Abstract
Identifying biomarkers able to discriminate individuals on different health trajectories is crucial to understand the molecular basis of age-related morbidity. We investigated multi-omics signatures of general health and organ-specific morbidity, as well as their interconnectivity. We examined cross-sectional metabolome and proteome data from 3,142 adults of the Cooperative Health Research in South Tyrol (CHRIS) study, an Alpine population study designed to investigate how human biology, environment, and lifestyle factors contribute to people’s health over time. We had 174 metabolites and 148 proteins quantified from fasting serum and plasma samples. We used the Cumulative Illness Rating Scale (CIRS) Comorbidity Index (CMI), which considers morbidity in 14 organ systems, to assess health status (any morbidity vs. healthy). Omics-signatures for health status were identified using random forest (RF) classifiers. Linear regression models were fitted to assess directionality of omics markers and health status associations, as well as to identify omics markers related to organ-specific morbidity. Next to age, we identified 21 metabolites and 10 proteins as relevant predictors of health status and results confirmed associations for serotonin and glutamate to be age-independent. Considering organ-specific morbidity, several metabolites and proteins were jointly related to endocrine, cardiovascular, and renal morbidity. To conclude, circulating serotonin was identified as a potential novel predictor for overall morbidity.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75627-3.
Keywords: Health status, Metabolomics, Proteomics, Comorbidity, Aging, CHRIS study
Subject terms: Computational models, Statistical methods, Computational biology and bioinformatics, Biomarkers, Diseases, Medical research
Introduction
Non-communicable diseases (NCDs) are the leading cause of morbidity and premature mortality globally1. Age itself is the leading predictor for most NCDs: NCD prevalence increases with age and multiple diseases tend to cluster among older individuals2. Common biological processes triggered by molecular damage and modified by cellular and systemic responses drive biological aging and modify risks for multiple diseases in a tissue-, organ- and system-specific manner3. In turn, these health outcomes feed back into the underlying biological processes impacting the rate of aging and enhancing the risk for further disease4. It is therefore important to enhance our understanding of the molecular basis of age-related diseases to improve measures for disease prevention and general health5. Omics-based biomarkers provide insights into the molecular processes driving functional decline, they also help monitoring health trajectories, age-related physiological decline and disease onset6. Such biomarkers can also support the development of prevention strategies targeting those processes and provide surrogate endpoints in intervention studies5. The goal is early identification of individuals at higher risk of diseases, who will benefit most from such preventive interventions7.
Protein biomarkers have the advantage of being direct biological effectors of the underlying genomic background7. Serum metabolomics on the other hand provides a snapshot of general physiological state of an organism, and is influenced by genetic, epigenetic, and environmental factors8,9. For instance, Tanaka et al. (2020)7 identified a proteomic signature of aging involving 76 proteins and predicting accumulation of chronic diseases and all-cause mortality. You et al. (2023)10 developed a disease specific proteomic risk score, which stratified the risk for 45 common disease conditions, resulting into an equivalent predictive performance over established clinical indicators for almost all endpoints. Similarly, Gadd et al. (2024)11 demonstrated the utility of proteomic scores in predicting several 10-year incident outcomes beyond factors, such as age, sex, lifestyle and clinically relevant biomarkers, showing the relevance of early proteomic contributions to major age-related diseases. Pietzner et al. (2021)12 used untargeted metabolomics to investigate signatures of multimorbidity and found that 420 metabolites are shared between at least two chronic diseases. A recent work demonstrated the potential of metabolomic profiles as a multi-disease assay to inform on the risk of many common diseases simultaneously. For 10-year outcome prediction of 15 selected endpoints a combination of age, sex and the metabolomic state was equal or outperformed established predictors13.
Advances in different omics technologies and computing capabilities have also enabled the integration of multi-omics data to capture the complex molecular interplay of health and disease14. In TwinsUK, using data from 510 women, Zierer et al. (2016)15 integrated four high-throughput omics datasets and demonstrated the interconnectivity of age-related diseases by highlighting molecular markers of the aging process, which might drive disease comorbidities.
Here, we aimed to identify multi-omics signatures of general health among adult individuals using cross-sectional data from the population-based Cooperative Health Research in South Tyrol (CHRIS) study, an Alpine population study designed to investigate how human biology, environment, and lifestyle factors contribute to people’s health over time. We specifically included targeted serum metabolomics16 and plasma proteomics data17. The Cumulative Illness Rating Scale (CIRS) based Comorbidity Index (CMI)18, reflecting disease status and severity in 14 relevant organ systems, was used to assess health status by classifying individuals into having any morbidity (CMI ≥ 1) or being healthy (CMI = 0). We then applied predictive models using a random forest classifier to determine metabolite and protein markers of health status and investigated differences in abundances of the relevant markers using linear regression models. Further, overall health status is a broader condition affecting multiple organ systems, for which the combination of systems affected by morbidity can vary across individuals. We therefore complemented our analysis by linear regression models investigating associations and inter-dependencies between CIRS organ-domain specific morbidity, such as the endocrine-metabolic or the renal domain, and individual proteins or metabolites .
Results
The main analytic sample consisted of n = 3,142 adult individuals from the CHRIS study with available metabolomics and proteomics data. The AbsoluteIDQ® p180 kit from Biocrates (Biocrates Life Sciences AG, Innsbruck, Austria) was used for metabolite quantification in fasting serum samples16. The high abundance plasma proteome was determined using the Scanning SWATH mass spectrometry-based approach17. We first present the study sample’s main characteristics by health status (any morbidity vs. healthy) and the relationships between the co-occurring comorbidities. Next, we provide findings from the random forest (RF) analysis, which was complemented by multiple linear regression analyses to better evaluate actual differences in abundances of each significant metabolite and protein from the RF analysis by health status, independent of age and sex. Finally, we investigated associations between organ specific morbidity and all available metabolites and proteins using linear regression models.
