Abstract
CRISPR-Cas, the adaptive immune system exclusive to prokaryotes, confers resistance against foreign mobile genetic elements. The CRISPR-Cas system is now being exploited by scientists in a diverse range of genome editing applications. CRISPR-Cas systems can be categorized into six different types based on their composition and mechanism, and there are also natural regulatory biomolecules in bacteria and bacteriophages that can either enhance or inhibit the immune function of CRISPR-Cas. The CRISPR-Cas systems are currently being trialed as a new tool for gene therapy to treat various human diseases, including cancers and genetic diseases, offering significant therapeutic potential. This paper comprehensively summarizes various aspects of the CRISPR-Cas system, encompassing its diversity, regulatory mechanisms, its clinical applications and the obstacles encountered.
Keywords: CRISPR-Cas, Genome editing, Regulatory mechanism, Clinical applications
1. Introduction
Given that it is estimated that 1023 phage infections may occur on earth every second [1,2], bacteria have ample opportunity to evolve a variety of defense mechanisms against such attacks. These defense mechanisms include the superinfection exclusion system [3,4], abortive infection [5,6], phage exclusion system [7,8], restriction-modification system [9,10], prokaryotic Argonautes (pAgos) [11], and the CRISPR-Cas system [12,13]. While all of these play crucial roles in bacterial defense against exogenous genetic elements, the CRISPR-Cas system is the only recognized adaptive genetic defense system, providing adaptive immunity to numerous bacteria and archaea [14].
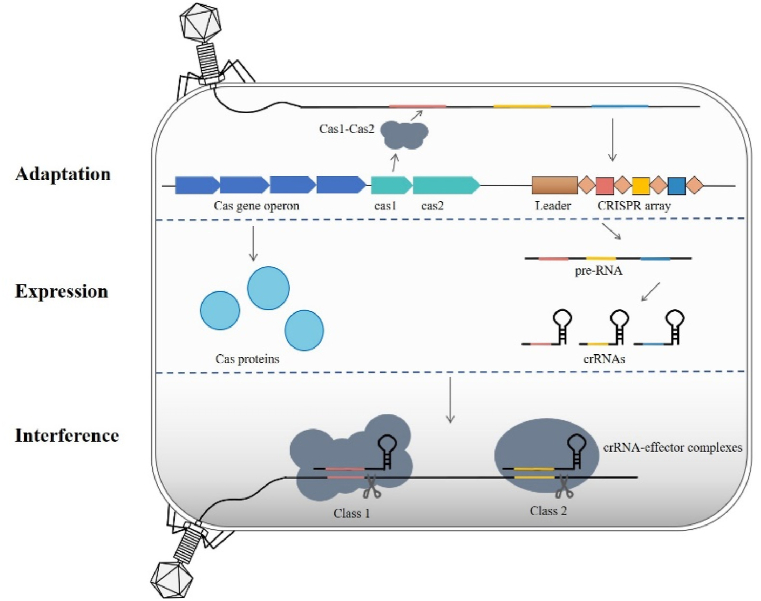
The CRISPR-Cas system is composed of two primary components: CRISPR (clustered regularly interspaced short palindromic repeats) and Cas (CRISPR-associated) proteins. CRISPR sequences consist of leader sequences, repetitive sequences and spacer sequences. The leader sequence is rich in AT bases and is located upstream of the CRISPR array, serving as the promoter of the CRISPR sequence. The repetitive sequence has a length of approximately 20-50bp bases and contains 5-7bp palindromic sequences. The spacer sequences are derived primarily from mobile genetic elements such as phages, and thus serve as a "memory bank" in which to store information about exogenous DNA, enabling targeted defense when the host is reinfected. Cas proteins are encoded by cas genes that are located in close proximity to CRISPR sequences and are essential for executing CRISPR-Cas immune responses [[15], [16], [17]]. In addition, protospacer adjacent motifs (PAMs) are also an important component of the CRISPR-Cas system. PAMs are short, conserved DNA sequences located next to one end of the protospacers. The presence of PAM sequences allows for self/nonself discrimination by preventing autoimmunity and self-cleavage in the absence within the CRISPR array, while also crucially enabling Cas proteins to recognize and bind to target DNA, facilitating accurate identification and cleavage of foreign genetic material by the CRISPR-Cas system. In general, CRISPR-Cas immunity encompasses three distinct phases: adaptation, expression, and interference (Fig. 1). During the adaptation process, fragments from the external piece of DNA, called protospacers, are incorporated into the CRISPR array as new spacers [18]. The Cas1 and Cas2 protein complexes present in almost all CRISPR-Cas systems play an important role in this process [19]. Moving on to the expression stage, the CRISPR array is transcribed and processed within the repeat region to produce an RNA consisting of a single spacer, flanked by (parts of) the repeat known as CRISPR RNAs (crRNAs). These crRNAs consist of a repeat sequence and a spacer sequence and acts as molecular guides to direct the Cas protein to the target site [20,21]. Finally, during interference, the crRNA directs the Cas protein to identify and target foreign nucleic acids corresponding to its spacer sequence and, once recognized, removes the foreign genetic material by recruiting nucleases or activating its own nuclease activity [22,23]. So overall, the three steps of CRISPR-Cas immunity work together to build a defense mechanism against invading genetic material, protecting the host organism from viral infection or other forms of foreign nucleic acids. The flexibility and efficiency of the CRISPR-Cas system has made it a widely used tool for genome editing and a key focus of research among scholars interested in new therapies for the prevention and treatment of human diseases.
Fig. 1.
The CRISPR-Cas immune system overview involves several steps. In the CRISPR-Cas system, Cas1 and Cas2 proteins capture and integrate invading genetic material into the CRISPR array, which is then transcribed into crRNAs. These crRNAs, along with Cas effector proteins, target and degrade matching sequences in the interference stage.
While the CRISPR-Cas adaptive immune system defends bacteria from phage infection it imposes a significant metabolic burden on the host [24]. Host regulatory factors therefore play a crucial role in controlling the CRISPR immune pathway so as to optimize immunity alongside fitness costs. Further exploration of CRISPR-Cas-related regulatory factors and their mechanisms of action will deepen our understanding of CRISPR biology. This knowledge will lay the groundwork to enhance the effectiveness of these widely-used genetic engineering tools thus opening the way for the wider clinical application of phage therapy.
