Abstract
Background and aim
Tacrolimus (TAC) has significantly improved kidney graft survival following transplantation, though it is associated with adverse side effects. The most prevalent complication resulting from excessive TAC exposure is the onset of de novo diabetes mellitus (DM), a condition that can negatively impact both renal graft function and patient outcomes. De novo DM is linked to an increased risk of chronic transplant dysfunction, as well as cardiovascular morbidity and mortality. Although the underlying mechanisms remain unclear, emerging research in the field of omics shows promise. The aim of this study was to investigate the metabolomic profile of kidney transplant patients who developed de novo DM, in comparison to those who did not, following TAC exposure, using untargeted metabolomic analysis through ultra-high-performance liquid chromatography–mass spectrometry (UHPLC-MS) and machine learning algorithms.
Methods
A cohort of 34 kidney transplant patients on a Tacrolimus regimen for at least 6 months was enrolled in the study, with serum samples collected from each patient. Comprehensive profiling of serum metabolites was performed, enabling the classification of patients into de novo diabetes mellitus and non diabetes groups. The metabolomic analysis of serum was conducted using UHPLC-MS.
Results
Of the 34 patients, 16 were diagnosed with TAC-induced diabetes. A total of 334 metabolites were identified in the serum samples, of which 10 demonstrated a significant correlation with the de novo diabetes mellitus group. Most of these metabolites were linked to alterations in lipid metabolism.
Conclusion
The application of metabolomics in kidney transplant patients undergoing a Tacrolimus regimen is both feasible and effective in identifying metabolites associated with de novo diabetes mellitus. This approach may provide valuable insights into the metabolic alterations underlying TAC-induced diabetes.
Keywords: Tacrolimus, diabetes mellitus, metabolomics, lipid metabolism
Background and aims
Tacrolimus, a potent immunosuppressive medication commonly used after kidney transplantation, has revolutionized the field by significantly improving graft survival rates. It belongs to the class of drugs known as calcineurin inhibitors and alongside mycophenolate mofetil (MMF) and prednisone plays a crucial role in suppressing the immune system, thereby ensuring the long-term success of the renal graft [1,2]. While tacrolimus has substantially improved transplant outcomes, it is characterized by substantial intra- and inter-individual pharmacokinetic variability and a narrow therapeutic window, causing numerous potential complications associated with its use [3–5].
One of the most common complications caused by TAC excessive exposure is represented by de novo diabetes mellitus (DNDM), a condition that can have detrimental effects on both renal graft and patient outcomes associating an increased risk of chronic transplant dysfunction and cardiovascular morbidity and mortality [6,7]. De novo diabetes mellitus refers to the onset of diabetes in individuals who did not have the condition prior to undergoing kidney transplantation [8].
The underlying mechanism by which tacrolimus contributes to the development of diabetes mellitus (DM) is not fully understood, but several factors have been proposed. Tacrolimus has been found to impair insulin secretion from pancreatic beta cells and increase insulin resistance or may produce a direct toxic effect on the beta cell, leading to a disruption in glucose metabolism [9–12]. Additionally, other factors such as genetic predisposition, obesity, and the use of other immunosuppressive medications may also contribute to the development of diabetes after kidney transplantation [13,14].
In order to mitigate the incidence of post-transplant diabetes mellitus after the administration of tacrolimus, the most effective approach entails the minimization of tacrolimus serum concentrations, albeit at the cost of potentially elevating the patient’s susceptibility to graft rejection [15–17]. The treatment of DM is complex and consists of a combination of lifestyle changes and medication [18,19]. Furthermore, managing diabetes in the post-transplant setting requires a multidisciplinary approach, involving close monitoring of blood glucose levels, adjustments in immunosuppressive regimens, and lifestyle modifications.
Metabolomics, a rapidly evolving field in medical research, offers a comprehensive approach of studying the metabolic profile of biological systems. It provides valuable insights into the complex interactions between genetic, environmental, and lifestyle factors that contribute to disease development and progression. In the context of tacrolimus therapy and the development of de novo diabetes mellitus after treatment in kidney transplant patients, metabolomics has emerged as a promising tool for understanding the underlying mechanisms and identifying potential biomarkers that may help in the early detection, prediction, and a personalized management of this complication [20–22].
In this article, we will explore the emerging role of metabolomics in understanding the metabolic changes induced by tacrolimus therapy and their association in de novo diabetes mellitus in kidney transplant patients. Thus, we aim to describe the serum metabolomic profile of kidney transplant patients that developed de novo diabetes mellitus compared to the ones that didn’t, using untargeted metabolomic investigation by UHPLC–MS and machine learning algorithms.
Methods
In this cross-sectional study, from a total of 135 consecutive patients who underwent kidney transplantation (KTx) at our hospital we recruited 34 patients, 16 that developed diabetes mellitus after the kidney transplant and a group of 18 patients age-gender matched that did not developed diabetes mellitus as the control group. These patients had stable creatinine levels, which we defined as a variation below 25% of the mean creatinine value. The study took place between May 2020 and July 2020 at the Clinical Institute of Urology and Renal Transplantation Cluj-Napoca. All enrolled patients were receiving a Tacrolimus (TAC)-based immunosuppressive therapy protocol (Advagraf 0.075–0.3 mg/kg/day) for at least six months after the surgery. We excluded patients who developed autoimmune diseases or lymphoproliferative disorders following the KTx procedure.
During the standard follow-up, we conducted clinical examinations, standard hematological and biochemical tests, as well as tacrolimus concentration analysis (tacrolinemia) for all patients. In accordance with the occurrence of de novo diabetes mellitus, the patient cohort was stratified into two distinct groups: individuals who manifested DNDM and those who remained unaffected. Subsequently, comprehensive profiling of serum metabolites was conducted for each respective group, facilitating the classification of patients into the DM group and the non-DM group.
Patients’ blood samples were obtained 24 hours after administering Advagraf, just before the next dose. Prior to collection, patients were required to fast for a minimum of 8 hours.
To collect the serum samples, vein puncture was performed using vacutainer tubes without anticoagulants. The collected blood was then subjected to centrifugation at 2000× g for 10 minutes to separate the blood serum. Aliquots of 1 mL were taken from the separated serum and stored at −80 °C until further analysis.
