Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and vaccination elicit both T cell and B cell immune responses in immunocompetent individuals. However, the mechanisms underlying the antiviral effects mediated by CD4+ T cells are not fully understood. In this study, we analyzed the culture supernatant (SN) from polyclonally stimulated human CD4+ T cells as a model for soluble mediators derived from SARS-CoV-2-stimulated CD4+ T cells. Interestingly, this SN inhibited SARS-CoV-2 propagation in a viral strain- and host cell type-dependent manner. The original wild-type showed the highest susceptibility, whereas the Delta variant exhibited resistance in the human monocyte cell line. In addition, antibody-dependent enhancement (ADE) of infection with the original strain was also abolished in the presence of the SN. The findings showed that the inhibitory effect on viral propagation by the SN was mostly attributed to interferon-γ (IFN-γ) that was present in the SN. These results highlight the potential role of IFN-γ as an anti-SARS-CoV-2 mediator derived from CD4+ T cells, and suggest that we need to understand the SARS-CoV-2 strain-dependent sensitivity to IFN-γ in controlling clinical outcomes. In addition, characterization of new SARS-CoV-2 variants in terms of IFN-γ-sensitivity will have important implications for selecting therapeutic strategies.
Subject terms: SARS-CoV-2, Infection
Introduction
Since the emergence of SARS-CoV-2, numerous studies have demonstrated the critical role of T cell responses in providing clinical protection against SARS-CoV-21,2. Experiments conducted on B-cell deficient mice have shown that vaccine-induced protective immunity against SARS-CoV-2 is mainly attributed to cellular immunity including T cells3. Similarly, studies using a macaque model of SARS-CoV-2 infection have highlighted the critical role of T cells and natural killer cells in controlling viral persistence4. In humans, studies examining infected or vaccinated individuals have shown that elicited T cells can cross-recognize various SARS-CoV-2 variants5,6. In addition, comparative studies of T cells derived from patients infected with SARS-CoV-2 and unexposed healthy donors have demonstrated the existence of pre-existing SARS-CoV-2 cross-reactive T cells in healthy donors, likely induced through previous encounters with endemic seasonal coronaviruses7–11. This T cell immunity, through both SARS-CoV-2 cross-reactive T cells and specific SARS-CoV-2 T cells, is important for maintaining a rapid defense against diverse SARS-CoV-2 variants and may result in asymptomatic or mild disease courses.
In our previous studies12,13, we demonstrated the potential of antibodies (Abs), induced after SARS-CoV-2 infection or vaccination, as well as some therapeutic Abs, to cause antibody-dependent enhancement (ADE) of infection. However, these studies used in vitro culture systems that did not include T cells; specifically, the co-culture involved SARS-CoV-2 and human myeloid cell lines (Mylc cell lines) as infection-recipient cells, with or without anti-SARS-CoV-2 Abs. Using these culture systems, it is possible to ascertain the potential for Abs themselves to cause ADE. However, we need to consider the involvement of many other cells, including T cells, in analyzing the possibility of ADE in vivo. In this study, we investigated the role of soluble factor(s) derived from human CD4+ T cells in modulating SARS-CoV-2 infection. Furthermore, we sought to re-evaluate the potential of Abs to cause ADE in the presence of factor(s) derived from T cells. As a model of soluble mediators derived from CD4+ T cells stimulated with SARS-CoV-2, we used culture supernatant (SN) from polyclonally stimulated human CD4+ T cell lines (hereafter referred to as stim-SN). During these studies, we unexpectedly discovered that stim-SNs could prevent the expansion of SARS-CoV-2, with their preventive effect varying across different SARS-CoV-2 strains. In addition, stim-SN was observed to inhibit ADE. Further analyses showed that the inhibitory effect of stim-SN was attributed to the presence of IFN-γ within it.
