Abstract
The study of whether life exists, is extinct, or not depends on various sophisticated experimental studies, as many different signatures of life can be used. The experimental procedures that can be performed to identify life can be further restricted by time, resources, and mobility constraints. Therefore, any research analyzing the presence of extraterrestrial life must be precise and unambiguous. This research focuses on the objective of the extraterrestrial life detection domain and seeks to provide an efficient protocol that can produce life detection decisions based on empirical data obtained through chemical analysis under time and resource-constrained conditions. While the majority of existing frameworks in this field are designed to identify biomolecules, our goal is to accomplish the same with minimal operational expense and mission complexity. We argue that the thoughtful integration of multiple biomolecular detections with lesser complexity and a robust framework can improve overall mission performance by satisfying the necessary time and resource constraints. In this study, a rapid multiple biomolecules-based life detection protocol (MBLDP-R) from soil samples is developed from scratch and embedded in an operational scientific rover subsystem targeted for planetary analysis missions. The study uses artificial biomolecule samples and simulated extraterrestrial environments to illustrate the suggested protocol’s end-to-end process. First, we list a few significant biomolecules, including lipids, proteins, carbohydrates, nucleic acids, ammonia, and pigments. Then, a weighted qualitative test scoring is applied to sort out the best test method for the finally selected biomolecules which are used as operational analogue to showcase the protocol’s in-situ analysis and decision-making capabilities. Based on the suitable biomolecules, a scientific exploration subsystem is developed, and the implemented protocol is built to perform onboard sample analysis. Evaluation results show that: (1) the proposed MBLDP-R protocol could effectively predict the classes with an average f1-score of 98.65% (macro) and 90.00% (micro), (2) the area under the Receiver Operating Characteristics (AUC-ROC) curve shows the sample categories to be correctly predicted 92% of the time (97% for Extant, 88% for Extinct, and 92% in the case of NPL), and (3) the protocol is time-efficient with an average completion time of 17.60 min, demonstrating the protocol’s rapid nature in detecting biosignatures in soil samples. The research outcome yields useful additional data for related future studies, particularly in the design of scientific frameworks for mission-specific requirements with limited resources while also serving as a reference point for constraint evaluation methods for similar systems.
Keywords: Life detection, Multiple biomolecule integration, Qualitative test-scoring, Life-detection rover subsystem, Computer-aided analysis
Subject terms: Astrobiology, Astrobiology
Introduction
Knowing whether there is life beyond Earth has been a vital question for humanity for a long time. Several scientific exploration missions have recently been set for Mars since it is most similar to the Earth in the Solar System1,2. Soil, also known as pedosphere, serves as a habitat for various life forms and is a rich source of organic matter3. Newer science missions4,5 to Mars are beginning to concentrate on soil and rock composition, in addition to examining the geology and environmental aspects of the red planet. And to do this, scientists choose to sample or analyse Martian soil and rocks. For instance, launched in July 2020, NASA’s Perseverance rover collects soil (regolith) and rock core samples and caches them in a specific coordinate that will be sent back to Earth for analysis in terrestrial labs. Simultaneously it searches for biosignatures in-situ using several onboard scientific equipment (PIXL, MEDA, SHERLOC)4. Similarly, the Tianwen-1 mission, launched in July 2020, aimed to learn more about the features of the surface and subsurface layers of Martian soil as well as the composition and types of rocks present there5.
Similarly, biomolecules are direct products of any cell or living organism and are essential components to test for the presence of life6. Studies7–10 demonstrated the effectiveness of using a single class of biomolecule, such as a protein, nucleic acid, or other biomolecules, as successful biosignature detection mechanisms. However, dependency on a single biomolecule detection has several limitations, including uncertainty of the presence of that particular biomolecule and generation of false positives while classifying the presence of life in a soil sample. We assume that a life-detection framework, containing a global decision mechanism based on the local detection results of multiple biomolecules might be useful to address this issue.
Interestingly, in the future human exploration mission (already planned by NASA in 2031)11,12, there might be instances where life detection will be necessary for a remote hazardous area where humans cannot explore physically. This type of exploration is expected to be conducted by a mobile rover capable of onboard evaluation and instant result generation since storing soil samples for a long time in an extraterrestrial environment can be difficult. To add on, the rover might not even make it out of the mission due to extreme environmental and geological challenges. In this type of use case, time will be a massive constraint along with the installation of lighter and simpler equipment, sacrificing the perks of installing heavy equipment for conducting complex experiments of detecting biomolecules like ATP, Nucleic acid, etc.
Thus, we contend that by meeting the requisite time and resource restrictions, a multi-biomolecule life detection approach with lighter assay and complexity can address the hypothetical corner cases where resource conservation and maneuverability are critical. Consequently, this research targets the extraplanetary life detection domain and aims to provide a simple yet effective operational framework to make life detection decisions from soil samples based on empirical data achieved through chemical analysis in less time and resources. To achieve the research’s objective, the methodological phases enumerated (a) Development of an efficient and rapid on-site life-detection protocol from soil samples integrating multiple biomolecules; (b) Development of a mechanical rover subsystem for soil sample collection and classification for implementing the proposed biosignature detection protocol; and (c) Implementation of a novel evaluation methodology to assess the performance of the protocols that are used for biosignature detection from soil samples.
In this study, we develop a Multiple Biomolecules-based Rapid Life Detection Protocol (MBLDP-R) from soil samples with the integration of protein, carbohydrate, and ammonium ions. Firstly, we compile a list of potentially significant biomolecules, including protein, carbohydrate, nucleic acid, ammonia, pigment, and lipid. Then, we determine the optimum test methods for the selected biomolecules using a weighted qualitative test-scoring approach. The selection of biomolecules and their chosen detection methods considered in this study are not constant in terms of all life detection strategies. Rather the study attempts to create a baseline procedure that demonstrates the identification of biomolecules and their detection mechanism selection criteria for any future development of similar mechanisms. Using information extracted from the designated biomolecules in the earlier phase, the structure of the proposed protocol is developed. A three-layered decision tree is at the heart of the protocol’s architecture. Based on the presence and absence of the selected biomolecules in the collected sample, the tree generates life presence results as outcomes. Furthermore, a subsystem for scientific investigation is constructed along with a fully functional Mars rover prototype13, and onboard sample analysis is implemented using the developed MBLDP-R to evaluate the results in real-life test cases. The artificial samples used in the evaluation process are modified adhering to the rules of the University Rover Challenge (URC), one of the premium global Robotics Challenges organized by the Mars Society at the Mars Desert Research Station (MDRS), Southern Utah, USA. We also have demonstrated the evaluation results in the Science mission of the URC. Evaluation results indicate that: (1) the proposed MBLDP-R protocol accurately predicts classes with an average f1-score of 98.65% (macro) and 90.00% (micro); (2) the sample categories are correctly predicted 92% of the time (97% Extant, 88% Extinct, and 92% in the case of NPL); and (3) the protocol is time-efficient with an average completion time of 17.60 min, demonstrating the rapid nature of the protocol to detect biosignatures in soil samples.
This paper is divided into five sections. The following section, section "Literature review", describes the prior work in this field. The research methodology is described in section "Methodology", which includes the protocol development and the rover subsystem for scientific exploration. Evaluation of the samples using real-life samples is described in section "Evaluation". Finally, section "Discussion and conclusion" summarizes the contribution of this work along with the limitations and future scopes.
Literature review
Several studies have been conducted to find the presence of life in soil samples. Some of these are directly intended for the exploration of Mars, and others have focused on finding a generic solution for the detection of life. A summary of the related works is included in Table 1.
Table 1.
Summary of the Related Works.
| Author | Type of research | Biomolecules used | Main methodology |
|---|---|---|---|
| Recko et al.14 (2017) | Soil sample collection platform | Not used | Soil sample collecting robotic platform with humidity and UV light observation-based suitability testing |
| Neveu et al.15 (2018) | Theoretical framework | Not used | A theoretical framework to guide the design of microbial life detection investigations. |
| Kite et al.16 (2018) | Not used | Framework in which life-detection missions are performed as hypothesis tests | |
| Sharukh et al.17 (2019) | Not used | A theoretical framework to highlight the significance of the companion mini-rover system for deep narrow scientific studies. | |
| Kiflen et al.9 (2020) | Biomolecule detection and Prototype development. | Nucleic Acid | UV-C spectrometer based on RNA/DNA extraction method to detect nucleic acid in the soil sample. |
| Goordial et al.8 (2017) | Nucleic Acid | Combination of numerous low-cost approaches to detect viable extant microorganisms and nucleic acid in soil samples. | |
| Mojarro et al.18 (2017) | Nucleic Acid | Automated in situ life detection instrument that uses nucleic acid extraction and nanopore sequencing on soil samples. | |
| Mora et al.10 (2020) | Amino acid | Microchip Electrophoresis Laser-Induced Fluorescence (ME-LIF) device receives and evaluates a liquid soil sample in an automated module for life detection. | |
| Abrahamsson et al.7 (2021) | Amino acid | Automated chiral amino acid analysis method for life detection. |
Recko et al.14 developed a robotic platform-based soil sample collection technique with a unique sample collection method and suitability test based on humidity and UV light observations before collecting the sample. However, the study did not cover the possible techniques of soil sample analysis for detecting biosignatures. Neveu et al.15 proposed a theoretical framework to guide the design of investigations to detect microbial life within the practical constraints of robotic space missions. They discussed extracting features related to life detection and the methodology for detecting them. However, this basic framework assumed the probability of microbial life being either above or below a rejection threshold. This measurement can be improved in a realistic scenario by quantifying a set of distinct numerical probability values. Kite et al.16 also described a framework with a proposition that flying life-detection missions as hypothesis tests will maximize scientific value. In contrast, a negative detection can also become scientifically important. Similar to this, a theoretical framework was proposed by Sharukh et al.17, highlighting the significance of the companion mini-rover system for deep, narrow scientific research. The outcomes of these proposed investigations are based on a theoretical framework or hypothesis that can be further investigated by creating robotic subsystems to evaluate any procedure using empirical data sets.
