Summary
The Arabidopsis megaspore mother cell (MMC) arises from somatic cells in the ovule primordium and enters meiosis to generate four megaspores. Only the most chalazal (functional megaspore, FM) survives, undergoing a series of mitoses to form the female gametophyte. We show that this commitment to the sexual germline requires spatial regulation of AUXINRESPONSEFACTOR10 (ARF10). GFP-fusion lines reveal ARF10 expression to be restricted to cells surrounding the MMC in wild type, but ectopically disseminated throughout the ovule in transgenic mARF10 lines insensitive to miR160, an ARF10 downregulator. Significantly, mARF10 ovules develop multiple FMs with differing ploidies, forming putative supernumerary gametophytes with altered polarity and cell identities – features of aposporous apomixis. Furthermore, we confirm the complexity of ovular ARF10 expression, being mediated by SEEDSTICK, ARGONAUTE1, and miR160. This work adds to our understanding of molecular switches possibly regulating aposporous apomixis, and may contribute the development of innovative plant breeding strategies.
Subject areas: Cell biology, Plant biology, Plant development
Graphical abstract
Highlights
-
•
Spatial regulation of ARF10 is essential for gametophyte commitment and development
-
•
mARF10 lines show multiple functional megaspores and altered gametophytes
-
•
ARF10 expression in ovules is controlled by SEEDSTICK, ARGONAUTE1, and miR160
-
•
This study could yield tools for advancing plant breeding
Cell biology; Plant biology; Plant development
Introduction
Flowering plants form seeds using two different mechanisms known as sexual and apomictic reproduction.1 In the sexual pathway, a meiotic division in the ovule primordia generates a reduced functional megaspore (FM) that, after a regulated series of mitoses, forms a female gametophyte containing two gametes: the haploid egg and the diploid central cell. Double-fertilization of these cells’ gametes by two pollen sperm nuclei then leads to production of the embryo and endosperm – key components of the developing seed. By contrast, apomixis (i.e., agamospermy) involves the formation of seeds without previous reductional division, with embryos spontaneously generated through parthenogenesis, and endosperm formed either autonomously, or after pseudogamy (fertilization of the central cell only).2 While sexuality leads to genetic variation and syngamy, apomixis generates clonal progeny genetically identical to the mother plant.3 Apomictic mechanisms are usually subcategorized as sporophytic, in which nucellar or integumental cells form maternal somatic embryos, or gametophytic, in which either the megaspore mother cell (MMC) itself (in the case of diplospory) or integumental/nucellar companion cells (in the case of apospory) differentiate in the ovule as non-reduced FMs, and form non-reduced megagametophytes by mitosis that generate parthenogenetic offspring.3
Apospory, the best characterized apomictic mechanism, has been reported in 110 genera belonging to all seven major clades of angiosperms and in most large orders.4 Contrary to adventitious embryony, which is predominant in fabids, malvids, and lamiids, clear phylogenetic tendencies are not apparent for apospory.4 It involves the development of extra non-reduced FMs (apospory initials or AIs) from the integuments and/or the nucellus, which are located around the legitimate MMC, the meiotic products and/or the reduced FM. Mature ovules typically feature one or more misoriented non-reduced embryo sacs of variable morphology, which can coexist with the legitimate sexually derived embryo sacs, or alternatively outcompete them.5
Female germline development in the model species Arabidopsis thaliana, which reproduces exclusively sexually, has been studied intensively in recent years.6,7,8,9 The process commences with the specification of a single subepidermal cell as the germline precursor, its enlargement to form the MMC, and its entry into meiosis. One of the four meiotic products, the FM, then initiates a controlled series of mitosis to form the mature gametophyte. A multitude of molecular mechanisms is required to establish MMC fate, and to regulate its subsequent differentiation. Importantly, canonical female germline commitment is dependent on a range of non-cell-autonomous and epigenetic pathways. For example, RNA-directed DNA methylation (RdDM) – a crucial epigenetic pathway in plants – has been shown to be required for the establishment of a single germline in the Arabidopsis pre-meiotic ovule.10 Significantly, SEEDSTICK (STK), a MADS-box transcription factor that acts as a master regulator of ovule identity, has recently been reported to directly control the transcription of two major RdDM components, AGO9 and RDR6.11
The phytohormone auxin has also been identified as playing a central part in ovule primordia. Polarity setting in the ovule involves localization of PIN auxin transporter proteins in the external cell layer (L1 layer) and an auxin maximum has been detected at the tip of the ovule primordia, directly above the site of MMC differentiation.12,13 Specificity of the auxin response is conferred by Auxin Response Factors (ARFs), transcriptional controllers that select target genes for regulation by the hormone. ARFs bind to auxin-response DNA elements (AuxRE) within the promoters of auxin-regulated genes, and either activate or repress transcription depending on a specific protein domain.14 ARF activity is controlled by IAA repressors and is promoted in the presence of auxin through AUX/IAA degradation. However, it has recently been shown that an ARF5/MP alternative isoform could function as a transcriptional activator within the ovule, even in regions of subthreshold auxin concentration.15 Some clades of the ARF family are modulated by the THO complex, for example THO/TREX was shown to repress the formation of extra MMCs, mediated by the TAS3-ARF3 module.16 Other ARFs, such as ARF6/ARF8 and ARF10/ARF16/ARF17, are regulated by miRNAs (miR167 and miR160, respectively).17,18,19,20,21 Huang et al.22 have recently characterized the expression of miR160 in Arabidopsis wild type ovules together with miR160-driven ARF17 modulation. Mature miR160 was detected not only in the chalaza and funiculus but also within the MMC, and one of its targets (ARF17) was shown to be a key determinant for promoting MMC specification.22 Auxin is not the only phytohormone involved in female germline acquisition for a recent study by Cai et al.23 showed that brassinosteroids biosynthesis and signaling components were also linked to female germline fate acquisition.
The study reported here is focused on the role of ARF10, another miR160 target, in the differentiation of the FM within the ovule. Our interest in this gene stemmed from recent comparative transcriptomic analyses intended to identify apomixis-candidate genes in the aposporous subtropical grass Paspalum notatum which revealed that both miR160 and ARF10 were differentially expressed in the apomictic genotypes.24,25 While current literature on ARF10 function is primarily focused on its activity in germination and somatic embryogenesis, blade outgrowth, leaf/leaflet initiation, and floral organ development,21,26,27,28,29 the detection of ARF10 differential expression in flowers of sexual and apomictic P. notatum plants at megasporogenesis/megagametogenesis stages suggested an additional role. By establishing the function of the miR160/ARF10 interaction in Arabidopsis sexual reproduction we have attempted to throw light on any role it may play in reproductive development. Our data reveal that strict spatial regulation of ARF10 by miR160 is required for the induction of a single female germline and, significantly, that ectopic expression of ARF10 throughout the ovule leads to the emergence of supernumerary FM, either reduced (surviving spores) or unreduced (originated from somatic cells). The formation of these extra FM leads to the development of misoriented, supernumerary, morphologically dissimilar female gametophytes, a phenotype mimicking aposporous apomixis. Furthermore, we also show that the regulation of ARF10 expression in the nucellus during ovule primordium development is mediated by a combination of (1) transcriptional regulation by the MADS-box transcription factor STK and (2) post-transcriptional control by AGO1 and miR160.
Results
ARF10 expression surrounds the MMC and its meiotic products
To study the expression pattern of ARF10 in wild type contexts, we used Arabidopsis lines expressing an ARF10_GREEN FLUORESCENT PROTEIN (GFP) fusion protein.21 No GFP signal was detected at early pre-meiotic stages when ovules are finger-like (stages 2-I, in Hernández-Lagana et al.30). At stage 2-II, when the MMC is clearly differentiated and immediately prior to meiosis, a weak but clear GFP signal was detected in the L1 and L2 layer, surrounding the MMC (Figure 1A). During meiotic divisions, the signal became restricted to a few cells surrounding the dividing MMC and, after meiosis, during the FG1 stage of gametogenesis, it appeared only at the top of the FM and started to be visible in the integuments (Figure S1A). At FG7, after three rounds of mitosis, the GFP signal remained in the outer integuments, being undetectable within the embryo sac (Figure S1B).
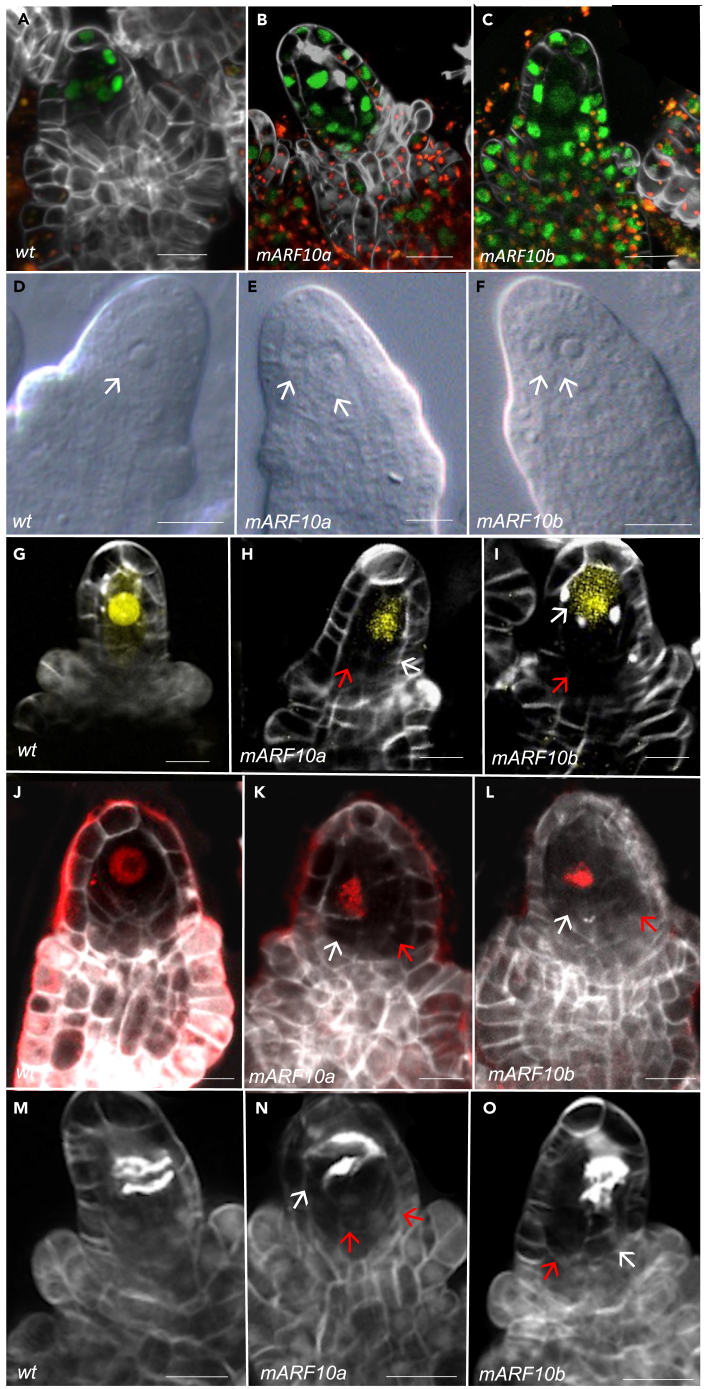
Figure 1.
Enlarged cells surrounding the MMC were identified in mARF10_GFP lines
(A) ARF10_GFP expression at pre-meiosis is detected in a few cells surrounding the MMC (L1 and L2 layers).
