Abstract
We have systematically investigated the duplex to hairpin conversion of oligoribonucleotides under the aspect of nucleobase methylation. The first part of our study refers to the self-complementary sequence rCGCGAAUUCGCGA, which forms a stable Watson–Crick base paired duplex under various buffer conditions. It is shown that this sequence is forced to adopt a hairpin conformation if one of the central 6 nt is replaced by the corresponding methylated nucleotide, such as 1-methylguanosine N2,N2-dimethylguanosine, N6,N6-dimethyladenosine (m62A) or 3-methyluridine. On the other hand, the duplex structure is retained and even stabilized by replacement of a central nucleotide with N2-methylguanosine (m2G) or N4-methylcytidine. A borderline case is represented by N6-methyladenosine (m6A). Although generally a duplex-preserving modification, our data indicate that m6A in specific strand positions and at low strand concentrations is able to effectuate duplex–hairpin conversion. Our studies also include the ssu ribosomal helix 45 sequence motif, rGACCm2GGm62Am62AGGUC. In analogy, it is demonstrated that the tandem m62A nucleobases of this oligoribonucleotide prevent duplex formation with complementary strands. Therefore, it can be concluded that nucleobase methylations at the Watson–Crick base pairing site provide the potential not only to modulate but to substantially affect RNA structure by formation of different secondary structure motifs.
INTRODUCTION
Up to now more than 90 differently modified nucleosides have been identified in cellular RNA (1). Modifications of high structural complexity mainly occur in tRNA (2,3). Structurally simple modifications, as represented by pseudouridine, 2′-O-ribose or base methylated nucleosides, are also found in rRNA, mRNA and snRNA (4). The function of the majority of nucleoside modifications is far from being fully understood. In recent years evidence has accumulated that important cellular processes are dependent on the presence of modified nucleosides. These processes are mRNA and rRNA maturation, ribosome assembly, rRNA processing, translation of the genetic code and recognition of tRNAs (5–7).
With respect to RNA secondary and tertiary structure, modified nucleotides are generally understood to modulate the physicochemical properties of an existing RNA fold (8–13). In our opinion, this strongly underestimates the structure-determining potential of modified nucleotides. Here we report that nucleosides that are methylated at the Watson–Crick base pairing site are capable of substantially affecting RNA structure by mediating folding into different secondary structure motifs. This is demonstrated by a comparison of selected sequences that exist in duplex conformations but change into hairpin conformations if single nucleobases are replaced by the corresponding methylated ones.
MATERIALS AND METHODS
RNA synthesis and purification
5′-O-DMT-2′-O-TOM-protected nucleoside cyanoethylphosphoramidites (A, C, G and U) were obtained from Xeragon AG (Switzerland).
Based on known procedures (14–21), we synthesized the methylated nucleosides 1-methylguanosine (m1G), N2-methylguanosine (m2G), N2,N2-dimethylguanosine (m22G), N6-methyladenosine (m6A), N6,N6-dimethyladenosine (m62A), N4-methylcytidine (m4C), 3-methyluridine (m3U) and 1-methylinosine (m1I) and transformed them into 5′-O-DMT-2′-O-TOM-protected nucleoside phosphoramidites (22,23); we will report on that in detail elsewhere.
All oligoribonucleotides were synthesized on CPG supports on a Pharmacia Gene Assembler Plus following slightly modified DNA standard methods: detritylation, dichloroacetic acid/1,2-dichloroethane (4:96, 2 min); coupling, phosphoramidites/acetonitrile (0.1 M, 120 µl, 2.5 min) were activated with benzyl thiotetrazole/acetonitrile (0.35 M, 360 µl); capping, (i) Ac2O/sym-collidine/THF (10:10:80), (ii) N-methylimidazole/THF (16:84); oxidation, I2/H2O/pyridine/THF (3:2:20:75, 45 s). Amidite solutions, tetrazole solutions and acetonitrile were dried over activated molecular sieves overnight. All sequences were synthesized trityl-off.
Deprotection and cleavage of oligonucleotides from the solid support were achieved with MeNH2 in EtOH (8 M, 0.75 ml) and MeNH2 in water (40%, 0.75 ml) for 3–6 h at 33°C. The solution was then evaporated to dryness. Removal of the 2′-O-silyl ethers was performed by treatment with tetrabutylammonium fluoride trihydrate (TBAF·3H2O) in THF (1 M, 0.90 ml) for at least 12 h at room temperature. The reaction was quenched by addition of Tris–HCl (1 M, pH 7.4, 0.95 ml). The volume of the solution was reduced to 1 ml and directly applied to a Sephadex G 10 column (30 × 1.5 cm) controlled by UV detection at 260 nm. The product was eluted with water and evaporated to dryness.
All oligoribonucleotides were purified by ion exchange chromatography on a semi-preparative Dionex DNAPac column at 80°C. Flow rate 2 ml/min; eluant A, 25 mM Tris–HCl, 6 M urea, in H2O, pH 8.0; eluant B, 25 mM Tris–HCl, 0.5 M NaOCl4, 6 M urea, in H2O, pH 8.0; detection at 265 nm; gradient I (to check the purity of the crude products after deprotection), 0–60% B in A over 45 min; gradient II (for purification), Δ20% B in A over 30 min. Fractions containing the purified oligonucleotide were desalted by loading onto a C18 SepPak cartridge (Waters/Millipore), followed by elution with 0.1–0.2 M (Et3NH)HCO3, water and then H2O/CH3CN (6:4). Combined fractions containing the oligonucleotide were lyophilized to dryness.
The expected masses were confirmed for all oligoribonucleotides by MALDI-TOF mass spectrometry.
Thermal denaturation studies
Absorbance versus temperature profiles were recorded at 250, 260, 265 and 270 nm on a Cary-1 spectrophotometer equipped with a multiple cell holder and a Peltier temperature control device. Each sequence was measured at five or six different concentrations ranging from ∼1 to 100 µM. Sequences 1–4 were measured in buffer solutions of 10 mM Na2HPO4, pH 7.0, containing either 150 mM or 1.0 M NaCl. All other sequences 5–21 were measured at a salt concentration of 150 mM NaCl. Data were collected after a complete cooling and heating cycle at a rate of 0.7°C/min. Melting transitions were reversible for all sequences and essentially the same with respect to the four different wavelengths.
