Abstract
Background/Objectives:
Pain is a challenging multifaceted symptom reported by most cancer patients. This systematic review aims to explore applications of artificial intelligence/machine learning (AI/ML) in predicting pain-related outcomes and pain management in cancer.
Methods:
A comprehensive search of Ovid MEDLINE, EMBASE and Web of Science databases was conducted using terms: “Cancer”, “Pain”, “Pain Management”, “Analgesics”, “Artificial Intelligence”, “Machine Learning”, and “Neural Networks” published up to September 7, 2023. AI/ML models, their validation and performance were summarized. Quality assessment was conducted using PROBAST risk-of-bias andadherence to TRIPOD guidelines.
Results:
Forty four studies from 2006-2023 were included. Nineteen studies used AI/ML for classifying pain after cancer therapy [median AUC 0.80 (range 0.76-0.94)]. Eighteen studies focused on cancer pain research [median AUC 0.86 (range 0.50-0.99)], and 7 focused on applying AI/ML for cancer pain management, [median AUC 0.71 (range 0.47-0.89)]. Median AUC (0.77) of models across all studies. Random forest models demonstrated the highest performance (median AUC 0.81), lasso models had the highest median sensitivity (1), while Support Vector Machine had the highest median specificity (0.74). Overall adherence to TRIPOD guidelines was 70.7%. Overall, high risk-of-bias (77.3%), lack of external validation (14%) and clinical application (23%) was detected. Reporting of model calibration was also missing (5%).
Conclusion:
Implementation of AI/ML tools promises significant advances in the classification, risk stratification, and management decisions for cancer pain. Further research focusing on quality improvement, model calibration, rigorous external clinical validation in real healthcare settings is imperative for ensuring its practical and reliable application in clinical practice.
Keywords: Cancer pain, Cancer pain management, Machine learning, Artificial intelligence
Introduction:
In recent years, the healthcare industry has witnessed a surge in the adoption of artificial intelligence (AI) and machine learning (ML) tools. These technologies have demonstrated their potential to transform healthcare delivery, diagnosis, and treatment decision-making [1]. AI/ML tools can analyze vast amounts of data, identify patterns, and provide predictive insights, thereby aiding healthcare professionals in making informed decisions [1, 2]. ML is a subfield of AI that includes systems that ‘learn from experience (E) with respect to some class of tasks (T) and performance measure (P), if its performance at tasks T, as measured by P, improves with experience E’ [3]. ML algorithms are essential in the clinical practice in medical imaging analysis, predicting patient outcomes, and personalizing treatment plans based on individual data profiles. They enhance diagnostic accuracy by analyzing radiological images and support clinical decision-making through evidence-based recommendations derived from large datasets [4, 5].
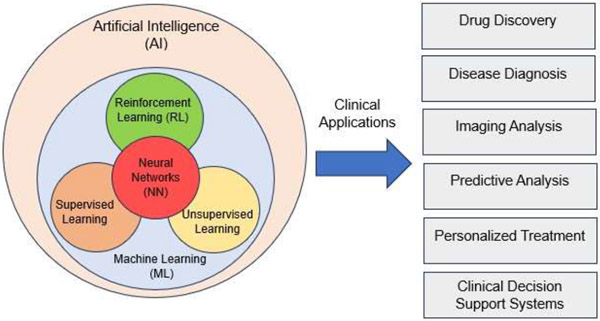
Neural Networks are a subtype of ML algorithms, which can have different degrees of complexity and evolve into the so-called deep learning (DL) algorithms when they involve a large number of layers. DL techniques enhance the clinical diagnosis and imaging analysis capabilities by leveraging complex neural network architectures like convolutional neural networks (CNNs) to analyze large-scale medical datasets. They also excel in image recognition tasks, such as identifying anomalies in medical imaging, which aids in precise disease diagnosis and monitoring. DL is crucial in medical image analysis, genomics, and healthcare operations optimization, predicting patient admission rates and resource utilization patterns [4]. Both ML and DL algorithms can be classed into supervised, unsupervised and reinforcement learning (RL) based on the way in which training data is presented to the model [14] (Figure 1).
Figure 1:
Types and clinical applications of Artificial Intelligence (AI) models.
RL in clinical applications can personalize treatment plans, optimize clinical decision support systems, enhance robotic surgery and rehabilitation, improve hospital resource management, aid in drug discovery, and refine health monitoring and medical imaging analysis [6, 7]. In supervised learning, the model is expected to learn the mapping between the training inputs and outputs presented during training to then be able to predict the outputs on an unseen input dataset. In unsupervised learning, the model is presented with unlabeled data and expected to learn the patterns and correlations by itself based on the input data only. In reinforcement learning, the algorithm learns to map inputs to actions by maximizing (or minimizing) a reward (or punishment) action evaluation signal. Additionally, AI algorithms can be grouped based on the task they are asked to perform: segmentation, regression or classification. Natural Language Processing (NLP) is an application of AI algorithms that is recently gaining interest in the clinical domain. NLP facilitates automated clinical documentation, enabling efficient automatic structured data extraction from clinical notes for billing, coding, and research purposes. Moreover, NLP-driven virtual assistants streamline patient interaction processes by scheduling appointments, providing medication reminders, and offering healthcare-related information promptly [5].
AI/ML applications have begun to make inroads into the field of cancer pain prediction and management. These technologies offer the promise of more accurate pain assessment, personalized treatment recommendations, and improved patient outcomes [8]. However, the extent of their utilization and their impact on cancer pain prediction and management is an area that requires further investigation, and the results of the best performing models in cancer induced pain remain controversial and disperse.
In the dynamic landscape of AI/ML, the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) guidelines stand as a crucial framework for ensuring the transparency and reproducibility of predictive models. Developed to enhance the reporting quality of studies involving prediction models, TRIPOD provides a structured approach to the design, analysis, and interpretation of such models [9]. Adhering to these guidelines is paramount as it not only facilitates the effective communication of research findings but also fosters trust in the outcomes of predictive models. In the realm of ML, where complex algorithms increasingly influence decision-making across various health domains, the importance of adhering to TRIPOD guidelines is accentuated by the critical need for model validation [10]. Lastly, the validation process plays a pivotal role in assessing a model's generalizability and reliability, ensuring that its predictive capabilities extend beyond the training data [9, 10].
The performance and generalizability of an AI/ML prediction model is assessed by testing the model on a data subset that has not been used during the model training process. The validation of a model can be internal or external, depending on whether the test dataset belongs to the same study cohort to that of the training dataset or is obtained from an entirely independent cohort. Both validation steps are crucial for the clinical implementation of a prediction model [10]. In particular, internal cross validation is a method to partition the data into training and testing subsets multiple times in order to provide a more accurate measure of model performance, especially with small datasets.
Comprehensive ML model performance assessment should include reporting of the model’s discrimination ability and model calibration as part of the internal and external validation process [11, 12]. Model discrimination is a measure of the model’s ability to differentiate between two classes (e.g., event vs. no event) given the available inputs [12]. Model calibration involves ensuring that the predicted probabilities align closely with the actual probabilities of events occurring [11].
Our scientific questions are:
Which AI/ML algorithms are used in cancer pain research and cancer pain management?
What are the applications of AI/ML models in cancer pain medicine?
Which are the best performing models for cancer pain prediction and opioids optimization?
To what degree do the existing AI/ML models in cancer pain prediction follow the TRIPOD guidelines with respect to model performance reporting?
What is the quality of the studies applied AI/ML in cancer pain medicine?
This systematic review aims to answer these questions and address the current gap in knowledge by analyzing existing literature on the role of AI/ML in cancer pain prediction and management decision-making. We also provide an overview of common supervised and unsupervised learning techniques, their general characteristics, and specific applications in cancer pain research, either in cancer pain prediction, cancer treatment related pain or pain management and opioids decisions.
Materials and Methods:
Protocol Registration:
Registration of this systematic review in the international prospective register database of systematic reviews (PROSPERO), was done on 16 October 2023 [ID number: CRD42023469865] in the context of human health care.
Search Strategy and Study Eligibility:
We conducted a systematic search of Ovid MEDLINE, Ovid EMBASE, and Clarivate Analytics Web of Science, for publications in English from the inception of databases to September 7, 2023. The concepts searched included “Cancer”, “Pain”, “Pain Management”, “Pain Measurement”, “Analgesics”, “Opioids”, “Artificial Intelligence”, “Machine Learning”, “Deep Learning”, “Expert System” and “Neural Networks”. Both subject headings and keywords were utilized. The terms were combined using AND/OR Boolean Operators. Animal studies, in vitro studies, and conference abstracts were excluded. The complete search strategy is detailed in Tables S1-S3.
Screening process:
Identified articles from the Search process were uploaded into the Covidence screening application [13] , and screening through Covidence was conducted by two independent reviewers. Screening of the titles and abstract was first conducted and then screening of the full text was done on the retrieved articles. Final included articles were extracted from Covidence for full-text review and data collection.
Inclusion Criteria:
Studies eligible for inclusion in this systematic review had the following criteria 1) be published in English, 2) investigate AI/ML applications in cancer pain medicine or cancer pain management, and 3) involve human subjects.
Exclusion Criteria:
Articles were excluded if they met any of the following criteria: 1) out of scope of our study 2) no AI/ML application, 3) non-cancer pain study, 3) not an original study (i.e., review article, letter, conference abstract), 4) duplicate publication or correction of an original article, 5) descriptive study that didn’t apply or test models.
Data Synthesis:
This study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines [14].
Data Collection Process:
The collected data included the following: AI/ML technique(s) used, input features and output variables, validation methods, and model performance metrics.
To evaluate the overall quality of the included articles, a rigorous assessment was undertaken, focusing on appraising both the risk of bias and adherence to reporting guidelines for each individual article. The risk of bias was evaluated using Prediction model Risk of Bias Assessment Tool (PROBAST) [15, 16], which examines four domains (participants, predictors, outcomes, and analysis) through 20 methodological questions to determine the overall risk of bias [16, 17].
Adherence to TRIPOD guidelines was analyzed using TRIPOD checklist [10]. Included articles were categorized according to the use of the AI/ML in cancer pain and grouped into the following groups: (1) models for prediction of cancer related pain intensity, cancer pain diagnosis or cancer pain initiation, (2) models for prediction of post cancer treatment pain (e.g., cancer surgery, RT or CT), (3) models for cancer pain management prediction or as a decision support system for cancer pain management.