(B and C) mARF10_GFP expression pattern (lines a and b), at pre-meiosis a strong GFP signal is detected in all ovule cells, including the MMC.
(D) DIC analysis in WT.
(E and F) DIC analysis in mARF10 lines a and b, respectively; white arrows indicate multiple enlarged cells.
(G) pKNU:nlsYFP expression in WT.
(H and I) pKNU:nlsYFP expression in mARF10 a and b, respectively; the YFP signal was detected only in one of the enlarged cells (white arrows), often displaced to one side by the presence of extra enlarged cells (red arrows).
(J–L) Meiotic marker ASY:RFP in WT and mARF10 lines. In the transgenic lines only one of the enlarged cells accumulated the meiotic precursor (white arrows), extra enlarged cell didn’t express it (red arrows).
(M–O) Renaissance staining in ovule primordia during meiosis; cell plates accumulating callose are detectable. In the WT context two central plates are identifiable; in the transgenic lines they are often displaced (white arrows), red arrows indicate the extra enlarged cells. Renaissance staining was used for the cell walls in all marker line backgrounds. GFP and YFP signals detects expression, RFP detects autofluorescence. Scale bars:10μm.
To determine ARF10 expression in the absence of miR160 regulation, we used two Arabidopsis lines21 in which the miR160-binding site was modified to make ARF10 insensitive to miR160 regulation, mARF10_GFP (mARF10a and mARF10b - Figures 1B and 1C, respectively, mARF10 stands for mutated ARF10). Confocal microscopy analyses revealed a strong GFP signal in the nuclei of all ovule primordia cells. A clear signal was observed just before meiosis - stage 2-II (Hernández-Lagana et al.30), with the fluorochrome being detectable in the whole ovule, even inside the MMC (Figures 1B and 1C). After meiosis (stage FG1), following FM differentiation, the signal remained visible in all cells (Figure S1C). During gametogenesis (FG2-FG6), it was particularly visible in all integument layers and in FG7 mature ovules (Figure S1D), where it was detected in the inner and outer integuments and even within the embryo sac. Taking our findings together with those Huang et al.,22 we conclude that, in wild type plants, ARF10 is transcribed throughout the ovule from stage FG0 (ovule primordia pre-meiosis) to FG7 (mature ovule), but the concomitant action of miR160 results in its silencing in all cells except for a few cells surrounding the MMC immediately before and during meiosis (from FG0 to FG1), and in the outermost cells of the external integuments at later stages (FG2-FG7).
mARF10_GFP lines exhibit supernumerary enlarged cells around the MMC
We then used the miR160-insensitive mARF10_GFP lines21 to study the impact of ARF10 expression repatterning on ovule development and fertility. Inflorescences from mARF10 lines a and b, and from wild type plants, were cleared and studied using Differential Interference Contrast (DIC) microscopy. At stage 2-I, FG0 (when the MMC becomes distinguishable from the surrounding nucellar cells owing to its increased size) a significant number of ovules were observed to contain multiple enlarged cells (Figures 1D–1F). The percentage of ovules with these multiple enlarged cells was 22.5% in mARF10a (n = 23/103), and 21.6% in mARF10b (n = 44/204), compared with only 7% in wild type ovules (n = 21/300).
To establish the identity of these supernumerary enlarged cells, we crossed the mARF10 lines with the MMC identity marker pKNU::nlsYFP31. Confocal analysis showed that, in the wild type context, the signal was clearly central and occupied all the available space (n = 318) (Figure 1G). In the mARF10 lines, only one cell expressed the YFP signal while the extra enlarged cells displayed no signal (n = 189/290) (Figures 1H and 1I); however, the cell generating the YFP signal was often displaced to the side of the ovule, presumably resulting from the expansion of one or more adjacent cells. YFP signal-counting showed no difference between the wild type and the mARF10 backgrounds. The commitment of these extra enlarged cells to meiotic entry was assessed using the meiotic precursor ASY3::RFP32 (Figure 1J). In the wild type context, the ASY3:RFP signal was detected only within the MMC (Figure 1J). When this marker line was introgressed in the mARF10 background, only one cell per ovule, often displaced to the side, expressed the RFP signal (n = 20/50) (Figures 1K and 1L), meaning that only one of the enlarged cells accumulates the meiotic precursor and is capable of entry into meiosis (Figures 1K and 1L). To further dissect meiotic progression, we have used Renaissance staining, which allows the detection of the callose meiotic septa11 (Figures 1M–1O). Again, only one cell was found to enter in meiosis (n = 100) in the mARF10 lines, often displaced to the side, when compared to wild-type ovules (n = 27/100). No alterations in the pattern of callose deposition around the MMC or in the meiotic septa were visible in the mARF10 lines. Taken together, these findings are consistent with the fact that only one of the enlarged cells have true MMC identity, since only one cell accumulates the meiotic precursor and forms callose septa, indicating meiotic entry and progression.
Seed set analysis of the two independent lines mARF10_GFPa (n = 350 ovules) and mARF10_GFPb (n = 186 ovules) revealed siliques with a significant percentage of unfertilized ovules when compared with wild type (n = 243 ovules) (28% for mARF10a and 27% for mARF10b) (Figures S2A–S2E). Moreover, the vegetative organs of mARF10_GFPa and mARF10_GFPb plants showed anomalies previously described by Liu et al.21 The most severe phenotype (seen in average in 1/20 plants) resulted in very small and completely sterile plants, i.e., siliques without any fertilized ovules. Other plants displayed more moderate phenotypes, with impaired development, less unfertilized ovules, and serrated leaves. After meiosis, at FG1 stage, mARF10 DIC analysis also showed extra enlarged cells around the legitimate FM in 25.8% of 348 ovules compared with 0% of 300 ovules in wild type (Figures S2F–S2H).
mARF10 lines present apospory-like phenotypes
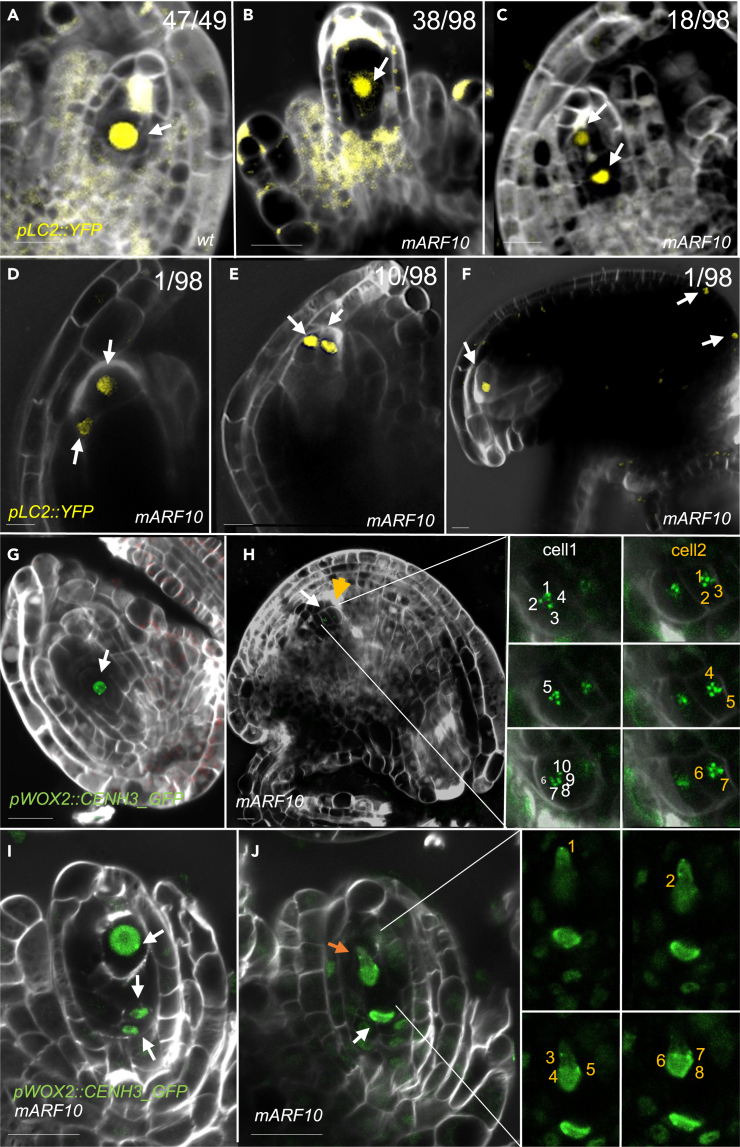
To further investigate the identity of the extra enlarged nucellar cells surrounding the FM, we crossed the FM identity marker pLC2:nlsYFP31 into the mARF10 lines (Figures 2A–2F). In the mARF10 sterile phenotype, only 5% of the ovules expressed the marker (n = 250). However, in mARF10 moderate phenotypes we detected several defects. In a considerable proportion of ovules, meiosis occurred when the integuments were just emerging, and the YFP signal was evident earlier than in wild type, i.e., before the integuments had surrounded the ovule (Figure 2B). Moreover, extra cells with FM identity appeared around the legitimate FM (Figures 2C–2F). These cells do not correspond to a 2n gametophyte since cell walls were clearly distinguishable between the marked cells (Figures 2C–2F). Some of them seem to be extra viable megaspores surviving from meiosis (i.e., they are located within the meiotic tetrad lineage) (Figure 2C), while others originate from surrounding non-reduced cells (i.e., they appear in the nucellus or the integuments) (Figures 2D and 2E). At ovule maturity, ectopically located signals were also detected in the integuments (Figure 2F). In mARF10 plants, the LC2 marker revealed 49% ovules with one FM, 19% with more than one FM and 32% without signal (n = 98). In a wild type background, we counted 98% ovules with canonical development and 2% ovules with altered development (possible evidence of 2 megaspores) (n = 49) (see statistical data in the legend of Figure 2). Our results therefore suggest in mARF10 lines there are extra enlarged nucellar cells that express the FM identity marker. By contrast, in pre-meiotic wild type ovules approximately 7% show enlarged cells surrounding the MMC, but they become no longer visible at FG1.To better understand whether these extra FM where reduced or unreduced, we used a second marker, pWOX2:CENH3_GFP, which is active specifically in the FM and clearly localized the chromosome centromeres, making it possible to count the number of chromosomes in each cell33 (Figures 2G–2J). In wild type backgrounds, five chromosomes were detected in the surviving FM after meiosis (Figure 2G). By contrast, in mARF10 backgrounds we found some cells expressing the pWOX2:CENH3_GFP marker that contain more than five chromosomes. From a total of n = 113 ovules analyzed, only n = 17 showed a clear signal and could be used for counting. Non-reduced cells expressing pWOX2:CENH3_GFP were observed after meiosis surrounding the reduced FM, and also in the mature ovule (Figures 2H–2J, video projections S1 and S2). These results suggest that some of the additional FM cells are non-reduced, a phenotype commonly detected in aposporous apomictic species.
Figure 2.
Extra functional megaspores detection in mARF10 background
(A) pLC2:YFP FM identity marker in the wild type background.
(B–F) pLC2:YFP in the mARF10 background: (B) YFP signal detects the functional megaspore, meiosis occurred early with respect to integument development; (C) extra YFP signal detected close to the degenerated megaspores; (D and E) extra YFP signals in the L1 layer; (F) YFP signal in sporophytic tissue at ovule mature stage. The LC2 FM marker signal was counted in a fluorescent microscope and a significance of statistical analysis was performed using 95% Confidence Interval (CI): 48.98% ovules with one functional megaspore (0.3882 < p < 0.5922), 19.39% with more than one functional megaspore (0.1236 < p < 0.2887) and 31.63% without signal (0.2281 < p < 0.4191) (n = 98 ovules). In a wild type background, 97.96% (0.8776 < p < 0.9989) showed one functional megaspore and 2.04% (0.0011 < p < 0.1224) two functional megaspores (n = 49).
