Abstract
The quality of Artemisia Argyi Folium (AAF), a traditional Chinese food ingredient, is intrinsically linked to its geographical origin, which this study explores through phenolic compounds and mineral elements. The contents of 17 phenols and 18 minerals differed significantly between geographically distinct samples according to UHPLC and ICP-MS, respectively. Chemometrics indicated that a supervised model, orthogonal partial least squares discriminant analysis (OPLS-DA), outperformed unsupervised methods at classifying AAF samples by their origins. Phenols were more effective at distinguishing samples from seven provinces, while minerals were adept at differentiating samples from the Dabie Mountain region (three provinces) and those from four other provinces. Six phenols and 10 minerals were important variables for discrimination. Complex correlations were observed between the contents of various phenols and minerals in AAF, with minerals possibly affecting the accumulation of phenols. This study provides an approach for distinguishing geographically distinct AAF samples and determining their geographical origins.
Keywords: Artemisia Argyi Folium, Phenolic compounds, Mineral elements, Geographical origin, Chemometrics
Highlights
-
•
Artemisia Argyi Folium (AAF) quality depends on the geographical origins.
-
•
Chemometric analysis distinguishes AAF origins based on phenolics and minerals.
-
•
Quantitatively analyzed 17 phenolics (9 phenolic acids, 8 flavonoids) by UHPLC.
-
•
Phenolic compounds were more useful for classifying samples from adjacent origins.
-
•
Observed complex correlations between phenolic compounds and mineral elements in AAF.
1. Introduction
Artemisia argyi Lévl. et Vant. is an edible and medicinal plant primarily distributed in China, Korea, Mongolia, and Japan. (Chang et al., 2021; Z. C. Wang et al., 2023). Artemisia Argyi Folium (AAF), which is commonly consumed as a traditional food ingredient, is used to prepare Chinese dishes such as qingtuan and dumplings (Cui et al., 2022; X. W. Song et al., 2019). AAF is composed of varied chemical constituents, including essential oils (Liu et al., 2021), flavonoids, phenolic acids (Li et al., 2019; W. Wu, Xiang, & He, 2021), terpenoids, and sterols (Zhan et al., 2022), which show antioxidant, hypoglycemic (Xiao et al., 2019), antimicrobial (H. B. Wang et al., 2020), anti-inflammatory (Chen et al., 2022), antitumor (Su et al., 2022), neuroprotective (L. K. Wu et al., 2022), and antiviral activities (X. W. Zhang et al., 2018).
The composition of food products is intricately linked to their provenance, and the variety and concentration of nutrients they contain are affected by crop soil quality, temperature, and other agricultural conditions (Jeong et al., 2023; Oliveira et al., 2019; Pacholczyk-Sienicka et al., 2024). Many different techniques have been proposed to determine the botanical and geographical origins of produce. Chemometrics combined with multi-component and multi-element analyses has proven efficacious when applied to diverse animal and plant products, such as honey, ginger, paprika, sesame, thyme, pork, and bovine bone (Campmajó et al., 2021; Qi et al., 2021; Wu, Xiang, & He, 2021; Wu, Zhao, et al., 2021; Yu et al., 2023).
A. argyi is cultivated in many regions of China, but Hubei Province is considered to provide one of the best sources of this plant's leaves (Liu et al., 2021). The quality and price of AAF from A. argyi plants of different geographical origins vary, but it is difficult to distinguish its origin by appearance only, leading to confusion about its provenance and limiting its commercial value. Hence, there is a need to develop a method of distinguishing AAF samples based on their components.
Phenolic compounds, such as phenolic acids and flavonoids, have demonstrated potential as pharmacological components of AAF (Li et al., 2019). Current research using phenolic compounds for fingerprint and quantitative analysis of non-volatile components of AAF still faces challenges such as long chromatographic separation times and poor separation of certain components(Chang et al., 2021; D. Zhang, Yao, et al., 2021). For example, cynarine and isochlorogenic acid A cannot be separated effectively. The method for quantitatively analyzing multi-index phenolic compounds requires further development. Alternatively, multi-isotopic and elemental signatures have proven to be effective for tracing the origins of foodstuffs. In contrast to stable isotope analysis, mineral element analysis has the advantages of rapidity, cost-effectiveness, and broad applicability (Nie et al., 2023; Qi et al., 2021). Moreover, chemometric methods can be utilized to analyze the phenolic compounds and mineral elements in AAF, enabling an examination and comparison of these components to trace the origins of AAF samples.
In this investigation, a comprehensive collection comprising 100 samples of AAF from A. argyi plants grown in seven provinces of China was amassed. Quantitative determination and comparison of 17 phenolic compounds (9 phenolic acids and 8 flavonoids) and 18 mineral elements from these samples was conducted. The components of the AAF samples were characterized using ultra-high performance liquid chromatography (UHPLC) with inductively coupled plasma mass spectrometry (ICP-MS) and chemometric techniques. Chemometrics, including hierarchical cluster analysis (HCA), principal component analysis (PCA), orthogonal partial least squares discriminant analysis (OPLS-DA), and Pearson's correlation analysis, were employed to analyze correlations between the components and classify AAF samples from various geographical origins.
The results indicated that the geographical origins of the samples could be distinguished by chemometric analysis of their contents of phenolic compounds and mineral elements, as these components were easy to classify and differentiate. This work contributes to a deeper and more comprehensive understanding of the characteristics of phenolic substances and mineral elements in AAF from plants of different geographical origins. The findings may also be used as a reference for AAF origin traceability analysis.
2. Material and methods
2.1. Reagents and standards
UHPLC-grade acetonitrile and formic acid were supplied by Fisher Chemical™ (Pittsburgh, PA, USA), and nitric acid and hydrochloric acid were obtained from Merck (Darmstadt, Germany). We sourced 95 % ethanol from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). A range of reference compounds was obtained from the National Institute for Food and Drug Control (Beijing, China), including chlorogenic acid, caffeic acid, schaftoside, isochlorogenic acid C, apigenin, vitexicarpin, and isoquercitrin. Additional compounds such as neochlorogenic acid, isochlorogenic acid B, isochlorogenic acid A, hispidulin, and cynarine were purchased from Chengdu Desite Biological Technology Co., Ltd. (Chengdu, China). Jaceosidin and eupatilin were acquired from Sinco Pharmachem Inc. (Delaware, USA). Cryptochlorogenic acid was provided by Chengdu Must Biological Technology Co., Ltd. (Chengdu, China), and 3,4,5-tricaffeoylquinic acid was sourced from Wuhan ChemFaces Biochemical Co., Ltd. (Wuhan, China). The compound 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone was secured from Shanghai Hongyong Biotechnology Co., Ltd. (Shanghai, China). The National Institute of Metrology (Beijing, China) provided the elemental standards. Purified water was obtained via an ELGA LabWater PURELAB Classic system (ELGA LabWater, UK) and underwent filtration using a 0.22 μm nylon membrane.