2. Regulation of the CRISPR-Cas system
In natural conditions, biomolecules regulate the transcriptional stage of the CRISPR-Cas system in bacteria. Examples include LeuO, Csa3a, Phrs and WYL1 proteins, all of which enhance immunity, as well as H-NS, LRP, and Csa3b, which negatively regulate the system. Furthermore, environmental stimuli like glucose and iron deficiency, quorum sensing, and bacteriostatic antibiotics can also affect the CRISPR-Cas system. In addition, accessory proteins in type III/VI CRISPR-Cas systems bolster immune activity, while bacteriophages have also developed anti-CRISPR proteins (Acrs) as a means to elude bacterial assaults. As Ning Jia et al. have already presented a comprehensive overview of the action mechanisms of anti-CRISPRcrs), this section will refrain from discussing Acrs [25].
2.1. Regulation of CRISPR-Cas system at the transcriptional level
Currently, several transcription factors have been recognized as active participants in the regulation of the CRISPR-Cas system. Transposon mutagenesis experiments have revealed that the nucleoid-associated protein H-NS, along with the leucine-reactive regulatory proteins LRP and LeuO, actively participate in modulating the expression of CRISPR-Cas components, thereby exerting control over their functionality. Among these, H-NS and LRP have the capability to bind upstream and downstream of the transcriptional start site of casA, thereby exerting a negative regulatory influence on the expression of this system. LeuO, meanwhile, exerts a positive regulatory effect on the expression stage of this system by binding to the promoter of the casA regulatory region, thus effectively counteracting the inhibitory effects induced by H-NS [26,27].
In the archaeon Sulfolobus islandicus, two transcriptional regulators, Csa3a and Csa3b, containing CARF domains, have been identified to regulate the expression of the type I-A CRISPR locus [28,29]. Specifically, while S. islandicus Csa3b acts as a transcriptional repressor for genes encoding the subtype I-A CRISPR spacer acquisition complex and subtype I-A target interference complex, which binding to the two regulation sites (RI and RII) upstream of the promoter Pcas. It also serves as a transcriptional activator for genes encoding the subtype III-B cmr complex. Conversely, S. islandicus Csa3a functions as a transcriptional activator for the expression of CRISPR arrays, the subtype I-A adaptation complex, and DNA repair proteins. Recent studies have revealed that the signal molecule cOAs can bind to the CARF domain of Csa3a and Csa3b proteins, triggering conformational changes in the C-terminal HTH domain and enhancing their regulatory functions. The signal molecule cOA is generated by the Type III system, while the effector Csa3a and Csa3b are encoded by the Type I system, indicating the ability of the CRISPR system to employ cOAs for regulating different CRISPR-Cas sites within the same organism [30].
Several studies have demonstrated that AlgU, AlgR, and AmrZ, which are alginate regulator proteins linked to biofilm production, play a role in repressing the expression of type I-F cas genes [31]. These transcriptional regulatory mechanisms exert their influence on CRISPR-Cas immunity during both the interference and adaptation stages by modulating the expression of multiple cas genes [32,33]. Additionally, RNA regulatory mechanisms are also significantly involved in promoting the expression of type I CRISPR loci and concurrently suppressing the expression of type II CRISPR loci. PhrS, which was discovered through a Small regulatory RNA (sRNA) library screen, enhances the transcription of CRISPR arrays by preventing the interaction between the type I-F CRISPR leaders of the CRISPR array and Rho protein, effectively putting an end to Rho-dependent transcriptional termination [34]. The WYL1 helper protein plays a crucial role in the CRISPR-Cas13d system by collaborating with the Cas13d enzyme to boost its RNA-targeting abilities. By guiding Cas13d to particular RNA sequences, WYL1 contributes to increasing the efficiency and accuracy of RNA editing and manipulation processes [35]. In type II CRISPR systems, trans-activating CRISPR RNA (tracrRNA) is transcribed from two distinct promoters, resulting in the production of both long and short forms. The long form of tracrRNA exerts a critical function by facilitating the recruitment of Cas9 to the cas gene promoter, which effectively suppresses the transcription of cas genes, thereby modulating the activity of the type II system [36].
2.2. Effects of external stimuli on the CRISPR-Cas system
The activity of the CRISPR-Cas system can also be influenced by external environmental stimuli [37,38]. In P. atrosepticum, the presence of glucose can lead to a reduction in cas gene expression by diminishing the levels of the transcriptional activator (CRP-cAMP complex). When exposed to elevated glucose levels, the adenylate cyclase gene cya1 exhibits reduced expression, resulting in a corresponding decrease in the abundance of CRP-cAMP complex. This decrease in the CRP-cAMP complex level resulted in a concomitant decrease in the expression of the adaptation genes cas1 and cas2-3. In addition, the expression of the CRISPR-Cas gene in P. aeruginosa type I-F was found to be significantly increased under low iron conditions; the ECF sigma factor PvdS actively promotes transcription of the CRISPR locus through an effective interaction with the promoter region of the cas1 gene, contributing to this increased expression. This adaptive response to glucose and iron limitation may be a strategy for bacteria to cope with the increased metabolic stress caused by phage infection.
The second environmental stimulus is high cell density, which is associated with an increased risk of phage infection. Under these conditions, cas gene expression increases by upregulating components of the quorum sensing pathway. For example, in Serratia cells, the quorum sensing autoinducer SmaR plays an important role in repressing CRISPR and cas gene expression (cas8e, cas1 and cas10). When cell density increases, N-acyl homoserine lactones (AHLs) accumulate and interact with SmaR, effectively blocking its ability to bind to DNA. This repression induces an increase in CRISPR and cas gene expression, which enhances the immune response mediated by the CRISPR-Cas system types I-E, I-F, and III-A [33]. More studies have shown that the LasIR/RhIIR autoinducer-receptor pair also regulates the expression of type I-E and I-F cas genes, suggesting that the expression of the CRISPR-Cas system in bacteria such as P. aeruginosa is controlled by a complex regulatory mechanism.
Recent studies have shown that bacteriostatic antibiotics enhance CRISPR immunity by slowing the production of mature phage particles. This delay maximises the time to conversion during phage infection. Bacteriostatic antibiotics are thus able to shift the balance in favour of the host immune system, allowing it to mount an immune response before the phage causes irreparable damage to the host [39].