Results:
Search and screening Results:
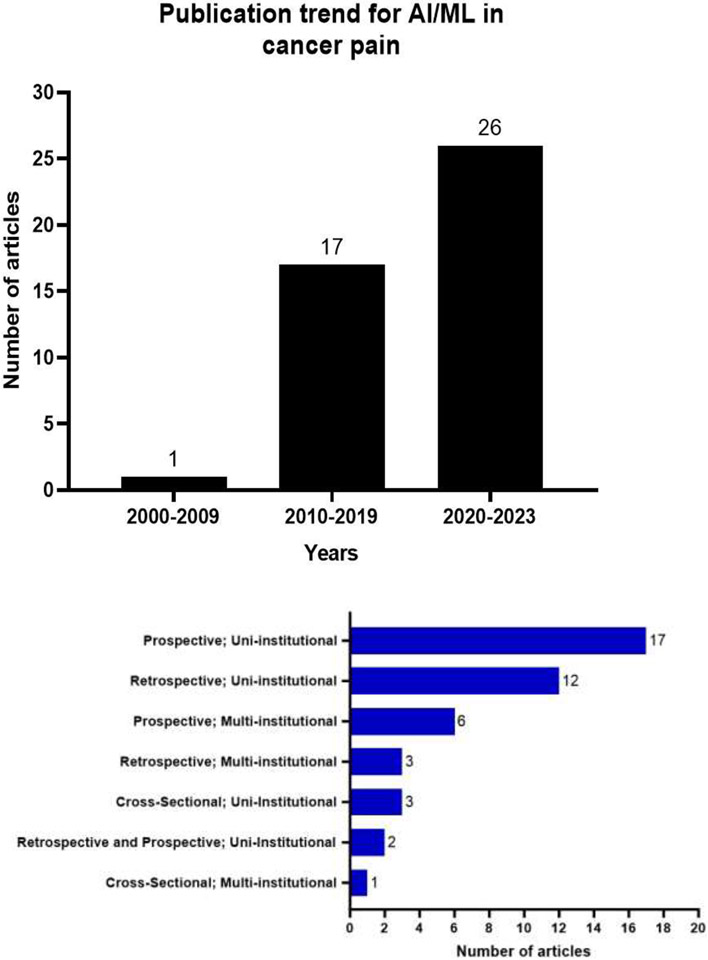
This comprehensive search resulted in identification of 436 studies through search of MEDLINE (n=189), EMBASE (n=189) and Web of Science (n=132). After screening and eligibility assessment, 283 articles were excluded. A total of 44 studies met the inclusion criteria and were included in this review. The search strategy process is illustrated in a PRISMA flow diagram (Figure 2). All included studies were published between 2006 and 2023, with an increase in the number of publications for AI/ML studies in cancer pain research from 1 (2006-2009) to 26 (2020-2023) (Figure 3.a).
Figure 2:
PRISMA Flow Diagram for systematic reviews of AI and ML in cancer pain research.
Figure 3:
a. Publications trends for AI/ML models used for cancer pain research between 2006-2023. b. Types of studies and the number of articles per each type.
Design and populations:
Most studies used a prospective uni-institutional cohort to develop their models (n=17 studies) while the least used approach involved the cross-sectional multi-institutional cohort design (n=1) (Figure 3.b). The median sample size to build the models was 320 (range: 21-46104, IQR 140-1000) 95% CI 156-900. Five studies (11%) did not specify the size of the study population. In 52% (n=23) of studies the cohort size was between 100-1000 patients, while 20% (n=9) of studies used >1000 patients and 16% (n=7) used <100 patients.
AI/ML algorithms used in cancer pain research:
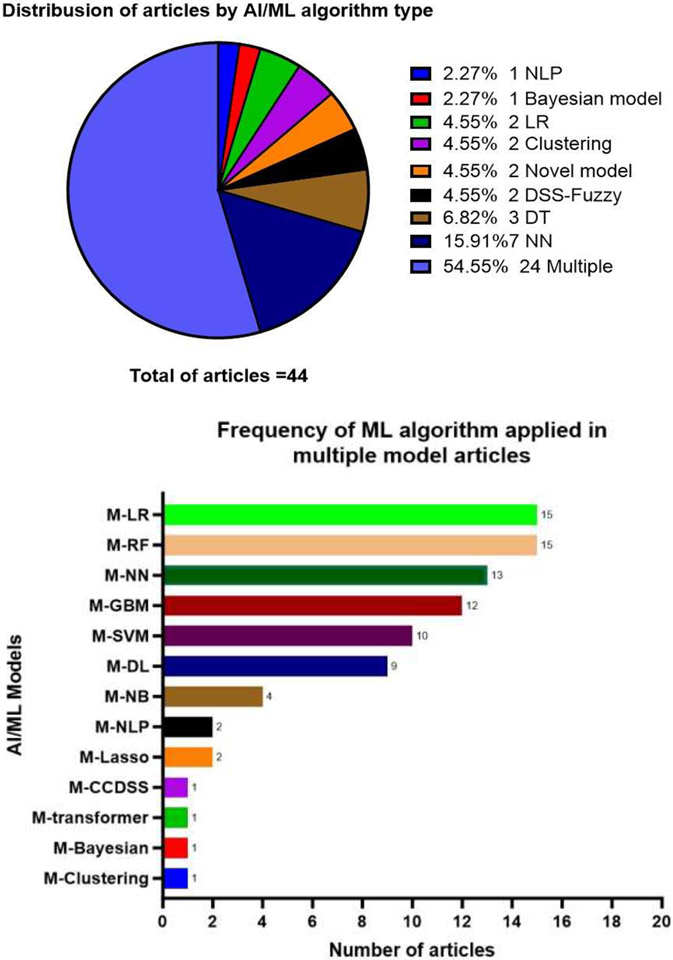
The most common AI/ML algorithms identified in this review, their general characteristics and specific use in cancer pain research are summarized in (Table 1). Some studies used a single model, while the majority of studies explored multiple models (each model was explored separately) (55%, n=24 articles) (Figure 4.a). The most common algorithms used in these multiple models studies were Random Forest (RF) and Logistic Regression (LR) (n=15 studies for each) (Figure 4.b). Other single model studies included Neural Networks (NN) (16% n=7), Decision Trees (DT) (7% n=3), Decision support system-Fuzzy (n=2), Clustering (n=2), LR (n=2), Bayesian networks (n=1), NLP (n=1) and 2 studies used novel models (Figure 4.a). Several other models used in multiple models studies (e.g., SVM, transformer, RF, GBM, Lasso, and NB) (Figure 4.b)
Table 1:
AI/ML model characteristics and applications in cancer pain research.
| Machine Learning Algorithm |
Characteristics | Cancer Pain Usage |
|---|---|---|
|
Supervised Learning [18] |
The output is labeled with the desired value (e.g., pain score or opioids dose) | |
| Classification | The output variable is a binary and/or categorical response | |
| Decision trees (DT) [19], [20], [21], [22], [23], [24], [8], [25], [26] |
Generates understandable rules with both categorical and continuous variables for prediction and classification purposes. |
|
| Random Forest (RF) [19], [27], [28], [29], [21], [30], [22], [31], [32], [8], [33], [34], [35], [36], [37] |
An ensemble approach that combines the output of multiple decision trees to reach a single high accurate result. It provides a good predictive performance, low overfitting, and easy interpretability. |
|
| Gradient boosting (GBM); extreme gradient boosting (XG Boost) [27], [28], [30], [22], [23], [32], [8], [34], [37] |
A robust ensemble boosting algorithm that trains the model sequentially for prediction and classification purposes. It combines several weak learners into strong learners. |
|
| Naïve Bayes (NB) Classifier [21], [32], [8] |
Used for binary or multi-class classification problems, based on Bayes rule and conditional independence assumption. |
|
| Regression | Predicts the probability using continuous data. | |
| Generalized linear mixed models (GLMMs), least absolute shrinkage and selection operator (Lasso), Linear regression (LR), Logistic regression (LR), Bayesian regression [38], [27], [39], [28], [29], [30], [22], [40], [41], [42], [35], [37], [26] |
Model that estimates the relationship between one dependent variable and one or more independent variables using a line. |
|
|
Support vector machine (SVM) [39], [29], [43], [21], [22], [23], [32], [36], [37] |
Type of supervised learning algorithm used to solve classification and regression tasks. Creates a hyperplane to separate two classes. |
|
|
Unsupervised Learning [31] |
Unlabeled and unclassified datasets used to train machines; Used to categorize unsorted data based on features, similarities and differences. |
|
|
Clustering [20], [44], [45] |
Machines divide the data into clusters based on features, similarities and differences. |
|
|
Association [46] |
Machines find interesting relations and connections among variables within large datasets that are input. |
|
|
Neural Networks (NN) [28], [20], [29], [21], [22], [23], [47], [48], [8], [33], [42], [34], [49], [50], [51], [52], [26] |
Composed of node layers, containing an input layer, one or multiple hidden layers and an output layer. Could be supervised, unsupervised or semi-supervised. |
|
| Deep Learning (DL) [53], [52] |
Subclass of NN that Includes many layers of the neural network and massive volumes of complex and disparate data. |
|
|
Natural Language Processing (NLP) [54], [55], [56] |
A component of AI that uses the ability of a computer program to understand human language either written or spoken, it could be supervised or unsupervised. |
|
|
Decision Support System [57], [58], [26] |
A subtype of AI using computerized programs to help decision making, judgment and actions in an organization. |
|
Figure 4:
a. Distribution of articles by AI/ML algorithm type. b. 'Frequency of ML algorithm applied in multiple model articles' (M; multiple).
AI/ML Models used for cancer related pain research:
This review identified 18 studies describing the implementation of AI/ML in cancer pain research (Table 2).
Table 2:
Characteristics of studies that used AI/ML algorithms in cancer pain research.