For sample preparation oligonucleotides were lyophilized to dryness, dissolved in the corresponding buffer from stock solutions and subsequently degassed. A layer of silicon oil was placed on the surface of the solution.
ΔH° and of ΔS° values for monomolecular melting transitions were derived from a two-state van’t Hoff analysis by fitting the shape of the individual α versus temperature curve; values of ΔH° and of ΔS° for bimolecular melting transitions were derived from 1/T versus lnc plots according to Marky and Breslauer (24) and Turner and co-workers (25,26). Errors for ΔH and ΔS arising from non-infinite cooperativity of two-state transitions and from the assumption of a temperature-independent enthalpy are typically 10–15%. An additional error is introduced when free energies are extrapolated far from the melting transitions; errors for ΔG° are typically 3–5%.
NMR spectroscopy
1H NMR NH spectra were acquired on a Bruker Avance 500 MHz DRX using excitation sculpting for suppression of the water signal (27) or, alternatively, on a Varian Unity 500, applying a selective excitation refocusing sequence employing selective pulses shaped according to the G4 [excitation (28); 2.62 ms, rf amplitude 1.74 kHz] or REBURP [refocusing (29); 1.4 ms, rf amplitude 4.47 kHz)] profile, respectively. Both shaped pulses were centered at 13 p.p.m.
Each of the sequences rCGCGAAUUCGCGA 3, rCGCm2GAAUUCGCGA 5 and rCGCm22GAAUUCGCGA 6 was measured at two different concentrations (0.30–0.40 mM and 0.07–0.10 mM) and at three different temperatures (40, 27 and 1°C) in 25 and 6 mM sodium arsenate buffer, pH 7.4, (Sigma) without further addition of sodium chloride. rCGCm1GAAUUCGCGA 4 was measured at concentrations of 2.2, 0.7 and 0.1 mM in 25, 12.5 and 6 mM arsenate buffer. rCGCGm62AAUUCGCGA 7, rCGCGAm62AUUCGCGA 8, rCGCGm6AAUUCGCGA 9, rCGCGAAm3UUCGCGA 11 and rCGCGAAUm3UCGCGA 12 were measured at concentrations of ∼0.2 mM at 26°C.
For sample preparation the oligoribonucleotides (triethyl ammonium salts) were lyophilized together with the corresponding amount of arsenate buffer, redissolved in water, lyophilized again and finally dissolved in H2O/D2O (9:1). The samples were equilibrated at room temperature; no heating and rapid cooling was performed.
Electrophoretic mobility shift assay
PAGE was performed using gels prepared from solutions of 19% acrylamide, 0.8% bis(acrylamide), 100 mM NaCl, 2 mM MgCl2, 100 mM Tris–HCl, 0.12% tetramethylethylenediamine and 0.06% ammonium persulfate, pH 7.4 (15 × 15 × 0.1 cm). For sample preparation 7 µl of loading buffer (100 mM NaCl, 2 mM MgCl2, 100 mM Tris–HCl and 6% glycerine) were added to the lyophilized oligonucleotides 1–6, 9, 10, 13 and 14 to obtain a final concentration of 45 µM. The samples were equilibrated at room temperature for 60 min. Electrophoresis was performed at room temperature. The electrophoresis buffer was 100 mM NaCl, 2 mM MgCl2, 100 mM Tris–HCl, pH 7.4. The gel was run at a constant voltage of 80 V, until the bromophenol blue marker had migrated ∼5 cm. The gels were visualized using Stains All dye in formamide.
RESULTS AND DISCUSSION
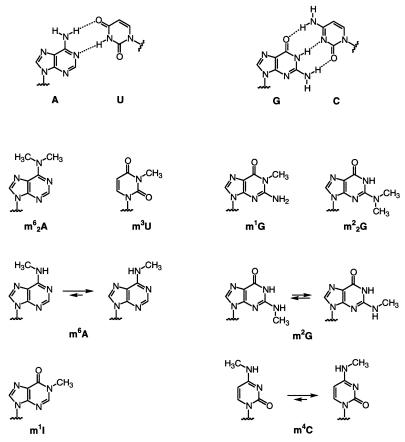
Among the naturally occurring methylated nucleosides we have focused on those that have a direct impact on the Watson–Crick base pairing pattern. The methyl groups of the nucleobases depicted in Figure 1 are located at sites that usually involve hydrogen bonding within a regular A-form duplex. Only in the cases of m2G, m6A and m4C rotamers exist that are capable of base pairing according to the Watson–Crick mode. For all other nucleobases shown (Fig. 1), namely m1G, m22G, m62A and m3U, a severe disturbance of the overall duplex structure would be expected if these nucleotides replaced the corresponding unmethylated nucleotide involved in a Watson–Crick base pair. Therefore, we asked whether a base pair disruption by methylation may also cause a change in secondary structure, provided the sequence itself allows this.
Figure 1.
Watson–Crick base pairing pattern of adenine-uracil (A-U) and guanine-cytosine (G-C) (above). Chemical constitution and conformation of methylated nucleotides encountered in cellular RNAs: 1-methylguanosine (m1G), N2,N2-dimethylguanosine (m22G), N6,N6-dimethyladenosine (m62A), 3-methyluridine (m3U), 1-methylinosine (m1I), N2-methylguanosine (m2G), N6-methyladenosine (m6A) and N4-methylcytidine (m4C). Preferred conformations of m4C, m6A and m2G are according to Engel and von Hippel (43).