(G) pWOX2:CENH3_GFP in wild type background, showing a single reduced FM with five chromosomes (green dots).
(H) mature ovule with two cells at the chalaza pole expressing the GFP marker (white and yellow arrow, respectively); the cells are non-reduced, as more than five green dots are detected per cell. In the right panels, different focal planes of each one of the cells are shown: 10 chromosomes are visible in cell 1 (white arrow), while 7 in cell 2 (for more details see Video S1).
(I and J) two different focal planes of the same ovule at FG1 stage: (I) three functional megaspores are detected (white arrows). The upper one display 5 chromosomes (five green dots) (for more details see Video S2), the bottom ones present more than 5 chromosomes. (J) in a different focal plane of the same ovule, new cells expressing the FM marker are visible: the upper one (yellow arrow) display at least 8 chromosomes (magnified panels at right, full view at Video S2). Scale bars:10μm.
mARF10 lines show gametophytic cell identity defects
To investigate female gametophyte development at later developmental stages (FG7), we used a combination of clearing and DIC microscopy of emasculated wild type and mARF10 pistils (Figures 3A and 3B, respectively). In the wild type line, a typical seven celled embryo sac was always observed (n = 300). In mARF10 lines we observed that 25% of the ovules presented misplaced supernumerary nuclei (Figure 3B, n = 370). Through Feulgen staining, we confirmed an abnormal cellular organization within mARF10 gametophytes, as illustrated in Figures 3C and 3D. Particularly noteworthy are several examples of aberrant development at the embryo sac seen in the mARF10 lines, which markedly differ from the canonical typical seven-celled embryo sacs observed in the wild type. Additional images of these atypical embryo sacs detected in mARF10 are shown in Figures S3A–S3E. These aberrant structures may indicate the presence of extra embryo sacs, a characteristic commonly observed in aposporous apomictic species like Paspalum notatum (for comparison, a DIC picture is provided) (Figure S3F).
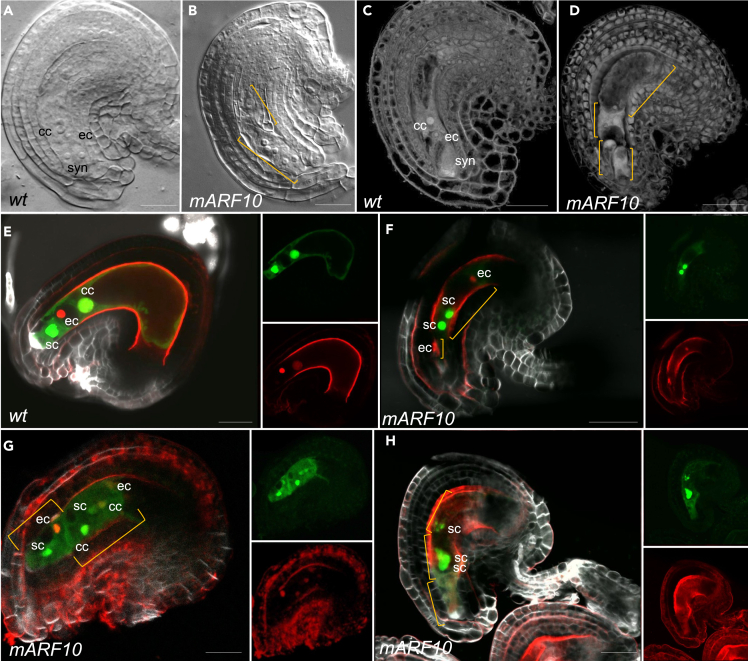
Figure 3.
mARF10 female gametophyte analysis revealed developmental alterations
(A and B) DIC images from wild type and mARF10 mature ovules, extra numerary nuclei were detected in the mARF10 mature embryo sacs.
(C and D) Feulgen staining of wild type and mARF10 mature ovules revealed different embryo sac limits when compared with wild type. Yellow bars mark different embryo sacs.
(E) Left panel: wild type mature ovule expressing FGR7.0; right panel: green and red channels of the same image, two synergids expressing the GFP marker are positioned in the micropillar pole, flanked by the egg cell (RFP red marker) and the central cell (yellowish color given by the red and green channel fusion).
(F–H) mARF10 ovules expressing FGR7.0. (F) two egg cells are detected in opposite poles of the embryo sac, and two synergids in the middle region (green and red channels are shown at right); (G) several cells showing synergid/central/egg cell identity.
(H) potentially three embryo sacs, in two of them groups of synergid cells are visible. Renaissance staining was used to mark cell walls. Green corresponds to GFP/YFP signal and red to RFP signal. Counting for the FGR7.0 marker crosses: wt-like ovules: 5; abnormal ovules: 37; ovules without signal: 74 (n = 116). For DIC imaging, the significance of statistical analysis was calculated by using 95% Confidence Intervals (CI), as follows: misplaced supernumerary nuclei surrounded by membranes 25% (0.2112 < p < 0.3022) (n = 370). In WT, no supernumerary nuclei were observed 0% (0 < p < 0.0158) (n = 300). sc: synergid cells; ec: egg cell; cc: central cell. Scale bars: 20μm.
However, it is essential to acknowledge the possibility that these additional cells may indeed belong to the same sac but are showing abnormal identities and positions. To determine whether several embryo sacs were present in the mature ovule and/or if the gametophyte cell-identity was compromised, the FGR7.0 female marker line34 was crossed into the mARF10_GFP background. FGR 7.0 combines marker genes that label synergid cells, the egg cell, and the central cell with different color (EC1:NLS_3xdsRed for egg cell, DD22:NLS_YFP for central cell/endosperm, and DD2:NLS_3xGFP for synergid cells/endosperm).34 In mature ovules from emasculated wild type flowers, the marker line showed normal embryo sacs (Figure 3E) but some ovules of the mARF10_GFP line (Figure 3F) were characterized by supernumerary egg cells situated at different poles of the embryo sac, distant from the correct location. The mARF10 lines also exhibited embryo sacs with an excessive number of cells, displaying concurrent identities of egg, synergid, and central cells (Figure 3G). Moreover, we also detected embryo sacs expressing only synergid identity at the middle of the sac (Figure 3H). Additional images depicting abnormal sacs with multiple identities and unusual locations can be seen in in Figure S3 (Figures S3G–S3O). The presence of cells expressing egg/synergid/central cell markers at abnormal locations suggests either the formation of supernumerary embryo sacs or cell identity misspecification.
As previously indicated, some mARF10 lines exhibited nearly sterile phenotypes. In these plants the increased presence of enlarged cells before meiosis and the occurrence of aberrant gametophytes were prominent, and these abnormalities appeared to be linked with impaired integument growth, suggesting a potential delay in integument formation (Figures S4A–S4D). More rarely, extreme examples of this defect were seen, where integuments remained undeveloped and gametophytes were directly expelled from the mature ovule or missing, or where an apparent aborted gametophyte was present adjacent to a normal gametophyte (Figures S4D–S4F). Taken together, our findings suggest the presence of one or several misoriented gametophytes with altered cell identity, a phenotype reminiscent of naturally-occurring aposporous apomictic plants.
arf10 defective mutants show aberrant reproductive phenotypes
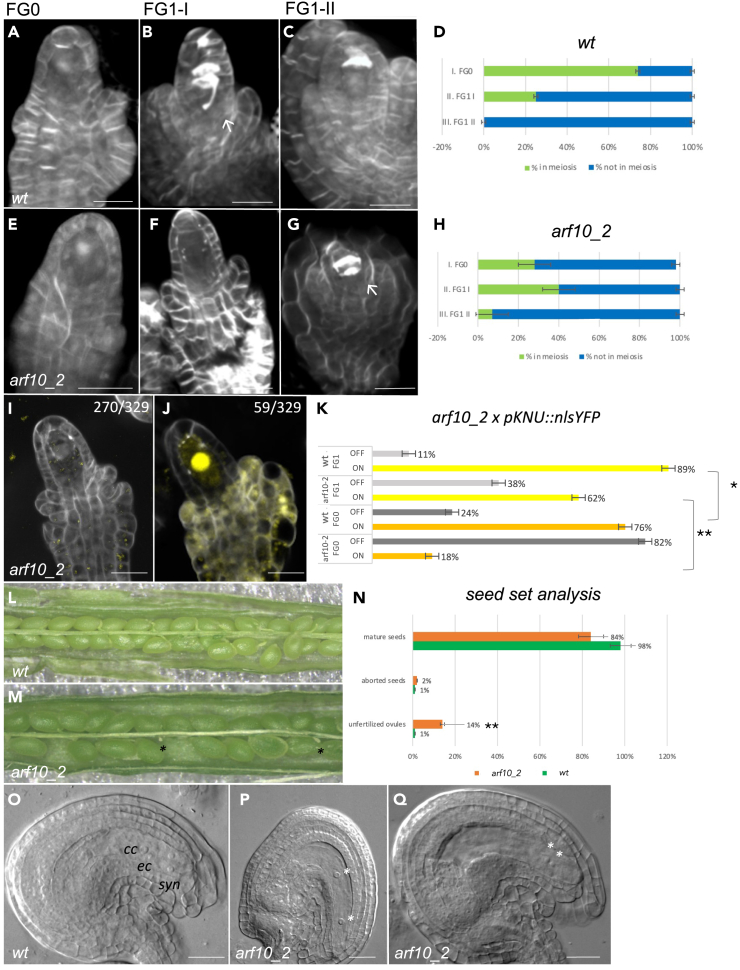
To gain deeper insights into the involvement of ARF10 in ovule development, we analyzed ovule development in a homozygous line carrying an insertional mutation, designated as arf10_2. To confirm the mutation site in the ARF10 sequence, PCR amplicons were sequenced. The ARF10 transcript representation was then quantified by qPCR, revealing a relative expression of 0.3 when compared with wild type plants (SD: 0.1569) (Figure S5C). Both cleared arf10_2 and wild type ovules were examined using DIC microscopy; at ovule pre-meiotic stage 2I some supernumerary enlarged cells were observed, but the numbers were not statistically significant in comparison with wild type (10.8% in arf10_2 n = 414 vs. 8.3% in wild type n = 336). Upon further examination of the ovule development, a potential delay in meiosis became evident. Notably, in a considerable proportion of ovules, the linear tetrad degeneration had not occurred by the time integuments enclosed the nucellus — 24.9% in arf10_2 (n = 334 ovules) compared to 0% in the wild type (n = 315 ovules) (Figures S5A and S5B). Renaissance staining was then used to follow meiotic progression in these ovules11 (Figures 4A–4H). To simplify statistical analysis, meiosis was divided into three FG stages with respect to integumental development: FG0, which corresponds to finger-like ovule prior to integument growth, FG1-I during which meiosis is taking place and before the ovular enclosure by the integuments and FG1-II, following meiosis when integuments completely enclose the nucellus. As seen in Figures 4A–4D, most wild type ovules entered meiosis at FG0 (74%, n = 239 ovules) and FG1-I (25%, n = 94 ovules), with meiosis being completed by FG1-II (0%, n = 145 ovules). By contrast, the proportion of arf10_2 ovules entering meiosis at FG0 was 28% (n = 298), while at FG1-I was 40% (n = 55) and 7% (n = 57) completed meiosis by FG1-II (Figures 4E–4H). According to these results, arf10_2 mutant ovules are delayed in entering meiosis, which extends into stage FG1-II. No alterations in the pattern of callose deposition around the MMC or in the meiotic septa were detected in the arf10_2 ovules (Figures 4E–4G). To further determine the identity of the cells delayed in meiotic entry, arf10_2 mutants were crossed with the pKNU:nlsYFP MMC marker line.31 Strikingly in FG0, 18% displayed signal (Figure 4J, n = 329), with 82% of the arf10_2 ovules showing no signal (Figure 4I, n = 329). In a wild type background, ovules with signal occurred in significantly higher numbers (Figure 4K, n = 318), pointing to either disruption or delay in the acquisition of MMC fate in the mutant. To further test the hypothesis that meiosis was delayed and probably also the acquisition of MMC identity, based on evidence from the KNU marker, we analyzed arf10_2 ovules closer to FG1 stage, with elongated integuments. Strikingly, 62% (n = 117/184) of the analyzed ovules displayed YFP signal, demonstrating that the acquisition of the identity was delayed as well as meiosis (Figure 4K). Seed set analysis showed a significant increase in the percentage of unfertilized ovules in mutant lines (14%, n = 792 ovules) when compared with wild type (1%, n = 738 ovules). Homozygous mutants produced only 84% mature seeds, a number significantly lower than wild type (98%) (Figures 4L–4N). Furthermore, we detected a lack of coordination between embryo sac and integument development, since in a substantial number of ovules progression of gametogenesis was delayed, with some ovules arrested at the two-nucleate stage (15.1% for arf10_2 ovules n = 396 and 2.3% for wild type ovules n = 300) (Figures 4O–4Q). Complementation tests demonstrated that in three plants of the cross arf10_2 x ARF10_GFP, there were 8% unfertilized ovules (n = 642) in average, which is statistically equivalent to the percentages detected in wild type plants. A second defective allele, arf10_ 3, was analyzed and the main phenotypes described above were confirmed (Figure S6G). Seed set was also compromised, as 21% of the ovules were unfertilized and an arrest in seed development was detected in 14% of the seeds (n = 831) in comparison with wild type plants, which presented only 5% unfertilized ovules and 1% of arrested seeds (n = 738) (Figures S5D–S5F). To confirm the mutation site in ARF10, PCR amplicons were sequenced and ARF10 transcript representation was quantified by qPCR. No expression was detected in the mutant (Figure S5G). A delay in entering meiosis was observed in 63% of the analyzed ovules (n = 305) which was partially reflected in a block of gametogenesis in 30% (n = 430), mainly at two-nucleate stage (Figures S5H and S5I). Taken together, these data indicate that, in the wild type, ARF10 expression in cells surrounding the MMC, and subsequently in the external integuments, is required for correct initiation of meiosis and progression of megagametogenesis.
