Abstract
Liaoning cashmere goat (LCG) is characterized by the highest individual cashmere yield, but its cashmere fineness tends to be coarse. Therefore, our research primarily focuses on reducing cashmere fineness. Through lipidomics screening and identification, we identified the crucial functional genes FA2H and ELOVL3 associated with cashmere fineness. Subsequently, using PCR-seq, we conducted gene typing and SNP analysis on the experimental population DNA, In the FA2H gene, a SNP locus T42443G was detected in LCG buck, with the TT genotype showing advantageous traits in cashmere fineness, meat quality, and body size, while the TG genotype demonstrated advantages in slaughter performance,In LCG doe, the TG genotype shows advantageous traits in cashmere fineness, milk production, and meat quality, while the TT genotype exhibits advantages in slaughter performance, lambing, and body size. In the ELOVL3 gene, a SNP locus C2133A was identified in LCG buck, where the CC genotype was advantageous for cashmere fineness, Only CA genotype was found in slaughter and meat quality. Additionally, and the CA genotype showed superiority in body size. On LCG doe, The CC genotype was the advantageous genotype in terms of cashmere fineness, milk production, slaughter performance, and meat quality. The CA genotype was the advantageous genotype in terms of lambing and body size. The dominant genotypes identified to influence both doe cashmere fineness and slaughter performance were TT and CC. The identified dominant haplotype combination for cashmere production performance in LCG was CCTG. The dominant haplotype combination for doe slaughter performance was the CCTT haplotype combination. The dominant haplotype combination for buck slaughter performance was the CATG haplotype combination. Therefore, the TT genotype of the FA2H gene and the CC genotype of the ELOVL3 gene in LCG buck, and the TG genotype of the FA2H gene and the CC genotype of the ELOVL3 gene in doe can be used as molecular markers for assisted selection of cashmere fineness. CCTG haplotype combination was the superior haplotype combinations for cashmere production performance. To provide a theoretical basis for the breeding and expansion of fine-fiber type new strains of LCG.
Keywords: SNP, Lipid-related genes, Slaughter performance, Gene substitution effect, Haplotype marker
1. Introduction
The cashmere goat is a valuable biological resource that plays a crucial role in Chinese animal husbandry,[1], [2], [3], [4] Thus, combined trials with emphasis on administration and genetic progress to improve animal outputs are of decisive significance,[5], [6], [7], [8] Economical and biological efficiency of small ruminant production enterprises generally improves by increasing productivity and reproductive performance of these animals.[9], [10], [11], [12], [13] The cashmere goat is a valuable biological resource that plays a crucial role in Chinese animal husbandry.14 China has over 20 native breeds of cashmere goats, which account for 75 % of the world's cashmere production. Among these breeds, LCG is renowned for its excellent cashmere yield.15 However, the growth of cashmere is influenced by many factors such as climate, breed, gender, age, body region, genetics, and nutrient absorption.16 In recent years, there has been increasing attention to the quality of cashmere products, with cashmere fineness being one of the key factors affecting cashmere quality.17 The LCG has a high cashmere yield, but the cashmere is moderately fine, with the Ministry of Agriculture requiring a fineness of 16 μm or less, and there is still room for decline.Therefore, methods need to be found to reduce its fiber diameter. Single nucleotide polymorphisms (SNPs) are widely used in animal genetic breeding research due to their numerous, widespread distribution, strong genetic stability, and ease of large-scale rapid detection.18 A number of genes related to cashmere fineness have been identified in the current study. The KIF-1 gene has been found to be associated with cashmere fineness in Xinjiang goats,19 Fu et al.,20 identified through high-throughput RNA sequencing that KRT26 and other genes are involved in hair follicle morphogenesis and skin development, potentially regulating cashmere fineness. On the basis of using expression profiling microarrays to detect candidate genes related to fine wool fibre diameter, Tian 21 established a real-time fluorescence quantitative PCR method and found that genes such as TXNIP and TFDP1 were related to cashmere fineness. Based on previous studies, this article found that during the growth and development of cashmere, in addition to the role played by the dermis of the skin, from September to December, the hair follicles extend into the subcutaneous adipose tissue to absorb nutrients. Therefore, through phenotypic omics screening of cashmere fineness differences, two genes, ELOVL3 and FA2H, were identified as significantly differentially expressed and potentially involved in regulating cashmere fineness.
The ELOVL3 gene is a member of the fatty acid elongase family.22 Anders Jacobson's team first discovered the very long-chain fatty acid elongase 3 (ELOVL3) in brown adipose tissue (BAT) exposed to cold temperatures.23 ELOVL3 is primarily expressed in the liver, brown adipose tissue, white adipose tissue, skin, and triglyceride-rich glands.24 Rolf Westerberg discovered that ELOVL3 is involved in the formation of specific neutral lipids essential for skin function. Mice lacking ELOVL3 exhibit hair loss, sebaceous gland hyperplasia, disrupted hair lipid content, and notably high levels of eicosenoic acid.25 Studies in some mammals have found that the ELOVL3 gene appears to play a role in the physiological processes of hair formation, follicle growth and development.26 Yu 27 conducted transcriptome sequencing on the shoulder blades of LCG and identified the FA2H gene as potentially influencing the fineness of cashmere. Zhou 28 used transcriptome sequencing to discover differential expression of the ELOVL3 gene in the skin tissues of Shanbei White Cashmere goats during their growth, quiescent, and regression periods. Wu 29 research on Nan jiang cashmere goats found that overexpression of the ELOVL3 gene can promote the growth of secondary hair follicle cells, further demonstrating the regulatory role of the ELOVL3 gene in cashmere goat traits.
FA2H is one of the metabolic enzymes for fatty acids. Research on this gene primarily focuses on human diseases such as cancer,30 hereditary spastic paraplegia,31 and others. Researchers have also found that this gene plays a crucial role in hair follicle growth. Wang et al. conducted weighted gene co-expression network analysis (WGCNA) on Inner mongolia cashmere goats and identified 12 candidate genes, including FA2H.32 Wu et al.,33 utilized transcriptome sequencing technology to study Nan jiang cashmere goats, identifying 7 candidate genes including FA2H. They found that overexpression of FA2H promotes proliferation of secondary hair follicle cells in cashmere goats.
This study was first carried out on LCG. The aim of this study is to investigate the effects of SNPs in the ELOVL3 and FA2H genes of LCG on their cashmere production performance, body size performance, milk production performance, lambing, slaughter performance, and meat quality performance. Through genetic diversity analysis and correlation analysis of six traits, we aim to identify the genotypes and haplotype combinations that affect these six traits.This will facilitate the breeding and improvement of LCG varieties, advance genetic breeding efforts for cashmere goats, and provide theoretical support for cultivating superior cashmere goats. Compared to other relevant studies, our study is more comprehensive, And for the first time, we investigated the effects of FA2H and ELOVL3 genes on the cashmere production performance of LCG.
2. Materials and methods
2.1. Experimental animals
At the Liaoning cashmere goat breeding center in Liaoning Province, China, 1,181 healthy LCG were selected, all of which were fed under consistent conditions. All animal handling procedures and protocols used in this study were approved in accordance with the guidelines of the Laboratory Animal Management Committee. (Animal Welfare Ethics Certificate Number: 2024.05.13), Blood samples for DNA extraction were collected under the guidance of a qualified veterinarian, obtaining 1 mL of blood from the jugular vein of each goat. After collection, the samples were placed into blood collection tubes containing EDTA and stored at −20°C.
2.2. Production performance phenotype data
The performance data of cashmere is measured by a portable all-weather cashmere fineness and length rapid testing machine. The cashmere is placed into the plate that comes with the analyser, the plate is placed into the analyser, after which the cashmere data is able to be obtained by clicking on Start Test.
The body size data is obtained from the intelligent body measurement system. The goat is driven into the instrument and its body measurements are automatically detected as it passes forward.
The lambing data is obtained through counting method. Counts were taken at lambing for each doe and summarised in the table.
The milk production data is obtained using a milk component analyzer. Prepare two sample bottles and fill them with the same goat's milk. Place one bottle with goat's milk under the test port of the milk analyzer, place the PH sensor in the other bottle with goat's milk, click on the test and wait for the test result.
Slaughter data is obtained according to the Operation Regulations for Slaughtering Poultry and Livestock (GB/T 43562–2023). All the data are obtained by weighing and calculating.
The quality of meat products is determined according to the Technical Specification for Meat Quality Determination (T/CAAA 102–2023).
2.3. DNA extraction
Take 200 μL of blood from the anticoagulant tube and transfer it to a centrifuge tube. Add 20 μL of Proteinase K and mix well. Then add Buffer DL, shake vigorously, and incubate at 56 °C in a water bath for 10 min. Next, add 200 μL of anhydrous ethanol to the centrifuge tube and mix well. Transfer the liquid to a DNA adsorption column and let it stand for two minutes. Centrifuge at 10,000 rpm at room temperature for 1 min and discard the waste liquid in the collection tube. Add 500 μL of GW Solution to the adsorption column, centrifuge at 10, 000 rpm for 30 s, and discard the waste liquid. Add 700 μL of Wash Solution to the adsorption column, centrifuge at 10, 000 rpm for 30 s, and discard the waste liquid. Repeat this step twice. Then centrifuge at 12,000 rpm at room temperature for 2 min to remove any remaining liquid. Remove the adsorption column and place it in a new centrifuge tube. Add 50 μL of CE Buffer, let it stand for 3 min, and centrifuge at 12,000 rpm at room temperature for 2 min. Collect the DNA solution and measure the sample's OD value using UV spectrophotometry. Store the qualified samples at −20 °C.