| Author, year |
Population | Cohort study |
AI/ML model (s) |
Input variables | Output variable |
Assessment tools |
Aim/ objective |
|---|---|---|---|---|---|---|---|
| Knudsen et al., 2011 [38] | 2278 Cancer patients | Cross-Sectional; Multi-institutional | LR | 46 variables -Functional status -Cognitive function |
1. Average pain 2. Worse pain 3. Pain relief |
11-point Numerical Rating Scales (0-10) | Identify variables associated with pain |
| Shimada et al, 2023 [24] | 213 patients with cancer | Retrospective; Multi-Institutional | DT | -Patient clinical and demographic characteristics - performance status | Pain in cancer patients | Characteristic visual information | Using ML to predict pain in End-of-Life cancer patients and at a wide range of times, including the "diagnostic stage of cancer" and the "treatment stage of cancer". |
| Cascella et al, 2023 [48] | Cancer patients | Prospective; Multi-Institutional | NN | -Facial expressions characteristic. - video recordings | Pain in cancer patients | video recordings. A set of 17 Action Units (AUs) was adopted. For each image | Constructed a ML model for discriminating between the absence and presence of pain in cancer patients using video analysis |
| Cascella et al., 2022 [33] | 158 cancer patients | Prospective; Uni-Institutional | RF GBM LASSO-RIDGE NN |
-Demographic -Clinical variables -Therapeutics Drugs for pain, |
Number of remote consultations due to pain | -Number of remote consultations -Pain severity assessment: Morphine dose |
Development of ML predictive models for identifying patients who may require more remote consultations due to pain. |
| DiMartin o et al., 2022 [54] | Cancer patients admitted to UNC Hospitals, a total of 1,644 hospitalizations were included. | Retrospective; Uni-institutional | NLP | -Clinical notes and records of symptom documentation-highest severity reported for pain | Pain severity labeled as: “controlled” (none, mild, not reported) or as “uncontrolled” (moderate or severe) | Common Terminology Criteria for Adverse Events (CTCAE) grade. | To evaluate NLP used to identify pain in EHR among hospitalized cancer patients. |
| Cascella et al., 2023 [34] | 226 cancer patients and 489 telemedicine visits for cancer pain management | Prospective; Multi-Institutional | DL (NN) RF GBM LASSO-RIDGE regression |
-Demographic -Clinical variables -Background pain (nociceptive, neuropathic) and breakthrough cancer pain (BTcP) |
The number of remote consultations” | The number of remote consultations recorded. | Investigate specific AI model efficacy for enhancing the telemedicine approach to cancer pain management. |
| Lou et al., 2022 [35] | Lung cancer patients with uncontrolled pain who participated in clinical consultations with physicians. | Retrospective; Uni-institutional | LR RF |
-Depth of prognosis discussion -Doctor's age, gender -Patient's race -Extent of shared decision making |
Patient Satisfaction with provider visit/treatment | Questions asked by physician and standardized patients (SPs) | Use ML to examine the association between patient satisfaction and physician factors in clinical consultations about cancer pain. |
| Lu et al., 2021 [55] | Child and adolescent survivors of cancer, aged 8 to 17 years, | Cross-Sectional; Uni-Institutional | Transformers (BERT) NLP SVM XGboost |
Meaning units in pain and fatigue as variables. | 1. Pain 2. Fatigue |
Standard surveys with prespecified content of PROs | Test the validity of NLP and ML algorithms in identifying different attributees of pain experienced by child and adolescent survivors of cancer. . |
| Moscato et al., 2022 [36] | 21 cancer patients | Prospective; Uni-institutional | SVM RF MP LR AdaBoost |
Physiological signals (e.g., photoplethysmo graphy, electrodermal activity) | Pain Experience (Pain ratings) | Edmonton Symptoms Assessment Scales (ESAS) for pain assessment. | Develop an automatic pain assessment method based on physiological signals recorded by wearable devices. |
| Heintzel man et al., 2013 [56] | 33 men with metastatic prostate cancer | Retrospective; Uni-institutional | NLP LR |
-Electronic, radiologic, RT, and pathology medical records Pain severity -Factors associated with pain experience (e.g., receipt of opioids and palliative radiation -Patient demographics (age, race/ethnicity). |
Pain Phenotype intensity: 1. Severe Pain 2. No Severe or controlled Pain |
Pain categorization model based on a conservative fourtiered pain scale: no pain (category 0); some pain (category 1); controlled pain (category 2); severe pain (category 3). | To test the feasibility of using text mining to predict pain in patients with metastatic prostate cancer. |
| Miettinen et al., 2021 [25] | 320 cancer patients with persistent pain | Prospective; Multi-institutional | DT (CART and PART) | -Pain Phenotype-Intensity related features -Pain etiology-related information -Psychological parameters -Demographic parameters -Lifestyle-related parameters, - Previous treatments -Comorbidities. |
1. Pain Intensity 2. Number of Pain areas 3. Pain Duration 4. Activity Pain Interference 5. Affective Pain interference |
The Brief Pain Inventory (BPI) for assessing pain intensity. | (1) identify patterns arising from different pain phenotypic parameters (2) selecting parameters in associating a patient with a particular pain phenotype. |
| Xuyi et al., 2021 [51] | Cancer patients between 2008 and 2015. [training and test cohorts consisted of 35,606 and 10,498 patients, respectively] | Retrospective; Uni-institutional | NN | 39 unique covariates: -Demographics, -Clinical features, -Treatment characteristics -Baseline PRO -Health care utilization measures. |
1. Severe Pain 2. Moderate to severe depression 3. Poor Well-being |
ESAS; and interRAI | Predict the risk of Severe Pain in cancer patients |
| Akshayaa et al., 2019 [53] | 900 Images of Cancer patients experiencing chronic pain and with ZFHX2 mutations. | Prospective; Uni-Institutional | Deep Convolutional Neural Network (CNN) | -Pathological images with ZFHX2 mutation-epigenomic alterations (DNA methylation), | Chronic pain | -Not specified | Designing an early mutation detection tool to identify the presence of pain inducing gene ZFHX2 using deep CNN. |
| Bang et al., 2023 [52] | 34,304 cancer patients. | Retrospective; Uni-institutional | Transformer CNN (DL) | -Pain intensity scores - lengths and time | Cancer Pain Exacerbation | 11-point Numerical Rating Scales (0-10) | Investigate the clinical relevance of DL models that predict the time of breakthrough pain onset in cancer patients. |
| Masukawa et al., 2022 [37] | 808 patients who died of cancer | Retrospective; Uni-Institutional | LR RF GBM SVM |
-Unstructured text data contained in EMRs. -Social distress -Spiritual pain Severe physical/psychological symptoms. |
1. Social Distress 2. Spiritual Pain 3. Severe Physical and Psychological Symptoms (e.g., pain) |
Japanese version of the Support Team Assessment Schedule (STAS-J). | Develop models to detect social distress, spiritual pain in terminally ill cancer patients. |
| Pombo et al., 2016 [26] | 32 cancer volunteers | Prospective; Uni-Institutional | CCDSS ANN Bayesian algorithms LR DT |
Clinical data, Treatment protocol Patient reported data | Mean pain intensity | -Not specified | To develop and validate a CCDSS as a method used pain evaluation system for pain intensity prediction. |
| Xu et al., 2020 [59] | Not specified | Retrospective; Multi-institutional | Herb Networks (SHN) and (THN) | -Pain-related herbs -Herbal molecules -Human and gut microorganism targets -Pathways Herb categories (HC1, HC2, HC3) |
1. Cancer pain 2. Chronic cough related neuropathic pain 3. Reproduction and autoimmune related pain |
Not specified | Understanding of the molecular mechanisms of pain subtypes that herbal drugs are participating |
| Pantano et al., 2020 [45] | 4,016 cancer patients suffering from BTcP. | Retrospective; Uni-institutional | Cluster analysis | Eight BTcP-defining variables (e.g., number of episodes, peaks duration, type, intensity, etc.) | Break through cancer pain (BTcP) | 11-point Numerical Rating Scales (NRS) (0-10) | Explore distinct subtypes of BTcP using an unsupervi sed learning algorithm. |
These studies employ various AI/ML models, showcasing their potential in understanding patient satisfaction, identifying pain-related attributes, and predicting pain in cancer patients across different age groups and healthcare settings. Most studies used clinical and demographic variables as input; however, some studies applied models utilizing innovative inputs such as physiological signals [18], facial expressions [19], textual data [20], and even herbal categories [21], indicating the exploration of novel data sources. This diversity in inputs strengthens the versatility of the models. Some studies showcase innovation by assessing pain through video analysis of facial expressions [19], wearable devices capturing physiological signals [18], and even employing NLP for identifying pain severity in unstructured medical records [22].
Most studies lacked detailed information on the extent of external validation of the AI models applied and the clinical application in real healthcare scenarios. Only 3 out of 18 studies performed external validation, and 7 studies discussed the clinical evaluation and application of the models used. Cascella et al, 2022 & 2023 [23] [24] applied external validation and clinical evaluation of the models for the remote consultation prediction in cancer patients. Additionally, Cascella et al, 2023 [19] performed a clinical trial to test the NN using video recording of facial expression for discriminating between the absence and presence of pain in cancer patients. Heintzelman et al., 2013 [25] proved the feasibility and generalizability of NLP through an external validation using separate data source, furthermore, they evaluated the clinical application of the NLP using text mining for pain prediction in patients with metastatic prostate cancer. Pombo et al., 2016 [26] clinically evaluated the CCDSS compared with clinical advice for pain intensity, however, there was a lack of generalizability of the decision model due to the uni-institutial cohort. Moscato et al., 2022 [18] applied a real-world context to develop an automatic pain assessment model, however, the small size cohort was a limitation with the need for bigger cohort for clinical validation.
AI/ML Models used for cancer treatment induced pain research:
Nineteen articles focused on implementing AI/ML algorithms in cancer treatment pain research were detected in our review. The characteristics of these studies are summarized in Table 3.
Table 3:
Characteristics of studies used AI/ML algorithms in cancer treatment related pain research.
| Author ,year |
Population | Cohort study |
AI/ML model (s) |
Input variables |
Output variable |
Assessment tool |
Aim/ objective |
|---|---|---|---|---|---|---|---|
| Chao et al., 2018 [19] | 197 cancer patients with Stage I NSCLC treated with SBRT | Prospective; Uni-institutional | DT RF |
25 patient, tumor and dosiomic features | Radiation induced chest wall pain, Chest wall syndrome (CWS) | CTCAE v4 grade ≥2 chest wall pain | Utilize ML algorithms to identify chest wall pain RT toxicity predictors to develop dose–volume constraints |
| Sun et al., 2023 [27] | 1152 patients with primary breast cancer undergoing mastectomy | Prospective; Multi-Institutional | RF GBM LR XGBoost |
6 leading predictors including (pain score, post-menopausal status, urban medical insurance, history of at least one operation, under fentanyl with sevoflurane general anesthesia, and received axillary lymph node dissection) | Chronic postsurgical pain (CPSP) at 12 months after surgery | - Modified Brief Pain Inventory > 0 -2016 International Association for the Study of Pain (IASP) criteria |
Develop prediction models for CPSP after breast cancer surgery using ML approaches |
| Olling et al., 2018 [39] | 131 NSCLC cases received RT | Retrospective; Uni-Institutional | LR SVM Generalized Linear Models (glmnet) |
Clinical, tumor and dose volume parameters. | Pain while swallowing (odynophagia) after RT in lung cancer | Semi-structured weekly in-person interviews reported by RT Technologist Nurse (RTN) | Generate predictive ML models for odynophagia needing prescription pain medication during lung RT lung cancer using nursing workflow |
| Juwara et al., 2020 [28] | 195 female patients scheduled to undergo breast cancer surgery | Prospective; Uni-institutional | Solitary Least square regression (OLS) Ridge regression (RR) Elasticnet (EN) RF GBM NN LR |
Demographic and Clinical features | Neuropathic pain after breast cancer surgery | 4 interview scores (DN4-interview; range: 0–7)- 3 months after surgery | Assess the utility of ML models developed as a tool to predict pain and identify specific pain characteristics after breast cancer surgery |
| Sipila et al., 2020 [20] | 337 women treated for breast cancer | Cross-Sectional; Uni-institutional | Cluster analysis NN DT |
- Psychological and sleep-related parameters - Parameters related to pain intensity and interference |