Health status and characteristics of the study sample
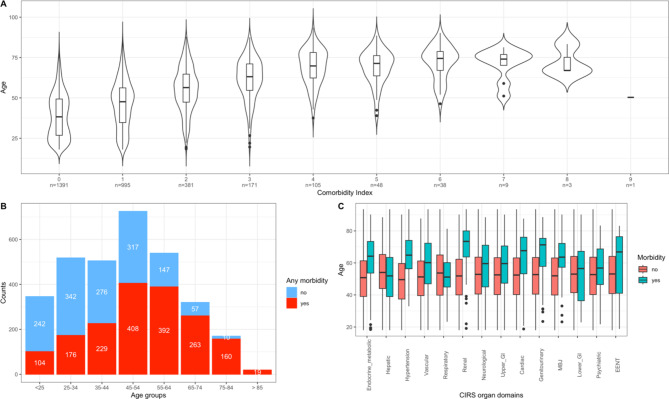
The characteristics for the main analytic sample are presented in Table 1. The CIRS organ domains most completely described by the available data (completeness ≥ 50%) were the hypertension, cardiac, respiratory, neurological, renal, vascular, endocrine-metabolic, hepatic and psychiatric/behavioral domains (Table 1). The remaining domains had < 50% completeness and in general a lower proportion of unhealthy individuals was observed for these domains (Table 1). Among all individuals, 56% (n = 1,751) were affected by at least one morbidity condition (CMI ≥ 1). As expected, these were on average older than healthy individuals (CMI = 0; Table 1; Fig. 1). The top five organ domains affected by health problems were the hepatic, vascular, hypertension, endocrine-metabolic and the respiratory domain, with 27.4%, 14.9%, 12.8%, 10.3%, and 8.8% of morbidity prevalence estimates, respectively. We additionally provide the characteristics for the CHRIS cohort, regardless of available omics data, to confirm the robustness of main characteristics and estimated disease prevalences in our analytic sample. These are reported in Supplementary Table S1 and Figure S1.
Table 1.
Characteristics of the main analytic sample with information on health status (any morbidity vs. healthy)a and CIRS domain specific morbidityb.
| Overall | Healthy | Any morbidity | Completeness c | |
|---|---|---|---|---|
| Sample size, n (%) | 3,142 | 1,391 (44%) | 1,751 (56%) | |
| Age, mean (SD) | 46.5 (16.7) | 39.0 (13.9) | 52.5 (16.3) | |
| Comorbidity Index, mean (SD) | 1.04 (1.34) | 0 | 1.86 (1.30) | |
| Sex, n (%) | ||||
| Females | 1,748 (55.6%) | 868 (62.4%) | 880 (50.3%) | |
| Males | 1,394 (44.4%) | 523 (37.6%) | 871 (49.7%) | |
| Morbidity in CIRS domains, yes (%) | ||||
| Hepatic | 861 (27.4%) | 861 (49.2%) | 59 | |
| Vascular | 467 (14.9%) | 467 (26.7%) | 62 | |
| Hypertension | 403 (12.8%) | 403 (23.0%) | 100 | |
| Endocrine-Metabolic | 325 (10.3%) | 325 (18.6%) | 74 | |
| Respiratory | 278 (8.8%) | 278 (15.9%) | 71 | |
| Upper Gastrointestinal | 168 (5.3%) | 168 (9.6%) | 44 | |
| Psychiatric and behavioral | 151 (4.8%) | 151 (8.6%) | 53 | |
| MBJd | 141 (4.5%) | 141 (8.1%) | 48 | |
| Renal | 132 (4.2%) | 132 (7.5%) | 59 | |
| Cardiac | 110 (3.5%) | 110 (6.3%) | 71 | |
| Neurological | 82 (2.6%) | 82 (4.7%) | 66 | |
| Lower Gastrointestinal | 67 (2.1%) | 67 (3.8%) | 44 | |
| Genitourinary | 58 (1.8%) | 58 (3.3%) | 33 | |
| EENTe | 11 (0.4%) | 8 (0.6%) | 32 |
aHealth status was assessed through the Cumulative Illness Rating Scale (CIRS) Comorbidity Index (CMI) by classifying individuals as having any morbidity (CMI ≥ 1) or being healthy (CMI = 0). bCIRS domain specific morbidity is defined as having a score ≥ 2 in the given domain. cCompleteness presents the coverage of the necessary information available in the CHRIS Study with regard to the CIRS guidelines expressed in percentages. dMBJ=Musculoskeletal, bones and joints. eEENT= Ears, eyes, nose and throat.
Fig. 1.
Distribution of health status and age in the main analytic sample. (A) Age distribution (y-axis) by Comorbidity Index values, which range from 0–9 (x-axis). The x-axis additionally provides information on the total number of participants with each corresponding index value. The Comorbidity Index value expresses the total number of CIRS organ domains scoring ≥ 2. (B) Absolute distribution of “Any morbidity” (no, yes) by age group. Red bars represent the number of participants with any morbidity in the respective age group, whereas the number of healthy subjects is shown in blue in this stacked bar plot (C) Age distribution by morbidity conditions (no, yes) in the specific CIRS domains.
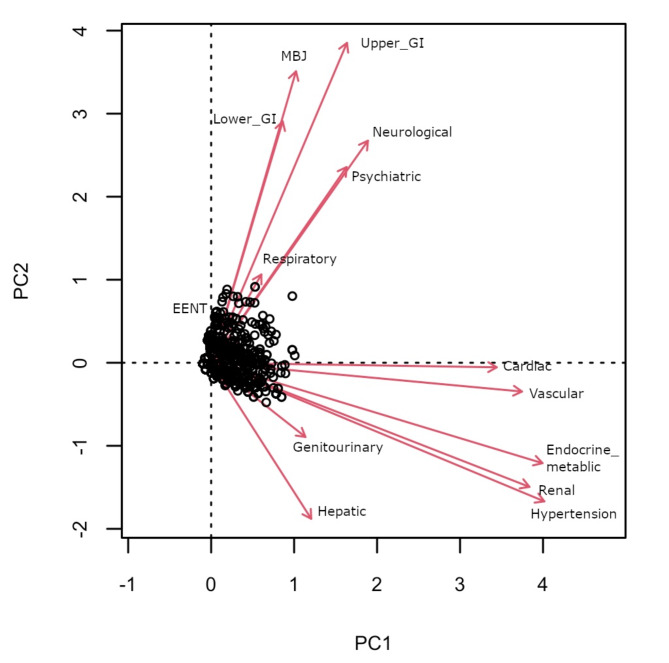
Next, we explored relationships between the 14 CIRS domains through ordinary correspondence analysis (OCA; Fig. 2), for which information was encoded as a binary variable in the analysis (no morbidity, morbidity). We observed proximity between the hypertension, renal and endocrine domains, the cardiac and the vascular domains, and between the neurological and the psychiatric domains. To compare the robustness of the comorbidity relationships we present the OCA analysis results for the CHRIS cohort in Supplementary Figure S2.