2.4. Primer design
The sequences of the FA2H gene (Reference number: NC_030825.1) and the ELOVL3 gene (Reference number: NC_030833.1) were obtained from the NCBI database. Specific primers were designed using Primer Premier 5 software 34 (Table 1).
Table 1.
Primer design of FA2H and ELOVL3 genes.
| Gene | Sense primer (Forward) | Anti-sense primer (Reverse) | TM(℃)F/R | Fragment size | Regions |
|---|---|---|---|---|---|
| FA2H | 5′GTTGGGATGAAGGGTTAG3′ | 5′CAGGAGGAGGAAAGAAGA3′ | 49.8 | 722 bp | 42249–42971 |
| ELOVL3 | 5′'ACCCCTATCCTGCCACCTGT3′ | 5′'GTGTTGGGACCACCCTCTGA3′ | 52.1 | 664 bp | 1681–2325 |
2.5. PCR amplification
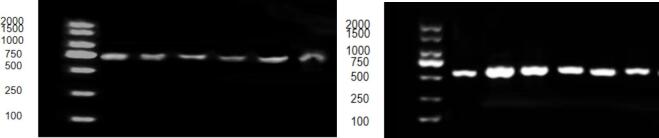
The PCR reaction system has a total volume of 50 μL, which includes 25 μL of 2x SanTaq PCR Mix solution, 1 μL of DNA template, 2 μL each of upstream and downstream primers, and 20 μL of ddH2O. Add these reagents to the PCR tube, mix thoroughly, and centrifuge. Perform the amplification in the PCR machine according to the PCR reaction conditions.The reaction conditions were pre-denaturation at 94℃ for 5 min, denaturation at 94℃ for 30 s, adjusting the temperature to 49.8℃-52.1℃ for annealing for 30 s, extension at 72℃ for 30 s, and finally keeping the extension at 72℃ for 10 min.Then electrophoresis was conducted at 130 V and 180 W for 20 min.After electrophoresis, observe whether the band of the electrophoresis result contains the target fragment (Fig. 1). If it is present, send the sample to Shanghai Biotechnology Co., Ltd. for sequencing.
Fig. 1.
Electrophoretic image FA2H (left), ELOVL3 (right).
2.6. Statistical analyses
Calculate genotype and allele frequencies, polymorphic information content (PIC), effective number of alleles (Ne), and heterozygosity (He). Perform single-factor analysis using SPSS software for the FA2H and ELOVL3 genes in relation to six traits of LCG. The integrity of the animal model was analyzed using Yijkl = μ + hi + pj + sk + ml + eijkl, Yijkl = observe value; μ = overall mean; hi = the effect of genotype or combined haplotype; pj = effect of season and farm; sk = effect of year; ml = effect of sire descent and eijkl = random error. Use Duncan's method for multiple comparisons. A P > 0.05 indicates no significant difference, <0.05 indicates a significant difference (marked with lowercase letters), and < 0.01 indicates a highly significant difference (marked with uppercase letters). Results should be presented as 'mean ± standard error.
3. Results
3.1. SNP locus sequencing map
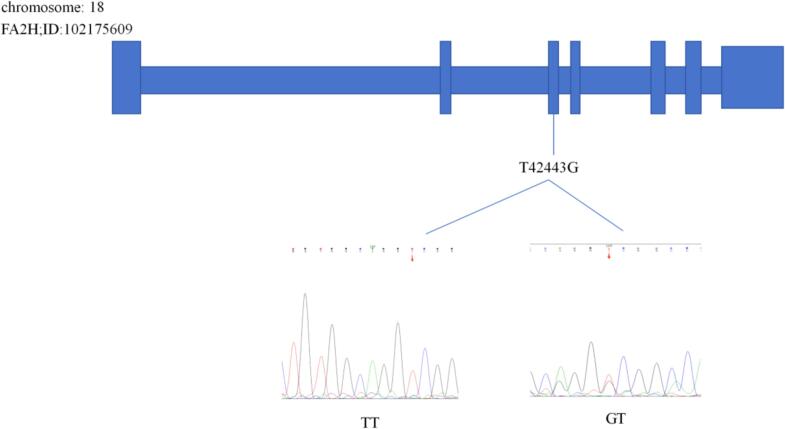
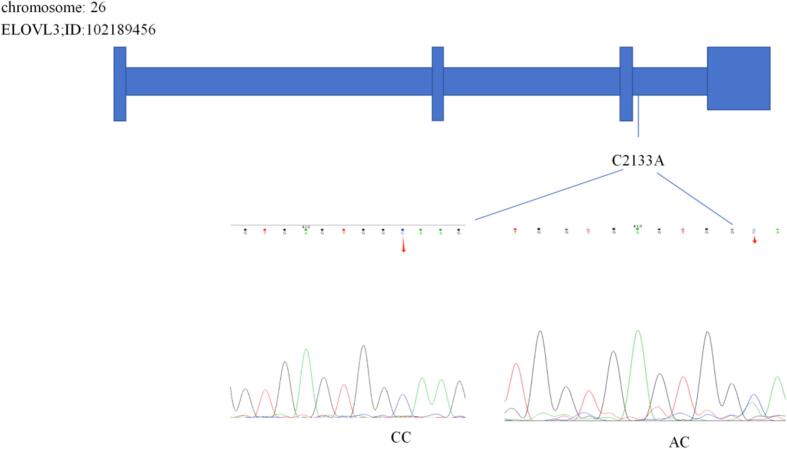
We compared the results and gene sequences of the FA2H and ELOVL3 genes. Using Chromas 2 and DNAMAN software, comparative analysis revealed one SNP (T42443G) was detected in the FA2H gene (Fig. 2) and one SNP (C2133A) was detected in the ELOVL3 gene (Fig. 3).
Fig. 2.
The T42443G locus of the FA2H gene.
Fig. 3.
The C2133A locus of the ELOVL3 gene.
3.2. Genetic diversity of the FA2H and ELOVL3 genes
The genotype and allele frequencies of SNP loci in the FA2H and ELOVL3 genes in LCG are presented in Table 2. Genes with frequencies greater than 0.5 are considered dominant. The polymorphism information content (PIC) values for the two loci range from 0.25 to 0.5, indicating moderate polymorphism. This suggests a significant genetic variation in these two genes in LCG, which could lead to substantial genetic progress.
Table 2.
Genetic diversity analysish of the FA2H and ELOVL3 genes in LCG.
| Name | loci | Genotype Frequency |
Allelic Frequencies | PIC | He | Ne | χ2 | P | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MM | Mm | mm | M | m | |||||||
| buck | T42443G | 0.43 | 0.57 | 0 | 0.72 | 0.28 | 0.32 | 0.41 | 1.68 | 3.57 | 0.06 |
| doe | T42443G | 0.37 | 0.63 | 0 | 0.69 | 0.31 | 0.34 | 0.43 | 1.76 | 16.36 | 5.24458E-05 |
| buck | C2133A | 0.08 | 0.92 | 0 | 0.54 | 0.46 | 0.37 | 0.50 | 1.99 | 18.14 | 2.05105E-05 |
| doe | C2133A | 0.24 | 0.76 | 0 | 0.62 | 0.38 | 0.36 | 0.47 | 1.89 | 30.30 | 3.70735E-08 |
3.3. Gene substitution effect analysis
The negative additive effect value at the T42443G locus of FA2H gene on LCG indicates that the substitution of the T42443G locus by T into G can improve the production performance, the positive additive effect value at the C2133A locus of ELOVL3 gene on LCG indicates that the substitution of the C2133A locus by C into A can reduce the production performance (Table 3).
Table 3.
Gene substitution effect analysis.
| Name | Loci | Dominant effect | Additive effect | Average effect of u gene | Average effect of U gene | The average effect of u instead of U |
|---|---|---|---|---|---|---|
| d | a | a1 | a2 | a | ||
| buck | T42443G | 8 | −5 | −1.09 | 0.43 | −1.52 |
| doe | T42443G | 34.5 | −14.5 | −1.15 | 0.53 | −1.67 |
| buck | C2133A | 22 | −1 | 0.41 | −0.35 | 0.76 |
| doe | C2133A | 52 | −10 | 1.67 | −1.01 | 2.68 |
3.4. Analysis of the relationship between SNPs and cashmere production performance
At the T42443G locus in buck the TT genotypes was highly significantly better than the TG genotypes in number of curls, significantly better than the TG genotypes in net cashmere rate, and the TT genotypes was also better in cashmere fineness. The CC genotypes was highly significantly better than the CA genotypes at the C2133A locus in terms of net cashmere rate, and the CA genotypes was highly significantly better than the CC genotypes in terms of cashmere yield. CC genotype was better in cashmere fineness.