3. CRISPR-cas system for clinical therapeutic applications
CRISPR-Cas systems have revolutionized the field of genetics and molecular biology by providing powerful genome editing tools that can precisely target and modify specific regions of the genome. The essential steps for genome editing with CRISPR-Cas involve designing a guide RNA (gRNA) that matches the target DNA sequence, delivering both the gRNA and Cas9 protein into cells, and allowing Cas9 to precisely cleave the DNA at the target site under the guidance of the gRNA. The cell's repair machinery then fixes the DNA break by introducing desired changes (such as gene knockout, insertion, or correction) or inducing mutations. This technology has enabled efficient genome editing in diverse organisms, from bacteria to plants and animals, and has found applications in gene therapy, disease modeling, and diagnostics [40].
Previously, Zahedipour et al. offered an overview of the most recent therapeutic and vaccine strategies based on CRISPR/Cas technology to combat human viral infections [41]. Similarly, Zhang et al. provided an overview of clinical trials that focus on utilizing CRISPR/Cas systems for the treatment of cancer, hematological disorders, endocrine diseases, immune system conditions [42]. Tianxiang Li et al. review the latest developments in gene-editing types, delivery vectors, and disease characteristics [43]. In this section, the latest references are summarized to offer an overview of CRISPR applications in genetic diseases, infectious diseases, cancers, and pathogen diagnostic platforms. Compared to previous reviews, this section concentrates on the mechanisms and impacts of CRISPR in these clinical scenarios.
3.1. CRISPR-based treatment for genetic diseases
Researchers are currently employing CRISPR-Cas9 technology to explore potential treatments for various rare genetic disorders, including hereditary hemoglobinopathies [44], Leber congenital amaurosis type 10 (LCA10) [45], and severe combined immunodeficiency (SCID) [46]. The outcomes of these endeavors have yielded promising results, fostering optimism within the scientific community.
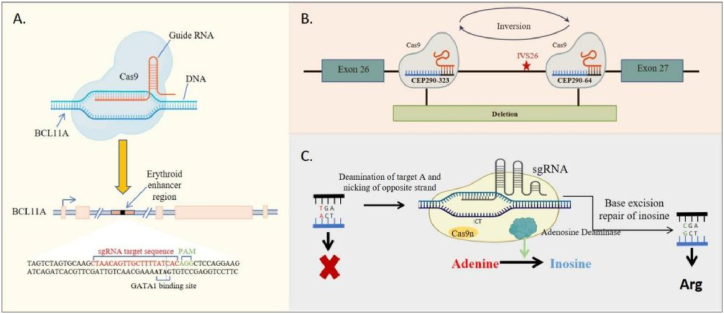
Transfusion-dependent β-thalassemia (TDT) and sickle cell disease (SCD) are severe monogenic disorders that can have life-threatening complications [[47], [48], [49]]. These conditions are characterized by the repression of γ-globin expression and fetal hemoglobin synthesis in erythroid cells, a process regulated by the transcription factor BCL11A [50]. Frangoul et al. employed an electroporation method to introduce CRISPR-Cas9 targeting the BCL11A erythroid-specific enhancer in CD34+ hematopoietic stem and progenitor cells derived from healthy donors (Fig. 2A). Patients who underwent treatment with autologous CD34+ cells edited using CRISPR-Cas9 demonstrated significant allelic editing in both bone marrow and blood, resulting in widespread increases in fetal hemoglobin levels and the complete elimination of vaso-occlusive episodes [44]. Similarly, LCA10 is a severe retinal dystrophy caused by mutations in the CEP290 gene [[51], [52], [53], [54]]. Morgan L. Maede et al. delivered S. aureus Cas9 protein (SaCas9) and two CEP290-specific guide RNAs (gRNAs) to photoreceptor cells by subretinal injection using an AAV5 vector (Fig. 2B). The patients receiving this treatment demonstrated mild but favorable clinical improvement, without any indication of toxicity or serious adverse effects [45].
Fig. 2.
(A) Schematic of the editing strategy for TDT and SCD; the sgRNA-targeted editing sites at the BCL11A erythroid-specific enhancer region, with the five exons of BCL11A labeled in gold boxes, GATA1 indicating the GATA1 transcription factor binding site, and PAM denoting the NGG DNA sequence after the Cas9 target DNA sequence; (B) The editing strategy for LCA10 involves flanking the IVS26 mutations with CEP290 gRNAs 323 and 64, leading to productive editing through the deletion or inversion of the intervening sequence; (C) The schematic of ABE for CD3d SCID shows that correcting the disease-causing defects is possible by developing ABE.
Building on advances in CRISPR-Cas technology, David Liu et al. successfully devised a single base editing technology called adenine base editing (ABE) [55]. This ABE approach involves the fusion of a catalytically impaired Cas9 nickase (Cas9n) with a DNA-modifying deaminase enzyme, facilitating the direct conversion of A·T-to-G·C base pairs (Fig. 2C). Notably, ABE circumvents the need to introduce double-stranded breaks (DSBs) and reduces the occurrence of indel byproducts [56]. The efficacy of this strategy has been substantiated in the treatment of various diseases in clinical trial, including SCID, demonstrating noteworthy therapeutic outcomes [46].
3.2. CRISPR-based treatment for infectious disease
The CRISPR-Cas system can also be used to treat infectious diseases such as cervical cancer caused by human papillomavirus, hepatocellular carcinoma caused by HBV infection, and acquired immunodeficiency syndrome (AIDS) caused by human immunodeficiency virus (HIV). CCR5, a cell membrane protein belonging to the superfamily of G protein-coupled receptors (GPCRs), is one of the key co-receptors involved in HIV-1 entry into living cells. Inactive mutations in the CCR5 protein can inhibit HIV-1 binding in vivo. The researchers used the CRISPR-Cas9 system to remove the CCR5Δ32 gene from donor CD34+ hematopoietic stem cells and then transplanted them into patients with AIDS-related leukemia, opening new frontiers in the treatment of AIDS and related diseases [57]. Similarly, in the process of using gene editing technology to treat HBV infection, the researchers produced four sgRNAs targeting conserved regions of HBV and further transfected the sgRNA-Cas9 plasmid into mice and found that the amount of HBV protein in the mice was significantly reduced, suggesting that CRISPR-Cas9 technology may offer a breakthrough in the treatment of HBV infection [58]. CRISPR-Cas9 technology has been used to inactivate the E6 and E7 genes of HPV to slow the progression of cervical cancer [59]. This report suggests that advances in genome editing using CRISPR-Cas technology may lay promising foundations for the treatment of infectious diseases.
