Nguyen et al. [20] |
| pTOP Blunt V2 | Integration/Deletion vector; AmpR, KanR | Invitrogen |
| pUC19 | Integration/Deletion vector; AmpR | This study |
| pTOP-ssadh | pTOP Blunt V2; ssadh deletion vector | This study |
| pTOP-ssadh-gltA | pTOP Blunt V2; ssadh deletion and gltA overexpression vector | This study |
| pUC19-pc | pUC19; pc promoter exchange vector | This study |
| pUC19-ppc | pUC19; ppc promoter exchange vector | This study |
| pTOP-GFP(Ppc) | pTOP-ssadh; GFP integration with the pc promoter | This study |
| pTOP-GFP(Pppc) | pTOP-ssadh; GFP integration with the ppc promoter | This study |
| pTOP-GFP(P18825) | pTOP-ssadh; GFP integration with the AYM39_18825 promoter | This study |
The following gene abbreviations were used: sdhB, succinate dehydrogenase, gltA, and citrate synthase. The following protein abbreviations were used: ms, malate synthase; il, isocitrate lyase; pc, pyruvate carboxylase; ppc, phosphoenolpyruvate carboxylase; ssadh, succinate semialdehyde dehydrogenase; AmpR, ampicillin resistance protein; KanR, kanamycin resistance protein
All mutant strains were constructed with vectors listed in Table 1 through electroporation and homologous recombination, following a previously described method [28]. To confirm whether it was correctly disrupted or integrated, PCR was performed using a primer pair that could specifically bind to the corresponding DNA region. By checking the band size of the amplified DNA, it was confirmed that the mutant strains were constructed correctly (Supplementary Fig. S6, Supplementary Fig. S7).
Media and culture conditions
All experiments on the cultivation of Methylomonas sp. DH-1 used nitrate mineral salt (NMS) medium containing 10 μM CuCl2, as described in a previous study [18]. Each culture was carried out in a 110 mL serum bottle with 10 mL of NMS medium and sealed with a butyl rubber stopper and aluminum cap. The headspace of the serum bottle was supplemented with 30% (v/v) of methane and 70% (v/v) of air using a disposable sterilized syringe. The cells were incubated in a shaking incubator at 30 °C with stirring at 200 rpm. The optical density of the cell culture (OD600) was measured at 600 nm using a UV-spectrophotometer (Shimadzu, Japan) and a 1.5 mL cuvette with a 1 cm path length. NMS agar plate containing 10 µg/ml kanamycin as antibiotic was used to select the mutant strains.
Antibiotic cassette removal
The kanamycin resistant gene between lox sequences was used as a selection marker for genetic engineering. After screening strains, containing the selection marker, the mutant strains were cultivated in 10 ml of NMS media containing 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 30 ℃ for 72 h to induce the expression of Cre recombinase for removal of the kanamycin resistant gene. Cells were spread onto the NMS agar plate containing 1 mM IPTG and cultivate 7 days to 10 days. The removal of antibiotic cassette gene was confirmed by PCR. [29].
Adaptive laboratory evolution (ALE)
To develop strains with improved performance, adaptive laboratory evolution was performed using CS strain. Cells were cultured in 55 mL serum bottles containing 5 mL NMS media with 10 µM CuCl2, and a mixture of 30% methane and 70% air was supplied to the headspaces. Every culture was inoculated at an initial OD600 of 0.1. The exponentially growing cells were repeatedly transferred to fresh culture medium for next round of growth.
Quantitative real-time PCR (qPCR)
Methylomonas sp. DH-1 and mutant cells were cultured in a 110 mL serum bottle with 10 mL of NMS medium. Cells were harvested in the exponential phase. RNA Protect® cell reagent (Qiagen, USA) was treated for 5 min at room temperature to prevent RNA degradation and centrifuged at 4 °C and 4,000 rpm. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, USA). Extracted total RNA was converted to cDNA by reverse transcription, performed in a total 20 µL reactions with GoScript™ reverse transcriptase (Promega, USA). 10 pmol of Random primer (Biorad, USA), 10 mM PCR nucleotide Mix (Promega, USA), 25 mM MgCl2 (Promega, USA), GoScript™ 5X Reaction buffer (Promega, USA), and 1 µg of RNA were used in one reaction. Samples were annealed at 25 °C for 5 min, extended at 42 °C for 1 h, and incubated at 70 °C for 15 min to inactivate reverse transcriptase using a Q-cycler 96 + (Hain Lifescience, Germany). qPCR analysis was performed with Quantstudio 6 flex (Applied Biosystems, USA) according to the manufacturer’s instructions (KAPA Biosystems, USA). The calculation method of RNA level quantification used Pfaffl method [31]. The quantified RNA level of each gene was normalized using the expression level of 16S rRNA gene. The primer sequences used are listed in Supplementary Table 1.
Bioreactor cultivation
Fermentation was carried out using a continuous stirred tank reactor (CNS, Korea) with 3.2 L working volume. Seed cultures for inoculation were prepared in 1 L baffled flasks containing 200 mL of NMS media. The flasks were sealed with a custom-made rubber stopper and a screw cap. The headspace of the flasks was purged using a mass flow controller (BROOKS, USA), and the gas composition was the same as that for serum bottle cultures. The bioreactor was inoculated at 0.1 of initial OD600. Filter (0.2 μm)-sterilized gas was supplied to the bioreactor continuously through a micro-gas sparger with mass flow controllers (BROOKS, USA) at a rate of 320 mL/min (0.1 vvm). The composition of the input gas followed a previously reported method (30% (v/v) of methane, 3.5% (v/v) of oxygen, and 66. 5% (v/v) of nitrogen) [28]. The pH was adjusted using a peristaltic pump with 2 N HCl and 5 N NaOH solution and was maintained between pH 6.85 and 6.95. The bioreactor was agitated at 800 rpm and maintained at 30 °C.
RNA sequencing data analysis
In the bioreactor, cell culture was started with a gas mixture of 30% (v/v) of methane, 3.5% (v/v) of oxygen, and 66.5% (v/v) of nitrogen. RNA samples were prepared from exponentially growing cells as described in the qPCR method. After RNA preparation, the supply gas composition was changed to 30% (v/v) of methane, 15% (v/v) of oxygen, and 55% (v/v) of nitrogen to increase the oxygen concentration. RNA samples were prepared 30 min after the oxygen composition increased. The integrity of the total RNA was assessed by the TapeStation RNA screentape (Agilent, #5067-5576). Only high-quality RNA preparations, with RNA integrity number (RIN) greater than 7.0, were used for RNA library construction. RNA libraries were prepared by Illumina TruSeq Stranded Total RNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA, #RS-122-2201) following the manufacturer’s protocol. The sequences of libraries were then submitted to an Illumina HiSeq 4000q (Illumina, Inc., San Diego, CA, USA). The paired-end (2 × 100 bp) sequencing was performed by the Macrogen Incorporated (Seoul, Korea). Quality-filtered reads were mapped to the reference genome sequence of Methylomonas sp. DH-1 (GenBank: CP014360.1). Based on the result, expression abundance of genes was calculated as transcripts per million (TPM). These sequence data have been submitted to the GenBank databases under BioProject accession number (PRJNA769841).
Whole genome sequencing
Whole genome sequencing was performed using Pacific Biosciences RS II and the Illumina NovaSeq sequencing platforms. Genomic DNA was extracted by Wizard Genomic DNA Purification Kit (Promega). A standard PacBio library with average 20-kb inserts was prepared using the RS II SMRTbell template preparation kit v1.0. PacBio long read data were generated with P6-C4 chemistry. The subreads were de novo assembled using the Hierarchical Genome Assembly Process (HGAP) pipeline (RS_HGAP_Assembly.2 protocol) in SMRT Analysis package v.2.3.0 [32]. The gaps between contigs in the draft genome were manually closed by in-house scripts using the greedy algorithm-based approach with pre-assembled reads from the HGAP pipeline. The subreads were mapped to the contigs using pbmm2 (v1.9.0) for error correction, and consensus sequences were generated from the alignments using the gcpp Arrow consensus caller (pbgcpp v2.0.2). In addition, to correct remaining errors with short read data, a paired end library for Illumina sequencing was constructed with TruSeq Nano DNA High Throughput Library Prep Kit and sequenced according to the manufacturer’s instructions (Illumina). The errors in assembled sequences were finally corrected using the Pilon v.1.22 program [33] with the preprocessed high-quality Illumina short read data which were quality-controlled using the Trimmomatic v.0.39 program [34]. For the visual validation and correction of the genome assembly, the mapped results with high-quality Illumina short read data were loaded and manually confirmed on the IGV viewer v.2.5.3 [35]. Protein-coding genes on the final complete genome sequence were predicted by Prodigal v.2.6.3 [36]. To functionally annotate the predicted genes, BLAST-searches were performed against PFAM database [37], COG database [35] and the annotated coding gene sequence set of the published DH-1 genome on GenBank database. To search the genomic variations on the final complete genome of CS40 strain compared to the published complete genome sequence of the DH-1 strain, genome sequence of DH-1, two inserted vector sequences and genome sequence of CS40 strain were analyzed using the Mauve program v2.4.0 [38]. For the case of the tandem repeats from the estimated variations, the sequences in the repeat region were extracted and precisely analyzed using Tandem repeats finder program [39]. The copy number of the repeat block in the tandem repeat regions on both strain genomes were compared with MEGA alignments. Additionally, the PacBio raw data were also mapped on DH-1 genome sequence using pbmm2 to confirm the large In/Del regions (more than 1 kb). The whole genome sequencing data of CS40 strain is submitted to the GenBank databases under BioProject accession number (PRJNA1066519).