To precipitate the protein content in the serum, a mixture of methanol and acetonitrile (in a ratio of 1:1) was added to 0.2 mL of serum, resulting in a total volume of 0.8 mL. The mixture was vortexed for 1 minute, kept at 4 °C for 6 hours, and vortexed again for 1 minute. After thorough mixing, the vials containing the mixture were centrifuged at 12,500× g for 5 minutes. The supernatant was then collected, and it underwent filtration through 0.2 μm nylon filters.
Tacrolinemia levels were measured using a semi-automated electrochemiluminescence immunoassay method on the ArchitectPlus CI4100 automatic analyzer [23,24]. Prior to the automated sequence on the Architect, a manual pretreatment step was conducted. This involved extracting the whole blood sample with a precipitation reagent and subjecting it to centrifugation. The resulting supernatant was carefully transferred to a Transplant Pretreatment Tube and then loaded onto the Architect iSystem for further analysis.
Moreover, the ArchitectPlus CI4100 automatic analyzer was employed for the standard laboratory assessment, encompassing a range of parameters. These included serum cholesterol, triglycerides, glycemia, aspartate aminotransferase (ASAT), alanine aminotransferase (ALAT), gamma-glutamyl transferase (GGT), amylases, total proteins (TP), potassium (K+), sodium (Na2+), chloride (Cl−), ionized calcium (Ca2+), magnesium (Mg2+), and uric acid (UA). The evaluation of renal function was accomplished by estimating the glomerular filtration rate (eGFR) through the application of the CKD-EPI equation, which is based on creatinine levels [25,26].
The metabolomic serum profile was analyzed using high-precision liquid chromatography (UHPLC)–mass spectrometry (MS) analysis.
For the UHPLC-MS analysis, a Bruker Daltonics MaXis Impact device (Bruker GmbH, Bremen, Germany) was utilized. The system consisted of a Thermo Scientific UHPLC UltiMate 3000 system with a Dionex Ultimate quaternary pump delivery and ESI+-QTOF-MS detection device. The analysis was performed on a C18 reverse-phase column (Acuity, UPLC C18 BEH, Dionex) with dimensions of 5 μm and 2.1 × 75 mm, maintained at a temperature of 25 °C. The flow rate was set at 0.3 mL/min, and the injection volume was 5.0 μL. The mobile phase used in the analysis was composed of eluent A (water containing 0.1% formic acid) and eluent B (methanol:acetonitrile 1:1, containing 0.1% formic acid). A gradient system was employed, starting with 99% A at minute 0, followed by 70% A at minute 1, 40% A at minute 2, 20% A at minute 6, and 100% B from minutes 9 to 10, followed by 5 minutes with 99% A. The total running time for the analysis was 15 minutes. The mass spectrometry parameters were set to analyze a mass range between 50 and 1000 Da. The nebulizing gas pressure was set at 2.8 bar, the drying gas flow was maintained at 12 L/min, and the drying gas temperature was set to 300 °C. Prior to each chromatographic run, a calibration with sodium formate was performed. Instrument control and data processing were carried out using the TofControl 3.2, Hystar 3.2, and Data Analysis 4.2 software packages provided by Bruker Daltonics.
The metabolites identified by UHPLC–MS were ranked based on their information gain, specifically in their capacity to distinguish between patients who developed de novo diabetes mellitus and those who did not. Utilizing the Kullback–Leibler divergence method, the top 10 metabolites were selected. A Student’s t-test was then performed for each of these metabolites, and the p-value was calculated. These 10 metabolites were subsequently chosen for further analysis. The classification accuracy for distinguishing between the two patient groups, based on each significant metabolite, was assessed using a receiver operating characteristic (ROC) curve, and the area under the curve (AUC) was calculated.
To quantitatively assess the multivariate classification efficacy of the 10 selected metabolites, two independent machine learning algorithms—naive Bayes and k-nearest neighbors (kNN)—were trained to discriminate between patients who developed de novo diabetes mellitus and those in the non-diabetic group. kNN was used as standard implementation in Quasar-Orange Software, using 5 as the number of neighbors, Euclidean as the metric parameter and uniform as the weight value. The cross-validation was done using the leave-one-out method. All models were rigorously cross-validated using the leave-one-out (LOO) method.
The inputs for the machine learning algorithms consisted of either the individual selected metabolites or all ten metabolites combined. Prior to classification, the data were normalized to unity. The classification performance was evaluated using several metrics: the area under the curve (AUC) derived from receiver operating characteristic (ROC) analysis, classification accuracy, F1 score, precision, and recall. These quality performance metrics were reported as the average values obtained from each iteration of the cross-validation process.
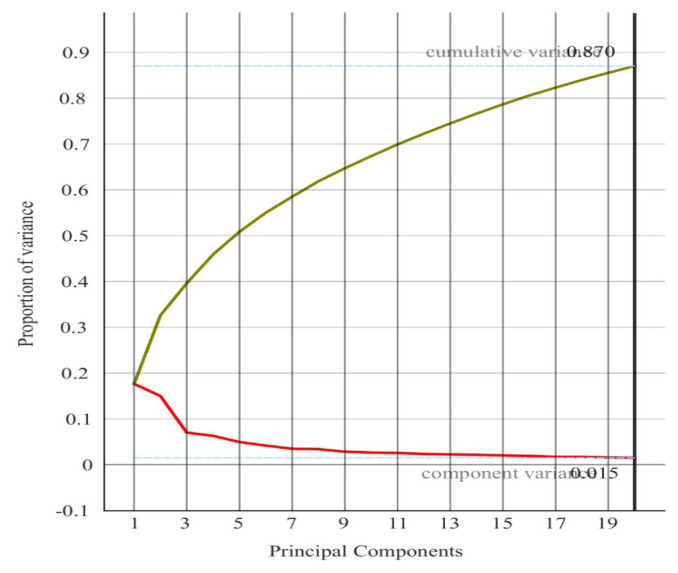
Subsequently, a principal component analysis (PCA) was conducted to explore the dataset, using the 10 selected metabolites as input variables. To enhance the representation of the model’s capacity to differentiate between the two groups, PCA was employed to reduce the dimensionality of the data. The relationship between the number of principal components and the explained variance in the original dataset is illustrated in figure 1.