2.2. Plant materials
One hundred dried AAF samples were supplied by Jing Brand Chizhengtang Pharmaceutical Co., Ltd. (Huangshi, China). All samples were composed of dried leaves of Artemisia argyi Lévl. et Vant. The samples originated from A. argyi plants grown in Hubei Province (HUB, n = 30), Henan Province (HN, n = 20), Anhui Province (AH, n = 10), Jiangxi Province (JX, n = 10), Guizhou Province (GZ, n = 10), Hebei Province (HEB, n = 10), and Liaoning Province (LN, n = 10) in China (Fig. 1). All voucher specimens were taxonomically identified by Prof. Daihua Sun (Hubei Provincial Engineering Technology Research Center of Traditional Chinese Medicine Formula Granules, Huangshi, China). Before analysis, the samples were pulverized into a powder and screened using 50-mesh sieves (355 μm ± 13 μm) to ensure their suitability for the experiments. The samples were collected in June 2023.
Fig. 1.
Geographical information of AAF samples from seven provinces in China.
2.3. Sample preparation
2.3.1. Preparation for UHPLC analysis
Precisely weighed quantities of 0.2 g of each powdered sample were placed into 50 mL conical flasks equipped with stoppers. Subsequently, 20 mL of 50 % ethanol was dispensed accurately into each flask, and the samples were subjected to extraction via ultrasonication (250 W, 53 kHz) for a duration of 30 min in an SK-8200HP ultrasonic water bath (Shanghai Kudos Ultrasonic Instrument Co., Ltd., Shanghai, China). Prior to injection, the ultimate extraction solution underwent filtration using a membrane filter with a pore size of 0.22 μm. The standard substances were precisely weighed and subsequently solubilized in 70 % methanol to create the stock solutions, all of which were maintained at 4 °C before use.
2.3.2. Preparation for ICP-MS analysis
Each powdered sample, weighing precisely 0.5 g, was meticulously added to pre-cleaned Teflon digestion vessels treated with nitric acid, after which 5 mL of concentrated nitric acid was added. Then, a Multiwave PRO microwave digestion system (Anton Paar, Graz, Austria) was used to digest the samples. The microwave temperature was increased gradually to 80 °C over 3 min, maintained for 10 min, increased to 130 °C over 3 min, maintained for 5 min, then increased to 180 °C over 15 min, and maintained for 8 min. Upon reaching ambient temperature, the digestion solution was diluted to volume in a volumetric flask with ultrapure water for elemental analysis. Blank solutions were formulated following the same procedure.
2.4. UHPLC analysis
Phenolic compounds (neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, caffeic acid, schaftoside, isoquercitrin, isochlorogenic acid B, cynarine, isochlorogenic acid A, isochlorogenic acid C, 3,4,5-tricaffeoylquinic acid, apigenin, hispidulin, jaceosidin, 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone, eupatilin, and vitexicarpin) were isolated on a 1290 Infinity II LC System connected to a Variable Wavelength Detector (VWD). The column was an ACQUITY UPLC HSS T3 column (150 mm × 2.1 mm, 1.8 μm, Waters, MA, USA). Utilizing a flow rate of 0.3 mL/min, a mobile phase consisting of 0.1 % aqueous formic acid (A) (v/v) and acetonitrile (B) was employed. The elution protocol was executed according to a predefined linear gradient, as follows: 10 % B at 0 min; 20 % B at 7 min; 27 % B at 17 min; 33 % B at 22 min; 38 % B at 27 min; 60 % B at 29 min; 70 % B at 32 min; 90 % B at 33–35 min. Following a 5-min interval, the mobile phase was returned to its initial parameters. The column oven was maintained at a temperature of 30 °C, and a 1 μL injection volume was employed. Phenolic compounds were detected at a wavelength of 330 nm. The quantitative analysis was carried out using external standards, demonstrating correlation coefficients surpassing 0.9990 across all calibration curves. Identification and quantification of 17 phenolic compounds were achieved through retention time analysis and calculation of individual standard linear equations.
2.5. ICP-MS analysis
Mineral element content analysis, encompassing Na, Mg, Al, K, Ca, Cr, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Cd, Ba, Hg, and Pb, was conducted utilizing ICAP RQ inductively coupled plasma mass spectrometry (Thermo Fisher Scientific, Bremen, Germany), following the guidelines stipulated in the Chinese Pharmacopeia (2020 Edition) (Commission, C. P., 2020). The instrument was typically operated under the following standard parameters: RF power: 1550 W, nebulizer pump: 40 rpm, sampling depth: 5.0 mm, carrier gas: 1.14 L/min, makeup gas: 0.80 L/min, CCT collision gas: 4.4 L/min, and cooling gas: 14.0 L/min, repeated two times. To correct for matrix effects and instrument drift, an internal standard containing Sc, Ge, Rh, In, Re, and Bi was employed. This internal standard (100 μg/L) was added to all samples and calibration standards. The R2 value of the calibration curve was 0.9990 or higher for all elements; the LOD, LOQ, RSD, and recovery rate of each element are shown in Table S1. The reported results represent the average of two replicate measurements.
2.6. Statistical analysis
The phenolic chromatographic fingerprint was constructed using the Similarity Evaluation System for Chromatographic Fingerprint of TCM (Chinese Pharmacopeia Commission, Beijing, China). One-way analysis of variance (ANOVA), HCA, PCA, and Pearson's correlation analysis were performed using OriginPro 2024b (OriginLab Corporation, Northampton, MA, USA); the orthogonal partial least squares discriminant analysis (OPLS-DA) and receiver operating characteristic (ROC) analysis were executed using SIMCA 14.1 software (Umetrics, Umea, Sweden).
3. Results and discussion
3.1. Phenolic compounds of AAF samples from different geographical origins
Phenolic compounds, such as phenolic acids and flavonoids, are the main non-volatile components of AAF and possess various biological activities (Chang et al., 2022; Z. C. Wang et al., 2023). Seventeen phenolic compounds, including nine phenolic acids and eight flavonoids, were determined by UHPLC (Table 1 and Fig. 2A). The chromatogram in Fig. 2B illustrates the peaks corresponding to the 17 phenolic compounds present in both the samples and the reference solution. Peaks 1–17 were characterized as neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, caffeic acid, schaftoside, isoquercitrin, isochlorogenic acid B, cynarine, isochlorogenic acid A, isochlorogenic acid c, 3,4,5-tricaffeoylquinic acid, apigenin, hispidulin, jaceosidin, 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone, eupatilin, and vitexicarpin, respectively. The mean concentrations of these 17 compounds were compared among AAF samples from different origins using analysis of variance (ANOVA), and the results from the analysis of these concentration differences are depicted using box plots (Fig. S1). The findings revealed a notable distinction (with a confidence level of 95 %) in the contents of phenolic compounds among all of the geographically distinct samples (P < 0.001), indicating that the origin impacts the levels of these constituents. Samples from AH exhibited the highest mean concentrations of isochlorogenic acid C, apigenin, hispidulin, jaceosidin, 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone, eupatilin, and vitexicarpin at 5.264 mg/g, 0.018 mg/g, 0.108 mg/g, 0.443 mg/g, 0.301 mg/g, 2.640 mg/g and 0.781 mg/g, respectively. The JX samples demonstrated higher mean values for most phenolic acids, while the concentrations of flavonoids in HEB and LN were comparatively lower.
Table 1.