Results
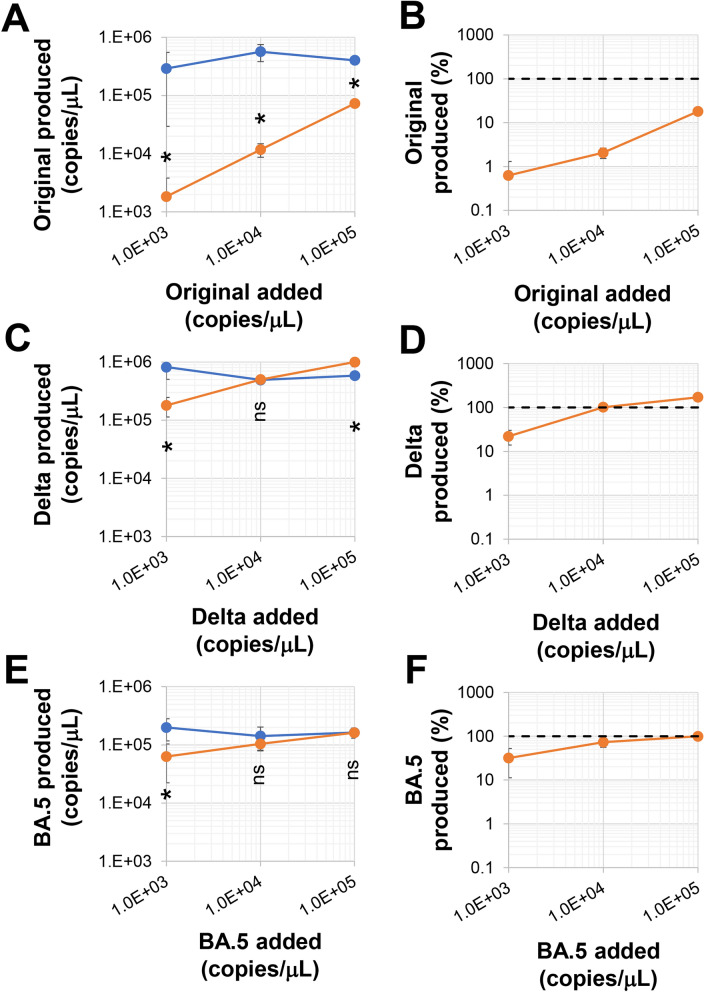
We used the Clone 35 cell line, a human iPS cell-derived, immortalized monocyte cell line that expresses both ACE2- and TMPRSS2, as the recipient cell line for SARS-CoV-2 infection12,13. Clone 35 cells were cultured with several SARS-CoV-2 strains in the presence of stim-SN (Fig. 1). Interestingly, stim-SN exhibited an obvious inhibitory effect on the replication of the original wild-type virus, but a minimal effect on the Delta and BA.5 strains. This inhibitory effect was also confirmed on day 2 culture and in the value of TCID50 (Supplementary Fig. 1). The variations in the inhibitory effect showed no obvious correlation with the magnitude of virus expansion (blue symbols in Fig. 1). We further characterized the inhibitory effects of stim-SN on the replication of the original SARS-CoV-2 strain. Stim-SN, prepared by culturing human CD4+ T cell lines with anti-CD3/CD28-beads in the presence or absence of exogenous IL-2, showed no obvious differences in inhibitory effects (Supplementary Fig. 2). Similarly, SN obtained from CD4+ T cell lines in a resting culture (rest-SN) showed no significant inhibitory effect. These findings suggest that soluble factor(s) derived specifically from stimulated CD4+ T cells are essential for achieving this inhibitory effect.
Fig. 1.
Inhibitory effect of supernatants from activated CD4+ T cells on SARS-CoV-2 variants. Clone 35 cells (2 × 104/well in a 96-well flat plate) were cultured with a titrated amount of SARS-CoV-2 (1 × 104copies/μL of viruses correspond to an MOI (multiplicity of infection) of approximately 0.5), both in the presence (orange symbols) or absence (blue symbols) (N = 4) of SNs (stim-SN, 10% final concentration) from activated CD4+ T cells. Three days post-inoculation, (A,C,E) the quantity of virus in the culture SNs was measured by qPCR. (B,D,F) The viral progeny levels in SNs in the presence of stim-SN are expressed as the percentage compared relative to the viral amounts in cultures without stim-SN. The control condition (viruses cultured in the absence of stim-SN) is represented by a dotted line. *p < 0.05. ns not significant.
We also used human iPS cell-derived alveolar epithelial cells as recipient cells for infection (Supplementary Fig. 3). Viral replication in these cells was also inhibited by stim-SN. However, in contrast to the results from Clone 35 cells, the inhibitory effect was comparable across both the original and Delta strains, showing no difference in efficiency. Taken together, these results demonstrate that soluble factor(s) derived from stimulated CD4+ T cells can produce anti-SARS-CoV-2 inhibitory effects, and that these inhibitory effects are exhibited in a virus strain- and host cell-dependent manner.
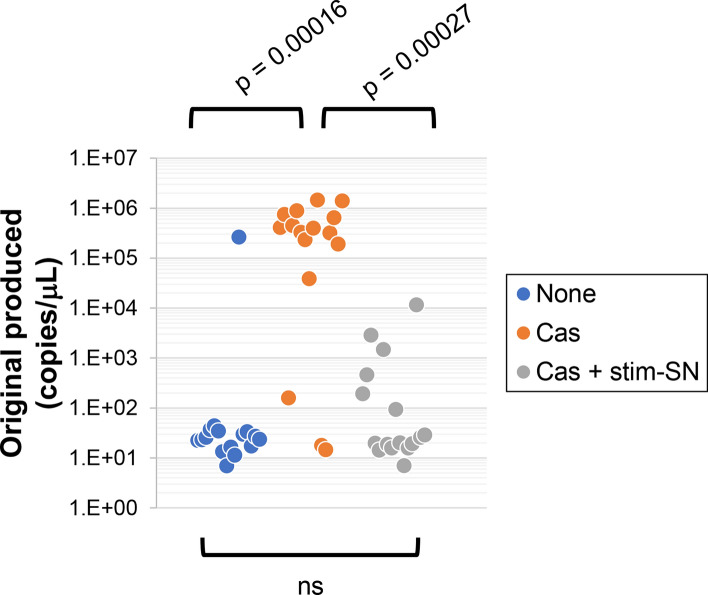
Next, we examined whether the inhibition of viral expansion by stim-SN was observed in another experimental setting. In a previous study12, we demonstrated that viral expansion does not occur in cultures of Clone 35 cells exposed to extremely low doses of the original SARS-CoV-2 strain (40 copies/μL, blue symbols in Fig. 2). However, expansion did occur in the presence of SARS-CoV-2 reactive Ab (for example, casirivimab at 1 ng/mL, orange symbols in Fig. 2), a phenomenon known as ADE. Under these experimental conditions, stim-SN exhibited a significant inhibitory effect against ADE (gray symbols), suggesting that it may play a role in maintaining antiviral homeostasis.
