Highlights
-
•
Twenty-eight novel actinobacterial type strains including 23 Streptomyces species.
-
•
Proposed species names in honour of female scientists.
-
•
Most of the studied strains showed antimicrobial activities against a range of Gram-positive and Gram-negative bacteria, and yeast.
-
•
Presence of numerous unique biosynthetic gene clusters (BGCs) encoding for potential novel bioactive compounds.
Keywords: Actinobacteria, Systematics, Drug discovery, Antibiotics
Abstract
Actinomycetes are a prolific source of bioactive natural compounds many of which are used as antibiotics or other drugs. In this study we investigated the genomic and biochemical diversity of 32 actinobacterial strains that had been deposited at the DSMZ–German Collection of Microorganisms and Cell Cultures decades ago. Genome-based phylogeny and in silico DNA-DNA hybridization supported the assignment of these strains to 26 novel species and two novel subspecies and a reclassification of a Streptomyces species. These results were consistent with the biochemical, enzymatic, and chemotaxonomic features of the strains. Most of the strains showed antimicrobial activities against a range of Gram-positive and Gram-negative bacteria, and against yeast. Genomic analysis revealed the presence of numerous unique biosynthetic gene clusters (BGCs) encoding for potential novel antibiotic and anti-cancer compounds. Strains DSM 41636T and DSM 61640T produced the antibiotic compounds A33853 and SF2768, respectively. Overall, this reflects the significant pharmaceutical and biotechnological potential of the proposed novel type strains and underlines the role of prokaryotic systematics for drug discovery. In order to compensate for the gender gap in naming prokaryotic species, we propose the eponyms for all newly described species to honour female scientists.
Graphical abstract

Introduction
The phylum Actinomycetota encompasses Gram-positive bacteria with a high G+C DNA content, grouped into six classes and 35 orders (https://lpsn.dsmz.de/). Strains of this phylum have different lifestyles, such as saprophytes, pathogens, opportunistic pathogens, symbionts, gastrointestinal and plant commensals etc., and are known for their wide range of microscopic forms, including rods and coccoid forms as well as branched filamentous mycelium that may separate to form spores. The latter are special cellular structures known for their resistance to various factors such as heat, desiccation and radiation (Bobek et al., 2017). Actinomycetota are widely distributed across diverse habitats, including terrestrial and aquatic ecosystems, as well as extreme environments from hot deserts to Arctic deep seas (Goodfellow et al., 2018). Thereby, the organisms are adapted to a wide range of different physico-chemical parameters, which shape their habitats, including climatic conditions, pH, temperature, moisture etc. (Mohammadipanah and Wink, 2015, Xie and Pathom-Aree, 2021).
The assignment of actinobacterial taxa was based on different phenotypic and genetic approaches that relied heavily on morphological, physiological, and biochemical properties in the early descriptions (Waksman and Curtis, 1916, Bergey et al., 1923, Sneath and Sokal, 1962, Sokal, 1966, Sneath, 1973, Stanier and Van Niel, 1941, Niel, 1946, Stanier and Niel, 1962) Since then, the prokaryotic systematics has evolved significantly to include polyphasic taxonomy, which is based on a combination of phenotypic (including chemotaxonomy), genomic (DNA:DNA hybridization), and genetic analyses (Vandamme et al., 1996, Kämpfer, 2012, Hugenholtz et al., 2021, Nouioui and Sangal, 2022). The latter was mainly based on the 16S rRNA gene, which has been considered as the gold standard marker used for comparative phylogenetic studies of bacteria. However, the resolution of the 16S rRNA gene for some taxa is limited and often not sufficient to distinguish closely related species (Labeda et al., 2012, Nouioui et al., 2018). Alternatively, multi-locus sequence analysis (MLSA), has emerged offering intermediate phylogenetic resolution (Thompson et al., 2005, Gevers et al., 2005). This approach involves phylogenetic comparisons based on a set of housekeeping genes and has clarified the phylogenetic relatedness of several complex taxa such as Streptomyces (Rong and Huang, 2014, Labeda et al., 2014, Labeda et al., 2017). Nevertheless, MLSA has shown certain limitations associated with the suitability and relevance of housekeeping genes selected on the basis of their degree of polymorphism (Hugenholtz et al., 2021). These constraints have been addressed by full-length genome sequence comparisons, which has become efficient and affordable over the last decade in the course of the progress of next generation sequencing technology and provide the most accurate phylogenetic resolution (Nouioui et al., 2018).
The systematics of the phylum Actinomycetota has undergone several changes over time due to advances in molecular biology and computational genomics. The techniques of DNA-DNA hybridisation (DDH) and DNA G+C content analysis, used since the 1960s for prokaryotic species delineation, had shifted from laborious and time-consuming wet-lab work to digital and more accurate data that have adjusted the taxonomic status of many misclassified taxa due to errors related to the sensitivity of the experiments (JP Meier-Kolthoff et al., 2013, JP Meier-Kolthoff et al., 2014). Other taxogenomic approaches, such as average nucleotide identity (ANI) and average amino acid identity (AAI) have been developed with cut-off values of 95–96% for species demarcation, which has further improved prokaryotic systematics (Goris et al., 2007, Richter and Rosselló-Móra, 2009). Given all these technological advances and the user-friendliness of taxonomic software and web servers (eg. GGDC (https://ggdc.dsmz.de/; (Auch et al., 2010)), the systematics of prokaryotes has been modernised by embracing the whole genome analysis (Hugenholtz et al., 2021, Nouioui and Sangal, 2022). The application of modern technology for bacterial systematic classification allowed to resolve the taxonomic status of several heterogenous complex taxa and helped to distinguish between closely related organisms leading to the reclassification and emendation of many orders, families, genera, species and subspecies. (Nouioui et al., 2018, Hahnke et al., 2016, Carro et al., 2018).
Strains of the phylum Actinomycetota are of agricultural, biotechnological, ecological, and medicinal interest (Goodfellow, 2012, Demain, 2014). Two-thirds of all clinically relevant antibiotics, as well as many anticancer, antifungal and immunosuppressive agents are derived from actinomycetes (Bérdy, 2012). This heterogenous group of actinobacterial organisms is the most promising source of new drugs and even though exploited for decades, actinomycetes still hold a huge genetic potential for the production of novel bioactive natural compounds (Gavriilidou et al., 2022). Thereby, natural products (NPs) and semi-synthetic substances derived from them still represent the majority of leads for drug developments compared to compounds obtained by combinatorial biosynthesis or synthetic biology approaches (Newman, 2008) the latter of which represent more modern technologies that still rely on a fundamental understanding of natural compounds biosynthetic pathways. According to the World Health Organisation (WHO) (WHO 2022) the discovery and production of new antibiotics by the pharmaceutical industry has been hampered by lengthy approval procedures, high costs and low success rates. Mining rare, understudied or novel actinomycetes species has proven to be a promising strategy for novel drug discovery (Bauman et al., 2021). The choice of the biological material can be crucial for the success rate of finding new NPs (Leopold-Messer et al., 2023) with especially new sources and/or biology that lead to new chemical entities (Goodfellow et al., 2018 Aug, Goodfellow and Fiedler, 2010).
The genus Streptomyces has been extensively studied, resulting in several taxonomic reclassifications and amendments due to advances in the taxonomic approach (Labeda et al., 2012, Labeda et al., 2014, Labeda et al., 2017, Witt and Stackebrandt, 1990, Pridham et al., 1965, Szabó and Marton, 1964). Strains of this taxon are known as producers of secondary metabolites such as antibiotics, anticancer, and antiviral bioactive compounds (Sanjivkumar et al., 2016, Sanjivkumar et al., 2018). However, several actinobacterial taxa are still understudied and little is known about their biotechnological potential in terms of bioactive compounds production. The following non-Streptomycetaceae genera are also the focus of this study: Nocardiopsis, Streptomonospora, Pseudonocardia, Blastococcus, and Jatrophihabitans.
Representatives of all these genera are present in the DSMZ strain collection. The German culture collection DSMZ was founded in 1969 as the national centre for culture collection in Germany and is renowned for its long-standing commitment to microbial taxonomic studies. It is nowadays one of the largest and most diverse culture collections worldwide, currently comprising more than 85.000 bioresources in total, including about 38.000 different bacterial strains. The bacterial diversity covers 80% of all microbial type strains, which come from more than 90 countries. The actinobacteria sub-collection contains more than 6.000 actinobacterial strains including many rare taxa, such as Actinoalloteichus, Kibdelosporangium, Tsukamurella, or Jiangella, as well as strains that are difficult to handle and/or to maintain, such as Frankia. Moreover, the culture collection contains ∼2.500 streptomycetes, many of which have been genome sequenced as part of the Genomic Encyclopedia of Bacteria and Archaea (GEBA) initiative (Seshadri et al., 2022).
In view of the continued emergence of drug-resistant bacteria associated with life-threatening infections, there is an urgent need for new antibiotics, and natural products from taxonomically unique actinobacterial strains are a promising source for novel antimicrobials. In this respect, a screening campaign for new species among a large number of actinobacterial strains deposited at the DSMZ decades ago, has been started focusing on the genus Streptomyces, which is the leading producer of antibiotics (Nouioui et al., 2024).
In this study, poorly characterized strains of Streptomyces, Nocardiopsis, Streptomonospora, Blastococcus, Pseudonocardia, and Jatrophihabitans, were subjected to polyphasic taxonomic characterisation including whole genome sequence analysis. The biotechnological potential of the strains was evaluated based on genome mining approaches and an antimicrobial activity screening. In total, 26 novel species, and two novel subspecies were identified and described and a reclassification of a Streptomyces species, which is so far the largest all-at-once description of that many actinobacterial strains. In order to compensate for the gender gap in names of prokaryotes honouring persons (Freese et al., 2023), we propose new taxon names to all newly described species to honour female scientists: Streptomyces hazeniae sp. nov., Streptomyces boetiae sp. nov., Streptomyces bugieae sp. nov., Streptomyces chisholmiae sp. nov., Streptomyces doebereineriae sp. nov., Streptomyces doudnae sp. nov., Streptomyces millisiae sp. nov., Streptomyces dubilierae sp. nov., Streptomyces edwardsiae sp. nov., Streptomyces evansiae sp. nov., Streptomyces gibsoniae sp. nov., Streptomyces gottesmaniae sp. nov., Streptomyces hesseae sp. nov., Streptomyces hintoniae sp. nov., Streptomyces mooreae sp. nov., Streptomyces johnsoniae sp. nov., Streptomyces litchfieldiae sp. nov., Streptomyces lonegramiae sp. nov., Streptomyces salyersiae sp. nov., Streptomyces lancefieldiae sp. nov., Streptomyces stephensoniae sp. nov., Blastococcus goldschmidtiae sp. nov., Jatrophihabitans lederbergiae sp. nov., Nocardiopsis lambiniae sp. nov., Pseudonocardia charpentierae sp. nov., and Streptomonospora wellingtoniae sp. nov. The diversity of the biosynthetic gene cluster families of the strains was comprehensively mapped and revealed the abundance of unique BGCs with the potential to encode novel secondary metabolites. For two of the strains, DSM 41636T and DSM 61640T, antibiotic substances (A33853 and SF2768, respectively) have been identified.
Materials and methods
Bacterial strains, source, maintenance and cultivation
Thirty-two actinobacterial strains were isolated from environmental samples (soil, marine sediments, water, plants, wasp) collected from different geographical locations worldwide and deposited at the DSMZ open culture collection years ago (Table 1). Strains were classified at genus level as Streptomyces sp. (n=27), Blastococcus sp. (n=1), Jatrophihabitans sp. (n=1), Nocardiopsis sp. (n=1), Pseudonocardiopsis sp. (n=1), and Streptomonospora sp. (n=1), based on their phenotypic and genetic data. All the reference strains used in this study are available in the DSMZ online catalogue (https://www.dsmz.de/collection/catalogue). Freeze-dried cells were used for chemotaxonomic analysis, with the exception of fatty acids analysis, for which fresh biomass was used. The growth conditions of the strains are listed in the DSMZ catalogue (https://www.dsmz.de/collection/catalogue).
Table 1.
Origin of the studied strains.
| The DSMZ strains | Habitat | Country | Strain designation and culture collection numbers | Proposed species name |
|---|---|---|---|---|
| Streptomyces strains | ||||
| DSM 3412T | soil | Tunisia | Tü 2253T, KCTC 59181T | Streptomyces gottesmaniae sp. nov. |
| DSM 40473T | soil | Unknown country | ATCC 12568T, BA-3572T, CBS 695.72T, IFO 13394T, IFO 13907T, ISP 5473T, KCC S-0694T, KCC S-0812T, NBRC 13394T, NBRC 13907T, RIA 1355T | Streptomyces hesseae sp. nov. |
| DSM 40712T | – | Unknown country | ETH 21066T, NRRL 2835T, Tü 41T | Streptomyces lancefieldiae sp. nov. |
| DSM 40932T | – | Unknown country | ATCC 13741T, ATCC 13793T, CBS 372.58T, ETH 24437T, NRRL B-1354T | Streptomyces stephensoniae sp. nov. |
| DSM 41014T | Sphagnum pots | Unknown country | IMRU 3065T, KCTC 59176T | Streptomyces hintoniae sp. nov. |
| DSM 41524T | soil | Unknown country | A10598T, ATCC 15166T | Streptomyces asiaticus subsp ignotus subsp. nov. |
| DSM 41527T | – | Unknown country | ATCC 21705T, SF-1293T | Streptomyces mooreae sp. nov. |
| DSM 41528T | – | Unknown country | ATCC 21722T, FERM-P 602T, SF-1084T | Streptomyces bugieae sp. nov. |
| DSM 41529T | soil | Japan | A-130T, ATCC 21840T, FERM-P 639T | Streptomyces lonegramiae sp. nov. |
| DSM 41602 | – | Unknown country | NRRL B-16257, U3S-25 | Streptomyces antimycoticus subsp. sporoclivatus subsp. nov. |
| DSM 41635 | rose root (rhizoplane, Rosa laxa) | Germany | 5-A4 | Streptomyces edwardsiae sp. nov. |
| DSM 41636T | rose root (rhizoplane, Rosa laxa) | Germany | 31-A2T, KCTC 59179T | Streptomyces edwardsiae sp. nov. |
| DSM 41640T | rose root (rhizoplane, Rosa laxa) | Germany | 13-A30T, KCTC 59177T | Streptomyces doebereineriae sp. nov. |
| DSM 41699T | soil sample from Tasek Bera | Malaysia | ATB-26T, KCTC 59183T | Streptomyces gibsoniae sp. nov. |
| DSM 41770T | water damaged gypsum liner in a children`s day care center | Finland | 157/96T, KCTC 59182T | Streptomyces salyersiae sp. nov. |
| DSM 41859 | agar plate culture of Aureobasidium pullulans | Germany | Agent 'Z' | Streptomyces evansiae sp. nov. |
| DSM 41886T | marine sediment | USA | CNB 984T, KCTC 59171T | Streptomyces johnsoniae sp. nov. |
| DSM 41921T | rhizospherical soil of resistant Vitis vinifera | Morocco | S6, KCTC 59180T | Streptomyces dubilierae sp. nov. |
| DSM 41972T | Atta colombica refuse dump | Panama | Av26–2T, DI-188T, KCTC 59178T | Streptomyces althioticus subsp. attaecolombicae subsp. nov. |
| DSM 41979T | sirex noctilio | USA | SA3-ActFT, KCTC 59185T | Streptomyces evansiae sp. nov. |
| DSM 41981T | solitary wasp | Panama | Sol5a-2T, KCTC 59175T | Streptomyces doudnae sp. nov. |
| DSM 41982 | solitary wasp | Panama | Sol7th | Streptomyces evansiae sp. nov. |
| DSM 42041T | gorgonian coral | China | SCSIO 10374T, KCTC 59186T | Streptomyces hazeniae sp. nov. |
| DSM 44915T | marine sediment | Palau | SCRIPP CNJ 962T, KCTC 59194T | Streptomyces chisholmiae sp. nov. |
| DSM 44917T | marine sediment | USA | SCRIPP CNQ 259T, KCTC 59173T | Streptomyces boetiae sp. nov. |
| DSM 44918T | marine sediment | Guam / USA | SCRIPP CNQ 703T, KCTC 59174T | Streptomyces millisiae sp. nov. |
| DSM 44938T | marine sediment | Bahamas | SCRIPP CNR 954T, KCTC 59172T | Streptomyces litchfieldiae sp. nov. |
| Actinobacterial strains | ||||
| Blastococcus sp. DSM 46792T | marble | Italy | 7CT, BC543T, KCTC 59190T | Blastococcus goldschmidtiae sp. nov. |
| Nocardiopsis sp. DSM 44743T | soil | China | DCDM15A35T, KCTC 59192T | Nocardiopsis lambiniae sp. nov. |
| Pseudonocardia sp. DSM 45834T | desert sand sample | China | CPCC 203558T, KCTC 59187T | Pseudonocardia charpentierae sp. nov. |
| Streptomonospora sp. DSM 45055T | marine sediment | USA | CNQ 327T, KCTC 59191T | Streptomonospora wellingtoniae sp. nov. |
| Jatrophihabitans sp. DSM 44399T | sandstone of Linnaeus Terrace (1600 m) | Antarctica | IFAM AA-499T, AA-499T | Jatrophihabitans lederbergiae sp. nov. |
Previously reported antibiotic production profile of the studied strains
Based on previous studies, six out of 27 Streptomyces strains are known for their ability to produce antibiotics, antifungals, and enzymes. Strains DSM 41524T, DSM 41527T, DSM 41528T, and DSM 41529T were deposited at the DSMZ collection on 19.05.1989 under the species name S. hygroscopicus subsp. hygroscopicus, while strain DSM 41602 was initially assigned to Streptomyces violaceusniger species. Strains DSM 41529T, DSM 41527T, DSM 41528T were known as producers of antibiotic A-130 (Oikawa et al., 1975), herbicide bialaphos (Murakami et al., 1986), and amylase (Koaze et al., 1974), respectively. Strains DSM 40932T and DSM 40712T, originally affiliated to Streptomyces griseus subsp. farinosus and Streptomyces albogriseolus, respectively. These strains were found to synthesize streptolins A, B, streptothricin, vitamin B12 and chalcomycin and echinomycin, respectively. The assignment of strains DSM 41524T, DSM 41527T, DSM 41528T, DSM 40932T, DSM 40712T, and DSM 41602T to the species listed above was based on numerical classification (Kämpfer et al., 1991). Strains DSM 41640T and DSM 3412T, deposited at the DSMZ culture collection between 1985–1994, were originally described as Streptomyces violaceusniger and Streptomyces galbus subsp. eurythermus, respectively. Strain DSM 3412T was known to produce antifungal macrolides called galbonolides A and B (Achenbach et al., 1988). Strain DSM 40473T was previously known as Streptoverticillium parvisporogenum (Locci, 1969), basonym of Streptomyces parvisporogenes that has been considered as a heterotypic synonym to Streptomyces abikoensis (Umezawa et al., 1951) Witt and Stackebrandt 1990. For this reason, this strain appears in different culture collections as Streptomyces abikoensis. Strain DSM 40473T was earlier described as the producer of the antibiotic PA-150 (Koe et al., 1957, English and McBride, 1957).
Strains DSM 41635 and DSM 41636T, and DSM 41770T were formerly assigned to Streptomyces griseoflavus and Streptomyces griseus, respectively. Strains DSM 41014T, DSM 41981T, DSM 41921T, DSM 41699 T, DSM 41982T, DSM 41859T, DSM 41972T, DSM 42041T, DSM 44917T, DSM 44915T, DSM 44938T, DSM 44918T, and DSM 41979T were accessed at the DSMZ between 1980 and 2011 as Streptomyces sp. Strain DSM 41886T was deposited in 2006 as ‘Marinispora sp.’; the genus Marinispora of the family Streptomycetaceae has never been validated. All the non-Streptomyces strains (Nocardiopsis sp. DSM 44743T, Blastococcus sp. DSM 46792T, Streptomonospora sp. DSM 45055T, Pseudonocardia sp. DSM 45834T) included in this study were deposited at the German collection between 2003–2013. All strain designations and culture accession numbers of the studied strains are provided in Table 1 and in the protologue for species description. No previous studies on the pharmaceutical biotechnological or ecological potential of these strains were performed, with the exception of those mentioned above.