On the contrary, Kiflen et al.9 developed a UV-C spectrometer based on Ribonucleic Acid (RNA)/ Deoxyribonucleic Acid (DNA) extraction method to detect nucleic acid in the soil sample. On a similar note, Goordial et al.8 used multiple low-cost techniques to detect viable extant microorganisms and nucleic acid from the soil sample in an environment. Similarly, Mojarro et al.18 developed the Search for Extra-Terrestrial Genomes (SETG) equipment to isolate and identify the sequence of nucleic acids from extant or preserved life on Mars. This instrument blends nucleic acid extraction and nanopore sequencing. However, automation of these multi-steps incorporating sophisticated critical steps for successful DNA extraction, such as desalting, competitive binding, DNA-protein separation, etc., is resource exhaustive and costly. In another study, Mora et al.10 created a Microchip Electrophoresis Laser-Induced Fluorescence (ME-LIF) instrument to receive and analyse a liquid soil sample in an automated module. This was a foundational development of microchip electrophoresis instruments for potential life-detection missions. However, this computerised module was developed to evaluate liquid soil samples based on protein (amino acid) detection. Similarly, Abrahamsson et al.7 prominently marked amino acid as a significant bio-molecule for life detection and proposed a novel automated chiral amino acid analysis method. These studies described various individual techniques of soil sample analysis.
Moreover, several missions and studies develop instruments and techniques to support future planetary expeditions by exploring Mars-like drilling tools for subsurface samples19, earth volcanic samples to model martian geology20, mission simulations in lava terrains to evaluate equipments21, and creating Mars analog environments to replicate surface condition22. Few potential future missions target biomolecule detection, such as the assessment of Enceladus’s23 (a moon of Saturn) ocean environment, plume composition, and biosignatures, as well as the exploration of metabolic products and signs of life while assessing the habitability and surface properties of Europa24 (a moon of Jupiter). Continuing the exploration for correct biomarker detection Aerts et al.25 proposed several techniques DNA, amino acids, lipids focusing on extracting the required biomolecule from samples and include High Performance Liquid Chromatography (HPLC), Gas Chromatography, Polymerase Chain Reaction (PCR), and acid digestion. Similarly, Gómez-Elvira et al.26 proposed detection protocols based on protein interactions and outlined a microarray-based instrument design that can operate in a space environment. While these studies contribute to scientific advancement, their computational complexity, time requirements, and financial demands constrain the rapid prototyping of in-field systems capable of delivering reasonable results under limited time and resource conditions. In this work, We aim to demonstrate a protocol for decision-making based on multiple assay outcomes, optimized for limited time and resources. We also propose a weight-based qualitative method to establish trade-offs among constraints (see section "Phase 4: qualitative test scoring for selection of test methods"), which can be extended to other situations by adjusting the weights accordingly. While not directly comparable to studies using realistic samples, weight-based qualitative analysis with synthetic samples serves as a valuable operational analogue to demonstrate our protocol for in-field decision-making based on multiple assay results.
Methodology
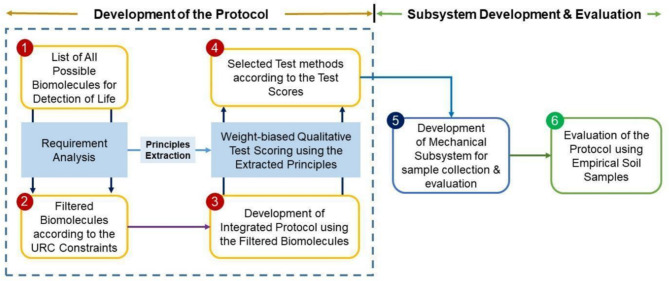
In this section, we explain the methodology of MBLDP-R and the phase-wise development procedure. Figure 1 illustrates the overview of the methodological framework. Firstly, a handful of important biomolecules such as protein, carbohydrate, nucleic acid, ammonia, pigment, and lipid are listed. The listed biomolecules are filtered to meet the URC 2021 guidelines. Then, we develop a multilevel decision tree-based protocol analysing the correlation among the selected biomolecules (Figs. 2 and 3). A weighted qualitative analysis is carried out across all the accepted techniques for identifying the chosen biomolecules in order to determine the test score for each method and identify which test method is optimal for each biomolecule. Then, the best test methods are implemented and integrated with the scientific exploration subsystem for the constructed rover PHOENIX (Fig. 4a). Finally, an exhaustive evaluation is carried out using empirical soil samples to evaluate the performance of the proposed protocol.
Fig. 1.
Research framework.
Fig. 2.
Architecture of the multiple biomolecule-based framework for soil sample detection.
Fig. 3.
Design of the rover subsystem.
Fig. 4.
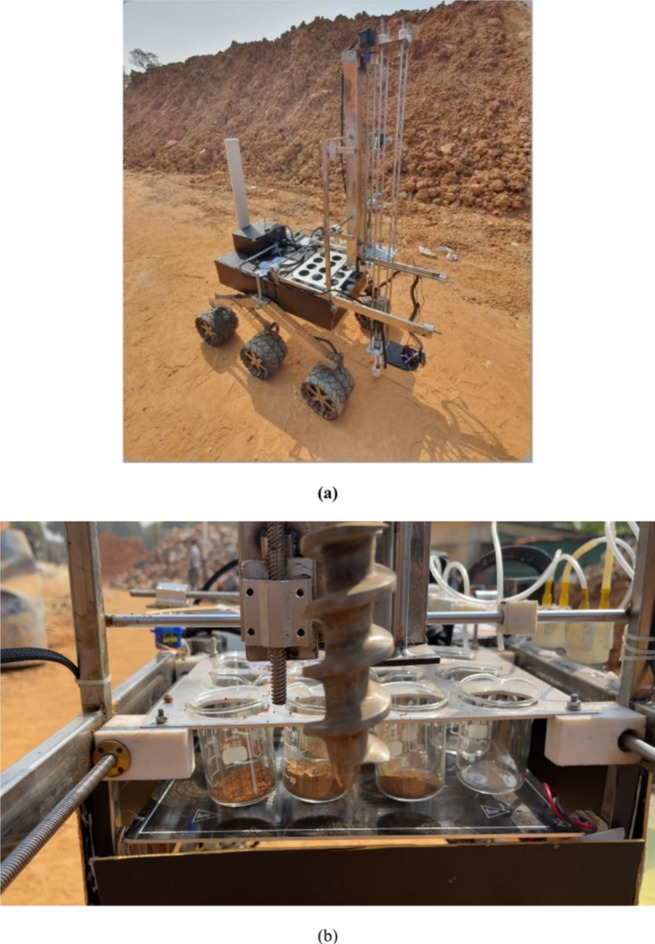
(a) Complete rover system (PHOENIX) with the attached scientific subsystem. (b) Collection of soil samples through the subsystem.
Development of MBLDP-R
The multiple biomolecules-based life detection protocol, MBLDP-R, is developed with the accumulation of works conducted in 4 phases (1) Potential list of biomolecules, (2) Selection of biomolecules based on requirement analysis, (3) Development of the protocol structure, and (4) Qualitative test scoring for the selection of the best test methods.
Phase 1: potential list of Biomolecules
Biomolecules are the chemical building blocks of living things, whereas biosignatures are observable signs of life in the environment. Despite some overlap, these two ideas are separate and have different functions in biology. A biosignature refers to any specific component, molecule, substance, or feature that can indicate past or present life and is distinct from an abiogenic (non-biological) background27. On the other hand, Biomolecules refer to essential organic compounds that play critical roles in various biological activities, including lipids, proteins, carbohydrates, and nucleic acids28. Making the distinction between possible and irrefutable biosignatures, the value of a biosignature is evaluated not only by the likelihood that life created it but also by the improbability of nonbiological processes producing it29. Biomolecules, when detected in specific contexts, serve as a subset of biosignatures, providing chemical evidence of life. For instance, chemical biosignatures such as, Molecular fossils provide evidence of past life as they are degradation products of biomolecules27. Major biomolecules have been considered a sign of extraterrestrial life, such as pigmented microorganisms30. However, since many biomolecules can be produced abiotically or due to other natural occurrences, one can argue against them being the concrete sign of life’s existence31.
In this work, we consider six major biomolecule protein, carbohydrate, ammonia, nucleic acid, lipid, and pigment as potential biomolecules for creating the multiple biomolecules-based biosignature detection protocol. Though not a conventional biomolecule, ammonia is considered here due to its derivation from the metabolism of amino acids and other biomolecules which contain nitrogen and is produced in soil from bacterial processes.
Protein. Extensive research circling archaeological remains dating from roughly 6000 years from today has pointed towards the correspondence between bimolecular preservation and the relationship between proteins, primarily collagen and mineral components, the durability of which depends chiefly on the elements of the soil32. If the thermal history of the burial environment is considered the deciding factor, relevant studies and models suggest that from warmer environments, proteins may provide opportunities to recover genetic information, as the diagenesis is controlled by slow, chemical decay of the organic phase33.
