Abstract
The maintenance of intestinal homeostasis is a fundamental process critical for organismal integrity. Sitting at the interface of the gut microbiome and mucosal immunity, adaptive and innate lymphoid populations regulate the balance between commensal micro-organisms and pathogens. Checkpoint inhibitors, particularly those targeting the CTLA-4 pathway, disrupt this fine balance and can lead to inflammatory bowel disease and immune checkpoint colitis. Here, we show that CTLA-4 is expressed by innate lymphoid cells and that its expression is regulated by ILC subset-specific cytokine cues in a microbiota-dependent manner. Genetic deletion or antibody blockade of CTLA-4 in multiple in vivo models of colitis demonstrates that this pathway plays a key role in intestinal homeostasis. Lastly, we have found that this observation is conserved in human IBD. We propose that this population of CTLA-4-positive ILC may serve as an important target for the treatment of idiopathic and iatrogenic intestinal inflammation.
Subject terms: Crohn's disease, Ulcerative colitis, Innate lymphoid cells, Mucosal immunology
At the mucosal interface of the gut and microbiome immune cells play pivotal roles to regulate between commensalism, colonisation and pathogenic invasion. Here, Lo et al. show CTLA-4 expression in innate lymphoid cells is linked to mucosal homeostasis in a microbiome dependent manner.
Introduction
Cytotoxic T lymphocyte–associated antigen 4 (CTLA-4 or CD152), is regarded as one of the key inhibitory molecules expressed by conventional CD4+ and CD8+ T cells1–6. CTLA-4 is homologous to CD28, a costimulatory molecule expressed on the surface of naïve CD4+ and CD8+ T cells that promotes T cell activation and proliferation7,8. Both CD28 and CTLA-4 bind to the ligands CD80/B7.1 and CD86/B7.2, which are usually found on the surface of antigen presenting cells (APC)9–11. CTLA-4 has a 10-100-fold greater affinity for both CD80 and CD86 and outcompetes CD28 binding and can also remove the ligands from cells by a process of transendocytosis12,13. Regulatory T cells (Treg) suppress aberrant and excessive immune responses via constitutive expression of CTLA-414–16. The importance of this is highlighted in Ctla4 knockout mice, which die prematurely from multi-organ failure due to excessive immune activation and lymphoproliferation17,18. Furthermore, in recent years, cancer patients undergoing treatment with anti-CTLA-4 and other immune checkpoint inhibitors (CPI) that block inhibitory molecules and enhance immune activation against tumours, have been found to develop autoimmune and immune-mediated inflammatory diseases19–27.
The discovery of innate lymphoid cells (ILCs) has been important for the understanding of immune responses, especially at barrier surfaces28–35. ILC1 can be activated by stimulation with the cytokines IL-12 and IL-18, express the transcription factor T-bet, and are potent producers of interferon-γ (IFNγ)28,36. ILC2 can be activated by alarmin cytokines, including IL-25 and IL-33, highly express the transcription factor GATA3 and are strong producers of interleukin (IL)−5 and IL-1337–39. ILC3 are activated by the cytokines IL-23, IL-1β, IL-6, IL-27 and TGF-β and express the transcription factor RORγt but can be further subdivided according to whether they express natural cytotoxicity receptors (NCRs) and the chemokine receptor CCR6. ILC3s are potent producers of IL-2240,41 and IL-17A42,43. Furthermore, CCR6+ embryonic lymphoid tissue inducer (LTi) cells and post-natal LTi-like ILC3s play important roles in lymphoid tissue organogenesis and are potent producers of IL-2244. ILCs can also transdifferentiate into other ILC lineages, with parallels to TH cell plasticity30,45–52. CTLA-4 and another inhibitory receptor pair, PD-1 and PD-L1, are expressed by ILC2 during Nippostrongylus brasiliensis infection, or stimulation with recombinant IL-2553. Transcripts encoding CTLA-4 are increased in intestinal inflammation in both bulk RNA-sequencing and single-cell RNA-sequencing (scRNA-seq) analyses54–57.
Despite recent discoveries and investigations into the immunopathology and biology of ILCs, there are still key limitations in the understanding of these immune cells and how they are regulated. In this study, we aim to further investigate and understand the role of surface checkpoint molecules, in particular CTLA-4, on ILCs and how their expression maintains homeostasis. We show that the checkpoint molecule CTLA-4 is expressed in intestinal lamina propria (LP) ILCs. Furthermore, our data highlight the importance of ILC expression of CTLA-4 for regulating immunological responsiveness in the colon, and identify roles for microbial and inflammatory cues in driving CTLA-4 expression by colonic ILCs.
Results
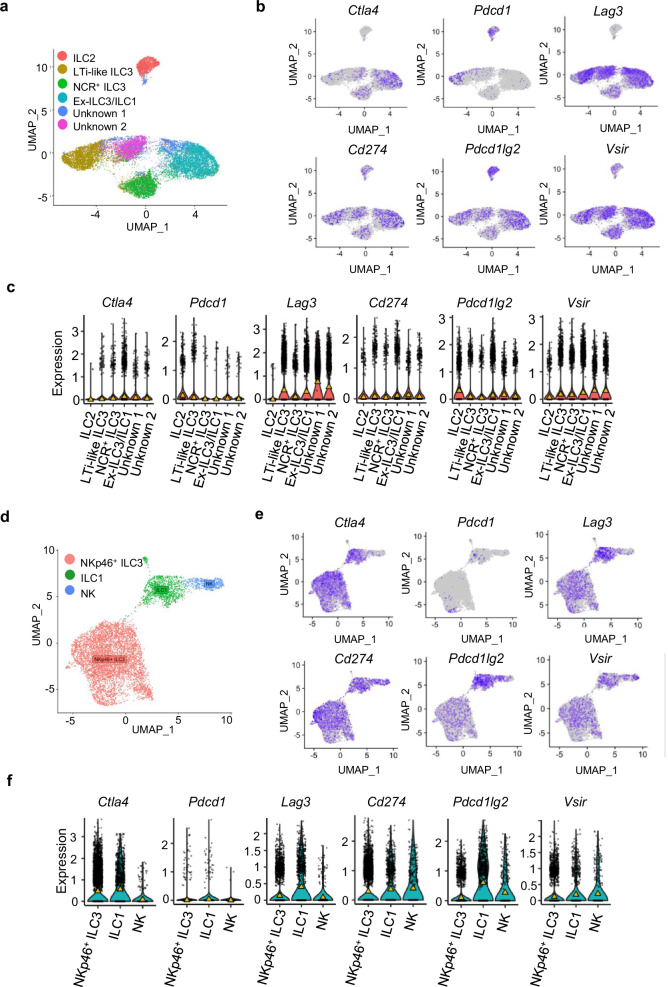
Immune checkpoint molecules are expressed across ILC populations in the colon
We have previously described the importance of ILCs in mediating colitis30,34,35,51. Moreover, we have found that ILC1 are a significant driver for the development of DSS-induced colitis and early IFNγ production is likely to be key in the induction of colitis by ILC152. We therefore performed a comprehensive analysis of the expression of well-characterised checkpoint molecules on murine ILCs at single-cell resolution. First, we utilised two publicly available scRNA-seq datasets of FACS-sorted ILCs from the small intestine lamina propria (SI LP) of wild-type (WT) mice57,58. The first dataset identified ex-ILC3/ILC1, ILC2, NCR+ ILC3s, LTi-like ILC3 and two undetermined ILC clusters (Fig. 1a). We found that Ctla4 expression was predominantly detected in ex-ILC3/ILC1, and to a lesser extent in NCR+ ILC3 (Fig. 1b, c). PD-1 (encoded by Pdcd1) was not highly expressed in NCR+ ILC3s and ex-ILC3/ILC1s subsets but was detected in ILC2 and LTi-like ILC3 (Fig. 1b, c). PD-L1 and PD-L2 (encoded by Cd274 and Pdcd1lg2, respectively) were detected more widely across all four clusters, but with ILC2 exhibiting the highest expression of PD-L2 (Fig. 1b, c). Interestingly, Lag3 and VISTA (encoded by Vsir) expression was also apparent in LTi-like ILC3, NCR+ ILC3 and ex-ILC3/ILC1 subsets, but less so in ILC2 (Fig. 1b, c). To confirm Ctla4 expression in NCR+ ILC subsets, we probed a second dataset from Krzywinska et al., which utilised sorted NKp46+ ILC and could, therefore, be used to investigate gene expression by NK cells, as well as ILC1 and NKp46+ ILC3 (Fig. 1d). In agreement with our initial findings, we detected transcripts encoding Ctla4 in NKp46+ ILC3 and ILC1 populations but not in the NK cells (Fig. 1e, f). Pdcd1 was not highly expressed in ILC1 and NKp46+ ILC3, but PD-L1 and PD-L2 expression was found across ILC1, NKp46+ ILC3 and NK cells (Fig. 1e, f). The distribution of Lag3 and Vsir expression was consistent with the first dataset, with high expression across NCR+ ILC3 and ILC1 subsets (Fig. 1e, f). Expression of Havcr2 could only be detected at very low levels across all three ILC subsets (Supplementary Figs. 1a–c).
Fig. 1. The immune checkpoint transcriptional landscape across intestinal ILC clusters.
a UMAP plot showing the six ILC populations previously identified from CD45+ Lin− CD127+ sorted cells by Fiancette et al. b UMAP plots and (c) violin plots showing the expression of canonical immune checkpoint inhibitory molecules across the six identified ILC subsets (median shown by the yellow triangle). Cell counts for each clusters are: ILC2 1468, LTi-like ILC3 3736, NCR+ ILC3 2984, Ex-ILC3/ILC1 5241, Unknown (1) 1115 and Unknown (2) 2412 (d) UMAP plot showing the three populations of NKp46+ sorted cells previously identified by Krzywinska et al. e UMAP plots and (f) violin plots showing the expression of canonical immune checkpoint inhibitory molecules across the three identified ILC populations (median shown by the yellow triangle). Cell counts for the clusters identified here were: NKp46+ ILC3 5739, ILC1 1261 and NK 462.
Blockade of CTLA-4 and PD-1 are linked to checkpoint inhibitor induced colitis in cancer patients59–61, and genetic variants in Ctla4 are linked to predisposition to IBD62,63. In our recently reported pre-clinical model of immune checkpoint colitis (CPI-C)64, we identified NCR+ ILC1/3 and ILC2 clusters using scRNA-seq from sorted CD45+ colonic lamina propria cells of healthy wildtype BALB/c mice. Here, we found similar expression of immune checkpoints on ILC subsets, as described above. Ctla4, Lag3, Pdcd1lg2 and Vsir exhibited higher expression in NCR+ ILC1/3 than ILC2, whereas Cd274 expression was higher in ILC2 (Supplementary Fig 1d). Thus, results from all these three independent murine scRNA-seq datasets indicate that Ctla4, as well as other immune checkpoint molecules, are expressed by certain subsets of ILC.