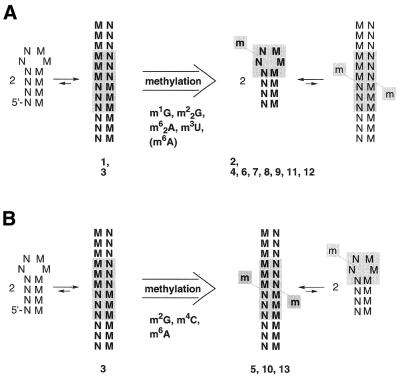
A straightforward approach to probe this involves duplexes of palindromic or self-complementary sequences. A self-complementary sequence system is experimentally advantageous over a non-self-complementary one with respect to our intended comparison of methylated versus unmethylated oligoribonucleotides (Scheme 1). In principle, both methylated and unmethylated self-complementary sequences should be able to form either a hairpin or a duplex. Simple criteria, such as dependence of the melting transition on strand concentration as well as the shape of the UV melting profiles and their hyperchromicity, should allow easy distinction between monomolecular and bimolecular interactions and, subsequently, determination of the thermodynamic pairing parameters. Furthermore, 1H NMR NH resonances aid in distinguishing hairpin from duplex structures. A combination of these tools was used in the study presented. Additionally, for selected sequences gel mobility experiments were performed.
Scheme 1. Site-specific methylation of a palindromic duplex. M, N, nucleotides capable of complementary Watson–Crick base pairing; mN, mM, methylated nucleotides. Depending on the kind of methylation the duplex is either converted into a hairpin (A) or the duplex is retained (B). The bold numbers refer to the sequences investigated (compare Table 1).
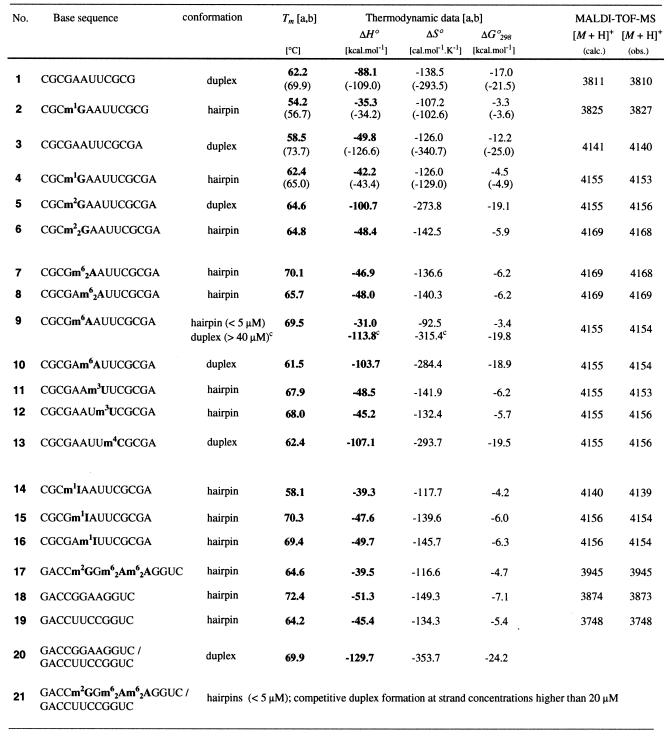
In general, hairpin formation is an intrinsic property of all palindromic sequences. In the DNA series the coexistence of hairpin and duplex structures is documented for several sequences, dCGCGAATTCGCG being the most famous among them (30–36). Low strand and low salt concentrations favor the hairpin, while high strand and high salt concentrations favor the duplex. Moreover, by heating and subsequent rapid cooling the hairpin conformations are kinetically trapped. Interestingly, in the RNA series palindromic sequences that establish a hairpin–duplex equilibrium are reported only for those that involve at least two mismatches within the corresponding duplex (37–41). This reflects the fact that RNA oligomers in micro- and millimolar concentrations thermodynamically prefer bimolecular over monomolecular interactions if formation of a fully Watson–Crick base paired duplex is possible. For example, this was observed for the Dickerson–Drew dodecamer analog, rCGCGAAUUCGCG 1 (Table 1). In buffer solutions containing 0.15 or 1.0 M NaCl this oligoribonucleotide showed a monophasic sigmoid melting profiles at neutral pH. A distinct dependence of the melting temperature on concentration (2–80 µM) provided evidence that this sequence existed as a bimolecular complex. Therefore, we considered this sequence type suitable for our comprehensive methylation study. As a first test we incorporated m1G instead of guanosine at strand position 4. Indeed, rCGCm1GAAUUCGCG 2 did show a monophasic sigmoid melting profile and, importantly, the independence of the melting temperature on concentration (2–90 µM) proved the existence of a monomolecular transition, which is only consistent with a hairpin structure. Moreover, the significantly lower hyperchromicity was in accord with the lower number of base pairs.
Table 1. Selection of methylated oligoribonucleotides and the corresponding reference sequences (5 µM total strand concentration, 150 mM NaCl, 10 mM Na2HPO4, pH 7.0); melting temperatures Tm, thermodynamic data of double helix formation and mass spectrometry dataa,b.
aTerms in parentheses display the thermodynamic data for buffer conditions of 1.0 M NaCl, 0.01 M Na2HPO4, pH 7.0.
bΔH° and ΔS° were determined from α versus T plots by curve fitting (hairpins) and from 1/T versus lnc plots (duplexes) (24–26).
cThermodynamic data for the duplex were derived from a 1/T versus lnc plot of the four concentrations 36, 88, 149 and 313 µM.
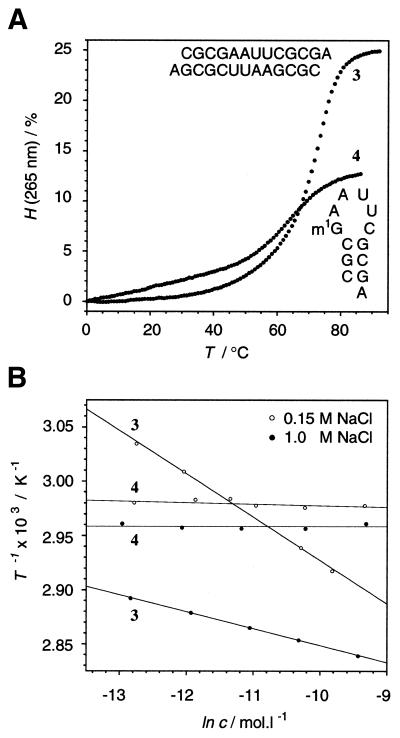
The same effect of methylation was found in a closely related sequence system having a 3′-dangling adenosine. rCGCGAAUUCGCGA 3 existed as a duplex, whereas rCGCm1GAAUUCGCGA 4 formed a hairpin structure in buffer solutions containing up to 1 M NaCl (Fig. 2). Surprisingly, and highly interestingly, in 150 mM NaCl buffer solutions a comparison of thermodynamic parameters of the unmethylated sequences rCGCGAAUUCGCG 1 and rCGCGAAUUCGCGA 3 showed that the latter sequence with a dangling nucleotide was more unstable. This is a rare example of a decrease in thermodynamic stability on attachment of a dangling nucleotide at the 3′-end of an oligoribonucleotide. Usually the attachment of a 3′-dangling nucleotide results in a profound increase in stability (25,26).