Ammonia. Mineralisation of nitrogen takes place in two steps, namely, ammonification and hydrolysis. In the first step, organic nitrogen is converted to ammonia, which in turn converts to ammonium ions in the presence of water in the second step. Ammonia, once formed, quickly escapes from the soil into the air. The positively charged ammonium ion is attracted to the negatively charged soil particles and thus acts as an exchangeable cation in soil34. The process is given below:
First step
Ammonification of organic nitrogen.
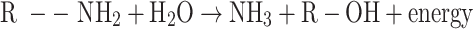
Second step
Hydrolysis of ammonia.
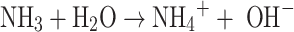
Carbohydrate. Carbohydrates play a key role in biological recognition processes35,36. Carbohydrates can be classified as monosaccharides, oligosaccharides and polysaccharides. Monosaccharides such as galactose and mannose are the chief forms of carbohydrates produced by microorganisms, while arabinose and xylose are formed by plant tissue and roots37,38. These carbohydrates are the byproducts of microbial metabolism and have vital roles to play in the formation and stabilisation of soil structure33.
Nucleic Acid. Nucleic acids are considered an important mark of living cells and are the basic molecules of life39. They are naturally made polymers consisting of nucleotides that can store, encode, transmit and express genetic information40,41. Deoxyribonucleic acid (DNA) from a mummified Egyptian kid was found in a paper by Paabo42, which provides the first evidence of the presence of DNA in archaeological remains (radiocarbon dated to nearly 2,500 years). This further strengthens the selection of nucleic acid for the consideration of extinct or extant life.
Pigments. Pigments are found in high amounts in plants43. The largest two sources of pigments are plants and microorganisms44,45. Bacterial pigments can be obtained from soils that are harvested commercially for a range of purposes, including their use in medical, food, cosmetic, and textile fields46. These pigments, which are responsible for the appearance of colours in higher plants, are classified into several groups: chlorophylls, flavonoids (chalcones, anthocyanins, flavonols, flavones), carotenoids (xanthophylls, carotenes) and betalains (betacyanin, betaxanthin)47. Moreover, prokaryotic (cyanobacteria) and eukaryotic cells (cyanelles, red algae, and cryptomonads) contain phycobilins, a class of water-soluble photosynthetic pigments, in the cytoplasm or the stroma of the chloroplast48.
Lipids. Lipid is one of the three most abundant biomolecules found in animal tissues, the other two being protein and carbohydrate43. Lipids also make up between 2 and 20% of dry weights of bacteria49. Up to 20% of soil humus exists in the form of lipids50.
Phase 2: selected biomolecules based on requirement analysis
Requirement analysis is conducted emphasising two factors: (a) rules of URC 2021 and (b) compatibility of the test equipment for the biomolecule with the rover subsystem.
In URC 20214851, the Science mission was designed to find out the presence of life in given soil samples. The goal was to categorise the samples into three significant classes (a) Extinct, (b) Extant and (c) No Presence of Life where:
Extinct: Fossilized life that is no longer metabolizing51.
Extant: Life that is metabolizing, or died recently enough for the biomolecules to still be intact51.
No Presence of life: No existence of life, inanimate/abiotic objects51.
URC 2021 set various constraints in the case of the detection of biomolecules. One of the foremost constraints was the use of hazardous substances like concentrated nitric acid (HNO3), sulphuric acid (H2SO4) etc., to conduct biomolecule detection tests. Moreover, the time limitation of 30 min for test completion played a major role in the case of selecting the biomolecules. Besides, the rover needs to traverse from the start gate to the sample location, collect the data from the samples, and pass it back to the base station. Thus, an onboard payload with the necessary testing capability (Fig. 5) is required to be developed, which will also be adept in sending necessary data back to the base station. Additionally, the weight of the total rover system with the scientific exploration subsystem must be less than 50 Kg, which restricts test designs based on heavy equipment. Finally, the budget limit for the whole system was US dollars 18,000. An analysis of all six potential biomolecules based on the requirements is made to find out suitable biomolecules for the protocol.
Fig. 5.
(a) Setup of test methods. (b) Onboard sample analysis in the developed subsystem.
Traditional spectrophotometric methods considered for detecting the likelihood of protein presence include the Biuret test, Ninhydrin test, Xanthoproteic test, Millon’s test and many more52. Although some of these traditional methods are not feasible considering time constraints and test preparations, some methods such as Biuret, Ninhydrin, and Xanthoproteic tests can be done under a time range of 1 to 10 min while applying a water bath as a heating source under a temperature range of 60 to 100 degrees52,53. More modern methods of protein detection include the use of various mass spectrometers such as IT-LIT, ToF-ToF, Q-Q-ToF, Q-Q-Q, FT-ICR, and QQ-LIT54. However, considering the factors of requirement analysis, such as the complexity of the equipment required for the tests, compatibility with the rover system and budget constraint of URC 2021, the traditional methods seems to be more suitable than the modern methods. On a similar note, methods of detecting ammonia from soil samples may include Schloesing’s methods, Baumann’s method or Russel’s methods55. Ammonia can also be detected by using a dilute sodium hydroxide solution and heating it. The resultant ammonia gas can be detected by its distinct pungent smell or by using a damp red litmus paper, which turns blue if the gas is present. This method is both simple and fast based on the type of heating source (less than a minute for direct heat). At present, various types of ammonia gas detecting sensors are available such as metal oxide-based sensors, conducting polymer sensors, tunable diode laser absorption spectroscopy (TDLAS), electrochemical sensors, surface acoustic wave sensors, field-effect transistor (FET) sensors, etc56. Easy detection procedures and readily available sensors make ammonia a potential biomolecule in these circumstances. On the other hand, for identifying the possibility of the presence of carbohydrates, several rapid tests based on specific colour reactions are available such as Molisch’s test, Anthrone test, Iodine test for glycans (starch, glycogen), Seliwanoff’s test for ketoses, Benedict’s test, Fehling’s test, and Picric acid test for reducing sugars Mucic acid test for galactose, Bial’s test for pentoses and Barfoed’s test to distinguish between monosaccharides from reducing disaccharides57. The majority of these test methods are fairly less complex and also time-efficient. Additionally, the equipment for conducting these tests is simple, lightweight and compatible with the rover system. Based on the conditions of requirement analysis, all three biomolecules: carbohydrates, ammonia and protein have the essential characteristics to be integrated as potential candidates for the protocol.
On the other hand, the nucleic acids or the microbial cells must be isolated from the soil particles and humic substances, which otherwise obscures nearly any observation, before the analysis of nucleic acid (DNA, RNA) from soil samples58. This multi-step approach of detection methods further complicates the implication of the nucleic acid detection system embedded in the rover. Moreover, automation of these multi-steps incorporating sophisticated key steps for successful DNA extraction, such as desalting, competitive binding, DNA-protein separation etc., is resource exhaustive and costly. In addition, extraction of DNA from different sources like a spore is generally a time-consuming process employed in the detection of nucleic acids from Martian or Mars-like soils, as well18 and biological reagents used in this process are a concern in the context of URC protection regulations. Moreover, 30 min, the time set for the competition, is not enough for the rover to manoeuvre, collect the sample and use the DNA extraction kits for sample analysis. In the case of pigment detection, paper chromatography59,60 is a prominent traditional method. Even though the technique can be applied to a variety of pigment detection tasks, its complexity as an onboard testing mechanism for the rover subsystem is increased because of its multi-step procedure. Moreover, modern techniques use High-Performance Liquid Chromatography (HPLC), a complex multi-step process that involves pigment extraction and HPLC analysis. The HPLC method can separate photosynthetic pigments like chlorophylls and carotenoids, etc61–63. The equipment needed for HPLC is sophisticated, non-modifiable and expensive. The equipment of HPLC is sophisticated, non-modifiable and very costly. A portable HPLC machine weighs up to 8 kg64, which is not very efficient considering the 50 Kg weight limit of the total rover system. Thus, it is certainly not possible to include pigment detection in the multiple biomolecule-based protocol. On the other hand, conventional methods for lipid quantification rely on solvent extraction and either gravimetric assays or chromatographic determination, which are time-consuming multi-step procedures that depend on the creation of the required test environments65. Thus, the complexity level of these methods is not suitable for the rover system and it is not convenient to execute the methods in an external environment. In contrast, colourimetric methods for lipid quantification constitute an attractive choice due to their fast response and simplified sample handling; however, they also usually require other preliminary steps, such as cell disruption and lipid extraction66, that are not convenient based on compatibility and complexity. Furthermore, other nonconventional spectroscopic methods include infrared spectroscopy, nuclear magnetic resonance spectroscopy, Raman spectroscopy, fluorescence spectroscopy and dielectric spectrometry66, which involve costly equipment. The above analysis shows that the tests of nucleic acid, pigment and lipid detection do not fulfil the minimum criteria of the requirement analysis. Thus, they are not considered for further analysis. Based on the above-mentioned discussions, protein, carbohydrate and ammonium ions are selected to develop a multiple biomolecule-based biosignature detection protocol for the soil samples.