Fig. 2.
Inhibition of antibody-dependent enhancement of infection by SNs from stimulation-culture of CD4+ T cells. Clone 35 cells were cultured with the original strain of SARS-CoV-2 (40 copies/μL) in the presence or absence of casirivimab (Cas, 1 ng/mL final concentration) or stim-SN from CD4+ T cells (10% final concentration) as indicated. Three days following inoculation, the amount of virus in the culture SNs was quantified by qPCR. Symbols represent individual wells (N = 16 per group). ns not significant.
We investigated whether stim-SN affects cells or the virus directly. The original SARS-CoV-2 strain (stim-SN-sensitive strain) and the Delta strain (stim-SN-insensitive strain in the case of Clone 35 cells) were cultured for 24 h in the presence or absence of stim-SN, after which the amount of virus was quantified using qRT-PCR (Fig. 3A). For both strains, the viral loads were comparable to those in control cultures, demonstrating that stim-SN does not directly affect the viruses themselves. To investigate another potential mechanism, we precultured Clone 35 cells with stim-SN for varying durations. We confirmed that cells were alive with more than 95% viability after this preculture. These pre-treated Clone 35 cells were washed three times to remove any residual stim-SN, and they were then cultured with SARS-CoV-2 viruses in the absence of stim-SN (Fig. 3B). Interestingly, pre-treated Clone 35 cells exhibited resistance against viral expansion of the original strain, but not the Delta strain, in a pre-treatment time-dependent manner. These results strongly suggest that stim-SN induces changes within or on the surface of Clone 35 cells, leading to the inhibition of viral replication. However, these changes selectively inhibit only specific strains (stim-SN-sensitive strains) in the case of Clone 35 cells.
Fig. 3.
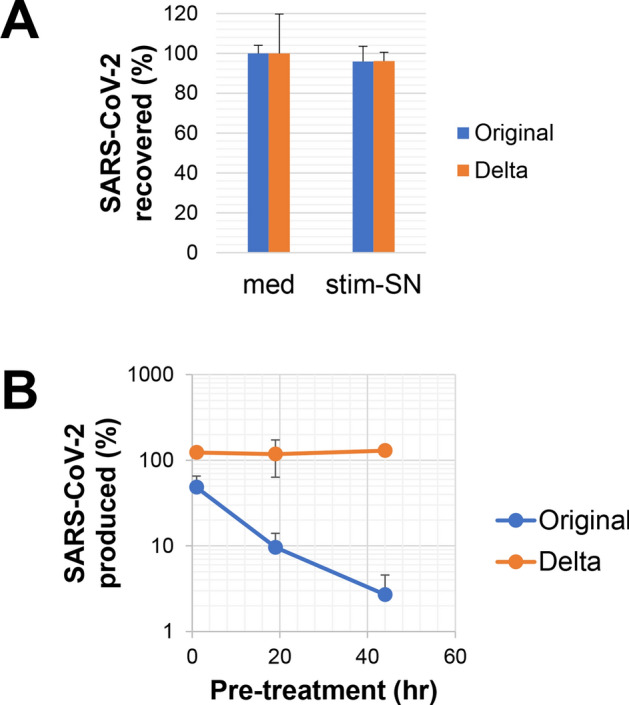
Influence of SNs from stimulation-culture on cells instead of viruses to exert an inhibitory effect against virus replication. (A) SARS-CoV-2 viruses (original or Delta strain) were cultured in the presence or absence of stim-SN at a final concentration of 10%. After 24 h, the amount of virus in the culture SNs was quantified by qPCR. The results are presented as the percentage of virus recovery compared to the control culture without stim-SN. (B) Clone 35 cells (5 × 105/well in a 6-well plate) were cultured in the presence or absence of stim-SN (10% final concentration) for 1, 19, or 44 h (pre-treatment). After pretreatment, the cells were harvested, washed three times, and counted. These pretreated cells were then co-cultured with SARS-CoV-2 viruses (original or Delta strain, 1 × 104 copies/μL). After 3 days, the amount of virus in culture SNs was quantified by qPCR. Viral production by cells pre-treated with stim-SN is shown as the percentage of production relative to cells that were pre-treated with medium (N = 4 per group).