Analytical methods
To analyze the amount of gas consumed, a quadrupole mass spectrometer (Hidden QGA, USA) was used for quantification. The off-gas composition was determined and quantified in real-time. Organic acids produced from bioreactor cultures were analyzed using high-performance liquid chromatography (HPLC). The culture was harvested periodically and centrifuged at 13,000 rpm at 4 °C for 10 min. The supernatants were filtrated by 0.2 μm of hydrophilic syringe filter (Hyundai Micro, Korea). Filtrated samples were analyzed using HPLC (Agilent, USA) with a UV detector at 230 nm and an Aminex HPX-87H organic acid column (300 7.8 mm; Bio-Rad, USA). The temperature was maintained at 50 ℃ Sulfuric acid 5 mM was used as the mobile phase and flown at 0.6 mL/min of 60 min each sample.
Green fluorescence analysis
One hundred milliliters of exponential phase cultures were harvested and centrifuged at 4,000 rpm and 4 °C for 10 min. The cell pellets were washed twice with deionized water, and the remaining liquid was removed entirely. Pellets were resuspended in 10 mL of lysis buffer containing 10 mM Tris, 1 mM EDTA, and 0.1% Triton-X 100. Then, ultrasonication was carried out at 4 °C for 15 min with a 30 s pulse and 15 s of repeated cooling using a VCX 750 (SONICS, USA). Cell extracts were created by incubating at 30 °C for 1 h and centrifuged at 4,000 rpm at 4 °C for 10 min. The concentration of total extracted protein was measured using bovine serum albumin (Protein Assay Standard II, BSA; Bio-Rad, USA) for normalization. Two hundred milliliters of Cell extracts (200 mL) were assayed in a 96-well plate (black sidewall and clear bottom; SPL Life science, Korea) by measuring fluorescence using a Synergy H1 Hybrid multi-mode microplate reader (Biotek, USA). The fluorescence signal was normalized to the protein concentration of the crude extract. Plate reader filters were set at excitation at 485 nm, emission at 512 nm, and gain 75.
Results
Construction of succinate producing DH-1 strain carrying Cre-lox system for multiple genetic manipulations
Previously, it was reported that succinate production by the DH-1 strain could be increased by integrating glyoxylate shunt genes (isocitrate lyase and malate synthase from E. coli) in place of sdhB (succinate dehydrogenase gene) [28]. However, the antibiotic cassette used for mutant selection cannot be removed from the genome. Therefore, an additional antibiotic cassette is necessary to manipulate the genome for further genetic engineering. The genetic engineering system for repeated use in DH-1 strain (DH-Cre strain) was recently published by Kang et al., [29]. Briefly, DH-Cre strain contains Cre recombinase whose expression can be induced by adding Isopropyl β-D-1-thiogalactopyranoside (IPTG). So, the integrated marker genes can be easily removed if it is flanked by lox sequences. After maker gene removal, the same antibiotic resistant gene can be used for the next engineering process. For convenient additional genetic engineering to improve succinate production, the strain (CS strain, Table 1) whose sdhB gene was replaced by glyoxylate shunt genes was constructed again based on the strain with the Cre-lox system (DH-Cre strain, Table 1). This newly created strain (CS strain) showed a succinate titer (218 mg/L) similar to that of the previously reported strain (DS-GL: 195 mg/L) [28]. Hereafter these performance values of CS strain were set as basal levels for comparison with other mutant strains constructed in this study. While maximum OD600 (3.08) decreased in comparison to DS-GL (6.3), its specific productivity (3.15 mg/g DCW/h) per dry cell weight (DCW) increased (DS-GL: 1.15 mg/g DCW/h) (Table 2).
Table 2.
Specific growth rate, succinate productivity, and titer of each mutant strain
| Strains | |||||||
|---|---|---|---|---|---|---|---|
| CS | CS40 | CS401 | CS402 | CS403 | CS404 | CS405 | |
| Specific productivity (mg/g DCW*/h) | 3.15 | 2.30 | 2.21 | 1.65 | 2.14 | 2.46 | 3.49 |
| Volumetric productivity (mg/L/h) | 1.80 | 2.60 | 3.51 | 3.45 | 3.92 | 3.98 | 5.84 |
| Specific titer (mg/g DCW*) | 382 | 286 | 279 | 208 | 274 | 306 | 419 |
| Volumetric titer (mg/L) | 218 | 323 | 433 | 435 | 502 | 496 | 702 |
All of these data are from batch cultivation in a 5L scale CSTR with continuous feeding of methane and oxygen
*DCW: dry cell weight. The highest value is marked as bold text
The effects of repetitive cultures of CS strain in NMS media
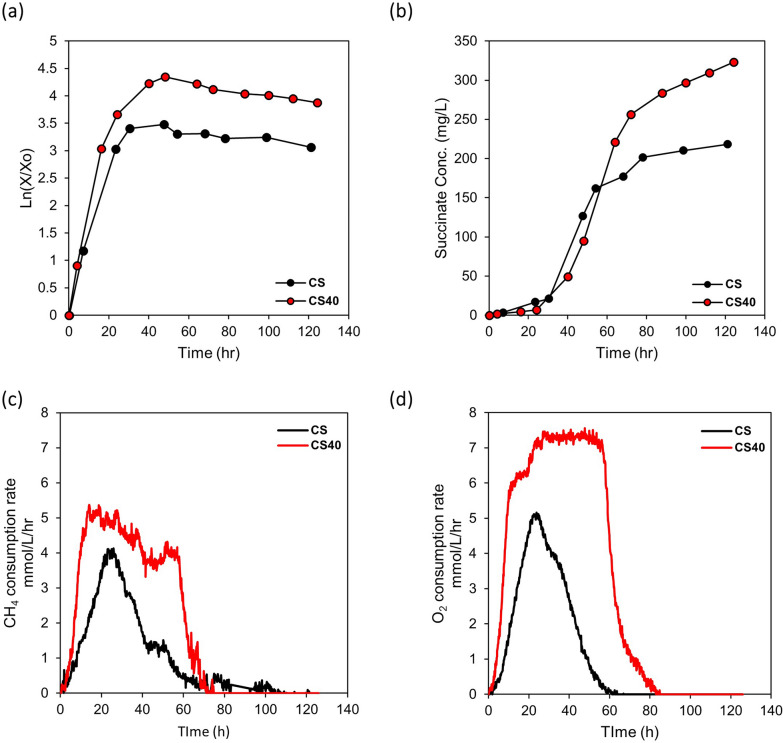
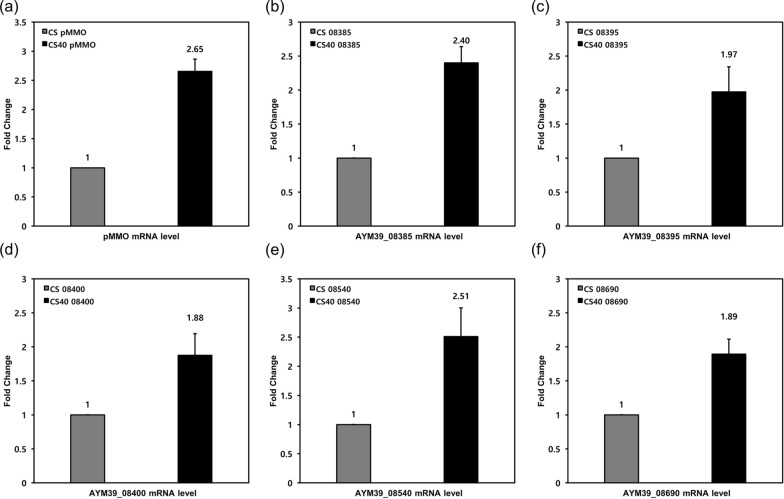
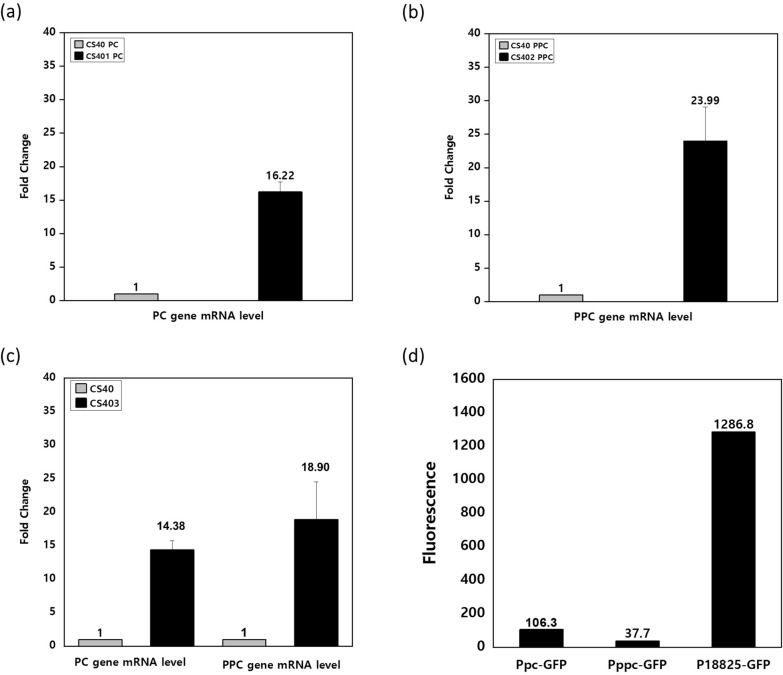
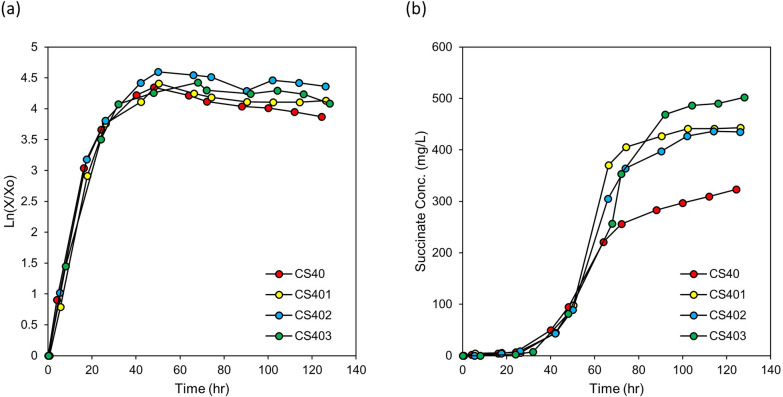
Adaptive laboratory evolution was conducted to improve cell performance. Cells were transferred to new NMS medium 40 times after their OD600 exceeded 1.0, as described in the Materials and Methods section. After 40 transfers (CS40 strain), the cell performance was tested in the bioreactor through batch cultivation where gaseous substrates (methane and oxygen) were continuously fed (Table 2, Fig. 2). The maximum cell concentration and succinate titer increased conspicuously with ALE. The volumetric succinate titer of CS40 strain was increased by 48%, from 218 mg/L to 323 mg/L, and its logarithmic growth showed improvement compared to the CS strain (Table 2, Fig. 2a). However, the specific titer of CS40 decreased from 382 mg/g DCW to 286 mg/g DCW compared to the CS strain due to the increase in cell concentration (Table 2). The productivity per media volume increased by 44% from 1.80 mg/L/h to 2.60 mg/L/h while the productivity per microbial dry cell weight decreased from 3.15 mg/g DCW/h to 2.30 mg/g DCW/h due to the increase in cell concentration (Table 2). Interestingly, the methane and oxygen consumption pattern of CS40 also changed with cell growth. Compared with CS strain, the maximum consumption rate of methane and oxygen increased and the prolonged maintenance of high gas consumption rate was observed (Fig. 2c, d). Through ALE, the maximum methane consumption rate of CS40 increased to 5.37 mmol/L/h, 30% higher than that of the CS