Persistent pain in breast cancer survivors | Brief Pain Inventory (BPI) for pain. 11-point Numerical Rating Scales (NRS) (0-10) | Identify psychological features that may influence poorer coping with persistent pain |
| Guan et al., 2023 [21] | Retrospective 857 patients and prospective 368 patients with HCC received TACE | Retrospective and Prospective; Uni-Institutional | RF SVM NN Naïve Bayes DT |
24 candidate variables, Demographics, clinical, tumor, and surgery data | Postoperative pain in HCC | NR: 0 points, no pain; 1-3 points, mild pain; 4-6, moderate pain; 7-10, severe pain. | To develop an early ML model for predicting pain after TACE in patients with HCC |
| Lotsch et al., 2017 [18] | 900 women who were treated for breast cancer | Retrospective; Uni-Institutional | Supervised classification ML techniques | Parameters acquired during the cold pain tolerance test | Presence or absence of persistent pain after breast cancer surgery, in 12-36 months | -11-point Numerical Rating Scales (NRS) (0-10) -Tonic cold pain test |
Use Supervised ML techniques to test how accurately the patients’ performance in a preoperatively performed tonic cold pain test could predict persistent post-surgery pain in breast cancer |
| Barber et al., 2022 [30] | 34 women after gynecologic cancer surgery | Prospective; Uni-institutional | LR RF GBM XGBoost |
-Clinical features -PRO responses -wearable device output |
The day of an unscheduled health care utilization event using PRO parameters (e.g., Pain intensity) | Patient - Reported Outcomes Measurement Information System (PROMIS) | Test the feasibility of implementing a postoperative monitoring program for women with gynecologic cancers composed of PROs and a wearable activity monitor. And predict the day of unscheduled health care utilization for pain |
| Reinbo lt et al., 2018 [46] | Patients with Breast Cancer received Aromatase inhibitors | Retrospective; Uni-Institutional | ML: novel analytic algorithm (NAA)-Correlation network | -Genetic SNPs -Demographics features. |
Aromatase inhibitor arthralgia (AIA) in breast cancer patients | AIA-positive and negative patients, we ranked based on their discriminatory power. | Evaluate the potential of a NAA to predict AIA using germline SNPs data obtained before treatment initiation |
| Im et al., 2006 [57] | 122 nurses working with cancer patients | Retrospective; Uni-institutional | Decision support computer program (DSCP) | Age, Ethnic and socioeconomic characteristics variables | Usage profile, accuracy, and acceptance of the DSCP | -Accuracy: measured by determining whether the decision support was appropriate and accurate -Satisfaction. (satisfaction score on a 110 scale) |
To evaluate a DSCP for cancer pain management |
| Wang et al., 2021 [23] | 746 lung cancer patients with bone metastasis | Retrospective; and prospective; Uni-Institutional | DT XGBoots SVM Bayesian NN (BNN) |
-Demographic and Clinical data. -The driver gene of lung cancer -Five differentially expressed proteins of bone metastases -VAS pain assessment |
Local treatment in lung cancer pain with bone metastasis to decrease pain. output | -Visual analog scale (VAS) -Quality of life (QoL) scores Bone Metastases Module (EORT C QLQ-BM22) |
Developed and validate ML models to predict patients who should receive local treatment to reduce pain in lung cancer patients with bone metastasis |
| Lotsch et al., 2018 [31] | 1000 breast cancer patients undergoing breast surgery | Prospective; Uni-Institutional | Both supervise and unsupervised mapping ML models 1.A supervised classification 2.A symbolic rulebased classifier |
542 different variables (clinical, demographics, medical history, medication, pre-oppain, opioids, tumor data) | Persistent pain after surgery in breast cancer patients | 11- point Numerical Rating Scales (NRS) (0-10) | Identify parameters that predict persistence of significant pain in breast cancer after surgery using ML models |
| Sipila et al., 2012 [40] | 489 breast cancer treated with surgery | Prospective; Uni-Institutional | Bayesian model | Demographics -Clinical -Vitals-treatment/Hormonal, Chemo, radio therapy -Type of surgery -Pre-surgery pain Psychological features |
Persistent pain after surgery in 6 months | 11-point Numerical Rating Scales (NRS) (0-10) | Develop a ML screening tool to identify presurgical factors that predict persistence of pain after 6 months from surgery in breast cancer |
| Lotsch et al, 2020 [44] | 763 women treated with surgery for breast cancer | Prospective; Uni-Institutional | Unsupervised automated evolutionary (genetic) algorithms Hierarchical clustering LR |
Patients’ temporal features of NRS ratings | Clusters or groups of patients sharing patterns in the time courses of pain between 6 and 36 months after breast cancer surgery. | 11-point Numerical Rating Scales (NRS) (0-10) | Identify subgroups of patients sharing similar time courses of postoperative persistent pain in breast cancer, using ML algorithms |
| Lotsch et al, 2018 [60] | 1000 Breast cancer received surgery | Prospective; Uni-Institutional | Supervised ML implemented as RF | Psychological factors (depressive symptoms, state and trait anxiety, and anger inhibition) | Persistent pain after breast cancer surgery (PPSP) | 11-point Numerical Rating Scales (NRS) (0-10) | Create a simple questionnaire with a good ML predictive power for persisting pain after surgery in breast cancer patients. |
| Kringel et al, 2019 [32] | 140 women undergoing breast cancer surgery | Prospective; Uni-institutional | RF Adaptive boosting kNN Naïve Bayes SVM LR |
Next-generation sequencing for selected 77 genes. | Persistent Pain vs Non-persistent pain after breast cancer surgery | 11-point Numerical Rating Scales (NRS) (0-10) | Assessment whether NGS-derived genotypes were associated with persistent pain in patients who were treated with breast cancer surgery. |
| Wang et al., 2021 [8] | 3489 Breast Cancer patients | Prospective; Uni-institutional | DT RF XGBoost Naïve Bayes (NB) CNN |
Age, endocrine therapy, radiotherapy, and chemotherapy | Pain in postoperative breast cancer patients | -Not reported | Investigate the factors influencing chronic pain after radical mastectomy in breast cancer patients, using AI/ML algorithms |
| Kringel et al., 2019 [42] | 140 women who had undergone breast cancer surgery | Prospective; Multi-Institutional | kNN SVM LR Naïve Bayes |
Global DNA methylation. DNA methylation status of TLR4, OPRM1, and LINE1 | Persistence of postoperative pain. | 11-point Numerical Rating Scales (NRS) (0-10) | Examine the association between the DNA methylation of two key genes of glial/opioid intersection and persistent postoperative pain. |
| Lotsch et al., 2022 [49] | 57 women who had undergone breast cancer surgery | Prospective; Uni-Institutional | Supervised and Unsupervised PCA NN RF |
Proteomics: patterns in 74 serum proteomic marker | Persistent postsurgical neuropathic pain (PPSNP) | -11-point Numerical Rating Scales (NRS) (0-10), sensory examination to diagnose PPSNP. -Brief Pain Inventory (BPI) |
Identify proteins that are most informative in identifying patients with and without PPSNP after breast cancer surgery |
As for the previous set of studies, most studies considered in this subgroup also lacked detailed information on the extent of external and clinical validation in real healthcare scenarios. Three out of 19 studies, used external clinical validation of the AI/ML models. Guan et al., 2023 and Wang et al., 2021 [27] [28] used an external prospective clinical cohort for clinical evaluation of the models. Additionally, Im et al., 2006 [29] used external cohorts of nurses from the internet for the clinical evaluation of the decision support computer program (DSCP) developed for pain prediction and management. Although studies used genomic, epigenomic and proteomic data to build the classification and clustering pain models, got the identified molecules from the clinical settings, were previously identified, however, no actual clinical validation and application were used, and the clinical evaluations of the models are still needed.
AI/ML Models used for cancer pain management prediction and decision:
Seven articles focused on implementing AI/ML algorithms in cancer pain management research were detected in our review. The characteristics of these studies are summarized in Table 4.
Table 4:
Characteristics of studies used AI/ML algorithms in cancer pain management prediction and decisions research.
| Author, year |
Population | Cohort study |
AI/ML model (s) |
Input variables |
Output variable(s) |
Assessment tool(s) |
Aims/ Objectives |
|---|---|---|---|---|---|---|---|
| Kumar et al., 2023 [29] | 257 cancer patients undergoing major breast surgery. | Cross-Sectional; Uni-institutional | Generalized linear regression model (GLM) SVM—Linear RF Bayesian regularized neural network (BRNN) |
-Nongenetic clinical and genetic factors (SNPs) -Cold pain test (CPT)scores -Pupillary response to fentanyl (PRF) |
1. 24-hour fentanyl requirement 2. 24-hour pain scores 3. Time for first analgesic (TFA) in the postoperative period |
-Cold pain testing -11-point Numerical Rating Scales (NRS) (0-10) -Pupillary response to fentanyl (PRF) using a linear Ultrasound probe (Sonosite, WA). |
Develop and validate robust AI/ML predictive models for postoperative fentanyl analgesic requireme nt and other related outcomes |
| Olesen et al., 2018 [43] | 1237 cancer pain patients | Retrospective; Multi-institutional | SVM | 18 SNPs within the μ and δ opioid receptor genes and the catechol-O-methyltransferase gene | Required opioid dose in cancer pain patients. in oral MED | Oral morphine equivalent dose (MED) | Predict required opioid dose in cancer pain patients, using genetic profiling |
| Bobrova et al., 2020 [22] | 90 pancreatic cancer patients received fentanyl TTS for pain relief | Prospective; Uni-institutional | LR k nearest neighbors’ algorithm (KNC) RF GBM DT NN SVM |
57 genetic and non-genetic factors: (13 genetics, 20 clinical, demographic, type of surgical treatment, 24 laboratories) | Development of Pharmaco-resistance of Fentanyl in pancreatic cancer patients with chronic pain | Pharmaco-resistance: according to the Naranjo scale Quality of life was assessed using the Palliative Medicine Symptom Rating Scale (ESAS), and cognitive functions were assessed using the Mental Status Assessment Scale (MMSE). |
Develop a calculator for personalized risk assessment of opioid-associated drug resistance in patients with pancreas cancer using fentanyl transdermal therapeutic system (TTS) as an example. |
| Facciorusso et al., 2019 [47] | 156 pancreatic cancer patients treated with repeat celiac plexus neurolysis | Retrospective; Uni-institutional | Artificial NN (ANN) LR |
Baseline demographics, clinical and treatment rCPN characteristics | Pain response after repeated celiac plexus neurolysis (rCPN) | Visual Analogue Scale (VAS), ranging from 0 (no pain) to 10 (maximal pain) | Build an artificial NN model to predict pain response in pancreatic cancer patients received rCPN |
| Dolendo et al., 2022 [41] | 148 cancer patients that underwent mastectomy | Retrospective; Multi-institutional | LR | Patient demographics, clinical and surgical characteristics | Total opioid use on postoperative day 1 | Oxycodone milligram equivalents (OME) | Identify risk factors and develop ML-based models to predict patients who are at higher risk for postoperative opioid use after mastectomy. |
| Im et al., 2011 [58] | 428 cancer patients (ethnic minority cancer patients) | Prospective; Uni-Institutional | Decision support computer system (DSCP) | -Demographics and socioeconomics (e.g., Ethnicity/race Sex) -Cancer pain experience |
Pain Treatment decisions | -5 cancer pain scales (the visual analog scale [VAS], the verbal descriptive scale [VDS], the Wong-Baker Faces Pain Scale [FS], the Brief Pain Index-Short Form [BPI-SF], and the McGill Pain Questionnaire-Short Form [MPQ-SF]) -The Memorial Symptom Assessment Scale [MSAS], and the Functional Assessment of Cancer Therapy Scale [FACT-G]. |
Develop a DSC to support nurses’ decisions about cancer pain management |
| Sokouti et al., 2014 [50] | 70 cancer patients | Prospective; Uni-Institutional | ANN | Thermal and electrical stimulations, and the time parameter | Total pain intensity management (Pain quality) | 11-point Numerical Rating Scales (NRS) (0-10) | Model an AI system to be more biologically efficient in pain quality modulation |
A lack of external validation and clinical evaluation of the models was also detected in all studies involved in this subgroup.
Performance of cancer related pain and pain management AI/ML models:
The main discrimination performance evaluation metrics of the AI/ML models were accuracy, receiver operating curve-area under the curve (ROC-AUC), sensitivity, specificity and root mean square errors. Several studies did not report any outcome performance of the models used. Table 5 summarizes the discrimination performance results and includes an assessment on the validation methods used and whether model calibration was assessed for the studies that applied AI/ML models in cancer pain studies.