Growth and morphological properties
The ability of the strains to grow in the presence of the following media was evaluated: International Streptomyces Project (ISP), ISP1 (DSMZ 1764), ISP2 (DSMZ 987) ISP3 (DSMZ 84), ISP4 (DSMZ 252), ISP5 (DSMZ 993), ISP6 (DSMZ 1269), ISP7 (DSMZ 1619), GYM (Glucose-Yeast extract-Malt extract = DSMZ 65) (Shirling and Gottlieb 1966), TSA (Trypticase Soy Agar = DSMZ 535), N-Z amine (DSMZ 554), GPHF (DSMZ 553), DSMZ 714, Luedemann (DSMZ 877), Bennett's (DSMZ 548), Gause synthetic media N°1, nutrient agar (DSMZ 1), Czapek peptone (DSMZ 83) agar media. Morphological traits, including the colour of the aerial and substrate mycelia of the strains, were recorded using the RAL colour chart. Moreover, the strains were subjected to a wide range of temperatures (4 °C, 10 °C, 15 °C, 25 °C, 28 °C, 37 °C, 42 °C, and 45 °C) and pH (5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0) tests. A bacterial suspension of 5 on the McFarland scale was used for inoculation of all duplicated tests (McFARLAND, 1907).
16S. rRNA gene-based identification and genome sequencing
Genomic DNA was extracted from active cultures of strains grown under optimal growth conditions (https://www.dsmz.de/collection/catalogue). DNA extraction was carried out by the microbial DNA service at the DSMZ as described before (Wright et al., 2023). The bacterial DNA was subjected to PCR-mediated amplification of a 16S rRNA gene and was sequenced using a 96-capillary-system from Applied Biosystems (ABI) as reported previously (Risdian et al., 2021).
For genome sequencing, a fresh biomass of 30–50 mg was harvested from the active culture of the strains and added to 500 µl of DNA/RNA Shield, a lysis buffer provided by the MicrobesNG service. Extraction, purification, quantitative and qualitative estimation of the DNA as well as sequencing on the Illumina platform were performed by MicrobesNG service (https://microbesng.com). 250 bp paired end reads and 30X depth of coverage for Illumina sequencing was used. MicrobesNG bioinformatic pipeline used for a final draft genome sequence includes Kraken (Wood and Salzberg, 2014), a system for taxonomic assignment, a software for mapping the reads, BWA mem (Li and Durbin, 2010, Li, 2013), and assembly programme, SPAdes (Bankevich et al., 2012). The genomes were annotated by NCBI, using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Li et al., 2021). The draft genome sequences of the studied strains have been deposited in GenBank. The genome accession numbers of the studied strains are listed in Table S1.
Phylogeny and comparative genomic studies
To confirm the authenticity of the strains, an almost complete 16S rRNA gene sequence (>1.400 bp) extracted from the draft genome sequence of the strains was aligned with that obtained from PCR using Basic Local Alignment Search Tool (BLASTN) available on the NCBI web server (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Zhang et al., 2000, Morgulis et al., 2008). BLAST comparative analysis of the 16S rRNA gene sequence of the strains of interest against those of the type strains of validly named species was carried out via the EZBioCloud server (https://www.ezbiocloud.net/) (Yoon et al., 2017). Maximum likelihood 16S rRNA gene and genome-based phylogenetic trees were constructed using the Type Strain Genome Server [TYGS, (https://tygs.dsmz.de/)] (Meier-Kolthoff and Göker, 2019, Meier-Kolthoff et al., 2022). Comparative genomic analysis for prokaryotic species and subspecies delineation based on digital DNA-DNA hybridization (dDDH) was performed between the genome of the studied strains and their closest phylogenomic neighbours using the recommended formula 4 provided by the TYGS (http://ggdc.dsmz.de) web server (Meier-Kolthoff et al., 2022). Genomic features of the strains and their close phylogenomic neighbours, such as genome size, G+C content, number of coding sequences, number of RNA genes, N50, were retrieved from GenBank.
Biochemical and chemotaxonomic analyses
The ability of the strains to metabolise a range of carbohydrates was examined using the API 50 CH (according to the manufacturer's instructions, Biomérieux, France). The enzymatic pattern of the strains was determined using the API ZYM kit (according to the manufacturor's instructions, Biomérieux, France). In addition, API 20 NE was used to determine the biochemical feature of Streptomonospora sp. DSM 45055T, Nocardiopsis sp. DSM 44743T, Blastococcus sp. DSM 46792T, Jatrophihabitans sp. DSM 44399T. Standard thin-layer chromatographic (TLC) procedures were used for analyses of polar lipids (Minnikin et al., 1984, Goodfellow and Kroppenstedt, 2006), isomers of diaminopimelic acid of the peptidoglycan (Schleifer and Kandler, 1972), and whole cell sugars (Lechevalier and Lechevalier, 1979). Fresh biomass of the strains was used for cellular fatty acid analysis. Fatty acids were converted into fatty acid methyl esters (FAMEs) following the protocol of saponification, methylation and extraction of (Sasser, 1990). FAMEs were analyzed by gas chromatography coupled to a flame ionization detector (Agilent instrument, model 6890N). A GCMS– run on an Agilent GC–MS 7000D instrument was used for identification of fatty acids (Vieira et al., 2021). Subsenquently, FAMEs were derivatized to dimethyl disulfide adducts to resolve the position of the single double bonds (Moss and Lambert-Fair, 1989) or to 4,4-dimethyloxazoline (DMOX) derivatives (Spitzer, 1996) to determine branched-chain fatty acid positions and cyclopostions, respectively.
Biosynthetic gene cluster and cluster similarity network analysis
A total of 76 genome sequences of the studied strains (n = 32) and their related phylogenomic relatives (n = 44) were subjected to antiSMASH analysis version 7.0.1 (Blin et al., 2023), using GenBank annotation files from the assemblies published with NCBI as input and relaxed strictness settings. The results were subsequently separated into all BGC regions, regions within contigs, and regions on contig edges. All sets of BGC regions (all regions, within contigs, on contig edges) were clustered into GCFs with BiG-SCAPE version 1.1.5 (Navarro-Muñoz et al., 2020) using the default cut-off of 0.3, including the MIBiG database, and allowing the mixing of all classes of natural products (–mix). The gene cluster network obtained from BiG-SCAPE was visualized with Cytoscape (version 3.10.2, (Shannon et al., 2003)). The strains were clustered according to their GCF coverage using the python function seaborn.clustermap with default settings (method=’average’, metric=’euclidean’).
Preparation of culture extracts and antimicrobial bioassay
The strains were cultivated in 50 mL of R5 medium at 28 °C (Nouioui et al., 2024). After three days, 5 mL of preculture were transferred in 50 mL of fresh R5 and NL800 medium, respectively, and the culture was grown for four days at 28 °C in 250 mL Erlenmeyer flasks on an orbital shaker (180 rpm) as described before (Nouioui et al., 2024). In cases where the strains did not grow in R5 or NL800, the optimal growth medium suggested for the strain was used, as listed on the DSMZ online catalogue. Organic compounds were extracted using 5 mL of ethyl acetate (EtAc) as described by Nouioui et al. (2024). Crude extracts were tested against reference strains of Gram-positive (Staphylococcus aureus DSM 18,827 (Multiple Antibiotic Resistant strain) and Enterococcus faecium DSM 20477T) and Gram-negative bacteria (Escherichia coli ΔtolC JW5503–1, Proteus vulgaris DSM 2140), as well as yeast (Candida albicans DSM 1386). All reference strains are available in the DSMZ online catalogue along with their respective growth conditions (https://www.dsmz.de/collection/catalogue). Antimicrobial tests were carried out according to Nouioui et al. (2024) using a 30 µl volume of methanolic crude extract applied in wells in agar plates inoculated with the test microorganisms listed above. The diameter of the inhibition zone was measured after incubation of the plates. Due to the large number of samples, only one biological replicate was prepared in each case.
SARP overexpression in strain DSM 41636T and DSM 61640T
To overexpress papR2 in streptomycetes, a conjugative, φBT1-based integrative construct was generated. The plasmid pIJ10257 (Hong et al., 2005) was used as a vector, which contains the ermE* promoter for the induction of gene transcription, a hygromycin resistance cassette (hygR) for selection, and oriT, required for the intergeneric conjugation from E. coli to Streptomyces. The papR2 gene was PCR amplified from the genomic DNA of Streptomyces pristinaespiralis PR11 using a pair of primers RM_papR2_exp_for (AAAAAACATATGAAGTTCCGCATTCTCGGTCC) and RM_papR2_exp_rev (AAAAAAGCTTCTAGTGGCCCGAGGCCG). The obtained 1 kb long DNA fragment was digested with HindIII and NdeI and then cloned into the respective sites of pIJ10257. This resulted in the plasmid pDS300, where papR2 transcription is under the control of the constitutive erythromycin promoter ermE*. The pDS300 plasmid was transferred to DSM 41636T and DSM 41640T by conjugation, as described in Kieser et al. (2000), resulting in the strain DSM 41636T::pDS300 and DSM 61640T::pDS300, respectively. As a control, strains carrying the empty vector pIJ10257 were used. Hygromycin B (50 µg/mL) was used for selection.
Antibiotic production conditions for DSM 41636T and DSM 61640T and preparation of culture extracts
The strains DSM 41636T and DSM 41640T were cultivated by inoculation from a GYM plate in 25 mL medium TSB at 28 °C in 250 mL Erlenmeyer flasks on an orbital shaker (180 rpm). After two days of cultivation, 5 mL of the preculture were used to inoculate 500 mL Erlenmeyer flasks with 50 mL of production media R5 or NL 19 as production cultures, that were incubated for four days at 28 °C on an orbital shaker at 180 rpm. For extraction of organic compounds, 20 mL of culture was extracted with 20 mL of ethyl acetate for 3 h at room temperature under constant vertical rotation. After centrifugation at 5.000 rpm for 10 min, the organic phase was completely dried using a centrifugal evaporator (SP Genevac EZ-2, “Low BP” program). The concentrated extracts were dissolved in 0.5 mL methanol (MeOH) and then used for LC-HRMS (liquid chromatography-high resolution mass spectrometry) analysis.
Chemical analyses for compound detection
Low resolution LC-MS data were recorded on a HPLC Agilent 1260 system connected to a G6125B mass spectrometer with a reversed-phase Poroshell C18 column (Agilent, 2.7 μm, 100 × 3 mm) using water as mobile phase A and acetonitrile as mobile phase B, both containing 0.1% formic acid (vol/vol) as a solvent modifier. Elution was carried out at 0.5 mL/min as follows: 0 min 95% A, 0.5 min 95% A, 18.5 min 5% A, 20.5 min 5% A, 20.8 min 95% A, 25 min 95% A. Full-scan mass spectra (m/z 200–2000) were collected in both positive and negative ESI modes. The following parameters were used: capillary voltage, 3000 V; nebulizer gas pressure, 35 psi; drying gas flow rate (N2), 10 L/min; drying gas temperature, 350 °C.
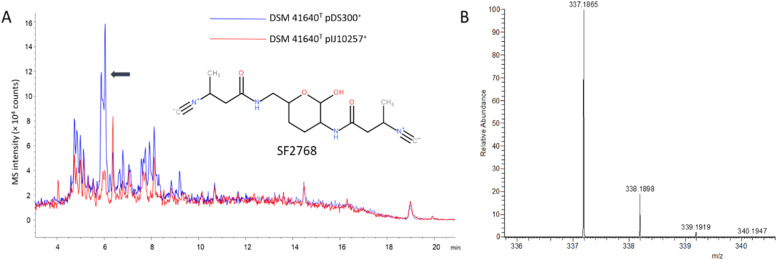
Identification of A33853 from DSM 41636T: A33853 was identified using a combination of its UV/vis spectrum and LC-HRMS. The UV/vis spectrum of the identified compound is virtually identical to the previously reported spectrum of A33853 (Michel et al., 1984). Its deprotonated molecule = m/z 390.0726 [M – H]– (calculated (calcd) for C20H13N3O6, 390.0732, Δ −1.54 ppm) yielded a formula (C20H13N3O6) matching that of A33853. Identification of SF2768 from DSM 41640T: SF2768 was identified using LC-HRMS. Its protonated molecule = m/z 337.1865 [M+H]+ (calculated (calcd) for C16H24N4O4, 337.187, Δ −1.48 ppm) yielded a formula (C16H24N4O4) matching that of SF2768.
Results and discussion
16S. phylogenetic analysis leads to the identification of new actinobacterial type strains
Phylogenetic analysis of Streptomyces strains based on 16S rRNA gene
Molecular identification based on the 16S rRNA gene sequence assign the strains to the class Actinomycetes and phylum Actinomycetota. Pairwise 16S rRNA gene sequence similarity confirms the affiliation of the strains to their respective genus. The authenticity of the strains was confirmed by comparing the 16S rRNA sequence obtained by PCR with that from the genome sequences of the strains. The 28 Streptomyces strains had 16S rRNA gene similarities between 98.2–100% with their closest phylogenetic relatives. The genus Streptomyces of the family Streptomycetaceae (Waksman and Henrici, 1943) and order Kitasatosporales (Kämpfer 2012) comprises more than 700 species with validly published names, with Streptomyces albus (Waksman and Henrici, 1943, Rossi, 1891) as the type species. Streptomycetes are present in different ecological niches (soil, sediment, plant, animal, human, etc.), though they predominate in soil. They are known for their saprophytic lifestyle. These filamentous actinomycetes are characterized by a classic sporulating life cycle consisting of spore germination and the development of primary substrate mycelia, followed by the formation of secondary aerial mycelia and production of spores.
A Maximum-likelihood (ML) phylogenetic tree based on 16S rRNA gene sequences showed that most of the Streptomyces strains had a distinct phylogenetic position within the evolutionary radiation of the genus Streptomyces (Figure S1). The phylogenetic distribution of the studied strains (DSM 41524T, DSM 41,602, DSM 41529T, DSM 41527T, DSM 42041T, DSM 41979T, DSM 41,982, DSM 41,859, DSM 40712T, DSM 41921T, DSM 41981T, DSM 41014T, DSM 41699T, DSM 44915T) in the ML tree is in concordance with the 16S rRNA gene sequence similarity (Table S2), as they were closely related to their reference strains which have the highest similarity values, unlike strains DSM 40473T, DSM 41528T, DSM 41,635, DSM 41636T, DSM 41770T, DSM 40932T DSM 3412T DSM 41640T, DSM 41640T, DSM 44938T, DSM 44917T, DSM 41886T, DSM 44918T (Figure S1, Table S2). The phylogenetic relationships of the strains were not well supported in the tree.
Phylogenetic analysis of non-Streptomyces strains based on 16S rRNA gene
The genera Nocardiopsis (Brocq-Rousseau, 1904, Meyer, 1976) and Streptomonospora (Cui et al., 2001, Li et al., 2003) belong to the family Nocardiopsidaceae (RAINEY et al., 1996, Zhi et al., 2009) and the order Streptosporangiales (Goodfellow, 2015), harbouring 47 and 10 validly named species, respectively. The type genus Pseudonocardia (Henssen, 1957) of the family Pseudonocardiaceae (Embley et al., 1988) and the order Pseudonocardiales (Labeda and Goodfellow, 2015) contains more than 65 validly named species. The genera Blastococcus (Ahrens and Moll, 1970) and Jatrophihabitans (Madhaiyan et al., 2013) of the families Geodermatophilaceae (Normand, 2006) and Jatrophihabitantaceae (Nouioui et al., 2018) are classified in the orders Geodermatophilales (Sen et al., 2014) and Jatrophihabitantales, which encompass 14 and 5 validly named species, respectively. The five non-Streptomyces strains had 16S rRNA gene similarities between 97.56–99.72% with their closest phylogenetic relatives.
Strains of the genus Blastococcus, which has Blastococcus aggregatus (Ahrens and Moll, 1970) as the type species, are characterized by the presence of rod and/or cocci-shaped cells that can be aggregated (Urzì et al., 2004, Hezbri et al., 2016). The cells have been shown to be motile rod-shaped cells and /or non-motile cocci (Lee, 2006). This group of microorganisms has been isolated from different habitats, such soil and sand from archaeological sites and deserts, marble, sediments, and medicinal plants (Montero-Calasanz et al., 2022). Strain DSM 46792T had 99.7% 16S rRNA gene similarity with the type strain of Blastococcus fimeti species. In the ML phylogenetic tree, the studied strain occupied a distinct branch closely associated with the type strains of B. fimeti and Blastococcus aggregatus species and next to Blastococcus aurantiacus DSM 44268T. It is evident from the low resolution of the 16S rRNA gene phylogeny that a more in-depth phylogenetic investigation is needed to clarify the taxonomic status of the strains. The genus Jatrophihabitans, type species Jatrophihabitans endophyticus (Madhaiyan et al., 2013), comprises non-spore forming strains with short rod cells. Jatrophihabitans strains have been isolated from plants and soils. Jatrophihabitans sp. DSM 44399T showed had 94.8% 16S rRNA gene similarity with the type strain of Jatrophihabitans telluris KCTC 39922T. These strains formed together a subclade closely related to Jatrophihabitans endophyticus DSM 45,627 T.
The genus Pseudonocardia, with Pseudonocardia thermophila (Henssen, 1957) as the type species, contains strains characterized by the presence of substrate mycelium with various degree of branching and thickness and an aerial mycelium generating square or oval elements or a chain of spores (>2 nm) when fragmented (Riahi et al., 2022). They have been isolated from soils, plants, sediments, wastewater, a gold mine cave, or molds (Riahi et al., 2022). As shown in Figure S2, Pseudonocardia sp. DSM 45834T is loosely associated with ‘Pseudonocardia humida’ strain S2–4T with 16S rRNA gene identity of 97.8%. However, Streptomonospora sp. DSM 45055T appeared in a distinct branch close to Streptomonospora litoralis DSM 106425T, though the highest 16S rRNA gene similarity of 99.5% was obtained with the type strain of Streptomonospora arabica S186T (Table S2).
Strains of the genus Nocardiopsis are widely distributed in the environment, including compost, animal, human clinical samples, plant material, the indoor environment and soil. They are fairly resistant to desiccation and play an important role in recycling organic substances due to their genetic ability to generate exoenzymes, surfactants, antibiotics that help them to survive under different conditions (Bennur et al., 2015). Nocardiopsis strains are filamentous bacteria with straight to flexuous or zigzag-shaped aerial mycelium and a smooth spore surface. Nocardiopsis sp. DSM 44743T was loosely associated with Nocardiopsis algeriensis CECT 8712T (99.3%) and adjacent to a poorly supported subcluster encompassing the type strain of Nocardiopsis flavescens (99.3%) and Nocardiopsis lucentensis (99.2%). Overall, it is clear from the 16S rRNA gene phylogeny that a single gene is not sufficient to distinguish between closely related species. Therefore, a genome-based tree was constructed.
Phylogenomic and comparative genomics analyses show the actual relationships of the identified actinobacterial strains
Phylogenomic and comparative genomic analysis of Streptomyces strains
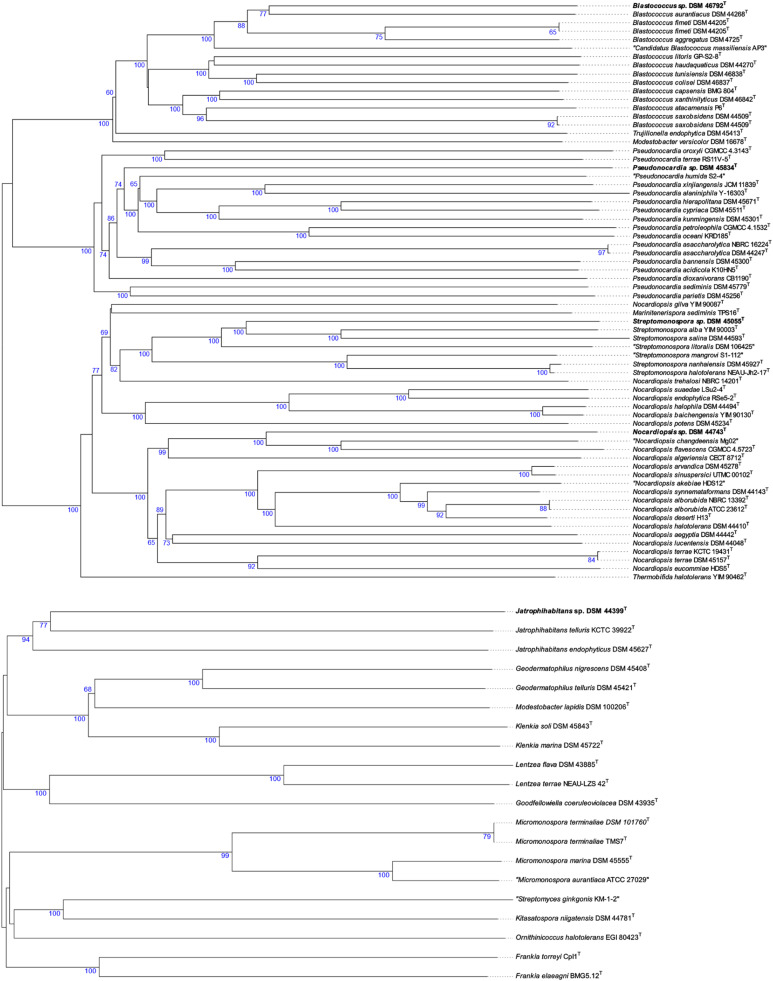
In order to assign all the strains to their correct taxonomic rank with high resolution and to have a clear overview of their evolutionary radiation with the corresponding genera, a genome-based phylogeny was constructed. The genome blast distance phylogenetic (GBDP) tree shown in Fig. 1 has highly supported branches. The taxonomic description given below is in the descending order of the strains in the genomic tree. Strain DSM 41524T was found in a distinct branch closely associated with S. asiaticus DSM 41761T, S. indonesiensis DSM 41759T, S. cangkringensis DSM 41769T, and S. rhizosphaericus DSM 41760T (Fig. 1). The branch lengths of S. indonesiensis, S. cangkringensis, and S. rhizosphaericus in the genome-based tree reflected their evolutionary convergence and call for revision of the taxonomic status of these validly named species. A narrow range of dDDH values ranged was obtained between strain DSM 41524T and S. asiaticus DSM 41761T (71.6%), S. indonesiensis DSM 41759T (71.5%), S. cangkringensis DSM 41769T (71.5%), and S. rhizosphaericus DSM 41760T (71.0%). However, the pairwise dDDH values between the genomic sequences of the type strains of S. asiaticus, S. indonesiensis, S. cangkringensis, and S. rhizosphaericus ranged between 96.2% to 98.4%, a value above the cut-off point of species demarcation. Therefore, S. indonesiensis, S. cangkringensis, and S. rhizosphaericus are heterotypic synonyms of S. asiaticus. These findings call for emending the description of the S. asiaticus species. The dDDH results between the genome sequence of strain DSM 41524T and its close neighbours showed that strain DSM 41524T forms a new subspecies within the Streptomyces asiaticus species for which the name Streptomyces asiaticus subsp. ignotus subsp. nov. is proposed.