The average concentrations of 17 phenolic compounds in AAF samples from various geographical origins.
| Components | Geographical origins (mg/g, mean ± SD) |
||||||
|---|---|---|---|---|---|---|---|
| HUB (n = 30) | HN (n = 20) | AH (n = 10) | JX (n = 10) | GZ (n = 10) | HEB (n = 10) | LN (n = 10) | |
| Neochlorogenic acid | 0.140f ± 0.055 | 0.370c ± 0.074 | 0.214e ± 0.011 | 0.544b ± 0.056 | 0.248de ± 0.019 | 0.287d ± 0.069 | 0.665a ± 0.023 |
| Chlorogenic acid | 0.855c ± 0.376 | 2.163b ± 0.669 | 1.067c ± 0.059 | 3.679a ± 0.285 | 2.306b ± 0.204 | 2.294b ± 0.156 | 4.182a ± 0.161 |
| Cryptochlorogenic acid | 0.161c ± 0.051 | 0.346b ± 0.085 | 0.298b ± 0.016 | 0.536a ± 0.039 | 0.214c ± 0.018 | 0.199c ± 0.015 | 0.356b ± 0.014 |
| Caffeic acid | 0.071c ± 0.066 | 0.091c ± 0.046 | 0.162b ± 0.011 | 0.234a ± 0.014 | 0.085c ± 0.01 | 0.121bc ± 0.043 | 0.112bc ± 0.013 |
| Schaftoside | 1.095ab ± 0.308 | 1.267a ± 0.459 | 0.541cd ± 0.060 | 0.607cd ± 0.392 | 0.336d ± 0.169 | 0.768bc ± 0.261 | 0.574cd ± 0.030 |
| Isoquercitrin | 0.425ab ± 0.168 | 0.363bc ± 0.174 | 0.407ab ± 0.042 | 0.112d ± 0.010 | 0.549a ± 0.052 | 0.225cd ± 0.010 | 0.173d ± 0.030 |
| Isochlorogenic acid B | 2.870bc ± 1.191 | 4.768a ± 1.512 | 3.091bc ± 0.141 | 5.297a ± 0.56 | 3.135bc ± 0.301 | 2.099c ± 0.198 | 3.615b ± 0.238 |
| Cynarine | 0.177e ± 0.071 | 0.420d ± 0.150 | 0.158e ± 0.012 | 0.345d ± 0.021 | 0.574c ± 0.076 | 1.467b ± 0.296 | 1.761a ± 0.082 |
| Isochlorogenic acid A | 5.737c ± 3.416 | 7.713b ± 1.711 | 6.280bc ± 0.157 | 11.378a ± 0.867 | 11.360a ± 1.198 | 7.297bc ± 0.906 | 11.883a ± 0.482 |
| Isochlorogenic acid C | 2.484d ± 1.083 | 3.657b ± 0.691 | 5.264a ± 0.170 | 5.214a ± 0.311 | 2.834cd ± 0.231 | 2.352d ± 0.184 | 3.397bc ± 0.147 |
| 3,4,5-Tricaffeoylquinic acid | 1.855a ± 1.272 | 1.783ab ± 1.057 | 0.813bc ± 0.060 | 0.405c ± 0.110 | 1.098abc ± 0.101 | 1.562abc ± 0.342 | 1.472abc ± 0.089 |
| Apigenin | 0.014ab ± 0.005 | 0.006c ± 0.004 | 0.018a ± 0.016 | 0.009bc ± 0.005 | 0.007bc ± 0.003 | 0.004c ± 0.001 | 0.004c ± 0.000 |
| Hispidulin | 0.056b ± 0.015 | 0.046c ± 0.007 | 0.108a ± 0.006 | 0.039cd ± 0.004 | 0.031d ± 0.012 | ND | 0.031d ± 0.002 |
| Jaceosidin | 0.304b ± 0.078 | 0.306b ± 0.037 | 0.443a ± 0.016 | 0.279bc ± 0.014 | 0.128d ± 0.017 | 0.048e ± 0.010 | 0.248c ± 0.030 |
| 5,7,3′-Trihydroxy-6,4′,5′-trimethoxyflavone | 0.118b ± 0.049 | 0.090c ± 0.023 | 0.301a ± 0.011 | 0.053d ± 0.022 | 0.053d ± 0.007 | 0.011e ± 0.015 | 0.027de ± 0.012 |
| Eupatilin | 1.568b ± 0.201 | 1.232c ± 0.171 | 2.640a ± 0.145 | 1.145c ± 0.044 | 0.572d ± 0.078 | 0.214e ± 0.038 | 0.705d ± 0.054 |
| Vitexicarpin | 0.637b ± 0.158 | 0.468c ± 0.069 | 0.781a ± 0.034 | 0.672ab ± 0.036 | 0.180e ± 0.0170 | 0.140e ± 0.016 | 0.352d ± 0.034 |
Samples originated from: Hubei Province (HUB), Henan Province (HN), Anhui Province (AH), Jiangxi Province (JX), Guizhou Province (GZ), Hebei Province (HEB), and Liaoning Province (LN). Note: Letters “a”–“f” within identical lines denote statistically significant differences (P < 0.05). ND denotes undetectable content. There was a notable difference in the contents of phenolic compounds among all of the geographically distinct samples (P < 0.001).
Fig. 2.
UHPLC analysis of 100 batches of AAF samples from seven provinces in China. (A) UHPLC fingerprints of 100 batches of AAF samples and the reference fingerprint (R); (B) UHPLC chromatograms of AAF sample (S1) and reference solution (S2). Peaks 1 to 17 represent neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, caffeic acid, schaftoside, isoquercitrin, isochlorogenic acid B, cynarine, isochlorogenic acid A, isochlorogenic acid C, 3,4,5-tricaffeoylquinic acid, apigenin, hispidulin, jaceosidin, 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone, eupatilin, and vitexicarpin.
3.2. Chemometric analysis of phenolic compounds in AAF samples from diverse geographical origins
3.2.1. HCA of phenolic compounds
The HCA of 17 phenolic compounds in 100 batches of AAF samples from different origins was carried out using a heat map (Fig. 3A), and the samples were found to be clustered into seven regions. The samples from the neighboring provinces of HUB and HN were similar, while the remaining samples could be distinguished by province. JX was quite different from samples originating in three other provinces in central China (HUB, HN, and AH), but GZ, JX, and those from northern China (HEB and LN) were similar.
Fig. 3.
Chemometric analysis of phenolic compounds in AAF samples from different geographical origins. (A) Pearson's correlation plot of the contents of 17 phenolic compounds. (A) Heat map analysis of the HCA classification of AAF samples and 17 phenolic compounds. (B) Biplot of the PCA of AAF samples from seven provinces and 17 phenolic compounds. (C) OPLS-DA score scatter plot. (D) Correlation circle between phenolic compounds and AAF samples. (E) VIP scores plot. (F) ROC curve of OPLS-DA (phenolic compounds).
The 17 phenolic compounds were clustered into four groups. The contents of group 1 (neochlorogenic acid, chlorogenic acid, isochlorogenic acid A, and cynarine) were higher in JX and LN; the contents of group 2 (cryptochlorogenic acid, caffeic acid, isochlorogenic acid B, and isochlorogenic acid C) were higher in JX; the contents of group 3 (schaftoside, 3,4,5-tricaffeoylquinic acid, and isoquercitrin) were higher in HUB and HN but lower in JX, LN, and AH; and the contents of group 4 (apigenin, hispidulin, eupatilin, 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone, jaceosidin, and vitexicarpin) were higher in AH. Most compounds in groups 1 and 2 were phenolic acids, while groups 3 and 4 were composed mainly of flavonoids. The clustering results were related to differences in the contents of each group of compounds.