Table 5:
Models performance, validation and calibration reported in included studies.
| Study | AI/ML model |
Accuracy | AUC | Sensitivity/ Recall |
Specificity/ Precision |
Root mean square error |
Validation | Calibration |
|---|---|---|---|---|---|---|---|---|
| AI/ML Models used for cancer related pain research | ||||||||
| Shimada et al, 2023 [24] | DT | 0.685 | 0.582 | 0.849 | 0.241 | Not reported | 10-fold Cross-Validation | Not Reported |
| Cascella et al, 2023 [48] | NN | 0.9448 | 0.98 | 0.968 | 0.9528 | Not reported | Train/test | Not Reported |
| Cascella et al., 2022 [33] | RF | 0.7 | 0.98 | 0.69 | 0.71 | Not reported | 8-fold cross-validation | Not Reported |
| GBM | 0.5 | 0.59 | 0.69 | 0.29 | Not reported | |||
| LASSO-RIDGE | 0.53 | 0.5 | 1 | 0 | Not reported | |||
| ANN | 0.57 | 0.95 | 0.5 | 0.64 | Not reported | |||
| DiMartino et al., 2022 [54] | NLP | 0.61 | Not reported | 0.69 | 0.46 | Not reported | 10-fold cross-validation | Not Reported |
| Cascella et al., 2023 [34] | RF | 0.8 | 0.99 | 0.69 | 0.71 | Not reported | K-fold cross-validation | Not Reported |
| GBM | 0.62 | 0.87 | 0.69 | 0.29 | Not reported | |||
| LASSO | 0.57 | 0.7 | 1 | 0 | Not reported | |||
| ANN | 0.71 | 0.92 | 0.5 | 0.64 | Not reported | |||
| Lou et al., 2022 [35] | RF | Not reported | 0.96 | Not reported | Not reported | Not reported | Not reported | Not Reported |
| LR | Not reported | 0.75 | Not reported | Not reported | Not reported | |||
| Lu et al., 2021 [55] | BERT | 0.870 | 0.875 | 0.507 | 0.950 | Not reported | 5-fold cross-validation | Not Reported |
| SVM | 0.859 | 0.868 | 0.366 | 0.969 | Not reported | |||
| XG Boost | 0.852 | 0.830 | 0.324 | 0.969 | Not reported | |||
| Moscato et al., 2022 [36] | SVM | 0.73 | 0.71 | 0.90 | 0.52 | Not reported | 10-fold cross-validation | Not Reported |
| RF | 0.64 | 0.65 | 0.72 | 0.54 | Not reported | |||
| MP | 0.61 | 0.59 | 0.68 | 0.52 | Not reported | |||
| LR | 0.65 | 0.66 | 0.73 | 0.49 | Not reported | |||
| Adaboost | 0.52 | 0.56 | 0.72 | 0.43 | Not reported | |||
| Heintzelm an et al., 2013 [56] | NLP | 0.95 | Not reported | Not reported | Not reported | Not reported | Blind test set | Not Reported |
| Miettinen et al., 2021 [25] | Cluster | 0.799 | Not reported | 0.741 | 0.877 | Not reported | 1000 Cross-Validation runs | Not Reported |
| DT | 0.67 | Not reported | Not reported | Not reported | Not reported | |||
| Akshayaa et al., 2019 [53] | CNN | 0.95 | Not reported | Not reported | Not reported | Not reported | Train/Test | Not reported |
| Masukaw a et al., 2022 [37] | RF | Not reported | 0.90 | Not reported | Not reported | Not reported | 5-fold cross-validation | Not reported |
| SVM | Not reported | 0.87 | Not reported | Not reported | Not reported | |||
| LR | Not reported | 0.86 | Not reported | Not reported | Not reported | |||
| GBM | Not reported | 0.89 | Not reported | Not reported | Not reported | |||
| AI/ML Models used for cancer treatment induced pain research | ||||||||
| Sun et al., 2023 [27] | RF | Not reported | Not reported | 0.362 | 0.914 | Not reported | 10-fold Cross-Validation |
XGBoost: 1. ICI; 0.050 (0.038-0.122) 2. E50; 0.046 (0.024-0.123) 3. E90; 0.071 (0.064-0.257) RF: 1.ICI; 0.093 (0.053-0.168) 2. E50; 0.109 (0.034-0.170) 3. E90; 0.129 (0.107-0.303) GBM: 1.ICI; 0.072 (0.049-0.156) 2. E50; 0.071(0.0 35-0.138) 3. E90; 0.114(0.083-0.286) Logistic Regression: 1. ICI; 0.070 (0.050 – 0.146) 2. E50; 0.070 (0.027 – 0.127) 3. E90; 0.111 (0.095 – 0.307) |
| LR | Not reported | Not reported | 0.769 | 0.622 | Not reported | |||
| GBM | Not reported | Not reported | 0.338 | 0.922 | Not reported | |||
| XG Boost | Not reported | Not reported | 0.339 | 0.907 | Not reported | |||
| Olling et al., 2018 [39] | LR | 0.83 | 0.84 | 0.95 | 0.61 | Not reported | Cross-Validation | Not Reported |
| Juwara et al., 2020 [28] | OLS | Not reported | Not reported | Not reported | Not reported | 1.43 | 10-fold cross-validation |
LR: 1. Unadjusted: Intercept: 0.02 (−0.50,0.47) and Slope 0.98 (0.42,1.58) 2. Adjusted: Intercept: 0.03 (−3.4,0.34) and Slope 1.00 (0.40, 1.60 |
| RR | Not reported | Not reported | Not reported | Not reported | 1.28 | |||
| EN | Not reported | Not reported | Not reported | Not reported | 1.31 | |||
| RF | Not reported | Not reported | Not reported | Not reported | 1.39 | |||
| GBM | Not reported | Not reported | Not reported | Not reported | 1.16 | |||
| NN | Not reported | Not reported | Not reported | Not reported | 1.50 | |||
| LR | Not reported | 0.68 | Not reported | Not reported | Not reported | |||
| Sipila et al., 2020 [20] | DT | 0.661 | Not reported | 0.583 | 0.636 | Not reported | Train/test | Not Reported |
| Guan et al., 2023 [21] | RF | Not reported | 0.871 | Not reported | Not reported | Not reported | Train/Test-Internal and external validation | Not Reported |
| DT | Not reported | 0.864 | Not reported | Not reported | Not reported | |||
| ANN | Not reported | 0.827 | Not reported | Not reported | Not reported | |||
| SVM | Not reported | 0.808 | Not reported | Not reported | Not reported | |||
| NB | Not reported | 0.803 | Not reported | Not reported | Not reported | |||
| Lotsch et al., 2017 [18] | Supervised classification ML techniques | 0.944 | Not reported | Not reported | Not reported | Not reported | Not Reported | Not Reported |
| Barber et al., 2022 [30] | RF | Not reported | 0.75 | Not reported | Not reported | Not reported | 5-fold cross-validation | Not Reported |
| Reinbolt et al., 2018 [46] | NAA | Not reported | 0.759 | Not reported | Not reported | Not reported | Leave-One-Out-Cross Validation | Not Reported |
| Wang et al., 2021 [23] | DT | 0.901 | 0.89 | 0.894 | 0.903 | Not reported | Train/test 10-fold cross validation | Not Reported |
| SVM | Not reported | 0.77 | Not reported | Not reported | Not reported | |||
| BNN | Not reported | 0.71 | Not reported | Not reported | Not reported | |||
| Lotsch et al., 2018 [31] | Supervised classification model | 0.86 | Not reported | Not reported | Not reported | Not reported | 100-fold cross-validation | Not Reported |
| Sipila et al., 2012 [40] | BM | 0.627 | Not reported | 0.81 | 0.44 | Not reported | Not Reported | Not Reported |
| Lotsch et al, 2018 [31] | Rule Based Classifier | 0.86 | 0.47 | 0.706 | 0.454 | Not reported | Cross-Validation | Not Reported |
| Kringel et al, 2019 [32] | RF | 0.71 | 0.71 | 0.74 | 0.74 | Not reported | Cross-Validation | Not Reported |
| GBM | 0.71 | 0.71 | 0.69 | 0.74 | Not reported | |||
| KNN | 0.65 | 0.65 | 0.61 | 0.696 | Not reported | |||
| NB | 0.67 | 0.67 | 0.696 | 0.696 | Not reported | |||
| SVM | 0.71 | 0.71 | 0.696 | 0.74 | Not reported | |||
| LR | 0.65 | 0.65 | 0.57 | 0.74 | Not reported | |||
| Wang et al., 2021 [8] | DT | 0.878 | 0.652 | 0.017 | 0.167 | Not reported | Train/Test-10-fold Cross validation | Not Reported |
| RF | 0.871 | 0.666 | 0.050 | 0.222 | ------ | |||
| XGB | 0.885 | 0.703 | 0.017 | 0.500 | Not reported | |||
| MLPC | 0.867 | 0.675 | 0.008 | 0.048 | Not reported | |||
| GNB | 0.721 | 0.685 | 0.500 | 0.205 | Not reported | |||
| CNN | 0.883 | 0.708 | 0.008 | 0.250 | Not reported | |||
| Kringel et al., 2019 [42] | CART | 0.717 | 0.794 | Not reported | Not reported | Not reported | Train/Test-Cross validation | Not Reported |
| kNN | 0.739 | 0.739 | Not reported | Not reported | Not reported | |||
| SVM | 0.804 | 0.826 | Not reported | Not reported | Not reported | |||
| Regression | 0.739 | 0.807 | Not reported | Not reported | Not report | |||
| NB | 0.739 | 0.802 | Not reported | Not reported | Not reported | |||
| Lotsch et al., 2022 [49] | RF | 0.58 | 0.58 | 0.65 | 0.50 | Not reported | Cross-Validation | Not Reported |
| Kumar et al., 2023 [29] | GLM | Not reported | Not reported | Not reported | Not reported | 0.130 | 10-fold cross validation | Not Reported |
| SVM | Not reported | Not reported | Not reported | Not reported | 0.127 | |||
| RF | Not reported | Not reported | Not reported | Not reported | 0.129 | |||
| NN | Not reported | Not reported | Not reported | Not reported | 0.132 | |||
| Facciorus so et al., 2019 [47] | ANN | Not reported | 0.94 | Not reported | Not reported | 0.057 | 10-fold Cross-Validation | Not Reported |
| LR | Not reported | 0.85 | Not reported | Not reported | 0.147 | |||
| Dolendo et al., 2022 [41] | LR | Not reported | 0.763 | Not reported | Not reported | Not reported | 10-Fold Cross-Validation | Not Reported |
| Ridge Regression | Not reported | 0.775 | Not reported | Not reported | Not reported | |||
| Lasso | Not reported | 0.799 | Not reported | Not reported | Not reported | |||
| Elastic Net Regression | Not reported | 0.801 | Not reported | Not reported | Not reported | |||
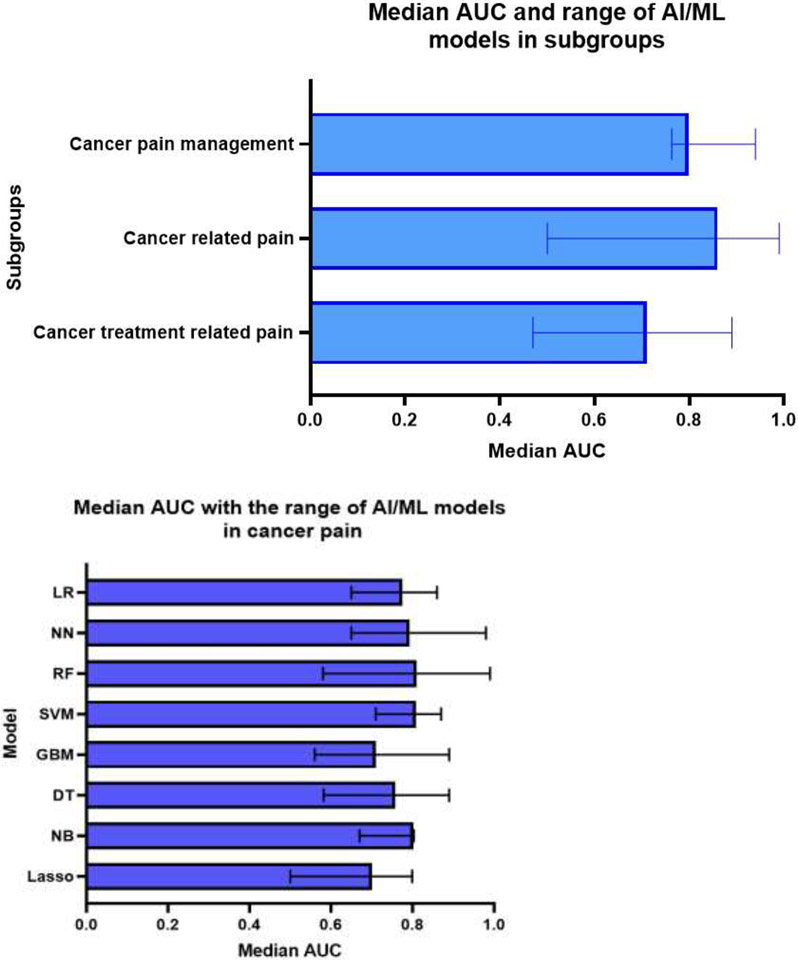
Median AUC across studies reported the models AUC performance was 0.77% (range 0.47-0.99). Models used for cancer related pain studies showed the highest AUC with median AUC 0.86 (range 0.50-0.99), while the median AUC across studies of AI/ML models for cancer treatment related pain research and cancer pain management was 0.71 (range 0.47-0.89) and 0.80 (range 0.76-0.94), respectively (Figure 5.a). The RF model showed the highest median AUC (0.81, range 0.58-0.99), while the Lasso model showed the lowest median AUC (0.70, range 0.50-0.79). Median AUC for SVM (0.808, range 0.71-0.87), NB (0.80, range 0.67-0.803), LR (0.78, range 0.65-0.86), NN (0.79, range 0.65-0.98), DT (0.76, range 0.58-0.89) and boosting (GBM) (0.71, range 0.56-0.89) (Figure 5.b). The Lasso model demonstrated the highest sensitivity (1.00), while the lowest specificity (0.00). SVM demonstrated the highest median specificity (0.74, range 0.52-0.97).