To clarify the changes induced by stim-SN on/within Clone 35 cells, we performed RNA sequencing (RNA-seq) of total RNA isolated from Clone 35 cells that were untreated, treated with rest-SN, or treated with stim-SN for 24 and 43 h (Supplementary Fig. 4A,B). During the differential expression analysis performed using iDEP.96 (http://bioinformatics.sdstate.edu/idep/), we observed increased expression of interferon-stimulated genes (ISGs) (Supplementary Table 1, with examples shown in Supplementary Fig. 4C). This prompted us to investigate whether type I (e.g., IFN-α2, IFN-β), II (IFN-γ), and III (IFN-λ) IFNs are present in stim-SN (Supplementary Fig. 5). The results showed that stim-SN contained background levels of IFN-α2, -β, and -λ and high levels of IFN-γ and that rest-SN contained a non-negligible amount of IFN-γ (for example, 62.9 ng/mL of IFN-γ in stim-SN, 1.3 ng/mL in rest-SN, and background < 0.015 ng/mL). To confirm the stim-SN-dependent transcriptional activation of ISGs, we examined the expression of representative ISGs (RSAD2, IFIT3, IFI35, ISG15, and ISG56)14–17 by qRT-PCR (Supplementary Fig. 6). We found that the expression of selected ISGs was elevated in the presence of stim-SN, a trend that was also apparent with IFN-γ itself.
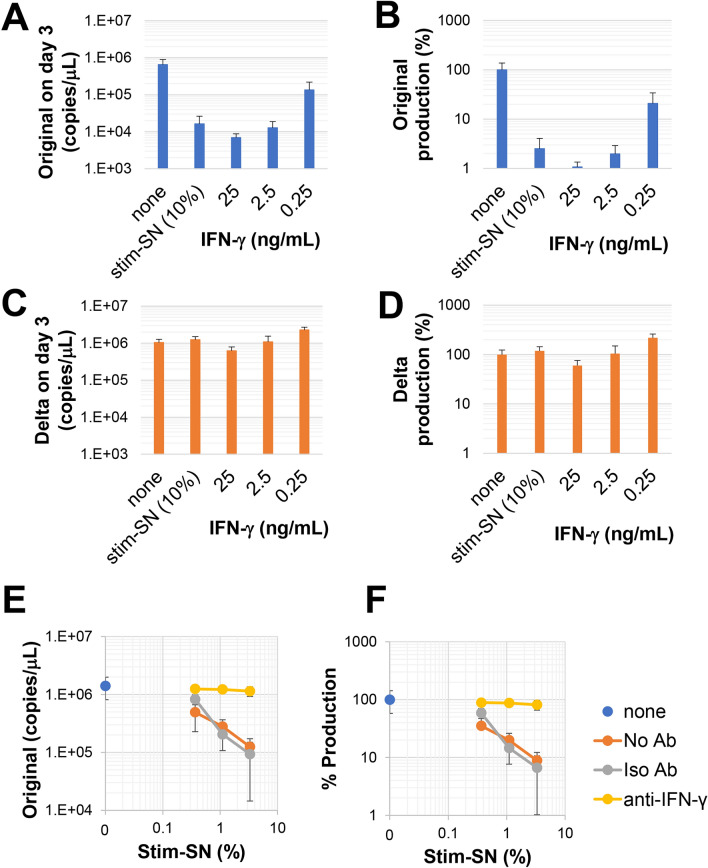
We investigated whether the observed antiviral effects of stim-SN on Clone 35 cells were attributed to the IFN-γ contained within the stim-SN. The addition of IFN-γ to the culture exhibited a dose-dependent inhibitory effect against the expansion of the original strain, but not against the Delta strain (Fig. 4A–D). The inhibitory effect of stim-SN on the original strain was blocked in the presence of anti-IFN-γ neutralizing Ab (Fig. 4E,F). In addition, as observed with stim-SN (Fig. 3B), the pre-treatment of Clone 35 cells with IFN-γ inhibited the expansion of the original strain, but had no effect on the Delta strain (Supplementary Fig. 7). Taken together, these results demonstrate that the IFN-γ present in stim-SN is critical for exerting an antiviral effect in different strains of SARS-CoV-2. Finally, we assessed whether the inhibitory effect mediated by stim-SN was dependent upon the induced amount of ISGs. To test this, Clone 35 cells were cultured in the presence or absence of stim-SN along with medium, in the presence of the original (sensitive) or Delta (insensitive) strain (1 × 104 copies/μL) (Supplementary Fig. 8). No clear difference was observed in the quantity of induced ISGs among the groups in the qPCR assay, indicating that the levels of ISGs examined in this study do not affect the sensitivity of SARS-CoV-2 strains to IFN-γ. This finding suggests that other critical factor(s) in Clone 35 cells induced by IFN-γ may be responsible for exerting the antiviral effect, or that intrinsic differences between the strains themselves could influence their responsiveness.