At the T42443G locus in doe the TT genotypes was highly significantly superior to the TG genotypes in cashmere yield and significantly superior to the TG genotypes in cashmere length, and the TG genotypes was highly significantly superior to the TT genotypes in number of curls and net cashmere rate. The TT genotypes was significantly better than the TG genotypes in terms of cashmere fineness. At the C2133A locus, the CC genotype shows highly significantly superior net cashmere rate compared to the CA genotype, while individuals with the CA genotype exhibit significantly better cashmere fineness and number of curls than those with the CC genotype (Table 4).
Table 4.
The cashmere production performance related to the FA2H and ELOVL3 genes in LCG.
| Name | Loci | Genotype | Quantities | Cashmere yield (g) | Cashmere fineness (μm) | Cashmere length (cm) | number of curls | Short cashmere rate (%) | Net cashmere rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| buck | T42443G | TT | 18 | 2000.00 ± 94.11 | 16.55 ± 0.26 | 9.73 ± 0.31 | 8.83 ± 0.19aA | 19.91 ± 3.15 | 78.20 ± 2.30a |
| buck | T42443G | TG | 26 | 1984.62 ± 60.56 | 16.80 ± 0.22 | 9.16 ± 0.52 | 5.29 ± 0.84bB | 16.92 ± 3.67 | 70.30 ± 2.72b |
| buck | C2133A | CC | 4 | 2025.00 ± 101.04 | 16.44±0.32 | 8.31 ± 1.40 | 4.10 ± 2.37ab | 26.97 ± 1.32a | 76.76 ± 0.22 |
| buck | C2133A | CA | 46 | 1904.35 ± 52.24 | 16.48 ± 0.16 | 9.44 ± 0.36 | 7.08 ± 0.57a | 17.35 ± 2.60ab | 76.32 ± 1.86 |
| doe | T42443G | TT | 261 | 1700.00 ± 20.57b | 16.95 ± 0.07a | 10.39 ± 0.17 | 5.71 ± 0.23b | 16.66 ± 0.79b | 70.57 ± 0.83bB |
| doe | T42443G | TG | 441 | 1836.73 ± 15.02a | 16.41 ± 0.06b | 9.55 ± 0.15 | 7.16 ± 0.19a | 18.01 ± 0.73a | 78.00 ± 0.69aA |
| doe | C2133A | CC | 180 | 1807.50 ± 26.98 | 16.32 ± 0.11b | 9.44 ± 0.236 | 8.69 ± 0.19aA | 15.32 ± 1.11 | 78.58 ± 1.13a |
| doe | C2133A | CA | 558 | 1770.97 ± 12.86 | 16.82 ± 0.05a | 9.96 ± 0.132 | 6.13 ± 0.17bB | 16.89 ± 0.59 | 72.44 ± 0.64b |
3.5. Analysis of the relationship between SNPs and milk production performance
The TG genotype at locus T42443G in LCG doe was highly significantly superior to the TT genotype at Fat, N, Cond. and Cru.Prot.The CC genotype was highly significantly superior to the CA genotype at the C2133A locus of the ELOVL3 gene in terms of Fat, Urea, and N (Table 5).
Table 5.
Analysis of the milk production performance of genes polymorphic loci.
| Name | Loci | Genotype | Quantities | Fat | Cru.Prot | Lactose | Urea | N | SnF | TS | Cond. | H.Index |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| doe | T42443G | TT | 128 | 7.22 ± 0.22 | 4.51 ± 0.03bB | 5.29 ± 0.02aA | 37.50 ± 0.26bB | 17.49 ± 0.12bB | 10.30 ± 0.04b | 17.88 ± 0.24 | 738.70 ± 5.84bB | 0.48 ± 0.01 |
| doe | T42443G | TG | 184 | 7.57 ± 0.23 | 5.29 ± 0.14aA | 5.02 ± 0.04bB | 41.34 ± 0.44aA | 19.29 ± 0.20aA | 10.92 ± 0.13a | 18.88 ± 0.25 | 784.57 ± 4.96aA | 0.49 ± 0.01 |
| doe | C2133A | CC | 84 | 7.96 ± 0.27aA | 5.31 ± 0.17 | 4.95 ± 0.06 | 42.51 ± 0.59aA | 19.84 ± 0.28aA | 10.85 ± 0.14 | 19.19 ± 0.33 | 789.90 ± 7.33 | 0.50 ± 0.01 |
| doe | C2133A | CA | 234 | 7.07 ± 0.18bB | 5.32 ± 0.18 | 5.09 ± 0.05 | 39.83 ± 0.57bB | 18.58 ± 0.26bB | 11.02 ± 0.18 | 18.48 ± 0.24 | 775.14 ± 7.50 | 0.47 ± 0.01 |
3.6. Analysis of the relationship between SNPs and slaughtering performance
At the T42443G locus in LCG buck, the TG genotype shows highly significantly superior traits in live weight before slaughter, carcass weight, net meat weight, slaughter rate, net meat rate, and GR compared to the TT genotype. At the C2133A locus, only the CA genotype was found.
At the T42443G locus in LCG doe, the TT genotype showshighly significantly superior traits in net meat weight and slaughter rate compared to the TG genotype. It was highly significantly superior to TG genotype in terms of net meat rate, carcass net meat rate. At the C2133A locus the CC genotype was significantly superior to the CA genotype in terms of carcass weight, net meat weight, slaughter rate, and net meat rate, It shows highly significant in eye muscle area (EMA) compared to the CA genotype (Table 6).
Table 6.
Analysis of the Slaughtering performance of genes polymorphic loci.
| Name | Loci | Genotype | Quantities | Live weight before slaughter(kg) | Carcass weight (kg) |
Net meat weight (kg) |
Slaughter rate (%) |
Net meat rate (%) |
Carcass net meat rate(%) | EMA(cm2) | GR(mm) | BFT(mm) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| buck | T42443G | TT | 15 | 45.02 ± 0.67bB | 22.48 ± 0.53bB | 17.52 ± 0.49bB | 49.95 ± 0.99bB | 38.90 ± 0.91bB | 77.82 ± 0.57 | 23.40 ± 1.53 | 4.73 ± 0.44bB | 1.62 ± 0.12 |
| buck | T42443G | TG | 12 | 49.75 ± 2.13aA | 26.20 ± 1.16aA | 20.63 ± 1.07aA | 52.65 ± 0.35aA | 41.30 ± 0.51aA | 78.42 ± 0.68 | 21.24 ± 0.51 | 7.73 ± 0.33aA | 1.92 ± 0.27 |
| buck | C2133A | CA | 26 | 46.30 ± 0.94 | 23.82 ± 0.64 | 18.60 ± 0.62 | 51.37 ± 0.66 | 40.01 ± 0.70 | 77.80 ± 0.59 | 22.41 ± 0.87 | 6.13 ± 0.38 | 2.09 ± 0.19 |
| doe | T42443G | TT | 42 | 48.94 ± 1.03 | 25.86 ± 0.67 | 21.59 ± 0.58a | 52.68 ± 0.43a | 43.94 ± 0.37aA | 83.40 ± 0.24aA | 20.45 ± 0.46 | 7.73 ± 0.27 | 2.89 ± 0.14 |
| doe | T42443G | TG | 18 | 43.30 ± 1.04 | 21.37 ± 0.46 | 17.07 ± 0.39b | 49.40 ± 0.17b | 39.46 ± 0.32bB | 79.86 ± 0.45bB | 19.07 ± 1.08 | 8.86 ± 0.46 | 2.98 ± 0.16 |
| doe | C2133A | CC | 5 | 55.00 ± 1.23 | 30.10 ± 0.78a | 25.40 ± 0.53a | 54.73 ± 0.76a | 46.18 ± 0.46a | 84.39 ± 0.31 | 26.95 ± 0.33aA | 9.37 ± 0.00 | 2.53 ± 0.00 |
| doe | C2133A | CA | 55 | 47.66 ± 0.83 | 24.11 ± 0.49b | 19.79 ± 0.44b | 50.58 ± 0.41b | 41.48 ± 0.41b | 81.97 ± 0.29 | 19.86 ± 0.29bB | 8.09 ± 0.25 | 2.81 ± 0.13 |
3.7. Analysis of the relationship between SNPs and meat quality performance
TT genotype was extremely significant to TG genotype in meat color b, dry matter and cooked meat rate at T42443G locus of FA2H gene of LCG buck. TG genotype was extremely significant to TT genotype in fat content. Only CA genotype was found at ELOVL3 gene.
TT genotype was significantly superior to TG genotype in meat color L and dry matter at T42443G locus of FA2H gene in LCG doe. TG genotype was extremely significant to TT genotype in meat color A, PH, protein content and fat content. CC genotype was extremely significant to CA genotype in meat color a, meat color b, fat content, cooked meat rate and shear force at C2133A locus of ELOVL3 gene (Table 7).
Table 7.
