Abstract
Imidazole Propionate (ImP), a gut-derived metabolite from histidine, affects insulin signaling in mice and is elevated in type 2 diabetes (T2D). However, the source of histidine and the role of the gut microbiota remain unclear. We conducted an intervention study in mice and humans, comparing ImP kinetics in mice on a high-fat diet with varying histidine levels and antibiotics, and assessed ImP levels in healthy and T2D subjects with histidine supplementation. Results show that dietary histidine is metabolized to ImP, with antibiotic-induced gut microbiota suppression reducing ImP levels in mice. In contrast, oral histidine supplementation resulted in increases in circulating ImP levels in humans, whereas antibiotic treatment increased ImP levels, which was associated with a bloom of several bacterial genera that have been associated with ImP production, such as Lactobacilli. Our findings highlight the gut microbiota’s crucial role in regulating ImP and the complexity of translating mouse models to humans.
Subject terms: Health care, Clinical microbiology
Introduction
Gut microbiota live in symbiosis with humans and affect human physiology1. In fact, gut microbiota modulate physiological processes through synthesis, extraction, and processing of nutrients that are important for the human body. Gut microbiota produce important metabolites, such as short-chain fatty acids (SCFA), bile salts, amino acid derivatives, and vitamins2. Microbial-derived metabolites have been shown to impact human metabolism, with both detrimental and beneficial effects described3,4. SCFA produced from indigestible fibers have been associated with reduced insulin resistance and improved blood pressure regulation5,6. In contrast, amino-acid and choline-related metabolites such as p-cresol sulfate7, indoxyl sulfate8, trimethylamine N-oxide (TMAO)9 and phenylacetyglutamine10 are linked to increased cardiovascular morbidity and mortality. Another amino acid-derived metabolite, imidazole propionate (ImP), has been shown to be increased in individuals with prediabetes and type 2 diabetes (T2D)11,12 and is associated with hypertension, heart failure and mortality13,14. ImP is a metabolite synthesized from histidine and is associated with low gut microbial gene richness12. In fact, ImP is produced from gut microbiota of individuals with T2D in a gut simulator following the addition of histidine11. Interestingly, ImP impairs insulin signaling by activating p38γ, followed by p62-mTORC-S6K1 and AKT-AMPK activation in vitro11,15, abolishing the effects of metformin in mice15.
Histidine is an essential amino acid, metabolized into either histamine or trans-urocanate. The latter is further metabolized into ImP, cis-urocanate, or glutamate16. Histidine deficiency can cause decreased mental alertness, anorexia, malaise, impaired erythropoiesis and skin lesions17, while excessive histidine is degraded in the liver and skin by histidine ammonia-lyase into trans-urocanate and cis-urocanate by exposure to sunlight18. Interestingly, cis-urocanate has been shown to have anti-inflammatory effects in multiple sclerosis19, intestinal inflammation20, and epithelial cells21. Although mechanistic studies have been performed in vitro or with ImP manipulation, the in vivo kinetics of ImP and urocanate production after oral histidine supplementation remain unknown.
Mice and humans share many similarities in terms of body composition, genetics, and physiology, which make them frequently used surrogates for humans in scientific research22. However, there are also significant differences between species, as mice have different energy metabolism23, bile acid24, and drug metabolism25. In addition, the microbiota of mice share similarities with humans in common phyla and genera, however, the abundances of phyla and species differ between mice and humans16,26. This may limit the generalizability of findings in murine studies and highlights the importance of both mice and human studies.
We, therefore, performed a dietary intervention study with orally administered histidine to study the systemic kinetics of the histidine-related metabolites urocanate and ImP in mice and humans. We also applied broad-spectrum antibiotics to suppress gut microbiota activity and determined the effect of histidine on metabolite kinetics as well as gut microbiota composition.
Results
Dietary histidine levels do not affect body weight or food intake in obese mice
Diet-induced obese (DIO) C57Bl/6J mice were stratified into a low histidine (1.28 g/kg), normal histidine (5.47 g/kg) or high histidine (25 g/kg) diet group for 7 weeks, Supplementary Table 1. To determine whether if dietary histidine affects body weight and food intake, weekly monitoring of these parameters was performed throughout the study.
We found that altering dietary histidine levels did not affect body weight significantly. However, mice receiving antibiotics had lower body weight, which started to manifest after 2 weeks of antibiotics use in the low and normal histidine groups (Supplementary fig. 1). Interestingly, this effect of antibiotic-induced weight loss was not observed in the high histidine group. Alterations in dietary histidine content did not lead to significant changes in food intake or body composition, as measured via an EchoMRI (Supplementary fig. 1).
Histidine intake leads to a dose-dependent increase in ImP in mice
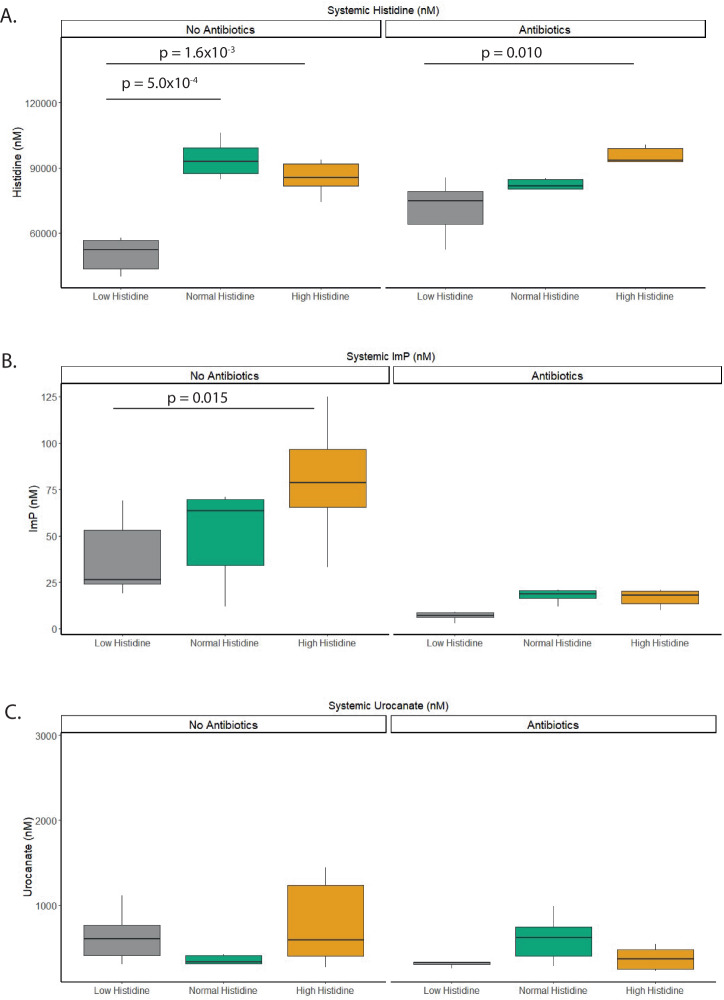
To determine whether dietary histidine intake increased circulating levels of the microbial-derived metabolite ImP, we measured fasting histidine, ImP, and urocanate levels in a subset of mice in systemic blood using LC-MS/MS (Fig. 1). Systemic histidine levels were significantly higher in the normal and high histidine group, compared to the low histidine group (Fig. 1a). In fact, systemic histidine levels increased from 52.2 µM (95% CI: 43.60–56.74 µM) in the low histidine group to 85.47 µM (81.60–91.74 µM, p = 0.0016) in the normal histidine group with no further significant increase in the high histidine group, indicating that the uptake of histidine in the systemic circulation is easily saturated. The addition of a broad spectrum of antibiotics did not mitigate this effect.