Figure 1.
The relationship between the number of principal components and the explained variance.
For the correlation analysis, we used the Pearson correlation coefficient.
All statistical analyses were performed using the Quasar-Orange software (Bioinformatics Laboratory of the University of Ljubljana) [27,28].
The study was approved by the Ethics Committee of the Clinical Institute of Urology and Kidney Transplantation, Cluj-Napoca, No. 2/2020, and by the Ethics Committee of the Iuliu Hatieganu University of Medicine and Pharmacy in Cluj-Napoca, No. 285/2020. Written informed consent was obtained from all patients following the rules and principles of the Helsinki Declaration.
Results
A total of 34 kidney transplant recipients were included in the study, comprising 16 patients who developed de novo diabetes mellitus following tacrolimus administration (DM group) and 18 patients who did not develop diabetes (non-DM group).
Table I presents the biochemistry panel, displaying the mean values and standard deviations for each parameter across both groups.
Table I.
Student’s t-test and info gain for the significantly different metabolites used to discriminate between patients that developed diabetes.
| Blood tests | DM group mean±SD |
non-DM group mean±SD |
|---|---|---|
| Cholesterol (mg/dl) | 176.7±30.5 | 212.3±52.4 |
| Triglycerides (mg/dl) | 166.7±92.4 | 149.1±65.6 |
| Potassium (mmol/L) | 4.6±0.5 | 4.4±0.7 |
| Amylases (U/L) | 91.3±39 | 102±50 |
| ASAT (U/L) | 18.5±5.3 | 19.6±7.7 |
| ALAT (U/L) | 23.7±14.5 | 27.2±26 |
| GGT (U/L) | 36±32 | 31.2±23 |
| TB (mg/dl) | 0.71±0.28 | 0.62±0.3 |
| Glycemia (mg/dl) | 125±21.7 | 94±10.2 |
| Total proteins(mg/dl) | 6.9±0.4 | 6.8±0.6 |
| Ca 2+ (mmol/L) | 4.5±0.3 | 4.7±0.5 |
| Cl − (mmol/L) | 107.1±3.8 | 107.3±3.9 |
| Na + (mmol/L) | 141.5±2.4 | 142.7±1.4 |
| Mg 2+ (mg/dL) | 1.68±0.2 | 1.71±0.2 |
| UA (mg/dl) | 7±1.3 | 6.8±1 |
Our dataset comprised 336 metabolites identified using ultra-high-performance liquid chromatography–mass spectrometry (UHPLC–MS). The metabolites were ranked based on their information gain, specifically their ability to discriminate between the DM and non-DM groups. The Kullback–Leibler divergence method was employed to select the top 10 metabolites. A Student’s t-test was performed for each of these metabolites, and p-values were calculated. These 10 metabolites, which showed significant differences between the two groups, were selected for further analysis (Table II).
Table II.
Student’s t-test and info gain for the significantly different metabolites. The mean levels of the metabolites represent peak UHPLC–MS intensities.
| Metabolite (counts) | DM Group Mean ± SD |
Non-DM Group Mean ± SD |
t-test Value | Info. gain | p-Value |
|---|---|---|---|---|---|
| Sphingomyelin (d18:1/18:1(9Z)) | 108369 ± 43377 | 198228 ± 143923 | 2.353 | 0.565 | 0.031 |
| Cer (t18:0/20:0(2OH)) | 148919 ± 91935 | 103460 ± 66817 | 2.727 | 0.553 | 0.015 |
| DG (18:1/22:4/0:0) | 67483 ± 41031 | 44790 ± 32875 | 2.960 | 0.519 | 0.011 |
| Eicosadienoyl-ethanolamine C22H41NO2 | 24001 ± 4531 | 31581 ± 29067 | 0.965 | 0.507 | 0.353 |
| PA (20:1/18:1) | 285202 ± 119440 | 100775 ± 52295 | 3.796 | 0.507 | 0.003 |
| N-oleoyl ethanolamine | 23802 ± 19994 | 25333 ± 22834 | 1.538 | 0.493 | 0.179 |
| PC (20:3/22:2) | 38371 ± 14542 | 22233 ± 14685 | 4.601 | 0.435 | 0.001 |
| PG (18:0/14:0) | 78710 ± 87800 | 70407 ± 76967 | 1.706 | 0.419 | 0.113 |
| LPI (18:0) | 49846 ± 33918 | 23393 ± 18312 | 2.595 | 0.403 | 0.067 |
| Tetrahydrocortisone | 32866 ± 18700 | 32158 ± 10496 | 0.173 | 0.386 | 0.865 |
Abbreviations: Cer – Ceramide; PA – phosphatidic acid; DG – diglyceride; PA - Phosphatidic acid; PC – phosphatidylcholine; PG – phosphatidylglycerol; LPI – lysophosphatidylinositols.