Analysis of the meat quality performance of genes polymorphic loci.
| Name | Loci | Genotype | Quantities | Meat color |
pH | Dry matter(%) | Protein content(%) | Fat content(%) | Drip loss(%) | Cooked meat rate(%) | Shear force(N) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | a | b | |||||||||||
| buck | T42443G | TT | 15 | 29.36 ± 0.23 | 14.86 ± 0.50 | 1.69 ± 0.03aA | 6.01 ± 0.02 | 26.72 ± 0.44aA | 21.66 ± 0.15 | 1.77 ± 0.16bB | 1.58 ± 0.07 | 67.06 ± 0.78aA | 73.80 ± 1.86 |
| buck | T42443G | TG | 12 | 29.60 ± 0.66 | 15.03 ± 0.59 | 1.56 ± 0.02bB | 6.05 ± 0.03 | 25.10 ± 0.21bB | 21.35 ± 0.25 | 2.39 ± 0.23aA | 1.65 ± 0.03 | 65.26 ± 0.20bB | 76.21 ± 2.25 |
| buck | C2133A | CA | 26 | 29.24 ± 0.32 | 14.94 ± 0.35 | 1.71 ± 0.07 | 6.06 ± 0.03 | 25.88 ± 0.30 | 21.39 ± 0.20 | 1.91 ± 0.14 | 1.65 ± 0.04 | 66.15 ± 0.75 | 75.61 ± 1.36 |
| doe | T42443G | TT | 42 | 30.69 ± 0.35a | 13.95 ± 0.18bB | 1.89 ± 0.11 | 5.95 ± 0.01bB | 28.37 ± 0.66a | 20.30 ± 0.20bB | 2.07 ± 0.13bB | 1.81 ± 0.04 | 67.99 ± 0.96 | 69.26 ± 1.87 |
| doe | T42443G | TG | 18 | 29.51 ± 0.25b | 15.03 ± 0.44aA | 2.00 ± 0.13 | 6.03 ± 0.02aA | 26.29 ± 0.24b | 21.44 ± 0.08aA | 2.79 ± 0.22aA | 1.85 ± 0.07 | 69.92 ± 0.24 | 69.83 ± 2.04 |
| doe | C2133A | CC | 5 | 31.17 ± 0.31 | 16.39 ± 0.00aA | 3.03 ± 0.02aA | 5.92 ± 0.01 | 24.43 ± 0.20bB | 20.71 ± 0.12 | 3.29 ± 0.11aA | 1.84 ± 0.03 | 77.99 ± 0.68aA | 86.87 ± 1.63aA |
| doe | C2133A | CA | 55 | 29.74 ± 0.30 | 14.13 ± 0.18bB | 1.80 ± 0.06bB | 5.96 ± 0.02 | 28.07 ± 0.47aA | 20.89 ± 0.18 | 2.28 ± 0.14bB | 1.78 ± 0.05 | 68.37 ± 0.60bB | 69.58 ± 1.43bB |
3.8. Analysis of the relationship between SNPs and lambing performance
At the T42443G locus in LCG doe, the TT genotype shows superiority in lambing compared to the TG genotype. At the C2133A locus, the CA genotype shows superiority in lambing compared to the CC genotype (Table 8).
Table 8.
Analysis of the lambing performance of genes polymorphic loci.
| Name | Loci | Genotype | Quantities | Number of kids |
|---|---|---|---|---|
| doe | T42443G | TT | 108 | 1.41 ± 0.5 |
| doe | T42443G | TG | 180 | 1.29 ± 0.04 |
| doe | C2133A | CC | 72 | 1.17 ± 0.04 |
| doe | C2133A | CA | 220 | 1.40 ± 0.03 |
3.9. Analysis of the relationship between SNPs and body size performance
At the T42443G locus in LCG buck, the TG genotype shows extremely significantly superior traits in sacral height compared to the TT genotype, The TT genotype shows significantly superior traits in body length and chest depth compared to the TG genotype. At the C2133A locus, the CA genotype shows significantly superior traits in chest width and chest circumference compared to the CC genotype.
At the T42443G locus in LCG doe, the TT genotype shows significantly superior traits in sacral height and body length compared to the TG genotype. At the C2133A locus, the CA genotype shows extremely significantly superior traits in body length and chest depth compared to the CC genotype (Table 9).
Table 9.
Analysis of the body size traits of genes polymorphic loci.
| Name | Loci | Genotype | Quantities | Body height (cm) | sacral height(cm) | Body oblique (cm) | Chest depth (cm) | Chest width (cm) | Waist width (cm) | Chest circumference(cm) |
tube circumference | Waist height (cm) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| buck | T42443G | TT | 28 | 74.66 ± 0.69 | 71.71 ± 0.72bB | 87.75 ± 0.92a | 36.14 ± 0.54a | 26.79 ± 0.92 | 21.43 ± 0.87 | 105.26 ± 1.00 | 12.57 ± 0.24 | 69.71 ± 0.73 |
| buck | T42443G | TG | 22 | 75.41 ± 0.78 | 74.05 ± 0.79aA | 81.41 ± 3.97b | 33.47 ± 1.72b | 26.98 ± 1.09 | 20.55 ± 0.63 | 105.71 ± 0.85 | 12.41 ± 0.20 | 69.18 ± 0.89 |
| buck | C2133A | CC | 20 | 74.90 ± 0.85 | 72.30 ± 1.04 | 87.10 ± 1.18 | 35.77 ± 0.52 | 25.70 ± 0.76b | 21.78 ± 0.75 | 103.27 ± 1.02b | 12.80 ± 0.31 | 68.00 ± 1.15 |
| buck | C2133A | CA | 30 | 75.82 ± 0.63 | 73.13 ± 0.53 | 88.33 ± 0.98 | 33.77 ± 1.42 | 28.53 ± 1.13a | 20.57 ± 0.59 | 105.96 ± 0.85a | 12.73 ± 0.18 | 70.43 ± 0.62 |
| doe | T42443G | TT | 282 | 64.05 ± 0.19 | 64.86 ± 0.19a | 79.99 ± 0.41a | 32.53 ± 0.13 | 23.54 ± 0.23 | 22.58 ± 0.27 | 99.29 ± 0.52 | 9.56 ± 0.06 | 63.79 ± 0.27 |
| doe | T42443G | TG | 444 | 63.87 ± 0.17 | 66.05 ± 0.15b | 77.67 ± 0.29b | 32.30 ± 0.14 | 23.47 ± 0.20 | 22.40 ± 0.23 | 99.51 ± 0.42 | 9.54 ± 0.05 | 63.80 ± 0.23 |
| doe | C2133A | CC | 210 | 63.50 ± 0.25 | 66.22 ± 0.25 | 76.07 ± 0.46bB | 31.82 ± 0.19bB | 23.67 ± 0.28 | 22.56 ± 0.34 | 100.95 ± 0.68 | 9.40 ± 0.06 | 64.64 ± 0.34 |
| doe | C2133A | CA | 594 | 64.27 ± 0.15 | 65.72 ± 0.14 | 78.86 ± 0.25aA | 32.90 ± 0.11aA | 23.79 ± 0.16 | 23.18 ± 0.19 | 99.64 ± 0.38 | 9.55 ± 0.04 | 64.03 ± 0.20 |
3.10. Analysis of the correlation between cashmere fineness and cashmere production performance in LCG
From Table 10, The length and net cashmere rate of doe was highly significantly correlated with cashmere fineness, cashmere yield, number of curls and short cashmere rate were significantly correlated with cashmere fineness, cashmere yield and net cashmere rate of buck was highly significantly correlated with cashmere fineness (Table 10).
Table 10.
Correlation coefficients between fineness of LCG and cashmere production performance.
| buck doe | Cashmere fineness (μm) | Cashmere yield (g) | Cashmere length (cm) | number of curls | Short cashmere rate (%) | Net cashmere rate (%) |
|---|---|---|---|---|---|---|
| Cashmere fineness (μm) | − | 0.278* | 0.322** | −0.246* | −0.238* | −0.936** |
| Cashmere yield (g) | 0.541** | − | 0.055 | −0.049 | −0.054 | −0.263* |
| Cashmere length (cm) | 0.084 | −0.266 | − | 0.078 | −0.309** | −0.293** |
| number of curls | −0.033 | 0.174 | 0.345* | − | −0.146 | 0.243* |
| Short cashmere rate (%) | −0.257 | 0.035 | −0.256 | −0.206 | − | 0.256* |
| Net cashmere rate (%) | −0.866** | −0.38** | 0.02 | 0.096 | 0.389** | − |
indicates highly significant correlation (P < 0.01), while.
indicates significant correlation (P < 0.05).
3.11. Path analysis results of fineness of LCG cashmere and cashmere production performance
From Table 11, it can be seen that the Net cashmere rate of LCG doe has the greatest direct effect on cashmere fineness (−0.905) followed by cashmere length (0.06), cashmere yield (0.036), number of curls (−0.028) and short cashmere rate (0.011), The maximum indirect effect of cashmere length on cashmere fineness is 0.262, followed by short cashmere rate (−0.248), cashmere yield (0.241),number of curls (−0.219) and net cashmere rate (−0.031). The direct effect of net cashmere rate on cashmere fineness is highest for LCG buck (−0.770), followed by cashmere yield (0.321). The largest indirect effect is cashmere yield (0.293), followed by net cashmere rate (−0.122) (Table 11).
Table 11.