Figure 2.
(A) Melting profiles of rCGCGAAUUCGCGA 3 and rCGCm1GAAUUCGCGA 4 (3 µM, 1.0 M NaCl, 10 mM Na2HPO4, pH 7.0). (B) Dependence of melting transitions on strand concentration c of sequences 3 and 4, (0.15 M or 1.0 M NaCl, 10 mM Na2HPO4, pH 7.0): 1/T versus lnc plots. H, hyperchromicity; T, temperature.
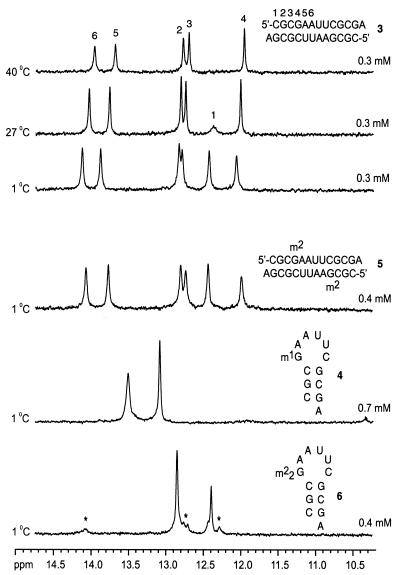
Although rCGCGAAUUCGCGA 3 did show a distinct dependence of melting temperature on concentration, we aimed at further proof of the exclusive occurrence of a duplex. In the 1H NMR proton spectra of rCGCGAAUUCGCGA 3 at a total strand concentration of 70 µM and in only 6 mM buffer solution without further addition of NaCl, six NH resonances were present, as expected for a symmetrical duplex structure of 12 bp (compare Fig. 3). One of the signals vanished at higher temperatures and was therefore assigned to the terminal base pair. Assignment of the remaining five NH protons was based on literature data for rCGCGAAUUCGCG (42).
Figure 3.
Selected 1H NMR (500 MHz) NH spectra: rCGCGAAUUCGCGA 3 (duplex, assignment according to 42), rCGCm2GAAUUCGCGA 5 (duplex), rCGCm1GAAUUCGCGA 4 (hairpin) and rCGCm22GAAUUCGCGA 6 (hairpin, asterisks indicate signals for competitive duplex formation to a minor extent). Total strand concentrations are as indicated. H2O/D2O, 9:1; sodium arsenate buffer, pH 7.4; 25 mM for 3, 5 and 6, 12.5 mM for 4)
The gel mobility data collected for sequences 1–4 were in accord with the observations described above (see Supplementary Material).
We considered the duplex of rCGCGAAUUCGCGA 3 at 150 mM NaCl as the most suitable sequence system for a comprehensive methylation study. On the one hand, with m1G we had a first example of a duplex–hairpin conversion. This conversion proceeded in up to 1 M NaCl concentrations. It was therefore not a specific property of a duplex with a small enthalpy value as observed for this sequence at 150 mM NaCl concentrations (Table 1). On the other hand, a duplex providing such a small enthalpy value enabled us to discover which methylated nucleosides cause significant stabilization of the duplex itself. This will be outlined in the following paragraph.
Methylated nucleotides that interfere with regular interbase Watson–Crick hydrogen bonding for conformational reasons (m6A, m2G and m4C)
The chemical constitution of the monomethylated nucleotides m2G, m6A and m4C allows the formation of a standard Watson–Crick base pair. However, the conformation of the methylated exocyclic amino functionality may interfere and has to be taken into account. In the case of m2G, studies at the monomer level demonstrated that rotation of the N2-methylamino moiety was not restricted at room temperature (43). Therefore, it can be assumed that formation of a Watson–Crick base pair is not significantly hindered. Indeed, when the guanosine at position 4 of rCGCGAAUUCGCGA was replaced by m2G we even found a significant stabilization by 7 kcal/mol of the corresponding duplex rCGCm2GAAUUCGCGA 5. Likewise, replacement of the adenosine at position 6 by m6A and replacement of the cytosine at position 9 by m4C resulted in a comparable stabilization of rCGCGAm6AUUCGCGA 10 and rCGCGAAUUm4CGCGA 13. This duplex stabilization by ribonucleotides monomethylated at their exocyclic amino functionality may well reflect a specific property of the rCGCGAAUUCGCGA system in 150 mM NaCl buffer solutions, not withstanding that Strobel and co-workers (44) recently acertained m2G to be iso-energetic to guanosine in other selected self-complementary RNA duplexes and in a GNRA hairpin loop.