Fig. 1. Histidine-related fasting metabolites in mice.
A Fasting Histidine serum levels increased with increasing dietary histidine concentrations. This effect is also visible when antibiotics are added to the die in the diet. B Fasting Imidazole propionate serum levels (ImP). ImP levels increase with increasing oral histidine load. This effect is abolished when antibiotics are added to the diet. C Fasting Urocanate serum levels. Oral histidine does not affect urocanate levels. Statistics performed with linear mixed effects model; FDR corrected. N = 6/group.
The production of the microbial-derived metabolite, ImP, showed a dose-dependently increase from 0.027 µM (CI:0.024–0.053 µM) in the low histidine group to 0.079 µM (0.066–0.097 µM) in the high histidine group (Fig. 1b). Antibiotics decreased the levels of ImP, and the dose-dependent response was no longer present. Urocanate levels did not significantly change after dietary histidine manipulation, and antibiotic usage did not affect its systemic concentrations (Fig. 1c).
Participant characteristics humans
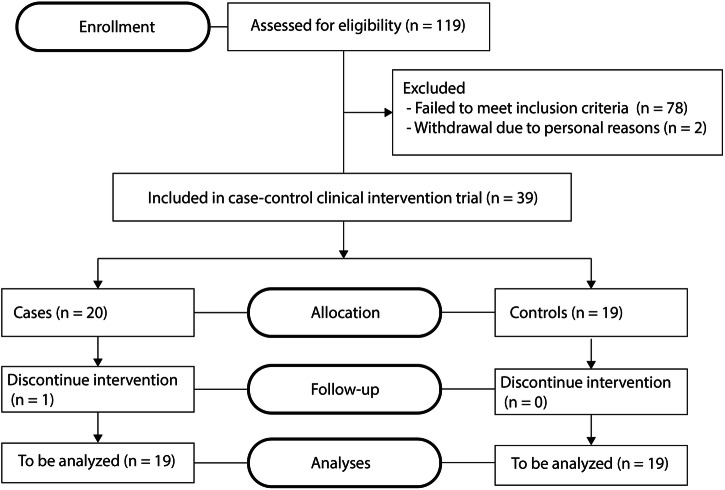
To understand how oral histidine affects ImP in humans, we performed an intervention study with oral histidine in 39 participants. We included participants with T2D (n = 20) and an age- and sex-matched control group (n = 19). Metabolic parameters such as BMI, weight, and total body fat differed between groups (Table 1). Importantly, resting energy expenditure measured by indirect calorimetry and caloric intake evaluated by food diaries and total body fat did not change during the intervention (Supplementary Table 2 and Supplementary fig. 3). Furthermore, individuals with T2D used oral diabetes medication. Side effects due to antibiotic treatment included gastro-intestinal complaints, and one case of skin desquamation was observed, which did not lead to discontinuation of the intervention. One participant was excluded before the final visit due to antibiotics use after a minor skin infection (Fig. 2). No serious adverse events occurred.
Table 1.
Baseline characterstics
| Control | Type 2 diabetes | p value | |
|---|---|---|---|
| n | 19 | 20 | – |
| Age (years) | 61.11 (7.69) | 59.95 (5.60) | 0.593 |
| Sex n (Female %) | 10 (52.6) | 10 (50.0) | 1.00 |
| BMI (kg/m2) | 25.49 (2.75) | 29.56 (2.87) | <0.001 |
| Waist/Hip ratio | 0.90 (0.05) | 0.98 (0.08) | 0.001 |
| Weight (kg) | 72.95 (13.13) | 87.08 (15.01) | 0.003 |
| Total body fat (%) | 27.16 (7.61) | 32.42 (7.18) | 0.035 |
| eGFR (mL/min/1.722) | 85.44 (5.55) | 81.80 (14.01) | 0.309 |
| Metformin use = Yes (%) | NA | 20 (100) | – |
| Statin use = Yes (%) | NA | 20 (100) | – |
| Sulfonylurea derivative use = Yes (%) | NA | 8 (40) | – |
| Antihypertensive drugs use = Yes (%) | NA | 9 (45) | – |
| Other drugs = Yes (%) | NA | 5 (25) | – |
Data displayed as mean ± SD or %. t-test for significance. Other = fibrates, antihistamines, thyroid supplementation, and antiplatelet therapy.
Fig. 2.
Flow diagram human intervention study.
Oral histidine supplementation increases ImP levels also after antibiotics
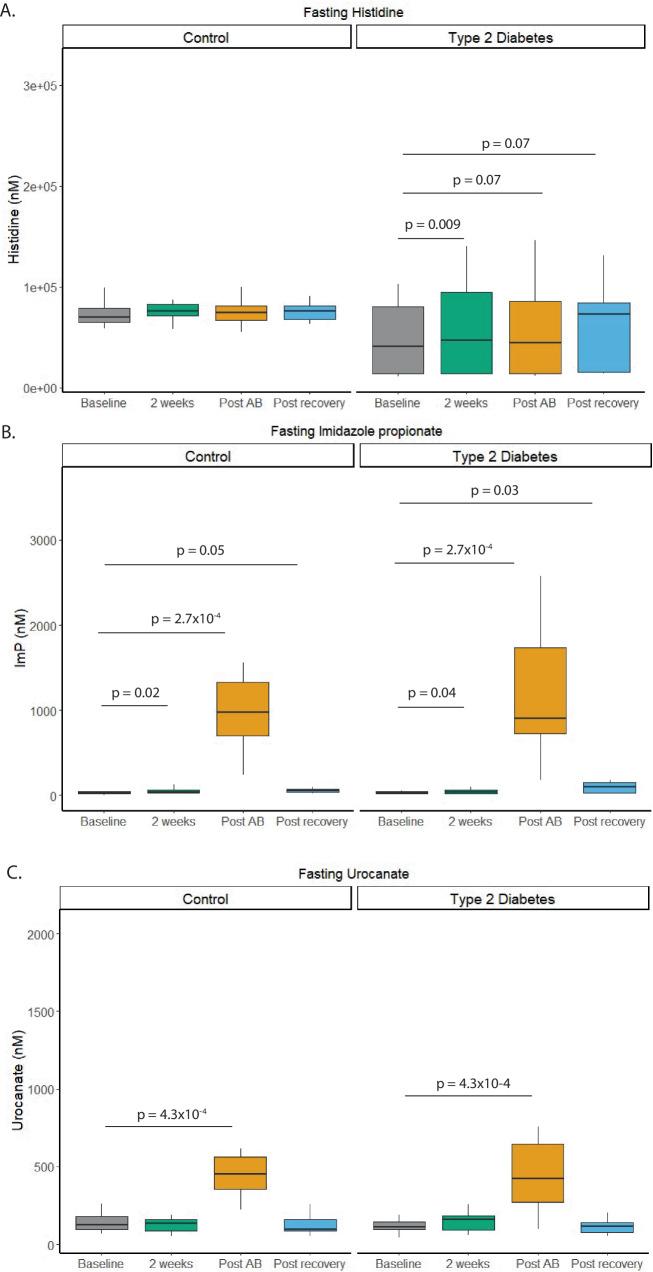
Oral histidine supplementation increased fasting serum histidine levels in participants with T2D (p = 0.009) but not in the control group (Fig. 3). After oral antibiotics and the subsequent recovery phase, a trend towards increased fasting histidine was observed (p = 0.07), for detailed results, see Supplementary Table 3. Importantly, ImP levels increased after antibiotic treatment compared to baseline in both non-diabetic and diabetic subjects (Supplementary Table 3). There was no difference between non-diabetic subjects and diabetic subjects when comparing ImP levels after antibiotics, possibly due to gut microbial changes in ImP-producing microbes.