CTLA-4 is expressed on certain ILC subsets constitutively and is increased in response to activation and subset-specific cytokine cues
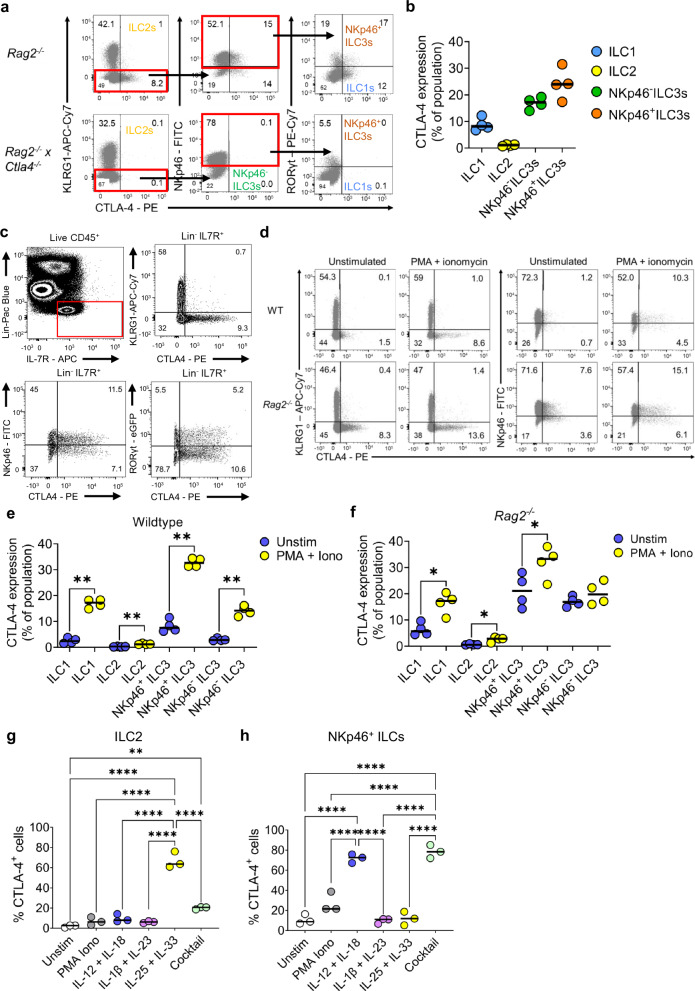
To validate these scRNA-seq data at the protein level, we used flow cytometry to measure CTLA-4 levels in Rag2−/− mice (lacking T cells) and, as a negative control, in Rag2−/− x Ctla4−/− double knockout mice. Unlike Ctla4−/− mice, which develop a spontaneous and fatal phenotype17,18, Rag2−/− x Ctla4−/− mice did not exhibit greater spleen and colon weights compared to Rag2−/− mice (Supplementary Fig. 2a and b). CTLA-4 staining was detected in ILC1s, NKp46+ ILC3s and NKp46- ILC3s from Rag2−/− mice, while expression of CTLA-4 in ILC2s at steady state was virtually undetectable (Fig. 2a, b).
Fig. 2. CTLA-4 is present on ILC2s and NCR+ ILCs.
a Representative flow cytometry plots and (b) summary dot plots from BALB/c Rag2−/− (n = 4) and BALB/c Rag2−/− x Ctla4−/− mice (n = 4) showing the proportion of CTLA-4-expressing cLP ILC1, ILC2, NKp46+ ILC3s and NKp46- ILC3 cells. c Flow cytometry plot showing CTLA-4 in the lamina propria of the colon of BALB/c WT mice and C57BL/6 RORγt-eGFP mice. LPMC were stimulated with PMA, ionomycin and monensin for 4 h. d Representative flow cytometry plots and (e) summary dot plots from BALB/c WT mice (n = 4) and (f) BALB/c Rag2−/− mice (n = 4) showing the proportion of CTLA-4-positive KLRG1 and NKp46 ILC upon stimulation with PMA and ionomycin. Both e and f used RM one-way ANOVA test with Holm-Sidak’s multiple comparison, where * P = 0.0215 and ** P = 0.0033 (g) KLRG1+ (n = 3) and (h) NKp46+ KLRG1- cLP ILC (n = 3) were FACS sorted and cultured with OP9-DL1 for 48 hrs in the presence of rIL7, rIL2 and rIL-15 (unstim), with the addition of either PMA and ionomycin, rIL-12 and rIL-18, rIL-1β and rIL-23, rIL-25 and rIL-33, or rIL-12, rIL-18, rIL-1β, rIL-6, rIL-27 and rTGF-β (cocktail). The proportion of CTLA-4-positive KLRG1+ CD90.2+ and KLRG1- NKp46+ CD90.2+ cLP ILC was measured upon seeding KLRG1+ and KLRG1- cLP ILC, respectively. Both g and h used one-way ANOVA test with Holm-Sidak’s multiple comparison * P = 0.0111 ** P = 0.0015 **** P < 0.0001. All experimental n in Fig. 2 are biologically independent mouse samples.
In PMA and ionomycin stimulated colonic lamina propria (cLP) CD127+ ILCs, from BALB/c mice, there was minimal CTLA-4 expression by ILCs containing Klrg1 (a marker of ILC2) (Fig. 2c). However, in Klrg1- ILCs, we observed CTLA-4 in subsets of both NKp46+ and NKp46- ILCs (Fig. 2c). Furthermore, utilising a C57BL/6 RORγt-eGFP reporter mouse line, we observed co-expression of CTLA-4 in both RORγt-eGFP+ and RORγt-eGFP- ILCs (Fig. 2c), further suggesting that CTLA-4 expression is present among NKp46+ ILC3 and NKp46- CCR6- ILC3. CTLA-4 staining was significantly increased upon stimulation with PMA and ionomycin compared to unstimulated ILC subsets isolated from both BALB/c mice and BALB/c WT animals (Fig. 2d–f). In order to validate the CTLA-4 expression on ILC, we also compared the expression of CTLA-4 on other immune cells that are known to express CTLA-4, like CD4+ T cells, especially TREGS, and myeloid cells, such as dendritic cells, using BALB/c wildtype mice (Supplementary Fig. 3a). As expected, we found that the major cell type expressing CTLA-4 was indeed CD4+ T cells, most likely due to including both conventional CD4+ T cells and TREGS within this population. However, our findings of the Ctla-4 expressing ILC1s, NKp46+ ILC3s and NKP46- ILC3s were still valid (Supplementary Figs. 3b and c). These data highlight that CTLA-4 is expressed on ILC subsets and that stimulation of ILCs can increase the expression of CTLA-4 in certain ILC subsets.
To further understand if CTLA-4 could be induced in a subset specific manner, we treated purified cLP ILCs (Supplementary Fig. 4a) with a selection of cytokines and assessed their CTLA-4 levels. Interestingly, we found that a combination of recombinant (r)IL-25 and rIL-33 was a potent inducer of CTLA-4 expression in ILC2 (Fig. 2g). In contrast, CTLA-4 expression by NKp46+ ILC was most strongly induced by rIL-12 + rIL-18 (Fig. 2h). CTLA-4 expression in NKp46+ ILCs and ILC2s was not further enhanced when a cocktail of rIL-1β, rIL-6, rIL-27 and rTGF-β was added to the medium (Fig. 2g, h). These in vitro data indicate that the canonical cytokines used to activate the respective ILC2 and NKp46+ ILC subsets can induce the expression of CTLA-4.
An IL-10-producing regulatory ILC subset in the intestine has been previously reported62, although more recent reports have challenged the validity of this finding65. Typically, CTLA-4 expressing Foxp3+ CD4+ TREG cells produce the anti-inflammatory cytokine IL-1066. Therefore, we sought evidence for a population of CD127+ ILC co-expressing IL-10 and CTLA-4 in the cLP and small intestine lamina propria of BALB/c mice. We detected no IL-10-producing ILCs upon stimulation with PMA and ionomycin in the cLP and minimal (4.7%) IL-10-producing ILCs in the small intestine, but none of these co-expressed CTLA-4 (Supplementary Fig. 5a). In contrast, we found a substantial CTLA-4+ ILC population among those stimulated ILCs, but IL-10 and CTLA-4 co-expression was not observed, indicating that CTLA-4 expression is separable from IL-10 production.
As CD28 and CTLA-4 bind to the same ligands (CD80/B7.1 and CD86/B7.2) and CTLA-4 is known to function by restricting CD28 engagement in T cells, we next investigated whether CD28 was expressed on ILCs at steady state. Analysis of the two publicly available scRNA-seq datasets used in Fig. 1 showed that Cd28 expression could be detected predominately in the ex-ILC3/ILC1 cluster and a small number of cells in the NCR+ ILC3 cluster (Supplementary Fig. 6a, b). In the second dataset by Krzywinska et al., Cd28 expression could be detected predominately in the ILC1 and NK cluster and a small number of cells in the NKp46+ ILC3 cluster (Supplementary Figs. 6c, d). In our own pre-clinical model of immune checkpoint colitis64, we also found a small number of NCR+ ILC1/3 cells which expressed Cd28 (Supplementary Fig. 6e). Consistent with the scRNA-seq data, we found that, in BALB/c wildtype mice, CD28 was highly expressed on ILC1s and NKp46+ ILC3 (Supplementary Fig. 6f). Therefore, ILC-expressed CTLA-4 could potentially alter ILC function by regulating CD28 engagement.
To investigate the role of CD28 on ILCs in mice, we generated a Rag2−/− x Cd28−/− mouse line (Supplementary Figs. 7a and b). At steady state, Rag2−/− x Cd28−/− mice had a slightly heavier colon, but no other macroscopic differences when compared to Rag2−/−Cd28+/+ littermates (Supplementary Figs. 7c–e). Remarkably, we found that the loss of Cd28 caused a significant loss of ILC1s, ILC2s and NKp46+ ILC3s in the colon (Supplementary Figs. 7f). The phenotype of ILCs was also markedly altered by Cd28 deficiency, with significantly increased CTLA-4 expression (Supplementary Figs. 8a and b) and IFNγ production (Supplementary Figs. 8c and d) in ILC1s and NKp46+ CCR6- ILC3s from the colon. These data indicate that CD28 co-stimulation can modulate ILC number and function, suggesting that CTLA-4 has the potential to alter ILC biology by regulating the CD28 pathway.