Figure 4.
arf10_2 defective mutant reproductive phenotype shows developmental delay
(A–C) wild type Renaissance staining highlighting meiotic progression at three different ovule stages; (A) FG0, finger-like ovule showing elongating integuments.
(B) FG1-I, meiosis ongoing or completed; the integuments have yet to cover the ovule.
(C) FG1-II after meiosis, integuments fully elongated, callose accumulation signifies tetrad degeneration.
(D) meiosis proportion in Renaissance-stained wild type ovules at three different stages.
(E–G) arf10-2 Renaissance staining highlighting meiotic progression at three different ovule stages (stages as in A–C). Meiosis is still in progress when integuments are fully elongated (white arrow).
(H) meiosis proportion in Renaissance-stained arf10_2 ovules at three different stages.
(I and J): arf10_2 ovules without (I) and with (J) pKNU:nlsYFP signal.
(K) Bar chart showing pKNU:nlsYFP expression in wild type and arf10-2 backgrounds at FG0 and FG1 stages: ON: YFP expression, OFF: no expression.
(L and M) seed set analysis in WT and arf10_2, black asterisks mark unfertilized ovules.
(N) proportion between mature seeds/aborted seeds/unfertilized ovules.
(O–Q) DIC imaging of wild type and arf10_2 mature ovules. Some arf10_2 ovules presented a block during gametogenesis; white asterisks indicate nuclei. The significance of differences between wild type and mutants for seed set and pKNU:YFP signal detection was evaluated by the Student’s t test (∗∗p < 0.05; ∗∗∗p < 0.001). For DIC imaging, the significance of statistical analyses was evaluated using the 95% Confidence Interval (CI): 15.15% vs. 2.33%; n = 396 and 300; 95% CI: 0.1184 < p < 0.1915 and 0.0102 < p < 0.0495 for arf10_2 and the wild type, respectively, Data are represented as mean ± SEM; scale bars: 20 μm; cc: central cell; ec: egg cell; syn: synergid cells.
ARGONAUTE 1 and SEEDSTICK regulate ARF10 nucellar silencing
To explore the mechanism underlying miR160-mediated ARF10 silencing, we attempted to identify the ARGONAUTE protein associated with this process. We first hypothesized that AGO1 played a role, since AGO1 transcripts had been reported as differentially expressed in pre-meiotic spikelets of aposporous apomictic and sexual Paspalum plants.25 Null alleles of AGO1 cause early lethality, so we analyzed a previously described viable but hypomorphic allele of ago1-2735 known to be defective in the RNaseH-like PIWI domain, which is involved in mRNA cleavage and translational repression.36
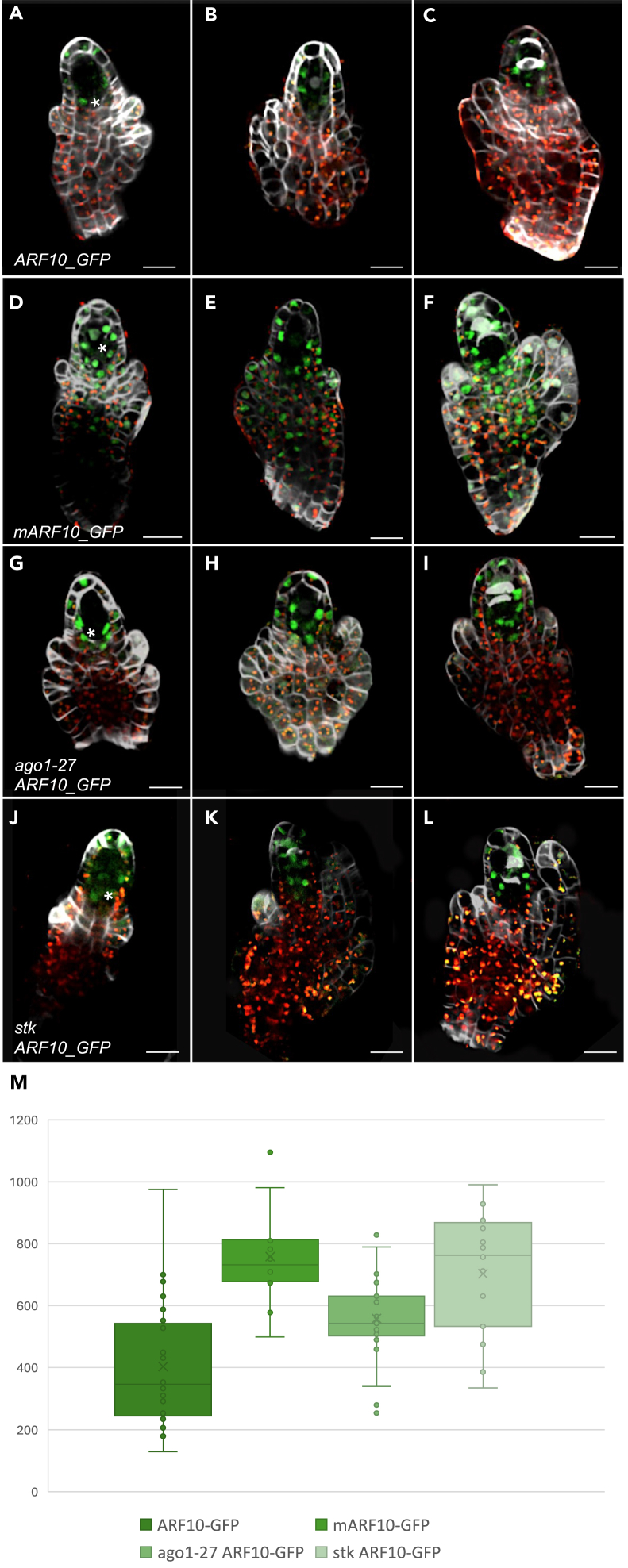
We first conducted an analysis of ARF10 expression in both wild type and ago1-27 backgrounds. In wild type samples (Figures 5A–5C), ARF10 exhibited expression primarily around the MMC, within the L1 and L2 layers, as mentioned before. Contrastingly, in backgrounds displaying miR160 insensitivity (mARF10_GFP) (Figures 5D–5F), expression was widespread throughout the ovule. Notably, in ago1-27 pre-meiotic/meiotic nucellar cells, a pronounced expression of ARF10 was observed within the nucellus (Figures 5G–5I), while no expression was detected in the MMC and chalaza.
Figure 5.
ARF10 expression patterns in different genetic contexts
(A and B) In wild type context, ARF10_GFP expression at pre-meiosis is detected in a few cells surrounding the MMC.
(C) in the post-meiotic ovule, ARF10_GFP expression is maintained in few L2 cells surrounding the meiocytes.
(D and E) mARF10_GFP expression pattern at pre-meiosis, a strong GFP signal is detected in all ovule cells, including the MMC.
(F) mARF10_GFP expression at post-meiosis, the GFP signal is detected in all ovule cells, including the funiculus and chalaza.
(G–I) ARF10_GFP protein accumulation in ago1-27 nucellar cells at three successive developmental stages, from MMC differentiation to meiosis. The expression is observed in the same domain as for the wild type context, but the signal is stronger (see quantification).
(J–L) ARF10_GFP protein accumulation in stk nucellar cells at three successive developmental stages, from MMC differentiation to meiosis. The expression is observed in the same domain as for the wild type context, but an increased signal is detected within the MMC. The signal is stronger (see quantification).
(M) mean GFP intensity detected in the nucellus for the different genotypes: mARF10, ago1 and stk backgrounds showed significantly higher signal levels with respect to wild type ones. Data are represented in a dot plot as mean ± SEM. Statistical analysis using one-way ANOVA and Tukey’s HSD test (p < 0.01) showed significant differences in GFP expression levels between WT and mARF10, ago1-27 ARF10_GFP, and stk ARF10_GFP. Scale bars: 20 μm. Cell walls were stained using Renaissance. Green signal corresponds to GFP.
GFP intensity variation among the different lines was quantified using ImageJ software, focusing exclusively on nucellar cells. Statistical analysis uncovered disparities in GFP intensity between ARF10_GFP and mARF10_GFP (Figure 5M). Significant differences were also observed between ARF10_GFP and ago1-27 ARF10_GFP, indicating a role for AGO1 in the nucellar regulation of ARF10 (Figure 5M). To better explore the connection between AGO1 and ARF10, we analyzed the phenotype of ago1-27 mutants during the early stages of ovule development. Approximately 25% (n = 203) of the ovules from ago1-27 mutants exhibited multiple enlarged cells during pre-meiotic stages, compared to only 4% (n = 435) in wild type samples. This observed phenotype bears a remarkable similarity to that observed in mARF10 lines. To further investigate potential disruptions in female gametophyte formation, we crossed ago1-27 with the FGR 7.0 marker (Figure S6). Mature ago1-27 ovules resulting from these crosses exhibited similar phenotypes to those observed in mARF10 lines (see Figures S6D–S6G). Notably, in many of these ovules, the positioning of the egg cell was significantly altered, being often found near the chalazal pole rather than the micropylar pole. Moreover, both synergid and central cells were frequently found to be misplaced - as seen for mARF10 in some cases.