Fig. 3. Overview fasting levels histidine related metabolites in humans.
A Fasting serum histidine levels increased in the type 2 diabetes group after oral histidine supplementation. B Fasting Imidazole propionate levels increased in T2D and controls after two weeks and further increased after antibiotics use. Also, after the recovery period ImP levels were still higher compared to baseline (C) Fasting Urocanate level increased after antibiotics use in both groups but not at other visits compared to baseline. Paired Wilcox test FDR corrected. Control n = 19, Type 2 Diabetes n = 20.
Kinetics of Urocanate and ImP changed after histidine intake and antibiotic use, see Supplementary figure 4. However, Cmax of histidine was increased in the control group (p = 0.04), after antibiotics and post recovery. In participants with T2D, the Cmax was increased after antibiotics and post recovery period but not after week 2 (Supplementary fig. 4 and Supplementary Table 4). The time point of maximal concentration (Tmax) was not affected (Supplementary fig. 5 and Supplementary Table 4). Fasting levels and area under the curve (AUC) of the downstream metabolite urocanate were only increased after antibiotics in both groups. However, the AUC was also increased after antibiotics in the group with type 2 diabetes (Supplementary fig. 5). Fasting levels of ImP, which is further downstream than urocanate in histidine metabolism16, were elevated after two weeks and after antibiotics in both groups. In the T2D group fasting ImP levels were also increased after the recovery period. A similar trend was observed in the AUC after antibiotics and after the recovery phase in participants with T2D. Urine ImP levels were increased after two weeks of histidine supplementation, which was further increased after antibiotics (Supplementary figs. 6 and 7 and Supplementary Table 5).
In concordance with significant increased fasting levels of urocanate and ImP after antibiotics, we observed different kinetic curves after antibiotics, which was also observed with increased Cmax post-antibiotics. Sub analysis of ethnic groups did not provide additional patterns.
Microbiota changes in mice
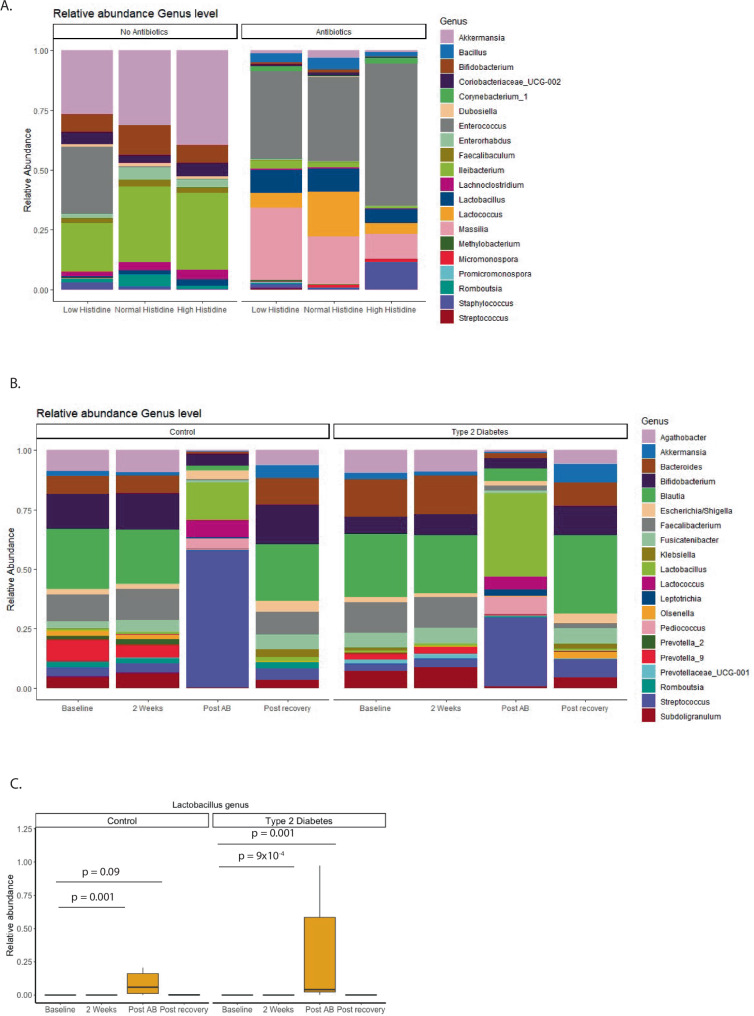
To evaluate the effects of histidine supplementation and antibiotic use on the gut microbiota profile, we first performed 16S rRNA analyses on the ceca of mice (Fig. 4a). The concentration of DNA and amplicon reads was significantly lower in the antibiotics group compared to the non-antibiotics group (Supplementary fig. 1 and Supplementary Table 6).
Fig. 4. Overview abundance in mice and humans.
A Stacked barplot relative abundance most prevalent genera in mice. B Stacked barplot relative abundance most prevalent genera in humans. C Relative abundance Lactobacillus genus in humans. Abundance is increased after antibiotics use and after the recovery period but not after 2 weeks of histidine use in both groups. Paired wilcox test with FDR correction. *FDR corrected p < 0.05. All data based on 16S rRNA Mice: all groups n = 10, Human: Control n = 19, Type 2 Diabetes n = 20.
In the non-antibiotic treated mice, we found that levels of Enterorhabdus genus were statistically increased in the normal histidine diet group, compared to the low histidine group (p = 0.0005), and high histidine group (p = 0.06). After FDR correction, only the increase in the normal group compared to the low histidine diet group remained statistically significant (p = 0.009). No other significant effects were observed, likely due to the small sample count (n = 10/group). Moreover, we found that the abundance of Enterococci (p = 0.04) and Bacilli (p = 0.04) decreased significantly following a high histidine diet in the non-antibiotic group, however, this significance was lost after correction for multiple comparisons. As previously reported, antibiotics had a strong effect on the microbiota, and we observed no significant effects between the different diet groups within the antibiotics group (Supplementary Table 7).
Overall, no genera were correlated with circulating histidine or ImP, with the exception of Faecalibaculum, which was associated with circulating histidine in the high histidine group (Supplementary Table 7). Interaction analysis showed that circulating levels of histidine were mediated by the genus Massilia. Urocanate levels were mediated by the genera Clostridium sensu stricto 1, Enterococcus, and Isobaculum. Interaction analysis for ImP did not show mediation by bacteria; however, a trend for Clostridium sensu stricto 12 was observed (Supplementary Table 8).
Microbiota changes in humans
After oral histidine intake, the abundance of several genera changed (Fig. 4b). Among the most abundant genera, Fusicatenibacter increased after 2 weeks in the T2D group but decreased after antibiotics in both groups. Similarly, Subdoligranulum increased in both groups after two weeks and decreased after antibiotics (Supplementary Table 9). Blautia increased in the control group.
Several bacteria increased after antibiotics, such as Streptococci in the control (p = 0.001) and T2D (p = 0.0008) groups. A trend toward increase was still visible in the post-recovery period of both groups (Supplementary Table 9). Another genus that increased after antibiotics in both groups was Lactobacillus (Fig. 4c).