ILCs upregulate CTLA-4 in a microbiota-dependent manner
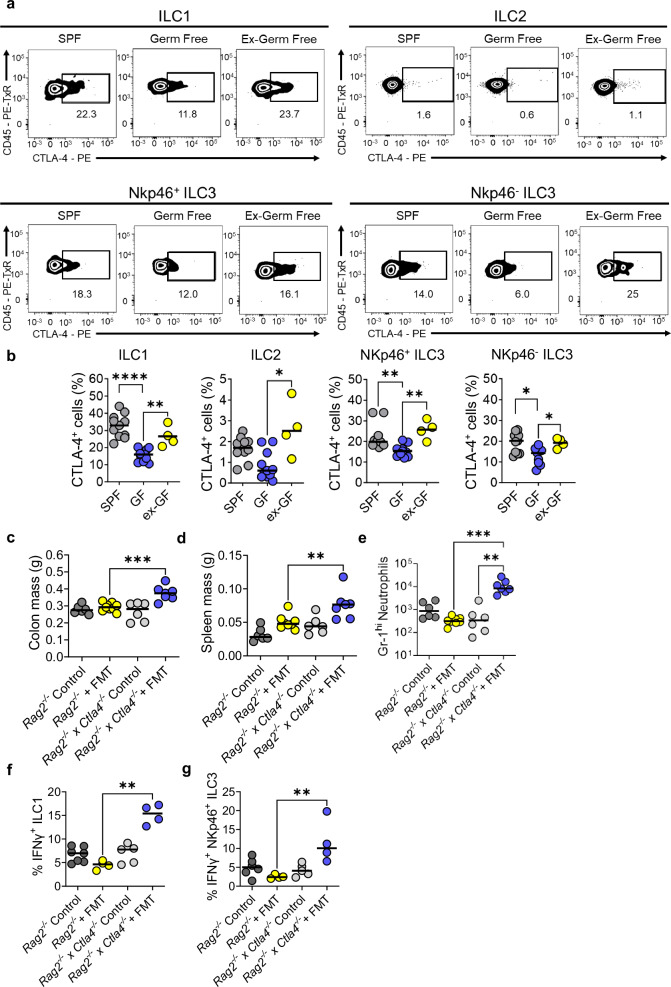
The composition of the intestinal microbiota has been shown to influence colitis susceptibility in both the general population and cancer patients treated with CPI67–70. Therefore, we investigated whether altering the intestinal microbiota would affect CTLA-4 expression in ILCs in the lamina propria of mice. To investigate this, we used C57BL/6 mice maintained under three housing conditions: germ free (GF), specific pathogen-free (SPF), and GF co-housed with SPF mice for 4 weeks prior, in order to recolonise GF mice with SPF microbiota (ex-GF). Altering the microbiota resulted in changes to the numbers of some ILC subsets; ILC1s and ILC2s were increased in GF mice and there was a trend for NKp46- ILC3s to be reduced in ex-GF mice, relative to SPF cell numbers (Supplementary Fig. 9a). Critically, CTLA-4 expression was significantly reduced in ILC1s and both NKp46+ and NKp46- ILC3s of GF mice when compared to SPF mice (Fig. 3a, b). Remarkably, the altered CTLA-4 expression in ILC from GF mice was restored in ex-GF mice, indicating that presence of the microbiota actively promotes CTLA-4 expression by colonic ILCs (Fig. 3a, b). Colonic explant biopsies from these mice were cultured for 24 h and analysis of the supernatant by ELISA showed that IL-18 production was significantly reduced in GF mice but restored in ex-GF mice, while IL-12 production was significantly higher in ex-GF mice (Supplementary Fig. 9b). These data highlight the importance of the microbiome in regulating CTLA-4 expression in ILCs and that key drivers of type 1 immunity were also dysregulated by the microbiome.
Fig. 3. ILC express CTLA-4 in a microbiota-dependent manner.
a Representative flow plots and (b) summary statistics showing the percentage of different ILC subsets (ILC1s, ILC2s, NCR+ ILC3s and NCR- ILC3s) that are positive for CTLA-4 in specific pathogen free (SPF) (n = 11), germ-free (GF) conditions (n = 11) and GF mice, which have been gavaged with SPF microbiota (ex-GF) (n = 4). One-sided Kruskal-Wallis Test with Dunn’s multiple comparison * P = 0.0106 ** P = 0.0048 **** P < 0.0001 (c) Colon, (d) spleen masses and (e) infiltrating Gr-1+ neutrophils between control BALB/c Rag2−/− (n = 6), FMT treated BALB/c Rag2−/− (n = 9), control BALB/c Rag2−/− x Ctla4−/− (n = 6) or FMT treated BALB/c Rag2−/− x Ctla4−/− (n = 7) ** P = 0.0037 *** P = 0.0007 One-sided Kruskal-Wallis Test. f IFNγ production from ILC1s (g) or NKp46+ ILC3s between control BALB/c Rag2−/− (n = 7), FMT treated BALB/c Rag2−/−(n = 4), control BALB/c Rag2−/− x Ctla4−/− (n = 5) or FMT treated BALB/c Rag2−/− x Ctla4−/− (n = 4) upon restimulation with PMA and ionomycin for 3 h. ** P = 0.0039 for f and P = 0.0052 for g using one-sided Kruskal-Wallis Test with Dunn’s multiple comparison. All experimental n in Fig. 3 are biologically independent mouse samples.
Next, we tested the impact of the microbiota on the expression of CTLA-4 on ILCs by giving C57BL/6 wild-type mice a cocktail of antibiotics (ampicillin, neomycin, vancomycin and metronidazole) in their drinking water over a period of two weeks. We found that cLP NKp46+ IFNγ+ ILCs (containing a mixed population of ILC1s and NCR+ ILC3s) from the antibiotic-treated mice had a lower per cell expression of CTLA-4 (Supplementary Figs. 9c and d). These data further illustrate that the microbiota play a role in regulating CTLA-4 expression in ILCs.
To further probe the impact of microbial regulation of CTLA-4 expression on ILCs, we transplanted a pro-inflammatory, colitogenic microbiota from TRUC mice71 to Rag2−/− and Rag2−/− x Ctla4−/− by oral gavage. In contrast to Ctla4 sufficient Rag2−/− mice, where faecal transplantation had no appreciable impact, in Rag2−/− x Ctla4−/− double knockout mice transplantation induced features of colitis, including increased colon mass, splenomegaly, increased infiltration of Gr-1hi neutrophils and expansion of IFNγ producing ILC1s and NKp46+ ILC3s (Fig. 3e–g and Supplementary Figs. 10a, 11a–e). There were no major changes in ILC subset numbers post FMT, indicating that it was the enhanced IFNγ production per ILC that was resulting in greater inflammation (Supplementary Figs. 11d and e). We conclude from these data that CTLA-4 is upregulated on ILCs in response to the microbiota and that the composition of the microbiota can induce an inflammatory response when regulation via CTLA-4 on ILCs is perturbed.
CTLA-4 restricts ILC mediated inflammation in the colon
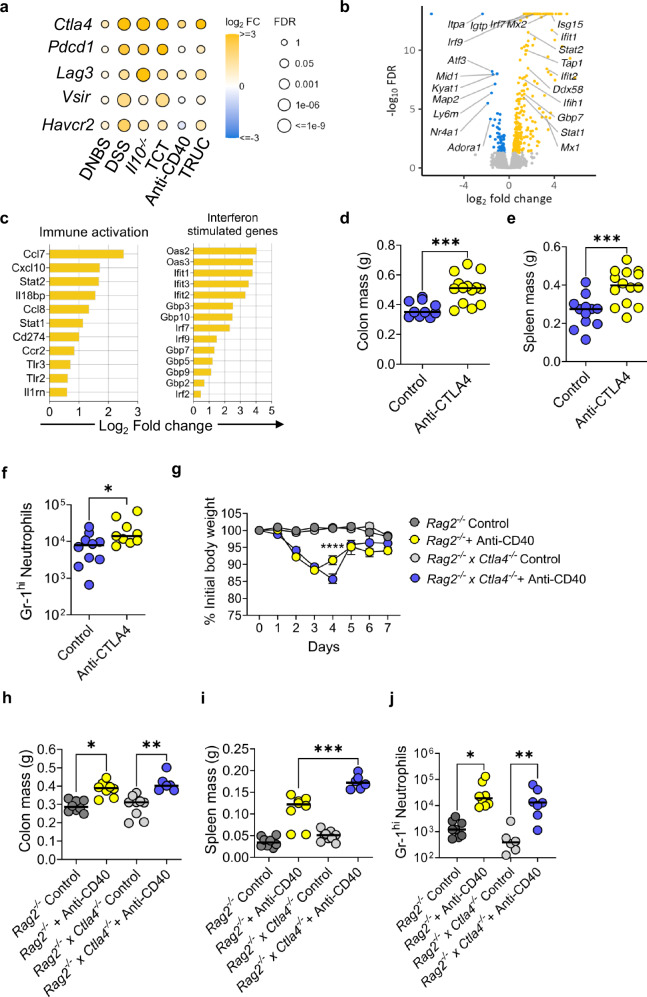
We next investigated whether inflammation is a driver of immune checkpoint expression on ILCs. We analysed the expression of checkpoint genes using RNA-seq from colonic tissue segments from six different pre-clinical models of colitis, including T cell-mediated models (Il10−/− mice and T cell transfer colitis), chemically induced models (Dinitrobenzene sulphonic acid (DNBS) induced colitis and dextran sulphate sodium (DSS) induced colitis) and two models of ILC-dependent colitis (Tbx21−/− xRag2−/− Ulcerative Colitis (TRUC) and anti-CD40 colitis in Rag−/− mice)35,72. Ctla4 was the most highly upregulated checkpoint gene in all the colitis models, with less pronounced changes in the expression of Pdcd1, Lag3, Vsir and Havcr2 (Fig. 4a). We observed induction of Ctla4 in the colon in both innate immune mediated models of colitis (Fig. 4a).
Fig. 4. CTLA-4 restrains innate immune activation in colitis.
a Heatmap of the changes in immune checkpoint expression in distal colon segments taken from different models of colitis (Il10−/− (n = 3), DSS (n = 4), T cell transfer (n = 4), DNBS (n = 4), TRUC (n = 4) and anti-CD40 (n = 4)) compared to control mice (controls were: WT mice (n = 4) for Il10−/−, DSS and DNBS models; Rag2−/− mice (n = 4) for T cell transfer, TRUC and anti-CD40 models. b Volcano plot showing the gene expression profile of distal colon segments taken from BALB/c Rag2−/− x Ctla4−/− mice (n = 4) compared to BALB/c Rag2−/− mice (n = 4). Positive log2 fold-changes indicate upregulation in Rag2−/− x Ctla4−/− mice, while negative log2 fold-changes indicate upregulation in Rag2−/− mice. c The most significantly upregulated differentially expressed genes involved in immune activation and interferon-stimulated genes from colon segments taken from Rag2−/− x Ctla4−/− mice (n = 4) compared to Rag2−/− mice (n = 4). d Colon mass, (e) spleen mass and (f) infiltrating Gr-1+ neutrophils between TRUC untreated mice (control) (n = 12) compared to TRUC mice treated with anti-CTLA-4 (n = 14) * P = 0.035 *** P = 0.0005 Two-tailed Mann Whitney U test. g weight change (with SEM) P < 0.0001 2-way ANOVA Test performed, (h) colon mass, (i) spleen mass and (j) infiltrating Gr-1+ neutrophils between control untreated BALB/c Rag2−/− mice (n = 8), BALB/c Rag2−/− mice treated with anti-CD40 (n = 8), untreated BALB/c Rag2−/− x Ctla4−/− mice (n = 9 for (h and i), n = 6 for (j) and BALB/c Rag2−/− x Ctla4−/− mice treated with anti-CD40 (n = 6 for (h and i,) n = 7 for (j) * P = 0.0165 ** P = 0.0036 *** P = 0.0005 One-sided Kruskal-Wallis Test with Dunn’s multiple comparison. All experimental n in Fig. 4 are biologically independent mouse samples.