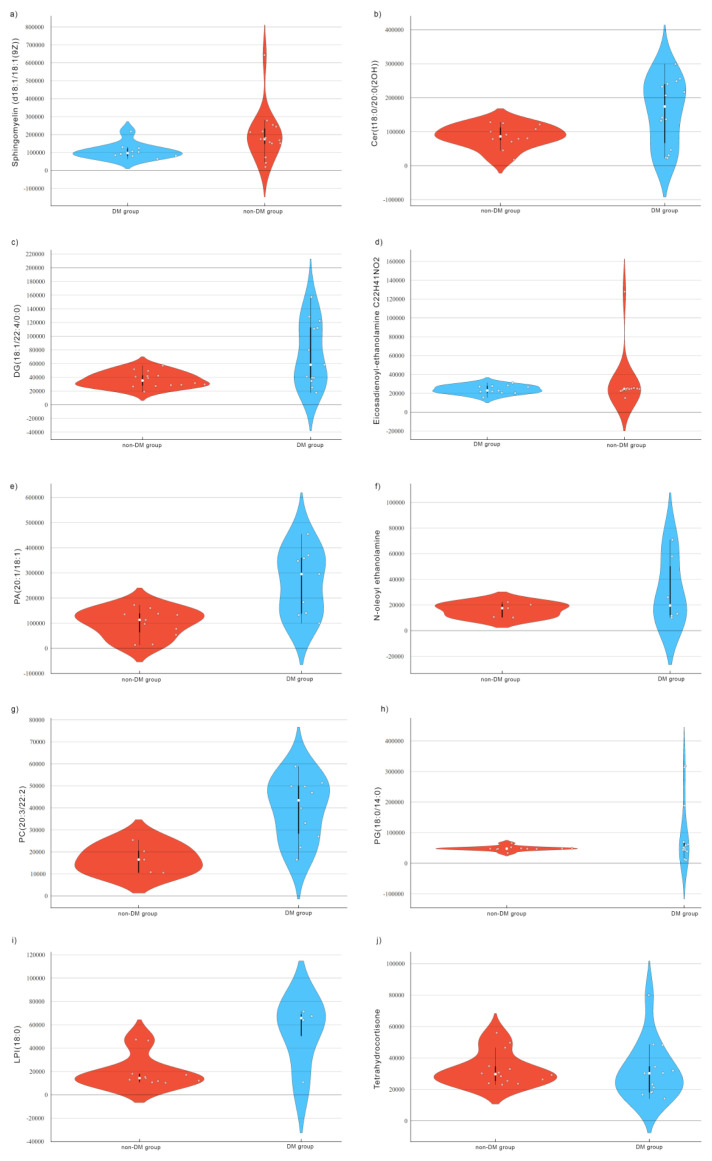
The violin plots derived from UHPLC-MS analysis for each of the 10 metabolites are presented in the figure below (Figure 2). These plots reveal a broader distribution with a higher density of data points in the DM group, compared to a narrower distribution and lower density of data points in the non-DM group for all ten metabolites. The wider plots indicate greater variability in the data for the metabolites. We can also observe a shift in the distribution of the violin plots between the two groups indicating a good discrimination between the patients that developed diabetes mellitus after TAC administration and the ones that did not.
Figure 2.
Violin plots of Sphingomyelin (d18:1/18:1(9Z)) (a), Cer(t18:0/20:0(2OH)) (b), DG(18:1/22:4/0:0) (c), Eicosadienoyl-ethanolamine C22H41NO2 (d), PA(20:1/18:1) (e), N-oleoyl ethanolamine (f), PC(20:3/22:2) (g), PG(18:0/14:0) (h), LPI(18:0) (i) Tetrahydrocortisone (j), for the DM group and non-DM group. Abbreviations: Cer – Ceramide; PA – phosphatidic acid; DG – diglyceride; PA - Phosphatidic acid; PC – phosphatidylcholine; PG – phosphatidylglycerol; LPI – lysophosphatidylinositols.
The ROC curves for the classification models demonstrated high discriminative power for both the naive Bayes and kNN algorithms. The AUC values were 0.902 and 0.816 for the naive Bayes and kNN models, respectively, indicating excellent classification performance (Table III).
Table III.
Head-to-head comparison of the area under the curves for the classification accuracy yielded by the ten metabolites using three supervised classification algorithms.
| Statistic model | AUC | CA | F1 | Precision | Recall |
|---|---|---|---|---|---|
| Naive Bayes | 0.902 | 0.647 | 0.607 | 0.798 | 0.647 |
| kNN | 0.816 | 0.676 | 0.657 | 0.705 | 0.676 |
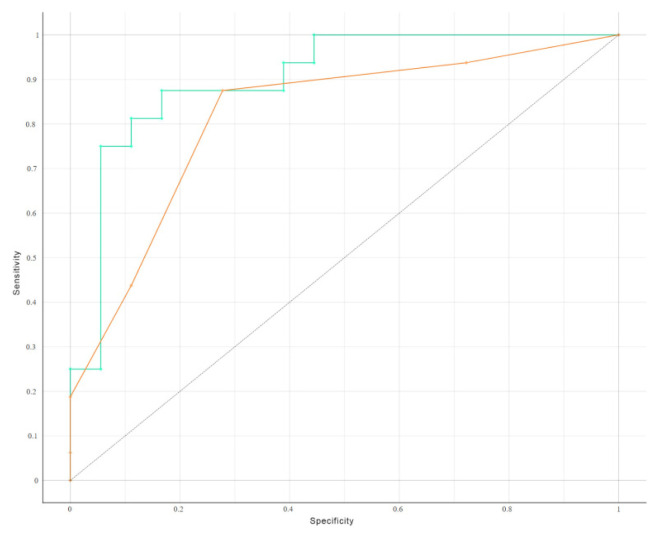
We conducted a head-to-head comparison of the receiver operating characteristic (ROC) curves to evaluate the classification accuracy of the 10 metabolites individually, as well as in combination, using Naïve Bayes analysis for supervised classification (Figure 3).
Figure 3.
Head-to-head comparison of the receiver operating characteristic curves (ROC) for the classification accuracy yielded by the ten metabolites using naïve Bayes analysis for supervised classification.
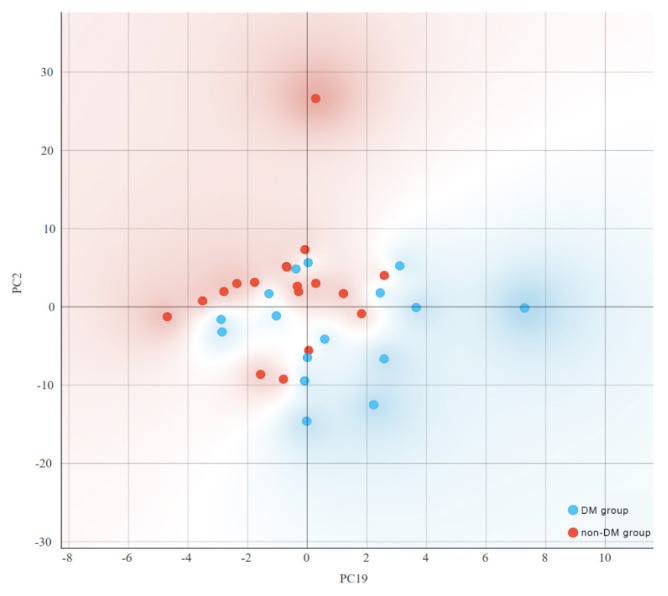
To more effectively represent the ability to distinguish between patients who developed DM following TAC administration and those who did not, we employed Principal Component Analysis (PCA) for dimensionality reduction. Figure 1 illustrates the relationship between the number of principal components (PCs) and the explained variability in the original dataset. The distribution of score values from the PCA of metabolic profiles for both DM and non-DM groups, specifically for PC2 and PC19, is depicted in figure 4. The distribution of score values following PCA of the two groups’ metabolic profiles (PC2 and PC19) show the clustering tendency of the patients that developed DM after TAC administration and the ones that did not.