3.2.2. PCA of phenolic compounds
A PCA was used to analyze the phenolic compound contents across samples from different origins. The cumulative variance contribution of the first three principal components (PCs) was 76.46 %. The initial principal component (PC1) elucidated 37.34 % of the overall variance, while the subsequent two PCs (PC2 and PC3) contributed 25.79 % and 13.33 %, respectively. As illustrated in Fig. 3B, the scores derived from PC1, PC2, and PC3, revealed seven distinct groups. The samples could, therefore, be distinguished effectively according to their province of origin. The confidence ellipses of HUB and HN samples partially overlapped, and the loadings of the phenolic compounds were divided into four groups. The comparison of the phenolic contents in samples from different provinces is displayed in a biplot—the results from the PCA were similar to those from the HCA, with geographically adjacent samples being relatively closer.
3.2.3. OPLS-DA and ROC of phenolic compounds
OPLS-DA is recognized as a highly effective approach for sample classification and the establishment of discriminant models, thus warranting its selection as a pivotal indicator in endeavors aimed at discriminating geographical origins (Du et al., 2021; Hye Hur et al., 2023). In the current study, the OPLS-DA was conducted on 17 phenolic compounds within a dataset comprising 80 AAF samples (80 % of the AAF samples from each geographical origin were chosen randomly). As illustrated in Fig. 3C, no overlap was observed among the seven groups of samples originating from distinct sources, indicating that the discrimination was excellent. The model exhibited favorable parameter values, with R2X, R2Y, and Q2Y values of 0.968, 0.843, and 0.800, respectively, indicating a robust fit and satisfactory predictive capability (Kang et al., 2022). After conducting 200 iterations of permutation tests (Fig. S2), the intercepts of R2 and Q2 were found to be below 0.3 and 0.05, respectively, signifying the absence of overfitting in the model (Long et al., 2023; Yu et al., 2023). The associations between the phenolic compounds and AAF samples are depicted in Fig. 3D. An inverse relationship between phenolic compound concentrations in the samples and the distances between their origins was observed (Mi et al., 2022). For example, the flavonoids including jaceosidin, 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone, hispidulin, apigenin, eupatilin, and vitexicarpin were distributed closely around the AAF samples from AH; the phenolic acids including cryptochlorogenic acid, caffeic acid, isochlorogenic acid B, and Isochlorogenic acid A were distributed closely around the AAF samples from JX; schaftoside, 3,4,5-tricaffeoylquinic acid, apigenin, and isoquercitrin were distributed closely around the AAF samples from HUB; and the maximum concentrations of these components were identified in each respective geographical origin.
Generally, variables with a variable importance in projection (VIP) > 1 are regarded as pivotal in discriminating between diverse sample groups (Lu et al., 2024; X. C. Song et al., 2021). Six phenolic components with VIP > 1 were subjected to screening by OPLS-DA (Fig. 3E): cynarine, vitexicarpin, schaftoside, caffeineic acid, isochlorogenic acid B, and 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone.
The accuracy of the OPLS-DA model at discriminating AAF samples from different geographical origins was evaluated using the ROC curve of the phenolic compounds (Mi et al., 2019; Mi et al., 2022). The areas under the ROC curve (AUC) for HUB and HN were 0.995 and 0.997, respectively, while the AUC for all other origins was 1 (Fig. 3F), indicating the robust discriminatory capacity of the phenolic compound contents in separating AAF samples from seven provinces (Yoon et al., 2022).
3.3. Mineral elements in AAF samples from different geographical origins
ICP-MS offers the benefits of heightened sensitivity, concurrent analysis of numerous elements, and expedited analysis. Such multi-element analyses hold promise for distinguishing geographical origins effectively (Mi et al., 2022; Yu et al., 2023; H. R. Zhang, Liu, et al., 2021). Eighteen chemical elements were quantitatively determined in AAF samples from different geographical origins. The most highly concentrated element in 100 batches of AAF samples was K, followed by Ca, Mg, Fe, Al, and Mn, while the contents of other elements remained relatively low (Table 2). ANOVA was utilized to assess the differences in the mean values of the 18 mineral elements among AAF samples from various origins. Comparations of the contents of mineral elements between samples of different geographical origins (seven provinces) are indicated in box plots (Fig. S3). The mean values of K, Mn, Ni, Zn, Cd, and Ba were highest in JX samples, whereas the mean values of Mg, Al, Cr, Co, Zn, and As in HUB were lower. The levels of the 18 mineral elements exhibited significant variation (at the 95 % confidence level, P < 0.001) across the different geographical origins, indicating that the environmental differences between the places of origin affected the contents of mineral elements in AAF.
Table 2.
The average concentrations of the 18 mineral elements in AAF samples originating from various geographical origins.
| Elements | Geographical origins (mg/g, mean ± SD) |
||||||
|---|---|---|---|---|---|---|---|
| HUB (n = 30) | HN (n = 20) | AH (n = 10) | JX (n = 10) | GZ (n = 10) | HEB (n = 10) | LN (n = 10) | |
| Na | 17.61b ± 13.36 | 52.44a ± 27.96 | 25.97b ± 7.79 | 11.3b ± 3.4 | 9.28b ± 5.59 | 48.68a ± 3.81 | 52.78a ± 8.39 |
| Mg | 2265.68d ± 367.74 | 2682.69c ± 227.48 | 3065.05b ± 169.16 | 1730.61e ± 74.11 | 3983.36a ± 198.19 | 2973.59bc ± 110.90 | 2828.44bc ± 139.78 |
| Al | 238.45d ± 67.55 | 425.41b ± 94.22 | 275.75cd ± 34.21 | 199.97d ± 23.32 | 356.07bc ± 27.10 | 782.19a ± 87.42 | 824.92a ± 48.62 |
| K | 30,682.70ab ± 4395.60 | 32,565.18a ± 4827.15 | 28,220.13b ± 1097.01 | 34,348.95a ± 2046.29 | 30,219.21ab ± 721.05 | 23,097.90c ± 1225.16 | 31,429.48ab ± 1233.38 |
| Ca | 10,657.84b ± 1282.71 | 10,049.97bc ± 789.17 | 9607.29c ± 374.02 | 6819.18d ± 401.09 | 10,605.88bc ± 150.62 | 12,029.27a ± 456.97 | 9978.58bc ± 227.28 |
| Cr | 3.34de ± 1.78 | 5.28c ± 1.40 | 4.58cd ± 0.89 | 2.86de ± 0.83 | 2.10e ± 0.13 | 11.39a ± 0.82 | 8.58b ± 0.73 |
| Mn | 186.37b ± 86.88 | 175.21b ± 60.73 | 125.53bc ± 6.80 | 594.83a ± 57.03 | 160.20bc ± 3.76 | 92.67c ± 6.51 | 155.93bc ± 16.06 |