Path coefficients of cashmere production Performance on fineness of LCG.
| Sex | Independent variable | Correlation coefficient | Direct action | Indirect effect |
||||
|---|---|---|---|---|---|---|---|---|
| Cashmere yield | Cashmere length | number of curls | Short cashmere rate | Net cashmere rate | ||||
| doe | Cashmere yield | 0.278 | 0.036 | 0.003 | 0.001 | −0.001 | 0.238 | |
| Cashmere length | 0.322 | 0.06 | 0.002 | −0.002 | −0.003 | 0.265 | ||
| Number of curls | −0.246 | −0.028 | −0.002 | 0.005 | −0.002 | −0.220 | ||
| Short cashmere rate | −0.238 | 0.011 | −0.002 | −0.019 | 0.004 | −0.231 | ||
| Net cashmere rate | −0.936 | −0.905 | −0.009 | −0.018 | −0.007 | 0.003 | ||
| buck | Cashmere yield | 0.541 | 0.321 | 0.293 | ||||
| Net cashmere rate | −0.866 | −0.770 | −0.122 | |||||
3.12. Results of stepwise multiple regression analysis of cashmere fineness and cashmere production performance in LCG
Table 12 shows that the optimal regression equation for doe is cashmere fineness = -0.077 net cashmere rate + 0.024 cashmere length + 0.0001 cashmere yield − 0.01 number of curls + 21.946.The optimal regression equation for buck is: fineness = -0.067 net cashmere rate + 0.001 cashmere yield + 0.016 cashmere length + 19.073. The multiple regression equations have final determination coefficients (R2) of 0.880 and 0.832, respectively. It means that the cashmere length, net cashmere rate, cashmere yield, number of curls, and short cashmere rate together can explain 88.0 % of the variation in cashmere fineness for doe. net cashmere rate, cashmere yield, and cashmere length explain 83.2 % of the variation, indicating a well-constructed model (Table 12).
Table 12.
Results of stepwise multiple regression analysis of cashmere production performance on cashmere fineness in LCG.
| Sex | Model | R2 | Adjusted R-squared | Standard error of estimate | F | P |
|---|---|---|---|---|---|---|
| doe | cashmere fineness = -0.08net cashmere rate + 22.586 | 0.876 | 0.876 | 0.459 | 5199.056 | 0.000 |
| cashmere fineness = -0.078net cashmere rate + 0.021cashmere length + 22.280 | 0.878 | 0.878 | 0.454 | 2655.831 | 0.000 | |
| cashmere fineness = -0.078net cashmere rate + 0.022cashmere length + 0.0001cashmere yield + 21.959 | 0.880 | 0.879 | 0.453 | 1787.580 | 0.000 | |
| cashmere fineness = -0.077net cashmere rate + 0.024cashmere length + 0.0001cashmere yield-0.01number of curls + 21.946 | 0.880 | 0.880 | 0.451 | 1349.376 | 0.000 | |
| buck | cashmere fineness = -0.077net cashmere rate + 22.374 | 0.750 | 0.745 | 0.543 | 144.052 | 0.000b |
| cashmere fineness = -0.069net cashmere rate + 0.001cashmere yield + 20.256 | 0.803 | 0.794 | 0.488 | 95.494 | 0.000c | |
| cashmere fineness = -0.067net cashmere rate + 0.001cashmere yield + 0.016 cashmere length + 19.073 | 0.832 | 0.821 | 0.455 | 76.117 | 0.000d |
3.13. Correlation analysis between cashmere fineness and slaughter performance of LCG
From Table 13, it is clear that carcass net meat rate and EMA of buck was highly significantly correlated with cashmere fineness and net meat rate was significantly correlated with cashmere fineness. Carcass weight, EMA, and BFT of doe was highly significantly correlated with cashmere fineness, and slaughter rate was significantly correlated with cashmere fineness (Table 13).
Table 13.
Correlation coefficients between cashmere fineness and slaughter performance of LCG.
| doe buck | Cashmere fineness | Live weight before slaughter | Carcass weight | Net meat weight | Slaughter rate | Net meat rate | Carcass net meat rate | EMA | GR | BFT |
|---|---|---|---|---|---|---|---|---|---|---|
| Cashmere fineness | − | −0.117 | −0.196 | −0.237 | −0.285 | −0.409* | −0.506** | −0.542** | 0.002 | 0.044 |
| Live weight before slaughter | 0.196 | − | 0.915** | 0.921** | 0.165 | 0.398* | 0.702** | −0.254** | 0.282** | 0.812** |
| Carcass weight | 0.277** | 0.96** | − | 0.995** | 0.548** | 0.716** | 0.723** | 0.05 | 0.421* | 0.781** |
| Net meat weight | 0.227 | 0.953** | 0.992** | − | 0.527** | 0.723** | 0.788** | 0.051 | 0.417* | 0.78** |
| Slaughter rate | 0.313* | 0.449** | 0.68** | 0.677** | − | 0.939** | 0.347 | 0.656** | 0.474* | 0.203* |
| Net meat rate | 0.174 | 0.54** | 0.73** | 0.769** | 0.93** | − | 0.647** | 0.567** | 0.509** | 0.368 |
| Carcass net meat rate | −0.173 | 0.465** | 0.49** | 0.593** | 0.374** | 0.688** | − | 0.098 | 0.361 | 0.521** |
| EMA | 0.367** | 0.474** | 0.432** | 0.393** | 0.137 | 0.086 | −0.071 | − | −0.167 | −0.082 |
| GR | 0.025 | 0.71** | 0.583** | 0.532** | −0.001 | −0.018 | −0.061 | 0.28** | − | 0.089 |
| BFT | −0.418** | 0.462** | 0.356** | 0.377** | −0.067 | 0.063 | 0.261** | −0.12 | 0.538** | − |
indicates highly significant correlation (P < 0.01), while.
indicates significant correlation (P < 0.05).
3.14. Results of through-traffic analysis of cashmere fineness and slaughter performance in LCG
From Table 14, it can be seen that the direct maximum of net meat rate and cashmere fineness of LCG buck is 5.718. Next is carcass net meat rate (−2.532) and EMA (−0.954). The maximum indirect path coefficient between carcass net meat rate and cashmere fineness is 3.577, followed by EMA (2.992) and net meat rate (−2.179). The direct throughput coefficient of BFT and cashmere fineness in doe is −0.711, Next is EMA (−0.159). The indirect flux coefficient of EMA versus cashmere fineness was 0.085, followed by BFT (0.019) (Table 14).
Table 14.
Path coefficients between slaughter performance and cashmere fineness in LCG.
| Sex | Independent variable | Correlation coefficient | Direct effect | Indirect effect |
|||
|---|---|---|---|---|---|---|---|
| Net meat rate | Carcass net meat rate | EMA | BFT | ||||
| buck | Net meat rate | −0.409* | 5.718 | −1.638 | −0.541 | ||
| Carcass net meat rate | −0.506** | −2.532 | 3.670 | −0.093 | |||
| EMA | −0.542** | −0.954 | 3.24 | −0.248 | |||
| doe | EMA | 0.367** | −0.159 | 0.085 | |||
| BFT | −0.418** | −0.711 | 0.019 | ||||
3.15. Results of stepwise multiple regression analysis of slaughter performance and cashmere fineness in LCG
From Table 15, the optimal regression equation for doe is cashmere fineness = -0.833 BFT + 0.19 Carcass weight − 0.187 carcass net meat rate + 29.921. For buck it is cashmere fineness = -0.183 EMA − 0.379 carcass net meat rate + 0.188 net meat rate + 42.595. The coefficients of determination (R-squared) for the multiple regression equation are 0.471 and 0.589, respectively (Table 15).
Table 15.
Path coefficients between slaughter performance and cashmere fineness in LCG.
| Sex | Model | R2 | Adjusted R-squared | Standard error of estimate | F | P |
|---|---|---|---|---|---|---|
| doe | Cashmere fineness = -0.625BFT + 18.549 | 0.175 | 0.161 | 1.16209 | 12.315 | 0.001b |
| Cashmere fineness = -0.884BFT + 0.144carcass weight + 15.773 | 0.383 | 0.361 | 1.01424 | 17.654 | 0.000c | |
| Cashmere fineness = -0.833BFT + 0.19carcass weight-0.187carcass net meat rate + 29.921 | 0.471 | 0.442 | 0.94750 | 16.590 | 0.000d | |
| buck | Cashmere fineness = -0.19EMA + 19.305 | 0.294 | 0.265 | 0.953 | 10.398 | 0.003b |
| Cashmere fineness = -0.12EMA-0.226carcass net meat rate + 36.728 | 0.501 | 0.460 | 0.818 | 12.064 | 0.000c | |
| Cashmere fineness = -0.183EMA-0.379carcass net meat rate + 0.188net meat rate + 42.595 | 0.589 | 0.535 | 0.758 | 10.988 | 0.000d |
3.16. The haplotype combinations of the FA2H gene T42443G and the ELOVL3 gene C2133A
Using SHEsis software, analysis of SNP loci in the FA2H and ELOVL3 genes revealed four haplotype combinations.(See Table 16)
Table 16.