Concurrently, we initiated a series of experiments aimed at unraveling the intricacies of STK regulation of ARF10, as STK has been shown to be involved in female germline development.11 To explore the relationship between STK, miR160 and ARF10, we examined ARF10 expression patterns in crosses between ARF10_GFP and stk-2 lines.37 As was observed in the ago1 background, in the stk mutant ARF10 was highly expressed in the nucellus (L1 and L2 layers), but in this case we detected an increment of the signal within the MMC during premeiotic/meiotic stages (Figures 5J–5L). This differential expression was confirmed by GFP quantification (Figure 5M). We also conducted chromatin immunoprecipitation (ChIP) followed by qPCR on pSTK:STK_GFP plants38 to explore the potential direct control of ARF10 expression by STK. Analysis of the ARF10 promoter region (2 kb upstream of the ATG starting site) revealed the presence of six possible STK binding sites, known as CARG boxes39 (Figure S7H). ChIP-qPCR results demonstrated that STK directly binds to the promoter region of ARF10 via CARG boxes 1 and 2 (Figure S6H), suggesting a direct repression of ARF10 expression by STK.
As STK was previously shown to be involved in the RNA directed DNA methylation (RdDM) pathway,11 we also investigated the possibility of a methylation control by performing Methylation Content Sensitive Enzyme ddRAD (MCSeEd) analysis40 in wild type plants versus stk mutants. Triplicate flower samples (up to and including stage11) were collected to minimize the impact of epigenetic variation between individuals.41,42 Eighteen (18) MCSeEd libraries were constructed by double restriction–ligations, using MseI in combination with one of three methylation-sensitive enzymes AciI, PstI, and EcoT22I for CG, CHG, and CHH contexts, respectively (Table S1). Exclusively mapped loci (MCSeEd loci) were normalized, filtered, and then analyzed with MethylKit to infer the number of differentially methylated positions (DMPs) between mutants and wild type plants, as either delta positive or delta negative. A total of 1,507 DMRs were scored in the stk mutants compared with the wild type plants (Table S1). The genomic location of each DMR was mapped onto Arabidopsis transcribed genic regions extended by 2.5 kb at both ends (extended gene bodies; EGBs, Table S2), and the genes belonging to these EGBs were defined as differentially methylated genes (DMGs). The list of DMGs included a CHH-DMR with negative delta methylation that intersected miR160 b at 109 bp from the Transcription Termination Site (TTS), while no methylation differences were encountered for the ARF10 genomic locus. To validate the differential methylation of the miR160-CHH-DMR, we employed the quantitative (q)MRE technique and, as expected, the digested samples of stk mutants showed a value lower than the wild-type ones, demonstrating that the positions belonging to the miR160-CHH-DMR were demethylated (Figure S6J; Table S3). Following this, we investigated whether STK influences the expression of miR160. We did a stem loop-PCR to quantify the levels of mature miRNA in the stk mutant background, revealing a significant downregulation of the mature form of miR160 (Figure S6I). Altogether, our findings suggest that STK may modulate ARF10 levels through two distinct mechanisms: (1) a direct negative regulation mechanism involving STK binding to the ARF10 promoter and (2) an indirect process involving methylation of the miR160b genomic locus, resulting in increased mature miR160 levels and subsequent ARF10 repression.
To investigate into cellular dynamics with greater precision, prompted by the distinct GFP pattern, we utilized both sense and antisense GFP probes via in situ hybridization. This approach enabled us to explore transcript accumulation at the cellular level in these lines. At pre-meiosis stages, a faint signal was detected across the ovule in ARF10_GFP lines (Figure S7A). However, in mARF10_GFP lines (Figure S7B) the accumulation of transcripts within the ovule was notably higher. Additionally, when comparing hybridization of ARF10_GFP with ago1-27 ARF10_GFP and stk ARF10_GFP, the signal was higher, but lower than that observed with mARF10_GFP (Figures S7C and S7D). In stk, a lower GFP hybridization signal was also detected within the MMC. In conclusion, the observed transcript levels suggest that both AGO1 and STK play roles in suppressing ARF10 expression within the nucellar layers, although their impact on transcript abundance appears to be minor. Conversely, in mARF10, the regulation by miR160 appears to have a significant impact, affecting transcript abundance in the nucellus, chalaza, and funiculus. It is noteworthy that transcripts were detected throughout the ovule rather than being confined to a specific cell type, suggesting potential involvement of post-transcriptional/translational mechanisms in ARF10 regulation. Finally, we tested the abundance of miR160 mature RNA by in situ hybridization, as demonstrated in Figure S8. In WT ovules, miR160 is distributed throughout the ovule primordia cells, except for the cells surrounding the MMC (Figure S8A). In stk mutants, the miR160 signal was markedly lower within the MMC (Figure S8B), suggesting that STK positively regulates miR160 expression, which agrees with the GFP hybridizations. Analysis of ago-1 mutants (Figure S8C) did not show a significant difference compared to the WT, indicating that AGO1 does not influence miR160 expression levels.
Discussion
Does ARF10 control a gametophytic factor?
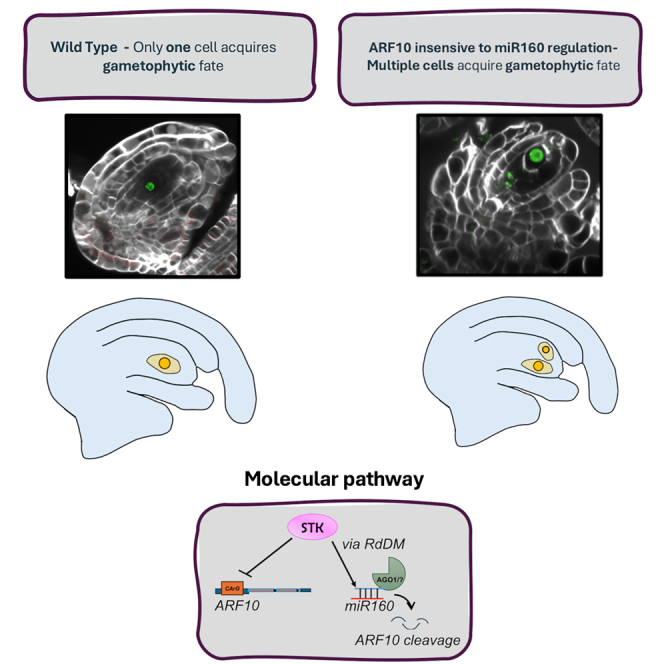
This study highlights the critical role of ARF10 during the regulation of gene expression in response to the plant hormone auxin. Our findings underscore the essential contribution of ARF10 to the development of both sexual and asexual female germlines. In a sexual wild-type plant, such as the model Arabidopsis, during the early stages of ovule development, we observed a highly specific expression pattern of ARF10 localized within the ovule nucellus, surrounding the MMC—the cell destined to undergo meiosis. Previous studies have demonstrated that ARF10 is regulated by miR160,21 but here we have analyzed the detailed dynamics of this regulation within the ovule, particularly in lines where ARF10 was insensitive to miR160 regulation (mARF10). GFP signal comparisons between ARF10-GFP and mARF10-GFP lines revealed that ARF10 transcription occurs ubiquitously across the ovule during all stages of development, but the expression is subsequently silenced by miR160 in most cells, except for those surrounding the MMC. Consequently, during the ovule primordia stage, the MMC appeared to be the only cell enveloped by ARF10-expressing cells.
The ectopic expression of ARF10 identified in the mARF10 lines induces a highly specific phenotype during ovule development: the emergence of multiple enlarged cells surrounding the MMC, lacking MMC identity initially but ultimately acquiring FM characteristics. As demonstrated through crosses with the FM-identity markers pLC2:nlsYFP and pWOX2:CENH3_GFP, in mARF10 lines, the additional enlarged cells originating from nucellar and integumental layers exhibit FM identity signal. Some of them presented a non-reduced status, characterized by possessing more than five chromosomes, while others were reduced, resulting directly from the meiotic events. Supernumerary surviving reduced megaspores that acquire an FM identity were also detected. At later developmental stages, we identified misoriented embryo sacs with anomalous morphology. A similar phenotype including the emergence of supernumerary reduced and unreduced FMs was recently reported in trimethylguanosine synthase 1 defective lines by Siena et al. (2023).43 Moreover, supernumerary reduced FM are also formed in Arabidopsis plants carrying loss-of-function mutations in the miR822.44
In the absence of ARF10 within the ovule in arf10 mutants, the progression of meiotic entry is compromised. Morphological identification of defective meiotic entry was achieved by staining division septa with the Renaissance stain, while observing the correlation between division progression and integument size. This observation was associated with the delay in the establishment of MMC identity, as revealed by the MMC marker pKNU:nlsYFP. The challenges observed in meiotic entry directly affect ovule development, resulting in arrest at the gametogenesis (FG2 stage) in a significant number of ovules, leading to abortion. The partial penetrance of this effect hints at the potential redundancy of ARF10 with another ARF, possibly ARF16, as indicated by Wójcick et al.26 Taking these findings into account collectively, we suggest that ARF10 plays a functional role in initiating the commitment to gametophyte formation. This process likely occurs before the acquisition of MMC identity, suggesting that early gametophyte commitment serves as a crucial checkpoint preceding reductional division entry. Since ARF10 is a transcriptional repressor, it is likely to be involved in the suppression of a repressor, allowing the release of a gametophytic signal of unknown nature to the MMC, inducing thereby a primary commitment to a gametophytic identity in a non-cell autonomous manner. This interpretation is further reinforced by the fact that the ectopic expression of ARF10 throughout the nucellus (in mARF10, stk and ago1 lines), which disrupts the ARF10 signal, fails to cause the development of multiple MMCs entering meiosis, but rather the emergence of supernumerary enlarged cells with FM identity, able to start gametogenesis. Recently, Huang et al.22 stated that pARF10:mARF10 and pARF16:mARF16, in contrast to pARF17:mARF17, fail to produce supernumerary MMC-like cells. Here, we confirmed the absence of legitimate supernumerary MMCs (cells expressing pKNU-Venus) in pARF10:mARF10 lines, but detected extra enlarged cells in the nucellus at pre-meiosis. However, any direct comparison between these two sets of results is necessarily complicated by the fact that different pARF10:mARF10 constructs were employed in the analyses (i.e., the mutated sites were different), and they were introduced in different genetic backgrounds (Ler vs. Col). Moreover, it is a well-known fact that environmental conditions influence the rate of formation of non-reduced FM in aposporous plants, which can have an effect on the detection of the phenotype.
ARF10 regulation in ovule nucellus is complex
Our data unveil a complex regulatory network governing ARF10 within the ovule primordia nucellus. This regulation involves components such as miR160, AGO1, and the MADS-box transcription factor STK. When ARF10 is active, it is primarily expressed in a single cell layer surrounding the MMC, detected in the L1 and L2 nucellus layers. In the absence of AGO1 or STK (in ago1-27 ARF10-GFP and stk ARF10-GFP mutants, respectively), ARF10 exhibits higher expression levels in the ovule primordia nucella with respect to WT ARF10-GFP). Furthermore, in stk mutants, a lower GFP signal is detected within the MMC, which is not observed in the ago-1 mutant background. However, when ARF10 is rendered insensitive to miR160 regulation (mARF10), its expression becomes ubiquitous, present in the MMC, as well as in all nucellar and chalazal cells. Thus, ARF10 appears to be transcribed in all ovule cells but silenced in the MMC, the chalaza, and part of the nucellar cells through a mechanism involving miR160.