| Fe | 314.06cd ± 93.91 | 515.15b ± 178.00 | 401.16bc ± 42.94 | 236.85d ± 22.28 | 366.56cd ± 19.21 | 995.55a ± 111.76 | 1114.56a ± 128.46 |
| Co | 0.20e ± 0.070 | 0.26d ± 0.04 | 0.30cd ± 0.03 | 0.26d ± 0.07 | 0.34c ± 0.02 | 0.45b ± 0.04 | 0.52a ± 0.04 |
| Ni | 2.66c ± 1.22 | 3.47b ± 0.57 | 2.76bcd ± 0.15 | 4.83a ± 0.22 | 0.97e ± 0.02 | 1.47e ± 0.13 | 1.84de ± 0.09 |
| Cu | 18.88a ± 4.91 | 15.28bc ± 1.06 | 13.21bcd ± 0.72 | 17.01ab ± 1.75 | 11.59d ± 0.32 | 13.17cd ± 0.39 | 13.02cd ± 0.44 |
| Zn | 38.82c ± 8.95 | 46.58ab ± 5.00 | 33.54cd ± 3.15 | 51.93a ± 3.76 | 31.87d ± 2.98 | 36.19cd ± 4.62 | 39.67bcd ± 4.65 |
| As | 0.22d ± 0.07 | 0.29c ± 0.05 | 0.20d ± 0.01 | 0.31c ± 0.06 | 0.65a ± 0.06 | 0.49b ± 0.06 | 0.65a ± 0.04 |
| Se | 0.91c ± 0.31 | 1.17b ± 0.31 | 0.67cd ± 0.05 | 1.43ab ± 0.16 | 0.58d ± 0.03 | 1.46a ± 0.21 | 1.43ab ± 0.1 |
| Cd | 0.58c ± 0.36 | 0.81b ± 0.24 | 0.28d ± 0.01 | 1.36a ± 0.07 | 0.49cd ± 0.03 | 0.22d ± 0.02 | 0.27d ± 0.02 |
| Ba | 47.55ab ± 11.42 | 41.55b ± 5.77 | 41.60ab ± 1.59 | 50.40a ± 8.67 | 9.06d ± 0.41 | 26.41c ± 2.17 | 27.24c ± 1.72 |
| Hg | 0.016b ± 0.004 | 0.022a ± 0.011 | 0.018ab ± 0.003 | 0.014b ± 0.003 | 0.018ab ± 0.002 | 0.020ab ± 0.003 | 0.013b ± 0.003 |
| Pb | 1.08cd ± 0.35 | 1.31bc ± 0.31 | 1.01cd ± 0.07 | 0.83d ± 0.07 | 1.10cd ± 0.06 | 1.45b ± 0.15 | 2.12a ± 0.23 |
Note: Letters “a”–“f” within lines denote statistically significant differences (P < 0.05). The levels of the 18 mineral elements exhibited significant variation (at the 95 % confidence level, P < 0.001) across the different geographical origins.
3.4. Chemometric analysis of mineral elements in AAF samples from different origins
3.4.1. HCA of mineral elements
The HCA of 18 mineral elements in 100 batches of AAF samples from different origins was carried out using a heat map (Fig. 4A), and the samples were clustered into seven groups. The samples from the Dabie Mountain region (HUB, HN, and AH) exhibited a relatively high degree of similarity, while the other samples could be distinguished according to the provinces from which they originated. Similar to the clustering of the phenolic compounds, the samples from JX were distinct from those of the three provinces surrounding the Dabie Mountains (HUB, HN, and AH).
Fig. 4.
Chemometric analysis of mineral elements in AAF samples from different geographical origins. (A) Heat map analysis of HCA classification of AAF samples and 18 mineral elements. (B) Biplot of PCA of AAF samples from 7 provinces and 18 mineral elements. (C) OPLS-DA score scatter plot (seven provinces). (D) OPLS-DA score scatter plot (five regions). Dabie Mountain region (DB). (E) Correlation circle between mineral elements and AAF samples. (F) VIP scores plot. (G) ROC curve of OPLS-DA (mineral elements).
The 18 mineral elements were grouped into five clusters. Group 1 elements (Na, Al, Fe, Co, Cr, and Pb) were found at higher concentrations in the HEB and LN samples, while HUB and JX contained lower levels. The group 2 (Mg and As) contents were found to be higher in GZ, LN, and HEB samples, and the contents of group 3 (Hg and Ca) were also found to be higher in HEB samples. Group 4 elements (K, Mn, Cd, Ni, Zn, and Se) were found at higher levels in JX but lower levels in GZ samples. Finally, the concentrations of group 5 elements (Cu and Ba) were higher in the HUB and JX samples.
The range of mineral elements found in samples from adjacent geographical origins was similar. However, the influence of rivers on element distribution was significant. For instance, although relatively close in distance, the sample collection site in JX Province was situated south of the Yangtze River, distinct from the other three provinces north of the Yangtze River (Dabie Mountains).
3.4.2. PCA of mineral elements
The cumulative variance contribution of the first four PCs was 79.35 %. The initial principal component (PC1) elucidated 39.91 % of the overall variance, while the subsequent three PCs (PC2, PC3, and PC4) contributed 21.69 %, 10.70 %, and 7.05 %, respectively. As illustrated in Fig. 4B, the comparison of mineral elements in samples from different provinces is directly displayed in biplots; the PC1, PC2, and PC3 scores indicated the presence of seven distinct groups. The samples from JX, GX, HEB, and AH were distinguished effectively according to their provinces of origin, while the scores and confidence ellipses of samples from the Dabie Mountain region (HUB, HN, and AH) partially overlapped. The loadings of the mineral elements were divided into five groups; the biplot visually presents the distribution of mineral elements, indicating that the geographical origin of the samples significantly influences their concentrations, as revealed by the PCA analysis.
3.4.3. OPLS-DA and ROC of mineral elements
The OPLS-DA was applied to 18 mineral elements within a dataset of 80 AAF samples (80 % of the AAF samples from each geographical origin were randomly chosen from the whole sample). The results from the samples originating in the Dabie Mountain region (HUB, HN, and AH) partially overlapped when analyzed according to the seven provinces (Fig. 4C). However, the OPLS-DA discriminated well between samples from five regions, the Dabie Mountain region, JX, GZ, HEB, and LN, indicating its effectiveness (Fig. 4D). Moreover, the values of the model parameters R2X, R2Y, and Q2Y were 0.928, 0.847, and 0.803, respectively, demonstrating that the model had strong explanatory and predictive capacity (Kang et al., 2022). After conducting 200 iterations of the permutation tests (Fig. S4), the intercepts of R2 and Q2 were found to be below 0.3 and 0.05, respectively, signifying the absence of overfitting in the model (Yu et al., 2023). The correlations between mineral elements and AAF samples are illustrated in Fig. 4E. For instance, elements including K, Mn, Cu, Zn, Ni, Ba, and Cd were distributed closely around the AAF samples from JX, with the maximum concentration levels observed in samples originating from this region. Ten mineral elements with VIP > 1 were screened out by OPLS-DA (Fig. 4F): Mn, As, Pb, Cr, Ba, Mg, Se, Ca, K, and Fe.
The accuracy of the OPLS-DA model at discriminating AAF samples from different geographical origins was evaluated using the mineral element ROC curve (Mi et al., 2019; Mi et al., 2022). The perfect classification performance, as evidenced by an AUC of 1 (Fig. 4G), underscored the robust discriminatory capacity of the mineral element content to distinguish between AAF samples from five regions.