Phase 3: development of the multilayered decision-tree Framework
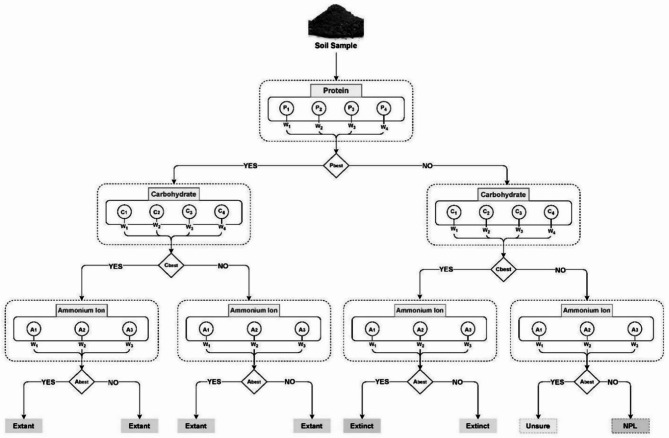
Using information compiled from the selected biomolecules (protein, carbohydrate and ammonium ion) mentioned in 3.1.2, a multilevel decision tree-based framework is developed to integrate the results of multiple biomolecular detection for classifying an unknown soil sample. A detailed architecture of the framework is illustrated in Fig. 2. The tree consists of 3 decision layers. Each of these layers contains unit blocks. This unit blocks work as decision breakpoints for the tree. From the topmost layer, the most weighted biomolecule unit blocks are present. In this research, the most weighted biomolecule for life presence making a decision is deemed to be protein. Then the carbohydrate and ammonia unit blocks are present in the later layers of the tree, respectively. Each layer of the tree is connected with a binary decision selector. These selectors help the protocol in branching out its decision from the topmost layer of the tree to the bottom. With each passing layer, the plausible decision-making from the protocol increases by an exponential of two.
Considering the structure of a unit block, each block contains a definite biomolecule as its domain, where a number of chemical analysis tests, related to the domain, are also present as the blocks cell. Each of these cells are weighted. The description of calculating the weight score based on a six-principle qualitative analysis can be found in the subsequent subsection "Phase 4: qualitative test scoring for selection of test methods" in detail. The weighted calculation of these cells determines which cell will be considered to be passed on to the binary decision selector for chemical analysis. Each unit block calculates the weight of each cell present within it and passes the most weighted cell to the binary decision selector. The binary decision selector then uses the chemical analysis test present in the past cell and based on its result passes a binary result (true/false) in the next layer. This unit block structure is followed for all the blocks of the tree until the leaf unit blocks of the last layer. The leaf unit blocks also pass their best weighted cell to binary decision selector but unlike passing it onto another layer, these selectors provide life expectancy decisions based on the flow of data on the tree.
Based on the presence and absence of the selected biomolecules in the collected sample, the tree generates life expectancy results as outcomes. The presence of protein is examined at the very first layer of the tree. Following that, the tree looks for carbohydrates, and lastly, ammonium ions. There are eight potential results of the proposed protocol. When all three types of biomolecules are present, the result is classified as Extant; if none are present, the classification is NPL. All living things have proteins, which cannot be found in an abiotic environment. Moreover, one important class of biomolecules that can be discovered in subsurface fossils is protein33. Similarly, microorganisms, plant tissue, or tree roots all contribute to the presence of carbohydrates in soil samples67. Fossilized samples of protozoa, mollusks, arthropods, and plants also contain carbohydrates68. Furthermore, ancient sedimentary rocks contain carbohydrates as well69. Even though both proteins and carbohydrates can be found as fossil molecules, carbohydrates exhibit greater resistance to decay than proteins, suggesting a greater likelihood of their retention in fossil records or absorption into fossil fuels70. Hence, the presence of protein always provides an Extant result for us independent of the other two sorts of molecules. The framework gives priority to the existence of carbohydrates in the absence of protein. Regardless of whether ammonium ion (NH4+) is present, the framework always produces the result “Extinct” if carbohydrates are present but protein is absent. Although ammonium ion (NH4+) can be generated abiotically or biotically, it can also be found in living things71. Because of this, any one of the three outcomes (Extant, Extinct, and NPL) cannot be determined by the presence of ammonium ion alone. This ambiguous outcome is treated as NPL by the framework to prevent difficulties.
Phase 4: qualitative test scoring for selection of test methods
Various tests exist for detecting proteins, carbohydrates, and ammonium ions. However, the requirement analysis discussed in 3.1.2 does not allow all the variants of the tests to be conducted in an onboard laboratory as a rover subsystem. Thus, six principles are extracted from the requirement analysis, and the principles are assigned a numerical weight according to priority based on requirement analysis factors. After that, a weighted test scoring based on these principles is conducted to select the best suitable test methods for the different biomolecules. Selected principles for qualitative analysis are as follows:
Hazardous Substances (HS). Any substance that can potentially affect safety and cause health hazards is deemed hazardous. Properties that can compromise the safety of the users and the components include flammability, explosiveness, toxicity, and the ability to oxidize. Concentrated H2SO4, HCl, NaOH, H2O2, Br are some examples of hazardous substances. For strict safety purposes, hazardous substances are not allowed in the on-site tests (Labels: Require HS-0, Don’t Require HS-1; Weight = 1).
Time (T). The amount of time required to conduct an experiment and to get the result (Labels: > than 25 min-0, <= 25 min − 1, Weight = 0.9).
Color Identification (CI). It defines the recognition of colours of the components of a chemical reaction once it reaches the desired stage. Although colour identification alone is not a reliable procedure in the identification of compounds due to the effect chemical impurities have on the colour of compounds, this technique essentially provides a head start to the identification process (Labels: Distinct colour-1, No Distinct Colour-0, weight = 0.8).
Unit Compound Identification (UCI). Unit compound (UC) identification such as amino acids for proteins and monosaccharides for carbohydrates is important as it ensures the feasibility of the test for all variants of that particular biomolecule (Labels: UC identified-1, UC Not Identified-0, weight = 0.7).
Direct Heat Contact (DHC). Direct heat contact refers to the process of test completion over a heating component rather than using a water bath. Although a water bath can provide more precise control over the heat generated for the tests, DHC is more suitable for a rapid and spill-free approach. Usage of heating components with direct contact of testing beakers also removes the complications of using water bath heating mechanism on a rover body. Regarding the safety of the rover, non-flame-generating heating components are considered for DHC. Thus, it is more convenient to use direct heat using a heating pad for a rover subsystem (Labels: Functional under DHC-1, Not Functional under DHC-0, weight = 0.5).
False Positive (FP). While identifying a specific compound, the presence of certain compounds can cause a positive result while the actual compound is absent. This positive result turns out to be a False Positive that is not desired (Labels: Doesn’t Give False Positive − 1, Gives False Positive – 0, weight = 0.3).
Modern instrumental methods are far more sensitive and accurate in terms of detecting or quantifying certain biomolecules than traditional spectrophotometric methods. However, these modern instruments can be expensive and difficult to manipulate for certain mission scenarios as discussed in the previous subsection. Considering the budget constraints given by the URC and the feasibility of detecting these biomolecules on rover bodies through on-board analysis, mostly traditional spectrophotometric detection methods have been considered in this research.
Some of the most popular test methods for the detection of proteins, carbohydrates and ammonium ions are considered for the scoring procedure. The tests are scored according to the characteristics of the test procedures in the six selected scoring principles. They are Biuret, Xanthoproteic, Millon’s and Ninhydrin test for protein detection; Benedict’s, Seliwanoff’s, Barfoed’s, Molisch’s and Iodine/Lugol’s tests for carbohydrate detection; and Schloesing’s method, direct distillation with magnesium ion and Litmus strip tests for ammonium detection. The above-mentioned tests are conducted in the Chemistry laboratory of MIST and the results are documented to determine the score of the principles. Test scoring results of the mentioned methods are shown in Table 2. The total score for any test has been calculated using the formula:
Table 2.
Weighted test scoring of the Biomolecule detection methods (X means insignificant).
| Biomolecule | Test name | HS (1) | T (0.9) |
CI (0.8) |
UCI (0.7) | DHC (0.5) |
FP (0.3) | Total Score |
|---|---|---|---|---|---|---|---|---|
| Protein | Biuret | 1 | 1 | 1 | 0 | 0 | 0 | 2.7 |
| Xanthoproteic | 0 | 1 | 1 | 1 | 1 | 1 | 3.2 | |
| Millon’s | 1 | 1 | 1 | 0 | 0 | 0 | 2.7 | |
| Ninhydrin | 1 | 1 | 1 | 1 | 1 | 1 | 4.2 | |
| Carbohydrate | Benedict Solution | 1 | 1 | 1 | 1 | 1 | 1 | 4.2 |
| Seliwanoff’s | 0 | 1 | 1 | 0 | 0 | 1 | 2.0 | |
| Barfoed’s | 1 | 1 | 1 | 1 | 0 | 1 | 3.7 | |
| Molisch’s | 0 | 1 | 1 | 0 | 0 | 0 | 1.7 | |
| Iodine/ Lugol’s Solution | 1 | 1 | 1 | 0 | 0 | 1 | 3 | |
| Ammonium | Schloesing Method | 0 | 1 | 0 | X | 0 | 1 | 1.2 |
| Direct Distillation with Magnesium ion | 1 | 0 | 0 | X | 1 | 1 | 1.8 | |
| Litmus Strip test | 1 | 1 | 1 | X | 1 | 1 | 3.5 |
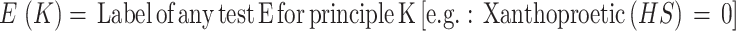 |
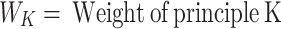 |
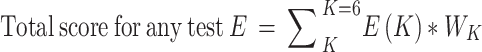 |
1 |
It is evident (Table 2) that although the Biuret test can detect the presence of peptide bonds in a substantial peptide bond concentrated protein sample, it fails to detect the unit compound amino acids (#4 UCI) in the soil specimen. Moreover, ammonium sulfate can often generate colored complexes or interfere with color development resulting in false positives (#6 FP). On a similar note, Millon’s test cannot identify the unit compound (#4), amino acids in the soil sample despite having a quick completion time of 2–3 min (#2 T). Moreover, compounds like salicylic acid and phenolic compounds can give rise to a false positive (#6) in this method. Both Biuret and Millon’s tests get a score of 2.7 (Table 2). In contrast, detection of amino acids by both Ninhydrin and Xanthoproteic tests can be stated as indirect protein assay methods. However, the prime component for the nitration reaction in the Xanthoproteic test is concentrated HNO3 which is a hazardous substance (#1 HS) and requires careful handling. The use of the Ninhydrin test is time-efficient with the detection time only between 3 and 4 min (#2). It can also easily detect amino acids (#4) indicating the likelihood of protein presence in the soil sample. Thus, the Ninhydrin test is selected with a total test score of 4.2 while the Xanthoproteic test is the nearest one with a test score of 3.2 (Table 2).