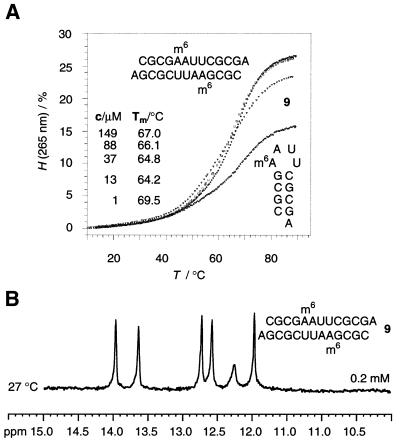
The higher duplex stability of rCGCGAm6AUUCGCGA 10 and rCGCGAAUUm4CGCGA 13 compared to that of rCGCGAAUUCGCGA 3 is not easily rationalized. From studies at the monomer level there is evidence that the methyl groups of m6A and m4C preferentially exist in the syn conformation (relative to N1 in the purine ring and relative to N3 in the pyrimidine ring) (43). Therefore, the formation of a Watson–Crick base pair involving m6A or m4C must be hindered to a small but definite extent and one is tempted to conclude that this hindrance would cause a decrease in pairing stability of the duplex. However, the preference for the syn conformation of the methyl group in m6A had an impact on the duplex–hairpin equilibrium when this nucleotide replaced the adenosine at position 5. At 1 and 3 µM concentrations rCGCGm6AAUUCGCGA 9 revealed monophasic melting profiles of low hyperchromicity with a melting transition at 69.5°C, indicating a monomolecular hairpin transition. At 13 µM strand concentrations we observed a sigmoid melting profile of significantly higher hyperchromicity with a transition temperature at only 64°C. At higher concentrations (up to 313 µM) the melting transition increased gradually and therefore reflected the occurrence of a bimolecular interaction, which is only compatible with a duplex structure. Accordingly, the six NH resonances found in the 1H NMR spectrum measured for a strand concentration of 0.2 mM corroborate the existance of a duplex (Fig. 4). The gel mobility assay of sequence 9 indicated predominantly duplex formation for a loading concentration of 45 µM in 100 mM NaCl, 2 mM MgCl2 and 100 mM Tris–HCl buffer, pH 7.4 (see Supplementary Material).
Figure 4.
(A) Examples of UV melting curves for the sequence rCGCGm6AAUUCGCGA 9. Strand concentrations <5 µM result in low hyperchromicites and invariable melting transitions of Tm = 69.5°C, indicating hairpin formation. Higher concentrations yield increased hyperchromicities and a linear 1/T versus lnc correlation (37–313 µM, Tm = 64.2–69.2°C), indicating duplex formation. 150 mM NaCl, 10 mM Na2HPO4, pH 7.0. (B) 1H NMR (500 MHz) NH spectrum of rCGCGm6AAUUCGCGA 9 at a concentration of 0.2 mM corroborates formation of a duplex (27°C, 25 mM arsenate buffer, pH 7.4).
However, the observation that at low strand concentrations (<5 µM) monomethylated adenosine was able to initiate the duplex–hairpin conversion is remarkable. To the best of our knowledge, this structure-determining feature of m6A has not been recognized so far and may enrich discussions about the reasons for frequent adenosine monomethylation in mRNA (45).
Methylated nucleotides that interfere with regular interbase Watson–Crick hydrogen bonding for constitutional reasons (m22G, m62A, m1G and m3U)
The chemical constitution of the methylated nucleotides m1G, m22G, m62A and m3U does not allow formation of a standard Watson–Crick base pair. For all nucleotide replacements with a representative of this nucleotide class we observed a duplex–hairpin conversion of our reference sequence rCGCGAAUUCGCGA 3. For instance, replacement of guanosine at position 4 by m22G, as well as replacement of adenosine at either position 5 or 6 by m62A resulted in the hairpin structures 6–8. A similar effect was observed for replacement of guanosine at position 4 by m1G and for replacement of uridine at either position 7 or 8 by m3U, as represented by oligoribonucleotides 4, 11 and 12. The number of NH resonances displayed in the 1H NMR spectra of 4, 6–8, 11 and 12 support the existence of hairpin structures (compare Fig. 3).
Two of the sequences mentioned in this paragraph, namely sequence 8 with m62A at strand position 6 and sequence 11 with m3U at strand position 7, showed a gradual appearance of a second melting transition at ∼35°C for strand concentrations >20 µM (see Supplementary Material). However, the 1H NMR spectra did not indicate formation of a duplex for these sequences. Interestingly, formation of duplex to a minor extent was observed in the case of rCGCm22GAAUUCGCGA 6 (Fig. 3).
Nucleotide replacement by 1-methylinosine (m1I)
Nucleotide replacement with m1I does not constitute a simple methylation of our reference sequence 3. Nevertheless, m1I was used in our study in order to shed light on the question of whether the hairpin rCGCm1GAAUUCGCGA is stabilized by a flipped base pair m1G-C within the hairpin loop involving N3 and N2-H of the purine and N4-H and N3 of the pyrimidine (46). Replacing the guanosine at position 4 with m1I closely resembled replacement with m1G but, due to the missing exocyclic amino group, a bidentate flipped base pair cannot be formed. These considerations may explain the observed lower stability of the hairpin rCGCm1IAAUUCGCGA 14 (Table 1). A NMR investigation of rCGCm1GAAUUCGCGA is currently underway in the laboratory of B. Jaun in order to obtain supporting structural information.
m1I can also be regarded as a model for m1A, a modification which is frequently found in cellular RNAs (4). Because of its reactivity and the easy hydrolyzation of the imino functionality, m1A is not suited to incorporation into oligoribonucleotides by the phosphoramidite approach and to subsequent standard deprotection procedures. Replacement of the adenosines at positions 6 and 7 by m1I did indeed result in hairpin structures, namely rCGCGm1IAUUCGCGA 15 and rCGCGAm1IUUCGCGA 16. It is tempting to postulate that m1A shows a similar behavior.
Comparison with DNA
Detailed structural investigations on alkylation at the nucleobases of dCGCGAATTCGCG and closely related sequences, namely dCGCGAm6ATTCGCG (47), dCGCm6GAATTCGCG (48), dCGCm6GAATTTCGCG (49) and dCGCm2IAATTCGCG (50), can be found in the literature. These sequences are observed as duplexes.
Nucleobase methylation versus mismatches in RNA
RNA shows great tolerance for mismatches within a double helical arrangement. Numerous reports provide detailed thermodynamic and structural data on mismatches, such as G·U, G·A, A·A, G·G, C·A, C·U and others. Two examples relevant to our studies are given here. Morse and Draper (41) investigated the sequences rCGCGGCGCG and rCGCGACGCG. These sequences have the possibility to form hairpins but exist as duplexes with a central mismatch. rGCGGCGC and rCGCGGCG, investigated by Kierzek et al. (51), also exist as duplexes.