The haplotype combinations of the FA2H gene T42443G and the ELOVL3 gene C2133A.
| Haplotype | H1:TT | H2:TG |
|---|---|---|
| H1:CC | CCTT | CCTG |
| H2:CA | CATT | CATG |
3.17. Correlation analysis of haplotype combinations of FA2H gene T42443G and ELOVL3 gene C2133A with cashmere production performance
A total of four haplotype combinations were found on LCG buck.The CCTT haplotype combination was highly significantly superior to the other haplotype combinations in terms of cashmere length, number of curls, and net cashmere rate.The CCTG haplotype combination was highly significantly superior to the other haplotype combinations in terms of cashmere yield, The CATG haplotype combination superior to other haplotype combinations in cashmere fineness.
A total of four haplotype combinations were found in LCG doe, The CCTG genotype was highly significantly superior to other haplotype combinations in terms of cashmere yield, number of curls, net cashmere rate and short cashmere rate. And CCTG haplotype combination of cashmere fineness was also the best (Table 17).
Table 17.
Haplotype combinations of two genes related to cashmere production performance in LCG.
| Name | Haplotype | Quantities | Cashmere yield(g) | Cashmere Fineness(μm) |
Cashmere length(cm) |
number of curls | Short cashmere rate (%) | Net cashmere Rate(%) |
|---|---|---|---|---|---|---|---|---|
| buck | CCTT | 2 | 1850.00 ± 34.72 | 16.99 ± 0.12 | 10.74 ± 0.08a | 8.20 ± 0.16aA | 19.68 ± 2.37 | 77.14 ± 2.54 |
| CCTG | 2 | 2200.00 ± 18.57 | 15.89 ± 0.16 | 8.58 ± 0.10b | 4.32 ± 0.17bB | 19.26 ± 1.37 | 76.37 ± 2.17 | |
| CATT | 14 | 1950.00 ± 108.18 | 16.39 ± 0.32 | 9.345 ± 0.17a | 9.10 ± 0.19aA | 19.67 ± 4.05 | 80.02 ± 2.68 | |
| CATG | 20 | 1900.00 ± 64.69 | 16.82 ± 0.21 | 9.60 ± 0.31a | 5.04 ± 0.95aAB | 17.37 ± 4.61 | 70.39 ± 2.36 | |
| doe | CCTT | 10 | 1700.00 ± 15.48cB | 18.10 ± 0.26aA | 9.12 ± 0.17 | 4.12 ± 0.20bC | 11.06 ± 1.26 | 56.94 ± 4.76bB |
| CCTG | 180 | 1866.67 ± 21.76aA | 16.20 ± 0.11cB | 9.54 ± 0.12 | 9.18 ± 0.13aA | 15.82 ± 1.17 | 79.72 ± 1.13aA | |
| CATT | 270 | 1722.22 ± 17.71bA | 16.86 ± 0.06bB | 10.33 ± 0.09 | 5.86 ± 0.23bA | 16.68 ± 0.80 | 71.82 ± 0.79bA | |
| CATG | 260 | 1788.46 ± 20.01bA | 16.49 ± 0.07bcB | 9.32 ± 0.10 | 6.14 ± 0.28bA | 18.55 ± 0.99 | 77.23 ± 0.83abA |
3.18. Correlation analysis of haplotype combinations of FA2H gene T42443G and ELOVL3 gene C2133A with slaughter performance
In the haplotype combinations of LCG doe, The CCTT haplotype combination shows highly significantly superior over other haplotype combinations in terms of live weight before slaughter, carcass weight, net meat weight, net meat rate, and EMA. The combination of CCTT and CATT haplotypes was highly significantly superior to the combination of CATG haplotypes in slaughtering rate, carcass net meat rate and GR. Therefore, CCTT was the dominant haplotype combination.
In the haplotype combinations of LCG buck, The CATG haplotype combination was significantly better than the CATT haplotype combination in GR, Therefore, CATG was the dominant haplotype combination (Table 18).
Table 18.
Haplotype combinations related to slaughter performance in LCG.
| Name | Haplotype | Quantities | Live weight before slaughter(kg) | Carcass weight (kg) |
Net meat weight (kg) |
Slaughter rate (%) |
Net meat rate (%) |
Carcass net meat rate(%) | EMA(cm2) | GR(mm) | BFT(mm) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| odoe | CCTT | 6 | 55.00 ± 0.76aA | 30.10 ± 0.54aA | 25.40 ± 0.56aA | 54.73 ± 0.32aA | 46.18 ± 0.63aA | 84.39 ± 0.23aA | 26.95 ± 0.48aA | 9.37 ± 0.39aA | 2.53 ± 0.15b |
| CATT | 36 | 47.93 ± 1.12bB | 25.15 ± 0.71bB | 20.95 ± 0.62bB | 52.34 ± 0.48bA | 43.56 ± 0.40bB | 83.24 ± 0.27aA | 19.37 ± 0.23cB | 7.46 ± 0.29bB | 2.95 ± 0.16ab | |
| CATG | 12 | 45.90 ± 0.81bB | 22.45 ± 0.41bB | 17.80 ± 0.45cB | 48.91 ± 0.02cB | 38.72 ± 0.30cC | 79.17 ± 0.58bB | 21.32 ± 1.15bB | 9.81 ± 0.49aA | 3.32 ± 0.16a | |
| buck | CATT | 15 | 45.02 ± 0.67 | 22.48 ± 0.53 | 17.52 ± 0.49 | 49.95 ± 0.99 | 38.90 ± 0.91 | 77.82 ± 0.57 | 23.40 ± 1.53 | 4.73 ± 0.44b | 1.62 ± 0.12 |
| CATG | 9 | 48.67 ± 2.79 | 25.93 ± 1.56 | 20.37 ± 1.44 | 53.22 ± 0.26 | 41.60 ± 0.66 | 78.14 ± 0.90 | 21.78 ± 0.58 | 7.41 ± 0.38a | 1.99 ± 0.36 |
4. Discussion
LCG is the world's highest yielding white cashmere goat breed, known for its long fiber length, high clean cashmere yield, moderate fineness of fiber, pure white coat, robust physique, strong adaptability, stable genetic performance, and effective improvement of medium to low yielding cashmere goats. It is hailed as a “Chinese national treasure”. Using LCG as the paternal line, five new local varieties have been cultivated, making outstanding contributions to the improvement and cultivation of Chinese cashmere goat breeds. Therefore, it is honored with the title “Father of Cashmere Goats”. This study primarily analyzed the effects of genotypes at different loci of the FA2H and ELOVL3 genes on six traits of LCG, as well as the influence of haplotype combinations of these two genes on cashmere production performance and slaughter performance. The SNP loci on the FA2H gene is T42443G, and on the ELOVL3 gene it is C2133A. The PIC values of the FA2H gene T42443G locus and the ELOVL3 gene C2133A locus in LCG range between 0.25 and 0.5, indicating moderate polymorphism. A higher HE value at these two loci indicates higher genetic variation, reflecting rich genetic resources and diversity. The Ne value at these two loci indicates a poor ability of the population to maintain allele stability during selection, mutation, or genetic variation. According to the X2 values, the T42443G and C2133A loci were not in Hardy-Weinberg equilibrium, likely due to high artificial selection intensity in the breeding farms, which might affect allele and genotype frequencies.
Based on the gene replacement effect, the T42443G locus showed a gene mutation with a negative additive effect, which has a positive impact. Conversely, the C2133A locus showed a gene mutation with a positive additive effect, which has a negative impact. In this study, it was found that in LCG buck, the TT genotype at the FA2H gene T42443G locus was the dominant genotype for cashmere production performance, while the TG genotype was dominant for slaughter performance, The CC genotype at locus C2133A of the ELOVL3 gene was the dominant genotype for cashmere producing performance, and only the CA genotype was found for slaughter performance. In LCG doe, the TG genotype at the FA2H gene T42443G locus was the dominant genotype for cashmere production performance, while the TT genotype was dominant for slaughter performance, The CC genotype at the C2133A locus of the ELOVL3 gene was the dominant genotype for cashmere production performance and slaughter performance. Haplotype combinations of the two genes revealed that the dominant haplotype combination for cashmeret production performance was CCTG. The dominant haplotype combination for buck in slaughter performance was CATG, and the dominant haplotype combination for doe in slaughter performance was CCTT. Reducing the fineness of LCG fibers has always been a concern for people, In previous studies, SNP analyses of the KRT26, TCHH,35 COL6A5, and LOC10218137436 genes were found to be associated with cashmere fineness. It has been suggested that the FA2H gene may be involved in the formation of myelin 2-hydroxygalactose cebuckide and sulpholipids.37 Deletion of the ELOVL3 gene leads to anti-obesity effects in mice, and the effect of the ELOVL3 gene in the liver on metabolic homeostasis and diet-induced metabolic diseases is dispensable.24 In medicine autosomal recessive mutations in the FA2H gene cause FAHN degeneration, which results in neurodegeneration.38 Wu et al.,33 conducted transcriptome sequencing of Jiang Nan cashmere goat skin tissues and speculated that the FA2H gene may be associated with cashmere fineness. The Xinjiang Academy of Animal Science, Institute of Animal Husbandry, has applied for a patent titled “Application, Regulation Methods, and Products of FA2H Gene in Preparation Controlling Cashmere Fineness,” indicating the correlation between the FA2H gene and cashmere fineness.