In the case of carbohydrate detection, Seliwanoff’s reagent contains highly concentrated HCl (#1) that may compromise the safety of the rover. Moreover, it detects aldose and ketose but is unable to detect monosaccharides (#4) and the use of a water bath (#5 DHC) is a must in this method. Similarly, Iodine/Lugol’s solution test can only detect polysaccharides (#4) and cannot identify monosaccharides. Although Barfoed’s method can successfully detect monosaccharides (#4), direct heat contact (#5) is not convenient for the test. Again, the concentrated sulfuric acid (#1) required to conduct Molisch’s test for the detection of carbohydrates is highly corrosive and is potentially explosive in its concentrated form. In addition, the presence of some organic acids (citric acid, lactic acid, oxalic acid, formic acid etc.) can give false-positive (#6) results in this method. Benedict’s test identifies monosaccharides (#4) in a soil sample and does not involve any corrosive ingredients (#1) in the process. Considering all the principles, Benedict’s test achieves the highest test score of 4.2 than Barfoed’s (3.7), Iodine/Lugol’s (3), Seliwanoff’s (2.0) or Molisch’s (1.7) test (Table 2). Thus, Benedict’s test is selected for the detection of carbohydrates. As previously mentioned, not all types of protein and carbohydrates are detected by the selected assays. The presence of these detection tests rather predicts the likelihood that the sample includes carbs and protein.
In the case of ammonium ion detection, the first Schloesing’s method requires strong sodium hydroxide. It, therefore, is discontinued because this strong alkali even at low temperatures gradually decomposes the organic nitrogen compounds giving rise to ammonium ions. Hydrochloric acid (#1) needed in the second Schloesing’s method removes approximately 60–70% of the added ammonium ions55. This method requires the distillation of the soil directly with magnesia, and the amount of ammonium obtained by distillation is dependent on the duration of the distillation, which is proportional to the amount of water and soil used and, on the amount of heat applied. Therefore, it takes more time (#2) to get the ammonium ions by this process. Moreover, these two methods do not give coloured identification (#3 CI) during the test. The use of hazardous substances and the length of time required to obtain the ammonium ions are drawbacks of the first two methods resulting in a test score of 1.2 and 1.8 respectively (Table 2). On the other hand, in the Litmus Strip test sodium hydroxide solution is mixed directly with the soil sample and then heated. The ammonium ions present in the sample then turns into ammonia gas and this gas turns the red litmus paper blue. With the highest score of 3.5 (Table 2) among all the ammonium ion detection methods, the litmus strip test method is selected for the detection.
The weighted analysis system for selecting assays is a dynamic component of the proposed protocol, as the weight criteria can be adjusted based on mission requirements and constraints to better align with a positive development stage later on. For instance, this study’s demonstrated case illustrates the protocol’s usage in URC. The URC mission criteria, as previously stated, required strict time and resource limits while discouraging the use of hazardous compounds. Therefore, the hazardous substance weight (HS) and time weight (T) are given higher priority than the others in this weighted analysis. The color weight is also emphasized due to the availability of color-detecting sensors, rather than more precise detectors that would reduce false positives. Similarly, in other extraterritorial mission scenarios, mission planners can dynamically adapt their in-situ experimentation plan using the weighted analysis even before the development phase begins. For example, in missions focused on rapid testing, the time weight (T) can be heavily prioritized to ensure speed, while exploratory missions requiring more precise results can reduce T and increase the false positive (FP) weight to enable more thorough analyses with fewer false positives. Similarly, in resource-limited missions, the color identification weight (CI) and the device heating control weight (DHC) can be elevated to favor simpler methods and heating mechanisms, whereas missions demanding precise control or advanced molecular detection can adjust these weights accordingly, ensuring the protocol aligns with varying mission priorities and constraints.
Development of the Rover Scientific Exploration Subsystem
The rover subsystem is developed to be attached to the main chassis to provide a tool suitable for collecting soil samples from remote and difficult terrains.
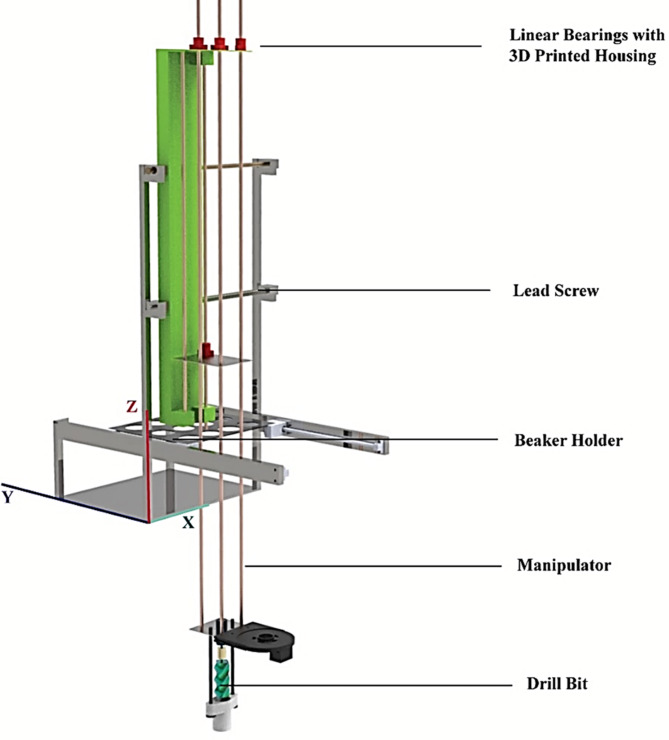
Design and fabrication
For evaluating the proposed protocol, a mechanical payload is designed as shown in (Fig. 3). The subsystem has four degrees of freedom (DOF) mechanism. There are three main parts of the developed subsystem such as (a) manipulator (b) sample collection chamber and (c) reagent mechanism. The body of the mechanical payload is made of stainless steel and aluminum sheets and is capable of collecting 4 soil samples at a time and conducting tests for protein (Ninhydrin), carbohydrate (Benedict), and ammonium ion (Litmus strip test) concurrently.
Manipulator
As depicted in Fig. 4b, the subsystem is able to gather soil samples using a three-degrees-of-freedom (DOF) manipulator to which a custom-made drill bit is mounted as the end effector. The drill bit is composed of mild steel and measures roughly 100 mm in length and 35 mm in diameter. The stainless steel plates are supported by linear rods with a diameter of 8 millimeters and linear bearings with a 3D-printed housing to ensure smooth motion. Two 8 mm-diameter square-threaded lead screws are used to drive the manipulator to collect samples of soil from the ground surface and then place them in the beaker for further processing. The lead screws are driven by two separate stepper motors (NEMA 17) A 5*8 coupler is used to connect lead screws to stepper motors. The manipulator may move along the X and Z axes using a lead screw mechanism, while the end effector spins along the Z axis to gather soil samples. A liquid sanitization mechanism is included to prevent cross-contamination. Following each sample collection, the end effector releases a flow of 3% hydrogen peroxide solution.
Sample collection chamber
The sample collection chamber comprises of 12 beakers made of borosilicate glass, a 2.5 mm thick aluminum frame, and a heat source. To gather samples, the frame adjacent to the beakers can move back and forth along the Y-axis. As illustrated in the diagram, samples are placed along the Y-axis and concurrent biomolecules testing are planned along the X-axis (Fig. 5b). As a heat source capable of generating 3,600 W/hour, a tailored heat pad is employed to provide the necessary heat for the testing.
Reagent flow mechanism
Three containers carrying the chemicals for the chosen tests, a litmus device for the ammonium test, and a framework for a single DOF tube make up the reagent mechanism. The containers are filled with Benedict’s reagent (carbohydrate test), Ninhydrin reagent (protein test), Sodium Hydroxide solution (ammonium ion test), and litmus paper strips in accordance with the selected tests (ammonium ion test). Reagents are delivered by three 12-volt DC Diaphragm Metering Micro Short Motor Water Pumps.
Detection mechanism
Four NEMA 17 stepper motors alongside the drivers are used to mobilize the motions of the subsystem. The whole rover system is controlled manually by a controller remote using a 915 MHz communication frequency, which has a range of more than 1 km. As shown in Fig. 6, the rover at first moves to the desired sample location and collects the sample using the manipulator. Using the motion of the beaker-holding frame along the y-axis, the samples collected are stored in three rows of a dedicated column of the collection chamber. Then, moving the manipulator along the x-axis the rover can collect up to four samples and store them in each column. After collecting samples for all four columns, the reagent flow mechanism adds the regents to the beakers by sliding down the frame along the x-axis to reach all the columns as shown in Fig. 5a. The heat source provides the required amount of heat. Then, by using a servomotor, a SS bar with three litmus strips is held on top of the beakers of row three as shown in Fig. 5b. The visual color change of rows one and two and the colour of litmus paper of row three gives the status of protein, carbohydrate and ammonium to the samples respectively. The observation is noted using USB camera feedback from the base station. Finally, the output of the three results is used as the input to the proposed MBLDP-R to classify the sample.