Figure 4.
The distribution of principal component (PC) score values (PC2 and PC19) of patients with metabolic profiles associated with the DM group and the non-DM group.
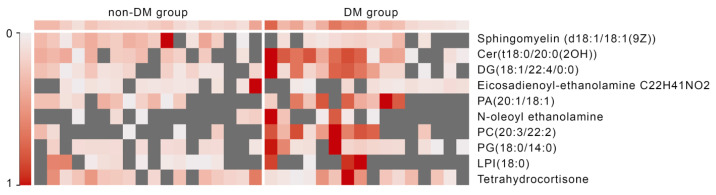
Figure 5 displays the heatmap of the ten serum metabolites identified through our UHPLC–MS analysis, demonstrating the clustering of metabolites based on the presence of diabetes mellitus.
Figure 5.
Heatmap of the levels of serum metabolites. Abbreviations: Cer – Ceramide; PA – phosphatidic acid; DG – diglyceride; PA - Phosphatidic acid; PC – phosphatidylcholine; PG – phosphatidylglycerol; LPI – lysophosphatidylinositols.
The left section of the heatmap represents patients who did not develop diabetes mellitus. This group exhibits a more variable expression pattern, with many gray areas indicating low or absent expression of certain metabolites. The right section corresponds to patients who developed diabetes mellitus. This group shows higher and more consistent levels of expression for most metabolites, as indicated by the predominance of red coloring. There appears to be a clear distinction in expression levels between the two groups. The DM group tends to have generally higher metabolite levels, suggesting that these metabolites may be associated with the development of diabetes mellitus following TAC administration.
Discussion
The primary objective of this study was to examine the relationship between kidney transplantation, tacrolimus treatment, and the development of de novo diabetes mellitus. Furthermore, the study aimed to investigate the potential of utilizing metabolomics biomarkers and machine learning mechanisms to predict and prevent the risk of DNDM in kidney transplant recipients undergoing tacrolimus treatment. The findings of this study suggest promising opportunities for incorporating personalized medicine strategies to optimize the selection and dosage of immunosuppressive therapies in order to prevent the occurrence of de novo diabetes mellitus.
Tacrolimus, a potent calcineurin inhibitor, is widely used in transplantation to prevent graft rejection. However, due to its narrow therapeutic window and high pharmacokinetic variability, patients are subjected to the diabetogenic effect of tacrolimus, which raises concerns among clinicians and researchers. The real challenge lies in finding the delicate balance of providing patients with the minimum required immunosuppression that yields maximum effectiveness. This balance is crucial to avoid complications arising from excessive exposure while also preventing graft rejection due to inadequate levels of immunosuppression [9,12,16,29].
Tacrolimus can contribute to the development of DNDM through various mechanisms, encompassing diminished insulin secretion, increased insulin resistance, or direct beta cell toxicity [9–12].
A recent study aimed to investigate the potential impact of diabetes mellitus and cytochrome P450 3A gene polymorphism on the concentrations of tacrolimus and its metabolites in kidney transplant recipients. The primary objective was to gain insights into the pharmacokinetics of tacrolimus in this patient population and explore the potential for personalized dosing strategies. The study findings present compelling evidence demonstrating that both diabetes mellitus and genetic polymorphism significantly influence the disposition of tacrolimus, as well as its metabolites, 13-DMT and 15-DMT. These results carry potential clinical implications in terms of interpreting therapeutic monitoring outcomes related to tacrolimus administration [30].
Other studies aimed to assess the potential of model-based follow-up dosing, incorporating patient characteristics and pharmacological data, to enhance the individualization of treatment in kidney transplant recipients. Specifically, those studies sought to determine whether model-based follow-up dosing could yield more precise tacrolimus exposure levels compared to the conventional approach of standard therapeutic drug monitoring following an initial algorithmic dosing regimen and revealed high rates of both graft survival and patient survival at 5 years post-transplant, with a high potential to minimize under- and overexposure to tacrolimus in the early posttransplant phase [31,32].
In our study, we identified 10 metabolites that tend to express in the group of patients diagnosed with diabetes post Tacrolimus exposure. The metabolism that is affected with a high predominance is the lipidic metabolism. As the majority of studies show, the long term administration of tacrolimus has effects on lipid metabolism with a high chance of developing diabetes, when compared with cyclosporine A, due to its effects on pancreatic beta cells. Sphingomyelin (d18:1/18:1(9Z)) is a cell membrane component, with an essential role in intracellular signal transduction, but also in cell maturation, apoptosis or proliferation. Elevated serum levels of sphingomyelin are associated with cardiovascular disease and metabolic syndrome [33].
Cer(t18:0/20:0(2OH)) is a very long chain ceramide that has been linked with comorbidities of obesity, steatosis, non-alcoholic steatohepatitis, and major adverse cardiac events. The latest studies make a strong difference between the 2 types of ceramiced, with long chain (C16 up to C20) and very long chain which are produced by acyl chain-specific ceramide synthases, CerS1–6. The ceramides that most tightly correlate with insulin resistance and hepatic steatosis are long-chain species, C16:0 or C18:0, ceramides produced predominantly by CerS6 in adipose and liver tissue [34].
From glycerolipids class is DG(18:1/22:4/0:0), a metabolite that showed important impact on insuline resistance. In a study conducted in a finish population, it showed that dysregulated levels of glycerolipids and sphingolipids are present in the serum plasma years before diabetes type 2 onset [35].
Eicosadienoyl-ethanolamine is a fatty amide (andanamides) obtained by the formal condensation of (11Z,14Z)-eicosadienoic acid with ethanolamine, the few studies that were addressed to this metabolite is showing that andanamides, are acting as an endogenous ligand of cannabinoids receptors in mammalian brain [36].