Fig. 1.

Phylogenomic tree showing the relationships between the Streptomyces strains and their closely related species. Tree inferred with FastME from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5.
In the same cluster of strain DSM 41524T, strain DSM 41602 formed a subcluster with S. mordarskii JCM 5052T placed next to S. antimycoticus NBRC 12839T and S. sporoclivatus NBRC 100767T forming together a well-supported cluster (Fig. 1). S. sporoclivatus species is a later heterotypic synonym of S. antimycoticus species. The dDDH values between strain DSM 41602 and S. mordarskii JCM 5052T was 93.5% while the scores obtained between the following pairs of strains DSM 41602 and S. sporoclivatus NBRC 100767T (76.0%); DSM 41602 and S. antimycoticus NBRC 12839T (75.6%); S. mordarskii JCM 5052T and S. antimycoticus NBRC 12839 T (75.6%) were between the threshold, 70–79%, for prokaryotic subspecies delineation. Furthermore, the in-silico G+C content of strain DSM 41602 is 70.9%, while the type strain of S. antimycoticus has 70.9 mol%; value within species variation of 1%. Therefore, strain DSM 41602, S. mordarskii JCM 5052T, S. sporoclivatus NBRC 100767T merit to be classified as a subspecies of S. antimycoticus for which the name Streptomyces antimycoticus subspecies sporoclivatus comb. nov. was proposed.
Fig. 1 showed that strain DSM 41529T formed a well-supported sub-cluster with the type strains of S. sparsogenes (Fig. 1). The phylogenomic position of the strain and its branch length clearly showed that it forms a new species within the evolutionary radiation of the genus Streptomyces. This conclusion agreed with the dDDH value below the threshold of 70% for species demarcation between the genome sequence of strain DSM 41529T and its close relative. Therefore, the strain DSM 41529T is considered as a type strain of a new species for which the name Streptomyces lonegramiae sp. nov., is proposed.
The phylogenomic tree showed that strain DSM 40473T was grouped with S. abikoensis JCM 4002T, forming a well-supported sub-cluster (Fig. 1). These results were in concordance with the dDDH value of 47.1% and justify the attribution of the strain to a new species, Streptomyces hesseae sp. nov.
In another phylogenetic lineage, strain DSM 41528T formed a well-supported sub-cluster with ‘S. endophytica HNM01140’ (Zhou et al., 2023) and next to a subclade housed strain DSM 41527T and its close relatives (Fig. 1). The dDDH value between strains DSM 41528T and HNM01140 was 70.8%, value slightly above the borderline of the 70% threshold for species demarcation. The strain has a G+C content of 71.7% while the type strain of Streptomyces endophytica has 72.2%, a value within species variation of 1% G+C (JP Meier-Kolthoff et al., 2014). At the time of writing, the name of S. endophytica species (Zhou et al., 2023) was not yet validly published and it is unclear how distant S. endophytica is phylogenetically from S. endophyticus (Li et al., 2013), as the genome of S. endophyticus is unavailable. In addition, the species epithet endophytica leads to confusion with the validly named S. endophyticus species. According to rule 53 of the International code of nomenclature of Prokaryotes (revision 2022) ‘an epithet is illegitimate if it duplicates a specific or subspecific epithet previously validly published for a species or subspecies’. As only legitimate names and epithets are taken into consideration, strain DSM 41528T cannot be affiliated to a new subspecies of ‘S. endophytica’, but it deserves to be considered as a novel species for which the name Streptomyces bugieae sp. nov. is proposed.
Strain DSM 41527T appeared in a distinct branch closely associated with S. caniferus NBRC 15389T and S. angustmyceticus JCM 4053T (Fig. 1). The dDDH values between the strain and its close relatives (41.7–42.5 < 70%) permit the assignment of strain DSM 41527T to a new Streptomyces species for which the name of Streptomyces mooreae sp. nov. is proposed.
In the next cluster, strain DSM 3412T had a phylogenetic position closely associated with ‘S. akebiae MG28T’, together forming a subclade next to S. deccanensis KCTC 19241T and S. caniscabiei NE06–02DT (Fig. 1). The dDDH between the genome sequence of strain DSM 3412T and its close relative ‘S. akebiae M G28T’ was between 64.7–46.8%, value below the 70% cut-off point for species demarcation (Table S3). Therefore, strains DSM 3412T should be considered as the type strain of new species for which the name Streptomyces gottesmaniae sp. nov., is proposed.
Strains DSM 41981T and DSM 41014T formed a well-supported distinct group next to the subcluster of strain DSM 41972T (Fig. 1) and showed a dDDH values below the 70% threshold for prokaryotic species affiliation. Therefore, strains DSM 41981T and DSM 41014T merit to be considered as novel species for which the names Streptomyces doudnae sp. nov., and Streptomyces hintoniae sp. nov. are proposed, respectively. Strain DSM 41972T was closely related to the type strains of S. griseorubens, S. althioticus, and S. matensis species (Fig. 1). S. griseorubens and S. matensis are heterotypic synonyms of S. althioticus. The dDDH values between the genomic sequences of strain DSM 41972T and those of S. althioticus JCM 4344T (72.3%) and S. matensis JCM 4277T (72.0%) fell between the established threshold for the prokaryotic subspecies demarcation, 70–79% (JP Meier-Kolthoff et al., 2013). Therefore, strain DSM 41972T merits to be considered as a new sub-species of S. althioticus for which the name Streptomyces althioticus subsp. attaecolombicae subsp. nov. is proposed.
Strains DSM 41635 and DSM 41636T formed a well-supported subclade closely related to the type strain of S. atrovirens JCM 6913T (Fig. 1). The close phylogenomic relationship of strains DSM 41635 and DSM 41636T was in line with a dDDH value of 82.1%, which is above the borderline of prokaryotic species delineation calling for affiliating them to the same species. The variation of the G+C content between strains DSM 41635 and DSM 41636T is within the range of 1% defined within a species (JP Meier-Kolthoff et al., 2014). Moreover, these strains have very similar genomic features as shown in Table S3. However, the dDDH values between strains DSM 41635 and DSM 41636T and validly named Streptomyces species were far below the 70% (Table S3). Thus, strains DSM 41635 and DSM 41636T merit to be considered as a new species for which the name Streptomyces edwardsiae sp. nov. is proposed with strain DSM 41636T as the type strain.
In a distant sub-clade from the strains listed above, strain DSM 41921T was placed in a distinct branch which reflected its genetic divergence from the validly named reference strains (Fig. 1). These findings were consistent with the dDDH score (36.2%) below 70%. Thus, strain DSM 41921T should be considered as new species within the genus Streptomyces for which the name Streptomyces dubilierae sp. nov. is proposed.
Strain DSM 40712T was closely related to S. ambofaciens ATCC 23877T and S. marokkonensis JCM 17027T, forming together a well-supported sub-group (Fig. 1). The genetic divergence of these strains was in line with the low dDDH values of 37.1–37.6%. Strain DSM 40712T is therefore a type strain of a new species for which the name Streptomyces lancefieldiae sp. nov., is proposed.
Strain DSM 41640T and its close relatives S. ciscaucasicus DSM 40275T and S. canus DSM 40017T (Fig. 1). However, S. ciscaucasicus is a later heterotypic synonym of S. canus. The divergent phylogenetic position of this strain within the evolution of the genus Streptomyces was justified by dDDH value below the cut-off point for species demarcation (Table S3). Thus, strain DSM 41640T forms a novel species for which the name Streptomyces doebereineriae sp. nov. is proposed.
Strain DSM 41699T was associated with S. glomeratus DSM 41457T and next to ‘S. cynarae HUAS 13–4′ and S. yaanensis CGMCC 4.7035T (Fig. 1). The dDDH values between the genome sequence of strain DSM 41699T and its closest relatives listed above ranged from 30.8–32.3%. Thus, strain DSM 41699T forms a new species within the genus Streptomyces for which the name Streptomyces gibsoniae sp. nov. is proposed.
Strain DSM 41770T was closely related to S. halstedii NRRL ISP-5068T, an earlier heterotypic synonym of S. griseolus, forming together a well-supported sub-group adjacent to the S. nitrosporeus JCM 4598T branch (Fig. 1). In another subcluster, DSM 40932T was placed in a distinct branch closely associated with S. cyaneofuscatus NRRL B-2570T, S. arboris TRM68085T, and the type strains of S. microflavus (Fig. 1). The divergence of the strains DSM 41770T and DSM 40932T from validly named Streptomyces species is evidenced by dDDH values below the 70% threshold defined for species demarcation (Wayne et al., 1987) as shown in Table S3. Therefore, strains DSM 41770T and DSM 40932T merit to be considered as novel species for which the names Streptomyces salyersiae sp. nov., and Streptomyces stephensoniae sp. nov., are proposed, respectively.
Strains DSM 41979T, DSM 41859, and DSM 41982 formed together a well-supported sub-cluster related to a sub-clade encompassing the type strains of S. diastaticus, S. rutgersensis, S. gourgerotii, S. coelicolor, S. limosus and S. albidoflavus (Fig. 1). However, S. rutgersensis, S. gourgerotii are a later heterotypic synonym of S. diastaticus while S. limosus is a later heterotypic synonym of S. albidoflavus. The dDDH between the genome sequence of the studied strains DSM 41979T, DSM 41859, and DSM 41982 ranged from 80.1% to 87.7%; which is coherent with their close phylogenetic relationships and their similar genomic features, including the G+C content (Table S1-S2). However, the highest dDDH value between these studied strains and Streptomyces species validly named was 23.4%, a value below the threshold for species demarcation. Consequently, strains DSM 41979T, DSM 41859, and DSM 41982 form a new species for which the name Streptomyces evansiae sp. nov. is proposed with strain DSM 41979T as the type strain.
In another phylogenetic lineage, the strain DSM 42041T was placed in a distinct branch associated with a sub-clade containing the type strains of S. qinglanensis, S. albus, S. diacarni, S. tubbatahanensis, and S. cacaoi, and next to the branch of S. grossypii N2–109T (Fig. 1). These results were consistent with the dDDH value below the cut-off point of 70% between the strain and all validly named Streptomyces species. Thus, strain DSM 42041T merits to be affiliated to a new species for which the name Streptomyces hazeniae sp. nov., is proposed.
The distinct branch of strain DSM 44938T was closely associated with a subclade that contained strains DSM 41886T formed with S. marincola SCSIO 64649T and next to S. radicis DS1–2T (Fig. 1). Strains DSM 44917T was found to be attached to this subclade with S. specialis GW41–1564T as the closest neighbour. In the same cluster, strain DSM 44918T was closely related to S. hainanensis DSM 41900T forming together a well-supported subclade next to Streptomyces hoynatensis KCTC 29097T. These strains were adjacent to a sub-cluster encompassed strain DSM 44915T and its closest phylogenetic neighbours S. zhaozhouensis CGMCC 4.7095T, S. sedi JCM 16909T, S. mimosae 3MP-10T, and S. triticirhizae NEAU-YY642T (Fig. 1). These phylogenetic relationships between the strains and their close phylogenomic neighbours were in line with the dDDH values below the threshold of 70% for prokaryotic species demarcation (Wayne et al., 1987) (Table S3). Therefore, strains DSM 41886T, DSM 44938T, DSM 44917T, DSM 44918T, and DSM 44915T form novel species for which the names Streptomyces johnsoniae sp. nov., Streptomyces litchfieldiae sp. nov., Streptomyces boetiae sp. nov., Streptomyces millisiae sp. nov., Streptomyces chisholmiae sp. nov. are proposed, respectively.
Phylogenomic and comparative genomic analysis of non-Streptomyces strains
The phylogenetic position of all the non-Streptomyces strains in the phylogenomic tree (Fig. 2) is well supported and a clear conclusion about the taxonomic status of the strains could be drawn. Blastococcus sp. DSM 46792T formed a well-supported subclade with Blastococcus aurantiacus DSM 44268T within the radiation of its respective genus (Fig. 2). These results were coherent with a dDDH value of 31.4% between strains DSM 46792T and DSM 44268T. Therefore, strain DSM 46792T deserves to be affiliated to a new species for which the name Blastococcus goldschmidtiae sp. nov. is proposed.
Fig. 2.
Phylogenomic tree showing the relationships between different actinobacterial strains and their closely related species. Tree inferred with FastME from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5.
Pseudonocardia sp. DSM 45834T was found to be a distinct branch closely related to a subcluster consisted of the type strains of ‘Pseudonocardia humida’, Pseudonocardia xinjiangensis, Pseudonocardia alaniniphila, Pseudonocardia cypriaca, Pseudonocardia kunmingensis, Pseudonocardia petroleophila, Pseudonocardia abyssalis, and Pseudonocardia oceani species (Fig. 2). The dDDH between strain DSM 45834T and its close phylogenomic relatives listed above (<22.0%) were far below the 70% threshold for species demarcation as shown in Table S3 and call for assigning strain DSM 45834T to a new Pseudonocardia species for which the name Pseudonocardia charpentierae sp. nov. is proposed. Streptomonospora sp. DSM 45055T appeared in a distinct branch close to the type strains of Streptomonospora alba and Streptomonospora salina species and phylogenetically distant from Streptomonospora arabica (Fig. 2). These outcomes were in line with the low dDDH values (28.1–26.8%) between the genome sequence of strain DSM 45055T and its close phylogenetic relatives. Thus, strain DSM 41055T merits to be assigned to a new species for which the name Streptomonospora wellingtoniae sp. nov. is proposed.
Nocardiopsis sp. DSM 44743T was placed in a well-supported distinct branch closely related to a subclade encompassed Nocardiopsis flavescens CGMCC 4.5723T and ‘Nocardiopsis changdeensis Mg02T’ and next to Nocardiopsis algeriensis CECT 8712T (Fig. 2). The dDDH between the genome of the studied strain and their close phylogenomic neighbours (24.2–29.1%) were below the 70% cut-off point for species demarcation. Therefore, strain DSM 44743T forms a new species for which the name Nocardiopsis lambiniae sp. nov., is proposed. Jatrophihabitans sp. DSM 44399T was grouped with Jatrophihabitans telluris KCTC 39922T and alongside the type strain of Jatrophihabitans endophyticus (Fig. 2). The phylogenetic relationship of strain DSM 44399T with its close relative is in line with the dDDH value of 20.4%, proving that the strain forms a new species for which the name Jatrophihabitans lederbergiae sp. nov. is proposed.
Genomic features including genome size, G+C content, number of RNA and coding sequences for all the studied strains and their closest phylogenomic neighbours are provided in Table S1.
Cultural and growth properties of the novel actinomycetes strains
Most of the Streptomyces strains were able to grow from 10°C to 37°C with optimal growth between 28°C and 37°C. Strains DSM 40932T, DSM 3412T, DSM 40434T, DSM 41979T, DSM 41982, DSM 41859, and DSM 41770T showed poor to moderate growth at 4°C and 10°C (Table S4). Only strains DSM 41981T and DSM 41921T showed a good growth at 45°C while strains DSM 44918T and DSM 44938T grew without aerial mycelia. All Streptomyces strains showed an optimal growth on DSMZ 65 medium and also on other media as shown in Table S3. A brown diffusible pigment was observed on ISP6 and ISP7 for strains DSM 41921T, DSM 3412T, DSM 41699T, while strains DSM 44917T and DSM 44915T produced a brown pigment on GYM and NZ amine media. In addition, strain DSM 42041T produced brown pigment on NZ amine. More detail about the growth properties and the morphological features of the strains are presented in Table S3 and Figure S3.
Strains DSM 41635 and DSM 41636T, which were found to belong to the same new species, were distinguished from each other on the basis of their morphology. Strain DSM 41636T developed lavender aerial mycelium after 7 days of incubation at 28°C on ISP2, while strain DSM 41635 had white-grey aerial mycelium under the same condition. Strains DSM 41979T, DSM 41859, and DSM 41982 had white aerial mycelium on ISP3, ISP5, ISP7 after 7 days of incubation at 28°C. Strain DSM 41859 showed white grey aerial mycelium on NZ amine medium unlike strains DSM 41979T and DSM 41982, which had no aerial mycelium, after 7 days of incubation at 28°C. Strain DSM 41528T showed a good growth on ISP1 to ISP7, DSMZ 535, GYM, and NZ amine media, on which it developed white aerial mycelium, which was greyish and pinkish on ISP1/ISP3 and NZ amine media, respectively. Moderate growth was observed at 25°C and 42°C, while good growth was recorded between 28°C and 37°C. However, the type strain of S. endophytica was not able to grow on ISP7 according to Zhou et al. 2023. Strain DSM 41972T, which forms a new subspecies within S. althioticus, had light grey (ISP1, ISP3, ISP6), white (ISP4, ISP5, ISP7), and light yellow (DSMZ 535, DSMZ 65, and NZ amine media) aerial mycelium after 7 days of incubation at 28°C. The strain showed weak growth at 17°C, 20°C, 42°C, and 45°C, while good growth was observed at 28°C, 35°C, and 37°C and at pH values between 5.0–9.0. Strain DSM 41602T showed good growth on ISP1 to ISP6, GYM, and NZ amine medium, on which the strain developed white-lavender (ISP2), grey (ISP3 and ISP4), and white (GYM) aerial mycelia. No aerial mycelium was observed on ISP1, ISP5, ISP6, and NZ amine medium despite the good growth of yellow brown substrate mycelium. A moderate growth of the strain was obtained on ISP7 and DSMZ 535 media. No diffusible pigment was observed on any of the tested media. Growth was detected between 17°C to 37°C. These growth properties were in line with those reported for its close relative S. antimycoticus species by Komaki and Tamura, 2020. Strain DSM 41524T showed a good growth on ISP1 to ISP7, DSMZ 535, DSMZ 65, and NZ amine agar plates on which aerial mycelium with grey (ISP1), white (ISP3, ISP4), and white greyish (ISP7) colour was detected. No aerial mycelium was observed on ISP2, ISP5, ISP6, DSMZ 535, and NZ amine, though a good development of yellow-brown substrate mycelium was obtained. A weak aerial mycelium was observed on medium DSMZ 65. No diffusible pigment was observed. The strain grew from 17°C to 42°C, whereby optimal growth was observed from 28°C to 37°C, unlike its close phylogenomic neighbour S. asiaticus DSM 1761T, which was able to grow at 45°C (Sembiring et al., 2000).
The growth properties for the non-Streptomyces strains were also determined and found to be in line with their respective genera. Blastococcus sp. DSM 46792T showed an optimal growth on GYM, DSMZ 553, DSMZ 714, and DSMZ 877 media at 28°C. A well grown culture of the strain was obtained at 28°C, 35°C, and 37°C while poor to moderate growth was observed at 17°C to 25°C, respectively. No growth was observed at 42°C and 45°C.