strain (4.13 mmol/L/h). And the maximum specific methane consumption rate of CS40 strain increased to 20.9 mmol/g DCW/h, 69.6% higher than that of the CS strain (12.32 mmol/g DCW/h), also (Supplementary Fig. S1). In addition, the duration of the high methane consumption rate in CS40 was extended up to 57 h, while the methane consumption rate of the CS strain reached its maximum at 24 h and started to decrease immediately after cell growth entered the early stationary phase (Fig. 2c). The total methane consumed by CS40 (3.93 g/L) during one batch culture also increased 1.96-fold compared to the CS strain (2.01 g/L). The volumetric oxygen consumption rate showed a similar pattern with volumetric methane consumption rate (Fig. 2d). This phenotype was also observed in other mutant strains that were constructed based on CS40 (Supplementary Fig. S8). It could be postulated that the improvement of methane and oxygen consumption might be caused by the increase of particulate methane monooxygenase (pMMO) expression. To confirm whether the expression level of pMMO changed or not, its transcription level was measured by qPCR. The mRNA level of pMMO gene in CS40 strain increased 2.65-fold compared to the CS strain (Fig. 4a).
Fig. 2.
Growth, succinate accumulation, and gas consumption rate of CS and CS40 strains during bioreactor culture. a The logarithmic graphs of CS and CS40 strains in the bioreactor. The logarithmic graphs of cell growth were included to show the changes in the growth phase clearly. b The changes of succinate concentration in media during cell growth in the bioreactor. c Methane consumption rate of mutant strains during fermentation. d Oxygen consumption rate of mutant strains during fermentation
Fig. 4.
Transcription level changes of duplicated genes and pMMO of CS40. a Transcription level change of pMMO in CS40. b, c, d, e, f The changes of transcription level of genes in duplication region. b AYM39_08385; Malyl-CoA lyase, c AYM39_08395; Malate thiokinase subunit beta, d AYM39_08400; Malate thiokinase subunit alpha, e AYM39_08540; Methenyltetrahydromethanopterin cyclohydrolase, f AYM39_08690; Peroxiredoxin. Transcription level of each gene in CS was set at one for comparison. Transcription level of each gene was normalized using transcription level of 16S rRNA gene. Each data set obtained from three separate experiments
To elucidate reasons for these phenotypical changes, the whole genome sequence of CS40 strain was completely analyzed using massive parallel sequencing technologies such as the Illumina sequencing technology and the PacBio single molecule real-time (SMRT) sequencing technology. The 147,671 subreads with a mean subread length of 7,880 bases generated by PacBio SMRT sequencing technology were de novo assembled using the Hierarchical Genome Assembly pipelines (HGAP) with sequencing depth of 134.4-fold. The 16,196,851 read pairs (150 bp X 2, sequencing depth > 800-fold) generated by Illumina sequencing technology were used to correct errors in the assembled contigs from HGAP. As results of genome assembly, the genome consists of a circular chromosome (4,937,345 bp with 56.48% G + C content) and a circular plasmid (274,155 bp with 51.55% G_C content).
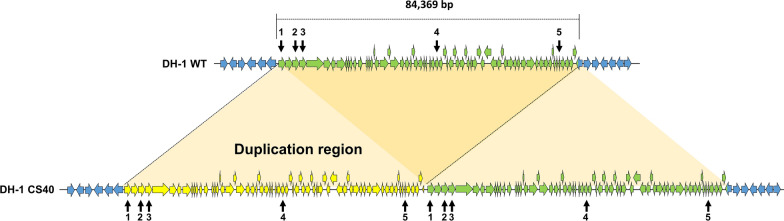
The chromosome of the CS40 strain increased by approximately 87 kb compared to the wild type DH-1 strain (chromosome, 4,849,532 bp; plasmid, 277,875 bp), whereas the plasmid size decreased by about 3.7 kb (Supplementary Fig. S2). Whole genome alignment between CS40 and DH-1 strain using Mauve indicated that the region containing the AYM39_08385 – AYM39_08720 genes (84.4 kb; 1,825,474–1,909,842 bp) on the chromosome of DH-1 strain was duplicated at the 1,909,843 bp on the chromosome of CS40 strain (Fig. 3, Table 3). Sequencing depth pattern of the CS40 strain’s PacBio raw data mapped on the chromosome sequence of DH-1 strain also revealed that the sequencing depth in the region increased approximately two-fold in chromosome of the CS40 strain (Supplementary Fig. S3). The duplicated region contains 68 genes (Table 4). To determine whether the duplication in the genome affected the expression of genes included in that region, the transcription levels at the beginning (AYM39_08385), middle (AYN39_08540), and end (AYM39_08690) of that region were measured using qPCR. The expression levels of these genes increased around twofold comparing to those of CS40 strain (Fig. 4b, e, f).
Fig. 3.
Synteny between the duplicated region in CS40 genome and the corresponding region in WT genome. Approximately 84 kb sequence including genes from AYM39_08385 to AYM39_08720 was duplicated in CS40 genome. Duplicated genes are indicated by yellow and green arrows in CS40 genome. The locus tag of genes in duplication region, with transcription levels confirmed by qPCR, is described as numbers (1, 2, 3, 4, 5). 1: AYM39_08385; 2: AYM39_08395; 3: AYM39_08400; 4: AYM39_08540; 5: AYM39_08690
Table 3.
Mutations of CS40 strain genome from whole genome shotgun sequencing data
| No | Loci | Mutation Type | WT | CS40 | Effects | Locus tag |
|---|---|---|---|---|---|---|
| 1 | aChromosome (261,875) | Point mutation | A | C | – | Intergenic region |
| 2 | Chromosome (1,110,051) | Insertion | – | G | – | Intergenic region |
| 3 | Chromosome (1,248,778) | Insertion | – | G | – | Intergenic region |
| 4 | Chromosome (1,909,843) | Insertion | – | 84,369 bp | Duplication | – |
| 5 | Chromosome (2,610,413–2,610,629) | Deletion | 217 bp | – | – | Intergenic region |
| 6 | Chromosome (2,805,889) | Point mutation | G | A | Missense (Ala175Val) | AYM39_12470 |
| 7 | Chromosome (4,271,282) | Deletion | C | – | Frame shift | AYM39_19030 |
| 8 | Chromosome (4,458,259) | Point mutation | C | A | – | Intergenic region |
| 9 | Chromosme (Integrated malate synthase) | Insertion | – | 132 bp | Fragmentation by stop codon | – |
| 10 | bPlasmid (16,199–19,918) | Deletion | 3720 bp | – | In frame deletion | AYM39_21655 |
| 11 | Plasmid (21,446–21,725) | Substitution | 280 bp | 280 bp | Missense | AYM39_21655 |
Table 4.