To determine whether loss of Ctla4 resulted in molecular perturbations in the colonic innate immune system, we using RNA-seq to analyse gene expression changes from a colon segment from the distal colon of Rag2−/− Ctla4−/− compared to Rag2−/− mice under homeostatic specific pathogen free (SPF) conditions. There were 344 differentially expressed genes (DEGs; FDR < 0.05) in the colon of Rag2−/− x Ctla4−/− in comparison with Rag2−/− mice, including 272 upregulated genes and 72 downregulated genes. Notably, many of the most upregulated genes were associated with immune activation (Ccl7, Cxcl10, Il18bp, Stat2, Stat1) and interferon stimulation (Gbp3, Gbp7, Irf7, Irf9, Ifit1) (Fig. 4b, c).
Pathway analysis demonstrated significant activation (FDR < 0.05) of multiple immune pathways, including interferon signalling (P < 4 × 10−10), dendritic cell maturation (P < 0.0006) and TREM1 signalling (P < 0.007) (Supplementary Fig. 12a). The most strongly predicted upstream activator of the gene expression changes observed in the colon of Rag2−/− Ctla4−/− mice was Ifng (Z-score= 8.5, P < 1.1 × 10−50) (Supplementary Figs. 12a-b). Other upstream regulators predicted to be activated included Stat1, the canonical signalling pathway triggered by Ifng (Z-score= 7.5, P < 6.9 × 10−66), Tnf (Z-score=5.5, P < 2 × 10−22) and type 1 interferon pathway components, including Ifna, Ifnar and Irf7 (Supplementary Fig. 12a–c). Together, these data indicate that when Ctla4 is deficient from innate cells, there is a transcriptional drive towards a more pro-inflammatory IFNγ-mediated transcriptional profile.
We, next, investigated whether this pro-inflammatory gene expression in the innate immune compartment, following genetic deletion or antibody blockade of CTLA-4, had a disease immunological phenotype by using three different colitis models. Previous data showed that the microbiota composition played a role in potentially driving disease, especially in the context of microbiota derived from TRUC mice (Fig. 3c–g). Therefore, we administered a CTLA-4 antagonist to TRUC mice and, when compared to control TRUC mice, we found that they suffered significantly more severe disease outcomes (based on either losing more than 15% weight loss), worse weight loss, significantly worse survival rates, heavier colon and spleen mass, and an increase in infiltrating neutrophils in the colonic lamina propria (Fig. 4d–f and Supplementary Figs. 13a–c). However, despite these clinical differences, there was no increase or changes observed in either IL-17A or IL-13 production from TRUC mice treated with anti-CTLA4 (Supplementary Figs. 13d–e).
Ctla4 expression was observed in the anti-CD40 model from the bulk RNA-seq data (Fig. 4a). Therefore, we applied the anti-CD40 model to Rag2−/− x Ctla4−/− in comparison with Rag2−/− mice to see if there would be a difference in colitis severity. We found that Rag2−/− x Ctla4−/− suffered more severe disease, lost more weight, and took longer to recover in this model and had significantly larger spleens, however colon masses and infiltrating neutrophils were the same in comparison to Rag2−/− treated with anti-CD40. (Fig. 4g–j and Supplementary Figs. 14a and b). The only significant difference in cytokine production came from IFNγ+ NKp46+ ILC3s (Supplementary Fig. 14c). However, Rag2−/− treated with anti-CD40 showed increased expression of CTLA-4 in all subsets of colonic ILCs (Supplementary Fig. 14d).
Lastly, administration of 5% DSS in the drinking water resulted in accelerated weight loss at day 2 and 3 and increased colon mass in Rag2−/− x Ctla4−/− mice in comparison with Rag2−/− mice, consistent with more severe colitis in the absence of CTLA-4, but there was no change in spleen mass or colon length (Supplementary Figs. 15a–d). Flow cytometry analysis of cytokines showed a slight increase in IFNγ-producing ILCs in Rag2−/− x Ctla4−/− mice in comparison to Rag2−/− mice after treatment with DSS but no change in IL-13/IL-5 producing ILC2 cells (Supplementary Figs. 15e and f).
Taken together, multiple colitis models show that CTLA-4 expressing ILCs play a role in regulating inflammation in the colon.
CTLA-4+ ILC are present in IBD patients
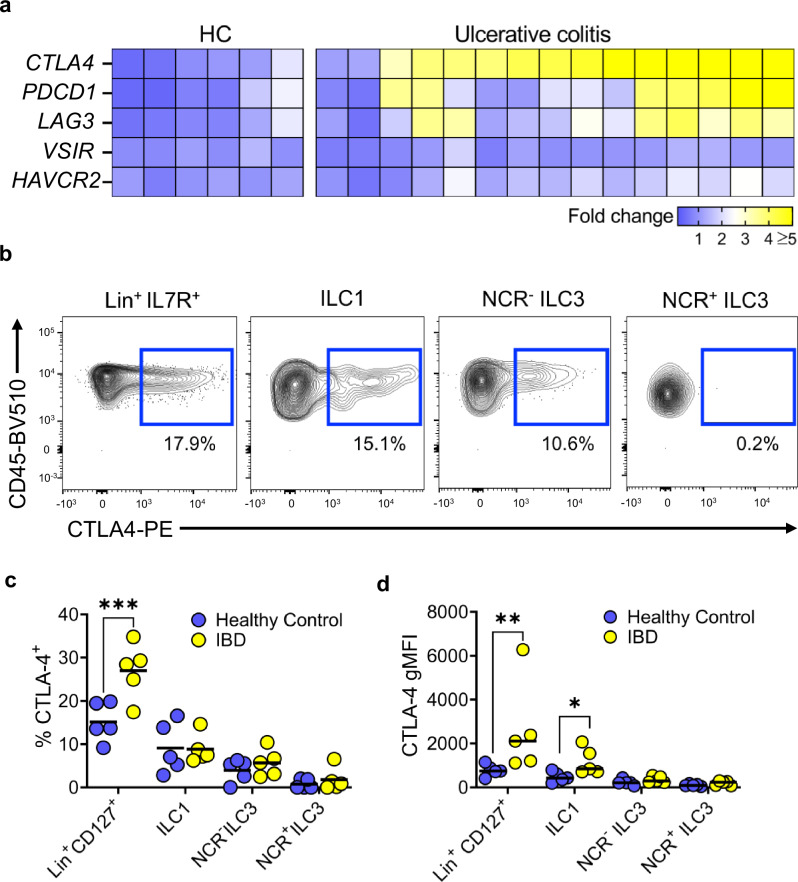
We sought to determine whether our findings in mice translated to patients with IBD. We, therefore, generated a dataset using colon biopsies from patients with active ulcerative colitis (UC) and healthy control subjects without intestinal inflammation to see if CTLA4 expression was upregulated under inflammatory conditions in the human colon (Supplementary Data 1). RNA-seq data analysis showed that CTLA4, and to a lesser extent PDCD1 and LAG3, were all upregulated in the colon of UC patients as compared to healthy control subjects (Fig. 5a). Analysis of a scRNA-seq dataset of human ILCs73 showed a similar pattern as previous murine scRNA-seq datasets; CTLA-4 and other checkpoint molecule gene expression could be identified on subsets of ILCs, mainly ILC1s and ILC3s, but there was very few cells expressing these in these clusters (Supplementary Figs. 16a-b). To determine whether CTLA-4 was expressed by human colonic ILCs, we extracted lamina propria mononuclear cells of patients undergoing colonoscopy and performed multiparameter flow cytometry. We found that CTLA-4 was present in human ILC1 and NCR- ILC3 cells, when comparing Lineage+ IL-7R+ shown as a positive control for CTLA-4 staining on bulk T cells among other immune cells (Fig. 5b and c and Supplementary Fig. 17a). CTLA-4 gMFI levels on ILC1 were also increased in the context of intestinal inflammation in IBD patients compared to healthy controls (Fig. 5d). Interestingly, we were also able to observe CD28 expression on ILC1s and NCR- ILC3s and there was also co-expression with CTLA-4 on these two subsets of ILCs (Supplementary Fig. 17b). These data indicate that CTLA-4 is upregulated on ILCs in IBD.
Fig. 5. CTLA-4+ ILC1 are present in IBD patients.
a Heatmap showing immune checkpoint molecule expression in the sigmoid colon of healthy patients (HC) (n = 6) and patients with ulcerative colitis (n = 15). b Representative flow plots and (c) summary statistics showing the percentage of different ILC subsets (ILC1s, NCR+ ILC3s and NCR- ILC3s) from patients with IBD (n = 5) and healthy controls (n = 5) that were positive for CTLA-4 after treatment with PMA, ionomycin and monensin for 3 h. *** P = 0.0002 2-way ANOVA with Šídák’s multiple comparisons Test. Lineage+ IL-7R+ shown as a positive control for CTLA-4 staining on bulk T cells. d Dot plot showing the CTLA-4 gMFI in different ILC subsets (ILC1, NCR+ ILC3 and NCR- ILC3) in patients with IBD (n = 5) compared with healthy controls (n = 5) * P = 0.0423 ** P = 0.0036 2-way ANOVA with Šídák’s multiple comparisons Test. All experimental n in Fig. 5 are biological independent human samples.
ILCs are expanded in, and mediate CPI-induced colitis
In recent years, blockade of immune checkpoints, in particular CTLA-4 and PD-1, has been found to cause a form of IBD, CPI-induced colitis (CPI-C). ILC clusters were identified in our pre-clinical model of CPI-C64, whereby BALB/c wildtype mice were given the same proinflammatory FMT and anti-CTLA4 and anti-PD1 intra-peritoneal injections to induce CPI-C. However, the role of immune checkpoint blockade in relation to ILCs is not well described in patients with CPI-induced colitis. Since expression of immune checkpoint proteins was observed in ILC populations in wild-type mice (Figs. 1 and 2 and Supplementary Fig. 1), and ILCs are important cells for the pathogenesis of IBD, we hypothesised that there may be a role for ILCs in CPI-induced colitis.