Streptomonospora sp. DSM 45055T showed a good growth on ISP1 (DSMZ 1764), DSMZ 514, and nutrient agar (DSMZ 1) media on which white-beige colonies were developed. The latter turned to have a dark brown colour on DSMZ 1159 medium. Optimal growth temperature was 28°C and at pH 6.0–7.0 on DSMZ 1159 medium. Moderate growth of this strain was obtained at 25°C, 35°C, and 37°C and on DSMZ 514 medium. Pseudonocardia sp. DSM 45834T showed optimal growth at 28°C on GYM and DSMZ 83 media. Poor and moderate growth was observed at 17°C and at 20°C, 25°C, 35°C, and 37°C, respectively. The growth of Nocardiopsis sp. DSM 44743T was poor at 4°C, 10°C, and 42°C, moderate at 17°C, 20°C, 25°C, and good at 28°C, 35°C, and 37°C (Table S1). Good growth was observed at pH 5.0 to 8.5 on GYM medium after 7 days of incubation. The strain showed white aerial mycelium at DSMZ 514 medium after 7 days of incubation at 28°C. Jatrophihabitans sp. DSM 44399T formed orange colonies on medium 621, ISP2, Bennet's media after incubation at 20°C. The strain was able to grow well at 20°C and 25°C and at pH 5.5 to 8.5.
Biochemical, enzymatic and chemotaxonomic analyses reveal the physiological properties of the strains
All the Streptomyces strains were distinguished from each other by their ability to metabolise a wide range of carbon and nitrogen sources. As shown in Table S5, most of the Streptomyces strains were unable to metabolise sugar alcohol substrates, such as L-arabitol, erythritol, xylitol, and D-sorbitol (except strain DSM 3412T). Strains DSM 41527T, DSM 41529T, DSM 41640T, DSM 41921T, DSM 44918T, DSM 44915T, and DSM 44938T oxidized D-adonitol, whereas strains DSM 40712T and DSM 41859 were able to use dulcitol.
The biochemical properties distinguished the studied strains, which belonged to the same new species (strains DSM 41635 and DSM 41636T; DSM 41859, DSM 41982, and DSM 41979T) from their close phylogenomic relatives (Table S5). The same applies to the studied strains DSM 41972T and DSM 41602T, which form new subspecies. Strains DSM 41635 and DSM 41636T were distinguished from each other by the ability of strain DSM 41635 to metabolise D-xylose, D-galactose, amygdalin, salicin, gentiobiose, and D-turanose and to produce ß-galactosidase and α-glucosidase, unlike strain DSM 41636T. The strains DSM 41979T, DSM 41859, and DSM 41982, which belong to the proposed new species, Streptomyces sirexnoctilio sp. nov. were differentiated from each other by the ability of strain DSM 41859T to degrade dulcitol and the inability of strain DSM 41982 to use arbutin and salicin. However, strain DSM 41979T and DSM 41859 were unable to oxidise D-melibiose and to produce α-galactosidase, respectively. More details about the biochemical and enzymatic profiles of the strains are shown in Table S5. The type strain of S. endophytica was able to use D-arabinose, D-xylose, L-rhamnose, D-mannitol, D-sorbitol (Zhou et al., 2023) unlike strain DSM 41528T. The culture of strain DSM 41528T was prepared following the same growth condition (ISP2 at 28°C for 7 days) of its close phylogenomic neighbour.
DSM 41602T forms a new subspecies within S. antimycoticus species. The strain was able to metabolise methyl-α-D-glucopyranoside unlike its close relative S. antimycoticus DSM 40284T. The latter oxidized dulcitol, inositol, D-sorbitol, methyl-α-S-mannopyranoside, amygdalin, D-saccharose, and inulin, unlike strain DSM 41602T. These strains had a very similar enzymatic profile though strain DSM 41602T was able to produce lipase (C 14), while strain DSM 42084T produced esterase (C 4) and esterase lipase (C 8). The two latter enzymes, cystine arylamidase and trypsin were produced by strain DSM 41972T unlike its relative S. althioticus DSM 40092T (Table S5).
For the non-Streptomyces strains, Pseudonocardia sp. DSM 45834T shared a similar enzymatic profile with its close phylogenomic neighbour Pseudonocardia kunmingensis DSM 45301T with the exception that it was unable to produce N-acetyl-ß-glucosaminidase, α-mannosidase, and α-fucosidase like its relative. Nocardiopsis sp. DSM 44743T was able to metabolise arbutin while its close phylogenomic relative Nocardiopsis flavescens DSM 45786T oxidized D-galactose, L-sorbose, dulcitol, D-sorbitol, starch, glycogen, D-tagatose, D-fucose, L-fucose, and potassium 5-ketogluconate. These strains had a similar enzymatic profile, though strain DSM 45786T produced α-mannosidase unlike strain DSM 44743T. More data are provided in Table S5.
Streptomonospora sp. DSM 45055T was differentiated from its close genomic neighbour Streptomonospora alba DSM 44588T by its ability to metabolise D-arabitol, D-adonitol, N-acetylglucosamine, glycerol, erythritol, D-ribose, L-rhamnose, and D-lyxose (Table S5).
Blastococcus sp. DSM 46792T was able to produce esterase C4, α-chymotrypsin and trypsin unlike its close neighbour Blastococcus aurantiacus DSM 44268T.
The chemotaxonomic traits of the genus Streptomyces are LL-diaminopimelic acid (LL-A2pm) in the cell wall peptidoglycan; a high amount of saturated isoand anteisofatty acids; and MK-9(H6) and MK-9(H8) as the predominant quinones. Streptomyces strains have a polar lipid pattern composed of diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphatidylinositol mannosides (PIM) (Kämpfer, 2012). The genus Nocardiopsis is characterized by the presence of meso-diaminopimelic acid (meso-A2pm) in its cell wall peptidoglycan and by the predominance of menaquinone MK-10 with various hydrogens. Quinone with 9 and 11 isoprene units have been detected in minor quantities. The major polar lipids are PI, phosphatidylcholine (PC), phosphatidylglyerol (PG); and phosphatidylmethylethanolamine (PME). The main fatty acids are branched (Hozzein et al., 2012). The chemotaxonomic traits of Pseudonocardia consist of meso-A2pm in the peptidoglycan and arabinose and galactose as cell wall sugars. Pseudonocardia strains have MK-8 (H4) as the predominant menaquinone; glucosamine-containing phospholipids, PC, PG, PE, PME, and glucosamine-phospholipids (PLs) as major polar lipids. The fatty acids pattern is predominated by isobranched fatty acids with a 16-carbon chain (Huang et al., 2012).
Blastococcus strains have cell wall peptidoglycan with meso-A2pm; galactose and arabinose as whole cell sugars; isobranched fatty acids with the presence of isoC16:1, C18:1 ω9c, C17:1 ω8c, isoC15:0, and C17:0; MK-9 (H4) as the predominant menaquinone. The main polar lipids are PC, PI, PE, DPG, and PG (Stackebrandt et al., 2012). The genus Jatrophihabitans is characterized by the presence of meso-A2pm in the cell wall peptidoglycan with acyl-type N-glycolylated; MK-9 (H4) as the predominant quinone; DPG and unidentified PL, as well as aminolipids; and isopalmitic acid, C18:1 ω9c, anteisoC17: 0 and C17:1 ω8c, as major fatty acid (Madhaiyan et al., 2013).
The chemotaxonomic features of all studied strains were coherent with those of their respective genera (Table S6). All the studied strains were distinguished from their closest neighbours by chemotaxonomic, biochemical and enzymatic traits.
New actinobacterial species and their antimicrobial activities
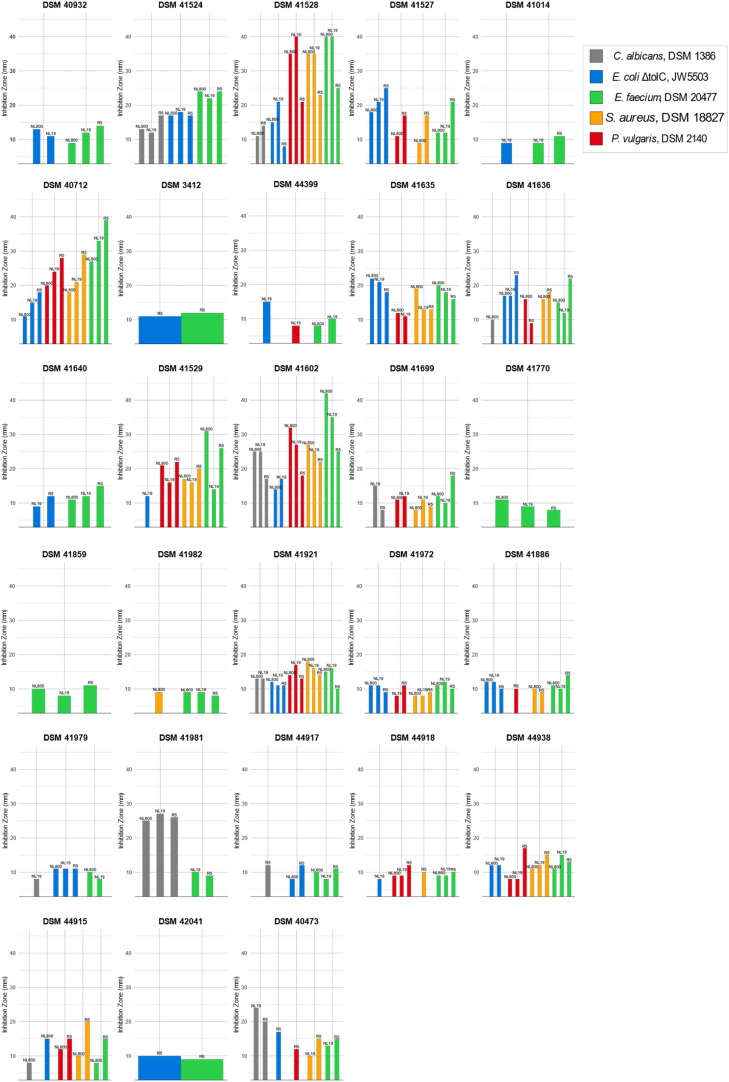
Streptomyces strains have been explored for new NPs and served as producers of not only hundreds of antibiotics but also enzymes, nanoparticles, or other industrially and pharmaceutically valuable biomolecules (Sanjivkumar et al., 2016, Sanjivkumar et al., 2018). In drug research, the discovery of the existence of numerous BGCs in the genomes of these strains has revived the research field (de Lima Júnior et al., 2023). The richness of the genetic composition of these filamentous bacteria in terms of specialized secondary metabolites, as well as the biotechnological advancements that have been successfully applied to Streptomyces, call for further exploiting the genetic diversity of this taxon, particularly after proving the effectiveness of taxonomy in drug discovery (Goodfellow and Fiedler, 2010, Thumar et al., 2010, Khodamoradi et al., 2021). It has been reported that Nocardiopsis strains produce toxins and immunomodulators, enzymes (amylases, cellulases, etc.) and bioactive compounds with antifungal, anticancer, and antimicrobial activities, such as pendolmycin or griseusins (Bennur et al., 2015). Streptomonospora strains are an interesting source of novel bioactive compounds, as shown by the discovery of two new anticancer compounds, named litoralimycin A1 and B2 from Streptomonospora litoralis (Khodamoradi et al., 2021), and an antibacterial compound named persiamycin A, which was isolated from Streptomonospora sp. PA3 (Matroodi et al., 2020). Some Pseudonocardia strains have been shown to be sources of interesting new bioactive compounds with antibacterial activity, such as branimycin and pseudonocardians A-C isolated from P. carboxydivorans M227 (Braña et al., 2017) and Pseudonocardia sp. SCSIO 01299 (Dekker et al., 1998, Li et al., 2011), respectively. Several anticancer compounds such as 4-(2-acetamidoethyl)-phenyl acetate and 4-((1,4-dioxooctahydropyrrolo [1,2-a] pyrazin-3-yl) methyl)-phenyl acetate were extracted from P. endophytica VUK-10 (Mangamuri et al., 2015, Mangamuri et al., 2016). Other strains (eg. Pseudonocardia sp. EC080529–01, Pseudonocardia autotrophic) produce bioactive compounds with antifungal activities, such as nystatin, garamycin, the polyene NPP A1, pseudonocardones A–C, 6-deoxy-8-O-methylrabelomycin (Mangamuri et al., 2016a, 2016b, Kim et al., 2017, Challinor and Bode, 2015, Barke et al., 2010, Sit et al., 2015), and the neuroprotective compound phenazostatin D, which was isolated from Pseudonocardia sp. B6273 (Maskey et al., 2003). Other strains appear to be more useful for bioremediation, such as Pseudonocardia sp. M43 (Lee et al., 2006), Pseudonocardia 1190 (Daye et al., 2003), Pseudonocardia sp. K1 (Kohlweyer et al., 2000), Pseudonocardia sp. KSF27 (Sakakibara et al., 2011), Pseudonocardia sp. RM423 (Apinya et al., 2015), Pseudonocardia alni AS4.1531 (Konkit and Lumyong, 2012), and Pseudonocardia sp. N23 (Derosa et al., 1996, Yamamoto et al., 2017). To assess the antibiotic potential of the strains, cell extracts were tested in bioassays against a panel of Gram-positive and Gram-negative bacterial test strains, as well as yeast, including clinically relevant pathogens, such as Staphylococcus aureus DSM 18,827, Enterococcus faecium DSM 20477T, Proteus vulgaris DSM 2140, E. coli ΔtolC JW5503–1, and Candida albicans DSM 1386, some of which are part of the recently established WHO Priority Pathogens list of the DSMZ (https://www.dsmz.de/collection/catalogue/microorganisms/special-groups-of-organisms/who-priority-pathogens-list). For some of the extracts, broad spectrum bioactivities were observed against all test strain. The vast majority of all Streptomyces strains exerted bioactivity against at least one microbial test strains as are summarized in Fig. 3.
Fig. 3.
Antimicrobial activities of the studied strains against Gram-positive and Gram-negative bacteria, and yeast.
However, no antimicrobial activities were detected from the crude extract of the non-Streptomyces strains.
Gene cluster network analyses depict biosynthetic potential
To investigate the BGC distribution between the identified strains (n=32) and their close phylogenomic neighbours (n=44), a gene cluster networking analysis was carried out using the BiG-SCAPE software with BGCs predicted by antiSMASH from seventy-six draft genome sequences in total. Additionally, the analysis included the MIBiG database to assess the novelty of the identified gene clusters. Due to the fragmented nature of the draft genomes, we separated the antiSMASH results into regions within contigs and on contig edges, and analyzed them separately. Out of 982 BGC regions predicted for the thirty-two identified strains (28 new type and 4 non-type strains), 407 were located on contig edges and 575 within contigs (Table S7). In the following, we will focus on the cluster networking analysis with BGC regions located within contigs to avoid overestimating biosynthetic diversity due to fragmented BGCs (Table S7). Together with the predicted BGC regions from the close phylogenetic neighbours and similar known BGCs from the MIBiG database (Terlouw et al., 2023), the BiG-SCAPE analysis of the regions inside contigs comprised 1508 BGC regions. BiG-SCAPE clustered these into 882 (excluding GCFs consisting only of MIBiG BGCs) gene cluster families (GCFs), of which 449 contained BGC regions from the newly identified strains (Fig. 4). A total of 265 out of these 449 GCFs were singletons. On the other hand, among the remaining 184 of the 449 GCFs, only 40 GCFs (comprising 65 BGCs from novel strains) were associated with a known BGC from MIBiG, indicating that few BGCs encode for the biosynthesis of known natural products (Table S7). For the other 144 GCFs (comprising 245 BGCs from novel strains), as well as the 265 singletons from novel strains, the associated NPs are unknown, showing a large potential of the respective strains for the production of novel compounds.
Fig. 4.
Gene cluster family network of GCFs as obtained from the BiG-SCAPE analysis and visualized with Cytoscape. Each node corresponds to a BGC. Green rectangles represent BGCs from the 32 studied strains, blue ovals are BGCs from reference strains, and purple triangles indicate known BGCs from the MIBiG database. Edges between two nodes represent a distance between the BGCs below the BiG-SCAPE default threshold of 0.3.
Among the 882 GCFs, 112 contained BGCs consisting of type I PKSs, 47 with PKS/NRPS-hybrid BGCs, and 259 with other PKS-related systems. Furthermore, 234 GCF contained NRP-related BGCs, 263 were associated with the production of RiPPs, and 149 with terpenes. Please note that some GCFs contain superclusters (BGC regions with multiple separate BGCs) and therefore fall into several of the above-mentioned classes.
In order to further analyse the relationship between the 28 novel strains and forty-four related strains, we performed a hierarchical clustering on the presence and absence of GCFs (only BGCs within contigs, singletons excluded). The resulting presence/absence map in combination with the dendrograms showed clearly distinguishable clusters (Figure S6). The grouping of these strains on the basis of GCFs is in line with their phylogenomic position within the genus Streptomyces. For instance, based on their GCFs, the novel strains DSM 41524T and DSM 41602T were grouped with the previously published strains Streptomyces asiaticus DSM 41761T and Streptomyces antimycoticus NBRC 12839T in a sub-cluster. This corresponds to their evolutionary relationship according to our phylogenomic analysis (Fig. 1). The cluster networking analysis revealed that these strains shared several GCFs, covering different types of BGCs (Figure S6) and it confirmed for some of the strains, such as DSM 41527T and DSM 3412T, the presence of BGCs encoding for the previously identified compounds. Strain DSM 41527T, known as bialaphos producer, contained a BGC similar to the bialaphos BGC0000406 deposited in MiBIG, whereas the galbonolid producer strain DSM 3412T, harboured a BGC similar to the galbonolid BGC0000065 deposited in MiBIG. However, for several strains previously reported as antibiotic producers, the respective BGCs could not be identified, as for example strains DSM 40932T (reported streptothricin producer), DSM 40712T (reported chalcomycin producer), and DSM 41529T (reported antibiotic A-130 producer). This could either be due to the fragmentation of the genomes and thus, missing BGCs, or indicate that the strains are not the original producers, but different strain deposits. The current standards for identifiying and confirming the authenticity of these old strains were not available at the time of deposition (between 1989–1993). Accordingly, it is worth analysing old strain deposits with new methods for their natural compound biosynthesis potential. In general, the current study can serve as preliminary data for further analytical chemistry research in the field of novel drug discovery.
Identification of compounds from new actinobacterial type strains
In addition to guiding novel drug discovery, our BiG-SCAPE analysis provided further insight on known natural compounds and their underlying BGCs. In the course of studies on the regulator-guided activation of BGCs, the SARP gene papR2 was heterologously expressed in strain DSM 41636T and DSM 41640T, as similarly reported in Nouioui et al. (2024). Bioassays with samples from the DSM 41636T::pDS300 and DSM 41640T::pDS300 heterologous expression strains resulted in increased bioactivity against K. rhizophila in comparison to the respective control samples, carrying an empty vector (data not shown). Extracts obtained from 4-day old cultures were analyzed with LC-MS. Comparison of metabolic profiles resulted in the identification of the benzoxazole antibiotic A33853 as a product from strain DSM 41636T::pDS300 (Fig. 5) and the diisonitrile natural product SF2768 from DSM 41640T::pDS300 (Fig. 6).
Fig. 5.
Detection of A33853 compound in extract samples of DSM 41636T::pDS300 by LC-MS analysis. (A) Overlaid chromatograms of ethyl acetate extracts derived from DSM 41636T strains grown in R5. A peak corresponding to A33853 is indicated by a black arrow. (B) UV–Vis profile of the identified compound that is virtually identical to the previously reported spectrum of A33853 (Michel et al., 1984). (C) HRMS data of the DSM 41636T::pDS300 sample with a deprotonated molecular ion of A33853 (m/z [M – H]– = 390.0726).
Fig. 6.
Detection of SF2768 in extract samples of DSM 41640T::pDS300 by LC-MS analysis. (A) Overlaid total ion chromatograms of ethyl acetate extracts derived from DSM 41640T strains grown in NL19. A peak corresponding to SF2768 is indicated by a black arrow. (B) HRMS data of the DSM 41640T::pDS300 sample with a protonated molecular ion of SF2768 (m/z [M + H]+ = 337.1865).
The benzoxazole compound A33853 was first isolated from Streptomyces sp., NRRL 12068 and shows anti-leishmania activity (Michel et al., 1984). A BGC was identified in DSM 41636T and the related strain DSM 41635, which showed 95% and 100% gene similarity in antiSMASH, respectively, to the A33853 BGC (MIBiG accession BGC0001292, (Lv et al., 2015)) (Figure S5 A-B).
SF2768 was first identified as a product from Streptomyces sp. (Tabata, 1995). The substance was originally isolated based on its fungicidal activity, but was also known for its bactericidal activity, later shown to be a copper-binding metallophore (Wang et al., 2017). A BGC was identified in DSM 41640T, which showed 66% gene similarity in antiSMASH to the SF2768 BGC (MIBiG accession BGC0001574) (Wang et al., 2017) (Figure S5 C) with all essential genes for SF2768 biosynthesis (SfaA-E from Wang et al, 2017) being present in the BGC of DSM 41640T. In accordance with the lower similarity, BiG-SCAPE clustered the BGC region from DSM 41640 with the published SF2768 BGC at a similarity threshold of 0.5, but not at the default threshold of 0.3.