PA(20:1/18:1) is a glycerolphospholipid found to have important parts in inflammation reactioA study conducted on mice population, both sexes, in thyroid, showed that the exposure to paclobutrazol and uniconazole significantly increased the biomarker levels of PG (12:0/15:0), PS (14:0/16:0), PA (20:1/15:0) and PG (13:0/17:0) in both sexes of rats [37].
N-oleoyl ethanolamine is one of the most studied metabolites in cardiovascular diseases, longevity and neurologic pathology, being linked also to the metabolic syndrome. There are studies showing this endogenous bioactive mediator of lipid homeostasis, who exerts vascular protection against intimal calcification, atherosclerosis; but his beneficial effect on vascular smooth muscle cell associated medial calcification has not been investigated in extension in humans, the majority of studies being conducted in mice models [38].
In a recent study, N-oleoyl ethanolamine was identified as a promising biomarker for bacterial infection diagnosis, where they studied the blood and the ascitis liquid from decompensated cirrhosis patients and for those with overlapping bacterial infections [39].
In what concernes PG(18:0/14:0), one of the most promising studies, presented the plasma lipidomics in patients with lung cancer and who underwent radiotherapy regimens. The authors aimed to identify biomarkers of diagnosis and prognosis, and also radiotherapy response in non-small-cell lung cancer (NSCLC) patients by plasma lipidomics analysis.
They compared lipid elements between weak and strong responses of NSCLC patients with radiotherapy, the obviously declined phosphatidylglycerol (PG 18: 0/14: 0, 18: 1/18: 3, and 18: 0/20: 1) elevated PI (20: 0/22: 5 and 18: 2/22: 4) and phosphatidic acid (PA 14: 0/20: 4, 14: 0/20: 3, and 18: 2/22: 4) could indicate poor therapeutic response for NSCLC patients. The results of ROC curve analysis suggested that PG (18: 0/20: 1 and 18: 0/14: 0) could clearly predict the radiotherapeutic response for NSCLC patients, and PS (18: 0/20: 0) and cholesterol were the first two lipid components with the most potential for the diagnosis of advanced NSCLC [40].
A study that observed the effects of dioxin on a cohort of male workers, showed the lipid metabolism dysregulation, especially the results revealed that dioxin exposure caused accumulations of triglyceride (TG), ceramide (Cer) and sphingoid (So), remodeling of glycerophospholipid, here being included phosphatidylcholine PC(20:3/22:2). Therefore, it is a promising area of research in what concernes inflammation, cardiovascular disease and hepatic diseas and their correlation with PC (20:3/22:2) [41].
Lysophosphatidylinositols (LPI) are bioactive lipids that are known to be involved in several pathophysiological processes such as cell proliferation, migration and tumorigenesis and were shown to play a role in metabolic disorders [42].
Studies suggest that LPI (18:0) plays a key role in several metabolic functions and possibly in metabolic disorders, while GPR55, the main LPI receptor, has been proposed as a metabolic regulator. Therefore, the LPI/GPR55 axis has been shown to be positively associated with obesity in humans and also altered levels of LPI have been linked with obesity and diabetes and they are also involved in inflammation and cancer. Further mechanistic studies are required to elucidate the potential role of LPI in the pathogenesis of metabolic disorders [43].
Tetrahydrocortisone is a corticosteroid hormone that is involved in the response to stress; it increases blood pressure and blood sugar levels and suppresses the immune system.
In a recent study, urinary steroid hormone profiles of patients with diabetic kidney disease were analyzed by gas chromatography–mass spectrometry and compared to an age and gender matched healthy control group taken out of a population study. They observed that the cohort with diabetic kidney disease excreted more tetrahydrosterone than the control group having a high significance in men, while the mineralo-corticoid receptor excretion was only increased for 18-hydroxytetrahydrocorticosterone in diabetic women. The excretion of most glucocorticoids was higher in the diabetic cohort compared with their healthy counterpart [44].
Our study evidenced that the most affected metabolism in diabetic patient group post-tacrolimus regimen was the lipidic metabolism, with a predilection on the phospholipids. Many other studies showed that after tacrolimus regimens, the lipidic metabolism is dysregulated, making many studies on lipidomics future keys of research in this area of transplantation.
One of the strengths of this study is the novelty in metabolomic approaches and the potential method for distinguishing between tacrolimus induced diabetes and non-diabetes patients after kidney transplant.
A future approach will involve the use of both urine and tissue metabolomics to validate our findings in a larger cohort of patients. Targeted metabolomics should be employed to confirm these results, alongside longitudinal monitoring of patients using metabolomic profiling, beginning prior to transplantation and continuing through the potential development of diabetes mellitus.
Personalised medicine is the key in transplant medicine and with the help of metabolomics, the tacrolimus regimens will find a way in helping patients without the cost of high comorbidity side effect.
Conclusions
Using serum metabolomics and machine learning algorithms we can find clear differences in the metabolic profile between patients that developed diabetes after TAC treatment and non-diabetic population, making it possible to identify future biomarkers. Future studies are required to confirm those metabolites through targeted metabolites.
Acknowledgement
This study was funded by a PCD nr 2461/7 2020–2021 grant from the Iuliu Haţieganu University of Medicine and Pharmacy, Cluj Napoca, Romania.