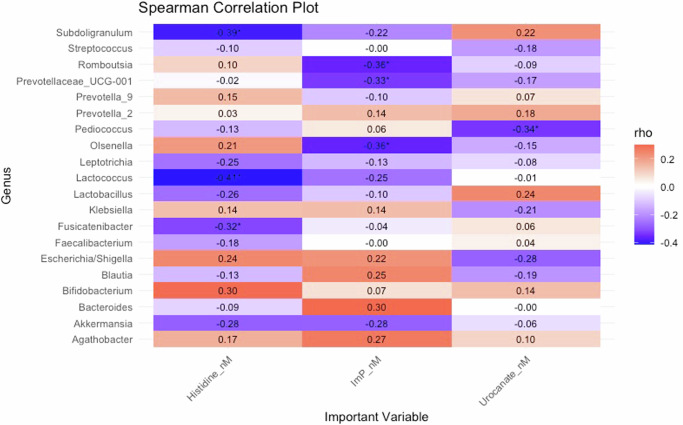
As several bacteria increased together with histidine-related plasma metabolites, we assessed the association between histidine-related metabolites and bacteria. Several bacteria were strongly associated with histidine, urocanate, and ImP. A trend toward an inverse association with fasting histidine was observed in five genera, namely Ruminococcus 1, Adlercreutzia, Lachnospiraceae ND3007 group, Ruminococcaceae_UCG-010 and Ruminococcaceae_UCG-007. A trend toward increased fasting histidine levels was observed in Rothia, Supplementary Table 10. Subdoligranulum (rho = −0.30, p = 0.0001, q = 0.004) and Fusicatenibacter (ρ = −0.26, p = 0.001, q = 0.02) were both negatively associated with histidine AUC. Some of the bacteria that increased after antibiotics, such as lactobacilli had a strong association with ImP (ρ = 0.49, p = 5.25 × 10−11, q = 5.98 × 10−10) and urocanate (ρ = 0.52, p = 3.14 × 10−12, q = 8.59 × 10−10) and other bacterial genera such as streptococci, pediococci, and lactococci were also significant. At baseline several bacteria were already associated with circulating metabolites (Fig. 5).
Fig. 5. Correlation heatmap prevalent genera and histidine-related metabolites in humans based on Spearman correlation at baseline.
*p < 0.05. All data is based on 16S rRNA. Human: Control n = 19, Type 2 Diabetes n = 20.
Interaction analyses revealed that 15 genera mediated changes in ImP concentration after two weeks; after antibiotics, 162 genera mediated the increase in ImP, and after the four weeks recovery phase, 23 genera mediated the ImP change (Supplementary Table 12). The genera Veillonella, Gemella, Actinomyces, Erysipelatoclostridium, Actinomyces, Romboutsia, and Graniculitella mediated ImP changes during all visits.
To better understand how antibiotics might impact the histidine transport system, we assessed the expression in peripheral blood monocytes of six amino acid transport that have a high affinity for histidine and are important for healthy metabolism. We found that in the control group SLC38A3, SLC7A5, SLC7A1, SLC7A8, and SLC43A2 were increased after antibiotics, and in the type 2 diabetes group, SLC28A3 and SLC7A5 were increased after antibiotics, see Supplementary fig. 12.
Discussion
In this study, we investigated the effects of oral histidine supplementation on circulating levels of histidine and related metabolites, urocanate and ImP, in both obese mice and humans with and without type 2 diabetes.
We found a dose-dependent increase in fasting serum ImP levels in mice that was completely abolished by the administration of antibiotics, clearly suggesting that ImP is produced by gut microbiota from orally supplemented histidine. In humans, we observed a relatively small increase in circulating histidine levels that, in contrast to the in mice, was dramatically increased after antibiotics.
ImP is produced from the histidine intermediate metabolite urocanate by the microbial enzyme urocanate reductase11,27,28. Consistent with our results, previous reports have demonstrated that ImP levels are associated with dietary histidine in mice29, but not in humans12, which may reflect differences in the gut transit. Furthermore, ImP levels are associated with reduced microbial gene richness12. To further investigate how depletion of the microbiota affects circulating ImP levels, we performed similar antibiotic treatments in mice and humans. As expected, the antibiotics reduced ImP levels in mice. However, ImP levels were dramatically increased in humans following antibiotics, where we observed increased ImP levels post antibiotics in both the T2D and the control group. Three processes are conceivable as an explanation for the increased ImP levels. First, oral antibiotics could remove competition from other bacteria, allowing potential ImP producers such as streptococci and lactobacilli to expand. This is supported by the finding that these bacteria were associated with ImP levels. Second, the antibiotics may have inhibited the growth of bacteria utilizing ImP as a substrate, although no such bacteria have been identified, thus contributing to increased circulating ImP levels. Third, the observed thinner stool and thereby increased transit time could result in more histidine being transported to the colon, which is usually mainly absorbed in the small intestine30, and thus increase the available pool for the colonic bacteria. This is consistent with the fact that ImP is produced by the colonic microbiota11. In addition to this, kinetic curves revealed that increased ImP concentrations were already observed two hours after the histidine challenge (Supplementary fig. 4), suggesting that absorption of ImP occurs in the small intestine.
In our study, we found lactobacilli to be significantly increased after antibiotic administration in humans but not in mice. In mice, a trend was visible in relative abundance, but this was not statistically significant.
However, it is important to note that lactobacilli are also more abundant in mice than in humans26, which limits the possibility to observe changes compared to humans. Due to the difference in gut microbiota composition, the effect of the antibiotics used may also result in metabolic changes in the microbiota of mice and humans following antibiotics. In humans, we observed that ImP levels were dramatically increased with antibiotic treatment and associated with increased abundance of lactobacilli and streptococci.
We included vancomycin in the antibiotic cocktail to suppress Gram-positive bacteria. However, vancomycin resistance in Gram-positive bacteria such as streptococci is emerging31, and they are likely to grow if there is no competition for substrates from other members of the microbiome. A similar argument can be made for other Gram-positive bacteria, which were increased in our study, such as lactobacilli32.
In addition to the increase in streptococci and lactobacilli in humans following antibiotic treatment, we observed increased abundances of the Subdoligranulum and Fusicatenibacter genera after 2 weeks of histidine supplementation, which then reduced in relative abundance following antibiotic administration. We also observed that Bifidobacterium increased after histidine supplementation in the control group and decreased in the T2D group post antibiotics. Furthermore, the genera Subdoligranulum, Fusicatenibacter, Bacteroides, and Bifidobacterium were inversely associated with systemic ImP levels in our study, which is consistent with previous findings identifying a negative correlation between the species Subdoligranulum variabile, and ImP but a positive correlation between ImP and Bifidobacterium longum and Bacteroides xylanisolvens in a European cross-sectional study12. However, in the previous study shotgun metagenomics was used to assess the microbiome, whereas we used 16S rRNA sequencing, which does not allow species level analyses to the same extent, which limits comparability between the studies.
We found several amino acid transporters that have a high affinity for histidine and are metabolically active in relation to glucose homeostasis30,33–36 to be upregulated after antibiotics in the control and type 2 diabetes group. As antibiotics increase transit time due to thinner stool, nutrient absorption is also decreased. The increased availability of histidine could thereby result in increased metabolization and higher levels of intermediate substrates such as ImP and urocanate, as we observed in our study. We observed a dose-dependent increase of serum histidine levels in our mice study, which was not abolished by antibiotic application. In contrast, ImP levels increase dose-dependently, but antibiotics mitigated this effect. ImP is most likely produced by bacteria from the metabolization of histidine11. However, histidine can be directly absorbed in the intestine by (sodium-dependent) amino acid transporters37. This could explain why histidine levels are not affected by antibiotic application in mice.