Fig. 2.
Biplot of ordinary correspondence analysis presenting relations between the 14 CIRS domains. Information on each domain was encoded as a binary variable (no morbidity, morbidity). Each data points represents one individual. CIRS domains with a stronger relation have longer (size consistency) and closer (direction consistency) loadings.
Multi omics signatures of health status
To avoid confounding of results due to the impact of medication17, we performed the analysis on metabolite and protein abundances adjusted for use of medications that were not considered in the CIRS definition (Supplementary Table S2).
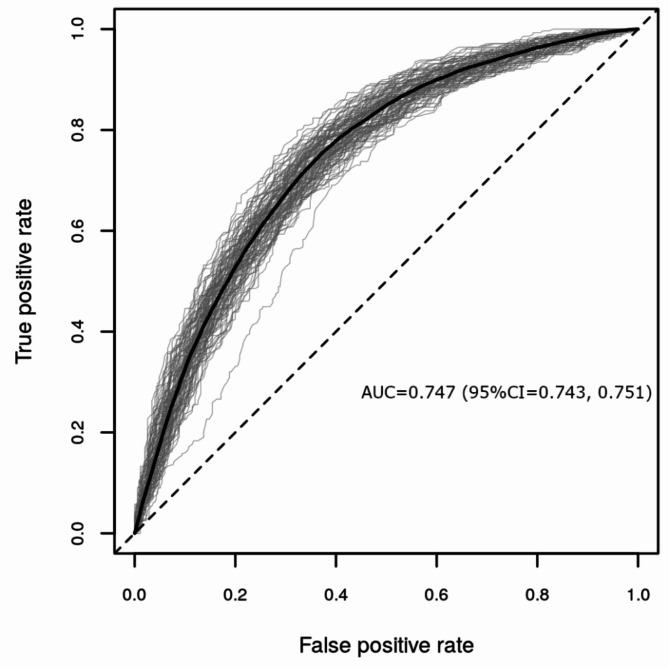
For the RF analysis we built a model including age, sex, 174 metabolites and 148 proteins as predictors. Overall, NModel = 100 RF models were generated, each containing NTree = 500 trees per model, using repeated random subsampling with 80% training and 20% validation set sizes, respectively. We compared the performance measures using the area under the receiver operating curve (ROC AUC), as well as the Matthew’s correlation coefficient (MCC) and MCC-F1. The RF model showed moderate performance (AUC = 0.747, 95% Confidence Interval (CI) = 0.743, 0.751; Fig. 3; MCC and MCC-F1 are presented in Supplementary Figure S3). In addition, we built models that included varying sets of the predictors, which were (a) age and sex, (b) age, sex and metabolites, (c) age, sex and proteins, and (d) age, sex, metabolites, proteins. When comparing differences in mean AUCs and mean MCCs, the metabolomics/proteomics-based models (b, c,d) generally showed greater performance over the model including age and sex only.When comparing the different omics-based models, statistically significant differences in performance were found, but these differences were not robust across the two performance measures AUC and MCC. A detailed performance comparison of these models is presented in Supplementary Text S1.
Fig. 3.
Performance evaluation of NModel = 100 random forest models including as predictors age, sex, 174 metabolites and 148 proteins to classify health status (any morbidity vs. healthy) represented as ROC AUCs. In the plot each validation run is shown as a gray line with the average curve shown in black.
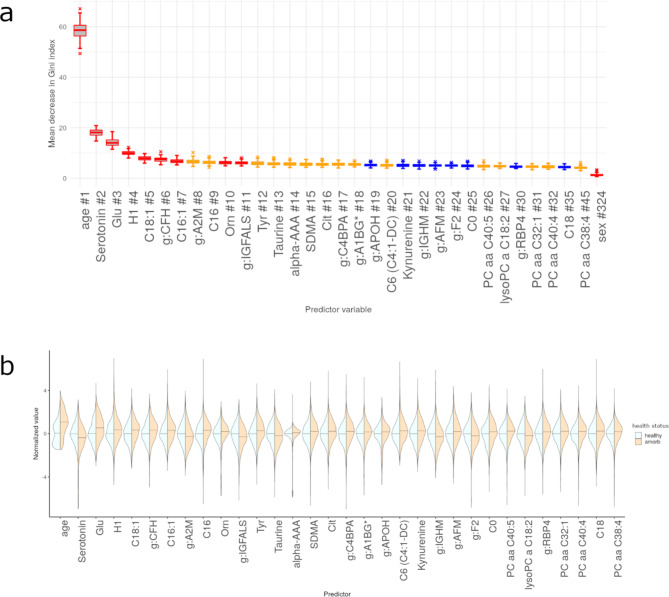
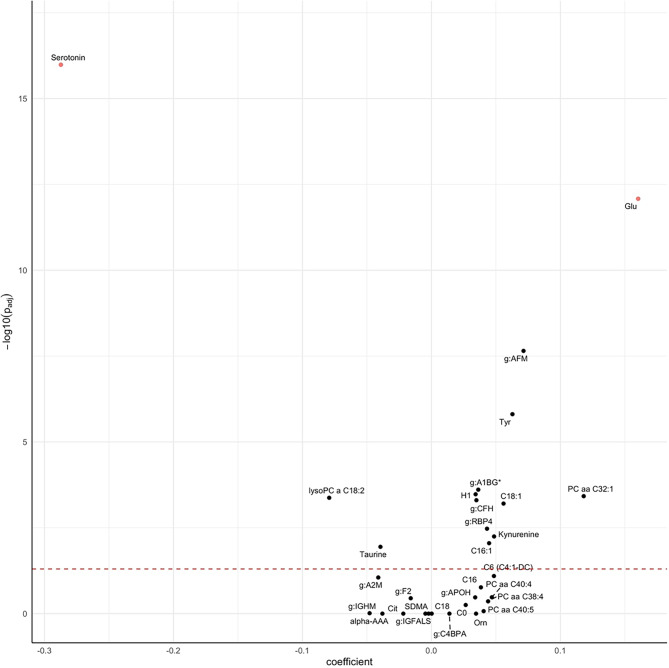
We next selected individual features (i.e., age, sex, metabolites or proteins) based on their importance (as expressed as mean decrease in Gini Index) and computed p-values for each feature’s Gini Index from an empirical background distribution. We selected a feature if it was at least 50 times significant at the level of α = 0.05 and obtained 33 features, including 21 metabolites and 10 proteins, with age being the most important and sex the least relevant for predicting health status (Fig. 4a). Among the top ten omics markers were the metabolites serotonin, glutamate, hexose, three acylcarnitines (C18:1, C16:1, C16), ornithine, and the proteins CFH, A2M and IGFALS. The medication-adjusted abundance distributions of these metabolites and proteins stratified by health status are presented in Fig. 4b. Individuals with any morbidity had lower mean abundance of serotonin, taurine and lysoPC C18:2, and higher mean abundance of all other metabolites. Regarding proteins, individuals with any morbidity had lower mean abundances of A2M, IGFALS, IGHM, and F2, and higher abundance of CFH, C4BPA, A1BG, APOH, AFM, and RBP4.