Furthermore, of particular interest is the direct regulation of ARF10 in the ovule primordia nucellus, which implies the existence of a complex mechanism likely orchestrated by the MADS box transcription factor STK and AGO1. Notably, our discovery reveals that STK can directly bind to the genomic region of ARF10, possibly repressing its expression. What is most intriguing is that in the nucellus, this repression operates in a dosage-dependent manner, as evidenced by our findings that ARF10 expression is higher in stk mutants, as observed through GFP quantification and in situ hybridization analysis. STK has been observed to interact with the RdDM pathway,11 prompting us to investigate whether it could potentially influence not only ARF10 but also miR160 expression through this epigenetic pathway. Previous studies have demonstrated that mutants within this pathway exhibit phenotypes associated with female germline fate acquisition, similar to what was described here for ARF10.10 To explore this further, we analyzed the methylation landscape of stk mutants compared to wild-type plants. We found that while the ARF10 locus remained unaffected, the miR160b locus displayed differential methylation. This discrepancy in the methylation (hypomethylation) status directly impacted miR160 expression levels, as evidenced by our quantification of mature miR160 using stem-loop PCR, which revealed a significant reduction in stk mutant backgrounds. We hypothesize that the regulation of ARF10 is intricately managed by STK, exerting masterful control through two distinct avenues: direct binding to its genomic region and indirect modulation via the RdDM pathway (methylation), which regulates the abundance of miR160 and consequently impacts ARF10 levels.
Another critical aspect of this puzzle involves AGO1. Its absence also results in an elevated expression of ARF10 in the ovule primordia nucelli, rather than a change in the spatial distribution. Considering the expression pattern of miR160, predominantly in the chalaza and funiculus, but also active in the ovule nucelli, we infer that mature miR160 likely has the ability to travel and silence its targets within the nucellus. This leads to the conclusion that AGO1 is the AGO protein facilitating the silencing of ARF10 in conjunction with miR160 in this context. Our findings underscore the previously documented specificity of AGO-mediated silencing. Specifically, AGO1 appears to regulate ARF10 processing exclusively in the nucellus, while not exhibiting such control in the chalaza or the MMC, where miR160 may be linked with other family members.
A model for the control of early female reproductive development
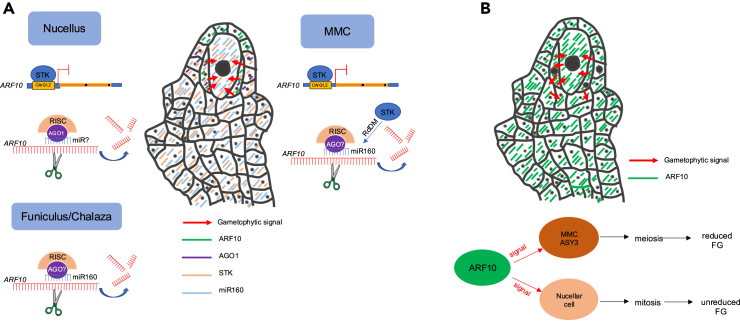
Based on the data provided here and elsewhere on miR160,22 AGO1,45 and STK38 on ovule expression domains, we propose a model for the STK- miR160/AGO1 control of ARF10 expression in female reproductive development (Figure 6). During Arabidopsis sexual wild type development, at late pre-meiosis (FG0), ARF10 is silenced by the action of miR160, STK and AGO1 in the nucellus (Figure 6A). Meanwhile, ARF10 accumulates in a restricted ring of L1/L2 nucellar cells surrounding the MMC. From this location, ARF10 induces the capacity for future gametophyte formation in the MMC cell and acts as a checkpoint for full MMC identity acquisition and progression into meiosis (Figure 6A). This ARF10 expression domain is maintained around the products of meiosis and is later relegated to the outer layer of the external integuments, where it may be necessary to promote normal gametophyte progression. In mARF10 lines, there is an increase of ARF10 levels in the nucellus, and an ectopic expression in the chalaza/funiculus and the MMC (Figure 6B), which induces: (1) an MMC premature entry into meiosis (note that the MMC is the only cell that accumulates meiotic precursors like ASY3) and (2) a commitment to form gametophytes from other nucellar cells (somatic). Both aspects of the reproductive phenotype might be related to the particularly high and generalized expression pattern of mARF10 in mARF10 lines, since numerous cells in the nucellus are surrounded by others expressing ARF10. Therefore, ARF10 expression possibly influences other nucellar cells, besides the MMC. However, since somatic nucellar cells fail to express other factors required for MMC identity and meiotic entry (perhaps elements of the THO/TREX complex/TAS3/ARF3, ARF17 or the RBR1 pathways), or even ASY3 meiotic precursors, they do not undergo megasporogenesis but bypass meiosis, acquire FM identity, and proceed directly into megagametogenesis.
Figure 6.
A proposed model for ARF10 function in the ovule
(A) In wild-type pre-meiotic ovule nucellus, ARF10 is partially and selectively silenced by STK in both the MMC and nucellus cells. STK binds to the genomic DNA at CArG boxes 1 and 2, repressing ARF10 expression. In the MMC, STK can also induce the expression of miR160 via the RdDM pathway. miR160 subsequently silences ARF10 in collaboration with an AGO protein of unknown identity. In the nucellus cells, AGO1 silences ARF10 in association with an unidentified miRNA. The regulation of ARF10 expression in the funiculus and chalaza regions involves miR160 in conjunction with an unknown AGO protein.
(B) In mARF10 lines, ARF10 is ectopically expressed in the nucellus cells, MMC, and funiculus/chalaza cells. This ectopic expression in the nucellus and MMC results in the widespread commitment of additional cells to form gametophytes. However, only the cell that accumulates meiotic precursors, such as ASY3 (i.e., the MMC), will undergo meiosis (sexual pathway), leading to the formation of a reduced female gametophyte. The generation of additional functional megaspores from somatic companion cells (non-reduced) eventually results in the development of supernumerary, anomalous gametophytes, similar to those observed in aposporous apomictic plants. Black dots represent the cell nuclei, question marks indicate unknown identity.
Conclusions
Here we present evidence that miR160 restricts the expression of ARF10 mainly to few cells around the MMC at pre-meiosis. ARF10 influences the entrance of the MMC into meiosis, by inducing commitment to form a gametophyte. Later, ARF10 appears to be implicated in the specification of megagametogenesis. Our experiments also indicate that the control of ARF10 by miR160 in the nucellus is mediated by AGO1, STK and RdDM. A higher accumulation of ARF10 in nucellar cells and the MMC induces phenotypes mimicking apospory (i.e., multiple non-reduced embryo sac formation with random orientation of cell-identity types and unusual embryo sac morphology). This study reveals a key mechanism involved in the germline fate acquisition in plant ovules and identifies a potential avenue for the development of molecular strategies for the induction of apomixis in sexual crop species – a long time target of the crop development industry. Considering that in natural apomicts like Paspalum notatum, ARF10 is overexpressed in florets at pre-meiosis and down-regulated during megagametogenesis,25 we speculate that full recreation of the apospory phenotype might require a more specific temporal control.
Limitations of the study
Current microscopy technologies, such as DIG or Feulgen confocal microscopy, do not allow for a conclusive assessment of the embryo sac border location in ovules carrying multiple megagametophytes. In the future, the development of markers that better reveal these limits might facilitate the identification of apospory-like phenotypes. Further experiments, such as Y1H or EMSA, should be conducted to confirm the repression of ARF10 by STK. Moreover, high-resolution in situ RNA sequencing will improve the definition of gene expression boundaries and refine the model presented here.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marta Mendes (marta.mendes@unimi.it).
Materials availability
This paper analyzes existing, publicly available plant materials (i.e., it did not generate new unique lines). The crosses between these materials are available from the lead contact upon request.
Data and code availability
-
•
McSeED sequencing data is available at NCBI Database: PRJNA750614.
-
•
Pipeline for methylation analysis from raw reads to DMR identification (https://bitbucket.org/capemaster/mcseed/src/master/).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
The project leading to this work has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No [645674], Project PROCROP and [872417], Project MAD; Agencia Nacional de Promoción Científica y Tecnológica, Argentina, Project PICT-2019-03414; Universidad Nacional de Rosario (UNR), Argentina, Project: 80020190300021UR; MAE, Italy/CONICET, Argentina cooperation project MAECI AR21GR03. Imaging analyses were carried out at NOLIMITS, an advanced imaging facility established by the University of Milan.
Author contributions
Conceptualization, S.P. and M.A.M.; Investigation, S.P., M.C., C.C., E.C., D.P., M.D.M., G.C.T., R.P., C.M., C.A., M.P., G.M, E.A., and M.A.M.; Methodology, S.P. and M.A.M.; Resources and Data Curation, M.P., E.A., and G.M.; Visualization: S.P., M.C., and M.A.M.; Writing – Original Draft, S.P. and M.A.M.; Writing – Review and Editing, S.P., E.A., H.D., L.C., and M.A.M., Funding Acquisition, S.P., L.C., and M.A.M.; Supervision: H.D., L.C., and M.A.M.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GFP polyclonal antibody | ChromoTek | RRID: AB_2749857 |
| Chemicals, peptides, and recombinant proteins | ||
| GoTaq DNA Polymerase | Promega | Cat #M3001 |
| Schiff reagent | Merck | 1090330500 |
| Renaissance 2200 staining | Renaissance Chemicals | https://www.renchem.co.uk/products/renaissance-SR-2200 |
| miRCURY LNA miRNA Detection Probes | Qiagen | Cat # 339111 YD00612441-BCD |
| Critical commercial assays | ||
| SV Total RNA Isolation Kit | Promega | Cat #Z3101 |
| Superscript II Reverse transcriptase | Invitrogen | Cat # 18064022 |
| Real Mix qPCR | Biodynamics | Cat # A6101 |
| iTaq SYBR green master mix | Bio-Rad | Cat#1725121 |
| DIG RNA Labeling Kit (SP6/T7) | Roche | Cat#11175025910 |
| Deposited data | ||
| Methylation raw sequence reads | this study | NCBI Database: PRJNA750614 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA750614 |
| Experimental models: Organisms/strains | ||
| Arabidopsis thaliana ARF10:mARF10-GFP line A | The Nottingham Arabidopsis Stock Centre (NASC) |
N16288 |
| Arabidopsis thaliana ARF10:mARF10-GFP line B | The Nottingham Arabidopsis Stock Centre (NASC) |
N16289 |
| Arabidopsis thaliana Col-0 | The Nottingham Arabidopsis Stock Centre (NASC) |
|
| Arabidopsis thaliana pKNU::nlsYFP | Tucker et al.,31 | |
| Arabidopsis thaliana pLC2::nlsYFP | Tucker et al.,31 | |
| Arabidopsis thaliana ASY3::RFP | Yang et al.,32 | |
| Arabidopsis thaliana pWOX2::CENH3_GFP | De Storme et al.,33 | |
| Arabidopsis thaliana FGR7.0 | Völz et al.,34 | |
| Arabidopsis thaliana arf10_2 | The Nottingham Arabidopsis Stock Centre (NASC) |
N655696 |
| Arabidopsis thaliana arf10_3 | The Nottingham Arabidopsis Stock Centre (NASC) |
N587560 |
| Arabidopsis thaliana stk-2 | Pinyopich et al.,37 | |
| Arabidopsis thaliana ago1-27 | Morel et al.,35 | |
| Oligonucleotides | ||
| Oligonucleotides for genotyping and expression analyses | See Table S4 | |
| Oligonucleotides for DNA methylation analysis | See Table S4 | |
| Oligonucleotides for Chromatin immunoprecipitation assay | See Table S4 | |
| Software and algorithms | ||
| Axiovision 4.1 | Carl Zeiss AG | https://www.micro-shop.zeiss.com/it/ch/ |
| LAS AF 2.2.0. A | Leica Microsystems Srl | https://www.leica-microsystems.com/ |
| R version 3.3.2 | R Core Team (2023) | www.r-project.org |
| Methylation Content Sensitive Enzyme Double-Digest Restriction-Site-Associated DNA (ddRAD) technique (MCSeEd) | Pipeline for methylation analysis from raw reads to DMR identification | https://bitbucket.org/capemaster/mcseed/src/master/ |
| REST-RG 2009 | QIAGEN | http://www.REST.de.com |
| QuantaSoft™ | BioRad |
https://www.bio-rad.com/en-it/SearchResults? Text=quantasoft |
Experimental model and study participant details
Plant material
For ARF10 expression analysis, we used an Arabidopsis SALK line (NASC ID N16287) transformed with a pARF10::ARF10_GFP construct.21 The ARF10 expression re-patterning in the absence of the miR160 function was studied in two different Arabidopsis NASC stocks (N16288, N16289) transformed with pARF10::mARF10_GFP fusion constructions, where ARF10 was mutated to be insensitive to miRNA160.21 These transgenic lines express a miR160-resistant form of ARF10 (mARF10), including four silent mutations in the miRNA target site in a Columbia background. During construction, the mARF10 gene, including the coding region (2.3 kb) and the 5'upstream region (3 kb), was fused to the green fluorescent protein.21 We also used specific cell-type reporter lines pKNU::nlsYFP,31 pLC2::nlsYFP,31 ASY3::RFP,32 pWOX2::CENH3_GFP33 and FGR7.0.34 Two defective arf10 mutant lines (NASC IDs N655696 - arf10_2 and N587560 - arf10_3) were employed to analyze ARF10 downregulation. The stk-237 and ago1-2735 mutants were used in crosses with the pARF10::ARF10_GFP line to carry on mechanistic studies. Plants were cultured on soil in growth chambers under a long-day photoperiod (16 h light 22°C/8 h dark 18°C) at 90 μmol photons m−2 s–1 light intensity using a mix of Sylvania 215-W cool white, fluorescent tubes and 60-W mate bulbs, at the University of Milan facilities.