This study could not identify one genus of bacteria that could explain the change in ImP, either in mice or humans. However, recent literature suggests that bacterial clusters interact with each other and depend on each other by cross-feeding, creating an ecosystem also described as a bacterial guild or trophic network38,39. This could explain the absence of strong correlations between abundant genera in mice in our study. Another reason for the absence of correlation could be the smaller sample size in mice compared to humans, which limits the statistical power of mice analyses.
However, a limitation in this study is that the administration of antibiotics does not only affect gut microbiota abundance per se, as the administration of this combination of antibiotics frequently resulted in diarrhea thereby potentially reducing gut transit time, which can affect nutritional status, bioavailability of nutrients, and thereby metabolism. Transit time was not measured in this study as a robust measurement (e.g. with the use of motility capsules) as the invasiveness would have been out of proportion for this study. However, we did collect Bristol stool scores40, but not all candidates properly filled out this information, and due to missing data and large variability, we could not incorporate this into our analyses, which is a limitation of this study. Furthermore, we used relative abundance, which can overestimate small changes in gut microbiota composition. In addition to this, absorption of histidine occurs mainly in the small intestine, and fecal microbiota composition as measured in our study is therefore not directly representative of the composition in the small intestine. Moreover, the high dose of histidine administered in our study will not be fully absorbed in the small intestine due to the saturation of histidine transporters. In the colon, the remaining histidine can still affect the growth of bacteria such as lactobacilli, which we measured with fecal microbiota. Thus, Fecal microbiota should therefore not be viewed as reflection of microbiota in the small intestine, but fecal microbiota can be affected by bacteria in the small intestine due to altered nutrient availability. Therefore, fecal microbiota can serve as surrogate markers for changes in small intestinal microbiota composition.
Furthermore, there are differences between how histidine was administered to mice (diet) and humans (capsules). However, we used the same antibiotics, and the exact same experimental and computational protocols were applied to analyze the gut microbiota composition. Furthermore, injection of ImP into mice increases fasting and postprandial glucose levels11, suggesting that ImP directly causes deterioration of glycemic control. However, oral supplementation of histidine in women with metabolic syndrome improves glycemic control41. Therefore, the effect of ImP on host metabolism could be dependent on the bioavailability, source, or microenvironment in the colon.
In conclusion, this study shows that systemic ImP is dose-dependently increased by dietary histidine supplementation in mice, and this increase can be abolished by gut microbiota suppression, via antibiotic treatment. Oral histidine supplementation resulted in significantly higher circulating histidine levels and slightly increased circulating ImP levels in humans. However, antibiotic treatment supports enhanced postprandial production of ImP. Future studies are needed to identify which bacteria, or bacterial consortia can produce ImP following antibiotic treatment in humans and the exact environmental conditions inducing ImP production. Experiments clarifying how and under which conditions ImP is produced should be performed in future studies. A better understanding of the complex interplay between diet, gut microbiota, and host metabolism will improve future dietary recommendations and will microbiota-tailored interventions.
Furthermore, we used relative abundance which can overestimate small changes in gut microbiota composition. Determining absolute abundances should give better insights into changes in the microbiome and could have been estimated using qPCR. Instead, as proxy qPCR quantification, we used the 16S amplicon library concentrations to estimate abundances. Even with qPCR data quantification would still be biased due to the extreme difference in feces matrix induced by antibiotics, which biases DNA extraction and PCR performance. Furthermore, we did not quantify total feces thus no absolute quantification could be performed.
Methods
Animal studies
Study design
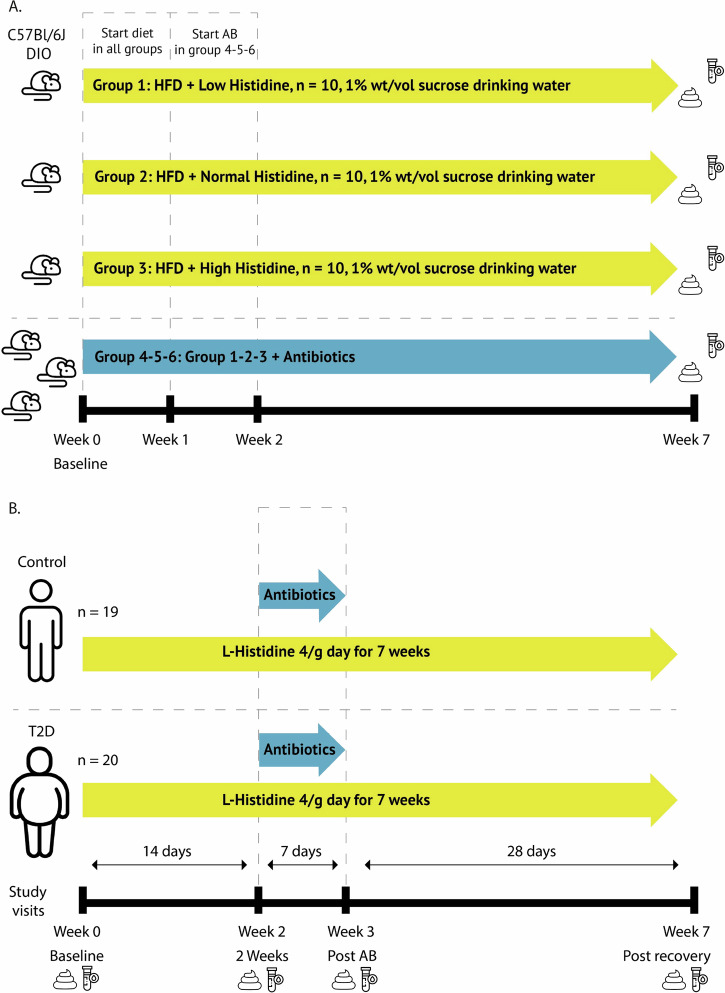
Sixty diet-induced obese (DIO) C57Bl/6J mice were purchased from Jackson laboratories (Bar Harbor, Maine, United States) with established obesity and insulin resistance at 14 weeks of age. Upon arrival in our facility, mice were switched from the standard high fat diet to chemically adjusted high fat diets with different levels of the essential amino acid histidine. In total, six diet groups (n = 10) were formed (Fig. 6a). Each diet group contained four cages, and at baseline, feces from all cages were collected, and sprinkled among the cages to correct for baseline microbiota status, as this was previously shown to reduce pre-study variability42. Mice followed the diet for 7 weeks, and food intake and body weight were tracked throughout the study by weighing the pellets and mice. At week 7, mice were killed. At killing, 4-h fasted mice were injected with a lethal dose of Ketamine/Xylazine (150 mg/kg and 15 mg/kg, respectively). Mice were fasted to reduce variability. Tissues were excised and snap frozen in liquid nitrogen. All mice were maintained in an internationally-approved animal facility. All experimental protocols were approved by the Institutional Animal Care and use Committee of the Cleveland Clinic.
Fig. 6. Study design.
A Mouse study with oral histidine supplementation in diet induced obese C57Bl/6J mice (DIO) during 7 weeks. Feces was collected from coecum. B Human study in type 2 diabetes (T2D) participants and controls during 7 weeks. Applied antibiotics were metronidazole, vancomycin and ciprofloxacin during 7 days. T2D: n = 20, n = 19. Blood and feces represent collection time point for specimen.