3.5. Prediction of geographical origin
OPLS-DA, utilizing phenolic compound and mineral element concentrations, successfully distinguished AAF samples from diverse geographical origins. To assess the robustness of the supervised models regarding phenolic compounds and mineral elements, a subset comprising 20 % of the samples was reserved for testing purposes to validate the accuracy of the models. Integration of the remaining 20 AAF samples with documented data (test set) and the 80 AAF samples from the discriminant models yielded a dataset of 100 samples (20 + 80) used to develop the OPLS-DA models. The developed model was subsequently tested using the test set to evaluate its robustness and predictive performance in classification. As illustrated in Table 3, all samples were accurately predicted, and the accuracy of the two constructed models was 100 %, exhibiting excellent predictability. Combining the quantitative analysis of phenolic compounds and mineral elements with chemometrics to classify the geographical origins of AAF may be a reliable method and has great application potential.
Table 3.
The prognostication of documented data regarding AAF samples through the developed OPLS-DA models of phenolic components (a) and mineral elements (b).
| (a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Test Set | Members | Accuracy (%) | HUB | HN | AH | JX | GZ | HEB | LN |
| HUB | 6 | 100 % | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| HN | 4 | 100 % | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| AH | 2 | 100 % | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| JX | 2 | 100 % | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| GZ | 2 | 100 % | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| HEB | 2 | 100 % | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| LN | 2 | 100 % | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Total | 20 | 100 % | 6 | 4 | 2 | 2 | 2 | 2 | 2 |
| (b) | |||||||||
| Test Set | Members | Accuracy (%) | DB | JX | GZ | HEB | LN | ||
| DB | 12 | 100 % | 12 | 0 | 0 | 0 | 0 | ||
| JX | 2 | 100 % | 0 | 2 | 0 | 0 | 0 | ||
| GZ | 2 | 100 % | 0 | 0 | 2 | 0 | 0 | ||
| HEB | 2 | 100 % | 0 | 0 | 0 | 2 | 0 | ||
| LN | 2 | 100 % | 0 | 0 | 0 | 0 | 2 | ||
| Total | 20 | 100 % | 12 | 2 | 2 | 2 | 2 | ||
3.6. Correlation analysis between phenolic compounds and mineral elements of AAF
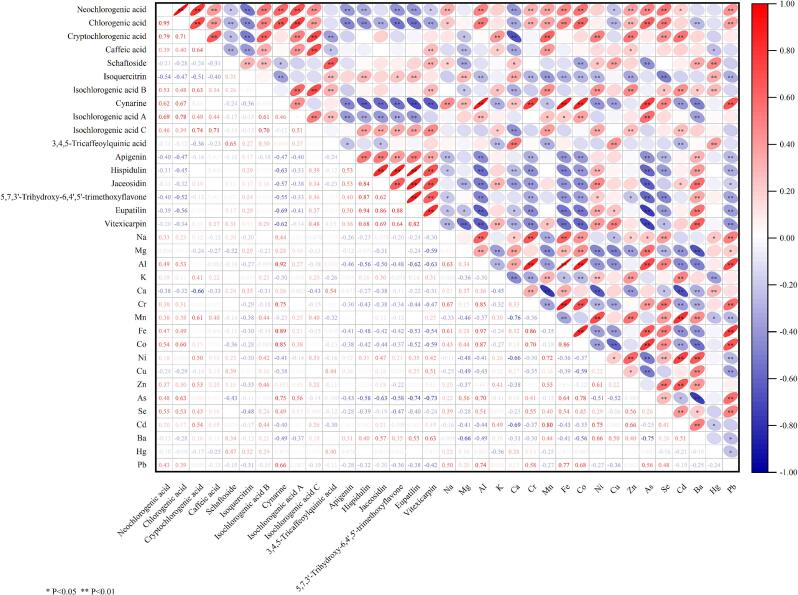
Pearson's correlation analysis was utilized to elucidate the intricate interrelations among the concentrations of the 17 phenolic compounds and 18 mineral elements. The correlation coefficients indicated that, of 595 pairs analyzed, 383 pairs met the threshold of P < 0.05 (Fig. 5). Within this subset, 308 pairs exhibited statistically significant positive correlations (R > 0), while 287 pairs displayed significant negative correlations (R < 0).
Fig. 5.
Correlation coefficients between the contents of phenolic compounds and mineral elements in AAF.
A significant positive relationship was observed between the concentrations of neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, caffeic acid, isochlorogenic acid B, isochlorogenic acid A and isochlorogenic acid C. Likewise, the contents of apigenin, hispidulin, jaceosidin, 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone, eupatilin, and vitexicarpin were significantly positively correlated. The 3,4,5-tricaffeoylquinic acid content was significantly positively correlated with schaftoside and isoquercitrin. The concentrations of neochlorogenic acid, chlorogenic acid, cynarine, and isochlorogenic acid A exhibited a negative correlation with the flavonoid content.
A positive relationship was observed between Na, Mg, Al, Ca, Cr, Fe, Co, and Pb contents. Likewise, the contents of K, Mn, Ni, Cu, Zn, Se, Cd, and Ba exhibited a positive correlation. The Se content was significantly positively correlated with Na, Al, Cr, Mn, Fe, Co, Ni, Zn, As, Cd, Ba, and Pb.
Furthermore, a significant correlation was observed between the contents of phenolic compounds and certain mineral elements in AAF. Each phenolic compound was found to be associated with 6 to 16 elements. For instance, the contents of five flavonoids (hispidulin, jaceosidin, 5,7,3′-trihydroxy-6,4′,5′-trimethoxyflavone, eupatilin, and vitexicarpin) exhibited a significantly positive correlation with Ni and Ba, while displaying a significantly negative correlation with Al, Cr, Fe, Co, As, and Pb. The contents of neochlorogenic acid and chlorogenic acid exhibited a significant positive correlation with Na, Al, Cr, Mn, Fe, Co, Zn, As, Se, and Pb, while exhibiting a significant negative correlation with Ca and Cu. Additionally, Se was negatively correlated with all flavonoids in AAF except for schaftoside. K demonstrated a significant positive correlation with jaceosidin and eupatilin, caffeic acid, cryptochlorogenic acid, isochlorogenic acid B, and isochlorogenic acid C.
The content of inorganic elements in plants has been shown to affect the accumulation of active components, thus influencing plant quality (Wei et al., 2024; Ye et al., 2023). Here, complex correlations were observed between the contents of various phenolic compounds and mineral elements, which may be attributed to environmental influences, such as differences in soil composition across distinct origins.
4. Conclusions
In this study, chemometric analyses and the quantification of phenolic compounds and mineral elements were used to distinguish AAF samples from different geographical origins. The results showed that the contents of phenolic compounds and mineral elements varied greatly among the samples. The two methods offer great potential for identifying and predicting the geographical origin of AAF samples and are suitable for tracing their source. OPLS-DA emphasized that the classification and discrimination performance of a supervised modeling method was better than that of unsupervised modeling methods, especially for samples from similar geographical origins. The phenolic constituents and mineral elements differed in their suitability for use in classifying and distinguishing samples. The phenolic constituents were more useful for classifying samples from adjacent origins. Analysis of the phenolic compounds enabled samples from seven provinces to be distinguished effectively, while analysis of the mineral element levels was useful for distinguishing between samples from the areas surrounding the Dabie Mountains (HUB, HN, and AH) and the other four provinces. Using the ROC curve, the classification accuracies for the two OPLS-DA models were 0.995 or higher. Furthermore, the developed OPLS-DA models were validated using an independent test set of 20 samples with 100 % accuracy. Complex correlations were observed between the contents of various phenolic compounds and mineral elements in AAF, with the mineral elements possibly affecting the accumulation of phenolic compounds. This study demonstrates the efficacy of chemometric analysis, in conjunction with quantitative determination of phenolic compounds and mineral elements, as a viable strategy for identifying the geographic origin of AAF samples and provides a basis for assessing the differences in the characteristics of AAF samples originating from different regions. It also provides some insights into mineral element application strategies in the cultivation of A. argyi. However, there are still some limitations to the current research. The mechanisms by which mineral elements influence the accumulation of phenolic components in AAF require further investigation. Additionally, there is a need to develop more effective methods of evaluating AAF quality and tracing its origin. Our research group is actively exploring the use of near-infrared spectroscopy (NIRS) for rapid qualitative and quantitative analysis of AAF.