Figure 5:
a. Median area under the receiver operating curve (AUC) across all included studies by subgroups. b. Median area under the receiver operating curve (AUC) across all included studies by AI/ML model.
Only 2 studies (Sun et al., 2023 [30] and Juwara et al., 2020 [31]), out of all articles reported the calibration of the AI/ML models (Table 5). Sun et al., used the integrated calibration index, E50, E90 and Hosmer-Lemeshow (H-L) test for calibration of the models used in the study, where XGBoost model showed better performance than multivariable LR model with ICI, 0.05 (95% CI (0.038-0.122)) which was lower than LR ICI, 0.07 (95% CI (0.050 – 0.146)) and the other models; RF and GBM didn’t show superiority in ICI and H-L P values reported for all models were (LR: 0.059, RF:0.429, GBM: 0.384 and XGBoost: 0.829) [30]. Jawara et al., generated the calibration plot of the predicted probability against the observed probabilities. The MCA adjusted model showed good calibration. The MCA unadjusted logistic classifier, the intercept: 0.02 (−0.50,0.47) and slope 0.98 (0.42,1.58), while for the MCA adjusted logistic classifier, the intercept: 0.03 (−3.4,0.34) and slope: 1.00 (0.40, 1.60) [31].
Adherence to TRIPOD Guidelines:
The overall compliance with the TRIPOD guidelines reporting checklist was 70.7%, revealing that 7 out of 31 domains fell below a 60% adherence rate. While reporting adherence surpassed 80% for components such as Study Design, Eligibility Criteria and Statistical Methods, it plummeted below 50% for crucial elements like blinding of outcomes/predictors, handling of missing data, model development, and identification of risk groups. Specifically, 75.4% of studies adequately specified their study population concerning inclusion/exclusion criteria and baseline characteristics. Lastly, 95.5% of abstracts provided adequate information on study methodology, and approximately 70% of studies disclosed funding sources (Figure 6).
Figure 6:
Frequency of adherence on included studies to reporting checklist of Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) guidelines.
Risk-of-bias assessment and quality of studies:
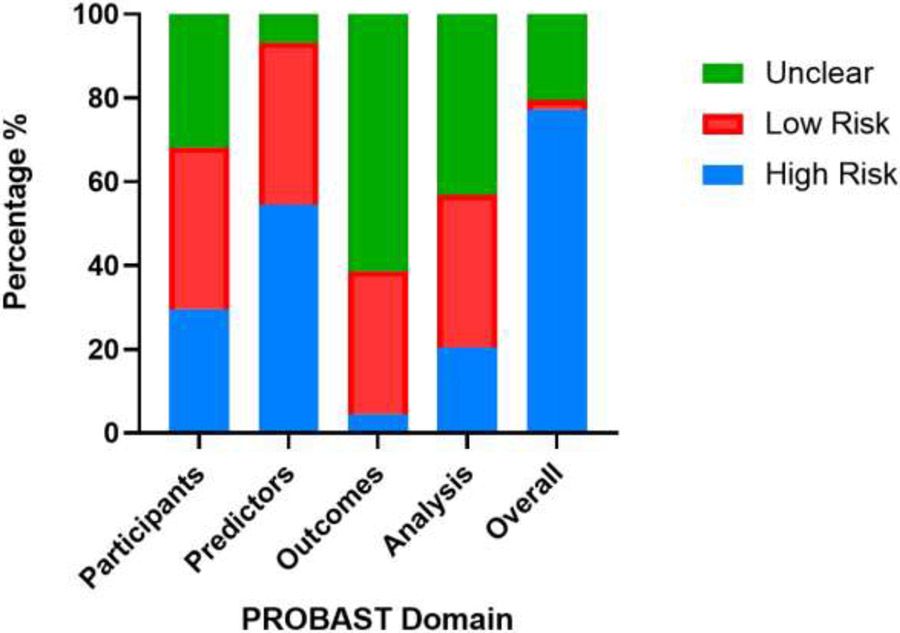
Of the 44 included studies, the overall risk of bias was high for 34 (77.3%), unclear for 9 (20.5%), and low for 1 (2.3%). High risk in the predictor’s domain (24/44) [54.5%] studies) was the main contributor to a study being overall high risk due to inadequate definition of predictors and unclear availability of predictor data at the time of model application. In the participants' domain, 13/44 (29.5%) were high risk mainly because inclusion and exclusion criteria for their study cohort were not described. Additionally, 27/44 (61.3%) were unclear in the outcomes domain because outcomes were vaguely defined, blinding was not performed, and the time interval between predictor assessment and outcome determination was not clear. In the analysis domain, some studies were high/unclear risk [28/44 (63.6%)] due to many studies did not report the number of participants with missing data, perform calibration to assess model performance, or account for overfitting. There was one study judged to be at low risk of bias: Xuyi et al. 2020 [32] (Figure 7).
Figure 7:
Frequency of risk-of-bias assessment of included studies using Prediction Model Risk of Bias Assessment Tool (PROBAST).
Discussion:
This comprehensive review delves into 44 studies that were identified through database searches, each utilizing distinct AI/ML methodologies within the field of cancer pain research published between 2006 and 2023. Most studies used prospective uni-institutial cohorts of cancer patients. In comparison to conventional statistical methods, AI/ML techniques have demonstrated superior performance, particularly in cancer pain prediction and pain management according to Sun et al. (2023) [30] and Juwara et al. (2020) [31]. The utilization of ML models, trained on big data sets of multiple features, has exhibited impressive mapping to select categories, classify patients and guide pain management decisions.
The results in this systematic review highlighted the substantial benefits of implementing AI/ML in cancer pain research, particularly in the context of predicting pain in cancer patients who have undergone cancer treatments. Out of the 44 studies, 19 focused on post-cancer treatment pain prediction, and 18 studies aimed at enhancing pain prediction and identifying high-risk factors associated with pain in cancer patients. Furthermore, our review pinpointed 7 studies that applied AI/ML for predicting cancer pain management and aiding in decisions regarding pain management (e.g., opioids usage).
Our comprehensive review illuminated the diverse array of AI/ML models deployed in the field of cancer pain research. Most of these models took the form of supervised classification or regression algorithms, including DT, RF, GBM, and LR. In addition, unsupervised clustering algorithms were applied to classify subgroups of pain patients and to identify key pain-related parameters. For making informed pain management decisions, advanced NNs were utilized and validated. AI-based decision support systems emerged as promising tools in guiding pain management choices. Most studies investigated multiple models (each model was tested separately) (55%), where RF and LR were the most common models used in these multiple models’ studies (n=15 studies for each). RF models demonstrated the highest performance across all studies (median AUC 81%), and Lasso models demonstrated the highest sensitivity (100%) while the lowest specificity (0%).
The expansion of clinical, genetic, and healthcare-related data has created an impetus for leveraging cutting-edge AI/ML techniques to harness this wealth of big data for the betterment of healthcare outcomes, especially for cancer patients contending with both the disease and its treatments. In our review, studies collectively demonstrated the potential of AI/ML models in predicting, assessing, and understanding pain among cancer patients, utilizing various inputs such as patient characteristics, imaging, video analysis, and clinical variables to improve pain management and remote consultations. While most studies used demographic and clinical data as input variables, only 4 studies included genetic data as inputs, Olesen et al., and Reinbolt et al., used single nucleotide polymorphisms (SNPs) data as inputs for the models [33]. More novel approaches were observed with studies including image data (pathology, radiological, dosiomic data) as model inputs related to cancer pain (Akshayaa et al.[34], and Chao et al., [35]). Our systematic review did not provide specific details on the evaluation process of these features or how they were selected for model input. Deep learning offers the potential of analyzing high dimensional data (e.g., images) in combination with non-image data. This is an opportunity that future studies should take towards more comprehensive models in cancer pain prediction. Five studies used video recording, machine signals or tests results as input data of the models [18, 19, 36]. Text mining and NLP open the opportunities for future studies in cancer pain using texts and different non-structured data,
Numerous studies have explored the power of AI and ML to predict pain in various medical diseases. For instance, Matsangidou et al., (2021) conducted a comprehensive investigation into the utilization of AI and ML techniques for pain prediction, showcasing the potential of these advanced technologies to enhance the accuracy and effectiveness of pain prediction and prognosis [37]. Such research exemplifies the growing significance of AI and ML in improving patient care and healthcare outcomes, However, studies specifically addressing cancer-related pain are limited, and there is a lack of data regarding the calibration of models and adherence to TRIPOD guidelines. To our knowledge, this is the first study to do a comprehensive investigation and analysis of all up-to-date studies focused on applying AI/ML models in cancer pain medicine and in cancer pain management decisions. We analyzed the different models applied in these studies and the median model performance. We categorized the studies according to the use of the models into three subgroups: cancer pain research, cancer treatment related pain, and cancer treatment (e.g., opioids) decisions. Our data analysis demonstrated an increase in the trend of AI/ML studies in cancer pain in the last few years, and that AI/ML models showed high performance in cancer pain prediction, classification and management decisions.
In our review, we extracted and analyzed data regarding the adherence to The Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) guidelines, models calibration, external validation, and clinical application of the AI/ML models. Interestingly, our results revealed 70.7% overall compliance with the TRIPOD reporting checklist. However, few studies tested models’ calibration (5%), performed external validation of internally tested models (n=6, 14%) or discussed the clinical application of the validated models (23%). According to Van Calster et. al (2019) [38], the performance assessment of AI/ML models that estimate disease risks or predict health outcomes for clinical decision-making should involve evaluating model discrimination (e.g., calculating ROC-AUC) and model calibration as essential elements in the evaluation process [38]. While most studies on AI/ML models focus on discrimination and classification performance, calibration of models is often overlooked.