Coronavirus disease 19 (COVID-19) is an acute respiratory infection caused by the SARS-CoV-2 virus, which was first reported in Wuhan, China in late 2019 and is the seventh coronavirus found to infect humans, posing a serious threat to humans [60]. Unfortunately, there are not many effective diagnostic technologies available to detect COVID-19, but CRISPR-Cas technology, which can accurately and rapidly identify pathogens with a fast detection speed of about 30 min from initial sample to final result [61], has been used to detect RNA viruses and identify bacterial pathogens, including influenza A virus and vesicular stomatitis virus [62,63]. Many scientists are developing CRISPR-based COVID-19 detection tools. Several Cas systems, such as Cas13a, Cas12a and Cas10, have been studied for successful nucleic acid detection.
3.3. CRISPR-based diagnosis and treatment for cancer
Given its renowned versatility in gene editing, the CRISPR-Cas9 system provides a strong framework for advances in cancer diagnosis and therapy. In that context, CRISPR-DS (CRISPR-Duplex Sequencing) is an innovative technique that utilizes Cas9-mediated digestion selectively to release megabase-sized fragments from genomic DNA [64]. This strategy offers numerous benefits, such as efficient target enrichment of small genomic regions, uniform sequencing coverage, ultra-precise readouts, and reduced DNA input requirements [64]. In a previous study, Julia Matas et al. utilized this strategy to investigate patients with colorectal cancer [65]. They identified a high prevalence of oncogenic KRAS and TP53 driver mutations in the normal colonic epithelium located away from the primary tumor in patients with colorectal cancer, highlighting the potential of these mutations as valuable markers for assessing colorectal cancer risk in clinical practice [65].
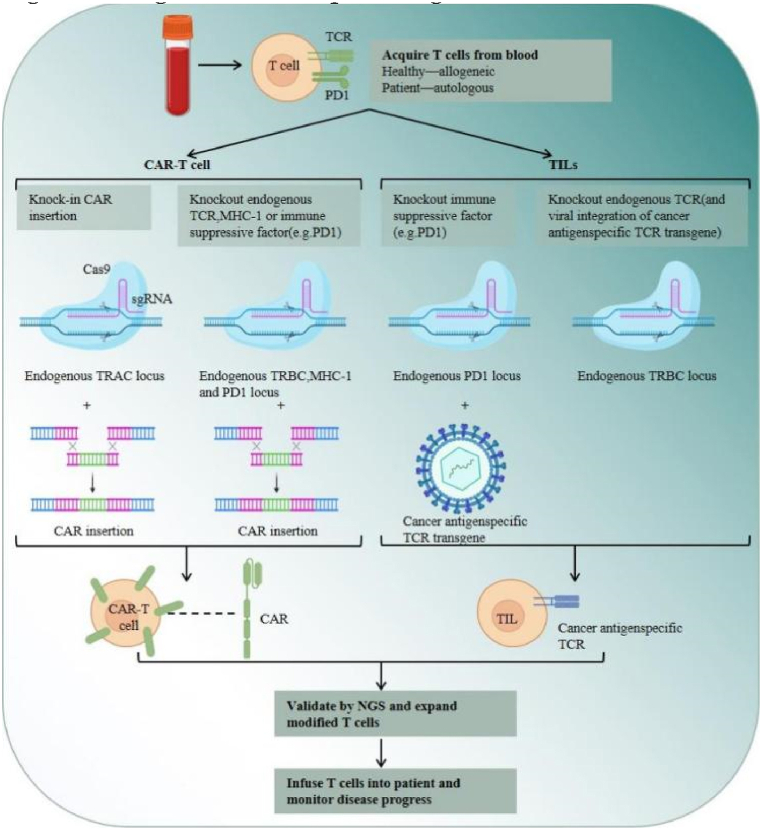
Chimeric Antigen Receptor (CAR) T-cells have undoubtedly revolutionized personalized cancer therapy [66]. While their success has been conspicuous in refractory B-cell malignancies, the optimization of CAR T-cell therapy for various other cancers, especially solid tumors, remains elusive. This can be attributed to challenges such as T-cell exhaustion, limited persistence of CAR T-cells, cytokine-related toxicities, and constraints associated with manufacturing autologous products [66]. In comparison, engineered T cell receptor-T (TCR-T) cell therapy, constructed using CRISPR-Cas9 technology, offers a more promising approach [67]. By introducing defective phenotypes of PD-1 [68], [69], TGF-β receptor II (TGFBR2) [70], CD7 and TRAC [71], the circulating number of TCR-T cells is increased. This gives this alternative therapy great potential to target a broad range of antigens, thus expanding the horizons for cancer treatment (Fig. 3).
Fig. 3.
Ex vivo CRISPR engineering of human T cells for adoptive T cell therapy; Allogeneic and autologous T cells can be tested to investigate the effect of CRISPR technology in tumor infiltrating lymphocytes (TILs) and chimeric antigen receptor (CAR) T cells.
3.4. CRISPR-based novel detection techniques
The CRISPR-Cas system has demonstrated significant potential in advancing the development of pathogen detection platforms. These platforms harness the specificity of the CRISPR-Cas system to target unique sequences in the pathogen's genome, facilitating precise and sensitive detection of these pathogens (Table 1).
Table 1.