Methylation of a nucleobase reduces the possibilities for hydrogen bonding and introduces a high steric demand. Both these features confine the number of energetically favorable mismatches between a methylated nucleobase and the former pairing partner. Consequently, the probability of retaining the duplex structure of a self-complementary sequence after methylation is decreased compared to after the introduction of a mismatch.
Methylation of a non-self-complementary duplex: the ribosomal helix 45 motif rGACCm2GGm62Am62AGGUC
The first part of our methylation study was based on a self-complementary sequence. Thereby, a single nucleotide replacement by a methylated counterpart inevitably generates a double interference in the corresponding duplex with methylated nucleobases located in opposite strands. This kind of methylation pattern is well known in DNA with respect to restriction–modification systems (52,53). In addition, we intended to expand our methylation studies to a non-self-complementary duplex.
Ribosomal helix 45 is the terminal helix at the 3′-end of the small subunit and is capped by a hypermethylated tetraloop, in which two successive m62A residues represent rRNA modifications that have been conserved from bacteria to eukaryotes. Choosing a helix 45 sequence analog of Bacillus stearothermophilus, rGACCm2GGm62Am62AGGUC 17, which has been structurally characterized by Rife and Moore (54), we were interested in the questions whether methylation helps to preserve this hairpin fold and whether the unmethylated sequence is forced into a duplex in the presence of the complementary strand. This situation mimics a folding trap that is avoided upon methylation. In this context, we also refer to recent computer simulations that demonstrate that RNA folding behavior is decisively improved by modifications (55,56).
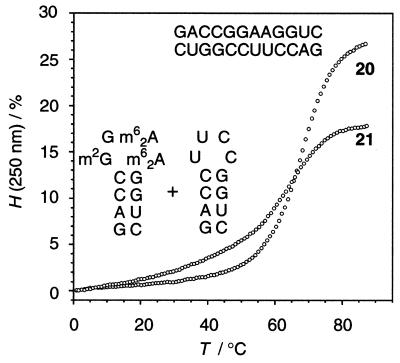
In solution the unmethylated sequence rGACCGGAAGGUC 18 formed an extra-stable stem–loop structure of the GNRA type. However, upon addition of the complementary dodecamer rGACCUUCCGGUC 19 formation of a duplex (20) was unequivocally ascertainable from the perfectly sigmoid shape of the melting profile and the concentration-dependent melting transition.
We then investigated mixtures of the methylated counterpart rGACCm2GGm62Am62AGGUC and rGACCUUCCGGUC 21. The low hyperchromicity and a melting transition at 64°C, comparable to that of the individual strands, support the assumption of the coexistence of two individual hairpins, at least for concentrations <5 µM. For concentrations >20 µM, the gradual occurrence of a second transition at ∼40°C indicated competitive duplex formation (Fig. 5).
Figure 5.
UV melting curves of rGACCGGAAGGUC/GACCUUCCGGUC 20 and rGACCm2GGm62Am62AGGUC/GACCUUCCGGUC 21. c = 3 µM. 150 mM NaCl, 10 mM Na2HPO4, pH 7.0.
Our current investigations focus on monomolecular helix 45 sequence constructs containing short complementary sequence partitions that are responsible for misfolds or two-state folding equilibria, as long as the sequences are unmethylated. These sequences fold into the correct hairpin structures upon methylation. We will report on that elsewhere.
CONCLUSIONS
In the present study we have investigated the effect of methylation on the self-complementary oligoribonucleotide duplex of rCGCGAAUUCGCGA. Site-specific single nucleotide replacements by the corresponding methylated counterparts, such as m22G, m62A, m1G and m3U, caused conversion of our reference duplex into a hairpin structure. In contrast, the monomethylated nucleotides m2G and m4C behaved as duplex-preserving modifications and even yielded higher duplex stabilities. Remarkably, the monomethylated nucleotide m6A demonstrated ambivalence with respect to duplex–hairpin conversion. At low strand concentrations our data indicated formation of a hairpin for the sequence rCGCGm6AAUUCGCGA, whereas at higher concentrations formation of a duplex was ascertained. Furthermore, the effects arising from the methylated ribonucleotides were not limited to a self-complementary sequence context. In analogy, we obtained experimental evidence that tandem methylation of two successive adenosine nucleotides in the ribosomal helix 45 motif, rGACCm2GGm62Am62AGGUC, prevents this hairpin structure from refolding into a duplex in the presence of the complementary sequence.
This basic study on RNA oligonucleotide methylation documents the potential of base-methylated nucleotides as primary determinants of RNA secondary structure.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This paper is dedicated to Prof. Heinz Falk (University of Linz) on the occasion of his 62nd birthday. R.M. thanks Prof. B. Kräutler (Innsbruck) for providing a new research home. We thank Dr R. Krishnamurthy (Scripps Institution, CA) for mass analyses, Prof. B. Auer and Prof. K. Bister (Innsbruck) for the HPLC system and C. Eichmüller and Prof. R. Konrat for their generous help concerning the NMR experiments in Innsbruck. Financial support from the Austrian Science Fund (P13216-CHE and P15042) and the Upper Austrian Government is gratefully acknowledged.
References
- 1.Crain P.F. and McCloskey,J.A. (1997) The RNA modification database. Nucleic Acids Res., 25, 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björk G.R. (1995) Biosynthesis and function of modified nucleosides. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 165–205.
- 3.Yokoyama S. and Nishimura,S. (1995) Modified nucleosides and codon recognition. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 207–223.
- 4.Motorin Y. and Grosjean,H. (1998) Appendix 1: Chemical structures and classification of post-transcriptionally modified nucleosides in RNA. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 543–549.
- 5.Grosjean H. and Benne,R. (eds) (1998) Modification and Editing of RNA. ASM Press, Washington, DC.
- 6.Simons R.W. and Grunberg-Manago,M. (eds) (1998) RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 7.Micura R., Pils,W. and Grubmayr,K. (2000) Bridged cyclic oligoribonucleotides as model compounds for codon-anticodon pairing. Angew. Chem. Int. Ed. Engl., 39, 922–926. [DOI] [PubMed] [Google Scholar]
- 8.Lane B.G. (1998) Historical perspectives on RNA nucleoside modifications. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 1–15.