Overall, genome-mining predicted two compounds from two strains of the set of ∼30 novel type strains from the DSMZ strain collection and SARP overexpression helped to confirm the production of these compounds.
Natural products have played an important role in a number of therapeutic areas such as cancer, infection, cardiovascular diseases (eg. statins), and sclerosis (eg. fingolimod). They are characterized by a wide range of molecular architecture and are liable to be improved, as illustrated by the RiPPs products, which can be predicted based on genome mining approaches using various computational tools and also can be modified by site-directed mutagenesis of their scaffolds (Delivoria et al., 2019, Zhong et al., 2020). NPs have more advantages than synthetic molecules, exemplified by their greater molecular rigidity, which can be very helpful in protein-protein interaction (Lawson et al., 2018, Atanasov et al., 2021). The large number of unknown BCGs suggest the genomic richness of the proposed novel type strains in producing novel secondary metabolites. These findings support previous findings that novel strains can lead to novel chemical entities and underline the importance of modern polyphasic taxonomy in the selection of biological material for novel drug discovery (Goodfellow et al., 2018 Aug, Goodfellow and Fiedler, 2010, Hoffmann et al., 2018). The strains included in this study were isolated decades ago and kept in a vital and stable form in the DSMZ's open collection. This report highlights the vital role of the culture collection in safeguarding microorganisms that can be very useful for novel drug discovery, health, food, and the environment, and therefore ensure the continuity of research studies. Conservation of microbial diversity and deposit in open culture collections should be essential for all beneficial strains and not only required for taxonomic description of novel type strain. This study provides an insight into the pharmaceutical and biotechnological potential of the proposed novel type strains, which are made available to the scientific communities (https://www.dsmz.de/collection/catalogue) for further research.
Taxonomic consequences
Description of Streptomyces gottesmaniae sp. nov
Streptomyces gottesmaniae (got.tes.ma'ni.ae. N.L. gen. fem. n. gottesmaniae, of Gottesman, named in honour of Susan Gottesman for her major contribution in the field of microbiology, biochemistry and molecular biology).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white to grey aerial mycelium on ISP2, ISP3, and GYM media. The strain grows well from 25 to 42 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D- and L-arabinose, D-arabitol, arbutin, D-cellobiose, D-fructose, L-fucose, D-galactose, gentiobiose, D-glucose, glycerol, glycogen, inositol, inulin, D-lactose (bovine origin), D-maltose, D-mannitol, D-mannose, D-melibiose, N-acetylglucosamine, potassium gluconate, potassium 5-ketogluconate, D-raffinose, L-rhamnose, D-ribose, D-saccharose (sucrose), D-sorbitol, starch (amidon), D-trehalose, and D-xylose. Positive for acid phosphatase, alkaline phosphatase, cystine arylamidase, esterase (C 4), esterase lipase (C 8), α-galactosidase, ß-galactosidase, α-glucosidase, ß-glucosidase, ß-glucuronidase, leucine arylamidase, lipase (C 14), α-mannosidase, N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, with traces of mannose, and galactose. The peptidoglycan contains LL-A2pm and trace of DL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, C15:0, C16:0, isoC15:0, isoC16:0, C16:1 cis9, anteisoC17:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, unidentified phospholipid, phosphoaminolipid, phosphoglycolipid. The strain was known to produce galbonolides A and B. The genome size of strain DSM 3412T is 11.2 Mb with DNA G+C content of 70.0%.
The type strain DSM 3412T (= Tü 2253T = KCTC 59181T) was isolated from soil sample collected in Tunisia. The GenBank accession number of the assembled draft genome is JAVRFJ000000000.1.
Description of Streptomyces hesseae sp. nov
Streptomyces hesseae (hes'se.ae. N.L. gen. fem. n. hesseae, of Hesse, in honour of Fanny Hesse (1850–1934) who introduced agar into microbiology).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white greyish to grey aerial mycelium on ISP1, ISP2, ISP5, ISP6, DSMZ 535, GYM, and NZ amine media that turn to have white beige and light green colour on ISP3, and ISP7 media, respectively. The strain grows well from 25 to 45°C, optimally at 28°C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-arabitol, D-glucose, glycerol, glycogen, D-mannose, N-acetylglucosamine, potassium gluconate, D-ribose, and starch (amidon). Positive for acid phosphatase, alkaline phosphatase, esterase (C 4), esterase lipase (C 8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, with trace of mannose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, isoC15:0, C16:0, isoC16:0, isoC17:0, anteisoC17:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phospholipid, phosphoglycolipid, and lipids. The genome size of strain DSM 40473T is 7.6 Mb with DNA G+C content of 71.7%.
The type strain DSM 40473T (= ATCC 12568T = CBS 695.72T = IFO 13394T = IFO 13907T = BA-3572T = ISP 5473T = KCC S-0694T = KCC S-0812T = NBRC 13394T = NBRC 13907T = RIA 1355T) was isolated from soil with unknown origin, but known as producer of antibiotic PA-150. The GenBank accession number of the assembled draft genome is JAVRFI000000000.1.
Description of Streptomyces lancefieldiae sp. nov
Streptomyces lancefieldiae (lan.ce.fi.el'di.ae. N.L. gen. fem. n. lancefieldiae, of Lancefield, in honour of Rebecca Lancefield (1895–1981) known for her significant contributions to microbiology, particularly bacteriology).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white aerial mycelium on ISP2, TSA, and NZ amine agar media that turn to have lavender colour on ISP3, ISP4, ISP5, ISP7, and GYM media. The strain grows well from 25 to 45°C, optimally at 28°C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate amygdalin, L-arabinose, D-arabitol, arbutin, D-cellobiose, dulcitol, D-fructose, D-galactose, D-glucose, glycerol, glycogen, inositol, inulin, D-lactose (bovine origin), D-maltose, D-mannitol, D-mannose, methyl-α-D-glucopyranoside, methyl-α-D-mannopyranoside, N-acetylglucosamine, potassium gluconate, L-rhamnose, D-ribose, salicin, and starch (amidon). Positive for acid phosphatase, alkaline phosphatase, α-chymotrypsin, esterase (C 4), esterase lipase (C 8), α-galactosidase, ß-galactosidase, α-glucosidase, leucine arylamidase, α-mannosidase, N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of ribose, with traces of glucose, mannose, and galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, isoC15:0, anteisoC17:0, C16:0, isoC16:0, C16:1 cis9. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, unknown lipid, phospholipid, aminolipid, phosphoglycolipid, and phosphoaminolipid. The genome size of strain DSM 40712T is 9.3 Mb with DNA G+C content of 71.4%.
The type strain DSM 40712T (= NRRL 2835T = Tü 41T = ETH 21066T) is with unknown isolation source. The GenBank accession number of the assembled draft genome is JAVRFH000000000.1.
Description of Streptomyces stephensoniae sp. nov
Streptomyces stephensoniae (ste.phen.so'ni.ae. N.L. gen. fem. n. stephensoniae, of Stephenson, named in honour of Marjory Stephenson (1885–1948) for her significant contributions to biochemistry and microbiology).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop orange aerial mycelium on ISP2 and GYM media that turn to have sandy yellow and white colour on ISP3 and DSMZ 535 media, respectively. The strain grows well from 20 to 37°C, optimally at 28°C, from pH 5.5 to 8.5, optimally at pH 7.0. The strain is able to assimilate amygdalin, D-arabitol, arbutin, D-cellobiose, gentiobiose, D-glucose, glycerol, glycogen, D-maltose, D-mannitol, D-mannose, N-acetylglucosamine, potassium gluconate, D-ribose, salicin, starch (amidon), and D-trehalose. Positive for acid phosphatase, alkaline phosphatase, α-chymotrypsin, cystine arylamidase, esterase lipase (C 8), ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, lipase (C 14), N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, trypsin, and valine arylamidase. Whole cell sugars consist of glucose, ribose with traces of mannose and galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC15:0, anteisoC15:0, anteisoC17:0, C16:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, unidentified phospholipids, aminolipid, phosphoglycolipid. The strain was known to produce streptolins A and B, streptothricin, vitamin B12. The genome size of strain DSM 40932T is 7.9 Mb with DNA G+C content of 72.0%.
The type strain DSM 40932T (= ATCC 13741T = ATCC 13793T = CBS 372.58T = ETH 24437T = NRRL B-1354T) is with unknown origin. The GenBank accession number of the assembled draft genome is JAVRFG000000000.1.
Description of Streptomyces hintoniae sp. nov
Streptomyces hintoniae (hin.to'ni.ae. N.L. gen. fem. n. hintoniae, of Hinton, named in honour of Jane Hinton for her contributions to microbiology, particularly for co-developing the Mueller-Hinton agar).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop beige aerial mycelium on DSMZ 535 and GYM media. The strain grows well from 25 to 28°C, optimally at 28°C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate amygdalin, L-arabinose, D-arabitol, arbutin, D-cellobiose, D-fructose, D-fucose, D-galactose, gentiobiose, D-glucose, glycerol, glycogen, inositol, D-lactose, D-maltose, D-mannitol, D-mannose, methyl-α-D-glucopyranoside, N-acetylglucosamine, potassium gluconate, potassium 2-ketogluconate, L-rhamnose, D-ribose, salicin, (bovine origin), starch (amidon), D-trehalose, D-turanose, and D-xylose. Positive for acid phosphatase, alkaline phosphatase, cystine arylamidase, esterase (C 4), esterase lipase (C 8), α-fucosidase, ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, lipase (C 14), α-mannosidase, N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose and ribose with trace of mannose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC14:0, anteisoC15:0, anteisoC17:0, isoC15:0, isoC16:0, C16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, lipid, phospholipids, phosphoglycolipid. The genome size of strain DSM 41014T is 9.0 Mb with DNA G+C content of 72.0%.
The type strain DSM 41014T (= IMRU 3066T = KCTC 59176T) was isolated from Sphagnum pots with unknown origin. The GenBank accession number of the assembled draft genome is JAVRFF000000000.1.
Description of Streptomyces asiaticus subsp. ignotus subsp. nov
Streptomyces asiaticus subsp. ignotus (i.gno'tus. L. masc. adj. ignotus, unknown, referring to the unknown origin of the strain).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop grey (on ISP1 medium), white- (ISP3, ISP4, DSMZ 65), and white grey aerial mycelium (on ISP7 medium) after 7 days of incubation at 28°C. The strain grows well at 25°C, 28°C, 35°C to 37°C, optimally at 28°C, from pH 5.5 to 9.0, optimally at pH 7.0. The strain is able to assimilate L-arabinose, glycerol, D-ribose, D-galactose, D-glucose, D-fructose, D-mannose, L-rhamnose, inositol, D-mannitol, methyl-a-D-mannopyranoside, methyl-a-D-glucopyranoside, N-acetylglucosamine, salicin, D-cellobiose, D-trehalose, D-raffinose, starch (amidon), glycogen, gentiobiose, potassium gluconate, and potassium 2-ketogluconate. The strain shows positive reactions for alkaline phosphatase, esterase lipase (C 8), lipase (C 14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, a-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, ß-galactosidase, ß-glucuronidase, and N-acetyl-ß-glucosaminidase. The whole cell sugars consist of glucose, mannose, and ribose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC14:0, anteisoC15:0, C16:0, isoC15:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phosphatidylmethylethanolamine, and unidentified lipids, phospholipids, and phosphoglycolipids. The genome size of strain DSM 41524T is 11.4 Mb with DNA G+C content of 75%.
The type strain DSM 41524T (= ATCC 15166T = A10598T) was isolated from soil of unknown origin. The GenBank accession number of the assembled draft genome is JAZBJO000000000.
Description of Streptomyces mooreae sp. nov
Streptomyces mooreae (moore'ae. N.L. gen. fem. n. mooreae, of Moore, in honour of Ruth Ella Moore (1903-1994) for her great work in the field of bacteriology).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white greyish to grey aerial mycelium on ISP1, ISP2, ISP3, ISP4, ISP5, ISP6, and ISP7 media that turn to have white to white beige colour on TSA, GYM, and NZ amine media. The strain grows well from 25 to 28°C, optimally at 28°C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-adonitol, D-arabitol, D-galactose, D-glucose, glycerol, glycogen, L-fucose, inositol, D-maltose, D-mannitol, D-mannose, D-melibiose, N-acetylglucosamine, potassium gluconate, D-raffinose, D-ribose, D-saccharose (sucrose), starch (amidon), D-trehalose, D-turanose, and D-xylose. Positive for acid phosphatase, alkaline phosphatase, esterase lipase (C 8), ß-galactosidase, leucine arylamidase, N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase.
Whole cell sugars consist of glucose, ribose, with traces of mannose and galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC15:0, anteisoC15:0, anteisoC17:0, isoC17:0, C16:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylmethylethanolamine, phosphatidylinositol, unidentified phospholipids, lipids, and phosphoglycolipid. The strain is known as producer of bialaphos. The genome size of strain DSM 41527T is 8.4 Mb with DNA G+C content of 71.1%.
The type strain DSM 41527T (= ATCC 21705T = SF-1293T) is with unknown origin. The GenBank accession number of the assembled draft genome is JAVRFE000000000.1.
Description of Streptomyces bugieae sp. nov
Streptomyces bugieae (bu'gie.ae. N.L. gen. fem. n. bugieae, of Bugie, in honour of Elizabeth Bugie for her role in the discovery of streptomycin, the first antibiotic effective against tuberculosis).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white-greyish (on ISP1, ISP2, ISP3 media), white (ISP4, ISP5, ISP6, ISP7, DSMZ 535, DSMZ 65), and white pinkish aerial mycelium (on DSMZ 553) after 7 days of incubation at 28 °C. The strain grows well at 28°C, 35 °C to 37 °C, optimally at 28 °C, from pH 5.5 to 9.0, optimally at pH 7.0. The strain is able to assimilate D-arabitol, glycerol, D-ribose, D-adonitol, D-galactose, D-glucose, D-fructose, D-mannose, D-mannitol, D-sorbitol, N-acetylglucosamine, D-maltose, D-melibiose, D-saccharose, D-trehalose, D-melezitose, D-raffinose, starch (amidon), glycogen, xylitol, D-turanose, and potassium gluconate. The strain shows positive reactions for alkaline phosphatase, esterase (C 4), esterase lipase (C 8), lipase (C 14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, a-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, ß-galactosidase, ß-glucuronidase, α-glucosidase, ß-glucosidase, and N-acetyl-ß-glucosaminidase. Whole cell sugars consist of galactose, glucose, mannose, and ribose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, anteisoC17:0, isoC17:0, C16:0, isoC15:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phosphatidylmethylethanolamine, unidentified lipids, phospholipid, and phosphoglycolipid, aminolipid. The strain was found to produce amylase. The genome size of strain DSM 41528T is 9.0 Mb with DNA G+C content of 71.7%.
The type strain, DSM 41528T (= ATCC 21722T = FERM-P 602T = SF-1084T) is with unknown origin. The GenBank accession number of the assembled draft genome is JAZBJP000000000.
Description of Streptomyces lonegramiae sp. nov
Streptomyces lonegramiae (lo.ne.gra'mi.ae. N.L. gen. fem. n. lonegramiae, of Lone Gram, in honour of Lone Gram for her significant contributions to microbial secondary metabolite research).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white aerial mycelium on ISP1, ISP2, ISP4, ISP7, TSA, and GYM media that turn to have light grey colour on ISP3 medium. The strain grows well from 25 to 28 °C, optimally at 28 °C, from pH 6.0 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-adonitol, amygdalin, D- and L-arabinose, D-arabitol, arbutin, D-cellobiose, D-fructose, L-fucose, D-galactose, gentiobiose, D-glucose, glycerol, glycogen, inositol, D-lactose (bovine origin), D-maltose, D-mannitol, D-mannose, D-melibiose, N-acetylglucosamine, potassium gluconate, potassium 2-ketogluconate, potassium 5-ketogluconate, D-raffinose, L-rhamnose, D-ribose, D-saccharose (sucrose), salicin, starch (amidon), and D-xylose. Positive for acid phosphatase, alkaline phosphatase, leucine arylamidase, N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, with traces of mannose, and galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, anteisoC17:0, C16:0, isoC15:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phosphatidylmethylethanolamine, unidentified phosphoglycolipid, and phospholipid. The strain was known to produce antibiotic A-130. The genome size of strain DSM 41529T is 11.9 Mb with DNA G+C content of 71.2%.
The type strain DSM 41529T (= FERM-P 639T = ATCC 21840T = A-130T) was isolated from a soil collected in Ikeda City in Japan. The GenBank accession number of the assembled draft genome is JAVRFD000000000.1.
Description of Streptomyces antimycoticus subsp. sporoclivatus (Preobrazhenskaya 1986 ex Krassilnikov 1970) comb. nov
Streptomyces antimycoticus subsp. sporoclivatus (spo.ro.cli'va.tus. N.L. masc. adj. clivatus; N.L. masc. adj. sporoclivatus).
Basonym: Streptomyces sporoclivatus (ex Krassilnikov 1970) Preobrazhenskaya 1986
The description is the same as provided by Gauze et al. 1983 and Komaki and Tamura (2020).
The genome size of strain NBRC 100767T is 11.0 Mb with DNA G+C content of 70.9%.
The type strain DSM 41461T = ATCC 43693T = INMI 97T = JCM 9094T = NBRC 100767T = VKM Ac-315T was isolated from soil. The GenBank accession number of the assembled genome is AP019620.1.
Description of Streptomyces edwardsiae sp. nov
Streptomyces edwardsiae (ed.ward'si.ae. N.L. gen. fem. n. edwardsiae, of Edwards, in honour of Katrina Jane Edwards for her significant contributions to soil microbiology).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop grey aerial mycelium on ISP1, ISP2, and GYM media. The strain grows well from 20 to 37 °C, optimally at 28 °C, from pH 5 to 8.5, optimally at pH 7.0. The strain is able to assimilate L-arabinose, D-arabitol, D-cellobiose, D-fructose, D-glucose, glycerol, glycogen, inositol, D-lactose (bovine origin), D-maltose, D-mannitol, methyl-α-D-glucopyranoside, N-acetylglucosamine, potassium gluconate, L-rhamnose, D-ribose, starch (amidon) and D-trehalose. Positive for alkaline phosphatase, esterase (C 4), esterase lipase (C 8), leucine arylamidase, lipase (C 14), and valine arylamidase.
Whole cell sugars consist of glucose, ribose with traces of galactose and mannose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, anteisoC17:0, C16:0, isoC16:0, isoC15:0, C16:1 cis9. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phospholipids, and phosphoglycolipid. The genome size of strain DSM 41636T is 7.9 Mb with DNA G+C content of 71.2%.
The type strain DSM 41636T (= 31-A2T = KCTC 59179T) was isolated from a rose root, Rosa laxa, collected in Germany. The GenBank accession number of the assembled draft genome is JAVRFA000000000.1.
Description of Streptomyces doebereineriae sp. nov
Streptomyces doebereineriae (doe.be.rei.ne'ri.ae. N.L. gen. fem. n. doebereineriae, of Döbereiner, in honour of Johanna Döbereiner (1924–2000) for her significant contributions in the field of plant-microbe interaction).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white aerial mycelium on ISP2, TSA, and NZ amine agar media that turn to have lavender colour on ISP3, ISP4, ISP5, ISP7, and GYM media. The strain grows well from 25 to 45 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-adonitol, L- and D-arabinose, D-arabitol, D-cellobiose, D-fructose, L-fucose, D-galactose, gentiobiose, D-glucose, glycerol, glycogen, inositol, inulin, D-lactose, D-maltose, D-mannitol, D-mannose, D-melibiose, N-acetylglucosamine, potassium gluconate, potassium 5-ketogluconate, D-raffinose, L-rhamnose, D-ribose, D-saccharose, starch, and D-xylose. Positive for acid phosphatase, alkaline phosphatase, cystine arylamidase, esterase (C 4), esterase lipase (C 8), ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, lipase (C 14), naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, rhamnose with traces of mannose and galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC15:0, anteisoC15:0, anteisoC17:0, C16:0, isoC16:0, C16:1 cis9, isoC17:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phospholipids, phosphoglycolipid. The genome size of strain DSM 41640T is 11.2 Mb with DNA G+C content of 70.0%.
The type strain DSM 41640T (= 13-A30T = KCTC 59177T) was isolated from a rose root, Rosa laxa, collected in Germany. The GenBank accession number of the assembled draft genome is JAVREZ000000000.1.