Technologies for detecting pathogens based on the CRISPR-Cas system.
| Types | Technical platform | Cas protein | Pre-amplification | Limit of detection | Pathogen | Year/Ref. |
|---|---|---|---|---|---|---|
| Type I-E | CONAN | Cas3 | RT-LAMP | 1 copy/μL | SARS-CoV-2, IAV |
2022 [78] |
| Type II | SCC-LFS | Cas9 | – | 1 CFU/mL | S. aureus | 2024 [79] |
| Type III | SCOPE | Cas10 | LAMP | 800aM | SARS-CoV-2 | 2021 [80] |
| CRISPR-Csm-based detection | Cas10 | RT-LAMP | 200 copies/mL | SARS-CoV-2 | 2021 [81] | |
| Type V | MoECS | Cas12 | – | fmol/L | SARS-CoV-2 Delta variant | 2022 [82] |
| CRISPR-ENHANCE | LbCas12a | RT-LAMP | – | SARS-CoV-2, HIV, HCV | 2022 [83] | |
| Cards-SSJ/K | Cas12a | RPA | 10 CFU/mL | S. suis serotype 2 | 2024 [84] | |
| RPA-CRISPR/Cas system | Cas12a | RPA | one parasite per milliliter | Neospora caninum | 2024 [85] | |
| CRISPR/Cas12a-SERS | Cas12a | – | 100 nM-10 fM | African swine fever virus | 2024 [86] | |
| RPA/CRISPR/Cas12a detection platform | Cas12a | RPA | 1 copy/μL | P. aeruginosa | 2024 [87] | |
| CBD platform | Cas12a | ERA | 1 CFU/mL |
S. aureus L.monocytogenes |
2024 [88] | |
| MP-RPA-CRISPR | Cas12b | RPA | 5 fg | M. pneumoniae | 2023 [89] | |
| sPAMC | LbCas12a | RPA | 1 copy/μL | SARS-CoV-2 | 2022 [90] | |
| CRISPR/Cas14a-G4 Biosensor | Cas14a | PCR | 5 copies/μL | African swine fever virus | 2024 [91] | |
| ACasB | Cas14a1 | – | 400 CFU/mL | S. aureus | 2022 [92] | |
| Type VI | mCARMEN | LwCas13a | PCR, RPA | 100 copies/μL | 21 types of human respiratory viruses | 2022 [93] |
| SCOPE | Cas13a | RPA | 0.5 copies/μL | Monkeypox virus | 2024 [94] | |
| CRISPR-Cas13a-based detection | LwaCas13a | RPA | 69 copies/μL | Avian influenza virus | 2023 [95] | |
| CRISPR/Ca13a-MTB | LwCas13a | PCR | 1 copy/μL | M. tuberculosis | 2023 [96] |
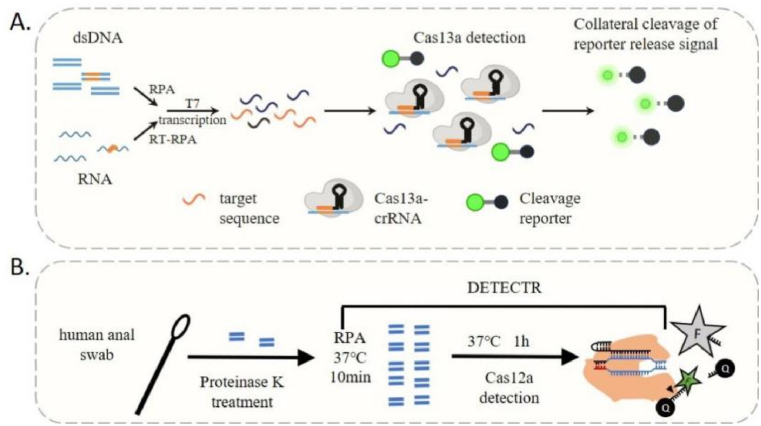
Kellner et al. have developed an innovative diagnostic tool based on CRISPR technology, combining nucleic acid pre-amplification with CRISPR-Cas enzymology to precisely detect and target desired DNA or RNA sequences. This novel strategy, known as Specific High Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK), enables the simultaneous detection of multiple RNA or DNA targets with exceptional sensitivity (Fig. 4A) [72,73]. SHERLOCK has demonstrated its ability to detect genotypes of bacterial and viral infectious disease agents, including SNP and antibiotic resistance genes [74]. Notably, SHERLOCK has successfully detected Zika and Dengue viruses directly from patient urine and serum samples, providing evidence of its potential in the field [73].
Fig. 4.
(A) The SHERLOCK assay steps consist of pre-amplifying the input DNA or RNA target, converting it to RNA through T7 transcription, and subsequently recognizing and binding by the Cas13-crRNA complex, which activates RNAse activity and results in the unquenching of the fluorescent RNA reporter; (B) the DETECTR assay involves extracting DNA, pre-amplifying it, and mixing it with CRISPR components, including Cas12 protein and a single-guide RNA (sgRNA), along with a fluorescent reporter. When the target DNA is introduced, Cas12 binds and activates, cleaving the reporter and releasing a fluorescent signal.
In a parallel endeavor, Janice S. Chen et al. ingeniously merged target-dependent Cas12a ssDNase activation with isothermal amplification, thereby giving birth to the innovative technique known as DNA Endonuclease Targeted CRISPR Trans Reporter (DETECTR) [75]. This approach not only achieves an unprecedented attomolar sensitivity for nucleic acid detection but also showcases a remarkable proficiency in rapidly and specifically detecting HPV and SARS-CoV-2 within human patient samples (Fig. 4B) [[75], [76], [77]]. Consequently, DETECTR has emerged as a simple platform for point-of-care diagnostics based on nucleic acid analysis.
4. Conclusions
In this review we have examined the intricate regulation of its activity by a diverse array of intracellular and extracellular factors, and illuminated its multifaceted application in the treatment of genetic diseases and tumors, and in the accurate detection of pathogens. There remain, however, several unresolved concerns regarding the use of the CRISPR-Cas system for gene editing. One of the major concerns with CRISPR-Cas is the potential for off-target effects, where the Cas enzyme may unintentionally edit DNA sequences leading to unintended consequences such as cancer or other genetic disorders. In addition, the body's immune system might recognize CRISPR-associated proteins, particularly Cas9, as foreign and initiate an immune response. This reduce the effectiveness of the therapy and potentially cause inflammation or other immune-related issues. Furthermore, viral vectors, nanoparticles, and other delivery technologies must be further optimized to accurately and specifically deliver CRISPR-Cas components to target cells or tissues. Inefficient delivery can result in suboptimal editing, while viral vectors pose risks of insertional mutagenesis and immune responses.