Fig. 6.
Evaluation using mock Science Mission setup.
Evaluation
The evaluation study is conducted in the chemistry laboratory of MIST and at a mock science mission field. The tests are conducted in a series of 10 events. At each event, 4 of the samples are placed in various locations of the mock science field. The gathered data from the field are transmitted back to the main base and the samples are predicted using the proposed protocol as a backbone algorithm. The field test videos are available at the deposited official System Acceptance Review (SAR) video72 of the team.
Preparation of samples
The soil samples are collected from a local site. To enhance or decrease the bioload, however, essential alteration is made. The samples are given the URC 2021 criteria labels of Extant, Extinct, and No Presence of Life (NPL). The “Extant” samples are made up of rich dirt. These samples have any unwanted rock particles removed. In addition, several of these samples have additional bioloads added to them (dextrose, albumin, etc.). According to the specific instructions of the URC science judges and organizing committeeg51, the No Presence of Life (NPL) samples are made by baking the soil for more than 24 h at 500. Two steps are taken to prepare the Extinct samples. Phase one involves heating the gathered samples at 500 degrees for more than 24 h to destroy any biosignatures that might have been present. Phase two involves creating an Extinct status by combining the samples with crushed fishbones.
Sensitivity analysis of the colorimetric reactions
An evaluation study is conducted to measure the sensitivity of the tests. The selected Ninhydrin test of this research is sensitive enough to detect up to 0.001 pmol of amines, tested from 15 sets of protein samples. This quantity yields a reading at 570 nm of 0.028 absorbance units and the typical reagent blank is 0.02 units using the standard assay parameters for the monitoring test. Amines present less than the lower limit in the soil sample will not produce any colors hence will not be detected by the rover scientific exploration subsystem and hence will produce a False Negative Output.
Study procedure
The evaluation is conducted in a mock science field of MIST. A total of 10 events are conducted where 4 samples are used in each event. In a single event, the rover has to traverse 4 sample locations (Fig. 6) and conduct the onboard sample analysis using the developed subsystem. The evaluation is conducted using 40 samples consisting of 18 Extant, 12 Extinct, and 10 NPL samples.
Results
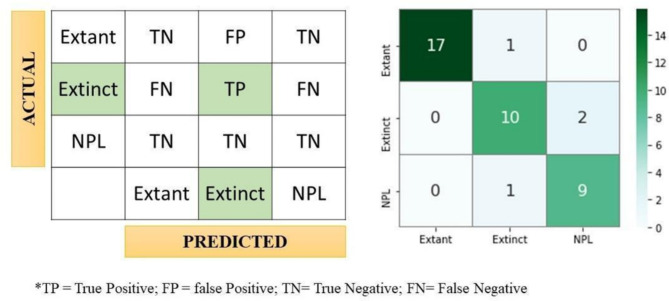
The study indicates that the methodology was 90% accurate (36 out of 40) in classifying samples into intended classes. In the case of Extant samples, the technique attains 94.44% accuracy (17 out of 18). Furthermore, the method’s accuracy is 83.33% (10 of 12) for Extinct samples and 90% (9 of 10) for NPL samples. However, accuracy is the most suited performance indicator for binary classes instead of numerous classes73. Thus, performance indicators like F1-score, precision, and recall are calculated per class to address this issue. Figure 7 depicts a confusion matrix used to evaluate class identification performance. Moreover, Micro and Macro averaging, widely acknowledged and regularly used in multiclass classification research, are utilized to assess the overall classification performance of MBLDP-R.
Fig. 7.
Confusion matrix of the evaluation study of the protocol.
Precision (See Eq. 2) is the ratio of true positive (TP) elements to positively predicted units (column sum of the predicted positives). It expresses the proportion of units deemed positive by the model, and, they are positive. In other words, precision reflects our confidence in the model’s accurate sample prediction. Recall is the ratio of True Positive elements to positively classified units (row sum of the actual positives). The prediction effectiveness of the model for the positive class is quantified by recall (see Eq. 3). Intuitively, it assesses the model’s capacity to locate all positive units in the dataset. F1-score evaluates the effectiveness of a classification model beginning with the confusion matrix; it combines precision and recalls’ metrics using the idea of harmonic mean. F1-score (See Eq. 4) is the weighted average of precision and recall, with a maximum score of 1 (100.00) and a minimum score of 0 (0.00). The proportional contribution of precision and recall equals the F1-score. The harmonic mean can determine the optimal trade-off between the two quantities.
Thus, for any class k among the three classes (Extant, Extinct and NPL):
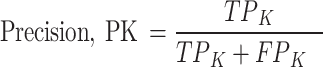 |
2 |
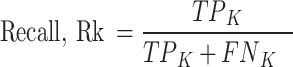 |
3 |
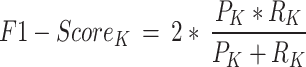 |
4 |
The macro-averaged measures of precision, recall, and F1-score are the simple average overall classes with equal weight to each incident type while micro average measures are based on the cumulative number of true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN) per studied type73.
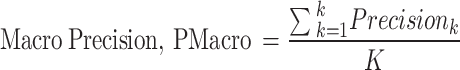 |
5 |
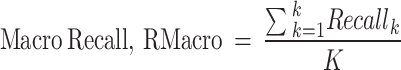 |
6 |
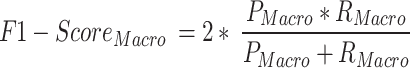 |
7 |
 |
8 |
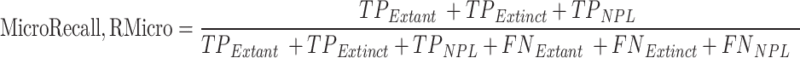 |
9 |
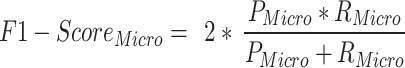 |
10 |
Class wise performance of the protocol is shown in Fig. 7. Among the three classes, Extant samples are better predicted by the protocol with an F1-score of 97.14. The protocol slightly outperforms in the case of Extinct Class prediction (F1-score: 83.33) than NPL class (F1-score: 85.71). The micro average for recall, precision, and F1-score in the proposed is 89.25, 88.38, and 88.72, respectively. Since this is a single labelled multi-classification problem, the micro average’s precision and recall are both equal to the protocol’s accuracy of 90%, from which the harmonic mean, or micro F1 score, is calculated as a similar value (Table 3).
Table 3.
Performance of MBLDP-R for different classes (Class Wise and in average measurements).
| Recall/sensitivity | Precision | F1-Score | |
|---|---|---|---|
| Extant | 94.44 | 100.00 | 97.14 |
| Extinct | 83.33 | 83.33 | 83.33 |
| NPL | 90.00 | 81.82 | 85.71 |
| ProtocolMacro | 89.25 | 88.38 | 88.72 |
| ProtocolMicro | 90.00 | 90.00 | 90.00 |
The area under the curve (AUC) of receiver operating characteristics (ROC) curve (AUC-ROC) is a performance metric that is based on a varying threshold value. ROC is a probability curve and the area under the curve (AUC) measures separability. In summary, the AUC metric announces the capability of the protocol in distinguishing the classes. AUC ranges from 0 to 1 and higher value of AUC depicts better model (1 depicts a perfect model). Mathematically, it can be created by plotting true positive rate (TPR) i.e., sensitivity or recall vs. false positive rate (FPR) (1- Specificity (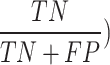 ) on varying threshold values74. AUC for the curve of the Extant class (0.97) demonstrates the highest score, followed by NPL (0.92) and Extinct (0.88) with a small margin between them. Overall, the micro and macro average of the protocol is 0.92 which depicts that MBLDP-R can correctly predict the class of an unknown sample in 92% of the instances.
) on varying threshold values74. AUC for the curve of the Extant class (0.97) demonstrates the highest score, followed by NPL (0.92) and Extinct (0.88) with a small margin between them. Overall, the micro and macro average of the protocol is 0.92 which depicts that MBLDP-R can correctly predict the class of an unknown sample in 92% of the instances.
Intriguingly, the average time to complete the tests is 17.6 min, whereas the least and maximum times are 15.20 and 19.45 min, respectively. Extant samples require 17.3 min, whereas Extinct samples require 16.8 min, and NPL samples require 18.2 min. In addition, negative tests require an average of 18.4 min longer. Positive protein tests require additional processing time. All three yes tests require the least time to complete, averaging 15.2 min.
Discussion and conclusion
In compliance with URC 2021 guidelines, a time-efficient, multiple biomolecules (protein, carbohydrate and ammonium ions) based life detection protocol from soil sample analysis (MBLDP-R) is established through the research work described here. One of the significant outcomes of this research is using a validation methodology for developing multiple biomolecule-based life detection protocols and proposing a functional protocol for classifying the samples which shows consistency and rigidity in mock in situ tests. MBLDP-R proposes a multiple biomolecule-based detection framework to be embedded in a rover’s scientific subsystem for planetary exploration. A weighted test scoring algorithm is designed to find the most suitable test method for the given situation. The implemented test-scoring scoring method can also be applied in different situations by setting the weights accordingly. A robust scientific exploration rover subsystem is developed to evaluate the proposed protocol. The evaluation study shows that the suggested protocol successfully identifies the biosignatures in soil samples and categorizes them into the three desired classifications Extant, Extinct, and No Presence of Life (NPL). Furthermore, the study provides evidence for the suggested protocol’s usability and functionality in real-world situations by showing how well it performs in simulated test environments. The protocol scores an adequate AUC value (0.92 Micro and 0.97 Macro) and excellent recall (89.25), precision (88.38), and F1-Score (88.72) on the experimental tests of identifying signs of life in mock soil samples. Moreover, an average time of 17.6 min for completing a full detection ensures that the requirement of the protocol to be rapid is met. Table 4 provides a summary of the several MBLDP-R feature factors as well as other well-known life detection approaches that were evaluated for this study.