References
- 1.Bamoulid J, Staeck O, Halleck F, Khadzhynov D, Paliege A, Brakemeier S, Dürr M, Budde K. Immunosuppression and results in renal transplantation. Eur Urol Suppl. 2016;15:415–429. Available from: https://daneshyari.com/article/preview/5694177.pdf. [Google Scholar]
- 2.Kidney Disease: Improving Global Outcomes (KDIGO) Transplant Work Group. KDIGO clinical practice guideline for the care of kidney transplant recipients. Am J Transplant. 2009;9(Suppl 3):S1–S155. doi: 10.1111/j.1600-6143.2009.02834.x. [DOI] [PubMed] [Google Scholar]
- 3.Thongprayoon C, Hansrivijit P, Kovvuru K, Kanduri SR, Bathini T, Pivovarova A, et al. Impacts of High Intra- and Inter-Individual Variability in Tacrolimus Pharmacokinetics and Fast Tacrolimus Metabolism on Outcomes of Solid Organ Transplant Recipients. J Clin Med. 2020;9:2193. doi: 10.3390/jcm9072193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degraeve AL, Moudio S, Haufroid V, Chaib Eddour D, Mourad M, Bindels LB, et al. Predictors of tacrolimus pharmacokinetic variability: current evidences and future perspectives. Expert Opin Drug Metab Toxicol. 2020;16:769–782. doi: 10.1080/17425255.2020.1803277. [DOI] [PubMed] [Google Scholar]
- 5.Yu M, Liu M, Zhang W, Ming Y. Pharmacokinetics, Pharmacodynamics and Pharmacogenetics of Tacrolimus in Kidney Transplantation. Curr Drug Metab. 2018;19:513–522. doi: 10.2174/1389200219666180129151948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez Cantarin MP. Diabetes in Kidney Transplantation. Adv Chronic Kidney Dis. 2021;28:596–605. doi: 10.1053/j.ackd.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Duijnhoven EM, Boots JM, Christiaans MH, van Hooff JP. Metabolic aspects of tacrolimus in renal transplantation. Consequences for the choice of an immunosuppressive regimen and for the management of post-transplant diabetes mellitus. Minerva Urol Nefrol. 2003;55:33–42. [PubMed] [Google Scholar]
- 8.Chowdhury TA. Post-transplant diabetes mellitus. Clin Med (Lond) 2019;19:392–395. doi: 10.7861/clinmed.2019-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jindal RM, Sidner RA, Milgrom ML. Post-transplant diabetes mellitus. The role of immunosuppression. Drug Saf. 1997;16:242–257. doi: 10.2165/00002018-199716040-00002. [DOI] [PubMed] [Google Scholar]
- 10.Zheng C, Wang C, Zhang T, Li D, Ni XF, Lin JH, et al. Exploring the Mechanism of Skeletal Muscle in a Tacrolimus-Induced Posttransplantation Diabetes Mellitus Model on Gene Expression Profiles. J Diabetes Res. 2020;2020:6542346. doi: 10.1155/2020/6542346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Zhao H, Chen B, Fan Z, Li N, Yue J, et al. FXR activation alleviates tacrolimus-induced post-transplant diabetes mellitus by regulating renal gluconeogenesis and glucose uptake. J Transl Med. 2019;17:418. doi: 10.1186/s12967-019-02170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Sun F, Zhang Y, Chen H, He N, Chen H, et al. Tacrolimus Induces Insulin Resistance and Increases the Glucose Absorption in the Jejunum: A Potential Mechanism of the Diabetogenic Effects. PLoS One. 2015;10:e0143405. doi: 10.1371/journal.pone.0143405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solhjoo M, Kumar SC. StatPearls [Internet] Treasure Island (FL): StatPearls Publishing; 2023. Aug 28, 2024. Jan 28, New Onset Diabetes After Transplant. [PubMed] [Google Scholar]
- 14.Smyrli M, Smyrlis A, Tsouka G, Apostolou T, Vougas V. Risk Factors of the Development of Diabetes Mellitus After Kidney Transplantation. Transplant Proc. 2021;53:2782–2785. doi: 10.1016/j.transproceed.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Song JL, Gao W, Zhong Y, Yan LN, Yang JY, Wen TF, et al. Minimizing tacrolimus decreases the risk of new-onset diabetes mellitus after liver transplantation. World J Gastroenterol. 2016;22:2133–2141. doi: 10.3748/wjg.v22.i6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ammari Z, Pak SC, Ruzieh M, Dasa O, Tiwari A, Jaume JC, et al. Posttransplant Tacrolimus-Induced Diabetic Ketoacidosis: Review of the Literature. Case Rep Endocrinol. 2018;2018:4606491. doi: 10.1155/2018/4606491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conte C, Secchi A. Post-transplantation diabetes in kidney transplant recipients: an update on management and prevention. Acta Diabetol. 2018;55:763–779. doi: 10.1007/s00592-018-1137-8. [DOI] [PubMed] [Google Scholar]
- 18.Jenssen T, Hartmann A. Emerging treatments for post-transplantation diabetes mellitus. Nat Rev Nephrol. 2015;11:465–477. doi: 10.1038/nrneph.2015.59. [DOI] [PubMed] [Google Scholar]
- 19.Montero N, Oliveras L, Soler MJ, Cruzado JM. Management of post-transplant diabetes mellitus: an opportunity for novel therapeutics. Clin Kidney J. 2021;15:5–13. doi: 10.1093/ckj/sfab131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pallares-Méndez R, Aguilar-Salinas CA, Cruz-Bautista I, Del Bosque-Plata L. Metabolomics in diabetes, a review. Ann Med. 2016;48:89–102. doi: 10.3109/07853890.2015.1137630. [DOI] [PubMed] [Google Scholar]
- 21.Jin Q, Ma RCW. Metabolomics in Diabetes and Diabetic Complications: Insights from Epidemiological Studies. Cells. 2021;10:2832. doi: 10.3390/cells10112832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen ZZ, Gerszten RE. Metabolomics and Proteomics in Type 2 Diabetes. Circ Res. 2020;126:1613–1627. doi: 10.1161/CIRCRESAHA.120.315898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toole B, Gechtman C, Dreier J, Kuhn J, Gutierrez MR, Barrett A, Niederau C. Evaluation of the New Cyclosporine and Tacrolimus Automated Electrochemiluminescence Immunoassays under Field Conditions. Clin Lab. 2015;61:1303–1315. doi: 10.7754/clin.lab.2015.150225. [DOI] [PubMed] [Google Scholar]