Poor calibration of predictive algorithms can be misleading, leading to incorrect and potentially harmful clinical decisions [38]. Furthermore, adhering to the TRIPOD guidelines is essential in the context of AI/ML models, these guidelines provide a structured framework for transparent and comprehensive reporting, ensuring the reliability and reproducibility of predictive models [9, 10]. Equally crucial is the process of external validation, where models are rigorously tested in diverse and independent datasets to assess their generalizability and reliability beyond the initial training data. This step is vital in affirming the robustness of the models and their applicability to real-world scenarios [39]. Addressing the lack of external evaluation and clinical application in studies on AI/ML for cancer pain prediction and management is crucial. This issue often arises due to challenges in accessing diverse and large datasets, concerns over patient privacy and ethical issues, resource and funding limitations for conducting comprehensive evaluations, and prioritization of initial model development over subsequent validation [40]. To mitigate these challenges and improve future research, it is essential to promote data sharing initiatives between research institutions and healthcare providers, establish a standardized benchmark datasets and data anonymization protocols, encourage replication studies, and integrate plans for external validation into study designs. By adopting these strategies, researchers can enhance the robustness and applicability of AI/ML models in clinical settings, thereby advancing the field of cancer pain management and ensuring the reliability of research findings.
Additionally, recurrent local validation should be considered in addition to external validation to test models’ reliability, safety and generalizability for clinical application, and Youssef et al., (2023) proposed the Machine Learning Operations-inspired paradigm for recurrent local validation of AI/ML models to maintain the validity of the models [41]. Furthermore, the clinical application of established ML models is of utmost importance to bridge the gap between research and practical healthcare settings. Understanding how these models perform in clinical practice enhances their utility and ensures informed decision-making. Several studies investigate robust AI/ML models in healthcare, however, few of these models have been clinically applied due to several limitations to translate AI/ML into clinical practices [4]. By prioritizing adherence to TRIPOD guidelines, conducting thorough repetitive local validations, external validations, and emphasizing the clinical application of ML models, the healthcare community can foster trust in AI-based tools, ultimately leading to improved patient outcomes and more effective healthcare interventions.
In our study, the risk-of-bias assessment of the 44 included studies reveals significant concerns about the quality and reliability of current research in the application of AI/ML for cancer pain prediction and management. With 77.3% of the studies categorized as having a high overall risk of bias. This analysis underscores the need for improved study design and reporting in AI and ML research for cancer pain prediction and management. Future research must focus on clearly defining predictors, establishing transparent inclusion and exclusion criteria, rigorously defining outcomes, and thoroughly reporting and analyzing data. There are considerable limitations that must be addressed to enhance the robustness and applicability of these models in clinical settings.
While deep reinforcement learning (RL) has exhibited remarkable efficacy in making morphine dosage decisions and optimizing pain management within the intensive care unit (ICU), as demonstrated by Lopez-Martinez et al. (2019) [7], our review, unfortunately, did not uncover any studies that had leveraged RL for the optimization of cancer pain management. This observation underscores a potential avenue for future research and development in the field, highlighting the need for exploring the application of RL techniques to enhance the management of pain in cancer patients.
Studies included in our review collectively demonstrate diverse applications of AI/ML models in cancer pain research, illuminating their potential in predicting, understanding patient satisfaction, identifying pain-related attributes, and managing pain in cancer patients across different age groups and healthcare settings. The identification of high-risk cancer pain patients and their associated risk factors is instrumental in stratifying patients according to their pain risk, which, in turn, informs the development of personalized pain management strategies. The integration of AI and ML in cancer pain prediction and patient risk stratification streamlines the analysis of complex big data with minimal human intervention, offering a promising path to improved outcomes and enhanced QOL for cancer patients suffering with pain. Furthermore, AI/ML techniques have proven effective in not only predicting but also diagnosing, categorizing, and recommending treatments for cancer pain. Although the clinical importance of applying these AI/ML models, several studies included in our review provided very limited details on the clinical validation of their AI/ML models in real healthcare settings, which is a crucial step in ensuring the applicability and reliability of these models in clinical practice. Clinical validation, in larger and more diverse cohorts, is vital to establish the predictive value of identified features and optimize pain treatment for cancer patients, enhancing the model's real-world applicability, generalizability and effectiveness in clinical settings.
Limitations:
While our diligent effort involved an extensive comprehensive exploration of the implementation of AI and ML in the domain of cancer pain medicine, it is important to have some caution in interpreting the results due to the limitations in the design of our study. Notably, we encountered substantial heterogeneity among the identified studies, encompassing variations in the models employed, diverse AI/ML performance metrics, and disparities in the outputs associated with pain. Additionally, there is a lack of some AI/ML performance metrics which may affect the overall models’ performance. Furthermore, it's essential to acknowledge that our search strategies did not explore other symptoms or cancer therapy toxicities that may have relevance to pain, such as mucositis, dysphagia, pneumonitis, and more. Our primary focus predominantly centered on patient-reported pain phenotypes, a deliberate choice that should be considered when evaluating the scope and implications of our findings. Very few data were available on the model calibration, external validation, and clinical application of the tested models in included articles which adds limitation of interpretation of the degree of biases, robustness, and clinical reliability of the applied models.
Future Directions:
Generalizability of the performance of the identified models that demonstrated high performance is required with external validation. Assessment of the robustness, clinical application of these models is needed to be conducted in the future for clinical use in real-world healthcare settings. The use of DL and RL models in the inclusion of imaging data for cancer pain medicine should be further explored.
Conclusion:
Recent advancements in AI and ML techniques have ushered in a new era of cancer pain research, with applications including cancer pain prediction, the anticipation of pain induced by cancer treatments, and aiding in pain management decisions. AI/ML models demonstrated strong performance in predicting cancer pain, risk stratification, and enabling personalized pain management. However, there is significant heterogeneity between the studies, which must be considered when evaluating model performance. Despite good adherence to TRIPOD guidelines, there were low rates of testing models’ calibration, external validation, and clinical application. The risk-of-bias analysis showed that many included studies had a high risk of bias, mainly due to poorly defined predictors and unclear data availability. There is an ongoing need for AL/ML models to be rigorously tested for calibration and externally clinically validated before their implementation in the real-world healthcare setting.
Supplementary Material
Key Message:
This systematic review examines the utilization of innovative artificial intelligence and machine learning tools for classifying, predicting and managing cancer pain. Results demonstrate the effectiveness of AI in these areas, highlighting the continued necessity for calibration testing and clinical validation of such models.
Acknowledgement:
We thank The American Legion Auxiliary (ALA) for the ALA Fellowship in Cancer Research, 2022-2024, through The University of Texas, MD Anderson Cancer Center, UTHealth Houston Graduate School of Biomedical Sciences (GSBS). We thank The McWilliams School of Biomedical Informatics at UTHealth Houston, The Student Governance Organization (SGO) Student Excellence award.
Funding Statement:
This work was supported by the following grants: Drs. Moreno and Fuller received project related grant support from the National Institutes of Health (NIH)/ National Institute of Dental and Craniofacial Research (NIDCR) (Grants R21DE031082); Dr. Moreno received salary support from NIDCR (K01DE03052) and the National Cancer Institute (K12CA088084) during the project period. Dr. Fuller receives grant and infrastructure support from MD Anderson Cancer Center via: the Charles and Daneen Stiefel Center for Head and Neck Cancer Oropharyngeal Cancer Research Program; the Program in Image-guided Cancer Therapy; and the NIH/NCI Cancer Center Support Grant (CCSG) Radiation Oncology and Cancer Imaging Program (P30CA016672). Dr. Fuller has received unrelated direct industry grant/in-kind support, honoraria, and travel funding from Elekta AB. Vivian Salama was funded by Dr. Moreno’s fund supported from Paul Calabresi K12CA088084 Scholars Program. Kareem Wahid was supported by an Image Guided Cancer Therapy (IGCT) T32 Training Program Fellowship from T32CA261856". Dr. Naser receives funding from NIH/NIDCR Grant (R03DE033550).
Footnotes
Conflict of interest:
Authors declare that they have no known competing commercial, financial interests or personal relationships that could be constructed as potential conflict of interest.
Declaration of using generative AI:
Authors declare the usage of Covidence software (with ML applied in it) as a screening tool in the screening phase. Authors used AI-assisted technologies for grammar correction in writing the manuscript.
References:
- 1.Char DS, Shah NH, Magnus D. Implementing Machine Learning in Health Care - Addressing Ethical Challenges. N Engl J Med. 2018;378(11):981–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sendak M, Gao M, Nichols M, Lin A, Balu S. Machine Learning in Health Care: A Critical Appraisal of Challenges and Opportunities. EGEMS (Wash DC). 2019;7(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell TM. Machine Learning 1997; Mitchell TM. Does machine learning really work? AI Magazine. 18(3):11. [Google Scholar]
- 4.Busnatu S, Niculescu AG, Bolocan A, Petrescu GED, Paduraru DN, Nastasa I, et al. Clinical Applications of Artificial Intelligence-An Updated Overview. J Clin Med. 2022;11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J. 2019;6(2):94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, See KC, Ngiam KY, Celi LA, Sun X, Feng M. Reinforcement Learning for Clinical Decision Support in Critical Care: Comprehensive Review. J Med Internet Res. 2020;22(7):e18477; [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang L, Zhang W, He XF, Zha HY. Supervised Reinforcement Learning with Recurrent Neural Network for Dynamic Treatment Recommendation. Kdd'18: Proceedings of the 24th Acm Sigkdd International Conference on Knowledge Discovery & Data Mining. 2018:2447–56. [Google Scholar]
- 7.Lopez-Martinez D, Eschenfeldt P, Ostvar S, Ingram M, Hur C, Picard R. Deep Reinforcement Learning for Optimal Critical Care Pain Management with Morphine using Dueling Double-Deep Q Networks. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:3960–3. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Zhu Y, Xue Q, Ji M, Tong J, Yang JJ, Zhou CM. Predicting chronic pain in postoperative breast cancer patients with multiple machine learning and deep learning models. J Clin Anesth. 2021;74:110423. [DOI] [PubMed] [Google Scholar]
- 9.Collins GS, Dhiman P, Andaur Navarro CL, Ma J, Hooft L, Reitsma JB, et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open. 2021;11(7):e048008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD). Ann Intern Med. 2015;162(10):735–6. [DOI] [PubMed] [Google Scholar]
- 11.Vaicenavicius J, Widmann D, Andersson C, Lindsten F, Roll J, Schön TB. Evaluating model calibration in classification. 22nd International Conference on Artificial Intelligence and Statistics, Vol 89. 2019;89. [Google Scholar]