Description of Streptomyces gibsoniae sp. nov
Streptomyces gibsoniae (gib.so'ni.ae. N.L. gen. fem. n. gibsoniae, of Gibson, named in honour of Jane Gibson for her great contributions in the field of microbial physiology and biochemistry).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop grey, white, and light grey aerial mycelium on ISP2, ISP3, and GYM media, respectively. The strain grows well from 25 °C to 37 °C, optimally at 28 °C, from pH 5.5 to 8.5, optimally at pH 7.0. The strain is able to assimilate D-arabitol, D-cellobiose, D-fructose, D-galactose, gentiobiose, D-glucose, glycerol, glycogen, inositol, D-mannitol, D-mannose, N-acetylglucosamine, potassium gluconate, potassium 5-ketogluconate, L-rhamnose, D-ribose, starch (amidon), and D-trehalose. Positive for acid phosphatase, alkaline phosphatase, cystine arylamidase, α-fucosidase, α-galactosidase, ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, α-mannosidase, N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, and mannose with traces of galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC15:0, anteisoC15:0, anteisoC17:0, isoC17:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, unidentified phospholipids, aminolipid, and phosphoglycolipid. The genome size of strain DSM 41699T is 10.2 Mb with DNA G+C content of 70.6%.
The type strain, DSM 41699T (= ATB-26T = KCTC 59183T) was isolated from soil sample collected in Tasek Bera, Malaysia. The GenBank accession number of the assembled draft genome is JAVREY000000000.1.
Description of Streptomyces salyersiae sp. nov
Streptomyces salyersiae (sa.ly.er'si.ae. N.L. gen. fem. n. salyersiae, of Salyers, in honour of Abigail A. Salyers (1942–2013) known as the mother of microbiome research).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop grey aerial mycelium on ISP1, ISP2, ISP5, ISP6, ISP7, DSMZ 535, GYM, and NZ amine media. The strain grows well from 20 to 37 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate amygdalin, L-arabinose, D-arabitol, D-cellobiose, D-fructose, gentiobiose, D-glucose, glycerol, glycogen, D-maltose, D-mannitol, D-mannose, N-acetylglucosamine, potassium gluconate, L-rhamnose, D-ribose, starch (amidon), D-trehalose, D-turanose, and D-xylose. Positive for esterase lipase (C 8), ß-galactosidase, α-glucosidase, leucine arylamidase, and lipase (C 14). Whole cell sugars consist of glucose, ribose, rhamnose with traces of mannose. The peptidoglycan contains LL-A2pm and trace of DL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, anteisoC17:0, C16:0, isoC15:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phospholipids, aminolipid, phosphoglycolipid. The genome size of strain DSM 41770T is 11.2 Mb with DNA G+C content of 70.0%.
The type strain DSM 41770T (= 157/96T = KCTC 59182T) was isolated from water collected from a damaged gypsum liner in a children`s day care centre in Finland. The GenBank accession number of the assembled draft genome is JAVREX000000000.1.
Description of Streptomyces evansiae sp. nov
Streptomyces evansiae (e.van'si.ae. N.L. gen. fem. n. evansiae, of Evans, in honour of Alice Catherine Evans, known for her great bacteriological work on milk and cheese and for discovering that Bacillus abortus caused Brucellosis in animals and humans, which resulted in the pasteurization of milk).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white aerial mycelium on ISP3, ISP5, and DSMZ 535 media. The strain grows well from 25 °C to 37 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate amygdalin, D-arabitol, arbutin, D-cellobiose, D-fructose, L-fucose, D-galactose, D-glucose, glycerol, D-lactose (bovine origin), D-mannitol, D-mannose, methyl-α-D-glucopyranoside, methyl-ß-D-xylopyranoside, N-acetylglucosamine, potassium gluconate, L-rhamnose, salicin, D-trehalose, and D-xylose. Positive for acid phosphatase, alkaline phosphatase, α-chymotrypsin, esterase (C 4), esterase lipase (C 8), ß-galactosidase, α-glucosidase, ß-glucosidase, ß-glucuronidase, leucine arylamidase, lipase (C 14), naphthol-AS-BI-phosphohydrolase, and valine arylamidase.
Whole cell sugars consist of glucose, ribose, with traces of mannose and galactose. The peptidoglycan contains LL-A2pm and trace of LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, isoC15:0, C16:0, isoC16:0, anteisoC17:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, unidentified phospholipids, phosphoaminolipid, phosphoglycolipid. The genome size of strain DSM 41979T is 7.5 Mb with DNA G+C content of 73.3%.
The type strain DSM 41979T (= SA3-ActFT = KCTC 59185T) was isolated from Sirex noctilio collected in the USA. The GenBank accession number of the assembled draft genome is JAVRET000000000.1.
Description of Streptomyces johnsoniae sp. nov
Streptomyces johnsoniae (john.so'ni.ae. N.L. gen. fem. n. johnsoniae, of Johnson, in honour of Mattiedna Johnson (1918–2003) for her significant contributions in antibiotic research).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop a weak white aerial mycelium on ISP1 agar media that is absent in NZ amine medium. The strain grows from 20 to 45 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is positive for acid phosphatase, alkaline phosphatase, ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, α-mannosidase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, with traces of mannose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC16:0, isoC16:1 cis9, anteisoC17:0, anteisoC17:1 cis9. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, unidentified phospholipids, and phosphoglycolipid. The genome size of strain DSM 41886T is 7.5 Mb with DNA G+C content of 72.5%.
The type strain DSM 41886T (= CNB 984T = KCTC 59171T) was isolated from marine sediment collected in the USA and known as cyclomarin A producer. The GenBank accession number of the assembled draft genome is JAVREV000000000.1
Description of Streptomyces dubilierae sp. nov
Streptomyces dubilierae (du.bi.li'er.ae. N.L. gen. fem. n. dubilierae, of Dubilier, in honour of Nicole Dubilier for her significant contributions to the ecological and evolutionary symbiosis of microbes).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop yellow grey aerial mycelium on ISP1 and ISP2 media that turn to have light brown colour on ISP3 and GYM media. The strain grows well from 25 to 50 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-adonitol, amygdalin, D- and L-arabinose, D-arabitol, arbutin, D-cellobiose, D-fructose, L-fucose, D-galactose, gentiobiose, D-glucose, glycerol, glycogen, inositol, D-maltose, D-mannitol, D-mannose, D-melibiose, methyl-α-D-glucopyranoside, methyl-α-D-mannopyranoside, N-acetylglucosamine, potassium gluconate, D-raffinose, L-rhamnose, D-ribose, D-saccharose (sucrose), salicin, starch (amidon), D-trehalose and D-xylose. Positive for acid phosphatase, alkaline phosphatase, esterase lipase (C 8), ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, lipase (C 14), N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, with traces of mannose and galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC14:0, isoC15:0, anteisoC15:0, anteisoC17:0, C16:0, isoC16:0, C16:1 cis9. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phospholipids, aminolipid, phosphoglycolipid. The genome size of strain DSM 41921T is 8.2 Mb with DNA G+C content of 72.4%.
The type strain DSM 41921T (= S6T = KCTC 59180T) was isolated from rhizospherical soil of Vitis vinifera, collected in Morocco. The GenBank accession number of the assembled draft genome is JAVREU000000000.1.
Description of Streptomyces althioticus subsp. attaecolombicae subsp. nov
Streptomyces althioticus subsp. attaecolombicae (at.tae.co.lom'bi.cae. N.L. gen, fem. n. attaecolombicae, of Atta colombica, the leafcutter ant species from which the strain was isolated).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium with well-developed light grey (ISP1, ISP3 media), white (on ISP4, ISP5, ISP7 medium), light grey (ISP6), and yellow (DSMZ 535 and 65, NZ-amine media) aerial after 7 days of incubation at 28 °C. The strain grows well at 28 °C, 35 °C to 37 °C, optimally at 28 °C, from pH 5.0 to 9.0, optimally at pH 7.0. The strain is able to assimilate a wide range of carbon source such as glycerol, L-arabinose, D-xylose, D-glucose, D-fructose, D-mannose, L-rhamnose, inositol, D-mannitol, N-acetylglucosamine, esculin / ferric citrate, salicin, D-cellobiose, D-maltose, D-trehalose, starch (amidon), glycogen, gentiobiose, L-arabitol, and potassium gluconate. The strain has enzymatic profile contains alkaline phosphatase, esterase (C 4), lipase (C 14), esterase lipase (C 8), leucine arylamidase, valine arylamidase, cystine arylamidase, and trypsin. The whole cell sugars consist of glucose and ribose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, anteisoC17:0, C16:0, isoC15:0, isoC16:0, C15:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phosphoglycolipid, unidentified aminolipid, phospholipid, and phosphoaminolipid. The genome size of strain DSM 41972T is 6.64 Mb with DNA G+C content of 72.2%.
The type strain DSM 41972T (= Av26–2T = DI-188T = KCTC 59178T) was isolated from Atta colombica refuse dump collected in Panama. The GenBank accession number of the assembled draft genome is JAVSGH000000000.
Description of Streptomyces doudnae sp. nov
Streptomyces doudnae (do.ud'nae. N.L. gen. fem. N. doudnae, of Doudna, in honour of Jennifer A. Doudna for her great contributions in biochemistry, genetics, and microbiology. She had a Nobel Prize (2020) in Chemistry for her pioneering work in the field of CRISPR genome editing).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white aerial mycelium on ISP2, TSA and NZ amine agar media that turn to have lavender colour on ISP3, ISP4, ISP5, ISP7, GYM media. The strain grows well from 25 to 45 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate L- and D-arabinose, D-arabitol, D-cellobiose, D-fructose, L-fucose, D-galactose, D-glucose, glycerol, glycogen, inositol, D-maltose, D-mannitol, D-mannose, N-acetylglucosamine, L-rhamnose, D-ribose, starch, D-trehalose, and D-xylose. Positive for acid phosphatase, alkaline phosphatase, cystine arylamidase, α-fucosidase, ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, lipase (C 14), α-mannosidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, and traces of mannose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) isoC14:0, isoC15:0, anteisoC15:0, anteisoC17:0, C16:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phospholipids, phosphoaminolipid, phosphoglycolipids. The genome size is 9.2 Mb with DNA G+C content of 72.9%.
The type strain DSM 41981T (= Sol5a-2T = KCTC 59175T) was isolated from a solitary wasp collected in Panama. The GenBank accession number of the assembled draft genome is JAVRES000000000.1.
Description of Streptomyces hazeniae sp. nov
Streptomyces hazeniae (ha.ze'ni.ae. N.L. gen. fem. n. hazeniae, of Hazen, in honour of Elizabeth Lee Hazen (1885–1975) for her great contributions to microbiology and the development of nystatin).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium with no aerial mycelium on ISP1, ISP2, ISP3, ISP4, ISP5, ISP7, and GYM media. The strain grows well from 17°C to 37 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-arabitol, D-cellobiose, D-fructose, gentiobiose, D-glucose, glycogen, D-maltose, D-mannitol, D-mannose, D-melibiose, D-raffinose, D-ribose, D-saccharose (sucrose), starch (amidon), D-turanose, and D-xylose. Positive for acid phosphatase, alkaline phosphatase, esterase (C 4), esterase lipase (C 8), α-glucosidase, leucine arylamidase, α-mannosidase, N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of glucose, ribose, galactose with trace of mannose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, isoC15:0, anteisoC17:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, and unidentified phospholipids. The genome size of strain DSM 42041T is 6.4 Mb with DNA G+C content of 72.8%.
The type strain DSM 42041T (= SCSIO 10374T = KCTC 59186T) was isolated from gorgonian coral in China. The GenBank accession number of the assembled draft genome is JAVREQ000000000.1.
Description of Streptomyces chisholmiae sp. nov
Streptomyces chisholmiae (chis.hol'mi.ae. N.L. gen. fem. n. chisholmiae, of Chisholm, in honour of Sallie ‘’Penny’’ W. Chisholm for her significant contributions to the ecology and evolution of ocean microbes.).
Gram-stain positive, aerobic actinobacterium with branched substrate mycelium with white and lavender aerial mycelium on ISP1, DSMZ 535, and ISP3 media, respectively. Red brown pigment was diffused produced on ISP7 medium. The strain grows well from 20 to 42 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-adonitol, amygdalin, L-arabinose, D-arabitol D-cellobiose, D-galactose, D-glucose, glycerol, D-lactose (bovine origin), D-maltose, D-mannitol, D-mannose, N-acetylglucosamine, L-rhamnose, D-ribose, starch (amidon), D-trehalose, and D-xylose. Positive for alkaline phosphatase, esterase lipase (C 8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, trypsin, and valine arylamidase. Whole cell sugars consist of glucose, ribose, with traces of mannose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC17:0, isoC16:0, isoC16:1 cis4, C15:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phospholipids, lipid, phosphoglycolipid. The genome size of strain DSM 44915T is 7.5 Mb with DNA G+C content of 73.7%.
The type strain DSM 44915T (= SCRIPP CNJ 962T = KCTC 59194T) was isolated from marine sediment collected in Palau. The GenBank accession number of the assembled draft genome is JAVREO000000000.1
Description of Streptomyces boetiae sp. nov
Streptomyces boetiae (bo.e'ti.ae. N.L. gen. fem. n. boetiae, of Boetius, in honour of Antje Boetius for her outstanding contributions to marine microbiology, formed under the assumption that Boetius is a Latin or Latinized name).
Gram-stain positive, aerobic actinobacterium with branched substrate mycelium with white and lavender aerial mycelium on ISP1, ISP2, TSBA and ISP3, ISP4, ISP7, and GYM media, respectively. White griseus aerial mycelium was developed on NZ amine agar medium. No diffusible pigment was produced. The strain grows well from 20 to 42 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-fructose, glycerol, glycogen, potassium gluconate, and D-trehalose. Positive for alkaline phosphatase, cystine arylamidase, esterase (C 4), esterase lipase (C 8), α-glucosidase, leucine arylamidase, lipase (C 14), and valine arylamidase. Whole cell sugars consist of glucose and ribose with traces of mannose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, isoC16:1 cis9, anteisoC17:0, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phosphatidylmethylethanolamine, unidentified phospholipids, phosphoglycolipid and phosphoaminolipid. The genome size of strain DSM 44917T is 6.0 Mb with DNA G+C content of 74.7%.
The type strain DSM 44917T (= SCRIPP CNQ 259T = KCTC 59173T) was isolated from marine sediment collected in the USA. The GenBank accession number of the assembled draft genome is JAVREN000000000.1.
Description of Streptomyces millisiae sp. nov
Streptomyces millisiae (mil.li'si.ae. N.L. gen. fem. n, millisiae. of Millis, in honour of Nancy F. Millis (1922–2012) who introduced applied microbiology and promoted wastewater microbiology in Australia).
Gram-stain positive, aerobic actinobacterium with branched substrate mycelium with grey brownish to brown colour on ISP2 and ISP3 media, respectively. Red brown pigment was diffused produced on ISP7 medium. The strain grows well from 25 to 45 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-adonitol, arbutin, D-maltose, D-mannitol, D-melibiose, methyl-α-D-glucopyranoside, L-rhamnose, salicin, and D-trehalose. Positive for acid phosphatase, alkaline phosphatase, α-galactosidase, ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, α-mannosidase, naphthol-AS-BI-phosphohydrolase, trypsin, and valine arylamidase. Whole cell sugars consist of glucose, ribose, with traces of mannose and galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, C16:1 cis9, C16:0, anteisoC17:0, anteisoC17:1 cis9, isoC16:0. Polar liprikhoid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, phosphatidylmethylethanolamine, unidentified phospholipids, and phosphoglycolipid. The genome size of strain DSM 44918T is 7.6 Mb with DNA G+C content of 72.4%.
The type strain DSM 44918T (= SCRIPP CNQ 703T = KCTC 59174T) was isolated from marine sediment collected in Guam, USA. The GenBank accession number of the assembled draft genome is JAVREM000000000.1
Description of Streptomyces litchfieldiae sp. nov
Streptomyces litchfieldiae (litch.fiel'di.ae. N.L. gen. fem. n. litchfieldiae, of Litchfield, in honour of Carol Ann Darlene Litchfield (1936–2012) for her great contributions to the microbiology of oceans and extreme environments).
Gram-stain positive, aerobic actinobacterium with branched substrate mycelium with lavender aerial mycelium on ISP3, ISP4, ISP5, ISP6, ISP7, and GYM media. White aerial mycelium develops on ISP2 and TSBA media. No diffusible pigment was produced on the tested media. The strain grows well from 20 to 37 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D-adonitol, D- and L-arabinose, D-arabitol, D-cellobiose, D-fructose, L-fucose, D-galactose, gentiobiose, D-glucose, glycerol, glycogen, inositol, D-lactose (bovine origin), D-maltose, D-mannitol, D-mannose, D-melibiose, N-acetylglucosamine, potassium gluconate, D-raffinose, L-rhamnose, D-saccharose (sucrose), salicin, starch (amidon), D-trehalose, and D-xylose. Positive for acid phosphatase, alkaline phosphatase, α-chymotrypsin, cystine arylamidase, esterase (C 4), esterase lipase (C 8), α-fucosidase, α-galactosidase, ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, α-mannosidase, N-acetyl-ß-glucosaminidase, naphthol-AS-BI-phosphohydrolase, trypsin, and valine arylamidase. Whole cell sugars consist of glucose, ribose, with traces of mannose and galactose. The peptidoglycan contains LL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, anteisoC17:0, anteisoC17:1 cis9, isoC16:1 cis9, isoC16:0. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, unidentified lipid, phospholipid, aminolipid, phosphoglycolipid. The genome size of strain DSM 44938T is 7.6 Mb with DNA G+C content of 72.4%.
The type strain DSM 44938T (= SCRIPP CNR954T = KCTC 59172T) was isolated from marine sediment collected in Bahamas. The GenBank accession number of the assembled draft genome is JAVREL000000000.1.
Description of Blastococcus goldschmidtiae sp. nov
Blastococcus goldschmidtiae (gold.schmid'ti.ae. N.L. gen. fem. n. goldschmidtiae, of Goldschmidt, in honour of Millicent C. Goldschmidt for her great contributions in the field of microbiology in particular in rapid testing methods for detecting microbial contaminants).
Gram-stain positive, aerobic actinobacterium with rod-, coccoid and non-motile cells forming orange colonies on ISP2, TSA, and GYM media. The strain grows well from 25 to 37 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. Positive for acid phosphatase, alkaline phosphatase, α-chymotrypsin, cystine arylamidase, esterase (C 4), esterase lipase (C 8), ß-glucosidase, leucine arylamidase, naphthol-AS-BI-phosphohydrolase, trypsin, and valine arylamidase. Whole cell sugars consist of glucose, mannose, and ribose. The fatty acids profile comprises (>5%) isoC16:1 cis9, isoC16:0, C17:1 cis9, C18:1 cis9. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylcholine, phosphatidylinositol, unidentified lipid, glycolipids. The genome size of strain DSM 46792T is 4.3 Mb with DNA G+C content of 73.3%.
The type strain DSM 46792T (= 7CT = BC543T = KCTC 59190T) was isolated from marble collected in Italy. The GenBank accession number of the assembled draft genome is JAVREI000000000.1.
Description of Nocardiopsis lambiniae sp. nov
Nocardiopsis lambiniae (lam.bi'ni.ae. N.L. gen. fem. n. lambiniae, of Lambin, in honour of Suzanne Lambin (1902–2008) for her great work on the evolution of bacterial cultures).
Gram-stain positive, aerobic actinobacterium with branched substrate mycelium with light brown and orange colonies on ISP1 and ISP2 media, respectively. White aerial mycelium develops on ISP6, TSBA, and DSMZ 83 media. No diffusible pigment was produced on the tested media. The strain grows well from 20 to 37 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. Positive for acid phosphatase, alkaline phosphatase, α-chymotrypsin, esterase (C 4), esterase lipase (C 8), α-glucosidase, ß-glucosidase, leucine arylamidase, lipase (C 14), N-acetyl-ß-glucosaminidase, and valine arylamidase. Whole cell sugars consist of ribose. The peptidoglycan contains DL-A2pm. The fatty acids profile comprises (>5%) anteisoC15:0, isoC16:0, anteisoC17:0, C17:1 cis9, C18:1 cis9, C18:2 cis9,12. Polar lipid pattern consists of phosphatidylcholine, phosphtidylglycerol, phosphatidylmethylethanolamine, phospholipids. The genome size of strain DSM 44743T is 6.2 Mb with DNA G+C content of 71.3%.
The type strain DSM 44743T (= DCDM15A35T = KCTC 59192T) was isolated from soil collected in China. The GenBank accession number of the assembled draft genome is JAVREP000000000.1.
Description of Pseudonocardia charpentierae sp. nov
Pseudonocardia charpentierae (char.pen'tie.rae. N.L. gen. fem. n. charpentierae, of Charpentier, in honour of Emmanuelle Charpentier for her great contributions to microbiology, genetics and biochemistry. She had a Nobel Prize (2020) in Chemistry for her pioneering work in the field of CRISPR genome editing).