The list of genes in the duplicated region of CS40
| No. | Locus tag | Product | No | Locus tag | Product |
|---|---|---|---|---|---|
| 1 | AYM39_08385 | Malyl-CoA lyase | 35 | AYM39_08555 | Aldehyde-activating protein |
| 2 | AYM39_08390 | Hypothetical protein | 36 | AYM39_08560 | Hypothetical protein |
| 3 | AYM39_08395 | Malate thiokinase subunit beta | 37 | AYM39_08565 | Hypothetical protein |
| 4 | AYM39_08400 | Malate thiokinase subunit alpha | 38 | AYM39_08570 | Methanofuran synthetase, MfnF |
| 5 | AYM39_08405 | Helicase SNF2 | 39 | AYM39_08575 | Aminodeoxychorismate synthase component I |
| 6 | AYM39_08410 | Transposase | 40 | AYM39_08580 | Uridylate kinase |
| 7 | AYM39_08415 | Hypothetical protein | 41 | AYM39_08585 | Hypothetical protein |
| 8 | AYM39_08420 | NrdJa | 42 | AYM39_08590 | Hypothetical protein |
| 9 | AYM39_08425 | Hypothetical protein | 43 | AYM39_08595 | GTP-binding protein |
| 10 | AYM39_08430 | Hypothetical protein | 44 | AYM39_08600 | GTP-binding protein |
| 11 | AYM39_08435 | NrdJb | 45 | AYM39_08605 | RND transporter |
| 12 | AYM39_08440 | Hypothetical protein | 46 | AYM39_08610 | Multidrug transporter |
| 13 | AYM39_08445 | Transposase | 47 | AYM39_08615 | Multidrug transporter |
| 14 | AYM39_08450 | Hypothetical protein | 48 | AYM39_08620 | Efflux transporter periplasmic adaptor subunit |
| 15 | AYM39_08455 | Hypothetical protein | 49 | AYM39_08625 | TetR family transcriptional regulator |
| 16 | AYM39_08460 | Hypothetical protein | 50 | AYM39_08630 | Hypothetical protein |
| 17 | AYM39_08465 | Hypothetical protein | 51 | AYM39_08635 | Multidrug transporter |
| 18 | AYM39_08470 | Hypothetical protein | 52 | AYM39_08640 | Transcription elongation factor GreB |
| 19 | AYM39_08475 | Integrase | 53 | AYM39_08645 | Peptidylprolyl isomerase |
| 20 | AYM39_08480 | Hypothetical protein | 54 | AYM39_08650 | Hypothetical protein |
| 21 | AYM39_08485 | Hypothetical protein | 55 | AYM39_08655 | Hypothetical protein |
| 22 | AYM39_08490 | Hypothetical protein | 56 | AYM39_08660 | Peptidase U6 |
| 23 | AYM39_08495 | Glutathionylspermidine synthase | 57 | AYM39_08665 | Sulfate transporter |
| 24 | AYM39_08500 | Hypothetical protein | 58 | AYM39_08670 | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase PgaB |
| 25 | AYM39_08505 | Two component system response regulator | 59 | AYM39_08675 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase |
| 26 | AYM39_08510 | Two component system sensor histidine kinase CreC | 60 | AYM39_08680 | Hypothetical protein |
| 27 | AYM39_08515 | Hypothetical protein | 61 | AYM39_08685 | Transcriptional regulator |
| 28 | AYM39_08520 | Nitrogen fixation protein (NifX) | 62 | AYM39_08690 | Peroxiredoxin |
| 29 | AYM39_08525 | Hypothetical protein | 63 | AYM39_08695 | Sodium pump decarboxylase subunit gamma |
| 30 | AYM39_08530 | tRNA-Ser | 64 | AYM39_08700 | Oxaloacetate decarboxylase |
| 31 | AYM39_08535 | Hypothetical protein | 65 | AYM39_08705 | Glutaconyl-CoA decarboxylase subunit beta |
| 32 | AYM39_08540 | Methenyltetrahydromethanopterin cyclohydrolase | 66 | AYM39_08710 | Hypothetical protein |
| 33 | AYM39_08545 | Alpha-L-glutamate ligase | 67 | AYM39_08715 | Hypothetical protein |
| 34 | AYM39_08550 | Triphosphoribosyl-dephospho-CoA synthase | 68 | AYM39_08720* | Transposase |
*This gene was partially duplicated within its sequence (The location of DH-1 WT chromosome from 1,909,795 to 1,909,843 bp; 49 bp)
Another insertion occurred in the heterologous sequence integrated to construct glyoxylate shunt. The nucleotide sequence of 132 bases containing two stop codons was inserted into the malate synthase gene integrated for glyoxylate shunt (Supplementary Fig. S4). So, the integrated malate synthase of 531 amino acid was fragmented to the smaller protein of 171 amino acid. This unexpected mutation might affect the enzyme activity severely. Intriguingly three genes (AYM39_08385: malyl-CoA lyase and AYM39_08395-08400: malate-CoA thiokinase) that had a potential to restore the missing link between glyoxylate and malate in the integrated glyoxylate shunt were found in the duplicated region of CS40 (Table 4, Fig. 1, Supplementary Fig. S5). Malyl-CoA lyase can mediate the reaction between glyoxylate and malyl-CoA. Malate thiokinase can convert malyl-CoA to malate. Actually, the transcription levels of these genes in CS40 strain increased approximately twofold comparing to those of CS strain, when measured by qPCR (Fig. 4b, c, d).
The gene encoding type II citrate synthase (AYM39_19030) was frameshifted by one base deletion, and potentially resulted in the loss of function. There is another gene (AYM39_09355) encoding the type II citrate synthase on the chromosome, so the complete loss of function is not expected. And 175th alanine of the helix-turn-helix domain-containing protein (AYM39_12470) was substituted to valine by point mutation. As results of Sanger sequencing, these two mutations were also found in the genome of CS strain (data not shown). So, these two mutations were not generated during ALE and could not be the reason for the improved performance of CS40 strain. The other mutations were found in non-coding regions. Additionally, sequence deletion and rearrangement happened in the natural plasmid (Table 3). The sequence of (AYM39_21655: hypothetical protein) has 9 times repetition of 744 bp. The sequence from 237 bp of 2nd repeat block to 236 bp of 7th repeat block was deleted. A portion of the 9th repeat block has been replaced with the sequence from base 276 to base 555 of the 4th repeat block.
The enhancement of succinate production by overexpressing pc and ppc genes
The activities of the pc and ppc genes produce oxaloacetate (OAA) from pyruvate or phosphoenolpyruvate by incorporating CO2. OAA is a substrate of citrate synthase, the first step enzyme in the TCA cycle. Therefore, the presence of both pc and ppc genes in DH-1 is advantageous for producing TCA cycle-related products such as succinate and 1,4-butanediol [28]. For overexpress the pc and ppc genes, a strong and constitutive promoter was needed. To select a strong promoter, the top 20 genes in the RNA relative abundance levels were screened (Table 5). RNA samples were obtained from cells growing under 3.5% and 15% O2. Among the top 20 genes, the cold-shock protein gene (AYM39_18825) exhibited relatively constant expression levels regardless of changes in O2 concentration, with the exception of transfer-messenger RNA. Its promoter region was selected for the next promoter exchange experiment. RNA-seq analysis was performed as described in the Materials and Methods section.
Table 5.
Top 20 RNA sequencing data of DH-1 genes for selection of promoter
| No | Locus tag | Type | Functions | TPM* (3.5% O2) | TPM* (15% O2) | Fold** |
|---|---|---|---|---|---|---|
| 1 | AYM39_17695 | tmRNA | Transfer-messenger RNA | 190,466 | 185,239 | 0.97 |
| 2 | AYM39_19775 | Protein | Methane monooxygenase subunit C | 96,356 | 27,806 | 0.29 |
| 3 | AYM39_20600 | Protein | RNase P RNA component class A | 60,979 | 50,865 | 0.83 |
| 4 | AYM39_14685 | Protein | Hypothetical protein | 27,449 | 3397 | 0.12 |
| 5 | AYM39_19765 | Protein | Methane monooxygenase subunit B | 27,086 | 8147 | 0.30 |
| 6 | AYM39_19770 | Protein | Methane monooxygenase subunit A | 26,255 | 5597 | 0.21 |
| 7 | AYM39_11910 | Protein | Hypothetical protein | 17,722 | 48 | 0.00 |
| 8 | AYM39_15615 | Protein | Methanol dehydrogenase | 13,706 | 4962 | 0.36 |
| 9 | AYM39_15600 | Protein | Methanol dehydrogenase | 11,638 | 6170 | 0.53 |
| 10 | AYM39_02740 | Protein | 3-hexulose-6-phosphate synthase | 8076 | 2515 | 0.31 |
| 11 | AYM39_13140 | Protein | hypothetical protein | 6170 | 3378 | 0.55 |
| 12 | AYM39_08555 | Protein | formaldehyde-activating enzyme | 6016 | 8798 | 1.46 |
| 13 | AYM39_18825 | Protein | Cold-shock protein | 5653 | 6162 | 1.09 |
| 14 | AYM39_01615 | Protein | Hypothetical protein | 5370 | 1018 | 0.19 |
| 15 | AYM39_15605 | Protein | Cytochrome c(L), periplasmic | 5200 | 3475 | 0.67 |
| 16 | AYM39_02470 | Protein | 3-hexulose-6-phosphate synthase | 5164 | 1902 | 0.37 |
| 17 | AYM39_02670 | Protein | Coenzyme PQQ precursor peptide PqqA | 5132 | 3324 | 0.65 |
| 18 | AYM39_15610 | Protein | Methanol oxidation system protein MoxJ | 4917 | 2890 | 0.59 |
| 19 | AYM39_02475 | Protein | 6-phospho-3-hexuloisomerase | 4729 | 1926 | 0.41 |
| 20 | AYM39_02725 | Protein | Hypothetical protein | 3871 | 2311 | 0.60 |
* TPM: Transcript per Million. ** Fold values represent ratio between TPM values of samples cultured under 3.5% and 15% O2 conditions. Genes showing relatively constant expression in response to oxygen composition change (fold: around 1.0) are indicated in bold letters
In the strains CS401(CS40 PAYM39_18825-pc), 402(CS40 PAYM39_18825-ppc), and 403 (CS401 PAYM39_18825-ppc), the upstream 400 bp region of the AYM39_18825 gene was inserted in front of the target genes (pc and ppc) in the genome of CS40 by homologous recombination (Supplementary Fig. S6 b, c, d, Table 1). Strength of this novel promoter was confirmed by qPCR (Fig. 5a, b, c). The transcription level of pc or ppc in the mutants (CS401, 402, 403) increased by 16 ~ 24-fold compared to their parent strain (CS40). Protein expression of this strong novel promoter was confirmed using green fluorescent protein (GFP). GFP gene sequences fused with the promoter regions of pc, ppc, and AYM39_18825 genes were integrated into the genome of CS40 (Supplementary Fig. S7). When GFP was expressed by the promoter of cold shock protein, its fluorescence was 12 ~ 34 times stronger than in other cases (Fig. 5 d).