Method details
Genotyping of arf10 defective mutants
Genomic DNA was extracted with Edwards Solution [200 mM Tris-HCl (pH 7.5), 250 mM NaCl, 25 mM EDTA, and 0.5% SDS] from one leaf of each plant, precipitated in isopropanol, resuspended, and used in genotyping experiments. Primers flanking both sides of the T insertion for arf10_2 and arf10_3 were designed at the T-DNA Primer design page of the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/tdnaprimers.2.html) and are described in Table S4. A primer located inside the T insertion, was used in combination with LP and RP to detect the presence of the insertion. Genomic DNA was amplified with the following primers combinations: 1) LP + LB: 2) RP + LB; 3) LP + RP (Table S4). Plants were classified as homozygous (Hm), heterozygous (Ht), or wild type based on the banding profile.
Microscopy
For DIC microscopy, ovules were clarified with chloral hydrate and examined in a Leica DM2500 microscope equipped with Nomarski differential interference contrast (DIC) optics. For fluorescent microscopy, fresh pistils were collected at different developmental stages for initial fluorescence evaluation, dissected in sterile H2O + 5% glycerol, and examined in a Nikon fluorescence microscope equipped with Axiovision software. For confocal laser scanning microscopy (CLSM), fresh material was collected, mounted in a 5% glycerol solution, and analysed immediately (to detect the fluorescence signal) or were processed with Feulgen staining (confocal analysis)46 to highlight nuclear DNA. The Feulgen technique first dissociate the two strands of DNA through hydrolysis by warm (60°C) 1 M hydrochloric acid, which liberates the hemiacetal functions of deoxyriboses. In an acidic environment, these hemiacetal functions are converted into aldehydic groups through a reversible reaction. Then, the Schiff reagent (i.e., fuchsin decolorated by insertion of a sulfonic group) reacts with the aldehydic groups. According to a possible interpretation, the carbonyl functions interact with the sulfonic groups included in the reagent to form alkyl sulfonic acids, while the chromophore function of the dye is re-established.46 Cell walls and meiotic septa were revealed using Renaissance 2200 staining.47 CLSM analysis was performed using a Leica TCS SPE microscope with a 488 nm argon laser line for excitation of GFP fluorescence. Emissions were detected between 505 and 580 nm. Images were collected in multi-channel mode, and overlay images were generated using Leica analysis software LAS AF 2.2.0.A.
DNA methylation analysis
Libraries were constructed according to Marconi et al.40 The methylation-sensitive enzymes chosen to infer CG, CHG, CHH methylation contexts were AciI, PstI, and EcoT22I, respectively, combined with MseI (Table S1). Pooled libraries were purified, size-selected for fragments in the range of 250 bp to 600 bp, quantified, normalized, and amplified with a primer that introduced an Illumina index (at the Y common adapter site) for demultiplexing (Table S4). Samples were then amplified using uniquely indexed primers, pooled, subjected to PCR-enrichment as described by Marconi et al.40 and Illumina-sequenced using 150 bp paired-end chemistry. High-quality raw reads from the Illumina sequencing of the CG, CHG, and CHH libraries were analyzed following the protocol and the pipeline described in Marconi et al.40 Reads were normalized and filtered, discarding all sites with a co-logarithm of the variation coefficient value higher than −0.35. The relative methylation levels at each site were calculated following the procedure in Marconi et al.,40 and the DMPs (Differentially Methylated Positions; only loci with a False Discovery Rate ≤ 0.05 were considered significant) were called following the MethylKit manual best practices.48 The mapping of the DMPs in the same scaffold and closer than a given threshold provided their clustering to identify the DMRs (Differentially Methylated Regions; only loci with a False Discovery Rate ≤ 0.05 were considered significant), based on the procedure reported in Marconi et al.40 Statistical analyses were performed in R version 3.3.2 (www.r-project.org) using the ‘stats’ and ‘gplots’ packages. The ‘stats’ package was used to estimate correlations and logistic regression. Complete linkage clustering was carried out using the ‘heatmap.2’ function of the ‘gplots’ package, combined with the ‘hclust’ and ‘dist’ functions, and with ‘ward.D2’ as the clustering method. Regarding CHH-DMR validation, as described by Hashimoto et al.,49 for each position of the CHH-DMR located 109 bp from the miR160 to be validated, the DNA was digested using EcoT22I (the methylation-sensitive enzyme for the corresponding methylcytosine context). The reaction mixture of 25 μL contained 100 ng DNA, 0.5 U of EcoT22I and its relative buffer. For the non-enzyme control (mock), distilled water was added instead of the enzyme. All the samples were then incubated at 37°C for 4 h, follow by heat inactivation at 65°C for 20 min. The Real-time PCR for the methylation status and the relative statistical analysis were performed as in Marconi et al.40 The sequence information of the primers that bracketed the enzyme site of each DMP are reported in Table S4. Data are available under the SRA accession number NCBI Database: PRJNA750614.
Silique analysis
The proportion of unfertilized ovules and seeds in the transgenic lines and in homozygous and heterozygous plants was counted with a Leica magnifying stereoscope equipped with a Leica Application Suites software.
qPCR analysis
Oligonucleotides for expression analysis of ARF10 (AT5G21150) for arf10_2: and arf10_3 are described in Table S4. To analyze ARF expression in the N655696 arf10 homozygous plants, total RNA was extracted from frozen flowers using the SV Total RNA Isolation Kit (PROMEGA) and reverse transcribed with Superscript II (INVITROGEN, Carlsbad, CA, USA) following manufacturer recommendations. Quantitative PCR reactions (final volume: 20 μL) included 0.5 μM gene-specific primers, 1X Real Mix qPCR (BIODYNAMICS, Buenos Aires, Argentina) and 20 ng of cDNA. In each experiment, two biological replicates were processed, including three technical replicates and negative control. Amplifications were performed in a Rotor-Gene Q thermocycler (QIAGEN, Hilden, Germany), as follows: 2 min at 94°C followed by 45 cycles of 94°C for 15 s, 57°C for 30 s, and 72°C for 17 s, and a final elongation step of 5 min at 72°C. The specificity of the PCR amplicons was checked by acquiring heat dissociation curves (from 60°C to 95°C). Relative quantitative expression levels were assessed using the REST-RG 2009 software (QIAGEN) with β-TUBULIN as an internal reference (Table S4). For the stem-loop PCR we followed the protocol used by Varkonyi-Gasic et al.50 and the are available in Table S4.
In situ hybridization
Samples were collected from ARF10_GFP, mARF10_GFP plants, homozygous ARF10_GFP stk-2 and homozygous ARF10_GFP ago1-27 plants. Arabidopsis thaliana inflorescences were fixed for 24h in FAA and embedded in paraffin wax as previously described by Galbiati et al.51 Sections of the embedded material were cut at a thickness of 10 μm using a microtome and mounted on poly-L-lysine slide (Bio-Optica). Sections were probed with digoxigenin-labeled for m5GFP antisense RNA. Hybridization and immunological detection were performed as previously described by Galbiati et al.51 The m5GFP specific antisense probes was amplified respectively with primers mGFP6 FW and RV (Table S4) and transcribed with the DIG RNA Labeling Kit (SP6/T7) by Roche. For the mature miR160 in situ hybridization protocol LNA-modified oligonucleotide-based probes were purchased from Qiagen. The slides were observed under a Zeiss Axiophot D1 microscope.
Chromatin immunoprecipitation assay followed by qRT-PCR (ChIP qPCR)
The promoter region of the ARF10 (2kb upstream to ATG) was analyzed to identify potential CArG boxes. The ChIP experiment was performed as previously described.52 One gram of inflorescence till anthesis was used for chromatin extraction from pSTK::STK-GFP38 and Col-0 plants. For immunoprecipitation we used 30 μl of GFP-trap for each sample (ChromoTek). Enrichment of the target regions was calculated by qPCR (iTaq Universal SYBR Green Supermix, Bio-Rad) using a Bio-Rad iCycler iQ optical system. The relative enrichment of the targets obtained from pSTK::STK-GFP inflorescences was compared with the enrichment obtained from WT. ACTIN 11 was used as reference gene. To establish the efficiency of the chromatin immunoprecipitation, we used the third CArG box of VDD as a positive control.11 Three independent ChIP experiments were performed, all the primers used are described in Table S4.
Quantification and statistical analysis
Statistical analyses of proportions
Part of the results was analysed using Student’s t-test that is performed with Excel (Microsoft), another part using the 95% confidence intervals of the true proportion (95% CI) were calculated according to the Newcombe method53 at the Vassarstats website (http://vassarstats.net/prop1.html) and other using one-way ANOVAs with post hoc Turkey test using an online tool (https://astatsa.com/OneWay_Anova_with_TukeyHSD/). Differentially methylated loci significance was calculated using MethylKit R package whereas differentially methylated regions were inferred by logistic regression using R version 3.3.2.
Published: October 9, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111115.
Supplemental information
Lists are separated for contexts and for the location of DMR (within gene sequence or regulatory region).