Fig. 4.
Differential inhibition of SARS-CoV-2 strains by IFN-γ. Clone 35 cells were cultured with either (A,B) the original SARS-CoV-2 strain or (C,D) the Delta strain in the presence or absence (N = 4) of stim-SN (10% final concentration) or a titrated amount of IFN-γ. After 3 days, the amount of virus in culture SNs was quantified by qPCR. The actual amount of viruses in SNs is shown in (A) and (C). (B,D) The amount of virus progeny in SNs, expressed as the percentage production relative to the amount of viruses cultured in the absence of stim-SN or IFN-γ. (E) Clone 35 cells were cultured with the original SARS-CoV-2 strain in the presence or absence (none) of titrated amounts of stim-SN, with medium (No Ab), isotype-matched Ab (Iso Ab) or anti-IFN-γ neutralizing Ab (anti-IFN-γ, 10 μg/mL final concentration). After 3 days, the amount of virus in the culture SNs was measured, shown in (E) and also expressed as the percentage production in (F) (N = 4 per group).
Discussion
Recent studies have shown the robust generation of T cell responses after SARS-CoV-2 infection or vaccination1,2. These T cell responses include the production of IFN-γ by both CD4+ T and CD8+ T cells18–22. However, IFN-γ is conventionally considered to be a signature cytokine of activated T cells. Type I (IFN-α, IFN-β) and III (IFN-λ) IFNs are recognized for their roles in antiviral defense23–25, and several in vitro experiments have shown that type I and III IFNs can suppress SARS-CoV-2 replication26–28. Consequently, the contribution of IFN-γ as an antiviral mediator in SARS-CoV-2 infection remains poorly understood29,30. Nonetheless, several recent reports have identified IFN-γ as a critical mediator in anti-SARS-CoV-2 responses3,4,15,31. In the present study using Clone 35 cells, we also found that IFN-γ produced from activated CD4+ T cells plays a potential role in the mediation of antiviral effects against SARS-CoV-2. It is important to note that different SARS-CoV-2 strains exhibit different sensitivities to IFN-γ in Clone 35 cells (Fig. 1). Interestingly, the IFN-γ-insensitive strains examined in this study, such as Delta and BA.5, were associated with peaks in SARS-CoV-2 pandemic infections. These strains demonstrated resistance to IFN-γ treatment in our experimental systems using Clone 35 cells, suggesting that these insensitive strains may possess a higher threshold for the antiviral effects induced by IFN-γ. Further research is required to identify the critical factor(s) that determine the differential sensitivity of SARS-CoV-2 strains to antiviral effects induced by IFN-γ. In addition, the effect of IFN-γ on other viruses including human coronaviruses also remains to be investigated. In contrast to Clone 35 cells, experiments using alveolar epithelial cells showed no difference in the sensitivity of the original and Delta strains to the antiviral effects of stim-SN, which includes IFN-γ (Supplementary Fig. 3). The experiments using human lung epithelial cells (Calu3) and ex vivo human lung tissues also showed the antiviral activity of IFN-γ.31 These results suggest that the quality and/or quantity of intracellular changes induced by IFN-γ may differ among Clone 35 cells and alveolar epithelial cells. Therefore, it remains to be investigated which specific factor(s) within these cells contribute to the antiviral effects induced by IFN-γ or ascribed to IFN-γ. Recent reports mentioned the involvement of nitric oxide32 or IDO131.
Several reports have demonstrated the existence of SARS-CoV-2 cross-reactive T cells in healthy donors who have not been exposed to SARS-CoV-28,10,11. These cross-reactive T cells may have been stimulated by endemic coronaviruses, leading to the production of various cytokines, including IFN-γ. In addition, there are numerous instances in an individual’s lifetime in which SARS-CoV-2-unrelated T cells can become activated and subsequently produce cytokines, including IFN-γ, similar to SARS-CoV-2-specific and/or cross-reactive T cells1,6–8. The IFN-γ produced incidentally by SARS-CoV-2-unrelated and activated T cells may also exert anti-SARS-CoV-2 replication effects as a bystander effector. This potential role of IFN-γ particularly in the early stages of SARS-CoV-2 infection, could partly influence the varying clinical outcomes observed, ranging from asymptomatic and mild to severe cases. However, it is also important to realize that high levels of IFN-γ in vivo may lead to a severe immune reaction referred to as cytokine storm33,34. These contrasting effects ascribed to IFN-γ highlight the importance of regulating its levels in vivo. Furthermore, it is essential to emphasize the importance of screening SARS-CoV-2 strains for their sensitivity to IFN-γ, since this is critical in determining and selecting strain-specific and effective therapies.