Fig. 5.
The increase in transcription and translation level of target genes by the promoter of AYM39_18825. a, b, c The changes of transcription level of pc and ppc genes by overexpression in the corresponding mutant strains. The expression level of each gene in CS40 was set at one for comparison. The variations in mutant strains (CS401, 402, and 403) were indicated as fold changes relative to the mRNA levels in the CS40 strain. Transcription level of each gene was normalized using transcription level of 16S rRNA gene. d The fluorescence signal intensity of GFP produced from the promoters of pc, ppc, and AYM39_18825 genes. The error bars mean standard deviations obtained from three separate experiments
The succinate production performance of these mutants with strengthened pc or ppc expression was tested in bioreactors through batch cultivations where gaseous substrates (methane and oxygen) were continuously fed (Fig. 6). The overexpression of pc or ppc was beneficial for increasing volumetric titer. When the pc gene was overexpressed (CS401), the titer increased to 443 mg/L at 126h cultivation time. CS402, in which the ppc gene was overexpressed, showed a similar maximum succinate titer (435 mg/L at 114h cultivation time). When the expression of these two genes was reinforced simultaneously (CS403), the succinate titer was improved further to 502 mg/L at 128h cultivation time (Fig. 6). The volumetric productivity of the CS401, CS402 and CS403 strains increased compared to the CS40 strain (2.60 mg/L/h), showing similar results of 3.51, 3.45 and 3.92 mg/L/h, respectively (Table 2). However, the specific productivity of these strains decreased compared to the CS40 strain (2.30 mg/g DCW/h), 2.21, 1.65 and 2.14 mg/g DCW/h, respectively.
Fig. 6.
Growth and succinate accumulation in mutants containing the overexpressed pc or ppc genes during fermentations. a The logarithmic cell growth graphs of mutant strains (CS40, 401, 402, and 403) in bioreactor. The logarithmic graphs of cell growth were included to clearly show the changes in the growth phase. b Accumulation of succinate during cell growth in the bioreactor. The graphs of CS40 strain are incorporated for comparison
The effects of succinate semialdehyde dehydrogenase gene knock out
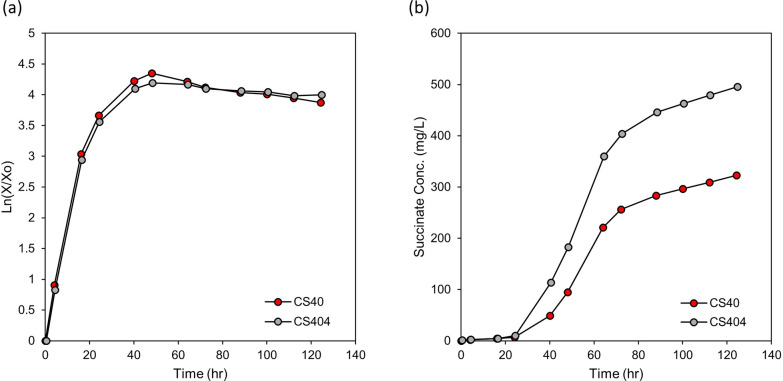
To further improve succinate production, genetic modifications for removing a possible succinate consumption route and enhancing the TCA cycle by increasing citrate synthase, the first enzyme of the TCA cycle, were attempted. A succinate semialdehyde dehydrogenase (ssadh) gene (AYM39_17625) was identified from the genome sequence of DH-1 (GenBank: CP014360.1). Considering that this enzyme mediates the conversion between succinate and succinate semialdehyde reversibly [40], it may consume succinate produced from the glyoxylate shunt and TCA cycle in CS40. Strain CS404, where the ssadh gene was deleted from the CS40 strain (Supplementary Fig. S6 e), was constructed. When CS404 was cultured in the bioreactor, its maximum volumetric succinate titer (496 mg/L at 124 h) increased by 53.5% compared with its parental strain CS40 (323 mg/L at 124 h; Fig. 7). The specific titer of CS404 (306 mg/g DCW) increased slightly by 7.06% compared with CS40 (286 mg/g DCW). The succinic acid production rate per volume of media also increased (3.98 mg/L/h, Table 2). The specific productivity also increased (2.46 mg/g DCW/h, Table 2).
Fig. 7.
Growth and succinate accumulation of mutants containing ssadh disruption. a The logarithmic cell growth graphs of mutant strains (CS40 and 404) in bioreactor. The logarithmic graphs of cell growth are included to show the changes in the growth phase clearly. b Accumulation of succinate during cell growth in the bioreactor. The graphs of CS40 strain are incorporated for comparison
Combination of genetic manipulations beneficial for succinate production
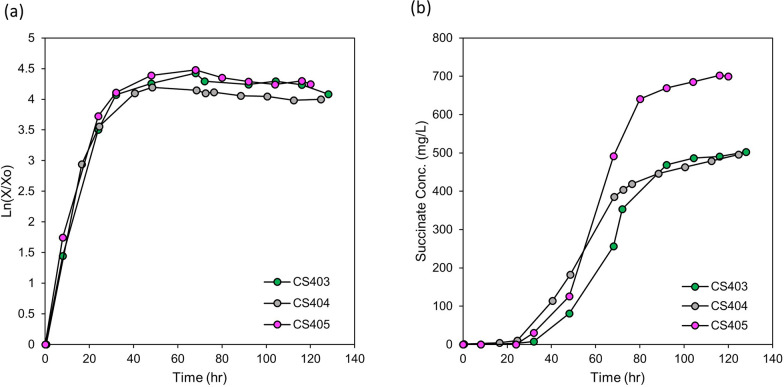
Genetic manipulations that improved succinate production performance were combined in one strain (CS405). Additionally, the effect of citrate synthase (gltA) was tested by transforming CS403 with the pTOP-ssadh-gltA vector. It was reported that citrate synthase overexpression could be beneficial for succinate production in C. glutamicum [41]. The ssadh gene was deleted and the gltA gene fused with the mxaF promoter was inserted simultaneously in the genome of CS403 containing overexpressed pc and ppc genes (Supplementary Fig. S6 f, Table 1). The performance of CS405 strain was tested in a bioreactor. It showed the best performance not only in succinate titer (702 mg/L) but also in specific productivity (3.49 mg/g DCW/h) and volumetric productivity (5.84 mg/L/h) when compared to other strains used in this study (Table 2, Fig. 8). The genetic variations that occurred during ALE of the CS strain and every additional rational genetic modification applied after ALE appeared to have synergistic effects on succinate production (Fig. 1).
Fig. 8.
Growth and succinate accumulation of the final mutant. a The logarithmic cell growth graphs of mutant strains (CS405) in the bioreactor. The logarithmic graphs of cell growth were included to show the changes in the growth phase clearly. b Accumulation of succinate during cell growth in the bioreactor. For comparison, the graphs of CS403 and 404 are included
Discussion
Methane gas composes large quantities of not only shale gas, but also biogas produced through anaerobic fermentation of organic waste. Unfortunately, it has higher global warming potential than carbon dioxide. Therefore, the production of industrially useful products with the ability to convert methane to chemicals have possibility for reducing atmospheric methane and anthropogenic methane emission [4]. The possibility of producing organic acids by methanotrophic bacteria was first suggested by Kalyuzhnaya et al., [10]. It was confirmed that succinate productivity could be increased through genetic engineering [28]. In this study, we attempted to further increase succinic acid production through a combination of ALE and rational genetic engineering.
To enable repetitive genetic engineering, the strain (DH-Cre) with the genome-integrated Cre-lox system, was used as a parental strain for the next steps. The succinate-producing strain in which the sdhB gene was replaced by glyoxylate shunt genes, was constructed again based on the DH-Cre strain. This newly created strain (CS strain) showed better performance than the previous strain, except for maximum cell growth. CS strain has the unexpected frame shift mutation in one of two citrate synthases present in the genome. It may negatively affect cell growth, considering that citrate synthase is the first enzyme of TCA cycle. Another difference between DS-GL and CS strains is the integrated Cre-lox system. The integration site of Cre-lox system was selected in the noncoding intergenic region with very low transcription level between AYM39_00230 and AYM39_00235. This site was chosen to prevent unexpected phenotypical change based on the RNA sequencing data. And the 450 bp region (59,550 bp to 59,101 bp in DH-1 genome) was deleted by homologous recombination [29]. In spite of very low transcription level, the deletion may affect cell growth negatively in an unknown way. Although the reason for the decrease in maximum OD600 wasn’t clearly elucidated in this study, a good strain for the subsequent developmental steps was constructed. The defect in maximum cell growth was overcome by ALE.
ALE with the CS strain caused interesting physiological changes in this strain. The methane and oxygen consumption speed in CS strain peaked at the late exponential phase in the CS strain and rapidly decreased as the early stationary phase began. Intriguingly, the CS40 strain subjected to the ALE process showed an extended period during which the high methane and oxygen consumption speed was maintained. These phenotypical changes can be explained by the increase of pMMO gene transcription in CS40 strain. What changes in CS40 induced the increase of pMMO gene expression is not clear yet. However, some duplicated genes in CS40 may be related to this phenotype. Among the 68 duplicated genes, there are three genes related to transcriptional regulation (AYM39_08505, 08625, 08685) and the increase of transcriptional regulator can shift the expression profile of many genes. There are some reports about reciprocal transcription regulation between sMMO and pMMO depending on copper concentration [42]. However, which protein can regulate or repress pMMO actually hasn’t been reported yet. Further genetic studies are needed to reveal how pMMO expression increased in CS40.