CRISPR interference (CRISPRi) uses an inactivated form of the Cas9 protein (dCas9) to bind to gRNA and target specific DNA sequences near the gene of interest. This binding prevents the transcription machinery from accessing the gene, effectively inhibiting the expression of that gene. CRISPR activation (CRISPRa) enhances the transcriptional activity of the target gene by fusing transcription activating factors (such as VP64, VPR, SAM, or SunTag) onto dCas9 and guiding this complex to the promoter region using sgRNA. CRISPRa provides a powerful tool for overexpression of specific genes, particularly suitable for studying gene function and disease mechanisms. By using specific guide RNAs that target the desired genes, CRISPRi and CRISPRa systems effectively promote gene silencing or activation, giving researchers the ability to precisely and reversibly manipulate gene expression [97,98].
In addition, as an intrinsic regulator of the CRISPR-Cas system, Acrs can precisely inhibit CRISPR-Cas system function. By introducing Acrs alongside CRISPR-Cas system components, researchers can fine-tune the timing and extent of gene editing to better control and reduce off-target effects. Timely delivery of AcrIIA4 to human cells has been shown to reduce off-target editing while maintaining desired on-target editing effects [99]. In particular, fusion of Acrs mutants with Cas9 has been shown to be an effective way to reduce unwanted off-target effects associated with gene editing [100]. According to the report, timed administration of AcrIIA2 and AcrIIA4 effectively regulated the duration of CRISPR-Cas9 activity, reducing cytotoxicity and increasing engraftment rates of human hematopoietic stem cells, with no effect on the rate of targeted genome editing [101].
While a few questions still linger, CRISPR continues to play a prominent role in the race to become the preferred tool for genome editing, poised to make a lasting impact in cancer treatment, drug design, and the management of genetic diseases. CRISPR-Cas based toolkits have demonstrated the potential for gene knockout, gene knockin, single-base and multi-base substitution, as well as transcriptional regulation. Our comprehension of biology and disease has greatly progressed thanks to this transformative technology.
CRediT authorship contribution statement
Hui Cheng: Writing – original draft. Haoyue Deng: Writing – review & editing, Writing – original draft. Dongdao Ma: Writing – review & editing. Mengyuan Gao: Writing – review & editing, Supervision. Zhihan Zhou: Investigation. Heng Li: Supervision. Shejuan Liu: Funding acquisition, Writing – review & editing. Tieshan Teng: Funding acquisition, Conceptualization.
Data availability statement
No data was used for the research described in the article.
Funding
This research was funded by The Key R&D and Promotion Projects of Henan Province (232102311139); China Postdoctoral Science Foundation (2021m690095); National Innovation and entrepreneurship training program for college students (20231022009 and 20231021006).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Montgomery M.T., et al. Yet more evidence of collusion: a new viral defense system encoded by gordonia phage CarolAnn. mBio. 2019;10 doi: 10.1128/mBio.02417-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duerkop B.A., et al. Murine colitis reveals a disease-associated bacteriophage community. Nature Microbiology. 2018;3:1023–1031. doi: 10.1038/s41564-018-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leavitt J.C., et al. Bacteriophage P22 SieA mediated superinfection exclusion. mBio. 2024;15 doi: 10.1128/mbio.02169-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domingo-Calap P., Mora-Quilis L., Sanjuán R. Social bacteriophages. Microorganisms. 2020;8 doi: 10.3390/microorganisms8040533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan S.-F., et al. Relationship between intrauterine bacterial infection and early embryonic developmental arrest. Chinese Med J. 2016;129:1455–1458. doi: 10.4103/0366-6999.183411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srinivas M., et al. Sexually transmitted infection rates and closure of family planning clinics because of abortion restrictions in Iowa. JAMA Netw. Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.39063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfarb T., et al. BREX is a novel phage resistance system widespread in microbial genomes. The EMBO journal. 2015;34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ofir G., et al. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nature microbiology. 2018;3:90–98. doi: 10.1038/s41564-017-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hille F., et al. The biology of CRISPR-cas: backward and forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 10.Birkholz N., Jackson S.A., Fagerlund R.D., Fineran Peter C. A mobile restriction–modification system provides phage defence and resolves an epigenetic conflict with an antagonistic endonuclease. Nucleic Acids Res. 2022;50:3348–3361. doi: 10.1093/nar/gkac147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esyunina D., et al. Specific targeting of plasmids with Argonaute enables genome editing. Nucleic Acids Res. 2023;51:4086–4099. doi: 10.1093/nar/gkad191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhokisham N., et al. CRISPR-cas system: the current and emerging translational landscape. Cells. 2023;12 doi: 10.3390/cells12081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y., et al. The CRISPR/cas system: a customizable toolbox for molecular detection. Genes. 2023;14 doi: 10.3390/genes14040850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burstein D., et al. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat. Commun. 2016;7 doi: 10.1038/ncomms10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shmakov S.A., et al. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio. 2017;8 doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X., et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol. Cell. 2021;81:4333–4345. doi: 10.1016/j.molcel.2021.08.008. e4334. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S., Stainer A., Dubrulle J., Simpkins C., Cooper J.A. Cas phosphorylation regulates focal adhesion assembly. Elife. 2023;12 doi: 10.7554/eLife.90234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiriaeva A., Fedorov I., Vyhovskyi D., Severinov K. Detection of CRISPR adaptation. Biochem. Soc. Trans. 2020;48:257–269. doi: 10.1042/BST20190662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jedrzejczyk D.J., et al. CRISPR-Cas12a nucleases function with structurally engineered crRNAs: SynThetic trAcrRNA. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-15388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao C., Beisel C.L. The tracrRNA in CRISPR biology and technologies. Annu. Rev. Genet. 2021;55:161–181. doi: 10.1146/annurev-genet-071719-022559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W., Samai P., Marraffini L.A. Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-cas immunity. Cell. 2016;164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sternberg S.H., LaFrance B., Kaplan M., Doudna J.A. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., Helal S.E., Peng N. CRISPR-Cas-Based engineering of probiotics. BioDesign Research. 2023;5 doi: 10.34133/bdr.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia N., Patel D.J. Structure-based functional mechanisms and biotechnology applications of anti-CRISPR proteins. Nat. Rev. Mol. Cell Biol. 2021;22:563–579. doi: 10.1038/s41580-021-00371-9. [DOI] [PubMed] [Google Scholar]