We have shown that the Clone 35 cells used in this study can be used to assess the potential of antibodies to cause ADE of infection12,13. Through these studies, we demonstrated that certain therapeutic Abs for SARS-CoV-2 might induce ADE within a narrow concentration range. These results were obtained from co-cultures of Clone 35 cells, SARS-CoV-2, and Abs. However, when considering the possibility of ADE occurring in vivo, it is necessary to account for the effects of other cell types. In this study, we showed that stim-SN containing IFN-γ can inhibit the induction of ADE (Fig. 2), demonstrating that IFN-γ is effective not only in suppressing viral replication, but also in preventing ADE. These results also suggest that the occurrence of ADE in vivo is less likely, even in the presence of Abs capable of causing ADE, because ADE occurs within a specific Ab concentration range12 and, as demonstrated in this study, under conditions where IFN-γ plays a minimal role.
Materials and methods
Viruses and reagents
SARS-CoV-2/Hu/DP/Kng/19-020 (original wild-type strain, GenBank accession number: LC528232.1) was obtained from the Kanagawa Prefectural Institute of Public Health, Kanagawa, Japan. The SARS-CoV-2 Delta strain was isolated from a patient (hCoV-19/Japan/RIMD-DVI-17/2021). The Omicron subvariant BA.5 (EPI_ISL_13241867) was obtained from the National Institute of Infectious Diseases, Japan. SARS-CoV-2 viruses were passaged twice in VeroE6-TMPRSS2 cells (purchased from the National Institutes of Biomedical Innovation, Health and Nutrition, JCRB Cell Bank, Japan). Virus stock was aliquoted, examined for the presence of virus RNA by quantitative RT-PCR, and tested for mycoplasma (the results were mycoplasma-free) before being used for the experiments in this study. All experiments using live SARS-CoV-2 were conducted in accordance with the approved operating procedures of the biosafety level 3 facility at the Research Institute for Microbial Diseases of Osaka University.
Peripheral blood mononuclear cell (PBMC) samples were obtained from volunteers. Informed consent was obtained from all subjects involved in this study. The Institutional Review Board of the Research Institute for Microbial Diseases, Osaka University, approved the collection and use of serum and PBMCs from volunteers in this study (Approval Number: RIMD-2021-9). All experiments using materials from humans were in accordance with the approved operating procedures and guidelines of the Research Institute for Microbial Diseases of Osaka University.
In some experiments, casirivimab (Cas) was used. Vials containing residual monoclonal antibody (mAb) following clinical use were stored at < 4 °C and kindly provided by Dr. Ryo Morita at Osaka City Juso Hospital, with the hospital’s authorization. Recombinant human IFN-γ and anti-IFN-γ Ab were purchased from Proteintech.
Preparation of human monocyte cell lines (Mylc) from human iPS cells
The procedure used to generate the human monocyte cell line has been described previously35,36. Briefly, immortalized myeloid cell lines were established through the lentivirus-mediated transduction of cMYC, BMI-1, GM-CSF, and M-CSF into human iPS cell-derived myeloid cells. These cell lines, referred to as Mylc, were further induced to express ACE2 and TMPRSS2 using lentiviral vectors. The established bulk cell lines were further cloned by limiting dilution. The resulting cloned cells (Clone 35 cells) maintained stable functional characteristics (specifically, susceptibility to infection) for at least 5 months. These clones were maintained for no longer than 2 months and used for all experiments. Clone 35 cells were cultured in MEMα (Gibco) supplemented with 10% (vol/vol) FBS at 37 °C under 5% CO2 and water-saturated humidity conditions.
Preparation of human alveolar epithelial cells from human iPS cells
The preparation of human alveolar epithelial cells from human induced pluripotent stem cells (iPSCs) was conducted using the air–liquid interface culture technique, as previously described37. Briefly, lung progenitor cells were stepwise induced from human iPSCs following a 21-day, 4-step protocol. At day 21, lung progenitor cells expressing the specific surface antigen carboxypeptidase M were isolated and seeded onto the upper chamber of a 24-well Cell Culture Insert (#353104, Falcon). This was followed by a 7-day differentiation period into alveolar epithelial cells. The alveolar differentiation medium was supplemented with dexamethasone (Cat# D4902, Sigma-Aldrich), KGF (Cat# 100-19, PeproTech), 8-Br-cAMP (Cat# B007, Biolog), 3-isobutyl 1-methylxanthine (IBMX) (Cat# 095-03413, FUJIFILM Wako), CHIR99021 (Cat# 1386, Axon Medchem), and SB431542 (Cat# 198-16543, FUJIFILM Wako) to induce the development of alveolar epithelial cells.