- 12.Roberta Pappadà FP. Discrimination in machine learning algorithms. arXiv:220700108 [statML]. 2022. [Google Scholar]
- 13.Covidence. Covidence systematic review software, Veritas Health Innovation, Melbourne, Australia. Available at www.covidence.org. [ [Google Scholar]
- 14.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moons KGM, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: A Tool to Assess Risk of Bias and Applicability of Prediction Model Studies: Explanation and Elaboration. Ann Intern Med. 2019;170(1):W1–W33. [DOI] [PubMed] [Google Scholar]
- 16.Wolff RF, Moons KGM, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann Intern Med. 2019;170(1):51–8. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Feridooni T, Cuen-Ojeda C, Kishibe T, de Mestral C, Mamdani M, Al-Omran M. Machine learning in vascular surgery: a systematic review and critical appraisal. NPJ Digit Med. 2022;5(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lotsch J, Ultsch A, Kalso E. Prediction of persistent post-surgery pain by preoperative cold pain sensitivity: biomarker development with machine-learning-derived analysis. Br J Anaesth. 2017;119(4):821–9. [DOI] [PubMed] [Google Scholar]
- 19.Chao HH, Valdes G, Luna JM, Heskel M, Berman AT, Solberg TD, Simone CB 2nd. Exploratory analysis using machine learning to predict for chest wall pain in patients with stage I non-small-cell lung cancer treated with stereotactic body radiation therapy. J Appl Clin Med Phys. 2018;19(5):539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sipila R, Kalso E, Lotsch J. Machine-learned identification of psychological subgroups with relation to pain interference in patients after breast cancer treatments. Breast. 2020;50:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan Y, Tian Y, Fan YW. Pain management in patients with hepatocellular carcinoma after transcatheter arterial chemoembolisation: A retrospective study. World J Gastrointest Surg. 2023;15(3):374–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bobrova O, Zyryanov S, Shnayder N, Petrova M. Personalized Calculator for Prediction of Opioid-Associated Pharmacoresistance in Patients with Pan Crease Cancer. Archiv Euromedica. 2020;10(4):20–2. [Google Scholar]
- 23.Wang Z, Sun J, Sun Y, Gu Y, Xu Y, Zhao B, et al. Machine Learning Algorithm Guiding Local Treatment Decisions to Reduce Pain for Lung Cancer Patients with Bone Metastases, a Prospective Cohort Study. Pain Ther. 2021;10(1):619–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimada K, Tsuneto S. Novel method for predicting nonvisible symptoms using machine learning in cancer palliative care. Sci Rep. 2023;13(1):12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miettinen T, Mantyselka P, Hagelberg N, Mustola S, Kalso E, Lotsch J. Machine learning suggests sleep as a core factor in chronic pain. Pain. 2021;162(1):109–23. [DOI] [PubMed] [Google Scholar]
- 26.Pombo N, Rebelo P, Araujo P, Viana J. Design and evaluation of a decision support system for pain management based on data imputation and statistical models. Measurement. 2016;93:480–9. [Google Scholar]
- 27.Sun C, Li M, Lan L, Pei L, Zhang Y, Tan G, et al. Prediction models for chronic postsurgical pain in patients with breast cancer based on machine learning approaches. Front Oncol. 2023;13:1096468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juwara L, Arora N, Gornitsky M, Saha-Chaudhuri P, Velly AM. Identifying predictive factors for neuropathic pain after breast cancer surgery using machine learning. Int J Med Inform. 2020;141:104170. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Kesavan R, Sistla SC, Penumadu P, Natarajan H, Chakradhara Rao US, et al. Predictive models for fentanyl dose requirement and postoperative pain using clinical and genetic factors in patients undergoing major breast surgery. Pain. 2023;164(6):1332–9. [DOI] [PubMed] [Google Scholar]
- 30.Barber EL, Garg R, Strohl A, Roque D, Tanner E. Feasibility and Prediction of Adverse Events in a Postoperative Monitoring Program of Patient-Reported Outcomes and a Wearable Device Among Gynecologic Oncology Patients. JCO Clin Cancer Inform. 2022;6(1):e2100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lotsch J, Sipila R, Tasmuth T, Kringel D, Estlander AM, Meretoja T, et al. Machine-learning-derived classifier predicts absence of persistent pain after breast cancer surgery with high accuracy. Breast Cancer Res Treat. 2018;171(2):399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kringel D, Kaunisto MA, Kalso E, Lotsch J. Machine-learned analysis of the association of next-generation sequencing-based genotypes with persistent pain after breast cancer surgery. Pain. 2019;160(10):2263–77. [DOI] [PubMed] [Google Scholar]
- 33.Cascella M, Coluccia S, Monaco F, Schiavo D, Nocerino D, Grizzuti M, et al. Different Machine Learning Approaches for Implementing Telehealth-Based Cancer Pain Management Strategies. J Clin Med. 2022;11(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cascella M, Scarpati G, Bignami EG, Cuomo A, Vittori A, Di Gennaro P, et al. Utilizing an artificial intelligence framework (conditional generative adversarial network) to enhance telemedicine strategies for cancer pain management. J Anesth Analg Crit Care. 2023;3(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lou Z, Vivas-Valencia C, Shields CG, Kong N. Examining how physician factors influence patient satisfaction during clinical consultations about cancer prognosis and pain. PEC Innov. 2022;1:100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moscato S, Orlandi S, Giannelli A, Ostan R, Chiari L. Automatic pain assessment on cancer patients using physiological signals recorded in real-world contexts. Annu Int Conf IEEE Eng Med Biol Soc. 2022;2022:1931–4. [DOI] [PubMed] [Google Scholar]
- 37.Masukawa K, Aoyama M, Yokota S, Nakamura J, Ishida R, Nakayama M, Miyashita M. Machine learning models to detect social distress, spiritual pain, and severe physical psychological symptoms in terminally ill patients with cancer from unstructured text data in electronic medical records. Palliative Medicine. 2022;36(8):1207–16. [DOI] [PubMed] [Google Scholar]
- 38.Knudsen AK, Brunelli C, Kaasa S, Apolone G, Corli O, Montanari M, et al. Which variables are associated with pain intensity and treatment response in advanced cancer patients? - Implications for a future classification system for cancer pain. Eur J Pain. 2011;15(3):320–7. [DOI] [PubMed] [Google Scholar]
- 39.Olling K, Nyeng DW, Wee L. Predicting acute odynophagia during lung cancer radiotherapy using observations derived from patient-centred nursing care. Tech Innov Patient Support Radiat Oncol. 2018;5:16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sipila R, Estlander AM, Tasmuth T, Kataja M, Kalso E. Development of a screening instrument for risk factors of persistent pain after breast cancer surgery. Br J Cancer. 2012;107(9):1459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolendo IM, Wallace AM, Armani A, Waterman RS, Said ET, Gabriel RA. Predictive Analytics for Inpatient Postoperative Opioid Use in Patients Undergoing Mastectomy. Cureus. 2022;14(3):e23079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kringel D, Kaunisto MA, Kalso E, Lotsch J. Machine-learned analysis of global and glial/opioid intersection-related DNA methylation in patients with persistent pain after breast cancer surgery. Clin Epigenetics. 2019;11(1):167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olesen AE, Gronlund D, Gram M, Skorpen F, Drewes AM, Klepstad P. Prediction of opioid dose in cancer pain patients using genetic profiling: not yet an option with support vector machine learning. BMC Res Notes. 2018;11(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lotsch J, Ultsch A, Kalso E. Data-science-based subgroup analysis of persistent pain during 3 years after breast cancer surgery: A prospective cohort study. Eur J Anaesthesiol. 2020;37(3):235–46. [DOI] [PubMed] [Google Scholar]
- 45.Pantano F, Manca P, Armento G, Zeppola T, Onorato A, Iuliani M, et al. Breakthrough Cancer Pain Clinical Features and Differential Opioids Response: A Machine Learning Approach in Patients With Cancer From the IOPS-MS Study. JCO Precis Oncol. 2020;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinbolt RE, Sonis S, Timmers CD, Fernandez-Martinez JL, Cernea A, de Andres-Galiana EJ, et al. Genomic risk prediction of aromatase inhibitor-related arthralgia in patients with breast cancer using a novel machine-learning algorithm. Cancer Med. 2018;7(1):240–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Facciorusso A, Del Prete V, Antonino M, Buccino VR, Muscatiello N. Response to repeat echoendoscopic celiac plexus neurolysis in pancreatic cancer patients: A machine learning approach. Pancreatology. 2019;19(6):866–72. [DOI] [PubMed] [Google Scholar]
- 48.Cascella M, Vitale VN, Mariani F, Iuorio M, Cutugno F. Development of a binary classifier model from extended facial codes toward video-based pain recognition in cancer patients. Scand J Pain. 2023. [DOI] [PubMed] [Google Scholar]
- 49.Lotsch J, Mustonen L, Harno H, Kalso E. Machine-Learning Analysis of Serum Proteomics in Neuropathic Pain after Nerve Injury in Breast Cancer Surgery Points at Chemokine Signaling via SIRT2 Regulation. Int J Mol Sci. 2022;23(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokouti B, Haghipour S. Pain Management Based on Spinal Cord Dorsal Horn System Response Identification Using Artificial Neural Networks. Biomedical Engineering-Applications Basis Communications. 2014;26(3). [Google Scholar]
- 51.Xuyi W, Seow H, Sutradhar R. Artificial neural networks for simultaneously predicting the risk of multiple co-occurring symptoms among patients with cancer. Cancer Med. 2021;10(3):989–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bang YH, Choi YH, Park M, Shin SY, Kim SJ. Clinical relevance of deep learning models in predicting the onset timing of cancer pain exacerbation. Sci Rep. 2023;13(1):11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akshayaa S, Vidhya R, Vyshnavi AMH, Namboori PKK. Exploring Pain Insensitivity Inducing Gene ZFHX2 by using Deep Convolutional Neural Network. Proceedings of the 2019 3rd International Conference on Computing Methodologies and Communication (Iccmc 2019). 2019:68–72. [Google Scholar]
- 54.DiMartino L, Miano T, Wessell K, Bohac B, Hanson LC. Identification of Uncontrolled Symptoms in Cancer Patients Using Natural Language Processing. J Pain Symptom Manage. 2022;63(4):610–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Z, Sim JA, Wang JX, Forrest CB, Krull KR, Srivastava D, et al. Natural Language Processing and Machine Learning Methods to Characterize Unstructured Patient-Reported Outcomes: Validation Study. J Med Internet Res. 2021;23(11):e26777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heintzelman NH, Taylor RJ, Simonsen L, Lustig R, Anderko D, Haythornthwaite JA, et al. Longitudinal analysis of pain in patients with metastatic prostate cancer using natural language processing of medical record text. J Am Med Inform Assoc. 2013;20(5):898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Im EO, Chee W. Evaluation of the decision support computer program for cancer pain management. Oncol Nurs Forum. 2006;33(5):977–82. [DOI] [PubMed] [Google Scholar]
- 58.Im EO, Chee W. The DSCP-CA: a decision support computer program--cancer pain management. Comput Inform Nurs. 2011;29(5):289–96. [DOI] [PubMed] [Google Scholar]
- 59.Xu X, Yang K, Zhang FL, Liu WW, Wang YY, Yu CY, et al. Identification of herbal categories active in pain disorder subtypes by machine learning help reveal novel molecular mechanisms of algesia. Pharmacological Research. 2020;156. [DOI] [PubMed] [Google Scholar]
- 60.Lotsch J, Sipila R, Dimova V, Kalso E. Machine-learned selection of psychological questionnaire items relevant to the development of persistent pain after breast cancer surgery. Br J Anaesth. 2018;121(5):1123–32. [DOI] [PubMed] [Google Scholar]
- 61.Matsangidou M, Liampas A, Pittara M, Pattichi CS, Zis P. Machine Learning in Pain Medicine: An Up-To-Date Systematic Review. Pain Ther. 2021;10(2):1067–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Calster B, McLernon DJ, van Smeden M, Wynants L, Steyerberg EW, Initiative S. Calibration: the Achilles heel of predictive analytics. Bmc Medicine. 2019;17(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramspek CL, Jager KJ, Dekker FW, Zoccali C, van Diepen M. External validation of prognostic models: what, why, how, when and where? Clin Kidney J. 2021;14(1):49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmed MI, Spooner B, Isherwood J, Lane M, Orrock E, Dennison A. A Systematic Review of the Barriers to the Implementation of Artificial Intelligence in Healthcare. Cureus. 2023;15(10):e46454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Youssef A, Pencina M, Thakur A, Zhu T, Clifton D, Shah NH. External validation of AI models in health should be replaced with recurring local validation. Nat Med. 2023;29(11):2686–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.