- 26.Dy R.L., Richter C., Salmond G.P., Fineran P.C. Remarkable mechanisms in microbes to resist phage infections. Annual review of virology. 2014;1:307–331. doi: 10.1146/annurev-virology-031413-085500. [DOI] [PubMed] [Google Scholar]
- 27.Fragel S.M., et al. Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli. Nucleic Acids Res. 2019;47:7363–7379. doi: 10.1093/nar/gkz506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charbonneau A.A., Eckert D.M., Gauvin C.C., Lintner N.G., Lawrence C.M. Cyclic tetra-adenylate (cA(4)) recognition by Csa3; implications for an integrated class 1 CRISPR-cas immune response in saccharolobus solfataricus. Biomolecules. 2021;11 doi: 10.3390/biom11121852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Q., et al. CRISPR-associated factor Csa3b regulates CRISPR adaptation and cmr-mediated RNA interference in Sulfolobus islandicus. Front. Microbiol. 2020;11:2038. doi: 10.3389/fmicb.2020.02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia P., Dutta A., Gupta K., Batish M., Parashar V. Structural basis of cyclic oligoadenylate binding to the transcription factor Csa3 outlines cross talk between type III and type I CRISPR systems. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borges A.L., et al. Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity. Nature microbiology. 2020;5:679–687. doi: 10.1038/s41564-020-0691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson A.G., Chang J.T., Taylor C., Fineran P.C. Regulation of the Type I-F CRISPR-Cas system by CRP-cAMP and GalM controls spacer acquisition and interference. Nucleic acids research. 2015;43:6038–6048. doi: 10.1093/nar/gkv517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson A.G., et al. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-cas systems. Mol. Cell. 2016;64:1102–1108. doi: 10.1016/j.molcel.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin P., et al. High-throughput screen reveals sRNAs regulating crRNA biogenesis by targeting CRISPR leader to repress Rho termination. Nat. Commun. 2019;10:3728. doi: 10.1038/s41467-019-11695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan W.X., et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell. 2018;70:327–339.e325. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Workman R.E., et al. A natural single-guide RNA repurposes Cas9 to autoregulate CRISPR-Cas expression. Cell. 2021;184:675–688.e619. doi: 10.1016/j.cell.2020.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Ivanova E., Khan S. How various drug delivery methods could aid in the translation of genome prime editing technologies. Genetics Research 2022. 2022;1–8 doi: 10.1155/2022/7301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deltcheva E., et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dimitriu T., et al. Bacteriostatic antibiotics promote CRISPR-Cas adaptive immunity by enabling increased spacer acquisition. Cell host & microbe. 2022;30:31–40.e35. doi: 10.1016/j.chom.2021.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 41.Zahedipour F., Zahedipour F., Zamani P., Jaafari M.R., Sahebkar A. Harnessing CRISPR technology for viral therapeutics and vaccines: from preclinical studies to clinical applications. Virus Res. 2024;341 doi: 10.1016/j.virusres.2024.199314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S., et al. Current trends of clinical trials involving CRISPR/Cas systems. Front. Med. 2023;10 doi: 10.3389/fmed.2023.1292452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li T., et al. CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduct. Targeted Ther. 2023;8:36. doi: 10.1038/s41392-023-01309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meisel R. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021;384:e91. doi: 10.1056/NEJMc2103481. [DOI] [PubMed] [Google Scholar]
- 45.Maeder M.L., et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 46.McAuley G.E., et al. Human T cell generation is restored in CD3δ severe combined immunodeficiency through adenine base editing. Cell. 2023;186:1398–1416.e1323. doi: 10.1016/j.cell.2023.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K., Doudna J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science (New York, N.Y.) 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandow A.M., Liem R.I. Advances in the diagnosis and treatment of sickle cell disease. J. Hematol. Oncol. 2022;15 doi: 10.1186/s13045-022-01237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan S.J., Zaidi S.A.T., Murtaza S.F., Asif M., Kumar V. Advancements in sickle cell disease (SCD) treatment: a review of novel pharmacotherapies and their impact on patient outcomes. Cureus. 2023;15 doi: 10.7759/cureus.42847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bauer D.E., Orkin S.H. Hemoglobin switching's surprise: the versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr. Opin. Genet. Dev. 2015;33:62–70. doi: 10.1016/j.gde.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corral-Serrano J.C., et al. Eupatilin improves cilia defects in human CEP290 ciliopathy models. Cells. 2023;12 doi: 10.3390/cells12121575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swarts D.C., et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westra E.R., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skorczyk-Werner A., Niedziela Z., Stopa M., Krawczyński M.R. Novel gene variants in Polish patients with Leber congenital amaurosis (LCA) Orphanet J. Rare Dis. 2020;15 doi: 10.1186/s13023-020-01634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landrum M.J., et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic acids research. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu C.L., Ruan M.Z.C., Mahajan V.B., Tsang S.H. Viral delivery systems for CRISPR. Viruses. 2019;11 doi: 10.3390/v11010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weissman J.L., Laljani R.M.R., Fagan W.F., Johnson P.L.F. Visualization and prediction of CRISPR incidence in microbial trait-space to identify drivers of antiviral immune strategy. The ISME journal. 2019;13:2589–2602. doi: 10.1038/s41396-019-0411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhen S., et al. In vitro and in vivo synergistic therapeutic effect of cisplatin with human Papillomavirus16 E6/E7 CRISPR/Cas9 on cervical cancer cell line. Translational oncology. 2016;9:498–504. doi: 10.1016/j.tranon.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding R., et al. CRISPR/Cas system: a potential technology for the prevention and control of COVID-19 and emerging infectious diseases. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.639108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rahimi H., et al. CRISPR systems for COVID-19 diagnosis. ACS Sens. 2021;6:1430–1445. doi: 10.1021/acssensors.0c02312. [DOI] [PubMed] [Google Scholar]