Preparation of culture supernatants
Human CD4+ T cells were purified from PBMCs using anti-human CD4 magnetic beads (Miltenyi Biotec). These CD4+ T cells (2 × 104/well) were cultured in 96-well round plates in the presence of anti-CD3/CD28-beads (Veritas, 1 × 104 beads/well) with or without IL-2 (10 U/mL) for 1–2 weeks (stimulation culture). Subsequently, cells were harvested, beads were removed, and the cells were washed and then cultured in the presence of IL-2 (resting culture). These stimulation and resting cultures were repeated every 1–2 weeks. Culture SNs from either the stimulation or resting cultures were harvested, filtered, and used as either stimulation (stim-SN) or resting (rest-SN) derived supernatants, respectively.
Viral infection
Clone 35 cells were cultured with SARS-CoV-2 in a 96-well format. The cytopathic effect on Clone 35 cells was observed after infection. Therefore, viral concentrations in the culture SNs after 3 days were quantified by qRT-PCR and used as an indicator of viral propagation. Total RNA from the culture SNs was extracted using a QIAamp Viral RNA Mini Kit (Qiagen) following the manufacturer’s instructions. The primers used for the qRT-PCR assays to quantify all SARS-CoV-2 were as follows: SARS-CoV-2 forward AGCCTCTTCTCGTTCCTCATCAC, reverse CCGCCATTGCCAGCCATTC. The fold-increase in viral quantity was calculated as follows: fold-increase = (virus concentration [copies/μL] in the experimental group with reagent)/(virus concentration [copies/μL] in the background control culture without reagent).
Quantitative RT-PCR
Total RNA was extracted using an RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. Following cDNA synthesis, gene expression levels were evaluated by qRT-PCR. The primers used for qRT-PCR were as follows: ISG15 forward TGGCGGGCAACGAATT, reverse GGGTGATCTGCGCCTTCA; IFI35 forward CACGATCAACATGGAGGAGTGC, reverse GGCAGGAAATCCAGTGACCAAC; RSAD2 forward CCAGTGCAACTACAAATGCGGC, reverse CGGTCTTGAAGAAATGGCTCTCC; IFIT3 forward CCTGGAATGCTTACGGCAAGCT, reverse GAGCATCTGAGAGTCTGCCCAA; ISG56 forward GCCTTGCTGAAGTGTGGAGGAA, reverse ATCCAGGCGATAGGCAGAGATC. Amplification signals were normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the housekeeping gene, with the following primers: forward GCAAATTCCATGGCACCGT, reverse TCGCCCCACTTGATTTTGG). Gene expression levels were then quantified relative to untreated controls using the delta-delta-(CT) method (2–ΔΔCT).
Measurement of IFNs
The concentration of IFNs in the SNs was measured using enzyme linked immunoassay kits for IFN-α2, IFN-β, and IFN-γ (BioLegend), as well as IFN-λ 3/1/2 (PBL Assay Science). The results were plotted as OD values.
RNA sequencing
Total RNA was extracted from cells using an miRNeasy Micro kit (Qiagen) following the manufacturer’s instructions. Library preparation was conducted according to the manufacturer’s instructions using a TruSeq Stranded mRNA Library Prep kit (Illumina). Sequencing was performed on an Illumina NovaSeq 6000 sequencer (Illumina) in a paired-end mode (2 × 101 nt). Following adapter sequence removal with Trimmomatic v0.38, the sequenced reads were mapped to the human reference genome (hg19) using TopHat (ver. 2.1.1). The number of fragments per kilobase of exon per million mapped fragments (FPKMs) was calculated using Cuffdiff (ver. 2.2.1). Data were analyzed using the online application iDEP (ver. 0.96, http://bioinformatics.sdstate.edu/idep/).
Statistical analysis
The significance of statistical differences was evaluated using the Mann–Whitney test. Probability (p) values < 0.05 were considered to be significant.
Supplementary Information
Acknowledgements
This work was supported by the Japan Agency for Medical Research and Development (AMED) (JP20he0822004). We thank the NGS core facility at the Research Institute for Microbial Diseases of Osaka University for the sequencing and data analysis. The authors thank FORTE Science Communications (https://www.forte-science.co.jp/) for English language editing.
Author contributions
J.S. planned and performed experiments, and analyzed the data with the assistance of T.Sasaki., G.H.O., R.K., Y.S., E.E.N., and K.M. Alveolar epithelial cells were provided by T.N. and Y.Y. The manuscript was written by J.S. and T.Shioda.
Data availability
All data generated or analyzed during this study are included in this article and the supplementary information files. The raw data have been deposited in the NCBI Gene Expression Omnibus database (GSE271988).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kazuo Miyazaki, Email: kmiyazaki@micantechnologies.com.
Tatsuo Shioda, Email: shioda@biken.osaka-u.ac.jp.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-77969-4.