Diet
Chemically modified high-fat diets, with different levels of histidine, were purchased from Research Diets, Inc (New Brunswick, United States) (Supplementary Table 1). The levels of free and total histidine in the diet were measured by Liquid chromatography–mass spectrometry (LC/MS) (details below, and Supplementary fig. 2).
After 2 weeks on the selected diets, an antibiotic cocktail (ciprofloxacin 1 g/L; metronidazole 1 g/L, and vancomycin 0.5 g/L) was added to the drinking water. Due to our previous experience with mice being reluctant to drink water with added metronidazole, we supplemented the drinking water with 1% (wt/vol) sucrose, as previously described43 (Fig. 6a).
Quantification of imidazole propionate, urocanate, and histidine in mouse plasma
Plasma ImP, urocanate, and histidine were quantified using ultraperformance liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Internal standards (IS; 13C3-ImP, 13C3-urocanate, and D3-histidine; 100 µl in methanol) and ice-cold methanol (800 µl) were added to the samples (20 µl). Samples were vortexed, spun down (21,000 x g at 4 °C for 15 min), and the supernatant (800 µl) was transferred into clean, labeled glass tubes (Borosilicate glass 12 × 75 mm, Cat # 14-961-26, Fisherbrand) and dried in a speed vacuum concentrator (Speed vac plus, SC210, Thermo Sevant). The dry residue was reconstituted in HPLC grade n-butanol (100 µl, Cat# A383-, Fisher Chemical) containing HCl (5%, w/v), sealed with safety caps (60828-726; VWR), vortexed, and heated (1 h at 70 °C) followed by drying under nitrogen. Dried residues were dissolved in a mixture of methanol: water (100 µl; 50:50 v/v with 0.1% acetic acid), tubes were vortexed, and liquid was transferred to glass vials with micro-insets and capped. Samples (3 µl for ImP and urocanate, and 0.2 µl for histidine) were subjected to the LC-MS/MS analysis on a system consisting of two Shimadzu LC-30 AD pumps (Nexera X2), a CTO 20AC oven operating at 30 °C, and a SIL-30 AC-MP autosampler in tandem with a 8050 triple quadruple mass spectrometer (Shimadzu Scientific Instruments, Inc., Columbia, MD, USA). For chromatographic separation, a Kinetex C18 column (50 mm × 2.1 mm; 2.6 μm) (Cat # 00B-4462-AN, Phenomenex, Torrance, CA) was used with solvent A (10 mM ammonium formate with 0.1% acetic acid in water) and B (0.1% acetic acid in methanol). Multiple reaction monitoring (MRM) transitions for the butyl ester derivatives were as follows: m/z 197/81 for ImP; m/z 200/81 for 13C3-ImP; m/z 195/121 for urocanate; m/z 198/124 for 13C3-urocanate; m/z 212/110 for histidine, and m/z 215/213 for D3-hisitidine. The following ion source parameters were applied: nebulizing gas flow, 3 l/min; heating gas flow, 10 l/min; interface temperature, 300 °C; desolvation line temperature, 250 °C; heat block temperature, 400 °C; and drying gas flow, 10 l/min. For data analysis software Lab Solution (version 5.89; Shimadzu) was used.
Quantification of free and total histidine in rodent diets
Free and total histidine in rodent diets were quantified using ultraperformance LC-MS/MS. Total histidine being the sum of protein bound histidine and free histidine. Free histidine was extracted by homogenizing food samples in 0.1 M HCl in methanol: water (1:1; v/v) in the presence of IS (D3-histidine). For total histidine, food samples were first hydrolyzed overnight in 6 M HCl in water (200 µl) under vacuum at 110 °C in the presence of IS (D3-histidine). After cooling down, water (2 ml) was added to the samples. Samples were then loaded onto Discovery DSC-SCX columns (1 ml; 100 mg, Supelco) previously washed with methanol and equilibrated with 0.2 M formic acid in water and eluted with 5% ammonia in 70% methanol in water. Eluates were first dried under a stream of nitrogen to remove ammonia, followed by drying in a speed vacuum concentrator (Speed vac plus, SC210, Thermo Sevant). The dry residue was reconstituted 0.1% acetic acid in water and analyzed on the same system as described above. Following MRM transitions were used: m/z 156/110 for histidine and m/z 159/113 for D3-hisitidine.
Human study
Study recruitment
We performed a non-randomized clinical trial by recruiting participants via online advertisements and advertisements in newspapers in the region of Amsterdam. Recruitment started in February 2020 and continued until December 2022. All participants were between 40–70 years of age. Participants in the control group had a BMI between 19–25 kg/m2. Participants with T2D had a BMI between 25–35 kg/m2, were on stable (oral anti-diabetic) drug treatment for three months, and had to use statins and metformin. Exclusion criteria included previous major cardiovascular events, proton pump inhibitor use, GLP-1 or insulin use, antibiotic use in the past three months, pregnancy, chronic illnesses (history of heart failure, eGFR < 30 ml/min, pulmonary disease, gastrointestinal disorders, or hematologic diseases) or other inflammatory diseases. Furthermore, patients were excluded with an active infection, previous intestinal surgery, smoking, vegetarian diet, >6 alcohol units per day or >14 alcohol units per week use, active malignancy, HbA1c > 9% (>75 mmol/mol) or if the subject was already involved in a clinical trial. The T2D and control group were age and sex matched.
Study design
After an initial screening visit, subjects had four study visits focusing on histidine and derived metabolite kinetics, during which blood samples were collected over six hours. Urine samples were also collected for 24 h as well as 24-hour feces. Subjects ingested 4 g of histidine capsules (Vital Cell Life l-Histidine 500 mg Capsules 100CP, Bunnik, The Netherlands) per day during 7 weeks (Fig. 6b). Compliance was evaluated by checking the medication container used as daily dispenser. After two weeks, subjects also started taking oral broad-spectrum antibiotics to suppress the gut microbiome (ciprofloxacin 500 mg once per day, metronidazole 500 mg twice per day, oral vancomycin 500 mg four times per day) for 7 days, along with the 4 g of l-histidine per day. This was followed by a study visit, and after a four-week reconstitution period, the final study visit took place. The study was conducted at the Amsterdam University Medical Centers location Academic Medical Center. All participants provided written informed consent, and the study was conducted in accordance with the Declaration of Helsinki (updated version 2013). The study was registered at the Dutch trial registry, registration number: NL8372 (https://www.onderzoekmetmensen.nl/en/trial/20133) and approved by the Ethical Review Board of Academic Medical Centers of the University of Amsterdam. The primary outcome was kinetics of histidine and histidine-related metabolites after oral histidine supplementation. Secondary outcomes including metabolic parameters will be published separately44. All authors had access to the study data and reviewed and approved the final manuscript.
Anthropometric measurements and food intake
Food intake was monitored three days before every study visit with a digital food diary app “Eetmeter” (v4.6.0 Stichting Voedingscentrum Nederland, The Netherlands). Energy expenditure was measured by carbon dioxide and oxygen productions during 20 min for indirect calorimetry using a ventilated hood system (Vmax Encore 29; SensorMedics, Anaheim, CA, United States) during every kinetic visit. Weight, body circumference and bioelectrical impedance analysis were also measured at every visit.