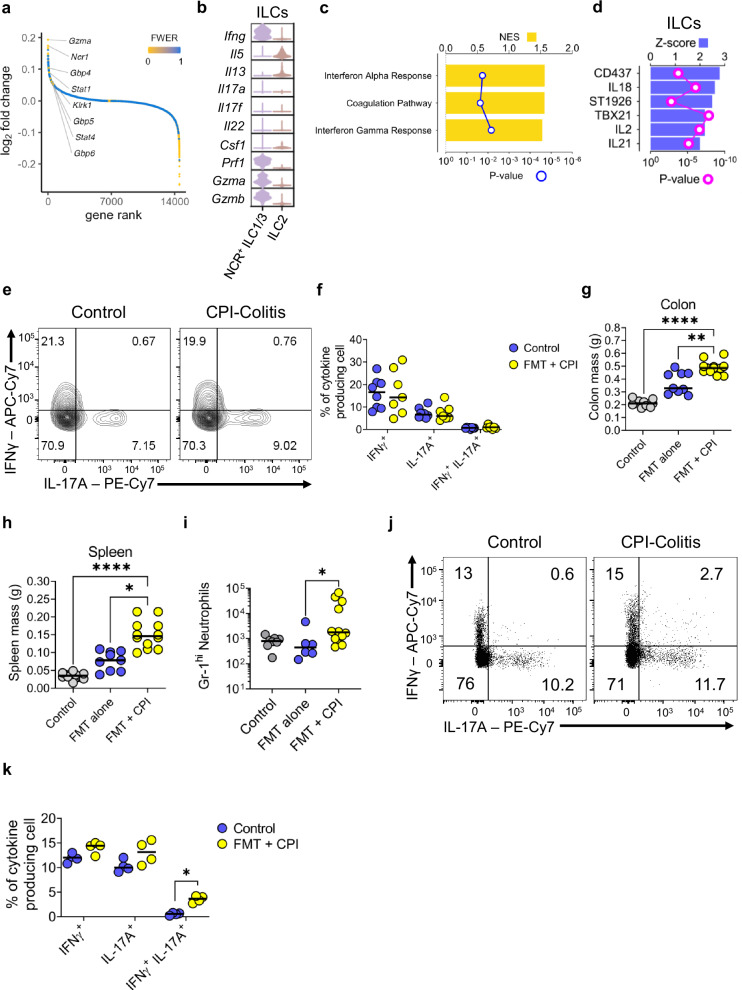
Analysis of the transcriptional changes of sorted live CD45+ lymphocytes from the colonic lamina propria (Supplementary Figs. 18a) of wild-type mice treated with combination CPI therapy revealed an increase in the expression of genes associated with interferon signalling, such as Stat1, Stat4, Gbp5 and Gbp6, as well as an increase in the expression of Ifng itself, in the cluster identified as NCR+ ILC1/3 in mice with CPI-C versus control healthy mice (Fig. 6a, b)64. Furthermore, there was an increase in the expression of genes encoding cytotoxic molecules, such as Gzmb, Gzma and Prf1 (Fig. 6a, b). Further analysis at pathway level using the Hallmark database showed increased activity of interferon alpha and interferon gamma pathways in the ILC clusters, indicating an IFNγ-related phenotype (Fig. 6c). Predicted upstream regulators of ILCs in CPI-C included Tbx21, encoding the transcription factor T-bet, and genes encoding the cytokines Il2, Il18 and Il21, suggesting that ILC1 may contribute to colitis in this model (Fig. 6d). The retinoic acid receptor (RAR)γ- agonist CD437 and the synthetic retinoid St1926 were also predicted to play a role in the gene expression changes observed in colonic ILC in CPI-induced colitis (Fig. 6d).
Fig. 6. NCR+ ILC are expanded in CPI-colitis.
a Distribution of gene expression changes in ILC clusters (ILC2s and NCR+ ILC1/3 s) from mice with CPI-induced colitis (n = 3) vs those from control wild-type BALB/c mice (n = 3). Gene were ranked by decreasing log fold changes and shown as circled coloured according to the associated false discovery rate. b Violin plots showing the expression levels of cytokines across the two ILC clusters in wild-type BALB/c control mice (n = 3) and mice with CPI-induced colitis (n = 3). c Pathways, identified by scanning the Hallmark gene signature dataset using GSEA, significantly upregulated (FDR < 0.05) in the two ILC clusters in mice with CPI-induced colitis (n = 3) vs control mice (n = 3). d Upstream regulators predicted to mediate the gene expression changes in CPI-induced colitis vs control samples from both ILC clusters. e Representative flow cytometry plots and (f) summary dot plot showing IFNγ and IL-17A cytokine production from Lin- IL-7R+ ILCs in wild-type mice treated with CPI-colitis (n = 7) compared to untreated mice (n = 8). The cells were restimulated with PMA and ionomycin for 3 h prior to analysis. g Colon and (h) spleen mass in untreated Rag2−/− mice (n = 12) and Rag2−/−mice given FMT only (n = 11) or FMT + CPI (n = 12). * P = 0.0413 ** P = 0.0022 **** P < 0.0001 Kruskal-Wallis Test with Dunn’s multiple comparison. i Gr-1+ neutrophil infiltration between untreated Rag2−/− mice (n = 8) and Rag2−/− mice given FMT only (n = 6) or FMT + CPI (n = 12). * P = 0.0366 Kruskal-Wallis Test with Dunn’s multiple comparison. j Representative flow cytometry plots and (k) summary dot plot showing IFNγ and IL-17A cytokine production from Lin- IL-7R+ ILCs in Rag2−/− mice treated with CPI-colitis (n = 4) compared to untreated control mice (n = 4). The cells were restimulated with PMA and ionomycin for 3 h prior to analysis. * P = 0.0286 Multiple Mann Whitney U Test. All experimental n in Fig. 6 are biologically independent mouse samples.
To validate this transcriptomic data at the protein level, we measured IFNγ and IL-17A production from IL-7R+ CD90+ ILCs in BALB/c wildtype mice with CPI-C compared to healthy control mice (for gating see Supplementary Fig. 18b). No differences were seen in cytokine production from ILCs in wild-type mice (Fig. 6e, f). However, in wild-type mice, disease is likely mostly driven by T cells, namely cytotoxic CD8+ tissue resident memory T cells60,61,64. To scrutinise the role of ILCs in the absence of T cells, we induced CPI-colitis in Rag2−/− mice. Using a similar methodology to our previous wild-type model64, Rag2−/− mice were treated with the same pro-inflammatory microbiota (FMT, deriving from the TRUC mice) as before and also injected weekly with combination anti-CTLA-4 and anti-PD1 for 3 weeks (Supplementary Fig. 19a). Notably, even in the absence of an adaptive immune system, we still observed induction of disease in Rag2−/− mice, including significantly increased colonic and splenic mass and an increase in the recruitment of neutrophils to the colon (Fig. 6g–i). Overall, the number of IFNγ producing ILCs was similar in CPI-C as compared to wildtype mice. However, we did observe a significant increase in polyfunctional IFNγ/IL-17A producing cells in the colon of mice with CPI-C (Fig. 6j, k), consistent with a pathogenic role for these cells74.
We also interrogated a publicly available scRNA-seq dataset from patients suffering with CPI-C61. Initial analysis of the normalized gene expression of CTLA4 (and other immune checkpoints) did not show expression of CTLA4 in the two ILC clusters identified by Luoma et al. (Supplementary Fig. 20a). This was most likely due to the very few cell numbers found in the clusters identified from the bulk CD45+ sorted lymphocytes and also normalizing the gene expression across all clusters, whereby the T cell clusters would have had much higher CTLA4 gene expression. Therefore, we looked at the actual proportional change in cellular abundance between these two clusters in healthy control compared to CPI-C patients. We found that the ILC1 population was expanded in patients suffering from CPI-C in comparison to healthy controls and when looking at the transcriptional changes between healthy compared to CPI-C that these ILCs were mainly cytotoxic and had a higher number of genes associated with the interferon signaling pathway (Supplementary Figs. 20b–d). These data suggest that the immunopathology of CPI-C might not solely be driven by adaptive immunity; ILCs may also play a role and could represent a potential therapeutic target in CPI-C too.
Discussion
This study provides insight into the regulation of CTLA-4 in ILCs, and how blockade or genetic deletion of this crucial regulatory molecule leads to exacerbated disease in mouse models of colitis and in patients with conventional IBD and CPI-C. The barrier surfaces, and the colon in particular, are challenged with maintaining immunological restraint against a multitude of different commensal microbes that vary across individuals and time, whilst remaining poised to repel invading pathogens. In humans and in mice, we identified CTLA-4 expression in colonic ILCs, a population of tissue-resident innate lymphocytes that have been implicated as important early effector cells in host defense against pathogens and restitution of the mucosal barrier, including in conventional inflammatory bowel diseases75–78. Our findings suggest that CTLA-4 is an important controller of the host response to the microbiome in the colon. These data are consistent with clinical data from CTLA4 heterozygote patients who develop life-threatening autoimmunity, including severe intestinal inflammation79,80.
We show that the ILC subsets predominantly expressing Ctla4 are ILC1, NCR- ILC3 and NCR+ ILC3. When Ctla4 is blocked or deleted, and inflammation is induced, these ILC subsets are responsible for making more proinflammatory cytokine. Notably, Ctla4 expression in ILCs was increased in Rag2−/− mice lacking adaptive immune cells and was upregulated following cell stimulation. Here, we show that CTLA-4 plays an important role in restraining innate immune activation in the colon in multiple colitis models. Genetic deletion of Ctla4 in the innate immune compartment triggered a pro-inflammatory transcriptional program in the colon and predisposed to more severe innate-immune mediated colitis, confirming that CTLA-4 curtails ILC-mediated mucosal inflammation.
We also found that Ctla4 expression was enhanced by ILC activation and regulated by microbial and inflammatory cytokines. Our data support a microbially-driven regulatory circuit of inducible Ctla4 expression in ILCs, as illustrated by the experimental recolonization of GF mice with SPF microbiota. Previous findings have suggested a reduction in NKp46+ ILC3s in GF mice78,81, however, we found that this was not the case in our study and found an increase in ILC1s and ILC2s in GF mice and no changes in ILC3 subsets. We also showed that the microbiota in mice did impact the expression of CTLA-4 on ILCs as seen when FMT was gavaged in the Rag2−/− mice and Rag2−/− mice lacking the Ctla4 gene. We propose that the composition of the bacteria within the gut can trigger upregulation of Ctla4 by ILC, leading in turn to suppression of microbially-driven immune activation. However, when the CTLA-4-driven regulatory circuit is lost, unopposed mucosal immune activation leads to colitis development. This is consistent with the shifts in the composition of the intestinal microbiota observed in immunotherapy-treated cancer patients who go on to develop CPI-induced colitis68–70. Moreover, manipulation of the intestinal microbiota looks to be a promising therapeutic option in the management of CPI-induced colitis82,83.
We also found that CTLA-4-expressing ILCs were present in patients with IBD. Based on the gene expression changes observed in colonic ILCs in our pre-clinical model of CPI-induced colitis, upstream analysis predicted significant activation of proximal mediators, such as cytokines that induce IFNγ production (Il18, Il21, Il27 and Il2), transcription factors (Tbx21), and MAPK kinases (Map2k1/2), flagging them as potentially druggable targets. The list of identified drug targets also includes the selective retinoic acid receptor (RAR)γ agonist CD437, and the retinoid ST1926, which were predicted regulators of colonic ILC transcriptomes in CPI-induced colitis. These ILC1 and IFNγ related genes and pathways were also corroborated by a recently published human CPI-induced colitis61 dataset and could be identified as a potential therapeutically tractable cellular target to this disease.
Lastly, as CTLA-4 and CD28 interact in a mechanistically relevant pathway, we examined CD28 expression on ILCs, revealing that CD28 was predominately expressed on ILC1s. CD28 has previously been noted on human peripheral blood ILCs84, but has not been well studied in murine ILCs. Mice deficient in both CD80 and CD86 were found to exhibit alterations in ILC subsets in the small intestine85, consistent with a role for CD28 and/or CTLA-4 on ILCs, however T cell/ILC crosstalk could not be excluded in these lymphoreplete animals. Here, we show in a T cell free setting, loss of CD28 impairs ILC homeostasis and function, potentially contributing to the significantly larger colons found in these Rag2−/− x Cd28−/− mice; most likely due to uncontrolled dysbiosis and inflammation. We further observed expression of CD28 on ILC1s and NCR- ILC3s in patients with IBD, and a proportion of these ILCs co-expressed CTLA-4. Since CD28 is present on ILC subsets, and appears to contribute to their homeostasis and function, it is possible that ILC-expressed CTLA-4 exerts its biological role by regulating CD28 engagement. CD28 signaling in ILC1 could play a significant role to promote the cellularity of ILC1 subsets. This could also explain why CTLA4 appears to restrain ILC1 cellularity in Rag2−/− mice. However, further work would be needed to fully investigate this possibility.