Table 4.
Comparison of MBLDP-R with earlier life detection protocols.
| Developed life detection protocols | Base theory of life detection | Soil sample collection subsystem | On-board analysis | Time efficiency | Evaluation using empirical samples | Resource/cost efficiency |
|---|---|---|---|---|---|---|
| (Mojarro et al. 2017)18 | Nucleic Acid | Yes | Yes | No | Yes | No |
| (Goordial et al. 2017)8 | Nucleic Acid | Yes | No | No | Yes | Yes |
| (Kiflen et al. 2020)9 | Nucleic Acid | Yes | Yes | Yes | No | Yes |
| (Mora et al. 2020)10 | Amino acid | Yes | Yes | No | Yes | No |
| MBLDP-R | Multiple Biomolecule Based | Yes | Yes | Yes | Yes | Yes |
Additionally, in contrast to other established techniques based on a single biomolecule, several biomolecule integrations reduce the likelihood of false negatives. For instance: single biomolecule-based approach of Mora et al.10 terms a sample as No Presence of Life (NPL) in the absence of amino acid in that sample. However, several other important biomolecules (lipid, carbohydrate, etc.) remain unidentified due to the architectural limitations of the protocol. Thus, many samples with the absence of amino acid but with the presence of other biomolecules are termed as NPL due to the lack of contribution of multiple biomolecules during the classification of the samples. A similar statement is also true for a handful of prevailing works8,9,18 where only nucleic acid has been used to detect the signature of life.
On the contrary, MBLDP-R aims to maximize the likelihood of detecting at least one biomolecule in the sample, thereby reducing the risk of obtaining a false negative result, by establishing a threshold value. For example: MBLDP-R uses the presence of both carbohydrates and ammonium ions or solely carbohydrates to identify the sample as Extant, making it capable of detecting life even in the absence of amino acids. However, our framework does not completely eliminate the possibility of receiving a false positive due to the inherent limitations of the individual biomolecule tests, which should be further explored in the future.
The research compares the weightage of different biomolecules found in nature in life detection decisions, the complexity of testing them on mobile robotic platforms, and how resource and time-consuming these tests might be. Following this comparison, the research provides MBLDP-R, which covers both the theoretical and technical aspects of life detection. Finally, with the on-field evaluations, the research provides a clear image of how MBLDP-R will operate in a time and resource-constrained environment, which can assist in the development of more resilient and efficient future protocols. Additionally, the developed subsystem with the implemented protocol is resource and time efficient unlike some of the prior life detection protocols8,10,18. Finally, starting with the development of the protocol, choosing the best tests for biomolecule detection, development of a robotic subsystem capable of sample collection and onboard analysis, and lastly, evaluation with the real-time samples demonstrate the holistic approach of this research that can be helpful for other researchers to explore multiple biomolecules-based life detection methods.
Even with its efficiency and consistent performance, the work has some limitations. Firstly, the initial filtration of the biomolecules is based on the guidelines of URC 2021. If there is no time limit, the selected biomolecules could be different and thereby the principles for test scoring would be different. As was previously noted in sub-subsection Phase 1: potential List of Biomolecules, the addition of time-consuming procedures like nucleic acid and pigment detection will have a greater influence on life detection decisions. So, adding these time-consuming but precise test techniques can increase operating time while improving the framework’s resilience and decision-making capability, paving a path toward claiming distinct science fidelity. This can be a potential extension of the research in the future. Additionally, there is also a chance of false positives because the selected analytes can be created abiotically; this can be mitigated in future extensions of this study by taking into account the investigation of biomolecules that cannot be made abiotically. Furthermore, due to limited publicly available information regarding materials widely recognized by experts as analogues to Mars regolith or potential biosignatures, we face challenges in making detailed distinctions between our samples in this work and the Martian regoliths, which can be potentially added in the future works. Secondly, the protocol is evaluated using a moderately sized sample of 40. There are future scopes to evaluate this protocol with a large sample size along with an exploration of variations of this protocol. Another limitation is the sensitivity of the protocol. For the detection of the biomolecules, the protocol ensured that the samples to be detected as Extant, Extinct or NPL are exactly that. Therefore, the protocol had no scope to test how sensitive it is in determining the different biomolecules. Whether the protocol would detect trace amounts of the biomolecules is not apparent in this study, which can be an interesting future scope of this research. Additionally, because the rover is anticipated to operate in an extraterrestrial environment, the soil sample that it has collected is taken into account as a potential test subject as such by the protocol. While measuring the desired biomolecules, the soil inorganic particles are not separated from the sample. It would take time to complete the separating process and would compromise the rapidness of the developed procedure. However, one intriguing extension of the research can be to separate the organic particles before analyzing a soil sample utilizing the proposed MBLDP-R.
Author contributions
AZ: Methodology, Evaluation Study, Writing - Original Draft; FA: Investigation, Software, Writing - Original Draft; HK: Supervision, Writing- Original Draft, Experiment Design FNA: Resources, Investigation, Writing; OS: Design, Fabrication, Writing; AMR: Design, Fabrication; SNN: Investigation, Validation, Writing; HMC: Investigation, Validation; MR: Supervision, Writing- Reviewing and Editing;
Funding
This research is funded by the Institute of Advanced Research (Grant No. IAR-2023-Pub-006), United International University, Bangladesh.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Akib Zaman, Email: akibzaman19@gmail.com.
Fardeen Ashraf, Email: fardeenashraf98@gmail.com.
References
- 1.Zorpette, G. Why go to Mars? Sci. Am.282, 40–43 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Zubrin, R. Why we earthlings should colonize mars! Theol. Sci.17, 305–316 (2019). [Google Scholar]
- 3.Speight, J. G. Water systems. in Natural Water Remediation (ed Speight, J. G.) 1–51 (Elsevier, (2020).
- 4.Li, C. et al. China’s Mars exploration mission and science investigation. Space Sci. Rev.217, 57 (2021). [Google Scholar]
- 5.Farley, K. A. et al. Mars 2020 mission overview. Space Sci. Rev.216, 1–41 (2020). [Google Scholar]
- 6.Aerts, J. W., Röling, W. F. M., Elsaesser, A. & Ehrenfreund, P. Biota and biomolecules in extreme environments on Earth: implications for life detection on Mars. Life. 4, 535–565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahamsson, V. et al. Extraction and separation of chiral amino acids for life detection on Ocean Worlds without using organic solvents or derivatization. Astrobiology. 21, 575–586 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Goordial, J. et al. In situ field sequencing and life detection in remote (79°26′N) Canadian high arctic permafrost ice wedge microbial communities. Front. Microbiol.8, (2017). [DOI] [PMC free article] [PubMed]
- 9.Kiflen, M., Shariff, O., Mahdi, H. & Balsara, F. Novel nucleic acid-based soil sample analysis system for planetary exploration. Preprint at (2020).
- 10.Mora, M. F., Kehl, F., da Costa, E., Bramall, N. & Willis, P. A. fully automated microchip electrophoresis analyzer for potential life detection missions. Anal. Chem.92, 12959–12966 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Wilson, J. NASA’s Orion flight test and the Journey to Mars. Preprint at (2017).
- 12.Wall, M. Astronauts will face many hazards on a journey to mars. Space Preprint at (2019).
- 13.Zaman, A. et al. Phoenix: towards Designing and developing a human Assistant Rover. IEEE Access.10, (2022).
- 14.Rećko, M., Tołstoj-Sienkiewicz, J. & Turycz, P. Versatile soil sampling system capable of collecting, transporting, storing and preliminary onboard analysis for Mars rover analogue. Solid State Phenom.260, 59–65 (2017). [Google Scholar]
- 15.Neveu, M., Hays, L. E., Voytek, M. A., New, M. H. & Schulte, M. D. The ladder of life detection. Astrobiology. 18, 1375–1402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kite, E. S., Gaidos, E. & Onstott, T. C. Valuing life-detection missions. Astrobiology. 18, 834–840 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Sharukh, M., Karim, B., Nv, S. & Thomas, J. Intelligent deployable mini rover from the mars rover for deep narrow scientific investigation. Int. J. Sci. Res. Comput. Sci. Eng. Inform. Technol. 174–184 (2019).
- 18.Mojarro, A., Ruvkun, G., Zuber, M. T. & Carr, C. E. Nucleic acid extraction from synthetic Mars analog soils for in situ life detection. Astrobiology. 17, 747–760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabor, A. ARADS project designs tools for finding signs of life. Preprint at (2019).
- 20.Gentry, D. et al. Field Exploration and Life Detection Sampling for Planetary Analogue Research (FELDSPAR): Variability and Correlation in Biomarker and Mineralogy Measurements from Icelandic Mars Analogues. in Lunar and Planetary Science Conference (2018).
- 21.Tabor, A. What is BASALT? - preparing for human deep space missions, on Earth. Preprint at (2019).