Metabolite analysis in human plasma
Human plasma metabolites were measured by stable isotope-dilution LC-MS/MS as previously described11, with some modifications. Briefly, 25 µL of plasma and urine samples were precipitated and diluted in glass vials using acetonitrile containing internal standards (D515N3-Histidine-, Cambridge Isotope Lab. Inc, 13C3-Imp and 13C3-urocanate, Astra Zeneca, Cambridge, UK). After vortexing and centrifugation, the supernatant was transferred to new glass vials and evaporated under a stream of nitrogen. The samples were then reconstituted in 5% HCl (37%) in n-butanol and placed in an oven at 70 °C for one hour allowing the n-butyl ester to be formed. After derivatization, the samples were evaporated and reconstituted in 150 µL of water:acetonitrile [90:10]. The samples were then analyzed using ultra-performance LC-MS/MS. The analytical system consisted of an Acquity UPLC I-class binary pump, sample manager and column manager coupled to a Xevo TQ-XS (Waters, Milford, MA, USA). The samples (2 µl) were injected onto a C18 BEH column (2.1 × 50 mm with 1.7 µm particles, Waters, Milford, MA, USA) and separated using a gradient consisting of water with 0.1% formic acid (A-phase) and acetonitrile with 0.1% formic acid (B-phase). The analytes were detected with multiple reaction monitoring using the transitions m/z 212/110 (histidine), 197/81 (ImP) and 195/93 (urocanate). For the internal standards, the transitions 220/118, 200/82 and 198/95 respectively were used. Calibration curves for histidine, ImP and urocanate were made in methanol and treated the same way as the samples.
PBMC isolation and storage
At each visit, blood was collected in EDTA tubes for PBMC isolation. Peripheral blood mononuclear cells (PBMC) were isolated from blood using Lymphoprep (GE Healthcare) and density gradient centrifugations according to the manufacturer’s protocol. After isolation, PBMCs were frozen in fetal bovine serum (FBS) containing 10% DMSO and stored in liquid nitrogen tanks.
RNA was isolated following standard RNA isolation protocol. PBMC were first thawed in 10 ml of 10%FBS-RPMI 1640 medium, centrifuged at 350 g for 10 min at 4 °C to remove DMSO. Afterwards, the pellet of PBMCs was resuspended in 1 ml Trizol (Sigma-Aldrich) to lyse cells. Afterwards, the RNA pellet was eluted in 20 µl RNAse-free water. RNA concentrations were measured using the NanoDrop 1000 (Thermo Scientific). 1μg of RNA was converted to cDNA with SensiFAST cDNA synthesis kit (Meridian Bioscience) according to the manufacturer’s instructions. qPCR was performed on a CFX Opus 384 PCR machine (BioRad) using SensiFAST SYBR No-ROX Green (Meridian Bioscience). RNA18S1 and HPRT genes were used as housekeeping genes, and the target gene expression was normalized to both housekeeping genes. Gene expression is shown as arbitrary units (AU) of 2^(-delta Ct).
Primers were manufactured by Sigma-Aldrich and the primers sequences are provided in Table 2.
Table 2.
Primers sequences qPCR
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| SLC38A3 | GCAGGCAACCAGAGGGTC | TCCCGAATGATGTCTTCCCC |
| SLC7A5 | GCCTGTGTTCTTCATCCTGG | GTGGAGAAGATGCCCTGGAG |
| SLC7A1 | TGCCTGGACAATAACAGCCC | TGGAACCTAGAAGACTGGCG |
| SLC7A8 | TGCCTGTCATATTTCCTGGGTG | AGGGTTAGCAGCTCAATGAAG |
| SLC43A2 | ACATGGACTACTCGGTGAAGA | CCGTGGTCACCTGCTTGTA |
| SLC15A4 | GGACAAACTGGTCGATCCCA | CCTTTTACTCTCCAAAATTCCTGCA |
| HPRT | TGACCTTGATTTATTTTGCATACC | CGAGCAAGACGTTCAGTCCT |
| RNA18S1 | GAGGGAGCCTGAGAAACGG | GTCGGGAGTGGGTAATTTGC |
Feces processing, sequencing, and bioinformatics
In the mice study, gut microbiota sequencing in the cohort was performed as previously described45. Briefly, mouse ceca were collected during necropsy, immediately snap frozen using liquid nitrogen, and stored at −80 °C.
For the human study, fresh stool samples were collected in the morning or evening before the study visit. Participants were asked to refrain from dietary and lifestyle changes during the study. Participants stored fecal samples directly after collection in a −20 °C freezer, transported them frozen to the research facility, where sampels were stored at −80 °C. Subsequently, mice and human feces were handled identically as following.
DNA from feces was extracted by application of a repeated bead beating protocol46. A single-step PCR protocol to target the V3-V4 region was used to generate barcoded 16S rRNA gene amplicons47. Ampure XP beads were used to purify PCR products, and the purified products were equimolarly pooled. The MiSeq platform was used to sequence the libraries by using 2 × 251 cycles of V3 chemistry. Forward and reverse reads were truncated to 240 and 210 bases, respectively, and merged with USEARCH48. Merged reads which did not pass the Illumina chastity filter with an expected error rate >2 or <380 bases were removed. For each individual sample amplicon sequence variants (ASV’s) were inferred with a minimum abundance ≥4 reads48. Taxonomical names were assigned to ASVs using the SILVA49 16S ribosomal database V132 and RDP classifier50. Subsequently, microbial counts per sample were rarefied to 30,000 counts. Data are available through the European Nucleotide Archive: ENA project ID: PRJEB71915.
Statistical analyses
Statistical analyses were performed in R (https://www.R-project.org/, v4.2.1) with libraries ‘tidyverse’ and ‘lme4’. Microbiome data were managed with the ‘phyloseq’ library. Libraries used for data visualization were ‘ggplot2’ or ‘corrplot’. Linear mixed effects models, Wilcoxon signed-rank test and Mann-Witney U tests were used when appropriate, with false discovery rate (FDR) correction, setting the alpha at 0.05 to minimize false positive rates. Interaction was test with comparison of linear mixed effects models with and without the interaction term, which were subsequently compared using an analysis of variance (ANOVA). Cmax represented the maximum concentration of the measured metabolite per kinetic test day and Tmax the time (in minutes) when the maximum concentration of the measured metabolite was reached during each kinetic test day. The area under the curve (AUC) was calculated according to the trapezoid method in R (version 4.2.1). To avoid bias introduced by comparing different groups, every participant functioned as their own control. Random effects at baseline were accounted for with linear mixed effects models. Missing data were omitted from the analysis.
Supplementary information
Acknowledgements
Mitch Aerts, Zeinab Maraha, Imane el Idrissi, and Nadia Mohamed are thanked for data gathering and curation for this study. Alinda Schimmel is acknowledged for helping with DNA isolation. Andrew Li Yim is acknowledged for input on epigenetics analyses. Images from Flaticon.com were used for Fig. 6.
Author contributions
Study conceptualization: M.V.W., I.A., A.H., R.B., W.J.M., V.V., S.D., I.N., J.A.W., K.W., A.K.G., A.K., D.R., H.H., F.B., J.M.B., S.H., and M.N. Statistical analysis, M.V.W., I.A., M.D., I.N., and P.O.B. data interpretation: M.V.W., I.A., I.N., P.O.B., M.D., and H.H. Manuscript drafting: M.V.W., I.A., I.N., H.H., F.B., M.N., and J.M.B. Funding: M.N., H.H., J.M.B., S.H., D.R., and F.B.
Data availability
Restrictions apply to the availability of some or data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. The corresponding author will, on request, detail the restrictions and any conditions under which access to data may be provided.