- 9.Grosjean H., Björk,G. and Maden,B.E.H. (1995) Nucleotide modification and base conversion of RNA: summary and outlook. Biochimie, 77, 3–6.7541253 [Google Scholar]
- 10.Agris P.F. (1995) The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol., 53, 79–129. [DOI] [PubMed] [Google Scholar]
- 11.Meroueh M., Grohar,P.J., Qui,J., SantaLucia,J.Jr, Scaringe,S.A. and Chow,C.S. (2000) Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res., 28, 2075–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundaram M., Crain,P.F. and Davis,D.R. (2000) Synthesis and characterization of the native anticodon domain of E. coli tRNALys: simultaneous incorporation of modified nucleosides mnm5s2U, t6A and pseudouridine using phosphoramidite chemistry. J. Org. Chem., 65, 5609–5614. [DOI] [PubMed] [Google Scholar]
- 13.Derreumaux S., Chaoui,M., Tevanian,G. and Fermandjian,S. (2001) Impact of CpG methylation on structure, dynamics and solvation of cAMP DNA responsive element. Nucleic Acids Res., 29, 2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekine M. and Satoh,T. (1991) A convenient method for the synthesis of N2,N2-dimethylguanosine by reductive carbon-sulfur bond cleavage with tributyltin hydride. J. Org. Chem., 56, 1224–1227. [Google Scholar]
- 15.Agris P.F., Malkiewicz,A., Kraszewski,A., Everett,K., Nawrot,B., Sochacka,E., Jankowska,J. and Guenther,R. (1995) Site-selected introduction of modified purine and pyrimidine ribonucleosides into RNA by automated phosphoramidite chemistry. Biochimie, 77, 125–134. [DOI] [PubMed] [Google Scholar]
- 16.Agris P.F., Guenther,R., Sochacka,E., Newman,W., Czerwinska,G., Liu,G., Ye,W. and Malkiewicz,A. (1999) Thermodynamic contribution of nucleoside modifications to yeast tRNAPhe anticodon stem loop analogs. Acta Biochim. Pol., 46, 263–172. [PubMed] [Google Scholar]
- 17.Rife J.P., Cheng,C.S., Moore,P.B. and Strobel,S.A. (1998) The synthesis of RNA containing the modified nucleotides N2-methyl-guanosine and N6,N6-dimethyl-adenosine. Nucl. Nucl., 17, 2281–2288. [Google Scholar]
- 18.Avino A.M., Mayordomo,A., Espuny,R., Bach,M. and Eritja,R. (1995) A convenient method for the preparation of N2,N2-dimethylguanosine. Nucl. Nucl., 14, 1613–1617. [Google Scholar]
- 19.Miah A., Reese,C.B. and Song,Q. (1997) Convenient intermediates for the preparation of C-4 modified derivatives of pyrimidine nucleosides. Nucl. Nucl., 16, 53–65. [Google Scholar]
- 20.Grasby J.A., Singh,M., Karn,J. and Gait,M. (1995) Synthesis and applications of oligoribonucleotides containing N4-methylcytosine. Nucl. Nucl., 14, 1129–1132. [Google Scholar]
- 21.Zemlicka J. (1970) Nucleic acid components and their analogs. CXXXII. Alkylation of some nuclei acid components and their analogs with dimethylformamide acetals. Collect. Czech. Chem. Commun., 35, 3572–3583. [Google Scholar]
- 22.Pitsch S., Weiss,P.A., Wu,X., Ackermann,D. and Honegger,T. (1999) Fast and reliable automated synthesis of RNA and partially 2′-O-protected precursors (‘caged RNA’) based on two novel, orthogonal 2′-O-protecting groups. Helv. Chim. Acta, 82, 1753–1761. [Google Scholar]
- 23.Wu X. and Pitsch,S. (1998) Synthesis and pairing properties of oligoribonucleotide analogues containing a metal-binding site attached to β-d-allofuranosyl cytosine. Nucleic Acids Res., 26, 4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marky L. and Breslauer,K. (1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers, 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- 25.Xia T., Mathews,D.H. and Turner,D.H. (1999) Thermodynamics of RNA secondary structure formation. In Söll,D., Nishimura,S. and Moore,P. (eds), Comprehensive Natural Product Chemistry. Elsevier, Oxford, UK, Vol. 8, pp. 21–47.