Ge et al.,39 conducted correlation regression analysis on body size and cashmere production performance in Yan Mountain cashmere goats, revealing that traits such as height, body length, and chest circumference are significantly positively correlated with cashmere yield. Gao et al.,40 conducted a correlation regression analysis on body measurements and body weight in Shaanbei White Cashmere goats, revealing significant or highly significant correlations between body length and body weight, Based on the literature above, we speculate whether there is a correlation between cashmere fineness and cashmere production performance as well as slaughter performance. Through correlation analysis, it is evident that in LCG, cashmere yield, cashmere length, net cashmere rate, were significantly or highly significantly correlated with cashmere fineness. Correlation analysis between cashmere fineness and slaughter performance showed that net meat rate and EMA were significantly or highly significantly correlated with fineness. Shi et al.,41 found a highly significant correlation between cashmere fineness and cashmere length in doe of northern Shaanxi white cashmere goats. Zhang et al.,42 found a highly significant correlation between cashmere fineness and cashmere yield in Yanshan cashmere goats. This is the same as the results analysed in this paper, suggesting that the analysis of correlations in this paper is of value.
LCG are important local breeds, and enhancing their production performance while reducing fineness has become a crucial research direction. In this study, we analysed the production performance of the SNP loci of FA2H and ELOVL3 genes from the genetic point of view to provide some help for the development of LCG breeding.
5. Conclusion
The Chinese Ministry of Agriculture and Rural Affairs has set forth core targets in the National goat Genetic Improvement Plan (2021–2035), requiring a 10 % increase in cashmere yield and cashmere fineness below 16 µm for Liaoning cashmere goat (LCG). LCG is characterized by the highest individual cashmere yield, but its cashmere fineness tends to be coarse. The findings of this study, In the FA2H gene, a SNP locus T42443G was detected in LCG buck, with the TT genotype showing advantageous traits in cashmere fineness, meat quality, and body size, while the TG genotype demonstrated advantages in slaughter performance,In LCG doe, the TG genotype shows advantageous traits in cashmere fineness, milk production, and meat quality, while the TT genotype exhibits advantages in slaughter performance, lambing, and body size. In the ELOVL3 gene, a SNP locus C2133A was identified in LCG buck, where the CC genotype was advantageous for cashmere fineness, Only CA genotype was found in slaughter and meat quality. Additionally, and the CA genotype showed superiority in body size. On LCG doe, The CC genotype was the advantageous genotype in terms of cashmere fineness, milk production, slaughter performance, and meat quality. The CA genotype was the advantageous genotype in terms of lambing and body size. Phenotypic correlation analysis of cashmere fineness with cashmere production performance and slaughter performance, Through multiple linear regression analysis, it was found that the trait positively correlated with cashmere fineness in buck' cashmere production performance was cashmere yield, the correlation coefficient was 0.541. The trait negatively correlated was net cashmere rate, the correlation coefficient was −0.866. The dominant genotypes identified to affect both cashmere fineness and cashmere production performance in buck was TT and CC. The traits positively correlated with the production performance of cashmere in doe was cashmere yield and cashmere length. The correlation coefficients were 0.278 and 0.322, The traits negatively correlated were number of curls, Short cashmere rate, and net cashmere rate, The correlation coefficients were −0.246, −0.238, and −0.936. The dominant genotypes identified to affect both the fineness of doe cashmere and cashmere production performance were TG and CC. Through multiple linear regression analysis, it was found that the traits negatively correlated with buck slaughter performance and cashmere fineness were net meat rate, carcass net meat rate, and EMA, The correlations were −0.409, −0.506, and −0.542. The dominant genotypes identified to affect both buck cashmere fineness and slaughter performance were TG and CA. The trait positively correlated with doe slaughter performance and cashmere fineness was EMA. The correlation coefficient was 0.367. The trait negatively correlated was backfat thickness, with a correlation coefficient of −0.418. The dominant genotypes identified to influence both doe cashmere fineness and slaughter performance were TT and CC. The identified dominant haplotype combination for cashmere production performance in LCG was CCTG. The dominant haplotype combination for doe slaughter performance was the CCTT haplotype combination. The dominant haplotype combination for buck slaughter performance was the CATG haplotype combination. Therefore, the TT genotype of the FA2H gene and the CC genotype of the ELOVL3 gene in LCG buck, and the TG genotype of the FA2H gene and the CC genotype of the ELOVL3 gene in doe can be used as molecular markers for assisted selection of cashmere fineness. It is hoped that this experiment can provide reference for future research.
Ethical approval
The goats all followed the guidelines of the journal Genetic Engineering and Biotechnology.
To ensure the ethical treatment of animals, all procedures and handling involving goat are carried out in accordance with the guidelines established by the Experimental Animal Management Committee of Shenyang Agricultural University.
Funding sources
This research was funded by National Natural Science Foundation of China (NO. 32272836).
CRediT authorship contribution statement
Shuaitong Li: Writing – original draft, Software. Lingchao Kong: Writing – review & editing. Siyi Li: Investigation. Yining Liu: Data curation. Yuan Pan: Methodology. Qingkun Liu: Data curation. Weihang Hong: Conceptualization. Hua Ma: Data curation. Qingyu Yuan: Formal analysis. Ran Duan: Formal analysis. Qiying Zhan: Formal analysis. Zeying Wang: Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Liaoning Cashmere Goat Breeding Center for providing experimental animals and related equipment to help this study.
This research was funded by National Natural Science Foundation of China (NO. 32272836).
References
- 1.Molaei Moghbeli S., Barazandeh A., Vatankhah M., et al. Genetics and non-genetics parameters of body weight for post-weaning traits in Raini Cashmere goats. Trop Anim Health Prod. 2013;45:1519–1524. doi: 10.1007/s11250-013-0393-4. [DOI] [PubMed] [Google Scholar]
- 2.Hajalizadeh Z., Dayani O., Khezri A., et al. The effect of adding fennel (Foeniculum vulgare) seed powder to the diet of fattening lambs on performance, carcass characteristics and liver enzymes. Small Rumin Res. 2019;175:72–77. doi: 10.1016/j.smallrumres.2019.04.011. [DOI] [Google Scholar]
- 3.Jafari Ahmadabadi S.A.A., Askari-Hemmat H., Mohammadabadi M., et al. The effect of Cannabis seed on DLK1 gene expression in heart tissue of Kermani lambs. Agricultural Biotechnology Journal. 2023;15(1):217–234. doi: 10.22103/jab.2023.21265.1471. [DOI] [Google Scholar]
- 4.Saadatabadi L.M., Mohammadabadi M., Nanaei H.A., et al. Unraveling candidate genes related to heat tolerance and immune response traits in some native sheep using whole genome sequencing data. Small Rumin Res. 2023;225 doi: 10.1016/j.smallrumres.2023.107018. [DOI] [Google Scholar]
- 5.Askari N., Mohammadabadi M., Baghizadeh A. ISSR markers for assessing DNA polymorphism and genetic characterization of cattle, goat and sheep populations. Iran J Biotechnol. 2011;9(3):222–229. [Google Scholar]
- 6.Vahabzadeh M., Chamani M., Dayani O., et al. Effect of Origanum majorana leaf (Sweet marjoram) feeding on lamb’s growth, carcass characteristics and blood biochemical parameters. Small Rumin Res. 2020;192 doi: 10.1016/j.smallrumres.2020.106233. [DOI] [Google Scholar]
- 7.Amirteymoori E., Khezri A., Dayani O., et al. Effects of linseed processing method (ground versus extruded) and dietary crude protein content on performance, digestibility, ruminal fermentation pattern, and rumen protozoa. Ital J Anim Sci. 2021;20(1):1506–1517. doi: 10.1080/1828051X.2021.1984324. [DOI] [Google Scholar]
- 8.Mohammadabadi M., Golkar A., Askari H.M. The effect of fennel (Foeniculum vulgare) on insulin-like growth factor 1 gene expression in the rumen tissue of Kermani sheep. Agricultural Biotechnology Journal. 2023;15(4):239–256. doi: 10.22103/jab.2023.22647.1530. [DOI] [Google Scholar]