Fig. 4.
Evaluation of importance for significant features identified through the random forest models including as predictors age, sex, 174 metabolites and 148 proteins, for health status (any morbidity vs. healthy). #: Rank of the specific feature. Predictor significance was estimated by permutation testing, which generates NModel = 100 background distributions, yielding 100 p-values for each predictor variable. Box plots are color-coded by the number of times a p-value was significant for such a variable (p < 0.05): red, all 100 runs returned a significant p-value; orange, between 80 and 99 runs returned a significant p-value; blue, between 50 and 79 runs returned a significant p-value. Panels are restricted to features that are significant in at least 50% of all runs. Density plots are scaled to have the same width, a median of zero (0) (horizontal bar) and standard deviation of one (1) for the healthy group. For comparability, the any morbidity group data have been scaled to the standard deviation of the healthy group. (a) Mean decrease in Gini Index for significant features. (b) Violin plots presenting scaled abundance distributions stratified by health status for significant features identified through RF, ordered by importance from left to right.
To better characterize the actual association between health status and each significant metabolite and protein from the RF analysis, we performed separate regression analyses with their abundance as the response variable and health status (any morbidity vs. healthy), age and sex as the explanatory variables, allowing us to evaluate the differences in abundances of these markers by health status independently of age or sex. Coefficients for health status were extracted from these models and are presented in Fig. 5 and Supplementary Table S3. In order to account for multiple hypothesis testing, we applied a Bonferroni correction by multiplying the p-values by the number of performed tests (n = 31 metabolites and proteins). Metabolites and proteins were considered relevant if their adjusted p-value was smaller than 0.05 and if the difference in abundance was larger than the data set-specific observed technical variance of that marker (see Methods for details). Individuals with any morbidity had, on average, 22% lower mean abundance of serotonin and 12% higher abundance of serum glutamate. Twelve other markers (C18:1, C16:1, lyso PC a C18:2, PC aa C32:1, tyrosine, taurine, hexose, kynurenine, AFM, CFH, RBP4, and A1BG) passed the multiple-testing correction, however, the observed differences in abundances did fall within the range of the technical variability and were thus not considered significant16. Given that age was the strongest predictor for health status in the RF model, we further investigated and compared the coefficients for age and health association from the regression models. For some markers, such as citrulline, abundances were almost entirely explained by age and the coefficient for the association with health status from the regression model was only very small, and not significant. For others, such as serotonin and glutamate, associations with health status were strong, even in these age-adjusted models, suggesting an age-independent association of these metabolites with health.
Fig. 5.
Volcano plot for the differential abundance of metabolomic and proteomic markers of health status (any morbidity vs. healthy). Coefficients represent the log2-difference in average concentrations between the groups.
Metabolomic and proteomic signatures related to CIRS domain specific morbidity
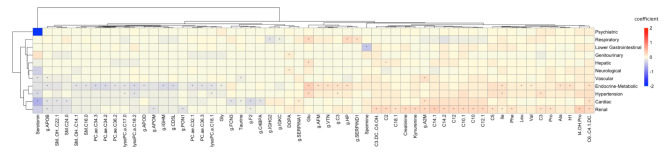
We additionally evaluated associations between CIRS organ-specific morbidity and all available metabolites and proteins. To do so, we implemented separate regression models for medication adjusted abundances of all 174 metabolites and 148 proteins as response variable and with each CIRS domain as well as age and sex as explanatory variables, and evaluated whether markers were shared across - or specific for any domain (Fig. 6; Supplementary Table S4). In total, 83 significant omics-disease associations were identified, passing both significance criteria (Bonferroni correction for multiple testing, consideration of technical variability), with 40 metabolites and 17 proteins being significant for ≥ 1 CIRS domain. Associations were observed with the cardiac, vascular, hypertension, endocrine-metabolic, renal, hepatic, psychiatric, neurological, respiratory and lower gastrointestinal and genitourinary domains (Fig. 6). Eleven metabolites (serotonin, glutamate, isoleucine, taurine, dihydroxyphenylalanine, several glycerophospholipids and acylcarnitines) and three proteins (F2, C3, A2M) were shared across multiple domains. For example, serotonin was related to the cardiac, vascular, hypertension and psychiatric systems, and glutamate to the hypertension, endocrine-metabolic, respiratory and hepatic domains. The proteins F2 and A2M were related to the cardiac, vascular, and the renal domains, and C3 to the hypertension and endocrine-metabolic domains. Overall, phosphatidylcholines and sphingolipids were all negatively associated with morbidity conditions in the related CIRS domain, whereas for acylcarnitines positive associations were observed. The directionality of biogenic amines and amino acids was not consistent across classes but remained consistent across CIRS domains. For proteins, negative associations were observed with APOB, APOD, APOM, IGHM, CD5L, PON1, FCN3, F2, C4BPA, IGHG2 and IGKC, whereas positive associations were found with SERPINA1, AFM, VTN, C3, HP, SERPIND1 and A2M. In general, all metabolites and proteins that were significantly associated with multiple domains showed consistent effect directions, being either negative or positive.
Fig. 6.
Heatmap presenting associations between metabolites, proteins and CIRS organ domains. Associations were obtained from multiple linear regression models adjusted for age and sex. Hierarchical clustering using the Euclidean distance as the similarity measure was applied by clustering rows and columns based on the coefficients describing the associations between metabolites and proteins with each corresponding CIRS organ domain. Only metabolites and proteins that were significantly associated with at least one specific CIRS domain are presented. Asterisks (*) highlight significant associations. β-coefficients are color-coded as follows: red hues represent positive relations and blue hues represent negative relations.