References
- 1.Moss, P. The T cell immune response against SARS-CoV-2. Nat. Immunol.23, 186–193 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Sette, A., Sidney, J. & Crotty, S. T cell responses to SARS-CoV-2. Annu. Rev. Immunol.41, 343–373 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Wang, X. et al. Vaccine-induced protection against SARS-CoV-2 requires IFN-γ-driven cellular immune response. Nat. Commun.14, 3440 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huot, N. et al. SARS-CoV-2 viral persistence in lung alveolar macrophages is controlled by IFN-γ and NK cells. Nat. Immunol.24, 2068–2079 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarke, A. et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell185, 847-859.e811 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarke, A. et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep. Med.2, 100355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loyal, L. et al. Cross-reactive CD4(+) T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science374, eabh1823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, J. et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature587, 270–274 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Grifoni, A. et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell181, 1489-1501.e1415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mateus, J. et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science370, 89–94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Bert, N. et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature584, 457–462 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Shimizu, J. et al. Reevaluation of antibody-dependent enhancement of infection in anti-SARS-CoV-2 therapeutic antibodies and mRNA-vaccine antisera using FcR- and ACE2-positive cells. Sci. Rep.12, 15612 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu, J. et al. The potential of COVID-19 patients’ sera to cause antibody-dependent enhancement of infection and IL-6 production. Sci. Rep.11, 23713 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol.32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busnadiego I, et al. Antiviral activity of type I, II, and III interferons counterbalances ACE2 inducibility and restricts SARS-CoV-2. mBio11, (2020). [DOI] [PMC free article] [PubMed]
- 16.Schoggins, J. W. Interferon-stimulated genes: what do they all do?. Annu. Rev. Virol.6, 567–584 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Saleiro D, et al. IFN-γ-inducible antiviral responses require ULK1-mediated activation of MLK3 and ERK5. Sci. Signal11, (2018). [DOI] [PMC free article] [PubMed]
- 18.Rydyznski Moderbacher C, et al. NVX-CoV2373 vaccination induces functional SARS-CoV-2-specific CD4+ and CD8+ T cell responses. J. Clin. Investig.132, (2022). [DOI] [PMC free article] [PubMed]
- 19.Barreiro, P. et al. A pilot study for the evaluation of an interferon gamma release assay (IGRA) to measure T-cell immune responses after SARS-CoV-2 infection or vaccination in a unique cloistered cohort. J. Clin. Microbiol.60, e0219921 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motozono, C. et al. The SARS-CoV-2 omicron BA.1 spike G446S mutation potentiates antiviral T-cell recognition. Nat. Commun.13, 5440 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gil-Bescós R, et al. Potency assessment of IFNγ-producing SARS-CoV-2-specific T cells from COVID-19 convalescent subjects. Life Sci. Alliance.6, (2023). [DOI] [PMC free article] [PubMed]
- 22.Krishna, B. A. et al. Spontaneous, persistent, T cell-dependent IFN-γ release in patients who progress to Long Covid. Sci. Adv.10, eadi9379 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, F. C. & Young, H. A. Interferons: Success in anti-viral immunotherapy. Cytokine Growth Factor Rev.25, 369–376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fensterl, V. & Sen, G. C. Interferons and viral infections. Biofactors35, 14–20 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Park, A. & Iwasaki, A. Type I and Type III interferons—induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe27, 870–878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudima, G., Kofiadi, I., Shilovskiy, I., Kudlay, D., Khaitov, M. Antiviral therapy of COVID-19. Int. J. Mol. Sci.24, (2023). [DOI] [PMC free article] [PubMed]
- 27.Lokugamage KG, et al. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol.94, (2020). [DOI] [PMC free article] [PubMed]
- 28.Vanderheiden A, et al. Type I and Type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol.94, (2020). [DOI] [PMC free article] [PubMed]
- 29.Kang, S., Brown, H. M. & Hwang, S. Direct antiviral mechanisms of interferon-gamma. Immune Netw.18, e33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akamatsu, M. A., de Castro, J. T., Takano, C. Y. & Ho, P. L. Off balance: Interferons in COVID-19 lung infections. EBioMedicine73, 103642 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, D. et al. Type-II IFN inhibits SARS-CoV-2 replication in human lung epithelial cells and ex vivo human lung tissues through indoleamine 2,3-dioxygenase-mediated pathways. J. Med. Virol.96, e29472 (2024). [DOI] [PubMed] [Google Scholar]
- 32.Silva, B. J. A. et al. IFN-γ-mediated control of SARS-CoV-2 infection through nitric oxide. Front. Immunol.14, 1284148 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karki, R. et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell184, 149-168.e117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadanandam, A. et al. A blood transcriptome-based analysis of disease progression, immune regulation, and symptoms in coronavirus-infected patients. Cell Death Discov.6, 141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haruta, M. et al. TAP-deficient human iPS cell-derived myeloid cell lines as unlimited cell source for dendritic cell-like antigen-presenting cells. Gene Ther.20, 504–513 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Imamura, Y. et al. Generation of large numbers of antigen-expressing human dendritic cells using CD14-ML technology. PLoS One11, e0152384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto, Y. et al. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat. Methods14, 1097–1106 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and the supplementary information files. The raw data have been deposited in the NCBI Gene Expression Omnibus database (GSE271988).