Gram-stain positive, aerobic non-motile actinobacterium with orange colonies that shows a good growth on GYM media. The strain grows well from 25 to 37 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The strain is able to assimilate D- and L-arabitol, gentiobiose, glycogen, D-lyxose, N-acetylglucosamine, D-melibiose, potassium gluconate, D-saccharose (sucrose), starch (amidon), and D-turanose. Positive for acid phosphatase, esterase (C 4), esterase lipase (C 8), α-galactosidase, ß-galactosidase, α-glucosidase, ß-glucosidase, leucine arylamidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase. Whole cell sugars consist of arabinose, galactose, glucose, mannose and ribose. The peptidoglycan contains DL-A2pm. The fatty acids profile comprises (>5%) C16:0, isoC16:0, C16:1 cis9, C16:0 10 methyl. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylcholine, phosphatidylinositol, phosphatidylethanolamine, phsophatidylmethylethanolamine, unidentified glycolipid, glycophospholipid, phospholipids, phosphoglycoaminolipid. The genome size of strain DSM 45834T is 6.7 Mb with DNA G+C content of 71.9%.
The type strain DSM 45834T (= CPCC 203558T = KCTC 59187T) was isolated from a desert sand, collected in China. The GenBank accession number of the assembled draft genome is JAVREJ000000000.1.
Description of Streptomonospora wellingtoniae sp. nov
Streptomonospora wellingtoniae (wel.ling.to'ni.ae. N.L. gen. fem. n. of wellingtoniae, in honour of Elizabeth Wellington for her great contributions to microbiomes and environmental microbiology).
Gram-stain positive, aerobic actinobacterium with white beige colonies that develop on ISP1 and nutrient agar media. Weak growth without aerial mycelium of the strain on ISP2, ISP3, and ISP4. The strain grows well from 25 to 37 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. Whole cell sugars consist of glucose, galactose, and ribose with traces of mannose. The peptidoglycan contains DL-A2pm. The fatty acids profile comprises (>5%) isoC16:1 cis4, isoC16:0, C16:1 cis9, C16:0, anteisoC17:0, TSBA C18:0 10 methyl. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylcholine, glycolipids, phospholipids, and phosphoglycolipid. The genome size of strain DSM 45055T is 5.5 Mb with DNA G+C content of 72.6%.
The type strain DSM 45055T (= CNQ 327T = KCTC 59191T) was isolated from marine sediment collected in the USA. The GenBank accession number of the assembled draft genome is JAVREK000000000.1.
Description of Jatrophihabitans lederbergiae sp. nov
Jatrophihabitans lederbergiae (le.der.ber'gi.ae. N.L. gen. fem. n. lederbergiae, of Lederberg, named in honour of Esther Miriam Zimmer Lederberg (1922–2006) for her great contributions to bacterial genetics and molecular biology).
Gram-stain positive, aerobic actinobacterium with extensively branched substrate mycelium that develop white aerial mycelium on ISP2, TSA, and NZ amine agar media that turn to have lavender colour on ISP3, ISP4, ISP5, ISP7, and GYM media. The strain grows well from 25 to 45 °C, optimally at 28 °C, from pH 5.5 to 8.0, optimally at pH 7.0. The enzymatic profile of the strain consists of leucine arylamidase, valine arylamidase, cystine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, ß-galactosidase, α-glucosidase, and ß-glucosidase. Whole cell sugars consist of glucose, ribose, rhamnose with traces of mannose and galactose. The peptidoglycan contains DL-A2pm. The fatty acids profile comprises (>5%) isoC16:1 cis9, isoC16:0, C17:1 cis9, C17:0 10 methyl, C18:1 cis9, TBSA C18:0 10 methyl. Polar lipid pattern consists of diphosphatidylglycerol, phosphatidylinositol, phosphatidylmethylethanolamine, glycoaminophosphatidylinositol, glycophospholipid, aminolipid, phosphoaminolipid, and phospholipids. The genome size of strain DSM 44399T is 5.2 Mb with DNA G+C content of 68.2%.
The type strain DSM 44399T (= AA-499T = IFAM AA-499T) was isolated from sandstone of Linnaeus Terrace (1600 m), Antarctica. The GenBank accession number of the assembled draft genome is JAVREH000000000.1.
Emended description of Streptomyces asiaticus Sembiring et al. 2001
The description is as stated by Sembiring et al. 2001 with the following modification.
The strain degrades adenine and pectin and grows at 10°C and 45°C. The type strain A14P1T (= DSM 41761T = JCM 11443T = NBRC 100774T = NCIMB 13675T) has a genome size 11.9 Mb with G+C content of 71.0%. The genome sequence is available in the GenBank under accession number NZ_JAGSHX000000000.1.
Author contributions
Phenotypic, genetic, genomic and phylogenetic analyses were carried out by IN, GP, MJ, CS, YM. Chemotaxonomic analyses were performed by MNS, SK, GP, IN. Genome assemblies were curated by IM and JPG-E. In silico and in vitro evaluation of the microbial potential of the strains were carried out by AZ, OH, MD, JPG-E, RM, YM. Cluster networking was performed by JB, UN. RM carried out genetic manipulation of strains and chemical analysis thereof. The manuscript was written by IN and YM. All authors read and approved the final version of the manuscript after contributing on previous versions.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to thank Sabine Gronow, Rüdiger Pukall, Christiane Baschien, and Andrey Yurkov for providing us with strains from the DSMZ culture collection for antimicrobial bioassays. We are grateful to Birgit Grün, Gesa Martens, and Anika Wasner (DSMZ-German culture collection) for excellent technical assistance. We thank Cathrin Spröer, Bettina Ehlert, Orsola Päuker, and Jolantha Swiderski for 16S gene sequencing and sequence analysis. We further thank Heike Freese for her help in naming the newly described type strains.
Funding
We gratefully acknowledge the funding received from the Leibniz Association “Leibniz Cooperative Excellence” programme (project ID: K445/2022).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2024.100290.
Contributor Information
Imen Nouioui, Email: imen.nouioui@dsmz.de.
Yvonne Mast, Email: yvonne.mast@dsmz.de.
Appendix. Supplementary materials
Data availability
The genome sequences of the 36 studied strains have been deposited at DDBJ/ENA/GenBank and the accessions numbers are provided in the manuscript (Table S1).
References
- Achenbach H, Mühlenfeld A, Fauth U, Zähner H. The galbonolides: novel, powerful antifungal macrolides from Streptomyces galbus ssp. eurythermus. Ann. N. Y. Acad. Sci. 1988;Vol. 544:128–140. doi: 10.1111/j.1749-6632.1988.tb40396.x. [DOI] [PubMed] [Google Scholar]
- Ahrens R, Moll G. A new budding bacterium from the Baltic Sea. Arch. Mikrobiol. 1970;70(3):243–265. [PubMed] [Google Scholar]
- Apinya T, Sombatsompop N, Prapagdee B. Selection of a Pseudonocardia sp. RM423 that accelerates the biodegradation of poly(lactic) acid in submerged cultures and in soil microcosms. Int. Biodeterior Biodegrad. 2015;99:23–30. Apr 1. [Google Scholar]
- Atanasov AG, Zotchev SB, Dirsch VM. International natural product sciences taskforce, Supuran CT. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 2021;20(3):200–216. doi: 10.1038/s41573-020-00114-z. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genomic. Sci. 2010;2(1):142–148. doi: 10.4056/sigs.541628. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barke J, Seipke RF, Grüschow S, Heavens D, Drou N, Bibb MJ, et al. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC. Biol. 2010;8:109. doi: 10.1186/1741-7007-8-109. Aug 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman KD, Butler KS, Moore BS, Chekan JR. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 2021;38(11):2100–2129. doi: 10.1039/d1np00032b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennur T, Kumar AR, Zinjarde S, Javdekar V. Nocardiopsis species: Incidence, ecological roles and adaptations. Microbiol. Res. 2015;174:33–47. doi: 10.1016/j.micres.2015.03.010. May. [DOI] [PubMed] [Google Scholar]
- Bergey D.H., Harrison F.C., Breed R.S., Hammer B.W., Huntoon F.M. Vol. 1923. The Williams & Wilkins Company; Baltimore: 1923. https://en.wikipedia.org/wiki/Bergey%27s_Manual_of_Systematic_Bacteriology (Bergey's Manual of Determinative Bacteriology [Internet]). Available from: [Google Scholar]
- Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, Alanjary M, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic. Acids. Res. 2023:gkad344. doi: 10.1093/nar/gkad344. May 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobek J, Šmídová K, Čihák M. A waking review: old and novel insights into the spore germination in Streptomyces. Front. Microbiol. 2017;8:2205. doi: 10.3389/fmicb.2017.02205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braña AF, Sarmiento-Vizcaíno A, Pérez-Victoria I, Otero L, Fernández J, Palacios JJ, et al. Branimycins B and C, antibiotics produced by the Abyssal Actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 2017;80(2):569–573. doi: 10.1021/acs.jnatprod.6b01107. Feb 24. [DOI] [PubMed] [Google Scholar]
- Brocq-Rousseau D. Sur un Streptothrix. Rev. Gen. Bot. 1904;16:219–230. [Google Scholar]
- Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. (Tokyo) 2012;65(8):385–395. doi: 10.1038/ja.2012.27. Aug. [DOI] [PubMed] [Google Scholar]
- Carro L, Nouioui I, Sangal V, Meier-Kolthoff JP, Trujillo ME, Montero-Calasanz M del C, et al. Genome-based classification of Micromonosporae with a focus on their biotechnological and ecological potential. Sci. Rep. 2018;8(1):525. doi: 10.1038/s41598-017-17392-0. Jan 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challinor VL, Bode HB. Bioactive natural products from novel microbial sources. Ann. N. Y. Acad. Sci. 2015;1354:82–97. doi: 10.1111/nyas.12954. Sep. [DOI] [PubMed] [Google Scholar]
- Cui XL, Mao PH, Zeng M, Li WJ, Zhang LP, Xu LH, et al. Streptimonospora salina gen. nov., sp. nov., a new member of the family Nocardiopsaceae. Int. J. Syst. Evol. Microbiol. 2001;51(Pt 2):357–363. doi: 10.1099/00207713-51-2-357. Mar. [DOI] [PubMed] [Google Scholar]
- Daye KJ, Groff JC, Kirpekar AC, Mazumder R. High efficiency degradation of tetrahydrofuran (THF) using a membrane bioreactor: identification of THF-degrading cultures of Pseudonocardia sp. strain M1 and Rhodococcus ruber isolate M2. J. Ind. Microbiol. Biotechnol. 2003;30(12):705–714. doi: 10.1007/s10295-003-0103-8. Dec. [DOI] [PubMed] [Google Scholar]
- de Lima Júnior AA, de Sousa EC, de Oliveira THB, de Santana RCF, da Silva SKR, Coelho LCBB. Genus Streptomyces: recent advances for biotechnological purposes. Biotechnol. Appl. Biochem. 2023;70(4):1504–1517. doi: 10.1002/bab.2455. Aug. [DOI] [PubMed] [Google Scholar]
- Dekker KA, Inagaki T, Gootz TD, Huang LH, Kojima Y, Kohlbrenner WE, et al. New quinolone compounds from Pseudonocardia sp. with selective and potent anti-Helicobacter pylori activity: taxonomy of producing strain, fermentation, isolation, structural elucidation and biological activities. J. Antibiot. (Tokyo) 1998;51(2):145–152. doi: 10.7164/antibiotics.51.145. Feb. [DOI] [PubMed] [Google Scholar]
- Delivoria DC, Chia S, Habchi J, Perni M, Matis I, Papaevgeniou N, et al. Bacterial production and direct functional screening of expanded molecular libraries for discovering inhibitors of protein aggregation. Sci. Adv. 2019;5(10):eaax5108. doi: 10.1126/sciadv.aax5108. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain AL. Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 2014;41(2):185–201. doi: 10.1007/s10295-013-1325-z. Feb. [DOI] [PubMed] [Google Scholar]
- Derosa CT, Wilbur S, Holler J, Richter P, Stevens YW. Health evaluation of 1,4-dioxane [Internet]. 1996 [cited 2024 Jun 11]. Available from: https://journals.sagepub.com/doi/abs/10.1177/074823379601200101. [DOI] [PubMed]
- Embley MT, Smida J, Stackebrandt E. The Phylogeny of Mycolate-less Wall Chemotype IV Actinomycetes and Description of Pseudonocardiaceae fam. nov. Syst Appl Microbiol. 1988;11(1):44–52. Nov 1. [Google Scholar]
- English AR, McBride TJ. PA 150, PA 153, and PA 166: new polyene antifungal antibiotics: biological studies. Antibiot. Annu. 1957;5:893–896. 1958. [PubMed] [Google Scholar]
- Freese HM, Giner-Pérez L, Oren A, Göker M, Arahal DR. The gender gap in names of prokaryotes honouring persons. Int. J. Syst. Evol. Microbiol. 2023;73(11) doi: 10.1099/ijsem.0.006115. [DOI] [PubMed] [Google Scholar]
- Gavriilidou A, Kautsar SA, Zaburannyi N, Krug D, Müller R, Medema MH, et al. Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes. Nat. Microbiol. 2022;7(5):726–735. doi: 10.1038/s41564-022-01110-2. May. [DOI] [PubMed] [Google Scholar]
- Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, et al. Opinion: Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005;3(9):733–739. doi: 10.1038/nrmicro1236. Sep. [DOI] [PubMed] [Google Scholar]
- Goodfellow M, Fiedler HP. A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek. 2010;98(2):119–142. doi: 10.1007/s10482-010-9460-2. Aug. [DOI] [PubMed] [Google Scholar]
- Goodfellow M, Kroppenstedt RM. The family Thermomonosporaceae: Actinomadura and Thermomonospora. Prokaryotes Vol 3 Archaea Bact Firmicutes Actinomycetes [Internet] 2006 https://eprints.ncl.ac.uk/2932 [cited 2024 Jun 10]; Available from: [Google Scholar]
- Goodfellow M, Nouioui I, Sanderson R, Xie F, Bull AT. Rare taxa and dark microbial matter: novel bioactive actinobacteria abound in Atacama Desert soils. Antonie Van Leeuwenhoek. 2018;111(8):1315–1332. doi: 10.1007/s10482-018-1088-7. Aug. [DOI] [PubMed] [Google Scholar]
- Goodfellow M. Bergey's manual® of systematic bacteriology. No Title. 2012;5:376. [Google Scholar]
- Goodfellow M. In: Bergey’s Manual of Systematics of Archaea and Bacteria [Internet] 1st ed. Whitman WB, editor. Wiley; 2015. Streptosporangiales ord. nov; pp. 1–3.https://onlinelibrary.wiley.com/doi/10.1002/9781118960608.obm00019 [cited 2024 Jun 10] Available from: [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57(1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Hahnke RL, Meier-Kolthoff JP, García-López M, Mukherjee S, Huntemann M, Ivanova NN, et al. Genome-based taxonomic classification of Bacteroidetes. Front. Microbiol. 2016;7:2003. doi: 10.3389/fmicb.2016.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssen A. Morphology and system of thermophilic actinomycetes. Arch. Mikrobiol. 1957;26(4):373–414. [PubMed] [Google Scholar]
- Hezbri K, Louati M, Nouioui I, Gtari M, Rohde M, Spröer C, et al. Blastococcus capsensis sp. nov., isolated from an archaeological Roman pool and emended description of the genus Blastococcus, B. aggregatus, B. saxobsidens, B. jejuensis and B. endophyticus. Int. J. Syst. Evol. Microbiol. 2016;66(11):4864–4872. doi: 10.1099/ijsem.0.001443. Nov. [DOI] [PubMed] [Google Scholar]
- Hoffmann T, Krug D, Bozkurt N, Duddela S, Jansen R, Garcia R, et al. Correlating chemical diversity with taxonomic distance for discovery of natural products in Myxobacteria. Nat. Commun. 2018;9(1):803. doi: 10.1038/s41467-018-03184-1. Feb 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong HJ, Hutchings MI, Hill LM, Buttner MJ. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J. Biol. Chem. 2005;280(13):13055–13061. doi: 10.1074/jbc.M413801200. Apr 1. [DOI] [PubMed] [Google Scholar]
- Hozzein WN, Tujillo ME., Genus I. Bergey’s Manual of Systematic bacteriology, The Actinobacteria, Parts A and B. 2nd edn. Springer; New York: 2012. Nocardiopsis Meyer 1976, 487AL; pp. 1–2083. [Google Scholar]
- Huang Y, Goodfellow M., Genus I. Pseudonocardia Henssen 1957, 408AL emend. Park, Park, Lee and Kim 2008, 2477. Bergey’s Man Syst. Bacteriol. 2012;5:1305–1323. [Google Scholar]
- Hugenholtz P, Chuvochina M, Oren A, Parks DH, Soo RM. Prokaryotic taxonomy and nomenclature in the age of big sequence data. ISMe J. 2021;15(7):1879–1892. doi: 10.1038/s41396-021-00941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodamoradi S, Hahnke RL, Mast Y, Schumann P, Kämpfer P, Steinert M, et al. Streptomonospora litoralis sp. nov., a halophilic thiopeptides producer isolated from sand collected at Cuxhaven beach. Antonie Van Leeuwenhoek. 2021;114(10):1483–1496. doi: 10.1007/s10482-021-01609-4. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Vol. 291. John Innes Foundation Norwich; 2000. (Practical Streptomyces Genetics). [Google Scholar]
- Kim HJ, Kang SH, Choi SS, Kim ES. Redesign of antifungal polyene glycosylation: engineered biosynthesis of disaccharide-modified NPP. Appl. Microbiol. Biotechnol. 2017;101(12):5131–5137. doi: 10.1007/s00253-017-8303-8. Jun. [DOI] [PubMed] [Google Scholar]
- Koaze Y, Nakajima Y, Hidaka H, Yoshida K, Ito J, Niwa T, et al. Production of new amylases by cultivation of Streptomyces and uses of these new amylases [Internet] US3804717A. 1974 https://patents.google.com/patent/US3804717A/en [cited 2024 Jun 10]. Available from: [Google Scholar]
- Koe BK, Tanner FW, Rao KV, Sobin BA, Celmer WD. Pa 150, PA 153, and PA 166: new polyene antifungal antibiotics. Antibiot. Annu. 1957;5:897–905. 1958. [PubMed] [Google Scholar]
- Kohlweyer U, Thiemer B, Schräder T, Andreesen JR. Tetrahydrofuran degradation by a newly isolated culture of Pseudonocardia sp. strain K1. FEMS Microbiol. Lett. 2000;186(2):301–306. doi: 10.1111/j.1574-6968.2000.tb09121.x. May 15. [DOI] [PubMed] [Google Scholar]
- Komaki H, Tamura T. Reclassification of Streptomyces fulvissimus as a later heterotypic synonym of Streptomyces microflavus. Int. J. Syst. Evol. Microbiol. 2020;70(9):5156–5162. doi: 10.1099/ijsem.0.004382. [DOI] [PubMed] [Google Scholar]
- Konkit M, Lumyong S. Poly(lactide) Degradation By Pseudonocardia alni. Chiang Mai J Sci. 2012 [Google Scholar]
- Kämpfer P, Kroppenstedt RM, Dott W. A numerical classification of the genera Streptomyces and Streptoverticillium using miniaturized physiological tests. Microbiology (N. Y) 1991;137(8):1831–1891. [Google Scholar]
- Kämpfer Genus I. The Actinobacteria, Part B Bergey’s Manual of Systematic Bacteriology. 2nd edn. Springer; New York: 2012. Streptomyces Waksman and Henrici 1943, 339AL emend. Rainey, Ward-Rainey and Stackebrandt, 1997, 486, emend. Kim, Lonsdale, Seong and Goodfellow 2003b, 113, emend. Zhi, Li and Stackebrandt 2009, 600; pp. 1455–1767. [Google Scholar]
- Labeda DP, Goodfellow M. Bergey's Manual of Systematics of Archaea and Bacteria [Internet] John Wiley & Sons, Ltd; 2015. Pseudonocardiaceae; pp. 1–7.https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118960608.fbm00044 [cited 2024 Jun 11]Available from: [Google Scholar]
- Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, et al. Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek. 2012;101(1):73–104. doi: 10.1007/s10482-011-9656-0. Jan 1. [DOI] [PubMed] [Google Scholar]
- Labeda DP, Doroghazi JR, Ju KS, Metcalf WW. Taxonomic evaluation of Streptomyces albus and related species using multilocus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int. J. Syst. Evol. Microbiol. 2014;64(Pt_3):894–900. doi: 10.1099/ijs.0.058107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeda DP, Dunlap CA, Rong X, Huang Y, Doroghazi JR, Ju KS, et al. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek. 2017;110(4):563–583. doi: 10.1007/s10482-016-0824-0. Apr 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ADG, MacCoss M, Heer JP. Importance of rigidity in designing small molecule drugs to tackle protein-protein interactions (PPIs) through stabilization of desired conformers. J. Med. Chem. 2018;61(10):4283–4289. doi: 10.1021/acs.jmedchem.7b01120. May 24. [DOI] [PubMed] [Google Scholar]
- Lechevalier MP, Lechevalier HA. The taxonomic position of the actinomycetic endophytes. 1979 [cited 2024 Jun 10]; Available from: https://www.cabidigitallibrary.org/doi/full/10.5555/19821971394.