In conclusion, this study demonstrates that the immune checkpoint protein CTLA-4 is expressed by mouse and human colonic ILCs in a manner that is regulated by the microbiota and specific inflammatory cues. Moreover, we show that CTLA-4 plays a key role in restraining innate immune activation during colonic inflammation in a variety of disease settings.
Methods
Animal husbandry
C57BL/6 and BALB/c wild-type mice (both Charles River) and BALB/c Rag2−/− mice and BALB/c Cd28−/− mice (Taconic) were sourced commercially. A colony of colitis-free Rag2−/− x Tbx21−/− (TRnUC) mice was generated as described previously. BALB/c Ctla4−/− mice were kindly provided by A. Sharpe (Harvard, Boston) and used to generate Rag2−/− x Ctla4−/− mice. RorcGFP mice were a kind gift of Dr Gérard Eberl. BALB/c Rag2−/− mice and BALB/c Cd28−/− mice were crossed to generate the Rag2−/− x Cd28−/− mice. All these mice were housed in specific pathogen-free facilities at King’s College London Biological Services Unit, Imperial College London Central Biomedical Services, University College London Biological Services Unit or Charles River Laboratories. C57BL/6 germ-free mice were housed in germ-free facilities at St. George’s University London and also at the University of Manchester. Breeding of Rag2−/− x Ctla4−/− mice, Rag2−/− x Cd28−/− mice and Rag2−/− mice was performed at University College London under Home Office Licences PPL PA8A94052 and PP5389651. These mice were housed in individually ventilated cages in a temperature- and humidity-controlled environment with a 14 h light and 10 h dark cycle and ad libitum feeding. Animals were provided with environmental enrichment including cardboard tunnels, paper houses, chewing blocks and aspen wood wool nesting material. All procedures were conducted under licenses (Home Office Licence Numbers PPL: 70/6792, 70/8127, 70/7869, P8999BD42 PA8A94052, PP5389651) from the United Kingdom (UK) Home Office in accordance with The Animals (Scientific Procedures) Act 1986 and licences were approved by each Animal Welfare and Ethical Review Body.
Cell isolation
cLP and Peyer’s Patch-free SI LP leucocytes were isolated using a published method84,85. Briefly, the epithelium was removed by incubation in HBSS lacking Mg2+ or Ca2+ (Invitrogen) supplemented with EDTA and HEPES. The tissue was further digested in HBSS supplemented with 2% foetal calf serum (FCS Gold, PAA Laboratories), 0.5 mg/ml collagenase D, 10 μg/ml DNase I and 1.5 mg/ml dispase II (all Roche). The LP lymphocyte-enriched population was harvested from a 40–80% Percoll (GE Healthcare) gradient interface. Leucocytes from human gut were isolated using a published protocol86.
Flow cytometry
Flow cytometry was performed as previously described51,85. LIVE/DEADTM stain (ThermoFisher Scientific Inc.) was used to determine cell viability. Lineage cocktails used included antibodies against CD3, CD45R, CD19, CD11b, TER-119, Gr-1, CD5 and FcεRI for murine ILC (ThermoFisher Scientific Inc.) and CD2, CD3, CD14, CD16, CD19, CD235a, and CD56 human ILC (ThermoFisher Scientific Inc.). The FoxP3 staining kit was used for intracellular staining of CTLA-4, transcription factors and cytokines. In case of cytokine analysis, cells were pre-stimulated with 100 ng/ml PMA and 2 µM ionomycin in the presence of 6 µM monensin for either 3 or 4 h prior to FACS analysis as indicated. Samples were acquired using an LSRFortessa™ cell analyzer (Becton Dickinson, USA) and data were analyzed using FlowJo software (Tree Star, USA). Details on the antibodies used in this study are listed in Supplementary Table 1.
ILC sorting and in vitro culture
Single-cell suspensions from colonic lamina propria were stained with fluorescently labelled antibodies as described and analysed and sorted using a BD FACSAria III cell sorter (BD Biosciences). Antibodies against CD45, lineage markers and IL-7Rα were used to separate CD45+ Lin- CD127+ cells. ILC were cultured in DMEM supplemented with 10% FCS, 1 x GlutaMax (Gibco), 50 U/ml penicillin, 50 µg/ml streptomycin, 10 mM HEPES, 1x non-essential amino acids (Gibco), 1 mM sodium pyruvate and 50 μM β-mercaptoethanol (Gibco). 20,000 sorted ILCs were plated per well of a flat-bottom 96-well plate pre-coated with 4000 OP9-DL1 following an established method. The medium was further supplemented with rmIL-7 and rhIL-2 (both at 10 ng/ml) and further cultures conditions as indicated. All cytokines were used at a final concentration of 10 ng/ ml. Cells were harvested and analysed by flow cytometry after 48 h in culture. In some conditions, the cells were stimulated with 100 ng/ml PMA and 2 µM ionomycin in the presence of 6 µM monensin for 4 h prior to cell harvest.
Faecal microbiota transplant treatment
Faecal content extracted from the caecum of colitis-confirmed TRUC mice was spun down at 600 g for 10 minutes and reconstituted in sterile PBS with 25% glycerol and then 200 µl was orally gavaged into mice at the beginning of FMT treatment. All mice used for these FMT experiments were 6 weeks of age at the start of experiments. All experimental mice were co-housed in specific pathogen–free facilities at King’s College London Biological Services Unit or Imperial Hammersmith CBS. Untreated non-FMT control mice were housed in a separate box to FMT treated mice.
In vivo murine antibody treatment
Mice used were 6-7 weeks old at the start of experiments and gender matched. Female mice were used in most experiments for this study. Mice treated with immune checkpoint blockade drugs were intraperitoneally (i.p) administered anti-CTLA-4 (9H10, BioXCell), using doses of 200 μg, and anti-PD-1, (RMP1-14, BioXCell) at a dose of 250 μg, once per week59. All experimental mice were co-housed in specific pathogen–free facilities at King’s College London Biological Services Unit or Imperial Hammersmith CBS. Untreated control mice were housed in a separate box to CPI + FMT treated mice. Mice were injected intraperitoneally with 100 μg anti-CD40, IgG2a monoclonal antibody rat anti-mouse FGK4.5 (BioXCell).
SPF recolonisation of GF mice and colonic biopsy explant culture
C57BL/6 mice were maintained under three separate housing conditions: germ free (GF), specific pathogen-free (SPF), and GF co-housed with SPF mice for 4 weeks prior, in order to recolonise GF mice with SPF microbiota (ex-GF). Three uniform colon explants, using a 3 mm biopsy punch (Instrapac), from the distal region of each of these mice were taken and cultured for 24 h in complete RPMI in 48well plates @ 37 °C and 5% CO2. After 24 h, supernatants from these wells were collected and spun down to remove any debris and stored at −80°C until required for further analysis.
ELISA
Cytokine concentrations were measured in culture supernatants by ELISA according to the manufacturers’ instructions for IL-12 (EMIL12B) and IL-18 (BMS618-3) (both kits from Thermo Fisher).
Antibiotic treatment
Cages of mice were treated for 2 weeks with their drinking water supplemented with 2% sucrose, ampicillin (0.5 g/l), vancomycin (0.5 g/l), neomycin (0.5 g/l) and metronidazole (0.5 g/l). Control mice only received 2% sucrose in the drinking water. Mice were sacrificed straight away after this 2-week treatment. Water bottles were monitored to ensure that mice were drinking the water mixture.
Single-cell RNA-seq
Colonic LPMCs from mice were initially sorted using a FACS Aria machine (BD Biosciences) based on live CD45+ gates and taken immediately to the 10X Chromium. Cells were suspended at 1 × 106/mL in PBS and 10,000 cells were loaded onto the ChromiumTM Controller instrument within 15 min after completion of the cell suspension preparation using GemCode Gel Bead and Chip, all from 10x Genomics (Pleasanton, CA), and following the manufacturer’s recommendations. Briefly, cells were partitioned into Gel Beads in Emulsion in the ChromiumTM Controller instrument where cell lysis and barcoded reverse transcription of RNA occurred. Libraries were prepared using 10x Genomics Library Kits (3’ end V3 kit) and sequenced on an Illumina HiSeq2500 according to the manufacturer’s recommendations. Read depth of more than 200 million reads per library, or an approximate average of 20,000 reads per cell was obtained with a recovery of 5000 cells.
The raw 10X Genomics sequencing libraries were processed using the Cell Ranger suite v.3.0.1 to demultiplex base call files, generate single cell feature counts for each library, and finally combine these data into one feature by barcode matrix. Read alignment and gene expression quantification made use of the CellRanger pre-built mouse (mm10 v. 3.0.0) reference data. The individual UMI count matrices were normalised to the same effective sequencing depth before they were aggregated. The merged UMI count matrix was imported in R v.3.6.1 and quality cheques were carried out to mitigate the effects of technical artefacts on downstream analyses. Filtering steps were taken to remove genes detected in less than 3 cells (barcodes), and barcodes with: 1) more than 12,400 UMIs (determined as three median absolute deviations above the median barcode library size); or 2) less than 198 detected genes (determined as three median absolute deviations below the median number of genes for all barcodes after log2 transformation); or 3) expression of the epithelial cell adhesion molecule (Epcam) or collagen alpha-1(I) chain (Col1a1) that would suggest contamination by epithelial cells or fibroblasts, respectively; or 4) co-expression of genes encoding chains of the CD3 complex (Cd3d, Cd3e and Cd3g) and those of the B cell antigen receptor (Cd79a, Cd79b) to limit further the impact of multiplets on downstream analyses. The filtered dataset was imported into Seurat v3.1.587, and the anchor-based integration workflow was followed to account for biological and technical batch differences between mice and sequencing libraries. For each sample, the UMI counts were normalised using the LogNormalize method and a scale factor equal to 10,000, and the top 10,000 most highly variable genes were identified using the vst method. Up to 10,000 integration anchor cells were identified for each pair of count matrices after dimensionality reduction to 20 coordinates via Canonical Component Analysis. These anchor sets guided the iterative process integrating the individual sample data into one shared space with all genes passing initial quality cheques. The expression data were scaled after regressing out the following sources of biological and technical variation: mouse id, sequencing library preparation and sequencing batches, number of UMIs detected for each cell and percent of UMIs for mitochondrial genes in each cell. After running Principal Component Analysis of the integrated expression data and inspecting the scree plot, 30 coordinates were used to represent the whole dataset into two dimensions using tSNE.
To cluster the integrated dataset, the k = 20 nearest neighbours of each cell were determined based the Euclidean distance of their expression profiles projected onto the first 30 principal components previously identified. A shared nearest neighbour graph was then built from these data to represent the neighbourhood overlap between pairs of cells using the Jaccard similarity index. From this graph, 24 cell clusters (identified with integers from 0 to 23) were computed by the Louvain algorithm for modularity optimisation with a resolution parameter equal to 0.8. Markers for each cluster were identified by differential expression analysis using MAST88. Only genes expressed in at least 5% of the cells in the cluster under consideration or the rest of the cell population were tested. Genes were considered differentially expressed if the absolute value of the natural log fold change was greater than 0.25 and the Bonferroni-adjusted P value was less than 0.001.