Discussion
In this cross-sectional analysis of CHRIS study data we identified age and 31 metabolites and proteins predicting overall health status (any morbidity vs. healthy) in adults using a random forest classifier. These markers included 21 metabolites (5 biogenic amines; 4 amino acids; 6 acylcarnitines; 5 glycerophospholipids; and hexose monosaccharides), and 10 plasma proteins. Subsequent regression analyses confirmed a sizeable association of health status with both serotonin and glutamate, which was independent of age and sex. Analyses on single CIRS domains further identified multiple metabolite- and protein-disease associations, most being related to cardiovascular, hypertension, endocrine-metabolic and renal morbidity, revealing strong molecular interconnectivity across these related domains.
Several studies have investigated omics markers of aging, longevity6,19, aging-related chronic diseases7,11–13,20,21, but only few have integrated multi-omics data with regards to general health assessment15,22. Independent of the approach used, great heterogeneity exists in the included omics technologies, protein and/or metabolite coverage, analytic tools as well as the outcome of interest, which makes comparison of studies challenging. In our study, multi-omics RF models identified several metabolites and proteins relevant for predicting health status (any morbidity vs. healthy).
Among the age-independent predictive metabolites was serotonin, which is involved in the regulation of energy, glucose and lipid metabolism. Changes in the serotonin system are known risk factors for many age-related diseases, such as diabetes and cardiovascular disease23,24, which was also observed in our study. Up to 95% of serotonin is produced in the gut, and only 5% of serotonin is synthesized by neurons, mainly in the central nervous system24. Although serotonin does not cross the blood-brain barrier, intestinal serotonin release causes neuronal activation in the brain stem, thus indirectly affecting the brain23. No other study has linked serotonin as a healthy aging marker per se. However, serotonin is a tryptophan derivative, and as inflammation and stress activate the tryptophan metabolism through the kynurenine pathway25, this consequently causes decreased production of serotonin26. The age associated upregulation of kynurenine and downregulation of serotonin therefore indicate a relevant role of tryptophan metabolism in inflammaging and aging27. Although tryptophan was included in the metabolomics panel in this study, it was not identified among the selected metabolites in the RF model. These results indicate a robust relation between health and circulating serotonin levels, but the role of tryptophan metabolism and related pathways, and the putative causal relationships deserves further investigation.
Glutamate was also identified as an age-independent predictor for health status. This amino acid has been previously associated with physical frailty in elderly individuals28, supporting our findings. It has further been linked to cardiovascular disease and has been suggested to be a potential biomarker of abdominal obesity and metabolic risk29. Glutamate is an important excitatory neurotransmitter in the brain, and although concentrations in the brain are much higher than in plasma, as the blood brain barrier is not very permeable to glutamate30, it is also one of the most abundant amino acids in the liver, kidney and skeletal muscle, showing great metabolic versatility31. Glutamate plays a key role in protein synthesis and degradation32 and is a by-product of the catabolism of branched chain amino acids29. By linking amino acid and carbohydrate metabolism glutamate supports energy production, which has further implications for insulin secretion32. More specifically, glutamate is also a source of alpha-ketoglutarate, which plays a key role in energy metabolism and aging processes and that has been implicated in improved life and health span33–36. In this study the morbidity group is associated with higher levels of circulating glutamate, which might reflect a depletion of alpha-ketoglutarate relative to healthy individuals.
Other relevant markers (metabolites hexose, C18:1, C16:1, lyso PC a C18:2, PC aa C32:1, tyrosine, taurine, kynurenine, and proteins AFM, CFH, RBP4, A1BG) were determined with the RF model, and the age independent associations confirmed by linear regression, but their difference in abundances was lower than the technical variance observed for these markers in the present data set. We provide further discussion of those markers in Supplementary Text S2.
When looking into organ-specific morbidity, most health conditions in our study were related to the hepatic domain, followed by the vascular, hypertension, endocrine-metabolic and the renal domains. Using OCA, we observed closer relatedness between the renal, hypertension and endocrine-metabolic, as well as the vascular and cardiac domains. Our analyses linking omics markers to specific CIRS domains supported such connections. For example, we observed the metabolites serotonin, glutamate, taurine, isoleucine, three glycerophospholipids (lysoPC a C17.0/18.1/18.2), and two acylcarnitines (C3, C6 (C4)-1-DC) and the proteins C3, APOB, F2, A2M and HP as common markers among the cardiac, vascular, hypertension, endocrine-metabolic, renal but also the respiratory domains. The co-occurrence of type-2 diabetes and cardiovascular diseases is very common, and its high degree of connectivity has been found with other diseases as well37. In addition, cardiovascular and kidney disease are closely interrelated and disease of one organ is known to cause dysfunction of the other38. Remaining molecular signatures were domain specific showing no connections among each other, such as the proteins VTN, APOD and a set of other amino acids (valine, leucine, alanine) being only related to the endocrine-metabolic domain. Such associations have also been reported previously in the literature39–41. Untargeted metabolomics analysis in the prospective, population-based EPIC-Norfolk study identified 420 metabolites that were shared among two or more chronic diseases12, further observing high connectivity among cardiometabolic and respiratory diseases across different biochemical classes of metabolites. Those findings highlight potential biological pathways related to the onset of multiple chronic diseases, such as liver and kidney function, lipid and glucose metabolism and low-grade inflammation, among others12.
Strengths of our investigation are the wealth of the available data resource, foremost with the availability of both metabolomics and proteomics data among the same participants. Additionally, available phenotypic parameters, both quantitative (such as blood parameters), self-reported or collected by trained study-nurses allowed a detailed characterization of the participants and enabled the assessment of the health status through the CIRS guidelines, which has been shown to be a useful tool to measure morbidity in clinical research42.