- Lee JJ, Yoon JH, Yang SY, ST Lee. Aerobic biodegradation of 4-methylpyridine and 4-ethylpyridine by newly isolated Pseudonocardia sp. strain M43. FEMS Microbiol. Lett. 2006;254(1):95–100. doi: 10.1111/j.1574-6968.2005.00019.x. Jan. [DOI] [PubMed] [Google Scholar]
- Lee SD. Blastococcus jejuensis sp. nov., an actinomycete from beach sediment, and emended description of the genus Blastococcus Ahrens and Moll 1970. Int. J. Syst. Evol. Microbiol. 2006;56(10):2391–2396. doi: 10.1099/ijs.0.64268-0. [DOI] [PubMed] [Google Scholar]
- Leopold-Messer S, Chepkirui C, Mabesoone MFJ, Meyer J, Paoli L, Sunagawa S, et al. Animal-associated marine Acidobacteria with a rich natural-product repertoire. Chem. 2023;9(12):3696–3713. Dec. [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WJ, Xu P, Zhang LP, Tang SK, Cui XL, Mao PH, et al. Streptomonospora alba sp. nov., a novel halophilic actinomycete, and emended description of the genus Streptomonospora Cui et al. 2001. Int. J. Syst. Evol. Microbiol. 2003;53(Pt 5):1421–1425. doi: 10.1099/ijs.0.02543-0. Sep. [DOI] [PubMed] [Google Scholar]
- Li S, Tian X, Niu S, Zhang W, Chen Y, Zhang H, et al. Pseudonocardians A–C, new diazaanthraquinone derivatives from a deap-sea actinomycete Pseudonocardia sp. SCSIO 01299. Mar Drugs. 2011;9(8):1428–1439. doi: 10.3390/md9081428. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao GZ, Zhu WY, Huang HY, Xu LH, Zhang S, et al. Streptomyces endophyticus sp. nov., an endophytic actinomycete isolated from Artemisia annua L. Int. J. Syst. Evol. Microbiol. 2013;63(Pt 1):224–229. doi: 10.1099/ijs.0.035725-0. Jan. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang M, Sun ZZ, Xie BB. Comparative genomic insights into the taxonomic classification, diversity, and secondary metabolic potentials of Kitasatospora, a genus closely related to Streptomyces. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.683814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM [Internet]. arXiv; 2013 [cited 2024 Aug 6]. Available from: http://arxiv.org/abs/1303.3997.
- Locci R. The genus Streptoveriticillium. A taxonomic study. Giorn Microbiol. 1969;17:1–60. [Google Scholar]
- Lv M, Zhao J, Deng Z, Yu Y. Characterization of the biosynthetic gene cluster for benzoxazole antibiotics A33853 reveals unusual assembly logic. Chem. Biol. 2015;22(10):1313–1324. doi: 10.1016/j.chembiol.2015.09.005. Oct 22. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Hu CJ, Kim SJ, Weon HY, Kwon SW, Ji L. Jatrophihabitans endophyticus gen. nov., sp. nov., an endophytic actinobacterium isolated from a surface-sterilized stem of Jatropha curcas L. Int. J. Syst. Evol. Microbiol. 2013;63(Pt 4):1241–1248. doi: 10.1099/ijs.0.039685-0. Apr. [DOI] [PubMed] [Google Scholar]
- Mangamuri UK, Vijayalakshmi M, Poda S, Manavathi B, Bhujangarao C, Venkateswarlu Y. Bioactive metabolites produced by Pseudonocardia endophytica VUK-10 from mangrove sediments: isolation, chemical structure determination and bioactivity. J. Microbiol. Biotechnol. 2015;25(5):629–636. doi: 10.4014/jmb.1407.07041. [DOI] [PubMed] [Google Scholar]
- Mangamuri UK, Muvva V, Poda S, Chitturi B, Yenamandra V. Bioactive natural products from Pseudonocardia endophytica VUK-10. J. Genet. Eng. Biotechnol. 2016;14(2):261–267. doi: 10.1016/j.jgeb.2016.10.002. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangamuri UK, Muvva V, Poda S, Manavathi B, Bhujangarao C, Yenamandra V. Chemical characterization & bioactivity of diketopiperazine derivatives from the mangrove derived Pseudonocardia endophytica. Egypt J. Aquat. Res. 2016;42(2):169–175. Jun 1. [Google Scholar]
- Maskey RP, Kock I, Helmke E, Laatsch H. Isolation and Structure Determination of Phenazostatin D, a New Phenazine froma Marine Actinomycete Isolate Pseudonocardia sp. B6273. Z Für Naturforschung B. 2003;58(7):692–694. Jul 1. [Google Scholar]
- Matroodi S, Siitonen V, Baral B, Yamada K, Akhgari A, Metsä-Ketelä M. Genotyping-guided discovery of Persiamycin A from sponge-associated halophilic Streptomonospora sp. PA3. Front Microbiol. 2020;11:1237. doi: 10.3389/fmicb.2020.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFARLAND J. The nephelometer:an instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J. Am. Med. Assoc. 1907;XLIX(14):1176–1178. Oct 5. [Google Scholar]
- Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019;10(1):2182. doi: 10.1038/s41467-019-10210-3. May 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M, Spröer C, Klenk HP. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013;195(6):413–418. doi: 10.1007/s00203-013-0888-4. Jun 1. [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC. Bioinformatics. 2013;14(1):60. doi: 10.1186/1471-2105-14-60. Feb 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, Fiebig A, et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genomic. Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Klenk HP, Göker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014;64(Pt 2):352–356. doi: 10.1099/ijs.0.056994-0. Feb. [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic. Acids. Res. 2022;50(D1):D801–D807. doi: 10.1093/nar/gkab902. Jan 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. Nocardiopsis, a New Genus of the Order Actinomycetales. Int. J. Syst. Evol. Microbiol. 1976;26(4):487–493. [Google Scholar]
- Michel KH, Boeck LD, Hoehn MM, Jones ND, Chaney MO. The discovery, fermentation, isolation, and structure of antibiotic A33853 and its tetraacetyl derivative. J. Antibiot. (Tokyo) 1984;37(5):441–445. doi: 10.7164/antibiotics.37.441. May. [DOI] [PubMed] [Google Scholar]
- Minnikin DE, O'Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods. 1984;2(5):233–241. Jul 1. [Google Scholar]
- Mohammadipanah F, Wink J. Actinobacteria from arid and desert habitats: diversity and biological activity. Front. Microbiol. 2015;6:1541. doi: 10.3389/fmicb.2015.01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Calasanz MDC, Yaramis A, Rohde M, Schumann P, Klenk HP, Meier-Kolthoff JP. Genotype-phenotype correlations within the Geodermatophilaceae. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.975365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24(16):1757–1764. doi: 10.1093/bioinformatics/btn322. Aug 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss CW, Lambert-Fair MA. Location of double bonds in monounsaturated fatty acids of Campylobacter cryaerophila with dimethyl disulfide derivatives and combined gas chromatography-mass spectrometry. J. Clin. Microbiol. 1989;27(7):1467–1470. doi: 10.1128/jcm.27.7.1467-1470.1989. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Anzai H, Imai S, Satoh A, Nagaoka K, Thompson CJ. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: molecular cloning and characterization of the gene cluster. Mol. Gen. Genet. MGG. 1986;205(1):42–53. Oct. [Google Scholar]
- Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, Parkinson EI, et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020;16(1):60–68. doi: 10.1038/s41589-019-0400-9. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ. Natural products as leads to potential drugs: an old process or the new hope for drug discovery? J. Med. Chem. 2008;51(9):2589–2599. doi: 10.1021/jm0704090. May 8. [DOI] [PubMed] [Google Scholar]
- Niel CB van. The classification and natural relationships of bacteria. Cold. Spring. Harb. Symp. Quant. Biol. 1946;11:285–301. Jan 1. [Google Scholar]
- Normand P. Geodermatophilaceae fam. nov., a formal description. Int. J. Syst. Evol. Microbiol. 2006;56(Pt 10):2277–2278. doi: 10.1099/ijs.0.64298-0. Oct. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Sangal V. Advanced prokaryotic systematics: the modern face of an ancient science. New. Microbes. New. Infect. 2022;49–50 doi: 10.1016/j.nmni.2022.101036. Nov 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, Kyrpides NC, et al. Genome-based taxonomic classification of the Phylum Actinobacteria. Front. Microbiol. 2018;9:2007. doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouioui I, Zimmermann A, Hennrich O, Xia S, Rössler O, Makitrynskyy R, et al. Challenging old microbiological treasures for natural compound biosynthesis capacity. Front. Bioeng. Biotechnol. 2024;12 doi: 10.3389/fbioe.2024.1255151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa H, Kawaguchi H, Kawamura Y. Antibiotic A-130-A and production thereof [Internet] US3903264A. 1975 https://patents.google.com/patent/US3903264A/en [cited 2024 Jun 10]. Available from: [Google Scholar]
- Pridham TG, Lyons AJ, Seckinger HL. Comparison of some dried holotype and neotype specimens of streptomycetes with their living counterparts1. Int. J. Syst. Evol. Microbiol. 1965;15(4):191–237. [Google Scholar]
- Rainey FA, Ward-Rainey N, Kroppenstedt RM, Stackebrandt E. The Genus Nocardiopsis Represents a Phylogenetically Coherent Taxon and a Distinct Actinomycete Lineage: Proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Evol. Microbiol. 1996;46(4):1088–1092. doi: 10.1099/00207713-46-4-1088. [DOI] [PubMed] [Google Scholar]
- Riahi HS, Heidarieh P, Fatahi-Bafghi M. Genus Pseudonocardia: What we know about its biological properties, abilities and current application in biotechnology. J. Appl. Microbiol. 2022;132(2):890–906. doi: 10.1111/jam.15271. Feb. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. Nov 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risdian C, Landwehr W, Rohde M, Schumann P, Hahnke RL, Spröer C, et al. Streptomyces bathyalis sp. nov., an actinobacterium isolated from the sponge in a deep sea. Antonie Van Leeuwenhoek. 2021;114(4):425–435. doi: 10.1007/s10482-021-01528-4. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong X, Huang Y. In: Methods in Microbiology [Internet] Goodfellow M, Sutcliffe I, Chun J, editors. Academic Press; 2014. Chapter 11 - Multi-locus Sequence Analysis: Taking Prokaryotic Systematics to the Next Level; pp. 221–251.https://www.sciencedirect.com/science/article/pii/S0580951714000178 [cited 2024 Jun 10](New Approaches to Prokaryotic Systematics; vol. 41). Available from: [Google Scholar]
- Rossi DT. Su di alcune specie di ‘Streptothrix’ trovate nell’aria studate in rapporto a quelle giá note a specialmente all’ ’Actinomyces’. Ann DellIstituto Ig Sper Univ Roma. 1891;1:399–438. [Google Scholar]
- Sakakibara F, Takagi K, Kataoka R, Kiyota H, Sato Y, Okada S. Isolation and identification of dieldrin-degrading Pseudonocardia sp. strain KSF27 using a soil–charcoal perfusion method with aldrin trans-diol as a structural analog of dieldrin. Biochem. Biophys. Res. Commun. 2011;411(1):76–81. doi: 10.1016/j.bbrc.2011.06.096. Jul 22. [DOI] [PubMed] [Google Scholar]
- Sanjivkumar M, Babu DR, Suganya AM, Silambarasan T, Balagurunathan R, Immanuel G. Investigation on pharmacological activities of secondary metabolite extracted from a mangrove associated actinobacterium Streptomyces olivaceus (MSU3) Biocatal. Agric. Biotechnol. 2016;6:82–90. Apr 1. [Google Scholar]
- Sanjivkumar M, Brindhashini A, Deivakumari M, Palavesam A, Immanuel G. Investigation on saccharification and bioethanol production from pretreated agro-residues using a mangrove associated actinobacterium Streptomyces variabilis (MAB3) Waste BioMass Valorization. 2018;9(6):969–984. Jun 1. [Google Scholar]
- Sasser M. MIDI technical note 101. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI, Newark},. 1990 May; (Technical Note #101).
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972;36(4):407–477. doi: 10.1128/br.36.4.407-477.1972. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sembiring L, Ward AC, Goodfellow M. Selective isolation and characterisation of members of the Streptomyces violaceusniger clade associated with the roots of Paraserianthes falcataria. Antonie Van Leeuwenhoek. 2000;78(3–4):353–366. doi: 10.1023/a:1010226515202. Dec. [DOI] [PubMed] [Google Scholar]
- Sen A, Daubin V, Abrouk D, Gifford I, Berry AM, Normand P. Phylogeny of the class Actinobacteria revisited in the light of complete genomes. The orders ‘Frankiales’ and Micrococcales should be split into coherent entities: proposal of Frankiales ord. nov., Geodermatophilales ord. nov., Acidothermales ord. nov. and Nakamurellales ord. nov. Int. J. Syst. Evol. Microbiol. 2014;64(Pt 11):3821–3832. doi: 10.1099/ijs.0.063966-0. Nov. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Roux S, Huber KJ, Wu D, Yu S, Udwary D, et al. Expanding the genomic encyclopedia of Actinobacteria with 824 isolate reference genomes. Cell Genom. 2022 doi: 10.1016/j.xgen.2022.100213. Nov 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit CS, Ruzzini AC, Van Arnam EB, Ramadhar TR, Currie CR, Clardy J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc. Natl. Acad. Sci. U S A. 2015;112(43):13150–13154. doi: 10.1073/pnas.1515348112. Oct 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath PH, Sokal RR. Numerical taxonomy. Nature. 1962;193:855–860. doi: 10.1038/193855a0. [DOI] [PubMed] [Google Scholar]
- Sneath PHA. The principles and practice of numerical classification. Numer. Taxon. [Internet] 1973;573 https://cir.nii.ac.jp/crid/1570572699884106752 [cited 2024 Jun 10] Available from: [Google Scholar]
- Sokal RR. Numerical taxonomy. Sci. Am. 1966;215(6):106–117. [Google Scholar]
- Spitzer V. Structure analysis of fatty acids by gas chromatography — low resolution electron impact mass spectrometry of their 4,4-dimethyloxazoline derivatives — A review. Prog. Lipid Res. 1996;35(4):387–408. doi: 10.1016/s0163-7827(96)00011-2. Dec 1. [DOI] [PubMed] [Google Scholar]
- Stackebrandt Schumann, Stackebrandt E, Schumann P. The Actinobacteria, Part B Bergey’s Manual of Systematic Bacteriology. 2nd edn. Springer; New York: 2012. Genus II. Blastococcus Ahrens and Moll 1970, 264AL emend. Urzì, Salamone, Schumann, Rohde and Stackebrandt 2004b, 257 emend. Lee 2006, 2394; pp. 1455–1767. 2012. [Google Scholar]
- Stanier RY, Niel CB. The concept of a bacterium. Arch Für Mikrobiol. 1962;42(1):17–35. doi: 10.1007/BF00425185. [DOI] [PubMed] [Google Scholar]
- Stanier RY, Van Niel CB. The main outlines of bacterial classification. J. Bacteriol. 1941;42(4):437–466. doi: 10.1128/jb.42.4.437-466.1941. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó I, Marton M. Comments on the first results of the international cooperative work on criteria used in characterization of Streptomycetes. Int. J. Syst. Evol. Microbiol. 1964;14(1):17–38. [Google Scholar]
- Tabata Y. SF2768, a new isonitrile antibiotic obtained from Streptomyces. Sci. Rep. Meiji Seika Kaisha. 1995;34:1–9. [Google Scholar]
- Terlouw BR, Blin K, Navarro-Muñoz JC, Avalon NE, Chevrette MG, Egbert S, et al. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic. Acids. Res. 2023;51(D1):D603–D610. doi: 10.1093/nar/gkac1049. Jan 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson FL, Gevers D, Thompson CC, Dawyndt P, Naser S, Hoste B, et al. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 2005;71(9):5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumar JT, Dhulia K, Singh SP. Isolation and partial purification of an antimicrobial agent from halotolerant alkaliphilic Streptomyces aburaviensis strain Kut-8. World J. Microbiol. Biotechnol. 2010;26(11):2081–2087. Nov 1. [Google Scholar]
- Umezawa H, Tazaki T, Fukuyama S. An antiviral substance, abikoviromycin, produced by Streptomyces species. Jpn. J. Med. 1951;4(5):331–346. doi: 10.7883/yoken1948.4.331. [DOI] [PubMed] [Google Scholar]
- Urzì C, Salamone P, Schumann P, Rohde M, Stackebrandt E. Blastococcus saxobsidens sp. nov., and emended descriptions of the genus Blastococcus Ahrens and Moll 1970 and Blastococcus aggregatus Ahrens and Moll 1970. Int. J. Syst. Evol. Microbiol. 2004;54(1):253–259. doi: 10.1099/ijs.0.02745-0. [DOI] [PubMed] [Google Scholar]
- Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60(2):407–438. doi: 10.1128/mr.60.2.407-438.1996. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira S, Huber KJ, Neumann-Schaal M, Geppert A, Luckner M, Wanner G, et al. Usitatibacter rugosus gen. nov., sp. nov. and Usitatibacter palustris sp. nov., novel members of Usitatibacteraceae fam. nov. within the order Nitrosomonadales isolated from soil. Int. J. Syst. Evol. Microbiol. 2021;71(2) doi: 10.1099/ijsem.0.004631. Feb. [DOI] [PubMed] [Google Scholar]
- Waksman SA, Curtis RE. The Actinomyces of the soil. Soil. Sci. 1916;1(2):99. Feb. [Google Scholar]
- Waksman SA, Henrici AT. The nomenclature and classification of the actinomycetes. J. Bacteriol. 1943;46(4):337–341. doi: 10.1128/jb.46.4.337-341.1943. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhu M, Zhang Q, Zhang X, Yang P, Liu Z, et al. Diisonitrile natural product SF2768 functions as a chalkophore that mediates copper acquisition in Streptomyces thioluteus. ACS. Chem. Biol. 2017;12(12):3067–3075. doi: 10.1021/acschembio.7b00897. Dec 15. [DOI] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, et al. Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 1987;37(4):463–464. [Google Scholar]
- WHO. Lack of innovation set to undermine antibiotic performance and health gains [Internet]. 2022 [cited 2024 Jun 10]. Available from: https://www.who.int/news/item/22-06-2022-22-06-2022-lack-of-innovation-set-to-undermine-antibiotic-performance-and-health-gains.
- Witt D, Stackebrandt E. Unification of the Genera Streptoverticillum and Streptomyces, and amendation of Streptomyces Waksman and Henrici 1943, 339AL. Syst. Appl. Microbiol. 1990;13(4):361–371. Oct 1. [Google Scholar]
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright L, Nouioui I, Mast Y, Bunk B, Spröer C, Neumann-Schaal M, et al. Nocardia australiensis sp. nov. and Nocardia spumae sp. nov., isolated from sea foam in Queensland, Australia. Int. J. Syst. Evol. Microbiol. 2023;73(8) doi: 10.1099/ijsem.0.005952. Aug. [DOI] [PubMed] [Google Scholar]
- Xie F, Pathom-Aree W. Actinobacteria from desert: diversity and biotechnological applications. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.765531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Inoue D, Kuroda M, Ike M. Draft genome sequence of Pseudonocardia sp. strain n23, a 1,4-dioxane-degrading bacterium. Genome Announc. 2017;5(44) doi: 10.1128/genomeA.01240-17. Nov 2e01240-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67(5):1613–1617. doi: 10.1099/ijsem.0.001755. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000;7(1–2):203–214. doi: 10.1089/10665270050081478. Feb. [DOI] [PubMed] [Google Scholar]
- Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int. J. Syst. Evol. Microbiol. 2009;59(3):589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- Zhong Z, He B, Li J, Li YX. Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs) Synth. Syst. Biotechnol. 2020;5(3):155–172. doi: 10.1016/j.synbio.2020.06.002. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Zhou Y, Li C, Wu W, Xu Y, Xia W, et al. Identification and genomic analyses of a novel endophytic actinobacterium Streptomyces endophytica sp. nov. with potential for biocontrol of yam anthracnose. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1139456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences of the 36 studied strains have been deposited at DDBJ/ENA/GenBank and the accessions numbers are provided in the manuscript (Table S1).