- 24.Qin X, Rui J, Xia Y, Mu H, Song SH, Raja Aziddin RE, et al. Multi-center Performance Evaluations of Tacrolimus and Cyclosporine Electrochemiluminescence Immunoassays in the Asia-Pacific Region. Ann Lab Med. 2018;38:85–94. doi: 10.3343/alm.2018.38.2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araújo NC, Rebelo MAP, da Silveira Rioja L, Suassuna JHR. Sonographically determined kidney measurements are better able to predict histological changes and a low CKD-EPI eGFR when weighted towards cortical echogenicity. BMC Nephrol. 2020;21:123. doi: 10.1186/s12882-020-01789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cusumano AM, Tzanno-Martins C, Rosa-Diez GJ. The Glomerular Filtration Rate: From the Diagnosis of Kidney Function to a Public Health Tool. Front Med (Lausanne) 2021;8:769335. doi: 10.3389/fmed.2021.769335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toplak M, Read ST, Sandt C, Borondics F. Quasar: Easy Machine Learning for Biospectroscopy. Cells. 2021;10:2300. doi: 10.3390/cells10092300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toplak M, Birarda G, Read S, Sandt C, Rosendahl SM, Vaccari L, et al. Infrared Orange: Connecting Hyperspectral Data with Machine Learning. Synchrotron Radiation News. 2017;30:40–45. [Google Scholar]
- 29.Cho YM, Park KS, Jung HS, Jeon HJ, Ahn C, Ha J, et al. High incidence of tacrolimus-associated posttransplantation diabetes in the Korean renal allograft recipients according to American Diabetes Association criteria. Diabetes Care. 2003;26:1123–1128. doi: 10.2337/diacare.26.4.1123. [DOI] [PubMed] [Google Scholar]
- 30.Chitnis SD, Ogasawara K, Schniedewind B, Gohh RY, Christians U, Akhlaghi F. Concentration of tacrolimus and major metabolites in kidney transplant recipients as a function of diabetes mellitus and cytochrome P450 3A gene polymorphism. Xenobiotica. 2013;43:641–649. doi: 10.3109/00498254.2012.752118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francke MI, Hesselink DA, Andrews LM, van Gelder T, Keizer RJ, de Winter BCM. Model-Based Tacrolimus Follow-up Dosing in Adult Renal Transplant Recipients: A Simulation Trial. Ther Drug Monit. 2022;44:606–614. doi: 10.1097/FTD.0000000000000979. [DOI] [PubMed] [Google Scholar]
- 32.Rummo O, Carmellini M, Kamar N, Durrbach A, Mousson C, Caputo F, et al. Long-term, prolonged-release tacrolimus-based immunosuppression in de novo kidney transplant recipients: 5-year prospective follow-up of the ADHERE study patients. Transpl Int. 2020;33:161–173. doi: 10.1111/tri.13527. [DOI] [PubMed] [Google Scholar]
- 33.Rhee J, Loftfield E, Albanes D, Layne TM, Stolzenberg-Solomon R, Liao LM, et al. A metabolomic investigation of serum perfluorooctane sulfonate and perfluorooctanoate. Environ Int. 2023;180:108198. doi: 10.1016/j.envint.2023.108198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruce CR, Risis S, Babb JR, Yang C, Lee-Young RS, Henstridge DC, et al. The sphingosine-1-phosphate analog FTY720 reduces muscle ceramide content and improves glucose tolerance in high fat-fed male mice. Endocrinology. 2013;154:65–76. doi: 10.1210/en.2012-1847. [DOI] [PubMed] [Google Scholar]
- 35.Suvitaival T, Bondia-Pons I, Yetukuri L, Pöhö P, Nolan JJ, Hyötyläinen T, et al. Lipidome as a predictive tool in progression to type 2 diabetes in Finnish men. Metabolism. 2018;78:1–12. doi: 10.1016/j.metabol.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Fontana A, Di Marzo V, Cadas H, Piomelli D. Analysis of anandamide, an endogenous cannabinoid substance, and of other natural N-acylethanolamines. Prostaglandins Leukot Essent Fatty Acids. 1995;53:301–308. doi: 10.1016/0952-3278(95)90130-2. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Xu Y, Wang Y, Liu C, Chen J, Fan S, et al. Study on endocrine disruption effect of paclobutrazol and uniconazole on the thyroid of male and female rats based on lipidomics. Ecotoxicol Environ Saf. 2022;234:113386. doi: 10.1016/j.ecoenv.2022.113386. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Sun X, Li X, Liu N. Oleoylethanolamide alleviates hyperlipidaemia-mediated vascular calcification via attenuating mitochondrial DNA stress triggered autophagy-dependent ferroptosis by activating PPARα. Biochem Pharmacol. 2023;208:115379. doi: 10.1016/j.bcp.2022.115379. [DOI] [PubMed] [Google Scholar]
- 39.Fischer P, Pandrea S, Grigoras C, Stefanescu H, Farcau O, Tefas C, et al. Blood Metabolomic Signatures to Identify Bacterial Infection in Patients with Decompensated Cirrhosis. J Gastrointestin Liver Dis. 2022;31:40–47. doi: 10.15403/jgld-4034. [DOI] [PubMed] [Google Scholar]
- 40.Lv M, Shao S, Du Y, Zhuang X, Wang X, Qiao T. Plasma Lipidomics Profiling to Identify the Biomarkers of Diagnosis and Radiotherapy Response for Advanced Non-Small-Cell Lung Cancer Patients. J Lipids. 2024;2024:6730504. doi: 10.1155/2024/6730504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang Y, Tang Z, Jiang Y, Ai C, Peng J, Liu Y, et al. Lipid metabolism disorders associated with dioxin exposure in a cohort of Chinese male workers revealed by a comprehensive lipidomics study. Environ Int. 2021;155:106665. doi: 10.1016/j.envint.2021.106665. [DOI] [PubMed] [Google Scholar]
- 42.Masquelier J, Alhouayek M, Terrasi R, Bottemanne P, Paquot A, Muccioli GG. Lysophosphatidylinositols in inflammation and macrophage activation: Altered levels and anti-inflammatory effects. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:1458–1468. doi: 10.1016/j.bbalip.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Arifin SA, Falasca M. Lysophosphatidylinositol Signalling and Metabolic Diseases. Metabolites. 2016;6:6. doi: 10.3390/metabo6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackermann D, Vogt B, Bochud M, Burnier M, Martin PY, Paccaud F, et al. Increased glucocorticoid metabolism in diabetic kidney disease. PLoS One. 2022;17:e0269920. doi: 10.1371/journal.pone.0269920. [DOI] [PMC free article] [PubMed] [Google Scholar]