- 22.Colen, J. Drilling for data: Simulating the Search for Life on mars. Preprint at (2017).
- 23.MacKenzie, S. M. et al. The Enceladus Orbilander mission concept: balancing return and resources in the search for life. Planet. Sci. J.2, 77 (2021). [Google Scholar]
- 24.Hand, K. P. et al. Science goals and mission architecture of the Europa lander mission concept. Planet. Sci. J.3, 22 (2022). [Google Scholar]
- 25.Aerts, J., Röling, W., Elsaesser, A. & Ehrenfreund, P. Biota and biomolecules in extreme environments on earth: implications for life detection on mars. Life (Basel). 4 (4), 535–565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gómez-Elvira, J. et al. Biology for Life Detection (MoBiLD) on Mars Authors (ESA Publications Division, 2003).
- 27.Domagal-Goldman, Shawn & Wright, Katherine & Adamala, Katarzyna & Rubia, Leigh & Bond, Jade & Dartnell, Lewis & Goldman, Aaron & Lynch, Kennda & Naud, Marie-Eve & Paulino Lima, Ivan Glaucio & Singer, Kelsi & Walter-Antonio, Marina & Abrevaya, Ximena & Anderson, Rika & Arney, Giada & Atri, Dimitra & Azua, Armando & Bowman, Jeff & Brazelton, William & Wong, Teresa. (2016). The Astrobiology Primer v2.0. Astrobiology. 16. 561–653. 10.1089/ast.2015.1460. [DOI] [PMC free article] [PubMed]
- 28.Zisk, R. Searching for biomolecules on Mars. Payload. (2022)., September 15 https://payloadspace.com/searching-for-biomolecules-on-mars/
- 29.Hays, L. E. et al. Biosignature preservation and detection in Mars analog environments. Astrobiology. 17, 363–400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poch, O. et al. Remote sensing of potential biosignatures from rocky, liquid, or icy (exo) planetary surfaces. Astrobiology. 17, 231–252 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Harman, C. E. et al. Abiotic O2 levels on planets around F, G, K, and M stars: effects of lightning-produced catalysts in eliminating oxygen false positives. Astrophys. J.866, 56 (2018). [Google Scholar]
- 32.Nielsen-Marsh, C. et al. Cambridge University Press,. The chemical degradation of bone. in Human Osteology: In Archaeology and Forensic Science (eds. Cox, M. & Mays, S.) 548 (2000).
- 33.Nielsen-Marsh, C. Biomolecules in fossil remains: multidisciplinary approach to endurance. Biochem. (Lond). 24, 12–14 (2002). [Google Scholar]
- 34.Walworth, J. Nitrogen in soil and the environment. Arizona.edu Preprint at (2017).
- 35.Dwek, R. A. & Glycobiology Toward understanding the function of sugars. Chem. Rev.96, 683–720 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Rudd, P. M. & Dwek, R. A. Glycosylation: heterogeneity and the {3D} structure of proteins. Crit. Rev. Biochem. Mol. Biol.32, 1–100 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Cheshire, M. V. Origins and stability of soil polysaccharide. J. Soil. Sci.28, 1–10 (1977). [Google Scholar]
- 38.Debosz, K., Vognsen, L. & Labouriau, R. Carbohydrates in hot water extracts of soil aggregates as influenced by long-term management. Commun. Soil. Sci. Plant. Anal.33, 623–634 (2002). [Google Scholar]
- 39.Singh, A., & Singh, R. DNA An Important Component of life. In Biochemistry, Biophysics, and Molecular Chemistry 195–207 (Apple Academic, 2020).
- 40.Seeman, N. C. & Sleiman, H. F. {DNA} nanotechnology. Nat. Rev. Mater.3, 17068 (2018). [Google Scholar]
- 41.Zhao, Y., Chen, F., Li, Q., Wang, L. & Fan, C. Isothermal amplification of nucleic acids. Chem. Rev.115, 12491–12545 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Pääbo, S. Molecular cloning of ancient Egyptian mummy {DNA}. Nature. 314, 644–645 (1985). [DOI] [PubMed] [Google Scholar]
- 43.Evershed, R. P. Biomolecular archaeology and lipids. World Archaeol.25, 74–93 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Cross, B. E., Edinberry, M. N. & Turner, W. B. Pigments of Gnomonia erythrostoma. I. The structures of erythrostominone, deoxyerythrostominone, and deoxyerythrostominol. J. Chem. Soc. Perkin. 1 (3), 380–390 (1972). [DOI] [PubMed] [Google Scholar]
- 45.Mizukami, H., Konoshima, M. & Tabata, M. Variation in pigment production in Lithospermum erythrorhizon callus cultures. Phytochemistry. 17, 95–97 (1978). [Google Scholar]
- 46.Mumtaz, R., Bashir, S., Numan, M., Shinwari, Z. K. & Ali, M. Pigments from soil bacteria and their therapeutic properties: a mini review. Curr. Microbiol.76, 783–790 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Młodzińska, E. Survey of plant pigments: Molecular and environmental determinants of plant colors. Abcbot pl (2009).
- 48.de Marsac, N. Phycobiliproteins and phycobilisomes: the early observations. Photosynth Res.76, 193–205 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Jones, J. G. Studies on lipids of soil micro-organisms with particular reference to hydrocarbons. J. Gen. Microbiol.59, 145–152 (1969). [DOI] [PubMed] [Google Scholar]
- 50.Stevenson, F. J. Lipids in soil. J. Am. Oil Chem. Soc.43, 203–210 (1966). [Google Scholar]
- 51.URC2020 Science Discussion. Youtube (2020). https://www.youtube.com/watch?v=jW21VtoHCag
- 52.Serra, J. A. Histochemical tests for proteins and amino acids; the characterization of basic proteins. Stain Technol.21, 5–18 (1946). [DOI] [PubMed] [Google Scholar]
- 53.Lubran, M. M. The measurement of total serum proteins by the Biuret method. Ann. Clin. Lab. Sci.8, 106–110 (1978). [PubMed] [Google Scholar]
- 54.Domon, B. & Aebersold, R. Mass spectrometry and protein analysis. Sci. (1979). 312, 212–217 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Potter, R. S. & Snyder, R. S. The determination of ammonia in soils. J. Ind. Eng. Chem.7, 221–226 (1915). [Google Scholar]
- 56.Kwak, D., Lei, Y. & Maric, R. Ammonia gas sensors: a comprehensive review. Talanta. 204, 713–730 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Katoch, R. Carbohydrate Estimations. in Analytical Techniques in Biochemistry and Molecular Biology 67–76Springer New York, (2011).
- 58.Bakken, L. R. & Frostegård, Å. Springer Berlin Heidelberg,. Nucleic acid extraction from soil. in Soil Biology 49–73 (2006).
- 59.Jensen, A. et al. Quantitative paper chromatography of carotenoids. Acta Chem. Scand.13, 1863–1868 (1959). [Google Scholar]
- 60.Strain, H. H., Sherma, J., Benton, F. L. & Katz, J. J. One-way paper chromatography of the chloroplast pigments of leaves. Biochim. Biophys. acta. 109, 1–15 (1965). [DOI] [PubMed] [Google Scholar]
- 61.Brotas, V. & Plante-Cuny, M. R. The use of {HPLC} pigment analysis to study microphytobenthos communities. Acta Oecol. (Montrouge). 24, S109–S115 (2003). [Google Scholar]
- 62.Suzuki, Y. & Shioi, Y. Identification of chlorophylls and carotenoids in major teas by high-performance liquid chromatography with photodiode array detection. J. Agric. Food Chem.51, 5307–5314 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Yuan, J., Zhang, Y., Shi, X., Gong, X. & Chen, F. Simultaneous determination of carotenoids and chlorophylls in algae by high performance liquid chromatography. Se Pu. 15, 133–135 (1997). [PubMed] [Google Scholar]
- 64.Portable high performance liquid chromatography. Axcendcorp.com Preprint at. (2020).
- 65.Mubarak, M., Shaija, A. & Suchithra, T. V. A review on the extraction of lipid from microalgae for biodiesel production. Algal Res.7, 117–123 (2015). [Google Scholar]
- 66.Patel, A. et al. Lipids detection and quantification in oleaginous microorganisms: an overview of the current state of the art. BMC Chem. Eng.1, (2019).
- 67.Oades, J. M. Soil organic matter and structural stability: mechanisms and implications for management. Plant. Soil.76, 319–337 (1984). [Google Scholar]
- 68.Swain, F. M. & Rogers, M. A. (2), (3),. Distribution of carbohydrate residues in some fossil specimens and associated sedimentary matrix and other geologic samples. J. Sediment. Res. 37, 12–24 (1967).
- 69.Palacas, J. G., Swain, F. M. & Smith, F. Presence of carbohydrates and other organic compounds in ancient sedimentary rocks. Nature. 185, 234 (1960). [Google Scholar]
- 70.Briggs, D. E. G. & Summons, R. E. Ancient biomolecules: their origins, fossilization, and role in revealing the history of life: prospects \& overviews. Bioessays. 36, 482–490 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Capone, D. G., Popa, R., Flood, B. & Nealson, K. H. Geochemistry. Follow the nitrogen. Sci. (1979). 312, 708–709 (2006). [DOI] [PubMed] [Google Scholar]
- 72.MIST Mongol Barota - URC SAR 2021. Youtube (2021). https://www.youtube.com/watch?v=0ezyJEdleQM
- 73.Sokolova, M. & Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 45, 427–437 (2009). [Google Scholar]
- 74.Narkhede, S. Understanding auc-roc curve. Towards Data Sci.26, 220–227 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.