CRediT authorship contribution statement
Lifei Hu: Writing – original draft, Software, Investigation, Conceptualization. Fengxiao Zhu: Writing – review & editing, Validation, Data curation. Yifan Wang: Writing – review & editing, Validation, Data curation. Tao Wu: Writing – review & editing, Methodology. Xin Wu: Writing – review & editing, Methodology. Zhian Huang: Writing – review & editing, Data curation. Daihua Sun: Supervision, Resources, Conceptualization. Mingxing Liu: Writing – review & editing, Validation, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Hubei Science and Technology Major Project, China(No. 2020ACA007, 2022ACA003)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2024.101909.
Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Campmajó G., Rodríguez-Javier L.R., Saurina J., Núñez O. Assessment of paprika geographical origin fraud by high-performance liquid chromatography with fluorescence detection (HPLC-FLD) fingerprinting. Food Chemistry. 2021;352 doi: 10.1016/j.foodchem.2021.129397. [DOI] [PubMed] [Google Scholar]
- Chang Y.Q., Fan W.X., Shi H., Feng X., Zhang D., Wang L.…Guo L. Characterization of phenolics and discovery of α-glucosidase inhibitors in Artemisia argyi leaves based on ultra-performance liquid chromatography-tandem mass spectrometry and relevance analysis. Journal of Pharmaceutical and Biomedical Analysis. 2022;220 doi: 10.1016/j.jpba.2022.114982. [DOI] [PubMed] [Google Scholar]
- Chang Y.Q., Zhang D., Yang G.Y., Zheng Y.G., Guo L. Screening of anti-lipase components of Artemisia argyi leaves based on Spectrum-effect relationships and HPLC-MS/MS. Frontiers in Pharmacology. 2021;12 doi: 10.3389/fphar.2021.675396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.K., Kuo C.H., Kuo W.W., Day C.H., Wang T.F., Ho T.J.…Lu C.Y. Artemisia argyi extract ameliorates IL-17A-induced inflammatory response by regulation of NF-κB and Nrf2 expression in HIG-82 synoviocytes. Environmental Toxicology. 2022;37(11):2793–2803. doi: 10.1002/tox.23637. [DOI] [PubMed] [Google Scholar]
- Commission, C. P. I. China Medical Science Press; Beijing: 2020. Pharmacopoeia of the People’s Republic of China. [Google Scholar]
- Cui Z.H., Li S.Q., Chang J.Y., Zang E.H., Liu Q., Zhou B.C.…Li M.H. The pharmacophylogenetic relationships of two edible medicinal plants in the genus Artemisia. Frontiers in Plant Science. 2022;13 doi: 10.3389/fpls.2022.949743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Chen W.L., Lei Y.T., Li F.C., Li H.M., Deng W., Jiang G.H. Discrimination of authenticity of Fritillariae Cirrhosae Bulbus based on terahertz spectroscopy and chemometric analysis. Microchemical Journal. 2021;168 doi: 10.1016/j.microc.2021.106440. [DOI] [Google Scholar]
- Hye Hur S., Kim H.Y., Kim Y.K., An J.M., Lee J.H., Kim H.J. Discrimination between Korean and Chinese kimchi using inductively coupled plasma-optical emission spectroscopy and mass spectrometry: A multivariate analysis of kimchi. Food Chemistry. 2023;423 doi: 10.1016/j.foodchem.2023.136235. [DOI] [PubMed] [Google Scholar]
- Jeong S., Seol D., Kim H., Lee Y., Nam S.H., An J.M., Chung H. Cooperative combination of LIBS-based elemental analysis and near-infrared molecular fingerprinting for enhanced discrimination of geographical origin of soybean paste. Food Chemistry. 2023;399 doi: 10.1016/j.foodchem.2022.133956. [DOI] [PubMed] [Google Scholar]
- Kang C.D., Zhang Y.Y., Zhang M.Y., Qi J., Zhao W.T., Gu J.…Li Y.Y. Screening of specific quantitative peptides of beef by LC–MS/MS coupled with OPLS-DA. Food Chemistry. 2022;387 doi: 10.1016/j.foodchem.2022.132932. [DOI] [PubMed] [Google Scholar]
- Li M., Chai X., Wang L.Y., Yang J., Wang Y.F. Study of the variation of phenolic acid and flavonoid content from fresh Artemisiae argyi folium to Moxa wool. Molecules. 2019;24(24):4603. doi: 10.3390/molecules24244603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He Y.N., Wang F., Xu R.C., Yang M., Ci Z.M.…Lin J.Z. From longevity grass to contemporary soft gold: Explore the chemical constituents, pharmacology, and toxicology of Artemisia argyi H.Lév. & vaniot essential oil. Journal of Ethnopharmacology. 2021;279 doi: 10.1016/j.jep.2021.114404. [DOI] [PubMed] [Google Scholar]
- Long Q.Q., Feng N., Gan H.X., Zhang X., Han Y.P., Gu J. Study on origin traceability of Amur cork-tree bark based on stable isotopes and multielements combined chemometrics. Industrial Crops and Products. 2023;194 doi: 10.1016/j.indcrop.2023.116380. [DOI] [Google Scholar]
- Lu L., Wang L., Liu R.Y., Zhang Y.B., Zheng X.Q., Lu J.L.…Ye J.H. An efficient artificial intelligence algorithm for predicting the sensory quality of green and black teas based on the key chemical indices. Food Chemistry. 2024;441 doi: 10.1016/j.foodchem.2023.138341. [DOI] [PubMed] [Google Scholar]
- Mi S., Shang K., Li X., Zhang C.H., Liu J.Q., Huang D.Q. Characterization and discrimination of selected China’s domestic pork using an LC-MS-based lipidomics approach. Food Control. 2019;100:305–314. doi: 10.1016/j.foodcont.2019.02.001. [DOI] [Google Scholar]
- Mi S., Wang Y.H., Zhang X.N., Sang Y.X., Wang X.H. Authentication of the geographical origin of sesame seeds based on proximate composition, multi-element and volatile fingerprinting combined with chemometrics. Food Chemistry. 2022;397 doi: 10.1016/j.foodchem.2022.133779. [DOI] [PubMed] [Google Scholar]
- Nie J., Yang J., Liu C.L., Li C.L., Shao S.Z., Yao C.X.…Yuan Y.W. Stable isotope and elemental profiles determine geographical origin of saffron from China and Iran. Food Chemistry. 2023;405 doi: 10.1016/j.foodchem.2022.134733. [DOI] [PubMed] [Google Scholar]
- Oliveira M.M., Cruz-Tirado J.P., Barbin D.F. Nontargeted analytical methods as a powerful tool for the authentication of spices and herbs: A review. Comprehensive Reviews in Food Science and Food Safety. 2019;18(3):670–689. doi: 10.1111/1541-4337.12436. [DOI] [PubMed] [Google Scholar]