The increased pMMO expression level of CS40 strain could enable a higher uptake of methane, leading to an increase in intracellular carbon substrates. It could lead to enhanced intracellular metabolism, subsequently resulting in cell growth improvement. Usually, methanotrophs have different carbon assimilation pathways depending on their type. The type I (gammaproteobacteria) assimilate carbon using RuMP cycle, and type II (alphaproteobacteria) assimilate carbon using the serine cycle [43]. In the case of DH-1, which is a type I methanotroph, it primarily uses the RuMP cycle for carbon assimilation, but it also possesses the serine cycle, providing multiple pathways for carbon assimilation [20]. So, when intracellular metabolism increases, it has a higher possibility of biomass accumulation than succinate. In methanotrophs, increased expression of pMMO by adding higher amounts of copper ions to the growth medium leads to increased growth yields [44]. As a result of increased cell growth, the volumetric succinate production of CS40, 401, 402, 403, 404 and 405 increased. However, the specific succinate production of CS40, 401, 402, 403 and 404 decreased comparing to CS strain, except for the final strain, CS405.
Unexpectedly, malate synthase integrated for building glyoxylate shunt, was fragmented in CS40 strain. Considering that nearly 2/3 of the entire protein sequence was deleted, this mutation may be critical to the protein activity. Interestingly, enzymes that were expected to complement the loss of malate synthase activity were found within the duplicated sequences of the CS40 strain. The genes of Malyl-CoA lyase and Malate-CoA ligase are included in the duplicated sequences of CS40, and it has been confirmed through qPCR that the transcription levels of these genes increased around twofold compared to the CS strain. Malyl-CoA lyase, mediating a reversible reaction between acetyl-CoA/glyoxylate and malyl-CoA, was reported to produce malyl-CoA using acetyl-CoA/glyoxylate in Rhodobacter spheroids. In R. spheroids, Malyl-CoA lyase can substitute for malate synthase activity along with malyl-CoA thioesterase [45]. Malate-CoA ligase, also known as malate thiokinase (mtk), is known to mediate a reversible reaction between malyl-CoA/ADP/Phosphate and malate/CoA/ATP [46]. Considering that these two reactions are reversible, there is a possibility that glyoxylate produced by isocitrate lyase can be converted into malate through these two enzymes complimenting the damaged malate synthase.
Based on NGS data, the most significant change during the adaptive laboratory evolution period is the duplication of the sequence spanning 84 kb. The 68 genes included in the duplicated region may have complex impacts on the physiology of the CS40 strain. The list of 68 genes includes proteins involved in nucleotide synthesis (NrdJ, uridylate kinase), transcription regulators (AYM39_08505, 08510, 08625, and 08685), oxidative stress response genes (glutathionylspermidine synthase, peroxiredoxin), transcription elongation factor GreB, proteins related to protein synthesis mechanisms (tRNA-Serine, Alpha-L-glutamate ligase, Peptidylprolyl isomerase), biofilm synthesis-related genes (Poly-beta-1,6-N-acetyl-D-glucosamine synthase), folate synthesis-associated genes (aminodeoxychorismate synthase component I), a nitrogen fixation-related gene (NifX), genes related to formaldehyde-oxidation to formate (methenyltetrahydromethanopterin (H4MPT) cyclohydrolase, aldehyde-activating protein, methanofuran synthetase MfnF), membrane transporters (AYM39_08605, 08610, 08615, 08620, 08635, and 08665), etc. [20, 47–58]. The higher cell growth of the CS40 strain compared to the CS strain may be related to the duplication of genes with various functions. For example, an increase in the expression of sulfate transporters could facilitate the influx of sulfate, a major component of the NMS medium. Oxidative stress is a stress which is inevitable to organisms growing in aerobic environments. An increase in oxidative stress response genes may positively affect the growth of methanotrophs that require oxygen for growth. NifX was reported to be related to the biosynthesis of Fe-Mo cofactor of nitrogenase [57]. Increased nitrogenase activity through enhanced cofactor synthesis may potentially facilitate a stable supply of nitrogen sources. Three genes related with formaldehyde oxidation pathway are included in the duplicated genes. Aldehyde-activating protein (AYM39_08555) can convert formaldehyde to 5,10-methylene tetrahydromethanopterin (H4MPT) which is transformed to 5,10-methenyl H4MPT by methylene H4MPT dehydrogenase (AYM39_08875). Methenyl H4MPT cyclohydrolase (AYM39_08540) converts 5,10-methenyl H4MPT into N5-formyl H4MPT. The formyl group of N5-formyl H4MPT is transferred to methanofuran, the cofactor of formyltransferase/hydrolase complex [58]. MfnF (AYM39_08570) was reported to be related to the biosynthesis of methanofuran [56]. More expression of genes related with formaldehyde oxidation may lead to more utilization of reducing power that can be generated by formaldehyde oxidation step. Additionally, the duplicated genes related to protein synthesis mechanisms may have positively influenced cell growth. In particular, the enzyme alpha-L-glutamate ligase, also known as glutamine synthetase, has been reported to exhibit high activity in the presence of nitrate, which is the nitrogen source in NMS [59]. An increase in translation-related genes may also have a positive impact on cell growth. Five more mutations were found in intergenic regions. They are either located in the upstream region (261,875 bp) of a hypothetical gene (AYM39_01125) or within sequences featuring 15–17 bp repetitive guanine nucleotide stretch (1,110,051 and 1,248,778 bp) or between two hypothetical genes (AYM39_05845 ~ 05850, 1,248,778 bp), or in the CRISPR region (2,610,413–2,610,629 deletion), or positioned in the intergenic region between 23S rRNA gene and tRNA-Ala (AYM39_19785-19,790, 4,458,259 bp). It seems that these mutations may have less significant effects on cell growth than 84 kb duplication.
To further improve succinate production, a new strong promoter applicable to DH-1 was screened using RNA sequencing data. The strong promoters that could maintain strong expression under both low and high O2 concentrations were searched because the novel promoter could be utilized for other studies not solely for this one. Previously it was reported that more organic acid could be produced under O2-limited condition and high O2 condition is favorable for biomass production in methanotrophs [10]. However, depending on targets, sometimes increasing cell concentration under high O2 concentration can be beneficial for titer improvement despite of some decrease in specific titer (target concentration per unit cell). So, it could be assumed that a promoter showing strong expression regardless of oxygen concentrations would have greater versatility. As a high O2 concentration, 15% O2 condition is similar to a typical methanotroph culture condition (30% methane, 70% air). For low O2 condition, 3.5% O2 was selected to make O2 limited condition easily. The promoter of cold shock protein showing a constant and strong expression even after O2 concentration change in the supplied gas was selected as a novel strong promoter (Table 5). Since the translation of cold shock protein is regulated in response to temperature change, the protein expression from the promoter sequence chosen was confirmed through GFP expression. When GFP was expressed from this cold shock protein promoter, 12 ~ 34 times stronger fluorescence signals than those from the pc or ppc promoters were obtained without any temperature change. Considering that the secondary structure of mRNA is important for cold shock protein translation, it can be postulated that the formation of the secondary structure was hindered by the sequences of other genes fused to the promoter.
The manipulated strains (CS401, 402, and 403) showed better volumetric titer and productivity than CS40 strain. However the specific titer and specific productivity reduced or kept similar to the levels of CS40 through pc and ppc overexpression due to increased cell growth. Methane taken by the mutant cells seems to be redirected for more cell growth rather than succinate production by pc and ppc overexpression. It can be assumed that the increase of volumetric parameters was driven by the increased cell growth. The overexpression of pc or ppc genes was expected to increase the OAA concentration. OAA can serve as a precursor or backbone for amino acids such as aspartate, isoleucine and lysine [60]. In C. glutamicum ATCC 21799 strain, it was reported that overexpression of native pyruvate carboxylase could lead to improved cell growth, compared to its parental strain [61].
The ssadh gene was deleted to prevent leakage of succinate. According to a previous studies, succinate semialdehyde dehydrogenase converts succinate into succinate semialdehyde in the presence of NADH [40]. During the stationary phase of microbial growth, the NADH/NAD+ ratio generally increases compared to the exponential growth phase. In this study, succinate production in all mutants began in the late exponential phase or early stationary phase. Considering the possibility of high intracellular NADH concentrations in these phases, it was postulated that the ssadh gene deletion could be beneficial for succinate production. In a previous study, the mutant of C. glutamicum ATCC13032, integrated with the transhydrogenase from E. coli, showed improved succinate production under aerobic conditions due to the transhydrogenase, which increases intracellular NADH [62]. Actually the mutant strain (CS404) without ssadh showed slight increases in specific titer, and specific productivity comparing to those of CS40. As for volumetric parameters, CS404 strain showed more significant improvement.
To further enhance succinate production, pc/ppc overexpression and ssadh disruption were integrated in one strain (CS405). At the same time, citrate synthase (gltA) was overexpressed by replacing ssadh gene with gltA fused to the mxaF promoter. CS405 strain showed the best performance among all other strains tested in this study. Not only volumetric parameters but also specific titer and productivity increased. Considering that OAA is a substrate of citrate synthase and that citrate synthase is the first enzyme of the TCA cycle, it can be inferred that the phenotypic changes are due to the increased carbon flow entering the TCA cycle. There was a report showing the possibility that the overexpression of ppc gene along with gltA gene could enhance cell growth by increasing the flux through the TCA cycle in E. coli [63].
All strains tested in this study showed a similar succinate production pattern. Succinate concentration started to increase after the early stationary phase began. Normally other succinate producers using liquid substrates as carbon sources are known to accumulate succinate while they are growing actively under anaerobic conditions [64]. Even in aerobic condition, C. glutamicum engineered for succinate production seemed to start making succinate at its exponential phase [65]. It can be assumed that the cells using gas as sole carbon source, cannot afford to accumulate succinate when they grow fast. This assumption is in good agreement with the previous report that showed enhanced organic acid production under the microaerobic condition where fast cell growth could be inhibited [10]. If cell growth and succinate production are competitive to each other, controlling cell growth by limiting nutrient source concentration would be a way to improve succinate production further. It was reported that TCA cycle flux could be reinforced by nitrogen starvation and mevalonate yield could be increased by sulfur starvation in E. coli [66]. Delicately controlling nutrient levels in the media to balance between cell growth and succinate production would be necessary to enhance succinate generation further.