- 26.Petersheim M. and Turner,D.H. (1983) Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp and ACCGGUp. Biochemistry, 22, 256–263. [DOI] [PubMed] [Google Scholar]
- 27.Hwang T.-L. and Shaka,A.J. (1995) Water suppression that works. Using excitation sculpting with gradients. J. Magn. Reson., 112A, 275–279. [Google Scholar]
- 28.Emsley L. and Bodenhausen,G. (1990) Gaussian pulse cascades: new analytical functions for rectangular selective inversion and in-phase excitation in NMR. Chem. Phys. Lett., 165, 469–476. [Google Scholar]
- 29.Geen H. and Freeman,R. (1991) Band-selective radiofrequency pulses. J. Magn. Reson., 93, 93–141. [Google Scholar]
- 30.Hald M., Pedersen,J.B., Stein,P.C., Kirpekar,F. and Jacobsen,J.P. (1995) A comparison of the hairpin stability of the palindromic d(CGCG(A/T)4CGCG) oligonucleotides. Nucleic Acids Res., 23, 4576–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xodo L.E., Manzini,G., Quadrifoglio,F., van der Marel,G.A. and van Boom,J.H. (1988) The duplex-hairpin conformational transition of d(CGCGCGATCGCGCG) and d(CGCGCGTACGCGCG): a thermodynamic and kinetic study. J. Biomol. Struct. Dyn., 6, 139–152. [DOI] [PubMed] [Google Scholar]
- 32.Kallick D.A. and Wemmer,D.E. (1991) 1H NMR of 5′CGCGTATATACGCG3′, a duplex and a four-membered loop. Nucleic Acids Res., 19, 6041–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh S., Patel,P.K. and Hosur,R.V. (1997) Structural polymorphism and dynamism in the DNA segment GATCTTCCCCCCGGAA: NMR investigations of hairpin, dumbell, nicked duplex, parallel strands and i-motif. Biochemistry, 36, 256–263. [DOI] [PubMed] [Google Scholar]
- 34.Pramanik P., Kanhouwa,N. and Kan,L. (1988) Hairpin and duplex formation in DNA fragments CCAATTTTGG, CCAATTTTTTGG and CCATTTTTGG: a proton NMR study. Biochemistry, 27, 3024–3031. [DOI] [PubMed] [Google Scholar]
- 35.Garcia A.E., Gupta,G., Soumpasis,D.M. and Tung,C.S. (1990) Energetics of the hairpin to mismatched duplex transition of d(GCCGCAGC) in NaCl solution. J. Biomol. Struct. Dyn., 8, 173–186. [DOI] [PubMed] [Google Scholar]
- 36.Lan T. and McLaughlin,L.W. (2001) Minor groove functional groups are critical for the B-form conformation of duplex DNA. Biochemistry, 40, 968–976. [DOI] [PubMed] [Google Scholar]
- 37.Kirchner R., Vogtherr,M., Limmer,S. and Sprinzl,M. (1998) Secondary structure dimorphism and interconversion between hairpin and duplex form of oligoribonucleotides. Antisense Nucleic Acid Drug Dev., 8, 507–516. [DOI] [PubMed] [Google Scholar]
- 38.Butcher S.E., Dieckmann,T. and Feigon,J. (1997) Solution structure of the conserved 16S-like ribosomal RNA UGAA tetraloop. J. Mol. Biol., 268, 348–358. [DOI] [PubMed] [Google Scholar]
- 39.Sich C., Ohlenschläger,O., Ramachandran,R., Görlach,M. and Brown,L.R. (1997) Structure of an RNA hairpin loop with a 5′-CGUUUCG-3′ loop motif by heteronuclear NMR spectroscopy and distance geometry. Biochemistry, 36, 13989–14002. [DOI] [PubMed] [Google Scholar]
- 40.Cabello-Villegas J. and Nikonowicz,E.P. (2000) Discriminating duplex and hairpin oligonucleotides using chemical shifts: application to the anticodon stem–loop of Escherichia coli tRNAPhe. Nucleic Acids Res., 28, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morse S.E. and Draper,D.E. (1995) Purine–purine mismatches in RNA helices: evidence for protonated G·A pairs and next-nearest neighbor effects. Nucleic Acids Res., 23, 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou S.H., Flynn,P. and Reid,B. (1989) Solid-phase synthesis and high-resolution NMR studies of two synthetic double-helical RNA dodecamers: r(CGCGAAUUCGCG) and r(CGCGUAUACGCG). Biochemistry, 28, 2422–2435. [DOI] [PubMed] [Google Scholar]
- 43.Engel J.D. and von Hippel,P.H. (1974) Effects of methylation on the stability of nucleic acid conformations: studies at the monomer level. Biochemistry, 13, 4143–4158. [DOI] [PubMed] [Google Scholar]
- 44.Rife J.P., Cheng,C.S., Moore,P.B. and Strobel,S.A. (1998) N2-methylguanosine is isoenergetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res., 26, 3640–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bokar J.A. and Rottman,F.M. (1998) Biosynthesis and functions of modified nucleosides in eucaryotic mRNA. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 183–200.
- 46.Murkard M.E., Turner,D.H. and Tinoco,I.Jr (1999) Appendix 1: Structures of base pairs involving at least two hydrogen bonds. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 675–680.
- 47.Frederick C.A., Quigley,G.J., van der Marel,G.A., van Boom,J.H., Wang,A.H.-J. and Rich,A. (1988) Methylation of the EcoRI recognition site does not alter DNA conformation: the crystal structure of d(CGCGAm6ATTCGCG) at 2.0-Å resolution. J. Biol. Chem., 263, 17872–17879. [DOI] [PubMed] [Google Scholar]
- 48.Sriram M., van der Marel,G.A., Roelen,H.L.P.F., van Boom,J.H. and Wang,A.H.-J. (1992) Structural consequences of a carcinogenic alkylation lesion on DNA: effect of O6-ethylguanine on molecular structure of the d(CGC[e6G]AATTCGCG)-netropsin complex. Biochemistry, 31, 11823–11834. [DOI] [PubMed] [Google Scholar]
- 49.Leonard G.A., Thomson,J., Watson,J.P. and Brown,T. (1990) High-resolution structure of a mutagenic lesion in DNA. Proc. Natl Acad. Sci. USA, 87, 9573–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang D., Gao,Y., Robinson,H., van der Marel,G.A., van Boom,J.H. and Wang,A.H. (1993) Structural effects of the C2-methylhypoxanthine:cytosine base pair in B-DNA: a combined NMR and X-ray diffraction study of d(CGC[m2I]AATTCGCG). Biochemistry, 32, 8672–8681. [DOI] [PubMed] [Google Scholar]
- 51.Kierzek R., Burkard,M.E. and Turner,D.H. (1999) Thermodynamics of single mismatches in RNA duplexes. Biochemistry, 38, 14214–14223. [DOI] [PubMed] [Google Scholar]
- 52.Leonhardt H., Rahn,H.-P. and Cardoso,M.C. (1999) Functional links between nuclear structure, gene expression, DNA replication and methylation. Crit. Rev. Eukaryot. Gene Expr., 9, 345–351. [DOI] [PubMed] [Google Scholar]
- 53.Tate P.H. and Bird,A.P. (1993) Effects of DNA methylation on DNA binding proteins and gene expression. Curr. Opin. Genet., 21, 163–167. [DOI] [PubMed] [Google Scholar]
- 54.Rife J.P. and Moore,P.B. (1998) The structure of a methylated tetraloop in 16S ribosomal RNA. Structure, 6, 747–756. [DOI] [PubMed] [Google Scholar]
- 55.Flamm C., Fontana,W., Hofacker,I.L. and Schuster,P. (2000) RNA folding at elementary step resolution. RNA, 6, 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wuchty S., Fontana,W., Hofacker,I.L. and Schuster,P. (1999) Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers, 49, 145–165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.