A first limitation is given by the study design, due to which participants with severe morbidity might have been underrepresented. In addition, for some of the CIRS domains only limited information in the CHRIS data was available, which led to a low completeness (< 50%) with respect to the CIRS guidelines for the EENT, genitourinary and the gastrointestinal domains. These domains were therefore affected by strong case-control imbalance, which might limit the power to detect any differences in mean abundances of metabolites and proteins. Moreover, given the low data completeness for these domains we cannot exclude any potential misclassification of cases and controls. In addition, metabolite and protein concentrations might be influenced by lifestyle, hormonal changes such as introduced by menopausal status and its treatment, as well as medication. Available information on medication use, irrelevant for the CIRS assessment, allowed adjustments to exclude spurious influence of this factor on the results. However, as the CIRS itself considers medication status to characterize certain diseases, it was not possible to distinguish whether the observed associations were driven by the disease itself or by the treatment for the given or related diseases. The present analyses are also limited to the set of metabolites and proteins that are possible to quantify by the analytical approaches used. Finally, given the cross-sectional design of the study we are only able to assess associations, hence no conclusions on temporal antecedence and causality can be drawn. Finally, we did not validate our models on an independent testing set. Despite these limitations, we were able to replicate several findings from previous studies, which supports reliable data and procedural quality within this study. Overall, using multi-omics data for profiling health status has great potential to identify changes in health trajectories at an earlier stage in life. This could help to develop new, effective target therapies for treating related as well as seemingly unrelated diseases occurring at the same time by uncovering common biological pathways connecting different underlying pathogenic mechanisms43.
To conclude, we identified several molecular signatures of overall health status. Specifically, circulating serotonin is suggested as a promising novel predictor for health and morbidity independent of age, implicating a potential key role of tryptophan metabolism and serotonin related pathways in sustaining health. The results also point to glutamate as another predictor for health and morbidity in adults, in agreement with previous studies relating this amino acid to frailty, metabolic and cardiovascular health. Future studies are needed to investigate the mediating role of these signatures in relation to lifestyle and the environment to promote healthy aging. In this regard the application of mendelian randomization approaches should be considered to further investigate causal links between circulating serotonin, serotonin metabolism and chronic disease.
Methods
Study cohort
The CHRIS study is a population-based cohort of 13,393 adults aged 18 and over recruited from 13 towns in the alpine Val Venosta/Vinschgau district in the Bolzano-South Tyrol province of northern Italy. The study was designed to investigate the genetic and molecular basis of age-related common chronic conditions and their interaction with lifestyle and environment in the general population44.
The study was approved by the Ethics Committee of the Healthcare System of the Autonomous Province of Bolzano. The study conforms to the Declaration of Helsinki, and with national and institutional legal and ethical requirements.
Metabolomics data were available for n = 6,415 individuals and proteomics data for n = 3,541. After excluding participants who were not fasting (n = 475) or had missing information on fasting status (n = 2), as well as women who were pregnant (n = 25) or unsure about pregnancy (n = 9), n = 3,142 individuals with overlapping omics data were available for analysis.
Data collection
Data, including collection of anthropometric measurements, blood and urine standard laboratory tests, blood pressure measurement and lifestyle information, were collected at the study center following overnight fasting44. Laboratory test data included all main cardiovascular and metabolic risk factors, and markers of iron metabolism, coagulation, renal damage, thyroid, and liver function. Blood (serum and plasma) and urine samples were collected and stored in a biobank.
To obtain information on disease history, participants were interviewed by trained study assistants. Specific clinical domains covered by the questionnaires were the circulatory and nervous system, as well as psychiatric disorders, cognition, autonomic and genitourinary function, endocrine, nutritional, and metabolic diseases. A detailed overview of the mode of assessment can be found elsewhere44. In addition, each questionnaire contained a section for “other diseases”, where participants could report any other condition not explicitly included in the domains as free text. Detailed medication information was collected by scanning the barcodes of the boxes of the medication used within the seven days prior to study center visit and brought by the study participants to the interview. The Anatomical Therapeutic Chemical (ATC) medicinal product classification coding system45 and the mode, frequency, and duration of drug administration was recorded for each scanned medication.
Assessment of the health status
To assess health in the CHRIS study sample, we used the Cumulative Illness Rating Scale (CIRS), which is a clinically relevant tool to measure the chronic medical illness burden by taking the number and the severity of chronic diseases into account46. For the purpose of this study we used the revised CIRS18, which assesses 14 organ related domains rating each of them according to the degree of severity ranging from: grade 0 (no impairment) to grade 4 (extremely severe impairment). Based on this guideline, we determined for all CHRIS participants the CIRS score for each domain by screening our questionnaire and interview data, as well as laboratory and clinical parameters for any given medical condition. We next derived the CIRS Comorbidity Index (CMI), which calculates the total number of CIRS organ domains with a score ≥ 2 (domains with moderate or severe morbidity). To define health status we classified individuals as being healthy if the CMI = 0 and as having any morbidity if CMI ≥ 1, which means that at least one CIRS organ domain was identified with moderate or severe morbidity. We additionally considered organ specific health by classifying individuals as being healthy if the CIRS organ domain scored < 2, or having morbidity if the respective domain scored ≥ 2.
We further assessed the completeness of the CIRS score with regard to the guidelines18 based on available CHRIS data for each domain. Detailed information on the completeness assessment is presented in Supplementary Text S3.
Metabolite and protein quantification
The AbsoluteIDQ® p180 kit from Biocrates (Biocrates Life Sciences AG, Innsbruck, Austria) was used for metabolite quantification in fasting serum samples. Details on data generation, quality assessment and normalization are provided in Verri Hernandes et al. (2022)16. In total, concentrations of 175 metabolites and lipids were quantified. Due to the large number of missing values, sarcosine was excluded from the present analysis. An overview of the included metabolites is presented in the Supplementary Table S5. According to common practice in quantitative proteomics/metabolomics analyses, abundances were log2-transformed prior to any data analysis to stabilize the variance and result in signal distributions that resemble more closely a normal distribution.
The high abundance plasma proteome was determined using the Scanning SWATH mass spectrometry-based approach. Details on sample processing, data acquisition and normalization are provided in Dordevic et al. (2024)17. In total 148 highly abundant proteins were quantified and included in this analysis. An overview of the included proteins is presented in Supplementary Table S6.
Metabolite and protein abundances were adjusted for frequent medication use with a linear model based approach: models were fitted separately for each metabolite or protein with their abundance as response, and medication (as individual binary variables) as explanatory variables. Only medications taken at least twice per week and not considered for CIRS scoring were used. The residuals from these models were used to construct medication-independent abundances for the RF and subsequent regression analyses. Supplementary Table S2 lists the medications for which abundances were adjusted.
Statistical analysis
Differences in characteristics were presented as mean (standard deviation, SD) for continuous variables and as percentages for categorical variables.