- Pacholczyk-Sienicka B., Modranka J., Ciepielowski G. Comparative analysis of bioactive compounds in garlic owing to the cultivar and origin. Food Chemistry. 2024;439 doi: 10.1016/j.foodchem.2023.138141. [DOI] [PubMed] [Google Scholar]
- Qi J., Li Y.Y., Zhang C., Wang C., Wang J.Q., Guo W.P., Wang S.W. Geographic origin discrimination of pork from different Chinese regions using mineral elements analysis assisted by machine learning techniques. Food Chemistry. 2021;337 doi: 10.1016/j.foodchem.2020.127779. [DOI] [PubMed] [Google Scholar]
- Song X.C., Canellas E., Nerin C. Screening of volatile decay markers of minced pork by headspace-solid phase microextraction–gas chromatography–mass spectrometry and chemometrics. Food Chemistry. 2021;342 doi: 10.1016/j.foodchem.2020.128341. [DOI] [PubMed] [Google Scholar]
- Song X.W., Wen X., He J.W., Zhao H., Li S.M., Wang M.Y. Phytochemical components and biological activities of Artemisia argyi. Journal of Functional Foods. 2019;52:648–662. doi: 10.1016/j.jff.2018.11.029. [DOI] [Google Scholar]
- Su S.H., Sundhar N., Kuo W.W., Lai S.C., Kuo C.H., Ho T.J.…Huang C.Y. Artemisia argyi extract induces apoptosis in human gemcitabine-resistant lung cancer cells via the PI3K/MAPK signaling pathway. Journal of Ethnopharmacology. 2022;299 doi: 10.1016/j.jep.2022.115658. [DOI] [PubMed] [Google Scholar]
- Wang H.B., Zhang M.L., Ma Y.R., Wang B., Shao M.W., Huang H.…Kang Z.H. Selective inactivation of gram-negative bacteria by carbon dots derived from natural biomass: Artemisia argyi leaves. Journal of Materials Chemistry B. 2020;8(13):2666–2672. doi: 10.1039/C9TB02735A. [DOI] [PubMed] [Google Scholar]
- Wang Z.C., Wang L., Huang H.T., Li Q.Y., Wang X.Y., Sun Q.…Li N. In vitro antioxidant analysis of flavonoids extracted from Artemisia argyi stem and their anti-inflammatory activity in lipopolysaccharide-stimulated RAW 264.7 macrophages. Food Chemistry. 2023;407 doi: 10.1016/j.foodchem.2022.135198. [DOI] [PubMed] [Google Scholar]
- Wei J., Wang Y., Tang X., Du Y., Bai Y., Deng Y.…Kang J. Correlation analysis between active components of Cornus officinalis and inorganic elements in rhizosphere soil and rapid analysis of origin quality by near-infrared spectroscopy combined with machine learning. Industrial Crops and Products. 2024;210 doi: 10.1016/j.indcrop.2024.118101. [DOI] [Google Scholar]
- Wu F.H., Zhao H.A., Sun J., Guo J.B., Wu L.M., Xue X.F., Cao W. ICP-MS-based ionomics method for discriminating the geographical origin of honey of Apis cerana Fabricius. Food Chemistry. 2021;354 doi: 10.1016/j.foodchem.2021.129568. [DOI] [PubMed] [Google Scholar]
- Wu L.K., Agarwal S., Kuo C.H., Kung Y.L., Day C.H., Lin P.Y.…Chiang C.Y. Artemisia leaf extract protects against neuron toxicity by TRPML1 activation and promoting autophagy/mitophagy clearance in both in vitro and in vivo models of MPP+/MPTP-induced Parkinson’s disease. Phytomedicine. 2022;104 doi: 10.1016/j.phymed.2022.154250. [DOI] [PubMed] [Google Scholar]
- Wu W., Xiang F., He F. Polyphenols from Artemisia argyi leaves: Environmentally friendly extraction under high hydrostatic pressure and biological activities. Industrial Crops and Products. 2021;171 doi: 10.1016/j.indcrop.2021.113951. [DOI] [Google Scholar]
- Xiao J.Q., Liu W.Y., Sun H.P., Li W., Koike K., Kikuchi T.…Zhang J. Bioactivity-based analysis and chemical characterization of hypoglycemic and antioxidant components from Artemisia argyi. Bioorganic Chemistry. 2019;92 doi: 10.1016/j.bioorg.2019.103268. [DOI] [PubMed] [Google Scholar]
- Ye Y.Y., Yan W., Peng L.J., He J.L., Zhang N., Zhou J.J.…Cai J. Minerals and bioactive components profiling in se-enriched green tea and the Pearson correlation with se. LWT. 2023;175 doi: 10.1016/j.lwt.2023.114470. [DOI] [Google Scholar]
- Yoon D., Shin W.C., Oh S.M., Choi B.R., Young Lee D. Integration of multiplatform metabolomics and multivariate analysis for geographical origin discrimination of Panax ginseng. Food Research International. 2022;159 doi: 10.1016/j.foodres.2022.111610. [DOI] [PubMed] [Google Scholar]
- Yu D.X., Guo S., Zhang X., Yan H., Mao S.W., Wang J.M.…Duan J.A. Combining stable isotope, multielement and untargeted metabolomics with chemometrics to discriminate the geographical origins of ginger (Zingiber officinale roscoe) Food Chemistry. 2023;426 doi: 10.1016/j.foodchem.2023.136577. [DOI] [PubMed] [Google Scholar]
- Zhan H., Xu W.F., Zhao X.X., Tian L.L., Zhang F., Wei H., Tao X.Y. Effects of Lactiplantibacillus plantarum WLPL01 fermentation on antioxidant activities, bioactive compounds, and flavor profile of Artemisia argyi. Food Bioscience. 2022;49 doi: 10.1016/j.fbio.2022.101908. [DOI] [Google Scholar]
- Zhang D., Yao L.M., Chang Y.Q., Yang G.Y., Xue Z.J., Wang L.…Guo L. Evaluation and comparison of bioactive constituents of Artemisiae argyi folium collected at different developmental stages. Journal of AOAC International. 2021;104(2):515–525. doi: 10.1093/jaoacint/qsaa105. [DOI] [PubMed] [Google Scholar]
- Zhang H.R., Liu W.Y., Shen Q.S., Zhao L.Y., Zhang C.H., Richel A. Discrimination of geographical origin and species of China’s cattle bones based on multi-element analyses by inductively coupled plasma mass spectrometry. Food Chemistry. 2021;356 doi: 10.1016/j.foodchem.2021.129619. [DOI] [PubMed] [Google Scholar]
- Zhang X.W., Wang S., Tu P.F., Zeng K.W. Sesquiterpene lactone from Artemisia argyi induces gastric carcinoma cell apoptosis via activating NADPH oxidase/reactive oxygen species/mitochondrial pathway. European Journal of Pharmacology. 2018;837:164–170. doi: 10.1016/j.ejphar.2018.07.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.