In the case of succinic acid production using microorganisms, feedstock costs account for about 20–30% of the total process cost, making it a critical factor [67, 68]. Using methane gas as a low-cost feedstock can provide the advantage of reducing process costs. However, for this advantage to be realized and for the methane-based succinic acid production process to be competitive in the market, the titer and productivity must reach levels comparable to those achieved with soluble substrates. There have been many studies describing microbial succinate production using soluble substrates. Some of them showed very high succinate production titers reaching around 100 g/L with Escherichia coli, Corynebacterium glutamicum and Mannheimia succiniciporducens [64, 69, 70]. Considering that studies on producing succinic acid using methanotrophs have been conducted more recently compared to studies utilizing other soluble substrates, there is still significant potential to enhance its performance through genetic engineering. Additionally, optimizing culture conditions for high cell density culture is also expected to improve its performances.
In summary, mutant strains with higher succinate production could be achieved by combining the overexpression of pc, ppc, and citrate synthase genes which could accelerate TCA cycle and the disruption of ssadh gene, an enzyme that could potentially consume succinic acid. The volumetric titer (702 mg/L) and volumetric productivity (5.84 mg/L/h) of succinate of final mutant (CS405) were successfully improved 3.22 and 3.24 times respectively, compared to their parent strain (CS; Table 2). The improved specific titer (419 mg/g DCW) and specific productivity (3.49 mg/g DCW/h) of final mutant proves that each cells produces succinate more and faster than parental strain. The final mutant showing the best performance (CS405) was created through three rounds of rational genetic engineering after ALE. This study confirmed that synergistic effects could be created by combining rational genetic engineering and ALE depending on spontaneous mutations.
Supplementary Information
Acknowledgements
This work was supported by the Technology Innovation Program (RS-2023-00265608, 1415188462, Development of production technology of value-added chemical using by-product gas in Naphtha Cracker) funded By the Ministry of Trade Industry & Energy (MOTIE, Korea). This research was supported by Korea Institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries, Korea (RS-2022-KS221653).
Biographies
Jae-Hwan Jo
A PhD. Student in Chonnam National University
Jeong-Ho Park
A Researcher in CJ CheilJedang Co.
Byung Kwon Kim
A Researcher in GI Biome Inc.
Seon Jeong Kim
A Master’s student in Chonnam National University
Chan Mi Park
A Post Doc. in Korea Institute of Energy Research
Chang Keun Kang
A Post Doc. in University of Seoul
Yong Jun Choi
A Professor in University of Seoul
Hyejin Kim
A graduate student in Kyung Hee University
Eun Yeol Lee
A Professor in Kyung Hee University
Myounghoon Moon
A Principal researcher in Korea Institute of Energy Research
Gwon Woo Park
A Senior researcher in Korea Institute of Energy Research
Sangmin Lee
A Professor in Chungnam National University
Soo Youn Lee
A Principal researcher in Korea Institute of Energy Research
Jin-Suk Lee
A Principal researcher in Korea Institute of Energy Research
Won-Heong Lee
A Professor in Chonnam National University
Jeong-Il Kim
A Professor in Chonnam National University
Min-Sik Kim
A Principal researcher in Korea Institute of Energy Research
Author contributions
J.-H. Jo: Writing—original draft, Investigation. J.-H. Park: Writing—original draft, Investigation. B. K. Kim: Writing—original draft, Data curation. S. J. Kim: Data curation. C. M. Park: Data curation. C. K. Kang: Investigation. Y. J. Choi: Conceptualization. H. Kim: Investigation
Funding
Financial supports were provided by the Ministry of Trade Industry & Energy (MOTIE, Korea) and the Ministry of Oceans and Fisheries, Korea.
Availability of data and materials
All data supporting the findings of this study are available within the paper and its Supplementary Information. RNA-seq data have been submitted to the GenBank databases under BioProject accession number (PRJNA769841). The whole genome sequencing data of CS40 strain is submitted to the GenBank databases under BioProject accession number (PRJNA1066519).
Declarations
Ethics approval and consent to participate
I (the corresponding author Min-Sik Kim) has read the journal policies and submit this manuscript in accordance with those policies. This study is not related with any clinical research. So ‘Ethics approval and consent to participate’ are not applicable for this study.
Consent for publication
By submitting my article, I (the corresponding author Min-Sik Kim) confirm that all the listed authors agreed to publish the data in Microbial Cell Factories and pay the APC in full if my article is accepted for publication. This article doesn’t include any details, images, or videos relating to an individual person. So ‘Consent for publication’ are not applicable for this study.
Competing interests
All authors of this paper, including Jae-Hwan Jo, Jeong-Ho Park, Byung Kwon Kim, Seon Jeong Kim, Chan Mi Park, Chang Keun Kang, Yong Jun Choi, Hyejin Kim, Eun Yeol Lee, Myounghoon Moon, Gwon Woo Park, Sangmin Lee, Soo Youn Lee, Jin-Suk Lee, Won-Heong Lee, Jeong-Il Kim, and Min-Sik Kim declare that the authors have no competing interests as defined by BMC, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jae-Hwan Jo and Jeong-Ho Park have contributed equally to this work.
Contributor Information
Jeong-Il Kim, Email: kimji@chonnam.ac.kr.
Min-Sik Kim, Email: kms0540@kier.re.kr.
References
- 1.Shindell DT, Faluvegi G, Koch DM, Schmidt GA, Unger N, Bauer SE. Improved attribution of climate forcing to emissions. Science. 2009;326:716–8. 10.1126/science.1174760. [DOI] [PubMed] [Google Scholar]
- 2.Dessus B, Laponche B, le Treut H. The importance of a methane reduction policy for the 21 ST century. 2009.
- 3.López JC, Rodríguez Y, Pérez V, Lebrero R, Muñoz R. CH4-based polyhydroxyalkanoate production: a step further towards a sustainable bioeconomy. In: López JC, editor. Biotechnological applications of polyhydroxyalkanoates. Singapore: Springer Singapore; 2019. p. 283–321. [Google Scholar]
- 4.Wang J, Salem DR, Sani RK. Microbial polymers produced from methane: overview of recent progress and new perspectives. In: Das S, Dash HR, editors. Microbial and natural macromolecules. Berlin: Academic Press; 2021. p. 117–42. [Google Scholar]
- 5.Clomburg JM, Crumbley AM, Gonzalez R. Industrial biomanufacturing: the future of chemical production. Science. 2017;355:25. [DOI] [PubMed] [Google Scholar]
- 6.Hwang IY, Hoon Hur D, Hoon Lee J, Park CH, Chang IS, Lee JW, et al. Batch conversion of methane to methanol using methylosinus trichosporium OB3B as biocatalyst. J Microbiol Biotechnol. 2015;25:375–80. [DOI] [PubMed] [Google Scholar]
- 7.Kalyuzhnaya MG, Puri AW, Lidstrom ME. Metabolic engineering in methanotrophic bacteria. Metab Eng. 2015;29:142–52. [DOI] [PubMed] [Google Scholar]
- 8.Conrado RJ, Gonzalez R. Envisioning the bioconversion of methane to liquid fuels. Science. 2014;343:621–3. [DOI] [PubMed] [Google Scholar]
- 9.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalyuzhnaya MG, Yang S, Rozova ON, Smalley NE, Clubb J, Lamb A, et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun. 2013;4:2785. [DOI] [PubMed] [Google Scholar]
- 11.Pham DN, Nguyen AD, Lee EY. Outlook on engineering methylotrophs for one-carbon-based industrial biotechnology. Chem Eng J. 2022;449:137769. [Google Scholar]
- 12.Lee OK, Hur DH, Nguyen DTN, Lee EY. Metabolic engineering of methanotrophs and its application to production of chemicals and biofuels from methane. Biofuels Bioprod Biorefin. 2016;10:848–63. [Google Scholar]
- 13.Strong PJ, Laycock B, Mahamud SNS, Jensen PD, Lant PA, Tyson G, et al. The opportunity for high-performance biomaterials from methane. Microorganisms. 2016;4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henard CA, Smith H, Dowe N, Kalyuzhnaya MG, Pienkos PT, Guarnieri MT. Bioconversion of methane to lactate by an obligate methanotrophic bacterium. Sci Rep. 2016;6:21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garg S, Wu H, Clomburg JM, Bennett GN. Bioconversion of methane to C-4 carboxylic acids using carbon flux through acetyl-CoA in engineered Methylomicrobium buryatense 5GB1C. Metab Eng. 2018;48:175–83. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen AD, Hwang IY, Lee OK, Kim D, Kalyuzhnaya MG, Mariyana R, et al. Systematic metabolic engineering of Methylomicrobium alcaliphilum 20Z for 2,3-butanediol production from methane. Metab Eng. 2018;47:323–33. [DOI] [PubMed] [Google Scholar]
- 17.Garg S, Clomburg JM, Gonzalez R. A modular approach for high-flux lactic acid production from methane in an industrial medium using engineered Methylomicrobium buryatense 5GB1. J Ind Microbiol Biotechnol. 2018;45:379–91. [DOI] [PubMed] [Google Scholar]
- 18.Hur DH, Na JG, Lee EY. Highly efficient bioconversion of methane to methanol using a novel type I Methylomonas sp. DH-1 newly isolated from brewery waste sludge. J Chem Technol Biotechnol. 2017;92:311–8. [Google Scholar]
- 19.Hur DH, Nguyen TT, Kim D, Lee EY. Selective bio-oxidation of propane to acetone using methane-oxidizing Methylomonas sp. DH-1. J Ind Microbiol Biotechnol. 2017;44:1097–105. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen AD, Hwang IY, Lee OK, Hur DH, Jeon YC, Hadiyati S, et al. Functional analysis of Methylomonas sp. DH-1 genome as a promising biocatalyst for bioconversion of methane to valuable chemicals. Catalysts. 2018;8:117. [Google Scholar]