To investigate the relations between CIRS-domain specific morbidity we encoded binary data for each domain as 0 (no morbidity) and 1 (morbidity) and performed ordinary correspondence analysis (OCA) using the rda function in the vegan package for R47. OCA is a technique used to visualize relationships between categorical variables creating a contingency table of their joint frequencies. Then, the chi-square distance between the row and column profiles is calculated to assess the association between categories. The chi-square distance matrix is decomposed using singular value decomposition, which is further used to create two-dimensional plots equivalent to PCA plots for continuous variables48.
Random forest and linear regression analysis
We used a random forest (RF) classification model to predict health status (any morbidity vs. healthy), including as predictors age, sex, 174 metabolites and 148 proteins, respectively. Overall, NModel = 100 RF models were generated, each containing NTree = 500 trees per model, considering 80% training set sizes. Predictions were validated by NModel = 100 times stratified repeated random subsampling with 20% test set sizes, such that the ratio between the healthy and the any morbidity group of the original set sizes remained the same for the subsampled sets. We used the randomForest R package49 to perform the RF analysis.
Model performance was measured based on the receiver operating characteristic (ROC) area under the curve (AUC), the Matthew’s correlation coefficient (MCC), and the unit-normalized Matthews correlation coefficient (MCC-F1).
The Gini Index was used to measure feature importance. We further estimated the significance of the Gini Indices for the random forest models by permuting the response variable using the rfPermute R package50. This generates a background distribution by calculating NPerm = 100 random forests, one for each permuted response variable, each with NTree = 500 trees. Usually, a background distribution is calculated only once; however, to increase the robustness of our results, we created a background distribution for each validation run, yielding Nmodel = 100 p-values for each predictor variable. We selected a predictor if in ≥ 50 out of the 100 models the predictor was significant at the level α = 0.05.
To further investigate associations between health status and the significant metabolites and proteins obtained from the RF analysis we fitted separate multiple linear regression models with abundances of each metabolite and protein as the response variable, and with health status, age and sex as the explanatory variables The resulting p-values were adjusted for multiple hypothesis testing using the Bonferroni method. After correction, results were considered statistically significant if the adjusted p-value remained below the conventional significance level of α = 0.05. For the present data sets we additionally had the advantage of repeated measurements of quality control (QC) samples, which allowed us to estimate the technical variance of each individual metabolite or protein in the analyzed data set. We therefore calculated the coefficient of variation (CV) for each metabolite and protein and used these as additional criteria in the selection of significant features: In addition to the statistical significance, we required for metabolites that the observed difference in abundances was at least two times larger than the technical variance and for proteins one time larger than the technical variance. While these thresholds were chosen arbitrarily, they reflect the difference in variability observed for the proteomics and metabolomics data sets, and this more stringent significance criterion ensured selection of the most reliable and consistent metabolites/proteins in the present data set.
Investigation of omics-markers related to CIRS domains
We additionally investigated associations between metabolites, proteins and the specific CIRS domains using again multiple linear regression models. We fitted separate linear regression models for each metabolite or protein using their abundances as response variable and each CIRS domain, age and sex as the explanatory variables.
Similarly to the previous analysis, we applied a Bonferroni correction by multiplying the p-values by the number of tests (metabolites:14 CIRS domains*174 metabolites; proteins: 14 CIRS domains*148 proteins), and considered results to be significant if the adjusted p-value remained below the conventional significance level of α = 0.05 and the average difference in abundance was larger than the technical variance, as described in the previous section.
All analyses were conducted using the R statistical software, version 4.1.0 (www.R-project.org).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
CHRIS study investigators thank all study participants, the Healthcare System of the Autonomous Province of Bolzano-South Tyrol, and all Eurac Research staff involved in the study. Bioresource Impact Factor Code: BRIF6107. The authors thank the Department of Innovation, Research University and Museums of the Autonomous Province of Bozen/Bolzano for covering the Open Access publication costs.
Author contributions
All authors contributed to the study conception and design. Material preparation and data collection were performed by J.R, V.H.H, F.D, P.P, M.G, L.F, C.P, F.A, V.F, M.R, M.M and E.H. Formal analysis and investigation was performed by C.W, E.H, N.D, J.R and F.D. All authors contributed to the interpretation of the findings. The first draft of the manuscript was written by E.H, C.W, N.D, J.R and F.D and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
The data that support the findings of this study are not openly available due to reasons of sensitivity. Individual level CHRIS study data can be requested for research purposes by submitting a dedicated request to the CHRIS Access Committee. Please visit https://chrisportal.eurac.edu/ for more information on the process or contact the corresponding author.
Declarations
Competing interests
The authors declare no competing interests.
Financial disclosure statement
The CHRIS study was funded by the Department of Innovation, Research and University of the Autonomous Province of Bolzano-South Tyrol and supported by the European Regional Development Fund (FESR1157). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Informed consent statement
Written informed consent was obtained from all subjects involved in the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.NCD Countdown 2030 collaborators. NCD countdown 2030: worldwide trends in non-communicable disease mortality and progress towards sustainable development goal target 3.4. Lancet Lond. Engl.392, 1072–1088 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Tchkonia, T. & Kirkland, J. L. Aging, cell senescence, and chronic disease: emerging therapeutic strategies. JAMA. 320, 1319 (2018). [DOI] [PubMed] [Google Scholar]
- 3.López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell. 186, 243–278 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Martin-Ruiz, C. & von Zglinicki, T. Biomarkers of healthy ageing: expectations and validation. Proc. Nutr. Soc.73, 422–429 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Deelen, J. Targeting multimorbidity: using healthspan and lifespan to identify biomarkers of ageing that pinpoint shared disease mechanisms. EBioMedicine. 67, 103364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menni, C. et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol.42, 1111–1119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka, T. et al. Plasma proteomic biomarker signature of age predicts health and life span. eLife. 9, e61073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaleckis, R., Murakami, I., Takada, J., Kondoh, H. & Yanagida, M. Individual variability in human blood metabolites identifies age-related differences. Proc. Natl. Acad. Sci. U S A. 113, 4252–4259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, C. H. & Gonzalez, F. J. Challenges and opportunities of metabolomics. J. Cell. Physiol.227, 2975–2981 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You, J. et al. Plasma proteomic profiles predict individual future health risk. Nat. Commun.14, 7817 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadd, D. A. et al. Blood protein assessment of leading incident diseases and mortality in the UK Biobank. Nat. Aging. 10.1038/s43587-024-00655-7 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietzner, M. et al. Plasma metabolites to profile pathways in noncommunicable disease multimorbidity. Nat. Med.27, 471–479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buergel, T. et al. Metabolomic profiles predict individual multidisease outcomes. Nat. Med.28, 2309–2320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babu, M. & Snyder, M. Multi-omics profiling for health. Mol. Cell. Proteom.22, 100561 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zierer, J. et al. Exploring the molecular basis of age-related disease comorbidities using a multi-omics graphical model. Sci. Rep.6, 37646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verri Hernandes, V. et al. Age, sex, body mass index, diet and menopause related metabolites in a large homogeneous alpine cohort. Metabolites12, 205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dordevic, N. et al. Hormonal Contraceptives Are Shaping the Human Plasma Proteome in a Large Population Cohort. (2023). 10.1101/2023.10.11.23296871.