- 62.Freije C.A., et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837.e811. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quan J., et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic acids research. 2019;47:e83. doi: 10.1093/nar/gkz418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nachmanson D., et al. Targeted genome fragmentation with CRISPR/Cas9 enables fast and efficient enrichment of small genomic regions and ultra-accurate sequencing with low DNA input (CRISPR-DS) Genome Res. 2018;28:1589–1599. doi: 10.1101/gr.235291.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matas J., et al. Colorectal cancer is associated with the presence of cancer driver mutations in normal colon. Cancer Res. 2022;82:1492–1502. doi: 10.1158/0008-5472.CAN-21-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ou X., Ma Q., Yin W., Ma X., He Z. CRISPR/Cas9 gene-editing in cancer immunotherapy: promoting the present revolution in cancer therapy and exploring more. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.674467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morris E.C., Stauss H.J. Optimizing T-cell receptor gene therapy for hematologic malignancies. Blood. 2016;127:3305–3311. doi: 10.1182/blood-2015-11-629071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rupp L.J., et al. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 2017;7:737. doi: 10.1038/s41598-017-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi B.D., et al. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. Journal for immunotherapy of cancer. 2019;7:304. doi: 10.1186/s40425-019-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang N., et al. TGF-β inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI insight. 2020;5 doi: 10.1172/jci.insight.133977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cooper M.L., et al. An "off-the-shelf" fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia. 2018;32:1970–1983. doi: 10.1038/s41375-018-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. Author Correction: SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2020;15:1311. doi: 10.1038/s41596-020-0302-z. [DOI] [PubMed] [Google Scholar]
- 73.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gootenberg J.S., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science (New York, N.Y.) 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen J.S., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science (New York, N.Y.) 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsou J.H., Leng Q., Jiang F. A CRISPR test for detection of circulating nuclei acids. Translational oncology. 2019;12:1566–1573. doi: 10.1016/j.tranon.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Broughton J.P., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshimi K., et al. CRISPR-Cas3-based diagnostics for SARS-CoV-2 and influenza virus. iScience. 2022;25 doi: 10.1016/j.isci.2022.103830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang J., et al. CRISPR/Cas9-mediated SERS/colorimetric dual-mode lateral flow platform combined with smartphone for rapid and sensitive detection of Staphylococcus aureus. Biosens. Bioelectron. 2024;249 doi: 10.1016/j.bios.2024.116046. [DOI] [PubMed] [Google Scholar]
- 80.Steens J.A., et al. SCOPE enables type III CRISPR-Cas diagnostics using flexible targeting and stringent CARF ribonuclease activation. Nat. Commun. 2021;12:5033. doi: 10.1038/s41467-021-25337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santiago-Frangos A., et al. Intrinsic signal amplification by type III CRISPR-Cas systems provides a sequence-specific SARS-CoV-2 diagnostic. Cell reports. Medicine. 2021;2 doi: 10.1016/j.xcrm.2021.100319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu C., et al. CRISPR-Cas12a-Empowered electrochemical biosensor for rapid and ultrasensitive detection of SARS-CoV-2 delta variant. Nano-Micro Lett. 2022;14:159. doi: 10.1007/s40820-022-00888-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen L.T., et al. CRISPR-ENHANCE: an enhanced nucleic acid detection platform using Cas12a. Methods (San Diego, Calif.) 2022;203:116–124. doi: 10.1016/j.ymeth.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 84.Wang L., et al. A CRISPR-Cas12a-based platform facilitates the detection and serotyping of Streptococcus suis serotype 2. Talanta. 2024;267 doi: 10.1016/j.talanta.2023.125202. [DOI] [PubMed] [Google Scholar]
- 85.Wang L., et al. Establishment of an ultrasensitive and visual detection platform for Neospora caninum based-on the RPA-CRISPR/Cas12a system. Talanta. 2024;269 doi: 10.1016/j.talanta.2023.125413. [DOI] [PubMed] [Google Scholar]
- 86.Wang H., et al. A CRISPR/Cas12a-SERS platform for amplification-free detection of African swine fever virus genes. Talanta. 2024;267 doi: 10.1016/j.talanta.2023.125225. [DOI] [PubMed] [Google Scholar]
- 87.Zhang W., Qu H., Wu X., Shi J., Wang X. Rapid, sensitive, and user-friendly detection of Pseudomonas aeruginosa using the RPA/CRISPR/Cas12a system. BMC Infect. Dis. 2024;24:458. doi: 10.1186/s12879-024-09348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu X., et al. A portable CRISPR-Cas12a triggered photothermal biosensor for sensitive and visual detection of Staphylococcus aureus and Listeria monocytogenes. Talanta. 2024;271 doi: 10.1016/j.talanta.2024.125678. [DOI] [PubMed] [Google Scholar]
- 89.Zhou J., et al. Rapid, ultrasensitive and highly specific diagnosis of Mycoplasma pneumoniae by a CRISPR-based detection platform. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1147142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu S., et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat. Biomed. Eng. 2022;6:286–297. doi: 10.1038/s41551-022-00861-x. [DOI] [PubMed] [Google Scholar]
- 91.Zhao X., et al. CRISPR/Cas14 and G-quadruplex DNAzyme-driven biosensor for paper-based colorimetric detection of african swine fever virus. ACS Sens. 2024;9:2413–2420. doi: 10.1021/acssensors.4c00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei Y., et al. Aptamer-based Cas14a1 biosensor for amplification-free live pathogenic detection. Biosens. Bioelectron. 2022;211 doi: 10.1016/j.bios.2022.114282. [DOI] [PubMed] [Google Scholar]
- 93.Welch N.L., et al. Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants. Nat. Med. 2022;28:1083–1094. doi: 10.1038/s41591-022-01734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Y., et al. Ultrasensitive single-step CRISPR detection of monkeypox virus in minutes with a vest-pocket diagnostic device. Nat. Commun. 2024;15:3279. doi: 10.1038/s41467-024-47518-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu Y., et al. CRISPR-Cas13a-based detection method for avian influenza virus. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1288951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ren W., et al. Development and clinical evaluation of a CRISPR/Cas13a-based diagnostic test to detect Mycobacterium tuberculosis in clinical specimens. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1117085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bendixen L., Jensen T.I., Bak R.O. CRISPR-Cas-mediated transcriptional modulation: the therapeutic promises of CRISPRa and CRISPRi. Mol. Ther. : the journal of the American Society of Gene Therapy. 2023;31:1920–1937. doi: 10.1016/j.ymthe.2023.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cai R., Lv R., Shi X., Yang G., Jin J. CRISPR/dCas9 tools: epigenetic mechanism and application in gene transcriptional regulation. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms241914865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shin J., et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aschenbrenner S., et al. Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aay0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X., Bai X.C., Chen Z.J. Structures and mechanisms in the cGAS-STING innate immunity pathway. Immunity. 2020;53:43–53. doi: 10.1016/j.immuni.2020.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.