Clusters were annotated to broad cell types using the SingleR (v.1.0.6) package89 and the Immunologic Genome Project (ImmGen) transcriptomic datasets for sorted populations of mouse immune cells90. To this end, SingleR was used to calculate cluster-level gene expression profiles from the individual cell’s data, and then to classify them using its correlation-based iterative algorithm with default parameter settings. The classification process yielded confident assignments based on the pruned scores for all but two sub-populations.
Analysis of publicly available datasets
Single cell count matrices were obtained from ArrayExpress accessions E-MTAB-979557, E-MTAB-1123858 and GEO accession GSE15005073. Count matrices were normalised using Seurat’s NormalizeData function before visualisation. UMAP co-ordinates and clustering metadata were as originally published. Therefore, downstream processing steps can be considered identical to those carried out as previously published. Each matrix was then normalised using SCTransform, followed by RunPCA (PCs=30) and RunUMAP (dims=30). Shared nearest neighbour and clustering were carried out using FindNeighbours (dims=30) and FindClusters respectively. NormalizeData was then ran, and this assay was used for downstream visualisation and differential expression analysis using the MAST algorithm88.
Processed data from droplet-based scRNA-seq profiles of CD45+ immune cells from healthy donors and patients with CPI colitis were downloaded from NCBI GEO (Series GSE144469)61. The UMI count matrices were filtered and normalised as described in the original publication using Seurat. The resulting dataset was integrated following the reciprocal PCA workflow and expression data were finally scaled by regressing out the percentage of mitochondrial transcripts and the number of UMIs detected in each cell. Cell-type assignments and UMAP coordinates were kindly provided by the authors. Average normalised gene expression for each population in CPI colitis was calculated and compared to equivalent data from the mouse data presented here by means of Spearman correlation. These expression profiles were also scanned for enrichment of hallmark signatures using single-sample GSEA. Spearman correlation between the output enrichment scores from the mouse and human data was also estimated.
Bulk RNA-seq analysis
For bulk RNAseq analysis, RNA was extracted from distal colon segments and purified cells with Qiazol reagent (Invitrogen). In this method, distilled solutions of RNA were purified using chloroform and isopropanol and eluted with ethanol before being resuspended in RNAse-free water. RNA was cleaned up using the Qiagen RNeasy Microkit and performing the DNA clean-up protocol as listed in the manufacturer’s guidelines. RNA samples were then checked for quality, contamination and concentration using a NanoDrop, Qubit spectrophotometer and Bioanalyzer. RNA with only a RIN score of above 7 were used for RNA sequencing. RNA was then stored at -80 °C prior to further analysis.
The quality of the raw library files was inspected with fastQC. Raw reads were trimmed and filtered to remove adaptor contamination and poor-quality bases using trimmomatic. Depending on the source organism, the resulting read files were mapped to the GRCm38 (mouse) genome assembly using Hisat2 with default parameters. The number of reads mapping to the genomic features annotated in Ensembl with a MAPQ score higher than or equal to 10 was calculated for all samples using htseq-count with default parameters.
Differential expression analysis between sample groups was performed in R using the Wald test as implemented in the DESeq2 package. P-values were adjusted for multiple testing according to the Benjamini and Hochberg procedure. The P- and corresponding FDR values were re-estimated empirically with fdrtool, when the histograms of the initial P-value distributions showed that the assumptions of the test were not met.
Pre-clinical colitis models
Dextran sulphate sodium (DSS) (36-50 Kda, MP Biomedicals, Ontario, USA) colitis was induced in mice by adding 3% DSS for wild-type model and 5% for Rag2−/− mice to the drinking water for a period of 5 days, after which DSS was removed. Mice were culled after 5 days of being on normal water. For the Rag2−/− x Ctla4−/− mouse related experiments, the relevant mice were sacrificed 2 days after the beginning of the treatment with 5% DSS in order to isolate cLP leucocytes. Anti-CD40 agonistic antibody induced colitis in Rag2−/− mice following i.p. injection of 100 μg of rat anti-mouse (clone: FGK4.5) (BioXCell, West Lebanon, USA). T cell transfer colitis was induced by injecting (i.p.) FACs sorted 0.5×106 live naïve CD4+CD25-CD62L+CD44- splenic T cells into Rag2−/− mice85. DNBS colitis was induced in C57BL/6 wild-type mice using DNBS (Sigma-Aldrich, France) resuspended in 50 μl of 30% ethanol (EtOH) in PBS. The DNBS solution was administered on day 1 by injection with a tuberculin syringe (Terumo, France) and a flexible plastic probe inserted 3.5 cm into the colon. TRUC colitis has been described elsewhere35,71. Immune checkpoint colitis was induced using our previously published method64, in brief, mice were given the FMT and combination anti-CTLA-4 and anti-PD-1, as described above, for 3 weeks.
Human samples
Studies in human tissues received ethical approval from Guy’s and St Thomas’s Trust (REC number: 15/LO/1998). Typically, 12-16 biopsies were taken from each patient. 1 of these biopsies would be taken in RNAlater for RNA extraction and bulk RNA-seq using the same above-described method. Colonic lamina propria mononuclear cells (cLPMCs) were isolated using either grids or by the digestion protocol described above from endoscopically acquired biopsy specimens. Isolated cLPMCs were stimulated with 100 ng/ml PMA and 2 µM ionomycin in the presence of 6 µM monensin for 3 h and then stained for flow cytometry as described above. Meta-data for patients used in this study are attached to Supplementary Data 1.
Statistics
Results are expressed as median ± IQR or mean ± SEM. Data were analysed using Two-way Student’s t-test, Two-way Mann-Whitney U test, Two-Way ANOVA Kruskal-Wallis test, as appropriate, using GraphPad Prism 10.0 (GraphPad Inc., USA). No data were excluded from the analyses. * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to acknowledge the BRC FlowCore at King’s College London and the MRC LMS FlowCore facility at Imperial College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, Imperial College London, King’s College London, or the Department of Health and Social Care. We acknowledge Novogene for the RNA sequencing and the BRC Genomics Facility at King’s College London for the 10X Genomics single-cell RNA sequencing. We would like to thank Dr Gérard Eberl (Institut Pasteur, Paris) for providing RorcGFP mice. We also would like to thank Professor Mike Curtis (King’s College London), Rene Ocho, Emma Mustafa and Robert Bond (all St. George’s University London) and Matthew Hepworth (University of Manchester) for generating and providing germ-free mice. We also thank Professor Zúñiga-Pflücker (Sunnybrook Research Institute, University of Toronto) for contributing OP9-DL1 cells. This study was supported by grants awarded by the Wellcome Trust (NP, WT101159 and WT225875, LSKW, 220772/Z/20/Z) and the UKRI Medical Research Council (GL and RGJ, MR/M003493/1 and MR/R001413/1; GL, MR/K002996/1; LSKW, MR/S0091401/1). NP was also funded by and the Imperial National Institute for Health Research (NIHR) Biomedical Research Centre (BRC). JFN acknowledges an RCUK/UKRI Rutherford Fund fellowship (MR/R024812/1), and GMJ (203757/Z/16/A) and ER (215027/Z/18/Z) were funded by a PhD fellowship from the Wellcome Trust. The research was also supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas and King’s College London (GL). The work of TK was supported by the UKRI BBSRC Gut Microbes and Health Institute Strategic Programme (BB/R012490/1 and its constituent projects BBS/E/F/000PR10353 and BBS/E/F/000PR10355) as well as a BBSRC Core Strategic Programme Grant for Genomes to Food Security (BB/CSP1720/1 and its constituent work packages, BBS/E/T/000PR9819 and BBS/E/T/000PR9817). TK was also supported by the UKRI BBSRC Institute Strategic Programme on Food Microbiome and Health BB/X011054/1 and its constituent project BBS/E/F/000PR13631. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. Work at the CRUK City of London Centre Single Cell Genomics Facility and Cancer Institute Genomics Translational Technology Platform was supported by the CRUK City of London Centre Award [C7893/A26233].
Author contributions
Study concept and design: G.M.L., N.P., and L.S.K.W. Acquisition of data: J.W.L., J.H.S., L.B.R., R.M., D.C., G.B., O.S.O., and M.M. Data analysis and interpretation: J.W.L., J.H.S., L.B.R., R.M., D.C., G.B., O.S.O., and M.M. Technical support: E.M.R., F.H., G.M.J. and E.R. Patient recruitment and consenting: O.S.O. and N.P. Obtained funding: G.M.L., L.S.K.W., N.P., J.F.N., R.G.J., T.K and R.M. Drafting of manuscript: J.W.L., J.H.S and R.M. Edits to manuscript: L.B.R., L.S.K.W., N.P., and G.M.L. Study supervision: G.M.L., and N.P.
Peer review
Peer review information
Nature Communications thanks Yingzi Cong and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All raw data in this study are available in the Source Data file, which has been uploaded with this manuscript. NGS data presented in the manuscript have been made publicly available through the Gene Expression Omnibus (GEO) database found at https://www.ncbi.nlm.nih.gov/geo/. GSE208395 “Transcriptome profiling of colon biopsies from pre-clinical models of colitis” for the bulk RNA-seq comparing different pre-clinical mouse models used for Fig. 4a (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE208395). GSE254247 for Fig. 4b-c comparing colons of Rag2−/− against Rag2−/− x Ctla4−/− titled “Transcriptomic profiling of the role of Ctla4 in the innate immune system” (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254247). GSE224758 for Fig. 5a comparing healthy controls with patients with ulcerative colitis titled “Gene expression profiling of colon biopsies from ulcerative colitis patients and healthy volunteers” (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE224758). GSE222959 titled “Single cell transcriptomics reveals colonic lymphocyte remodelling and emergence of polyfunctional, cytolytic lymphocyte responses in CPI-induced colitis” for the single cell RNA-seq data comparing the wildtype CPI + FMT treated BALB/c CPI-C mice to healthy untreated wildtype BALB/c mice used in Fig. 6 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE222959) and has previously been published64. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jonathan W. Lo, Jan-Hendrik Schroeder, Luke B. Roberts, Rami Mohamed.
Contributor Information
Nick Powell, Email: nicholas.powell@imperial.ac.uk.
Graham M. Lord, Email: graham.lord@manchester.ac.uk
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51719-6.