- 9.Zamani P., Akhondi M., Mohammadabadi M.R., et al. Genetic variation of Mehraban sheep using two intersimple sequence repeat (ISSR) markers. Afr J Biotechnol. 2011;10(10):1812–1817. doi: 10.4314/AJB.V10I10. [DOI] [Google Scholar]
- 10.Safaei S.M.H., Dadpasand M., Mohammadabadi M., et al. An origanum majorana leaf diet influences myogenin gene expression, performance, and carcass characteristics in lambs. Animals. 2022;13(1):14. doi: 10.3390/ani13010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohammadabadi M.R., Tohidinejad F. Charachteristics determination of Rheb gene and protein in Raini Cashmere goat. Iranian Journal of Applied Animal Science. 2017;7(2):289–295. [Google Scholar]
- 12.Mohammadinejad F., Mohammadabadi M., Roudbari Z., et al. Identification of Key Genes and Biological Pathways Associated with Skeletal Muscle Maturation and Hypertrophy in Bos taurus, Ovis aries, and Sus scrofa. Animals. 2022;12(24):3471. doi: 10.3390/ani12243471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shokri S., Khezri A., Mohammadabadi M., et al. The expression of MYH7 gene in femur, humeral muscle and back muscle tissues of fattening lambs of the Kermani breed. Agricultural Biotechnology Journal. 2023;15(2):217–236. doi: 10.22103/jab.2023.21524.1486. [DOI] [Google Scholar]
- 14.Liu F., Wamg J., Yue Y., et al. Derivation of arbas cashmere goat induced pluripotent stem cells in LCDM with trophectoderm lineage differentiation and interspecies chimeric abilities. Int J Mol Sci. 2023;24:14728. doi: 10.3390/ijms241914728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin M., Lu J., Fei X., et al. Genetic signatures of selection for cashmere traits in chinese goats. Animals (basel). 2020;10:1905. doi: 10.3390/ani10101905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S., Ge W., Luo Z., et al. Integrated analysis of coding genes and non-coding RNAs during hair follicle cycle of cashmere goat (Capra hircus) BMC Genomics. 2017;18:767. doi: 10.1186/s12864-017-4145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y., Cai W., Chen R., et al. Metabolomic analysis and MRM verification of coarse and fine skin tissues of liaoning cashmere goat. Molecules. 2022;27:5483. doi: 10.3390/molecules27175483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su R., Lin J., Chen L., et al. Research progress of high-throughput automated SNP detection technology. Chinese Journal of Cell Biology. 2019;41:1412–1422. https://link.cnki.net/urlid/31.2035.Q.20190812.1508.030 [Google Scholar]
- 19.Fang G., Fang Y., Li L., et al. Correlation between exon 6 of goat KIF-Ⅰ gene and cashmere traits. Journal of Northwest a&f University (natural Science Edition). 2010;38:47–52. doi: 10.13207/j.cnki.jnwafu.2010.04.033. [DOI] [Google Scholar]
- 20.Fu X., Zhao B., Tian K., et al. Integrated analysis of lncRNA and mRNA reveals novel insights into cashmere fineness in Tibetan cashmere goats. PeerJ. 2020;8:e10217. doi: 10.7717/peerj.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Y. Xinjiang Agricultural University; 2012. Screening of candidate genes related to fine wool fiber diameter of chinese merino sheep (Xinjiang Type) and their association analysis. [Google Scholar]
- 22.Jakobsson A., Jörgensen J.A., Jacobsson A. Differential regulation of fatty acid elongation enzymes in brown adipocytes implies a unique role for Elovl3 during increased fatty acid oxidation. Am J Physiol Endocrinol Metab. 2005;289:E517–E526. doi: 10.1152/ajpendo.00045.2005. [DOI] [PubMed] [Google Scholar]
- 23.Tvrdik P., Asadi A., Kozak L.P., et al. Cig30, a mouse member of a novel membrane protein gene family, is involved in the recruitment of brown adipose tissue. J Biol Chem. 1997;272:31738–31746. doi: 10.1074/jbc.272.50.31738. [DOI] [PubMed] [Google Scholar]
- 24.Qin Z., Wang P., Chen W., et al. Hepatic ELOVL3 is dispensable for lipid metabolism in mice. Biochem Biophys Res Commun. 2023;658:128–135. doi: 10.1016/j.bbrc.2023.03.075. [DOI] [PubMed] [Google Scholar]
- 25.Westerberg R., Tvrdik P., Undén A.B., et al. Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem. 2004;279:5621–5629. doi: 10.1074/jbc.M310529200. [DOI] [PubMed] [Google Scholar]
- 26.Guillou H., Zadravec D., Martin P.G., et al. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Yu Y., Cao Y., Piao Q., et al. Screening of genes related to diameter of cashmere fibers in Liaoning cashmere goats. Chinese Herbivore Science. 2014:147–149. https://kns.cnki.net/kcms2/article/abstract?v=G7eKQ1uxNXfVbASWgGDsoaxTdSvb5UK118QGHwQERFgm8T8JqIBHOntRsxFfARESK7tmDa2JT-HVqB0VXWIGV0AKBeBydIW6VdiEeaKHiDm5XyUqt70Av9gogaExgP_5GAwbdqhYHaS-gxrvBs3khZFgg-PAL7iIUpAx62hjENJUXRLk1u1q-YjjR5TOcuMf&uniplatform=NZKPT&language=CHS [Google Scholar]
- 28.Zhou G. Northwest A&F University; 2018. Screening of key genes involved in the secondary hair follicle cycle of shaanbei cashmere goats and functional verification of LHX2 and miR-144. [Google Scholar]
- 29.Wu C. Screening of genes related to cashmere traits in Jiangnan cashmere goats and functional validation of ELOVL3 and FA2H genes. Xinjiang Agricultural University. 2022 doi: 10.27431/d.cnki.gxnyu.2022.000175. [DOI] [Google Scholar]
- 30.Hirao-Suzuki M., Koga T., Sakai G., et al. Fatty acid 2-hydroxylase (FA2H) as a stimulatory molecule responsible for breast cancer cell migration. Biochem Biophys Res Commun. 2020;531:215–222. doi: 10.1016/j.bbrc.2020.07.043. [DOI] [PubMed] [Google Scholar]
- 31.Cao L., Huang X.J., Chen C.J., et al. A rare family with Hereditary Spastic Paraplegia Type 35 due to novel FA2H mutations: a case report with literature review. J Neurol Sci. 2013;329:1–5. doi: 10.1016/j.jns.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 32.Wang J., Sui J., Mao C., et al. Identification of key pathways and genes related to the development of hair follicle cycle in cashmere goats. Genes (Basel) 2021;12:180. doi: 10.3390/genes12020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C., Li J., Xu X., et al. Effect of the FA2H Gene on cashmere fineness of Jiangnan cashmere goats based on transcriptome sequencing. BMC Genomics. 2022;23:527. doi: 10.1186/s12864-022-08763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai Z., Chen X., Wang J. Primer design using Primer Premier 5.0. Northwest Med Educ. 2008;(04):695–698. https://kns.cnki.net/kcms2/article/abstract?v=cF0fONyw0YL9P2Z-vLt9eh74jIN0KP8L3dJh61XHTUTdsghhD1-D038ACd2L3wav0XWohhLJ_CrSFbGnCht06FjR5-E-0qQGbrrOEH6AjOB38BmvyUimDssL72APF9JpWzs39BZr6Yvt6CRN0ndcsVbkNyEKK2OTf8uif-IVS3_KAy8QezJhMGPpxxX0rAHZ&uniplatform=NZKPT&language=CHS [Google Scholar]
- 35.Xu Y., Zhang X., Hui T., et al. Association analysis for SNPs of KRT26 and TCHH genes with cashmere production performance, body measurement traits and milk production traits in Liaoning cashmere goats. Anim Biotechnol. 2021;34:698–708. doi: 10.1080/10495398.2021.1996386. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Qin Y., Gu M., et al. Association between the cashmere production performance, milk production performance, and body size traits and polymorphism of COL6A5 and LOC102181374 genes in Liaoning cashmere goats. Anim Biotechnol. 2023;34:4415–4429. doi: 10.1080/10495398.2022.2155177. [DOI] [PubMed] [Google Scholar]
- 37.Alderson N.L., Rembiesa B.M., Walla M.D., et al. The human FA2H gene encodes a fatty acid 2-hydroxylase. J Biol Chem. 2004;279:48562–48568. doi: 10.1074/jbc.M406649200. [DOI] [PubMed] [Google Scholar]
- 38.Hashemi N., Abadi N.R.S., Alavi A., et al. The first reports of FA2H-associated neurodegeneration from two unrelated Iranian families. Neurol Sci. 2023;44:4359–4362. doi: 10.1007/s10072-023-06932-4. [DOI] [PubMed] [Google Scholar]
- 39.Ge C., Liu H., Li H., et al. Correlation regression analysis of body size, body weight, cashmere thickness, and cashmere yield in Yan mountain cashmere goats. J Anim Ecol. 2021;42:22–25. doi: 10.3969/j.issn.1673-1182.2021.05.005. [DOI] [Google Scholar]
- 40.Gao Y., Yan H., Feng P., et al. Regression analysis of body measurements and body weight of Shaanbei white cashmere goat. Chinese Herbivores. 2010;30:30–32. https://qikan.cqvip.com/Qikan/Article/Detail?id=35363638 [Google Scholar]
- 41.Shi L., Cao D., Zhang Q., et al. Analysis of coefficient of variation of fineness and length of Shaanbei white cashmere goat fiber. Heilongjiang Animal Husbandry and Veterinary Medicine. 2022;43:45–49. doi: 10.13881/j.cnki.hljxmsy.2021.07.0198. [DOI] [Google Scholar]
- 42.Zhang Y., Han C., Hong Z., et al. Correlation, regression, and path analysis of fineness and hair and down characteristics of Yan mountain cashmere goats. Chinese Journal of Animal Husbandry. 2024;60:183–187. doi: 10.19556/j.0258-7033.20221129-01. [DOI] [Google Scholar]