- 21.Zeikus JG, Jain MK, Elankovan P. Biotechnology of succinic acid production and markets for derived industrial products. Appl Microbiol Biotechnol. 1999;51:545–52. [Google Scholar]
- 22.Song H, Lee SY. Production of succinic acid by bacterial fermentation. Enzyme Microb Technol. 2006;39:352–61. [Google Scholar]
- 23.McKinlay JB, Vieille C, Zeikus JG. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol. 2007;76:727–40. [DOI] [PubMed] [Google Scholar]
- 24.Bechthold I, Bretz K, Kabasci S, Kopitzky R, Springer A. Succinic acid: a new platform chemical for biobased polymers from renewable resources. Chem Eng Technol. 2008;31:647–54. [Google Scholar]
- 25.Samuelov NS, Lamed R, Lowe S, Zeikus IG. Influence of CO2-HCO3- levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl Environ Microbiol. 1991;57:3013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol. 2004;7:182–96. [DOI] [PubMed] [Google Scholar]
- 27.Sánchez AM, Bennett GN, San KY. Efficient succinic acid production from glucose through overexpression of pyruvate carboxylase in an Escherichia coli alcohol dehydrogenase and lactate dehydrogenase mutant. Biotechnol Prog. 2005;21:358–65. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen DTN, Lee OK, Hadiyati S, Affifah AN, Kim MS, Lee EY. Metabolic engineering of the type I methanotroph Methylomonas sp. DH-1 for production of succinate from methane. Metab Eng. 2019;54:170–9. [DOI] [PubMed] [Google Scholar]
- 29.Kang CK, Jeong SW, Jo JH, Park JH, Kim MS, Yang JE, et al. High-level squalene production from methane using a metabolically engineered Methylomonas sp. DH-1 strain. ACS Sustain Chem Eng. 2021;9:16485–93. [Google Scholar]
- 30.Choi YJ. Mutant strain for improving genome of microorganism, method for preparing same, and method for improving genome of microorganism by using same. Republic of Korea: KIPRIS; 2020. [Google Scholar]
- 31.Pfaffl MW. Relative quantification. In: Pfaffl MW, editor. Real-time PCR. Milton Park: Taylor & Francis; 2007. p. 89–108. [Google Scholar]
- 32.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 33.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinf. 2003;4:41. 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014. 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999. 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alhasawi AA, Thomas SC, Tharmalingam S, Legendre F, Appanna VD. Isocitrate lyase and succinate semialdehyde dehydrogenase mediate the synthesis of α-ketoglutarate in pseudomonas fluorescens. Front Microbiol. 2019;10:1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu N, Xia H, Wang Z, Zhao X, Chen T. Engineering of acetate recycling and citrate synthase to improve aerobic succinate production in Corynebacterium glutamicum. PLoS ONE. 2013;8:e60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen AK, Gerdes K, Murrell JC. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol Microbiol. 1997;25:399–409. [DOI] [PubMed] [Google Scholar]
- 43.Tucci FJ, Rosenzweig AC. Direct methane oxidation by copper- and iron-dependent methane monooxygenases. Chem Rev. 2024;124:1288–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu SS, Chen KH, Tseng MY, Wang YS, Tseng CF, Chen YJ, et al. Production of high-quality particulate methane monooxygenase in high yields from Methylococcus capsulatus (Bath) with a hollow-fiber membrane bioreactor. J Bacteriol. 2003;185:5915–24. 10.1128/jb.185.20.5915-5924.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erb TJ, Frerichs-Revermann L, Fuchs G, Alber BE. The apparent malate synthase activity of Rhodobacter sphaeroides is due to two paralogous enzymes, (3S)-malyl-coenzyme A (CoA)/β-methylmalyl-CoA lyase and (3S)-malyl-CoA thioesterase. J Bacteriol. 2010;192:1249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoenke S, Wild MR, Dimroth P. Biosynthesis of triphosphoribosyl-dephospho-coenzyme A, the precursor of the prosthetic group of malonate decarboxylase. Biochemistry. 2000;39:13223–32. [DOI] [PubMed] [Google Scholar]
- 47.Fischer G, Bang H, Mech C. Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed Biochim Acta. 1984;43:1101–11. [PubMed] [Google Scholar]
- 48.Kang W-K, Icho T, Isono S, Kitakawa M, Isono K. Characterization of the gene rimK responsible for the addition of glutamic acid residues to the C-terminus of ribosomal protein $6 in Escherichia coli KI2. Mol Gen Gen. 1989. 10.1007/BF02464894. [DOI] [PubMed] [Google Scholar]
- 49.Ye QZ, Liu J, Walsh CT. p-Aminobenzoate synthesis in Escherichia coli: purification and characterization of PabB as aminodeoxychorismate synthase and enzyme X as aminodeoxychorismate lyase. Proc Natl Acad Sci USA. 1990;87:9391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bollinger JM, Kwon DS, Huisman GW, Kolter R, Walsh CT. Glutathionylspermidine metabolism in Escherichia coli purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase*. J Biol Chem. 1995. 10.1074/jbc.270.23.14031. [DOI] [PubMed] [Google Scholar]
- 51.Bucurenci N, Serina L, Zaharia C, Landais SP, Danchin A, Baˆrzu O, et al. Mutational analysis of UMP kinase from Escherichia coli. J Bacteriol. 1998. 10.1128/JB.180.3.473-477.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Preston JF, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torrents E. Ribonucleotide reductases: essential enzymes for bacterial life. Front Cell Infect Microbiol. 2014. 10.3389/fcimb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci. 2015;40:435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalyuzhnaya MG, Nercessian O, Lidstrom ME, Chistoserdova L. Development and application of polymerase chain reaction primers based on fhcD for environmental detection of methanopterin-linked C1-metabolism in bacteria. Environ Microbiol. 2005;7:1269–74. 10.1111/j.1462-2920.2004.00831.x. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Xu H, Jones MK, White RH. Identification of the final two genes functioning in methanofuran biosynthesis in Methanocaldococcus jannaschii. J Bacteriol. 2015;197:2850–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rangaraj P, Rüttimann-Johnson C, Shah VK, Ludden PW. Accumulation of 55Fe-labeled precursors of the iron-molybdenum cofactor of nitrogenase on NifH and NifX of Azotobacter vinelandii. J Biol Chem. 2001;276:15968–74. [DOI] [PubMed] [Google Scholar]
- 58.Chistoserdova L, Laukel M, Portais JC, Vorholt JA, Lidstrom ME. Multiple Formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J Bacteriol. 2004;186:22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murrell JC, Dalton H. Ammonia assimilation in Methylococcus capsulatus (Bath) and other obligate methanotrophs. Microbiology. 1983;129:1197–206. 10.1099/00221287-129-4-1197. [Google Scholar]
- 60.Scott JH, O’Brien DM, Emerson D, Sun H, McDonald GD, Salgado A, et al. An examination of the carbon isotope effects associated with amino acid biosynthesis. Astrobiology. 2006;6:867–80. 10.1089/ast.2006.6.867. [DOI] [PubMed] [Google Scholar]
- 61.Koffas MAG, Jung GY, Aon JC, Stephanopoulos G. Effect of pyruvate carboxylase overexpression on the physiology of Corynebacterium glutamicum. Appl Environ Microbiol. 2002;68:5422–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamauchi Y, Hirasawa T, Nishii M, Furusawa C, Shimizu H. Enhanced acetic acid and succinic acid production under microaerobic conditions by Corynebacterium glutamicum harboring Escherichia coli transhydrogenase gene pntAB. J Gen Appl Microbiol. 2014;60:112–8. [DOI] [PubMed] [Google Scholar]
- 63.De Maeseneire SL, De Mey M, Vandedrinck S, Vandamme EJ. Metabolic characterisation of E. coli citrate synthase and phosphoenolpyruvate carboxylase mutants in aerobic cultures. Biotechnol Lett. 2006;28:1945–53. [DOI] [PubMed] [Google Scholar]
- 64.Litsanov B, Brocker M, Bott M. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol. 2012;78:3325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Litsanov B, Kabus A, Brocker M, Bott M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb Biotechnol. 2012;5:116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masuda A, Toya Y, Shimizu H. Metabolic impact of nutrient starvation in mevalonate-producing Escherichia coli. Bioresour Technol. 2017;245:1634–40. [DOI] [PubMed] [Google Scholar]
- 67.Diwekar U, Xiang D, Abdul Shaik M, Shaikh F. The design and techno economic analysis of a succinic acid production facility.
- 68.Efe Ç, van der Wielen LAM, Straathof AJJ. Techno-economic analysis of succinic acid production using adsorption from fermentation medium. Biomass Bioenerg. 2013;56:479–92. [Google Scholar]
- 69.Vemuri GN, Eiteman MA, Altman E. Succinate production in dual-phase Escherichia coli fermentations depends on the time of transition from aerobic to anaerobic conditions. J Ind Microbiol Biotechnol. 2002;28:325–32. 10.1038/sj/jim/7000250. [DOI] [PubMed] [Google Scholar]
- 70.Ahn JH, Seo H, Park W, Seok J, Lee JA, Kim WJ, et al. Enhanced succinic acid production by Mannheimia employing optimal malate dehydrogenase. Nat Commun. 2020. 10.1038/s41467-020-15839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information. RNA-seq data have been submitted to the GenBank databases under BioProject accession number (PRJNA769841). The whole genome sequencing data of CS40 strain is submitted to the GenBank databases under BioProject accession number (PRJNA1066519).