Competing interests
M.N. is a member of the scientific advisory board of Caelus Health and all honoraria are paid to the employer Amsterdam University Medical Centres. F.B. receives research funding from Biogaia AB, is co-founder and shareholder of Roxbiosens Inc and Implexion AB, and is on the scientific advisory board of Bactolife A/S. K.C. has held a collaborative research contract with Danone Research in the context of the MetaCardis project. D.H.v.R. has participated in advisory boards for AstraZeneca, Boehringer Ingelheim‐Eli Lilly Alliance, MSD, Novo Nordisk and Sanofi, and has received research grants from AstraZeneca, Boehringer Ingelheim‐Eli Lilly Alliance, MSD and Sanofi. All honoraria are paid to the employer of Amsterdam University Medical Centres. All other authors declare to have no related conflict of interest.
Ethics approval
The study was conducted in accordance with the Declaration of Helsinki (updated version 2013).
Informed consent
All participants provided written informed consent.
Consent for publication
All coauthors read and approved the manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Moritz V. Warmbrunn, Ilias Attaye.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00592-8.
References
- 1.Chakaroun, R. M., Olsson, L. M. & Backhed, F. The potential of tailoring the gut microbiome to prevent and treat cardiometabolic disease. Nat. Rev. Cardiol.20, 217–235 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Rowland, I. et al. Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr.57, 1–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrema, H., IJzerman, R. G. & Nieuwdorp, M. Emerging role of intestinal microbiota and microbial metabolites in metabolic control. Diabetologia60, 613–617 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Fromentin, S. et al. Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat. Med.28, 303–314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu, J., Moore, B. N. & Pluznick, J. L. Short-Chain Fatty Acid Receptors And Blood Pressure Regulation: Council On Hypertension Mid-career Award For Research Excellence 2021. Hypertension79, 2127–2137 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Backhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell165, 1332–1345 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Vanholder, R. et al. The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: a systematic review. J. Am. Soc. Nephrol.25, 1897–1907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sallee, M. et al. The aryl hydrocarbon receptor-activating effect of uremic toxins from tryptophan metabolism: a new concept to understand cardiovascular complications of chronic kidney disease. Toxins (Basel)6, 934–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang, W. H. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med.368, 1575–1584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemet, I. et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell180, 862–877.e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh, A. et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell175, 947–961.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Molinaro, A. et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat. Commun.11, 5881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molinaro, A. et al. Microbially produced imidazole propionate is associated with heart failure and mortality. JACC Heart Fail11, 810–821 (2023). [DOI] [PubMed] [Google Scholar]
- 14.van Son, J. et al. Plasma imidazole propionate is positively correlated with blood pressure in overweight and obese humans. Nutrients13, 2706 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koh, A. et al. Microbial imidazole propionate affects responses to metformin through p38gamma-dependent inhibitory AMPK phosphorylation. Cell Metab.32, 643–653.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acuna, I. et al. Rapid and simultaneous determination of histidine metabolism intermediates in human and mouse microbiota and biomatrices. Biofactors48, 315–328 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Kopple, J. D. & Swendseid, M. E. Evidence that histidine is an essential amino acid in normal and chronically uremic man. J. Clin. Invest.55, 881–891 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart, P. H. & Norval, M. The multiple roles of urocanic acid in health and disease. J. Invest. Dermatol.141, 496–502 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Correale, J. & Farez, M. F. Modulation of multiple sclerosis by sunlight exposure: role of cis-urocanic acid. J. Neuroimmunol.261, 134–140 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Albert, E. et al. cis-Urocanic acid attenuates acute dextran sodium sulphate-induced intestinal inflammation. PLoS ONE5, e13676 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korhonen, E. et al. UV-B-induced inflammasome activation can be prevented by cis-Urocanic acid in human corneal epithelial cells. Invest. Ophthalmol. Vis. Sci.61, 7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampton, T. How useful are mouse models for understanding human atherosclerosis? Review examines the available evidence. Circulation135, 1757–1758 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Tschop, M. H. et al. A guide to analysis of mouse energy metabolism. Nat. Methods9, 57–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J. & Dawson, P. A. Animal models to study bile acid metabolism. Biochim. Biophys. Acta Mol. Basis Dis.1865, 895–911 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martignoni, M., Groothuis, G. M. & de Kanter, R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Exp. Opin. Drug Metab. Toxicol.2, 875–894 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, T. L., Vieira-Silva, S., Liston, A. & Raes, J. How informative is the mouse for human gut microbiota research? Dis. Model Mech.8, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venskutonyte, R. et al. Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production. Nat. Commun.12, 1347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bogachev, A. V., Bertsova, Y. V., Bloch, D. A. & Verkhovsky, M. I. Urocanate reductase: identification of a novel anaerobic respiratory pathway in Shewanella oneidensis MR-1. Mol. Microbiol.86, 1452–1463 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Zeng, X. et al. Gut bacterial nutrient preferences quantified in vivo. Cell185, 3441–3456.e19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broer, S. Intestinal amino acid transport and metabolic health. Annu. Rev. Nutr.43, 73–99 (2023). [DOI] [PubMed] [Google Scholar]
- 31.Poyart, C. et al. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother.41, 24–29 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abriouel, H. et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res. Int.78, 465–481 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi, T. et al. The histidine transporter SLC15A4 coordinates mTOR-dependent inflammatory responses and pathogenic antibody production. Immunity41, 375–388 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Fotiadis, D., Kanai, Y. & Palacin, M. The SLC3 and SLC7 families of amino acid transporters. Mol. Asp. Med.34, 139–158 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Javed, K. & Fairweather, S. J. Amino acid transporters in the regulation of insulin secretion and signalling. Biochem. Soc. Trans.47, 571–590 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Broer, S. Amino acid transporters as modulators of glucose homeostasis. Trends Endocrinol. Metab.33, 120–135 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Scalise, M. et al. The human SLC7A5 (LAT1): the intriguing histidine/large neutral amino acid transporter and its relevance to human health. Front Chem.6, 243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, G. et al. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med.13, 22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Goffau, M. C. et al. Gut microbiomes from Gambian infants reveal the development of a non-industrialized Prevotella-based trophic network. Nat. Microbiol.7, 132–144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blake, M. R., Raker, J. M. & Whelan, K. Validity and reliability of the Bristol Stool Form Scale in healthy adults and patients with diarrhoea-predominant irritable bowel syndrome. Aliment. Pharmacol. Ther.44, 693–703 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Feng, R. N. et al. Histidine supplementation improves insulin resistance through suppressed inflammation in obese women with the metabolic syndrome: a randomised controlled trial. Diabetologia56, 985–994 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Miyoshi, J. et al. Minimizing confounders and increasing data quality in murine models for studies of the gut microbiome. PeerJ6, e5166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goulding, D. R. et al. Comparative efficacy of two types of antibiotic mixtures in gut flora depletion in female C57BL/6 mice. Comp. Med.71, 203–209 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warmbrunn, M. V. et al. Oral histidine affects gut microbiota and MAIT cells improving glycemic control in type 2 diabetes patients. Gut Microbes16, 2370616 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Attaye, I. et al. Protein supplementation changes gut microbial diversity and derived metabolites in subjects with type 2 diabetes. iScience26, 107471 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costea, P. I. et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol.35, 1069–1076 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Kozich, J. J. et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl.Environ. Microbiol.79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res.41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol.73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Restrictions apply to the availability of some or data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. The corresponding author will, on request, detail the restrictions and any conditions under which access to data may be provided.