References
- 1.Guntermann, C. & Alexander, D. R. CTLA-4 suppresses proximal TCR signaling in resting human CD4+ T cells by inhibiting ZAP-70 Tyr319 phosphorylation: a potential role for tyrosine phosphatases. J. Immunol.168, 4420–4429 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Kong, K.-F. et al. Protein kinase C-η controls CTLA-4–mediated regulatory T cell function. Nat. Immunol.15, 465–472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krummel, M. F. & Allison, J. P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med.182, 459–465 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qureshi, O. S. et al. Constitutive Clathrin-mediated endocytosis of CTLA-4 persists during T cell activation. J. Biol. Chem.287, 9429–9440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchbinder, E. I. & Desai, A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am. J. Clin. Oncol.39, 98–106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowshanravan, B., Halliday, N. & Sansom, D. M. CTLA-4: a moving target in immunotherapy. Blood131, 58–67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linsley, P. S. et al. Binding of the B cell activation antigen B7 to CD28 costimulates T cell proliferation and interleukin 2 mRNA accumulation. J. Exp. Med.173, 721–730 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turka, L. A., Ledbetter, J. A., Lee, K., June, C. H. & Thompson, C. B. CD28 is an inducible T cell surface antigen that transduces a proliferative signal in CD3+ mature thymocytes. J. Immunol.144, 1646–1653 (1990). [PubMed] [Google Scholar]
- 9.Azuma, M. et al. B70 antigen is a second ligand for CTLA-4 and CD28. Nature366, 76–79 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Linsley, P. S. et al. CTLA-4 is a second receptor for the B cell activation antigen B7. J. Exp. Med.174, 561–569 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pentcheva-Hoang, T., Egen, J. G., Wojnoonski, K. & Allison, J. P. B7-1 and B7-2 selectively recruit CTLA-4 and CD28 to the immunological synapse. Immunity21, 401–413 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Collins, A. V. et al. The interaction properties of costimulatory molecules revisited. Immunity17, 201–210 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Qureshi, O. S. et al. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science332, 600–603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi, T. et al. Immunologic self-tolerance maintained by Cd25+Cd4+regulatory T cells constitutively expressing cytotoxic T lymphocyte–associated antigen 4. J. Exp. Med.192, 303–310 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Read, S. et al. Blockade of CTLA-4 on CD4+CD25+; regulatory T cells abrogates their function in vivo. J. Immunol.177, 4376 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wing, K. et al. CTLA-4 control over foxp3+ regulatory T cell function. Science322, 271–275 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Tivol, E. A. et al. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity3, 541–547 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Waterhouse, P. et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla4. Science270, 985–988 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Khoja, L., Day, D., Wei-Wu Chen, T., Siu, L. L. & Hansen, A. R. Tumour- and class-specific patterns of immune-related adverse events of immune checkpoint inhibitors: a systematic review. Ann. Oncol.: Off. J. Eur. Soc. Med. Oncol.28, 2377–2385 (2017). [DOI] [PubMed] [Google Scholar]
- 20.De Velasco, G. et al. Comprehensive meta-analysis of key immune-related adverse events from CTLA-4 and PD-1/PD-L1 inhibitors in cancer patients. Cancer Immunol. Res.5, 312–318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassel, J. C. et al. Combined immune checkpoint blockade (anti-PD-1/anti-CTLA-4): Evaluation and management of adverse drug reactions. Cancer Treat. Rev.57, 36–49 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Phan, G. Q. et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc. Natl Acad. Sci. USA100, 8372–8377 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med.363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robert, C. et al. Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med.372, 2521–2532 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Antonia, S. J. et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol.17, 883–895 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Cohen, E. E. W. et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet (Lond., Engl.)393, 156–167 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Motzer, R. J. et al. Nivolumab plus Ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med.378, 1277–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernink, J. H., Mjösberg, J. & Spits, H. Human ILC1: to be or not to be. Immunity46, 756–757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs, A. ILC1s in tissue inflammation and infection. Front. Immunol.7, 104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrido-Mesa, N. et al. T-bet controls intestinal mucosa immune responses via repression of type 2 innate lymphoid cell function. Mucosal Immunol.12, 51–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klose, C. S. N. et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell157, 340–356 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Spits, H. et al. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol.13, 145 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Spits, H. & Cupedo, T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol.30, 647–675 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Powell, N. et al. Interleukin 6 increases production of cytokines by colonic innate lymphoid cells in mice and patients with chronic intestinal inflammation. Gastroenterology149, 456–467 e415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell, N. et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity37, 674–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernink, J. H. et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol.14, 221 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Licona-Limón, P., Kim, L. K., Palm, N. W. & Flavell, R. A. TH2, allergy and group 2 innate lymphoid cells. Nat. Immunol.14, 536 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Mjösberg, J. M. et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol.12, 1055 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Price, A. E. et al. Systemically dispersed innate IL-13–expressing cells in type 2 immunity. Proc. Natl Acad. Sci.107, 11489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cella, M. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature457, 722 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanos, S. L. et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22–producing NKp46+ cells. Nat. Immunol.10, 83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eberl, G. Development and evolution of RORγt+ cells in a microbe’s world. Immunological Rev.245, 177–188 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Luci, C. et al. Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat. Immunol.10, 75 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Tachó-Piñot R., et al. Bcl6 is a subset-defining transcription factor of lymphoid tissue inducer-like ILC3. Cell Rep.42, 113425 (2023). [DOI] [PMC free article] [PubMed]
- 45.Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature517, 293 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Klose, C. S. N. et al. A T-bet gradient controls the fate and function of CCR6−RORγt+ innate lymphoid cells. Nature494, 261 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Koch, M. A. et al. T-bet controls regulatory T cell homeostasis and function during type-1 inflammation. Nat. Immunol.10, 595–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lazarevic, V., Glimcher, L. H. & Lord, G. M. T-bet: a bridge between innate and adaptive immunity. Nat. Rev. Immunol.13, 777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenzie Andrew, N. J., Spits, H. & Eberl, G. Innate lymphoid cells in inflammation and immunity. Immunity41, 366–374 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Melo‐Gonzalez, F. & Hepworth, M. R. Functional and phenotypic heterogeneity of group 3 innate lymphoid cells. Immunology150, 265–275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroeder, J. H. et al. T-bet controls cellularity of intestinal group 3 innate lymphoid cells. Front Immunol.11, 623324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schroeder J.-H., et al. Sustained post-developmental t-bet expression is critical for the maintenance of type one innate lymphoid cells in vivo. Front. Immunol.12, 760198 (2021). [DOI] [PMC free article] [PubMed]
- 53.Schwartz, C. et al. ILC2s regulate adaptive Th2 cell functions via PD-L1 checkpoint control. J. Exp. Med. 214, 2507–2521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeis, P. et al. In Situ maturation and tissue adaptation of type 2 innate lymphoid cell progenitors. Immunity53, 775–792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flamar, A. L. et al. Interleukin-33 induces the enzyme tryptophan hydroxylase 1 to promote inflammatory group 2 innate lymphoid cell-mediated immunity. Immunity52, 606–619.e606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ricardo-Gonzalez, R. R. et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol.19, 1093–1099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiancette, R. et al. Reciprocal transcription factor networks govern tissue-resident ILC3 subset function and identity. Nat. Immunol.22, 1245–1255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krzywinska E., et al. The transcription factor HIF-1alpha mediates plasticity of NKp46+ innate lymphoid cells in the gut. J. Exp. Med.219, e20210909 (2022). [DOI] [PMC free article] [PubMed]
- 59.Wei, S. C. et al. Distinct cellular mechanisms underlie anti-ctla-4 and anti-pd-1 checkpoint blockade. Cell170, 1120–1133.e1117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sasson, S. C. et al. Interferon-gamma-producing cd8(+) tissue resident memory t cells are a targetable hallmark of immune checkpoint inhibitor-colitis. Gastroenterology161, 1229–1244 e1229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luoma, A. M. et al. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell182, 655–671 e622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, S. et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell171, 201–216 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Zeissig, S. et al. Early-onset Crohn’s disease and autoimmunity associated with a variant in CTLA-4. Gut64, 1889–1897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lo, J. W. et al. Immune checkpoint inhibitor-induced colitis is mediated by polyfunctional lymphocytes and is dependent on an IL23/IFNγ axis. Nat. Commun.14, 6719 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bando J. K., et al. ILC2s are the predominant source of intestinal ILC-derived IL-10. J. Exp. Med.217, e20191520 (2020). [DOI] [PMC free article] [PubMed]
- 66.Walker, L. S. K. CTLA-4 and autoimmunity: new twists in the tale. Trends Immunol.36, 760–762 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaput, N. et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol.: Off. J. Eur. Soc. Med. Oncol.28, 1368–1379 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Vetizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dubin, K. et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun.7, 10391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell131, 33–45 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uhlig, H. H. et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity25, 309–318 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Mazzurana, L. et al. Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res. 31, 554–568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morrison, P. J. et al. Th17-cell plasticity in <em>Helicobacter hepaticus</em>–induced intestinal inflammation. Mucosal Immunol.6, 1143–1156 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Buonocore, S. et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature464, 1371–1375 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diefenbach, A., Gnafakis, S. & Shomrat, O. Innate lymphoid cell-epithelial cell modules sustain intestinal homeostasis. Immunity52, 452–463 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Goldberg, R., Prescott, N., Lord, G. M., MacDonald, T. T. & Powell, N. The unusual suspects-innate lymphoid cells as novel therapeutic targets in IBD. Nat. Rev. Gastroenterol. Hepatol.12, 271–283 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Satoh-Takayama, N. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity29, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Kuehn, H. S. et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science345, 1623–1627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schubert, D. et al. Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat. Med.20, 1410–1416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanos, S. L. et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol.10, 83–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, Y. et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med.24, 1804–1808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang, F., Yin, Q., Chen, L. & Davis, M. M. Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc. Natl Acad. Sci. USA115, 157–161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanos S. L., Diefenbach A. Isolation of NK Cells and NK-Like Cells from the Intestinal Lamina Propria. In: Natural Killer Cell Protocols: Cellular and Molecular Methods (ed Campbell K. S.). (Humana Press, 2010). [DOI] [PubMed]
- 85.Lo, J. W. et al. A population of naive-like CD4(+) T cells stably polarized to the T(H) 1 lineage. Eur. J. Immunol.52, 566–581 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Omer, O. S., Powell, N. & Lord, G. M. Characterizing innate lymphoid cell phenotype and function in human inflammatory bowel disease. Methods Mol. Biol.2121, 199–211 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol.16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol.20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heng, T. S. & Painter, M. W. The immunological genome project: networks of gene expression in immune cells. Nat. Immunol.9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All raw data in this study are available in the Source Data file, which has been uploaded with this manuscript. NGS data presented in the manuscript have been made publicly available through the Gene Expression Omnibus (GEO) database found at https://www.ncbi.nlm.nih.gov/geo/. GSE208395 “Transcriptome profiling of colon biopsies from pre-clinical models of colitis” for the bulk RNA-seq comparing different pre-clinical mouse models used for Fig. 4a (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE208395). GSE254247 for Fig. 4b-c comparing colons of Rag2−/− against Rag2−/− x Ctla4−/− titled “Transcriptomic profiling of the role of Ctla4 in the innate immune system” (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE254247). GSE224758 for Fig. 5a comparing healthy controls with patients with ulcerative colitis titled “Gene expression profiling of colon biopsies from ulcerative colitis patients and healthy volunteers” (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE224758). GSE222959 titled “Single cell transcriptomics reveals colonic lymphocyte remodelling and emergence of polyfunctional, cytolytic lymphocyte responses in CPI-induced colitis” for the single cell RNA-seq data comparing the wildtype CPI + FMT treated BALB/c CPI-C mice to healthy untreated wildtype BALB/c mice used in Fig. 6 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE222959) and has previously been published64. Source data are provided with this paper.