References
- 1.Nogler G.A. In: Embryology of Angiosperms. Johri B.M., editor. Springer-Verlag; 1984. Gametophytic apomixis; pp. 475–518. [Google Scholar]
- 2.Koltunow A.M., Grossniklaus U. Apomixis: a developmental perspective. Annu. Rev. Plant Biol. 2003;54:547–574. doi: 10.1146/annurev.arplant.54.110901.160842. [DOI] [PubMed] [Google Scholar]
- 3.Hand M.L., Koltunow A.M.G. The genetic control of apomixis: asexual seed formation. Genetics. 2014;197:441–450. doi: 10.1534/genetics.114.163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hojsgaard D., Klatt S., Baier R., Carman J.G., Hörandl E. Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. Crit. Rev. Plant Sci. 2014;33:414–427. doi: 10.1080/07352689.2014.898488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crane C.F. In: The Flowering of Apomixis: From Mechanisms to Genetic Engineering. Savidan Y., Carman J.G., Dresselhaus T., editors. CIMMYT, IRD, European Commission DG VI FAIR; 2001. Classification of apomictic mechanisms; pp. 24–43. [Google Scholar]
- 6.Pinto S.C., Mendes M.A., Coimbra S., Tucker M.R. Revisiting the female germline and its expanding toolbox. Trends Plant Sci. 2019;24:455–467. doi: 10.1016/j.tplants.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Petrella R., Caselli F., Roig-Villanova I., Vignati V., Chiara M., Ezquer I., Tadini L., Kater M.M., Gregis V. BPC transcription factors and a Polycomb Group protein confine the expression of the ovule identity gene SEEDSTICK in Arabidopsis. Plant J. 2020;102:582–599. doi: 10.1111/tpj.14673. [DOI] [PubMed] [Google Scholar]
- 8.Hater F., Nakel T., Groß-Hardt R. Reproductive Multitasking: The Female Gametophyte. Annu. Rev. Plant Biol. 2020;71:517–546. doi: 10.1146/annurev-arplant-081519-035943. [DOI] [PubMed] [Google Scholar]
- 9.Yu S.X., Jiang Y.T., Lin W.H. Ovule initiation: the essential step controlling offspring number in Arabidopsis. J. Integr. Plant Biol. 2022;64:1469–1486. doi: 10.1111/jipb.13314. [DOI] [PubMed] [Google Scholar]
- 10.Olmedo-Monfil V., Durán-Figueroa N., Arteaga-Vázquez M., Demesa-Arévalo E., Autran D., Grimanelli D., Slotkin R.K., Martienssen R.A., Vielle-Calzada J.-P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendes M.A., Petrella R., Cucinotta M., Vignati E., Gatti S., Pinto S.C., Bird D.C., Gregis V., Dickinson H., Tucker M.R., Colombo L. The RNA-dependent DNA methylation pathway is required to restrict SPOROCYTELESS/NOZZLE expression to specify a single female germ cell precursor in Arabidopsis. Development. 2020;147 doi: 10.1242/dev.194274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 13.Ceccato L., Masiero S., Sinha Roy D., Bencivenga S., Roig-Villanova I., Ditengou F.A., Palme K., Simon R., Colombo L. Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S.-B., Xie Z.-Z., Hu C.G., Zhang J.-Z. A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016;7:47. doi: 10.3389/fpls.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cucinotta M., Cavalleri A., Guazzotti A., Astori C., Manrique S., Bombarely A., Oliveto S., Biffo S., Weijers D., Kater M.M., Colombo L. Alternative splicing generates a MONOPTEROS isoform required for ovule development. Curr. Biol. 2021;31:892–899.e3. doi: 10.1016/j.cub.2020.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Su Z., Zhao L., Zhao Y., Li S., Won S., Cai H., Wang L., Li Z., Chen P., Qin Y., Chen X. The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module. Curr. Biol. 2017;27:1597–1609.e2. doi: 10.1016/j.cub.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mallory A.C., Bartel D.P., Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR 17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J.W., Wang L.J., Mao Y.B., Cai W.J., Xue H.W., Chen X.Y. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J.H., Han S.J., Yoon E.K., Lee W.S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006;34:1892–1899. doi: 10.1093/nar/gkl118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu M.F., Tian Q., Reed J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- 21.Liu P.-P., Montgomery T.A., Fahlgren N., Kasschau K.D., Nonogaki H., Carrington J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 22.Huang J., Zhao L., Malik S., Gentile B.R., Xiong V., Arazi T., Owen H.A., Friml J., Zhao D. Specification of female germline by microRNA orchestrated auxin signaling in Arabidopsis. Nat. Commun. 2022;13:6960. doi: 10.1038/s41467-022-34723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai H., Liu L., Huang Y., Zhu W., Qi J., Xi X., Aslam M., Dresselhaus T., Qin Y. Brassinosteroid signaling regulates female germline specification in Arabidopsis. Curr. Biol. 2022;32:1102–1114.e5. doi: 10.1016/j.cub.2022.01.022. [DOI] [PubMed] [Google Scholar]
- 24.Ortiz J.P.A., Leblanc O., Rohr C., Grisolia M., Siena L.A., Podio M., Colono C., Azzaro C., Pessino S.C. Small RNA-seq reveals novel regulatory components for apomixis in Paspalum notatum. BMC Genom. 2019;20:487. doi: 10.1186/s12864-019-5881-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podio M., Colono C., Siena L., Ortiz J.P.A., Pessino S.C. A study of the heterochronic sense/antisense RNA representation in florets of sexual and apomictic Paspalum notatum. BMC Genom. 2021;22:185. doi: 10.1186/s12864-021-07450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wójcik A.N., Nodine M.D., Gaj M.D. miR160 and miR166/165 contribute to the LEC2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Front. Plant Sci. 2017;8:2024. doi: 10.3389/fpls.2017.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wójcikowska B., Gaj M.D. Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep. 2017;36:843–858. doi: 10.1007/s00299-017-2114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damodharan S., Corem S., Gupta S.K., Arazi T. Tuning of SlARF10A dosage by sly-miR160a is critical for auxin-mediated compound leaf and flower development. Plant J. 2018;96:855–868. doi: 10.1111/tpj.14073. [DOI] [PubMed] [Google Scholar]
- 29.Mei S., Zhang M., Ye J., Du J., Jiang Y., Hu Y. Auxin contributes to jasmonate-mediated regulation of abscisic acid signaling during seed germination in Arabidopsis. Plant Cell. 2023;35:1110–1133. doi: 10.1093/plcell/koac362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez-Lagana E., Mosca G., Mendocilla-Sato E., Pires N., Frey A., Giraldo-Fonseca A., Michaud C., Grossniklaus U., Hamant O., Godin C., et al. Organ geometry channels reproductive cell fate in the Arabidopsis ovule primordium. Elife. 2021;10 doi: 10.7554/eLife.66031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucker M.R., Okada T., Hu Y., Scholefield A., Taylor J.M., Koltunow A.M.G. Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development. 2012;139:1399–1404. doi: 10.1242/dev.075390. [DOI] [PubMed] [Google Scholar]
- 32.Yang C., Hu B., Portheine S.M., Chuenban P., Schnittger A. State changes of the HORMA protein ASY1 are mediated by an interplay between its closure motif and PCH2. Nucleic Acids Res. 2020;48:11521–11535. doi: 10.1093/nar/gkaa527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Storme N., Keçeli B.N., Zamariola L., Angenon G., Geelen D. CENH3-GFP: a visual marker for gametophytic and somatic ploidy determination in Arabidopsis thaliana. BMC Plant Biol. 2016;16:1. doi: 10.1186/s12870-015-0700-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Völz R., Heydlauff J., Ripper D., von Lyncker L., Groß-Hardt R. Ethylene signaling is required for synergid degeneration and the establishment of a pollen tube block. Dev. Cell. 2013;25:310–316. doi: 10.1016/j.devcel.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Morel J.B., Godon C., Mourrain P., Béclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song J.J., Liu J., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 37.Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- 38.Mizzotti C., Ezquer I., Paolo D., Rueda-Romero P., Guerra R.F., Battaglia R., Rogachev I., Aharoni A., Kater M.M., Caporali E., Colombo L. SEEDSTICK is a master regulator of development and metabolism in the Arabidopsis seed coat. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendes M.A., Guerra R.F., Berns M.C., Manzo C., Masiero S., Finzi L., Kater M.M., Colombo L. MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell. 2013;25:2560–2572. doi: 10.1105/tpc.112.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marconi G., Capomaccio S., Comino C., Acquadro A., Portis E., Porceddu A., Albertini E. Methylation content sensitive enzyme ddRAD (MCSeEd): a reference-free, whole genome profiling system to address cytosine/adenine methylation changes. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-51423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker C., Hagmann J., Müller J., Koenig D., Stegle O., Borgwardt K., Weigel D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz R.J., Schultz M.D., Lewsey M.G., O′Malley R., Urich M.A., Libiger O., Schork N.J., Ecker J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siena L.A., Michaud C., Selles B., Vega J.M., Pessino S.C., Ingouff M., Ortiz J.P.A., Leblanc O. TRIMETHYLGUANOSINE SYNTHASE1 mutations decanalize female germline development in Arabidopsis. New Phytol. 2023;240:597–612. doi: 10.1111/nph.19179. [DOI] [PubMed] [Google Scholar]
- 44.Tovar-Aguilar A., Grimanelli D., Acosta-García G., Vielle-Calzada J.-P., Badillo-Corona J.A., Durán-Figueroa N. The miRNA822 loaded by ARGONAUTE9 modulates the monosporic female gametogenesis in Arabidopsis thaliana. Plant Reprod. 2024;37:243–258. doi: 10.1007/s00497-023-00487-2. [DOI] [PubMed] [Google Scholar]
- 45.Su Z., Wang N., Hou Z., Li B., Li D., Liu Y., Cai H., Qin Y., Chen X. Regulation of female germline specification via small RNA mobility in Arabidopsis. Plant Cell. 2020;32:2842–2854. doi: 10.1105/tpc.20.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braselton J.P., Wilkinson M.J., Clulow S.A. Feulgen staining of intact plant tissues for confocal microscopy. Biotech. Histochem. 1996;71:84–87. doi: 10.3109/10520299609117139. [DOI] [PubMed] [Google Scholar]
- 47.Musielak T.J., Slane D., Liebig C., Bayer M. Versatile optical clearing protocol for deep tissue imaging of fluorescent proteins in Arabidopsis thaliana. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. MethylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashimoto K., Kokubun S., Itoi E., Roach H.I. Improved quantification of DNA methylation using methylation-sensitive restriction enzymes and real-time PCR. Epigenetics. 2007;2:86–91. doi: 10.4161/epi.2.2.4203. [DOI] [PubMed] [Google Scholar]
- 50.Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galbiati F., Sinha Roy D., Simonini S., Cucinotta M., Ceccato L., Cuesta C., Simaskova M., Benkova E., Kamiuchi Y., Aida M., et al. An integrative model of the control of ovule primordia formation. Plant J. 2013;76:446–455. doi: 10.1111/tpj.12309. [DOI] [PubMed] [Google Scholar]
- 52.Ezquer I., Mizzotti C., Nguema-On E., Gotté M., Beauzamy L., Ebeling Viana V., Dubrulle N., Costa de Oliveira A., Caporali E., Koroney A.-S., et al. The developmental regulator SEEDSTICK controls structural and mechanical properties of the Arabidopsis seed coat. Plant Cell. 2016;28:2478–2492. doi: 10.1105/tpc.16.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newcombe R.G. Two-Sided Confidence Intervals for the Single Proportion: Comparison of Seven Methods. Stat. Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lists are separated for contexts and for the location of DMR (within gene sequence or regulatory region).
Data Availability Statement
-
•
McSeED sequencing data is available at NCBI Database: PRJNA750614.
-
•
Pipeline for methylation analysis from raw reads to DMR identification (https://bitbucket.org/capemaster/mcseed/src/master/).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.