- 18.Salvi, F. et al. A manual of guidelines to score the modified cumulative illness rating scale and its validation in acute hospitalized elderly patients. J. Am. Geriatr. Soc.56, 1926–1931 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Menni, C. et al. Circulating proteomic signatures of chronological age. J. Gerontol. Ser. A. 70, 809–816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orwoll, E. S. et al. Proteomic assessment of serum biomarkers of longevity in older men. Aging Cell.19:e13253 10.1111/acel.13253 (2020). [DOI] [PMC free article] [PubMed]
- 21.Santos-Lozano, A. et al. Successful aging: insights from proteome analyses of healthy centenarians. Aging. 12, 3502–3515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, W. et al. A population-based study of precision health assessments using multi-omics network-derived biological functional modules. Cell. Rep. Med.3, 100847 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fidalgo, S., Ivanov, D. K. & Wood, S. H. Serotonin: from top to bottom. Biogerontology. 14, 21–45 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Martin, A. M. et al. The diverse metabolic roles of peripheral serotonin. Endocrinology. 158, 1049–1063 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Calder, P. C. et al. Health relevance of the modification of low grade inflammation in ageing (inflammageing) and the role of nutrition. Ageing Res. Rev.40, 95–119 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Kanova, M. & Kohout, P. Tryptophan: a unique role in the critically ill. Int. J. Mol. Sci.22, 11714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lassen, J. K. et al. Large-scale metabolomics: predicting biological age using 10,133 routine untargeted LC–MS measurements. Aging Cell. 10.1111/acel.13813 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calvani, R. et al. A distinct pattern of circulating amino acids characterizes older persons with physical frailty and sarcopenia: results from the BIOSPHERE Study. Nutrients. 10, 1691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maltais-Payette, I., Allam-Ndoul, B., Pérusse, L., Vohl, M. C. & Tchernof, A. Circulating glutamate level as a potential biomarker for abdominal obesity and metabolic risk. Nutr. Metab. Cardiovasc. Dis.29, 1353–1360 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Hawkins, R. A. The blood-brain barrier and glutamate. Am. J. Clin. Nutr.90, 867S–874S (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brosnan, J. T. Glutamate, at the interface between amino acid and carbohydrate metabolism. J. Nutr.130, 988S–990S (2000). [DOI] [PubMed] [Google Scholar]
- 32.Kelly, A. & Stanley, C. A. Disorders of glutamate metabolism. Ment Retard. Dev. Disabil. Res. Rev.7, 287–295 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Treberg, J. R., Banh, S., Pandey, U. & Weihrauch, D. Intertissue differences for the role of glutamate dehydrogenase in metabolism. Neurochem Res.39, 516–526 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Canfield, C. A. & Bradshaw, P. C. Amino acids in the regulation of aging and aging-related diseases. Transl Med. Aging. 3, 70–89 (2019). [Google Scholar]
- 35.Nassar, K. et al. The significance of caloric restriction mimetics as anti-aging drugs. Biochem. Biophys. Res. Commun.692, 149354 (2024). [DOI] [PubMed] [Google Scholar]
- 36.Kurhaluk, N. Tricarboxylic acid cycle intermediates and individual ageing. Biomolecules. 14, 260 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guasch-Ferré, M. et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care. 39, 833–846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, M. et al. Cardiovascular disease and its relationship with chronic kidney disease. Eur. Rev. Med. Pharmacol. Sci.18, 2918–2926 (2014). [PubMed] [Google Scholar]
- 39.Cai, X. et al. Population serum proteomics uncovers a prognostic protein classifier for metabolic syndrome. Cell. Rep. Med.4, 101172 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coan, P. M. et al. Complement factor B is a determinant of both metabolic and cardiovascular features of metabolic syndrome. Hypertension. 70, 624–633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kollerits, B. et al. Plasma concentrations of afamin are associated with prevalent and incident type 2 diabetes: a pooled analysis in more than 20,000 individuals. Diabetes Care. 40, 1386–1393 (2017). [DOI] [PubMed] [Google Scholar]
- 42.de Groot, V., Beckerman, H., Lankhorst, G. & Bouter, L. How to measure comorbiditya critical review of available methods. J. Clin. Epidemiol.56, 221–229 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Dash, P., Mohapatra, S. R. & Pati, S. Metabolomics of multimorbidity: could it be the Quo Vadis? Front. Mol. Biosci.9, 848971 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattaro, C. et al. The Cooperative Health Research in South Tyrol (CHRIS) study: rationale, objectives, and preliminary results. J. Transl Med.13, 348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.WHO Collaborating Centre for Drug Statistics Methodology, ATC classification index with DDDs. Oslo, Norway 2021. (2022).
- 46.Linn, B. S., Linn, M. W. & Gurel, L. Cumulative illness rating scale. J. Am. Geriatr. Soc.16, 622–626 (1968). [DOI] [PubMed] [Google Scholar]
- 47.Oksanen, J. et al. _vegan: Community Ecology Package_. R package version 2.6-4. (2022).
- 48.Legendre, P. & Legendre, L. Numerical Ecology. 3rd English Ed. (Elsevier).
- 49.Liaw, A. & Wiener, M. Classification and regression by randomForest. R News. 2 (3), 18–22 (2002). https://CRAN.R-project.org/doc/Rnews/ [Google Scholar]
- 50.Archer, E. & rfPermute estimate permutation p-values for random forest importance metrics. 2.5.2 (2011). 10.32614/CRAN.package.rfPermute
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are not openly available due to reasons of sensitivity. Individual level CHRIS study data can be requested for research purposes by submitting a dedicated request to the CHRIS Access Committee. Please visit https://chrisportal.eurac.edu/ for more information on the process or contact the corresponding author.